Abstract
Polyadenylate-binding protein cytoplasmic 1 (PABPC1) is dysregulated in malignancies, which is considered as a potential therapeutic target for many cancer types. By alternative splicing (AS) for gastric cancer (GC), we described PABPC1-modulated AS events in this study. PABPC1 expression was analyzed in 408 GC tissues from The Cancer Genome Altas (TCGA) database. Human gastric adenocarcinoma (AGS) cells were transfected with PABPC1-specific small interfering RNA (siPABPC1) with siCtrl as a negative control. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was done for the determination of transcripts. To detect the differentially expressed genes (DEGs) and 10 different types of AS events, RNA sequencing (RNA-seq) was performed. DEGs were analyzed for functional categories including gene ontology, and the Kyoto encyclopedia of genes and genomes pathway were analyzed for DEGs. GC displayed an elevated expression of PABPC1. PABPC1 was efficiently knocked down in AGS cells. Here, we excavated 1234 PABPC1-regulated DEGs, among which 502 were down-regulated and 732 were up-regulated compared to the siCtrl group. A total of 94 DEGs were involved in inflammation and immune response. Results from qRT-PCR validated the up-regulation of 10 immune and inflammation-related DEGs in the siPABPC1 group. PABPC1 deficiency causes 1304 AS events differentially occurred in AGS cells. The most common type of AS events regulated by PABPC2 is alternative 5′ splice sites. qRT-PCR confirmed the transcription level of five immune-related genes, in which AS events were detected in the siPABPC1 group. PABPC1 knockdown mediates AS events and thus the transcript level of immune and inflammation-related genes in AGS cells. PABPC1-regulated oncogenic AS events display potential as targets for therapeutic development.
Keywords: PABPC1, alternative splicing events, gastric cancer, differentially expressed genes, immune pathway, RNA sequencing
Impact Statement
This study elucidates a novel molecular regulatory mechanism involving PABPC1-mediated alternative splicing events in gastric cancer, which is based on RNA sequencing technology and bioinformatic analysis. These differentially expressed genes (DEGs) were predicted to participate in regulating the immune pathway, as well as alternative splicing events, which are candidates for interfering with PABPC1’s oncogenic functions in human gastric adenocarcinoma cells. The obtained results highlight a comprehensive link between dysregulation of alternative splicing events and gastric cancer, specifically those regulated by PABPC1. PABPC1-regulated oncogenic alternative splicing events display potentials as targets for therapeutic development. The screened DEGs and signaling pathways can be studied as drug targets and can be used for medicine design.
Introduction
As a multifactorial disease, gastric cancer (GC) is affected by Helicobacter pylori infection, smoking, alcohol consumption, and a high salt diet.1,2 GC was the fifth most commonly diagnosed cancer with 1,000,000 new cases according to global cancer statistics by the International Agency for Research on Cancer. 3 GC was estimated to cause 769,000 deaths globally, which was the fourth leading cause of cancer-related death worldwide in 2020. 3 Recently, notable evidence suggested that the incidence of GC is increasing among young adults aged less than 50 years. 4 Emerging findings postulated that the frequent utilization of antibiotics and acid suppressants may contribute to the rising prevalence of autoimmune gastritis and dysbiosis of the gastric microbiome, causing an increased incidence of GC among younger individuals.5,6 To date, the treatment of GC is still largely based on surgery, chemotherapy, and radiotherapy. Recently, some new individualized regimens have been proposed for treating GC. Of particular interest, splicing regulatory networks-mediated alternative splicing (AS) events are related to immune features, tumor microenvironment, and clinical characteristics in patients with H. pylori-negative GC, suggesting that these may be promising therapeutic candidates.7,8,9
There are many transcripts from human protein-coding genes generated by AS events of different types, including A3SS&ES, A5SS&ES, exon skipping (ES), mutually exclusive 3′ untranslated regions (3pMXE), mutually exclusive 5′ untranslated regions (5pMXE), intron retention (IntronR), alternative 3′ splice site (A3SS), alternative 5′ splice site (A5SS), mutually exclusive exon (MXE), and cassette exon (CassetteExon). These events have been discovered in the context of physiological diseases and some biological processes.10,11 Splice variants are responsible for diversifying gene expression regulation, which functionally result in the diversity of proteomes and contribute to cell proliferation, migration, or immune escape.12 –14 According to the previous description, some compounds (e.g. rapamycin, metformin, dorsomorphin, enzastaurin, and rigosertib) have been reported to alter AS events and regulate signaling pathways such as mTOR, AMPK, PKC, and PI3K/AKT.15 –18 These dysregulated AS events may provide promising targets in colorectal cancer, 13 breast cancer, 19 and lung cancer. 14 Genome-wide transcriptome data have focused on illustrating the relationship between survival of cancer patients and AS events, which demonstrated the potential mechanisms for survival-related AS events. 8 Furthermore, the alterations in splicing factors such as abnormal expression, mutation, or post-transcriptional modification provide genetic evidence for a straightforward correlation between cancer pathogenesis and splicing dysregulation.20 –23
PABPC1 encodes polyadenylate-binding protein cytoplasmic 1 protein that is a binding partner of heterogeneous nuclear ribonucleoprotein L-like protein (hnRNPLL). 24 PABPC1 can bind the poly(A) tail of mRNA, and then mediate pre-mRNA splicing and mRNA stability.25,26 Some efforts have been made to confirm the correlation between clinicopathological factors and PABPC1 expression in malignancies including esophageal cancer, 27 ovarian cancer, 28 glioblastoma,29,30 and GC. 31 However, little is known whether the pathogenesis and progression of GC are affected by PABPC1-mediated AS events. Here, this study investigated the differentially expressed genes (DEGs) in PABPC1-mediated AS events and analyzed the signal pathways regulated by AS-related DEGs in GC.
Materials and methods
Data collection and analysis
RNA sequencing (RNA-seq) data of GC were collected from The Cancer Genome Altas-Stomach Adenocarcinoma (TCGA-STAD), which is available at https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga.
Transcriptome data of GC were analyzed for PABPC1 expression. GEPIA (available at http://gepia.cancer-pku.cn/detail.php###) was then used for the analysis of PABPC1 expression in 408 tumor tissues and 36 normal tissues, according to Tang’s method. 32
Cell culture and transfection
Human gastric adenocarcinoma (AGS) cells with CDH1, CTNNB1, KRAS, and PIK3CA mutations were provided by Procell Life Science & Technology (Item No. CL-0022; Wuhan, China).
AGS cells were seeded and incubated in Ham’s F12 medium. The medium was added with penicillin (100 U/mL) and streptomycin (100 µg/mL), and supplemented with 10% fetal bovine serum (all from Sigma-Aldrich, St. Louis, MO, USA). The cell culture was maintained at 37°C in a humidified incubator containing 5% CO2.
The effects of PABPC1 on AS events were assayed after treatment with PABPC1-specific small interfering RNA (siPABPC1) (sense, 5′-GCUCCUAAAUGAUCGCAAATT-3′; Gemma, Suzhou, China). A scrambled siRNA (sense, 5′-UUCUCCGAACGUGUCACGUTT-3′) served as a silencer negative control (siCtrl, Gemma). For transfection, AGS cells were seeded in 24-well plates (6.0 × 105 cells in each well). AGS cells were transfected with siPABPC1 (10 ng) or siCtrol (10 ng) using Lipofectamine™ 3000 transfection reagent (Thermo Fisher Scientific) after 24 h of pre-incubation.
Quantitative reverse-transcription polymerase chain reaction for gene expression
Total RNA was extracted with TRIZOL (Ambion, Austin, TX, USA). HifairTM II first strand cDNA synthesis kit (Low Rox Plus; YEASEN, Shanghai, China) was used for cDNA synthesis. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) was performed with Hieff qPCR SYBR Green Master Mix (Low Rox Plus) with the specific primers for PABPC1, CD55, HLA-F, IRF7, and STAT2 (Table 1), on the Bio-Rad S1000 (Bio-Rad, Hercules, CA, USA). For the normalization of gene expression, GAPDH was used as an internal reference, and the transcript levels were calculated using 2−ΔΔCT method. 33
Table 1.
qRT-PCR primers for the detected genes.
| Genes | Forward primer | Reverse primer |
|---|---|---|
| C4BPB | 5′-GAACCTCTGCGAAGCCATG-3′ | 5′-TTGCCTTCAACTCAGCTTTCTT-3′ |
| CD55 | 5′-TTTGCTTGGGACGCTAGTAAC-3′ | 5′-AGAGGTGTAGGTGTGCTAAGAA-3′ |
| HLA-F | 5′-GGAATGAATGGCTGCGACAT-3′ | 5′-GGTAGGTCCTGAACTCCTCTG-3′ |
| IRF7 | 5′-TGGAGTTCTCATTAGACTGGGT-3′ | 5′-TGGATAGCAGCAGCCTCAG-3′ |
| STAT2 | 5′-ATGGAGTCCTATCCTGTGTCTG-3′ | 5′-CTGCTGTGCTGGGAGGTATA-3′ |
| PABPC1 | 5′-ACTGACATTCTGAGCTATTCCA-3′ | 5′-TTGAACCTTATGTACCGAGCAA-3′ |
| GAPDH | 5′-GGTCGGAGTCAACGGATTTG-3′ | 5′-GGAAGATGGTGATGGGATTTC-3′ |
qRT-PCR: quantitative reverse-transcription polymerase chain reaction.
RNA-Seq
DEGs and AS events were determined by RNA-seq. The extracted RNA was subjected to electrophoresis using 1.5% agarose gel for assaying RNA integrity. The mRNAs were isolated from RNA extracts using VAHTS mRNA capture Beads (Vazyme, Nanjing, China).
RNA-seq library was constructed with KAPA Stranded mRNA-Seq Kit (KAPA Biosystems, Wilmington, MA, USA) and 1 μg of total RNA. The enriched polyadenylated mRNAs were then converted into double-strand cDNA. DNA was ligated to diluted Roche Adaptor after end repairing and A-tail processing. Products with 300–500 bps were selected for amplification, purification, quantification, and sequencing on a Novaseq 6000 system (Illumina, CA, USA).
RNA-Seq raw data alignment
For quality filtering, raw reads were filtered using FASTX-Toolkit Version 0.013 (available at http://hannonlab.cshl.edu/fastx_toolkit). After discarding raw reads involving 2-N bases as the first two nucleotides, adaptors and low-quality bases were trimmed. Short reads less than 16 nts were removed. TopHat2 was used to align the collected clean reads to the GRch38 Genome, with four mismatches allowed. 34 Uniquely mapped reads were retained, followed by gene reads number counting. Then fragments per kilobase of transcript per million mapped reads (FPKM) were calculated. 35
DEGs analysis in siPABPC1-transfected AGS cells
To screen DEGs between siCtrl and siPABPC1 groups, the R Bioconductor package edger was employed. 36 DEGs between siPABPC1 and siCtrl groups were screened with the following cut-off criteria: a false discovery rate (FDR) of <0.05 and a fold change of >2 or ⩽0.5. Student’s t-test was used for multiple hypothesis testing. A significant difference was indicated by P < 0.05.
AS events analysis
ABLas-v0.2 (available at https://github.com/ablifedev/ABLas/) was used for the definition and quantification of AS events and PABPC1-regulated AS events (RASEs). In brief, 10 types of AS events classified on the basis of the splice junction reads were detected using ABLas, including CassetteExon, MXE, A5SS, A3SS, IntronR, 5pMXE, 3pMXE, ES, A5SS&ES, and A3SS&ES.
Function prediction of DEGs
Functional categories were implemented with KOBAS 2.0 Server, 37 which involved gene ontology (GO) terms and the Kyoto encyclopedia of genes and genomes (KEGG) pathways. The potential targets of DEGs regulated by PABPC1 were analyzed. P < 0.05 indicates that DEG-involved pathways were significantly mediated by PABPC1.
Statistical analysis
Gene expression levels are shown as the mean ± standard deviation. Student’s t-test was performed for the comparison of the differences in gene expression between the siCtrl and siPABPC1 groups. P values < 0.05 were set as the indication of significant differences in gene expression.
Results
PABPC1 expression in GC tissues and construction of AGS cell model low expressing PABPC1
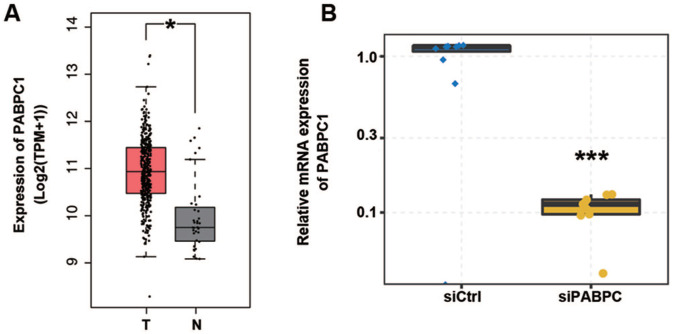
Based on transcriptome data from the TCGA database, PABPC1 was detected up-regulated in 408 tumor tissues of GC patients compared to 36 normal para-carcinoma tissues (P < 0.05, Figure 1(A)). Hence, it was assumed that PABPC1 plays roles in the regulation of gene expression and AS events associated with GC pathogenesis. To prove this hypothesis, we knocked down PABPC1 in AGS cells using siRNA. We detected PABPC1 mRNA expression in siPABPC1- and siCtrl-transfected AGS cells. Data of qRT-PCR showed that PABPC1 was efficiently knocked down with an approximately 75% reduction in siPABPC1 cells compared with siCtrl cells (P < 0.001), as shown in Figure 1(B).
Figure 1.
PABPC1 was down-regulated in gastric cancer tissue and PABPC1 mRNA expression was altered in AGS cells after transfection. (A) Data extracted from TCGA database suggest PABPC1 expression in gastric cancer tissues; boxplots show the median (center line), first quartile (Q1, lower quartile), third quartile (Q3, upper quartile), maximum (the highest data point in the data set excluding any outliers), and minimum (the lowest data point in the data set excluding any outliers); *P < 0.05 versus normal tissues (Student’s t-test). (B) AGS cells transfected with siRNA for PABPC1 or negative control were assayed by qRT-PCR for PABPC1 quantification. Data represent three independent technical repetitions and three independent biological repetitions; ***P < 0.001 versus siCtrl (Student’s t-test). Data are expressed as mean ± standard deviation, where the standard deviation is indicated as error bars. (A color version of this figure is available in the online journal.)
PABPC1 deficiency contributed to DEGs in AGS cells
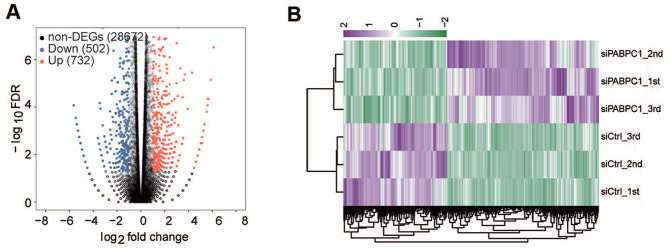
RNA-seq was performed to confirm the effects of PABPC1 down-regulation on gene expression. The edge R package was used for the analysis of digital gene expression data. We excavated a total of 1234 PABPC1-regulated DEGs and 28,672 non-DEGs in AGS cells. Of note, PABPC1 deficiency resulted in down-regulation of 502 DEGs and up-regulation of 732 DEGs in AGS cells at a cutoff of FDR < 0.05 and fold change ⩾ 2 or ⩽ 0.5 (Figure 2(A)). Hierarchical clustering analysis showed a high consistency of PABPC1-mediated transcription in the siPABPC1 groups or siCtrl groups (Figure 2(B)).
Figure 2.
Identification of PABPC1-regulated DEGs by RNA-seq. (A) Volcano plots presenting differentially expressed genes in AGS cells after transfection with siPABPC1 or siCtrl. The colored dots denote significantly down-regulated (blue), up-regulated (red), or non-differentially expressed genes (gray) in siPABPC1-transfected cells compared to siCtrl-treated cells. (B) Clustering analysis of siCtrl and siPABPC1 groups based on the differentially expressed genes. The top bar values represent the relative expression of the differentially expressed genes. (A color version of this figure is available in the online journal.)
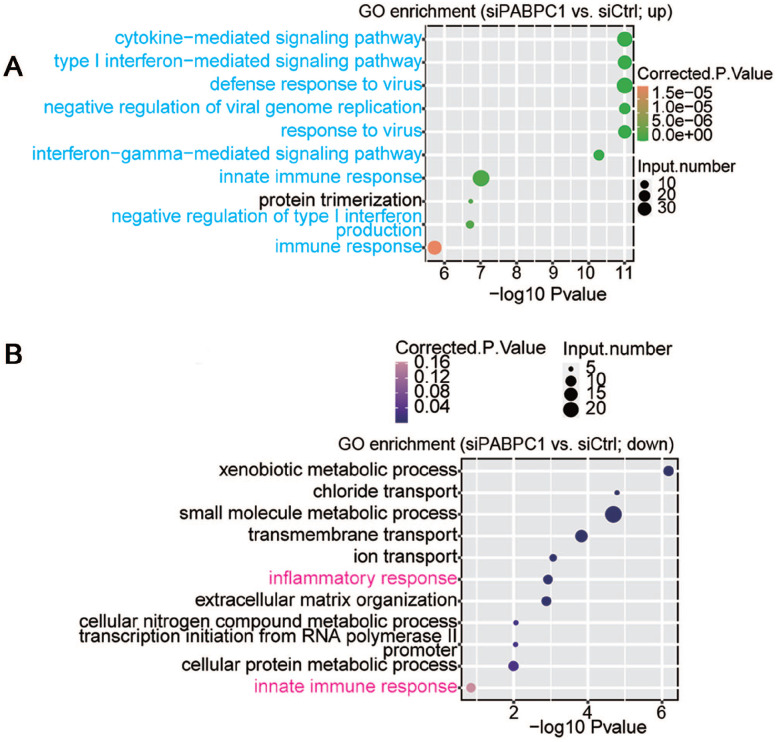
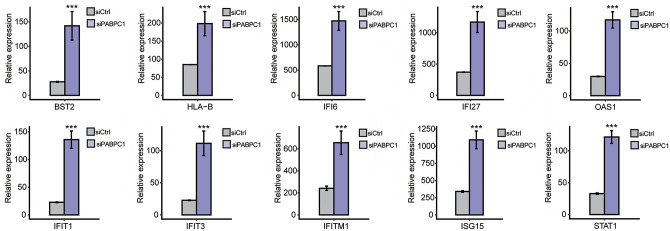
The potential biological roles of the 732 DEGs were analyzed by functional clustering. As shown in Figure 3(A), these genes were enriched in immuno-regulatory biological processes involving cytokines, type I interferon, interferon gamma, and immune response. These significantly down-regulated genes were enriched in inflammatory response, innate immune response, metabolic processes of xenobiotic, small molecules, cellular nitrogen compound and cellular protein, chloride and ion transport, transmembrane transport, extracellular matrix organization, and transcription initiation from RNA polymerase II promoter (Figure 3(B)). Among the PABPC1-mediated DEGs, we analyzed 94 DEGs associated with inflammation and immune response. Hierarchical clustering analysis indicated high consistency in PABPC1-mediated transcription of immune- and inflammation-related genes between siPABPC1 and siCtrl groups (Figure (4)). RNA-Seq data revealed increased transcriptions of BST2, HLA-B, IFI6, IFI27, IFIT1, IFIT3, IFITM1, ISG15, OAS1, and STAT1 in siPABPC1-treated AGS cells (P < 0.001, Figure (5)).
Figure 3.
(A) Up-regulated and (B) down-regulated biological processes were compared between siPABPC1-treated AGS cells and siCtrl-treated cells after GO enrichment analysis. (A color version of this figure is available in the online journal.)
Figure 4.
Heatmap of PABPC1-regulated genes involved in immune response and inflammation. The right bar values represent the relative expression of the indicated genes. (A color version of this figure is available in the online journal.)
Figure 5.
RNA-seq quantification method for analyzing the transcript abundance of BST2, HLA-B, IFI6, IFI27, OAS1, IFIT1, IFIT3, IFITM1, ISG15, and STAT1 in siCtrl (gray) and siPABPC1 (purple) groups. ***P < 0.001 versus siCtrl (Student’s t-test). Data are expressed as mean ± standard deviation, where the standard deviation is indicated as error bars. (A color version of this figure is available in the online journal.)
Analysis of PABPC1-regulated AS events in each class
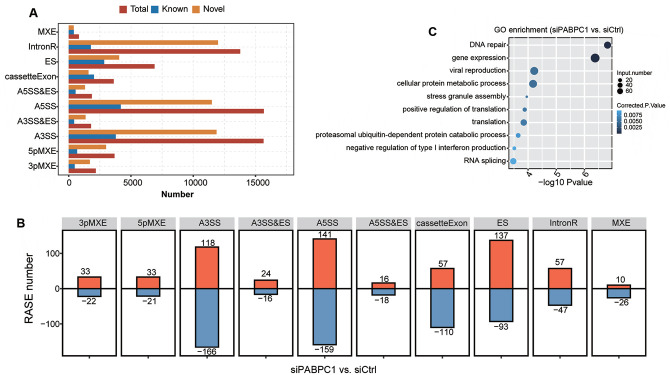
To gain further insights into the roles of PABPC1, transcriptome data were utilized to explore PABPC1-related AS events in AGS cell lines. In total, 17,167 known AS events and 48,748 non-known AS events were screened. These AS events were associated with 3pMXE, 5pMXE, A3SS, A3SS&ES, A5SS, A5SS&ES, cassetteExon, ES, IntronR, and MXE (Figure 6(A)). PABPC1 down-regulation triggered the decline of 1304 AS events differentially expressed in AGS cells (Figure 6(B)). A5SS was the most common class of PABPC1-regulated AS events up-regulated in AGS cells, followed by ES and A3SS. The top 3 down-regulated AS events were A3SS, A5SS, and ES in siPABPC1-treated cells. Functional enrichment analysis of RASE showed that PABPC1-regulated AS events involved in biological processes associated with DNA repair, gene expression, viral production, cellular protein metabolism, stress granule assembly, translation, negative regulation of type I interferon production, proteasomal ubiquitin-dependent protein catabolic process, and RNA splicing (Figure 6(C)). To validate PABPC1-regulated AS events, we detected five cancer-associated RASEs using RNA-Seq technique. PABPC1-induced A5SS-mediated A5SS events in POLR2J (P < 0.001, Supplementary Figure S1(A)). IntronR event was affected by PABPC1 down-regulation, which may cause an alteration in NXF1 expression (P < 0.05, Supplementary Figure S1(B)). PABPC1 altered ES event of KARS (P < 0.05, Supplementary Figure S1(C)), IntronR event of ADARB1 (P < 0.05, Supplementary Figure S1(D)), and A5SS event of GTF3C1 (P < 0.01, Supplementary Figure S1(E)), suggesting the regulatory roles of PABPC1 in gene expression.
Figure 6.
PABPC1-regulated AS events in siCtrl and siPABPC1 groups. (A) Number of AS events in each category including A3SS&ES, A5SS&ES, 3pMXE, 5pMXE, A3SS, A5SS, cassetteExon, ES, IntronR, and MXE. (B) Number of PABP1-regulated AS events in the indicated categories. (C) Gene ontology analysis for biological processes of PABP1-regulated AS events in siPABPC1-treated AGS cells compared with siCtrl-transfected cells. (A color version of this figure is available in the online journal.)
Correlation analysis of PABPC1-mediated AS events and DEGs in AGS cells
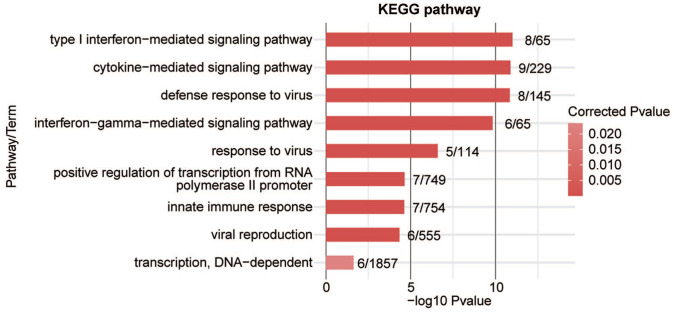
To analyze whether AS events contribute to alterations in transcriptional levels, we overlapped PABPC1-regulated DEGs with AS events. It was noticed that PABPC1 simultaneously mediated the transcription and AS process of 57 genes (Figure 7). These genes may participate in cytokine-mediated signal pathway, type I interferon-mediated signal pathway, interferon gamma-mediated signal pathway, innate immune response, transcription from RNA polymerase II promoter, as well as defense and response to the virus (Figure 8). Finally, five immune-related genes (e.g. C4BPB, CD55, HLA-F, IRF7, and STAT2) were screened, and results from RNA-seq showed that PABPC1 silence induced the down-regulation of C4BPB and CD55 and the transcription of HLA-F, IRF7, and STAT2 (P < 0.001, Figure 9). RNA-Seq analysis showed that PABPC1 mediated A3SS event of C4BPB (P < 0.05, Supplementary Figure S2(A)), 3pMXE event of CD55 (P < 0.05, Supplementary Figure S2(B)), IntronR of HLA-F (P < 0.05, Supplementary Figure S2(C)), ES event of IRF7 (P < 0.05, Supplementary Figure S2(D)), and IntronR of STAT2 (P < 0.05, Supplementary Figure S2(E)). qRT-PCR confirmed that the changed AS events altered mRNA expression of C4BPB, CD55, HLA-F, IRF7, and STAT2 (P < 0.05 and P < 0.001, Supplementary Figure S2).
Figure 7.

A total of 57 overlapped AS events were obtained between RASG (1241) and DEGs (1177). The differentially expressed AS events and DEGs were compared between siPABPC1-treated AGS cells and siCtrl cells. (A color version of this figure is available in the online journal.)
Figure 8.
Top 9 KEGG pathway items of PABPC1-regulated DEGs with alteration in AS events. (A color version of this figure is available in the online journal.)
Figure 9.
Effects of PABPC1 on DEGs with changes in AS events. Relative expression of immune-related genes (C4BPB, CD55, HLA-F, IRF7, and STAT2) in siCtrl (gray) and siPABPC1 (purple) groups was determined by RNA-seq. Data represent three independent biological repetitions. ***P < 0.001 versus siCtrl (Student’s t-test). Data are expressed as mean ± standard deviation, where the standard deviation is indicated as error bars. (A color version of this figure is available in the online journal.)
Discussion
The dysregulation of RNA splicing patterns has been associated with the modification of cellular transcripts in GC.38 –40 Peng et al. 24 revealed that PABPC1 interacts with hnRNPLL and facilitates the binding of hnRNPLL to the target mRNAs. However, it has not been well defined that PABPC1-mediated AS events play roles in the pathogenesis of GC. Based on RNA-Seq data, we performed bioinformatics analysis and found that PABPC1 knockdown altered gene transcription and AS events in AGS cells. PABPC1-regulated genes engaged in the regulation of immune pathways. Therefore, we speculated that the knockdown of PABPC1 in AGS cells could affect the expression of immune-related genes and AS events.
PABPC1 is abnormally expressed in several malignancies including glioblastoma, 29 hepatocellular carcinoma, 41 and gastric carcinoma. 42 qRT-PCR results showed that PABPC1 mRNA was enriched in GC tissues compared to the corresponding para-carcinoma tissues. Of note, PABPC1 binding sites within genetically encoded mRNA sequences affect gene expression, which has been disclosed by cross-linking immunoprecipitation coupled with high-throughput sequencing. 43 In addition, RNA-Seq analysis was carried out and 502 down-regulated and 732 up-regulated DEGs were identified in PABPC1-silenced AGS cells. GO and KEGG analysis indicated that the down-regulated and up-regulated DEGs were associated with the regulation of immune-related pathways.
It has been proposed that immune escape is one of the important causes for tumor progression and treatment failure. 44 The cancer cells can metastasize through evading recognition and destruction of the immune system. 45 Primary tumor-secreted factors, affecting the intra-tumor microenvironment and systemic immune environment, play an important role in this process. 46 After silencing PABPC1, the expression of mRNA of several genes (i.e. BST2, HLA-B. IFI1, IFI6, IFI27, IFIT1, IFIT3, IFITM1, ISG15, OAS1, and STAT1) was increased. Among these genes, ISG15 is reported to trigger an antitumor immune response against breast cancer upon secretion into the extracellular milieu. 47 In addition, STAT1 transcription factor mediates the surface expression of interferon-γ-induced programmed death ligand-1, which forms an inhibitory checkpoint with PD-1 acceptor to induce immune escape of tumor. 48 Moreover, HLA-B expression is related to biomarkers of T-cell activation (e.g. GZMA, GZMB, and PRF1) in basal-like breast cancers. 49 Therefore, we suggested that PABPC1 in GC may regulate the GC phenotype by mediating immune-related genes expression.
It has been recognized about the roles of AS events in cancer development, and the splicing process induces the general, cancer type-specific and subtype-specific changes in cancer cells. 50 Cancer cells can utilize AS events to evade anticancer therapy.51,52 At the present time, it remains unknown whether PABPC1-mediated carcinogenesis is associated with AS events in GC. We found that knockdown of PABPC1 significantly altered AS events in POLR2J, KARS, ADARB1, NXF1, and GTF3C1. POLR2J subunit isoform functions with translation initiation factor eIF3 involved in regulating downstream gene expression. 53 For the biological roles of this protein, KARAS mutations contribute to defective aminoacylation of mitochondrial and cytoplasmic tRNALys. 54 In addition, genomics screening has validated ADARB1 as putative cancer targets for identifying novel candidates. 55 In mice, RNA metabolism is disturbed by heterozygous mutations of NXF1, which induces lymphopenia. 56 Moreover, GTF3C1 is required for RNA polymerase III-mediated transcription, and its prognostic significance has been reported in colorectal cancer. 57 Furthermore, PABPC1 may influence the development of GC by regulating AS events, affecting gene expression and protein function, which in turn changes the expression signature of downstream genes.
In this study, we demonstrated that PABPC1 regulated both transcription and AS events in AGS cells. PABPC1-regulated DEGs and AS events were partially overlapping, including C4BPB, CD55, HLA-F, IRF7, and STAT2. KEGG pathway analysis suggested that PABPC1-regulated DEGs and AS events may affect immune-related signaling pathways involving type I interferon, cytokine, and RNA polymerase II promoter, viral multiplication, and DNA-dependent transcription. It has been shown that tumor cells generate membrane-bound complement regulatory proteins to induce immune escape. CD55 emerges as an important target for immune escape, anti-apoptosis, and targeted therapy of cancer cells. 58 Indeed, it has been shown that CD55 is involved in tumor metastasis and dedifferentiation and protects cells from complement-mediated attacks. 59 High expression of CD55 in GC is related to GC progress and displays prognostic value. 60 Different types of AS events have been found in C4BPB and IRF7 and play important roles in immune regulation.61,62 The expression of IRF7-targeting genes was inhibited in bone metastasis of breast cancer, which is associated with immune escape. 63 AS events in STAT2 mediate anti-apoptosis and drug resistance in cancer cells. 64 Thus, high expression of PABPC1 affects protein function by regulating gene expression and AS events, which in turn regulates the immune response of cancer cells and influences the development of GC.
Infiltration level of specific immune cells is related to the pathological progression of GC. 65 Analysis has elucidated that AS events-mediated specific protein modification might reshape the cancer immune microenvironment, suggesting a relationship between infiltration and AS events.66,67 CD55 is the membrane-bound complement regulatory protein, which is widely distributed on human lymphoid and non-lymphoid cells. 67 In this study, PABPC1 silence down-regulated the expression of CD55, implying that PABPC1-mediated changes in CD55 expression may be involved in the immune infiltration of GC. Evidence revealed that IRF7 is strongly associated with the abundance of immune cells like dendritic cells, B cells, and neutrophil cells, and IRF7 plays critical roles in immune escape in cancer. 68 These results suggested that PABPC1 may participate in modulating AS events that alter protein modification and then affect immune infiltration in GC.
Conclusions
In summary, this study validated the high expression of PABPC1 in GC. Our comprehensive association analysis of DEGs with AS events confirmed the effects of PABPC1-mediated AS events on gene expression. These DEGs and AS events were predicted to participate in regulating immune pathways, which are candidates for interfering PABPC1’s oncogenic functions in AGS cells. This study still has some limitations. For instance, although we uncovered PABPC1-regulated DEGs and AS events, PABPC1-mediated positive and negative regulatory mechanisms are unclear. In addition, further studies about the detailed contribution of PABPC1-mediated AS events to disease progression should be experimentally confirmed.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221121631 for Polyadenylate-binding protein cytoplasmic 1 mediates alternative splicing events of immune-related genes in gastric cancer cells by Xincai Xu, Wenbin Zhang, Hua Gao, Yi Tan, Yangchao Guo and Tiehan He in Experimental Biology and Medicine
Footnotes
Authors’ Contributions: XX collected and analyzed the data. WZ and HG contributed to the experiment. YT and YG performed the statistical analysis. XX, WZ, and YT wrote the manuscript. TH designed the research and revised the manuscript. All authors read and approved the final manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Tiehan He  https://orcid.org/0000-0002-6658-2803
https://orcid.org/0000-0002-6658-2803
Supplemental Material: Supplemental material for this article is available online.
References
- 1. IARC working group on the evaluation of carcinogenic risks to humans. Infection with Helicobacter pylori. Schistosomes, Liver Flukes and Helicobacter pylori. IARC Monogr Eval Carcinog Risks Hum 1994;61:177–2407715070 [Google Scholar]
- 2. Plummer M, Franceschi S, Vignat J, Forman D, de Martel C. Global burden of gastric cancer attributable to Helicobacter pylori. Int J Cancer 2015;136:487–90 [DOI] [PubMed] [Google Scholar]
- 3. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49 [DOI] [PubMed] [Google Scholar]
- 4. National Cancer Institute’s PROSPR Consortium, Corley DA, Sedki M, Ritzwoller DP, Greenlee RT, Neslund-Dudas C, Rendle KA, Honda SA, Schottinger JE, Udaltsova N, Vachani A, Kobrin S, Li CI, Haas JS. Cancer screening during the coronavirus disease-2019 pandemic: a perspective from the national cancer institute’s PROSPR consortium. Gastroenterology 2021;160:999–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Anderson WF, Rabkin CS, Turner N, Fraumeni JF, Jr, Rosenberg PS, Camargo MC. The changing face of Noncardia gastric cancer incidence among US non-Hispanic whites. J Natl Cancer Inst 2018;110:608–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Camargo MC, Anderson WF, King JB, Correa P, Thomas CC, Rosenberg PS, Eheman CR, Rabkin CS. Divergent trends for gastric cancer incidence by anatomical subsite in US adults. Gut 2011;60:1644–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu C, Hu C, Li Z, Feng J, Huang J, Yang B, Wen T. Systematic profiling of alternative splicing in Helicobacter pylori-negative gastric cancer and their clinical significance. Cancer Cell Int 2020;20:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang S, Hu Z, Lan Y, Long J, Wang Y, Chen X, Xu X, Zeng Z, Ouyang Y. Prognostic significance of survival-associated alternative splicing events in gastric cancer. Aging 2020;12:21923–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feng H, Jin Z, Liu K, Peng Y, Jiang S, Wang C, Hu J, Shen X, Qiu W, Cheng X, Zhao R. Identification and validation of critical alternative splicing events and splicing factors in gastric cancer progression. J Cell Mol Med 2020;24:12667–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ule J, Blencowe BJ. Alternative splicing regulatory networks: functions, mechanisms, and evolution. Molecular Cell 2019;76:329–45 [DOI] [PubMed] [Google Scholar]
- 11. Lin C, Yu B, Zhang M, Chen Y, Li L, Zhao D. Systematic analyses of the differentially expressed alternative splicing events in gastric cancer and its clinical significance. Front Genet 2020;11:522831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dvinge H, Guenthoer J, Porter PL, Bradley RK. RNA components of the spliceosome regulate tissue- and cancer-specific alternative splicing. Genome Res 2019;29:1591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wan L, Yu W, Shen E, Sun W, Liu Y, Kong J, Wu Y, Han F, Zhang L, Yu T, Zhou Y, Xie S, Xu E, Zhang H, Lai M. SRSF6-regulated alternative splicing that promotes tumour progression offers a therapy target for colorectal cancer. Gut 2019;68:118–29 [DOI] [PubMed] [Google Scholar]
- 14. Sheng J, Zhao Q, Zhao J, Zhang W, Sun Y, Qin P, Lv Y, Bai L, Yang Q, Chen L, Qi Y, Zhang G, Zhang L, Gu C, Deng X, Liu H, Meng S, Gu H, Liu Q, Coulson JM, Li X, Sun B, Wang Y. SRSF1 modulates PTPMT1 alternative splicing to regulate lung cancer cell radioresistance. Ebiomedicine 2018;38:113–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Passacantilli I, Frisone P, De Paola E, Fidaleo M, Paronetto MP. hnRNPM guides an alternative splicing program in response to inhibition of the PI3K/AKT/mTOR pathway in Ewing sarcoma cells. Nucleic Acids Res 2017;45:12270–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brockhoff M, Rion N, Chojnowska K, Wiktorowicz T, Eickhorst C, Erne B, Frank S, Angelini C, Furling D, Rüegg MA, Sinnreich M, Castets P. Targeting deregulated AMPK/mTORC1 pathways improves muscle function in myotonic dystrophy type I. J Clin Invest 2017;127:549–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McCuaig RD, Dunn J, Li J, Masch A, Knaute T, Schutkowski M, Zerweck J, Rao S. PKC-Theta is a novel SC35 splicing factor regulator in response to T cell activation. Front Immunol 2015;6:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sanidas I, Polytarchou C, Hatziapostolou M, Ezell SA, Kottakis F, Hu L, Guo A, Xie J, Comb MJ, Iliopoulos D, Tsichlis PN. Phosphoproteomics screen reveals akt isoform-specific signals linking RNA processing to lung cancer. Mol Cell 2014;53:577–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Anczuków O, Akerman M, Cléry A, Wu J, Shen C, Shirole NH, Raimer A, Sun S, Jensen MA, Hua Y, Allain FH, Krainer AR. SRSF1-regulated alternative splicing in breast cancer. Mol Cell 2015;60:105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Popli P, Richters MM, Chadchan SB, Kim TH, Tycksen E, Griffith O, Thaker PH, Griffith M, Kommagani R. Splicing factor SF3B1 promotes endometrial cancer progression via regulating KSR2 RNA maturation. Cell Death Dis 2020;11:842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gökmen-Polar Y, Neelamraju Y, Goswami CP, Gu Y, Gu X, Nallamothu G, Vieth E, Janga SC, Ryan M, Badve SS. Splicing factor ESRP1 controls ER-positive breast cancer by altering metabolic pathways. EMBO Reports 2019;20:e46078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Palangat M, Anastasakis DG, Fei DL, Lindblad KE, Bradley R, Hourigan CS, Hafner M, Larson DR. The splicing factor U2AF1 contributes to cancer progression through a noncanonical role in translation regulation. Genes Develop 2019;33:482–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seiler M, Peng S, Agrawal AA, Palacino J, Teng T, Zhu P, Smith PG, Buonamici S, Yu L. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Reports 2018;23:282–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peng Y, Yuan J, Zhang Z, Chang X. Cytoplasmic poly(A)-binding protein 1 (PABPC1) interacts with the RNA-binding protein hnRNPLL and thereby regulates immunoglobulin secretion in plasma cells. J Biol Chem 2017;292:12285–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patel GP, Bag J. IMP1 interacts with poly(A)-binding protein (PABP) and the autoregulatory translational control element of PABP-mRNA through the KH III-IV domain. FEBS J 2006;273:5678–90 [DOI] [PubMed] [Google Scholar]
- 26. Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 2014;159:1365–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takashima N, Ishiguro H, Kuwabara Y, Kimura M, Haruki N, Ando T, Kurehara H, Sugito N, Mori R, Fujii Y. Expression and prognostic roles of PABPC1 in esophageal cancer: correlation with tumor progression and postoperative survival. Oncol Rep 2006;15:667–71 [PubMed] [Google Scholar]
- 28. Feng C, Han Y-H, Qi N, Li J, Sheng Q-H, Liu Y, Yang L-L. Functional implications of PABPC1 in the development of ovarian cancer. J Open Med 2021;16:805–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Su R, Ma J, Zheng J, Liu X, Liu Y, Ruan X, Shen S, Yang C, Wang D, Cai H, Li Z, Xue Y. PABPC1-induced stabilization of BDNF-AS inhibits malignant progression of glioblastoma cells through STAU1-mediated decay. Cell Death Dis 2020;11:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang Q, Wang Z, Bao Z, Zhang C, Wang Z, Jiang T. PABPC1 relevant bioinformatic profiling and prognostic value in gliomas. Future Oncol 2020;16:4279–88 [DOI] [PubMed] [Google Scholar]
- 31. An T, Deng L, Yang Z, Chai C, Wang Y, Ouyang J, Lu X, Zhang C. High expression of PABPC1 predicts worse survival of gastric cancer patients. 2020, https://www.researchgate.net/publication/346634672_High_Expression_of_PABPC1_Predicts_Worse_Survival_of_Gastric_Cancer_Patients/fulltext/5fca94cea6fdcc697be0298c/High-Expression-of-PABPC1-Predicts-Worse-Survival-of-Gastric-Cancer-Patients.pdf
- 32. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. O’Connell GC, Treadway MB, Petrone AB, Tennant CS, Lucke-Wold N, Chantler PD, Barr TL. Leukocyte dynamics influence reference gene stability in whole blood: data-driven qRT-PCR normalization is a robust alternative for measurement of transcriptional biomarkers. Lab Med 2017;48:346–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26:139–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 2011;39:W316–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Armero VES, Tremblay MP, Allaire A, Boudreault S, Martenon-Brodeur C, Duval C, Durand M, Lapointe E, Thibault P, Tremblay-Létourneau M, Perreault JP, Scott MS, Bisaillon M. Transcriptome-wide analysis of alternative RNA splicing events in Epstein-Barr virus-associated gastric carcinomas. PLoS ONE 2017;12:e0176880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shi Y, Chen Z, Gao J, Wu S, Gao H, Feng G. Transcriptome-wide analysis of alternative mRNA splicing signature in the diagnosis and prognosis of stomach adenocarcinoma. Oncol Rep 2018;40:2014–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takeda J, Suzuki Y, Sakate R, Sato Y, Gojobori T, Imanishi T, Sugano S. H-DBAS: human-transcriptome database for alternative splicing: update 2010. Nucleic Acids Res 2010;38:D86–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhang H, Xu H-B, Kurban E, Luo H-W. LncRNA SNHG14 promotes hepatocellular carcinoma progression via H3K27 acetylation activated PABPC1 by PTEN signaling. Cell Death Dis 2020;11:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhu J, Ding H, Wang X, Lu Q. PABPC1 exerts carcinogenesis in gastric carcinoma by targeting miR-34c. Int J Clin Exp Pathol 2015;8:3794–802 [PMC free article] [PubMed] [Google Scholar]
- 43. Kini HK, Silverman IM, Ji X, Gregory BD, Liebhaber SA. Cytoplasmic poly(A) binding protein-1 binds to genomically encoded sequences within mammalian mRNAs. RNA 2016;22:61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kim JM, Chen DS. Immune escape to PD-L1/PD-1 blockade: seven steps to success (or failure). Ann Oncol 2016;27:1492–504 [DOI] [PubMed] [Google Scholar]
- 45. Gonzalez H, Hagerling C, Werb Z. Roles of the immune system in cancer: from tumor initiation to metastatic progression. Genes Develop 2018;32:1267–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Albini A, Bruno A, Noonan DM, Mortara L. Contribution to tumor angiogenesis from innate immune cells within the tumor microenvironment: implications for immunotherapy. Front Immunol 2018;9:527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Burks J, Reed RE, Desai SD. Free ISG15 triggers an antitumor immune response against breast cancer: a new perspective. Oncotarget 2015;6: 7221–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cerezo M, Guemiri R, Druillennec S, Girault I, Malka-Mahieu H, Shen S, Allard D, Martineau S, Welsch C, Agoussi S, Estrada C, Adam J, Libenciuc C, Routier E, Roy S, Désaubry L, Eggermont AM, Sonenberg N, Scoazec JY, Eychène A, Vagner S, Robert C. Translational control of tumor immune escape via the eIF4F-STAT1-PD-L1 axis in melanoma. Nat Med 2018;24:1877–86 [DOI] [PubMed] [Google Scholar]
- 49. Noblejas-López MDM, Nieto-Jiménez C, Morcillo García S, Pérez-Peña J, Nuncia-Cantarero M, Andrés-Pretel F, Galán-Moya EM, Amir E, Pandiella A, Győrffy B, Ocana A. Expression of MHC class I, HLA-A and HLA-B identifies immune-activated breast tumors with favorable outcome. Oncoimmunology 2019;8:e1629780–162980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang Y, Qian J, Gu C, Yang Y. Alternative splicing and cancer: a systematic review. Signal Trans Target Therapy 2021;6:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bonnal SC, López-Oreja I, Valcárcel J. Roles and mechanisms of alternative splicing in cancer – implications for care. Nat Rev Clin Oncol 2020;17:457–74 [DOI] [PubMed] [Google Scholar]
- 52. Sciarrillo R, Wojtuszkiewicz A, Assaraf YG, Jansen G, Kaspers GJL, Giovannetti E, Cloos J. The role of alternative splicing in cancer: from oncogenesis to drug resistance. Drug Resist Updat 2020;53:100728. [DOI] [PubMed] [Google Scholar]
- 53. Proshkin SA, Shematorova EK, Souslova EA, Proshkina GM, Shpakovski GV. A minor isoform of the human RNA polymerase II subunit hRPB11 (POLR2J) interacts with several components of the translation initiation factor eIF3. Biochemistry 2011;76:976–80 [DOI] [PubMed] [Google Scholar]
- 54. Wang Y, Zhou JB, Zeng QY, Wu S, Xue MQ, Fang P, Wang ED, Zhou XL. Hearing impairment-associated KARS mutations lead to defects in aminoacylation of both cytoplasmic and mitochondrial tRNA(Lys). Sci China Life Sci 2020;63:1227–39 [DOI] [PubMed] [Google Scholar]
- 55. Flanagan JM, Funes JM, Henderson S, Wild L, Carey N, Boshoff C. Genomics screen in transformed stem cells reveals RNASEH2A, PPAP2C, and ADARB1 as putative anticancer drug targets. Mol Cancer Ther 2009;8:249–60 [DOI] [PubMed] [Google Scholar]
- 56. Chappaz S, Law CW, Dowling MR, Carey KT, Lane RM, Ngo LH, Wickramasinghe VO, Smyth GK, Ritchie ME, Kile BT. Germline heterozygous mutations in Nxf1 perturb RNA metabolism and trigger thrombocytopenia and lymphopenia in mice. Blood Advances 2020;4:1270–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Anuraga G, Tang WC, Phan NN, Ta HDK, Liu YH, Wu YF, Lee KH, Wang CY. Comprehensive analysis of prognostic and genetic signatures for general transcription factor III (GTF3) in clinical colorectal cancer patients using bioinformatics approaches. Current Iss Mol Biol 2021;43:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cimmino F, Avitabile M, Pezone L, Scalia G, Montanaro D, Andreozzi M, Terracciano L, Iolascon A, Capasso M. CD55 is a HIF-2α marker with anti-adhesive and pro-invading properties in neuroblastoma. Oncogenesis 2016;5:e212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dho SH, Lim JC, Kim LK. Beyond the role of CD55 as a complement component. Immune Netw 2018;18:e11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Yin X, Shen C, Yin Y, Cai Z, Wang J, Zhao Z, Chen X, Chen Z, Chen H, Zhang B. Overexpression of CD55 correlates with tumor progression and poor prognosis in gastric stromal tumors. Onco Targets Ther 2019; 12:4703–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Criado García O, Sánchez-Corral P, Rodríguez de Córdoba S. Isoforms of human C4b-binding protein. II. Differential modulation of the C4BPA and C4BPB genes by acute phase cytokines. J Immunol 1955;155:4037–43 [PubMed] [Google Scholar]
- 62. Schultz KL, Vernon PS, Griffin DE. Differentiation of neurons restricts Arbovirus replication and increases expression of the alpha isoform of IRF-7. J Virol 2015;89:48–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Bidwell BN, Slaney CY, Withana NP, Forster S, Cao Y, Loi S, Andrews D, Mikeska T, Mangan NE, Samarajiwa SA, de Weerd NA, Gould J, Argani P, Möller A, Smyth MJ, Anderson RL, Hertzog PJ, Parker BS. Silencing of Irf7 pathways in breast cancer cells promotes bone metastasis through immune escape. Nat Med 2012;18:1224–31 [DOI] [PubMed] [Google Scholar]
- 64. Du Z, Fan M, Kim JG, Eckerle D, Lothstein L, Wei L, Pfeffer LM. Interferon-resistant Daudi cell line with a Stat2 defect is resistant to apoptosis induced by chemotherapeutic agents. J Biol Chem 2009;284: 27808–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ren F, Zhao Q, Zhao M, Zhu S, Liu B, Bukhari I, Zhang K, Wu W, Fu Y, Yu Y, Tang Y, Zheng P, Mi Y. Immune infiltration profiling in gastric cancer and their clinical implications. Cancer Sci 2021;112:3569–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang Z, Zhu L, Li K, Sun Y, Giamas G, Stebbing J, Peng L, Yu Z. Alternative splicing events in tumor immune infiltration in renal clear cell carcinomas. Cancer Gene Ther. Epub ahead of print 4 April 2022. DOI: 10.1038/s41417-022-00426-9 [DOI] [PubMed] [Google Scholar]
- 67. Shi JY, Bi YY, Yu BF, Wang QF, Teng D, Wu DN. Alternative splicing events in tumor immune infiltration in colorectal cancer. Front Oncol 2021;11:583547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Guo L, Fang T, Jiang Y, Liu D. IRF7 is a prognostic biomarker and associated with immune infiltration in stomach adenocarcinoma. Int J Gen Med 2021;14:9887–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_15353702221121631 for Polyadenylate-binding protein cytoplasmic 1 mediates alternative splicing events of immune-related genes in gastric cancer cells by Xincai Xu, Wenbin Zhang, Hua Gao, Yi Tan, Yangchao Guo and Tiehan He in Experimental Biology and Medicine