Abstract
Positive effects of alcohol drinking such as anxiolysis and euphoria appear to be a crucial factor in the initiation and maintenance of alcohol use disorder (AUD). However, the mechanisms that lead from chromatin reorganization to transcriptomic changes after acute ethanol exposure remain unknown. Here, we used Assay for Transposase-Accessible Chromatin followed by high throughput sequencing (ATAC-seq) and RNA-seq to investigate epigenomic and transcriptomic changes that underlie anxiolytic effects of acute ethanol using an animal model. Analysis of ATAC-seq data revealed an overall open or permissive chromatin state that was associated with transcriptomic changes in the amygdala after acute ethanol exposure. We identified a candidate gene, Hif3a (Hypoxia-inducible factor 3, alpha subunit), that had ‘open’ chromatin regions (ATAC-seq peaks), associated with significantly increased active epigenetic histone acetylation marks and decreased DNA methylation at these regions. The mRNA levels of Hif3a were increased by acute ethanol exposure, but decreased in the amygdala during withdrawal after chronic ethanol exposure. Knockdown of Hif3a expression in the central nucleus of amygdala attenuated acute ethanol-induced increases in Hif3a mRNA levels and blocked anxiolysis in rats. These data indicate that chromatin accessibility and transcriptomic signatures in the amygdala after acute ethanol exposure underlie anxiolysis and possibly prime the chromatin for the development of AUD.
Subject terms: Neuroscience, Addiction
Introduction
Clinical and preclinical studies have shown that anxiolytic effects of acute ethanol promote higher alcohol consumption and are crucial for the development of alcohol use disorder (AUD) [1–4]. Humans with anxiety disorders have the tendency to transition from occasional drinking to dependence more rapidly than those without [5, 6]. The extended amygdaloid circuitry is implicated in the regulation of anxiety behavior and serves as a critical hub in mediating acute and chronic ethanol’s behavioral and molecular effects [7–9]. Alcohol’s actions on modulating the excitability of the extended amygdaloid circuitry may play an important role in anxiety behaviors [10]. It is well established that the behavioral effects of acute and chronic alcohol exposure are under complex epigenetic control [2, 9].
We recently showed acute ethanol-induced anxiolysis is dependent on microRNA-494 (miR-494) signaling, which impacts regulation of CBP [CREB (cyclic adenosine monophosphate response element binding protein) binding protein], p300, and Cbp/p300-interacting transactivator 2 (CITED2) in the amygdala [11]. Acute ethanol exposure is associated with increases in histone H3K9 and H4K8 acetylation, via inhibition of histone deacetylases (HDACs), in addition to decreases in H3K9me2 and G9a (histone methyltransferase) protein levels in the amygdala of rats [12–15]. Recent studies have shown similar increases in histone acetylation in mouse hippocampus, prefrontal cortex and liver after acute ethanol exposure [16]. Furthermore, the increased histone acetylation in the amygdala and associated behavioral changes such as decreased anxiety undergo neuroadaptation (tolerance) and are reversed following withdrawal from chronic ethanol exposure [2, 12, 13]. However, in the amygdala, the genome-wide loci where these epigenomic and associated transcriptomic changes occur following a low dose of ethanol are currently unknown.
In this study, we sought to investigate this question by utilizing two genome-wide approaches: Assay for Transposase-Accessible Chromatin with high throughput sequencing (ATAC-seq), which identifies changes in genome accessibility, and RNA-seq, which assesses changes in global gene expression following acute ethanol [17, 18]. ATAC-seq makes use of the integrative capabilities of the hyperactive Tn5 transposase and next-generation sequencing to identify more accessible (open) versus less accessible (closed) regions across the genome and also provides a wealth of information regarding protein-DNA interactions [17, 19]. By functionally linking this with data from RNA-seq, which provides a global transcriptomic readout, we identified candidate gene targets containing differential chromatin modifications at their transcriptional control regions that correlated with differential expression in the amygdala, likely contributing to the anxiolytic effects of acute ethanol.
Materials and methods
Acute ethanol exposure paradigm
Adult male and female Sprague–Dawley rats were purchased from Harlan (Indianapolis, IN) and group housed with ad libitum food and water access in a temperature-controlled room on a 12/12-h light/dark cycle. All procedures were performed in accordance with the National Institute of Health Guidelines for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee. Animals were randomly assigned to various groups and the number of rats in each group was based on our previous publications [11–13]. Acute ethanol (1 g/kg, intraperitoneal) or n-saline injections and anxiety behavioral measurements [elevated plus-maze (EPM) exploration test] were performed as described previously [13]. Rats were subjected to explore EPM for 5 min 1 h after injections. Immediately after behavioral measurements or 1 h after ethanol injections (without behavioral measurements), animals were anesthetized with isoflurane, and brain regions [amygdala (central, medial, and basolateral nuclei), bed nucleus of the stria terminalis (BNST) and dorsal hippocampus] were dissected (within 5 min) and stored at −80 °C until processed for various biochemical studies as described below. The Analox Alcohol Analyzer (Lunenburg, MA) was used to measure blood alcohol levels at the time of brain collection. The average blood ethanol level (mg%) of ethanol-treated male rats was 89.3 ± 2.9 (n = 40; mean ± SEM), as we have previously observed (11, 13–15). The average blood ethanol level (mg%) of ethanol-treated female rats was 74.5 ± 7.6 (n = 8; mean ± SEM), The mean ± SEM (n = 41) body weights (gm) of both treatment groups were not significantly different from each other in males (Control = 304.6 ± 4.3; Ethanol = 302.8 ± 3.6). Similarly, in females, the mean ± SEM (n = 8) body weights (gm) among both groups were not different from each other (Control = 204 ± 2.8; Ethanol = 200.6 ± 3.1). All data presented here were generated in adult male rats except Fig. S3, which was performed in adult female rats. Experiments were not performed blindly.
ATAC-sequencing
Following acute ethanol treatment, frozen amygdala samples (six controls and six ethanol treated) were tagmented with Illumina’s Nextera DNA sample prep kit (Illumina®, San Diego, CA). Following purification, DNA was amplified using custom Nextera library-specific primers. The resulting libraries were assessed for quality nucleosome ladder profiles and quantification on the Agilent Bioanalyzer with the Agilent DNA HS assay kit (Agilent Technologies, Santa Clara, CA). Libraries were paired-end sequenced using the Illumina® platform. The bioinformatic processing of ATAC-seq data is described in supplementary methods.
RNA-sequencing
Total RNA was extracted from amygdala tissues obtained from a separate cohort of rats, using the RNeasy® kit (QIAGEN, Germantown, MD) following the manufacturer’s protocol. A total of six controls (saline-treated) and six ethanol-treated rats were used. Agilent Tapestation (Agilent Technologies, Santa Clara, CA) was used to assess RNA quality using the RNA Integrity Number (RIN) assessment, and the average RIN value for the samples indicate high quality and integrity of isolated RNA (Control = 8.7 ± 0.118; Ethanol = 8.9 ± 0.119). Total RNA from 2 animals was pooled, resulting in n = 3 in each group for RNA-seq. The concentrations were determined by fluorometric analysis, and 1 µg of RNA in 20 µl of RNAse-Free water was used for library preparation. The libraries were prepared and sequenced in the DNA Services laboratory of the Roy J. Carver Biotechnology Center at the University of Illinois at Urbana-Champaign. The Illumina® platform was used for library preparation and single-end sequencing using the TruSeq Stranded mRNAseq Sample Prep kit (Illumina®, San Diego, CA). Libraries were sequenced for 100nt on three lanes of a HiSeq2500 (Illumina® platform). The bioinformatic analysis of RNA-seq data is described in supplementary methods.
Integration of ATAC-seq with RNA-seq data
ATAC peaks were annotated to nearby genes on the basis of close proximity (promoter association, 2 kb ± from transcription start site [TSS]) or potential long-range interactions (distal association, up to 200 kb from TSS). The genes in the 2 kb list (31 genes, Table 1, FDR < 0.2) were cross-referenced for their presence in the RNA-seq data (FDR < 0.2) for mRNA changes.
Table 1.
Genes that are associated with ATAC-seq peaks at their promoters (TSS ± 2 kb).
| Peak ID | Chromosome | Start | End | Gene name | TSS ± 2 kb | Ethanol/Control: logFC | Ethanol/Control: Q Value |
|---|---|---|---|---|---|---|---|
| peak_3217 | 1 | 78998888 | 79004845 | Hif3a | −1019 | 0.739398 | 1.51668E−10 |
| peak_68740 | 20 | 8090968 | 8098770 | Fkbp5 | 0 | 0.563998 | 1.33121E−05 |
| peak_40629 | 15 | 30183087 | 30184570 | LOC290156 | 0 | 0.613106 | 0.000106 |
| peak_40797 | 15 | 31685828 | 31687526 | LOC108353020 | 148 | 0.775034 | 0.000129 |
| peak_108576 | 8 | 53142419 | 53148464 | Zbtb16 | 0 | 0.419321 | 0.002019 |
| peak_40843 | 15 | 32135389 | 32137992 | LOC102556508 | 0 | 0.482184 | 0.002740 |
| peak_40647 | 15 | 30553539 | 30555161 | Trav14s1 | 677 | 0.779843 | 0.003097 |
| peak_40834 | 15 | 32042566 | 32045051 | LOC103693854 | 0 | 0.460100 | 0.005350 |
| peak_40541 | 15 | 29366687 | 29370177 | LOC684995 | 16 | 0.555889 | 0.006129 |
| peak_63413 | 2 | 140705353 | 140708314 | Mgst2 | −82 | 0.536870 | 0.009204 |
| peak_3216 | 1 | 78995896 | 78998702 | Hif3a | 0 | 0.431469 | 0.010197 |
| peak_96228 | 6 | 76175484 | 76176788 | Psma6 | 1024 | 0.605321 | 0.012949 |
| peak_40796 | 15 | 31675907 | 31679085 | LOC102555014 | 0 | 0.393897 | 0.014117 |
| peak_10302 | 1 | 221753875 | 221755151 | Pygm | −1174 | 0.685527 | 0.037892 |
| peak_44676 | 16 | 8497159 | 8500489 | Ogdhl | 0 | 0.378384 | 0.073691 |
| peak_67100 | 2 | 239413152 | 239414771 | Cxxc4 | −111 | −0.417057 | 0.078753 |
| peak_86677 | 5 | 33180626 | 33182976 | Maged2 | 0 | −0.317445 | 0.082602 |
| peak_62601 | 2 | 105000505 | 105002148 | LOC682402 | 0 | −0.552217 | 0.100118 |
| peak_105292 | 7 | 130540544 | 130541153 | Acr | −167 | 0.764952 | 0.100712 |
| peak_40773 | 15 | 31510679 | 31512130 | LOC682708 | 0 | 0.482470 | 0.112276 |
| peak_87750 | 5 | 69832439 | 69834831 | Nipsnap3b | 0 | −0.451526 | 0.112958 |
| peak_44673 | 16 | 8466095 | 8467877 | LOC102550503 | 0 | 0.482126 | 0.123118 |
| peak_9080 | 1 | 201498140 | 201500195 | Htra1 | 0 | 0.371520 | 0.134251 |
| peak_21401 | 11 | 30362069 | 30365302 | Sod1 | 0 | −0.284925 | 0.161426 |
| peak_92245 | 5 | 165026989 | 165029576 | LOC108351059 | 0 | 0.393443 | 0.165461 |
| peak_97599 | 6 | 111475449 | 111477839 | Slirp | 0 | −0.353192 | 0.165461 |
| peak_97599 | 6 | 111475449 | 111477839 | Alkbh1 | 0 | −0.353192 | 0.165461 |
| peak_40642 | 15 | 30533803 | 30535711 | LOC102552674 | 0 | 0.512589 | 0.170042 |
| peak_103519 | 7 | 109203060 | 109206986 | Zfat | 525 | 0.314836 | 0.181991 |
| peak_1056 | 1 | 31493765 | 31495003 | LOC100911237 | 0 | −0.690268 | 0.185516 |
| peak_34139 | 14 | 7617155 | 7618444 | Slc10a6 | 0 | 0.602676 | 0.190299 |
Candidate gene analysis for mRNA, histone acetylation, and DNA methylation fold changes
Total RNA was extracted, as described above, from the amygdala, BNST, and dorsal hippocampus. Reverse transcription followed by candidate gene qPCR to evaluate mRNA fold changes were performed, as described previously [11]. The occupancy of acetylated histones (H3K9/14ac, Cat. No. 06-599 MilliporeSigma, Burlington, MA; H3K27ac, Cat. No. 39133, Active Motif, Carlsbad, CA) and HIF3A (Cat. No. ab10134, Abcam, Waltham, MA) was evaluated at specific sites of genes using chromatin immunoprecipitation (ChIP) assay in the amygdala followed by qPCR and relative fold changes were determined, as described previously [20, 21]. DNA methylation fold changes were analyzed at the promoter sites of Hif3a and Slc10a6 genes in the amygdala using MethylMiner™ assay for 5-methylcytosine (5mC) followed by qPCR and determination of relative fold changes, as published by us [22]. For all the qPCR assays, relative fold changes were determined using the ΔΔCt method [23]. Primer sequences are included in Table S4.
Stereotaxic surgery and infusion of Hif3a siRNA into CeA
Animals were bilaterally cannulated with CMA/11 guide cannulae (CMA Microdialysis, North Chelmsford, MA) targeting the central nucleus of amygdala (CeA) using coordinates from bregma (posterior −2.5 mm, medial-lateral ±4.2 mm, ventral −5.1 mm) and siRNA infusion procedures as described previously by us [21, 24]. One week after surgery, they were infused with either negative control siRNA (Cat. No. 1027310) or Hif3a siRNA (Cat. No. SI01521604) (1 µg in 0.5 µl/side) (QIAGEN, Germantown, MD) using probes that delivered siRNA into the CeA, 3 mm beyond the guided cannulas [24]. The sequences of the Hif3a siRNA were: sense, 5′-CGACGAGAGGAUUGCAGAATT-3′; antisense 5′-UUCUGCAAUCCUCUCGUCGCA-3′. The siRNAs were dissolved in iFect solution (Neuromics, Edina, MN). Post-infusion (~23 h), rats were treated with ethanol (1 g/kg) or saline, and after 1 h, anxiety measures were determined using EPM [13]. Following behavioral measurements, rats were anesthetized with isoflurane, and amygdaloid tissues (predominantly CeA but also including surrounding medial and basolateral amygdala) were dissected and stored at −80 °C for mRNA measurements as described above.
Chronic ethanol treatment
Adult male rats received liquid Lieber–DeCarli ethanol or control diets for 15 or 16 days as described previously [13, 14]. Ethanol-fed rats were withdrawn for 0 or 24 h, anesthetized, and sacrificed to dissect out the amygdala for the measurement of mRNA levels of Hif3a using qPCR, as described above.
Statistical Analysis
Statistical analysis was performed using SigmaStat software suite (Systat Software Inc., San Jose, CA). Differences between control and ethanol groups were analyzed using Student’s unpaired two-tailed t test. For Hif3a siRNA experiments, two-way ANOVA followed by Tukey’s post-hoc test was performed. For the Hif3a chronic ethanol and withdrawal experiment, Kruskal–Wallis one-way analysis of variance followed by Tukey’s post-hoc test was used. Inclusion/exclusion criteria were not preestablished. Outliers were determined using interquartile range (IQR) method. For whole genome approaches (ATAC-seq and RNA-seq), details of statistical analysis are provided in supplementary methods.
Results
Acute ethanol produces an anxiolytic-like response
We measured anxiety-like behaviors 1 h after acute ethanol by using the elevated plus maze exploration (EPM) test. The results confirmed our earlier findings [11, 13–15] that acute ethanol significantly increases the percentage of open-arm entries and percentage of time spent in the open arms (Fig. S1), which are indicators of the anxiolytic effects of acute ethanol in male rats. Similar findings were also observed in female rats (Fig. S3A).
ATAC-seq analysis in the amygdala after acute ethanol exposure
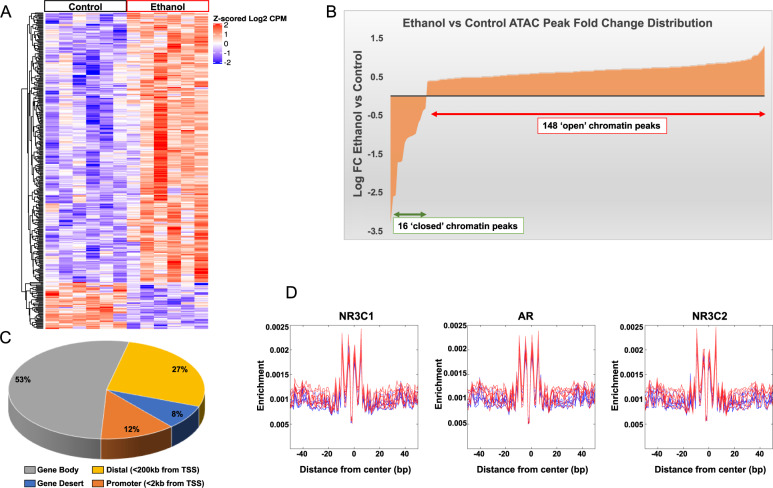
We used ATAC-seq to assess chromatin accessibility in the amygdala after acute ethanol exposure and obtained a total of 118,836 peaks (Table S1). We observed a differential pattern of distribution (Heatmap, Fig. 1A) with an overall increase in genome-wide chromatin accessibility after acute ethanol exposure (Fig. 1A, B). The majority of peaks were localized to intergenic and distal regulatory regions, while about 12% of these peaks (14,707) were in promoter proximal regions (±2 kb of TSS) (Fig. 1C). We observed 164 significant peaks that were differentially altered in the amygdala between ethanol-treated and control rats at FDR < 0.05: 148 were ‘open’-chromatin peaks in ethanol-treated versus saline-treated controls and 16 were ‘closed’ (Fig. 1B). We identified 31 genes (FDR < 0.2) that had peaks at their promoters (±2 kb of TSS) (Table 1). Ingenuity Pathway Analysis (IPA®) of these genes resulted in two gene networks (Fig. S4) containing Hif3a (hypoxia-inducible factor 3 subunit alpha) and Slc10a6 (solute carrier family 10 member 6). Hif3a promoter contained two peaks (peak_3217 and peak_3216) and Slc10a6 contained one peak (peak_34139) that were upregulated after acute ethanol exposure (Table 1). Interestingly, the most significant differential promoter peak was associated with Hif3a, a transcription factor involved in adaptive responses to hypoxic stress impacting diverse regulatory biological pathways [25, 26]. These data suggest that expression of these two genes may be altered in the amygdala, most likely induced by acute ethanol exposure.
Fig. 1. ATAC-seq data analysis in the amygdala of acute ethanol exposed rats.
A Heatmap showing differential ATAC peaks (FDR < 0.2; 345 peaks) and chromatin accessibility in the amygdala after acute ethanol exposure. Red represents increases in peaks and blue represents decreases in peaks. Individual peaks are represented in rows and individual animals from control and ethanol groups (n = 6 animal per treatment) are shown in columns. B Distribution of ATAC peak fold changes indicating that out of the 164 peaks at FDR < 0.05, 148 peaks were associated with ‘open’ chromatin regions in the amygdala of acute ethanol-treated compared to 16 peaks in the control rats. C ATAC-seq data analysis in the amygdala of acute ethanol exposed rats showing distribution of ATAC peaks across genomic features. D Footprinting images for the top 3 (out of 41) motifs derived from FIMO analysis of 572 vertebrate motifs from the JASPAR database. The motifs satisfied the following criteria: q < 0.01 and >50% enrichment (log2 ratio >0.585). The blue lines represent controls and the red lines represent ethanol-treated samples. [(NR3C1- Glucocorticoid Receptor; Nuclear Receptor Subfamily 3 Group C Member 1), (AR- Androgen Receptor), NR3C2 (Mineralocorticoid Receptor; Nuclear Receptor Subfamily 3 Group C Member 2)].
We further searched for putative transcription factor (TF) binding motifs in the differential ATAC peak regions (FDR < 0.2; 345 peaks) to identify potential regulators of differential chromatin accessibility in the amygdala, and we also generated footprinting images to supplement these data. True TF-DNA binding should prevent integration of the transposase at the binding site, which is reflected in these footprints as a drop-in enrichment or a pattern change around the motif that matches the size of the binding site. The observed reduction in transposon binding at the site of the motif then implies occupancy of those sites by another molecule, which, based on the motif specificity, is most likely the TF [17] (Fig. 1D and Table S2). Predicted transcription factors that bind these motifs include those that have been implicated in alcohol responses, such as the glucocorticoid receptor, NR3C1 [27–29].
RNA-seq analysis and validations of gene expression in the amygdala after acute ethanol exposure
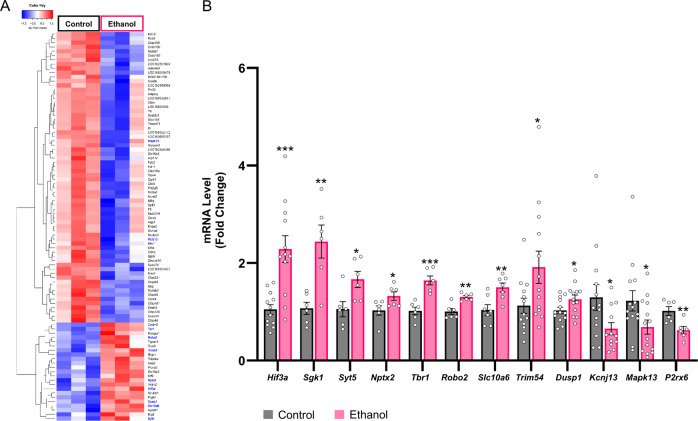
To evaluate whether acute ethanol-induced chromatin accessibility is responsible for transcriptomic changes in the amygdala of male rats, we performed RNA-seq using the Illumina® platform. The high sequencing depth resulted in 62.58–70.76 million reads per sample (Fig. S5). After calculating fold changes and FDR corrected values (Table S3), we used an FDR < 0.2, to identify 91 genes that exhibited significant differential fold changes in the ethanol group compared to controls (Heatmap, Fig. 2A). Pathway analysis using IPA® revealed several networks involved in critical cellular processes including organismal injury, cell death and survival, cellular organization and tissue development. The top 4 networks are shown in Fig. S6. For mRNA validations, we selected candidate genes from the IPA® networks that had increased and decreased transcript levels after acute ethanol compared to controls: Increased—Hif3a, Syt5 (Synaptotagmin 5), Tbr1 (T-box, Brain, 1), Trim54 (Tripartite Motif Containing 54), Robo2 (Roundabout Guidance Receptor 2), Nptx2 (Neuronal Pentraxin 2), Slc10a6 and Dusp1 (Dual Specificity Phosphatase 1); Decreased—Kcnj13 (Potassium Inwardly Rectifying Channel Subfamily J Member 13) and Mapk13 (Mitogen-Activated Protein Kinase 13). Quantitative RT-PCR confirmed that the expression changes were similar to the changes seen in the RNA-seq data, thus validating the findings (Fig. 2B). We further validated two genes (FDR > 0.2), Sgk1 (Serum/Glucocorticoid Regulated Kinase 1) and P2rx6 (Purinergic Receptor P2X 6) (Fig. 2B) that exhibited increased and decreased expression, respectively, in ethanol-treated rats compared to controls. Sgk1 is a known glucocorticoid response gene, and since our ATAC peak motif analysis identified putative NR3C1 (glucocorticoid receptor) binding motifs to be enriched, we chose this gene. Furthermore, Sgk1 functions as an important modulator of ion channel function and neuronal plasticity and plays a role in regulating acute and chronic ethanol phenotypes via its actions in mouse prefrontal cortex [27]. In addition, we measured Hif3a mRNA in the amygdala of female rats and found that acute ethanol exposure significantly increased Hif3a mRNA levels (Fig. S3B), similar to male rats (Fig. 2B).
Fig. 2. RNA-seq data analysis in the amygdala of acute ethanol-exposed rats.
A HeatMap of RNA-seq differentially expressed transcripts at FDR < 0.2.Hierarchical clustering and heatmap of differentially expressed mRNAs (FDR < 0.2) from acute ethanol and normal saline (control) treated rats. Red represents increases in overall expression and blue represents decreases in expression. Individual mRNAs are represented in rows and individual experiments (n = 3 in each treatment; RNA from two rats are pooled for each treatment) are shown in columns. Gene names highlighted in blue were used for the validations. B Validations of mRNA levels of selected candidates from RNA-seq data in the amygdala of acute ethanol exposed rats. Expression levels of mRNAs were significantly different in the amygdala of acute ethanol-exposed rats compared to saline-treated control rats, as measured by qPCR. Data represents the mean ± SEM and individual values are shown on bar diagrams with circle dots for control and ethanol groups (n = 6–14; Two-tailed Student’s t test; *p < 0.05; **p < 0.01; ***p < 0.001).
Integration of ATAC-seq and RNA-seq analysis and validations
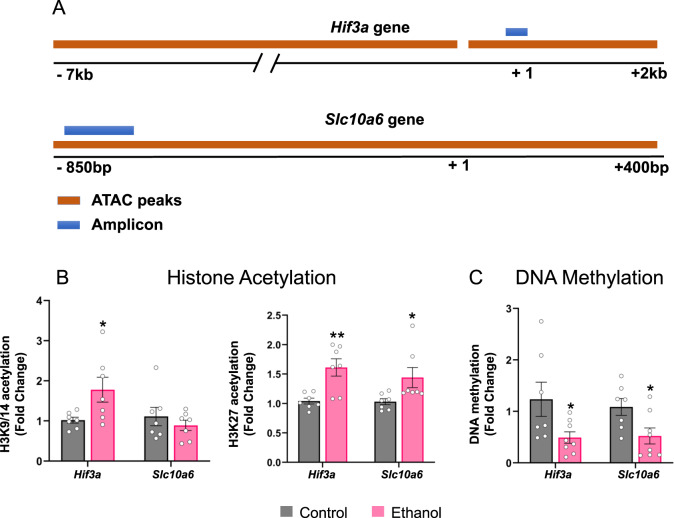
Amongst the 31 genes with differential chromatin peaks in their promoters (Table 1), two candidates that emerged from IPA, Hif3a and Slc10a6, also exhibited increased mRNA expression following acute ethanol compared to controls in both the RNA-seq data (FDR < 0.2) and qPCR validations (Fig. 2B). We used ChIP and DNA methylation assays to investigate the open-chromatin peak regions at the promoters of Hif3a and Slc10a6 by designing primers within the peak locations for both Hif3a (peak that overlaps TSS) and Slc10a6 (Table 1; Fig. 3A). For ChIP, we used antibodies against two well-characterized activating histone modifications, H3K9/14ac and H3K27ac [2, 9]. We observed an increase in H3K27ac occupancy at both Hif3a and Slc10a6 (Fig. 3B) and an increase in H3K9/14ac occupancy at the Hif3a site, but no change in H3K9/14ac occupancy at the Slc10a6 site (Fig. 3B). In addition, we observed a decrease in 5-methylcytosine (5mC) levels (Fig. 3C) at these locations of Hif3a and Slc10a6. These findings are suggestive of an “open” or permissive chromatin structure, as reflected by ATAC-seq data. These data suggest that increased histone acetylation and decreased DNA methylation at Hif3a and Slc10a6 promoters in the amygdala may be associated with their observed increased mRNA levels after acute ethanol exposure.
Fig. 3. Histone acetylation (H3K9/14ac; H3K27ac) and DNA methylation (5-methylcytosine, 5mC) changes reveal relaxed chromatin architecture at the ATAC-seq peak regions near the transcription start site (TSS) of Hif3a and Slc10a6 genes.
A Schematic showing the location of ATAC-seq peaks and location of primers for qPCR validation for Hif3a and Slc10a6. B The “open peak” regions at the transcriptional control regions of Hif3a and Slc10a6 were evaluated by chromatin immunoprecipitation (ChIP) assay representing fold changes of H3K27ac and H3K9/14ac occupancy at selected regions of Hif3a and Slc10a6 in ethanol-treated and control rats. C Changes in DNA methylation were also evaluated at the same loci of Hif3a and Slc10a6 in amygdala of ethanol-treated and control rats and represented as fold changes in DNA methylation (5mC levels). Values are represented as the mean ± SEM and individual values are shown on bar diagrams with circle dots for control and ethanol groups (n = 7–8; two-tailed Student’s t test *p < 0.05; **p < 0.01).
Hif3a gene knockdown in the CeA promotes anxiety and attenuates anxiolytic effects of acute ethanol
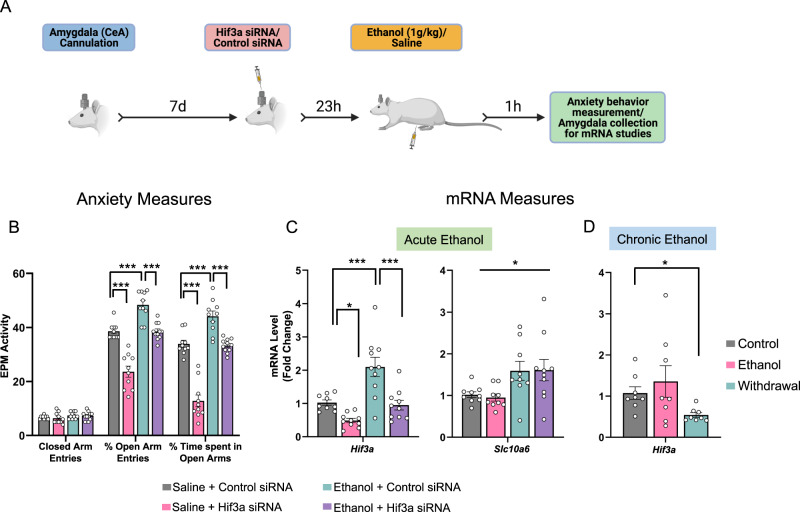
Results from merging the two big data sets suggest that ‘open’ ATAC-seq peaks (chromatin accessibility) may be responsible for increased mRNA levels of Hif3a in the amygdala after acute ethanol exposure. In addition, we observed that Hif3a mRNA levels were increased in BNST and hippocampal brain regions after acute ethanol exposure (Fig S2). We investigated whether induction of Hif3a mRNA in the adult male CeA is involved in the ethanol-induced anxiolysis (Fig. 4A), as this brain region is implicated in anxiety and alcohol-related behaviors [2, 8, 30]. It was found that Hif3a small interfering RNA (siRNA) infusion into the CeA significantly attenuated ethanol-induced increases in Hif3a mRNA levels and associated anxiolysis in rats (Fig. 4B, C). Interestingly, Hif3a siRNA infusion in ethanol-naïve control rats decreased Hif3a mRNA levels in the amygdala and provoked anxiety-like behaviors. The differential reduction in anxiety by Hif3a siRNA in control and alcohol-treated animals may be related to differential baseline anxiety levels. In order to ascertain whether these changes were specific to Hif3a mRNA expression, we also measured mRNA levels of Slc10a6 in these rats and observed no changes in mRNA levels of Slc10a6 in Hif3a siRNA infused rats (Fig. 4C). Taken together, these results suggest that increased expression of Hif3a in the CeA mediates acute ethanol-induced anxiolysis.
Fig. 4. Hif3a siRNA infusion into the central nucleus of amygdala (CeA) blocks ethanol-induced anxiolysis and reduces mRNA levels of Hif3a, but not Slc10a6. Hif3a mRNA is decreased during withdrawal after chronic ethanol exposure.
A Schematic representation of the experimental design for the siRNA infusion and behavioral measurement. B Bar diagram showing anxiety-like behaviors following Hif3a siRNA infusion into the CeA with or without acute ethanol treatment (1 g/kg, intraperitoneal) in rats. Hif3a siRNA infusion into CeA (24 h prior) prevented anxiolytic effects of acute ethanol in rats. Values are represented as mean ± SEM (n = 10–11; two-way analysis of variance (Percent open arm entries: treatment effect, F1,37 = 72.397, p < 0.001, group effect F1,37 = 71.445, p < 0.001, treatment × group interaction, F1,37 = 3.078, p = 0.088; Percent time spent in open arms: treatment effect, F1,37 = 97.836, p < 0.001, group effect, F1,37 = 90.159, p < 0.001, treatment × group interaction, F1,37 = 10.175, p < 0.01) followed by post-hoc Tukey’s test: ***p < 0.001). C The mRNA levels of Hif3a and Slc10a6 were also measured in the amygdala of rats infused with control siRNA or Hif3a siRNA into CeA then treated with either saline or ethanol. Values are represented as mean ± SEM [n = 9–10; Two-way analysis of variance (Hif3a: treatment effect, F1,35 = 23.54, p < 0.001; group effect, F1,35 = 19.366, p < 0.001; Slc10a6: group effect, F1,33 = 11.003, p < 0.01) followed by post-hoc Tukey’s test: *p < 0.05; ***p < 0.001]. D Bar diagram showing changes in Hif3a mRNA levels in the amygdala of control, chronic ethanol-treated and ethanol-withdrawn (24 h) rats. Values are represented as mean ± SEM (n = 8; Kruskal–Wallis one-way analysis of variance on ranks (H2 = 7.3, p < 0.05) followed by Tukey’s test, *p < 0.05). Individual values are shown on bar diagrams with circle dots for various groups.
Chronic ethanol exposure and withdrawal decrease Hif3a expression in the amygdala
We used an established animal model of alcohol dependence, wherein rats exhibit anxiety-like behaviors during withdrawal following chronic ethanol exposure, and measured Hif3a expression in these amygdala tissues [13, 14]. It was found that Hif3a mRNA levels were significantly decreased during withdrawal (24 h) compared to control animals, but that they were unchanged when ethanol was still in the system (0 h withdrawal; Fig. 4D). These data suggest that decreased Hif3a expression in the amygdala may be involved in development of ethanol withdrawal-related anxiety-like behaviors. This is mechanistically supported by the fact that infusion of Hif3a siRNA into the CeA provoked anxiety-like behaviors in alcohol naïve male control rats (Fig. 4B, C).
Acute ethanol-induced HIF3A binding to Npy1r promoter induces Npy1r mRNA levels
Next we examined the molecular mechanisms by which HIF3A might be involved in the regulation of acute ethanol-induced anxiolysis. Recently, a HIF3A ChIP-seq study in human cell lines identified genome-wide HIF3A binding sites [31]. Based on this observation, we utilized the JASPAR database [32] to identify putative hypoxia response element (HRE) sites at the rat neuropeptide Y receptor Y1 (Npy1r) gene promoter (Fig. S7A). It has been shown that activation of Npy1r in the amygdala, particularly in the CeA, produces anxiolytic effects [33, 34]. Using an antibody against HIF3A, we performed the ChIP assay and observed increased HIF3A binding following acute ethanol in the vicinity of a predicted HRE site at the Npy1r promoter region in the amygdala (Fig. S7A, B). Expression levels for this gene trended to increase after acute ethanol exposure in RNA-seq data (FDR < 0.475; Table S3). We confirmed these results by measuring Npy1r mRNA levels following acute ethanol exposure and observed a significant increase in the amygdala (Fig. S7C). These data suggest the possibility that a HIF3A mediated increase in Npy1r mRNA levels in the amygdala could be involved in alcohol-induced anxiolysis.
Discussion
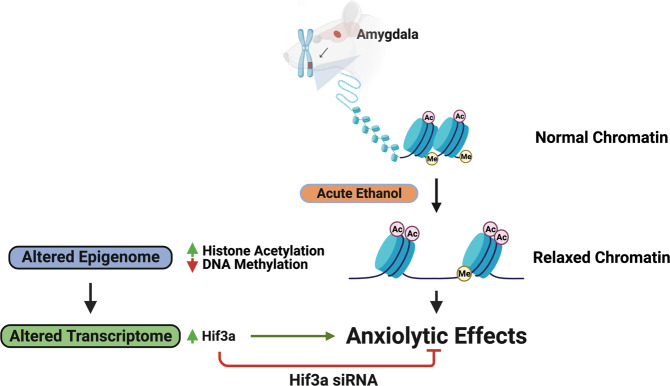
The present study has identified epigenomic and transcriptomic changes in the amygdala produced by acute ethanol exposure as well as a candidate gene, Hif3a, that is mechanistically linked to the anxiolytic effects of ethanol. Here, we used next-generation sequencing approaches, ATAC-seq and RNA-seq, to demonstrate that acute ethanol has the ability to induce a predominantly ‘open’ chromatin structure in the amygdala, thereby facilitating DNA-protein interactions and altering the transcriptome (Fig. 5). Our earlier findings indicate that acute ethanol decreases HDAC activity and increases CBP levels with associated increases in H3K9ac and H4K8ac levels in rat amygdala [13, 15]. Corroborating these findings, our ATAC-seq analysis revealed an overall “open” or permissive chromatin state, in the amygdala of acute ethanol-exposed rats. Footprinting analysis further identified important ethanol-regulated transcription factor motifs such as the glucocorticoid receptor, NR3C1 [27–29]. It is known that TF motifs occur in intergenic regions, which interact with specific core promoter elements via chromatin looping [35], as revealed by approaches such as chromosome conformation capture (3C) and its derivatives [36]. RNA-seq revealed several candidate genes involved in critical cellular functions such as mediation of stress responses (Hif3a, Sgk1), synaptic signaling and communication (Syt5, Nptx2), and regulation of intracellular MAP kinase signaling (Dusp1).
Fig. 5. This model depicts the ability of acute ethanol to rapidly alter the epigenome in the amygdala and produce transcriptomic changes.
These genome-wide chromatin accessibility and transcriptomic signatures in the amygdala are associated with anti-anxiety effects of ethanol in rats. One such candidate gene is hypoxia-induced gene transcription factor, Hif3a, which is epigenetically induced by acute ethanol, and anxiolytic effects associated with acute ethanol are prevented by inhibiting Hif3a expression in the central nucleus of amygdala of rats. The rapid and dynamic epigenomic modifications from a low dose of ethanol clearly suggest that these molecular processes may prime the amygdala to the emotional negative consequences that are most commonly associated with and promote alcohol use disorder.
Comparing ATAC-seq with RNA-seq data revealed two promising candidates, Hif3a (Hypoxia-inducible factor 3, alpha subunit) and Slc10a6 (solute carrier family 10 member 6), which had increased chromatin accessibility around their TSS’ along with up-regulation of both transcripts after acute ethanol exposure in the amygdala. The ‘open’ chromatin regions at these promoters had differential histone modifications associated with them. Combinatorial histone modifications are typically loci specific [37], and here we observed that acute ethanol increased H3K27ac and H3K9/14ac levels at the promoter of Hif3a and H3K27ac but not H3K9/14ac at the promoter of Slc10a6. DNA methylation (5mC) was decreased at both gene promoters. SLC10A6 also known as sodium-dependent organic anion transporter (SOAT), is part of the SLC family of transporters, which are involved in active and passive transport of various inorganic ions, neurotransmitters, amino acids and various other substrates, and plays a role in the transport of sulfate steroids and associated signaling pathways [38]. Interestingly, other members of this family, such as the glutamate transporter Slc17a7 and the GABA transporter Slc6a11, have been implicated in mediating alcohol responses [39, 40]. HIF3A is a transcription factor that is involved in adaptive responses to hypoxic stress and impacts multiple regulatory pathways [26]. Other members of this family, such as HIF1A, are known to interact with CITED2 (cAMP-responsive element-binding protein [CBP]/p300-interacting transcriptional modulator), which has been linked to the anxiolytic response to acute ethanol [11, 41]. Hypoxia response element (HIF binding) or HRE sites have been shown to occur in conjunction with cAMP response element-binding (CREB) sites, activator protein 1 (AP1), CCAAT-enhancer binding protein (CEBP) and ATF3 sites and TF’s binding these sites are thought to functionally interact and co-operate to affect hypoxia-induced changes in gene expression [41–43]. CREB signaling, as has been shown in various studies, is crucial for anxiety and alcohol comorbid phenotypes in rats via chromatin remodeling mechanisms [2, 9]. Here, we observed that Hif3a expression is increased in the amygdala during acute ethanol but is normalized after chronic ethanol exposure and significantly decreased during withdrawal. Similarly, we have previously reported that in the amygdala, acute ethanol exposure increased histone acetylation, CBP levels, and inhibited HDAC activity, which is normalized after chronic ethanol exposure. During withdrawal after chronic ethanol exposure, however, HDAC activity is increased and CBP levels are decreased, and histone acetylation also is decreased [11, 13, 15]. These data suggest that Hif3a as well as other transcriptomic changes that occur during acute ethanol exposure may undergo neuroadaptive processes [8] most likely involving dynamic epigenetic changes [2, 11, 13] after chronic ethanol exposure and subsequent withdrawal.
Inhibition of Hif3a expression in the CeA was able to attenuate acute ethanol-induced anxiolysis. A recent study [31] revealed HIF3A occupancy at transcriptional control regions of critical genes implicated in regulating alcohol and anxiety-related comorbid behaviors, such as neuropeptide Y (Npy) and activity-regulated cytoskeletal associated protein (Arc). Acute ethanol exposure has been shown to increase expression of Npy and Arc in the amygdala that is possibly involved in the anxiolytic actions of alcohol [13, 14]. The receptor for Npy is the Npy1r gene, and activation of NPY receptor Y1 with a NPY1R agonist produces anxiolysis [33, 34]. Furthermore, it has been shown that NPY via NPY1R activation suppresses the corticotropin-releasing factor (CRF) and modulates GABAergic pathways in the CeA to regulate anxiety and alcohol-related behaviors [8, 34, 44, 45]. We identified HRE sites at Npy1r control regions and tested this relationship. We observed increased HIF3A binding at the Npy1r promoter following acute ethanol exposure. Furthermore, this was associated with increased expression of Npy1r in the amygdala. These data suggest that HIF3A may regulate alcohol-induced anxiolysis via activating NPY1R signaling in the amygdala.
Data collected here suggest that inhibition of Hif3a expression in the CeA is sufficient to attenuate ethanol-induced anxiolysis, but involvement of other brain circuitry cannot be ruled out, as we observed that acute ethanol exposure increased Hif3a expression in BNST and hippocampus. These brain circuitries also play an important role in alcohol-related anxiety and emotional behaviors [8, 46, 47]. It is well established that there are sex differences in acute ethanol sensitivity [48–50], so we tested this idea and similar to male rats, acute ethanol exposure produced anxiolytic effects and increased Hif3a expression in the amygdala of female rats. These findings suggest that HIF3A may be a common target in mediating acute ethanol-induced anxiolysis in both sexes.
In summary, the combined use of two powerful genome-wide approaches, ATAC-seq and RNA-seq identified genomic loci that may be involved in the rapid epigenetic regulation of the transcriptome in the amygdala after acute ethanol exposure, suggesting that even a low dose of ethanol may impact the epigenome and drive processes responsible for addictive behaviors (Fig. 5). Clinically, it has been shown that acute alcohol priming causes high-risk social drinkers to demonstrate binge consumption behavior similar to heavy drinkers [51]. We identified an epigenetic target, Hif3a, in the amygdala that regulates anxiety-related behaviors after ethanol exposure. Hif3a is induced following acute ethanol, which is associated with anxiolytic-like behavior [11], and decreased during withdrawal after chronic ethanol exposure, which is associated with increased anxiety-like behavior [13, 14]. Interestingly, knockdown of Hif3a in the CeA induced anxiety-like behaviors in ethanol-naïve control rats, mimicking the ethanol withdrawal effects on these measures. These data provide the important link between Hif3a and ethanol-related anxiety behaviors (Fig. 5). This study also reiterates the fact that acute ethanol-induced epigenomic changes can serve as a molecular indicator of genes that could potentially become dysregulated upon chronic exposure and subsequent withdrawal. AUD comorbid with anxiety has a complex multifactorial etiology, and these data make a compelling argument to target HIF3A signaling in the amygdala for the development of pharmacological interventions in the treatment of AUD.
Supplementary information
Acknowledgements
The authors would like to thank, Dr. Alvaro Hernandez from University of Illinois at Urbana Champaign for performing RNA-sequencing and Dr. Kevin White from University of Chicago for performing ATAC-sequencing. We also thank Ms. Gina Giase for her help with ATAC-seq-related experiments and Dr. Tara Teppen for providing help in animal handling and brain dissections. The schematic design of the experiment in Fig. 4 and models (Fig. 5, Fig. S7) were prepared using BioRender.
Author contributions
SCP conceived the idea and conceptual framework, designed, supervised, and received funding for the study. HRK, HZ, and JPB generated animals, performed behavioral experiments, and collected brains. HRK, YC, and HK performed biochemical experiments. MM-C, AWS, and CL performed bioinformatics of ATAC-seq data. JD performed bioinformatics of RNA-seq data. HRK, DRG, CL, and MM-C further evaluated and analyzed these data sets. SCP and HRK acquired all data and performed further analyses and interpretations. HRK and SCP wrote the manuscript. All authors reviewed and edited the manuscript and approved the final version.
Funding
This work was supported by National Institute on Alcohol Abuse and Alcoholism (NIAAA) grants, P50AA-022538 (Center for Alcohol Research in Epigenetics), U01AA-019971, U24AA-024605 [Neurobiology of adolescent Drinking in Adulthood (NADIA) project], RO1AA-010005, and by the Department of Veterans Affairs I01BX004517 (VA Merit Grant) and Senior Research Career Scientist Award (IK6BX006030) to SCP. The authors are solely responsible for the content of the article and do not represent the official views of the National Institutes of Health or the U.S. Department of Veterans Affairs.
Data availability
All the data on which conclusions are based are included either in the main manuscript or in supplemental materials. The ATAC-seq (accession number GSE 171399) and RNA-seq (accession number GSE 171400) data sets have been deposited at gene expression omnibus (GEO) of NCBI (SuperSeries GSE accession number 171412).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41380-022-01732-2.
References
- 1.Gilman JM, Ramchandani VA, Davis MB, Bjork JM, Hommer DW. Why we like to drink: a functional magnetic resonance imaging study of the rewarding and anxiolytic effects of alcohol. J Neurosci. 2008;28:4583–91. doi: 10.1523/JNEUROSCI.0086-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pandey SC, Kyzar EJ, Zhang H. Epigenetic basis of the dark side of alcohol addiction. Neuropharmacology. 2017;122:74–84. doi: 10.1016/j.neuropharm.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sayette MA. The effects of alcohol on emotion in social drinkers. Behav Res Ther. 2017;88:76–89. doi: 10.1016/j.brat.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spanagel R, Montkowski A, Allingham K, Stöhr T, Shoaib M, Holsboer F, et al. Anxiety: a potential predictor of vulnerability to the initiation of ethanol self-administration in rats. Psychopharmacology (Berl) 1995;122:369–73. doi: 10.1007/BF02246268. [DOI] [PubMed] [Google Scholar]
- 5.Anker JJ, Kushner MG. Co-occurring alcohol use disorder and anxiety: bridging psychiatric, psychological, and neurobiological perspectives. Alcohol Res. 2019;40:arcr.v40.1.03. [DOI] [PMC free article] [PubMed]
- 6.Kushner MG, Maurer E, Menary K, Thuras P. Vulnerability to the rapid (”telescoped”) development of alcohol dependence in individuals with anxiety disorder. J Stud Alcohol Drugs. 2011;72:1019–27. doi: 10.15288/jsad.2011.72.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hur J, Kaplan CM, Smith JF, Bradford DE, Fox AS, Curtin JJ, et al. Acute alcohol administration dampens central extended amygdala reactivity. Sci Rep. 2018;8:16702. doi: 10.1038/s41598-018-34987-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–38. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishnan HR, Sakharkar AJ, Teppen TL, Berkel TD, Pandey SC. The epigenetic landscape of alcoholism. Int Rev Neurobiol. 2014;115:75–116. doi: 10.1016/B978-0-12-801311-3.00003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harrison NL, Skelly MJ, Grosserode EK, Lowes DC, Zeric T, Phister S, et al. Effects of acute alcohol on excitability in the CNS. Neuropharmacology. 2017;122:36–45. doi: 10.1016/j.neuropharm.2017.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teppen TL, Krishnan HR, Zhang H, Sakharkar AJ, Pandey SC. The potential role of amygdaloid microRNA-494 in alcohol-induced anxiolysis. Biol Psychiatry. 2016;80:711–9. doi: 10.1016/j.biopsych.2015.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berkel TDM, Zhang H, Teppen T, Sakharkar AJ, Pandey SC. Essential role of histone methyltransferase G9a in rapid tolerance to the anxiolytic effects of ethanol. Int J Neuropsychopharmacol. 2019;22:292–302. doi: 10.1093/ijnp/pyy102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pandey SC, Ugale R, Zhang H, Tang L, Prakash A. Brain chromatin remodeling: a novel mechanism of alcoholism. J Neurosci. 2008;28:3729–37. doi: 10.1523/JNEUROSCI.5731-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pandey SC, Zhang H, Ugale R, Prakash A, Xu T, Misra K. Effector immediate-early gene arc in the amygdala plays a critical role in alcoholism. J Neurosci. 2008;28:2589–2600. doi: 10.1523/JNEUROSCI.4752-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakharkar AJ, Zhang H, Tang L, Shi G, Pandey SC. Histone deacetylases (HDAC)-induced histone modifications in the amygdala: a role in rapid tolerance to the anxiolytic effects of ethanol. Alcohol Clin Exp Res. 2012;36:61–71. doi: 10.1111/j.1530-0277.2011.01581.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mews P, Egervari G, Nativio R, Sidoli S, Donahue G, Lombroso SI, et al. Alcohol metabolism contributes to brain histone acetylation. Nature. 2019;574:717–21. doi: 10.1038/s41586-019-1700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol. 2015;109:21.29.1–29.9. doi: 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Kyzar EJ, Bohnsack JP, Kokare DM, Teppen T, Pandey SC. Adolescent alcohol exposure epigenetically regulates CREB signaling in the adult amygdala. Sci Rep. 2018;8:10376. doi: 10.1038/s41598-018-28415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kyzar EJ, Zhang H, Pandey SC. Adolescent alcohol exposure epigenetically suppresses amygdala Arc enhancer RNA expression to confer adult anxiety susceptibility. Biol Psychiatry. 2019;85:904–14. doi: 10.1016/j.biopsych.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sakharkar AJ, Kyzar EJ, Gavin DP, Zhang H, Chen Y, Krishnan HR, et al. Altered amygdala DNA methylation mechanisms after adolescent alcohol exposure contribute to adult anxiety and alcohol drinking. Neuropharmacology. 2019;157:107679. doi: 10.1016/j.neuropharm.2019.107679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Zhang H, Sakharkar AJ, Shi G, Ugale R, Prakash A, Pandey SC. Neuropeptide Y signaling in the central nucleus of amygdala regulates alcohol-drinking and anxiety-like behaviors of alcohol-preferring rats. Alcohol Clin Exp Res. 2010;34:451–61. doi: 10.1111/j.1530-0277.2009.01109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safran M, Kaelin WG. HIF hydroxylation and the mammalian oxygen-sensing pathway. J Clin Invest. 2003;111:779–83. doi: 10.1172/JCI18181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Costin BN, Dever SM, Miles MF. Ethanol regulation of serum glucocorticoid kinase 1 expression in DBA2/J mouse prefrontal cortex. PLoS ONE. 2013;8:e72979. doi: 10.1371/journal.pone.0072979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gatta E, Grayson DR, Auta J, Saudagar V, Dong E, Chen Y, et al. Genome-wide methylation in alcohol use disorder subjects: implications for an epigenetic regulation of the cortico-limbic glucocorticoid receptors (NR3C1) Mol Psychiatry. 2021;26:1029–41. doi: 10.1038/s41380-019-0449-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy A, Mittal N, Zhang H, Pandey SC. Modulation of cellular expression of glucocorticoid receptor and glucocorticoid response element-DNA binding in rat brain during alcohol drinking and withdrawal. J Pharmcol Exp Ther. 2002;301:774–84. doi: 10.1124/jpet.301.2.774. [DOI] [PubMed] [Google Scholar]
- 30.Gilpin NW, Melissa A, Herman MA, Roberto M. The central amygdala as an integrative hub for anxiety and alcohol use disorders. Biol Psychiatry. 2015;77:859–69. doi: 10.1016/j.biopsych.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolonen JP, Heikkilä M, Malinen M, Lee HM, Palvimo JJ, Wei GH, et al. A long hypoxia-inducible factor 3 isoform 2 is a transcription activator that regulates erythropoietin. Cell Mol Life Sci. 2020;77:3627–42. doi: 10.1007/s00018-019-03387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46:D260–D266. doi: 10.1093/nar/gkx1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eva C, Serra M, Paolo Mele P, Carlo Panzica G, Oberto A. Physiology and gene regulation of the brain NPY Y1 receptor. Front Neuroendocrinol. 2006;27:308–39. doi: 10.1016/j.yfrne.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 34.Gilpin NW. Neuropeptides in central amygdala: role in anxiety- and alcohol-related behaviors. Alcohol. 2012;46:329–37. doi: 10.1016/j.alcohol.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–39. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denker A, de Laat W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 2016;30:1357–82. doi: 10.1101/gad.281964.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Södersten E, Toskas K, Rraklli V, Tiklova K, Björklund AK, Ringnér M, et al. A comprehensive map coupling histone modifications with gene regulation in adult dopaminergic and serotonergic neurons. Nat Commun. 2018;9:1226. doi: 10.1038/s41467-018-03538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grosser G, Bennien J, Sánchez-Guijo A, Bakhaus K, Döring B, Hartmann M, et al. Transport of steroid 3-sulfates and steroid 17-sulfates by the sodium-dependent organic anion transporter SOAT (SLC10A6) J Steroid Biochem Mol Biol. 2018;179:20–25. doi: 10.1016/j.jsbmb.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 39.Augier E, Barbier E, Dulman RS, Licheri V, Augier G, Domi E, et al. A molecular mechanism for choosing alcohol over an alternative reward. Science. 2018;360:1321–6. doi: 10.1126/science.aao1157. [DOI] [PubMed] [Google Scholar]
- 40.Ponomarev I, Wang S, Zhang L, Harris RA, Mayfield RD. Gene coexpression networks in human brain identify epigenetic modifications in alcohol dependence. J Neurosci. 2012;32:1884–97. doi: 10.1523/JNEUROSCI.3136-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dengler VL, Galbraith M, Espinosa JM. Transcriptional regulation by hypoxia inducible factors. Crit Rev Biochem Mol Biol. 2014;49:1–15. doi: 10.3109/10409238.2013.838205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Firth JD, Ebert BL, Ratcliffe PJ. Hypoxic regulation of lactate dehydrogenase A. Interaction between hypoxia-inducible factor 1 and cAMP response elements. J Biol Chem. 1995;270:21021–7. doi: 10.1074/jbc.270.36.21021. [DOI] [PubMed] [Google Scholar]
- 43.Villar D, Ortiz-Barahona A, Gómez-Maldonado L, Pescador N, Sánchez-Cabo F, Hackl H, et al. Cooperativity of stress-responsive transcription factors in core hypoxia-inducible factor binding regions. PLoS ONE. 2012;7:e45708. doi: 10.1371/journal.pone.0045708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heilig M, Koob GF, Ekman R, Britton KT. Corticotropin- releasing factor and neuropeptide Y: role in emotional integration. Trends Neurosci. 1994;17:80–85. doi: 10.1016/0166-2236(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 45.Gilpin NW, Roberto M. Neuropeptide modulation of central amygdala neuroplasticity is a key mediator of alcohol dependence. Neurosci Biobehav Rev. 2012;36:873–88. doi: 10.1016/j.neubiorev.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ferreira VMM, Takahashi RN, Morato GS. Dexamethasone reverses the ethanol-induced anxiolytic effect in rats. Pharm Biochem Behav. 2000;66:585–90. doi: 10.1016/s0091-3057(00)00255-0. [DOI] [PubMed] [Google Scholar]
- 47.Centanni SW, Bedse G, Patel S, Winder DG. Driving the downward spiral: alcohol-induced dysregulation of extended amygdala circuits and negative affect. Alcohol Clin Exp Res. 2019;43:2000–13. doi: 10.1111/acer.14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grant BF, Goldstein RB, Saha TD, Chou SP, Jung J, Zhang H, et al. Epidemiology of DSM-5 alcohol use disorder: results from the national epidemiologic survey on alcohol and related conditions III. JAMA Psychiatry. 2015;72:757–66. doi: 10.1001/jamapsychiatry.2015.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirson D, Khom S, Rodriguez L, Wolfe SA, Varodayan FP, Gandhi PJ, et al. Sex differences in acute alcohol sensitivity of naïve and alcohol dependent central amygdala GABA synapses. Alcohol Alcohol. 2021;56:581–8. doi: 10.1093/alcalc/agab034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Witt ED. Puberty, hormones, and sex differences in alcohol abuse and dependence. Neurotoxicology Teratol. 2007;29:81–95. doi: 10.1016/j.ntt.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 51.Sloan ME, Gowin JL, Janakiraman R, Ester CD, Stoddard J, Stangl B, et al. High-risk social drinkers and heavy drinkers display similar rates of alcohol consumption. Addict Biol. 2020;25:e12734. doi: 10.1111/adb.12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data on which conclusions are based are included either in the main manuscript or in supplemental materials. The ATAC-seq (accession number GSE 171399) and RNA-seq (accession number GSE 171400) data sets have been deposited at gene expression omnibus (GEO) of NCBI (SuperSeries GSE accession number 171412).