Abstract
Automatic detection of lung diseases using AI-based tools became very much necessary to handle the huge number of cases occurring across the globe and support the doctors. This paper proposed a novel deep learning architecture named LWSNet (Light Weight Stacking Network) to separate Covid-19, cold pneumonia, and normal chest x-ray images. This framework is based on single, double, triple, and quadruple stack mechanisms to address the above-mentioned tri-class problem. In this framework, a truncated version of standard deep learning models and a lightweight CNN model was considered to conviniently deploy in resource-constraint devices. An evaluation was conducted on three publicly available datasets alongwith their combination. We received 97.28%, 96.50%, 97.41%, and 98.54% highest classification accuracies using quadruple stack. On further investigation, we found, using LWSNet, the average accuracy got improved from individual model to quadruple model by 2.31%, 2.55%, 2.88%, and 2.26% on four respective datasets.
Keywords: Stack ensemble technique, Deep neural network, Chest radiography, Lung diseases, Pneumonia, Covid-19
Introduction
Coronavirus disease often referred to as Covid-19, is a submicroscopic pathogen resulting from worldwide pandemics. Coronavirus causes rapid bronchial distress disorder, a severe type of asthma. It is a kind of atypical, human-to-human transmissible pneumonia. Due to its enormous adverse influence on the healthcare of the general community, the Covid-19 pandemic is currently the most serious issue affecting our entire world. To fight Covid-19, governments and rulers imposed a variety of different strategies, policies, and lifestyles [38]. Science and technology had significantly impacted the implementation of these new ideas and techniques.
Reverse transcriptase-polymerase chain reaction (RT-PCR) [21] and enzyme-linked immunosorbent assay (ELISA) [63] are two of the most commonly used methods for detecting Covid-19 viruses. The most effective screening tool for finding Covid-19 patients is RT-PCR, which can locate the virus’s RNA from lower respiratory tract samples. The whole testing method for identifying pathogens using RT-PCR is manual and time-consuming, with a high risk of false negatives of 39–61%. In any event, significant clinical development leading to pneumonia, chest imaging studies are regularly conducted in suspected or confirmed Covid-19 patients, by the recommendation of WHO [43]. Antibodies, antigens, proteins, and glycoproteins are routinely measured in biological samples using an immunological test known as ELISA.
According to preliminary research, people with Covid-19 or pneumonia infection show anomalies in their chest radiographs. It was suggested that radiography examination might be used as a critical means of pneumonia-based disease screening in epidemic areas [4]. Radiography analysis is an excellent complement to the RT-PCR test and, in some instances, even provides a positive index. Accommodations for chest imaging are easily accessible in modern medical systems, even though radiographic images cannot simply and rapidly solve our purposes. There was a high demand for expert radiologists in this epidemic. The healthcare industry needs a solution to this problem. The field of computer vision and image processing could manage this problem with the aid of advanced tools and techniques [17]. Machine learning and deep learning were extremely promising solutions for managing these issues.
Artificial intelligence is widely being applied in medical imaging identification, and analysis [39]. The advantage of deep learning-based techniques such as CNN, RNN, LSTM-RNN, etc., in the field of computer-vision, outperformed the work of professional radiologists [29]. Those CNNs-based framework extract feature and classification prediction capabilities are quite a height, but due to a tremendous amount of data required for the training purpose, authors [6] used pre-trained models to save CPU power and calculation time.
The chest X-ray image is readily available due to its low-cost compared to other tests; the correct diagnosis from only these images is an immediate need for mankind. Since it is difficult to diagnose COVID-19 and pneumonia from X-ray images manually by medical practitioners, an automated system is required to correctly diagnose these diseases from the pool of normal, pneumonia, and COVID-19 x-ray images.
In this research, we experimented with one of the largest datasets for COVID-19 identification, comprised of 20738 images in three classes. Here, a hybrid deep learning architecture is designed by the stack-ensemble method and tuned lightweight version of the three standard deep learning frameworks and a custom CNN framework. The proposed architecture is developed to consider the issue of deploying the system in low-configuration devices to make the diagnosis faster with high performance.
The key contributions of the present work are as follows:
A novel stack ensemble deep learning framework namely LWSNet is proposed to segregate Covid-19, Pneumonia and Normal from x-ray imagery.
During stack ensemble process, truncated versions of MobileNetV2, VGG19 and InceptionResNetV2 were considered, making the final model lightweight for running at resource-constrained environments.
Three distinct public datasets were used to evaluate the performance of the proposed system.
Review of state-of-the-arts
It is essential to quarantine patients as soon as possible in order to control this infectious disease. Available resources are fast running out due to a constantly rising number of patients and lengthy treatment time [21]. Researchers from many disciplines and policymakers urgently need to develop a strategy to control the unwanted situation. We are attempting to concentrate on numerous computer vision areas to speed up the entire procedure. We explored some previously mentioned research areas on computer vision intelligent systems for automatic diagnosis. Researchers are using radiographic images to apply machine learning, and deep learning algorithms as the main categories to classify Covid-19 automatically and pneumonia disease [19, 20, 28, 55]. Other subcategories, such as multi-layer perceptron [62], ensemble [57], LSTM [22], fusion [53], and fuzzy [62], were applied for categorization. Those categories can implement different imaging modalities like X-ray, CT, etc.
Stack ensemble classification
The method of increasing the performance of the classifier by aggregating the already learned sub-models to tackle the same classification task is known as ensemble learning [36]. Each base learner takes a vote, and the meta-learner, a model that learns to improve the base learners’ predictions, receives the final prediction. Tang et al. [57] suggested ensemble learning can solve deep learning’s drawbacks by making predictions with several models rather than as a single model. Their experiment showed the results with good accuracy of 95%. Saha et al. [50] proposed a model that extracted deep features from X-ray images, then used an ensemble classifier. They obtained individual scores before implementing an ensemble classifier, which provides better accuracy from 1320 images. The highest accuracy, precision, recall, and F1-score were 98.91%, 100%, 97.28%, and 98.89%. Li et al. [31] combined ensemble with VGG16 as a base model, and they used cascade classifier from multiple training sets.
Annavarapu et al. [5] introduced a new ensemble technique for reducing the computationally learning cost. They named this approach the snapshot ensemble technique. Snapshot Ensembling’s adaptability with a wide range of network models and learning tasks was verified by its cyclic learning rate scheduling. This snapshot ensemble approach saved the local minima parameter and changed the model during runtime. They used ResNet50 as a base architecture. Chowdhury et al. [9] used the snapshot ensemble model, which is based on EfficientNet model. To identify and predict a critical section, they used the Grad-CAM model to visualize a CAP. They published their overall accuracy of 96.07% for Covid-19 detection.
Upadhyay et al. [60] implemented a fusion-based model to categorize three distinct classes. They used the handcrafted color-space method to collect specific features from X-ray images and then applied a stacked ensemble. Abdar et al. [1] proposed novel model named Uncertainty FuseNet. It is based on the fusion method using ensemble dropout currently. This model produced good results in noisy data.
Gifani et al. [52] implemented a pre-trained model using a total of 15 pre-trained architectures and fine-tuned the target classes. Among the 15 pre-trained architectures, the majority of voting was applied only on 5 architectures.
Deep transfer learning
Deep Transfer Learning is a method of deep learning where knowledge is transmitted from one model to the other. Zhu et al. [64] used traditional CNN and VGG16 net models. They optimized both the models and evaluated the predictions. But a weakness of their work was the selection of fewer chest images. Gupta et al. [18] worked with a five pre-trained integrated stack model called InstaCovNet-19. For boosting classification, various training and pre-processing techniques were applied. They benchmarked their model on other state-of-the-art approaches. Fan et al. [12] experimented with five pre-trained models and applied three optimizers using different learning rates. In addition, they used a 10-fold cross-validation approach. They achieved an average accuracy of 97%. Mohammadi et al. [35] discussed four popular pre-trained models with binary classification and obtained 99.1% accuracy using MobileNet for identifying Covid-19 disease from a chest X-ray. Niu et al. [42] proposed a new technique named distant domain transfer learning (DDTL). They used two models, namely reduced-size U-net Segmentation and Distant Feature Fusion. Their models worked on unstructured data and efficiently handled the variation in distribution during the training and testing data. Rezaee et al. [48] introduced a pre-trained deep learning architecture-based hybrid deep transfer learning technique. They utilized feature extraction, feature selection, and a support vector machine to classify. Other researchers presented their approach in aiming for pneumonia-based disease as an exception to these limited categories of deep learning and machine learning model. Saha et al. [49] used a pre-processing method to transform image data into an undirected graph such that simply the edges of the image are considered rather than the entire image. These networks showed impressive accuracy of 99% for the limited dataset. Another effective approach utilized by researchers for classification is the segmentation method. Munusamy et al. [41] proposed an architecture using segmentation of CT images and segregation on X-ray images. They used the U-net model for segmentation purposes and combined the location information. They compared their classification model with different pre-trained standard architecture, and overall accuracy of 99% was reported.
Lightweight CNN
In lightweight CNN, layers of the network are made reduced considering the system’s complexity, and accuracy trade-off [40]. Karakanis et al. [26] implemented a lightweight model in addition to a conditional generative adversarial network to develop synthetic images to replace the minimal data set. They began with a two-class dataset but later switched to a three-class dataset, which performed better. Paluru et al. [44] proposed 7.8 times fewer lightweight parameters using U-net architecture. They showed that their model performed well compared to the other six standard models. They claimed that their model could also run on a low configuration system. Ter-Sarkisov et al. [58] discussed lightweight segmentation models based on R-CNN masks with different standard architecture. The lightweight model isn’t limited to working with fixed images or structural data. Trivedi et al. [59] proposed a lightweight architecture based on MobileNet that can identify pneumonia from chest x-ray images. A benchmark dataset containing 5856 images in two classes was used. Additionally, they discussed the total training time for preparing the lightweight model and represented the accuracy at 97.09%.
The state-of-the-art research dealt with available datasets ignoring the performance over the conglomeration effects of these datasets. The system deployment issue on the resource-constraint devices is also a significant research gap to the best of our knowledge.
Proposed method
In this work, we considered deep learning-based models. Medical fields were significantly benefited from deep learning, including the detection of lung infection. In recent years, due to their impressive classification capabilities, deep learning methods were gained popularity on COVID-19. It was also observed that deep learning models outperform handcrafted feature-based models [14]. To train a deep learning model, a huge amount of data and a good amount of training time are required. So to abstain from these issues, here, transfer learning-based models were considered.
Transfer learning [15] is a machine training strategy where a system is built utilizing many training samples for a specific assignment and then used as the starting solution in another study. Rather than constructing the whole architecture initially, a pre-trained framework was used since this learning technique assures how an architecture trained for one problem may perform on another issue. As a result, the learning process is more resilient and adaptable. In addition, building a deep neural framework from scratch necessitates a high volume of samples and a significant quantity of cost time. Transfer learning allows one to focus on the beginning efforts rather than developing an entire deep architecture. Through the use of the transfer learning technique, three well-established deep learning architectures are transformed into lightweight models that are fine-tuned, as discussed in Section 3.1.
Amongst different deep learning frameworks [32], CNN is a very useful technique for processing spatial data. It is generally made up of three tiers: convolution, pooling, and dense. The design of such networks allows us to learn a wide range of complicated patterns that a basic neural network often fails to perform. CNN-based frameworks are a wide variety of uses, including self-driving cars, robots, surgical operations, etc. The convolutional layer transfers the presence of features observed in individual portions of the input images into a feature map. The process of creating a feature map from this layer is as follows:
| 1 |
Where, represents the kernal for the jth,kth pixel in cth layer over the instance ρm,n and * represent the convolution operator.
| 2 |
Here, λ denotes ReLU (rectified linear unit) activation function, which can be expressed as
| 3 |
if(z < 0);Re(z) = 0otherwise,Re(z) = z,where,zdenotestheneuroninput.
The softmax activation function is utilized in the network’s last dense layer, can be represented as
| 4 |
Here, n signifies the source vector, which has a length of m.
Proposed lightweight models
In this work, we proposed lightweight versions of existing deep CNN models: MobileNetV2, VGG19, and InceptionResNetV2, and designed a custom lightweight CNN (CLCNN) architecture. Developing lightweight stack ensemble models aims to deploy the system into resource-constraint devices. The number of parameters generated in the lightweight models is significantly less than the original counterpart.
Lightweight MobileNetV2
MobileNet-V2 is built on the principles of MobileNet-V1, which uses depth-wise separable convolution as a robust building component. They introduced an inverted residual block and a linear bottleneck framework in this version [51]. The original MovibleNetV2 architecture has fifty-three levels and 3.4 million parameters in its final version. In contrast, the proposed lightweight version consists of only the top twenty-five layers and utilizes 0.139331 M parameters. The structure and parameter details of this architecture are shown in Table 1.
Table 1.
Layer-wise parameter details for the lightweight models
| Layers | Network Input Dimension (# Parameter) | |||
|---|---|---|---|---|
| MobileNetV2 | InceptionResNetV2 | VGG19 | Proposed | |
| Conv1 (Conv2D) | 112×112×32 (864) | 111×111×32 (864) | 224×224×64 (1792) | 222×222×64 (1792) |
| BatchNormalization_Conv1 | 112×112×32 (128) | 111×111×32 (96) | 224×224×64 (36928) | 220×220×32 (18464) |
| Activation | – | 111×111×32 (0) | – | – |
| Conv1_relu (ReLu) | 112×112×32 (0) | – | – | 108×108×16 (2320) |
| Expanded_conv_depthwise | 112×112×32 (288) | 112×112×64 (0) | 112×112×64 (0) | – |
| BatchNormalization_3 | 112×112×32 (128) | – | 112×112×128 (73856) | – |
| Expanded_conv_depthwise_relu | 112×112×32 (0) | – | – | – |
| Expanded_conv_project | 112×112×16 (512) | – | 112×112×128 (147584) | – |
| Expanded_conv_project_BN | 112×112×16 (64) | – | – | – |
| Block_1_expand (Conv2D) | 112×112×96 (1536) | – | – | – |
| Block_1_expand_BN | 112×112×96 (384) | – | – | – |
| Block_1_expand_relu (ReLu) | 112×112×96 (0) | – | – | – |
| Block_1_pad (ZeroPadding2D) | 113×113×96 (0) | – | – | – |
| BatchNormalization_3 | – | 109×109×32 (96) | – | – |
| Activation_1 | – | 109×109×32 (0) | – | – |
| Conv2d_4 (Conv2D) | – | 109×109×64 (18432) | – | 106x106x8 (1160) |
| BatchNormalization_4 | – | 109×109×64 (192) | – | – |
| Expanded_conv_depthwise | 56×56×96 (864) | – | – | – |
| Expanded_conv_depthwise_BN | 56×56×96(384) | – | – | |
| Expanded_conv_depthwise_relu | 56×56×96 (0) | – | – | – |
| Block_1_expand (Conv2D) | 56×56×24 (2304) | – | – | – |
| Block_1_expand_BN | 56×56×24 (96) | – | – | – |
| Block_2_expand (Conv2D) | 56×56×144 (3456) | – | – | – |
| Block_2_expand_BN | 56×56×144 (576) | – | – | – |
| Block_2_expand_relu (ReLu) | 56×56×114 (0) | – | – | – |
| Block_2_depthwise | 56×56×144 (1296) | – | – | – |
| Block_2_expand_BN | 56×56×144(576) | – | – | – |
| Activation_2 (Conv2D) | – | 109×109×64 (0) | – | – |
| Max_pooling2d_2 | – | 54×54×80 (0) | – | – |
| Conv2d_5 (Conv2D) | – | 54×54×80 (5120) | – | 106×106×16 (2320) |
| BatchNormalization_5 | – | 54×54×80 (240) | – | – |
| Activation_3 (Conv2D) | – | 54×54×80 (0) | – | – |
| Conv2d_6 (Conv2D) | – | 52×52×192 (138240) | – | – |
| BatchNormalization_6 | – | 52×52×192 (576) | – | – |
| Activation_4 (Conv2D) | – | 52×52×192 (0) | – | – |
| Max_pooling2d_1 | – | 25×25×192 (0) | 56×56×128 (0) | – |
| Conv2d_10 (Conv2D) | – | 25×25×64 (12288) | 27×27×32 (36896) | 51×51×8 (1160) |
| BatchNormalization_10 | – | 25×25×64 (192) | – | – |
| Block_2_expand_relu (ReLu) | 56×56×144 (0) | – | - | – |
| Block_2_project (Conv2D) | 56×56×24 (3456) | – | – | – |
| Conv2d_1 (Conv2D) | 27×27×32 (6944) | 12×12×32 (18464) | – | 49×498 (584) |
| Droupout_1 (Dropout) | 23328 (0) | 4608 (0) | 23328 (0) | – |
| Flatten_1 (Flatten) | 23328 (0) | 4608 (0) | 23328 (0) | – |
| Droupout_2 (Dropout) | – | 4608 (0) | – | – |
| Batch_normalization | 23328 (93312) | 4608 (18432) | 23328 (93312) | – |
| Dense | 3 (69987) | 3 (9218) | 3 (69987) | 3 (13827) |
| Trainable parameters | 139,331 | 226,131 | 413,699 | 42,835 |
Lightweight VGG19
Simonyan and Zisserman [54] at the University of Oxford, UK, in early 2014, designed a CNN model named VGG network. VGG (Visual Geometry Group) was trained using the ImageNet ILSVRC dataset, consisting of more than 1 million pictures. According to these picture patterns, pictures are divided into 1000 categories and utilize more than 100 thousand images for training and 50 thousand images for validation. VGG-19 is a VGG variation with 19 densely linked layers that routinely outperform other state-of-the-art models. The model is made up of convolutional and fully connected layers that allow for enhanced feature extraction and the use of maxpooling instead of average pooling for downsampling, and modifying the linear unit (ReLU) as the activation function, selecting the largest value in the image area as the pooled value.
Lightweight InceptionResNetV2
The Inception-ResNet architecture [56] combines Inception block with residual module. In each Inception component, there is a filter expansion layer, which is used to scale up the depth of the filter bank to match the depth of the input. We made a lightweight version of this architecture by considering only the top twenty layers out of one hundred sixty-four. There are 0.2261 M parameters generated in this pre-trained lightweight architecture, whereas 55 M parameters are in the original architecture. In Table 1 the structure and parameter details of the lightweight InceptionResNetV2 architecture model are presented.
Custom lightweight CNN (CLCNN)
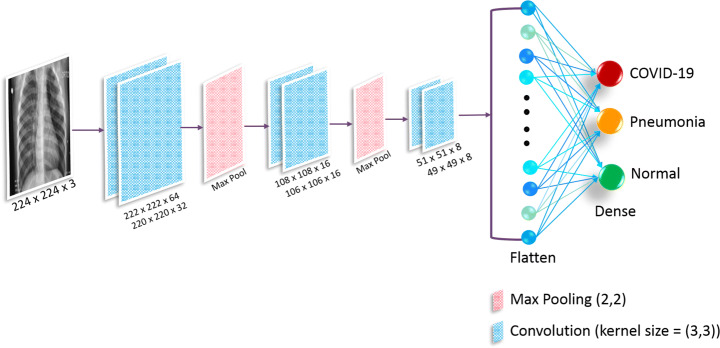
In this CLCNN framework (shown in Fig. 1), six convolution layers with 64, 32, 16, 16, 8, and 8 filters accompanied by two max-pool layers having a pooling size of 2 × 2 were considered. One max-pool layer is placed after the first two convolutions layers, and the second one is employed at the end of the fourth convolution layer. A dense/output layer of size 3 was used. In ablation study, we obtained the accuracies of 96.25%, 96.45%, 96.42%, and 96.35%, using 0.3, 0.4, 0.5, and 0.6 dropout values, respectively. Since a dropout of 0.4 yielded the best accuracy, we considered this value throughout the rest of the experiments. The layer-wise number of trainable parameters generated is shown in Table 1. The number of trainable parameters in original and lightweight versions is tabulated in Table 2.
Fig. 1.
CLCNN architecture for Covid-19 and pneumonia classification
Table 2.
The comparison of number of trainable parameters (Million) between original and lightweight versions
| Architecture Name | Trainable Parameter | Less parameter (%) of lightweight model | |
|---|---|---|---|
| Original Model | Lightweight Model | ||
| MobileNetV2 | 3.4 M | 0.139331 M | 96.03 |
| InceptionResNetV2 | 55 M | 0.226131 M | 99.59 |
| VGG19 | 144 M | 0.413699 M | 99.71 |
| Customize CNN | – | 0.042835 M | – |
Lightweight stack ensemble learning
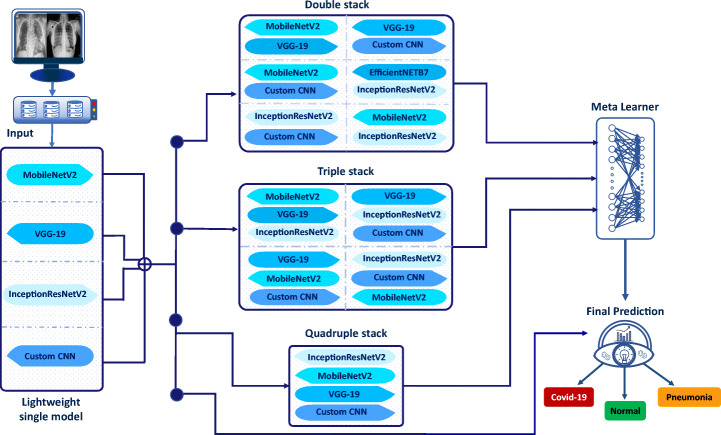
The prediction outcome suffers from high variance, and generalization problems [24] occur due to noise in the training data and the unpredictability of the deep learning models. We considered a stack ensemble approach to boost efficiency. It uses deep learning architecture for non-linear integration of predictors to increase prediction accuracy while reducing training errors. Specifically, several ensemble techniques are available, but among all the technique stacking generalization, [61] method is most effective in terms of accuracy. Before obtaining the final prediction, passing information through one group of classifiers to other sets of classifiers is known as stacking generalization. The information in the classifier network originates from several subgroups of the training set. The original training set is split into numerous subgroups of training sets, a distinguishing property of stacking generalization. Each sub-training group is utilized to gather biased information on the dataset’s generalization behavior, which is then used to populate the classifier network. This study developed an ensemble-based stacked framework for improving classification by combining predictions of different models and obtaining the highest positive true predictive values.
Figure 2 depicts the architecture of the proposed lightweight stack ensemble learning framework. In this architecture, we considered lightweight CNN models as single models. Then the stacked ensemble was designed considering double stack, triple stack, and quadruple stack. The performance of the single models was evaluated after classification from the final predictor. The double stack consists of, MobileNetV2 & VGG19, MobileNetV2 & InceptionResNetV2, MobileNetV2 & CLCNN, VGG19 & InceptionResNetV2, VGG19 & CLCNN, and InceptionResNetV2 & CLCNN. Similarly, a triple stack consists of three base architecture such as InceptionResNetV2 & VGG19 & MobileNetV2, InceptionResNetV2 & VGG19 & CLCNN, and MobileNetV2 & VGG19 & CLCNN. This quadruple stack comprises MobileNetV2, VGG19, InceptionResNetV2, and CLCNN. This multi-stack prediction is fed to the meta-learner. In the meta-learner, three dense layers having sizes 512, 256, and 128 were deployed. The meta-learner predictions are fed to the final prediction block, which comprises a dense layer of dimension 3.
Fig. 2.
Four-step experiments of the proposed lightweight stack ensemble learning architecture
Experiment
The LWSNet is a classification problem to detect Covid-19 from chest X-ray images. Since the infection is unusual and has pneumonia-like characteristics, we included pneumonia images along with Covid-19 and normal images in our experiment.
Setup
We performed training and evaluation of our proposed system on a GPU machine consists of two core Intel(R) Core(TM) i5-10400 H CPU @ 2.60 GHz 2.59 GHz, 32 GB of RAM, and two NVIDIA Quadro RTX 5000. It runs on Windows 10 Pro OS version 20H2 with TensorFlow 2.0.0 installed for deep learning model training and inference, where cuDNN is enabled to speed up the training computation on a GPU device.
Datasets
The datasets used in this study were obtained from three different public sources and contained three classes. As a result, since Covid-19 is a novel disease, only a limited number of benchmark datasets are available for studies. The first datasets were collected from Kaggle repositories, which contained 6432 samples divided into three classes. The second dataset was constructed using 50% of chest X-ray images from Kaggle repositories with three classes. This dataset contains 21165 chest X-ray images from four classes. The third dataset is mixed with two public repositories; one is the GitHub repository with Covid-19 images, which is constantly updated by a researcher named Cohen et al. [11] at the Montreal University, and the other datasets are taken from the Kaggle repository [37] with two classes: Normal and Pneumonia. These mixed datasets are also available in the Kaggle repository. In order to conduct further experiments, we prepared a combined dataset. Merge three datasets with their respective classes to create a combined dataset. Table 3 represents the number of images containing three categories: Covid-19, Pneumonia, and Normal, from four different datasets. In Fig. 4 the sample sizes using an 8:2 train-set of three datasets are shown. The sample images of three classes corresponding to the three datasets are presented in Fig. 3.
Table 3.
Details of the databses used in the current experiment
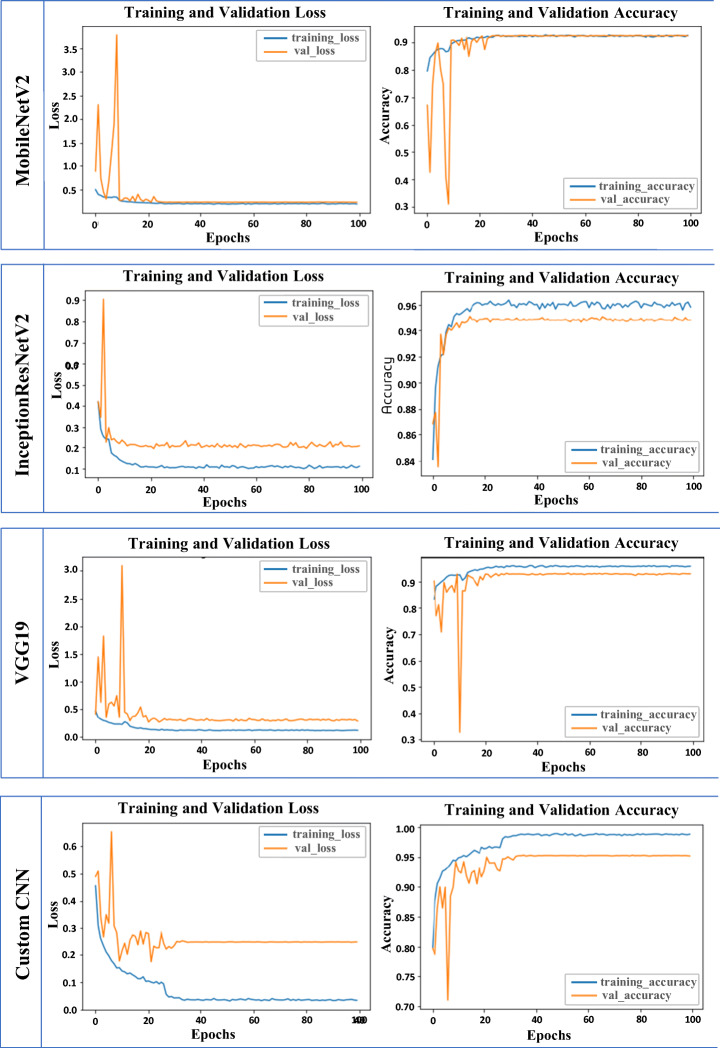
Fig. 4.
Training accuracy and training loss, validation accuracy and validation loss for individual lightweight models on dataset 1
Fig. 3.
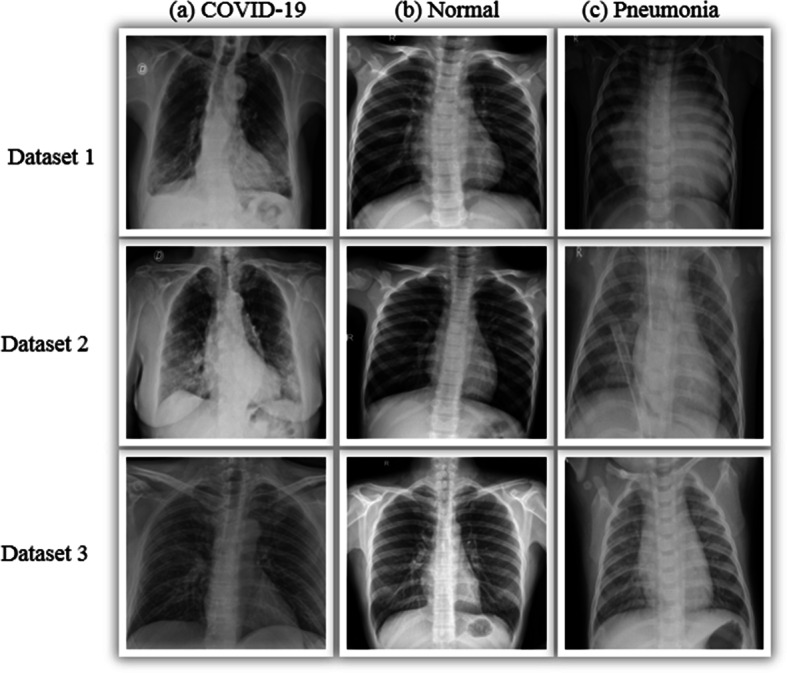
Organize the three datasets Dataset 1, Dataset 2, and Dataset 3 with their corresponding classes of images in three columns: (a) Covid-19, (b) normal, and (c) pneumonia
Evaluation protocol
To evaluate the performance of our LWSNet, we used different evaluation metrics since accuracy is not sufficient [13] to justify the performance of the proposed framework, especially in disease detection cases. The classification of diseases is based directly on the number of True Positives, False Negatives, True Negatives, and False Positives. Using these values, the following matrices are used to evaluate model performance from different perspectives:
Sensitivity
Sensitivity is a measure that evaluates the number of patients who was detected as positive in a scenario when the patient is genuinely effected.
Specificity
Specificity is a measure that evaluates the number of patients who was not been detected as negative in a scenario when the patient is genuinely not effected.
F1-Score
Statistics use F1 scores to determine how accurate a test is when analyzing classification data. During the computation of the F1 score, the precision and recall of the test are both considered. The F1 score is the harmonic average of precision and recall, where 1 is the best and 0 is the worst score.
Precision
Precision is the positive prediction value for the corresponding diseases. A predicted value of this disease is calculated based on true and false positives.
Negative predictive value
The negative predicted value is calculated based on the true, and false negative values.
Miss rate
Miss rate refers to the result of a test which suggests that a person does not have a specific disease when, in reality, that person does in fact, have that disease.
Fall-out
One of the most significant risks of receiving a false positive result is when the person is cohorted with other patients who are suffering with Covid-19 and is thus exposed to the virus.
False discovery rate
When making multiple comparisons, the false discovery rate is a means of conceptualizing the rate of type I error in the null hypothesis testing that is often used. Statistical approaches for regulating false discovery rates are intended to keep the predicted percentage of rejected null hypotheses) from exceeding a certain threshold.
False omission rate
It is The statistical technique, which is the complement of negative predictive value, is employed in multiple hypothesis testing to account for numerous comparisons. It calculates the percentage of false negatives that are wrongfully rejected.
Training regime
The datasets are of different sizes, so the images were resized into dimensions of 224 × 224 as a pre-processing step. The datasets were split into 8:2 train-test sets. The reason for the 8:2 division is that it was observed that the 8:2 train-test set gave better results compared to other ratios [64]. By this split, there are 5144 trains and 1288 tests for dataset 1. For dataset 2, 6061 images were considered in training, while 1515 images were for testing. Similarly, 5384 images were used in training, and the rest, 1346, were kept for testing dataset 3. To show the robustness of LWSNet, we combined all three datasets with their respective classes. In this combination, there are 16592 trained images and 4146 test images. At the beginning of the experiment, the learning rate was set to 0.001. The performance was evaluated at the interval of 0.001 learning rate. A dropout value was set at 0.5 throughout our experiment. Accordingly, the initial and secondary momentum exponential decay rates were fixed to 0.9 and 0.999. The epsilon level was set to 0.0000001. The value of the AMSGrad Boolean optimization parameter variable was set to false.
Results & analysis
Several levels of experiments were conducted to build the LWSNet model. The experiment was performed on single as well as double, triple, and quadruple stacking experiments. A total of 60 experiments were conducted during the experimentation period. To demonstrate the performance of LWSNet, we used different statistical approaches.
To test the underfit and overfit of single architectures, the accuracies and corresponding losses were presented in Fig. 4. It is seen that using mobileNetV2 training and validation loss were almost null, whereas, in InceptionResNetV2, there is a validation loss of 0.17%. Similarly, VGG19 and Custom CNN loss were generated at 0.08 and 0.27. In Table 4 the accuracies and losses corresponding to the learning rates are presented for the quadruple stack model on four datasets. Changing the learning rate is very significant in building a better DL model. We observed that accuracy gradually increased and loss decreased when learning rates were decreased.
Table 4.
Accuracy and loss values for the three datasets and the combination of three datasets in the quadruple stack model for diverse learning rates
| Learning Rate | Dataset 1 | Dataset 2 | Dataset 3 | Combined | ||||
|---|---|---|---|---|---|---|---|---|
| Accuracy | Loss | Accuracy | Loss | Accuracy | Loss | Accuracy | Loss | |
| 0.001 | 0.9805 | 0.0912 | 0.9658 | 0.2430 | 0.9651 | 0.3751 | 0.9786 | 0.1283 |
| 0.0001 | 0.9719 | 0.1785 | 0.9643 | 0.2186 | 0.9641 | 0.3660 | 0.9786 | 0.1085 |
| 1.00E-05 | 0.9727 | 0.1829 | 0.9741 | 0.2196 | 0.9638 | 0.3613 | 0.9854 | 0.1101 |
| 1.00E-06 | 0.9728 | 0.1852 | 0.9658 | 0.2241 | 0.9634 | 0.3630 | 0.9779 | 0.1136 |
| 1.00E-07 | 0.9719 | 0.1826 | 0.9643 | 0.2216 | 0.9638 | 0.3623 | 0.9767 | 0.1126 |
| 1.00E-08 | 0.9727 | 0.1833 | 0.9658 | 0.2245 | 0.9634 | 0.3616 | 0.9774 | 0.1131 |
| 1.00E-09 | 0.9727 | 0.1848 | 0.9658 | 0.2235 | 0.9634 | 0.3615 | 0.9777 | 0.1116 |
| 1.00E-10 | 0.9727 | 0.1848 | 0.9643 | 0.2234 | 0.9638 | 0.3601 | 0.9779 | 0.1128 |
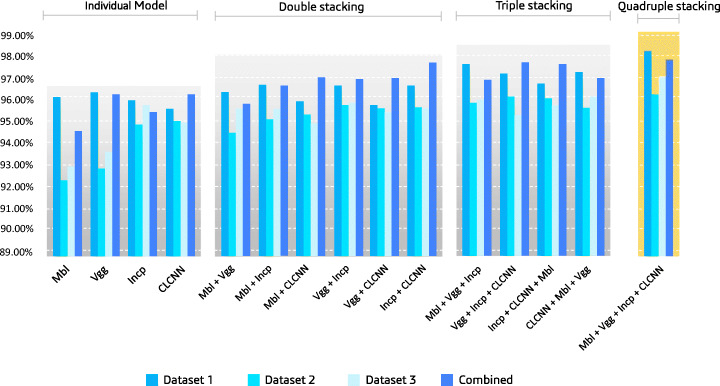
The results of individual lightweight architecture: MobileNetV2 (Mbl), VGG-19 (Vgg), InceptionResNetV2 (Incp) and Custom lightweight CNN (CLCNN) are depicted in Fig. 5, also represent other three stacking experiment results. In the four different individual lightweight models, the average accuracy of the datasets 1, 2, 3, and combined is 96.22%, 93.95%, 94.52%, and 95.84%, respectively. The average values of MobileNetV2, VGG-19, InceptionResNetV2, and CLCNN for those four datasets are 94.00%, 94.48%, 95.74%, and 95.39%, respectively.
Fig. 5.
Consolidated accuracy graph to single architecture to quad stack architecture. The individual model’s abbreviation within brakets is MobileNetV2(Mbl), VGG19(Vgg), InceptionResNetV2(Incp), and custom lightweight CNN (CLCNN) and ‘+’ sign indicate that models are concatenated for ensemble stacking
In this experiment, the batch size of 32, initial learning rate 0.001, optimizer RMSprop, activation function ReLu, and epoch size 100 were considered. We utilized these parameters for training four individual architectures. The results of four datasets showed that our CLCNN architecture performed well as compared to the pre-trained lightweight architecture. In the CLCNN architecture, only 42,899 trainable parameters were used, which were 2.24, 4.27, and 8.64 times less than mobileNetV2, InceptionResNetV2, and VGG19, respectively. The parameters are tabulated in Table 2. Stacking approaches were divided into three phases according to the proposed lightweight stack ensemble architecture. The stacking process in the first phase was done by selecting two lightweight models among the 4 variants. Using these selection criteria, a total of six combinations are there in a dataset. As a result, using the double-stack ensemble technique, a total of 24 experiments were carried out on four datasets. Compared to individual models, in the double-stack ensemble model, the accuracies improved by 0.32%, 1.54%, 1.14%, and 1.22% for dataset 1, dataset 2, dataset 3, combined, respectively. For dataset 2, the double-stacked architecture performed better than the single-lightweight architecture in terms of average accuracy. In the second step, we took three distinct models and combined them into a single stack. Three lightweight models were selected from four types of the triple stack model. The average accuracy gained by the triple stack model compared with the double stack was 0.88%, 0.68%, 0.36%, and 0.51% for datasets 1, 2, 3, and combined, respectively.
Testing was conducted further by changing the number of epochs from 50 to 300 with 50 epoch intervals considering the batch size of 32. But, the system’s performance didn’t increase for the increasing number of an epoch. Considering 100 epochs, we experimented again by changing the batch size by 64, 128, and 256. For 128 batch size, the quadruple stack block returns the highest accuracies of 98.54%, 96.50%, 97.41%, and 98.10%, on dataset 1, 2, 3 and combined, respectively.
The accuracy value of a system cannot be the only measure of its performance. We further calculated other statistical measures to check the architecture performance. We used nine statistical metrics to evaluate the LWSNet architecture’s efficiency in terms of correctly and incorrectly identified X-ray images with their respective diseases. From the Table 5 we observed that false positive rate is 0.0009, 0.0181, 0.0026 and 0.0010 for Covid-19 classes in Dataset 1, 2, 3, and Combined, respectively.
Table 5.
Quad stack performance evaluation for different datasets
| Rate | Dataset 1 | Dataset 2 | Dataset 3 | Combined | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | N | P | C | N | P | C | N | P | C | N | P | |
| Sensitivity | 0.9661 | 0.9650 | 0.9848 | 0.9632 | 0.9708 | 0.9148 | 0.9716 | 0.9678 | 0.9847 | 0.9652 | 0.9739 | 0.9839 |
| Specificity | 0.9991 | 0.9866 | 0.9676 | 0.9819 | 0.9539 | 0.9920 | 0.9854 | 0.9795 | 0.9886 | 0.9820 | 0.9956 | |
| F1-score | 0.9785 | 0.9619 | 0.9842 | 0.9524 | 0.9743 | 0.9165 | 0.9771 | 0.9601 | 0.9864 | 0.9517 | 0.9733 | 0.9892 |
| Precision | 0.9913 | 0.9589 | 0.9836 | 0.9419 | 0.9779 | 0.9182 | 0.9828 | 0.9525 | 0.9882 | 0.9386 | 0.9728 | 0.9946 |
| Negative predictive | 0.9966 | 0.9886 | 0.9698 | 0.9887 | 0.9395 | 0.9917 | 0.9957 | 0.9902 | 0.9735 | 0.9937 | 0.9828 | 0.9870 |
| Miss rate | 0.0339 | 0.0339 | 0.0152 | 0.0368 | 0.0368 | 0.0852 | 0.0284 | 0.0284 | 0.0153 | 0.0348 | 0.0348 | 0.0161 |
| Fall-out | 0.0009 | 0.0134 | 0.0324 | 0.0181 | 0.0461 | 0.0080 | 0.0026 | 0.0146 | 0.0205 | 0.0114 | 0.0180 | 0.0044 |
| False discovery | 0.0087 | 0.0411 | 0.0164 | 0.0581 | 0.0221 | 0.0818 | 0.0172 | 0.0475 | 0.0118 | 0.0614 | 0.0272 | 0.0054 |
| False omission | 0.0034 | 0.0114 | 0.0302 | 0.0113 | 0.0605 | 0.0083 | 0.0043 | 0.0098 | 0.0265 | 0.0063 | 0.0172 | 0.0130 |
Error analysis
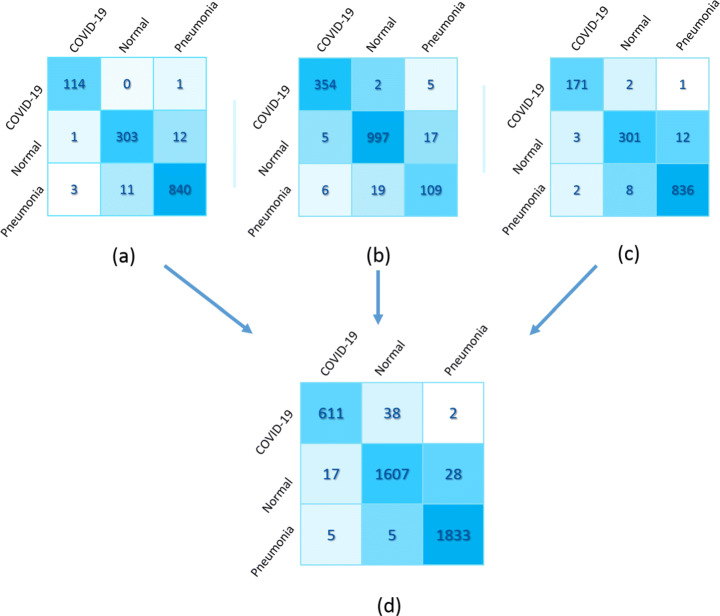
It was observed from Table 4 that changing the learning rates 0.001, 1.00e-06, and 1.00e-05, the lowest error rate of 1.46%, 2.59%, 3.49%, and 1.90% were generated for a quadruple stack using dataset 1, 2, 3, and combined, respectively. The correctly and incorrectly classified samples for each respective dataset can be understood easily from the confusion matrices as shown in Fig. 6. It is observed that for dataset 1 in Fig. 6(a) the lowest error was generated, i.e., 26 samples out of 1285 sample size were misclassified. As a result of this sample size, one instance of Covid-19 was misclassified as pneumonia, and one instance of normal was misinterpreted as Covid-19, while three instances of pneumonia were wrongly classified as Covid-19. Similarly, in the combined dataset, 22 samples were misclassified to detect Covid-19, while 30 samples were misclassified to detect pneumonia (Fig. 6(d)).
Fig. 6.
Confusion matrix of quad stack architecture of (a) Dataset 1, (b) Dataset 2, (c) Dataset 3, and (d) Combined dataset
In Fig. 7, the misclassified instances of images are shown. The possible reasons of misclassification are noisy, blur, and opaque images.
Fig. 7.
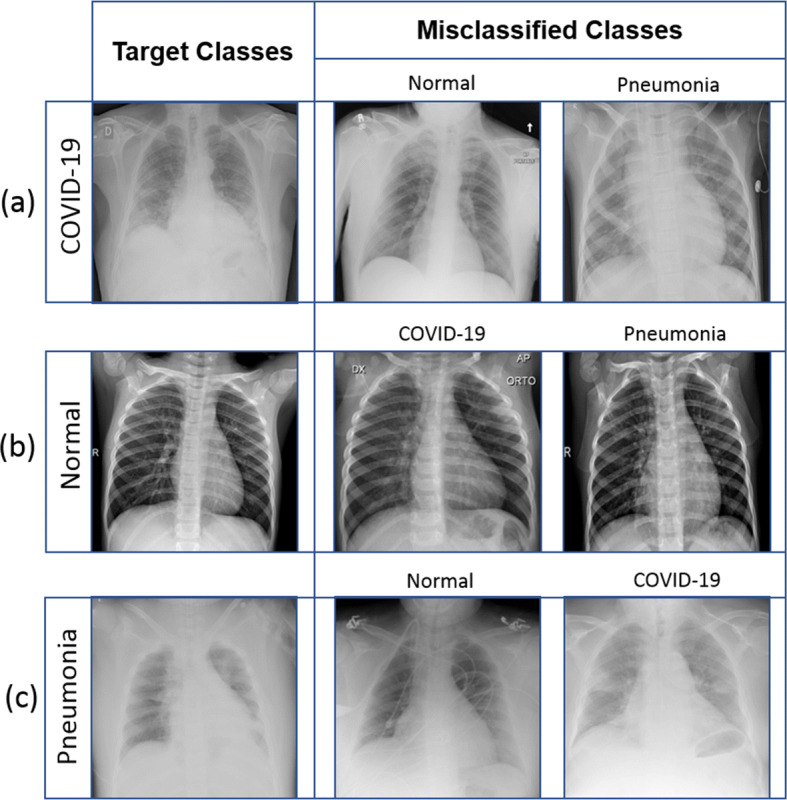
The first column represents the target class, and misclassified samples of the target class are represented by the second and third columns. Here, the first row and the first column represent the target class of Covid-19. In the same row, the second and third columns represent misclassified samples that are normal and pneumonia, similarly in the second and third rows
Comparative study
We compared LWSNet with the standard lightweight deep learning models for dataset 1, 2, 3, and combined dataset. The results indicate that the proposed LWSNet model is most effective in four datasets, particularly in the combined dataset. The accuracy of LSWNet was improved by 3.77%, 2.07%, 2.88%, and 2.09% when compared with lightweight MobileNet, InceptionResNetV2, and CLCNN in the combined dataset, respectively. In Table 6, along with accuracy, precision, sensitivity, and f1-score, the inference time for a batch size of 128 is also presented for single models and LSWNet.
Table 6.
Evaluation metrics of lightweight deep learning models with LWSNet on given X-ray images datasets
| Datasets | Models Name | Precision | Sensitivity | f1-score | Accuracy(%) | Time/step |
|---|---|---|---|---|---|---|
| Dataset 1 | MobileNet | 0.9520 | 0.9563 | 0.9540 | 96.34 | 694 ms |
| InceptionResNetV2 | 0.9508 | 0.9529 | 0.9512 | 96.19 | 695 ms | |
| VGG19 | 0.9498 | 0.9588 | 0.9541 | 96.58 | 693 ms | |
| Custom CNN | 0.9550 | 0.9447 | 0.9498 | 95.80 | 692 ms | |
| LWSNet | 0.9632 | 0.9765 | 0.9697 | 97.28 | 690 ms | |
| Dataset 2 | MobileNet | 0.8979 | 0.8992 | 0.8982 | 92.51 | 85 ms |
| InceptionResNetV2 | 0.9350 | 0.9350 | 0.9350 | 95.05 | 86 ms | |
| VGG19 | 0.8870 | 0.9101 | 0.8978 | 93.04 | 84 ms | |
| Custom CNN | 0.9349 | 0.9310 | 0.9334 | 95.21 | 84 ms | |
| LWSNet | 0.9512 | 0.9489 | 0.9500 | 96.50 | 80 ms | |
| Dataset 3 | MobileNet | 0.9142 | 0.9205 | 0.9178 | 93.15 | 421 ms |
| InceptionResNetV2 | 0.9494 | 0.9572 | 0.9512 | 95.98 | 419 ms | |
| VGG19 | 0.9165 | 0.9318 | 0.9236 | 93.82 | 420 ms | |
| Custom CNN | 0.9412 | 0.9438 | 0.9425 | 95.16 | 419 ms | |
| LWSNet | 0.9745 | 0.9747 | 9746 | 97.41 | 417 ms | |
| Combined Dataset | MobileNet | 0.9440 | 0.9294 | 0.9362 | 94.77 | 383 ms |
| InceptionResNetV2 | 0.9633 | 0.9565 | 0.9598 | 96.48 | 384 ms | |
| VGG19 | 0.9515 | 0.9522 | 0.9517 | 95.66 | 386 ms | |
| Custom CNN | 0.961 | 0.9477 | 0.9540 | 96.45 | 385 ms | |
| LWSNet | 0.9779 | 0.9719 | 0.9748 | 98.54 | 384 ms |
The proposed architecture was compared with other state-of-the-art architecture. In Table 7, it is seen that the accuracies were improved by 4.54%, 0.93%, and 0.28% and the F1-score on dataset 1 were improved by 4.48%, 1.12%, and 1.48% in comparison with the article of Gour et al. [16], Abdar et al. [1], and Jain et al. [23], respectively. Also, comparing the techniques [3, 27, 33, 46], with this proposed method for dataset 2, the accuracies got improved by 0.73%, 5.86%, 1.39%, and 0.50%. But, compared with the work of Aggarwal et al. [2] a loss of 0.50% of accuracy was also observed. Using the same dataset, the f1-scores of our method comparing with Rahman [46] and Lafraxocite [33] were improved by 1.11% and 1.97%, respectively. For dataset 3, the f1-scores in our technique were improved by 0.48%, 5.49%, 0.66%, and 4.46%, and the accuracies were gained by 0.40%, 6.91%, 8.41%, and 0.98% comparing the results of the articles of [7, 8, 30, 34], respectively.
Table 7.
Comparison with same public X-ray images dataset by using different evaluation metrics
| Datasets | Author’s | Precision | Sensitivity | f1-score | Accuracy(%) |
|---|---|---|---|---|---|
| Dataset 1 | Gour et al. [16] | 0.9581 | 0.9333 | 0.9300 | 92.74 |
| Abdar et al. [1] | 0.9635 | 0.9637 | 0.9636 | 96.35 | |
| Jain et al. [23] | 0.9800 | 0.9466 | 0.9600 | 97.00 | |
| Proposed LWSNet | 0.9632 | 0.9765 | 0.9697 | 97.28 | |
| Dataset 2 | Lafraxo et al. [27] | 0.9369 | 0.9366 | 0.9367 | 95.77 |
| Ahmed et al. [3] | 0.9200 | 0.8900 | 0.8980 | 90.64 | |
| Rahman et al. [46] | 0.9455 | 0.9456 | 0.9453 | 95.11 | |
| Loey et al. [33] | 0.9608 | 0.9604 | 0.9605 | 96.00 | |
| Aggarwal et al. [2] | 0.9766 | 0.9733 | 0.9733 | 97.00 | |
| Proposed LWSNet | 0.9512 | 0.9489 | 0.9500 | 96.50 | |
| Dataset 3 | Li et al. [30] | 0.9700 | 0.9709 | 0.9698 | 97.01 |
| Mangal et al. [34] | 0.9128 | 0.9414 | 0.9198 | 90.50 | |
| Chatterjee et al. [8] | 0.8500 | 0.9350 | 0.9680 | 89.00 | |
| Kanwal et al. [25] | 0.1000 | 0.1000 | 0.1000 | 93.00 | |
| Chakraborty et al. [7] | 0.9900 | 0.9368 | 0.9300 | 96.43 | |
| Proposed LWSNet | 0.9745 | 0.9747 | 0.9746 | 97.41 |
Conclusion
In this work, we presented LWSNet, a lightweight stack ensemble architecture to segregate Covid and non-Covid pneumonia. We explored truncated versions of three state-of-the-art networks, namely MobileNetV2, VGG19, InceptionResNetV2, and CLCNN architecture for the said problem. Further, the stack ensemble technique was employed on these four tailors-made models. Three different stacking techniques, namely double, triple and quadruple stacking, were experimented. Among all, the quadruple stack ensemble produced the highest accuracy of 97.28%, 96.50%, 97.41%, and 98.54%, using datasets 1, 2, 3, and combined, respectively, which outperform the state-of-the-art.
Our plan for the future is threefold: (i) the experiments will be carried out on other radiological images such as CT and MRI, (ii) different advanced deep learning architectures such as generative adversarial network, attention-based encoder-decoder, zero-shot learning, etc., will also be explored, and (iii) fusion of deep and handcrafted features, and clinical information (upon availability) will also be evaluated.
Acknowledgement
The fourth author would like to acknowledge Indian Council of Medical Research, Govt of India [Ref. No. BMI/12(81)/2021] for the research work.
Data Availability
This study used a secondary dataset as described by Rahman et al. in [10, 37, 45, 47]. The dataset can be obtained from the https://www.kaggle.com repository using this direct links: https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database, https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumoniahttps://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia, https://www.kaggle.com/datasets/prashant268/chest-xray-covid19-pneumoniahttps://www.kaggle.com/datasets/prashant268/chest-xray-covid19-pneumonia.
Declarations
Conflict of Interests
The authors declare that they have no conflict of interest.
Footnotes
Asifuzzaman Lasker and Mridul Ghosh contributed equally to this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Asifuzzaman Lasker, Email: asifuzzaman.lasker@gmail.com.
Mridul Ghosh, Email: mridulxyz@gmail.com.
Sk Md Obaidullah, Email: sk.obaidullah@gmail.com.
Chandan Chakraborty, Email: chakraborty.nitttrk@gmail.com.
Kaushik Roy, Email: kaushik.mrg@gmail.com.
References
- 1.Abdar M, Salari S, Qahremani S, Lam H-K, Karray F, Hussain S, Khosravi A, Acharya U R, Nahavandi S (2021) Uncertaintyfusenet: robust uncertainty-aware hierarchical feature fusion with ensemble monte carlo dropout for covid-19 detection. arXiv:2105.08590 [DOI] [PMC free article] [PubMed]
- 2.Aggarwal S, Gupta S, Alhudhaif A, Koundal D, Gupta R, Polat K. Automated covid-19 detection in chest x-ray images using fine-tuned deep learning architectures. Expert Syst. 2022;39(3):12749. doi: 10.1111/exsy.12749. [DOI] [Google Scholar]
- 3.Ahmed F, Bukhari SAC, Keshtkar F. A deep learning approach for covid-19 8 viral pneumonia screening with x-ray images. Digital Government: Research and Practice. 2021;2(2):1–12. doi: 10.1145/3431804. [DOI] [Google Scholar]
- 4.Ai T, Yang Z, Hou H, Zhan C, Chen C, Lv W, Tao Q, Sun Z, Xia L. Correlation of chest ct and rt-pcr testing for coronavirus disease 2019 (covid-19) in China: a report of 1014 cases. Radiology. 2020;296(2):32–40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Annavarapu CSR, et al. Deep learning-based improved snapshot ensemble technique for covid-19 chest x-ray classification. Appl Intell. 2021;51(5):3104–3120. doi: 10.1007/s10489-021-02199-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apostolopoulos ID, Mpesiana TA. Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakraborty S, Murali B, Mitra AK. An efficient deep learning model to detect covid-19 using chest x-ray images. Int J Environ Res Public Health. 2022;19(4):2013. doi: 10.3390/ijerph19042013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee S, Saad F, Sarasaen C, Ghosh S, Khatun R, Radeva P, Rose G, Stober S, Speck O, Nürnberger A (2020) Exploration of interpretability techniques for deep covid-19 classification using chest x-ray images. arXiv:2006.02570 [DOI] [PMC free article] [PubMed]
- 9.Chowdhury NK, Kabir MA, Rahman MM, Rezoana N. Ecovnet: a highly effective ensemble based deep learning model for detecting covid-19. PeerJ Comput Sci. 2021;7:551. doi: 10.7717/peerj-cs.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury ME, Rahman T, Khandakar A (2022) Covid-19 radiography database. https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database. Accessed 10 May 2022
- 11.Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M (2020) Covid-19 image data collection: prospective predictions are the future. arXiv:2006.11988
- 12.Fan Z, Jamil M, Sadiq M T, Huang X, Yu X (2020) Exploiting multiple optimizers with transfer learning techniques for the identification of covid-19 patients. J Healthc Eng 2020 [DOI] [PMC free article] [PubMed]
- 13.Ghosh M, Mukherjee H, Obaidullah SM, Santosh K, Das N, Roy K. Lwsinet: a deep learning-based approach towards video script identification. Multimed Tools Appl. 2021;80(19):29095–29128. doi: 10.1007/s11042-021-11103-8. [DOI] [Google Scholar]
- 14.Ghosh M, Baidya G, Mukherjee H, Obaidullah S M, Roy K (2022) A deep learning-based approach to single/mixed script-type identification, pp 121–132
- 15.Ghosh M, Roy SS, Mukherjee H, Obaidullah SM, Santosh K, Roy K. Understanding movie poster: transfer-deep learning approach for graphic-rich text recognition. Vis Comput. 2022;38(5):1645–1664. doi: 10.1007/s00371-021-02094-6. [DOI] [Google Scholar]
- 16.Gour M, Jain S (2020) Stacked convolutional neural network for diagnosis of covid-19 disease from x-ray images. arXiv:2006.13817
- 17.Gupta A (2019) Current research opportunities for image processing and computer vision. Comput Sci 20
- 18.Gupta A, Gupta S et al, katarya R (2021) Instacovnet-19: a deep learning classification model for the detection of covid-19 patients using chest x-ray. Appl Soft Comput 99:106859 [DOI] [PMC free article] [PubMed]
- 19.Gupta RK, Sahu Y, Kunhare N, Gupta A, Prakash D (2021) Deep learning based mathematical model for feature extraction to detect corona virus disease using chest x-ray images. Int J Uncertain, Fuzziness Knowl-Based Syst:921–947
- 20.Hou J, Gao T. Explainable dcnn based chest x-ray image analysis and classification for covid-19 pneumonia detection. Sci Rep. 2021;11(1):1–15. doi: 10.1038/s41598-018-33214-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Islam MZ, Islam MM, Asraf A. A combined deep cnn-lstm network for the detection of novel coronavirus (covid-19) using x-ray images. Informat Med Unlocked. 2020;20:100412. doi: 10.1016/j.imu.2020.100412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jain R, Gupta M, Taneja S, Hemanth DJ. Deep learning based detection and analysis of covid-19 on chest x-ray images. Appl Intell. 2021;51(3):1690–1700. doi: 10.1007/s10489-020-01902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jakubovitz D, Giryes R, Rodrigues MRD (2018) Generalization error in deep learning. arXiv:1808.01174
- 25.Kanwal A, Chandrasekaran S. 2dcnn-bicudnnlstm: hybrid deep-learning-based approach for classification of covid-19 x-ray images. Sustainability. 2022;14(11):6785. doi: 10.3390/su14116785. [DOI] [Google Scholar]
- 26.Karakanis S, Leontidis G. Lightweight deep learning models for detecting covid-19 from chest x-ray images. Comput Biol Med. 2021;130:104181. doi: 10.1016/j.compbiomed.2020.104181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lafraxo S, El Ansari M (2021) Covinet: automated covid-19 detection from x-rays using deep learning techniques. In: 2020 6th IEEE congress on information science and technology (CiSt). IEEE, pp 489–494
- 28.Lasker A, Ghosh M, Obaidullah SM, Chakraborty C, Roy K (2022) Deep features for covid-19 detection: performance evaluation on multiple classifiers. In: International conference on computational intelligence in pattern recognition. Springer, pp 313–325
- 29.LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 30.Li T, Han Z, Wei B, Zheng Y, Hong Y, Cong J (2020) Robust screening of covid-19 from chest x-ray via discriminative cost-sensitive learning. arXiv:2004.12592
- 31.Li X, Tan W, Liu P, Zhou Q, Yang J (2021) Classification of covid-19 chest ct images based on ensemble deep learning. J Healthc Eng [DOI] [PMC free article] [PubMed]
- 32.Liu W, Wang Z, Liu X, Zeng N, Liu Y, Alsaadi FE. A survey of deep neural network architectures and their applications. Neurocomputing. 2017;234:11–26. doi: 10.1016/j.neucom.2016.12.038. [DOI] [Google Scholar]
- 33.Loey M, El-Sappagh S, Mirjalili S. Bayesian-based optimized deep learning model to detect covid-19 patients using chest x-ray image data. Comput Biol Med. 2022;142:105213. doi: 10.1016/j.compbiomed.2022.105213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mangal A, Kalia S, Rajgopal H, Rangarajan K, Namboodiri V, Banerjee S, Arora C (2020) Covidaid: covid-19 detection using chest x-ray. arXiv:2004.09803
- 35.Mohammadi R, Salehi M, Ghaffari H, Rohani A, Reiazi R. Transfer learning-based automatic detection of coronavirus disease 2019 (covid-19) from chest x-ray images. J Biomed Phys Eng. 2020;10(5):559. doi: 10.31661/jbpe.v0i0.2008-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohammed M, Mwambi H, Omolo B, Elbashir MK (2018) Using stacking ensemble for microarray-based cancer classification. In: 2018 International conference on computer, control, electrical, and electronics engineering (ICCCEEE). IEEE, pp 1–8
- 37.Mooney P (2020) Chest X-ray images (pneumonia). www.kaggle.com. https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia. Accessed 10 May 2022
- 38.Mukherjee H, Dhar A, Obaidullah S, Santosh K, Roy K et al (2021) Covid-19: a necessity for changes and innovations. In: COVID-19: prediction decision-making, and its impacts, pp 99–105
- 39.Mukherjee H, Ghosh S, Dhar A, Obaidullah SM, Santosh K, Roy K. Deep neural network to detect covid-19: one architecture for both ct scans and chest x-rays. Appl Intell. 2021;51(5):2777–2789. doi: 10.1007/s10489-020-01943-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mukherjee H, Ghosh S, Dhar A, Obaidullah S M, Santosh K, Roy K (2021) Shallow convolutional neural network for covid-19 outbreak screening using chest x-rays. Cogn Comput:1–14 [DOI] [PMC free article] [PubMed]
- 41.Munusamy H, Muthukumar KJ, Gnanaprakasam S, Shanmugakani TR, Sekar A. Fractalcovnet architecture for covid-19 chest x-ray image classification and ct-scan image segmentation. Biocybernetics Biomed Eng. 2021;41(3):1025–1038. doi: 10.1016/j.bbe.2021.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu S, Liu M, Liu Y, Wang J, Song H. Distant domain transfer learning for medical imaging. IEEE J Biomed Health Inform. 2021;25(10):3784–3793. doi: 10.1109/JBHI.2021.3051470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Organization WH et al (2020) Clinical management of severe acute respiratory infection (sari) when Covid-19 disease is suspected: interim guidance, 13 March. Technical report, World Health Organization
- 44.Paluru N, Dayal A, Jenssen HB, Sakinis T, Cenkeramaddi LR, Prakash J, Yalavarthy PK. Anam-net: anamorphic depth embedding-based lightweight cnn for segmentation of anomalies in covid-19 chest ct images. IEEE Trans on Neural Netw and Learn Syst. 2021;32(3):932–946. doi: 10.1109/TNNLS.2021.3054746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prashant P (2020) Covid-19 diagnosis using X-ray images. Kaggle. https://www.kaggle.com/code/prashant268/covid-19-diagnosis-using-x-ray-images. Accessed 10 May 2022
- 46.Rahman T, Khandakar A, Qiblawey Y, Tahir A, Kiranyaz S, Kashem SBA, Islam MT, Al Maadeed S, Zughaier SM, khan MS, et al. Exploring the effect of image enhancement techniques on covid-19 detection using chest x-ray images. Comput Biol Med. 2021;132:104319. doi: 10.1016/j.compbiomed.2021.104319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Refat CMM (2020) Chest X-ray images pneumonia and covid-19. https://www.kaggle.com/masumrefat/chest-xray-images-pneumonia-and-covid19. Accessed 10 May 2022
- 48.Rezaee K, Badiei A, Meshgini S (2020) A hybrid deep transfer learning based approach for covid-19 classification in chest x-ray images. In: 2020 27th national and 5th international Iranian conference on biomedical engineering (ICBME). IEEE, pp 234–241
- 49.Saha P, Mukherjee D, Singh PK, Ahmadian A, Ferrara M, Sarkar R. Retracted article: graphcovidnet: a graph neural network based model for detecting covid-19 from ct scans and x-rays of chest. Sci Rep. 2021;11(1):1–16. doi: 10.1038/s41598-021-87523-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Saha P, Sadi MS, Islam MM. Emcnet: automated covid-19 diagnosis from x-ray images using convolutional neural network and ensemble of machine learning classifiers. Informat Med Unlocked. 2021;22:100505. doi: 10.1016/j.imu.2020.100505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C. (2018) Mobilenetv2: inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4510–4520
- 52.Shalbaf A, Vafaeezadeh M. Automated detection of covid-19 using ensemble of transfer learning with deep convolutional neural network based on ct scans. Int J Comput Assist Radiol Surg. 2021;16(1):115–123. doi: 10.1007/s11548-020-02286-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharifrazi D, Alizadehsani R, Roshanzamir M, Joloudari JH, Shoeibi A, Jafari M, Hussain S, Sani ZA, Hasanzadeh F, khozeimeh F, et al. Fusion of convolution neural network, support vector machine and sobel filter for accurate detection of Covid-19 patients using x-ray images. Biomed Sig Process Conrol. 2021;68:102622. doi: 10.1016/j.bspc.2021.102622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556
- 55.Singh D, Kumar V, Yadav V, Kaur M. Deep neural network-based screening model for Covid-19-infected patients using chest x-ray images. Int J Pattern Recognit Artif Intell. 2021;35(03):2151004. doi: 10.1142/S0218001421510046. [DOI] [Google Scholar]
- 56.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI conference on artificial intelligence
- 57.Tang S, Wang C, Nie J, Kumar N, Zhang Y, Xiong Z, Barnawi A. Edl-covid: ensemble deep learning for Covid-19 case detection from chest x-ray images. IEEE Trans Ind Informat. 2021;17(9):6539–6549. doi: 10.1109/TII.2021.3057683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ter-Sarkisov A. Lightweight model for the prediction of Covid-19 through the detection and segmentation of lesions in chest ct scans. Int J Autom Comput Artif Intell Mach Learn. 2021;2(1):01–15. [Google Scholar]
- 59.Trivedi M, Gupta A. A lightweight deep learning architecture for the automatic detection of pneumonia using chest X-ray images. Multimed Tools Appl. 2022;81(4):5515–5536. doi: 10.1007/s11042-021-11807-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Upadhyay K, Agrawal M, Deepak D. Ensemble learning-based Covid-19 detection by feature boosting in chest X-ray images. IET Image Process. 2020;14(16):4059–4066. doi: 10.1049/iet-ipr.2020.1127. [DOI] [Google Scholar]
- 61.Wolpert DH. Stacked generalization. Neural Netw. 1992;5(2):241–259. doi: 10.1016/S0893-6080(05)80023-1. [DOI] [Google Scholar]
- 62.Zhang X, Wang D, Shao J, Tian S, Tan W, Ma Y, Xu Q, Ma X, Li D, Chai J, et al. A deep learning integrated radiomics model for identification of coronavirus disease 2019 using computed tomography. Sci Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-83237-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou C, Bu G, Sun Y, Ren C, Qu M, Gao Y, Zhu Y, Wang L, Sun L, Liu Y. Evaluation of serum igm and igg antibodies in Covid-19 patients by enzyme linked immunosorbent assay. J Med Virol. 2021;93(5):2857–2866. doi: 10.1002/jmv.26741. [DOI] [PubMed] [Google Scholar]
- 64.Zhu J, Shen B, Abbasi A, Hoshmand-Kochi M, Li H, Duong TQ. Deep transfer learning artificial intelligence accurately stages Covid-19 lung disease severity on portable chest radiographs. Plos One. 2020;15(7):0236621. doi: 10.1371/journal.pone.0236621. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study used a secondary dataset as described by Rahman et al. in [10, 37, 45, 47]. The dataset can be obtained from the https://www.kaggle.com repository using this direct links: https://www.kaggle.com/datasets/tawsifurrahman/covid19-radiography-database, https://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumoniahttps://www.kaggle.com/datasets/paultimothymooney/chest-xray-pneumonia, https://www.kaggle.com/datasets/prashant268/chest-xray-covid19-pneumoniahttps://www.kaggle.com/datasets/prashant268/chest-xray-covid19-pneumonia.