Summary
The TERT/CLPTM1L risk locus on chromosome 5p15.33 is a pleiotropic cancer risk locus in which multiple independent risk alleles have been identified, across well over ten cancer types. We previously conducted a genome-wide association study in uveal melanoma (UM), which uncovered a role for the TERT/CLPTM1L risk locus in this intraocular tumor and identified multiple highly correlated risk alleles. Aiming to unravel the biological mechanisms in UM of this locus, which contains a domain enriched in active chromatin marks and enhancer elements, we demonstrated the allele-specific enhancer activity of this risk region using reporter assays. In UM, we identified the functional variant rs452384, of which the C risk allele is associated with higher gene expression, increased CLPTM1L expression in UM tumors, and a longer telomere length in peripheral blood mononuclear cells. Electrophoretic mobility shift assays and quantitative mass spectrometry identified NKX2.4 as an rs452384-T-specific binding protein, whereas GATA4 preferentially interacted with rs452384-C. Knockdown of NKX2.4 but not GATA4 resulted in increased TERT and CLPTM1L expression. In summary, the UM risk conferred by the 5p locus is at least partly due to rs452384, for which NKX2.4 presents strong differential binding activity and regulates CLPTM1L and TERT expression. Altogether, our work unraveled some of the complex regulatory mechanisms at the 5p15.33 susceptibility region in UM, and this might also shed light on shared mechanisms with other tumor types affected by this susceptibility region.
Keywords: cancer genetics, functional genomics, TERT/CLPTM1L, uveal melanoma
Graphical abstract

Derrien et al. identified rs452384 as a functional variant implicated in uveal melanoma risk at the multi-cancer 5p15.33 risk locus, mediating allele-specific expression and differentially binding NKX2.4 and GATA4 transcription factors. These findings unravel some of the complex regulatory mechanisms in uveal melanoma susceptibility.
Introduction
Although a rare cancer, uveal melanoma (UM) is the most frequent primary intraocular malignant tumor in adults. It arises from the malignant transformation of melanocytes from the uveal tract composed of the choroid, ciliary body, and iris. In up to 50% of individuals with UM, the disease metastasizes, almost invariably to the liver,1,2 and is associated with poor prognosis and a median survival of 12 months. UM is a genetically simple tumor, with two main oncogenic driver events characterized by the constitutive activation of the Gαq pathway through mutually exclusive mutations of GNA11, GNAQ, or more rarely CYSLTR2 or PLCB4,3,4 and a second event that involves almost mutually exclusive mutations of BAP1, SF3B1, or EIF1AX, defining UM subgroups with high, middle, and low metastatic risk, respectively.5,6,7
UM has an incidence rate of 5.6 persons per million persons per year in the US and, strikingly, is more frequent (10- to 20-fold higher incidence) in populations of European ancestry compared with those of Asian or African-American origins.8,9 Other important risk factors for UM include light iris color and fair skin;10 however a role for pigmentation protecting against ultra-violet radiation to explain this epidemiology was excluded by sequencing studies.6,11 Both familial occurrences of UM and population studies suggest the implication of genetic risk factors in UM. To date, BAP1 is the only known highly penetrant predisposition gene in UM,12,13 with pathogenic germline mutations occurring in 1.6% of all individuals with UM.14 Recently, we identified MBD4 as a UM predisposition gene with moderate penetrance, conferring a relative risk of 9.2 of developing a UM.15 However, these genes explain only a fraction of familial UM cases, suggesting the presence of additional genetic risk factors in UM.
We recently conducted a genome-wide association study (GWAS) in UM with genetic imputation, including a total of 1,142 individuals with UM and 882 healthy controls.16 In addition to identifying two risk regions involved in eye pigmentation determination, at the IRF4 (chr6) and HERC2/OCA2 (chr15) loci, we also confirmed the previously identified association signal on the TERT/CLPTM1L locus on chromosome 5p15.33, comprising multiple risk variants in high linkage disequilibrium (LD) spanning CLPTM1L introns.16,17 The lack of functional evidence explaining the mechanisms underlying this 5p15.33 genomic region in UM prompted us to investigate the biological consequences of the TERT/CLPTM1L risk locus in UM.
The 5p15.33 TERT/CLPTM1L locus is an extensively characterized multi-cancer risk locus,18,19,20,21 and over 10 tumor types are associated with this risk region, including carcinomas from ER-negative breast, colon, lung, pancreas, prostate, kidney, ovary, head and neck, esophagus, and endometrium, as well as germ cell tumor, cutaneous melanoma, and glioma. Importantly, up to ten independent risk loci have now been identified within this genomic region, encompassing both CLPTM1L and TERT.18,20 In addition, some alleles have different directionalities of effect (risk or protective, depending on the tumor type),18,20,22 suggesting tissue-specific regulation and oncogenic consequences. The association signal found by our GWAS, led by rs370348 (odds ratio OR [CI 95%] = 1.59 [1.35; 1.86]) and corresponding to the region marked by rs465498 in the metanalysis of Chen and colleagues, was also found in cutaneous melanoma, lung, and pancreatic cancers.18,23,24,25 Conditional analyses in our GWAS revealed that no independent variant in this region was associated with UM risk other than the identified locus marked by rs370348.
Although the TERT/CLPTM1L locus has been extensively characterized in some tumor types, much of the underlying biological mechanisms remain to be deciphered. To investigate the underlying biological mechanisms of UM predisposition linked to the 5p15.33 TERT/CLPTM1L locus, we functionally characterized the susceptibility region and in UM identified rs452384 as a functional variant that mediates allele-specific binding of the NKX2.4 nuclear factor and the transcriptional activity of the region, including TERT and CLPTM1L.
Materials/subjects and methods
Additional detailed methods are provided in the supplemental information.
Subjects
Subjects included in the study of telomere length are a subpopulation (N = 326) of our GWAS,16 and include germline DNA of 118 European controls and 208 individuals with UM (age, sex, and genotype of individuals are provided in Table S1). Since the European control KIDRISK cohort is strongly biased for male individuals (to match kidney cancer population), the telomere analysis was restricted to male individuals in order to avoid sex as a confounding factor. The study was approved by the Ethical Committee and Internal Review Board at the Institut Curie, and all individuals consented to participate in the study.
Cell lines and culture
All cell lines described in this study are derived from UM. The MP41 and MM66 cell lines were derived at Institut Curie.26 The Mel202 cell line was purchased from the European Searchable Tumour Line Database (Tubingen University, Germany). The OMM1 and OMM2.5 cell lines were kindly provided by P.A. van der Velden (Leiden University Medical Center, the Netherlands). Cell lines were cultured in RPMI 1640 (Gibco) supplemented with 10% fetal bovine serum (FBS) (MP41, Mel202, OMM1, and OMM2.5) or 20% FBS (MM66). Authentication of cell lines was verified by short tandem repeat (STR) testing. Cells were regularly tested for Mycoplasma and found to be negative each time.
In silico characterization of the TERT/CLPTM1L region and functional variant prioritization
All combinations of inter-variant allele count squared correlation (r2) were calculated with plink 1.9 (https://www.cog-genomics.org/plink/1.9) using all available genotypes from our previous study16 that are localized on CLPTM1L (chr5:1317867-1345214, hg19). To prioritize probable functional variants and narrow down the number of candidate risk SNPs kept for functional analysis, ENCODE annotations (H3K27ac, DNase I hypersensitivity (DHS)-sequencing, transcription factor (TF) chromatin immunoprecipitation sequencing (ChIP-seq), and DNA repeated regions) from the CLPTM1L region were downloaded using UCSC genome browser and plots were done with R (v4.1.1). H3K27ac and DHS annotations were generated from ENCODE sequencing data from all available cell lines: GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK, NHLF (for H3K27ac annotations) and K562, NHLF, HSMM, HUVEC, NHEK, HepG2, GM12878, H7-ES, and HeLa-S3 cell lines (for DHS annotations).
RNA and DNA extraction
RNA and genomic DNA extraction from UM cell lines was performed using Qiagen’s RNeasy Plus Mini Kit and DNeasy Blood & Tissue kit respectively, following the manufacturer’s instructions. RNA and DNA quality and yield were obtained using a Nanodrop 2000 spectrophotometer. Tumor DNA and RNA samples from individuals with UM were further purified from melanin and other contaminants using the OneStep PCR Inhibitor Removal Kit (Zymo Research).
Vector constructs and luciferase assay
The 3kb genomic region surrounding UM risk SNPs was PCR amplified from the MP41 cell line, heterozygous for the 5p UM risk haplotype.17 HD Cloning Plus kit was used for PCR amplification of the 3kb fragment and to subclone the insert into a pGL3-Promoter Firefly luciferase reporter vector (Promega). The same technique was used in the fragment deletion analysis (fragments A−G). All primers are listed in Table S2. Plasmids were amplified and verified at the insert position by Sanger sequencing, and clones of both haplotypes (high-risk and low-risk) were selected. Sanger sequencing of high-risk and low-risk plasmids and previously performed whole-genome sequencing of the MP41 cell line confirmed the absence of SNPs (other than the three tested SNPs) and of somatic mutations differing between the two haplotypes. The only exception is a small repetitive sequence upstream of fragment F (Figure 1A, chr5: 1,129,600–1,130,200) in which the absence of sequence difference could not be verified due to the poor mappability and alignment of the sequence – potential SNPs in this region were thus excluded from the analysis. Plasmids were co-transfected with pRL-CMV Renilla luciferase control vector (Promega) in UM cell lines using Lipofectamine 2000 reagent (ThermoFisher Scientific) following the manufacturers’ instructions. Cells were collected 48h post-transfection, and luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega). Luminescence signal from Firefly luciferase was normalized to that of Renilla luciferase, and normalized luminescence was compared with that of transfection with empty vector.
Figure 1.
Allele-specific regulatory activity conferred by rs452384 at the TERT/CLPTM1L chr5p15.33 uveal melanoma risk locus
(A) A sub-region at the TERT/CLPTM1L risk locus is enriched in marks of open chromatin and behaves as an enhancer. Top panel: Map of the TERT/CLPTM1L genomic region along with highly correlated variants associated with risk of uveal melanoma (UM) identified in our initial GWAS study, aligned on hg19 genome. Chromosomal positions of TERT and CLPTM1L are shown, where vertical bars represent exons and arrows indicate transcriptional directionality. Significant UM risk SNPs are shown in red bars, and other SNPs appear in grey. LD pairwise correlation structure for SNPs in this region (zoom-in of CLPTM1L) is shown by shadings of grey according to r2 values. Layered ENCODE chromatin marks (H3K27ac, DNase I hypersensitivity sequencing, transcription factor ChIP-seq -TF - and DNA repeated regions) were downloaded from UCSC genome browser and were generated from ENCODE using sequencing data from all available cell lines (GM12878 in red, H1-hESC in yellow, HSMM in turquoise, HUVEC in light blue, K562 in blue, NHEK in violet, and NHLF in pink). Finally, genomic positions of amplicons used for ChIP experiments are shown by orange circles (ChIP-H3K27ac) and green squares (ChIP-NKX2.4 and GATA4). Bottom panel: Fragment deletion analysis strategy to narrow down the sub-region from the full haploblock tested (“full vector”; represented by a red rectangle), mediating the most differential luciferase activity between the high-risk (HR) and low-risk (LR) UM haplotypes. The full vector insert was trimmed from its 5' end or 3' end to generate smaller fragments A to G, represented by grey bars and subsequently used for luciferase assays (Figure 1C). UM risk variants within the region tested are shown in black and red.
(B) Allele-specific luciferase activity of the HR and LR haploblocks of the CLPTM1L locus enriched in active histone marks (red fragment in Figure 1A) in two UM cell lines, MP41 (Figure 1) and OMM2.5 (Figure S2). The enhancer region (“full vector”) containing either protective or risk UM variants was cloned upstream of a pGL3-promoter reporter vector, followed by transfection and luciferase assay. Luciferase activity was normalized to Renilla luciferase levels and to empty vector (EV). Mean relative luciferase activity ± standard deviation (SD) is shown (left); experiments were performed in triplicate and repeated independently in n = 6 experiments. Mean HR/LR luciferase ratios ± SD are also shown (right); the hypothetical ratio of HR/LR = 1, representing equal luciferase activity between the two haplotypes, is indicated by a black dashed line.
(C) Fragment deletion analysis by luciferase activity assay. Individual fragments A to G (Figure 1A) of both haplotypes (HR and LR) were tested for luciferase activity as described in 1B, in MP41 (Figure 1) and OMM2.5 (Figure S2). Graphs represent the differential luciferase ratio (HR/LR) observed in each fragment, compared with vector containing the full insert initially tested (“full vector”). Mean luciferase ratios ± SD are shown for n = 3 independent experiments.
(D) Site-directed mutagenesis in reporter vectors harboring Fragment F (“Fragment F vector”) of rs452932, rs452384, and rs370348, the three variants associated with UM risk within the tested region, from their protective to risk allele in MP41 (Figure 1) and OMM2.5 (Figure S2). Resulting vectors were tested in luciferase activity assays and compared to vectors containing the full LR or HR haplotypes (i.e., Fragment F vector harboring all three variants with either their protective allele or risk allele). Mean luciferase ratios ± SD are shown for n = 3 independent experiments.
(E) Double site-directed mutagenesis at rs452932 and rs452384 positions (both SNPs T>C) in MP41, compared with “single” mutants described in 1D by luciferase activity assays. The same process was subsequently repeated starting from the HR haplotype vector and mutating tested variants to their protective allele (C>T). Graphs represent mean ± SD for experiments carried out n = 3 independent times. Unpaired two-sided t test p values: ns for p > 0.05, ∗ for p < 0.05, ∗∗ for p < 0.005, and ∗∗∗ for p < 0.001.
Site-directed mutagenesis
Site-directed mutagenesis was performed on the full-length pGL3-P-CLPTM1L vector (CLPTM1L intronic enhancer sequence described above) to evaluate the individual effects of the three SNPs, rs452932T/C, rs452384T/C, and rs370348A/G, identified in the UM GWAS within the insert sequence. The QuickChange II Site-directed mutagenesis kit (Agilent) was used to specifically mutate the full-length pGL3-P-CLPTM1L vector at the three individual positions, using 35 to 45bp mutagenic primers (Table S2). After mutant strand synthesis using PfuUltra HF DNA polymerase, resulting DpnI-treated DNA was transformed into XL10-Gold ultracompetent cells (Agilent). This process was repeated on both genotypes (high- and low-risk) of the pGL3-P-CLPTM1L vector to obtain clones with all possible combinations of the three SNPs’ genotypes and validate in both directions (high-to-low risk and low-to-high risk) the effect of changing the SNPs’ alleles. Correct sequences were verified by Sanger sequencing of the mutant clones. Resulting vectors were assessed for luciferase activity described above.
Electrophoretic mobility shift assay (EMSA)
The detailed procedure is available in the supplemental information. 25bp biotinylated DNA double-stranded oligos centered around rs452384 alleles are described in Table S2. EMSAs were performed using the LightShift Chemiluminescent EMSA Kit (20148 Thermo Scientific). For supershift assays using FLAG-tagged proteins, MP41 cells were transfected with pcDNA3.1(+)C-(K)DYK (C terminus FLAG tag) vectors expressing candidate proteins (NKX2.4, GATA4, DLX6, or PITX2) or empty vector (GenScript). Protein overexpression was verified by western blotting using monoclonal anti-Flag M2 antibody (Sigma F1804), anti-NKX2.4 antibody (abcam ab189202), anti-GATA4 antibody (Invitrogen MA5-15532), and anti-Histone H3 antibody (abcam ab1791) as a nuclear loading control. Supershift experiments were carried out by adding 2μg monoclonal anti-Flag M2 antibody (Sigma F1804) or 2μg normal mouse IgG (Santa Cruz sc2025) antibody as a negative control.
DNA pulldown and mass spectrometry
The full procedure (pulldown, mass spectrometry, data analysis) is described in the supplemental information. Proteins bound to rs452384-C or -T alleles were obtained by DNA pulldown assay using 25bp biotinylated double-stranded probes (including a negative control probe, Table S2) and Dynabeads M−280 Streptavidin (11205D Invitrogen), performed in n = 5 replicates. Peptides bound to beads were identified by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
ChIP
ChIP experiments were carried out using the iDeal ChIP-Seq kit for Transcription Factors (Diagenode C01010055) according to the manufacturer’s instructions, using MP41 (rs452384-TC), Mel202 (rs452384-TT), and OMM1 (rs452384-CC) cell lines. Due to the absence of ChIP-grade antibody for NKX2.4, cells were first transfected with a pcDNA3.1(+)NKX2.4-HA expression vector (GenScript) using lipofectamine 2000 (ThermoFisher Scientific) and following the manufacturer’s instructions. ChIP experiments were carried out using 12 million cells per condition, 48h after transfection for NKX2.4 ChIP. 4μg of anti-HA tag antibody (ChIP grade, Abcam ab9110), 4 μg of anti-GATA4 antibody (Invitrogen MA5-15532), or 4 μg of rabbit IgG as a nonspecific control (Diagenode C15410206) were used for individual ChIP experiments on sheared chromatin from 4 million cells. For Mel202 and OMM1 ChIP, IP’d DNA was then analyzed by qPCR using SYBR Green PCR Master Mix (Applied Biosystems 4309155) and qPCR primers designed to amplify the rs452384 target region in both rs452384-TT and -CC genotype contexts compared with other sites within the CLPTM1L gene (Figure 1A and Table S2). For MP41, input and IP’d DNA was assessed by droplet digital PCR (ddPCR) for analysis of allelic enrichment (supplemental methods). For H3K27ac ChIP on MP41 and OMM2.5 cell lines, essentially the same protocol was used except that the iDeal ChIP-Seq kit for histones (Diagenode C01010051) was used, with 4μg of H3K27ac antibody (Diagenode C15410196) and 4μg of negative control IgG antibody (described earlier).
End-point droplet digital PCR
Following NKX2.4 and/or GATA4 knockdowns in Mel202 and OMM1 cell lines (supplemental methods), expression levels of TERT and CLPTM1L were assessed. Due to the very low expression of TERT in UM, ddPCR was used to assess expression levels of TERT and CLPTM1L normalized to that of housekeeping gene GUSB using Bio-Rad ddPCR technology and following manufacturer’s instructions (Mel202: n = 5 independent experiments for siNKX2.4, n = 3 for siGATA4; OMM1: n = 3 independent experiments for siNKX2.4). Taqman Gene Expression assays targeting either TERT (FAM-labeled, Cat# Hs00972650_m1) or CLPTM1L (FAM-labeled, Cat# Hs00363947_m1) in combination with GUSB (VIC-labeled, Cat# Hs00939627_m1) were used in PCR reactions containing ddPCR Supermix for probes (No dUTP) (Bio-Rad). Water-emulsion droplets were generated using Bio-Rad QX100 Droplet Generator, and resulting mixes were read in a QX100 Droplet Reader after PCR on a C1000 Touch thermocycler. ddPCR data were analyzed with Bio-Rad QuantaSoft™ software, reporting fluorescence from channel 1 (FAM) and channel 2 (VIC) on the basis of the number of FAM- and VIC- single-positive droplets (fitted to a Poisson distribution). Relative expression ratios of TERT/GUSB and CLPTM1L/GUSB represent the ratio of concentration of target DNA molecule (in units of copies/mL input) for each fluorophore/target molecule. To compare relative gene expression (expression ratios) between conditions (siCTRL, siNKX2.4, and siGATA4), unpaired two-sided t tests with pooled biological variances were used.
Telomere length measurement
Cohorts used for the analysis are described in Subjects. The full procedure is available in the supplemental information. Relative telomere length was measured by quantitative PCR using Cawthon’s gold standard method,27 with primers targeting TERT and HMBS (single copy gene) (Table S2), adapted to correct amplification efficiency using the TeloPCR method.28 Samples were stratified according to their rs452384 genotype in order to assess correlation with telomere length.
Statistical analysis
For luciferase and qPCR assays, unpaired two-sided t tests were used to assess statistical significance between measures. For enrichment of proteins in the mass spectrometry analysis and to determine the significance of changes in protein abundance between two conditions, a linear model (adjusted on peptides and biological replicates) was performed, and p values were adjusted using the Benjamini–Hochberg false discovery rate (FDR) procedure. In ddPCR analyses, t tests with pooled biological variances were used to assess significance of differences in gene expression between groups and differences in allelic enrichment ratios in ChIP experiments (see supplemental methods). For telomere length analysis, difference in telomere length between the three rs452384 genotypes was measured using an age-adjusted generalized linear additive model (GLM). To assess pairwise differences in telomere length, non-parametric tests (3-way non-parametric Kruskal-Wallis Rank Sum test and pairwise Wilcoxon Rank Sum test) were used. P values < 0.05 were considered statistically significant. Representation of significance on graphs is as follow: ns for p > 0.05, ∗ for p < 0.05, ∗∗ for p < 0.005, and ∗∗∗ for p < 0.001.
Results
The 5p15.33 UM risk region mediates allele-specific regulatory effects
Our GWAS in UM identified three susceptibility regions, including one at chromosome 5p15.33 at the TERT/CLPTM1L locus, marked by rs370348 and containing multiple genetically linked SNPs spanning CLPTM1L introns (Figure 1A).16 To prioritize probable functional variants mediating regulatory activity, we searched the regulatory marks surrounding UM risk SNPs annotated by the ENCODE project on the UCSC Genome Browser. A 3kb sub-region (hg19; chr5:1,329,015–1,332,000) was enriched in acetylated H3K27 (H3K27ac), a major histone mark flanking enhancer elements (Figure 1A).29 This region is also associated with DNase I hypersensitivity clusters, indicating a region of open chromatin, in agreement with the presence of TF ChIP-seq clusters at this genomic region (Figure 1A). Of note, the directly upstream region (hg19; chr5:1,326,000–1,329,000) also displayed moderate H3K27ac enrichment. However, this region consists of unmappable highly repetitive minisatellite sequences (Figure 1A), making both in silico and in vitro functional characterization challenging, and thus was not kept for further analysis.
The 3kb region retained for analysis contained three SNPs (rs452932, rs452384, and rs370348) that were significantly associated with UM risk (red bars in Figure 1A) and enriched in H3K27ac (Figure 1A). Furthermore, these SNPs were among variants with highest OR and most significant p values.16 These observations prompted us to further characterize this 3kb CLPTM1L sub-region.
We first assessed whether this region also harbored active chromatin elements in a UM context. To do so, we performed ChIP for H3K27ac mark using UM cell lines MP41 and OMM2.5, followed by quantitative PCR on the region surrounding rs452384. This SNP was chosen because it is directly flanked by two strong H3K27ac signals and because it lies within a DNase I cluster in the ENCODE database (Figure 1A). In MP41, the UM risk region surrounding rs452384 (CLPTM1L intron 8) indeed displayed a 3.5- to 4-fold higher enrichment over input DNA compared with two other CLPTM1L regions devoid of the regulatory marks, close to exons 3 and 4 (Figure S1A). In OMM2.5, the rs452384 region was also highly enriched in HK27ac (Figure S1B) compared with IgG and to the exon 4 region, and unlike in MP41, the exon 3 region was also enriched. To further characterize this region and search for an allele-specific differential enhancer activity, we subcloned the 3kb intronic regulatory region of both the low-risk (LR) and high-risk (HR) haplotypes 5' to the promoter of a pGL3-promoter reporter vector driving luciferase expression. This CLPTM1L regulatory region induced a strong increase in luciferase signal in two UM cell lines compared with empty vector (EV), characterized in MP41 by a 7.79-fold ±3.20 (mean ± SD over EV) enrichment for LR haplotype and a 21.01-fold ±4.73 for HR haplotype; in the OMM2.5 cell line, the enrichment was 3.38-fold ±0.83 for LR versus 6.37-fold ±1.56 for HR (Figures 1B and S2A). The average HR/LR luciferase ratio in MP41 was 2.85 ± 0.62 (mean ± SD, unpaired two-tailed t test pLRvsHR = 0.0002) and 1.90 ± 0.18 in OMM2.5 (pLRvsHR = 0.002) (Figure 1B). Taken together, these results confirmed that the CLPTM1L genomic risk region is part of an enhancer region, with a haplotype-specific transcriptional activity in a UM context characterized by a significant 2- to 3-fold higher increase for the HR haplotype compared with the LR.
Allele-specific regulatory effects at the chr5p UM risk locus are mediated by rs452384
We next performed a deletion analysis to further define the sub-region conferring the most differential haplotype-specific transcriptional activity (HR/LR ratio). We generated smaller fragments (A to G) from the full 3kb region (Figure 1A) and repeated the luciferase assay comparing the HR/LR ratio. Construct F mediated the most haplotype-specific luciferase activity in MP41 cells, and there were similar results in OMM2.5 (Figures 1C and S2B). Within construct F (and within the full region), three SNPs, rs452932, rs452384, and rs370348, are associated with UM risk. To test the individual effect of each SNP on luciferase activity, we performed site-directed mutagenesis of the full-length LR haplotype vector at the position of each variant to its HR allele (rs452932-C, rs452384-C, and rs370348-G). In both cell lines, although luciferase levels in the rs370348A>G mutant vector were close to levels in the LR vector, rs452932T>C resulted in a significant increase in luciferase activity toward the levels seen in the HR vector, and rs452384T>C caused an even more significant increase (Figures 1D and S2C). Transfection with a T>C double mutant (rs452932T>C; rs452384T>C) resulted in comparable luciferase levels between the T>C double mutant, rs452384T>C single mutant, and the HR vector (Figure 1E), ruling out a synergistic effect of the two SNPs. Reverse experiment mutating the HR vector to LR alleles confirmed these findings (Figure 1E).
Taken together, our data indicate that rs452384 is the variant within the tested region exhibiting the highest allele-specific gene regulation and that it might have a functional role in UM.
The rs452384 genotype is associated with CLPTM1L expression and telomere length
The two gene candidates most likely to be regulated in an allele-specific manner, driven by the rs452384 surrounding region, are CLPTM1L and TERT (Figure 1A). We previously reported an expression quantitative trait loci (eQTL) analysis showing that the rs421284 genotype at 5p15.33 (r2 > 0.9 with rs452384) is associated with CLPTM1L expression in UM tumors,17 and there is a positive correlation with the risk (C) allele. TERT expression in UM tumor samples is barely detectable, making eQTL analyses challenging. We thus performed a colocalization analysis based only on CLPTM1L eQTL. Using eCAVIAR30 and expression datasets from UM, normal cutaneous melanocytes, and GTEX tissues, we calculated the CoLocalization Posterior Probability (CLPP) of the SNPs in the candidate region, and none reached the proposed threshold of 0.01. Although this analysis failed to further pinpoint a potential causal SNP, it confirmed the same directionality of rs452384/rs465498 in CLPTM1L eQTL with primary melanocytes (Curie UVM: prs452384 = 9.07 x 10−3; prs465498 = 9.07 x 10−3; normal melanocytes: prs452384 = 2.46 x 10−4; prs465498 = 3.47 x 10−4),31 but an opposite direction in other tissues included in GTEx (Figure S3).
As an alternative approach for evaluating a potential role of rs452384 in TERT expression, we sought to use telomere length as a surrogate marker of TERT expression. We measured relative telomere length by qPCR in germline DNA samples obtained from peripheral blood mononuclear cells (PBMCs) of 326 males with UM and healthy controls (KIDRISK study) from our GWAS in UM,16 for which the rs452384 genotype was known. We observed a significant increase in telomere length in the CC (risk) genotype group (median relative length = 27.6) compared with CT (length = 21.8) and TT (length = 20.3); this was a 36% increase in CC compared with TT (GLM p value = 9.78 x 10−3; Figures 2 and S4). These results indicate that the chr5p region influences telomere length, where rs452384-T alleles are associated with shorter telomeres than rs452384-C, and this could reflect a potential regulatory effect of rs452384 on TERT expression (in the same direction of effect as for CLPTM1L). These findings indicate that both CLPTM1L and TERT should be considered as candidate target genes downstream off rs452384 at the 5p UM risk locus.
Figure 2.
Relative telomere length in peripheral blood mononuclear cells (PBMCs) of UM-affected individuals and controls according to rs452384 genotype
Relative telomere length was measured by qPCR using primers for telomeric repeats and normalized to expression of a single copy gene, HMBS. Germline DNA samples (dots) from PBMCs of N = 208 male UM-affected individuals and N = 118 male healthy individuals from the GWAS cohort were used, stratified according to their rs452384 genotype. Number of individuals in each genotype group is indicated in brackets. Differences in telomere length were assessed using an age-adjusted generalized linear additive model (GLM) p value. Pairwise comparisons of telomere length (between two genotypes) are shown in Figure S4.
NKX2.4 is an rs452384-T allele-specific interactor whereas GATA4 preferentially binds rs452384-C
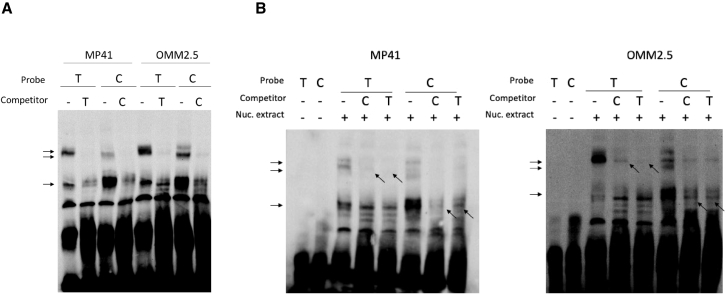
The fact that rs452384 alleles affect CLPTM1L expression and telomere length prompted us to further investigate the mechanisms underlying the biological regulation induced by this functional variant in UM. Hypothesizing that rs452384 could result in allele-specific gene regulation through the differential binding of nuclear factors, we performed electrophoretic mobility shift assays (EMSA) in UM cell lines. Rs452384-T and rs452384-C probes exhibited distinct shift patterns, which were consistent across 5 UM cell lines, MP41, OMM2.5, OMM1, MM66, and Mel202 (Figures 3A and S5A). Addition of excess specific unlabeled competitor probes confirmed the specificity of the shifts (Figures 3A, 3B, and S5B). These results suggest that rs452384 exhibits allele-specific binding of nuclear complexes, prompting us to further investigate the transcription factors mediating these effects.
Figure 3.
rs452384 preferentially binds nuclear proteins
Electrophoretic mobility shift assays (EMSAs) using double-stranded biotinylated DNA oligonucleotides containing either rs452384-T (protective allele) or rs452384-C (risk allele) in the MP41 and OMM2.5 UM cell lines.
(A) Nuclear complexes preferentially bind to the protective allele (T) or to the risk allele (C) (black arrows). Upon addition of specific unlabeled competitor probes of the same allele in 100X molar excess, these complexes disappear or are strongly attenuated.
(B) Nuclear complex that is preferentially bound to C probe (bottom horizontal arrow) is more displaced with unlabeled C probe compared with unlabeled T probe added in 100X excess (bottom angled arrows), and vice versa for complex preferentially bound to T probe.
To identify proteins bound to rs452384 alleles, we performed DNA-pulldown assays using 25bp biotin-labeled double-stranded oligonucleotides surrounding rs452384-T (“T probe”) and -C (“C probe”) alleles, as well as a nonrelevant biotinylated probe as a negative control (“NEG”),32 followed by quantitative mass spectrometry. Several TFs were found to be preferentially enriched with one allele, and NKX2.4 and GATA4 showed the highest fold changes (T- and C-enriched, respectively) and most significant p values compared with other candidates (Figure 4A, Tables 1 and S3). Filtering out proteins not significantly enriched against NEG, 6 TFs, NKX2.4, HOXC9 and DLX6 (T-enriched), and GATA4, TRPS1, and PITX2 (C-enriched), remained enriched in rs452384 probes (Figures 4B, S6A, and S6B). Unlike the other TFs for which both T and C probes were enriched compared with NEG, NKX2.4 was only enriched in T (but not C) probe compared with NEG (Figure 4B), suggesting that its interaction is specific to the T allele. To further select direct interactors of rs452384, only NKX2.4, GATA4, DLX6, and PITX2 were kept for further analyses because their predicted binding motif perfectly or closely matched the genomic sequence surrounding rs452384 (Figure S7).
Figure 4.
NKX2.4 is an allele-specific interactor of rs452384-T regulating TERT and CLPTM1L, whereas GATA4 is enriched in rs452384-C
(A and B) Quantitative mass spectrometry analysis after DNA pulldown using either rs452384-T, rs452384-C, or negative control (NEG) biotinylated probes, showing enrichment of proteins with C or T alleles. Each condition was performed in n = 5 independent biological replicates, and proteins with at least three distinct peptides and a Benjamini-Hochberg adjusted p value of ≤ 0.05 (see Methods) were kept for analysis.
(A) Volcano plot of all proteins enriched in rs452384-T or -C probes, denoted by arrows pointing left and right, respectively, represented as log2(fold change C/T) on the x-axis and –log10(p value) (y-axis). A horizontal red line represents the significant p value threshold of ≤ 0.05, whereas the vertical green lines indicate the absolute fold change threshold ≥ 1.5 of C/T and T/C enrichments. Enriched transcription factors are shown; only those written in bold are also significantly enriched against NEG probe.
(B) Correlation plot comparing enrichment ratios T/NEG (y-axis) to C/NEG (x-axis) on a log2 scale. Green lines indicate an absolute fold change threshold of ≥1.5. Proteins that specifically bind to T or C alleles are found in the top left and bottom right squares, respectively, whereas those preferentially enriched in one allele (but still enriched in both alleles vs NEG) are found in the top right corner, where the black dashed diagonal deliminates the T (left)- or C (right)-enriched interactors.
(C) Supershift EMSAs using rs452384-T or -C probes, nuclear extracts of MP41 cells transfected with either NKX2.4-flag-tag (left) or GATA4-flag-tag (right) expression vectors, and 2μg of anti-flag antibody (Ab) or negative anti-IgG control. Results show an allele-specific supershift (top arrows) of NKX2.4-flag with rs452384-T probe, whereas a preferential supershift in rs452384-C probe compared with -T is seen with GATA4-flag. Both complexes without addition of antibody (bottom arrows) disappear or greatly diminish upon addition of 100X excess of unlabeled competitor C and T probes.
(D) rs452384T>C on chr5p15.33 disrupts a predicted DNA binding motif for NKX2.4 (JASPAR database),33 whereas the rs452384 C allele favors GATA4 binding compared with rs452384 T (the core binding motif itself remains unaltered, but C is preferred over T at the 10th position).
(E and F) Chromatin immunoprecipitation (ChIP) experiments of Mel202 (E) and MP41 cells (F) using an anti-HA antibody (after transfection with NKX2.4-HA expression vector) or an anti-GATA4 antibody. In (E) enrichment at the rs452384 genomic position was measured by qPCR, compared with two other regions within CLPTM1L exons 3 and intron 4 (green squares in Figure 1A and Table S2) and compared with IP with empty vector (EV) (for NKX2.4-HA) or IgG (for GATA4). Graphs represent mean ± SD for a representative experiment (n = 3 independent experiments in total). In (F), allele-specific enrichment of MP41 immunoprecipitated DNA at rs452384 was measured by ddPCR, relative to input DNA allelic ratio. Graph represents T/C allelic ratios ± 68% Poisson confidence intervals for n = 3 independent experiments; an unpaired two-sided t test was used to compare allelic enrichment in each group compared with the hypothetical ratio of 1 (dashed line) representing equal enrichment of C and T alleles.
(G) siRNA-mediated knockdown of NKX2.4 and GATA4. The Mel202 UM cell line homozygous for rs452384 protective T allele was transfected with siRNAs targeting either NKX2.4 or GATA4, or with a negative siRNA control (siCTRL). Knockdown efficiencies of both genes are shown in Figure S11A. Resulting expression of TERT and CLPTM1L relative to that of the GUSB housekeeping gene was measured by ddPCR using Taqman gene expression assays and compared with siCTRL. Independent experiments were performed n = 5 times for siNKX2.4 and n = 3 times for siGATA4. Graphs represent relative gene expression ± 68% Poisson confidence intervals for each biological replicate. Unpaired two-sided t tests were used to compare expression levels between conditions. P values: ns for p > 0.05, ∗ for p < 0.05, ∗∗ for p < 0.005, and ∗∗∗ for p < 0.001.
Table 1.
Protein candidates identified by quantitative mass spectrometry to be significantly enriched in rs452384-T or -C alleles
| Protein | rs452384 allele specificitya | T/C fold change ratiob | Adj. p-valc | T/NEGdfold change ratio | Adj. p valc |
|---|---|---|---|---|---|
| NKX2.4 | T-specific | 13.05 | 3.1 x 10−31 | 6.40 | 2.0 x 10−28 |
| DLX6 | T-enriched | 2.96 | 1.3 x 10−8 | 8.89 | 4.7 x 10−19 |
| Protein | rs452384 allele specificitya | C/T fold change ratiob | Adj. p valc | C/NEGd fold change ratio | Adj. p valc |
| GATA4 | C-enriched | 2.98 | 4.4 x 10−23 | 36.32 | 3.5 x 10−18 |
| PITX2 | C-enriched | 1.73 | 4.4 x 10−5 | 4.90 | 2.9 x 10−9 |
“C-” or “T-enriched” proteins refer to proteins enriched in both (C vs NEG) and (T vs NEG) quantifications that were more enriched in one allele over the other; “T-specific” proteins refers to proteins only enriched in (T vs NEG) quantifications but not enriched in (C vs NEG), suggesting clear specificity for the T allele.
Fold change ratio represents the relative enrichment of candidate proteins between the two conditions tested (T/C or C/T).
Adj. p val: Benjamini-Hochberg adjusted p value.
NEG: negative control probe.
To determine whether any of these four TFs preferentially bound rs452384-T (NKX2.4, DLX6) or -C (GATA4, PITX2), we next performed supershift EMSA using FLAG-TFs. Unlike for DLX6 and PITX2 (Figure S8), expression of exogenous NKX2.4-FLAG resulted in a supershift specific to rs452384-T probe upon addition of anti-FLAG. Conversely for GATA4-FLAG, a supershift was predominantly observed with rs452384-C, but it was also detected in rs452384-T (Figure 4C). These results are consistent with mass spectrometry data that identified NKX2.4 as an rs452384-T specific interactor and GATA4 as preferentially enriched in rs452384-C compared with -T. Indeed, rs452384-C completely disrupts NKX2.4 binding, affecting a highly conserved T residue of the core binding motif TNAAGTG,33,34 whereas rs452384 alleles only marginally affect the GATA4 binding site, altering a less conserved residue (Figure 4D).
To further validate genomic enrichment and allele-specificity of NKX2.4 and/or GATA4, we performed ChIP of over-expressed NKX2.4-HA-tagged protein and endogenous GATA4 in the Mel202, MP41, and OMM1 cell lines (rs452384-TT, -CT, and -CC, respectively) using HA-tag and GATA4 antibodies. By performing a qPCR targeting the genomic region around rs452384 and other nearby regions within CLPTM1L (exon 3 and intron 4, Figure 1A), we observed in Mel202 (rs452384-TT) a specific enrichment of NKX2.4-HA at the genomic position of rs452384 compared with other regions, as well as a slight enrichment for GATA4 (Figure 4E). In the OMM1 cell line, both NKX2.4 and GATA4 were enriched at the rs452384 genomic locus (Figure S9). To address whether these enrichments were specific to one allele, we then assessed in the MP41 heterozygous cell line the allele-specific enrichment of NKX2.4 and GATA4 on immunoprecipitated DNA by ddPCR using an rs452384 Taqman genotyping probe. This indicated a significant increase in the rs452384 T/C enrichment ratio for NKX2.4, compared with the hypothetical T/C ratio of 1 representing an absence of allelic enrichment (two-sided t test, p = 0.007), and a non-significant trend toward a decrease in the T/C ratio for GATA4 (p = 0.053) (Figure 4F). Taken together, these results confirm that NKX2.4 is the strongest and most specific rs452384 interactor, preferentially binding rs452384-T, whereas GATA4 is enriched in rs452384-C.
NKX2.4 knockdown increases TERT and CLPTM1L mRNA levels
Given the genomic position of the enhancer element encompassing rs452384 within CLPTM1L and near the TERT promoter (Figure 1A), we tested whether siRNA-mediated depletion of NKX2.4 could impact the expressions of CLPTM1L and/or TERT in the Mel202 and OMM1 cell lines. The Mel202 cell line was chosen for its homozygous rs452384-TT status, the genotype preferentially bound by NKX2.4, whereas OMM1 served as comparison with the opposite genotype (rs452384-CC). Due to the very low expression levels of TERT in UM tumors and cell lines, we assessed TERT and CLPTM1L expression by ddPCR. In Mel202 (rs452384-TT), NKX2.4 knockdown resulted in a small yet consistent increase in CLPTM1L expression and a more pronounced increase in TERT expression compared with the negative control (“siCTRL”), resulting in significant fold changes of 1.54 for TERT and 1.24 for CLPTM1L (TERT relative expression: siCTRL = 0.020 ± 0.004; siNKX2.4 = 0.031 ± 0.006; p = 7.32 x 10−3; CLPTM1L: siCTRL = 2.19 ± 0.20, siNKX2.4 = 2.71 ± 0.35, t test p = 1.28 x 10−2) (Figure 4G). GATA4 knockdown, however, did not alter CLPTM1L or TERT expression significantly, although there was a trend toward decreased expression for both genes. By comparison, in OMM1 (rs452384-CC), NKX2.4 knockdown did not lead to any clear change in either CLPTM1L or TERT expression levels (Figure S10). These results suggest that NKX2.4 might act as a repressor of CLPTM1L and TERT in an rs452384-T context. Of note, overexpression of NKX2.4 and/or GATA4 did not induce changes in CLPTM1L or TERT expression in Mel202 cells (Figure S11).
Taken together, our results show that NKX2.4 regulates TERT and CLPTM1L expression and they corroborate our previous findings indicating that rs452384 influences both CLPTM1L and telomere length, itself regulated by TERT. These findings link rs452384 to NKX2.4 through a common functional role in gene regulation, further supporting their DNA-TF interaction demonstrated by mass spectrometry.
Discussion
In this study, we functionally characterized within the 5p15.33 UM risk locus identified by GWAS a regulatory region16 harboring active chromatin marks and three of the most significant UM risk variants (rs421284, rs452932, and rs370348). We identified rs452384 as a functional SNP at this locus, which induces allele-specific gene regulation, and further evidenced NKX2.4 differential binding to this variant. We finally suggest that UM risk conferred by these regulatory elements acts through transcriptional regulation of CLPTM1L and TERT, the latter influencing telomere length.
We identified NKX2.4 and GATA4 as TFs enriched with rs452384-T and -C respectively, where NKX2.4 is a T-specific interactor (UM protective allele) and GATA4 is preferentially enriched with the C (risk) allele, as shown by in vitro mass spectrometry experiments and confirmed by in vivo ChIP approaches. The preferential binding of NKX2.4 to rs452384-T over -C in allele-specific ChIP experiments, along with the weak but significant increase in CLPTM1L and TERT expression levels upon NKX2.4 knockdown, is in accordance with the low penetrance of this predisposing locus over a person’s lifetime.
NKX2.4 belongs to the NKX2 homeobox family of transcription factors, which recognize the core consensus DNA sequence 5'-TNAAGTG-3',35 have highly tissue-dependent expression patterns, and play a critical role in lineage-specific regulation of organ development.36,37,38 NKX2.4 is poorly characterized, probably because of its low RNA expression levels in normal tissues except in the hypothalamus and the pituitary gland (The Human Protein Atlas).39 It shares high homology with NKX2.1, a driver of thyroid- and lung-specific gene expression.40 Interestingly, rs452384, identified here as a functional variant in UM, is also a risk variant in lung adenocarcinoma (LUAD) and has been reported to bind to NKX2.1 in an allele-specific manner in LUAD that carries the rs452384-T and not -C genotype.41 Another NKX2 member, NKX2.5, has a C-terminal inhibitory (repressor) domain that is removed upon GATA4 co-binding, resulting in a sharp increase in transcriptional activity.34,42 This result is in agreement with our ChIP experiments in OMM1 (rs452384-CC), in which both GATA4 and NKX2.4 are enriched, and our siRNA knockdown experiments, where repressor activity for NKX2.4 (but not GATA4) binding to rs452384-T (protective) allele is demonstrated by the observed increase of CLPTM1L and TERT upon NKX2.4 knockdown. Similarly, we showed here that NKX2.4 binds a DNA motif disrupted by rs452384-C, whereas GATA4 binds slightly upstream of rs452384, and its core consensus sequences is only marginally affected by the SNP. A hypothesis is that NKX2.4 and GATA4 compete for a partially overlapping binding site with a common -AAG- core sequence (Figure 4D), and that NKX2.4 T-allele binding might displace or prevent GATA4 binding. Conversely, one could also hypothesize that GATA4 binding prevents NKX2.4 from exerting its repressor activity, which would be consistent with dual knockdown of NKX2.4 and GATA4 in Mel202, attenuating the effects of individual knockdowns (Figure S11).
Interestingly, although they do not function as TFs, we found in our mass spectrometry experiment proteins belonging to the high mobility group (HMG) superfamily of non-histone chromatin proteins, such as HMGB2 and HMGA1, to be significantly enriched with the IP sample containing rs452384-C (risk) probe (Table S3). The observed binding of TFs to rs452384 described here might thus also be accompanied by allele-specific chromatin remodelling complexes, as suggested by the preferential binding of HMG proteins to rs452384-C.
TERT and CLPTM1L, which both lie in the 5p15.33 risk locus, remain the most plausible genes regulated by rs452384, although we cannot exclude a role of more distant genes. Our previous work showed that rs421284 (highly linked to rs452384, r2 > 0.9) risk (C) genotype is positively correlated with CLPTM1L expression, in the same direction of effect as rs465498 in cutaneous melanoma (r2 > 0.9).17 Strikingly, the GTEx database indicates an inverse directionality of effect for rs452384 genotype in skin tissues (whether sun-exposed or not), where the C (risk) allele is negatively correlated with CLPTM1L expression. A potential explanation is illustrated by the study of Zhang and colleagues,31 who pointed out the heterogeneity of skin samples from the GTEx database, in which primary melanocytes – cell-of-origin of both UM and cutaneous melanoma – only represent a minor fraction of the skin tissue, resulting in major differences in eQTL directionality of effect between skin tissues and melanocytes. Indeed, our reanalysis of these datasets for the 5p15.33 locus showed that rs452384 and other linked risk alleles are negatively associated with CLPTM1L in skin tissues, but, as in primary UM, positively associated in primary melanocytes (to a higher extent than in the UM TCGA dataset, highlighting the importance of cell-type-specific eQTLs). The basis of this cell-type-specific gene regulation of the CLPTM1L locus is presently unknown.
The function of CLPTM1L and its implication in tumorigenesis has not yet been studied in UM to our knowledge, but the protein is overexpressed in pancreatic, ovarian, and lung tumor cells, and it was shown to contribute to RAS-dependent transformation.43 CLPTM1L has also been described to act as an anti-apoptotic factor upon endoplasmic reticulum stress, to promote growth, and to confer resistance to chemotherapy.44,45,46,47 More studies, such as CLPTM1L-targeted inhibition and its effect on cell growth and survival, will be needed to decipher any tumorigenic role of CLPTM1L in UM.
On the other hand, TERT, encoding the catalytic component of the telomerase enzyme, maintains genome integrity by elongating telomeres at chromosome ends, preventing chromosomal fusions and rearrangements that might induce cell senescence.48 Telomere maintenance has been extensively linked to cancer, and TERT reactivation is a possible mechanism by which tumor cells could maintain abnormal cell survival and promote cancer development.49 Several lines of evidence support a role of TERT in rs452384 allele-specific UM risk: (1) the GTEx database indicates a correlation between rs452384 and TERT expression in oesophagus; (2) 5p15.33 risk SNPs (including some within CLPTM1L introns) in lung carcinomas and others are associated with TERT, but not CLPTM1L, expression;19,50 and (3) TERT SNPs play a significant role in predisposition to many solid tumors, including cutaneous melanoma.20,21,51 As previously mentioned, TERT mRNA is barely detectable in UM tumors,17,52 making it challenging to assess for genotype-expression correlations. Because TERT is a key regulator of telomere length in normal tissues, we measured telomere length in PBMC of individuals with UM and healthy controls as a potential surrogate marker of TERT expression. We showed longer telomeres within the rs452384-CC (UM risk) genotype, corroborating a potential role for this SNP in the differential regulation of TERT (and thus of telomere length) in UM risk at the 5p15.33 locus, although we cannot rule out that telomere length is also affected by mechanisms independent of TERT-driven regulation at the 5p locus. The fact that rs452384-T alleles are associated with shorter telomeres than rs452384-C alleles in PMBC is consistent with a possible long-term effect of a decrease in TERT expression (not directly observable) in an rs452384-T context, downregulated by NKX2.4. The 1.5-fold increase in TERT expression upon NKX2.4 knockdown might be enough to mediate changes in telomere length, as the telomerase enzyme is usually found in low abundance and its activity is tightly regulated, making telomere length sensitive to even very subtle changes in TERT expression.53 Thus, the absence of detectable TERT expression in UM tumors does not rule out a role of the regulation of this gene by risk haplotype in UM development prior to tumorigenesis, such as in normal uveal melanocytes or during neural crest differentiation and migration. Because the telomere length analysis was done in the hematopoietic tissue, studies in the poorly accessible healthy uveal melanocytes would nevertheless be required for definitive conclusions.
While no other risk signal independent from the peak marked by rs370348 has been identified by our GWAS,16 we cannot rule out that other untested variants also participate in mediating UM risk in the 5p15.33 region. This analysis prioritized UM risk variants that are enriched in H3K27ac marks based on ENCODE data, and therefore noteworthy limitations to this study are that (1) ENCODE annotations used here are based on cell lines other than melanocytes, and thus cell-type-specific regulatory annotations such as melanocyte-specific enhancers might have been missed; and (2) we cannot exclude that other candidate (and untested) SNPs within the CLPTM1L region might also increase UM risk, as part of the same risk signal as rs452384. Of note, the candidate region harboring regulatory elements marks contains a large tandem-repetitive minisatellite 3kb region lying next to, and for some repeats within, the tested region (Figure 1A). This poorly mappable region precludes assessment by standard techniques, both in silico, due to poorly defined polymorphisms in this region, and in vitro, due to its sequence instability in cloning approaches. Therefore, although thorough functional analysis of the tested region definitely points toward a role for rs452384 in mediating UM risk, long-read sequencing approaches and synthetic DNA fragments represent the next steps for assessing the presence of additional causal variants in this minisatellite region.
In summary, this work elucidates some of the biological mechanisms underlying the 5p15.33 TERT/CLPTM1L susceptibility region in UM, one of the three major independent low-penetrant risk loci in UM. We identified rs452384 as a functional variant, located in a region of active chromatin elements behaving as an enhancer and implicated in allele-specific gene regulation. Using different approaches (EMSA, mass spectrometry, siRNA-mediated silencing, and ChIP) we showed that NKX2.4 preferentially binds to the low-risk (T) allele of rs452384 and represses the transcription of the surrounding region, which contains two genes already involved in tumorigenesis. Our results implicate CLPTM1L and TERT as target genes and indicate a correlation between rs452384 genotype and telomere length. Future work will be required in order to establish a causal link between rs452384 alleles and CLPTM1L and TERT expression levels, such as through chromatin interaction experiments (3C and derived techniques) and rs452384 isogenic cell line models. Altogether, our work unraveled some of the complex regulatory mechanisms at the 5p15.33 susceptibility region in UM, paving the way for a thorough assessment of TERT, CLPTM1L, and telomere length in UM tumorigenesis.
Acknowledgments
A.-C.D. was supported by the Horizon 2020 program and innovation program under the Marie Skłodowska-Curie grant agreement No. 666003 and the Ligue Nationale Contre le Cancer. A.H. was supported by the Site de Recherche Intégrée sur le Cancer (SiRIC) of Institut Curie. L.M. was supported by the Horizon 2020 program UM Cure (No. 667787). D.L. was supported by “Région Ile-de-France” and Fondation pour la Recherche Médicale. M.R. was supported by the «Interface Inserm » grant. This study was funded by the Institut National de la Santé et de la Recherche Médicale (Inserm), the Institut Curie, and the Ligue Nationale Contre le Cancer (Labellisation).
Author contributions
A.-C.D. and M.-H.S. conceived the project and designed and developed its methodology. A.-C.D, O.G., F.D., M.N., L.M., and M.I.E.D. performed experiments and/or data acquisition, and A-C.D. and A.H. performed data presentation and statistical analyses. J.N. provided help for statistical analysis. N.C., O.C., G.C.-T., and J.Z.-C. provided resources and/or analytic tools. R.M. provided insightful advice in experiment conceptualization and analysis. A.-C.D. and M.-H.S. wrote and edited the manuscript. O.G., D.L., M.R., and R.M. reviewed the manuscript. M.-H.S. supervised the study. All authors reviewed and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: December 1, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.11.004.
Web resources
HOMOCOCO motif collection, https://hocomoco11.autosome.ru/
JASPAR database, https://jaspar.genereg.net/
PLINK v1.9, https://www.cog-genomics.org/plink/1.9
UCSC genome browser, https://genome.ucsc.edu/
Supplemental information
These include a subset of patients and controls from our prior UM GWAS study, including 208 UM patients (UM_1-208) and 118 European controls (KIDRISK_1-118) from the UM GWAS
Data availability
Genotyping data of uveal-melanoma-affected individuals included in the GWAS have been deposited on the European Genome-Phenome Archive (EGA) under accession numbers EGAS00001002334 (UM-affected individuals from dataset 1) and EGAS0000100233 (UM-affected individuals from datasets 2 and 3). Genotyping data of French controls (KIDRISK consortium US NCI U01CA155309; G. Scelo) are accessible from dbGAP under study accession number phs001271.v1.p1 (European controls from KIDRISK study). The accession number for the proteomics data reported in this paper is PXD028954, deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository.54
References
- 1.Singh M., Durairaj P., Yeung J. Uveal melanoma: a review of the literature. Oncol. Ther. 2018;6:87–104. doi: 10.1007/s40487-018-0056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jager M.J., Shields C.L., Cebulla C.M., Abdel-Rahman M.H., Grossniklaus H.E., Stern M.H., Carvajal R.D., Belfort R.N., Jia R., Shields J.A., Damato B.E. Uveal melanoma. Nat. Rev. Dis. Primers. 2020;6:24. doi: 10.1038/s41572-020-0158-0. [DOI] [PubMed] [Google Scholar]
- 3.Van Raamsdonk C.D., Bezrookove V., Green G., Bauer J., Gaugler L., O'Brien J.M., Simpson E.M., Barsh G.S., Bastian B.C. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009;457:599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johansson P., Aoude L.G., Wadt K., Glasson W.J., Warrier S.K., Hewitt A.W., Kiilgaard J.F., Heegaard S., Isaacs T., Franchina M., et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016;7:4624–4631. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harbour J.W., Onken M.D., Roberson E.D.O., Duan S., Cao L., Worley L.A., Council M.L., Matatall K.A., Helms C., Bowcock A.M. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010;330:1410–1413. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furney S.J., Pedersen M., Gentien D., Dumont A.G., Rapinat A., Desjardins L., Turajlic S., Piperno-Neumann S., de la Grange P., Roman-Roman S., et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov. 2013;3:1122–1129. doi: 10.1158/2159-8290.CD-13-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin M., Maßhöfer L., Temming P., Rahmann S., Metz C., Bornfeld N., van de Nes J., Klein-Hitpass L., Hinnebusch A.G., Horsthemke B., et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat. Genet. 2013;45:933–936. doi: 10.1038/ng.2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aronow M.E., Topham A.K., Singh A.D. Uveal melanoma: 5-year update on incidence, treatment, and survival (SEER 1973-2013) Ocul. Oncol. Pathol. 2018;4:145–151. doi: 10.1159/000480640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bishop K.D., Olszewski A.J. Epidemiology and survival outcomes of ocular and mucosal melanomas: a population-based analysis. Int. J. Cancer. 2014;134:2961–2971. doi: 10.1002/ijc.28625. [DOI] [PubMed] [Google Scholar]
- 10.Weis E., Shah C.P., Lajous M., Shields J.A., Shields C.L. The association between host susceptibility factors and uveal melanoma: a meta-analysis. Arch. Ophthalmol. 2006;124:54–60. doi: 10.1001/archopht.124.1.54. [DOI] [PubMed] [Google Scholar]
- 11.Royer-Bertrand B., Torsello M., Rimoldi D., El Zaoui I., Cisarova K., Pescini-Gobert R., Raynaud F., Zografos L., Schalenbourg A., Speiser D., et al. Comprehensive genetic landscape of uveal melanoma by whole-genome sequencing. Am. J. Hum. Genet. 2016;99:1190–1198. doi: 10.1016/j.ajhg.2016.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiesner T., Obenauf A.C., Murali R., Fried I., Griewank K.G., Ulz P., Windpassinger C., Wackernagel W., Loy S., Wolf I., et al. Germline mutations in BAP1 predispose to melanocytic tumors. Nat. Genet. 2011;43:1018–1021. doi: 10.1038/ng.910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdel-Rahman M.H., Pilarski R., Cebulla C.M., Massengill J.B., Christopher B.N., Boru G., Hovland P., Davidorf F.H. Germline BAP1 mutation predisposes to uveal melanoma, lung adenocarcinoma, meningioma, and other cancers. J. Med. Genet. 2011;48:856–859. doi: 10.1136/jmedgenet-2011-100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta M.P., Lane A.M., DeAngelis M.M., Mayne K., Crabtree M., Gragoudas E.S., Kim I.K. Clinical characteristics of uveal melanoma in patients with germline BAP1 mutations. JAMA Ophthalmol. 2015;133:881–887. doi: 10.1001/jamaophthalmol.2015.1119. [DOI] [PubMed] [Google Scholar]
- 15.Derrien A.C., Rodrigues M., Eeckhoutte A., Dayot S., Houy A., Mobuchon L., Gardrat S., Lequin D., Ballet S., Pierron G., et al. Germline MBD4 mutations and predisposition to uveal melanoma. J. Natl. Cancer Inst. 2021;113:80–87. doi: 10.1093/jnci/djaa047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mobuchon L., Derrien A.C., Houy A., Verrier T., Pierron G., Cassoux N., Milder M., Deleuze J.F., Boland A., Scelo G., et al. Different pigmentation risk loci for high-risk monosomy 3 and low-risk disomy 3 uveal melanomas. J Natl Cancer Inst. 2021 doi: 10.1093/jnci/djab167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mobuchon L., Battistella A., Bardel C., Scelo G., Renoud A., Houy A., Cassoux N., Milder M., Cancel-Tassin G., Cussenot O., et al. A GWAS in uveal melanoma identifies risk polymorphisms in the CLPTM1L locus. NPJ Genom. Med. 2017;2:5. doi: 10.1038/s41525-017-0008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen H., Majumdar A., Wang L., Kar S., Brown K.M., Feng H., Turman C., Dennis J., Easton D., Michailidou K., et al. Large-scale cross-cancer fine-mapping of the 5p15.33 region reveals multiple independent signals. HGG Adv. 2021;2:100041. doi: 10.1016/j.xhgg.2021.100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang J., Jia J., Makowski M., Xu M., Wang Z., Zhang T., Hoskins J.W., Choi J., Han Y., Zhang M., et al. Functional characterization of a multi-cancer risk locus on chr5p15.33 reveals regulation of TERT by ZNF148. Nat. Commun. 2017;8:16159. doi: 10.1038/ncomms15034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z., Zhu B., Zhang M., Parikh H., Jia J., Chung C.C., Sampson J.N., Hoskins J.W., Hutchinson A., Burdette L., et al. Imputation and subset-based association analysis across different cancer types identifies multiple independent risk loci in the TERT-CLPTM1L region on chromosome 5p15.33. Hum. Mol. Genet. 2014;23:6616–6633. doi: 10.1093/hmg/ddu363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rafnar T., Sulem P., Stacey S.N., Geller F., Gudmundsson J., Sigurdsson A., Jakobsdottir M., Helgadottir H., Thorlacius S., Aben K.K.H., et al. Sequence variants at the TERT-CLPTM1L locus associate with many cancer types. Nat. Genet. 2009;41:221–227. doi: 10.1038/ng.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bojesen S.E., Pooley K.A., Johnatty S.E., Beesley J., Michailidou K., Tyrer J.P., Edwards S.L., Pickett H.A., Shen H.C., Smart C.E., et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat. Genet. 2013;45:371–384. doi: 10.1038/ng.2566. 384e371-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McKay J.D., Hung R.J., Han Y., Zong X., Carreras-Torres R., Christiani D.C., Caporaso N.E., Johansson M., Xiao X., Li Y., et al. Large-scale association analysis identifies new lung cancer susceptibility loci and heterogeneity in genetic susceptibility across histological subtypes. Nat. Genet. 2017;49:1126–1132. doi: 10.1038/ng.3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Law M.H., Bishop D.T., Lee J.E., Brossard M., Martin N.G., Moses E.K., Song F., Barrett J.H., Kumar R., Easton D.F., et al. Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat. Genet. 2015;47:987–995. doi: 10.1038/ng.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein A.P., Wolpin B.M., Risch H.A., Stolzenberg-Solomon R.Z., Mocci E., Zhang M., Canzian F., Childs E.J., Hoskins J.W., Jermusyk A., et al. Genome-wide meta-analysis identifies five new susceptibility loci for pancreatic cancer. Nat. Commun. 2018;9:556. doi: 10.1038/s41467-018-02942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amirouchene-Angelozzi N., Nemati F., Gentien D., Nicolas A., Dumont A., Carita G., Camonis J., Desjardins L., Cassoux N., Piperno-Neumann S., et al. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Mol. Oncol. 2014;8:1508–1520. doi: 10.1016/j.molonc.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cawthon R.M. Telomere measurement by quantitative PCR. Nucleic Acids Res. 2002;30:e47. doi: 10.1093/nar/30.10.e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ningarhari M., Caruso S., Hirsch T.Z., Bayard Q., Franconi A., Védie A.L., Noblet B., Blanc J.F., Amaddeo G., Ganne N., et al. Telomere length is key to hepatocellular carcinoma diversity and telomerase addiction is an actionable therapeutic target. J. Hepatol. 2021;74:1155–1166. doi: 10.1016/j.jhep.2020.11.052. [DOI] [PubMed] [Google Scholar]
- 29.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A., et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hormozdiari F., van de Bunt M., Segrè A.V., Li X., Joo J.W.J., Bilow M., Sul J.H., Sankararaman S., Pasaniuc B., Eskin E. Colocalization of GWAS and eQTL signals detects target genes. Am. J. Hum. Genet. 2016;99:1245–1260. doi: 10.1016/j.ajhg.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang T., Choi J., Kovacs M.A., Shi J., Xu M., NISC Comparative Sequencing Program, Melanoma Meta-Analysis Consortium. Goldstein A.M., Trower A.J., Bishop D.T., et al. Cell-type-specific eQTL of primary melanocytes facilitates identification of melanoma susceptibility genes. Genome Res. 2018;28:1621–1635. doi: 10.1101/gr.233304.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Redondo-Muñoz J., Ugarte-Berzal E., Terol M.J., Van den Steen P.E., Hernández del Cerro M., Roderfeld M., Roeb E., Opdenakker G., García-Marco J.A., García-Pardo A. Matrix metalloproteinase-9 promotes chronic lymphocytic leukemia b cell survival through its hemopexin domain. Cancer Cell. 2010;17:160–172. doi: 10.1016/j.ccr.2009.12.044. [DOI] [PubMed] [Google Scholar]
- 33.Fornes O., Castro-Mondragon J.A., Khan A., van der Lee R., Zhang X., Richmond P.A., Modi B.P., Correard S., Gheorghe M., Baranašić D., et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48:D87–D92. doi: 10.1093/nar/gkz1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sepulveda J.L., Belaguli N., Nigam V., Chen C.Y., Nemer M., Schwartz R.J. GATA-4 and Nkx-2.5 coactivate Nkx-2 DNA binding targets: role for regulating early cardiac gene expression. Mol. Cell Biol. 1998;18:3405–3415. doi: 10.1128/mcb.18.6.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watada H., Mirmira R.G., Kalamaras J., German M.S. Intramolecular control of transcriptional activity by the NK2-specific domain in NK-2 homeodomain proteins. Proc. Natl. Acad. Sci. USA. 2000;97:9443–9448. doi: 10.1073/pnas.97.17.9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanfel M.N., Moses K.A., Schwartz R.J., Zimmer W.E. Regulation of organ development by the NKX-homeodomain factors: an NKX code. Cell. Mol. Biol. 2005;51 OL785-799. [PubMed] [Google Scholar]
- 37.He Y., Liu X.Y., Gong R., Peng K.W., Liu R.B., Wang F. NK homeobox 2.2 functions as tumor suppressor in colorectal cancer due to DNA methylation. J. Cancer. 2020;11:4791–4800. doi: 10.7150/jca.43665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorsi B., Mosbruger T.L., Smith M., Hill J.T., Yost H.J. Nkx2.5-dependent alterations of the embryonic heart DNA methylome identify novel cis-regulatory elements in cardiac development. bioRxiv. 2017 doi: 10.1101/186395. Preprint at. [DOI] [Google Scholar]
- 39.Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 40.Boggaram V. Thyroid transcription factor-1 (TTF-1/Nkx2.1/TITF1) gene regulation in the lung. Clin. Sci. 2009;116:27–35. doi: 10.1042/CS20080068. [DOI] [PubMed] [Google Scholar]
- 41.Yang C., Stueve T.R., Yan C., Rhie S.K., Mullen D.J., Luo J., Zhou B., Borok Z., Marconett C.N., Offringa I.A. Positional integration of lung adenocarcinoma susceptibility loci with primary human alveolar epithelial cell epigenomes. Epigenomics. 2018;10:1167–1187. doi: 10.2217/epi-2018-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benson D.W., Silberbach G.M., Kavanaugh-McHugh A., Cottrill C., Zhang Y., Riggs S., Smalls O., Johnson M.C., Watson M.S., Seidman J.G., et al. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J. Clin. Invest. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.James M.A., Vikis H.G., Tate E., Rymaszewski A.L., You M. CRR9/CLPTM1L regulates cell survival signaling and is required for Ras transformation and lung tumorigenesis. Cancer Res. 2014;74:1116–1127. doi: 10.1158/0008-5472.CAN-13-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke W.R., Amundadottir L., James M.A. CLPTM1L/CRR9 ectodomain interaction with GRP78 at the cell surface signals for survival and chemoresistance upon ER stress in pancreatic adenocarcinoma cells. Int. J. Cancer. 2019;144:1367–1378. doi: 10.1002/ijc.32012. [DOI] [PubMed] [Google Scholar]
- 45.Jia J., Bosley A.D., Thompson A., Hoskins J.W., Cheuk A., Collins I., Parikh H., Xiao Z., Ylaya K., Dzyadyk M., et al. CLPTM1L promotes growth and enhances aneuploidy in pancreatic cancer cells. Cancer Res. 2014;74:2785–2795. doi: 10.1158/0008-5472.CAN-13-3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parashar D., Geethadevi A., McAllister D., Ebben J., Peterson F.C., Jensen D.R., Bishop E., Pradeep S., Volkman B.F., Dwinell M.B., et al. Targeted biologic inhibition of both tumor cell-intrinsic and intercellular CLPTM1L/CRR9-mediated chemotherapeutic drug resistance. NPJ Precis. Oncol. 2021;5:16. doi: 10.1038/s41698-021-00152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.James M.A., Wen W., Wang Y., Byers L.A., Heymach J.V., Coombes K.R., Girard L., Minna J., You M. Functional characterization of CLPTM1L as a lung cancer risk candidate gene in the 5p15.33 locus. PLoS One. 2012;7:e36116. doi: 10.1371/journal.pone.0036116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheung A.L.M., Deng W. Telomere dysfunction, genome instability and cancer. Front. Biosci. 2008;13:2075–2090. doi: 10.2741/2825. [DOI] [PubMed] [Google Scholar]
- 49.Maciejowski J., de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017;18:175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang Y.C., Fu W.P., Zhang J., Zhong L., Cai S.X., Sun C. rs401681 and rs402710 confer lung cancer susceptibility by regulating TERT expression instead of CLPTM1L in East Asian populations. Carcinogenesis. 2018;39:1216–1221. doi: 10.1093/carcin/bgy084. [DOI] [PubMed] [Google Scholar]
- 51.Huang F.W., Hodis E., Xu M.J., Kryukov G.V., Chin L., Garraway L.A. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barthel F.P., Wei W., Tang M., Martinez-Ledesma E., Hu X., Amin S.B., Akdemir K.C., Seth S., Song X., Wang Q., et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat. Genet. 2017;49:349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNally E.J., Luncsford P.J., Armanios M. Long telomeres and cancer risk: the price of cellular immortality. J. Clin. Invest. 2019;129:3474–3481. doi: 10.1172/JCI120851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
These include a subset of patients and controls from our prior UM GWAS study, including 208 UM patients (UM_1-208) and 118 European controls (KIDRISK_1-118) from the UM GWAS
Data Availability Statement
Genotyping data of uveal-melanoma-affected individuals included in the GWAS have been deposited on the European Genome-Phenome Archive (EGA) under accession numbers EGAS00001002334 (UM-affected individuals from dataset 1) and EGAS0000100233 (UM-affected individuals from datasets 2 and 3). Genotyping data of French controls (KIDRISK consortium US NCI U01CA155309; G. Scelo) are accessible from dbGAP under study accession number phs001271.v1.p1 (European controls from KIDRISK study). The accession number for the proteomics data reported in this paper is PXD028954, deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository.54