Abstract
Mass spectrometry imaging (MSI) is emerging as a powerful analytical tool for detection, quantification, and simultaneous spatial molecular imaging of endogenous and exogenous molecules via in situ mass spectrometry analysis of thin tissue sections without the requirement of chemical labeling. The MSI generates chemically specific and spatially resolved ion distribution information for administered drugs and metabolites, which allows numerous applications for studies involving various stages of drug absorption, distribution, metabolism, excretion, and toxicity (ADMET). MSI-based pharmacokinetic imaging analysis provides a histological context and cellular environment regarding dynamic drug distribution and metabolism processes, and facilitates the understanding of the spatial pharmacokinetics and pharmacodynamic properties of drugs. Herein, we discuss the MSI’s current technological developments that offer qualitative, quantitative, and spatial location information of small molecule drugs, antibody, and oligonucleotides macromolecule drugs, and their metabolites in preclinical and clinical tissue specimens. We highlight the macro and micro drug-distribution in the whole-body, brain, lung, liver, kidney, stomach, intestine tissue sections, organoids, and the latest applications of MSI in pharmaceutical ADMET studies.
Keywords: Mass spectrometry imaging, MSI, Drug absorption, Drug distribution, Xenobiotic metabolism, Pharmacokinetics, Toxicology
1. Mass spectrometry imaging in modern drug development and toxicology studies
Modern drug discovery and development requires a comprehensive assessment of drug candidates’ absorption, distribution, metabolism, excretion, and toxicity (ADMET). Refinement of analytical techniques that detect and quantify drugs, metabolites, relevant biomarkers and gain knowledge regarding distribution and delivery of a therapeutic agent to a target organ or tissue, has been fundamental to the drug development process. Small molecule or biologic pharmaceutical agents can only elicit the desired pharmacological activities when they are present in the vicinity of the therapeutic target sites at effective concentrations and for an appropriate duration of time [1]. The failure of many preclinical or clinical drug candidates could be related to insufficient drug exposure at the disease site or unacceptable toxicity [2]. Nonspecific accumulation of parent drug or toxic metabolites in unexpected tissues could lead to off-target toxicity. Thus, identifying the all-relevant molecular entities and quantifying their abundance, spatial distribution, penetration, and interaction with their targets in different tissues is important for understanding the pharmacology and toxicology of the pharmaceutical agents and aids in selection of new drug candidates.
The quantitative ADMET studies, especially tissue distribution analysis, are traditionally derived from autoradiography analysis, high-performance liquid chromatography (HPLC), or liquid chromatography tandem mass spectrometry (LC-MS/MS) based tissue homogenate analysis. Autoradiography is a molecular imaging technique that records the spatial distribution of radiolabeled pharmaceuticals in whole or dissected animals’ organs and tissues [3,4]. Autoradiography requires radiolabeled compounds and a long exposure time (days or weeks) to obtain high-resolution results [4]. While autoradiography is highly sensitive, it relies solely on radiolabeled tracers, which are difficult to prepare and environmentally unfriendly. Additionally, autoradiography lacks the ability to differentiate the labeled parent compound from their degradation products or metabolites that retain the tracer, thus making the actual molecular identity of the radioactivity it tracks and/or compound quantitation unavailable. Conventional tissue homogenate-based LC-MS/MS analysis does not provide spatial information of drug distribution. The immunohistochemical (IHC) method has been routinely used in toxicological pathology study of tissues; however, it is a low-throughput technique which also requires target-specific reagents [5]. Other imaging methods, like positron emission tomography (PET) and single-photon emission computed tomography (SPECT), require radioactive tracers, offer only millimeter spatial resolution, and provide limited molecular information. Fluorescence microscopy generally requires tagging of a drug with a fluorescent probe, which could impact the physiocochemical properties, uptake, and distribution of drug within the body.
Mass spectrometry imaging (MSI) is emerging as a powerful alternative analytical tool for determining spatial distribution and quantification of endogenous biomolecules and exogenous drug molecules by directly analyzing thin tissue sections based on their mass-to-charge (m/z) ratio [6]. MSI is a molecular imaging technique capable of multiplex imaging analysis, allowing simultaneous detection of thousands of molecules including endogenous biomolecules such as proteins, peptides, lipids, small metabolites and exogenous molecules such as drugs and drug metabolites in specific tissue microstructures at cellular and subcellular spatial resolution in a single experiment [6]. MSI can provide spatial distribution information that the conventional LC-MS/MS method lacks. MSI avoids the use of radio-labeling agents required by autoradiography method and determines chemical specificity of both drugs and their metabolites [7]. Compared to the conventional histologic techniques, MSI is a label-free and high-throughput technique that can assess a comprehensive array of molecular species without a priori knowledge of the targeted molecule of interest or target-specific reagents. It therefore, promotes the discovery of toxic biomarkers and facilitates mechanistic insights into toxicological pathways [8]. MSI technology offers powerful new dimensions and capabilities to aid in drug development.
MSI is considered as a “molecular microscope” for biology and medicine [9,10,11], and has broad applications for molecular pathology, tissue/cellular biology, drug discovery and development, and forensic science [12]. In this review, we focus on the current applications of MSI in pharmaceutical ADMET and site-specific pharmacokinetics (PK) studies and discuss MSI’s recent technical advances in qualitative and quantitative molecular ion imaging of small and large drug molecules in various tissues (brain, lung, liver, kidney, stomach, intestine, tumors, whole body) and brain organoids in preclinical and clinical models. Particularly, we highlight MSI advances in ADMET studies from the last 5 years with sophisticated instrumentation, enhanced spatial resolution, absolute quantification of detected drug molecules, and improved sample preparation methods.
2. Introduction of MSI
2.1. Principles and fundamentals of MSI
Mass spectrometry (MS) analysis is an analytical technique that identifies ionized molecules based on the m/z, which is generated from different ionization technologies including electrospray ionization (ESI), electron impact ionization (EI), fast atom bombardment (FAB), atmospheric pressure chemical ionization (APCI), and matrix-assisted laser desorption/ionization (MALDI). MS instruments necessarily have the following components: 1) a sample introduction system that either introduces sample molecules directly into the instrument via an insertion probe/plate or direct infusion using a sample capillary inlet or follow online chromatographic separation; 2) an ionization source like ESI to generate the ionized molecules; 3) a mass analyzer to detect ions according to the m/z; and 4) a mass detector to quantify the total number of ions of each mass present [13]. Sample ionization may take place either in high vacuum or at atmospheric pressure, but ion detection and analysis require a high vacuum. The commonly employed mass analyzers include triple quadrupole (QqQ), quadrupole ion trap (QTRAP), time-of-flight (TOF), or high-mass accuracy and high-resolution mass analyzers such as orbitrap, Fourier transform ion cyclotron resonance (FTICR), quadrupole-TOF (QTOF), or TOF/TOF. Each of these mass analyzers has its own advantages and limitations in terms of mass resolution, transmission, duty cycle, and quantitative abilities [14,15].
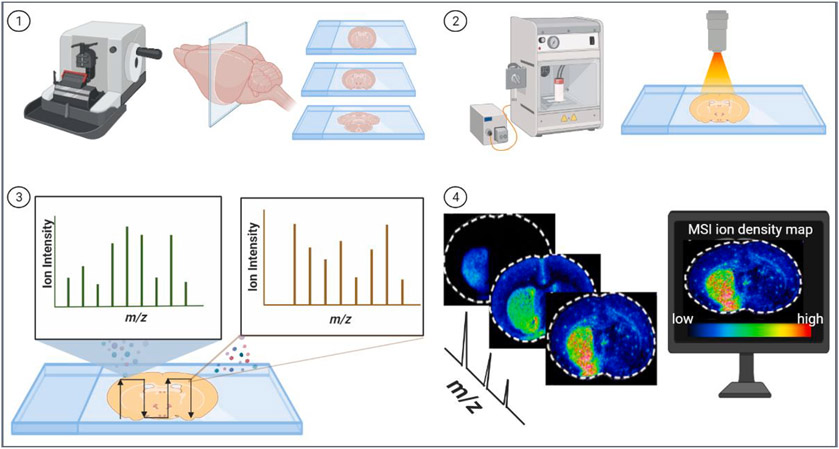
MSI adds an additional layer of information to traditional MS technologies. MSI not only identifies and quantifies each mass present in a tissue section, but also provides un-biased spatial distribution of each molecule [5,6]. MSI technologies were pioneered by Richard Caprioli. His team successfully demonstrated the use of MALDI-MSI to visualize large biomolecules with insulin in a rat pancreas section, hormone peptide in a rat pituitary section, and a membrane protein in cells in 1997 [16]. In a typical workflow for spatial analysis of tissue section using MALDI-MSI (Fig. 1), frozen or formalin fixed paraffin embedded (FFPE) tissues are sliced into thin tissue sections and sprayed with a layer of energy-absorbing matrix to form uniform microcrystals. A pulsed laser then irradiates the matrix-coated tissue, desorbs the tissue and ionizes molecules in different spots on the defined region of tissues [17]. Ion signals from an ordered array of all laser shots produce the m/z mass spectrum, individual ion m/z values in each spectrum that correspond to the individual compound’s molecular weights are assembled to produce selected images of the areas within the tissue in which the molecules are located. Summing all the ions in each spectrum produces a total ion density map [18]. The distribution maps of multiple analytes can be correlated with tissue histological features. Instrument software is used to set the data acquisition grid pattern with a predetermined number of laser shots per grid coordinate, and the grid pattern has a fixed center with the size depending on the image resolution required, usually between 25 and 250 μm [19].
Fig. 1.
A typical workflow for spatial analysis of tissue section using matrix-assisted laser desorption/ionization (MALDI) mass spectrometry imaging (MSI). 1). Frozen or FFPE tissue sections are cut into thin sections on glass slides by microtome or cryostat. 2). Tissues sections are coated with an energy-absorbing matrix via a matrix sprayer and placed into the mass spectrometer. 3). A pulsed laser desorbs the tissue and ionizes molecules at different spots on the defined region of tissues. The m/z values of the ionized molecules are determined by mass analyzer. 4). The localization pattern of individual molecular species present on the tissue surface is extracted and positioned on the histological image of the tissue. MS images are generated by computer software based on molecules’ m/z ion intensities. The color reference bar represents the relative intensity of different ions. Each m/z image has its own intensity scale encoded with the blue (low intensity) to red (high intensity) color gradient.
Since the initial demonstration of the imaging capability for MALDI-MSI directly from tissue sections, several innovative ionization technologies and sample preparations have been developed to allow unbiased visualization of biomolecular spatial arrangements in tissue sections. Historically, such a view was only available using traditional histology in tandem with molecular identification via mass spectrometry. Selecting the appropriate combination of ionization methods and MS components to pair with MSI technology is crucial for optimal imaging.
2.2. Ionization methods
2.2.1. MALDI
Several ionization methods are commonly employed in MSI, each with their own strengths and limitations (Table 1). The MALDI technique is the most popular and versatile soft ionization method that allows the analysis of large intact biomolecules that would normally fragment under hard ionization conditions. MALDI-MSI also enables analysis of a wider mass range (0–50,000 Da) than other MSI technologies [20]. Tissue sections analyzed by MALDI-MSI must be uniformly coated with a chemical matrix (section 2.4.2) (Fig. 1). During analysis, a pulsed ultraviolet (UV) or infrared (IR) laser beam irradiates the sample surface in an array of discrete points at a pre-defined pixel size, triggering ablation and desorption of the molecules from the sample surface [21]. The matrix molecules absorb the laser energy and transfer energy to the analyte, and the analyte molecules are generally ionized by being protonated or deprotonated with surrounding matrix molecules; the resulting ions are accelerated into the mass analyzer for detection (Fig. 2). The most common mass analyzer that can be interfaced with MALDI include TOF, QTRAP, Orbitrap, and FTICR. Matrix-assisted laser desorption electrospray ionization (MALDESI) paired with a FTICR can be used as an effective analytical method in analysis of peptides and proteins in a single acquisition [22]. MALDI can be operated under high vacuum or atmospheric pressure. Spatial resolution on MALDI-MSI can range from 30 to 50 μm on commercial instruments, a spatial resolution of up to 10 μm is achievable by decreasing the pre-defined pixel size, which can cause an increase in sample run time and make high resolution images rather time consuming [23,24]. A molecule’s ionization efficiency depends on the wavelength of the laser. Traditional MALDI has sensitivity issues due to limited ionization and ion suppression. In a typical MALDI analysis of tissues, molecules that are desorbed from the surface by the laser as neutral species are normally not detected. MALDI-2 is an effective laser-based post-ionization technique introduced in 2015 by Soltwisch et al. [25]. MALDI-2 utilizes a second orthogonal laser beam to interact with the analyte desorbed from the first laser pulse to allow a charge transfer from post-ionized matrix molecules to neutral analyte molecules [25]. MALDI-2 can improve sensitivity by up to 1–3 orders of magnitude depending on samples and matrix [25-27]. The secondary laser in MALDI-2 can ionize neutral species microseconds after they desorb, enabling their detection. This enhances the signal, dramatically shifts the limit of detection, and permits the use of smaller spot sizes for higher spatial resolution. The mechanism of improved ionization by proton transfer in MALDI-2 was postulated to involve resonant two-photon ionization of the matrix by the post-ionization laser [25]. MALDI-MSI is applicable to molecules of wide mass range including both low (<1 kDa) and high molecular mass compounds (~100 kDa) [28,5]. MALDI-MSI is used in a variety of research areas and clinical applications to provide spatial distribution information alongside quantitative analysis.
Table 1.
Comparison of different MSI ionization methods.
| Ionization | Advantages | Disadvantages |
|---|---|---|
| MALDI | 10–100 μm spatial resolution, fmol-zmol sensitivity, mass resolution up to 40,000 Allows for analysis for a large mass range m/z 0 – 50,000 Da, allows for the generation of ions of small and large molecules. Has been used for quantitative analysis on tissue |
Sample preparation and sample matrix required Matrix signals may interfere with the signal of analyte Low sensitivity for low molecular weight compounds Sample damage depending on the laser frequency |
| DESI | 40–200 μm spatial resolution for DESI, 10–100 μm spatial resolution for nanoDESI, Ambient conditions, no vacuum required. Minimal sample preparation, no matrix required, faster analysis time |
Low sensitivity for high m/z (>2000) ions At present, limited sample surface can be analyzed |
| SIMS | 0.5–1 μm spatial resolution Static and dynamic ion modes provide surface and depth profile respectively |
Low sensitivity for high m/z (>1000) ions due to fragmentation. Quantification is difficult |
| LA-ICP | Spatial resolutions < 1 μm possible Elemental imaging, trace metal detection and quantification Quantification of metal-labelled antibodies |
Complex isotopic fractionation, isobaric interference lack of matrix matched standards for quantification Destructive action on the specimen |
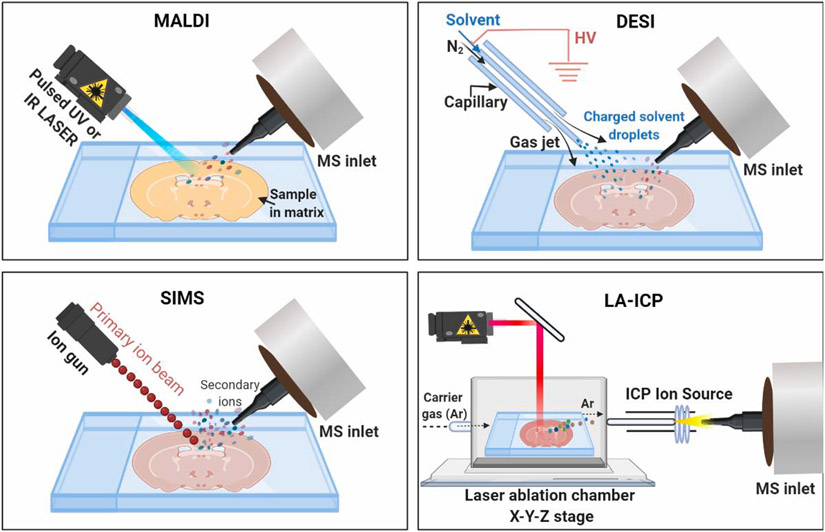
Fig. 2.
Schematic drawing of ion source of matrix-assisted laser desorption/ionization (MALDI), desorption electrospray ionization (DESI), secondary ion mass spectrometry (SIMS), and laser ablation inductively coupled plasma (LA-ICP) MSI. For MALDI-MSI analysis, the tissue section is coated with a thin layer of energy-absorbing matrix and irradiated by UV or IR laser pulses to desorb and release the ions of analyte. DESI-MSI is carried out by directing electrosprayed charged droplets and ions of solvent onto the surface of the tissue section; the impact of the charged particles produces gaseous ions of analyte. SIMS uses a beam of primary ions to irradiate the tissue surface and facilitate ionization by transferring energy from primary ions to the secondary ions of analyte. For LA-ICP-MSI analysis, tissue is placed in a chamber and ablated with laser; the laser induced sample particles are transported by a carrier gas and ionized in the plasma. After different ionization steps, the resulting ions are guided into the mass spectrometers for m/z analysis.
2.2.2. DESI
Desorption electrospray ionization (DESI) was first introduced in 2004 by Graham Cooks’ group in an attempt to eliminate the need for a vacuum system to analyze samples [29]. DESI is an ambient ionization technique that does not require a matrix or sample preparations (Table 1). DESI employs charged solvent droplets sprayed onto the analyte surface from a pneumatically assisted electrospray source. The microdroplets act as projectiles and desorb ions from the sample surface as a result of electrostatic and pneumatic forces; the resulting ions are accelerated into the mass analyzer for detection (Fig. 2) [30,31,32]. The desorbed secondary droplets undergo electrospray-like solvent evaporation and ionization, and these ions are transferred to the MS via an atmospheric pressure ion-transfer line [30,31,32]. For low molecular weight molecules, ionization occurs by charge transfer of an electron or a proton. For high molecular weight molecules, multiple charges in the droplet can be transferred to the analyte. This allows for the acquisition of electrospray-like mass spectrum information with multiple charged ions and adducts. Nitrogen is the most commonly used nebulizing gas. Solvent systems with or without an acid modifier are most commonly used for DESI. A 1:1 methanol:water solution solvent system has proven to have a high signal intensity for molecules with m/z range of 700–1000 [33,34,35]. Small molecule analysis with a m/z < 700 shows increased sensitivity when a 1:1 dimethylformamide: water solution is used [34]. Analytes that do not undergo ESI typically have a low detection sensitivity [31]. This problem can be overcome by the use of reactive DESI methodology which employs a derivatization agent in the DESI solvent [36]. Another drawback to DESI technology is that, while it can easily analyze compounds < m/z 2000, compounds with higher masses require a more sophisticated system or pre-scanning the surface [37]. In addition, DESI-MSI coupled with ion mobility (IM) separation, an ion separation technique used in gaseous phases based on molecular differences in size, shape and charge when an electric field is applied, has been utilized to add an additional dimension of resolution [38]. Ion mobility separation can be highly beneficial when analyzing complex samples as it enables detection and isolation of various ions with overlapping signals which would otherwise remain buried in the noise and thus be undetectable without this additional mode of separation [39]. While the spatial resolution of initial DESI-MSI (<500 μm) is not as good as in MALDI-MSI [40], a spatial resolution of 20–100 μm pixel size can be achieved by optimizing the DESI spray’s geometry [41]. Nanospray DESI (nanoDESI) uses a self-aspirating nanospray capillary to directly transport and ionize analyte that is desorbed from the surface [42]. NanoDESI-MSI was shown to have a lateral resolution of 10 μm on tissue sections [43]. The MSI analytical capabilities can be further extended when coupling nanoDESI with IM separation for spatially-resolved analysis of complex tissue samples [44]. The DESI technique is applicable to solids, liquids, adsorbed gases, tissues sections, and live cells [45]. DESI-MSI can therefore be utilized in several pharmaceutical development activities including formulation and in vitro release method development [38].
2.2.3. SIMS
Secondary ion mass spectrometry (SIMS) was originally used for inorganic materials but is now aiding the analysis of biological materials (Table 1). SIMS utilizes a highly focused primary ion beam under high-vacuum to bombard the sample surface and produce atomic collisions, some of these collisions result in the release of secondary ion particles (Fig. 2) [46,47]. These charged ions are transmitted through a MS analyzer, where the m/z is measured. SIMS has the advantage of high lateral resolution at micrometer or submicrometer range. SIMS technology has been expanded to include TOF-SIMS, FTICR-SIMS, and NanoSIMS. In TOF-SIMS, the secondary ions created by the primary ion beam are extracted into an analyzer using high voltage potential; the molecular mass is determined by measuring the time-of-flight from the sample’s surface to the detector [47]. TOF-SIMS provides analysis of a large mass range and improves mass resolution and sensitivity. The FTICR-SIMS platform uses a C60 primary ion gun source. The C60 powder is heated under a vacuum and the evaporated molecules are ionized and focused through several ion optics while moving towards the sample. C60 FTICR-SIMS can achieve high mass accuracy and high mass resolving power at a spatial resolution of 40 μm range [48,49,50]. NanoSIMS uses a reactive primary ion beam like Cesium+ (Cs+) or Oxygen− (O−) beam for positive or negative secondary ion analysis, respectively [46]. The beam size can be adjusted below 50 nm for the Cs+ ion gun and below 200 nm for the O− ion gun, which provides high lateral resolution and allows the visualization of elemental and isotopic composition of the analyzed sample at nanometer scale [51]. The primary ion source for SIMS methodology is limited to small molecule analysis (<1000 Da); this is due to the high energy levels required to fragment large molecules [20]. SIMS methodology is typically applied to analysis of elements, atomic clusters, drugs, and other low molecular mass compounds within tissue sections.
2.2.4. LA-ICP-MSI
Laser ablation inductively coupled plasma mass spectrometry imaging (LA-ICP-MSI) is used to conduct elemental analysis of solid samples and tissues (Table 1). Laser ablation removes material from a solid surface by irradiating it with a laser beam. The laser induced sample particles are transported via an argon carrier gas into the ICP ionization source where the high-temperature (up to 10,000 K) plasma (ionized gas) fully decomposes a sample into its constituent elements and transforms those elements into ions by removing loosely held electrons [52,53] (Fig. 2). The ICP torch pushes the analyte aerosols to the MS creating m/z and MSI data [54]. LA-ICP-MSI can use metal-labelled antibodies to detect and quantify proteins [55]. A distinct disadvantage to using LA-ICP-MSI is its destructive action on the specimen; post laser ablation, a small pit encompassing the size of the analytical volume remains [56]. In addition, LA-ICP-MSI can have difficulties in quantification due to matrix effects and molecular ion formation [55]. Coupling the LA-ICP to a TOF has aided detection of proteins labeled with rare earth elements in biological samples [57]. Resolution for elemental analysis using LA-ICP-MSI can be as low as 5 μm [58], although it is possible to achieve a resolution of 500 nm using a dual-pulse laser under vacuum conditions [59]. LA-ICP-MSI technology primarily produces atomic information and detects different isotopes of the same element at very low concentrations. LA-ICP-MSI shows great promise in quantitative metal imaging in biological tissue samples and metal-containing drug analysis [60].
2.3. MSI for quantitative analysis
Absolute quantification using MSI is still challenging due to ion suppression by endogenous molecules, heterogeneity of sample surface, matrix effects, and fluctuations in MS detectors. Different tissue quantification approaches have been explored. The dilution series model uses calibration standards spotted onto a control tissue. The drug concentration is estimated by comparing the control ion intensities to those of the dosed sample. The mimetic tissue model uses tissue homogenates spiked with calibration standards to quantify target compounds in tissue sections; this method requires more sample preparation but closely simulate drug “in-tissue” [61]. The tissue extinction coefficient method refers to a regional correction factor calculated by the intensity of the standard on tissue divided by the intensity of the standard on the glass slide [62]. Normalizing drug or metabolite ion signals against a stable isotope-labeled internal standard compensates for ion suppression and matrix effects. Drug quantification on tissues is done ex vivo and, in some cases, after the tissue has been formalin fixed. This process would stop all biological cell functions and limit or halt additional drug migration, allowing for precise drug distribution imaging. As with traditional mass spectrometry systems, a system suitability test should be performed when using MSI to confirm accurate quantification. This can be accomplished by using the MSI to analyze a defined spot on the glass of the tissue slide that has been spiked at known concentration with the mass range of interest. This will evaluate accurate mass identification and accurate quantification prior to sample analysis. MSI also suffers from relatively low sensitivity compared to LC-MS/MS. Multiple reaction monitoring (MRM) combined with MSI can selectively detect and quantify specific precursor ion to product ion transitions and provide a high-throughput analysis of multiple known analytes on tissue sections [61]. The analytical validation of the quantitative MSI is still challenging. Several groups compared different quantitative MSI platforms with respect to limit of detection (LOD), linearity, sensitivity, reproducibility, and precision and accuracy of high and low quality control (QC) samples of drugs in dog liver samples [63,61,64]. Recent reviews present detailed description of MSI relative quantitation, absolute quantitation, calibration curve construction, and normalization strategies [65;66]. High selectivity of analyte can be further achieved by using the ion mobility separation or high resolving power FTICR-MSI. Derivatization strategies, matrix optimization, and improved tissue processing methods can also help to develop a robust and validated quantitative MSI method for drug analysis.
2.4. Sample preparation and data acquisition
2.4.1. Sample preparation
MSI is commonly performed on flash-frozen tissue samples. Tissue perfusion prior to tissue harvest can be performed to avoid detecting drug and metabolites in capillaries that supply blood to the organs such as brain. It is essential to stop any cellular reactions and enzyme-mediated degradation of molecules and reduce molecule diffusion across the tissues when harvesting tissue samples. Thus, tissues should be frozen quickly in liquid nitrogen to minimize these variables and to preserve tissue morphology. Thin tissue sections for MSI typically range from 5 to 20 μm thickness. FFPE tissue, which is routinely used in histological analysis and diagnosis, can also be used for MSI. Originally tissues for MSI analysis could not have extra additives like optimal cutting temperature (OCT) compound to minimize ion suppression. Recent sample preparation has been optimized to allow for additives like OCT, although additional sample preparation steps to properly remove OCT compound or paraffin from the tissues while not disrupting the key tissue components being analyzed are needed [67]. In contrast to fresh frozen tissues, the use of antigen retrieval technique and sample enzyme digestion are performed for FFPE samples to obtain a partial reversal of the protein crosslinking caused by the FFPE preparation [68,69]. FFPE tissue sample preparation has been optimized by various labs to improve MALDI-MSI performance [67,70]. Carter et al. used FFPE to inflate and fix lung tissue samples from rhesus macaques and this sample preparation improved MSI sensitivity and resolution without significant signal loss [71]. The chemical structures of drug and drug metabolites should not be modified in the FFPE tissue; some very specific metabolites could be modified in theory (e.g., primary amine may react with formalin), but they should be readily recognized based on the exact mass. In general, both DESI and SIMS can be performed directly after sectioning. Unlike MALDI-MSI samples, SIMS and DESI sample preparation processes typically consist of freeze or vacuum drying of samples using a vacuum desiccator [72,39]. Optimization of thin tissue sections can include washing the sample to remove cell debris, excess salts, or lipids in order to prevent ion suppression, or adding compounds to perform on-sample enzymatic digestion or derivatization [69]. Histochemical staining using compatible dyes like methylene blue or cresyl violet can be performed on the same tissue section before or after an MSI experiment or on the consecutive tissue section to allow correlation of MSI images with histological information [73].
2.4.2. Matrix chemistry
A matrix coating is required to aid in MALDI ionization of compounds. This coating is typically applied with a matrix sprayer to evenly coat the entire tissue surface and avoid regions of high and low ionization (Fig. 1). Selection of the proper sample matrix is critical to achieving reliable imaging results. The matrix is usually a small organic, aromatic molecule that has an absorbance matching the wavelength of the laser, which allows for optimum absorption of energy from the laser to avoid substantial fragmentation [74]. The benefits of applying the matrix on the sample to MALDI-MSI analysis are: (1) minimizing fragmentation of the analyte ions by absorbing the majority of the laser beam’s energy; (2) inducing collisional cooling during the adiabatic expansion of the cloud of material moving into the vacuum; and (3) separating the analyte crystals from other interfering substances like salts and large molecules [75]. As the matrix crystalizes, analytes are extracted from the tissue section and co-crystalized. The frequently used matrixes for MALDI-MSI and their structures are listed in Table 2. The most common matrixes for small molecule analysis include 2,5-dihydroxybenzoic acid (DHB) and α-cyano-4-hydroxycinnamic acid (CHCA). For additional information on MSI matrices, we refer the reader to the available comprehensive review papers [69,74,76]. Matrix application methods include manual droplet disposition, airbrushing using pneumatic nebulization, an acoustic reagent multi-spotter, electrospray disposition, or a pneumatic sprayer [74,77]. The quality of matrix application can impact MSI’s mass and spatial resolution and sensitivity.
Table 2.
Frequently used MALDI matrixes to assist desorption and ionization of analyte.
| Matrix | Abbreviation | Structures | Applications | References |
|---|---|---|---|---|
| 2,6-Dihydroxyacetophenone | DHA |
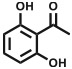
|
lipids, proteins | [77] |
| 2,5-Dihydroxybenzoic acid | DHB |
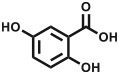
|
small molecules, lipids, oligosaccharides | [92,94] |
| 1,5-Diaminonaphthalene | DAN |
|
lipids | [158] |
| α-Cyano-4-hydroxycinnamic acid | CHCA |
|
small molecules, peptide, and proteins | [159] |
| Sinapinic acid | SA |
|
peptide and proteins | [77,159] |
2.4.3. Data acquisition, processing, and normalization
Data acquisition, processing, and analysis for MSI are much more complex than in traditional MS data analysis due to large data sets and a high degree of dimensionality. Traditional MS data acquisition and analysis create a total ion count (TIC), which is a chromatogram that sums up the intensities of all mass spectral peaks along the same scan, providing information as to how multiple analytes elute in a single injection. Traditional MS also provides qualitative and quantitative detection of an analyte by comparing detected m/z ratios to isotopically labelled internal standards. MSI adds an important layer of complexity to this already complicated and detailed sample analysis by providing spatial distribution information for each compound. Each pixel created in the MSI experiment contains the information of an entire mass spectrum [65]. As with traditional MS, there are several preprocessing steps that can reduce experimental variance in the data set; these include normalization, baseline correction, spectral realignment, and smoothing [78]. Analyte intensities and data reproducibility can be negatively affected by sample preparation, matrix selection, ion suppression, and ionization inefficiencies in a complex sample matrix. Normalization to the TIC is the most common method of avoiding misleading artifacts and improper analysis. Normalization ensures that all spectra obtained have the same integrated area and it is assumed that there is a comparable number of signals in each spectrum [65,79,80]. However, it should be noted that normalization to the TIC is not applicable when comparing different tissue types. For samples containing different tissue types like a whole-body section, a reference compound should be added and normalization should be performed to the reference compound [65,79]. As with traditional MS data, smoothing is an integral preprocessing step. Smoothing allows for mitigation of background noise and overall data improvement. Smoothing is traditionally used when the parent spectrum is difficult to distinguish due to intense background noise; however, peak smoothing should be used with caution to avoid misrepresentation of data. This is equally important in MSI to avoid unexpected variability between pixels that may misrepresent the in vivo data distributions [65]. A more detailed description of MSI data processing, visualization, and statistical analysis of MSI data can be found in an excellent review from Buchberger et al. [65]. The reproducibility of MSI data analysis is critical as the regulatory agencies generally expect the co-efficient of variance (CV) of reproducibility should not exceed 20% for analytical techniques [81]. Several labs have addressed the issue of MSI site-to-site reproducibility [82,83,84] and multicenter studies were carried out to study the MSI reproducibility and data processing guidelines to insure the reliability and transparency of MSI data [85,86,87]. Several commercial software programs and tools such as MSiReader (a free open source MSI processing and visualization tool), SCiLS from Bruker, ImageQuest from Thermo, and High-Definition Imaging (HDI) from Waters are available for advanced data processing and statistical analysis. To overcome problems with MSI data standardization and reproducibility, an open-source computational platform Galaxy framework was developed to link 18 different MSI data analysis tools to enable all major MSI analysis steps such as visualization, preprocessing, statistical analysis, and image co-registration [88]. The MSI data analysis in Galaxy allows accessible and transparent data analysis.
3. MSI applications in visualization and quantification of drug absorption, distribution, and excretion in preclinical tissues
3.1. MSI monitors drug absorption in gastrointestinal tract and PK imaging
In drug development, a drug’s plasma concentration is commonly used to determine systemic exposure, but sometimes it does not correlate with the quantity of the drug distributed in the target tissue. For locally delivered drugs to the distal gastrointestinal (GI) tract to treat intestinal diseases, systemic plasma concentrations do not accurately represent the tissue exposure of these GI-targeted agents. Knowledge on the region-specific drug absorption profiles from the GI tract of orally administered drugs can facilitate drug design and selection of the optimal dosing regimen. In this regard, MSI offers a complementary analytical method to enable visualization of the quantity and distribution of drugs in different GI tract regions and provides a longitudinal absorption profile at different time points.
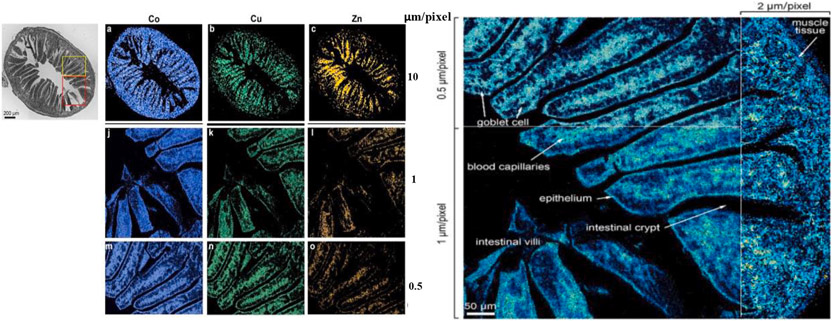
In 2021, Meng et al. used a LA-ICP-MSI platform to investigate the distributions of three metal drugs used in photodynamic therapy after ingestion by oral gavage (CuPc, ZnPc and CoTsPc) in the small intestine of mice [60]. The overlay MS image of 59Co, 63Cu and 67Zn distribution in the small intestine at different spatial resolutions (10, 1, 0.5 μm/pixel) is presented in Fig. 3 (left panel). All three drugs had wide distribution in the intestinal epithelia and blood capillaries of the intestine, indicating these drugs are absorbed by the epithelium and transported to other parts of body by the blood capillaries. The zoom-in MSI image showed the goblet cells in the small intestine at 0.5 μm spatial resolution (Fig. 3, right panel). This study thus demonstrates the power of high-resolution MSI in revealing vital information about drug absorption and transportation processes in the intestinal villi.
Fig. 3.
High resolution LA-ICP-MSI distribution analysis of photodynamic therapy drugs (CuPc, ZnPc, and CoTsPc) in the small intestine of mouse after oral administration. Left panel shows the optical image of the intestine slice and the MS images of 59Co, 63Cu and 67Zn distribution in a same intestine slice with spatial resolutions at 10, 1, and 0.5 μm/pixel. Images (a-c) in the left panel are MSI of Co, Cu and Zn of the small intestine at 10 μm/pixel. Images (j-l) are MSI of Co, Cu and Zn of the red region outlined in the optic image of the small intestine at 1 μm/pixel. Images (m-o) are MSI of Co, Cu and Zn of the yellow region outlined in the optic image of the small intestine at 0.5 μm/pixel. The right panel shows the zoomed-in MS image of the intestine to identify the cells within the intestine at different spatial resolutions (0.5, 1 and 2 μm/pixel). Printed with permission from [60]. Copyright 2022, ACS.
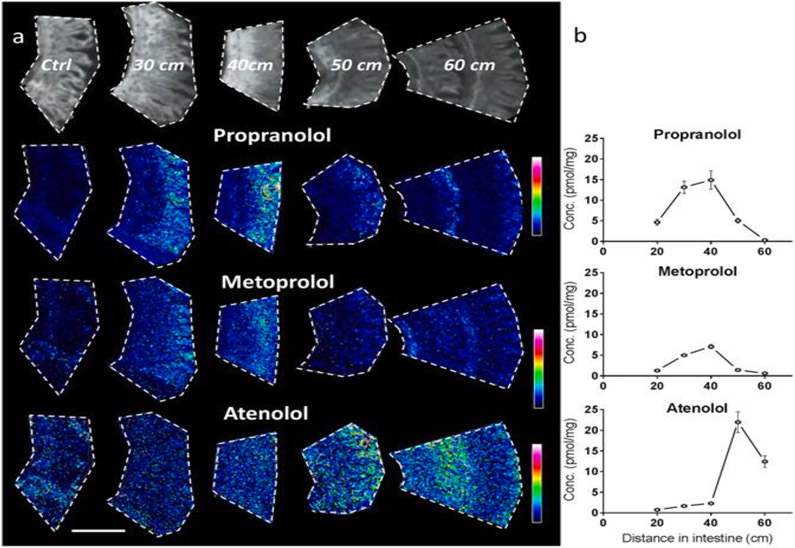
Nilsson et al. used MALDI-MSI to study the oral absorption of three FDA-approved, well-characterized permeability and absorption marker drugs (two high permeable drugs, propranolol and metoprolol, and a low-moderate permeable drug, atenolol) in rat GI tissues at 15 μm spatial resolution [89]. The propranolol and metoprolol MALDI-MSI images showed the highest relative ion abundance 30–40 cm downstream from the stomach into the small intestine, which gradually decreased by 50–60 cm, while atenolol demonstrated the highest abundance at a 50–60 cm distance from the stomach (Fig. 4a). The MSI ion images also showed that propranolol and metoprolol were mostly located around the upper part of the villi; in contrast, atenolol was more evenly distributed along the whole crypt-villus axis. The MSI findings were confirmed by the LC-MS/MS measurement of intestinal homogenates derived from matched tissue regions (Fig. 4b). These tissue absorption profiles are in agreement with published oral PK for all three compounds, supporting that high permeability drugs are associated with rapid oral absorption [89].
Fig. 4.
MALDI-MSI study of longitudinal (proximal to distal) and crypt-villus distribution of oral propranolol, metoprolol, and atenolol in rat small intestine. Images and concentration determinations from tissues collected 20 min post oral administration (1 μmol/kg) at different distances distal from the pyloro-duodenal transition. (a) MALDI-MSI ion distribution images of three drugs propranolol, metoprolol, and atenolol in intestine, scale bar is 1 mm. All tissue sections were analyzed at the same occasion and each compound is displayed with its own relative intensity scale (rainbow) with blue representing lower intensity and white representing highest intensity. (b) Homogenate concentrations (pmol/mg) measured by LC-MS/MS for the three drugs. X-error bars indicate position of the sampling ± 1 cm distal from the pyloro-duodenal transition. Printed with permission from [89]. Copyright 2022.
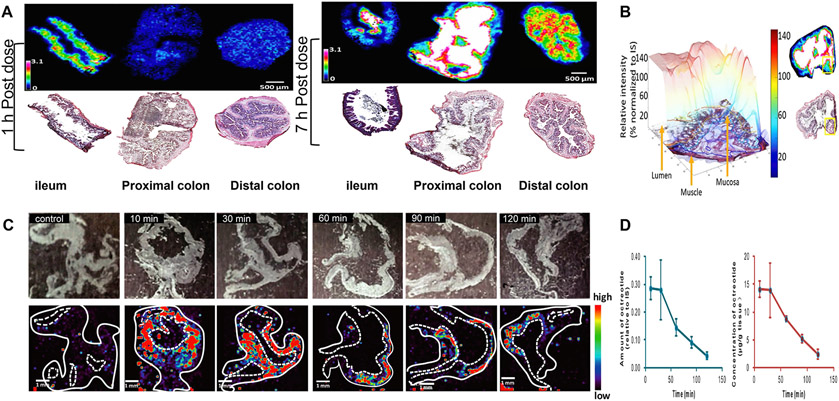
In 2021, Huizing et al. studied the distribution of tofacitinib, a FDA approved oral drug to treat rheumatoid arthritis and ulcerative colitis, in the GI tract (ileum, proximal colon, and distal colon) at 1 h and 7 h post single oral dose in rats using a quantitative MALDI-FTICR-MSI method [90]. Quantification was achieved using the 13C and 15N stable isotope-labeled tofacitinib as the internal standard (IS). A fixed concentration of the 13C,15N-tofacitinib was sprayed over the entire tissue section prior to coating it with 2,5-dihydroxybenzoic acid (DHB) MALDI matrix. The ratio of the drug/IS signal intensity was measured for three regions (the lumen, mucosa, and muscle layer) per intestinal section and in three different areas similar in size and pathological homogeneity. At 1 h post oral dosing (systemic Tmax), tofacitinib distributed highest in the ileum, whereas tofacitinib mainly distributed in the colon at 7 h post dosing (Fig. 5A). A 3D intensity plot coupled with hematoxylin & eosin (H&E) stains showed that MSI can differentiate at 7 h between the lumen and intestinal wall layers with a high exposure of drug in the muscular layer of the proximal colon (Fig. 5B). The study therefore demonstrates that MSI, coupled with classic H&E staining and stable isotope IS, provides a quantitative description of drug absorption and the passage of drugs across different regions of the intestinal wall [90].
Fig. 5.
MALDI-MSI analysis of distribution of oral drugs in the intestine of rats following oral administration. (A) Distribution of oral tofacitinib in the ileum, proximal colon, and distal colon with corresponding Hematoxylin & Eosin (H&E) stains beneath MSI ion distribution images at 1 h and 7 h post-dose. The ion intensity signal was normalized to the signal of the isotope-labeled standard. (B) 3D intensity plot of the tofacitinib of the selected region in the proximal colon 7 h post-dose. Right-side images show the intensity distribution of tofacitinib and the H&E image of the entire section with the yellow box corresponding to the region used for 3D visualization. Arrows point to the muscle layer, mucosa, and lumen areas. Adapted with permission from [90]. Copyright 2022, ACS. (C) Representative optical and MALDI-MSI images of octreotide in mouse stomach section collected at various time points ranging from 10 min to 120 min after oral administration of octreotide. (D) Tissue concentration-time curve of octreotide in mouse stomach determined by MALDI-MSI (left panel, green) and by LC-MS/MS (right panel, red) to show similar pharmacokinetic curves using the different technologies (tissue imaging via MSI and tissue homogenate using LC-MS/MS). Adapted with permission from [91]. Copyright 2022.
Rao et al. performed a MALDI-TOF-MSI-based PK imaging of octreotide, an octapeptide drug used to treat neuroendocrine tumors, in mouse stomach, intestine, and liver sections after oral administration [91]. In this assay, a structurally similar IS was spotted onto tissue sections together with DHB matrix solution. The distribution of octreotide in the stomach section was clearly visualized at different time points from 10 to 120 min (Fig. 5C). The drug concentration was much higher in the corpora ventriculi as compared to cardia, fundus, and pylorus regions, reflecting the heterogeneity of drug distribution in tissue. The most exciting observation was that the tissue concentration-time curve of octreotide in the stomach obtained via MSI was consistent with findings using the well-established LC-MS/MS method (Tmax at 30 min and a stomach elimination half-life t½ of 34.1 min) (Fig. 5D). The quantitative performance of the MALDI-TOF-MSI octreotide assay was validated for specificity, selectivity, linearity, sensitivity, accuracy, and precision [91]. Together, these studies suggest that MSI techniques can not only visualize the spatial distribution of a drug in tissues, but also generate meaningful data for construction of the tissue PK profiles and calculation of key PK parameters.
3.2. MSI analysis of drug distribution in central nervous system tissues
3.2.1. Distribution of antipsychotics and Alzheimer’s disease drugs in brain tissue
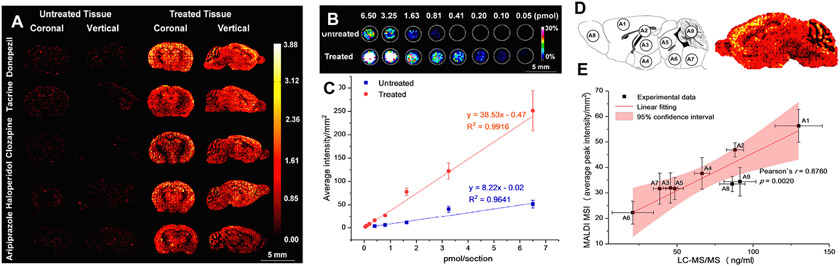
The blood-brain barrier (BBB) and blood-cerebrospinal fluid barrier can significantly limit a drug’s permeability to the central nervous system (CNS). MSI technologies have been successfully used to detect and quantify spatial distribution of CNS-targeted drugs in preclinical brain tissue sections [92]. Using a MALDI TOF-TOF MSI method, Chen at al. simultaneously detected and quantified five different intraperitoneally injected CNS drugs (three antipsychotics: clozapine, haloperidol, and aripiprazole; and two Alzheimer’s disease drugs: donepezil and tacrine) in mouse brain tissue sections at a 150 μm resolution (Fig. 6A) [92]. The brain tissues were sectioned, vacuum dried, washed with ammonium acetate solution, incubated with trifluoroacetic acid vaper, then washed with n-hexane before MALDI-MSI analysis [92]. Relatively high abundance of the five CNS drugs was observed in the cerebral cortex, followed by the hippocampus, thalamus, and stratum regions. This type of tissue preparation enhanced drug detection and decreased ion suppression across all five CNS drugs and allowed for improved quantification (regression values R2 > 0.9) (Fig. 6B, C) [92]. This MALDI-MSI quantification method was validated by conventional LC-MS/MS analysis of tissue punches from the nine brain regions of the same mouse brain prepared for MALDI MSI, with the LC-MS/MS data finding similar region average peak intensities (Fig. 6D, E) [92]. These results demonstrate that the integrated tissue pretreatment protocol coupled with MALDI-MSI technology allow reliable quantitative analysis of the spatial distribution of drugs in the brain.
Fig. 6.
MALDI-MSI analysis of coronal and sagittal brain sections from mice dosed intraperitoneally with CNS drugs donepezil, tacrine, clozapine, haloperidol, and aripiprazole. (A) MALDI-MSI images of mouse brain sections treated with five drugs at 150 μm lateral resolution in positive ion mode, comparing the different intensities in untreated tissue versus pretreated tissues. (B–C) The quantitative calibration curves of donepezil obtained from MALDI-MSI for untreated (R2 = 0.9641) and pretreated (R2 = 0.9916) mouse tissue sections. (D) LC – MS/MS validation of the tissue-specific distribution of donepezil in brain tissues, tissue punches collected from different brain regions used for the LC – MS/MS analyses and MALDI imaging of donepezil. A1 (cortex), A2 (hippocampus), A3 (thalamus), A4 (hypothalamus), A5 (midbrain), A6 (pons), A7 (medulla), A8 (cerebellum), and A9 (olfactory bulb). (E) Correlation of the amount of donepezil from different cerebrum regions determined by MALDI-MSI and LC – MS/MS. Adapted with permission from [92]. Copyright 2022 ACS.
3.2.2. Anticancer drug distribution in CNS tissue
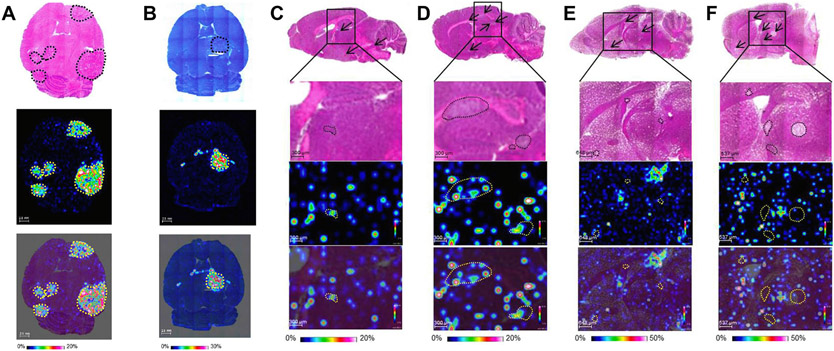
When developing drugs to treat brain tumor and brain metastases, it is always challenging to ensure a drug’s ability to cross the BBB and blood-tumor barrier (BTB) to reach the intended site of action. Tanaka et al. used MALDI-MSI to quantitatively evaluate the spatial distribution of two oral tyrosine kinase inhibitors, epertinib and lapatinib, in mouse models of HER2-positive breast cancer brain metastases and T790M-EGFR-expressing lung cancer brain metastases [93]. Fig. 7A, B show the histological H&E and Giemsa staining of tumor-bearing brain tissue sections and the MALDI-MSI ion distribution of epertinib and lapatinib in the lung cancer brain metastases mouse model [94]. Quantitative MSI confirmed that brain tumor concentrations of both epertinib and lapatinib were equivalent, while plasma concentrations were significantly different at 4 h after oral administration [94]. In a separate BR3 breast cancer brain metastases mouse brain sections, epertinib and lapatinib distribution in tumors was visualized by MALDI-MSI at 4 and 8 h after oral drug administration (Fig. 7C-F). Quantitative MALDI-MSI analysis using epertinib-d9 and lapatinib-d7 as the internal standard showed that epertinib concentration was 10 times higher than that of lapatinib in tumors [94]. These findings indicate that drug distribution to the CNS site does not necessarily correspond to the physiochemical properties of the drug or to systemic exposure, making the addition of spatial drug distribution information especially valuable in CNS drug discovery research.
Fig. 7.
Quantitative analysis and distribution of anticancer drug epertinib and lapatinib in brain metastases using MALDI-MSI. Representative images of horizontal brain sections of mice with lung cancer brain metastases developed by intraventricular injection of NCI-H1975-luc cancer cells and stained with H&E (A top row) and giemesa staining (B top row) with the black dashed lines indicating tumor position. The ion images of epertinib (A, middle row) and lapatinib (B, middle row) in brain section at 4 h after oral administration of epertinib hydrochloride or lapatinib ditosylate monohydrate at a dose of 100 mg/kg. The overlay images of MSI of epertinib (A, lower row) and lapatinib (B, lower row) on the stained images. Distribution of epertinib and lapatinib in BR3 breast cancer brain metastases shown at 4 h (C) and 8 h (D) post dose of epterinib hydrochloride (50mg/kg) and 4 h (E) and 8 h (F) post dose of lapatinib ditosylate monohydrate (50mg/kg). (C-F) top row showing H&E staining, middle row is the ion images of epertinib and lapatinib and the bottom row is the overlay of the ion images on the H&E stain. Arrows indicate tumor position of the choroid plexus, black and yellow dashed lined indicate tumor region. Adapted with permission from [94]. Copyright 2022.
Another study combined MALDI-MSI with LC-MS/MS methodologies to compare the effect of the multidrug resistance protein 1 (MDR1) on the intra-brain accumulation of an oral kinase inhibitor, alectinib, in Mdr1a/b knockout (KO) mice [95]. MSI images showed a significant increase in alectinib exposure and distribution in the brain sections of Mdr1a/b KO mice as compared to that in brain section of FVB wild-type mice at 1 and 4 h post oral drug administration [95]. These data indicate that brain distribution of alectinib was affected by efflux transporter MDR1 and was not associated with blood concentration [95,94]. These studies thus reiterate the value of being able to visualize and quantitate spatial distribution of drugs in drug discovery and development and highlights the importance of understanding the role of transporters in drug distribution.
3.2.3. Antiretroviral drug distribution in CNS tissue
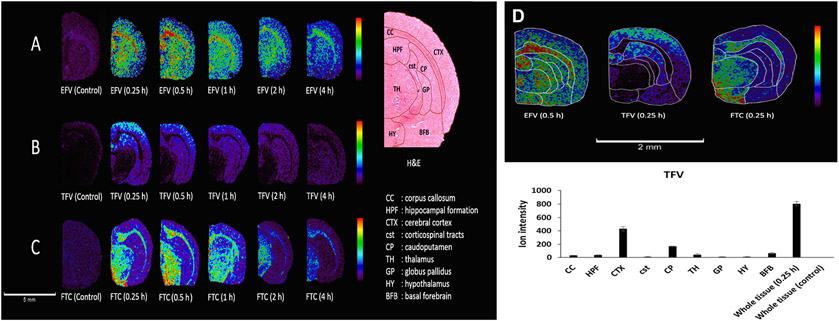
CNS distribution of antiretroviral drugs has been evaluated using MALDI-MSI to determine the regional drug localization patterns and PK in rat brains [96]. In this study, the antiretroviral drugs efavirenz, tenofovir, and emtricitabine were dosed in healthy rats at 50 mg/kg via intraperitoneal injection. A LC-MS/MS method was developed to quantify the antiretroviral drugs in both plasma and brain homogenate samples in tandem with a MALDI-MSI method used to map the drugs’ time dependent CNS exposure. MALDI-MSI of coronal rat brain sections showed a time-dependent spatial distribution of all three antiretroviral drugs at a lateral resolution of 100 μm. Efavirenz displayed a high degree localization across the entire brain, tenofovir localized mainly in the cortex, and emtricitabine distributed heterogeneously in the thalamus, corpus callosum, and hypothalamus (Fig. 8A-C). The relative ion abundance of the three antivirals in different brain regions can be quantified (Fig. 8D). It was evident that tenofovir had the highest intensity in the cerebral cortex (CTX), followed by the caudoputamen (CP) at 0.25 h post-dose. Tenofovir had the lowest brain concentration compared to the other two drugs using the LC-MS/MS analysis of whole brain homogenates [96]. These findings make a clear case for the advantages of MSI in monitoring drug delivery. While the homogenized tissue analysis did not provide enough information about drug penetration across the BBB, the MALDI-MSI images show drug localization at different brain regions at different levels.
Fig. 8.
MALDI-MSI analysis of regional brain distribution of three antiretroviral drugs efavirenz (EFV), tenofovir (TFV), and emtricitabine (FTC) in the rat brain after intraperitoneal administration. MSI images of time dependent spatial distribution of EFV (A), IFV (B), and FTC (C) in coronal rat brain sections with a spatial resolution of 100 μm. H&E staining with regional labels to define regions of interest. (D) Relative ion abundance of the three drugs in different brain regions (top panel) and a representative TFV ion intensity plot at 0.5 h post-dose. Scale bar: 2 mm. CC: corpus callosum; HPF: hippocampal formation; CTX: cerebral cortex; cst: corticospinal tracts; CP: caudoputamen; TH: thalamus; GP: globus pallidus; HY: hypothalamus; BFB: basal forebrain. Adapted with permission from [96]. Copyright 2022 ACS.
3.2.4. MSI imaging of L-DOPA-treated Parkinson’s disease brain sections
Shariatgorji et al. recently developed a reactive matrix designed to selectively target primary amine groups on neurotransmitters. Using a fluoromethylpyridinium-based reactive matrix and MALDI-MSI, they visualized distinct alterations of neurotransmitters and their metabolites in brain tissue sections from rat models of Parkinsonism [97]. Specifically, the authors showed the MALDI-MSI detection of endogenous dopamine and various neurotransmitters and metabolites acquired from the brain tissue section of a unilateral 6-hydroxydopamine lesioned rat model of Parkinson’s disease treated with L-3-4-dihydroxyphenylalanine (L-DOPA) for three weeks, followed by a final dose of deuterated L-DOPA-d3. Derivatized dopamine was predominantly detected in the striatal region of rat brain sections. Tracing the metabolism of the isotopically labelled L-DOPA-d3, this group was able to identify brain areas that uptake the dopamine precursor and simultaneously obtain distinct localization patterns for dopamine metabolites [97]. This study demonstrates that MALDI-MSI methodology can enable the simultaneous detection and mapping of comprehensive pathways of biochemical and pharmacological pathways of drug-induced neurotransmitters from a brain tissue section.
3.2.5. Mapping drug-induced change in metabolome in human brain organoids
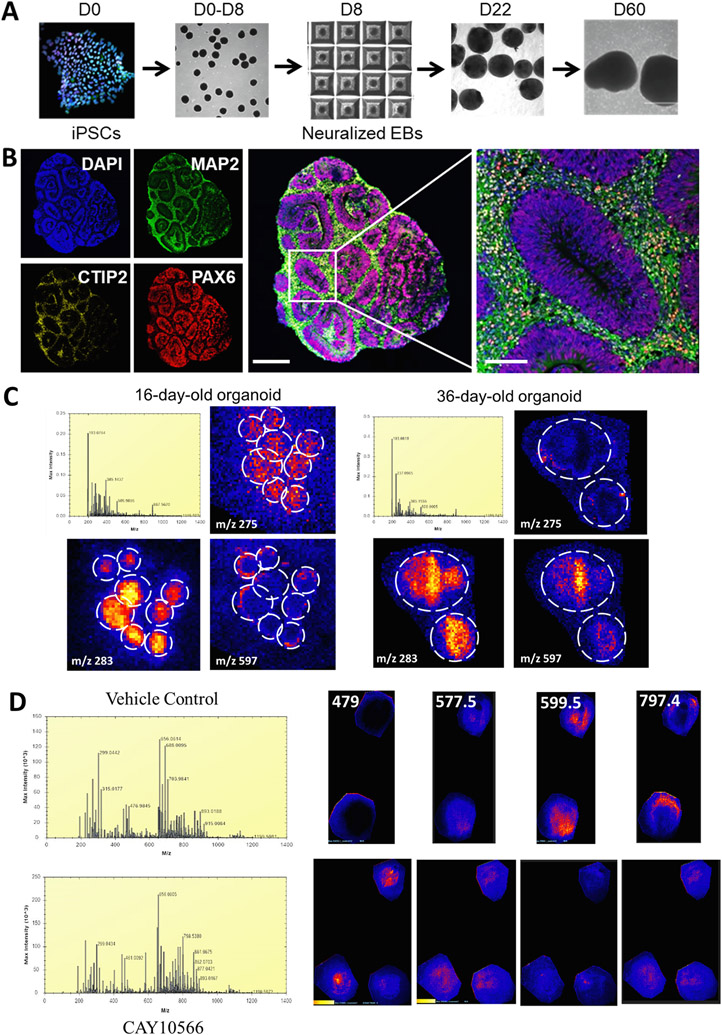
Human brain organoids are three-dimensional self-organizing, stem-cell-derived structures that resemble in vivo tissue counterparts in both cell composition and organ architecture [98]. They have quickly become an important tool for basic and translational research with wide applications for disease modeling using human cells as primary source, drug development, screening, optimization, and personalized medicine [99,100,101]. Because organoids are produced from patient samples, the patient-specific genetic background is preserved, making them an ideal model system to study genetic disorders and develop gene or other therapies to counteract them. However, before these models are fully utilized in the medical, biology research, and pharmaceutical industries, more characterization needs to be undertaken. In our study, we aimed to examine whether MSI could be an informative tool to study changes in the organoid metabolome and its response to treatments. We used forebrain organoids generated from induced pluripotent stem cells (iPSCs) by modified Pasca protocol [102]. These organoids have well characterized neuroprogenitor zones (rosettes), neurons and astroglia of different maturation stages (Fig. 9). To map selected metabolites, we used MALDI-TOF (Synapt 2G-Si HDMS, Waters) calibrated for the mass range from m/z 50–2000 [103]. We first examined whether MSI can detect any metabolome differences in organoids of different age. Indeed, at 16 days, when organoids are populated solely with neuroprogenitors (organoid size < 0.5 mm), we could detect an abundance of small molecules (<400 Da) concentrated either in the center of an organoid (Fig. 9C, left panel m/z 275 and 283) or outlining the periphery of the organoid (Fig. 9C, left panel, m/z 597). In contrast, at 36 days, when organoids have immature neurons that migrate toward the periphery (organoid size is about 0.5 mm), we saw differences in the same small molecules – some maintained their localization in the center of the organoid (Fig. 9C, right panel, m/z 283), some were subsequently not detected (m/z 275), and some moved from the periphery to the center of the organoid (m/z 597). We then examined whether we could detect any differences if the organoids were treated with the fatty acid desaturase inhibitor, CAY10566, as neuroprogenitors are highly enriched with fatty acids and depend on monounsaturated fatty acids for their survival [104]. After 3 days of treatment, we could detect the presence and distribution of some metabolites in the organoid (Fig. 9D). These data demonstrate that MSI could be used to study human brain organoids, although more optimization needed to ensure proper tissue fixation and sectioning to achieve the best results.
Fig. 9.
MSI can be used to characterize small molecule/metabolome changes in human brain organoids. A. iPSCs grown on feeders are transferred into low-attachment AggreWell 800 to develop about 300 embryoid bodies (EBs) per batch. To achieve rapid and efficient neural induction, both the bone morphogenetic protein (BMP) and transforming growth factor beta (TGF-β) signaling pathways are inhibited with small molecules, dorsomorphin and SB-431542, respectively. On day 8, the floating spheroids were moved to suspension dishes and serum-free media containing fibroblast growth factor-2 (FGF2) and ascorbic acid. To promote differentiation of neuro-progenitors into neurons, FGF2 was replaced with brain-derived neurotrophic factor (BDNF) and neurotrophic factor 3 (NT3) starting on day 22 to 60, after which we used only neural medium without growth factors. B. These organoids contain ventricular zone-like regions consisting of PAX6 + cells surrounded by MAP2 + and CTIP2 + neurons. Scale bars: low mag 500 μm, high mag 100 μm. C. MSI of 16- and 36-day old organoids detects differences in the abundance and distribution of metabolites. We used MALDI-TOF (Synapt 2G-Si HDMS, Waters) calibrated for the mass range from m/z 50–2000 Da. Laser power was set to 250 (arb units) with 300 laser shots per pixel. The laser raster step was set to 60 μm to match the laser spot size of 30 μm. Tissue was processed as published [103] and sections (14 μm) were coated with the MALDI matrix 9-aminoacridine. The coated slides were rehydrated in a heated humidifying chamber for 2 min. Before loading the slide into the MS, the slide was scanned with an EPSON scanner to map the areas of interest into the High Definition Imaging (HDI) 1.4 software and the data were acquired in negative ionization mode. 16d and 36d organoids have different metabolome profiles, as seen by the mass spectrometry data (upper panels). Individual metabolites (red-yellow signal) were visualized in the organoids (m/z listed in the corner; yellow indicates higher concentration than red). Blue signal originates from the matrix. In the 16 day sample, 8 organoids are seen (they were small and more can fit on the slide), while in the 36 day sample, 2 organoids occupied the same area. D. 60-day-old organoids were treated with CAY10566, an inhibitor of the stearoyl-CoA desaturases that catalyze the conversion of saturated fatty acids into monounsaturated fatty acids, for 3 days. Tissue was processed and MSI performed as in (C). We could observe differences in the content, abundance, and distribution of metabolites (m/z listed in the corner).
3.3. MSI imaging of drug localization in lung tissues
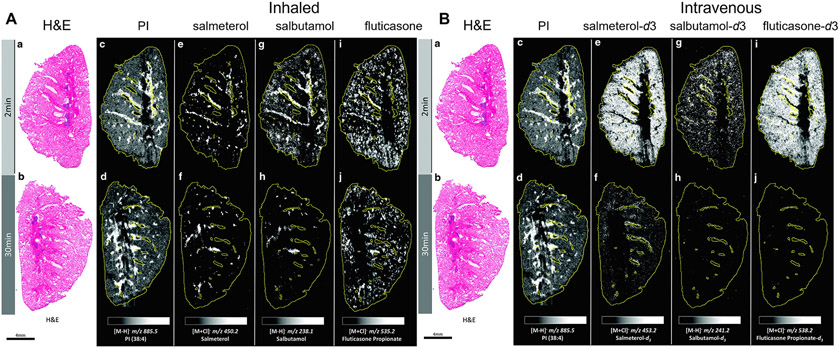
The lung is a highly heterogenous organ, and there is only limited information on the details of lung tissue drug distribution after inhalation therapy. Due to high pulmonary pressures and gravity, the blood flow in the lungs is unevenly distributed [105], which may impact drug transport in the lung. In 2020, Hamm and colleagues applied a DESI-MSI method to determine the lung distribution and retention of slowly dissolving neutral fluticasone propionate (a glucocorticoid receptor agonist) and the soluble bases salmeterol (a long-acting β-adrenoreceptor-agonist) and salbutamol (a short-acting β-adrenoreceptor-agonist), which were simultaneously delivered to rats by inhaled nebulization [106]. The three deuterated drugs were also delivered to rats by intravenous injection which resulted in homogenous lung distribution for all drugs. In contrast, inhaled salmeterol and salbutamol were preferentially retained in bronchiolar tissue, while inhalation delivered fluticasone propionate was retained in all regions of the lungs (Fig. 10). These MSI results demonstrated that inhaled small molecule chemotypes are differentially distributed in the lung tissue after inhalation; the regional localization and retention of drugs are highly dependent on the drugs’ structure and physicochemical properties.
Fig. 10.
Whole-lung imaging by DESI-MSI in negative detection mode at 70 μm spatial resolution. (A) H&E-stained images from 2 (A-a) and 30 (A-b) minutes after administration through inhaled nebulization. Molecular images of endogenous (A-c, A-d) phosphatidylinositol (PI), inhaled (A-e, A-f) salmeterol, (A-g, A-h) salbutamol, and (A-i, A-j) fluticasone propionate. (B) H&E-stained images from 2 (B-a) and 30 (B-b) minutes after intravenous administration. Molecular images of endogenous (B-c, B-d) PI, intravenous-dosed (B-e, B-f) salmeterol-d3, (B-g, B-h) salbutamol-d3, and (B-i, B-j) fluticasone- d3 propionate. Intensity scale is fixed for each molecular species independently; monochromatic lighter colors correspond to higher relative abundance and darker to lower abundance. Tissue area is outlined in yellow line. Adapted with permission from [106]. Copyright 2020, Mary Ann Liebert, Inc.
Similarly, Yamamoto et al. used a DESI-TOF-MS method to visualize the spatial localization of inhaled ciclesonide, a corticosteroid, and its two metabolites in rat lung sections after administration of a single dose of 1-μm ciclesonide aerosol particles [107]. The MSI data revealed that ciclesonide was localized in the airway epithelium, while the two metabolites were much more homogenously distributed throughout the lung tissue, including the alveoli and bronchi. These data demonstrated the efficient delivery of 1-μm aerosol particles to the deep portions of the rat lungs, although a portion of the particles was deposited at the airway epithelium [107].
Nilsson et al. used MALDI-MSI to track and quantify the distribution of inhaled tiotropium, a bronchodilator, within lungs of dosed rats, in both MS and MS/MS mode at 200 μm spatial resolution [108]. Tiotropium was administered as an aerosol of a nebulized solution to rats (1.1 mg/kg) by exposing the rats in an inhalation chamber for 15 min. The quantification of tiotropium levels in lung tissue was achieved by comparison to drug standard samples spotted on a control tissue using MALDI-MSI. Tiotropium precursor MS ions and fragmented product MS/MS ions were dispersed in a concentration gradient (80 fmol – 5 pmol) away from the central airways into the lung parenchyma and pleura based on a MSI ion map of lung tissue sections; the finding was consistent with a rapid absorption of tiotropium [108].
In addition to inhaled drugs, Treu et al. developed a MALDI-MSI workflow to image orally administered anti-tuberculosis (TB) antibiotics in the mouse lung with high mass resolution and accuracy (<1.5 ppm) and high spatial resolution (10 μm pixel size) [109]. The detection of antibiotics (pyrazinamide, rifampicin, ethambutol, isoniazid, moxifloxacin, and clofazimme) was accomplished in mouse lung sections from the same treated animals using alternating scan mode and the molecular identity of the antibiotics was confirmed by on-tissue MS/MS analysis. The selected ion monitoring (SIM) acquisition increased the sensitivity of drug detection in MALDI-MSI. Clofazimine was imaged with 10 μm pixel size revealing for the first time the clofazimine accumulation in lipid deposits around airways and the non-homogenous distribution of clofazimine across all lung tissues [109]. Together, MSI techniques enable the tracking of drug transport within the lung tissue compartment and provide evidence for the efficient design and delivery of inhaled and ingested drugs to target lung tissues.
3.4. MSI imaging of drug distribution and elimination in kidney tissues
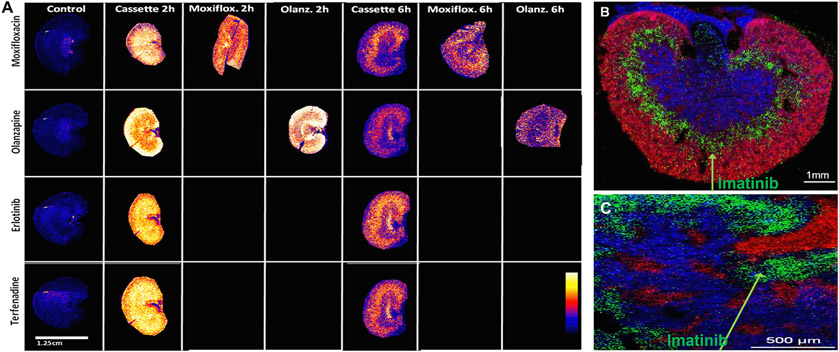
Kidney drug clearance plays an important role in the drug elimination process. Several recent reports highlight the use of MSI for spatial distribution of drugs in the kidney. Swales et al. used MALDI-MSI to examine the spatial distribution of eight drugs (haloperidol, bufuralol, midazolam, clozapine, terfenadine, erlotinib, olanzapine, and moxfloxacin) simultaneously in multiple tissues after oral or intravenous cassette dosing in rats (four compounds per dose route), thus increasing throughput while decreasing the number of animals used [110]. Fig. 11A displays the MSI ion abundance distributions of cassette- and discrete-dosed compounds (moxifloxacin, olanzapine, erlotinib, and terfenadine) in rat kidney sections at 2- and 6-h post dose. The drug distribution was homogenous throughout the kidney tissues at 2 h post dose, and reductions of the drugs in the kidneys were observed at 6 h post dosing. Erlotinib and terfenadine were distributed in both the cortex and medulla of the kidney, with greater intensity in the medullary region. In contrast, moxifloxacin and olanzapine were more localized in the medulla of the kidney [110]. These results suggest that a combination of MALDI-MSI and cassette dosing approach can provide label-free PK and drug distribution data at high throughput in organs at the early stage of the drug discovery and development process.
Fig. 11.
(A) MALDI-MSI analysis of distributions of cassette- and discrete-dosed drugs (moxifloxacin, olanzapine, erlotinib, and terfenadine) in rat kidney sections (14 μm) at 2 and 6 h post dose. Moxifloxacin and olanzapine distributions are comparable at the two time points validating the cassette dosing approach. Printed with permission from [110]. Copyright 2022, ACS. (B) MALDI-MSI ion map of selected ions including imatinib (green), endogenous phospholipid PC (34:1) (blue), and PC (32:0) (red) in kidney section at 35 μm resolution. Adapted with permission from [111]. Copyright 2022, Springer. (C) Overlay MSI image of selected ions including imatinib (green), endogenous phospholipid PC (38:5) (blue), and heme (red) in kidney section at 10 μm resolution. Adapted with permission from [111]. Copyright 2022, Springer.
Rompp et al. described a high mass resolution and high spatial resolution MALDI-Orbitrap MSI method to analyze the spatial distribution of the anticancer drugs imatinib in mouse kidneys [111]. Imatinib showed a well-defined distribution in the stripe of the outer medulla region of the kidney (green color, Fig. 11B). The identity of imatinib was verified via on-tissue MS/MS measurements. Selected ion images of endogenous phospholipid PC (32:0) (red color, Fig. 11B) provided additional information on the structure of the kidney sections in the overlay of selected ion images at 35 μm spatial resolution. The ion image at 10 μm lateral resolution confirmed the specific location of imatinib in the outer medulla (green color, Fig. 11C), and the selected ion images of phospholipid PC (38:5) (blue color) and heme (red color) gave a good indication of the histological features of the kidney section (Fig. 11C) [111].
Nilsson and colleagues used MALDI-MSI to measure the distribution of nephrotoxic antibiotics polymyxin B1 and colistin in rat kidney tissue sections [112]. Polymyxins are cationic polypeptide drugs that are highly potent against Gram-negative resistant bacteria but are used as a last-line therapy due to their dose-limiting toxicity. MALDI-MSI was performed at 100 μm in rat kidney tissues obtained after 1, 2, and 7 day subcutaneous injection of the drugs at their maximum tolerated doses. Polymyxin B1 displayed greatest abundance in the kidney cortex, and the cortical accumulation of the drug increased with increased number of days dosed. A high resolution MALDI-MSI at 20 μm spatial resolution showed the distribution of colistin ([M + H]+ at m/z 1170) in relation to the heme (m/z 616) in the cortex region on day 7 post dosing [112]. The observed high accumulation of polymyxin B1 and colistin in the rat renal cortex provides valuable evidence in characterizing the nephrotoxicity of these drugs.
3.5. MSI analysis of the drug distribution in whole-body animal tissue sections
In addition to individual organs, MSI can be applied to whole-body sections of an intact animal carcass. MSI is capable of differentiating molecular species of drugs and metabolites at a spatial resolution similar to the quantitative whole-body autoradiography (QWBA) technique (typically 50–100 μm resolution). Caprioli and colleagues were the first to report direct molecular analysis of whole-body rat tissue sections using MALDI-MSI [113]. After a single oral dose of the antipsychotic drug olanzapine (8 mg/kg), 20 μm-thick whole-body sagittal rat tissue sections were sprayed with DHB matrix. The drug, N-desmethyl metabolite, and 2-hydroxymethyl metabolite were detected by MALDI-MSI in measurable amounts in almost all tissues at 2 h post-dose (Fig. 12A). The highest olanzapine ion signal was detected in the lung, followed by the spleen, bladder, kidney, liver, thymus, brain and spinal cord (the target organ), and testis. In contrast, the metabolites were mainly detected in the liver, kidney, and bladder. These results were in full agreement with previously published quantification data [113].
Fig. 12.
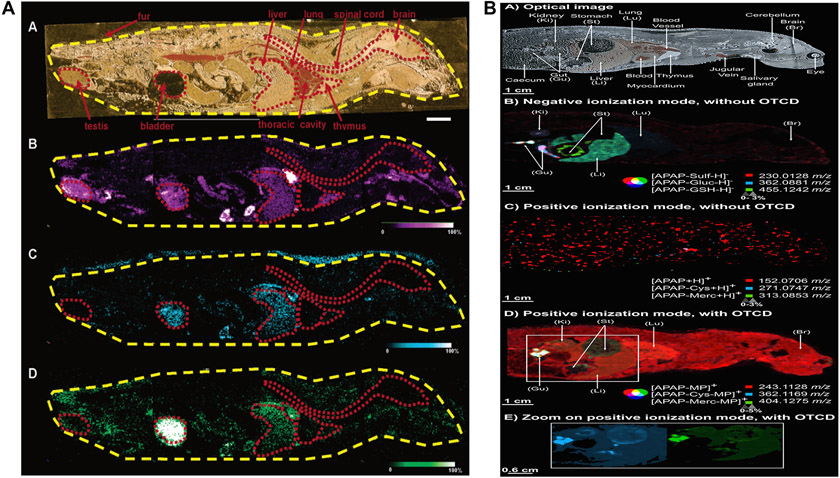
Whole-body spatially resolved MSI analysis of drug and metabolites. (A). MALDI-MSI determining olanzapine and metabolite distribution at 2 h post oral administration of 8 mg/kg olanzapine in a whole rat sagittal tissue section. Optical image of a dosed rat tissue section, organs are outlined in red (A-A). MS ion images of olanzapine (m/z 256) (A-B), N-desmethyl metabolite (m/z 256) (A-C), and 2-hydroxymethyl metabolite (m/z 272) (A-D) are displayed. Bar, 1 cm. Ion intensity: blue being low intensity and white being high intensity. Reprinted with permission from [113]. Copyright 2022 ACS. (B). Detection of acetaminophen and metabolite distribution under different conditions by MALDI-MSI on whole-body sections from rats after intravenous administration of 300 mg/kg acetaminophen. Spatial resolution: 400 μm. (B-A) Optical image of the section. (B-B) Negative ionization mode without on-tissue chemical derivatization (OTCD). (B-C) Positive ionization mode without OTCD. (B-D) Positive ionization mode with OTCD. (E) Zoom on positive ionization mode with OTCD. Reprinted with permission from [114]. Copyright 2022 ACS.
Merdas and colleagues used a MALDI-MSI and on-tissue chemical derivatization (OTCD) protocol to detect acetaminophen (APAP) and metabolites in whole-body rat tissue sections treated with a single intravenous dose of 300 mg/kg of APAP [114]. As shown in Fig. 12B, without derivatization, MALDI-MSI did not detect the drug in the whole-body tissue section. When 2-fluoro-1-methylpyridinium p-toluene sulfonate was used as a derivatization reagent, the OTCD procedure enhanced the ionization of APAP and metabolites and allowed direct visualization and quantification of APAP by MALDI-MSI in the whole-body tissues. The quantification of APAP in tissue was achieved by direct comparison to calibration curves constructed based on manually spotted standards on tissues, and the accuracy of the MALDI-MSI quantification was confirmed by conventional LC-MS method [114]. The successful application of MSI to the detection of drugs and metabolites in whole-body tissue sections will advance our understanding of drug PK/PD and efficacy relationship in drug discovery and development.
4. MSI applications in drug metabolism and toxicity
Accurately understanding the biodistribution, accumulation, and metabolism of administered drugs is required in drug development [115,116]. For therapeutic agents to produce the desired pharmacological effect, they must be well distributed to the intended target site [117,118]. The accumulation of a parent drug or metabolites in untargeted tissues may cause unexpected secondary pharmacological activity or toxicity [119,120]. Spatial drug concentrations in tissues based only on information from tissue homogenates cannot capture the heterogeneity of compound distributions that are influenced by the tissue microenvironment. Detailed visualization of drug metabolism and distribution in tissues, and within discrete tissue regions, could provide valuable information when interpreting pharmacological effects and investigating the mechanisms of drug toxicity. Both MALDI and DESI have been frequently used to image drugs and their metabolites in sectioned tissues to study drug metabolism and toxicity [121,15]. Although the use of MSI in biomedical and basic molecular biology research has steadily increased in recent years, only a limited number of MSI studies have focused on drug metabolism, drug safety, and toxicological studies because of challenges such as matrix interference, isobaric interferences, poor sensitivity, and limited sample throughput. In this section, we review MSI applications in studies of drug distribution in the liver and drug-induced liver injury (DILI).
4.1. Hepatic metabolism
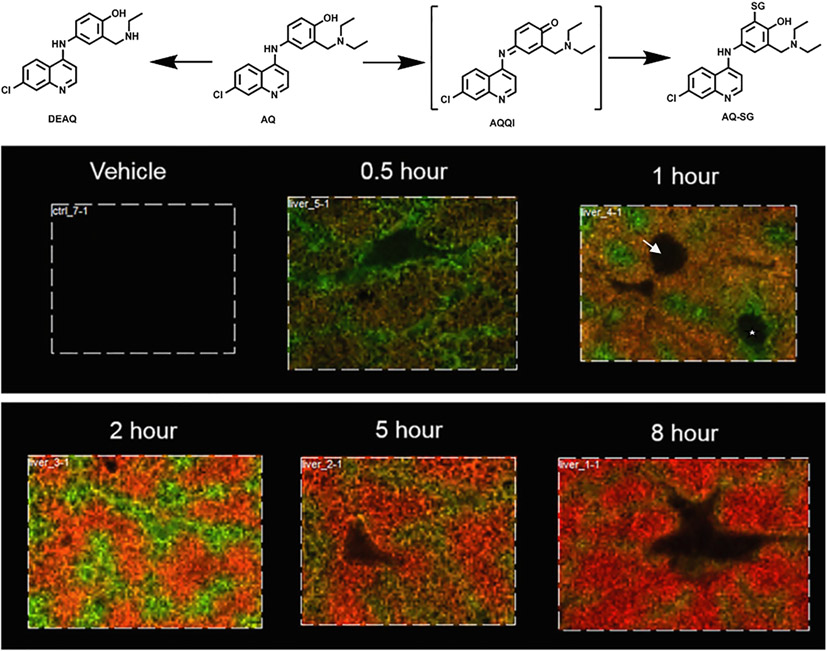
It is well appreciated that drug metabolism plays important roles in drug safety and efficacy [122]. As the primary site for drug metabolism [123], the liver is crucial for converting prodrugs to active metabolites or active drugs to inactive or toxic forms. Metabolic activation of drugs can result in reactive metabolites, which may lead to drug toxicity by covalently binding to biomacromolecules [124,125]. The role of liver as the major site in drug metabolism makes it a target of drug toxicity. Combined with histology studies, understanding the temporal and spatial distribution of drugs and metabolites in the liver may help us to understand the relationship between reactive metabolites and DILI. Serious idiosyncratic DILI and agranulocytosis have been associated with amodiaquine (AQ), a medication used to treat malaria [126,127]. The exact mechanism by which AQ caused hepatotoxicity remains unclear, but it is generally thought that the metabolic activation of AQ to a reactive quinoneimine (AQQI; Fig. 13), which binds irreversibly to proteins, contributes to its toxicity. Grove et al. employed MALDI-MSI with a high resolution and mass accuracy to study the spatial and temporal distribution of AQ stable metabolites such as de-ethyl-AQ (DEAQ) and the glutathione-adduct of AQ (AQ-SG) in rat livers (Fig. 13) [128].
Fig. 13.
Amodiaquine (AQ) was metabolized to de-ethyl-amodiaquine (DEAQ) or the glutathione adduct (AQ-SG) in rat liver. MALDI-MSI with a 25 μm spatial resolution reveals zonation patterns in the liver lobule at different time points after a single 100 mg/kg oral dose. The distribution of AQ (green) and DEAQ (red) at 0.5, 1, 2, 5, and 8 h time points varied. Arrow indicates the central veins; * indicates a portal triad region. Adapted with permission from Grove, et al. 2018. Copyright 2022.
In Fig. 13, an overlay of the molecular ion images of AQ (green) and DEAQ (red) at various time points shows that AQ was rapidly absorbed and reached maximum absorption at 2 h post-dose, followed by its clearance; whereas DEAQ abundance lagged behind AQ (from which it was produced via metabolism) but remained in the liver much longer. Additionally, the images show that AQ and DEAQ were not evenly distributed in each tissue section (Figs. 13, 2 h time point). In this study, drug and metabolite localization in the livers were also assessed by comparing images of H&E staining with MALDI ion images. Higher levels of AQ were detected near the portal triad and a lower intensity was seen near the central veins, while DEAQ had the reverse distribution (Figs. 13, 1 h time point). These findings correspond with the zonation of cytochrome P450 enzymes in liver lobules: P450 enzymes are in greater abundance near the central vein than near the portal triad area [129]. More extensive conversion of AQ to DEAQ by P450s may explain the observed distribution of DEAQ and AQ intensities across the tissue. MSI of glutathione (GSH) in liver sections revealed depletion of GSH in the centrilobular region and the formation of AQ-SG, suggesting that AQ is bioactivated to quinoneimine (AQQI) in the centrilobular region and that it depletes the GSH to form AQ-SG. AQ-SG formation is an alternative explanation for the lower observed abundance of AQ in this region. Histology studies indicated that AQ-induced liver injury also occurred mainly around the central vein. The MSI of liver sections, together with histology, demonstrated that the bioactivation of AQ is an initial step in AQ-induced liver injury by depleting GSH. Similar temporally resolved spatial imaging of the distribution of acetaminophen, its metabolites, and GSH adducts in mouse livers has been investigated using MALDI-MSI [130]. Recently, MSI was used to spatially profile diclofenac and its metabolites in various tissues [131]. Together, these studies demonstrate that integrating MSI and histology can provide vital information for understanding the role of drug metabolism in hepatoxicity.
4.2. Drug-induced toxicity
DILI is a life-threatening adverse effect that has been reported in rare instances for many drugs in a small fraction of patients, but few mechanisms of DILI have been elucidated [132]. Although substantial progress in understanding DILI has been made by employing genomics and metabolomics of humans and transgenic animal models, mechanistic studies of drug toxicity and the development of suitable animal models remain challenging [132]. In addition to its role in the metabolism of xenobiotics and thus a broad range of drugs, the liver plays a vital role in the catabolism and anabolism of lipids, carbohydrates, proteins [133], and sterols (e.g., primary bile acids) [134]. Using high mass accuracy and high spatial resolution MS, it is now possible to determine the distribution of endogenous metabolites and lipids and explore the interrelationships between drug distribution and metabolomic or lipidomic changes in tissues. With this capability, pharmaceutical scientists are starting to employ MALDI-MSI to investigate spatial biochemical changes in tissues as they relate to drug efficacy and toxicity. Kampa et al. studied the underlying mechanisms of amitriptyline-induced liver injury in rats by monitoring site-specific metabolic alterations, including phospholipidosis, using MSI and histology [135]. MSI detected lipids that showed distinct distribution patterns based on overlay imaging of MSI and liver histology. The lipid with m/z 794.51, putatively identified as PC (34:3), accumulated in the periportal zone whereas m/z 810.60, putatively identified as PC (38:4), accumulated in the centrilobular zone. Other lipid species also displayed zonal distribution patterns. Although no specific pattern was found for the spatial abundance of amitriptyline and its metabolites, their spatial abundances were non-uniform throughout the liver. This work extends our understanding of how amitriptyline treatment impacts lipid production and zonation pattern in the hepatic lobule pertaining to phospholipidosis [136]. Further investigations are needed to identify how amitriptyline disturbs phospholipid homeostasis in rat livers. In analogous work, MALDI-MSI integrated with omics analysis revealed that lipid homeostasis, oxidative stress, mitochondria damage and energy expenditure are closely associated with cadmium toxicity in mouse models, including liver toxicity [135]. Overall, MSI is becoming an effective tool to study the mechanism of DILI in combination with histology and other advanced technologies such as metabolomics.
5. MSI study of the drug distribution in clinical human tissues
In the past few years, MSI has gained popularity for its utility in analyzing drug and metabolite distribution in surgically dissected human tissue sections. In 2022, Ferey et al. reported a MALDI-FTICR-MSI analysis of platinum anticancer drug oxaliplatin in 12 ovary sections from women with peritoneal metastases before and after hyperthermic intraperitoneal chemotherapy [137]. The mass spectrum of a platinum-containing oxaliplatin has characteristic isotopic m/z values (m/z 397.0653, 398.0675, and 399.0679) due to the presence of three most abundant platinum isotopes (194Pt, 195Pt, and 196Pt). The high magnetic field (12 T) of the FTICR MSI provides ultra-high resolving power (>220,000) and high mass accuracy (<1 ppm), enabling the separation of the most isobaric species and allowing the accurate detection and non-ambiguous molecular formula assignments of oxaliplatin and its metabolites in human tissue sections. Interestingly, the original oxaliplatin compound was not detected in the ovary sections. Instead, a discriminative metabolite was seen at the peripheral location of tumoral peritoneum tissue which corresponded to the oxaliplatin-methionine complex based on accurate mass analysis [137]. This clinical result of detecting metabolites of Pt-species in ovary tissues after chemotherapy provides new mechanistic insights into the pharmacology of platinum chemotherapy.
Nishimura et al. conducted a prospective clinical study to visualize the distribution of the molecular targeted anticancer drug erlotinib in the drug-related skin dermatitis/eruption of patients with advanced pancreatic cancer (skin dermatitis/eruption being the most common side effect of erlotinib) [138]. The difference in erlotinib accumulation in the tissues of normal skin and dermatitis-affected skin was investigated using MALDI-MSI in combination with laser microdissection. The MSI data showed that erlotinib distribution in the dermatitis-affected skin was more heterogenous than that in the normal skin, and the dermatitis-affected skin contained statistically higher concentrations of the drug than in the superficial skin layer of adjacent normal skin. The focal distribution of erlotinib was further validated by regional laser microdissection and LC-MS/MS analysis [138]. Combined LC-MS/MS and MSI PK tissue analysis are useful for evaluating clinical samples and elucidating the pharmacological and toxicological mechanisms of drug actions.
Prieaux and colleagues recruited a cohort of 15 tuberculosis-infected patients who received a single dose of a TB cocktail containing key drugs (isoniazid, rifampicin, pyrazinamide, and moxifloxacin) before lung resection surgery, and then conducted MALDI-MSI to study the spatial distribution of TB drugs in intact TB lesions [139]. The MSI ion maps showed differences in the penetration of each TB drug or metabolite in caseous foci and cellular layers of the lesions. Pyrazinamide and isoniazid metabolite diffused favorably and rapidly into the necrotic cores and cellular compartments, thus reaching two critical persister populations/regions, including extracellular anaerobic bacilli in caseum and intracellular bacteria in acidic phagolysosomes. Rifampicin accumulated in necrotic foci where extracellular anaerobic bacteria reside. In contrast, the MALDI-MSI spatial distribution of moxifloxacin was different; whereas the drug accumulated in cellular regions, it did not diffuse well into the acellular caseum. The striking drug distribution and kinetics of accumulation of TB drugs across TB lesions provide a spatial PK and PD explanation for the treatment properties (or lack thereof) of current TB drugs [139], highlighting the importance of MSI in the decision-making process of pharmaceutical drug discovery and development.
6. MSI analysis of tissue distribution of macromolecule therapeutics
Over the past two decades, biologics such as proteins, antibodies, antibody-drug conjugates, and oligonucleotides have been granted FDA approvals at a rapid pace. In situ tissue distribution of complex modalities of antibody-based therapeutics has been investigated using radio- or fluorescent dye-labeled probes and relies on analytical approaches such as fluorescence microscopy, autoradiography, electron microscopy, and PET [140,141]. However, these assessments may not always be appropriate because of labeling complexity and lack of antibody cross-reactivity to different species. Biotransformation is an additional concern as these methods are not suitable for assessing the “intactness” of an antibody. MSI offers a complementary assay to detect and quantify macromolecules. To overcome the difficulty of ionizing high molecular weight macromolecules and the mass range limitation of MS, Ait-Belkacem et al. developed a MALDI in-source decay (ISD) MSI method to detect the fragmentation of bevacizumab and palivizumab, two FDA approved humanized IgG1 therapeutic antibodies, on glioblastoma-bearing mouse brain tissue sections at 80 μm lateral resolution [142]. ISD is a “top-down” approach that fragments ions directly in the MS ion source, rather than after the laser shot, allowing for the identification of major proteins without further treatment [143]. Using the MALDI-ISD-MSI technique and 1,5-diaminonaphthalene (DAN) as a matrix, ISD fragmentation of the immunoglobulin amine and carboxyl terminal parts generated a long series of characteristic ions after cleavage of N-Cα bonds on the peptide backbone. A sequence of 30 residues corresponding to the variable domain of the heavy and light chains of bevacizumab and palivizumab was obtained, which provided information for protein identification. The intensities of three major characteristic ions were summed to provide the image of antibodies distribution within the brain slices derived from mouse brain tissue sections bearing intracranial U87 glioblastoma tumors and treated with an intraperitoneal injection of bevacizumab and palivizumab. Bevacizumab was detected primarily in the tumor area, whereas palivizumab was distributed throughout the brain tissues. These results support the clinical application of bevacizumab for glioblastoma treatment [142].
MALDI-MSI has been used to detect the release and distribution of drug payload from antibody-drug conjugate (ADC) in tumor tissues [144]. Fujitwara et al. prepared an ADC by conjugating anti-human tissue factor antibody and monomethyl auristatin E (MMAE), a highly toxic anticancer agent, via a linker to direct the drug payload to cancer cells specifically. Although they attempted to ionize the ADC directly into the tumor tissue by selective fragmentation with different proteases, it was difficult to differentiate the ADC, antibody, and MMAE. The MALDI-MSI visualized only free drug payload MMAE without interference from MMAE in ADC based on the MMAE-specific MS/MS fragment. The in-situ detection of the accumulation of MMAE released from ADC in the tumor tissues at 3, 24, and 75 h after the administration of ADC was further validated by LC-MS/MS analysis [144]. These results suggest that MSI is a powerful tool for monitoring ADC disposition and payload release in ADC development at the preclinical stage.
Oligonucleotide-based therapeutics have been developed extensively in recent years. As of 2020, ten oligonucleotide drugs have received regulatory approval [145]. Hybridization methods such as the enzyme-linked immunosorbent assay (ELISA) are generally used for PK evaluation of oligonucleotides [146]. Nakashima et al. developed a new MALDI-TOF-MSI method to detect phosphonothioate antisense oligonucleotide ASO-2 (21 nucleotide units) in rat eyeballs [147]. ASO-2 oligonucleotide, a sequence of the FDA-approved oligonucleotide therapeutic fomivirsen, was observed in rat eye sections 30 min post-intravitreous administration in rats. The ion signal of [M─H]− at m/z 6683 corresponding to ASO-2 was localized in the central part of the vitreous body immediately after drug administration (Fig. 14A-b), and later localized more in the retina of eye 30 min after administration (Fig. 14A-c, 14A-e,) [147].
Fig. 14.
MALDI-MSI detection of oligonucleotide-based therapy. (A) The ion image of control rat eye section (A-a) and antisense oligonucleotide ASO-2 in rat eye sections immediately (A-b) or 30 min (A-c) after intravitreous injection of ASO-2 (500 μg). The [M─H]− of the ASO-2 at m/z 6683 was monitored. The eye section was stained with H&E (A-d) and merged with the ion image (A-e). Scan resolution: 150 μm. Printed with permission from [147]. Copyright 2022. (B, C) MALDI-MSI detection of an antisense oligonucleotide containing locked nucleic acids (LNA-A) in mouse kidney section after intravenous administration of LNA-A (1 mmol). (B) Mass spectra of control and 0.5, 2, and 4 h post-dose. (C) MS ion image of LNA-A (m/z 4959) and its metabolite (m/z 4614). Scale bar: 5 mm. Adapted with permission from [148]. Copyright 2022, Wiley.
In 2018, Yokoi et al. reported using MALDI-MSI to detec antisense oligonucleotide containing locked nucleic acids (LNA-A) in mouse kidneys [148]. To remove most of the salts, lipids and other interfering compounds from the tissue to enhance the detection sensitivity of the LNA-A, a series of washing steps with 70% ethanol (EtOH) followed by EtOH and Carnoy’s solution (EtOH/chloroform/acetic acid mixture) and a final acetone wash were implemented. The [M─H]− ion (m/z 4959) in the LNA-A spectrum was detected in kidney tissues at 0.5, 2 and 4 h after 1 nmol of LNA-A intravenous injection in mice (Fig. 14B, C). Importantly, the [M─H]− (m/z 4614), a metabolite ion which represents a removal of terminal deoxyguanosine thiophosphate in LNA-A, was detected in all treated mouse kidneys (Fig. 14C). These exciting results demonstrate the potential of MSI-based drug distribution analysis of oligonucleotide therapeutics and metabolites in tissues.
7. Conclusions and future perspective
Comprehensive knowledge of PK properties (absorption, distribution, metabolism, and excretion) and toxicity is key to understanding how effectively therapeutic agents reach their targets. MSI offers the unique ability to provide in situ imaging and quantification of small molecule and macromolecule drugs and their metabolites in preclinical and clinical tissue studies with high spatial resolution. MSI also provides unique information about macro and micro distribution of drugs in the context of the anatomic and metabolic heterogeneity of tissues, offers potential insights into impediments to drug access, and provides opportunities to guide drug dosing and delivery in drug development. The advantages of MSI techniques (such as high molecular selectivity and parallel acquisition of multiple analytes) over conventional methods such as autoradiography and immunohistochemistry technologies will allow for higher throughput pharmaceutical ADMET analysis.
While absolute quantitation using MSI still face challenges such as variation of signal (pixel-to-pixel), ion-suppression effects and other confounding factors (such as geometric set-up of instrument, tissue heterogeneity, etc.), the incorporation of MSI-based qualitative and quantitative studies in preclinical and clinical drug development is becoming more common. The precision, repeatability, and reproducibility of MSI studies need to be verified and well documented to accelerate the translation of the technique to routine clinical applications. Currently there are very few studies using MSI to exam the impact of active transporters in tissue drug uptake and efflux, intracellular biotransformation of prodrugs, receptor-mediated drug uptake, or the impact of these factors on the ratio of drug concentration between tissue and plasma (Kp) at steady state. It would be interesting to apply the MSI technique to understand how transporters play a role in various drug distribution processes; for instance, the spatial distribution of drug in hepatocytes versus bile canaliculi in biliary excretion or renal tubular cells versus tubular lumen in renal excretion of a given drug. In the future, we expect that improved MSI resolution coupled with immunohistochemical studies will improve MSI’s quantitative ability to differentiate drug distribution at the cellular and subcellular level and allow direct quantification of important PK parameters such as Kp value of drug distribution in a given organ.
Current commercially available MSI instruments offer spatial resolution at 5–10 μm. Technical advances in MSI resolution approaching the sub-micrometer range will be capable of addressing many pharmacological questions related to the mechanism of action of a drug and its metabolites in target tissues and the therapeutic or toxic effects of these compounds [149,150]. Exciting research opportunities in the biochemical and pharmacology areas include the following.
Single-cell and subcellular drug distribution by MSI analysis.
PK and biodistribution analysis of drugs at the organ level provide insight into how drugs are distributed throughout the body, but single-cell and subcellular PK imaging studies at single-cell resolution provide insight into drug action at the cellular level and offer understanding of cellular pharmacology [151]. Single-cell PK imaging has been conducted on drugs with intrinsic fluorescence using the fluorescence microscopy imaging technique [152,151]. The application of high resolution MSI to provide information about a drug and its metabolite distribution at single-cell and subcellular resolution, along with 3D imaging will aid our understanding of cellular pharmacology and the toxicology of drug molecules.
Multimodality paradigm for drug distribution analysis.
While MSI technology can generate a wealth of chemical information via mapping the spatial distribution of hundreds of biomolecules, drugs, and metabolites throughout a tissue section, MSI spatial resolution can be further improved by combining it with various anatomical mapping imaging methods that have relatively low chemical specificity but high spatial resolution. Caprioli and colleagues described a predictive imaging modality created by “fusing” MSI and microscopy technologies. MSI-microscopy fusion analysis creates sharper MSI images, predicts ion distribution in areas that were not measured by MSI, enriches biological signals, and attenuates instrument artifacts [153].
Several research groups have reported fusing MSI and magnetic resonance imaging (MRI) techniques [154,155,156]. Because MRI scans visualize the anatomy of the tissue and the distribution of water, the MSI-MRI fusion workflow merges information from MRI and MSI and establishes the co-localization of ionized molecules with water distribution in tissues [155]. Agar et al. reported using an automatic method to nonlinearly align hyperspectral molecular MALDI-MSI images with anatomical information derived from 7 T in vivo MRI scans [154]. Verbeeck et al developed a computational approach to integrate the MRI-based rat brain atlas with the FTICR-MSI data from coronal brain sections of a Parkinson’s disease rat model [156]. Additionally, the coupling of MSI with various vibrational spectroscopic imaging techniques, such as Roman spectroscopy and Fourier transform infrared spectroscopy, has been explored [157]. The advantages of combining multimodal imaging technologies (high spatial resolution and high chemical specificity) with MSI will undoubtedly improve drug distribution analysis, allow fine mapping of drug molecules with their therapeutic target in the native tissue, and further support drug discovery and toxicological studies.
MSI technology is an important tool that can be used to assess drug distribution, target engagement, and pharmacodynamics biomarkers; explore in situ pharmaco-metabolome changes in response to drug administration; promote the discovery of toxic biomarkers and mechanistic insights into toxicological pathways; and to provide key insights into the PK/PD and ADMET properties of drugs. Further development of MSI-related instrumentation and absolute quantification methods, including coupling MSI with multimodal imaging system and “omics” technologies (such as proteomics, lipidomics, and metabolomics on tissues), will provide significant benefits to pharmacological research. As a result, preclinical and clinical drug development research will utilize MSI in ever-broadening applications.
Acknowledgement
This work was supported by the National Institute of Diabetes and Digestive and Kidney (R01-DK121970) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (R61HD099995) to Feng Li. Feng Li was also partially supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development (P01 HD087157) and Bill and Melinda Gates Foundation (INV-001902) to Dr. Martin M. Matzuk. Xinli Liu acknowledges funding support from the National Cancer Institute (NIH R15CA182769, P20CA221731, P20CA221696), the Cancer Prevention and Research Institute of Texas (CPRIT RP150656), and the University of Texas MD Anderson Cancer Center Duncan Family Institute for Cancer Prevention and Risk Assessment and UHAND Partnership Cancer Prevention Disparities Seed Funding Research Program. The human brain organoid work was partially supported by the BCM IDDRC Grant (P50HD10355) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development for use of the Microscopy Core facilities, the RNA In Situ Hybridization Core facility, and the Human Neuronal Differentiation Core facility, and NIGMS R01 GM120033, Cynthia and Antony Petrello Endowment, Mark A. Wallace Endowment to M. Maletic-Savatic. We are grateful to Dr. Sivan Osenberg who generated the organoids, Dr. Prasanna Kandel for the drug treatment, Dr. Dodge Baluya for MSI, and Victoria Jialu Huang for assistance of graphics.
Abbreviations:
- MSI
Mass Spectrometry Imaging
- ADMET
Absorption, Distribution, Metabolism, Excretion, Toxicity
- HPLC
High Performance Liquid Chromatography
- LC-MS/MS
Liquid Chromatography Tandem Mass Spectrometry
- IHC
Immunohistochemical
- MS
Mass Spectrometry
- MALDI
Matrix-Assisted Laser Desorption/Ionization
- ESI
Electrospray Ionization
- FAB
Fast Atom Bombardment
- APCI
Atmospheric Pressure Chemical Ionization
- QqQ
Triple Quadrupole
- QTRAP
Quadrupole Ion Trap
- TOF
Time-of-flight
- FTICR
Fourier Transform Ion Cyclotron Resonance
- QTOF
Quadrupole Time-of-flight
- PET
Positron Emission Tomography
- SPECT
Single-Photon Emission Computed Tomography
- MRM
Multiple Reaction Monitoring
- DESI
Desorption Electrospray Ionization
- IM
Ion Mobility
- UV
Ultraviolet
- m/z
Mass-to-charge ratio
- SIMS
Secondary Ion Mass Spectrometry
- LA-ICP-MSI
Laser ablation inductively coupled plasma mass spectrometry imaging
- OCT
Optimal Cutting Temperature
- FFPE
Formalin Fixed Paraffin Embedded
- SA
Sinapinic Acid
- DHB
2,5-Dihydroxybenzoic Acid
- DAN
1,5-Diaminonaphthalene
- CHCA
α-Cyano-4-Hydroxycinnamic Acid
- DHA
2,6-Dihydroxyacetophenone
- TIC
Total Ion Count
- GI
Gastrointestinal
- PK
Pharmacokinetics
- IS
Internal Standard
- CNS
Central Nervous System
- BBB
Blood-Brain Barrier
- BTB
Blood-Tumor Barrier
- HER2
Human Epidermal Growth Factor Receptor 2
- EGFR
Epidermal Growth Factor Receptor
- MDR1
Multidrug Resistance Protein 1
- KO
Knockout
- CP
Caudoputamen
- CTX
Cerebral Cortex
- L-DOPA
L-3–4-dihydroxyphenylalanine
- iPSCs
Pluripotent stem cells
- OTCD
On-tissue Chemical Derivatization
- QWBA
Quantitative Whole-Body Autoradiography
- DILI
Drug-Induced Liver Injury
- AQ
Amodiaquine
- AQQI
Amodiaquine Quinoeimine
- AQ-SG
Amodiaquine-Glutathione-Adduct
- DEAQ
De-ethyl-Amodiaquine
- ISD
In-source Decay
- ADC
Antibody-drug Conjugate
- MMAE
Monomethyl Auristatin E
- ELISA
Enzyme-linked Immunosorbent Assay
- LNA
Locked Nucleic Acids
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Waring MJ, Arrowsmith J, Leach AR, Leeson PD, Mandrell S, Owen RM, Pairaudeau G, Pennie WD, Pickett SD, Wang J, Wallace O, Weir A, An analysis of the attrition of drag candidates from four major pharmaceutical companies, Nat. Rev. Drug Discov 14 (7) (2015) 475–486, 10.1038/nrd4609. [DOI] [PubMed] [Google Scholar]
- [2].Cook D, Brown D, Alexander R, March R, Morgan P, Satterthwaite G, Pangalos MN, Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework, Nat. Rev. Drug Discov 13 (6) (2014) 419–431, 10.1038/nrd4309. [DOI] [PubMed] [Google Scholar]
- [3].Solon EG, Schweitzer A, Stoeckli M, Prideaux B, Autoradiography, MALDI-MS, and SIMS-MS imaging in pharmaceutical discovery and development, AAPS J. 12 (1) (2010) 11–26, 10.1208/s12248-009-9158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Solon EG, Autoradiography techniques and quantification of drug distribution, Cell Tissue Res. 360 (1) (2015) 87–107, 10.1007/s00441-014-2093-4. [DOI] [PubMed] [Google Scholar]
- [5].Schwamborn K, Caprioli RM, Molecular imaging by mass spectrometry–looking beyond classical histology, Nat. Rev. Cancer 10 (9) (2010) 639–646, 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- [6].Chughtai K, Heeren RM, Mass spectrometric imaging for biomedical tissue analysis, Chem. Rev 110 (5) (2010) 3237–3277, 10.1021/cr100012c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Prideaux B, Stoeckli M, Mass spectrometry imaging for drug distribution studies, J. Proteomics 75 (16) (2012) 4999–5013, 10.1016/j.jprot.2012.07.028. [DOI] [PubMed] [Google Scholar]
- [8].Jiang H, Gao S, Hu G, He J, Jin H, Innovation in drug toxicology: Application of mass spectrometry imaging technology, Toxicology 464 (2021), 153000, 10.1016/j.tox.2021.153000. [DOI] [PubMed] [Google Scholar]
- [9].Prentice BM, Caprioli RM, Vuiblet V, Label-free molecular imaging of the kidney, Kidney Int. 92 (3) (2017) 580–598, 10.10l6/j.kint.2017.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Seeley EH, Schwamborn K, Caprioli RM, Imaging of intact tissue sections: moving beyond the microscope, J. Biol. Chem 286 (29) (2011) 25459–25466, 10.1074/jbc.R111.225854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Setou M, Kurabe N, Mass microscopy: high-resolution imaging mass spectrometry, J. Electron. Microsc. (Tokyo) 60 (1) (2011) 47–56, 10.1093/jmicro/dfq079. [DOI] [PubMed] [Google Scholar]
- [12].Zhou LY, Wang X, Xiang P, Mass spectrometry imaging and its application in forensic toxicology, Fa Yi Xue Za Zhi 37 (3) (2021) 402–1401, 10.12116/j.issn.1004-5619.2020.300502. [DOI] [PubMed] [Google Scholar]
- [13].Siuzdak G, An Introduction to Mass Spectrometry Ionization: An Excerpt from The Expanding Role of Mass Spectrometry in Biotechnology, 2nd ed.; MCC Press: San Diego, 2005. 10.1016/j.jala.2004.01.004. [DOI] [Google Scholar]
- [14].Li C, Chu S, Tan S, Yin X, Jiang Y, Dai X, Gong X, Fang X, Tian D, Towards higher sensitivity of mass spectrometry: a perspective from the mass analyzers, Front. Chem 9 (2021), 813359, 10.3389/fchem.2021.813359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Swales JG, Hamm G, Clench MR, Goodwin RJA, Mass spectrometry imaging and its application in pharmaceutical research and development: a concise review, Int. J. Mass Spectrom 437 (2019) 99–112, 10.1016/j.ijms.2018.02.007. [DOI] [Google Scholar]
- [16].Caprioli RM, Farmer TB, Gile J, Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS, Anal. Chem 69 (23) (1997) 4751–4760, 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- [17].Murayama C, Kimura Y, Setou M, Imaging mass spectrometry: principle and application, Biophys. Rev 1 (3) (2009) 131, 10.1007/s12551-009-0015-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chaurand P, Caprioli RM, Direct profiling and imaging of peptides and proteins from mammalian cells and tissue sections by mass spectrometry, Electrophoresis 23 (18) (2002) 3125–3135, . [DOI] [PubMed] [Google Scholar]
- [19].Stoeckli M, Farmer TB, Caprioli RM, Automated mass spectrometry imaging with a matrix-assisted laser desorption ionization time-of-flight instrument, J. Am. Soc. Mass Spectrom 10 (1) (1999) 67–71, 10.1016/S1044-0305(98)00126-3. [DOI] [PubMed] [Google Scholar]
- [20].Gemperline E, Chen B, Li L, Challenges and recent advances in mass spectrometric imaging of neurotransmitters, Bioanalysis 6 (4) (2014) 525–540, 10.4155/bio.13.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cole R, Electrospray and MALDI Mass Spectrometry Fundamentals, Instrumentation, Practicalities, and Biological Applications, 2nd edition, Wiley, 2010. [Google Scholar]
- [22].Sampson JS, Hawkridge AM, Muddiman DC, Generation and detection of multiply-charged peptides and proteins by matrix-assisted laser desorption electrospray ionization (MALDESI) Fourier transform ion cyclotron resonance mass spectrometry, J. Am. Soc. Mass Spectrom 17 (12) (2006) 1712–1716, 10.1016/j.jasms.2006.08.003. [DOI] [PubMed] [Google Scholar]
- [23].Gemoll T, Roblick UJ, Habermann JK, MALDI mass spectrometry imaging in oncology (Review), Mol. Med. Rep 4 (6) (2011) 1045–1051, 10.3892/mmr.2011.566. [DOI] [PubMed] [Google Scholar]
- [24].DeLaney K, Phetsanthad A, Li L, Advances in high-resolution maldi mass spectrometry for neurobiology, Mass Spectrom. Rev 41 (2) (2022) 194–214, 10.1002/mas.21661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Soltwisch J, Kettling H, Vens-Cappell S, Wiegelmann M, Muthing J, Dreisewerd K, Mass spectrometry imaging with laser-induced postionization, Science 348 (6231) (2015) 211–215, 10.1126/science.aaa1051. [DOI] [PubMed] [Google Scholar]
- [26].Ellis SR, Soltwisch J, Paine MRL, Dreisewerd K, Heeren RMA, Laser post-ionisation combined with a high resolving power orbitrap mass spectrometer for enhanced MALDI-MS imaging of lipids, Chem. Commun. (Camb.) 53 (53) (2017) 7246–7249, 10.1039/c7cc02325a. [DOI] [PubMed] [Google Scholar]
- [27].Potthoff A, Dreisewerd K, Soltwisch J, Detailed characterization of the postionization efficiencies in MALDI-2 as a function of relevant input parameters, J. Am. Soc. Mass Spectrom 31 (9) (2020) 1844–1853, 10.1021/jasms.0c00072. [DOI] [PubMed] [Google Scholar]
- [28].Schwamborn K, The importance of histology and pathology in mass spectrometry imaging, Adv. Cancer Res 134 (2017) 1–26, 10.1016/bs.acr.2016.11.001. [DOI] [PubMed] [Google Scholar]
- [29].Takats Z, Wiseman JM, Gologan B, Cooks RG, Mass spectrometry sampling under ambient conditions with desorption electrospray ionization, Science 306 (5695) (2004) 471–473, 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- [30].Ifa DR, Wu C, Ouyang Z, Cooks RG, Desorption electrospray ionization and other ambient ionization methods: current progress and preview, Analyst 135 (4) (2010) 669–681, 10.1039/b925257f. [DOI] [PubMed] [Google Scholar]
- [31].Takats Z, Strittmatter N, McKenzie JS, Ambient mass spectrometry in cancer research, In Adv. Cancer Res Vol. 134 (2017) 231–256, 10.1016/bs.acr.2016.11.011. [DOI] [PubMed] [Google Scholar]
- [32].Zoltan Takats JMW, Bogdan Gologan R, Cooks G, Mass_spectrometry sampling under ambient conditions with desorption electrospray ionization, Science 306 (5695) (2004) 471–473, 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- [33].Eberlin LS, Ferreira CR, Dill AL, Ifa DR, Cooks RG, Desorption electrospray ionization mass spectrometry for lipid characterization and biological tissue imaging, Biochim. Biophys. Acta 1811 (11) (2011) 946–960, 10.1016/j.bbalip.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Eberlin LS, Ifa DR, Wu C, Cooks RG, Three-dimensional vizualization of mouse brain by lipid analysis using ambient ionization mass spectrometry, Angew. Chem. Int. Ed. Engl 49 (5) (2010) 873–876, 10.1002/anie.200906283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Manicke NE, Wiseman JM, Ifa DR, Cooks RG, Desorption electrospray ionization (DESI) mass spectrometry and tandem mass spectrometry (MS/MS) of phospholipids and sphingolipids: ionization, adduct formation, and fragmentation, J. Am. Soc. Mass Spectrom 19 (4) (2008) 531–543, 10.1016/j.jasms.2007.12.003. [DOI] [PubMed] [Google Scholar]
- [36].Cotte-Rodriguez I, Takats Z, Talaty N, Chen H, Cooks RG, Desorption electrospray ionization of explosives on surfaces: sensitivity and selectivity enhancement by reactive desorption electrospray ionization, Anal. Chem 77 (21) (2005) 6755–6764, 10.1021/ac050995+. [DOI] [PubMed] [Google Scholar]
- [37].Bodzon-Kulakowska A, Cichon T, Golec A, Drabik A, Ner J, Suder P, DESI-MS as a tool for direct lipid analysis in cultured cells, Cytotechnology 67 (6) (2015) 1085–1091, 10.1007/s10616-014-9734-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Pierson EE, Midey AJ, Forrest WP, Shah V, Olivos HJ, Shrestha B, Teller R, Forster S, Bensussan A, Helmy R, Direct drug analysis in polymeric implants using desorption electrospray ionization - mass spectrometry imaging (DESI-MSI), Pharm. Res 37 (6) (2020) 107, 10.1007/s11095-020-02823-x. [DOI] [PubMed] [Google Scholar]
- [39].Towers MW, Karancsi T, Jones EA, Pringle SD, Claude E, Optimised desorption electrospray ionisation mass spectrometry imaging (DESI-MSI) for the analysis of proteins/peptides directly from tissue sections on a travelling wave ion mobility Q-ToF, J. Am. Soc. Mass Spectrom 29 (12) (2018) 2456–2466, 10.1007/s13361-018-2049-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wiseman JM, Ifa DR, Song Q, Cooks RG, Tissue imaging at atmospheric pressure using desorption electrospray ionization (DESI) mass spectrometry, Angew. Chem. Int. Ed. Engl 45 (43) (2006) 7188–7192, 10.1002/anie.200602449. [DOI] [PubMed] [Google Scholar]
- [41].Tillner J, Wu V, Jones EA, Pringle SD, Karancsi T, Dannhorn A, Veselkov K, McKenzie JS, Takats Z, Faster, more reproducible DESI-MS for biological tissue imaging, J. Am. Soc. Mass Spectrom 28 (10) (2017) 2090–2098, 10.1007/s13361-017-1714-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Roach PJ, Laskin J, Laskin A, Nanospray desorption electrospray ionization: an ambient method for liquid-extraction surface sampling in mass spectrometry, Analyst 135 (9) (2010) 2233–2236, 10.1039/c0an00312c. [DOI] [PubMed] [Google Scholar]
- [43].Yin R, Burnum-Johnson KE, Sun X, Dey SK, Laskin J, High spatial resolution imaging of biological tissues using nanospray desorption electrospray ionization mass spectrometry, Nat. Protoc 14 (12) (2019) 3445–3470, 10.1038/s41596-019-0237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Unsihuay D, Mesa Sanchez D, Laskin J, Quantitative mass spectrometry imaging of biological systems, Annu. Rev. Phys. Chem 72 (2021) 307–329, 10.1146/annurev-physchem-061020-053416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Takats Z, Wiseman JM, Cooks RG, Ambient mass spectrometry using desorption electrospray ionization (DESI): instrumentation, mechanisms and applications in forensics, chemistry, and biology, J. Mass Spectrom 40 (10) (2005) 1261–1275, 10.1002/jms.922. [DOI] [PubMed] [Google Scholar]
- [46].Nunez J, Renslow R, Cliff JB 3rd, Anderton CR, NanoSIMS for biological applications: current practices and analyses, Biointerphases 13 (3) (2017)03B301, 10.1116/1.4993628. [DOI] [PubMed] [Google Scholar]
- [47].Sodhi RN, Time-of-flight secondary ion mass spectrometry (TOF-SIMS):– versatility in chemical and imaging surface analysis, Analyst 129 (6) (2004) 483–487, 10.1039/b402607c. [DOI] [PubMed] [Google Scholar]
- [48].DeBord JD, Smith DF, Anderton CR, Heeren RMA, Paša-Tolić L, Gomer RH, Fernandez-Lima FA, Williams JG, Secondary ion mass spectrometry imaging of Dictyostelium discoideum aggregation streams, PLoS ONE 9 (6) (2014) e99319, 10.1371/journal.pone.0099319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Smith DF, Kiss A, Leach FE 3rd, Robinson EW, Pasa-Tolic L, Heeren RM, High mass accuracy and high mass resolving power FT-ICR secondary ion mass spectrometry for biological tissue imaging, Anal. Bioanal. Chem 405 (18) (2013) 6069–6076, 10.1007/s00216-013-7048-1. [DOI] [PubMed] [Google Scholar]
- [50].Smith DF, Robinson EW, Tolmachev AV, Heeren RM, Pasa-Tolic L, C60 secondary ion Fourier transform ion cyclotron resonance mass spectrometry, Anal Chem 83 (24) (2011) 9552–9556, 10.1021/ac2023348. [DOI] [PubMed] [Google Scholar]
- [51].Musat N, Foster R, Vagner T, Adam B, Kuypers MM, Detecting metabolic activities in single cells, with emphasis on nanoSIMS, FEMS Microbiol. Rev 36 (2) (2012) 486–511, 10.1111/j.1574-6976.2011.00303.x. [DOI] [PubMed] [Google Scholar]
- [52].Watrous JD, Dorrestein PC, Imaging mass spectrometry in microbiology, Nat. Rev. Microbiol 9 (9) (2011) 683–694, 10.1038/nrmicro2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Wilschefski SC, Baxter MR, Inductively coupled plasma mass spectrometry: introduction to analytical aspects, Clin. Biochem. Rev 40 (3) (2019) 115–133, 10.33176/AACB-19-00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Pozebon D, Scheffler GL, Dressler VL, Nunes MAG, Review of the applications of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) to the analysis of biological samples, J. Anal. At. Spectrom 29 (12) (2014) 2204–2228, 10.1039/c4ja00250d. [DOI] [Google Scholar]
- [55].Sussulini A, Becker JS, Becker JS, Laser ablation ICP-MS: application in biomedical research, Mass Spectrom. Rev 36 (1) (2017) 47–57, 10.1002/mas.21481. [DOI] [PubMed] [Google Scholar]
- [56].Weiskirchen SK, Kim P, Weiskirchen R, Laser ablation inductively coupled plasma spectrometry: metal imaging in experimental and clinical wilson disease, Inorganics 7(54) (2019), 10.3390/inorganics7040054. [DOI] [Google Scholar]
- [57].Giesen C, Wang HA, Schapiro D, Zivanovic N, Jacobs A, Hattendorf B, Schuffler PJ, Grolimund D, Buhmann JM, Brandt S, Varga Z, Wild PJ, Gunther D, Bodenmiller B, Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry, Nat. Methods 11 (4) (2014) 417–422, 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- [58].Gimenez Y, Busser B, Trichard F, Kulesza A, Laurent JM, Zaun V, Lux F, Benoit JM, Panczer G, Dugourd P, Tillement O, Pelascini F, Sancey L, Motto-Ros V, 3D imaging of nanoparticle distribution in biological tissue by laser-induced breakdown spectroscopy, Sci. Rep 6 (2016) 29936, 10.1038/srep29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Meng Y, Gao C, Lin Z, Hang W, Huang B, Nanoscale laser-induced breakdown spectroscopy imaging reveals chemical distribution with subcellular resolution, Nanoscale Adv. 2 (9) (2020) 3983–3990, 10.1039/d0na00380h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Meng Y, Gao C, Lu Q, Ma S, Hang W, Single-cell mass spectrometry imaging of multiple drugs and nanomaterials at organelle level, ACS Nano 15 (8) (2021) 13220–13229, 10.1021/acsnano.1c02922. [DOI] [PubMed] [Google Scholar]
- [61].Lamont L, Hadavi D, Viehmann B, Flinders B, Heeren RMA, Vreeken RJ, Porta Siegel T, Quantitative mass spectrometry imaging of drugs and metabolites: a multiplatform comparison, Anal. Bioanal. Chem 413 (10) (2021) 2779–2791, 10.1007/s00216-021-03210-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hamm G, Bonnel D, Legouffe R, Pamelard F, Delbos JM, Bouzom F, Stauber J, Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor, J. Proteomics 75 (16) (2012) 4952–4961, 10.1016/j.jprot.2012.07.035. [DOI] [PubMed] [Google Scholar]
- [63].Barry JA, Ait-Belkacem R, Hardesty WM, Benakli L, Andonian C, Licea-Perez H, Stauber J, Castellino S, Multicenter validation study of quantitative imaging mass spectrometry, Anal. Chem 91 (9) (2019) 6266–6274, 10.1021/acs.analchem.9b01016. [DOI] [PubMed] [Google Scholar]
- [64].Porta T, Lesur A, Varesio E, Hopfgartner G, Quantification in MALDI-MS imaging: what can we learn from MALDI-selected reaction monitoring and what can we expect for imaging? Anal. Bioanal. Chem 407 (8) (2015) 2177–2187, 10.1007/s00216-014-8315-5. [DOI] [PubMed] [Google Scholar]
- [65].Buchberger AR, DeLaney K, Johnson J, Li L, Mass spectrometry imaging: a review of emerging advancements and future insights, Anal. Chem 90 (1) (2018) 240–265, 10.1021/acs.analchem.7b04733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Unsihuay D, Yin R, Sanchez DM, Yang M, Li Y, Sun X, Dey SK, Laskin J, High-resolution imaging and identification of biomolecules using Nano-DESI coupled to ion mobility spectrometry, Anal. Chim. Acta 1186 (2021), 339085, 10.1016/j.aca.2021.339085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Hermann J, Noels H, Theelen W, Lellig M, Orth-Alampour S, Boor P, Jankowski V, Jankowski J, Sample preparation of formalin-fixed paraffin-embedded tissue sections for MALDI-mass spectrometry imaging, Anal. Bioanal. Chem 412 (6) (2020) 1263–1275, 10.1007/s00216-019-02296-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Casadonte R, Caprioli RM, Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry, Nat. Protoc 6 (11) (2011) 1695–1709, 10.1038/nprot.2011.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Goodwin RJA, Sample preparation for mass spectrometry imaging: small mistakes can lead to big consequences, J. Proteomics 75 (16) (2012) 4893–4911, 10.1016/j.jprot.2012.04.012. [DOI] [PubMed] [Google Scholar]
- [70].O’Rourke MB, Padula MP, Smith C, Youssef P, Cordwell S, Witting P, Sutherland G, Crossett B, Optimal preparation of formalin fixed samples for peptide based matrix assisted laser desorption/ionization mass spectrometry imaging workflows, J. Vis. Exp 131 (2018), 10.3791/56778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Carter CL, Jones JW, Farese AM, MacVittie TJ, Kane MA, Inflation-fixation method for lipidomic mapping of lung biopsies by matrix assisted laser desorption/ionization-mass spectrometry imaging, Anal. Chem 88 (9) (2016) 4788–4794, 10.1021/acs.analchem.6b00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Amaya KR, Sweedler JV, Clayton DF, Small molecule analysis and imaging of fatty acids in the zebra finch song system using time-of-flight-secondary ion mass spectrometry, J. Neurochem 118 (4) (2011) 499–511, 10.1111/j.1471-4159.2011.07274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Rocha B, Ruiz-Romero C, Blanco FJ, Mass spectrometry imaging: a novel technology in rheumatology, Nat. Rev. Rheumatol 13 (1) (2017) 52–63, 10.1038/nrrheum.2016.184. [DOI] [PubMed] [Google Scholar]
- [74].Kaletas BK, van der Wiel IM, Stauber J, Lennard JD, Guzel C, Kros JM, Luider TM, Heeren RM, Sample preparation issues for tissue imaging by imaging MS, Proteomics 9 (10) (2009) 2622–2633, 10.1002/pmic.200800364. [DOI] [PubMed] [Google Scholar]
- [75].Zenobi R, Knochenmuss R, Ion formation in MALDI mass spectrometry, Mass Spectrom. Rev 17 (1998) 337–366, . [DOI] [Google Scholar]
- [76].MacAleese L, Stauber J, Heeren RM, Perspectives for imaging mass spectrometry in the proteomics landscape, Proteomics 9 (4) (2009) 819–834, 10.1002/pmic.200800363. [DOI] [PubMed] [Google Scholar]
- [77].Seeley EH, Oppenheimer SR, Mi D, Chaurand P, Caprioli RM, Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections, J. Am. Soc. Mass Spectrom 19 (8) (2008) 1069–1077, 10.1016/j.jasms.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Norris JL, Cornett DS, Mobley JA, Andersson M, Seeley EH, Chaurand P, Caprioli RM, Processing MALDI mass spectra to improve mass spectral direct tissue analysis, Int. J. Mass. Spectrom 260 (2–3) (2007) 212–221, 10.1016/j.ijms.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Deininger SO, Cornett DS, Paape R, Becker M, Pineau C, Rauser S, Walch A, Wolski E, Normalization in MALDI-TOF imaging datasets of proteins: practical considerations, Anal. Bioanal. Chem 401 (1) (2011) 167–181, 10.1007/s00216-011-4929-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Fonville JM, Carter C, Cloarec O, Nicholson JK, Lindon JC, Bunch J, Holmes E, Robust data processing and normalization strategy for MALDI mass spectrometric imaging, Anal. Chem 84 (3) (2012) 1310–1319, 10.1021/ac201767g. [DOI] [PubMed] [Google Scholar]
- [81].Guidance for industry: bioanalytical method validation. (2013). https://www.regulations.gov/docket/FDA-2013-D-1020. [Google Scholar]
- [82].Boskamp T, Casadonte R, Hauberg-Lotte L, Deininger S, Kriegsmann J, Maass P, Cross-normalization of MALDI mass spectrometry imaging data improves site-to-site reproducibility, Anal. Chem 93 (30) (2021) 10584–10592, 10.1021/acs.analchem.1c01792. [DOI] [PubMed] [Google Scholar]
- [83].Ly A, Longuespee R, Casadonte R, Wandernoth P, Schwamborn K, Bollwein C, Marsching C, Kriegsmann K, Hopf C, Weichert W, Kriegsmann J, Schirmacher P, Kriegsmann M, Deininger SO, Site-to-site reproducibility and spatial resolution in MALDI-MSI of peptides from formalin-fixed paraffin-embedded samples, Proteomics Clin. Appl 13 (1) (2019), e1800029, 10.1002/prca.201800029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Morikawa-Ichinose T, Fujimura Y, Murayama F, Yamazaki Y, Yamamoto T, Wariishi H, Miura D, Improvement of sensitivity and reproducibility for imaging of endogenous metabolites by matrix-assisted laser desorption/ionization-mass spectrometry, J. Am. Soc. Mass Spectrom 30 (8) (2019) 1512–1520, 10.1007/s13361-019-02221-7. [DOI] [PubMed] [Google Scholar]
- [85].Dekker TJ, Balluff BD, Jones EA, Schone CD, Schmitt M, Aubele M, Kroep JR, Smit VT, Tollenaar RA, Mesker WE, Walch A, McDonnell LA, Multicenter matrix-assisted laser desorption/ionization mass spectrometry imaging (MALDI MSI) identifies proteomic differences in breast-cancer-associated stroma, J. Proteome Res 13 (11) (2014) 4730–4738, 10.1021/pr500253j. [DOI] [PubMed] [Google Scholar]
- [86].McDonnell LA, Rompp A, Balluff B, Heeren RM, Albar JP, Andren PE, Corthals GL, Walch A, Stoeckli M, Discussion point: reporting guidelines for mass spectrometry imaging, Anal. Bioanal. Chem 407 (8) (2015) 2035–2045, 10.1007/s00216-014-8322-6. [DOI] [PubMed] [Google Scholar]
- [87].Rompp A, Both JP, Brunelle A, Heeren RM, Laprevote O, Prideaux B, Seyer A, Spengler B, Stoeckli M, Smith DF, Mass spectrometry imaging of biological tissue: an approach for multicenter studies, Anal. Bioanal. Chem 407 (8) (2015) 2329–2335, 10.1007/s00216-014-8410-7. [DOI] [PubMed] [Google Scholar]
- [88].Foll MC, Moritz L, Wollmann T, Stillger MN, Vockert N, Werner M, Bronsert P, Rohr K, Gruning BA, Schilling O, Accessible and reproducible mass spectrometry imaging data analysis in Galaxy, GigaScience 8 (12) (2019), 10.1093/gigascience/giz143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Nilsson A, Peric A, Strimfors M, Goodwin RJA, Hayes MA, Andren PE, Hilgendorf C, Mass Spectrometry Imaging proves differential absorption profiles of well-characterised permeability markers along the crypt-villus axis, Sci. Rep 7 (1) (2017) 6352, 10.1038/s41598-017-06583-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Huizing LRS, McDuffie J, Cuyckens F, van Heerden M, Koudriakova T, Heeren RMA, Vreeken RJ, Quantitative mass spectrometry imaging to study drug distribution in the intestine following oral dosing, Anal. Chem 93 (4) (2021) 2144–2151, 10.1021/acs.analchem.0c03956. [DOI] [PubMed] [Google Scholar]
- [91].Rao T, Shen B, Zhu Z, Shao Y, Kang D, Li X, Yin X, Li H, Xie L, Wang G, Liang Y, Optimization and evaluation of MALDI TOF mass spectrometric imaging for quantification of orally dosed octreotide in mouse tissues, Talanta 165 (2017) 128–135, 10.1016/j.talanta.2016.12.049. [DOI] [PubMed] [Google Scholar]
- [92].Chen Y, Tang W, Gordon A, Li B, Development of an integrated tissue pretreatment protocol for enhanced MALDI MS imaging of drug distribution in the brain, J. Am. Soc. Mass Spectrom 31 (5) (2020) 1066–1073, 10.1021/jasms.0c00003. [DOI] [PubMed] [Google Scholar]
- [93].Tanaka H, Hirata M, Shinonome S, Wada T, Iguchi M, Dohi K, Inoue M, Ishioka Y, Hojo K, Yamada T, Sugimoto T, Masuno K, Nezasa K, Sato N, Matsuo K, Yonezawa S, Frenkel EP, Shichijo M, Preclinical antitumor activity of S-222611, an oral reversible tyrosine kinase inhibitor of epidermal growth factor receptor and human epidermal growth factor receptor 2, Cancer Sci. 105 (8) (2014) 1040–1048, 10.1111/cas.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Tanaka Y, Hirata M, Shinonome S, Torii M, Nezasa KI, Tanaka H, Distribution analysis of epertinib in brain metastasis of HER2-positive breast cancer by imaging mass spectrometry and prospect for antitumor activity, Sci. Rep 8 (1) (2018) 343, 10.1038/s41598-017-18702-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Aikawa H, Hayashi M, Ryu S, Yamashita M, Ohtsuka N, Nishidate M, Fujiwara Y, Hamada A, Visualizing spatial distribution of alectinib in murine brain using quantitative mass spectrometry imaging, Sci. Rep 6 (2016) 23749, 10.1038/srep23749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Ntshangase S, Mdanda S, Singh SD, Naicker T, Kruger HG, Baijnath S, Govender T, Mass spectrometry imaging demonstrates the regional brain distribution patterns of three first-line antiretroviral drugs, ACS Omega 4 (25) (2019) 21169–21177, 10.1021/acsomega.9b02582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Shariatgorji M, Nilsson A, Fridjonsdottir E, Vallianatou T, Kallback P, Katan L, Savmarker J, Mantas I, Zhang X, Bezard E, Svenningsson P, Odell LR, Andren PE, Comprehensive mapping of neurotransmitter networks by MALDI-MS imaging, Nat. Methods 16 (10) (2019) 1021–1028, 10.1038/s41592-019-0551-3. [DOI] [PubMed] [Google Scholar]
- [98].Qian X, Song H, Ming GL, Brain organoids: advances, applications and challenges, Development 146 (8) (2019), 10.1242/dev.166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Hoffman GE, Schrode N, Flaherty E, Brennand KJ, New considerations for hiPSC-based models of neuropsychiatric disorders, Mol. Psychiatry 24 (1) (2019) 49–66, 10.1038/s41380-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Mead BE, Karp JM, All models are wrong, but some organoids may be useful, Genome Biol. 20 (1) (2019), 10.1186/s13059-019-1677-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Quadrato G, Brown J, Arlotta P, The promises and challenges of human brain organoids as models of neuropsychiatric disease, Nat. Med 22 (11) (2016) 1220–1228, 10.1038/nm.4214. [DOI] [PubMed] [Google Scholar]
- [102].Manganas LN, Dura I, Osenberg S, Semerci F, Tosun M, Mishra R, Parkitny L, Encinas JM, Maletic-Savatic M, BASP1 labels neural stem cells in the neurogenic niches of mammalian brain, Sci. Rep 11 (1) (2021) 5546, 10.1038/s41598-021-85129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Chen K, Baluya D, Tosun M, Li F, Maletic-Savatic M, Imaging mass spectrometry: a new tool to assess molecular underpinnings of neurodegeneration, Metabolites 9 (7) (2019), 10.3390/metabo9070135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kandel P, Semerci F, Mishra R, Choi W, Bajic A, Baluya D, Ma LiHua, Chen K, Cao AC, Phongmekhin T, Matinyan N, Jiménez-Panizo A, Chamakuri S, Raji IO, Chang L, Fuentes-Prior P, MacKenzie KR, Benn CL, Estébanez-Perpiñá E, Venken K, Moore DD, Young DW, Maletic-Savatic M, Oleic acid is an endogenous ligand of TLX/NR2E1 that triggers hippocampal neurogenesis, Proc. Natl. Acad. Sci. U. S. A 119 (13) (2022), e2023784119, 10.1073/pnas.2023784119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Nayek S, De Silva IW, Aguilar R, Lund AK, Verbeck GF, Toxicological alterations induced by subacute exposure of silver nanoparticles in Wistar rats, J. Appl. Toxicol 41 (6) (2021) 972–986, 10.1002/jat.4086. [DOI] [PubMed] [Google Scholar]
- [106].Hamm GR, Backstrom E, Brulls M, Nilsson A, Strittmatter N, Andren PE, Grime K, Friden M, Goodwin RJA, Revealing the regional localization and differential lung retention of inhaled compounds by mass spectrometry imaging, J Aerosol. Med. Pulm Drug Deliv 33 (1) (2020) 43–53, 10.1089/jamp.2019.1536. [DOI] [PubMed] [Google Scholar]
- [107].Yamamoto E, Taquahashi Y, Kuwagata M, Saito H, Matsushita K, Toyoda T, Sato F, Kitajima S, Ogawa K, Izutsu KI, Saito Y, Hirabayashi Y, Iimura Y, Honma M, Okuda H, Goda Y, Visualizing the spatial localization of ciclesonide and its metabolites in rat lungs after inhalation of 1-mum aerosol of ciclesonide by desorption electrospray ionization-time of flight mass spectrometry imaging, Int. J. Pharm, 595 (2021) 120241, 10.1016/j.ijpharm.2021.120241. [DOI] [PubMed] [Google Scholar]
- [108].Nilsson A, Fehniger TE, Gustavsson L, Andersson M, Kenne K, Marko-Varga G, Andrén PE, Morty RE, Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry, PLoS ONE 5 (7) (2010) e11411, 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Treu A, Kokesch-Himmelreich J, Walter K, Holscher C, Rompp A, Integrating high-resolution MALDI imaging into the development pipeline of anti-tuberculosis drugs, J. Am. Soc. Mass Spectrom 31 (11) (2020) 2277–2286, 10.1021/jasms.0c00235. [DOI] [PubMed] [Google Scholar]
- [110].Swales JG, Tucker JW, Strittmatter N, Nilsson A, Cobice D, Clench MR, Mackay CL, Andren PE, Takats Z, Webborn PJ, Goodwin RJ, Mass spectrometry imaging of cassette-dosed drugs for higher throughput pharmacokinetic and biodistribution analysis, Anal. Chem 86 (16) (2014) 8473–8480, 10.1021/ac502217r. [DOI] [PubMed] [Google Scholar]
- [111].Rompp A, Guenther S, Takats Z, Spengler B, Mass spectrometry imaging with high resolution in mass and space (HR(2) MSI) for reliable investigation of drug compound distributions on the cellular level, Anal. Bioanal. Chem 401 (1) (2011) 65–73, 10.1007/s00216-011-4990-7. [DOI] [PubMed] [Google Scholar]
- [112].Nilsson A, Goodwin RJ, Swales JG, Gallagher R, Shankaran H, Sathe A, Pradeepan S, Xue A, Keirstead N, Sasaki JC, Andren PE, Gupta A, Investigating nephrotoxicity of polymyxin derivatives by mapping renal distribution using mass spectrometry imaging, Chem. Res. Toxicol 28 (9) (2015) 1823–1830, 10.1021/acs.chemrestox.5b00262. [DOI] [PubMed] [Google Scholar]
- [113].Khatib-Shahidi S, Andersson M, Herman JL, Gillespie TA, Caprioli RM, Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry, Anal. Chem 78 (18) (2006) 6448–6456, 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- [114].Merdas M, Lagarrigue M, Umbdenstock T, Lhumeau A, Dartiguelongue F, Vanbellingen Q, Da Violante G, Pineau C, Study of the distribution of acetaminophen and its metabolites in rats, from the whole-body to isolated organ levels, by matrix-assisted laser desorption/ionization mass spectrometry imaging after on-tissue chemical derivatization, Anal. Chem 93 (39) (2021) 13242–13250, 10.1021/acs.analchem.1c02487. [DOI] [PubMed] [Google Scholar]
- [115].Reichel A, Lienau P, Pharmacokinetics in drug discovery: an exposure-centred approach to optimising and predicting drug efficacy and safety, Handb. Exp. Pharmacol 232 (2016) 235–260, 10.1007/164_2015_26. [DOI] [PubMed] [Google Scholar]
- [116].Zhang Z, Tang W, Drug metabolism in drug discovery and development, Acta Pharm. Sin. B 8 (5) (2018) 721–732, 10.1016/j.apsb.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Lanao JM, Fraile MA, Drug tissue distribution: study methods and therapeutic implications, Curr. Pharm. Des 11 (29) (2005) 3829–3845, 10.2174/138161205774580679. [DOI] [PubMed] [Google Scholar]
- [118].Mouton JW, Theuretzbacher U, Craig WA, Tulkens PM, Derendorf H, Cars O, Tissue concentrations: do we ever learn? J. Antimicrob. Chemother 61 (2) (2008) 235–237, 10.1093/jac/dkm476. [DOI] [PubMed] [Google Scholar]
- [119].Castellino S, Groseclose MR, Wagner D, MALDI imaging mass spectrometry: bridging biology and chemistry in drug development, Bioanalysis 3 (21) (2011) 2427–2441, 10.4155/bio.11.232. [DOI] [PubMed] [Google Scholar]
- [120].Pellegatti M, Pagliarusco S, Drug and metabolite concentrations in tissues in relationship to tissue adverse findings: a review, Expert. Opin. Drug Metab. Toxicol 7 (2) (2011) 137–146, 10.1517/17425255.2011.545053. [DOI] [PubMed] [Google Scholar]
- [121].Cornett DS, Reyzer ML, Chaurand P, Caprioli RM, MALDI imaging mass spectrometry: molecular snapshots of biochemical systems, Nat. Methods 4 (10) (2007) 828–833, 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- [122].Ganesh S, Almazroo OA, Tevar A, Humar A, Venkataramanan R, Drug metabolism, drug interactions, and drug-induced liver injury in living donor liver transplant patients, Clin. Liver. Dis 21 (1) (2017) 181–196, 10.1016/j.cld.2016.08.011. [DOI] [PubMed] [Google Scholar]
- [123].Almazroo OA, Miah MK, Venkataramanan R, Drug metabolism in the liver, Clin. Liver Dis 21 (1) (2017) 1–20, 10.1016/j.cld.2016.08.001. [DOI] [PubMed] [Google Scholar]
- [124].Baillie TA, Future of toxicology-metabolic activation and drug design: challenges and opportunities in chemical toxicology, Chem Res Toxicol 19 (7) (2006) 889–893, 10.1021/tx060062o. [DOI] [PubMed] [Google Scholar]
- [125].Kalgutkar AS, Dalvie D, Predicting toxicities of reactive metabolite-positive drug candidates, Annu. Rev. Pharmacol. Toxicol 55 (2015) 35–54, 10.1146/annurev-pharmtox-010814-124720. [DOI] [PubMed] [Google Scholar]
- [126].Neftel KA, Woodtly W, Schmid M, Frick PG, Fehr J, Amodiaquine induced agranulocytosis and liver damage, Br. Med. J. (Clin. Res. Ed.) 292 (6522) (1986) 721–723, 10.1136/bmj.292.6522.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Olliaro P, Nevill C, LeBras J, Ringwald P, Mussano P, Garner P, Brasseur P, Systematic review of amodiaquine treatment in uncomplicated malaria, Lancet 348 (9036) (1996) 1196–1201, 10.1016/S0140-6736(96)06217-4. [DOI] [PubMed] [Google Scholar]
- [128].Grove KJ, Hoque S, Rudewicz PJ, Investigation of amodiaquine localization in liver lobules using matrix-assisted laser desorption/ionization imaging mass spectrometry, Rapid Commun. Mass Spectrom 33 (3) (2019) 252–258, 10.1002/rcm.8339. [DOI] [PubMed] [Google Scholar]
- [129].Lindros KO, Zonation of cytochrome P450 expression, drug metabolism and toxicity in liver, Gen. Pharmacol 28 (2) (1997) 191–196, 10.1016/s0306-3623(96)00183-8. [DOI] [PubMed] [Google Scholar]
- [130].Sezgin S, Hassan R, Zuhlke S, Kuepfer L, Hengstler JG, Spiteller M, Ghallab A, Spatio-temporal visualization of the distribution of acetaminophen as well as its metabolites and adducts in mouse livers by MALDI MSI, Arch. Toxicol 92 (9) (2018) 2963–2977, 10.1007/s00204-018-2271-3. [DOI] [PubMed] [Google Scholar]
- [131].Brown H, Mesa Sanchez D, Yin R, Chen B, Vavrek M, Cancilla M, Zhong W, Shyong B, Zhang R, Li F, Laskin J, Mass spectrometry imaging of diclofenac and its metabolites in tissues using nanospray desorption electrospray ionization, ChemRxiv, 2020. Doi: 10.26434/chemrxiv.13194422.v1. [DOI] [PubMed] [Google Scholar]
- [132].Lu Y, Zhao X-M, Hu Z, Wang L, Li F, LC–MS-based metabolomics in the study of drug-induced liver injury, Curr. Pharmacol. Rep 5 (1) (2019) 56–67, 10.1007/s40495-018-0144-3. [DOI] [Google Scholar]
- [133].Malarkey DE, Johnson K, Ryan L, Boorman G, Maronpot RR, New insights into functional aspects of liver morphology, Toxicol. Pathol 33 (1) (2005) 27–34, 10.1080/01926230590881826. [DOI] [PubMed] [Google Scholar]
- [134].Planas-Paz L, Orsini V, Boulter L, Calabrese D, Pikiolek M, Nigsch F, Xie Y, Roma G, Donovan A, Marti P, Beckmann N, Dill MT, Carbone W, Bergling S, Isken A, Mueller M, Kinzel B, Yang YI, Mao X, Nicholson TB, Zamponi R, Capodieci P, Valdez R, Rivera D, Loew A, Ukomadu C, Terracciano LM, Bouwmeester T, Cong F, Heim MH, Forbes SJ, Ruffner H, Tchorz JS, The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver zonation and size, Nat. Cell Biol, 18 (5) (2016) 467–479, 10.1038/ncb3337. [DOI] [PubMed] [Google Scholar]
- [135].Zeng T, Guo W, Jiang L, Luo Q, Shi Z, Lei B, Zhang J, Cai Z, Integration of omics analysis and atmospheric pressure MALDI mass spectrometry imaging reveals the cadmium toxicity on female ICR mouse, Sci. Total Environ 801 (2021) 149803, 10.1016/j.scitotenv.2021.149803. [DOI] [PubMed] [Google Scholar]
- [136].Bhandari N, Figueroa DJ, Lawrence JW, Gerhold DL, Phospholipidosis assay in HepG2 cells and rat or rhesus hepatocytes using phospholipid probe NBD-PE, Assay Drug Dev. Technol 6 (3) (2008) 407–419, 10.1089/adt.2007.119. [DOI] [PubMed] [Google Scholar]
- [137].Ferey J, Larroque M, Schmitz-Afonso I, Le Maitre J, Sgarbura O, Carrere S, Quenet F, Bouyssiere B, Enjalbal C, Mounicou S, Afonso C, Imaging matrix-assisted laser desorption/ionization fourier transform ion cyclotron resonance mass spectrometry of oxaliplatin derivatives in human tissue sections, Talanta 237 (2022) 122915, 10.1016/j.talanta.2021.122915. [DOI] [PubMed] [Google Scholar]
- [138].Nishimura M, Hayashi M, Mizutani Y, Takenaka K, Imamura Y, Chayahara N, Toyoda M, Kiyota N, Mukohara T, Aikawa H, Fujiwara Y, Hamada A, Minami H, Distribution of erlotinib in rash and normal skin in cancer patients receiving erlotinib visualized by matrix assisted laser desorption/ionization mass spectrometry imaging, Oncotarget 9 (26) (2018) 18540–18547, 10.18632/oncotarget.24928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Prideaux B, Via LE, Zimmerman MD, Eum S, Sarathy J, O’Brien P, Chen C, Kaya F, Weiner DM, Chen PY, Song T, Lee M, Shim TS, Cho JS, Kim W, Cho SN, Olivier KN, Barry CE 3rd, Dartois V, The association between sterilizing activity and drug distribution into tuberculosis lesions, Nat. Med 21 (10) (2015) 1223–1227, 10.1038/nm.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Chakravarty R, Goel S, Valdovinos HF, Hernandez R, Hong H, Nickles RJ, Cai W, Matching the decay half-life with the biological half-life: ImmunoPET imaging with (44)Sc-labeled cetuximab Fab fragment, Bioconjug. Chem 25 (12) (2014) 2197–2204, 10.1021/bc500415x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Zhu L, Glick J, Flarakos J, Bioanalytical challenges in support of complex modalities of antibody-based therapeutics, AAPS J. 22 (6) (2020) 130, 10.1208/s12248-020-00517-1. [DOI] [PubMed] [Google Scholar]
- [142].Ait-Belkacem R, Berenguer C, Villard C, Ouafik L, Figarella-Branger D, Beck A, Chinot O, Lafitte D, Monitoring therapeutic monoclonal antibodies in brain tumor, MAbs 6 (6) (2014) 1385–1393, 10.4161/mabs.34405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Debois D, Bertrand V, Quinton L, De Pauw-Gillet MC, De Pauw E, MALDI-in source decay applied to mass spectrometry imaging: a new tool for protein identification, Anal. Chem 82 (10) (2010) 4036–4045, 10.1021/ac902875q. [DOI] [PubMed] [Google Scholar]
- [144].Fujiwara Y, Furuta M, Manabe S, Koga Y, Yasunaga M, Matsumura Y, Imaging mass spectrometry for the precise design of antibody-drug conjugates, Sci. Rep 6 (2016) 24954, 10.1038/srep24954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Roberts TC, Langer R, Wood MJA, Advances in oligonucleotide drug delivery, Nat. Rev. Drug. Discov 19 (10) (2020) 673–694, 10.1038/s41573-020-0075-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Zanardi TA, Han SC, Jeong EJ, Rime S, Yu RZ, Chakravarty K, Henry SP, Pharmacodynamics and subchronic toxicity in mice and monkeys of ISIS 388626, a second-generation antisense oligonucleotide that targets human sodium glucose cotransporter 2, J. Pharmacol. Exp. Ther 343 (2) (2012) 489–496, 10.1124/jpet.112.197426. [DOI] [PubMed] [Google Scholar]
- [147].Nakashima Y, Setou M, Distribution of antisense oligonucleotides in rat eyeballs using MALDI imaging mass spectrometry, Mass Spectrom. (Tokyo) 7 (1) (2018) A0070, 10.5702/massspectrometry.A0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Yokoi H, Kasahara Y, Obika S, Doi T, Kamada H, Development of a detection method for antisense oligonucleotides in mouse kidneys by matrix-assisted laser desorption/ionization imaging mass spectrometry, Rapid Commun. Mass Spectrom 32 (23) (2018) 1984–1990, 10.1002/rcm.8274. [DOI] [PubMed] [Google Scholar]
- [149].Zavalin A, Todd EM, Rawhouser PD, Yang J, Norris JL, Caprioli RM, Direct imaging of single cells and tissue at sub-cellular spatial resolution using transmission geometry MALDI MS, J. Mass Spectrom 47 (11) (2012) i, 10.1002/jms.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Zavalin A, Yang J, Hayden K, Vestal M, Caprioli RM, Tissue protein imaging at 1 mum laser spot diameter for high spatial resolution and high imaging speed using transmission geometry MALDI TOF MS, Anal. Bioanal. Chem 407 (8) (2015) 2337–2342, 10.1007/s00216-015-8532-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Thurber GM, Yang KS, Reiner T, Kohler RH, Sorger P, Mitchison T, Weissleder R, Single-cell and subcellular pharmacokinetic imaging allows insight into drug action in vivo, Nat. Commun 4 (2013) 1504, 10.1038/ncomms2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Laughney AM, Kim E, Sprachman MM, Miller MA, Kohler RH, Yang KS, Orth JD, Mitchison TJ, Weissleder R, Single-cell pharmacokinetic imaging reveals a therapeutic strategy to overcome drug resistance to the microtubule inhibitor eribulin, Sci. Transl. Med 6 (261) (2014) 261ra152, 10.1126/scitranslmed.3009318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Van de Plas R, Yang J, Spraggins J, Caprioli RM, Image fusion of mass spectrometry and microscopy: a multimodality paradigm for molecular tissue mapping, Nat. Methods 12 (4) (2015) 366–372, 10.1038/nmeth.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Abdelmoula WM, Regan MS, Lopez BGC, Randall EC, Lawler S, Mladek AC, Nowicki MO, Marin BM, Agar JN, Swanson KR, Kapur T, Sarkaria JN, Wells W, Agar NYR, Automatic 3D nonlinear registration of mass spectrometry imaging and magnetic resonance imaging data, Anal. Chem 91 (9) (2019) 6206–6216, 10.1021/acs.analchem.9b00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Grelard F, Legland D, Fanuel M, Arnaud B, Foucat L, Rogniaux H, Esmraldi: efficient methods for the fusion of mass spectrometry and magnetic resonance images, BMC Bioinf. 22 (1) (2021) 56, 10.1186/s12859-020-03954-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Verbeeck N, Spraggins JM, Murphy MJM, Wang HD, Deutch AY, Caprioli RM, Van de Plas R, Connecting imaging mass spectrometry and magnetic resonance imaging-based anatomical atlases for automated anatomical interpretation and differential analysis, Biochim. Biophys. Acta Proteins Proteom 1865 (7) (2017) 967–977, 10.1016/j.bbapap.2017.02.016. [DOI] [PubMed] [Google Scholar]
- [157].Tuck M, Blanc L, Touti R, Patterson NH, Van Nuffel S, Villette S, Taveau JC, Rompp A, Brunelle A, Lecomte S, Desbenoit N, Multimodal imaging based on vibrational spectroscopies and mass spectrometry imaging applied to biological tissue: a multiscale and multiomics review, Anal. Chem 93 (1) (2021) 445–477, 10.1021/acs.analchem.0c04595. [DOI] [PubMed] [Google Scholar]
- [158].Strnad S, Prazienkova V, Sykora D, Cvacka J, Maletinska L, Popelova A, Vrkoslav V, The use of 1,5-diaminonaphthalene for matrix-assisted laser desorption/ionization mass spectrometry imaging of brain in neurodegenerative disorders, Talanta 201 (2019) 364–372, 10.1016/j.talanta.2019.03.117. [DOI] [PubMed] [Google Scholar]
- [159].van Hove ER, Smith DF, Fornai L, Glunde K, Heeren RM, An alternative paper based tissue washing method for mass spectrometry imaging: localized washing and fragile tissue analysis, J. Am. Soc. Mass Spectrom 22 (10) (2011) 1885–1890, 10.1007/s13361-011-0203-z. [DOI] [PMC free article] [PubMed] [Google Scholar]