Abstract
Galactose is a secondary fermentable sugar that requires specific regulatory and structural genes for its assimilation, which are under catabolite repression by glucose. When glucose is absent, the catabolic repression is attenuated, and the structural GAL genes are fully activated. In Saccharomyces cerevisiae, the GAL pathway is under selection in environments where galactose is present. However, it is unclear the adaptive strategies in response to long-term propagation in galactose as a sole carbon source in laboratory evolution experiments. Here, we performed a 4,000-generation evolution experiment using 48 diploid Saccharomyces cerevisiae populations to study adaptation in galactose. We show that fitness gains were greater in the galactose-evolved population than in identically evolved populations with glucose as a sole carbon source. Whole-genome sequencing of 96 evolved clones revealed recurrent de novo single nucleotide mutations in candidate targets of selection, copy number variations, and ploidy changes. We find that most mutations that improve fitness in galactose lie outside of the canonical GAL pathway. Reconstruction of specific evolved alleles in candidate target of selection, SEC23 and IRA1, showed a significant increase in fitness in galactose compared to glucose. In addition, most of our evolved populations (28/46; 61%) fixed aneuploidies on Chromosome VIII, suggesting a parallel adaptive amplification. Finally, we show greater loss of extrachromosomal elements in our glucose-evolved lineages compared with previous glucose evolution. Broadly, these data further our understanding of the evolutionary pressures that drive adaptation to less-preferred carbon sources.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00239-022-10079-9.
Keywords: Experimental evolution, Galactose, Saccharomyces cerevisiae
Introduction
Catabolic repression–the inability to utilize a less-preferred carbon source when a more favorable one is present–is a core principle of metabolic gene regulation. The standard example is the lac operon in E. coli where expression of the structural genes needed to assimilate alternative carbon sources require that two conditions be met: the absence of the preferred carbon source and the presence of the alternative carbon source (Jacob and Monod 1961). This logic is used to regulate the assimilation of a variety of secondary fermentable (e.g., sucrose, maltose, and galactose) and non-fermentable (e.g., acetate, glycerol, and ethanol) carbon sources (Zaman et al. 2008; Fendt and Sauer 2010).
In the yeast, Saccharomyces cerevisiae, galactose is a widely used secondary carbon source. Galactose is a common sugar found in dairy products, fruits, grains, and vegetables (Acosta and Gross 1995; Lee et al. 2011). Galactose assimilation requires three regulatory genes and four structural genes, which are expressed only when glucose is absent and galactose is present. In the absence of glucose, the Snf1 complex attenuates catabolite repression by phosphorylating the Mig1 transcriptional repressor, leading to its inactivation (Conrad et al. 2014; Nair and Sarma 2021). Full activation of galactose structural genes is mediated by a transcription factor (Gal4p), a repressor (Gal80p), and a co-inducer (Gal3p). In the absence of galactose, Gal80p sequesters Gal4p in the cytoplasm. When present, galactose binds to Gal3p, which in turn binds to Gal80p, releasing Gal4p. Once released, Gal4p relocalizes to the nucleus and increases up to ~ 1000-fold the transcription of the GAL structural genes, which are clustered in a 7 kb region on Chromosome II (Sellick et al. 2008).
Galactose initially enters the cell through low-affinity hexose transporters (Escalante-Chong et al. 2015). However, once the switch occurs, galactose enters the cell predominantly through the high-affinity galactose transporter Gal2 (Conrad et al. 2014). Once in the cell, three main enzymes are necessary to catalyze four sequential steps in galactose assimilation. Gal1p is the galactokinase. Gal10p contains two catalytic domains: a mutarotase that interconverts galactose enantiomers and an epimerase that converts UDP-galactose to UDP-glucose. Gal7p is the galactose-1-phosphate uridylyltransferase. These enzymes are tightly regulated to reduce their costly expression and avoid accumulation of the toxic intermediate galactose-1-phosphate (Slepak et al. 2005; Wang et al. 2015).
The genetic switch from glucose to galactose utilization has been extensively studied as a model system to understand the evolution of gene clustering (Hittinger and Carroll 2007, Lang and Botstein 2011, Harrison et al. 2021), transcriptional regulation (van denBrink et al. 2009; New et al. 2014; Peng et al. 2015), nutrient-sensing metabolism (Escalante-Chong et al. 2015; Lee et al. 2017), and cellular memory (Acar et al. 2005; Sood and Brickner 2017). The clustering and regulation of the GAL pathway has evolved independently multiple times in fungi. Relative to other yeast, S. cerevisiae evolved strong repression and slow induction of the GAL genes, which confers a fitness advantage in environments where glucose is in excess (Harrison et al. 2021). However, studies on natural environments identified novel strategies in S. cerevisiae isolates that are associated with improved growth on galactose. For example, natural variation in catabolite repression varies with the length of diauxic lag such that galactose is only assimilated once glucose drops below a certain threshold (Wang et al. 2015). Lineages isolated from dairy products harbor GAL variation, such as GAL3 polymorphisms, GAL2 amplification, and GAL cluster introgressions from S. paradoxus reflecting adaptation to a galactose-rich environment (Duan et al. 2018; Legras et al. 2018; Boocock et al. 2021). These suggest that the structural and regulatory genes in the GAL pathway are under selection in environments where galactose is present. However, natural environments are complex and variable environments; therefore, it is unclear how selection acts to improve the growth in a simple environment containing galactose as the sole carbon source.
Experimental evolution of microbes is a powerful tool to identify genomic changes during adaptation in different environments (Voordeckers et al. 2015; Swamy and Zhou 2019). The molecular targets of selection can be informative on the pressure that drive adaptation in each system. For example, in glucose-limitation laboratory evolution, recurrent mutations arise in nutrient signaling pathways, such as the Ras/PKA pathway and TOR pathway (Venkataram et al. 2016). In chemostat, amplification of glucose transporters is another recurrent adaptive mechanism (Kao and Sherlock 2008; Wenger et al. 2011). In glucose-rich laboratory evolution, genetic targets of selection are involved in cell wall biogenesis, assembly, and cytokinesis, as well as nutrient sensing and signaling (Lang et al. 2013; Fisher et al. 2018; Marad et al. 2018; Johnson et al. 2021). Previous laboratory evolution experiments in galactose have identified mutations in genes encoding the transcriptional repressor GAL80 (Quarterman et al. 2016), proteins involved in polarized growth (Jerison et al. 2020), and regulators of the Ras/PKA pathway (Hong et al. 2011; Hong and Nielsen 2013). In contrast to variation found in galactose-rich natural environments, mutations identified in laboratory evolution are predominantly outside the canonical GAL genes.
Previously, we evolved 48 diploid populations in glucose over 4,000 generations (Marad et al. 2018). Here, we perform an identical 4,000-generation diploid evolution experiment using galactose as the sole carbon source. We directly compare the rates of adaptation and genetic targets of selection between glucose and galactose evolution. Consistent with previous work, we find that most mutations that improve fitness in galactose lie outside of the canonical GAL pathway. In addition, we identify recurrent copy number variants, including aneuploidies on Chromosome VIII. Reconstruction of specific evolved alleles in SEC23 and IRA1 showed significant increases in fitness under galactose conditions. Finally, we show multiple losses of extrachromosomal elements, two of which (2-micron plasmid and Killer phenotypes) were also observed in galactose-evolved populations, and one of which (mitochondrial genome) was not.
Results
Higher Rate of Adaptation in Rich Galactose Medium Compared to Rich Glucose Medium
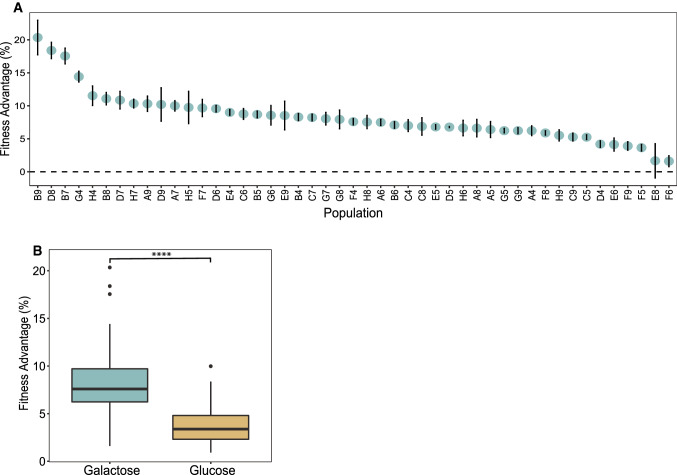
To determine the evolutionary response to constant growth in an alternative carbon source, we evolved 48 replicate populations of the same diploid ancestor in a rich medium with galactose as the sole carbon source (YPGal). After 2,000 generations, all populations increased in fitness compared to the ancestor (Fig. 1A) with a mean fitness gain of 8.2 ± 0.1% (α = 0.05). This is significantly greater than the fitness gains for the identically evolved populations using glucose as a sole carbon source (p < 0.0001, Wilcoxon rank-sum test; Fig. 1B), which showed mean fitness gains of 3.7 ± 0.06% (α = 0.05) in the first 2,000 generations of evolution.
Fig. 1.
Galactose-evolved populations have a higher fitness advantage after 2,000 generations of laboratory evolution. A Relative fitness advantage of 48 evolved populations on galactose. Vertical error bars reflect ± standard error of the regression. B Adaptation in galactose has a significantly higher fitness advantage compared with glucose-evolved populations (p < 0.0001, Wilcoxon rank-sum test). Box plot reflects the mean fitness of 48 populations in both conditions. Galactose populations have a mean fitness advantage of 8.2%, whereas glucose populations have a mean of 3.7%. Glucose fitness data are from Marad et al. 2018
To identify mutations that arose in the galactose-evolved populations, we sequenced two clones from each of the 48 populations at Generation 4,000. We found 2,406 de novo mutations across 96 evolved clones distributed across all 16 chromosomes with a mean of 25 ± 6 mutations per clone. Of these mutations, 794 are present in both clones from the same population, and 735 mutations are only found in one clone.
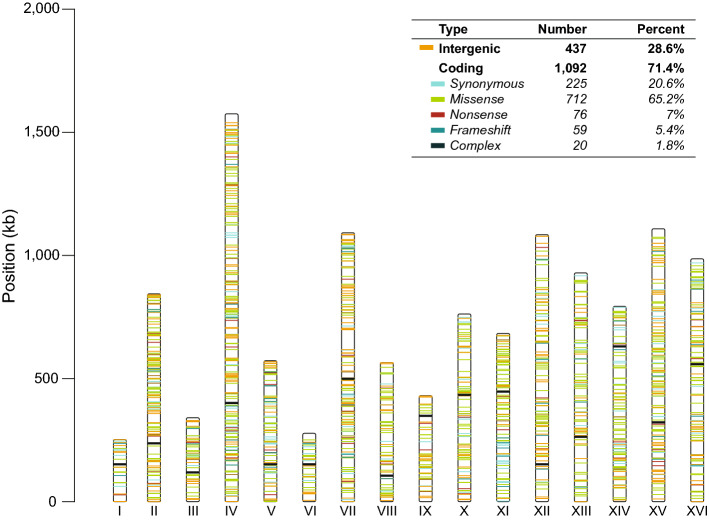
Of the 1,529 unique mutations, 91% are single nucleotide polymorphisms, most of which are in coding genes (1092 out of 1529; 71.4%). 712 of these mutations are mostly missense (46.6%), 225 are synonymous (14.7%), 76 are nonsense (5%), 59 are frameshift (3.9%), and 20 are complex mutations (1.3%) (Fig. 2 and Supplementary Data). Consistent with our previous work (Fisher et al. 2018; Marad et al. 2018), we find that heterozygous mutations outnumber homozygous mutations ~ 19:1 (Supplementary Fig. 1A), and that homozygous mutations are clustered on the right arm of Chromosome XII (Supplementary Fig. 1B). This region is a hotspot for loss of heterozygosity (LOH) due to the rDNA locus: approximately one third of the homozygous mutations are located on this chromosome.
Fig. 2.
The spectrum of mutations in 96 evolved clones under galactose evolution. Distribution of 1,529 unique evolved mutations across 16 Chromosomes. The vertical bars represent each Chromosome labeled by Roman numeral. The horizontal lines reflect each mutation colored based upon their protein-coding effect (top box). Other mutations group conservative inframes, disruptive inframes, and complex mutations. Centromers are represented with a black horizontal line
Beneficial Mutations are Commonly Found Outside the Canonical GAL Pathway
To identify the pathways under selection in galactose medium we performed Gene Ontology (GO) enrichment using all 860 coding genes that were mutated across all populations. This analysis showed, at a broad level, an enrichment for genes involved in: positive regulation of the metabolic process, cellular response to stimulus, intracellular signal transduction, and cellular component organization (biological process enrichment, p < 0.001). We next restricted our GO-term enrichment analysis to the 24 genes that acquired mutations in three or more populations. This showed enrichment for genes involved in the negative regulation of Ras signal transduction including IRA1, IRA2, and BEM2 (p < 0.005). In addition, we found mutations in GPB1, PDE2, and CDC25 in one or two populations. All the mutations found in Ras pathway genes are nonsynonymous.
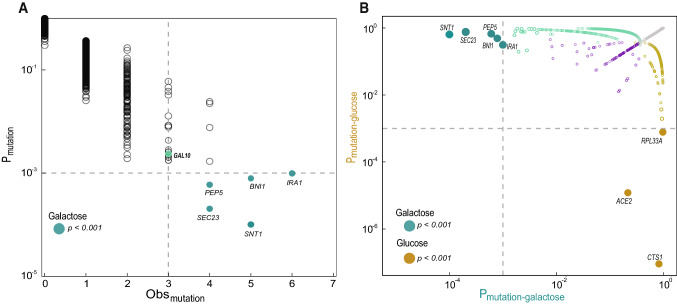
We identified individual genes that are targets of selection by determining which genes are mutated more often than expected by chance across replicate populations. Using this approach, we identified five targets of selection with a p < 0.001 (Fig. 3A). These five genes are involved in the Ras/PKA pathway (IRA1), histone deacetylation (SNT1), endosome organization/vacuolar biogenesis (PEP5), secretory pathway (SEC23), and actin filament (BNI1). Notably, we did not find a GO enrichment or a significant number of mutations in genes with an obvious connection to galactose metabolism. We found two missense and one conservative inframe in GAL10, two missense mutations in GAL2, one missense mutation in GAL4, one missense mutation in GAL7, one nonsense and one synonymous in GAL1. None of these genes, however, were acquired more mutations than expected by chance at p < 0.001. None of the candidate targets of selection in galactose are significantly overrepresented in glucose evolution experiment and vice versa (Fig. 3B). The most common targets of selection in glucose-evolved populations are cell wall-associated genes, CTS1, ACE2, and RPL33A (p < 0.001, Poisson distribution; Fig. 3B). Of these, only one mutation in ACE2 was observed in one galactose-evolved population.
Fig. 3.
Common targets of selection in galactose-evolved populations. A Plotted on the x-axis is the observed number of coding sequence (CDS) mutations in each of the 5800 genes. The y-axis is the probability that the observed number of CDS mutations in each gene (without mutation type distinction) occurred by chance. We used a Poisson distribution weighted for the length of each gene (Fisher et al. 2018). The gray dotted line indicates the cut-off of genes with 3 or more CDS mutations with corresponding p-value < 0.001 (solid dark cyan). Genes in this area are common targets of selection. We also denoted gal10 with p-value = 0.002 (solid light green). B Common targets of selection in galactose-evolved populations differ from glucose-evolved populations. Plotted is the ratio of the probability that the observed number of CDS mutations in a gene occurred by chance in galactose populations (cyan circles) versus glucose populations (gold circles). The gray dotted line indicates the cut-off genes with a p-value < 0.001. Targets of selection were labeled with the gene name and bigger solid dark cyan circles. Open purple circles indicate CDS mutated in both conditions. Gray circles indicate CDS that were not mutated under any conditions. Glucose data are from Marad et al. 2018
Based on the types of mutations observed, we can determine how selection is acting on the candidate targets of selection (Table 1). We observed enrichment for nonsense and frameshift mutations (high impact mutations) in SEC23, PEP5, and IRA1, suggesting selection for loss-of-function. In contrast, we only observe missense mutations in SNT1. Out of the five mutations found in BNI1, three are synonymous and conservative inframe mutations, suggesting that this locus may not have a strong effect in the adaptation of galactose. Though there are a few notable exceptions of beneficial synonymous mutations in other systems (Bailey et al. 2014; Kristofich et al. 2018), most synonymous mutations in our system are neutral (Buskirk et al. 2017) and reach high frequency by genetic hitchhiking (Lang et al. 2013). Ignoring synonymous mutations removes a large background of hitchhiker mutations and produces more robust predictions of true targets of selection. If we restrict our analysis to nonsynonymous mutations, PEP5 and BNI1 are no longer significant at p < 0.001 (Supplementary Data).
Table 1.
Common targets of selection in galactose-evolved populations
| Gene | p-valuea | Amino acid change | Zygosity | Cloneb | Biological process (SGD) |
|---|---|---|---|---|---|
|
SNT1 (Chr III) |
4.55–04 |
Ala168Serd Val237Ile Val374Leu Arg1009Lysd |
Heterozygous Heterozygous Heterozygous Heterozygous |
F51, F52 B42 E81, E82 A91, A92 |
Subunit of the Set3C deacetylase complex |
|
SEC23 (Chr XVI) |
8.01–05 |
Gln167c Gln494c Arg592Iled Leu701fs |
Heterozygous Heterozygous Heterozygous Heterozygous |
F41, F42 A91, A92 G61, G62 H61, H62 |
COPII vesicle coating |
|
IRA1 (Chr II) |
2.84–04 |
Ser700Prod Gln933c Pro1013fs Leu1402c Gln2083c Ala2292Ser |
Homozygous Homozygous Heterozygous Heterozygous Heterozygous Heterozygous |
C41, C42 H81, H82 A91, A92 G91, G92 A61, A62 B51, B52 |
Inhibitory Regulator of the RAS-cAMP pathway |
Bolded clones indicate the SNP was found in all clones of the population
aStatistical analysis focus on nonsynonymous mutations
bClones indicate the well ID followed by the clone number
cNonsense
dReconstructed mutations
fs Frameshift
Galactose-Evolved Alleles Show Carbon Source Dependence Effect
To quantify the fitness effect of individual mutations, we reconstructed evolved mutations in putative targets of selection (snt1-A168S, snt1-R1009K, sec23-R592I, ira1-S700P, Table 1) as well as evolved mutations in the galactose pathway (gal10-D302N, gal10-R433P, gal7-Q265E, and gal1-G347*, Supplementary Fig. 3B) even though our statistical analysis did not identify them as candidate targets of selection. Each mutation was reconstructed in the ancestral background as haploids, heterozygous diploids, and homozygous diploids. As a control, we constructed two previously identified adaptive mutations, hsl1-A262P (Buskirk et al. 2017; Vignogna et al. 2021), and ace2-R669* (Marad et al. 2018) from haploid-glucose evolution and diploid-glucose evolution, respectively.
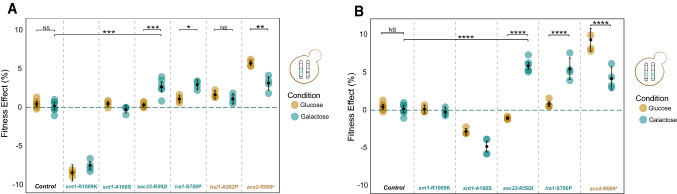
We performed competitive fitness assays of the reconstructed strains against a fluorescently labeled ancestral reference in both galactose and glucose. Reconstruction of heterozygous evolved mutations in the GAL pathway does not show fitness advantage in galactose (Supplementary Fig. 3B). We observed the same pattern for the homozygous reconstruction. In contrast, for all evolved mutations identified in our candidate targets of selection, we find that the fitness effects in glucose and galactose are highly correlated (r = 0.69, p = 0.0122, Pearson correlation coefficient; Supplementary Fig. 2). Nevertheless, the magnitude of the fitness effect was greater in the condition in which the specific mutation arose, and this difference is more exaggerated for homozygous mutations. We found a fitness advantage of the heterozygous sec23-R592I mutation of 3.9 ± 0.4% (α = 0.05; p < 10−6 one-way ANOVA, Tukey's HSD test) in galactose than glucose and a larger fitness advantage of 6.2 ± 0.5% (α = 0.05; p < 10−8, one-way ANOVA, Tukey's HSD test) as homozygous in galactose compared to glucose. We observed similar fitness advantage for ira1-S700P mutation; 3.3 ± 0.3% (α = 0.05; p < 10−3, one-way ANOVA, Tukey’s HSD test) as heterozygous and 5.3 ± 0.3% (α = 0.05; p < 10−8, one-way ANOVA, Tukey's HSD test) as homozygous (Fig. 4B). Despite the greater fitness benefit of the homozygote mutations, only ira1-S700P lost heterozygosity in the evolution experiment. Surprisingly, neither of the two SNT1 alleles provided a selective benefit in galactose or glucose. The heterozygous snt1-R1009K mutation is strongly deleterious, but it is neutral as a homozygote. The snt1-A592S shows the opposite pattern: the heterozygous mutation is neutral but homozygous mutation is deleterious (Fig. 4). As expected, the ploidy, glucose-evolved alleles of hsl1-A262P and ace2-R669* are more beneficial in glucose although both alleles also improved fitness in galactose (Fig. 4 and Supplementary Fig. 3). Our results show that evolved mutations on SEC23 and IRA1 are beneficial only in the condition in which they arose.
Fig. 4.
Galactose-evolved mutations show variation in fitness effect and carbon source dependence. Average fitness effects of the reconstructed mutations in snt1 (n = 4), sec23 (n = 6), and ira1 (n = 4) in both zygosities; A heterozygous and B homozygous mutations. Fitness effect were compared with wild-type version (n = 6), hsl1 mutation (n = 4) from glucose, haploid evolution (Fisher et al. 2018), and ace2 mutation (n = 4) from glucose, diploid-evolution (Marad et al. 2018). We were not able to generate hsl1 homozygous mutation. The sec23-R592I has a significant fitness gain in galactose compare to glucose. Asterisk (****) indicate p < 10–8, (***) p < 10–5, (**) p < 10–4, (*) p < 10–3, and NS: not significant; one-way ANOVA, tukey's HSD test. Error bars are the s.e.m
Aneuploidies and CNVs are Common in Galactose-Evolved Populations
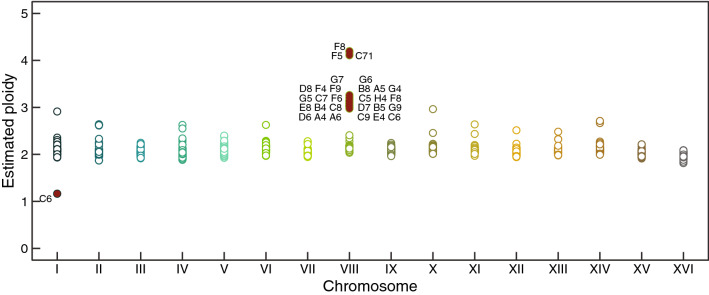
In addition to point mutations, copy number variants (CNVs) and aneuploidies can contribute to adaptation (Gresham et al. 2008; Yona et al. 2012; Sunshine et al. 2015). To identify CNVs and aneuploidies in our galactose-evolved populations, we calculated the relative coverage across all 16 chromosomes. We find that most of our evolved clones (28/46; 61%) have one or two extra copies of Chromosome VIII. In addition, we find one population with monosomy for Chromosome I (Fig. 5). We validated these findings by sequencing eight random clones by long-read sequencing (Supplementary Fig. 4A). Although the high occurrence of additional copies for Chromosome VIII strongly suggests an adaptive strategy under galactose, populations with extra copies do not correlate with higher fitness than euploid populations (p = 0.56, Wilcoxon rank-sum test; Supplementary Fig. 4B).
Fig. 5.
Aneuploidy on Chromosome VIII appears to be a parallel evolutionary adaptation in the galactose condition. To estimate the ploidy of 96 clones, we calculated the median coverage across each chromosome compared to genome-wide coverage. Baseline ploidy is 2 N. Aneuploidies are shown as filled red circles and labeled by name population. Empty circles indicate euploidy. Notably, duplication of Chromosome VIII occurred in 28 populations out of 46 populations
In addition to loss or gain of entire chromosomes, we also detect large (> 40 kb) CNVs in our evolved clones, including eight unique amplifications and two deletions (Table 2 and Supplementary Fig. 5). We identified several populations with CNVs in the right arm of Chromosome II; however, in only population does the amplification include the GAL1-GAL10-GAL7 gene cluster (Population A4, Chromosome II: 268,984–275,164). No other recurrent CNVs were identified that overlap with GAL genes (Table 2 and Supplementary Fig. 5). In Populations A4 and B9, we identified duplication of genes associated with Ras/PKA signaling pathway; IRA1 (Chromosome II: 534,751–544,029) and RAS2 (Chromosome XIV: 440,898–441,866), respectively. Additionally, we found amplifications of regions containing hexose transporters genes; HXT14 (Population B9), HXT9, HTX8, HXT16 (Population G4), HXT4, HXT1, and HXT5 (populations with aneuploidies for Chromosome VIII). These data demonstrate recurrent amplifications of regions that contain genes encoding hexose transporters, suggesting a potential adaptive mechanism in our galactose evolution. We do not, however, find a significant correlation between higher fitness and CNVs (p = 0.89, Wilcoxon rank-sum test; Supplementary Fig. 4B).
Table 2.
Structural variants in galactose-evolved populations
| Chr | Start (kb) | Stop (kb) | Length (kb) | Copy number | Class | Cloneb |
|---|---|---|---|---|---|---|
| I | 0 | 161,000 | 161 | 1 N | Aneuploidy | C61, C71c |
| II1,3 | 226,000 | 844,051 | 618 | 3 N | CNV | A41, A42c |
| II | 666,000 | 836,000 | 170 | 3 N | CNV | B61, B62 |
| II3 | 354,000 | 836,000 | 482 | 3 N | CNV | H71, H72 |
| II | 737,000 | 844,051 | 107 | 1 N | CNV | G71, G72 |
| VII | 749,000 | 1,092,105 | 343 | 3 N | CNV | D51, D52 |
| VIII2 | 0 | 844,051 | 844 | 3 N & 4 N | Aneuploidy | G71, G72, G61, G62, D81, D82, F41, F42, F91, F92, B81, B82, A51, A52, G41, G42, G51, G52, C71c, C61c, F61c, F62c, F71c, C51, C52, H41, H42, F81, F82, E81, E82, B41, B42, C81, C82, D71, D72, B51, B52, G91, G92, D61, D62, A41c, A42c, E71c, E72c, A61, A62, C91, C92, E41, E42, C61c, C721c, D91, D92 |
| X2 | 502,000 | 762,303 | 260 | 4 N | CNV | G41,G42 |
| XI | 0 | 297,000 | 297 | 3 N & 4 N | CNV | H91, H92, H82 |
| XIII | 379,000 | 750,000 | 371 | 3 N | CNV | F52 |
| XIII | 0 | 44,000 | 44 | 3 N | CNV | D41, D42 |
| XIV2,3 | 0 | 479,001 | 479 | 3 N | CNV | B91, B92 |
Bolded population indicates that the CNV was found in all clones of the population
bClones indicate the well ID followed by the clone number
cPopulations with cross-contamination (See Supplementary Data)
1Duplication of the GAL cluster. Coordinates: chrII. 268,984–275,164, W303 ref
2Duplication of hexose transporters
3Duplication of components of the Ras/PKA pathway
Changes in Copy Number of Extrachromosomal Elements and Reduction of the Killer Phenotype in Galactose-Evolved Populations
In addition to evolution in the nuclear genome, we determined the extent to which cytoplasmic elements (the mitochondrial genome, the 2-micron plasmid, and the yeast dsRNA Killer virus) changed during evolution in galactose. Using read depth as a proxy, we estimated the copy number of mitochondrial DNA for each clone according to Chiara et al. 2020. Estimates of mitochondrial copy number is highly correlated between clones from the same population (r = 0.928, p < 0.0001, Pearson correlation; Supplementary Fig. 7A). Most of the populations show a significant decrease in the mitochondrial copy number (p < 0.0001, Wilcoxon rank-sum test), with a global mean of ~ 8:1 mitochondrial to nuclear genomes (Supplementary Fig. 6A) and with only six galactose-evolved populations maintaining or increasing mitochondrial copy number (19:1 ratio between mitochondrial and nuclear genomes). In contrast mitochondrial copy number is maintained throughout evolution in glucose-evolved populations (Supplementary Fig. 7C).
Like mitochondrial copy number, our estimates of the 2-micron plasmid copy number are highly correlated between clones from the same population (r = 0.66, p < 0.0001, Pearson correlation; Supplementary Fig. 7B). Our galactose-evolved populations show a drastic loss of 2-micron copy number (from 133 initial copies to an average of 57 copies per every copy of the nuclear genome). Only three populations showed a higher 2-micron copy number than the ancestor (Supplementary Fig. 6B). We did not observe differences in reduction of 2-micron copy number between galactose and glucose-evolved populations (p = 0.43, Wilcoxon rank-sum test; Supplementary Fig. 7D). We also did not find correlation between reduction of mitochondrial DNA and 2-micron (r = -0.07, p = 0.48, Pearson correlation).
As a proxy for the loss of the killer toxin, we quantified killing ability in our populations (Buskirk et al. 2020). Similar with glucose evolution, we find that the majority of the populations (42/48) lost totally or partially the killer-associated phenotype (Supplementary Fig. 6C, Supplementary Fig. 8). We did not observe a significant difference of the loss of the killer phenotype between both conditions (p = 0.17, Wilcoxon rank-sum test). Consistent with previous glucose evolution, reduction of the 2-micron and killing ability is also a common mechanism in our galactose evolution adaptation. In contract, mitochondrial DNA has recurrent losses only in galactose evolution.
Discussion
The genetic switch from glucose to galactose utilization in S. cerevisiae has been extensively studied (Acar et al. 2005, Ronen and Botstein 2006, van den Brink et al. 2009, Escalante-Chong et al. 2015). However, the strategies for long-term adaptation to galactose as a sole carbon source are not well understood. To determine how galactose affects adaptation in budding yeast, we evolved 48 diploid populations in rich galactose medium under identical conditions to our previous glucose evolution experiment (Marad et al. 2018). We show that fitness gains over the first 2,000 generations were greater in the galactose medium compared to glucose. We find that beneficial mutations in the galactose-evolved populations are beneficial in glucose medium but that the magnitude of the benefit is smaller. Similarly, glucose-evolved mutations are strongly beneficial in glucose, but beneficial in galactose, suggesting that environment-specific selective pressures drive the fixation of specific mutations in each environment. These results are consistent with findings of others showing a positive correlation between fitness in glucose and fitness in galactose (Chen and Zhang 2020; Jerison et al. 2020).
To identify the targets of selection during long-term growth on galactose, we sequenced two clones from each population after 4,000 generations. Several studies have identified functional variants in the GAL pathway that tune the response of the galactose pathway in natural populations (Roop et al. 2016; Lee et al. 2017; Boocock et al. 2021). It is, therefore, reasonable to expect laboratory adaptation to constitutive galactose to select for mutations in these same genes. While we did observe several mutations in the GAL pathway genes, we do not observe more mutations than expected by chance, and reconstruction experiments failed to identify a fitness advantage (Supplementary Fig. 3B).
We identify candidate targets of selection outside of the canonical galactose pathways, with SNT1, SEC23, and IRA1 as the most significant hits. IRA1 is repeatedly observed as a target of selection across evolution experiments (Lang et al. 2013; Fisher et al. 2018; Li et al. 2018; Johnson et al. 2021), whereas SNT1 is rarely observed (Fisher et al. 2018). SEC23 has not been previously identified as recurrent mutated gene in glucose. These three genes show enrichment of missense and nonsense mutations consistent with being under selection in galactose. In diploid evolution experiments, beneficial mutations are partially dominant or overdominant (Aggeli et al. 2022). Reconstruction of evolved alleles shows that ira1-S700P and sec23-R592I are partially dominant. Consistent with being partially dominant, we find that the allele ira1-S700P fixed as a homozygote. It is not surprising that sec23-R592I did not undergo loss of heterozygosity since the majority of mutations in the evolved clones are heterozygous (1,444 of 1,529 mutations).
IRA1 is a negative regulator of the Ras/PKA signaling pathway, one of the best-known glucose-triggered signaling cascades (Tamanoi 2011; Broach 2012). The Ras pathway is a hotspot for functional variation owing to its role in regulating metabolic, transcriptional, and physiological responses to nutrient availability. Because of this, genes in the Ras pathway are common targets of selection in nearly all yeast evolution experiments including rich glucose medium (Lang et al. 2013), defined low glucose medium (Venkataram et al. 2016), as well as glucose-limited (Kvitek and Sherlock 2013) and nitrogen-limited (Hong and Gresham 2014) chemostats.
Mutations identified in experimental evolution are often pleiotropic, affecting fitness (either positively or negatively) in other environments (Ostrowski et al. 2005; Jerison et al. 2020; Bakerlee et al. 2021). Mutations in the Ras pathway have been shown to result in fitness tradeoffs under alternative carbon sources (Hong et al. 2011; Hong and Nielsen 2013) or prolonged starvation (Li et al. 2018). Here, we find that the galactose-evolved mutation, ira1-S700P, has a greater fitness benefit in galactose compared in glucose. These results suggest that though the Ras/PKA pathway is a general hub for adaptive mutations in all laboratory evolution experiments, individual mutations are condition specific, adjusting cell physiology match the environment.
Outside of the Ras/PKA pathway, the secretory pathway is another common target of selection in galactose. Apart from recurrent mutations in SEC23, we observe independent moderate-impact mutations in six more genes that encode secretory proteins. The endoplasmic reticulum has a role in maintaining metabolic homeostasis by sensing nutrient availability (Fu et al. 2012). Another common target of selection is SNT1, which encodes a histone deacetylase. However, despite SNT1 being a common target of selection, neither of our reconstructed SNT1 mutations are beneficial on their own (Fig. 4). It is possible the presence of epistasis interactions between SNT1 mutations and the genetic background. The snt1-A1009S and snt1-V237I mutations arose in populations with mutations in ira1 and ira2, respectively. Genetic interaction among co-evolved mutations is common in experimental evolution (Fisher et al. 2019; Vignogna et al. 2021).
Increasing the copy number of specific genes whose expression level is limiting is another common mode of adaptive evolution. For example, the amplification of transporter and specific permeases are a recurrent adaptive mechanism to selection in medium limited for glucose (Gresham et al. 2008; Kao and Sherlock 2008), nitrogen (Hong and Gresham 2014), sulfur (Sunshine et al. 2015), or raffinose (Scott et al. 2017). An essential step in utilizing galactose is its transport achieved by Gal2p, a closely related hexose transporter (Boles and Hollenberg 1997). Notably, we did not detect any duplications of the transporter GAL2 located on Chromosome XII. Of the four CNVs on Chromosome II, only the largest one encompasses the GAL1-GAL10-GAL7 gene cluster (Supplementary Fig. 5). We do, however, find that 61% of our populations have fixed aneuploidies on Chromosome VIII. We suggest that a strong adaptive advantage in our populations can be explained by the amplification of hexose transporters on Chromosome VIII: HXT4, HXT1, and HXT5. Under galactose conditions, selection for spontaneous duplication of Chromosome VIII may favor nutrient transporters, leading to more efficient nutrient uptake into central metabolism (Torres et al. 2007). We only detected one point mutation in any of the HXT genes (a single missense mutation in HXT3).
Most of the observed adaptive processes have focused on the nuclear genome. Little is known about the evolution of extrachromosomal elements in different environments. Changes in copy number can give us a first light of whether these elements are under selection or not. Recently, Johnson et al. 2021 showed the recurrent loss of 2-micron plasmids and reduction of the killer phenotype of the Killer virus under rich glucose and synthetic complete media. Here we find a similar pattern in 2-micron plasmids and Killer-associated phenotypes in galactose-evolved populations (Supplementary Fig. 6). The selective benefit decreasing 2-micron copy number are unclear; however, we previously showed that the loss of Killer virus occurs due to an intracellular fitness advantage of viruses that do not produce toxin (Buskirk et al. 2020).
We observe multiple losses of mitochondrial DNA (Supplementary Fig. 6). When growing on galactose, S. cerevisiae does not exhibit a Kluyver effect (inability to grow on and ferment sugars under anaerobic conditions). Therefore, it can consume galactose under respiration and fermentation (van den Brink et al. 2009). The existence of this respiration-dependent assimilation of sugars is an essential link between mitochondria and sugar utilization (Quarterman et al. 2016). The reduction of mitochondrial genome copy number may indicate that the evolved populations are increasing fermentation relative to respiration. Mitochondrial genome copy number, however, is dynamic with considerable strain-to-strain variation (Galeota-Sprung et al. 2022). We, therefore, cannot determine if the changes we observe are adaptive or are a secondary response to changes in growth.
Overall, we find that long-term adaptation to growth on galactose is driven mainly by mutations outside of the canonical GAL pathway. Specifically, we identify Ras/PKA signaling as a major target of selection, as it is in nearly all environments. We find that glucose and galactose-evolved alleles are more beneficial in the condition in which they arose. Unlike in glucose-evolved populations, we find recurrent aneuploidy for Chromosome VIII suggestion a potential role in galactose adaptation by increasing the copy number of HXT genes. We observe a reduction in 2-micron copy number, mitochondrial copy number, and killing ability across our evolved populations.
Methods
Strains Construction
All strains used in this study are derived from the W303 background (ade2-1, CAN1, his3-11, leu2-3,112, trp1-1, URA3, bar1Δ::ADE2, hmlαΔ::LEU2, GPA1::NatMX, ura3Δ::pFUS1-yEVenus). All the derivative strains are identified by their yGIL prefix and the number in the Lang Lab yeast collection. Mutant strains were constructed using CRISPR-Cas9 as described previously (Fisher et al. 2019). Repair oligonucleotide templates were generated by amplifying 500 bp gBlocks (IDT) that contained the point mutation and a synonymous PAM site substitution. We co-transformed a high-copy plasmid that contains the Cas9 gene and the guide RNA expression cassette (pML104; Addgene #67,638) (Laughery et al. 2015) and the linear repair template into yGIL432 (MATa) and yGIL646 (MATα) strains. We designed the following sgRNA; snt1-A168S (5ʹ-TGGAGCGTTT GATAATGCCG AGG-3ʹ), snt1-R1009K (5ʹ-ATGGCTCTAT AAGACCATTT GGG-3ʹ), sec23-R592I (5ʹ-TTGGGATCTT CTTAAATAAT AGG-3ʹ), and ira1-S700P (5ʹ-GAAGAGATTC TACAACTTGT TGG-3ʹ). After editing, we removed the plasmid on media containing 5-FOA. Diploid mutants were generated by crossing each mutant strain with the opposite mating type mutant strain or wild-type strain to produce homozygotes and heterozygotes, respectively. Crosses were sporulated to confirm diploid strains. All plasmids and strain construction were confirmed by Sanger sequencing (Genscript).
Evolution Experiment
Long-term evolution was performed as described previously (Marad et al. 2018). Briefly, the ancestral diploid strain, yGIL672, was grown to saturation in YPD (yeast extract, peptone, dextrose) medium. A dilution of 1:210 was used to seed 48 replicate populations in a single round bottom 96-well plate. The cultures were propagated for 4,000 generations in YPGal medium (yeast extract, peptone, 2% galactose) containing ampicillin (100 mg/mL) and tetracycline (25 mg/mL) to prevent bacterial contamination. The cultures were incubated at 30 °C in an unshaken 96-well plate. Every 24 h, the populations were diluted 1:1,024 by serial dilution 1:32 (4 μl into 125 μl) × 1:32 (4 μl into 125 μl) into a new YPGal 96-well plate. This regime corresponds to 10 generations of growth per day at an effective population size of ~ 105. The long-term experimental evolution was performed using the Biomek Liquid Handler. Approximately every 50 generations, populations were cryo-archived in 15% glycerol at − 80 °C.
Competitive Fitness Assays
To measure the effect of evolved mutations we used flow-cytometry-based competitive fitness assay as described previously (Buskirk et al. 2017). We used for the competition the reference strains; yGIL519 (MATa), yGIL699 (MATα), and yGIL702 (MATa/α). These strains have the ancestral background with constitutive ymCitrine integrated at the URA3 locus. For all the evolved alleles, we performed the competition in YPD and YPGal under identical conditions. Diploid and haploid strains were competed in independent 96-well plates in an identical fashion to the evolution experiment. The reference and the experimental strains were mixed 1:1 at Generation 0 using a Biomek Liquid handler. The assays were performed for 50 generations and sampled every 10 generations. To measure density using flow-cytometry (BD FACSCanto II), we transferred 4 µL of each sample into 60µL of PBS and stored at 4 °C for 1 day. Data were analyzed in FlowJo 10.3. The selective coefficient was calculated as the slope of the change in the natural log ratio between query and reference strains. For each fitness measurement, we performed between 4–6 independent biological replicates and one technical replicate.
Whole-Genome Sequencing
Each population, from generation 4,000, was struck to singles colonies on YPGal and two clones were isolated for sequencing. Clones were grown to saturation in YPGal and total genomic DNA was isolated for each sample using phenol–chloroform extraction and ethanol precipitation. We used the Nextera sequencing library preparation kit as described previously (Buskirk et al. 2017). Sequencing was performed on an Illumina HiSeq 2500 sequencer with 150-nucleotide paired-end reads by the Sequencing Core Facility within the Lewis-Sigler Institute for Integrative Genomics at Princeton University.
The Oxford Nanopore MinION Genomic DNA Sequencing Kit (SQK-LSK109) was used to prepare the DNA libraries of 8 evolved clones. Briefly, we performed DNA extraction using a QIAGEN 100/G. One microgram of DNA per sample was diluted in 48µL of water. End-repair was performed using NEBNext FFPE Repair Mix and the product was purified using 60 μl of Agencourt AMPure XP beads. End-prepped gDNA was quantified using the Qubit High Sensitivity assay. dA-tailing was performed using NEBNext dA-tailing module. A ligation reaction was then performed by adding 2.5µL of the Native Barcode (EXP-PBC001 and EXP-PBC096) and 25µL of Blunt/TA Ligase Master Mix. The adapter-ligated DNA was purified using Agencourt AMPure XP beads and washed with long fragment buffer. DNA was quantified using a Qubit and then loaded on to Minion R9.4.1 (product code FLO-MIN106D R9) flow cells.
Sequencing Analysis Pipeline
Raw Illumina sequencing data were concatenated and demultiplexed using barcode_splitter 0.18.6 from L. Parsons (Princeton University). Adapter sequences were trimmed using trimmomatic/0.36 (Bolger et al. 2014) using PE -phred33 parameter. Each sample was aligned to the complete and annotated W303 genome (Matheson et al. 2017) using BWA-MEM, v.0.7.15 (Li and Durbin 2009). Each clone was sequenced to an average depth of approximately 50X coverage (Supplementary Fig. 1). Common variants were called using FreeBayes/1.1.0 (Garrison and Marth 2012), using default parameters. Variants common were filtered using the VCFtools/0.1.15 vcf-isec function (v.0.1.12b). Individual VCF files were annotated using SnpEff/5.0 using -formatEff flag (Cingolani et al. 2012). Manually we did a more punctual filtering of common variants by viewing BAM files using Integrated Genome Viewer (Broad Institute). We removed the variant calls found in low complexity regions (TY elements, centromeres, and telomeric regions) and with less than 5X coverage. Only nuclear mutations were analyzed. We used R package idiogramFISH to plot the distribution of our evolved mutations. Zygosity was determined by establishing a cut-off of mutation frequency values above 0.9 and p-values < 0.001. If both parameters were satisfied, we called those mutations homozygous. Copy Number Variants (CNVs) were identified using ControlFREEC version 11.5 (Boeva et al. 2012). All the.txt outputs were merged into one data frame to remove all the common calls. We applied the following criteria to filtered CNVs: the variant is calling more than 10 times, had length less than 5 kb, and variants that were located close to telomeres and centromeres (5 kb of distance) regions. CNVs and aneuploidies were corroborated by visual inspection of chromosome coverage plots created in R. Briefly, we used samtools-depth to calculate per-site depth from the sorted-bam files. We divided the median chromosome coverage by the median genome-wide coverage using non-overlapping 1000 bp window size and 500 nt step size. The total coverage of the genome was normalized to 2 (diploid population). The same analysis was used to estimate copy number of the 2-micron plasmid. Copy number of mitochondrial DNA was estimated using the same sliding window approach but only from the ATP6, COX2, and COX3 regions (De Chiara et al. 2020) to avoid overcounting the highly repetitive sequences in the mitochondrial genome. For Nanopore sequencing, we performed basecalling and demultiplex barcode using guppy/3.1.5. We aligned reads to the S288C reference using the long-read mapper ngmlr/0.2.7 and identified structural variants using Sniffles (Sedlazeck et al. 2018). To visualize chromosome coverage, we used the same methodology described for Illumina data. For glucose-evolved populations, we performed the same analysis as galactose. However, the whole-genome sequencing was performed at Generation 2,000.
Gene Ontology analysis was conducted using Gene Ontology Term Finder (https://www.yeastgenome.org/) and PANTHER (http://pantherdb.org/), both analysis were conducted on Sep, 2022.
Identification of Common Targets
The p-values of coding genes hit by a mutation just by chance were calculated using the Poisson distribution (Fisher et al. 2018). We determined the probability that chance alone explains the observed number of mutations across all 5,800 genes by assuming a random Poisson distribution of CDS mutations weighted by the length of the gene across the 8,453,525 bp genome-wide CDS. First, we determined p-values of the 1,040 CDS mutations without mutation type distinction. However, to increase the statistical power, we also performed the same statistical analysis but focus on nonsynonymous CDS mutation. Final common targets of selection were defined as genes with three or more nonsynonymous CDS mutations and a corresponding probability of less than 0.1%. To determine if the targets of selection are unique to galactose, we compare the p-values with prior glucose evolution experiments (Marad et al. 2018). To determine the targets of selection for each condition, we calculated the ratio between probabilities (Supplementary Data).
Halo Assay
Killer phenotype was performed using the protocol described previously (Buskirk et al. 2020). Assay was performed using YPD agar that had been buffered to pH 4.5 (citrate–phosphate buffer), dyed with methylene blue (0.003%), and poured into a 1-well rectangular cell culture plate. Killing ability was assayed against a hypersensitive tester strain (yGIL1097). The hypersensitive tester was grown to saturation, diluted 1:10, and spread (150 ml) evenly on the buffered agar. Glucose and galactose-evolved populations were grown to saturation, concentrated five-fold, and spotted (2 µL) using liquid handler (Biomek FX). Plates were incubated at room temperature for 3 days before assessment. Killer phenotype was scored according to the scale as shown in Buskirk et al. 2020.
Statistical Analyses
All statistical analyses reported were performed using tools in the R Stats package in R v.4.0.2. All plots were produced in R using the ggplot2 package (Wickham et al. 2016).
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the members of the Lang Lab for comments on the manuscript. This study was supported by a grant from the National Institutes of Health (R01GM127420). Portions of this research were conducted on Lehigh University’s Research Computing infrastructure partially supported by the National Science Foundation (Award 2019035).
Author Contributions
GIL contributed to conceptualization, supervision, and funding acquisition. AAM and, GIL contributed to formal analysis, writing–original draft preparation, writing–review & editing, and visualization. AAM, AC, DM, and SWB contributed to investigation.
Data Availability
The short-read sequencing data reported in this study have been deposited to the NCBI BioProject database, accession number PRJNA835840. The previously published dataset from Marad et al. 2018 is available under NCBI BioProject PRJNA418180.
Declarations
Conflict of interest
The authors declare no competing interests.
References
- Acar M, Becskei A, Van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435(7039):228–232. doi: 10.1038/nature03524. [DOI] [PubMed] [Google Scholar]
- Acosta PB, Gross KC. Hidden sources of galactose in the environment. Eur J Pediatr. 1995;154(2):S87–S92. doi: 10.1007/BF02143811. [DOI] [PubMed] [Google Scholar]
- Aggeli D, Marad DA, Liu X, Buskirk SW, Levy SF, Lang GI. "Overdominant and partially dominant mutations drive clonal adaptation in diploid Saccharomyces cerevisiae". Genetics. 2022;221(2):iyac061. doi: 10.1093/genetics/iyac061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey SF, Hinz A, Kassen R. Adaptive synonymous mutations in an experimentally evolved Pseudomonas fluorescens population. Nat Commun. 2014;5(1):1–7. doi: 10.1038/ncomms5076. [DOI] [PubMed] [Google Scholar]
- Bakerlee CW, Phillips AM, Ba ANN, Desai MM. Dynamics and variability in the pleiotropic effects of adaptation in laboratory budding yeast populations. Elife. 2021;10:e70918. doi: 10.7554/eLife.70918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O, Barillot E. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28(3):423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles E, Hollenberg CP. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev. 1997;21(1):85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boocock J, Sadhu MJ, Durvasula A, Bloom JS, Kruglyak L. Ancient balancing selection maintains incompatible versions of the galactose pathway in yeast. Science. 2021;371(6527):415–419. doi: 10.1126/science.aba0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broach JR. Nutritional control of growth and development in yeast. Genetics. 2012;192(1):73–105. doi: 10.1534/genetics.111.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk SW, Peace RE, Lang GI. Hitchhiking and epistasis give rise to cohort dynamics in adapting populations. Proc Natl Acad Sci U S A. 2017;114(31):8330–8335. doi: 10.1073/pnas.1702314114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buskirk SW, Rokes AB, Lang GI. Adaptive evolution of nontransitive fitness in yeast. Elife. 2020;9:e62238. doi: 10.7554/eLife.62238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Zhang J. Antagonistic pleiotropy conceals molecular adaptations in changing environments. Nat Ecol Evol. 2020;4(3):461–469. doi: 10.1038/s41559-020-1107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2014;38(2):254–299. doi: 10.1111/1574-6976.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Chiara M, Friedrich A, Barré B, Breitenbach M, Schacherer J, Liti G. Discordant evolution of mitochondrial and nuclear yeast genomes at population level. BMC Biol. 2020;18(1):1–15. doi: 10.1186/s12915-020-00786-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan S-F, Han P-J, Wang Q-M, Liu W-Q, Shi J-Y, Li K, Zhang X-L, Bai F-Y. The origin and adaptive evolution of domesticated populations of yeast from Far East Asia. Nat Commun. 2018;9(1):1–13. doi: 10.1038/s41467-018-05106-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante-Chong R, Savir Y, Carroll SM, Ingraham JB, Wang J, Marx CJ, Springer M. Galactose metabolic genes in yeast respond to a ratio of galactose and glucose. Proc Natl Acad Sci. 2015;112(5):1636–1641. doi: 10.1073/pnas.1418058112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt S-M, Sauer U. Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst Biol. 2010;4(1):1–11. doi: 10.1186/1752-0509-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Buskirk SW, Vignogna RC, Marad DA, Lang GI. Adaptive genome duplication affects patterns of molecular evolution in Saccharomyces cerevisiae. PLoS Genet. 2018;14(5):e1007396. doi: 10.1371/journal.pgen.1007396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher KJ, Kryazhimskiy S, Lang GI. Detecting genetic interactions using parallel evolution in experimental populations. Philos Trans R Soc Lond B Biol Sci. 2019;374(1777):20180237. doi: 10.1098/rstb.2018.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu S, Watkins SM, Hotamisligil GS. The role of endoplasmic reticulum in hepatic lipid homeostasis and stress signaling. Cell Metab. 2012;15(5):623–634. doi: 10.1016/j.cmet.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Galeota-Sprung B, Fernandez A, Sniegowski P. Changes to the mtDNA copy number during yeast culture growth. Royal Soc Open Sci. 2022;9(7):211842. doi: 10.1098/rsos.211842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison, E. and G. Marth (2012). "Haplotype-based variant detection from short-read sequencing." arXiv preprint arXiv:1207.3907.
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4(12):e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison M-C, LaBella AL, Hittinger CT, Rokas A. The evolution of the GALactose utilization pathway in budding yeasts. Trends Genet. 2021;38:97–106. doi: 10.1016/j.tig.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hittinger CT, Carroll SB. Gene duplication and the adaptive evolution of a classic genetic switch. Nature. 2007;449(7163):677–681. doi: 10.1038/nature06151. [DOI] [PubMed] [Google Scholar]
- Hong J, Gresham D. Molecular specificity, convergence and constraint shape adaptive evolution in nutrient-poor environments. PLoS Genet. 2014;10(1):e1004041. doi: 10.1371/journal.pgen.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong K-K, Nielsen J. Adaptively evolved yeast mutants on galactose show trade-offs in carbon utilization on glucose. Metab Eng. 2013;16:78–86. doi: 10.1016/j.ymben.2013.01.007. [DOI] [PubMed] [Google Scholar]
- Hong K-K, Vongsangnak W, Vemuri GN, Nielsen J. Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc Natl Acad Sci. 2011;108(29):12179–12184. doi: 10.1073/pnas.1103219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3(3):318–356. doi: 10.1016/S0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jerison ER, Nguyen Ba AN, Desai MM, Kryazhimskiy S. Chance and necessity in the pleiotropic consequences of adaptation for budding yeast. Nature Ecol Evol. 2020;4(4):601–611. doi: 10.1038/s41559-020-1128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MS, Gopalakrishnan S, Goyal J, Dillingham ME, Bakerlee CW, Humphrey PT, Jagdish T, Jerison ER, Kosheleva K, Lawrence KR, Min J, Moulana A, Phillips AM, Piper JC, Purkanti R, Rego-Costa A, McDonald MJ, Nguyen Ba AN, Desai MM. Phenotypic and molecular evolution across 10,000 generations in laboratory budding yeast populations. Elife. 2021 doi: 10.7554/eLife.63910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet. 2008;40(12):1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristofich J, Morgenthaler AB, Kinney WR, Ebmeier CC, Snyder DJ, Old WM, Cooper VS, Copley SD. Synonymous mutations make dramatic contributions to fitness when growth is limited by a weak-link enzyme. PLoS Genet. 2018;14(8):e1007615. doi: 10.1371/journal.pgen.1007615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvitek DJ, Sherlock G. Whole genome, whole population sequencing reveals that loss of signaling networks is the major adaptive strategy in a constant environment. PLoS Genet. 2013;9(11):e1003972. doi: 10.1371/journal.pgen.1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Botstein D. A test of the coordinated expression hypothesis for the origin and maintenance of the GAL cluster in yeast. PLoS ONE. 2011;6(9):e25290. doi: 10.1371/journal.pone.0025290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, Desai MM. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013;500(7464):571–574. doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughery MF, Hunter T, Brown A, Hoopes J, Ostbye T, Shumaker T, Wyrick JJ. New vectors for simple and streamlined CRISPR–Cas9 genome editing in Saccharomyces cerevisiae. Yeast. 2015;32(12):711–720. doi: 10.1002/yea.3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KS, Hong ME, Jung SC, Ha SJ, Yu BJ, Koo HM, Park SM, Seo JH, Kweon DH, Park JC. Improved galactose fermentation of Saccharomyces cerevisiae through inverse metabolic engineering. Biotechnol Bioeng. 2011;108(3):621–631. doi: 10.1002/bit.22988. [DOI] [PubMed] [Google Scholar]
- Lee KB, Wang J, Palme J, Escalante-Chong R, Hua B, Springer M. Polymorphisms in the yeast galactose sensor underlie a natural continuum of nutrient-decision phenotypes. PLoS Genet. 2017;13(5):e1006766. doi: 10.1371/journal.pgen.1006766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legras J-L, Galeote V, Bigey F, Camarasa C, Marsit S, Nidelet T, Sanchez I, Couloux A, Guy J, Franco-Duarte R. Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol Biol Evol. 2018;35(7):1712–1727. doi: 10.1093/molbev/msy066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Venkataram S, Agarwala A, Dunn B, Petrov DA, Sherlock G, Fisher DS. Hidden complexity of yeast adaptation under simple evolutionary conditions. Curr Biol. 2018;28(4):515–525.e516. doi: 10.1016/j.cub.2018.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marad DA, Buskirk SW, Lang GI. Altered access to beneficial mutations slows adaptation and biases fixed mutations in diploids. Nat Ecol Evol. 2018;2(5):882–889. doi: 10.1038/s41559-018-0503-9. [DOI] [PubMed] [Google Scholar]
- Matheson K, Parsons L, Gammie A. "Whole-genome sequence and variant analysis of w303, a widely-used strain of saccharomyces cerevisiae". G3 (bethesda) 2017;7(7):2219–2226. doi: 10.1534/g3.117.040022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair A, Sarma SJ. The impact of carbon and nitrogen catabolite repression in microorganisms. Microbiol Res. 2021;251:126831. doi: 10.1016/j.micres.2021.126831. [DOI] [PubMed] [Google Scholar]
- New AM, Cerulus B, Govers SK, Perez-Samper G, Zhu B, Boogmans S, Xavier JB, Verstrepen KJ. Different levels of catabolite repression optimize growth in stable and variable environments. PLoS Biol. 2014;12(1):e1001764. doi: 10.1371/journal.pbio.1001764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski EA, Rozen DE, Lenski RE. Pleiotropic effects of beneficial mutations in Escherichia coli. Evolution. 2005;59(11):2343–2352. [PubMed] [Google Scholar]
- Peng W, Liu P, Xue Y, Acar M. Evolution of gene network activity by tuning the strength of negative-feedback regulation. Nat Commun. 2015;6(1):1–9. doi: 10.1038/ncomms7226. [DOI] [PubMed] [Google Scholar]
- Quarterman J, Skerker JM, Feng X, Liu IY, Zhao H, Arkin AP, Jin Y-S. Rapid and efficient galactose fermentation by engineered Saccharomyces cerevisiae. J Biotechnol. 2016;229:13–21. doi: 10.1016/j.jbiotec.2016.04.041. [DOI] [PubMed] [Google Scholar]
- Ronen M, Botstein D. Transcriptional response of steady-state yeast cultures to transient perturbations in carbon source. Proc Natl Acad Sci. 2006;103(2):389–394. doi: 10.1073/pnas.0509978103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roop JI, Chang KC, Brem RB. Polygenic evolution of a sugar specialization trade-off in yeast. Nature. 2016;530(7590):336–339. doi: 10.1038/nature16938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott AL, Richmond PA, Dowell RD, Selmecki AM. The influence of polyploidy on the evolution of yeast grown in a sub-optimal carbon source. Mol Biol Evol. 2017;34(10):2690–2703. doi: 10.1093/molbev/msx205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, Von Haeseler A, Schatz MC. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15(6):461–468. doi: 10.1038/s41592-018-0001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellick CA, Campbell RN, Reece RJ. Galactose metabolism in yeast—structure and regulation of the Leloir pathway enzymes and the genes encoding them. Int Rev Cell Mol Biol. 2008;269:111–150. doi: 10.1016/S1937-6448(08)01003-4. [DOI] [PubMed] [Google Scholar]
- Slepak T, Tang M, Addo F, Lai K. Intracellular galactose-1-phosphate accumulation leads to environmental stress response in yeast model. Mol Genet Metab. 2005;86(3):360–371. doi: 10.1016/j.ymgme.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Sood V, Brickner JH. Genetic and epigenetic strategies potentiate Gal4 activation to enhance fitness in recently diverged yeast species. Curr Biol. 2017;27(23):3591–3602.e3593. doi: 10.1016/j.cub.2017.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunshine AB, Payen C, Ong GT, Liachko I, Tan KM, Dunham MJ. The fitness consequences of aneuploidy are driven by condition-dependent gene effects. PLoS Biol. 2015;13(5):e1002155. doi: 10.1371/journal.pbio.1002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swamy K, Zhou N. Experimental evolution: its principles and applications in developing stress-tolerant yeasts. Appl Microbiol Biotechnol. 2019;103(5):2067–2077. doi: 10.1007/s00253-019-09616-2. [DOI] [PubMed] [Google Scholar]
- Tamanoi F. Ras signaling in yeast. Genes. Cancer. 2011;2:210–215. doi: 10.1177/1947601911407322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317(5840):916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- van den Brink J, Akeroyd M, van der Hoeven R, Pronk J, De Winde J, Daran-Lapujade P. Energetic limits to metabolic flexibility: responses of Saccharomyces cerevisiae to glucose–galactose transitions. Microbiology. 2009;155(4):1340–1350. doi: 10.1099/mic.0.025775-0. [DOI] [PubMed] [Google Scholar]
- Venkataram S, Dunn B, Li Y, Agarwala A, Chang J, Ebel ER, Geiler-Samerotte K, Herissant L, Blundell JR, Levy SF, Fisher DS, Sherlock G, Petrov DA. Development of a comprehensive genotype-to-fitness map of adaptation-driving mutations in yeast. Cell. 2016;166(6):1585–1596.e1522. doi: 10.1016/j.cell.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignogna RC, Buskirk SW, Lang GI. Exploring a local genetic interaction network using evolutionary replay experiments. Mol Biol Evol. 2021;38(8):3144–3152. doi: 10.1093/molbev/msab087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers K, Kominek J, Das A, Espinosa-Cantu A, De Maeyer D, Arslan A, Van Pee M, van der Zande E, Meert W, Yang Y. Adaptation to high ethanol reveals complex evolutionary pathways. PLoS Genet. 2015;11(11):e1005635. doi: 10.1371/journal.pgen.1005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Atolia E, Hua B, Savir Y, Escalante-Chong R, Springer M. Natural variation in preparation for nutrient depletion reveals a cost–benefit tradeoff. PLoS Biol. 2015;13(1):e1002041. doi: 10.1371/journal.pbio.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger JW, Piotrowski J, Nagarajan S, Chiotti K, Sherlock G, Rosenzweig F. Hunger artists: yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 2011;7(8):e1002202. doi: 10.1371/journal.pgen.1002202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H, Chang W, Wickham MH. "Package ‘ggplot2’". Create elegant data visualisations using the grammar of graphics. Version. 2016;2(1):1–189. [Google Scholar]
- Yona AH, Manor YS, Herbst RH, Romano GH, Mitchell A, Kupiec M, Pilpel Y, Dahan O. Chromosomal duplication is a transient evolutionary solution to stress. Proc Natl Acad Sci. 2012;109(51):21010–21015. doi: 10.1073/pnas.1211150109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Im Lippman S, Zhao X, Broach JR. How saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The short-read sequencing data reported in this study have been deposited to the NCBI BioProject database, accession number PRJNA835840. The previously published dataset from Marad et al. 2018 is available under NCBI BioProject PRJNA418180.