Abstract
The biofilm state is the preferred lifestyle of bacteria in nature. Within a biofilm, the resident bacteria are protected from environmental stresses, antibiotics and other antimicrobials, including those due to multiple immune effectors of their host during conditions of disease. Thereby, biofilms contribute significantly to pathogenicity, recalcitrance to clearance and chronicity/recurrence of bacterial diseases, including diseases of the respiratory tract. In the absence of highly effective, biofilm-targeted therapeutics, antibiotics are commonly prescribed to attempt to treat these diseases, however, in light of the canonical resistance of biofilm-resident bacteria to antibiotic-mediated killing, this ineffectual practice often fails to resolve the diseased condition and contributes significantly to the global threat of rising antimicrobial resistance.
Nontypeable Haemophilus influenzae is a common respiratory tract disease co-pathogen, often present in partnership with other airway pathogens. Herein we aspired to determine whether either of two monoclonal antibodies we developed, one specific for NTHI [directed against the majority subunit (PilA) of the type IV pilus (T4P) of NTHI] and the other able to act agnostically on all bacteria tested to date (directed against a structural protein of the biofilm matrix, a DNABII protein), were able to disrupt 2-genera biofilms wherein NTHI co-partnered with another respiratory tract pathogen. These monoclonals were tested singly as well as when within an antibody cocktail.
The monoclonal directed against the NTHI antigen PilA was only effective on single species NTHI biofilms and not on single species biofilms formed by other unrelated species. However, when NTHI co-partnered with any of 5 respiratory tract pathogens tested here (Burkholderia cenocepacia, Staphylococcus aureus, Pseudomonas aeruginosa, Streptococcus pneumoniae or Moraxella catarrhalis), this exclusively NTHI-directed monoclonal was able to disrupt these 2-genera biofilms. Conversely, the monoclonal antibody directed against protective epitopes of a DNABII protein, significantly disrupted all single species and 2-genera biofilms, which reflected the universal presence of this structural protein in all tested biofilm matrices. However, greatest release of both pathogens from a 2-genera biofilm was uniformly achieved by incubation with a 1:1 cocktail of both monoclonals. These data support the use of an approach wherein patients with respiratory tract disease could be treated with a therapeutic monoclonal antibody cocktail to release NTHI and its common co-pathogens from the protective biofilm to be killed by either traditional antibiotics and/or host immune effectors.
Keywords: S. aureus, S. pneumoniae, M. catarrhalis, B. cenocepacia, P. aeruginosa, rsPilA, IHF, HU
Highlights
-
•
NTHI forms unique spatial relationships with airway pathogens in 2 genera biofilms.
-
•
Close integration within biofilm enabled effectiveness of NTHI-specific monoclonal.
-
•
Monoclonal antibody mediated biofilm dispersal + disruption yielded additive benefit.
1. Introduction
Biofilms are increasingly recognized as major contributors to the pathogenicity, recurrence, chronicity and recalcitrance-to-treatment of respiratory tract infections, as well as many other diseases beyond the airway [4,5,15,16,21,36,47,71]. Bacteria resident within a biofilm are canonically highly resistant to antibiotic-mediated killing and overall antimicrobial resistance (AMR), which is often to multiple classes of antibiotics. AMR is an urgent and rapidly increasing worldwide public health problem responsible for over 2.8 million infections and more than 35,000 deaths annually in the United States alone [2], whereas the global burden of AMR is far greater with at least 700,000 deaths globally per year, a number expected to increase to 10 million deaths globally per year by 2050) [48]. Socioeconomically, biofilm diseases present a burden to the U.S. economy that is estimated to be $20-$55 billion per year [68] and is anticipated to cost between $300 billion to $1 trillion globally by 2050 [11,57].
While the definition of a biofilm continues to evolve, it is widely accepted that these are communities of bacteria encased in a polymeric matrix that protects resident bacteria from both antibiotics and host immune effectors, amongst many other stressors and potential means of eradication [1,46,63,65,66,71,73]. The biofilm matrix is highly variable but commonly comprised of proteins, carbohydrates, and extracellular DNA (eDNA). Whereas much elegant work to characterize single-species biofilms has been conducted and has contributed significantly to our understanding of both biofilm biology and the role of biofilms in disease, it is increasingly evident that biofilms present within sites of chronic and recurrent diseases are typically comprised of multiple species and/or several genera of bacteria.
Chronic and recurrent diseases of the airway wherein biofilms play an important role include chronic obstructive pulmonary disease (COPD), otitis media (OM), chronic rhinosinusitis (CRS), and cystic fibrosis (CF). Importantly, these biofilms are indeed often polymicrobial, however commonly nontypeable Haemophilus influenzae (NTHI) is identified as either a primary or secondary co-pathogen of the disease course [8,21,28,38,41,58,59,72]. The role of NTHI as a predominant contributor to multiple upper and lower respiratory tract infections is evidenced by its’ co-culture from disease sites along with numerous other bacterial genera that include Streptococcus pneumoniae, Moraxella catarrhalis, Pseudomonas aeruginosa, Staphylococcus aureus and Burkholderia cenocepacia [12,23,33,34,41,47,62,67,69,71].
The combination of the: 1) propensity for all bacteria in nature to reside within a biofilm; 2) role of bacterial biofilms in chronic and recurrent diseases; 3) inherent resistance of biofilm-resident bacteria to antimicrobials; 4) global incidence of AMR, 5) our limited repertoire of effective biofilm-mediated disease treatment or prevention strategies to date and 6) the polymicrobial nature of biofilms within sites of chronic/recurrent bacterial disease, mandates development of novel approaches. Herein, whereas we too have conducted many evaluations of single-species biofilms to date, we now begin to ascertain our ability to disrupt polymicrobial biofilms. In this study, we evaluated our ability to disrupt 2-genera biofilms wherein NTHI is resident along with another clinically important respiratory tract pathogen. To do so, we use two distinct monoclonal antibodies, both singly and when combined in a 1:1 cocktail, to determine their relative abilities to disrupt single-species and 2-genera biofilms. The first monoclonal antibody targets the majority subunit of the type IV twitching pilus of NTHI [52], and thus could be expected to directly act only upon this member of any 2-genera biofilm. The second monoclonal antibody targets a universal structural element of the biofilm matrix, protective epitopes of the two bacterial DNA-binding proteins of the DNABII family [10], which we’ve shown is capable of disrupting biofilms formed by diverse pathogens of 23 genera tested to date [10,18,26,45,53,60].
We hypothesize that the relative effectiveness of the two monoclonal antibodies, or a cocktail of both, will be dictated by not only the molecular targets of these antibodies, but also by the spatial relationships between NTHI and each of the five additional airway pathogens selected for assay when they are allowed to form a 2-genera biofilm in vitro. Here we test the potential for these distinctly directed monoclonal antibodies to work additively or synergistically as we continue our efforts to optimize development of a novel therapeutic approach for diseases of the respiratory tract wherein NTHI commonly contributes to the disease course.
2. Materials and methods
2.1. Respiratory tract pathogens
Bacterial pathogens used, their source and appropriate growth medium are listed in Table 1. For this study, we chose Staphylococcus aureus, Burkholderia cenocepacia, Streptococcus pneumoniae, Pseudomonas aeruginosa and Moraxella catarrhalis as co-partners for NTHI due to their predominance as airway disease co-pathogens.
Table 1.
Bacterial strains used in this study.
| Bacterial Strains Used | Strain | Agar | Source |
|---|---|---|---|
| Nontypeable Haemophilus influenzae | 86-028NP | Chocolate agar | Pediatric otitis media patient |
| Sirakova et. al. [64]. | |||
| Staphylococcus aureus | 29213 | Tryptic Soy agar | ATCC |
| Burkholderia cenocepacia | K56-2 | Luria-Bertani agar | Mahenthiralingam et al. [37] |
| Streptococcus pneumoniae | 1121 | TSA II + 5% sheep blood | McCool et. al. [42] |
| Pseudomonas aeruginosa | 142–1 | Tryptic Soy agar | University of South Texas |
| Moraxella catarrhalis | 7169 | BHI agar | Luke et. al. [35]. |
| Nontypeable Haemophilus influenzae GFP reporter | 86-028NP/pRSM2211 | Chocolate agar | Mason et. al. [40] |
| Pseudomonas aeruginosa mCherry reporter | PAO1/pCJ5.2 | Tryptic Soy agar | Gift from Daniel J Wozniak Ph.D. (unpublished to date) |
To conduct studies of 2-genera biofilms wherein NTHI was one of the co-pathogens, we first needed to determine the optimal ratios of each species to inoculate into our assay system that would allow NTHI to survive as well within a 2-genera biofilm as it would within an NTHI single species biofilm when recovered under the same conditions of testing [e.g. biofilm age, treatment duration]. This was important as whereas one monoclonal targets NTHI specifically, the other is species-agnostic; thereby to be able to assay for any potential additive and/or synergistic effectiveness when the antibodies were tested at one universal dose either singly or as part of a 1:1 cocktail, we needed to be able to control the relative contribution of NTHI to any 2-genera biofilm.
Bacteria were grown from frozen stocks on appropriate agar (Table 1) at 37°C, in a humidified atmosphere of 5% CO2 for 16-20 h. One or 2 well-isolated colonies were then picked, suspended in BHI supplemented with 2 μg each of heme and β-NAD per ml (sBHI), then adjusted to achieve the appropriate concentration for each strain in a 100 μl volume. Prior to initiating this study, we empirically tested various ratios of each species in combination with NTHI to pre-determine the inoculum of each to admix and allow to form a 2-genera biofilm wherein we could recover approximately 4.5E7-7E7 NTHI from control wells that contained sterile sBHI only. These inoculum ratios of NTHI:co-partnered pathogen varied between 1:3 when NTHI formed a biofilm with M. catarrhalis to 2E7:1 when NTHI formed a biofilm with P. aeruginosa (Table 2).
Table 2.
Ratio of inocula used in NTHI : co-pathogen 2-genera biofilms.
| NTHI | Inoculum (cfu/well) | Co-Pathogen | Inoculum (cfu/well) | Ratio NTHI: Co-Pathogen | Age of Biofilm at Treatment | Treatment Duration | Mean cfu NTHI Recovered from sBHI Wells |
|---|---|---|---|---|---|---|---|
| NTHI | 2.00E+04 | S. aureus | 2.50E+03 | 8 : 1 | 24 h | 2 h | 4.4e7 |
| NTHI | 2.00E+04 | B. cenocepacia | 6.00E+03 | 3 : 1 | 24 h | 2 h | 4.9e7 |
| NTHI | 1.50E+08 | S. pneumoniae | 5.00E+04 | 3E3 : 1 | 24 h | 2 h | 4.5e7 |
| NTHI | 1.00E+08 | P. aeruginosa | 5 | 2E7 : 1 | 16 h | 2 h | 7e7 |
| NTHI | 2.00E+04 | M. catarrhalis | 6.00E+04 | 1 : 3 | 24 h | 16 h | 1.8e8 |
2.2. Biofilm formation
One hundred microliters of each bacterial suspension were added to wells of an 8-well chambered coverglass (Fisher Scientific), either alone or in combination. To maintain equivalent volumes in each chamber, 100 μl sBHI was added to any well that contained a single species, so that all wells contained 200 μl total final volume prior to incubation. Chambered coverglasses were incubated at 37°C, 5% CO2 for 24 h to permit biofilm formation. For the study of NTHI co-partnered with P. aeruginosa, each single species biofilm, as well as these 2-genera biofilms, were incubated for only 16 h because P. aeruginosa would outgrow any size tested inoculum of NTHI by 24 h in this assay system.
2.3. Biofilm disruption assay
Murine monoclonal antibodies directed against either recombinant, soluble PilA (α-rsPilA) [31,43,44,49,50], or one directed against a chimeric synthetic peptide designed to mimic the protective epitopes of the alpha- and beta-subunits of the DNABII protein IHF (α-DNABII) [55], were diluted in sBHI, then pre-warmed to 37°C. Medium was removed from wells prior to a gentle wash with 200 μl of sterile sBHI to remove non-biofilm resident bacteria. Two hundred microliters of each antibody (to deliver 2.5 μg) or a 1:1 cocktail of both (to deliver 2.5 μg of each antibody) was added and the chambered coverglasses were returned to the 37°C, 5% CO2 incubator for 2 h. Due to the very unique spatial relationship between NTHI + M. catarrhalis within a 2-genera biofilm as we described previously [44], these cultures were incubated for up to 16 h after addition of the monoclonal antibodies.
2.4. Collection of bacteria newly released from biofilm-residence (NRel) by the action of the monoclonal antibodies
To collect bacteria that had been newly released from biofilm residence (NRel) by either α-rsPilA or α-DNABII, we used a previously described methodology [32,45]. Briefly, 150 μl of the antibody and NRel-containing medium was carefully removed from the well so as not to disturb any remaining biofilm. Then, 200 μl of sterile DPBS was slowly added to facilitate recovery of any remaining NRel (wash). 150 ul of the DPBS wash was combined with the recovered antibody/NRel-containing medium to generate the final NRel suspension. As a control, the same process was used to collect bacteria present in the medium above biofilms when treated with sBHI only. These final suspensions were gently sonicated (2.8L Ultrasonic bath, FisherSci) for 2 min to dissociate any aggregated bacteria prior to serially dilution and plating on to appropriate agar to calculate the relative disruptive capability of each antibody for single species and 2-genera biofilms. Each assay was conducted a minimum of 3 times with assays run on separate days.
2.5. Validation of biofilm disruption by CSLM imaging
Following treatments and recovery of NRel as described above, residual biofilms were stained with BacLightTM Bacterial Viability Kit (Molecular Probes, Eugene OR) according to manufacturer’s protocol, and fixed in a solution of 1.6% paraformaldehyde, 2.5% glutaraldehyde and 4.0% acetic acid in 0.1 M phosphate buffer. Wells were viewed by confocal scanning laser microscopy (CSLM) on a Zeiss LSM 800 confocal microscope, and images were rendered with Zeiss Zen software. Biofilm biomass was calculated by COMSTAT2 analysis. Each assay was repeated three times on separate days.
2.6. Examination of biofilm architecture and spatial relationships of single species and 2-genera biofilms by scanning electron microscopy (SEM)
For all single species biofilms, as well as any 2-genera biofilms wherein each pathogen had a distinct cellular morphology, we determined biofilm architecture and spatial relationships by SEM, as we could easily identify each pathogen visually. As such, single species and 2-genera biofilms were allowed to form as described above in 8-well chambered coverglass culture vessel into which a sterile 12 mm glass coverslip had been inserted prior to incubation. Biofilms were allowed to form for 16 h prior to overnight fixation at 4°C in 2.5% glutaraldehyde (Electron Microscopy Sciences, Hatfield, Pa) in 0.1 M phosphate buffer pH 7.4. Sixteen hours was chosen as the incubation period for all SEM imaging to allow ready discrimination of each bacterium and their relationship to each other before increased biofilm density obscured this detail. Samples were post-fixed with 1% osmium tetroxide in 0.1 M phosphate buffer, pH 7.2 (Electron Microscopy Sciences), dehydrated slowly through a series of graded ethanol solutions, then dried overnight using hexamethyldisilazane (HMDS) (Electron Microscopy Sciences). Coverslips were mounted onto 15 mm aluminum stubs, sealed with colloidal silver (Electron Microscopy Sciences) and air dried. Dried stubs were sputter coated with 2 nm gold/palladium and imaged using a Hitachi S-4800 SEM/TEM.
2.7. Examination of biofilm architecture and spatial relationships of single species and 2-genera biofilms by confocal microscopy
For those 2-genera biofilms wherein both pathogens were Gram negative rods, we relied upon use of CSLM and one reporter isolate to discriminate biofilm architecture and spatial relationships. To this end, selected single species and 2-genera biofilms were allowed to form in 8-well chambered coverglass as described, a P. aeruginosa PAO1 reporter that incorporated an mCherry-expressing plasmid pCJ5.2 and NTHI #86-028NP which expresses GFP under the control of the ompP2 promoter [40] were used. After 16 h, biofilms were gently washed with DPBS to remove non-adherent bacteria, fixed with 1.6% paraformaldehyde, 2.5% glutaraldehyde, 4% acetic acid in 0.1 M phosphate buffer, pH 7.4, [19,32,43,53,55], then imaged with a Zeiss 800 CLSM. To calculate co-localization in the CLSM images, a multi-step process with BiofilmQ v0.2.2 [20,29] was used. Briefly, each Z-stack was subjected to image preparation, then segmentation which involved separation using an intensity threshold filter to isolate individual bacteria followed by object declumping by cube segmentation (26 vox or 1.11 μ m) and finally parameter calculation to determine 3-D overlap. This feature quantifies the volume overlap or co-localization of two fluorescent channels in a given space.
2.8. Statistical analysis
Two-way ANOVA models were used to assess the effect of α-rsPilA and α-DNABII on single species or 2-genera biofilms. P-values were adjusted for multiple comparisons where appropriate, using the Tukey method and those wherein a p-value of ≤0.05 were considered statistically significant. Statistical analyses were completed using GraphPad Prism, version 9 (GraphPad Software, San Diego, California).
3. Results
3.1. SEM or CSLM imaging of single species and 2-genera biofilms
To determine how each of the 5 respiratory tract pathogens was interacting, or not, with NTHI when they formed a 2-genera biofilm, we imaged each single species and 2-genera biofilm when formed at the concentration used to inoculate a 2-genera biofilm (Table 2). However, as mentioned, due to the fact that by 24 h, all biofilms were too dense to readily discern each pathogen, these were imaged at 16 h. Representative images of each single species and 2-genera biofilm at 16 h are shown in Fig. 1&2.
Fig. 1.
Scanning electron micrographs of 16 h biofilms formed by NTHI, S. aureus, M. catarrhalis, and S.pneumoniae either as a single species biofilm or within a 2-genera biofilm. Panel A - 16 h NTHI biofilm pseudo-colored green. Panel B - 16 h S. aureus biofilm pseudo-colored gold. Panel C - 16 h NTHI + S. aureus 2-genera biofilm demonstrates close interrelationship between the two bacterial species. Panel D - 16 h S.pneumoniae biofilm (pseudo-colored red). Note diplococcal morphology. INSET - Magnified image of diplococcus in Panel D. Panel E - 16 h NTHI + S.pneumoniae biofilm. Note both close interrelationship between these two bacterial species and also the long chaining morphology of S.pneumoniae. Panel F - 16 h M. catarrhalis biofilm (pseudo-colored purple). Panel G - 16 h NTHI + M. catarrhalis biofilm. Note that NTHI surrounds and grows over spherical aggregate M. catarrhalis biofilms. Scale bars in Panels A–G = 5 μm, scale bar INSET = 1 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
By cellular morphology, we were able to readily distinguish NTHI from either S. aureus, S. pneumoniae or M. catarrhalis by SEM. As such, we pseudocolored each pathogen to better illustrate their spatial relationship with NTHI within 2-genera biofilms. When grown as a single species for 16 h, both NTHI (regardless of the starting inoculum) and S. aureus were typically organized in clusters, some denser than others (Fig. 1A&B), whereas S. pneumoniae appeared in small clusters of diplococci (Fig. 1D), and M. catarrhalis was arranged exclusively in dense spherical aggregates (Fig. 1F), as we have reported previously [44]. When identical inocula of both species were used for 2-genera biofilms, we observed that S. aureus and NTHI did not form separate biofilm structures and instead were intermingled (Fig. 1C), as were NTHI and S. pneumoniae when co-partnered (Fig. 1E). Interestingly however, when S. pneumoniae formed a biofilm with NTHI, it appeared to adopt a nearly exclusively long chain morphology instead of the traditional diplococcus morphology we observed in S. pneumoniae single species biofilms (see Fig. 1D). When co-partnered with M. catarrhalis, NTHI densely surrounded M. catarrhalis biofilm structures and grew up and over these spherical aggregates (Fig. 1G).
For P. aeruginosa and B. cenocepacia, which have cellular morphologies similar to NTHI, to understand the spatial relationship between two pathogens within a 2-genera biofilm, we visualized these biofilms by fluorescent confocal microscopy (Fig. 2). NTHI formed tall biofilms with what appeared to be obvious water channels at 16 h (Fig. 2A&C), with relative overall biomass densities that reflected the different inocula of NTHI used (e.g. 2e4 to partner with B. cenocepacia or 1e8 to partner with P. aeruginosa). Both B. cenocepacia and P. aeruginosa formed substantial single species biofilms with distinct characteristics (Fig. 2B&D). When identical inocula as used for single species biofilms were used, we observed that when co-partnered with NTHI, both B. cenocepacia (Fig. 2E–G) and P. aeruginosa (Fig. 2H–J) were well-integrated with NTHI throughout the 2-genera biofilms (see merged panels, Fig. 2G&J). The fraction of the biomasses that overlap in the 2-genera biofilms formed by NTHI with either B. cenocepacia or P. aeruginosa, based on analysis of co-localization of the two fluorescent signals, were 27% and 39%, respectively. These values appeared to align well with what could be discerned visibly in the merged images in Fig. 2G&J.
Fig. 2.
3D-reconstructed CSLM micrographs of 16 h or 24 h single species or 2-genera biofilms formed by NTHI, B. cenocepacia and/or P. aeruginosa, respectively. Panel A&C - Single species 24 h (panel A) or 16 h (panel C) biofilms formed by an NTHI reporter wherein biofilms were initiated at the same seeding density as used in the associated 2-genera biofilms formed when NTHI co-partnered with either B. cenocepacia or P. aeruginosa, respectively. Panel B - Phase contrast micrograph of single species 24 h B. cenocepacia biofilm (pseudo-colored violet). Panel D - Single species 16 h biofilm formed by a P. aeruginosa reporter (pseudo-colored orange). Panel E-G – 24 h 2-genera biofilm formed by NTHI + B. cenocepacia wherein the contribution by NTHI (Panel E) and that of B. cenocepacia (Panel F) are shown both individually, as well as within the merged image in Panel G. Note the close spatial relationship between these 2 bacterial species within the 2-genera biofilm. Panel H-J - 16 h 2-genera biofilm formed by NTHI + P. aeruginosa wherein the contribution by NTHI (Panel H) and that of P. aeruginosa (Panel I) are shown both individually, as well as within the merged image in Panel J. Note again the close spatial relationship between these two bacterial species within the 2-genera biofilm. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Biofilm disruption assays
3.2.1. Disruption of single species NTHI biofilms
Assay of single species NTHI biofilms served as both a comparator to 2-genera biofilms and as a bridge to an earlier study [45]. For all studies herein, we used α-rsPilA at approximately 1/4 the concentration (2.5 μg) of prior work [[43], [44], [45],52], whereas α-DNABII was used at approximately ½ the concentration of prior work (5 μg) [9,10,26,45,51,[53], [54], [55]] as a means to be able to demonstrate any additive or synergistic effect when α-rsPilA was prepared in a 1:1 cocktail with α-DNABII.
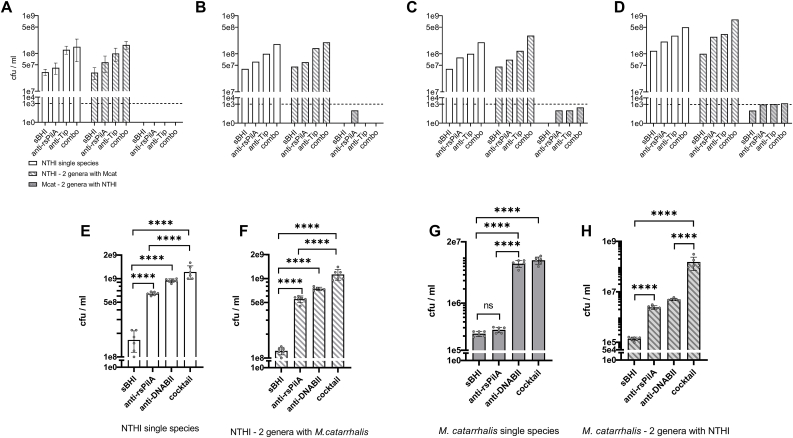
For all NTHI single species biofilm assays, we recovered a largely consistent concentration of NTHI from wells incubated with sterile sBHI only that reflected bacteria present within the medium above the biofilm (Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7C). These bacteria reflected those that had either never been biofilm resident and were perhaps simply growing in the culture medium or were those that had come off the biofilm due to natural remodeling during the 2 h incubation period. Due to use of a longer incubation time during treatment (increase from 2 h to 16 h), the concentration of NTHI recovered from sBHI wells was greater for studies of partnerships with M. catarrhalis (Fig. 7D&E). NTHI that had been newly released from biofilm residence by either α-rsPilA or α-DNABII were significantly greater than that recovered from sBHI only wells (3A, 4A, 5A, 6A & 7E) (p ≤ 0.05 – p ≤ 0.0001).
Fig. 3.
Relative release of biofilm-resident NTHI and B. cenocepacia from single species and 2-genera biofilms due to the action of monoclonal antibodies directed against either the T4P of NTHI or a DNABII structural biofilm matrix protein used alone as well as within a 1:1 monoclonal antibody cocktail. Panel A - Recovery of NTHI from a 24 h single species biofilm after incubation with sterile sBHI, anti-rsPilA, anti-DNABII or a cocktail of both monoclonal antibodies for 2 h with any of these agents. Panel B - Recovery of NTHI from a 24 h 2-genera biofilm when co-partnered with B. cenocepacia after incubation with the same agents. Panel C - Recovery of B. cenocepacia from a 24 h single species biofilm after incubation with sterile sBHI, anti-rsPilA, anti-DNABII or a cocktail of both monoclonal antibodies for 2 h with any of these agents. Panel D - Recovery of B. cenocepacia from a 24 h 2-genera biofilm when co-partnered with NTHI after incubation with the same agents. Data are presented as mean ± sd based on n = 6 (*p< 0.05, **p≤ 0.01, ***p≤0.001, ****p≤0.0001). In addition to overall trends as detailed in text, note both the ability of anti-rsPilA to release B. cenocepacia from biofilm residence when within a 2-genera biofilm with NTHI and the additive benefit derived from use of the antibody cocktail which induced a 3-fold greater increase in release of both NTHI and B. cenocepacia from biofilm residence compared to incubation in sBHI.
Fig. 4.
Relative release of biofilm-resident NTHI and S. aureus from single species and 2-genera biofilms due to the action of monoclonal antibodies directed against either the T4P of NTHI or a DNABII structural biofilm matrix protein used alone as well as within a 1:1 monoclonal antibody cocktail. Panel A - Recovery of NTHI from a 24 h single species biofilm after incubation with sterile sBHI, anti-rsPilA, anti-DNABII or a cocktail of both monoclonal antibodies for 2 h with any of these agents. Panel B - Recovery of NTHI from a 24 h 2-genera biofilm when co-partnered with S. aureus after incubation with the same agents. Panel C - Recovery of S. aureus from a 24 h single species biofilm after incubation with sterile sBHI, anti-rsPilA, anti-DNABII or a cocktail of both monoclonal antibodies for 2 h with any of these agents. Panel D - Recovery of S. aureus from a 24 h 2-genera biofilm when co-partnered with NTHI after incubation with the same agents. Data are presented as mean ± sd based on n = 6 (*p< 0.05, **p≤ 0.01, ***p≤0.001, ****p≤0.0001). In addition to overall trends as detailed in text, note both the ability of anti-rsPilA to release S. aureus from biofilm residence when within a 2-genera biofilm with NTHI and the additive benefit derived from use of the antibody cocktail which induced a 2- or 4-fold greater increase in release of NTHI and S. aureus, respectively from biofilm residence compared to incubation in sBHI.
Fig. 5.
Relative release of biofilm-resident NTHI and P. aeruginosa from single species and 2-genera biofilms due to the action of monoclonal antibodies directed against either the T4P of NTHI or a DNABII structural biofilm matrix protein used alone as well as within a 1:1 monoclonal antibody cocktail. Panel A - Recovery of NTHI from a 16 h single species biofilm after incubation with sterile sBHI, anti-rsPilA, anti-DNABII or a cocktail of both monoclonal antibodies for 2 h with any of these agents. Panel B - Recovery of NTHI from a 16 h 2-genera biofilm when co-partnered with P. aeruginosa after incubation with the same agents. Panel C - Recovery of P. aeruginosa from a 16 h single species biofilm after incubation with sterile sBHI, anti-rsPilA, anti-DNABII or a cocktail of both monoclonal antibodies for 2 h with any of these agents. Panel D - Recovery of P. aeruginosa from a 16 h 2-genera biofilm when co-partnered with NTHI after incubation with the same agents. Data are presented as mean ± sd based on n = 6 (*p< 0.05, **p≤ 0.01, ***p≤0.001, ****p≤0.0001). In addition to overall trends as detailed in text, note both the ability of anti-rsPilA to release P. aeruginosa from biofilm residence when within a 2-genera biofilm with NTHI and the additive benefit derived from use of the antibody cocktail which induced a 3- or 4-fold greater increase in release of NTHI and P. aeruginosa, respectively from biofilm residence compared to incubation in sBHI.
Fig. 6.
Relative release of biofilm-resident NTHI and S.pneumoniae from single species and 2-genera biofilms due to the action of monoclonal antibodies directed against either the T4P of NTHI or a DNABII structural biofilm matrix protein used alone as well as within a 1:1 monoclonal antibody cocktail. Panel A - Recovery of NTHI from a 24 h single species biofilm after incubation with sterile sBHI, anti-rsPilA, anti-DNABII or a cocktail of both monoclonal antibodies for 2 h with any of these agents. Panel B - Recovery of NTHI from a 24 h 2-genera biofilm when co-partnered with S.pneumoniae after incubation with the same agents. Panel C - Recovery of S.pneumoniae from a 24 h single species biofilm after incubation with sterile sBHI, anti-rsPilA, anti-DNABII or a cocktail of both monoclonal antibodies for 2 h with any of these agents. Panel D - Recovery of S.pneumoniae from a 24 h 2-genera biofilm when co-partnered with NTHI after incubation with the same agents. Data are presented as mean ± sd based on n = 6 (*p< 0.05, **p≤ 0.01, ***p≤0.001, ****p≤0.0001). In addition to overall trends as detailed in text, note both the ability of anti-rsPilA to release S.pneumoniae from biofilm residence when within a 2-genera biofilm with NTHI and the additive benefit derived from use of the antibody cocktail which induced a 3- or 7-fold greater increase in release of NTHI and S. pneumoniae, respectively from biofilm residence compared to incubation in sBHI.
Fig. 7.
Relative release of biofilm-resident NTHI and M. catarrhalis from single species and 2-genera biofilms due to the action of monoclonal antibodies directed against either the T4P of NTHI or a DNABII structural biofilm matrix protein used alone as well as within a 1:1 monoclonal antibody cocktail. Panels A-D - We were unable to recover M. catarrhalis either consistently or at a level that exceeded the minimal level of detection (dashed horizontal line) when we used the same concentration of monoclonal antibodies or time period of exposure as used with all other single or 2-genera biofilms (e.g. 2.5 μg/2 h shown in Panel A). Various combinations of increased antibody concentration (from 2.5 μg to 10 μg) or increased incubation period (from 2 h to 16 h) did not fully mitigate inability to release of M. catarrhalis to a level above the limit of detection for this assay (dotted line) despite observing a dose- and time-dependent outcome (Panel B, 2.5 μg/6 h; Panel C, 2.5 μg/16 h; Panel D, 10 μg/6 h). When conditions were adjusted to use of 10 μg of each antibody either alone or as a cocktail for 16 h on a 24 h biofilm, consistent recovery of M. catarrhalis was achieved. Panels E - Recovery of NTHI from a 24 h single species biofilm after incubation with sterile sBHI, anti-rsPilA, anti-DNABII or a cocktail of both monoclonal antibodies for 16 h with any of these agents. Panel F - Recovery of NTHI from a 24 h 2-genera biofilm when co-partnered with M. catarrhalis after incubation with the same agents. Panel G - Recovery of M. catarrhalis from a 24 h single species biofilm after incubation with sterile sBHI, anti-rsPilA, anti-DNABII or a cocktail of both monoclonal antibodies for 16 h with any of these agents. Panel H - Recovery of M. catarrhalis from a 24 h 2-genera biofilm when co-partnered with NTHI after incubation with the same agents. Dotted line in panels A, B, C, & D indicates the level of detection in these assays. Data are presented as mean ± sd based on n = 6 (*p< 0.05, **p≤ 0.01, ***p≤0.001, ****p≤0.0001). In addition to overall trends as detailed in text, note both the ability of anti-rsPilA to release M. catarrhalis from biofilm residence when within a 2-genera biofilm with NTHI and the additive benefit derived from use of the antibody cocktail which induced a 9- or >1000-fold greater increase in release of NTHI and M. catarrhalis, respectively from biofilm residence compared to incubation in sBHI.
Treatment of single species NTHI biofilms with a 1:1 cocktail of both monoclonals (2.5 μg α-rsPilA + 2.5 μg α-DNABII), each of which were capable of releasing NTHI from biofilm-residence, consistently induced a significant release of NTHI NRel (p ≤ 0.0001) that typically exceeded that observed when either monoclonal was used alone at the reduced doses used here. This latter observation demonstrated an overall additive disruptive benefit conferred by the antibody cocktail for single species NTHI biofilms. Highly similar results were obtained for recovery of NTHI from any of the five 2-genera biofilms following treatment with either sBHI only, either of the two monoclonals singly or the 1:1 monoclonal antibody cocktail (Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7F), although there were some slight differences in p-values obtained.
3.2.2. Disruption of B. cenocepacia, S. aureus, P. aeruginosa, S.pneumoniae or M. catarrhalis within single species biofilms
For each single species biofilm built by any of the selected five airway pathogens, we did not observe any release from biofilm-residence by incubation with α-rsPilA as the concentrations of bacteria recovered within the medium above the biofilms was equivalent to that of wells that contained sBHI only. This outcome was expected given the NTHI-specific nature of α-rsPilA (Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7G). Moreover, this outcome indicated that addition of non-specific serum to the culture medium did not induce heightened growth of any of the 5 tested co-pathogens due to this serum serving as a food source. However, for 2 of these 5 pathogens (B. cenocepacia and S. pneumoniae), incubation with the reduced dose of α-DNABII antibody, used here for 2 h, induced significant release of NRel compared to incubation with sBHI only (p ≤ 0.01) (Fig. 3, Fig. 6C). This latter observation was not observed for single species S. aureus or P. aeruginosa biofilms (Fig. 4, Fig. 5C) under the conditions tested.
For all 5 single species biofilms (B. cenocepacia, S. aureus, P. aeruginosa S. pneumoniae and M. catarrhalis), greatest overall release from biofilm residence was achieved by incubation with the cocktail of both monoclonal antibodies (Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7G. There were no significant differences between use of α-DNABII or the cocktail for any of these single species biofilms, which thus reflected that no additional benefit was achieved via inclusion of α-rsPilA on single species biofilms formed by these unrelated respiratory tract pathogens, again as anticipated due to the NTHI-specific nature of α-rsPilA.
3.2.3. Disruption of B. cenocepacia, S. aureus, P. aeruginosa, S.pneumoniae or M. catarrhalis within 2-genera biofilms when co-partnered with NTHI
For all 2-genera biofilms, incubation with sBHI continued to provide the targeted baseline number of non-biofilm resident bacteria recoverable in the medium above the biofilms during the 2 h incubation period. Intriguingly however, now incubation with α-rsPilA released a significant number of all 5 bacterial co-pathogens (B. cenocepacia, S. aureus, P. aeruginosa, S. pneumoniae and M. catarrhalis) from biofilm-residence when partnered with NTHI (Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7H) (p ≤ 0.05 to 0.01). This outcome is likely due to the close physical association between NTHI and each of these airway pathogen co-partners as shown in Figs. 1 and 2. For all of these respiratory tract co-pathogens, when partnered with NTHI in a 2-genera biofilm, greatest release from biofilm residence was achieved via use of the 1:1 cocktail of α-rsPilA admixed with α-DNABII (Fig. 3, Fig. 4, Fig. 5, Fig. 6D) (p ≤ 0.001 to 0.0001). This degree of release was significantly greater than that obtained by incubation with any single monoclonal compared to sBHI. These data again demonstrated the additive disruptive benefit of incubation of these 2-genera biofilms with a cocktail of the two monoclonal antibodies.
Two-genera biofilms of NTHI co-partnered with M. catarrhalis (Fig. 7), presented a unique relationship, as when we used the same assay conditions as that used for all other 2-genera biofilms tested here (e.g. incubation with 2.5 μg of each monoclonal for 2 h) we did not observe the same pattern of disruption as described above. Whereas NTHI consistently behaved as described and expected, we were unable to recover a reliable or consistent count of M. catarrhalis from any of the control or treated chamberglass wells (Fig. 7A). As such, we then conducted a small proof-of-concept trial (n = 1) wherein we screened the potential to use increased incubation periods (from 2h to either 6h or 16 h) to mediate release of M. catarrhalis, however these longer incubation periods did not consistently allow us to recover measurable quantities of M. catarrhalis (Fig. 7B&C). Thereby, we attempted to mediate release of M. catarrhalis from biofilm residence via use of a 4- or 2-fold fold greater concentration of each monoclonal (e.g. 10 μg of both) for a 6 h incubation period (Fig. 7D) and while this did slightly improve our ability to recover M. catarrhalis, these values barely reached the limit of detection for this assay (dashed horizontal line). However, when we assessed use of 10 μg of each monoclonal for 16 h [44] on each single species biofilms of NTHI or M. catarrhalis (Fig. 7E & H), we achieved an outcome highly similar to that described above for both NTHI and the other 4 pathogen co-partners. As indicated earlier, the concentration of NTHI recovered from sBHI wells was greater than that for other 2 genera assays conducted here and reflected the increased period of incubation during treatment (from 2h to 16 h). We’ve previously shown that this dose and time period was permissive for NTHI to express both PilA and AI-2 [44], both of which are needed for active dispersal of NTHI from biofilm residence when treated with α-rsPilA.
Again however, here we saw that wherein incubation with α-rsPilA was effective on single species NTHI biofilms (Fig. 7E), but not on single species M. catarrhalis biofilms (Fig. G), this NTHI-specific antibody did mediate release of both species from a 2-genera biofilm (Fig. 7F&H). In this co-partnership, wherein NTHI largely surrounds and grows over M. catarrhalis aggregates, release of M. catarrhalis is associated with expression of AI-2 by NTHI [44], with this latter quorum sensing molecule important given the discovery by Armbruster et al. [3] who showed that M. catarrhalis can eavesdrop on expression of AI-2 by NTHI. Greatest release from biofilm residence for both pathogens was achieved by incubation with the monoclonal antibody cocktail (p ≤ 0.001 to 0.0001).
The above described commonly observed outcome wherein α-rsPilA mediates release of a non-NTHI co-partner pathogen is likely due to the close intermingled spatial relationship between NTHI and B. cenocepacia, S. aureus, P. aeruginosa and S. pneumoniae, as well as the very unique relationship between NTHI and M. catarrhalis when they form a 2-genera biofilm in this assay system.
3.3. Validation of biofilm disruption with release of NRel by CSLM
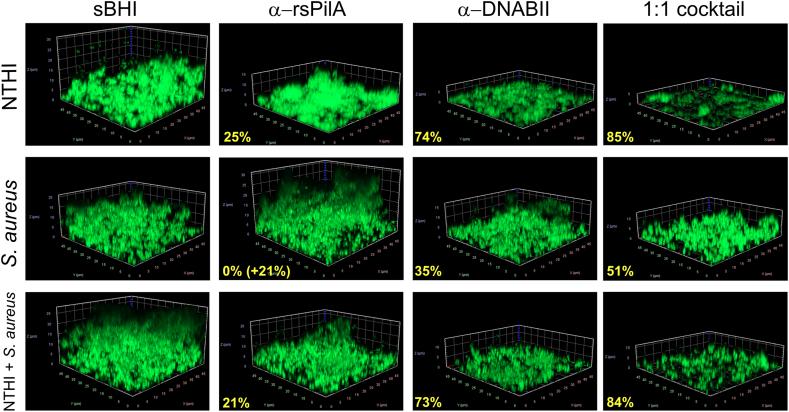
To provide additional supportive evidence of the relative disruption of each single species and 2-genera biofilm, after recovery of any NRel within each chamberslide, we subjected the residual biofilm to vital staining for imaging by CSLM. Representative images for NTHI single species biofilms following treatment with either sBHI, α-rsPilA, α-DNABII or a 1:1 cocktail of both monoclonal antibodies are shown in Fig. 8, as are those for single species S. aureus biofilms and 2-genera biofilms formed by NTHI + S. aureus. Note relative disruption compared to treatment with sBHI by each monoclonal antibody when used alone as well as within the cocktail. Greatest disruption was achieved by the 1:1 cocktail for NTHI, S.aureus and the 2-genera NTHI + S. aureus biofilm (85%, 51% and 84% respectively). Further, note that incubation of S. aureus single species biofilms with α-rsPilA resulted in an increase in biomass as in the absence of expression of the targeted antigen (e.g. NTHI PilA) by S. aureus, this serum likely served as an additional food source during the 2 h incubation period.
Fig. 8.
Percent reduction of biofilm biomass after exposure to various treatments and removal of newly released bacteria. Representative images of the residual biofilms formed by either NTHI, S. aureus or the 2-genera biofilm of NTHI + S. aureus after exposure to either media alone (sBHI), α-rsPilA, α-DNABII or the 1:1 cocktail of both monoclonal antibodies. After recovery of NRel the residual biofilm was stained with Live/Dead stain, imaged by CSLM and the biomass calculated by COMSTAT software. The percent reduction of the biomass compared to media alone wells are indicated in bold yellow font. Note that when S. aureus single species biofilms are incubated with α-rsPilA, which does not target an S. aureus antigen, there was an increase of resulting biomass as this non effective serum can serve as a food source. These data provide additional support to those data which indicated relative release of NRel following treatment with each of these agents as shown in Fig. 4. Representative images for the remaining four co-pathogens both as single species and within 2-genera biofilms with NTHI are shown in Supplemental Fig. 1. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Representative images of biofilms formed by B. cenocepacia, S. pneumoniae, P. aeruginosa or M. catarrhalis both as single species biofilms as well as when within a 2-genera biofilm with NTHI are shown in Suppl. Fig. 1 where we again saw a similar pattern of relative disruptive capability. Note again that incubation of both S. pneumoniae or P. aeruginosa with α-rsPilA resulted in an increase in biomass, likely due to this non-effective serum serving as a food source for these two pathogens which similarly do not express the antigenic target of this monoclonal. Whereas there was no increase in biomass for either B. cenocepacia or M. catarrhalis when incubated with α-rsPilA, any observed disruption was minor (e.g. 17% or 12%, respectively) and may indicate limited biofilm remodeling during the 2 h incubation period.
4. Discussion
Due to the multiple mechanisms, both physical and physiological, by which bacteria resident within a biofilm are protected from antimicrobials and immune effectors, it is imperative to be able to first disrupt the biofilm in order to expose the resident bacteria for eradication during a disease state. We, and others, show that bacteria released from a biofilm have a transient unique phenotype that is commonly one of greater sensitivity to the killing action of select antibiotics. This disruption can be due to exposure to nitric oxide, pyruvate, physical disruption or antibody-mediated, among others [7,10,13,14,27,44,45].
We have developed two disruption strategies that result from incubation of biofilms with monoclonal antibodies specifically directed against protective epitopes of two distinct bacterial proteins. One monoclonal antibody targets the protective epitope of the majority subunit of the type IV pilus of NTHI (PilA). This monoclonal mediates active dispersal of NTHI from biofilm residence due to upregulated expression of both the quorum sensing molecule AI-2 and its T4P [52]. Thus, α-rsPilA is a species-specific therapeutic biological, specifically developed for those diseases of the respiratory tract induced by NTHI. The second monoclonal antibody targets the combined protective domains of essential structural bacterial proteins (e.g. HU and IHF) that stabilize the biofilm matrix by crosslinking strands of extracellular DNA (eDNA). e-DNA is a major universal structural component of the bacterial biofilm matrix and this lattice-like scaffold is stabilized by proteins of the DNABII family [26]. Thereby, incubation with a monoclonal antibody that targets the DNABII proteins induces an equilibrium shift that rapidly destabilizes the eDNA lattice, resulting in collapse of the biofilm matrix with concomitant release of the resident bacteria. This anti-DNABII monoclonal antibody has been effective against biofilms built by many diverse pathogens we’ve tested to date, and thus we consider it to be more of a species-agnostic therapeutic [10,18,26,32,55,60]. Both monoclonals release bacteria from biofilm-residence, by either active dispersal or passive disruption [10,44,45,53,55].
NTHI is a common co-pathogen of multiple diseases of the upper and lower airway, and thus is cultured along with one or more of several other primary respiratory tract pathogens and rarely, if ever, is present as a single pathogen [8,28,38,58,59,72]. We were thus curious as to the potential additive or synergistic biofilm dispersal/disruption capability if we used these two monoclonals as a new therapeutic cocktail approach for polymicrobial biofilms that included NTHI. To do so, we generated 2-genera biofilms that incorporated NTHI as one of the bacterial co-partners with which to assess the biofilm-resident bacterial release potential of these two monoclonals when used alone as well as when in a 1:1 cocktail. Despite the species specificity of α-rsPilA, when NTHI was resident within a biofilm with either B. cenocepacia, S. aureus, P. aeruginosa or S. pneumoniae, its close physical relationship with these other pathogens appeared to facilitate their release from biofilm-residence by this monoclonal. As NTHI dispersed itself from these 2-genera biofilms, this activity likely also physically disrupted its co-partner from the 2-genera biofilm.
In the unique partnership between NTHI and M. catarrhalis, NTHI surrounds and ultimately largely encases the large spherical aggregate M. catarrhalis biofilm structures, as we reported earlier [44]. In this earlier study, when we exposed an NTHI + M. catarrhalis biofilm to only α-rsPilA, NTHI upregulates expression of both its type IV pilus and AI-2. As originally described by Armbruster et al. [3], M. catarrhalis can then eavesdrop on this expression of AI-2 by NTHI, so that when NTHI disperses from biofilm residence, M. catarrhalis disperses as well [44]. Here however, we expanded upon these early observations to now determine the potential additive and/or synergistic efficacy of α-rsPilA when admixed with α-DNABII to mediate release of M. catarrhalis from a 2-genera biofilm formed in partnership with NTHI. Whereas the unique spatial relationship between NTHI and M. catarrhalis in a 2-genera biofilm required the use of conditions that allowed time for upregulated expression of both PilA and AI-2 by NTHI [44], this was not required for any other 2-genera biofilm assayed here. This latter observation suggested that for the other 4 co-partnerships, release from biofilm residence by α-rsPilA, when used alone as a treatment, was primarily due to physical disruption mechanisms, but does not rule out any contribution from quorum sensing, particularly given the well-established role of AI-2 in interspecies crosstalk [6,24,30,70].
As anticipated, use of the species-agnostic α-DNABII was universally disruptive against biofilms formed by all six tested pathogens, unlike α-rsPilA, and effectively released these bacteria from both single species and 2-genera biofilms. In all cases however, the use of the cocktail of both monoclonals mediated the greatest release of both pathogens from residence within a 2-genera biofilm. While a noted change in cellular morphology of S. pneumoniae from diplococcus (when within a single species biofilm), to long chains when co-partnered with NTHI had no effect on relative release of S. pneumoniae from either single species or 2-genera biofilm residence in this study, it was nonetheless an observation of interest. While this may have reflected the need to limit the inoculum size for S. pneumoniae in single species biofilms in order to facilitate development of a 2-genera biofilm when partnered with NTHI, the chaining morphotype for S. pneumoniae has also been associated with enhanced adherence and colonization capabilities [61], but can also promote complement-dependent killing [17]. Each of these parameters could have consequences for successful colonization of its human host and/or disease induction/clearance when S. pneumoniae is within a polymicrobial biofilm with NTHI.
Release of bacteria from biofilm residence results in a transient phenotype wherein these bacteria are more readily killed by traditional antibiotics that are ineffective when they are resident within a biofilm [10,44,45] [14,7,13,22,27,39], an outcome often achievable at a greatly reduced dose. A combinatorial approach wherein the described monoclonal cocktail is delivered along with a reduced dose of appropriate antibiotic would likely be necessary if, for example, release from biofilm residence results in a sub-population with an enhanced pathogenicity phenotype as has been shown for S. pneumoniae in murine models when this pathogen was released from biofilm residence by influenza A virus [56], or under conditions wherein any pathogen is released from a biofilm in an immunocompromised patient. In other cases, as we have now shown in three distinct animal models of: 1) NTHI-induced experimental otitis media in the chinchilla [53,55]; 2) P. aeruginosa-induced lung infection in mice [53] and 3) Aggregatibacter actinomycetemcomitans-induced periimplantitis in rats [25], rapid clearance of bacteria newly released by treatment with either of these monoclonals and resolution of disease was effectively mediated by innate immune effectors in the absence of any co-delivered antibiotic.
Collectively, these new data support continued development of a combinatorial therapeutic approach for respiratory tract infections wherein NTHI is resident within a pathogenic polymicrobial biofilm.
CRediT authorship contribution statement
Joseph A. Jurcisek: Investigation, Methodology, Visualization, Formal analysis, Writing – review & editing. Llwyatt K. Hofer: Methodology, Visualization, Formal analysis. Steven D. Goodman: Conceptualization, Formal analysis, Writing – review & editing, Supervision. Lauren O. Bakaletz: Conceptualization, Resources, Formal analysis, Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Lauren O. Bakaletz reports financial support was provided by National Institutes of Health. Steven D. Goodman reports financial support was provided by National Institutes of Health. L.O.B. is an inventor of technology related to PilA-derived immunogens that is licensed to GlaxoSmithKline Biologicals. L.O.B. and S.D.G. are inventors of technology related to the DNABII-directed approach, rights to which have been licensed to Clarametyx Biosciences, Inc.
Acknowledgments & funding
This work was supported by the National Institutes of Health [NIH/NIDCD R01 DC003915 to LOB and NIH/NIDCD R01 DC011818 to LOB & SDG]. The authors are grateful to the Biostatistics Resource at Nationwide Children’s Hospital (BRANCH) and particularly Joe Stanek for statistical consultation and analysis. The authors also thank Dr. Danial Wozniak for generously providing the Pseudomonas aeruginosa strain PA01 reporter isolate that expresses mCherry and to Jennifer Neelans for outstanding assistance with manuscript preparation.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2022.100096.
Contributor Information
Joseph A. Jurcisek, Email: Joseph.Jurcisek@nationwidechildrens.org.
Llwyatt K. Hofer, Email: Llwyatt.Hofer@nationwidechildrens.org.
Steven D. Goodman, Email: Steven.Goodman@nationwidechildrens.org.
Lauren O. Bakaletz, Email: Lauren.Bakaletz@nationwidechildrens.org.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Supplemental Fig. 1.
Representative images of residual biofilms that remained after exposure to various treatments and removal of newly released bacteria. Representative images of the residual biofilms after exposure to either media (sBHI), α-rsPilA, α-DNABII or the 1:1 cocktail of both monoclonals for the co-pathogens B. cenocepacia, S. pneumoniae, P, aeruginosa and M. catarrhalis both as single species biofilms as well as within a 2-genrea biofilm with NTHI are shown. The percent reduction of biomass compared to media alone are shown in bold yellow font. Note that once again incubation with α-rsPilA resulted in an increase in biomass for both S. pneumoniae and P. aeruginosa.
Data availability
Data will be made available on request.
References
- 1.Ahearn C.P., Gallo M.C., Murphy T.F. Insights on persistent airway infection by non-typeable Haemophilus influenzae in chronic obstructive pulmonary disease. Pathog Dis. 2017;75 doi: 10.1093/femspd/ftx042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antibiotic resistance threats in the United States, 2019. 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf Atlanta, GA.
- 3.Armbruster C.E., Hong W., Pang B., Weimer K.E., Juneau R.A., Turner J., Swords W.E. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. mBio. 2010;1 doi: 10.1128/mBio.00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Azimi S., Lewin G.R., Whiteley M. The biogeography of infection revisited. Nat Rev Microbiol. 2022;20:579–592. doi: 10.1038/s41579-022-00683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakaletz L.O. Bacterial biofilms in the upper airway - evidence for role in pathology and implications for treatment of otitis media. Paediatr Respir Rev. 2012;13:154–159. doi: 10.1016/j.prrv.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Banerji R., Kanojiya P., Saroj S.D. Role of interspecies bacterial communication in the virulence of pathogenic bacteria. Crit Rev Microbiol. 2020;46:136–146. doi: 10.1080/1040841X.2020.1735991. [DOI] [PubMed] [Google Scholar]
- 7.Berlanga M., Gomez-Perez L., Guerrero R. Biofilm formation and antibiotic susceptibility in dispersed cells versus planktonic cells from clinical, industry and environmental origins. Antonie Leeuwenhoek. 2017;110:1691–1704. doi: 10.1007/s10482-017-0919-2. [DOI] [PubMed] [Google Scholar]
- 8.Bose S., Grammer L.C., Peters A.T. Infectious chronic rhinosinusitis. J Allergy Clin Immunol Pract. 2016;4:584–589. doi: 10.1016/j.jaip.2016.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandstetter K.A., Jurcisek J.A., Goodman S.D., Bakaletz L.O., Das S. Antibodies directed against integration host factor mediate biofilm clearance from Nasopore. Laryngoscope. 2013;123:2626–2632. doi: 10.1002/lary.24183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brockson M.E., Novotny L.A., Mokrzan E.M., Malhotra S., Jurcisek J.A., Akbar R., Devaraj A., Goodman S.D., Bakaletz L.O. Evaluation of the kinetics and mechanism of action of anti-integration host factor-mediated disruption of bacterial biofilms. Mol Microbiol. 2014;93:1246–1258. doi: 10.1111/mmi.12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burki T.K. Superbugs: an arms race against bacteria. Lancet Respir Med. 2018;6:668. doi: 10.1016/S2213-2600(18)30271-6. [DOI] [PubMed] [Google Scholar]
- 12.Cappelletty D. Microbiology of bacterial respiratory infections. Pediatr Infect Dis J. 1998;17:S55–S61. doi: 10.1097/00006454-199808001-00002. [DOI] [PubMed] [Google Scholar]
- 13.Chambers J.R., Cherny K.E., Sauer K. Susceptibility of Pseudomonas aeruginosa dispersed cells to antimicrobial agents is dependent on the dispersion cue and class of the antimicrobial agent used. Antimicrob Agents Chemother. 2017;61 doi: 10.1128/AAC.00846-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chua S.L., Liu Y., Yam J.K., Chen Y., Vejborg R.M., Tan B.G., Kjelleberg S., Tolker-Nielsen T., Givskov M., Yang L. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun. 2014;5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 15.Ciofu O., Moser C., Jensen P.O., Hoiby N. Tolerance and resistance of microbial biofilms. Nat Rev Microbiol. 2022;20:621–635. doi: 10.1038/s41579-022-00682-4. [DOI] [PubMed] [Google Scholar]
- 16.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 17.Dalia A.B., Weiser J.N. Minimization of bacterial size allows for complement evasion and is overcome by the agglutinating effect of antibody. Cell Host Microbe. 2011;10:486–496. doi: 10.1016/j.chom.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Devaraj A., Justice S.S., Bakaletz L.O., Goodman S.D. DNABII proteins play a central role in UPEC biofilm structure. Mol Microbiol. 2015;96:1119–1135. doi: 10.1111/mmi.12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devaraj A., Novotny L.A., Robledo-Avila F.H., Buzzo J.R., Mashburn-Warren L., Jurcisek J.A., Tjokro N.O., Partida-Sanchez S., Bakaletz L.O., Goodman S.D. The extracellular innate-immune effector HMGB1 limits pathogenic bacterial biofilm proliferation. J Clin Invest. 2021;131 doi: 10.1172/JCI140527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dhekane R., Mhade S., Kaushik K.S. Adding a new dimension: multi-level structure and organization of mixed-species Pseudomonas aeruginosa and Staphylococcus aureus biofilms in a 4-D wound microenvironment. Biofilms. 2022;4 doi: 10.1016/j.bioflm.2022.100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dicker A.J., Huang J.T.J., Lonergan M., Keir H.R., Fong C.J., Tan B., Cassidy A.J., Finch S., Mullerova H., Miller B.E., Tal-Singer R., Chalmers J.D. The sputum microbiome, airway inflammation, and mortality in chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2021;147:158–167. doi: 10.1016/j.jaci.2020.02.040. [DOI] [PubMed] [Google Scholar]
- 22.Duguid I.G., Evans E., Brown M.R., Gilbert P. Effect of biofilm culture upon the susceptibility of Staphylococcus epidermidis to tobramycin. J Antimicrob Chemother. 1992;30:803–810. doi: 10.1093/jac/30.6.803. [DOI] [PubMed] [Google Scholar]
- 23.Escribano Montaner A., Garcia de Lomas J., Villa Asensi J.R., Asensio de la Cruz O., de la Serna Blazquez O., Santiago Burruchaga M., Mondejar Lopez P., Torrent Vernetta A., Feng Y., Van Dyke M.K., Reyes J., Garcia-Corbeira P., Talarico C.A., group E.P.-S.-s. Bacteria from bronchoalveolar lavage fluid from children with suspected chronic lower respiratory tract infection: results from a multi-center, cross-sectional study in Spain. Eur J Pediatr. 2018;177:181–192. doi: 10.1007/s00431-017-3044-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan Q., Wang H., Mao C., Li J., Zhang X., Grenier D., Yi L., Wang Y. Structure and signal regulation mechanism of interspecies and interkingdom quorum sensing system receptors. J Agric Food Chem. 2022;70:429–445. doi: 10.1021/acs.jafc.1c04751. [DOI] [PubMed] [Google Scholar]
- 25.Freire M.O., Devaraj A., Young A., Navarro J.B., Downey J.S., Chen C., Bakaletz L.O., Zadeh H.H., Goodman S.D. A bacterial-biofilm-induced oral osteolytic infection can be successfully treated by immuno-targeting an extracellular nucleoid-associated protein. Mol Oral Microbiol. 2017;32:74–88. doi: 10.1111/omi.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodman S.D., Obergfell K.P., Jurcisek J.A., Novotny L.A., Downey J.S., Ayala E.A., Tjokro N., Li B., Justice S.S., Bakaletz L.O. Biofilms can be dispersed by focusing the immune system on a common family of bacterial nucleoid-associated proteins. Mucosal Immunol. 2011;4:625–637. doi: 10.1038/mi.2011.27. [DOI] [PubMed] [Google Scholar]
- 27.Goodwine J., Gil J., Doiron A., Valdes J., Solis M., Higa A., Davis S., Sauer K. Pyruvate-depleting conditions induce biofilm dispersion and enhance the efficacy of antibiotics in killing biofilms in vitro and in vivo. Sci Rep. 2019;9:3763. doi: 10.1038/s41598-019-40378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Green B.J., Wiriyachaiporn S., Grainge C., Rogers G.B., Kehagia V., Lau L., Carroll M.P., Bruce K.D., Howarth P.H. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartmann R., Jeckel H., Jelli E., Singh P.K., Vaidya S., Bayer M., Rode D.K.H., Vidakovic L., Diaz-Pascual F., Fong J.C.N., Dragos A., Lamprecht O., Thoming J.G., Netter N., Haussler S., Nadell C.D., Sourjik V., Kovacs A.T., Yildiz F.H., Drescher K. Quantitative image analysis of microbial communities with BiofilmQ. Nat Microbiol. 2021;6:151–156. doi: 10.1038/s41564-020-00817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang K., Xu Y., Yuan B., Yue Y., Zhao M., Luo R., Wu H., Wang L., Zhang Y., Xiao J., Lin F. Effect of autoinducer-2 quorum sensing inhibitor on interspecies quorum sensing. Front Microbiol. 2022;13 doi: 10.3389/fmicb.2022.791802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jurcisek J.A., Bakaletz L.O. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J Bacteriol. 2007;189:3868–3875. doi: 10.1128/JB.01935-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurbatfinski N., Goodman S.D., Bakaletz L.O. A humanized monoclonal antibody potentiates killing of diverse biofilm-forming respiratory tract pathogens by antibiotics. Antimicrob Agents Chemother. 2022;66 doi: 10.1128/AAC.01877-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewnard J.A., Givon-Lavi N., Dagan R. Interaction with nontypeable Haemophilus influenzae alters progression of Streptococcus pneumoniae from colonization to disease in a site-specific manner. J Infect Dis. 2019;220:1367–1376. doi: 10.1093/infdis/jiz312. [DOI] [PubMed] [Google Scholar]
- 34.Lim J.H., Ha U., Sakai A., Woo C.H., Kweon S.M., Xu H., Li J.D. Streptococcus pneumoniae synergizes with nontypeable Haemophilus influenzae to induce inflammation via upregulating TLR2. BMC Immunol. 2008;9:40. doi: 10.1186/1471-2172-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luke N.R., Jurcisek J.A., Bakaletz L.O., Campagnari A.A. Contribution of Moraxella catarrhalis type IV pili to nasopharyngeal colonization and biofilm formation. Infect Immun. 2007;75:5559–5564. doi: 10.1128/IAI.00946-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magalhaes A.P., Franca A., Pereira M.O., Cerca N. Unveiling co-infection in cystic fibrosis airways: transcriptomic analysis of Pseudomonas aeruginosa and Staphylococcus aureus dual-species biofilms. Front Genet. 2022;13 doi: 10.3389/fgene.2022.883199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahenthiralingam E., Bischof J., Byrne S.K., Radomski C., Davies J.E., Av-Gay Y., Vandamme P. DNA-Based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol. 2000;38:3165–3173. doi: 10.1128/JCM.38.9.3165-3173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Margolis E., Yates A., Levin B.R. The ecology of nasal colonization of Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus: the role of competition and interactions with host's immune response. BMC Microbiol. 2010;10:59. doi: 10.1186/1471-2180-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marks L.R., Davidson B.A., Knight P.R., Hakansson A.P. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio. 2013;4 doi: 10.1128/mBio.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mason K.M., Munson R.S., Jr., Bakaletz L.O. Nontypeable Haemophilus influenzae gene expression induced in vivo in a chinchilla model of otitis media. Infect Immun. 2003;71:3454–3462. doi: 10.1128/IAI.71.6.3454-3462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mayhew D., Devos N., Lambert C., Brown J.R., Clarke S.C., Kim V.L., Magid-Slav M., Miller B.E., Ostridge K.K., Patel R., Sathe G., Simola D.F., Staples K.J., Sung R., Tal-Singer R., Tuck A.C., Van Horn S., Weynants V., Williams N.P., Devaster J.M., Wilkinson T.M.A., Group A.S. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax. 2018;73:422–430. doi: 10.1136/thoraxjnl-2017-210408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCool T.L., Cate T.R., Moy G., Weiser J.N. The immune response to pneumococcal proteins during experimental human carriage. J Exp Med. 2002;195:359–365. doi: 10.1084/jem.20011576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mokrzan E.M., Ward M.O., Bakaletz L.O. Type IV pilus expression is upregulated in nontypeable Haemophilus influenzae biofilms formed at the temperature of the human nasopharynx. J Bacteriol. 2016;198:2619–2630. doi: 10.1128/JB.01022-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mokrzan E.M., Novotny L.A., Brockman K.L., Bakaletz L.O. Antibodies against the majority subunit (PilA) of the type IV pilus of nontypeable Haemophilus influenzae disperse Moraxella catarrhalis from a dual-species biofilm. mBio. 2018;9 doi: 10.1128/mBio.02423-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mokrzan E.M., Ahearn C.P., Buzzo J.R., Novotny L.A., Zhang Y., Goodman S.D., Bakaletz L.O. Nontypeable Haemophilus influenzae newly released (NRel) from biofilms by antibody-mediated dispersal versus antibody-mediated disruption are phenotypically distinct. Biofilms. 2020;2 doi: 10.1016/j.bioflm.2020.100039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moscoso M., Garcia E., Lopez R. Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J Bacteriol. 2006;188:7785–7795. doi: 10.1128/JB.00673-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murphy T.F., Faden H., Bakaletz L.O., Kyd J.M., Forsgren A., Campos J., Virji M., Pelton S.I. Nontypeable Haemophilus influenzae as a pathogen in children. Pediatr Infect Dis J. 2009;28:43–48. doi: 10.1097/INF.0b013e318184dba2. [DOI] [PubMed] [Google Scholar]
- 48.No time to wait: securing the future from drug-resistant infections. 2019. https://www.who.int/publications/i/item/no-time-to-wait-securing-the-future-from-drug-resistant-infections
- 49.Novotny L.A., Adams L.D., Kang D.R., Wiet G.J., Cai X., Sethi S., Murphy T.F., Bakaletz L.O. Epitope mapping immunodominant regions of the PilA protein of nontypeable Haemophilus influenzae (NTHI) to facilitate the design of two novel chimeric vaccine candidates. Vaccine. 2009;28:279–289. doi: 10.1016/j.vaccine.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novotny L.A., Clements J.D., Bakaletz L.O. Transcutaneous immunization as preventative and therapeutic regimens to protect against experimental otitis media due to nontypeable Haemophilus influenzae. Mucosal Immunol. 2011;4:456–467. doi: 10.1038/mi.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novotny L.A., Amer A.O., Brockson M.E., Goodman S.D., Bakaletz L.O. Structural stability of Burkholderia cenocepacia biofilms is reliant on eDNA structure and presence of a bacterial nucleic acid binding protein. PLoS One. 2013;8 doi: 10.1371/journal.pone.0067629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Novotny L.A., Jurcisek J.A., Ward M.O., Jr., Jordan Z.B., Goodman S.D., Bakaletz L.O. Antibodies against the majority subunit of type IV pili disperse nontypeable Haemophilus influenzae biofilms in a LuxS-dependent manner and confer therapeutic resolution of experimental otitis media. Mol Microbiol. 2015;96:276–292. doi: 10.1111/mmi.12934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novotny L.A., Jurcisek J.A., Goodman S.D., Bakaletz L.O. Monoclonal antibodies against DNA-binding tips of DNABII proteins disrupt biofilms in vitro and induce bacterial clearance in vivo. EBioMedicine. 2016;10:33–44. doi: 10.1016/j.ebiom.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Novotny L.A., Goodman S.D., Bakaletz L.O. Redirecting the immune response towards immunoprotective domains of a DNABII protein resolves experimental otitis media. NPJ Vaccines. 2019;4:43. doi: 10.1038/s41541-019-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Novotny L.A., Goodman S.D., Bakaletz L.O. Targeting a bacterial DNABII protein with a chimeric peptide immunogen or humanised monoclonal antibody to prevent or treat recalcitrant biofilm-mediated infections. EBioMedicine. 2020;59 doi: 10.1016/j.ebiom.2020.102867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettigrew M.M., Marks L.R., Kong Y., Gent J.F., Roche-Hakansson H., Hakansson A.P. Dynamic changes in the Streptococcus pneumoniae transcriptome during transition from biofilm formation to invasive disease upon influenza A virus infection. Infect Immun. 2014;82:4607–4619. doi: 10.1128/IAI.02225-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pulingam T., Parumasivam T., Gazzali A.M., Sulaiman A.M., Chee J.Y., Lakshmanan M., Chin C.F., Sudesh K. Antimicrobial resistance: prevalence, economic burden, mechanisms of resistance and strategies to overcome. Eur J Pharmaceut Sci. 2022;170 doi: 10.1016/j.ejps.2021.106103. [DOI] [PubMed] [Google Scholar]
- 58.Ramakrishnan V.R., Hauser L.J., Feazel L.M., Ir D., Robertson C.E., Frank D.N. Sinus microbiota varies among chronic rhinosinusitis phenotypes and predicts surgical outcome. J Allergy Clin Immunol. 2015;136:334–342 e331. doi: 10.1016/j.jaci.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 59.Rickard A.H., Gilbert P., High N.J., Kolenbrander P.E., Handley P.S. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/s0966-842x(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 60.Rocco C.J., Bakaletz L.O., Goodman S.D. Targeting the HUbeta protein prevents Porphyromonas gingivalis from entering into preexisting biofilms. J Bacteriol. 2018;200 doi: 10.1128/JB.00790-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez J.L., Dalia A.B., Weiser J.N. Increased chain length promotes pneumococcal adherence and colonization. Infect Immun. 2012;80:3454–3459. doi: 10.1128/IAI.00587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rotta Detto Loria J., Rohmann K., Droemann D., Kujath P., Rupp J., Goldmann T., Dalhoff K. Nontypeable Haemophilus influenzae infection upregulates the NLRP3 inflammasome and leads to caspase-1-dependent secretion of Interleukin-1beta - a possible pathway of exacerbations in COPD. PLoS One. 2013;8 doi: 10.1371/journal.pone.0066818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sauer K., Stoodley P., Goeres D.M., Hall-Stoodley L., Burmolle M., Stewart P.S., Bjarnsholt T. The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat Rev Microbiol. 2022;20:608–620. doi: 10.1038/s41579-022-00767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sirakova T., Kolattukudy P.E., Murwin D., Billy J., Leake E., Lim D., DeMaria T., Bakaletz L. Role of fimbriae expressed by nontypeable Haemophilus influenzae in pathogenesis of and protection against otitis media and relatedness of the fimbrin subunit to outer membrane protein A. Infect Immun. 1994;62:2002–2020. doi: 10.1128/iai.62.5.2002-2020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Slinger R., Chan F., Ferris W., Yeung S.W., St Denis M., Gaboury I., Aaron S.D. Multiple combination antibiotic susceptibility testing of nontypeable Haemophilus influenzae biofilms. Diagn Microbiol Infect Dis. 2006;56:247–253. doi: 10.1016/j.diagmicrobio.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 66.Starner T.D., Shrout J.D., Parsek M.R., Appelbaum P.C., Kim G. Subinhibitory concentrations of azithromycin decrease nontypeable Haemophilus influenzae biofilm formation and diminish established biofilms. Antimicrob Agents Chemother. 2008;52:137–145. doi: 10.1128/AAC.00607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Eldere J., Slack M.P., Ladhani S., Cripps A.W. Non-typeable Haemophilus influenzae, an under-recognised pathogen. Lancet Infect Dis. 2014;14:1281–1292. doi: 10.1016/S1473-3099(14)70734-0. [DOI] [PubMed] [Google Scholar]
- 68.Ventola C.L. The antibiotic resistance crisis: part 1: causes and threats. P T. 2015;40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 69.Wang Z., Maschera B., Lea S., Kolsum U., Michalovich D., Van Horn S., Traini C., Brown J.R., Hessel E.M., Singh D. Airway host-microbiome interactions in chronic obstructive pulmonary disease. Respir Res. 2019;20:113. doi: 10.1186/s12931-019-1085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waters C.M., Bassler B.L. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 71.Weeks J.R., Staples K.J., Spalluto C.M., Watson A., Wilkinson T.M.A. The role of non-typeable Haemophilus influenzae biofilms in chronic obstructive pulmonary disease. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.720742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Welp A.L., Bomberger J.M. Bacterial community interactions during chronic respiratory disease. Front Cell Infect Microbiol. 2020;10:213. doi: 10.3389/fcimb.2020.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Whitchurch C.B., Tolker-Nielsen T., Ragas P.C., Mattick J.S. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.