Abstract
Background
Sarcopenia is defined as an age‐related progressive loss of muscle mass and/or strength. Although different factors can contribute to this disease, the underlying mechanisms remain unclear. We assessed transcriptional heterogeneity in skeletal muscles from sarcopenic and control mice at single‐cell resolution.
Methods
A mouse model was established to study sarcopenic skeletal muscles. Single‐cell RNA‐seq was performed on tibialis anterior (TA) muscle cells collected from sarcopenic and control mice. A series of bioinformatic analyses were carried out to identify and compare different cell types under different conditions. Immunofluorescence staining and western blotting were used to validate the findings from single‐cell experiments. Tube formation assays were conducted to further evaluate the effects of Gbp2 on endothelial cells during angiogenesis.
Results
A murine sarcopenia model was successfully established using a senescence‐accelerated mouse strain (SAMP6, n = 5). Sarcopenia phenotype was induced by administration of dexamethasone (20 mg/kg) and reduced physical activity. Senescence‐resistant mice strain (SAMR1) and SAMP6 strain with similar activity but injected with PBS were recruited as two control groups. As signs of sarcopenia, body weight, muscle cell counts and cross‐sectional fibre area were all significantly decreased in sarcopenic mice (P value = 0.004, 0.03 and 0.035, respectively). After quality control, 13 612 TA muscle single‐cell transcriptomes were retained for analysis. Fourteen cell clusters were identified from the profiled cells. Among them, two distinct endothelial subtypes were found to be dominant in the sarcopenia group (42.2% cells) and in the two control groups (59.1% and 47.9% cells), respectively. 191 differentially expressed genes were detected between the two endothelial subtypes. Sarcopenia‐specific endothelial cell subtype exhibited a dramatic increase in the interferon family genes and the interferon‐inducible guanylate‐binding protein (GBP) family gene expressions. For example, Igtp and Gbp2 in sarcopenic endothelial cells were 5.4 and 13.3 times higher than those in the control groups, respectively. We further validated our findings in muscle specimens of sarcopenia patients and observed that GBP2 levels were increased in endothelial cells of a subset of patients (11 of 40 patients, 27.5%), and we identified significantly higher CD31 and GBP2 co‐localization (P value = 0.001128). Finally, we overexpressed Gbp2 in human umbilical vein endothelial cells in vitro. The endothelial cells with elevated Gbp2 expression displayed compromised tube formation.
Conclusions
Our single‐cell‐based results suggested that endothelial cells may play critical roles in sarcopenia development through interferon‐GBP signalling pathways, highlighting new therapeutic directions to slow down or even reverse age‐related sarcopenia.
Keywords: Sarcopenia, Skeletal muscle, Single‐cell RNA‐seq, Endothelial cell, Interferon, Guanylate‐binding protein
Introduction
Sarcopenia is a condition described by progressive decline in skeletal muscle mass and/or muscle strength. The clinical consequences of sarcopenia include falls, fractures, decreased mobility, or even death. 1 As age is one of the key risk factors for this syndrome, sarcopenia is increasingly considered to be a critical public health challenge in the ageing population. 2 The estimated direct healthcare cost for sarcopenia was $18.5 billion in the USA in 2000 3 and increased to $40.4 billion in a 2019 estimate, 4 amounting to a considerable economic burden for the healthcare system. Therefore, there is an urgent need to understand this disease and develop novel preventive and therapeutic approaches.
Many efforts have been devoted to understand the mechanisms driving sarcopenia. Age‐related changes have been observed to be involved in sarcopenia pathogenesis, such as impaired muscle cell regeneration, chronic inflammatory microenvironment in skeletal muscles, altered cell–cell communication and mitochondrial dysfunction. 5 , 6 Transcriptome studies have been performed to elucidate the molecular mechanisms underlying sarcopenia. For example, Giresi et al. identified the increasing of genes which were involved in mediating cellular responses to inflammation and apoptosis in skeletal muscles of sarcopenia patients. 7 Borsch et al. found that a rodent sarcopenia model recapitulated mitochondrial changes and inflammatory responses observed in human sarcopenia. 8 However, most of these studies used microarray or bulk RNA‐seq, which can only reflect the average gene expression of a mixture of various cell types.
Single‐cell RNA‐seq (scRNA‐seq) technology makes it possible to quantify gene expression at single‐cell resolution, allowing us to study transcriptome changes in different cell types from a single sample without experimentally sorting the cell types. Several pioneering studies have been conducted to use single‐cell technology in sarcopenia‐related research. Zhang et al. identified that expression of genes related to interferon‐gamma responses is decreased during muscle regeneration after tissue injury in aged mice. Following scRNA‐seq, these authors discovered that a novel macrophage subset might be responsible for such changes. 5 In another study, researchers used single‐nuclei sequencing to investigate age‐related alterations in muscle cell composition. 9 However, a direct comparison between sarcopenic and normal skeletal muscle at single‐cell resolution is still lacking.
In this study, we established a mouse model of sarcopenia and studied the molecular profiling differences between sarcopenic skeletal muscle and control samples using scRNA‐seq. We revealed extensive cellular heterogeneity in skeletal muscles and found that endothelial cells exhibited the greatest difference between sarcopenic and control biopsies. Expression of interferon signalling pathway genes and interferon‐inducible guanylate‐binding protein (GBP) family genes was drastically increased in sarcopenic‐specific endothelial cells. We then validated these findings using muscle specimens from human sarcopenia patients. The Gbp2 gene was further investigated for its function in endothelial cell tube formation. Our work provided the first reference single‐cell transcriptomic atlas for murine skeletal muscle under sarcopenia as well as steady conditions and suggested potential new therapeutic targets for age‐related muscle loss and sarcopenia.
Methods
Animal model and clinical samples
The mice were handled according to European Community guidelines. The senescence accelerated mouse prone 6 (SAMP6) and senescence mouse resistant 1 (SAMR1) were purchased from the Department of Experimental Animal Science, Peking University Medical Center. All experimental animal protocols were approved by the Kunming Medical University Ethics Committee for Animal Experimentation. In this study, 5‐month‐old SAMP6 and SAMR1 mice were used. Animals were exposed to a standard 12:12‐h light/dark cycle with reduced activity space and normal chow. Ten SAMP6 mice were divided into a sham control group (P6Sham) and an experimental group (P6DX), with five mice in each group. Five SAMR1 mice were used as a control group (control). To induce sarcopenia phenotypes, all mice were housed in limited space to reduce their physical activities. Dexamethasone (20 mg/kg) was injected intraperitoneally into the P6DX group, whereas PBS was injected into the P6Sham and control groups. All mice were injected for 7 consecutive days and weighed at the same time each day.
All clinical specimens were collected at the Sixth Affiliated Hospital of Kunming Medical University and were approved by the Ethics Committee of the Sixth Affiliated Hospital of Kunming Medical University. All patients signed an informed consent form. All samples were obtained from discarded muscle tissue that had been removed during joint replacement or lumbar surgery.
HE staining, muscle cell counting and cross‐sectional area quantification
Muscle tissues were immersed in 4% paraformaldehyde for 7 days, dehydrated using a sucrose gradient solution (20%, 30% and 40% sucrose/4% paraformaldehyde) and then immersed in optimal cutting temperature (OCT) medium for 12 h for embedding. After embedding, the transverse sections (8–12 μm thick) were cut and used for haematoxylin and eosin (H&E) staining.
Acquired images were analysed using Image‐Pro Plus v.6.0 (Media Cybernetics, USA). The numbers of myofibres in each field of view were counted separately using microns as the standard unit. The total area of myofibres in each field of view was measured, and the cross‐sectional area of was calculated as the total area of myofibres/number of myofibres.
Western blotting
Nuclear and cytoplasmic fractions were extracted from tissues and cells using the Nuclear and Cytoplasmic Protein Extraction Kit (Solarbio, R0010), subjected to 12% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes. After blocking, membranes were incubated overnight with antibodies against GBP2 (Proteintech, SA00001‐2) (diluted 1:2000) and GAPDH (Cell Signaling, 97166) (diluted 1:2000). The membranes were then incubated with HPR‐conjugated goat anti‐rabbit IgG (Proteintech, SA00001‐2) (diluted 1:2000) for 2 h at room temperature. The blots were developed using an enhanced chemiluminescence kit (Millipore, Billerica, MA, USA), and bands were quantified using a Bio‐Rad imaging system (Hercules, CA, USA).
Immunofluorescent staining
Similar to H&E staining, muscle tissues of mice and humans were immersed in paraformaldehyde for 7 days with sucrose gradient dehydration and then embedded in OCT medium. The tissues were cut into 10‐μm sections using a frozen microtome, and immunofluorescent staining was performed after preparation. Briefly, the samples were washed three times with 0.1% Triton X‐100 in PBS (0.1% PBST) for 10 min each. The samples were permeabilized using 500 μL of 0.3% Triton X‐100 in PBS for 20 min at room temperature. The samples were again washed three times with 0.1% PBST for 30 min each. Next, 10% goat serum in 0.1% PBST was added to the samples for 2 h at room temperature, followed by 200 μL primary antibody cocktail, which was incubated overnight at room temperature. The following primary antibody cocktails in 10% goat serum in 0.1% PBST were used: Gbp2 (Proteintech, 11854‐1‐AP) (diluted 1:500) and CD31 (Abcam, ab24590). The samples were washed three times with 0.1% PBST for 10 min each. The samples were then incubated with the secondary antibodies for 2 h at room temperature. The secondary antibodies, each used at 1:1000 dilution, were as follows: goat anti‐mouse IgG AF555 (Abcam, ab150118) and goat anti‐rabbit IgG AF488 (Abcam, ab150081). After incubation for 2 h, the samples were washed four times with 0.1% PBST for 10 min each. Finally, a drop of mounting medium with DAPI (Abcam, ab104139) was added to seal the samples. The samples were stored at 4°C away from light until imaging. Samples were imaged using a confocal microscope (LSM880, ZEISS) and SP5 (LEICA).
GBP2 over‐expression in human endothelial cells and tube formation assay
Human umbilical vein endothelial cells (HUVECs) were donated by the Science and Technology Achievements Incubation Center of Kunming Medical University. HUVECs were cultured up to passage 4 (P4) in basal medium: high‐glucose Dulbecco's modified Eagle's medium (DMEM; Gibco, 11995073) supplemented with 10% fetal bovine serum (FBS; VWR, 142D19) and 1% penicillin/streptomycin (Sigma‐Aldrich, P4458) at 37°C in humidified air with 5% CO2. A plasmid over‐expression system (GenePharma, China) was used for Gbp2 over‐expression (Figure S1). HUVECs were transfected with GP‐transfected Mate (GenePharma). Expression of GBP2 protein was verified via western blotting.
In tube formation assays, endothelial cells were cultured in a gel of membrane extract, and their ability to form tube‐like structures was assessed. The growth factor‐reduced basement membrane extract Matrigel matrix (Corning, 356234) was used as a culture substratum. Briefly, a thin layer of Matrigel (50 μL) was placed over pre‐chilled 96‐well culture plates until completely solidified. Trypsinized HUVECs resuspended in DMEM medium with only PBS were added to the preconditioned culture plates at a density of 2 2*10^4cells/well. At 8 h, randomly chosen visual fields per well were photographed, and the length and area of the network of connected cells (tube formation) were measured using ImageJ 2.0 software. Tube (capillary‐like) formation was defined as a ring‐like structure with a length/width ratio of 4.
Single‐cell RNA‐seq and analysis
Tibialis anterior (TA) muscles were dissected from each mouse; single‐cell suspensions were obtained by enzymatic digestion. Briefly, muscle biopsies were minced and digested with Collagenase II, Dispase II, DNAnase I solution at 37°C; dissociated cells were filtered and resuspended in PBS. Red Blood Cell Lysis Solution (Miltenyi Biotec, Germany) was applied to further remove erythrocytes. After removal, cells were washed and resuspended in PBS. The cell suspension was loaded onto the Chromium single‐cell controller (10X Genomics) to generate single‐cell gel beads in the emulsion according to the manufacturer's protocol. Single‐cell 3′ libraries were prepared for captured cells using Gel Bead Kit V3. The libraries were finally sequenced using an Illumina Novaseq 6000 sequencer with a sequencing depth of at least 100 000 reads per cell with pair‐end 150‐bp reading strategy.
Raw sequencing data were processed by Cell Ranger version 4.0 (10X Genomics). Alignment (to mouse mm10 reference genome), filtering, barcode counting and UMI counting were performed with cellranger count module to generate gene expression matrix. scRNA‐seq data analysis was performed using Seurat R package (Version 3.2.0). 10 In quality control step, cells were filter by the following criteria: (1) mitochondrial abundance <15%, (2) minimum gene detected >800 and (3) maximum gene detected <99 percentile among all cells (18 730 genes). Principal component analysis was applied on the highly variable genes and cells were projected and visualized on 2D dimensions using uniform manifold approximation and projection (UMAP) on the first 20 PCs. Cells were clustered by Louvain algorithm implemented in Seurat. Gene set enrichment analysis (GSEA) were performed using fgsea R package. 11 Cell–cell interaction was inferred using CellChat R package. 12
Results
Successful establishment of a murine sarcopenia model
We first established a mouse sarcopenia model according to previous reports 13 , 14 , 15 (Figure 1A, left). Briefly, the SAMP6 mice with accelerated ageing phenotypes were induced sarcopenia by reduced physical activity and dexamethasone injection (P6DX group). SAMR1 and SAMP6 mice with similar physical activity strength and PBS injection were used as two control groups (R1 control and P6Sham, respectively). The body weights of P6DX sarcopenia mice were significantly reduced after induction (Figure 1B; P value = 0.004, one‐way ANOVA). The muscle cell counts and cross‐sectional fibre area were also both decreased in P6DX group (Figure 1C and 1D; P value = 0.03 and 0.035, respectively, one‐way ANOVA). H&E staining on skeletal muscle presented clearly loosed muscle cell structure in P6DX group (Figure 1E). By immunofluorescence staining, we confirmed that type1 slow muscle fibres were enriched in P6DX (Figure 1F), indirectly indicating the loss of type 2 fibres, which was a signature of aged skeletal muscle. Taking these phenotype evaluations together, we successfully established a mice model that recapitulates sarcopenia.
Figure 1.
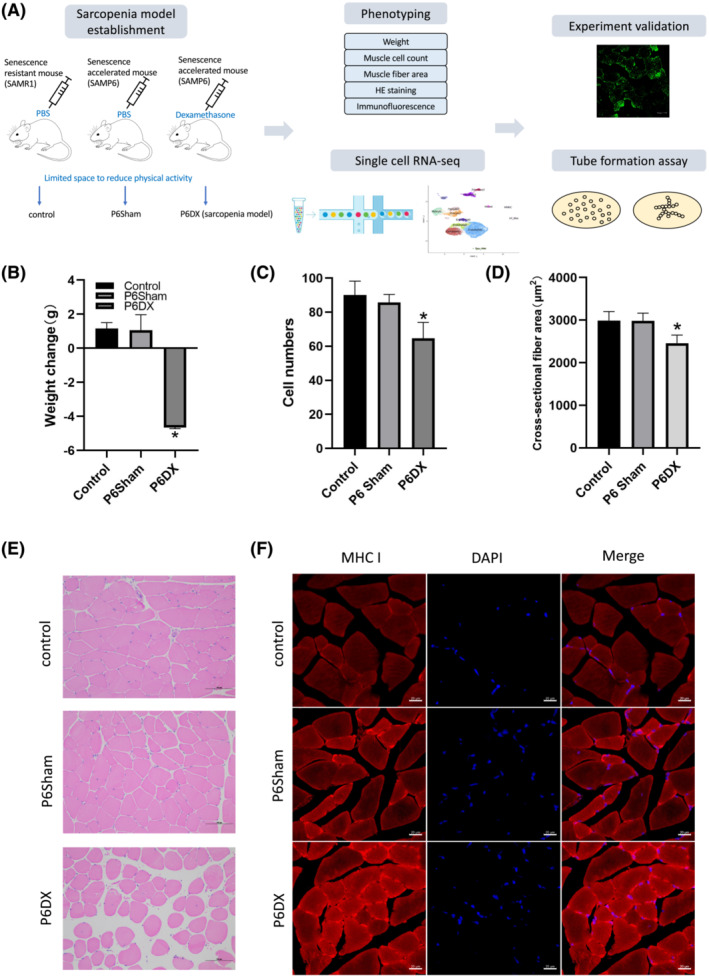
Establishment and evaluation of a murine sarcopenia model. (A) A scheme to illustrate the workflow of this study. A mouse model for sarcopenia is established and evaluated by different criteria. TA biopsies from sarcopenia mice (n = 5) as well as from mice in two other control groups (n = 5 each group) are subject to single‐cell transcriptome analysis. Findings from scRNA‐seq are validated by in vivo and in vitro experiments. (B–D) Criteria to evaluate sarcopenia mice model. The body weights, muscle cell counts and cross‐sectional fibre area are significantly decreased in P6DX sarcopenia mice group during the sarcopenia establishment procedure. (E) HE‐stained TA muscle of R1 control, P6Sham and P6DX groups; the muscle cell structures are loose in P6DX sarcopenia sample. Scale bar represents 100 μm. (F) Immunofluorescence staining of the three experimental groups. Myosin heavy chain I (MCH I) is increased in P6DX sarcopenia sample. Scale bar represents 20 μm.
Single‐cell transcriptome analysis revealed diverse cellular composition in murine skeletal muscle
We performed scRNA‐seq on mice skeletal muscle biopsies collected from three experiment groups: (1) R1 control group (control): SAMR1 + PBS, (2) sham group (P6Sham): SAMP6 + PBS and (3) sarcopenia group (P6DX): SAMP6 + dexamethasone. Each group contained five mice. We first established a cell type atlas for mouse skeletal muscles (Figure 2A and 2B). After quality control and data pre‐processing, 13,612 cells (4,762 control, 4,250 P6Sham, 4,600 P6DX) were retained for downstream analysis. In total, 14 clusters were identified from the cell population; each corresponded to a unique cell type or cell subtype. Cell type identities were assigned to each cluster by canonical markers from previous publications (Figure 2C and Table S1).
Figure 2.
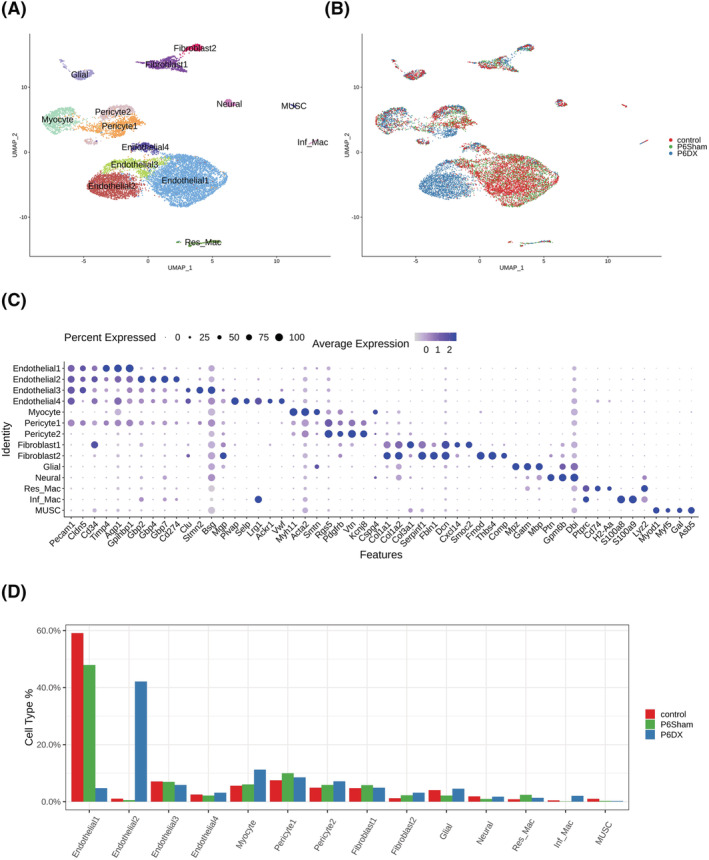
Cell types identified from skeletal muscle biopsies by scRNA‐seq. (A) Cell atlas of skeletal muscle from sarcopenia and control mice presented by UMAP. Fourteen cell types are revealed from single‐cell transcriptome data. (B) Transcriptomic atlas coloured by experiment groups. Except endothelial1 and endothethlial2, different experiment groups largely overlap in different cell clusters. (C) Marker genes used to annotate cell types. Dot size represents % of cells of that cluster expressing the given gene, whereas colour indicates the expression level of that cluster. (D) Bar plot depicting the % of each cell type in each group; endothelial1 is enriched control and P6Sham groups, whereas endothelial2 is enriched in P6DX group.
Four distinct endothelial cell subtypes were revealed from the profiled cells. Endothelial1 expressed pan‐endothelial cell markers Pecam1, Cldn5 and Cd34 16 , 17 ( Figure S2A). Endothelial2, in addition to pan‐endothelial markers, expressed high levels of GBP family genes, such as Gbp2, Gbp4 and Gbp7, and interferon signalling pathway genes, such as Ifit3, Ifi47 and Ifit1 ( Figure S2B), suggesting an active interplay between GBP and interferons in Endothelial2. Endothelial3 expressed Clu and Stmn2 ( Figure S2C), hinting their arterial origins. 18 , 19 Endothelial4 cells were marked by Plvap, Selp and Vwf expressions ( Figure S2D), indicating that they were from Weibel–Palade bodies (WPBs), the storage granules of endothelial cells. 18
Two pericyte and two fibroblast subtypes were also identified, respectively. Pericyte1 and Pericyte2 both expressed canonical pericyte markers Pdgfrb, Rgs5 and Vtn. 20 Pericyte1 also had high expression level of Pecam1 (Cd31), which was largely absent in Pericyte2, whereas Pericyte2 had slightly higher expression of Cspg4 (Ng2) ( Figure S2E). Pan‐fibroblast markers such as collagen genes (Col1a1, Col1a2, Col3a1) and Serpinf1, Fbln1 and DcnP were highly expressed in Fibroblast1 and Fibroblast2. 21 Fibroblast1 can be further characterized by expression of Cxcl14 and Smoc2. Autocrine CXCL14 can stimulate fibroblasts migration and ERK‐dependent proliferation. 22 Fibroblast2 was likely to be skeletal muscle perimysial cells, marked by expression of Fmod, Thbs4 and Comp ( Figure S2F).
A cluster of muscle cells (Myocyte) was identified by myosin gene Myh11 and alpha smooth muscle actin Acta2 gene ( Figure S2G). Glial cell and Neural cell were annotated by Mpz, Gatm, Mbp and Ptn, Gpm6b, Dbi, respectively ( Figure S2H and S2I). Two macrophage groups were also observed in the cell population. Resident Macrophage (Res_Mac) expressed Cd74 and histocompatibility complex HLA genes (H2‐Aa), and inflammatory macrophage (Inf_Mac) expressed pro‐inflammatory molecules S100a8 and S100a9 ( Figure S2J). Finally, we recognized a small cluster of muscle stem cells (MUSC), with unique expression of Myod1 and Myf5 ( Figure S2K).
Among the 14 cell clusters, Endothelial1 and Endothelial2 were the most abundant cell types, Endothelial1 constituted 59.1% and 47.9% of control and P6Sham group cells, respectively, and Endothelial2 constituted 42.2% of P6DX group cells. Endothelial1 and Endothelial2 showed high sample origin bias, 96.1% Endothelial1 cells were from control and P6Sham groups, and 96.5% Endothelial2 cells were from P6DX group (Figure 2B and 2D). Both Endothelial1 and Endothelial2 expressed canonical endothelial markers but formed clearly separated cell clusters, suggesting that endothelial cells from sarcopenia samples had distinct transcriptome profiles from homoeostatic status. More systemic comparison of these two endothelial groups will be conducted in the next section. Except Endothelial1 and Endothelial2, cells from the different experiment groups were largely overlapped in each of the cell clusters (Figure 2B). We compared the cell type composition among the three experiment groups (Figure 2D). The % of myocyte in P6DX group (11.3%) was ~2‐fold of that in control (5.6%) and P6Sham (6.1%) group. But the % of muscle stem cells was much lower in P6Sham (0.3%) and P6DX (0.2%) groups compared with control group (1.0%). Moreover, inflammatory macrophages were mostly from P6DX group.
Distinct endothelial cell subtypes were observed from different experiment groups
In the previous section, we observed that the major Endothelial cell cluster of P6DX group (Endothelial2) was distinct from that in control and P6Sham groups (Endothelial1), suggesting that endothelial cells may play an important role in sarcopenia. To further compare the difference between Endothelial1 and Endothelial2, we assessed the differentially expressed (DE) genes (|Fold Change| > 1.5 and adjusted P value < 0.05) between Endothelial1 and Endothelial2 (Figure 3A). Totally, 191 DE genes were identified; compared with Endothelial1, 141 genes were increased in Endothelial2; and 50 genes were decreased (Table S2). The top10 increased and decreased genes were visualized in Figure 3B. Among the top 20 increased genes in Endothelial2, six were GBPs (Gbp2, Gbp4, Gbp7, Gbp6, Gbp3 and Gbp5), and six were interferon signalling pathway related genes (Igtp, Ifit3, Ifi47, Ifit1, Iigp1, Ifitm3). For example, the expression level of Gbp2 in Endothelial2 was 13.3‐fold of that in Endothelial1; 89% cells in Endothelial2 were detected to express Gbp2 versus only 30% in Endothelial1. GBPs are an interferon inducible subfamily of guanosine triphosphatases (GTPases); previously, they were recognized to serve as major protective hubs both inside and outside of the immune system. 23 Our result suggested that interferon‐induced GBPs may have a novel role in sarcopenia pathogenesis.
Figure 3.
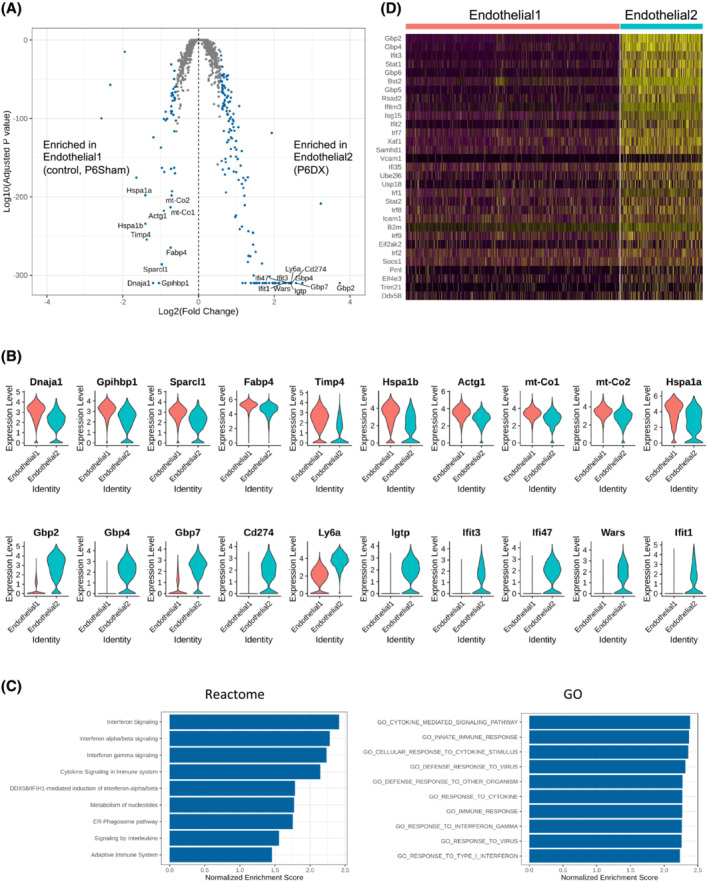
Distinct endothelial cell clusters between experiment groups. (A) Volcano plot showing differentially expressed genes between endothelial1 and endothelial2 cell clusters. Differentially expressed (DE) genes (|fold change| > 1.5 and adjusted P value < 0.05) are coloured blue. Top 10 DE genes by adjusted P value are labelled. (B) Violin plot to compare expression level of top 10 DE genes between endothelial1 and endothelial2. Upper panel is the top 10 genes enriched in endothelial1, and the bottom panel is the top 10 genes enriched in endothelial2. (C) Bar plot to present Reactome pathways (left) and GO terms (right) positively enriched in endothelial2 compared with endothelial1. (D) Heatmap to show the leading genes in interferon signalling pathway in endothelial1 and endothelial2.
We next performed GSEA to investigate pathways that positively or negatively enriched in Endothelial2 compared with Endothelial1. In Reactome 24 pathway collections, interferon signalling‐related pathways were among the top positive enriched pathways by normalized enrichment scores (NES). Cytokine‐related pathways were also positively enriched in Endothelial2 (Figure 3C, left). In gene ontology (GO) terms, ‘Cytokine mediated signaling pathway’, ‘Cellular response to cytokine stimulus’ and ‘Cellular response to cytokine stimulus’ were the top 3 positively enrich terms (Figure 3C, right). ‘Response to interferon gamma’ was also among the top positively enriched terms. We further surveyed the expression of the leading genes in interferon signalling pathway in Endothelial1 and Endothelial2, which showed significant elevation in Enodothelial2 (Figure 3D).
Decreased cell–cell interactions in sarcopenia skeletal muscle cells
Cell–cell communications through ligand–receptor pairs is critical for the crosstalk between different cell types and coordinating various biological processes. To investigate if the cell–cell communications were disturbed in sarcopenia samples, we inferred cell–cell interactions using CellChat 12 and compared the interaction network among the three experiment conditions. Because Endothelial1 was the major endothelial cell cluster for control and P6Sham group and Endothelial2 was the major one for P6DX, we used Endothelial_major to refer to Endothelial1 in control and P6Sham and to Endothelial2 in P6DX, respectively. To make reliable inference, we also excluded cell types with small cell numbers (<50 cells in one condition). Overall, there are more cell–cell interactions inferred from control and P6Sham cells (Figure 4A and 4B); the total interaction counts decreased by 22.0% in P6DX cells compared with control cells. Moreover, the interaction strength was also lowest in P6DX cells (Figure 4B). We next compared the differential number of interactions and interaction strength between P6DX cells and control cells in different cell type pairs (Figure 4C). In most cell type pairs, P6DX had less interactions and lower interaction strength. For example, in Pericyte2 (sender)–Myocyte (receiver) pair, there was 46 interactions in control cells, but only 23 in P6DX cells. These results together suggested that the cell–cell communications were compromised in P6DX cells and may attribute to the sarcopenia process.
Figure 4.
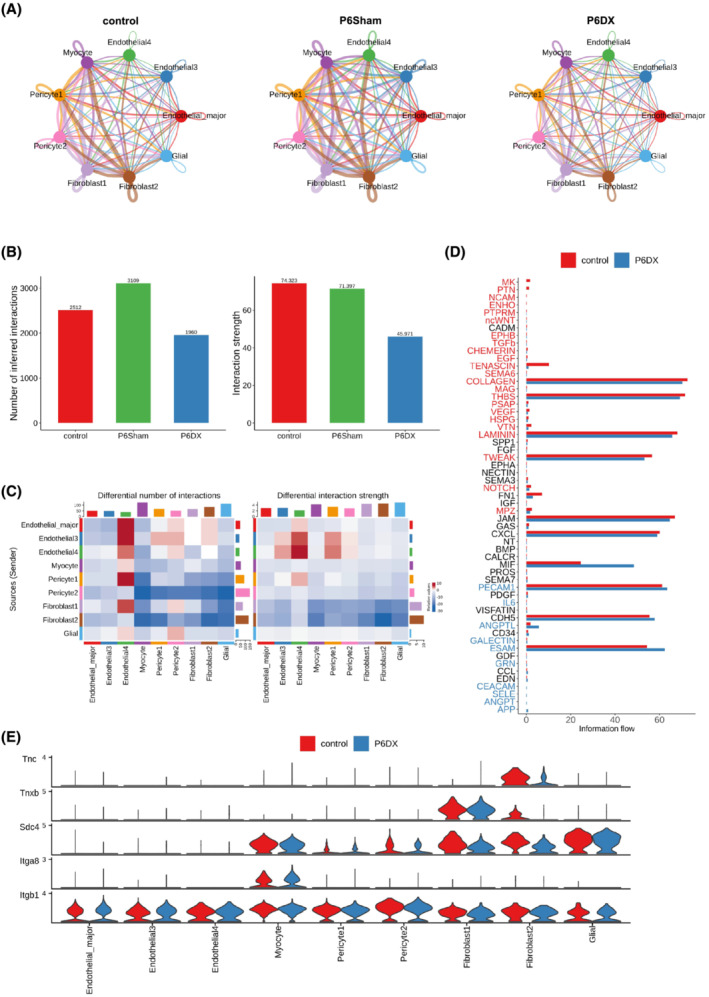
Cell–cell interactions inferred from scRNA‐seq data. (A) Cell–cell interaction network among the major cell types in each experiment group. Edge width represents number of interactions between the two connecting cell types. P6DX group has a less dense network comparing to control and P6Sham groups. (B) Bar plot to compare number of interactions and interaction strength among the three groups. P6DX has fewest interactions and lowest interaction strength. (C) Heatmap to compare differential number of interactions and interaction strength between P6DX and control group. P6DX has fewer interactions and lower interaction strength in most cell type pairs. (D) Bar plot to compare the overall information flow of each signalling pathway. The signalling pathways coloured red are enriched in control, and these coloured blue are enriched in the P6DX. (E) Expression of signalling genes related to TENASCIN signalling pathway in control and P6DX groups.
We also investigated the signalling pathways that were changed between P6DX cells and control cells by comparing the information flow (interaction strength). The signalling pathways with increased interaction strength in control were coloured as red, and that in P6DX were coloured as blue (Figure 4D). For example, TENASCIN signalling pathway had much weaker interaction strength in P6DX. We further evaluated the expressions of ligand receptors involving in this signalling pathway, showing that Tnc, Tnxb and Sdc4 had lower expression Fibroblast2 in P6DX compared with control (Figure 4E).
Validation of Gbp2 expression in mice and human sarcopenia muscle biopsies
From scRNA‐seq data, we observed unusual high Gbp2 expression in endothelial cells in P6DX group, and we next validated this result using experimental approaches. We first assessed the Gbp2 expression by western blot (Figure 5A and 5B). We showed that the protein expression of Gbp2 was significantly higher in P6DX samples compared with the other two groups (P value = 0.01699 in P6DX vs. control and 0.02943 in P6DX vs. P6Sham, Student's t‐test). We further inspected Gbp2 expression using immunofluorescence staining. Muscle sections from the three groups were stained with Gbp2 and Cd31 to identify Gbp2‐expressing endothelial cells. Whereas Gpb2 was detected broadly in endothelial cells in P6DX group, Gbp2 was mostly absent in R1 control and P6Sham groups (Figure 5C, left panel). The Cd31 and Gbp2 co‐localization rate was significantly higher in P6DX group than the other two groups (Figure 5C, right panel; P value = 1.57e‐05 in P6DX vs. control and 1.592e‐05 in P6DX vs. P6Sham, Student's t‐test).
Figure 5.
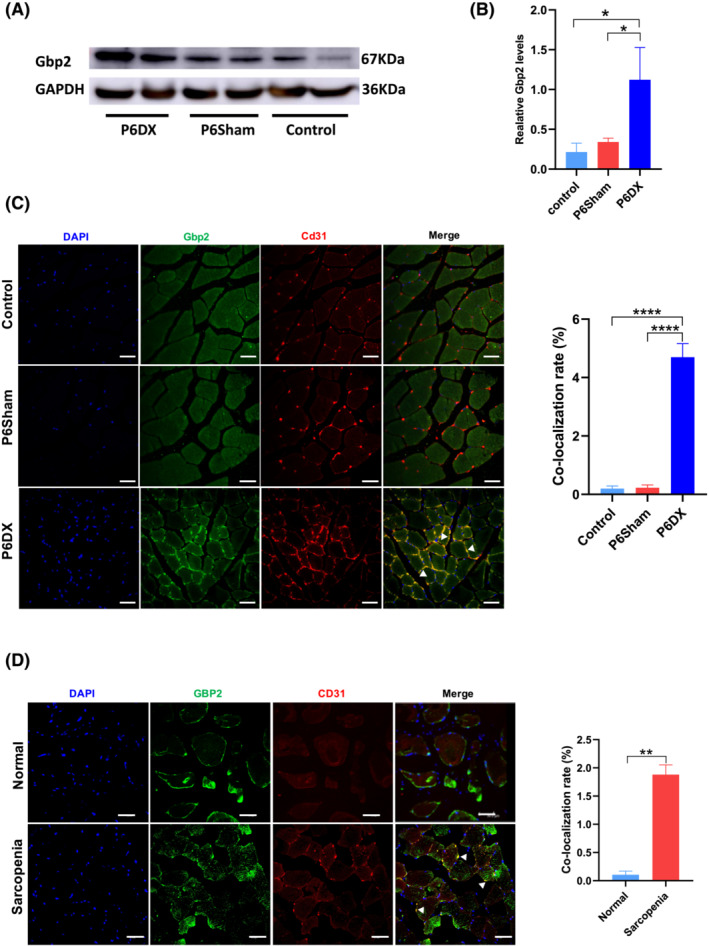
Gbp2 increased in mice and human sarcopenia muscle biopsies. (A) Western blot image to show that Gbp2 expression is increased in P6DX group. (B) Quantification of western blot results. Gbp2 expression is significantly higher than the other two groups (* indicates P value < 0.05). (C) Left: Immunofluorescence of mouse TA; expression of Gbp2 is identified in endothelial cells in the P6DX group (white arrows). Blue: DAPI, green: Gbp2, red: Cd31. Scale bar represents 20 μm. Right: Co‐localization rate of Cd31 and Gbp2 (**** indicates P value < 0.0001) (D) Left: Expression of GBP2 in human muscle tissue; expression of GBP2 in endothelial cells is observed in sarcopenia patients (white arrows). Blue: DAPI, green: GBP2, red: CD31. Scale bar represents 20 μm. Right: Co‐localization rate of CD31 and GBP2, (** indicates P value < 0.01).
We next investigated whether GBP2 was also present in human sarcopenia patients. We recruited 40 sarcopenia patients diagnosed by recommendations of the European Working Group on Sarcopenia in Older People. 25 Among the 40 patients, 22 were women, aged from 60 to 87 years, and none of them had used any glucocorticoids in the recent past. During joint replacement or lumbar fusion surgery, we collected muscle tissues associated with the surgical unavoidably removal. We then made frozen muscle specimen sections for immunofluorescence according to the instructions. We identified GBP2 expression in endothelial cells from a subset of sarcopenia patients (Figure 5D, left panel). Overall, 27.5% (11 out of 40) patients were found to exhibit significant GBP2 expression in the collected specimens. In those patients, we identified significantly higher CD31 and GBP2 co‐localization rate than non‐sarcopenia controls (Figure 5D, right panel; P value = 0.001128, Student's t‐test).
Over‐expression of GBP2 led to compromised endothelial cell tube formation
Skeletal muscle is highly vascularized; many studies have reported the importance of vascularization in skeletal muscle function for the physiological demand such as restoring oxygen and nutrient supply. 26 The endothelial tube formation assay provides a tool for the assessment of angiogenesis in vitro. In this study, we successfully constructed HUVEC‐GBP2 over‐expression cells by plasmid over‐expression system (‘Methods’). The GBP2 over‐expression was validated by Western blot (Figure 6A and 6B); the GBP2 over‐expression cells had significantly higher GBP2 (P value = 0.006532 in GBP2 Over‐expression vs. Empty Plasmid and P value = 0.008091 in GBP2 Over‐expression vs. Control). In endothelial cell tube formation assay, we found that the tubule formation ability was significantly decreased in the GBP2 over‐expression group by microscopy inspection. No clear cell aggregation was observed to form tubules in over‐expression cells, whereas obvious tubule formation was identified in the control group (Figure 6C).
Figure 6.
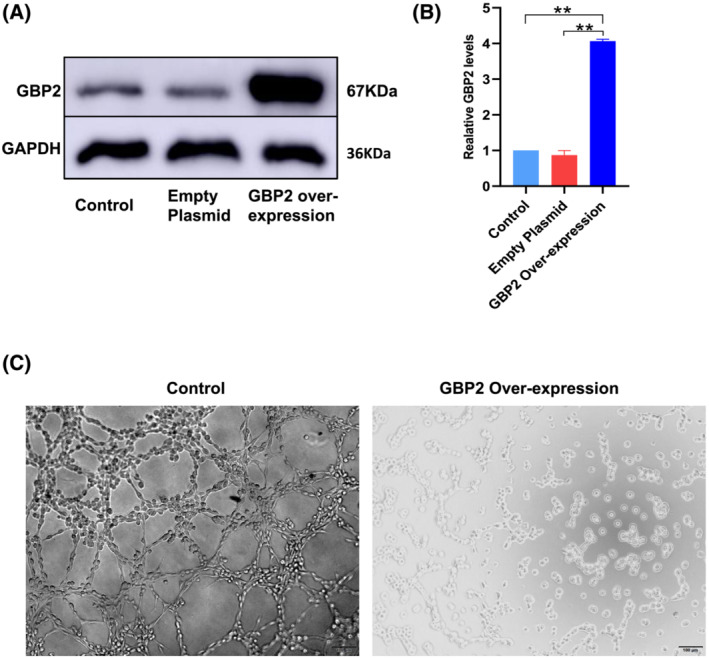
Over‐expression of Gbp2 in endothelial cells inhibited the tube formation. (A and B) Western blot to verify GBP2 over‐expression in HUVEC (** indicates P value < 0.01). (C) Endothelial cell tube formation assay result. Left: Control group; robust tubule formation is observed; right: HUVEC has reduced tube formation ability in GBP2 over‐expression environment. Images were taken at 8 h.
Discussion
Single‐cell technologies are becoming increasingly important in biological researches; however, a single‐cell transcriptomic reference for sarcopenia is still lacking. In this study, we constructed a mouse model for sarcopenia and applied scRNA‐seq to provide a comprehensive cell type landscape for skeletal muscle under sarcopenic as well as steady conditions. We revealed 14 cell types/subtypes from our single‐cell datasets. To our surprise, we found that endothelial cells made the largest proportion among the cell population. In addition, distinct endothelial cell subtypes have been identified between sarcopenic and control samples. Drastically high expression of interferon genes and the resulting interferon‐induced GBP genes were identified in the sarcopenia‐specific endothelial cells. Elevated Gbp2 gene expression was also observed in endothelial cells from human sarcopenia patients' skeletal muscle biopsies, suggesting that our findings may indicate new therapeutic directions for sarcopenia.
Supported by several previous studies, endothelial cells have recently been acknowledged to involve in mechanistic pathways of skeletal muscle dysfunctions. A recent in vitro study demonstrated that the high glucose‐treated endothelial cells secreted higher levels of inflammatory cytokines and the damaging free radical superoxide, which consequently affects skeletal muscle satellite cell growth and differentiation. 27 Another study reported that apoptosis of capillary endothelial cells accounts for more than 75% of apoptosis in the old mouse muscle. Dysfunction in endothelial cell could disturb the integrity of the endothelial monolayer and the delivery of nutrients and oxygen as well as the removal of toxic metabolic products, which finally affect muscular homoeostasis. 28 In our study, we found the major endothelial cells from sarcopenia samples were clearly separated from control samples in the UMAP two‐dimensional representations, suggesting they had distinct transcriptional profiles. From our gene‐level and pathway‐level analysis, we created a gene signature list for sarcopenia endothelial cells. Interferon signalling pathway genes were among the top of the gene list, indicating that endothelial cells may contribute to sarcopenia progression through an interferon‐dependent pathway. This result is consistent with the enhanced gamma interferon signalling during ageing. 29 It has been proposed that chronic inflammation makes substantial contribution to ageing and ageing‐related muscle dysfunction. 6 Our study provided new evidence to support this hypothesis. In a recent study, it has been reported that responses to interferon‐gamma were significantly down‐regulated during muscle regeneration in aged mice, which differed from our increased interferon observations. The authors further applied single‐cell sequencing and discovered that a novel population of interferon‐responsive macrophages attributed to the down‐regulation. These controversial results highlighted the importance to discuss the molecular changes under specified cell type context, which can be achieved by single‐cell technologies.
GBP family genes were another cluster of genes highly increased in sarcopenic endothelial cells in our study, which is a gene family that can be highly upregulated by interferon signalling. GBP genes were originally identified by their unusual ability to bind guanosine monophosphate (GMP) nucleotide agarose and were later emerging as central orchestrators of innate immunity to infection and inflammation. 23 Although this interferon‐induced gene family assembles functions to protection against invading pathogens, a growing number of evidences suggest their new functions that are not directly related to defending invaders. 30 , 31 , 32 A recent study uncovered that GBP2 and GBP5 can both negatively regulate osteoclast differentiation in vitro. Moreover, GBP5 knockout mice showed greater age‐associated bone loss, which was mediated by inflammation. 32 In this study, we presented that GBP genes may also be positively associated with senescence of skeletal muscle, the neighbours of bones with close ties, playing a multifarious role in the musculoskeletal system. In another study, after inducing by interferon, GBP1 was shown to restrain cellular proliferation in endothelial cells through an undefined mechanism. 31 Increased expression of GBP1 also correlated with reduced blood vessel density formed by endothelial cells. 33 In our study, after overexpressing GBP2 in human umbilical vein endothelial cells, the tube formation was significantly inhibited, suggesting that increased GBP genes could lead to compromised angiogenesis in skeletal muscles. The blood vessels not only provides oxygen and nutrients supplies to skeletal muscles but also plays a key role in muscle development regeneration. 26 Future studies can be conducted to further elucidate how GBP genes can disturb vascular function in skeletal muscle remodelling and senescence to establish promising therapeutic strategies.
A limitation of our study is that we recruited mice with accelerated ageing rather than natural ageing mice, which may not reflect the exact ageing procedure. However, senescence‐accelerated mouse prone (SAMP) mice strains have been successfully be used in several osteoporosis and sarcopenia studies. 34 , 35 , 36 Our study can build basis for future studies recruiting natural ageing mice. In our human clinical samples, we observed that a subset of sarcopenia patients also showing pronounced GBP2 expression in endothelial cells, which was consistent with our animal model, although we did not detect high GBP2 expression by immunofluorescence in other patients. This result indicated that the disease heterogeneity for sarcopenia. Future clinical studies will be needed to stratify the patient population, and personalized therapeutic approaches based on diagnostics at molecular level can be beneficial for patients.
In summary, our study provided the first single‐cell RNA‐seq reference dataset for sarcopenia mouse model. We demonstrated that endothelial cell may play an overlooked role in sarcopenia, potentially through an interferon‐induced GBP signalling pathway. Interferon–GBP interplay may be a new promising therapeutic target for age‐related sarcopenia.
Conflict of interest
Zhi Peng, Xiaolin Kuang, Chen Yu, Shiwei Niu, Yongjun Du, Di Lu, Shaobo Li, Zhaowei Teng and Sheng Lu declare that they have no conflict of interest. Ruoyu Zhang is a paid consultant of InnoVec Biotherapeutics. The manuscript has been approved by all authors for publication.
Funding
This work was funded by National Natural Science Foundation of China (Nos. 81960268 and 31960136); the Joint special fund of Applied Fundamental Research of Kunming Medical University granted by the Science and Technology Office of Yunnan (Nos. 202001AY070001‐172 and 2018FE001(‐174)); Joint Project of Yunnan Provincial Science and Technology Department‐Local Universities (2018FH001‐084 and 2019FH001‐015).
Supporting information
Figure S1. GBP2 expression plasmid structure
Figure S2. Marker gene expression in each cell type. (A) Marker gene expression of Endothelial 1. (B) Marker gene expression of Endothelial 2. (C) Marker gene expression of Endothelial 3. (D) Marker gene expression of Endothelial 4. (E) Marker gene expression of Pericyte. (F) Marker gene expression of Fibroblast. (G) Marker gene expression of Myocyte. (H) Marker gene expression of Glial. (I) Marker gene expression of Neural. (J) Marker gene expression of Macrophage. (K) Marker gene expression of MUSC.
Table S1. Supporting Information
Table S2. Supporting Information
Acknowledgements
We thank Mr Wei Song and our colleagues from Yunnan Provincial Key Laboratory of Digital Orthopedics, Yunnan Provincial Lv‐Weijia Academician Workstation for their discussion and comments on the manuscript. The authors of this manuscript certify that they comply with the ethical guidelines for authorship and publishing in the Journal of Cachexia, Sarcopenia and Muscle. 37
Peng Z., Zhang R., Kuang X., Yu C., Niu S., Du Y., Lu D., Li S., Teng Z., and Lu S. (2022) Single‐cell RNA‐seq reveals interferon‐induced guanylate‐binding proteins are linked with sarcopenia, Journal of Cachexia, Sarcopenia and Muscle, 13, 2985–2998, doi: 10.1002/jcsm.13091
Contributor Information
Zhaowei Teng, Email: tengzhaowei2003@163.com.
Sheng Lu, Email: drlusheng@163.com.
References
- 1. Santilli V, Bernetti A, Mangone M, Paoloni M. Clinical definition of sarcopenia. Clin Cases Miner Bone Metab 2014;11:177–180. [PMC free article] [PubMed] [Google Scholar]
- 2. Therakomen V, Petchlorlian A, Lakananurak N. Prevalence and risk factors of primary sarcopenia in community‐dwelling outpatient elderly: A cross‐sectional study. Sci Rep 2020;10:19551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Janssen I, Shepard DS, Katzmarzyk PT, Roubenoff R. The healthcare costs of sarcopenia in the United States. J Am Geriatr Soc 2004;52:80–85. [DOI] [PubMed] [Google Scholar]
- 4. Goates S, Du K, Arensberg MB, Gaillard T, Guralnik J, Pereira SL. Economic impact of hospitalizations in US adults with sarcopenia. J Frailty Aging 2019;8:93–99. [DOI] [PubMed] [Google Scholar]
- 5. Zhang C, Cheng N, Qiao B, Zhang F, Wu J, Liu C, et al. Age‐related decline of interferon‐gamma responses in macrophage impairs satellite cell proliferation and regeneration. J Cachexia Sarcopenia Muscle 2020;11:1291–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang J, Leung K‐S, Chow SKH, Cheung WH. Inflammation and age‐associated skeletal muscle deterioration (sarcopaenia). J Orthop Transl 2017;10:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giresi PG, Stevenson EJ, Theilhaber J, Koncarevic A, Parkington J, Fielding RA, et al. Identification of a molecular signature of sarcopenia. Physiol Genomics 2005;21:253–263. [DOI] [PubMed] [Google Scholar]
- 8. Börsch A, Ham DJ, Mittal N, Tintignac LA, Migliavacca E, Feige JN, et al. Molecular and phenotypic analysis of rodent models reveals conserved and species‐specific modulators of human sarcopenia. Commun Biol 2021;4:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perez K, McGirr J, Limbad C, Doi R, Nederveen JP, Nilsson MI, Tarnopolsky M, Campisi J, Melov S. Single nuclei profiling identifies cell specific markers of skeletal muscle aging, sarcopenia and senescence. medRxiv 2021:2021.2001.2022.21250336. [DOI] [PMC free article] [PubMed]
- 10. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM III, et al. Comprehensive integration of single‐cell data. Cell 2019;177:1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. bioRxiv 2021:060012.
- 12. Jin S, Guerrero‐Juarez CF, Zhang L, Chang I, Ramos R, Kuan C‐H, et al. Inference and analysis of cell‐cell communication using CellChat. Nat Commun 2021;12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen H, Yao X, Emura S, Shoumura S. Morphological changes of skeletal muscle, tendon and periosteum in the senescence‐accelerated mouse (SAMP6): A murine model for senile osteoporosis. Tissue Cell 2006;38:325–335. [DOI] [PubMed] [Google Scholar]
- 14. Xie W‐q, He M, Yu DJ, Wu YX, Wang XH, Lv S, et al. Mouse models of sarcopenia: classification and evaluation. J Cachexia Sarcopenia Muscle 2021;12:538–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chiu CS, Weber H, Adamski S, Rauch A, Gentile MA, Alves SE, et al. Non‐invasive muscle contraction assay to study rodent models of sarcopenia. BMC Musculoskelet Disord 2011;12:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Goncharov NV, Nadeev AD, Jenkins RO, Avdonin PV. Markers and biomarkers of endothelium: When something is rotten in the state. Oxid Med Cell Longev 2017;2017:9759735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kalucka J, de Rooij LPMH, Goveia J, Rohlenova K, Dumas SJ, Meta E, et al. Single‐cell transcriptome atlas of murine endothelial cells. Cell 2020;180:764–779.e20. [DOI] [PubMed] [Google Scholar]
- 18. Zhao Q, Eichten A, Parveen A, Adler C, Huang Y, Wang W, et al. Single‐cell transcriptome analyses reveal endothelial cell heterogeneity in tumors and changes following antiangiogenic treatment. Cancer Res 2018;78:2370–2382. [DOI] [PubMed] [Google Scholar]
- 19. Chen MB, Yang AC, Yousef H, Lee D, Chen W, Schaum N, et al. Brain endothelial cells are exquisite sensors of age‐related circulatory cues. Cell Rep 2020;30:4418–4432.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamazaki T, Mukouyama Y‐s. Tissue specific origin, development, and pathological perspectives of pericytes. Front Cardiovasc Med 2018;5:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Muhl L, Genové G, Leptidis S, Liu J, He L, Mocci G, et al. Single‐cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun 2020;11:3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Augsten M, Hägglöf C, Olsson E, Stolz C, Tsagozis P, Levchenko T, et al. CXCL14 is an autocrine growth factor for fibroblasts and acts as a multi‐modal stimulator of prostate tumor growth. Proc Natl Acad Sci U S A 2009;106:3414–3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tretina K, Park E‐S, Maminska A, MacMicking JD. Interferon‐induced guanylate‐binding proteins: Guardians of host defense in health and disease. J Exp Med 2019;216:482–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2020;48:D498–d503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cruz‐Jentoft AJ, Bahat G, Bauer J, Boirie Y, Bruyère O, Cederholm T, et al. Sarcopenia: revised European consensus on definition and diagnosis. Age Ageing 2018;48:16–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Latroche C, Gitiaux C, Chrétien F, Desguerre I, Mounier R, Chazaud B. Skeletal muscle microvasculature: A highly dynamic lifeline. Phys Ther 2015;30:417–427. [DOI] [PubMed] [Google Scholar]
- 27. Kargl CK, Nie Y, Evans S, Stout J, Shannahan JH, Kuang S, et al. Factors secreted from high glucose treated endothelial cells impair expansion and differentiation of human skeletal muscle satellite cells. J Physiol 2019;597:5109–5124. [DOI] [PubMed] [Google Scholar]
- 28. Wang H, Listrat A, Meunier B, Gueugneau M, Coudy‐Gandilhon C, Combaret L, et al. Apoptosis in capillary endothelial cells in ageing skeletal muscle. Aging Cell 2014;13:254–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bandrés E, Merino J, Vázquez B, Inogés S, Moreno C, Subirá ML, et al. The increase of IFN‐γ production through aging correlates with the expanded CD8+highCD28−CD57+ subpopulation. Clin Immunol 2000;96:230–235. [DOI] [PubMed] [Google Scholar]
- 30. Vestal DJ, Jeyaratnam JA. The guanylate‐binding proteins: Emerging insights into the biochemical properties and functions of this family of large interferon‐induced guanosine triphosphatase. J Interferon Cytokine Res 2011;31:89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Honkala AT, Tailor D, Malhotra SV. Guanylate‐binding protein 1: An emerging target in inflammation and cancer. Front Immunol 2020;10:3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Place DE, Malireddi RKS, Kim J, Vogel P, Yamamoto M, Kanneganti T‐D. Osteoclast fusion and bone loss are restricted by interferon inducible guanylate binding proteins. Nat Commun 2021;12:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bleiziffer O, Hammon M, Arkudas A, Taeger CD, Beier JP, Amann K, et al. Guanylate‐binding protein 1 expression from embryonal endothelial progenitor cells reduces blood vessel density and cellular apoptosis in an axially vascularised tissue‐engineered construct. BMC Biotechnol 2012;12:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dry‐fermented soybean food (Cheonggukjang) ameliorates senile osteoporosis in the senescence‐accelerated mouse prone 6 model. J Med Food 2019;22:1047–1057. [DOI] [PubMed] [Google Scholar]
- 35. Zhang N, Chow SKH, Leung KS, Lee HH, Cheung WH. An animal model of co‐existing sarcopenia and osteoporotic fracture in senescence accelerated mouse prone 8 (SAMP8). Exp Gerontol 2017;97:1–8. [DOI] [PubMed] [Google Scholar]
- 36. Romanick M, Thompson LV, Brown‐Borg HM. Murine models of atrophy, cachexia, and sarcopenia in skeletal muscle. Biochim Biophys Acta 2013;1832:1410–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. von Haehling S, Morley JE, Coats AJS, Anker SD. Ethical guidelines for publishing in the Journal of Cachexia, Sarcopenia and Muscle: Update 2021. J Cachexia Sarcopenia Muscle 2021;12:2259–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. GBP2 expression plasmid structure
Figure S2. Marker gene expression in each cell type. (A) Marker gene expression of Endothelial 1. (B) Marker gene expression of Endothelial 2. (C) Marker gene expression of Endothelial 3. (D) Marker gene expression of Endothelial 4. (E) Marker gene expression of Pericyte. (F) Marker gene expression of Fibroblast. (G) Marker gene expression of Myocyte. (H) Marker gene expression of Glial. (I) Marker gene expression of Neural. (J) Marker gene expression of Macrophage. (K) Marker gene expression of MUSC.
Table S1. Supporting Information
Table S2. Supporting Information


