SUMMARY
Tendon injuries heal via a scar-mediated response, and there are no biological approaches to promote more regenerative healing. Mouse flexor tendons heal through the formation of spatially distinct tissue areas: a highly aligned tissue bridge between the native tendon stubs that is enriched for adult Scleraxis-lineage cells and a disorganized outer shell associated with peri-tendinous scar formation. However, the specific molecular programs that underpin these spatially distinct tissue profiles are poorly defined. In the present study, we combine lineage tracing of adult Scleraxis-lineage cells with spatial transcriptomic profiling to define the overarching molecular programs that govern tendon healing and cell-fate decisions. Pseudotime analysis identified three fibroblast trajectories (synthetic, fibrotic, and reactive) and key transcription factors regulating these fate-switching decisions, including the progression of adult Scleraxis-lineage cells through the reactive trajectory. Collectively, this resource defines the molecular mechanisms that coordinate the temporo-spatial healing phenotype, which can be leveraged to inform therapeutic candidate selection.
Graphical abstract
In brief
Ackerman et al. integrated spatial transcriptomic profiling and fate mapping of adult Scleraxis-lineage cells during murine tendon healing to define key regulators of the healing process. This study provides important insights regarding the mechanisms that dictate cell fate in a spatially dependent manner.
INTRODUCTION
Tendons are responsible for facilitating skeletal locomotion throughout the body. Disruptions in tendon structure-function result in a dramatic reduction in quality of life, and these disruptions can occur with both acute traumatic injuries and as a result of chronic degeneration. Upon injury, tendons heal via a fibrotic scar-tissue response rather than regeneration of the native tendon structure, with this fibrotic response leading to chronic deficits in tendon function. Despite the frequency of tendon injuries (greater than 300,000 per year in the United States),1 the massive socioeconomic cost (estimated at >$400 million per year for flexor tendon injuries)2 and the complications associated with the fibrotic healing response, there are currently no consensus biological or pharmacological approaches to improve the healing process. A significant barrier to identifying promising therapeutics to enhance tendon healing is the limited understanding of the cellular and molecular programs that govern the fibrotic tendon healing process, particularly in the context of the temporal and spatial regulators of healing. For example, the tendon heals with spatially distinct patterns including the formation of highly organized bridging tissue between the native tendon ends, as well as the formation of disorganized scar tissue at the periphery of the injury site. Thus, understanding the specific molecular programs that dictate these spatially distinct tissue programs is key to harnessing the intrinsic regenerative potential and blunting the fibrotic tendency of the healing tendon.
In addition to broadly defining the spatial-molecular map of fibrotic tendon, delineating how different cell populations contribute to the healing process is a key step in further informing therapeutic candidate selection. Recent work has demonstrated substantial heterogeneity of the basal tendon cell environment,3–5 with different populations making distinct contributions to the healing process.6,7 Scleraxis (Scx), a basic helix-loop-helix transcription factor, is a critical regulator of tendon formation,8,9 with the loss of Scx resulting in developmental deficits in tendon. In addition, Scx-lineage (ScxLin) cells are the predominant cell population in the adult tendon and play a critical role in tendon healing,6,10,11 particularly in the formation of an organized cellular bridge between the injured tendon stubs.3 However, defining the function of ScxLin cells and their associated molecular programs is complicated by the inherent heterogeneity of this lineage: the ScxLin includes those cells that expressed Scx during embryonic development, the progeny of these cells, as well as any cell that expressed Scx during postnatal growth, adult homeostasis, and in response to injury. While comprehensive definition of the functions of ScxLin cells is important, we are particularly interested in adult ScxLin cells (ScxAi9), the cells that actively express Scx in the adult tendon during homeostasis. Adult ScxLin cells can become myofibroblasts during tendon healing, with myofibroblasts mediating multiple aspects of tissue repair including deposition, contraction, and remodeling of a new extracellular matrix (ECM).12 However, persistent or dysregulated myofibroblast function can promote a fibrotic healing response due to exuberant and sustained ECM deposition.12 In many tissues, the initiating step of myofibroblast differentiation is activation of quiescent, tissue-resident fibroblasts. In the current study, we take advantage of spatial transcriptomic profiling to examine the temporospatial activation and myofibroblast differentiation profile of adult ScxAi9 cells and to elucidate the regulation and function of these cells. Collectively, these data provide critical insights into the spatial-molecular regulation of fibrotic tendon healing and identify key intervention points to modulate cell fate, fibrosis, and healing potential.
RESULTS
Adult ScxAi9 cells undergo progressive and spatially dependent activation and myofibroblast differentiation to mediate both ECM elaboration and fibrosis
Tenocytes are largely quiescent during homeostasis,13 but there is evidence that injury can induce fibroblast proliferative expansion and activation during healing.14,15 Adult ScxAi9 cells were labeled prior to acute flexor tendon injury and repair, including a washout period to ensure that any cells that turned on expression of Scx in response to injury were not labeled (Figure 1A). While there are proliferating cell nuclear antigen (PCNA)+ ScxAi9 cells in both the tendon stub and bridging scar tissue at day 8 post-repair, proliferation of ScxAi9 cells peaks between days 10 and 14 in both the stub and bridging regions. While many ScxAi9 cells within the stub remained PCNA+ at day 21, ScxAi9 cells in the bridging tissue were almost exclusively PCNA−. By day 28 post-repair, only a few ScxAi9 cells in both the tendon stubs and bridging tissue were PCNA+ (white arrows, Figure 1B).
Figure 1. Adult ScxAi9 cells undergo progressive and spatially dependent activation and myofibroblast differentiation to mediate both ECM elaboration and fibrosis.
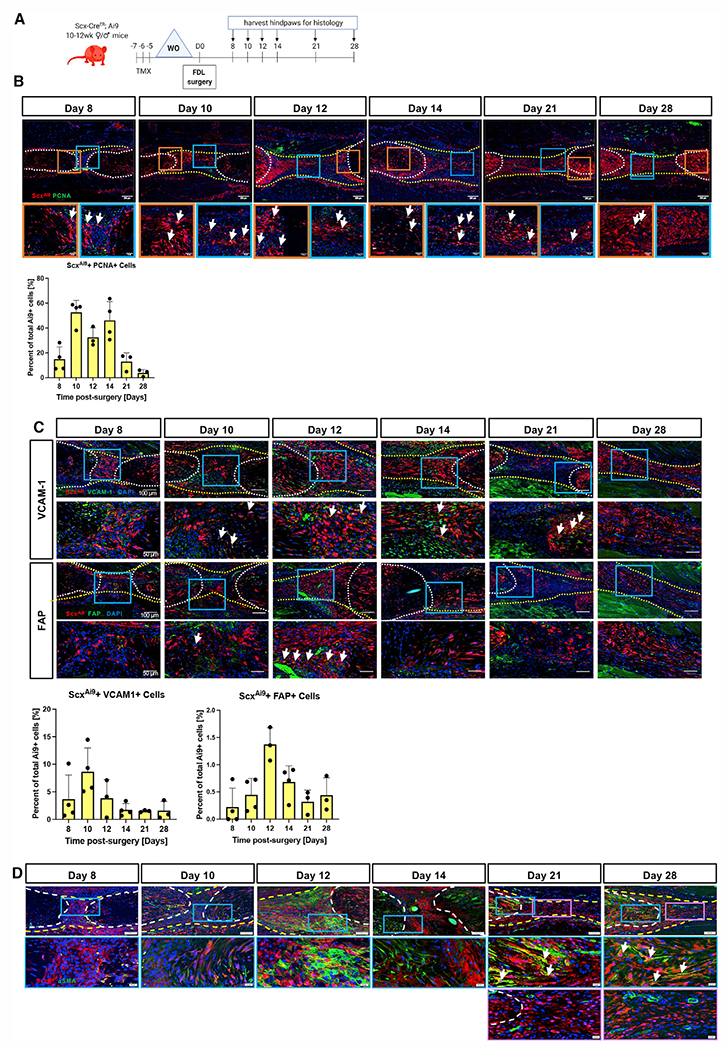
(A) ScxAi9 mice were injected with tamoxifen (Tmx) for 3 days, followed by a 4-day washout (WO) period prior to tendon repair surgery, and tendons were harvested between 8 and 28 days post-repair. Scale bars on low-magnification images represent 200 μm and 50 μm on higher-magnification images.
(B and C) (B) Co-immunofluorescence of ScxAi9 (red) and PCNA (green), (C) ScxAi9 (red) and VCAM-1 (green) (top), or ScxAi9 (red) and FAP (green) (bottom) between 8 and 28 days post-repair. Nuclei are stained with DAPI. Tendon is outlined by white dotted lines and scar tissue by yellow dotted lines. Examples of co-localization indicated by white arrows. Scale bars on low-magnification images represent 100 μm and 50 μm on higher-magnification images.
(D) Co-immunofluorescence between ScxAi9 (red) and αSMA+ myofibroblasts (green) between 8 and 28 days post-repair. Scale bars on low-magnification images represent 200 μm and 20 μm on higher-magnification images. Blue and purple boxes indicate locations of higher magnification images. White arrows indicate examples of co-localization.
Quantitative data are plotted mean ± standard deviation. n = 3–4 per time point.
See also Figure S1.
To further determine the spatial-temporal patterns of fibroblast activation of adult ScxAi9 cells during tendon healing, de novo expression of vascular cell adhesion protein 1 (VCAM-1) and fibroblast activation protein (FAP) were evaluated in these cells. At day 8, several ScxAi9+ cells co-expressed VCAM-1, while some VCAM-1+ cells were ScxAi9− (Figure 1C). While overall VCAM-1 staining increased through day 14, the proportion of ScxAi9+ VCAM-1+ cells progressively decreased after day 10. A substantial decrease in overall VCAM-1 expression was observed at days 21 and 28. At day 8, there was minimal expression of FAP, and a few FAP+ ScxAi9 cells were seen at day 10 (white arrows, Figure 1C); however, most FAP expression was adjacent to ScxAi9 cells (Figure 1C). By day 12, many ScxAi9 cells in the bridging tissue were FAP+; however, by day 14, FAP expression had dramatically decreased in both the bridging tissue and tendon stubs, with only a few FAP+ cells remaining in the tendon stubs. These low levels of FAP expression in the bridging tissue persisted through day 28. Collectively, these data suggest that fibroblast activation of ScxAi9 cells takes place between approximately day 10 and 14 and that cell activation is largely restricted to the bridging tissue.
Fibroblast activation is associated with both ECM production and myofibroblast differentiation. We have previously shown that adult ScxAi9 cells form a highly aligned cellular bridge between the native tendon ends,3 yet it is unclear whether these cells elaborate this new ECM or simply use an existing ECM scaffold as a bridge during tendon repair. To address this, we examined the relationship between ScxAi9 cells and ECM deposition using sequential immunofluorescence and Masson’s trichrome staining on the same samples. As we have previously demonstrated a lack of both ScxAi9 cells and new collagen ECM between the native tendon ends at day 7,3 we analyzed samples starting at day 8 post-surgery (Figure S1). At day 8, a few ScxAi9 cells had egressed from the tendon stubs, accompanied by modest collagen fiber presence in the healing tissue (Figure S1). At day 10, a substantial increase in the extent of both ScxAi9 and collagen ECM was observed, with strong overlap between the cells and ECM, and this tandem elaboration of the cell/ECM bridge continued through day 12 (Figure S1). These data demonstrate concomitant development of the ScxAi9 cellular bridge and the collagen ECM bridge, suggesting a key role for ScxAi9 cells in elaboration of this new organized matrix. Interestingly, there was a clear demarcation between ScxAi9+ cells (white arrows) and ScxAi9− cells (yellow arrows) at the periphery of the new ECM bridge (Figure S1), suggesting the presence of discrete molecular programs and cell populations in this area. We next examined expression of the collagen chaperone protein Hsp47 and observed abundant co-localization of Hsp47 in ScxAi9 cells as the new ECM was developed from day 8 to 12 (Figure S1), further supporting ScxAi9 cells as the primary producers of this new ECM bridge.
We have previously shown that adult ScxAi9 cells are not a primary contributor to myofibroblast fate at day 14;3 however, based on the transient “activated” state of ScxAi9 cells and their ECM elaboration phenotype, we examined myofibroblast fate over the course of healing more comprehensively (Figure 1D). At days 8 and 10, minimal α smooth muscle actin (αSMA)+ myofibroblasts were observed. However, by day 12, many cells in the expanded αSMA+ population were derived from adult ScxAi9 cells. Moreover, consistent with our previous data, at day 14, αSMA+ ScxAi9-derived myofibroblasts were primarily restricted to the tendon stubs and not in the bridging granulation tissue (Figure 1D), with this expression pattern continuing through day 28, indicating spatially dependent myofibroblast differentiation and persistence. Interestingly, these data suggest that not all adult ScxAi9 cells that undergo fibroblast activation continue to a mature myofibroblast phenotype and that the contribution to myofibroblast fate is both time and location dependent.
Integration of multiple spatial RNA sequencing (RNA-seq) datasets to elucidate transcriptional changes in cellular/molecular programs over time
The clear temporal and spatial dependence of the adult ScxAi9 contribution to ECM organization and both transient and persistent myofibroblast populations suggest that there are discrete molecular programs underlying these fate-function dynamics. Therefore, we utilized spatial transcriptomics to define and spatially resolve these distinct molecular programs during tendon healing.
Adult ScxAi9 cells were labeled prior to injury, and hind paws were harvested from uninjured controls, and at days 14, 21, and 28 post-surgery, to encompass different phases of tendon healing and adult ScxAi9 function. For each section, a region of interest (ROI) including the healthy tendon (i.e., uninjured [UI]) or the entire area of the healing tendon (tendon and scar tissue) were selected for subsequent integration and analysis (Figures 2A and S2). Integrating the selected ROI from each time point allows identification of heterogeneous molecular programs at a higher resolution than analysis of a single time point and facilitates the identification of transcriptional dynamics over time. Integrated analysis identified six conserved molecular programs (i.e., clusters) during tendon healing (Figure 2B), which were then annotated via analysis of differentially expressed genes (DEGs), biological Gene Ontology (GO) terms, and spatial localization (Figures 2C–2E).
Figure 2. Integration of tendon and scar ROI over time defines transcriptome dynamics during healing.
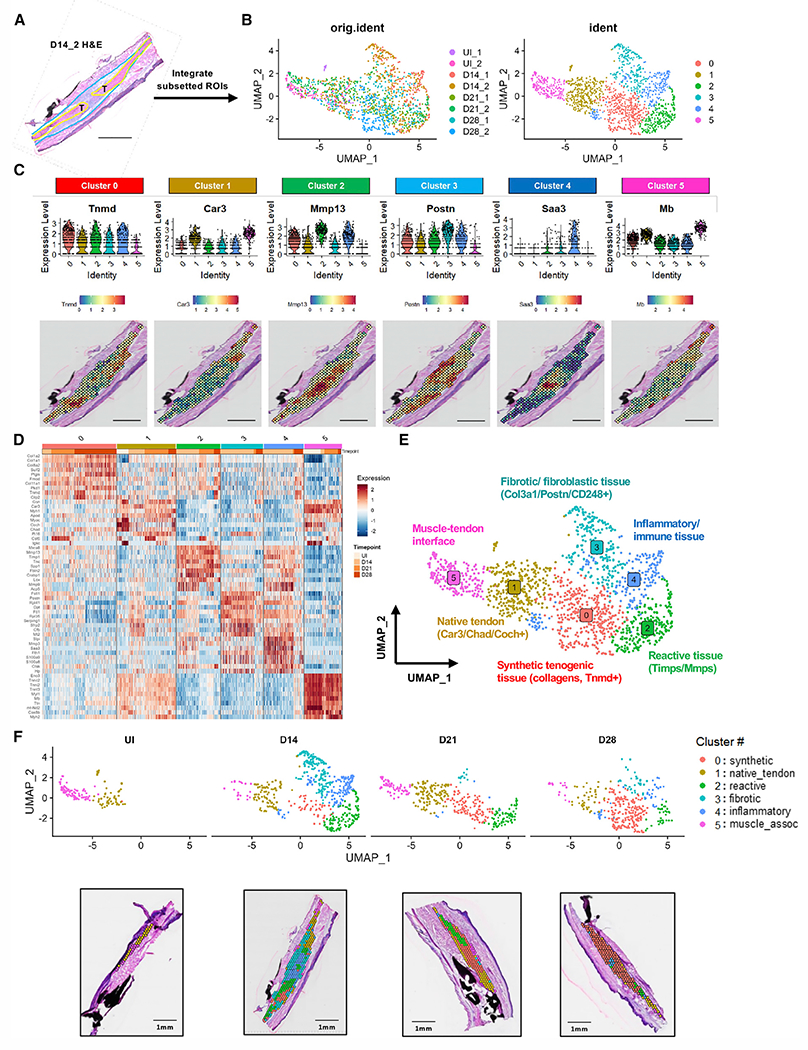
(A) Representative H&E image from day 14_2 sample; ROI outlined in yellow (tendon ends) and blue (scar tissue) was subsetted for integration.
(B) Uniform manifold approximation and projection (UMAP) of integrated and batch-corrected dataset demonstrates (left) distribution of spots from each individual sample and (right) six distinct clusters.
(C) Violin plots and spatial mapping of highly expressed genes within each integrated cluster.
(D) Heatmap of top 10 DEGs from each cluster.
(E) Annotated UMAP of integrated dataset.
(F) Distribution and localization of integrated clusters over time represented by both UMAP (top) and spatial plot (bottom, one representative sample at each time point).
UI indicates uninjured healthy tendon. Scale bars represent 1 mm.
See also Figures S2 and S3.
Cluster 0, which is predominantly located in less reactive areas of native tendon further from the repair site, represents a highly synthetic tenogenic program, expressing predominantly Tnmd, Col1a1, and Fmod (C0synthetic; Figures 2C–2E and S3). GO term analysis reveals that this population is involved in ECM organization (GO: 0030198), collagen fibril organization (GO: 0030199), and wound healing (GO: 0042060), reinforcing this annotation.
Cluster 1, enriched in Coch, Chad, and Car3, most closely approximates native tendon-like tissue (C1native_tendon) and is found abundantly in the UI tendon samples and areas distant from the injury site (Figures 1C and 1F). GO analysis indicates upregulation of numerous contractile/muscle-related processes (Figure S3), potentially a byproduct of the lower resolution of this spatial technique or indicative of the lack of tendon-specific curated processes.
Cluster 2 represents reactive tissue found immediately adjacent to the repair site (C2reactive) and is characterized by high expression of Mmp13/Lox/Fbln2 (Figures 2C–2E). Consistent with its annotation, many of the enriched pathways in C2reactive relate to ECM organization: collagen fibril organization (GO: 0030199), ECM organization (GO: 0030198), supramolecular fiber organization (GO: 0097435), collagen catabolic processes (GO: 0030574), and positive regulation of many cellular activities (cell adhesion, migration, proliferation, collagen biosynthesis) (Figure S3).
Cluster 3 defines a fibrotic/fibroblastic program (C3fibrotic) at the peripheral shell of the healing tendon and is enriched in pro-fibrotic markers Col3a1, Postn, and Thbs3 (Figures 2C–2E and S3). The top GO terms found in this cluster are all related to translation (translation, GO: 0006412; cytoplasmic translation, GO: 0002181; rRNA processing, GO: 0006364), indicating high levels of ECM synthesis.
Cluster 4, located immediately at the repair site and adjacent to C2reactive, identifies an inflammatory program (C4inflammatory) with high expression of Saa3, alarmins S100a8 and S100a9, and markers consistent with a variety of immune cells such as neutrophils (e.g., Lcn2) and monocytes (Figures 2C–2E and S3). In addition to GO terms associated with immune system processes (GO: 0002376) and neutrophil chemotaxis (GO: 0030593), this cluster also has GO terms associated with ECM organization (e.g., collagen fibril organization, cell-matrix adhesion).
Cluster 5 exhibits features of both muscle and native tendon tissue, representing a muscle-tendon interface (C5muscle-assoc) (Figures 2C–2E and S3).
Distribution of clusters over time supports these annotations, with only C1native_tendon and C5muscle-assoc present in UI tendons, expansion of C4inflammatory at day 14, and enrichment of C0synthetic through day 28. Interestingly, the presence of C2reactive persists from day 14 to 28 (Figure 2F), suggesting that this sustained reactive response may hinder a complete return to a normal tenogenic program.
Identification of fibroblastic differentiation trajectories and key transcriptional regulators
The nature of this comprehensive spatiotemporal dataset allows us to gain important insights into the potential differentiation pathways of tendon-resident fibroblasts over the course of healing. First, C5muscle-assoc and C4inflammatory were removed from subsequent analysis, retaining only those clusters that could participate in tendon-associated differentiation trajectories.
We then conducted trajectory analysis, using C1native_tendon as the root for all lineage trajectories, and found trifurcations toward C3fibrotic (blue line, Figure 3A), C2reactive (green line, Figure 3A), and C0synthetic (red line, Figure 3A), suggesting that tendon cells may differentiate to one of these three fates following activation. Given the clear role for adult ScxAi9 cells in multiple aspects of tendon healing, and taking advantage of the built-in lineage tracing of ScxAi9 (tdTomato expression) in our experimental design, we subset the Tdtomato.pos (ScxAi9+) and Tdtomato.-neg (ScxAi9−) populations within each cluster using expression tdTomato >1 (Figures 3B and 3C). Interestingly, while C2reactive comprised the highest percentage of tdTomato+ cells (82%), a relatively high percentage of spots within C4inflammatory were also tdTomato+ (64%; Figure 3D). To better understand the relationship between ScxAi9 and C4inflammatory, we examined the expression of ScxAi9 (RFP) and CD45. Surprisingly, many ScxAi9+ cells co-expressed CD45 (Figure S4). While CD45 is commonly used to define cells of hematopoietic origin, recent studies have identified CD45 as a possible marker of circulating fibrocytes.16,17 As such, future work will be needed to address the surprising possibility that there may be a non-tendon derived ScxAi9+ CD45+ population during healing. To confirm fibroblast activation in our spatial dataset, we examined expression of activation markers within each cluster and observed increased Fap and Acta2 expression in the tdTomato+ subset of C2reactive, relative to the tdTomato subset of this cluster, suggesting a functional difference compared with non-ScxAi9 cells (Figure S5).
Figure 3. ScxAi9 pseudotime trajectory analysis from native tendon to the reactive cellular/molecular program.
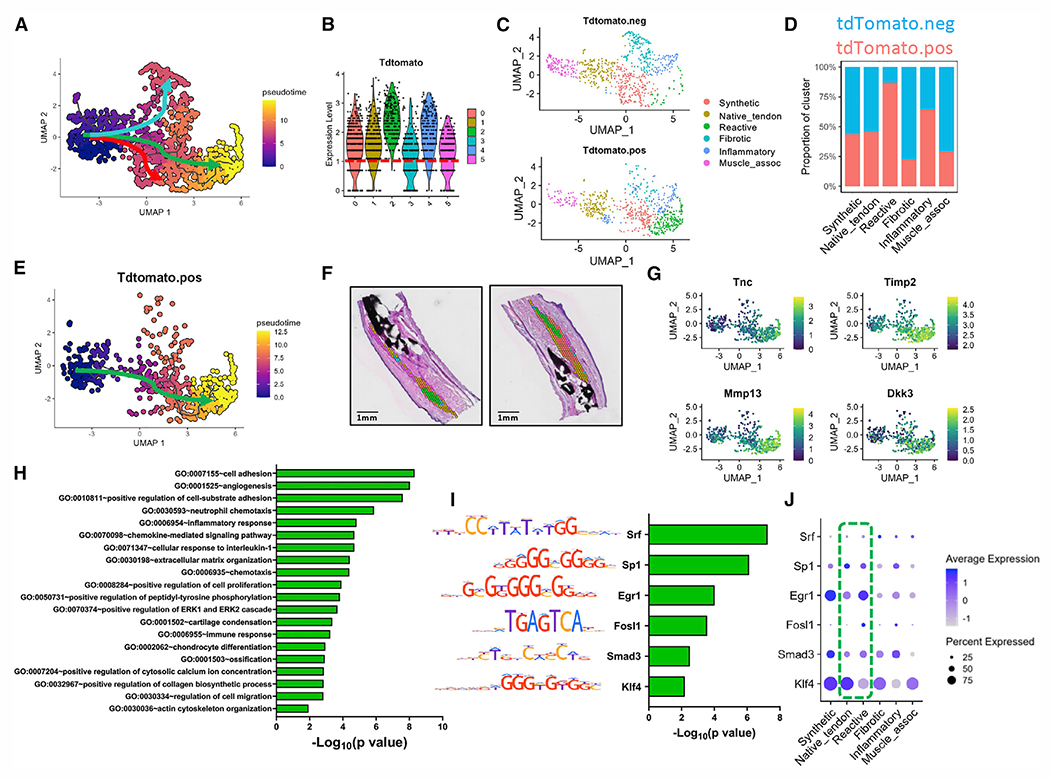
(A) UMAP depicting differentiation pathways from C1native_tendon to C3fibrotic (blue arrow), C2reactive (green), and C0synthetic (red).
(B and C) Violin plot depicting expression pattern of (B) tdTomato, indicative of ScxAi9 cells, across all clusters and (C) tdTomato+/− subsets based on a cutoff of tdTomato >1 (red dotted line).
(D) Quantification of ScxAi9 spots per cluster.
(E) Pseudotime trajectory analysis for the tdTomato+ subset follows the reactive module pathway (green arrow).
(F and G) Spatial localization of C2reactive at day 21 post-injury (F), along with feature plots of representative genes enriched along the reactive differentiation route, including Timp2, Tnc, Mmp13, and Dkk3 (G).
(H) Identification of GO terms enriched the reactive module.
(I and J) TF binding motif analysis predicts transcription factors Srf, Sp1, Egr1, Fosl1, Smad3, and Klf4 as regulators of the reactive program trajectory (I), which are also shown with a dot plot (J) to demonstrate expression level and localization within clusters.
Green box highlights expression levels in native tendon and reactive tissue. Scale bars represent 1 mm.
See also Figure S5.
To determine which genes may be governing fate decisions, we analyzed gene modules contributing to fate specification of ScxAi9 from C1native_tendon to C2reactive. Genes upregulated along this trajectory included Tnc, Timp2, Mmp13, and Dkk3 (Figure 3G). GO analysis revealed that upregulated pathways along this differentiation route were consistent with an actively remodeling state: increased angiogenesis (cell adhesion, GO: 0007155; angiogenesis, GO: 0001525; positive regulation of cell proliferation, GO: 0008284), inflammatory processes (neutrophil chemotaxis, GO: 0030593; inflammatory response, GO: 0006954), and matrix organization (ECM organization, GO: 0030198; positive regulation fo collagen biosynthetic process, GO: 0032967) (Figure 3H). Transcription factor analysis predicts that that Srf, Sp1, Egr1, Fosl1, Smad3, and Klf4 are likely to regulate this differentiation trajectory (Figure 3I). Notably, Egr1, Smad3, and Klf4 are highly expressed in C0synthetic, through which the reactive trajectory passes, suggesting thatthese represent key fate-switching transcription factors from C0synthetic to C2reactive (Figure 3J).
We then examined the trajectories of tdTomato−, with paths from C1native_tendon into C3fibrotic (Figure 4), and C0synthetic (Figure 5). The C1native_tendon to C3fibrotic path is described by the fibrotic module and upregulation of markers such as Adamts15, Ccl6, Col14a1, and Dpt (Figures 4A–4C). The majority of the highly enriched pathways in this trajectory indicate involvement of a variety of immune cells and pathways (neutrophil chemotaxis, GO: 0030593; immune system process, GO: 0002376; innate immune response, GO: 0045087) as well as inflammation (inflammatory response, GO: 0006954; cellular response to tumor necrosis factor, GO: 0071356) (Figure 4D). These pathways, combined with the early and transient presence of this cluster in proximity to C4inflammatory (Figures 2 and 4B), suggests that C3fibrotic is highly immunoresponsive and may interact with infiltrating immune cells to initiate abundant ECM production. Transcription factor (TF) binding motif analysis further confirms the inflammatory-immune bias of this trajectory, with high expression of Nfkb1 and Rela (a binding partner in the nuclear factor κB [NF-κB] complex), two proteins known to act as main regulators of inflammatory signaling cascades, as well as interferon regulatory factor 8 (Irf8), involved in major histocompatibility (MHC) complex I expression (Figure 4E). Several other notable pathways are implicated along this trajectory route: Jun is broadly expressed among clusters and is shown to interact with Fosb, with which it forms the dimerized TF AP-1 to control cell differentiation, proliferation, and apoptosis. Interestingly, Tcf4 has been implicated in neuronal differentiation18 and is widely expressed in all clusters, hinting at nerve ingrowth processes following tendon injury (Figures 4E and 4F).
Figure 4. Pseudotime trajectory analysis from native tendon to the fibrotic cellular/molecular program.
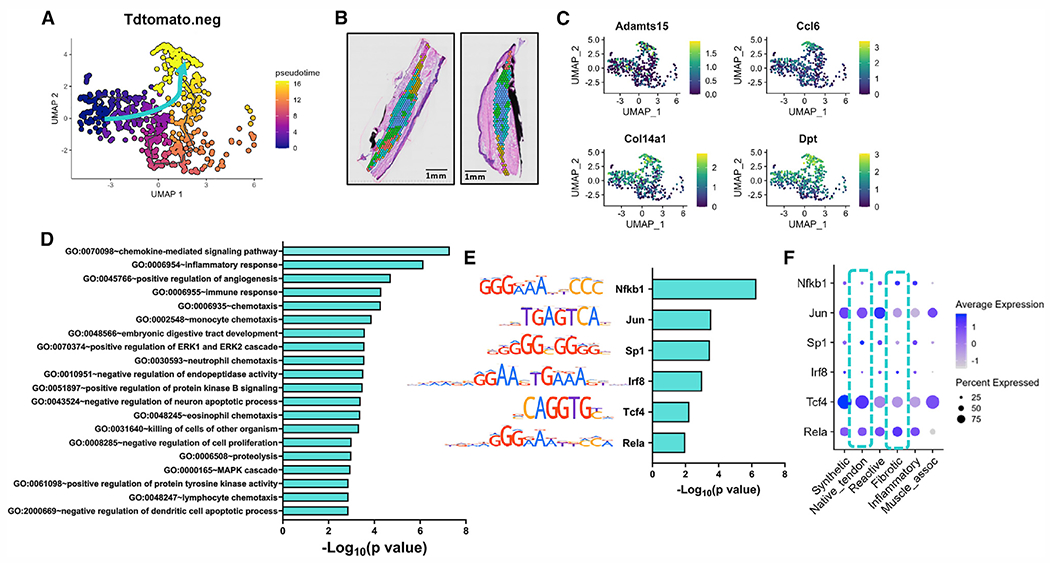
(A–C) UMAP depicting the specific pseudotime trajectory chosen for defining the differentiation path (blue arrow) (A), spatial localization of C3fibrotic at day 14 post-injury (B), and feature plots of representative genes enriched along this route, including Adamts15, Ccl6, Col14a1, and Dpt (C).
(D) Identification of GO terms enriched in the fibrotic module.
(E and F) TF binding motif analysis predicts transcription factors Nfkb1, Jun, Sp1, Irf8, Tcf4, and Rela as key regulators of the trajectory on this differentiation route (E), along with dot plot to demonstrate expression level and localization within clusters (F).
Blue box highlights expression levels in native tendon and fibrotic tissue. Scale bars represent 1 mm.
See also Figure S5.
Figure 5. Pseudotime trajectory analysis from native tendon to the synthetic cellular/molecular program.
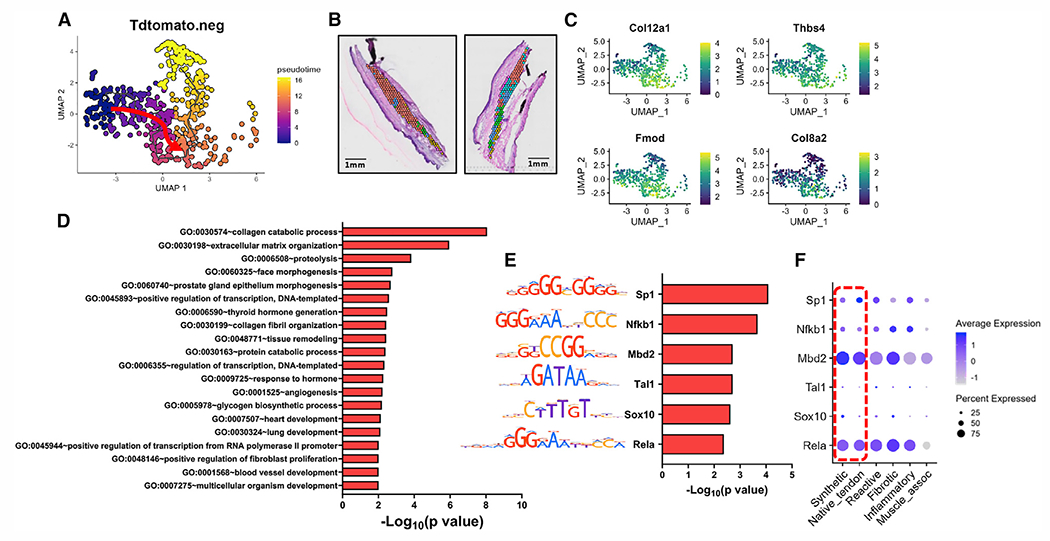
(A–C) UMAP depicting the specific pseudotime trajectory chosen for defining the differentiation path (red arrow) (A), spatial localization of C0synthetic at day 28 post-injury (B), and feature plots of representative genes enriched along this route, including Col12a1, Thbs4, Fmod, and Col8a2 (C).
(D) Identification of GO terms enriched the synthetic module.
(E) TF binding motif analysis predicts transcription factors Sp1, Nfkb1, Mbd2, Tal1, Sox10, and Rela to be key regulators of the trajectory in this synthetic module.
(F) Dot plot of TFs demonstrates expression level and localization within clusters.
Red box highlights expression levels in native tendon and synthetic tissue. Scale bars represent 1 mm.
See also Figure S5.
Finally, the differentiation pathway to C0synthetic is defined by the trajectory from C1native_tendon to C0synthetic, with C0synthetic being the predominant cluster at day 28 (Figures 5A and 5B). Upregulated along this route are genes indicative of increased collagen deposition (Col12a1, Col8a2), along with proteoglycans (Thbs4, Fmod), which are commonly found in mature connective tissues. Consistent with this, highly upregulated pathways involve collagen organization (collagen catabolic process, GO: 00305074; ECM organization, GO: 0030198; tissue remodeling, GO: 0048771) and several related to developmental processes (face morphogenesis, GO: 0060325; heart development, GO: 0007507; lung development, GO: 0030324), suggesting a developmentally guided remodeling program (Figure 5D). Transcriptional regulation includes factors Sp1, Nfkb1, Mbd2, Tal1, Sox10, and Rela. The repeated presence of Nfkb1 and Rela as top TFs regulating differentiation further underlines the important role of NF-κB signaling during tendon healing.19 Interestingly, while Sox10 is commonly studied in neural crest development, it has also been implicated in tendon progenitor migration and development, potentially supporting a more regenerative mechanism underlying C0synthetic differentiation.20,21
Cellular interactome analysis defines C2reactlve as the most talkative cluster
In addition to understanding the signaling pathways and programs that are specific to each molecular program, an important strength of spatial transcriptomics is the ability to define the interactions between physically proximate clusters. Therefore, we next established the patterns of cell-cell crosstalk both within a given molecular cluster (i.e., autocrine signaling) and between molecular clusters that are in proximity to each other (i.e., paracrine signaling). Inferring the relative strength of cellular communication using CellChat, we observed that C2reactive both qualitatively and quantitatively makes up the majority of interactions and exhibits the highest strength of interaction in both the tdTomato+ and tdTomato− subsets (Figure 6A). Relative information flow analysis reveals significant signaling heterogeneity between the ScxAi9+ and ScxAi9− subsets, with several pathways mainly enriched in the ScxAi9+ subset (Notch, Epha, App, Ncam, Cdh5, Sema7, Ptprm, Visfatin) (Figures 6B and 6C). The overall signaling patterns indicate that C2reactive most often participates in autocrine signaling. Therefore, we defined the specific signaling pathways between ScxAi9+ and ScxAi9− populations within this cluster. Thrombospondin signaling (Thbs4-Cd47) and Spp1 signaling (Itgav-Itgb1 and Itga5−+Itgb1) were the predominant different signaling pathways between the ScxAi9+ and ScxAi9− subsets in C2reactive. In addition, several signaling pairs indicated receptors that are only present in the C2reactiVe/ScxAi9+ population, including TGFbR2, Cd47, and Epha2/3 (Figure 6D). We then examined paracrine signaling from C0synthetic, C3fibrotic, and C4inflammatory clusters to the ScxAi9+ subset in C2reactive. Consistent with the signaling communication within C2reactive, thrombospondin signaling (Thbs4-Cd47) had the highest predicted communication probability from C4inflammatory to the ScxAi9+ subset in C2reactive (Figure 6E). Interestingly, there was substantial heterogeneity in communication between the ScxAi9+ subset in C2reactive and the ScxAi9+ and ScxAi9− subsets in the remaining clusters (Figure 6E), suggesting that spatial communication is far more specialized than has previously been appreciated.
Figure 6. Identification of the interactome in relation to the ScxLin+ population.
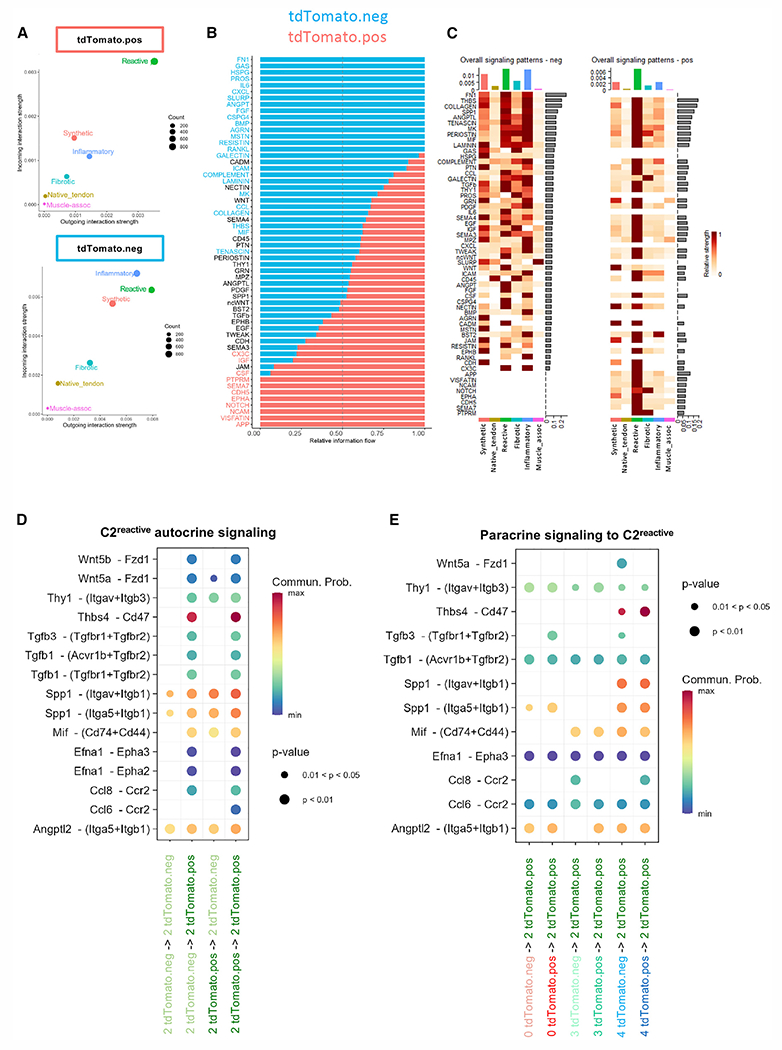
(A) Overall interaction strength of each cluster in the tdTomato+ subset (top) and tdTomato− subset (bottom) demonstrates differential interaction strength for C2reactive.
(B) Relative information flow between subpopulations; top signaling pathways colored red are enriched in the tdTomato+ subset, and those colored blue enriched in the tdTomato− subset.
(C) Overall signaling patterns for each subset breaks down the overall information flow to demonstrate which integrated clusters participate in the top signaling pathways.
(D) Dot plot of the identified ligand-receptor interactions within and between the tdTomato+ and tdTomato− subsets in C2reactive.
(E) Dot plot of ligand-receptor interactions between the tdTomato+ and tdTomato− subsets of C0synthetic, C3fibrotic, and C4inflammatory to the tdTomato+ subset of C2reactive. Dot size and color indicate the significance of the interaction (given as a range of p values), and the probably of communication, respectively.
See also Figures S6 and S7.
Finally, we looked at overall signaling patterns independent of ScxAi9 (Figures S6 and S7). As noted above, C2reactive was the most communicative cluster (Figures S6A and S6B). In line with our unbiased clustering (Figures 2B and 2C), Periostin (Postn) signaling was most likely to be outgoing from C3fibrotic, and complement signaling originated mainly from C3fibrotic and C4inflammatory (Figure S6C). We also conducted more in-depth interrogation of the predominant signaling pathways to better define key ligand-receptor interactions within and between C2reactive and C4inflammatory clusters. Thrombospondin signaling (Thbs4-Sdc1 and Thbs4-Cd47) were the most probable interactions within both C2reactive and C4inflammatory clusters, as well as between C2reactive and C4inflammatory clusters (Figures S6D and S7).
DISCUSSION
Dissecting the molecular programs that underlie the physiological and pathological aspects of tendon healing is critical for the development and translation of biological and pharmacological approaches to improve tendon healing. ScxLin cells are critical for both tendon homeostasis and repair, and a subset of these cells is found to contribute to the myofibroblast fate following acute injury. Here, we have demonstrated the time- and location-dependent proliferation, activation, and myofibroblast fate of adult ScxLin cells (ScxAi9) during tendon healing. These data demonstrated a spatial response in which ScxAi9 cells initially contribute to an organized cellular bridge at the repair site without progression to a myofibroblast phenotype, followed by progressive differentiation to αSMA+ myofibroblasts, with these cells primarily restricted to the remodeling stubs of the native tendon. Given the importance of the myofibroblast fate to both successful wound healing and fibrotic progression, we sought to define the specific spatial-molecular programs underlying the overall healing process, and the distinct responses of adult ScxAi9 cells, using spatial transcriptomics.
During normal wound healing, “fibroblast activation,” in which tissue-resident fibroblast populations break quiescence, proliferate, and contribute to the healing, is typically initiated via production of damage-associated molecular proteins (DAMPs),22 and activation is strongly associated with subsequent myofibroblast differentiation.23,24 While fibroblast activation has been observed in tenocytes,25,26 the cell lineage of myofibroblast origin can also impact myofibroblast function.27,28 Thus, we have focused specifically on the activation profile and myofibroblast differentiation of adult ScxLin (ScxAi9) cells. Somewhat surprisingly, we found that activation markers were predominantly associated with ScxAi9 cells in the organized bridging tissue between the native tendon ends, with these cells undergoing a transient differentiation to αSMA+ myofibroblasts, followed by loss of αSMA staining in these cells by day 14. In contrast, minimal expression of activation markers was observed in ScxAi9 in the native tendon stubs even though these cells progress to a more persistent myofibroblast fate through day 28. The differences in activation profile between transient and persistent myofibroblasts suggest that distinct molecular programs likely underlie these fate decisions and, as such, represent potentially important intervention opportunities to modulate myofibroblast activity. While the resolution of current spatial transcriptomics technology does not allow us to precisely resolve these spatially distinct processes at the single-cell level, pseudotime analysis provides key insights into the regulation of distinct tissue features.
Pseudotime analysis using C1native_tendon as the root identified fate trifurcation into (1) synthetic/tenogenic, (2) fibrotic/fibroblastic tissue, and (3) reactive tissue. The adult ScxAi9 population was shown to exclusively follow a differentiation trajectory from native tendon to reactive tissue. Given the approximate co-localization of C2reactive and C4inflammatory clusters at the site of tendon injury, this trajectory is likely to be heavily influenced by ongoing interaction with immune cells or inflammatory mediators. This resident fibroblast-immune cell interaction has previously been shown to be a key driver of both fibroblast activation and myofibroblast differentiation.29–32 While this process is key to proper initiation of wound healing, sustained interaction can lead to a chronically activated tissue state and fibrosis.33 Consistent with this, we observe both the persistence of C4inflammatory through day 28 and a sustained myofibroblast presence in the native tendon stubs. Normal resolution of wound healing is characterized by clearance of immune cells and myofibroblasts; as such, disruption of this chronically reactive state may promote restoration of a normal tenogenic program. Indeed, several of the transcriptions factors that are predicted to mediate the “reactive” pseudotime module are strongly associated with myofibroblast differentiation and tissue fibrosis. For example, aberrant Egr1 expression is associated with scleroderma34,35 and idiopathic pulmonary fibrosis,36 while Egr1−/− attenuates myofibroblast differentiation.37 However, Egr1 is also a highly mechanosensitive TF expressed during tendon homeostasis,38–40 suggesting potential context-dependent functions that may vary between tendon homeostasis and healing.
Consistent with only a minor subpopulation of tdTomato+ cells (ScxAi9+) in both C0synthetic and C3fibrotic, the ScxAi9 population did not take part in either of these differentiation trajectories from C1native_tendon. Based on the related biological processes, the localization of C0synthetic further away from the injury site, and a shift toward enrichment for C0synthetic by day 28 post-healing, we suggest that this trajectory defines the restoration of injured tendon back to its native structure. As such, stimulation of signaling pathways specific to this molecular cluster may be a means to restore native tendon structure and function by supporting pro-tenogenic programming. While the TFs identified as regulating this process have not been extensively studied in the context of tendon, this module represents a potentially important approach to restore normal tenogenic function after injury.
In contrast to C2reactive and C0synthetic, C3fibrotic is located within a peripheral capsule surrounding the primary repair area. Importantly, this exterior scar tissue shell is likely a critical driver of peritendinous adhesion formation. In the murine model, which forms transient peritendinous adhesions, the C3fibrotic cluster is relatively transient, with predominant enrichment at day 14 and a subsequent decrease through day 28. Thus, future work is needed to establish whether the sustained presence of this tissue program may be working to promote persistent adhesion formation in the context of clinical flexor tendon injuries. Accordingly, several TFs predicted to regulate this fibrotic module are strongly associated with fibrosis, such as Rela and Nfkb1.41–44 RelA/NF-κB inhibition has been shown to reduce tendon scarring,45 suggesting that inhibition of the fibrotic module drives more regenerative healing.
In addition to those TFs specific to individual modules, common TFs were also identified between modules, suggesting broader regulation of certain aspects of the healing process. For example, Srf1 was predicted to regulate both the reactive and synthetic modules. Srf1 is involved in cell growth and differentiation,46,47 while Sp1 was predicted to regulate all three pseudotime modules, consistent with its known functions relating to collagen biosynthesis.48,49
While spatial transcriptomics does not provide single-cell resolution, a major benefit is the ability to interrogate the cross-talk between clusters in physical proximity, thereby allowing identification of potential key interactions underpinning these molecular programs. Our cellular interactome data derived from the integrated dataset focused primarily on the reactive population (C2reactive), as this cluster represented the bulk of both outgoing and incoming signaling patterns, consistent with a sustained “reactive” phenotype. Thbs4-Sdc1 and Thbs4-Cd47 were the main ligand-receptor interactions both within C2reactive autocrine signaling and between C2reactive and C4inflammatory. Thrombospondin signaling, and more specifically the Thbs4 ligand, seems to exert broad effects throughout healing, consistent with a requirement for Thbs4 in the normal formation of tendon and myotendinous junction.50,51 However, an interesting restriction of Thbs4-Cd47 signaling was observed: the tdTomato+ subset of C2reactive was exclusively and consistently identified as expressing Cd47 receptor component of this ligand-receptor pair. Within the C2reactive cluster, this interaction is only observed in tdTomato+ autocrine signaling and tdTomato− signaling to tdTomato+. Moreover, Thbs4-Cd47 was identified only in C4inflammatory (tdTomato+ and tdTomato− subsets) signaling to the tdTomato+ subset of C2reactive. Given the role of Cd47 functioning as a “do not eat me” signal to patrolling macrophages,52,53 this may be an important mechanism by which adult ScxAi9 cells maintain a persistent reactive myofibroblast phenotype in the native tendon stubs, thereby preventing restoration of a normal tenogenic profile.
In addition to defining the key programs and events that dictate the spatial map of tendon healing, these data shed important light on potential regulators of tendon homeostasis that have not previously been appreciated. For example, in the integrated analysis, Cochlin (Coch) was identified as a defining gene for the native tendon cluster. Cochlin is an ECM protein that comprises the predominant non-collagen ECM component of the cochlea and vestibule of the inner ear,54 and Coch mutations lead to sensineuronal healing loss.55 While the role of Coch in tendon is not clear, recent work from Wunderli et al. demonstrates downregulation of Coch in an ex vivo model of hyper vascular/matrix unloading-mediated tendinopathy.56 Moreover, Wang et al. demonstrated that Scx is required for the formation of tendon in the middle ear,57 and Coch expression is observed in these structures.58 Finally, proteomic analysis of aged tendons identified Coch as a predominant protein that is lost during natural aging,59 suggesting a potential role for tenocyte-derived Cochlin in tendon homeostasis.
In summary, these data provide unprecedented temporal-spatial molecular profiling of the tendon healing process, define the molecular programs that underly distinct tissue features, and establish potential transcriptional regulators of different aspects of cell fate and function. Defining the distinct molecular programs that predominate during different phases of healing in a spatially dependent way will substantially enhance our understanding of the complex cellular and molecular milieu of tendon healing and will facilitate identification of more targeted therapeutic approaches to improve tendon healing.
Limitations of the study
One of the main limitations of this study is the resolution (55 μm per spot) of the current spatial transcriptomics technology, which is not capable of capturing single-cell information. However, the ability to define the predominant molecular programs of fibrotic tendon healing in an unbiased way, and subsequently map these programs to specific tissue features, provides an unparalleled level of spatial-molecular definition of the tendon healing process. In addition, spatial data can subsequently be integrated with single-cell RNA-seq datasets to provide more granular definition of the healing process. Related to the limited resolution, we are currently unable to distinguish between the potential contribution of the epitenon relative to the main body of the tendon. While this study provides significant information regarding spatially resolved transcriptomic profiles of tendon healing in young, metabolically normal mice, future work will be necessary to define likely alterations in the spatial-molecular map of tendon healing due to aging or other comorbidities.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Alayna Loiselle (alayna_loiselle@urmc.rochester.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Spatial transcriptomics data have been deposited at Gene Expression Omnibus (GEO: GSE216214) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Polyclonal Goat Anti-tdTomato | SICGEN | AB8181-200; RRID: AB_2722750 |
| Monoclonal Anti-Actin, alpha smooth muscle -CY3 | Sigma Life Sciences | Cat: C6198; RRID: AB_476856 |
| Monoclonal Anti-Actin, alpha smooth muscle -FITC | Sigma Life Sciences | Cat: F3777; RRID: AB_476977 |
| Polyclonal Rabbit Anti-Fibroblast Activation Protein, alpha | Abcam | Cat: ab53066; RRID: AB_880077 |
| Monoclonal Rabbit Anti-Hsp47 | Abcam | Cat: ab109117; RRID: AB_10888995 |
| Monoclonal Mouse Anti-PCNA | Abcam | Cat: ab29; RRID: AB_303394 |
| Monoclonal Rabbit Anti-VCAM1 | Abcam | Cat: ab134047; RRID: AB_2721053 |
| Polyclonal Rabbit Anti-CD45 | Abcam | Cat: ab10558; RRID: AB_442810 |
| Alexa Fluor 488-AffiniPure F(ab’)2 Fragment Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | Cat: 715-546-150; RRID: AB_2340849 |
| Rhodamine Red-X-AffiniPure F(ab’)2 Fragment Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat: 711-296-152; RRID: AB_2340614 |
| Alexa Fluor 488-AffiniPure F(ab’)2 Fragment Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch | Cat: 705-546-147; RRID: AB_2340430 |
| Rhodamine Red-X-AffiniPure F(ab’)2 Fragment Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch | Cat: 705-296-147; RRID: AB_2340424 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Tamoxifen | Sigma Life Sciences | Cat: T5648 |
| NucBlue Live Cell Stain ReadyProbes reagent | Invitrogen | Cat: R37605 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Visium Spatial Gene Expression Slide and Reagents Kit | 10x Genomics | Cat: 1000187 |
|
| ||
| Deposited data | ||
|
| ||
| Spatial transcriptomic dataset | Gene Expression Omnibus | GSE216214 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Scx-CreERT2 | Dr. Ronen Schweitzer | N/A |
| Mouse: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | Cat: #007909; RRID: IMSR_JAX:007909 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ Image Processing and Analysis in Java | ImageJ | https://imagej.nih.gov/ij/ |
| Cell Ranger v.3.0.2 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| R studio | R Studio | https://www.rstudio.com |
| GraphPad Prism version 9.0 | Dotmatics | https://www.graphpad.com |
| Monocle3 R package | Trapnell et al.60 Qiu et al.61 Traag et al.62 |
https://cole-trapnell-lab.github.io/monocle3/ |
| RcisTarget R package | Aibar et al.63 | https://bioconductor.org/packages/release/bioc/html/RcisTarget.html |
| Batchelor R package | Haghverdi et al.64 | https://bioconductor.org/packages/release/bioc/html/batchelor.html |
| DAVID Gene Functional Classification Tool version 6.8 | Huang et al.65 | https://david.ncifcrf.gov/gene2gene.jsp |
| CellChat v.1.1.3 | Jin et al.66 | http://www.cellchat.org |
| Seurat V4.0 R package | Hao et al.67 | https://www.rdocumentation.org/packages/Seurat/versions/4.1.0 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Mouse models
Scx-CreERT2 mice were generously provided by Dr. Ronen Schweitzer. ROSA-Ai9 (#007909) mice were obtained from the Jackson Laboratory (Bar Harbor, ME, USA). ROSA-Ai9 mice express the red fluorescent protein variant tdTomato following Cre-mediated recombination. Scx-CreERT2 mice were crossed to the ROSA-Ai9 strain to trace adult Scx-lineage cells (ScxAi9). Cre+ males and females were used for all studies and underwent tendon surgery at 10–12 weeks of age. For all experiments, ScxAi9 animals received three 100mg/kg i.p. tamoxifen (Tmx; #T5648, Sigma Life Sciences) injections beginning seven days prior to flexor tendon repair surgery to trace and assess the fate of adult ScxAi9 cells, while also ensuring that no subsequent labelling occurred during tendon healing by allowing for a washout period of four days. All mice were maintained under pathogen-free conditions and housed with no more than five animals per cage under standardized light-dark cycle conditions with ad libitum access to food and water. The vivarium was maintained under controlled temperature and humidity.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. All animal procedures were approved by the University Committee on Animal Research (UCAR) at the University of Rochester.
METHOD DETAILS
Flexor tendon repair
At 10–12 weeks of age, male and female mice underwent complete transection and repair of the flexor digitorum longus (FDL) tendon in the hind paw as previously described.6,68,69 Mice were injected prior to surgery with 15–20μg of sustained-release buprenorphine and then anesthetized with Ketamine (100mg/kg) and Xylazine (10mg/kg). Following cleaning and sterilization of the surgery area, the FDL tendon was transected at the myotendinous junction to reduce strain-induced rupture of the repair site and the skin was closed with a 5-0 suture. This myotendinous junction injury naturally heals throughout the first week of healing, re-introducing strain to the tendon. A small incision was then made on the posterior surface of the hind paw, the FDL tendon was located and completely transected. The tendon was repaired using an 8-0 suture and the skin was closed with a 5-0 suture.
Histology and immunofluorescence
Hind paws were harvested (n = 3–5 per time-point) from both uninjured ScxAi9 mice, and ScxAi9 mice at post-operative days 8, 10, 12, 14, 21, and 28 for paraffin sectioning. Hind paws were fixed in 10% formalin for 72 hours at room temperature, decalcified in Webb-Jee EDTA for two weeks, processed and embedded in paraffin. Three-micron sagittal sections were cut, de-waxed, rehydrated, and probed with antibodies for tdTomato (1:500, AB8181, SICGEN), αSMA-CY3 (1:200, C6198, Sigma Life Sciences, St. Louis, MO, USA), αSMA-FITC (1:500, F3777, Sigma Life Sciences), HSP47 (1:250, ab109117, Abcam), PCNA (1:100, ab29, Abcam, Cambridge, MA), FAP (1:500, ab53066, Abcam), VCAM1 (1:1000, ab134047, Abcam) CD45 (1:250, ab10558, Abcam). The following secondary antibodies were used: Donkey anti-mouse 488 (for PCNA) (1:200, #715-546-150, Jackson Immuno), Donkey anti-rabbit Rhodamine-Red-X (for FAP, Hsp47, VCAM-1, CD45) (1:200, #711-296-152, Jackson Immuno), Donkey anti-goat Rhodamine-Red-X (for tdTomato) (1:200, #705-296-147, Jackson Immuno), Donkey anti-goat 488 (for tdTomato) (1:200, #705-546-147, Jackson Immuno).
Nuclei were counterstained with NucBlue (#R37605, Invitrogen), and imaging was performed using a VS120 Virtual Slide Microscope (Olympus, Waltham, MA, USA). For Masson’s Trichrome staining of ScxAi9, following imaging of immunofluorescent staining, coverslips were gently removed in PBS and samples subsequently stained with Masson’s Trichrome to visualize collagen content. Figures 1B and 1D were pseudo-colored using Image J (https://imagej.nih.gov/ij/) for ease of interpretation.
Spatial transcriptomics (spatial RNA-sequencing)
Hind paws from ScxAi9 mice were harvested at 14-, 21-, and 28-days post-surgery, and from un-injured ScxAi9 mice (n = 2 per time-point), embedded in cryomatrix and snap-frozen in isopentane pre-cooled with liquid nitrogen. Ten-micron frozen sections were cut through the sagittal plane of the tendon and mounted on spatially barcoded capture areas of a Gene Expression slide (#1000187, Visium Spatial Gene Expression, 10X Genomics). Sections were fixed and stained with H&E for digital imaging, permeabilized and then a reverse transcription mix was added to each section. The bound cDNA was then released from the slide and used to prepare a library for sequencing following the 10X Genomics protocol. Paired-end sequencing was performed on the Illumina NextSeq 550 sequencer (Illumina Inc, San Diego CA) to obtain approximately 50,000 reads per spot covered with tissue within each capture area. Sequence data was then overlaid on to the H&E-stained digital image based on spatial orientation of the barcodes. The 10X gene-expression data were first processed using CellRanger (v.3.0.2, 10X Genomics). Normalized feature-barcode matrices were then used for downstream analysis.
Spatial RNA-seq data processing, integration, and visualization
Spatial RNA-seq datasets of the samples from different timepoints were processed in R Studio using Seurat (V4.1).67 Each sample was subsetted with barcodes obtained from Loupe to encompass the region of interest (tendon ends and associated scar tissue, Figure S1), and then underwent quality control to filter out low-quality reads and spots with high mitochondrial content. SCTransform was used to normalize the data, and dimensionality reduction performed with 15 principal components.
After each replicate was processed in this way, samples were then integrated with an anchor-based approach implemented in Seurat, and batch-corrected with the Fastmnn function from batchelor R package.64 Unsupervised clustering at 0.5 resolution identified six distinct clusters representing cellular/molecular programs present throughout fibrotic tendon healing. Annotation of each cluster was performed through examination of top differentially expressed genes (DEG) between clusters, gene ontology (GO) analysis, and spatial localization of markers (Figures 2 and S3). DEGs were genes expressed in at least 25% of cells within the cluster and with a fold change of more than 0.25 in natural log scale. GO analysis to determine the biological functions of a given cluster was completed by submitting the top 100 DEGs of each cluster to the Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/) bioinformatics resource.65
Pseudotemporal ordering and lineage trajectories
Monocle360–62 was used to reconstruct differentiation trajectories from native tendon tissue cells by computing and ordering the sequence of gene expression changes of the cells collected from different time points.60–62 First, both the muscle-tendon interface and inflammatory clusters from the integrated spatial RNA-seq dataset were removed, as we are primarily focused on investigating differentiation trajectories of tendon cells. Next, the integrated spatial RNA-seq dataset created in Seurat was converted into a Monocle3 object using the SeuratWrappers R package to maintain cluster embeddings. We used the learn_graph and order_cells functions to identify potential differentiation trajectories in our integrated spatial-seq dataset with the non-reactive/native tendon-like cell cluster as the root of lineage trajectories.
To determine sets of genes (i.e., gene modules) governing the differentiation of native tendon-like cells into peripheral fibroblastic tissue, active remodeling tissue, or synthetic fibroblast tissue during wound healing, we first subset each trajectory individually using choose_graph_segments, then used the graph_test and find_gene_modules (resolution = 0.001) to define specific gene modules using hierarchical cluster analysis. Additionally, GO term analysis was performed on each gene module to determine their associated biological functions.
Transcription factor binding motif and protein-protein network analysis
To obtain transcription factor (TF) scores and TF binding motifs of the genes within modules that control lineage specification of native tendon tissue during healing, we entered gene lists from each synthetic, remodeling, and fibrotic gene modules in RcisTarget R package63 using default parameters and mm9-tss-centered-10kb-7species.mc9nr.feather as the database. Furthermore, gene lists from each module were submitted to Transcriptional Regulatory Relationships Unrevealed by Sentence-based Text (TRRUST) mining, a manually curated database of human and mouse transcriptional regulatory networks, to compare and combine the results obtained from RcisTarget analysis.63 For each gene module, top six TFs with corresponding binding motif were selected and visualized.
Cell-cell interactome analysis
To identify cell-cell crosstalk between each cluster in the integrated data, as well as that between tdTomato+ (ScxAi9+) and tdTomato− (ScxAi9−) populations, we used CellChat (v.1.1.3),66 an open source R package that uses a manually curated signaling molecule interaction database to comprehensively analyze intercellular communications from seq data. The advantage of using this method is that CellChat takes multimeric ligand-receptor complexes into account to infer signaling patterns with mass action models. Quantitative communication analysis is performed via social network analysis tools, pattern recognition models, and manifold learning approaches, allowing identification of the signaling role for each specified population.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of the percent of ScxAi9+ cells that were also positive for either PCNA, FAP, or VCAM-1 expression (Figure 1) was conducted in a blinded semi-automated manner using Visiopharm image analysis software v.6.7.9.2590 (Visiopharm, Horsholm, Denmark). A region of interest (ROI) was drawn to include the tendon stubs and bridging scar tissue for each sample, and automatic segmentation using a threshold classifier was used to define discrete cell populations based on fluorescent intensity. Data are presented as the percent of ScxAi9+ that are also positive for the marker specified. An n = 3–4 independent biological samples per time-point were analyzed, and data normality was confirmed using GraphPad Prism Version 9.4.1. Data are presented ± standard deviation (SD).
Supplementary Material
Highlights.
Fibroblastic tissue demonstrates molecular trifurcation during tendon healing
Adult Scleraxis-lineage cells are enriched in the reactive tissue trajectory
The reactive cluster demonstrates high autocrine and paracrine communication
ACKNOWLEDGMENTS
We would like to thank the Histology, Biochemistry and Molecular Imaging (HBMI) Core for technical assistance with the histology. We would also like to thank the UR Genomics Research Core for assistance with spatial transcriptomics. This work was supported in part by National Institutes of Health/ NIAMS F31 AR074815 (to K.T.B.), F31 AR077398 (to J.E.A.), K99 AR080757 (A.E.C.N.), R00 AR075899 (to C.-L.W.), and R01 AR073169 and R01 AR077527 (to A.E.L.). The HBMI and BBMTI Cores were supported by NIH/NIAMS P30 AR069655. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111706.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Pennisi E (2002). Tending tender tendons. Science 295, 1011. 10.1126/science.295.5557.1011. [DOI] [PubMed] [Google Scholar]
- 2.Mehrzad R, Mookerjee V, Schmidt S, Jehle CC, Kiwanuka E, and Liu PY (2019). The economic impact of flexor tendon lacerations of the hand in the United States. Ann. Plast. Surg S3, 419–423. 10.1097/SAP.0000000000001950. [DOI] [PubMed] [Google Scholar]
- 3.Best KT, and Loiselle AE (2019). Scleraxis lineage cells contribute to organized bridging tissue during tendon healing and identify a subpopulation of resident tendon cells. FASEB J. 33, 8578–8587. 10.1096/fj.201900130RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Micheli AJ, Swanson JB, Disser NP, Martinez LM, Walker NR, Oliver DJ, Cosgrove BD, and Mendias CL (2020). Single-cell transcriptomic analysis identifies extensive heterogeneity in the cellular composition of mouse Achilles tendons. Am. J. Physiol. Cell Physiol 319, C885–C894. 10.1152/ajpcell.00372.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kendal AR, Layton T, Al-Mossawi H, Appleton L, Dakin S, Brown R, Loizou C, Rogers M, Sharp R, and Carr A (2020). Multi-omic single cell analysis resolves novel stromal cell populations in healthy and diseased human tendon. Sci. Rep 10, 13939. 10.1038/s41598-020-70786-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Best KT, Korcari A, Mora KE, Nichols AE, Muscat SN, Knapp E, Buckley MR, and Loiselle AE (2021). Scleraxis-lineage cell depletion improves tendon healing and disrupts adult tendon homeostasis. Elife 10, e62203. 10.7554/eLife.62203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ackerman JE, Nichols AE, Studentsova V, Best KT, Knapp E, and Loiselle AE (2019). Cell non-autonomous functions of S100a4 drive fibrotic tendon healing. Elife 8, e45342. 10.7554/eLife.45342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murchison ND, Price BA, Conner DA, Keene DR, Olson EN, Tabin CJ, and Schweitzer R (2007). Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 134, 2697–2708. [DOI] [PubMed] [Google Scholar]
- 9.Yoshimoto Y, Takimoto A, Watanabe H, Hiraki Y, Kondoh G, and Shukunami C (2017). Scleraxis is required for maturation of tissue domains for proper integration of the musculoskeletal system. Sci. Rep 7, 45010. 10.1038/srep45010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dyment NA, Hagiwara Y, Matthews BG, Li Y, Kalajzic I, and Rowe DW (2014). Lineage tracing of resident tendon progenitor cells during growth and natural healing. PLoS One 9, e96113. 10.1371/journal.pone.0096113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Howell K, Chien C, Bell R, Laudier D, Tufa SF, Keene DR, Andarawis-Puri N, and Huang AH (2017). Novel model of tendon regeneration reveals distinct cell mechanisms underlying regenerative and fibrotic tendon healing. Sci. Rep 7, 45238. 10.1038/srep45238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pakshir P, and Hinz B (2018). The big five in fibrosis: macrophages, myofibroblasts, matrix, mechanics, and miscommunication. Matrix Biol. 68-69, 81–93. 10.1016/j.matbio.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Grinstein M, Dingwall HL, O’Connor LD, Zou K, Capellini TD, and Galloway JL (2019). A distinct transition from cell growth to physiological homeostasis in the tendon. Elife 8, e48689. 10.7554/eLife.48689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sakabe T, Sakai K, Maeda T, Sunaga A, Furuta N, Schweitzer R, Sasaki T, and Sakai T (2018). Transcription factor scleraxis vitally contributes to progenitor lineage direction in wound healing of adult tendon in mice. J. Biol. Chem 293, 5766–5780. 10.1074/jbc.RA118.001987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyment NA, Liu CF, Kazemi N, Aschbacher-Smith LE, Kenter K, Breidenbach AP, Shearn JT, Wylie C, Rowe DW, and Butler DL (2013). The paratenon contributes to scleraxis-expressing cells during patellar tendon healing. PLoS One 8, e59944. 10.1371/journal.pone.0059944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LeBleu VS, and Neilson EG (2020). Origin and functional heterogeneity of fibroblasts. FASEB J. 34, 3519–3536. 10.1096/fj.201903188R. [DOI] [PubMed] [Google Scholar]
- 17.Reilkoff RA, Bucala R, and Herzog EL (2011). Fibrocytes: emerging effector cells in chronic inflammation. Nat. Rev. Immunol 11, 427–435. 10.1038/nri2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischer B, Azim K, Hurtado-Chong A, Ramelli S, Fernández M, and Raineteau O (2014). E-proteins orchestrate the progression of neural stem cell differentiation in the postnatal forebrain. Neural Dev. 9, 23. 10.1186/1749-8104-9-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Best KT, Nichols AEC, Knapp E, Hammert WC, Ketonis C, Jonason JH, Awad HA, and Loiselle AE (2020). NF-kappaB activation persists into the remodeling phase of tendon healing and promotes myofibroblast survival. Sci. Signal 13, eabb7209. 10.1126/scisignal.abb7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu K, Pan X, Qiu X, Wang D, Dong N, Yang L, and Li S (2018). Neural crest-derived cells migrate from nerve to participate in Achilles tendon remodeling. Wound Repair Regen. 26, 54–63. 10.1111/wrr.12614. [DOI] [PubMed] [Google Scholar]
- 21.Xu K, Shao Y, Xia Y, Qian Y, Jiang N, Liu X, Yang L, and Wang C (2021). Tenascin-C regulates migration of SOX10 tendon stem cells via integrin-α9 for promoting patellar tendon remodeling. Biofactors 47, 768–777. 10.1002/biof.1759. [DOI] [PubMed] [Google Scholar]
- 22.Turner NA (2016). Inflammatory and fibrotic responses of cardiac fibroblasts to myocardial damage associated molecular patterns (DAMPs). J. Mol. Cell. Cardiol 94, 189–200. 10.1016/j.yjmcc.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, and Gabbiani G (2007). The myofibroblast: one function, multiple origins. Am. J. Pathol 170, 1807–1816. 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hinz B (2007). Formation and function of the myofibroblast during tissue repair. J. Invest. Dermatol 127, 526–537. 10.1038/sj.jid.5700613. [DOI] [PubMed] [Google Scholar]
- 25.Dakin SG, Buckley CD, Al-Mossawi MH, Hedley R, Martinez FO, Wheway K, Watkins B, and Carr AJ (2017). Persistent stromal fibroblast activation is present in chronic tendinopathy. Arthritis Res. Ther 19, 16. 10.1186/s13075-016-1218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akbar M, McLean M, Garcia-Melchor E, Crowe LA, McMillan P, Fazzi UG, Martin D, Arthur A, Reilly JH, Mclnnes IB, and Millar NL (2019). Fibroblast activation and inflammation in frozen shoulder. PLoS One 14, e0215301. 10.1371/journal.pone.0215301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shook BA, Wasko RR, Rivera-Gonzalez GC, Salazar-Gatzimas E, López-Giráldez F, Dash BC, Muñoz-Rojas AR, Aultman KD, Zwick RK, Lei V, et al. (2018). Myofibroblast proliferation and heterogeneityare supported by macrophages during skin repair. Science 362, eaar2971. 10.1126/science.aar2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lynch MD, and Watt FM (2018). Fibroblast heterogeneity: implications for human disease. J. Clin. Invest 128, 26–35. 10.1172/JCI93555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou J, Shi J, Chen L, Lv Z, Chen X, Cao H, Xiang Z, and Han X (2018). M2 macrophages promote myofibroblast differentiation of LR-MSCs and are associated with pulmonary fibrogenesis. Cell Commun. Signal 16, 89. 10.1186/s12964-018-0300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ploeger DT, Hosper NA, Schipper M, Koerts JA, de Rond S, and Bank RA (2013). Cell plasticity in wound healing: paracrine factors of M1/M2 polarized macrophages influencethe phenotypical state ofdermal fibroblasts. Cell Commun. Signal 11, 29. 10.1186/1478-811X-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mewhort HEM, Lipon BD, Svystonyuk DA, Teng G, Guzzardi DG, Silva C, Yong VW, and Fedak PWM (2016). Monocytes increase human cardiac myofibroblast-mediated extracellular matrix remodeling through TGF-beta1. Am. J. Physiol. Heart Circ. Physiol 310, H716–H724. 10.1152/ajpheart.00309.2015. [DOI] [PubMed] [Google Scholar]
- 32.Glim JE, Niessen FB, Everts V, van Egmond M, and Beelen RHJ (2013). Platelet derived growth factor-CC secreted by M2 macrophages induces alpha-smooth muscle actin expression by dermal and gingival fibroblasts. Immunobiology 218, 924–929. 10.1016/j.imbio.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Braga TT, Agudelo JSH, and Camara NOS (2015). Macrophages during the fibrotic process: M2 as friend and foe. Front. Immunol 6, 602. 10.3389/fimmu.2015.00602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhattacharyya S, Chen SJ, Wu M, Warner-Blankenship M, Ning H, Lakos G, Mori Y, Chang E, Nihijima C, Takehara K, et al. (2008). Smad-independent transforming growth factor-beta regulation of early growth response-1 and sustained expression in fibrosis: implications for scleroderma. Am. J. Pathol 173, 1085–1099. 10.2353/aj-path.2008.080382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharyya S, Wu M, Fang F, Tourtellotte W, Feghali-Bostwick C, and Varga J (2011). Early growth response transcription factors: key mediators of fibrosis and novel targets foranti-fibrotic therapy. Matrix Biol. 30, 235–242. 10.1016/j.matbio.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yasuoka H, Hsu E, Ruiz XD, Steinman RA, Choi AMK, and Feghali-Bostwick CA (2009). The fibrotic phenotype induced by IGFBP-5 is regulated by MAPK activation and egr-1-dependent and -independent mechanisms. Am. J. Pathol 175, 605–615. 10.2353/aj-path.2009.080991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu M, Melichian DS, de la Garza M, Gruner K, Bhattacharyya S, Barr L, Nair A, Shahrara S, Sporn PHS, Mustoe TA, et al. (2009). Essential roles for early growth response transcription factor Egr-1 in tissue fibrosis and wound healing. Am. J. Pathol 175, 1041–1055. 10.2353/ajpath.2009.090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F, Zhang A, and Richardson DW (2019). Regulation of the tenogenic gene expression in equine tenocyte-derived induced pluripotent stem cells by mechanical loading and Mohawk. Stem Cell Res. 39, 101489. 10.1016/j.scr.2019.101489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaut L, Robert N, Delalande A, Bonnin MA, Pichon C, and Duprez D (2016). EGR1 regulates transcription downstream of mechanical signals during tendon formation and healing. PLoS One 11, e0166237. 10.1371/journal.pone.0166237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Havis E, and Duprez D (2020). EGR1 transcription factor is a multifaceted regulator of matrix production in tendons and other connective tissues. Int. J. Mol. Sci 21, E1664. 10.3390/ijms21051664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moles A, Sanchez AM, Banks PS, Murphy LB, Luli S, Borthwick L, Fisher A, O’Reilly S, van Laar JM, White SA, et al. (2013). Inhibition of RelA-Ser536 phosphorylation by a competing peptide reduces mouse liver fibrosis without blocking the innate immune response. Hepatology 57, 817–828. 10.1002/hep.26068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Treiber M, Neuhöfer P, Anetsberger E, Einwächter H, Lesina M, Rickmann M, Liang S, Kehl T, Nakhai H, Schmid RM, and Algl H (2011). Myeloid, but not pancreatic, RelA/p65 is required for fibrosis in a mouse model of chronic pancreatitis. Gastroenterology 141, 1473–1485. e1–7. 10.1053/j.gastro.2011.06.087. [DOI] [PubMed] [Google Scholar]
- 43.Luedde T, and Schwabe RF (2011). NF-kappaB in the liver–linking injury, fibrosis and hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol 8, 108–118. 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hou J, Ma T, Cao H, Chen Y, Wang C, Chen X, Xiang Z, and Han X (2018). TNF-alpha-induced NF-kappaB activation promotes myofibroblast differentiation of LR-MSCs and exacerbates bleomycin-induced pulmonary fibrosis. J. Cell. Physiol 233, 2409–2419. 10.1002/jcp.26112. [DOI] [PubMed] [Google Scholar]
- 45.Chen S, Jiang S, Zheng W, Tu B, Liu S, Ruan H, and Fan C (2017). RelA/p65 inhibition prevents tendon adhesion by modulating inflammation, cell proliferation, and apoptosis. Cell Death Dis. 8, e2710. 10.1038/cddis.2017.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Posern G, and Treisman R (2006). Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 16, 588–596. 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 47.Gualdrini F, Esnault C, Horswell S, Stewart A, Matthews N, and Treisman R (2016). SRF Co-factors control the balance between cell proliferation and contractility. Mol. Cell 64, 1048–1061. 10.1016/j.molcel.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenwel P, Inagaki Y, Hu W, Walsh M, and Ramirez F (1997). Sp1 is required for the early response of alpha2(I) collagen to transforming growth factor-beta1. J. Biol. Chem 272, 19738–19745. 10.1074/jbc.272.32.19738. [DOI] [PubMed] [Google Scholar]
- 49.Verrecchia F, Rossert J, and Mauviel A (2001). Blocking sp1 transcription factor broadly inhibits extracellular matrix gene expression in vitro and in vivo: implications for the treatment of tissue fibrosis. J. Invest. Dermatol 116, 755–763. 10.1046/j.1523-1747.2001.01326.x. [DOI] [PubMed] [Google Scholar]
- 50.Subramanian A, and Schilling TF (2014). Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions. Elife 3, e02372. 10.7554/eLife.02372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frolova EG, Drazba J, Krukovets I, Kostenko V, Blech L, Harry C, Vasanji A, Drumm C, Sul P, Jenniskens GJ, et al. (2014). Control of organization and function of muscle and tendon by thrombospondin-4. Matrix biology. Matrix Biol. 37, 35–48. 10.1016/j.matbio.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai RK, and Discher DE (2008). Inhibition of “self” engulfment through deactivation of myosin-II at the phagocytic synapse between human cells. J. Cell Biol 180, 989–1003. 10.1083/jcb.200708043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, and Lindberg FP (2000). Role of CD47 as a marker of self on red blood cells. Science 288, 2051–2054. 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- 54.Robertson NG, Skvorak AB, Yin Y, Weremowicz S, Johnson KR, Kovatch KA, Battey JF, Bieber FR, and Morton CC (1997). Mapping and characterization of a novel cochlear gene in human and in mouse: a positional candidate gene for a deafness disorder, DFNA9. Genomics 46, 345–354. 10.1006/geno.1997.5067. [DOI] [PubMed] [Google Scholar]
- 55.Robertson NG, O’Malley JT, Ong CA, Giersch ABS, Shen J, Stankovic KM, and Morton CC (2014). Cochlin in normal middle ear and abnormal middle ear deposits in DFNA9 and Coch (G88E/G88E) mice. J. Assoc. Res. Otolaryngol 15, 961–974. 10.1007/s10162-014-0481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wunderli SL, Blache U, Beretta Piccoli A, Niederöst B, Holenstein CN, Passini FS, Silván U, Bundgaard L, Snedeker JG, and Auf dem Keller U (2020). Tendon responseto matrix unloading is determined by the patho-physiological niche. Matrix Biol. 89, 11–26. 10.1016/j.matbio.2019.12.003. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, Bresee CS, Jiang H, He W, Ren T, Schweitzer R, and Brigande JV (2011). Scleraxis is required for differentiation of the stapedius and tensor tympani tendons of the middle ear. J. Assoc. Res. Otolaryngol 12, 407–421. 10.1007/s10162-011-0264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robertson NG, Jones SM, Sivakumaran TA, Giersch ABS, Jurado SA, Call LM, Miller CE, Maison SF, Liberman MC, and Morton CC (2008).A targeted Coch missense mutation: a knock-in mouse model for DFNA9 late-onset hearing loss and vestibular dysfunction. Hum. Mol. Genet 17, 3426–3434. 10.1093/hmg/ddn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korcari A, Nichols AEC, Buckley MR, and Loiselle AE (2022). Depletion of Scleraxis-lineage cells accelerates tendon ECM aging and promotes retention of a specialized remodeling tenocyte population that drives enhanced tendon healing. Preprint at bioRxiv. 10.1101/2022.01.20.477119. [DOI] [Google Scholar]
- 60.Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, and Rinn JL (2014). The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol 32, 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, and Trapnell C (2017). Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Traag VA, Waltman L, and van Eck NJ (2019). From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep 9, 5233. 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, Rambow F, Marine JC, Geurts P, Aerts J, et al. (2017). SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Haghverdi L, Lun ATL, Morgan MD, and Marioni JC (2018). Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol 36, 421–427. 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huang DW, Sherman BT, Tan Q, Collins JR, Alvord WG, Roayaei J, Stephens R, Baseler MW, Lane HC, and Lempicki RA (2007). The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 8, R183. 10.1186/gb-2007-8-9-r183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, and Nie Q (2021). Inference and analysis of cell-cell communication using CellChat. Nat. Commun 12, 1088. 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Loiselle AE, Bragdon GA, Jacobson JA, Hasslund S, Cortes ZE, Schwarz EM, Mitten DJ, Awad HA, and O’Keefe RJ (2009). Remodeling of murine intrasynovial tendon adhesions following injury: MMP and neotendon gene expression. J. Orthop. Res 27, 833–840. 10.1002/jor.20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ackerman JE, and Loiselle AE (2016). Murine flexor tendon injury and repair surgery. J. Vis. Exp 10.3791/54433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Spatial transcriptomics data have been deposited at Gene Expression Omnibus (GEO: GSE216214) and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| Polyclonal Goat Anti-tdTomato | SICGEN | AB8181-200; RRID: AB_2722750 |
| Monoclonal Anti-Actin, alpha smooth muscle -CY3 | Sigma Life Sciences | Cat: C6198; RRID: AB_476856 |
| Monoclonal Anti-Actin, alpha smooth muscle -FITC | Sigma Life Sciences | Cat: F3777; RRID: AB_476977 |
| Polyclonal Rabbit Anti-Fibroblast Activation Protein, alpha | Abcam | Cat: ab53066; RRID: AB_880077 |
| Monoclonal Rabbit Anti-Hsp47 | Abcam | Cat: ab109117; RRID: AB_10888995 |
| Monoclonal Mouse Anti-PCNA | Abcam | Cat: ab29; RRID: AB_303394 |
| Monoclonal Rabbit Anti-VCAM1 | Abcam | Cat: ab134047; RRID: AB_2721053 |
| Polyclonal Rabbit Anti-CD45 | Abcam | Cat: ab10558; RRID: AB_442810 |
| Alexa Fluor 488-AffiniPure F(ab’)2 Fragment Donkey Anti-Mouse IgG (H+L) | Jackson ImmunoResearch | Cat: 715-546-150; RRID: AB_2340849 |
| Rhodamine Red-X-AffiniPure F(ab’)2 Fragment Donkey Anti-Rabbit IgG (H+L) | Jackson ImmunoResearch | Cat: 711-296-152; RRID: AB_2340614 |
| Alexa Fluor 488-AffiniPure F(ab’)2 Fragment Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch | Cat: 705-546-147; RRID: AB_2340430 |
| Rhodamine Red-X-AffiniPure F(ab’)2 Fragment Donkey Anti-Goat IgG (H+L) | Jackson ImmunoResearch | Cat: 705-296-147; RRID: AB_2340424 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Tamoxifen | Sigma Life Sciences | Cat: T5648 |
| NucBlue Live Cell Stain ReadyProbes reagent | Invitrogen | Cat: R37605 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Visium Spatial Gene Expression Slide and Reagents Kit | 10x Genomics | Cat: 1000187 |
|
| ||
| Deposited data | ||
|
| ||
| Spatial transcriptomic dataset | Gene Expression Omnibus | GSE216214 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| Mouse: Scx-CreERT2 | Dr. Ronen Schweitzer | N/A |
| Mouse: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | Cat: #007909; RRID: IMSR_JAX:007909 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ Image Processing and Analysis in Java | ImageJ | https://imagej.nih.gov/ij/ |
| Cell Ranger v.3.0.2 | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest |
| R studio | R Studio | https://www.rstudio.com |
| GraphPad Prism version 9.0 | Dotmatics | https://www.graphpad.com |
| Monocle3 R package | Trapnell et al.60 Qiu et al.61 Traag et al.62 |
https://cole-trapnell-lab.github.io/monocle3/ |
| RcisTarget R package | Aibar et al.63 | https://bioconductor.org/packages/release/bioc/html/RcisTarget.html |
| Batchelor R package | Haghverdi et al.64 | https://bioconductor.org/packages/release/bioc/html/batchelor.html |
| DAVID Gene Functional Classification Tool version 6.8 | Huang et al.65 | https://david.ncifcrf.gov/gene2gene.jsp |
| CellChat v.1.1.3 | Jin et al.66 | http://www.cellchat.org |
| Seurat V4.0 R package | Hao et al.67 | https://www.rdocumentation.org/packages/Seurat/versions/4.1.0 |


