Summary
By combining data from 160,500 individuals with breast cancer and 226,196 controls of Asian and European ancestry, we conducted genome- and transcriptome-wide association studies of breast cancer. We identified 222 genetic risk loci and 137 genes that were associated with breast cancer risk at a p < 5.0 × 10−8 and a Bonferroni-corrected p < 4.6 × 10−6, respectively. Of them, 32 loci and 15 genes showed a significantly different association between ER-positive and ER-negative breast cancer after Bonferroni correction. Significant ancestral differences in risk variant allele frequencies and their association strengths with breast cancer risk were identified. Of the significant associations identified in this study, 17 loci and 14 genes are located 1Mb away from any of the previously reported breast cancer risk variants. Pathways analyses including 221 putative risk genes identified multiple signaling pathways that may play a significant role in the development of breast cancer. Our study provides a comprehensive understanding of and new biological insights into the genetics of this common malignancy.
Keywords: breast cancer, multi-ancestry meta-analysis, transcriptome-wide association study
Using data from 386,000 Asian- and European-ancestry women, we conducted extensive genome- and transcriptome-wide association studies that identified 222 risk loci and 137 genes in association with breast cancer risk. These studies, along with pathway analyses, provide a comprehensive understanding of and new biological insights into the genetics of breast cancer.
Introduction
Breast cancer is the most commonly diagnosed cancer in women worldwide, with an estimated 2.3 million new cases in 2020.1 Genetic factors play a critical role in the etiology of both familial and sporadic breast cancers. In addition to breast cancer predisposition genes, such as BRCA1 and BRCA2,2,3,4 common genetic variants in approximately 200 loci have been identified in genome-wide association studies (GWASs).5,6,7 However, most GWASs of breast cancer have been conducted among women of European ancestry,8 and GWASs conducted among women of Asian ancestry have had relatively smaller sample sizes.9,10 Although most susceptibility loci have been shown to be shared across European and Asian populations, the lead variants at some susceptibility loci can be different between these two populations given their differences in genetic architecture.11,12 To identify additional genetic risk loci and provide a more comprehensive understanding of breast cancer genetics, we conducted cross-ancestry meta-analyses of data from the Asia Breast Cancer Consortium (ABCC) and the Breast Cancer Association Consortium (BCAC), including 386,696 women (139,523 of Asian ancestry and 247,173 of European ancestry). Furthermore, we performed a transcriptome-wide association study (TWAS) to uncover putative breast cancer susceptibility genes and gain biological insights into the genetics of this common malignancy.
Subjects and methods
Study population
In this study, we conducted a cross-ancestry meta-analysis using data from two large breast cancer genetic research consortia: ABCC and BCAC. All studies were approved by relevant institutional ethical committees. The detailed descriptions of participating studies are described in the supplemental information. In brief, the 133,384 individuals with breast cancer and 113,789 controls of European ancestry included in this analysis were from BCAC, which consisted of three datasets: iCOGS (38,349 individuals with breast cancer and 37,818 controls), OncoArray (80,125 individuals with breast cancer and 58,383 controls), and other GWASs (14,910 individuals with breast cancer and 17,588 controls).6 For European-ancestry participants, we used summary statistics data generated in BCAC, following the data use agreements. Individuals of Asian ancestry included in this analysis were 27,116 individuals with breast cancer and 112,407 controls recruited by studies in AABC and BCAC (Table S1). Proper informed consent was obtained from all study participants.
Genotyping and quality control
Genotyping and quality control procedures for the contributing studies have been described previously.5,6,7,9,10,11,13,14,15,16,17,18,19 After quality control, we imputed all datasets using the 1000 Genomes Project Phase 3 and excluded variants with an imputation quality score (R2) <0.3. Variants with a minor allele frequency (MAF) of >0.01 in Asian-ancestry datasets or >0.005 in European-ancestry datasets were included for association analyses.
Statistical meta-analyses
Analyses using logistic regression models were performed within each of the ABCC studies, except Biobank Japan project (BBJ2), to estimate the per-allele odds ratio (OR) for each variant using PLINK 2.0.20 Age and the top two principal components (PCs) were adjusted as covariates. The number of PCs included in the regression was determined by evaluating the Scree plot. Summary statistics were acquired for BBJ2 and BCAC-European dataset. Age and top five PCs were adjusted in BBJ as covariates.13 The country of contributing studies and the first ten PCs were adjusted in the BCAC-European dataset.6 A fixed-effects model was used for ancestry-specific meta-analyses and cross-ancestry meta-analyses for risk of overall breast cancer and estrogen receptor (ER) subtypes using METAL.21 The heterogeneity of risk estimates was evaluated using Cochran’s Q statistic and I2. We estimated the statistical power of our cross-ancestry meta-analyses with at 5 × 10−8 (Figure S1). We had 80% power to detect a minimum per-allele OR of 1.07, 1.05, 1.04, and 1.03 for variants with a MAF of 0.05, 0.15, 0.20, and 0.30, respectively. In order to take into account of the population heterogeneity, we also used the meta-regression approach implemented in MR-MEGA22 in cross-ancestry meta-analyses for overall breast cancer. At each risk locus, we performed fine-mapping analysis using SuSiE23 and constructed a 95% credible set for the lead variant at the locus (detailed methods in supplemental information). We investigated the ancestral heterogeneity of the lead variants and all variants in the credible sets.
Novel risk loci were defined as loci with the sentinel variants located at least 1 Mb away from any of the risk variants identified by previous GWASs included in the NHGRI-EBI GWAS Catalog.24 For each novel locus, we conducted conditional analyses to identify additional independent signals located flanking ± 500 kb from the lead variant. The GCTA-COJO was used for the conditional analyses. In each iteration of the stepwise conditional analysis, we conducted ancestry-specific conditional analyses and combined the results by a fixed-effects model using METAL. Asian samples (N = 20,554) genotyped by Multi-Ethnic Genotyping Array (MEGA) chips were used as a reference panel for linkage disequilibrium (LD) estimation among women of Asian ancestry. For women of European ancestry, we used 5,000 samples from the Vanderbilt University Medical Center biobank (BioVU) genotyped by MEGA as a reference panel for LD estimation.25,26 Since the conditional analyses were restricted to local regions of the novel loci identified at genome-wide significance, we used 1 × 10−4 as significance level (adjusting for ∼500 comparisons in each locus). If the variant with the lowest conditional p was lower than 1 × 10−4, it was considered an independent signal at that locus, and it was subsequently adjusted, along with the lead variant, from cross-ancestry meta-analyses in later iterations. This process was repeated until there were no variants with a cross-ancestry conditional p < 1 × 10−4.
Genetic variance explained by novel risk variants
We estimated the genetic variance explained by novel risk variants identified in this study using a log-additive model:
where n is the total number of novel risk variants, is the MAF of the ith variant, is the log-OR for the ith variant and is the standard error of . The explained genetic variance was estimated for overall breast cancer and by ER subtypes for Asian- and European-ancestry populations, respectively.
Transcriptome-wide association analysis
We used RNA sequencing data from 115 samples collected from European-ancestry women from the Genotype-Tissue Expression Project (GTEx, version 8) to build prediction models for each gene expressed in normal breast tissue. Germline genotyping data were obtained using whole-genome sequencing (WGS) of genomic DNA extracted from blood samples. The details of data processing are described in the supplemental information. We used a cross-tissue approach, joint-tissue imputation (JTI), to build prediction models for gene-expression levels in normal breast tissue.27 Besides breast tissue, data from all 31 other tissues were borrowed in the JTI approach to leverage shared genetic regulation and improve prediction performance in a tissue-dependent manner (Table S10). Prediction models were built using genetic variants within flanking +/− 500 kb from the respective gene boundaries. Five-fold cross-validation was conducted to validate the models internally. Genes with a model prediction R > 0.1 were included for association analyses.
To evaluate the performance of prediction models, we performed an external validation using 86 tumor-adjacent normal breast tissue samples from European-ancestry females with breast cancer in The Cancer Genome Atlas (TCGA). We calculated the Spearman’s correlation between the prediction performance (R2) in GTEx and TCGA.
We conducted association analyses of predicted gene expression with breast cancer risk with S-PrediXcan tool,28 using the summary statistics from our ancestry-specific and cross-ancestry meta-analyses of GWASs for breast cancer. For genes identified at Bonferroni correction in the association analyses, we also conducted TWAS fine-mapping analyses and colocalization analyses. Pathway analyses were conducted for protein-coding genes. The details of statistical analyses were described in supplemental information.
Results
By cross-ancestry meta-analyzing GWAS data from 160,500 individuals with breast cancer and 226,196 controls of Asian and European ancestry using fixed-effects models, we identified 23,461 variants in 184 regions that were associated with overall breast cancer risk at genome-wide significance level (p < 5.00 × 10−8; Table S2). Twenty-seven additional risk loci were uncovered in population-specific analyses, including 25 loci identified in European-specific GWASs and two in Asian-specific GWASs. In total, we identified 211 loci showing a significant association with risk of overall breast cancer. Of them, 16 loci are novel, with the sentinel variants located at least 1 Mb away from any of the risk variants identified by previous GWASs (Table 1).
Table 1.
Results for the lead risk variants at 17 novel loci identified in cross-ancestry meta-analyses of GWAS data
| Variants | Loci | Nearest gene | Gene region | Allelesa | EAFb | OR (95% CI) | pc | I2, % | p_het |
|---|---|---|---|---|---|---|---|---|---|
| Overall | |||||||||
| rs727477 | 2p22.1 | SLC8A1 | Intron | G/T | 0.36 | 0.97 (0.96, 0.98) | 2.85 × 10−8 | 52.1 | 0.03 |
| rs3010266 | 5q13.2 | LINC02056 | 8.5 kb from 5′ | A/G | 0.24 | 0.96 (0.95, 0.98) | 3.56 × 10−8 | 0 | 0.83 |
| rs6890591d | 5q35.2 | CPEB4 | 3.3 kb from 3′ | A/T | 0.38 | 0.97 (0.96, 0.98) | 3.25 × 10−8 | 50.5 | 0.04 |
| rs3829964 | 6p21.2 | CDKN1A | Intron | T/C | 0.47 | 0.97 (0.96, 0.98) | 4.61 × 10−9 | 0 | 0.46 |
| rs74392007 | 6q22.31 | HSF2 | 5.4 kb from 5′ | T/C | 0.12 | 1.05 (1.03, 1.07) | 1.55 × 10−8 | 0 | 0.93 |
| rs3778663 | 6q27 | AFDN | Intron | A/G | 0.13 | 1.06 (1.04, 1.07) | 8.51 × 10−9 | 0 | 0.69 |
| rs17167576 | 7p21.2 | AC005019.3e | 5.5 kb from 3′ | A/T | 0.37 | 1.03 (1.02, 1.04) | 6.93 × 10−9 | 47.2 | 0.05 |
| rs3988353 | 8p22 | PCM1 | Intron | CT/C | 0.42 | 1.03 (1.02, 1.04) | 4.32 × 10−8 | 0 | 0.81 |
| rs1937680 | 10q21.1 | PRKG1 | Intron | C/A | 0.36 | 1.03 (1.02, 1.04) | 8.18 × 10−9 | 1.3 | 0.42 |
| rs11354045 | 11q23.1 | ALG9 | Intron | CT/C | 0.35 | 1.03 (1.02, 1.04) | 2.68 × 10−8 | 22.3 | 0.25 |
| rs36028244 | 11q23.3 | PCSK7 | Intron | C/CTTA | 0.07 | 1.06 (1.04, 1.08) | 1.77 × 10−8 | 0 | 1.00 |
| rs3809114 | 12q13.3 | INHBE | 5′ UTRf | G/A | 0.47 | 0.97 (0.96, 0.98) | 2.33 × 10−8 | 37.8 | 0.12 |
| rs956006 | 15q22.2 | TLN2 | Intron | T/C | 0.32 | 1.03 (1.02, 1.05) | 3.54 × 10−8 | 1.7 | 0.42 |
| rs4797754 | 18p11.21 | LDLRAD4 | Intron | G/C | 0.31 | 1.03 (1.02, 1.05) | 2.08 × 10−8 | 0 | 0.50 |
| rs112208395 | 20q11.23 | PHF20 | Intron | C/CT | 0.14 | 1.05 (1.03, 1.07) | 4.11 × 10−8 | 0 | 0.96 |
| rs74157632g | 10q26.11 | DENND10 | Missense | G/A | 0.05 | 0.86 (0.81, 0.90) | 1.41 × 10−8 | 0 | 1.00 |
| ER-negative | |||||||||
| rs2123844 | 17p13.2 | ZZEF1 | Intron | A/C | 0.07 | 1.13 (1.09, 1.18) | 2.81 × 10−10 | 37.4 | 0.16 |
Effect allele/reference allele.
Effect allele frequency.
Unless otherwise specified, p derived from meta-analyses using fixed-effects model.
Identified using cross-ancestry meta-regression (Table S6). The p derived from cross-ancestry fixed-effects model is (Table S2).
AC005019.3 (ENSG00000224330) does not have a gene symbol in HUGO yet.
UTR, untranslated region.
Identified in Asian-specific GWASs. The p for cross-ancestry fixed-effects model is (Table S2).
Analyses by ER status identified 13,392 variants in 100 loci and 2,425 variants in 34 loci that were associated with ER-positive and ER-negative breast cancer, respectively, at the genome-wide significance level (Tables S3 and S4). Two loci for ER-positive and nine loci for ER-negative breast cancer did not overlap with any of the loci identified for overall breast cancer. Of them, 17p13.2, associated with ER-negative breast cancer risk, has not yet been reported in previous GWASs (Table 1).
Of the 222 lead risk variants identified in our study that were associated with the risk of either overall breast cancer (n = 211) or exclusively ER-positive (n = 2) or ER-negative (n = 9) breast cancer, 68 variants showed a significantly different association by ER status at a false discovery rate (FDR) <0.05 in heterogeneity tests (Table S7). Among them, eight risk loci were not reported previously. Except for rs12335941 at 9p21.3, all other seven variants had a stronger association with ER-positive than ER-negative breast cancer. Of the 32 variants showing a different association at a Bonferroni-corrected p < 2.25 × 10−4 (0.05/222, Table 2), five lead variants showed an opposite direction of the association by ER status.
Table 2.
Results for breast cancer risk loci showing different associations by estrogen receptor status
| Variants | Loci | Allelea | EAFb |
ER-Positive |
ER-Negative |
p for ER heterogeneity | ||
|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | p | OR (95% CI) | p | |||||
| rs2506885 | 1p36.22 | T/A | 0.34 | 0.95 (0.94, 0.97) | 5.91 × 10−10 | 0.88 (0.86, 0.90) | 3.68 × 10−27 | 2.63 × 10−8 |
| rs11249433 | 1p11.2 | G/A | 0.39 | 1.13 (1.11, 1.15) | 3.45 × 10−59 | 1.01 (0.99, 1.04) | 0.29 | 1.01 × 10−15 |
| rs12129456 | 1q32.1 | G/T | 0.38 | 1.02 (1.00, 1.03) | 0.03 | 0.92 (0.90, 0.94) | 1.52 × 10−13 | 2.00 × 10−13 |
| rs2169137 | 1q32.1 | G/C | 0.25 | 1.00 (0.98, 1.02) | 0.9 | 1.13 (1.11, 1.16) | 4.03 × 10−24 | 2.30 × 10−17 |
| rs56158184 | 2p23.2 | C/T | 0.09 | 1.03 (1.00, 1.05) | 0.02 | 0.89 (0.86, 0.92) | 1.01 × 10−9 | 1.60 × 10−10 |
| rs2016394 | 2q31.1 | A/G | 0.44 | 0.94 (0.93, 0.96) | 1.05 × 10−16 | 1.00 (0.98, 1.02) | 0.91 | 2.51 × 10−6 |
| rs4442975 | 2q35 | G/T | 0.46 | 1.15 (1.14, 1.17) | 1.42 × 10−92 | 1.05 (1.03, 1.07) | 1.12 × 10−5 | 3.72 × 10−14 |
| rs552647 | 3p24.1 | A/C | 0.48 | 1.12 (1.10, 1.14) | 6.35 × 10−60 | 1.05 (1.03, 1.07) | 4.89 × 10−6 | 1.06 × 10−7 |
| rs7697216 | 4q34.1 | T/C | 0.15 | 0.89 (0.87, 0.91) | 1.17 × 10−30 | 0.98 (0.96, 1.01) | 0.24 | 1.49 × 10−8 |
| rs2853669 | 5p15.33 | G/A | 0.31 | 0.96 (0.95, 0.97) | 3.29 × 10−8 | 0.89 (0.87, 0.91) | 3.03 × 10−24 | 4.32 × 10−8 |
| rs7710996 | 5p12 | A/G | 0.25 | 1.00 (0.98, 1.02) | 0.97 | 1.07 (1.04, 1.09) | 1.50 × 10−8 | 3.84 × 10−6 |
| rs10941679 | 5p12 | G/A | 0.31 | 1.16 (1.14, 1.18) | 5.38 × 10−86 | 1.02 (1.00, 1.05) | 0.04 | 1.45 × 10−20 |
| rs59957907 | 5q11.2 | G/A | 0.22 | 1.19 (1.17, 1.21) | 2.95 × 10−90 | 1.06 (1.04, 1.09) | 2.09 × 10−6 | 2.46 × 10−13 |
| rs60954078 | 6q25.1 | G/A | 0.17 | 1.16 (1.14, 1.19) | 1.75 × 10−41 | 1.33 (1.29, 1.37) | 6.92 × 10−76 | 2.18 × 10−12 |
| rs910416 | 6q25.1 | C/T | 0.46 | 0.95 (0.94, 0.96) | 3.23 × 10−13 | 0.91 (0.89, 0.93) | 1.08 × 10−21 | 1.02 × 10−4 |
| rs116426014 | 8p23.3 | G/A | 0.26 | 1.03 (1.01, 1.04) | 0.01 | 1.09 (1.06, 1.12) | 1.83 × 10−10 | 1.68 × 10−4 |
| rs60037937 | 9q31.2 | T/TAA | 0.26 | 1.10 (1.08, 1.11) | 7.92 × 10−28 | 1.03 (1.00, 1.05) | 0.04 | 1.57 × 10−5 |
| rs7862747 | 9q31.2 | C/A | 0.36 | 0.88 (0.87, 0.90) | 1.89 × 10−58 | 0.98 (0.96, 1.00) | 0.05 | 4.49 × 10−13 |
| rs7098100 | 10p12.31 | A/G | 0.34 | 1.07 (1.06, 1.09) | 9.46 × 10−21 | 0.97 (0.95, 1.00) | 0.02 | 1.42 × 10−12 |
| rs9420318 | 10q26.12 | A/G | 0.33 | 0.94 (0.93, 0.95) | 2.55 × 10−17 | 1.00 (0.98, 1.02) | 0.74 | 6.53 × 10−6 |
| rs2981579 | 10q26.13 | A/G | 0.41 | 1.32 (1.31, 1.34) | 3.72 × 10−359 | 1.06 (1.04, 1.08) | 4.23 × 10−8 | 5.37 × 10−74 |
| rs78540526 | 11q13.3 | T/C | 0.07 | 1.39 (1.35, 1.42) | 3.11 × 10−137 | 1.01 (0.97, 1.05) | 0.73 | 1.67 × 10−36 |
| rs199504893 | 11q22.3 | CA/C | 0.41 | 1.02 (1.00, 1.03) | 0.01 | 0.94 (0.92, 0.96) | 3.31 × 10−9 | 1.56 × 10−10 |
| rs1292011 | 12q24.21 | G/A | 0.39 | 0.90 (0.89, 0.92) | 3.34 × 10−47 | 0.97 (0.95, 0.99) | 0 | 1.05 × 10−7 |
| rs1744947 | 14q24.1 | T/C | 0.15 | 1.08 (1.06, 1.10) | 8.58 × 10−14 | 1.00 (0.97, 1.03) | 0.82 | 2.26 × 10−5 |
| rs4784227 | 16q12.1 | T/C | 0.24 | 1.26 (1.25, 1.28) | 1.03 × 10−202 | 1.15 (1.13, 1.18) | 3.57 × 10−36 | 3.21 × 10−11 |
| rs2123844 | 17p13.2 | A/C | 0.07 | 1.03 (1.00, 1.06) | 0.03 | 1.13 (1.09, 1.18) | 2.81 × 10−10 | 6.69 × 10−5 |
| rs745983748 | 18q11.2 | A/AAGTGTT | 0.32 | 0.93 (0.91, 0.94) | 6.12 × 10−24 | 1.01 (0.99, 1.03) | 0.44 | 3.07 × 10−10 |
| rs4609972 | 19p13.11 | C/G | 0.48 | 1.00 (0.98, 1.01) | 0.80 | 0.88 (0.86, 0.90) | 6.13 × 10−35 | 6.60 × 10−24 |
| rs34753522 | 20q12 | C/T | 0.35 | 0.96 (0.94, 0.97) | 3.21 × 10−8 | 1.02 (1.00, 1.04) | 0.1 | 8.07 × 10−6 |
| rs2403907 | 21q21.1 | A/C | 0.29 | 0.91 (0.90, 0.93) | 1.09 × 10−32 | 0.97 (0.95, 1.00) | 0.02 | 3.14 × 10−6 |
| rs4822992 | 22q12.1 | A/G | 0.02 | 1.25 (1.19, 1.31) | 7.16 × 10−19 | 1.00 (0.93, 1.09) | 0.91 | 6.23 × 10−6 |
Effect allele/reference allele.
Effect allele frequency.
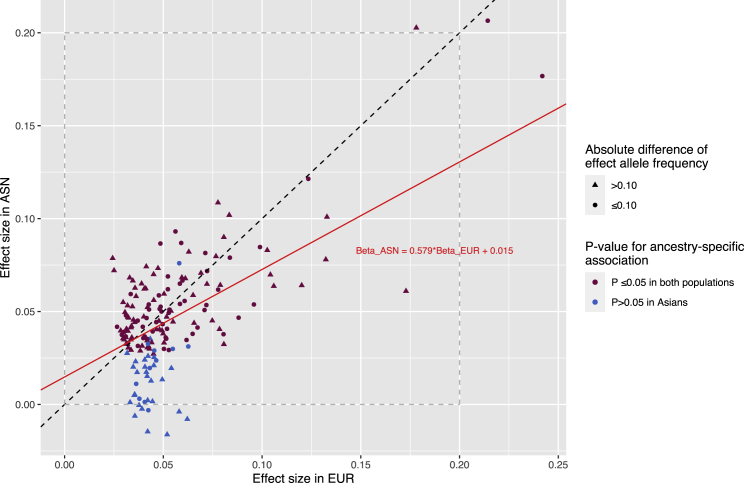
Of the 211 lead risk variants for overall breast cancer, 166 variants had a >25% difference in the effect allele frequency between Asian-ancestry and European-ancestry women (Figure S2). Seventeen lead variants, all identified from ancestry-specific GWASs, are rare (a MAF of <0.01) in one population but common in the other population. For nine of these lead variants, all variants included in their 95% credible sets were rare in one population but common in the other population (Table S2). Of the 194 common risk variants in both populations, 36 showed a significant difference in risk estimates between Asian- and European-ancestry populations at p < 0.05, including 31 lead variants with the entire credible sets showing ancestral heterogeneity in risk estimates (p < 0.05). Three variants showed ancestral heterogeneity with a p < 2.58 × 10−4, the significance level after adjusting for multiple comparisons (0.05/194) (Table S2). In particular, variant rs59957907 showed a highly significant ancestral difference in risk estimate with a p for heterogeneity of 1.27 × 10−104. Overall, risks estimated in European-ancestry populations are larger than those estimated in Asian-ancestry populations with a regression beta coefficient of 0.579 derived from linear regression (Figure 1, Table S2). The ancestral difference observed in our study could be underestimated, as variants with similar risk estimates were more likely to be identified by cross-ancestry meta-analyses.
Figure 1.
Comparison of risk estimates for lead risk variants between Asian- and European-ancestry women
The red regression line shows the trend of risk estimates in both ancestry groups. To be conservative, the regression was performed excluding four variants with risk estimates >0.15 in European-ancestry women, which could be outliers or with a high leverage. The black dashed diagonal line shows where risk estimates are the same in both ancestries.
Twenty-three previously reported index variants are not located at the regions identified at genome-wide significance in our meta-analyses. However, 16 of them were associated with breast cancer risk at p < 2.04 × 10−4, a significant level with Bonferroni correction for comparisons of 245 index variants. Of the remaining seven index risk variants, four were previously identified in a GWAS by breast cancer intrinsic subtypes6 (Table S8). Two index variants showed a nominally significant association with breast cancer in cross-ancestry and European-ancestry meta-analyses (p < 0.05). Only variant rs9348512 showed a null association with overall breast cancer risk (p = 0.505). The association with this variant was originally reported in a GWAS conducted among individuals with BRCA2 mutation29 but was not replicated in subsequent studies.5,6
The sentinel variants at all 17 newly identified risk loci showed the same association direction in both Asian- and European-ancestry populations (Tables S2 and S4). Except for the Asian-specific risk variant rs74157632, all other lead variants are common, with a MAF >0.01 in both populations. Significant ancestral heterogeneity was observed for rs6890591 (identified by meta-regression) and rs74157632 (identified as Asian specific). The estimated ORs for these 17 lead variants in the BCAC and AABC studies are shown in Table S5. The proportion of variance explained by the 17 novel loci identified in our study was 1.15% for overall breast cancer, 1.07% for ER-positive breast cancer, and 1.03% for ER-negative breast cancer in Asian-ancestry populations. The corresponding numbers are 0.74%, 0.61%, and 1.03% for European-ancestry populations. The higher percentage of genetic variation explained by these new loci in Asian- compared to European-ancestry populations was because of the population differences in the risk estimates at the new loci. Of the 17 novel loci, one locus was specific to the Asian populations. For the remaining 16 loci, the effect size, as measured using OR, was larger in Asian- than in European-ancestry populations for nine loci, including two loci showing a significant difference (p for heterogeneity <0.05). In only two loci, the OR for the lead variant was larger in European- than in Asian-ancestry populations, but no significant heterogeneity was found in either locus. The Asian-specific lead variant rs74157632 (GenBank: NM_207009.4; c.658A>G; p.Asn220Asp) is a missense variant of protein-coding gene DENND10, which has been shown to regulate the progression of epidermal growth factor receptor (EGFR) trafficking.30 Eleven lead variants are located in the intronic regions of genes. Some of these genes have been reported to be involved in breast cancer cell migration and invasion (SLC8A1,31 CDKN1A,32 AFDN,33 TLN234), resistance to radiotherapy (ALG935), and TGF-β (LDLRAD436) or p53 (PHF2037) signaling pathways.
For each of the novel loci identified in this study, we performed conditional analyses for variants located within 500 kb of the lead variant, adjusted for the lead variant separately for Asian and European descendants, to identify potential secondary association signals. These results were then combined by meta-analyses. We found eight independent association signals (conditional p < 1.0 × 10−4) at six loci: 2p22.1, 6q22.31, 6q27, 8p22, 15q22.2, and 18p11.21 (Table S9). There were two additional independent association signals found at loci 8p22 and 18p11.21.
To identify putative breast cancer susceptibility genes, we conducted a transcriptome-wide association analysis (TWAS). We used whole-genome sequencing data generated in genomic DNA samples and RNA sequencing data generated in normal tissues obtained from 115 individuals included in the GTEx project (version 8) to build genetic models to predict gene expression across the transcriptome (Material and methods, Table S10). Of the 30,362 genes evaluated, models were successfully built for 17,127 genes, in which 10,820 genes could be predicted with R > 0.1. The performance of the models was evaluated using the adjacent normal breast tissue samples from TCGA. Overall, genes that were predicted with R > 0.1 in GTEx data were also predicted well in TCGA tumor-adjacent normal tissue data (correlation coefficient of 0.69; Figure S3).
Of the 10,820 genes evaluated using GWAS data from 160,500 individuals with breast cancer and 226,196 controls, we identified 137 genes in association with risk of breast cancer at the Bonferroni-corrected threshold of p < 4.62 × 10−6, including 76 protein-coding genes (Tables S11 and S18). Of them, 14 genes at 13 loci are located at least 1 Mb away from any of the previous GWAS-identified risk variants for breast cancer (Table 3), including 11 genes associated with overall breast cancer risk and three additional genes associated with ER-positive breast cancer. CPNE1 is located at a novel risk locus identified in our cross-ancestry meta-analyses. CPNE1 has been reported to be overexpressed in triple-negative breast cancer and promotes tumorigenesis and radio-resistance by the AKT signaling pathway.38 In addition, we also identified 87 genes (including 39 protein-coding genes) that are located in known risk loci but have not yet been reported in previous TWASs39,40,41,42 (Table S11).
Table 3.
Genes identified in TWASs in novel loci in association with breast cancer risk
| Locia | Gene | Gene type | Z score | p | R2b |
|---|---|---|---|---|---|
| Overall | |||||
| 1p11.2 | NBPF8 | Pseudogene | 7.05 | 1.76 × 10−12 | 0.23 |
| 1p11.2 | PFN1P2 | Pseudogene | 9.22 | 2.87 × 10−20 | 0.22 |
| 3p21.31 | RNF123 | Protein coding | 4.63 | 3.62 × 10−6 | 0.26 |
| 5p15.31 | NSUN2 | Protein coding | −4.89 | 1.01 × 10−6 | 0.37 |
| 10q26.13 | EEF1AKMT2 | Protein coding | −4.70 | 2.63 × 10−6 | 0.34 |
| 15q15.1 | SRP14-DT | LincRNA | −4.80 | 1.55 × 10−6 | 0.29 |
| 15q15.3 | STRCP1 | Pseudogene | −4.66 | 3.18 × 10−6 | 0.12 |
| 17p12 | MAP2K4 | Protein coding | 4.99 | 6.06 × 10−7 | 0.02 |
| 19q13.12 | ZNF793-AS1 | Antisense RNA | −4.94 | 7.64 × 10−7 | 0.10 |
| 20q11.22 | CPNE1 | Protein coding | −4.68 | 2.88 × 10−6 | 0.38 |
| 20q13.33c | RGS19 | Protein coding | 4.64 | 3.47 × 10−6 | 0.07 |
| ER-positive | |||||
| 6p22.1 | H4C12 | Protein coding | 5.01 | 5.54 × 10−7 | 0.07 |
| 11q13.2 | RHOD | Protein coding | 4.78 | 1.73 × 10−6 | 0.19 |
| 5q13.2c | GUSBP14 | Pseudogene | 5.08 | 3.73 × 10−7 | 0.08 |
Unless otherwise specified, results are based on TWAS analyses using cross-ancestry GWAS data.
Prediction performance derived using GTEx data.
Genes identified from association analysis using European-ancestry GWAS data.
Of the 137 genes identified by TWAS, 15 genes showed different associations with ER-positive and ER-negative breast cancer, with a p for heterogeneity <3.65 × 10−4 (0.05/137; Tables 4 and S12). Of them, protein-coding genes ABHD8 and ANKLE1 at 19p13.11 showed an exclusive association with ER-negative breast cancer, and similar heterogeneity also was found for the lead variant rs4808616 at this risk locus. These findings were supported by a previous study, which identified ABHD8 and ANKLE1 as potential target genes at the risk locus 19p13.11.43
Table 4.
TWAS-identified breast cancer risk genes showing a significantly different association by estrogen receptor status
| Loci | Gene | Gene type |
ER-Positive |
ER-Negative |
p for ER heterogeneity | ||
|---|---|---|---|---|---|---|---|
| Z score | P | Z score | p | ||||
| 1p11.2 | SRGAP2C | Protein coding | −9.45 | 3.32′10−21 | −1.47 | 0.14 | 6.99′10−5 |
| 1p11.2 | H3P4 | Pseudogene | 8.89 | 6.05′10−19 | 1.10 | 0.27 | 1.72′10−4 |
| 1p11.2 | RP11-343N15.2a | LincRNA | −8.74 | 2.27′10−18 | −1.00 | 0.32 | 3.35′10−5 |
| 1p11.2 | EMBP1 | Pseudogene | −8.38 | 5.23′10−17 | −0.27 | 0.78 | 9.32′10−6 |
| 1p36.13 | KLHDC7A | Protein coding | −7.10 | 1.27′10−12 | 0.10 | 0.92 | 5.79′10−6 |
| 1p36.22 | DFFA | Protein coding | 4.37 | 1.26′10−5 | 7.60 | 2.96′10−14 | 9.54′10−5 |
| 1q22 | GBAP1 | Pseudogene | −6.66 | 2.73′10−11 | 0.59 | 0.56 | 2.54′10−5 |
| 1q22 | THBS3 | Protein coding | 5.72 | 1.07′10−8 | −0.89 | 0.38 | 8.72′10−5 |
| 1q32.1 | PTPRVP | Pseudogene | −1.50 | 0.14 | 6.67 | 2.52′10−11 | 1.36′10−10 |
| 2q35 | TNP1 | Protein coding | 5.85 | 5.04′10−9 | −0.37 | 0.71 | 5.44′10−5 |
| 5p12 | MRPS30-DT | Antisense RNA | 16.38 | 2.48′10−60 | −0.15 | 0.88 | 4.20′10−21 |
| 5q11.2 | CTD-2310F14.1a | Antisense RNA | 14.50 | 1.17′10−47 | 3.73 | 1.90′10−4 | 4.24′10−7 |
| 8p23.3 | SEPT14P8 | Pseudogene | −2.29 | 0.02 | −6.00 | 1.98′10−9 | 2.53′10−4 |
| 19p13.11 | ABHD8 | Protein coding | −0.51 | 0.61 | 9.64 | 5.25′10−22 | 2.39′10−15 |
| 19p13.11 | ANKLE1 | Protein coding | −0.24 | 0.81 | 6.74 | 1.62′10−11 | 8.17′10−9 |
RP11-343N15.2 (ENSG00000231429) and CTD-2310F14.1 (ENSG00000271828) do not have gene symbols in HUGO yet.
In addition, 16 genes showed a significantly different association between Asian- and European-ancestry women at the Bonferroni-corrected threshold p for heterogeneity <3.65 × 10−4, including seven protein-coding genes (Table S13). Of them, CASP8 and ALS2CR12 at 2q33.1 and HLA-F at 6p22.1 showed a stronger association with breast cancer risk in Asian-ancestry women than in European-ancestry women. The CASP8 gene plays a central role in extrinsic apoptosis44 and has been reported to be associated with breast cancer risk in previous TWASs among European-ancestry women.39,40,41,42
To identify the most likely target genes in the locus in which multiple genes were found to be associated with breast cancer risk in TWASs, we performed fine-mapping analyses using FOCUS.45 In total, we identified 69 genes showing significant posterior inclusion probability and thus included them in the credible target gene sets (Table S14). In addition, we identified 50 genes that were colocalized with both GWASs and eQTL signals from colocalization analyses using COLOC46 (Table S15), including 28 genes included in the credible target gene sets from TWAS fine-mapping analyses.
We performed pathway analyses to identify biological pathways that may play a role in breast cancer etiology. Of the 137 genes identified in our TWASs in association with breast cancer risk, 76 located in 53 genomic regions are protein-coding genes. In 47 regions, we were able to identify 53 genes as putative target genes with supporting evidence from either fine-mapping analyses (n = 25), colocalization analyses (n = 10), or both (n = 18). Additionally, for the remaining 152 loci, in which no target genes were identified in TWASs, we selected 89 protein-coding genes previously reported as putative target genes47 and 79 protein-coding genes located nearby the lead variants identified in our GWAS. In total, 221 putative risk genes for breast cancer were included in our pathway analysis (supplemental methods and Table S16). We identified multiple signaling pathways that were significantly associated with breast cancer risk at FDR <0.05, including p53, cGMP-PKG, TNF, and MAPK signaling pathways, as well as pathways of DNA-binding transcription activator activity and cell cycle phase transition48,49,50 (Table S17).
Discussion
We conducted a large GWAS and TWAS of breast cancer, including 386,696 women of Asian and European ancestry. In total, 222 genetic risk loci and 137 genes were identified by GWAS and TWAS, respectively, in association with breast cancer risk after adjusting for multiple comparisons.
Our pathway analyses identified multiple biological pathways that have been implicated in the development of breast and other cancers. For example, CACNA1A, DUSP4, FGFR2, MAP2K4, MAP3K1, MYC, NF1, PLA2G6, TAB2, TGFBR2, and TP53 are involved in mitogen-activated protein kinase (MAPK) signaling pathway.48,51 ATG10, CDKAL1, KLF4, MAF8, and MAP3K1 are regulated by the activation of KRAS.51 KRAS is a proto-oncogene from the RAS family and a part of the RAS/MAPK pathway. Although the RAS signaling pathway is commonly activated in breast cancer, somatic mutations of RAS are not common in individuals with breast cancer.52 Our findings indicate that the germline alternation of genes involved in the RAS signaling pathway could play a role in the development and progression of breast cancer.
Although the p53 pathway is often altered in breast cancer tissues, particularly those from ER-negative and triple-negative cancer, germline mutations of TP53 are detected only in less than 1% of individuals with breast cancer.53 In this study, we found that 15 genes (CASP8, CCND1, CCNE1, CDKN1A, CHEK2, MDM4, INHBB, KLF4, MXD1, PHLDA3, PIDD1, TNNI1, TP53, ZFP36L1, ZNF365) are involved in the p53 signaling pathway,48,51 providing support that germline alterations of this pathway could play a more significant etiologic role than what is appreciated based on analyzing TP53 alone. Intriguingly, the MDM4 and CCNE1 are located at risk loci with a stronger association with ER-negative than ER-positive breast cancer. Our TWAS also found that the expression of MDM4 was exclusively associated with an increased risk of ER-negative breast cancer. These findings suggest that the p53 signaling pathway plays an important role in the risk of breast cancer, especially ER-negative breast cancer.
By increasing the sample size and incorporating transcriptome data, we were able to identify 30 novel associations in loci and genes that are located >1 Mb away from any of the previously reported breast cancer risk variants. The discovery of these novel associations further expanded our understanding of the genetic and biological mechanism of breast cancer development. For example, the lead variant at the novel risk locus 6p21.2 is located at the intronic region of CDKN1A. CDKN1A regulates cell-cycle progression as a cyclin-dependent kinase inhibitor32 and plays an important role in both PI3K/AKT signaling pathway and p53 pathway.51
MAP2K4 at 17p12 is a novel target gene identified by our TWAS. This gene encodes a member of the mitogen-activated protein kinase and it is involved in multiple signaling pathways, including MAPK pathway, EGF pathway, FAS signaling pathway,51 and PI3K/AKT signaling pathway.54 In addition, our TWAS identified 39 protein-coding genes that are located in known risk loci but have not yet been reported in previous TWAS. Of them, MDM4, PLA2G6, and RIT1 are involved in the p53 pathway, RAS/MAPK pathway, and PI3K/AKT pathway, respectively. These newly identified putative breast cancer risk genes could be potential targets for therapies.
Given the much larger sample size for GWASs conducted in European descendants compared to those conducted in East Asians, many of the associations were driven by data from European-ancestry GWASs. Increasing the sample size for GWASs of non-European populations will be valuable to fully uncover the genetic basis for breast cancer. In our TWAS, we built gene prediction models using European-ancestry samples from GTEx. Given the difference in genetic architectures between Asian and European descendants, some of these models may not perform well in TWASs in Asian populations, affecting the detection of significant association signals, particularly in regions where significant ancestral differences exist. Using Asian-specific gene prediction models in future studies should help to identify additional genes associated with breast cancer risk.
In summary, in this large GWAS and TWAS for breast cancer, we uncovered a large number of genetic variants associated with breast cancer risk and identified potential target genes for this common cancer. We discovered significant differences for many of these variants and genes in association with breast cancer risk by ER status and ancestry. We identified multiple signaling pathways that play an etiologic role in breast cancer risk and propose that germline alterations in TP53, RAS, and MAPK pathways may play a more significant role in the etiology of breast cancer than what is currently appreciated. Our study provides substantial insights into the genetics and biology of breast cancer.
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agents. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. This research was supported in part by U.S. National Institutes of Health grants R01CA235553, R01CA202981, R01CA124558, R01CA148667, R01CA158473, R01CA064277, R37CA070867, and UM1CA182910 (to W.Z.), R01CA118229 and R01CA092585 (to X.-O.S.), R01CA122756 (to Q.C.), and R01CA137013 (to J. Long); Department of Defense Idea Awards BC011118 (to X.-O.S.) and BC050791 (to Q.C.); and Ingram Professor and Anne Potter Wilson Chair and Research Reward funds (to W.Z.). Sample preparation and genotyping assays at Vanderbilt were conducted at the Survey and Biospecimen Shared Resources and Vanderbilt Microarray Shared Resource, which are supported in part by the Vanderbilt-Ingram Cancer Center (P30CA068485). Data analyses were conducted using the Advanced Computing Center for Research and Education (ACCRE) at Vanderbilt University. Additional information is provided in the supplemental information.
Declaration of interests
The authors declare no competing interests.
Published: November 9, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2022.10.011.
Supplemental information
Data and code availability
Access to the ABCC data can be requested by submission of an inquiry to Dr. Wei Zheng (wei.zheng@vanderbilt.edu). Request for access to the BCAC data can be submitted directly to BCAC (http://bcac.ccge.medschl.cam.ac.uk/). All GTEx data are publicly available through dbGaP: phs000424.v8.p2. TCGA data are publicly available through National Cancer Institute’s Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). Access to the custom code: https://github.com/pingjie/EURASN_GWAS/.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Hu C., Hart S.N., Gnanaolivu R., Huang H., Lee K.Y., Na J., Gao C., Lilyquist J., Yadav S., Boddicker N.J., et al. A population-based study of genes previously implicated in breast cancer. N. Engl. J. Med. 2021;384:440–451. doi: 10.1056/NEJMoa2005936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breast Cancer Association Consortium. Dorling L., Carvalho S., Allen J., González-Neira A., Luccarini C., Wahlström C., Pooley K.A., Parsons M.T., Fortuno C., et al. Breast cancer risk genes - association analysis in more than 113, 000 women. N. Engl. J. Med. 2021;384:428–439. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Narod S.A. Which genes for hereditary breast cancer? N. Engl. J. Med. 2021;384:471–473. doi: 10.1056/NEJMe2035083. [DOI] [PubMed] [Google Scholar]
- 5.Michailidou K., Lindström S., Dennis J., Beesley J., Hui S., Kar S., Lemaçon A., Soucy P., Glubb D., Rostamianfar A., et al. Association analysis identifies 65 new breast cancer risk loci. Nature. 2017;551:92–94. doi: 10.1038/nature24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H., Ahearn T.U., Lecarpentier J., Barnes D., Beesley J., Qi G., Jiang X., O’Mara T.A., Zhao N., Bolla M.K., et al. Genome-wide association study identifies 32 novel breast cancer susceptibility loci from overall and subtype-specific analyses. Nat. Genet. 2020;52:572–581. doi: 10.1038/s41588-020-0609-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shu X., Long J., Cai Q., Kweon S.-S., Choi J.-Y., Kubo M., Park S.K., Bolla M.K., Dennis J., Wang Q., et al. Identification of novel breast cancer susceptibility loci in meta-analyses conducted among Asian and European descendants. Nat. Commun. 2020;11:1217. doi: 10.1038/s41467-020-15046-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin A.R., Kanai M., Kamatani Y., Okada Y., Neale B.M., Daly M.J. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 2019;51:584–591. doi: 10.1038/s41588-019-0379-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng W., Long J., Gao Y.-T., Li C., Zheng Y., Xiang Y.-B., Wen W., Levy S., Deming S.L., Haines J.L., et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat. Genet. 2009;41:324–328. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai Q., Zhang B., Sung H., Low S.-K., Kweon S.-S., Lu W., Shi J., Long J., Wen W., Choi J.-Y., et al. Genome-wide association analysis in East Asians identifies breast cancer susceptibility loci at 1q32.1, 5q14.3 and 15q26.1. Nat. Genet. 2014;46:886–890. doi: 10.1038/ng.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng W., Zhang B., Cai Q., Sung H., Michailidou K., Shi J., Choi J.-Y., Long J., Dennis J., Humphreys M.K., et al. Common genetic determinants of breast-cancer risk in East Asian women: a collaborative study of 23 637 breast cancer cases and 25 579 controls. Hum. Mol. Genet. 2013;22:2539–2550. doi: 10.1093/hmg/ddt089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y., Tao R., Shu X., Cai Q., Wen W., Gu K., Gao Y.-T., Zheng Y., Kweon S.-S., Shin M.-H., et al. Incorporating polygenic risk scores and nongenetic risk factors for breast cancer risk prediction among asian women. JAMA Netw. Open. 2022;5 doi: 10.1001/jamanetworkopen.2021.49030. e2149030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ishigaki K., Akiyama M., Kanai M., Takahashi A., Kawakami E., Sugishita H., Sakaue S., Matoba N., Low S.-K., Okada Y., et al. Large-scale genome-wide association study in a Japanese population identifies novel susceptibility loci across different diseases. Nat. Genet. 2020;52:669–679. doi: 10.1038/s41588-020-0640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michailidou K., Beesley J., Lindstrom S., Canisius S., Dennis J., Lush M.J., Maranian M.J., Bolla M.K., Wang Q., Shah M., et al. Genome-wide association analysis of more than 120, 000 individuals identifies 15 new susceptibility loci for breast cancer. Nat. Genet. 2015;47:373–380. doi: 10.1038/ng.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Q., Long J., Lu W., Qu S., Wen W., Kang D., Lee J.-Y., Chen K., Shen H., Shen C.-Y., et al. Genome-wide association study identifies breast cancer risk variant at 10q21.2: results from the Asia Breast Cancer Consortium. Hum. Mol. Genet. 2011;20:4991–4999. doi: 10.1093/hmg/ddr405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Long J., Cai Q., Sung H., Shi J., Zhang B., Choi J.-Y., Wen W., Delahanty R.J., Lu W., Gao Y.-T., et al. Genome-wide association study in east Asians identifies novel susceptibility loci for breast cancer. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002532. e1002532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han M.-R., Long J., Choi J.-Y., Low S.-K., Kweon S.-S., Zheng Y., Cai Q., Shi J., Guo X., Matsuo K., et al. Genome-wide association study in East Asians identifies two novel breast cancer susceptibility loci. Hum. Mol. Genet. 2016;25:3361–3371. doi: 10.1093/hmg/ddw164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y., Long J., Lu W., Shu X.-O., Cai Q., Zheng Y., Li C., Li B., Gao Y.-T., Zheng W. Rare coding variants and breast cancer risk: evaluation of susceptibility Loci identified in genome-wide association studies. Cancer Epidemiol. Biomarkers Prev. 2014;23:622–628. doi: 10.1158/1055-9965.EPI-13-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim H.c., Lee J.-Y., Sung H., Choi J.-Y., Park S.K., Lee K.-M., Kim Y.J., Go M.J., Li L., Cho Y.S., et al. A genome-wide association study identifies a breast cancer risk variant in ERBB4 at 2q34: results from the Seoul breast cancer study. Breast Cancer Res. 2012;14:R56. doi: 10.1186/bcr3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang C.C., Chow C.C., Tellier L.C., Vattikuti S., Purcell S.M., Lee J.J. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Willer C.J., Li Y., Abecasis G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mägi R., Horikoshi M., Sofer T., Mahajan A., Kitajima H., Franceschini N., McCarthy M.I., COGENT-Kidney Consortium T2D-GENES Consortium. Morris A.P., Morris A.P. Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum. Mol. Genet. 2017;26:3639–3650. doi: 10.1093/hmg/ddx280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G., Sarkar A., Carbonetto P., Stephens M. A simple new approach to variable selection in regression, with application to genetic fine mapping. J. Roy. Stat. Soc. B. 2020;82:1273–1300. doi: 10.1111/rssb.12388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E., et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019;47:D1005–D1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roden D.M., Pulley J.M., Basford M.A., Bernard G.R., Clayton E.W., Balser J.R., Masys D.R. Development of a large-scale De-identified DNA biobank to enable personalized medicine. Clin. Pharmacol. Ther. 2008;84:362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasimatis K.R., Abraham A., Ralph P.L., Kern A.D., Capra J.A., Phillips P.C. Evaluating human autosomal loci for sexually antagonistic viability selection in two large biobanks. Genetics. 2021;217:1–10. doi: 10.1093/genetics/iyaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou D., Jiang Y., Zhong X., Cox N.J., Liu C., Gamazon E.R. A unified framework for joint-tissue transcriptome-wide association and Mendelian randomization analysis. Nat. Genet. 2020;52:1239–1246. doi: 10.1038/s41588-020-0706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barbeira A.N., Dickinson S.P., Bonazzola R., Zheng J., Wheeler H.E., Torres J.M., Torstenson E.S., Shah K.P., Garcia T., Edwards T.L., et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gaudet M.M., Kuchenbaecker K.B., Vijai J., Klein R.J., Kirchhoff T., McGuffog L., Barrowdale D., Dunning A.M., Lee A., Dennis J., et al. Identification of a BRCA2-specific modifier locus at 6p24 related to breast cancer risk. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003173. e1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Zhang K., Qi L., Hu Q., Shen Z., Liu B., Deng J., Zhang C., Zhang Y. DENN domain-containing protein FAM45A regulates the homeostasis of late/multivesicular endosomes. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:916–929. doi: 10.1016/j.bbamcr.2019.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Zhu Q., Zhang X., Zai H.-Y., Jiang W., Zhang K.-J., He Y.-Q., Hu Y. circSLC8A1 sponges miR-671 to regulate breast cancer tumorigenesis via PTEN/PI3k/Akt pathway. Genomics. 2021;113:398–410. doi: 10.1016/j.ygeno.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 32.Zaremba-Czogalla M., Hryniewicz-Jankowska A., Tabola R., Nienartowicz M., Stach K., Wierzbicki J., Cirocchi R., Ziolkowski P., Tabaczar S., Augoff K. A novel regulatory function of CDKN1A/p21 in TNFα-induced matrix metalloproteinase 9-dependent migration and invasion of triple-negative breast cancer cells. Cell. Signal. 2018;47:27–36. doi: 10.1016/j.cellsig.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Fournier G., Cabaud O., Josselin E., Chaix A., Adélaïde J., Isnardon D., Restouin A., Castellano R., Dubreuil P., Chaffanet M., et al. Loss of AF6/afadin, a marker of poor outcome in breast cancer, induces cell migration, invasiveness and tumor growth. Oncogene. 2011;30:3862–3874. doi: 10.1038/onc.2011.106. [DOI] [PubMed] [Google Scholar]
- 34.Li L., Li X., Qi L., Rychahou P., Jafari N., Huang C. The role of talin2 in breast cancer tumorigenesis and metastasis. Oncotarget. 2017;8:106876–106887. doi: 10.18632/oncotarget.22449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X., He Z., Guo L., Wang C., Lin C., Ye L., Wang X., Li Y., Yang M., Liu S., et al. ALG3 contributes to stemness and radioresistance through regulating glycosylation of TGF-β receptor II in breast cancer. J. Exp. Clin. Cancer Res. 2021;40:149. doi: 10.1186/s13046-021-01932-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakano N., Maeyama K., Sakata N., Itoh F., Akatsu R., Nakata M., Katsu Y., Ikeno S., Togawa Y., Vo Nguyen T.T., et al. C18 ORF1, a novel negative regulator of transforming growth factor-β signaling. J. Biol. Chem. 2014;289:12680–12692. doi: 10.1074/jbc.M114.558981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cui G., Park S., Badeaux A.I., Kim D., Lee J., Thompson J.R., Yan F., Kaneko S., Yuan Z., Botuyan M.V., et al. PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53. Nat. Struct. Mol. Biol. 2012;19:916–924. doi: 10.1038/nsmb.2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao Z., Ma X., Zhang Y., Sun Y., Lv W., He K., Xia R., Wang P., Gao X. CPNE1 predicts poor prognosis and promotes tumorigenesis and radioresistance via the AKT singling pathway in triple-negative breast cancer. Mol. Carcinog. 2020;59:533–544. doi: 10.1002/mc.23177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu L., Shi W., Long J., Guo X., Michailidou K., Beesley J., Bolla M.K., Shu X.-O., Lu Y., Cai Q., et al. A transcriptome-wide association study of 229, 000 women identifies new candidate susceptibility genes for breast cancer. Nat. Genet. 2018;50:968–978. doi: 10.1038/s41588-018-0132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferreira M.A., Gamazon E.R., Al-Ejeh F., Aittomäki K., Andrulis I.L., Anton-Culver H., Arason A., Arndt V., Aronson K.J., Arun B.K., et al. Genome-wide association and transcriptome studies identify target genes and risk loci for breast cancer. Nat. Commun. 2019;10:1741. doi: 10.1038/s41467-018-08053-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrenson K., Kar S., McCue K., Kuchenbaeker K., Michailidou K., Tyrer J., Beesley J., Ramus S.J., Li Q., Delgado M.K., et al. Functional mechanisms underlying pleiotropic risk alleles at the 19p13.1 breast-ovarian cancer susceptibility locus. Nat. Commun. 2016;7:12675. doi: 10.1038/ncomms12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fritsch M., Günther S.D., Schwarzer R., Albert M.-C., Schorn F., Werthenbach J.P., Schiffmann L.M., Stair N., Stocks H., Seeger J.M., et al. Caspase-8 is the molecular switch for apoptosis, necroptosis and pyroptosis. Nature. 2019;575:683–687. doi: 10.1038/s41586-019-1770-6. [DOI] [PubMed] [Google Scholar]
- 43.Wen W., Chen Z., Bao J., Long Q., Shu X.-O., Zheng W., Guo X. Genetic variations of DNA bindings of FOXA1 and co-factors in breast cancer susceptibility. Nat. Commun. 2021;12:5318. doi: 10.1038/s41467-021-25670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng H., Gusev A., Pasaniuc B., Wu L., Long J., Abu-Full Z., Aittomäki K., Andrulis I.L., Anton-Culver H., Antoniou A.C., et al. Transcriptome-wide association study of breast cancer risk by estrogen-receptor status. Genet. Epidemiol. 2020;44:442–468. doi: 10.1002/gepi.22288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mancuso N., Freund M.K., Johnson R., Shi H., Kichaev G., Gusev A., Pasaniuc B. Probabilistic fine-mapping of transcriptome-wide association studies. Nat. Genet. 2019;51:675–682. doi: 10.1038/s41588-019-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giambartolomei C., Vukcevic D., Schadt E.E., Franke L., Hingorani A.D., Wallace C., Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004383. e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fachal L., Aschard H., Beesley J., Barnes D.R., Allen J., Kar S., Pooley K.A., Dennis J., Michailidou K., Turman C., et al. Fine-mapping of 150 breast cancer risk regions identifies 191 likely target genes. Nat. Genet. 2020;52:56–73. doi: 10.1038/s41588-019-0537-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gene Ontology Consortium The Gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–D334. doi: 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Galiè M. RAS as supporting actor in breast cancer. Front. Oncol. 2019;9:1199. doi: 10.3389/fonc.2019.01199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schon K., Tischkowitz M. Clinical implications of germline mutations in breast cancer: TP53. Breast Cancer Res. Treat. 2018;167:417–423. doi: 10.1007/s10549-017-4531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu S., Huang J., Zhang Y., Liu Y., Zuo S., Li R. MAP2K4 interacts with Vimentin to activate the PI3K/AKT pathway and promotes breast cancer pathogenesis. Aging (Albany NY) 2019;11:10697–10710. doi: 10.18632/aging.102485. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Access to the ABCC data can be requested by submission of an inquiry to Dr. Wei Zheng (wei.zheng@vanderbilt.edu). Request for access to the BCAC data can be submitted directly to BCAC (http://bcac.ccge.medschl.cam.ac.uk/). All GTEx data are publicly available through dbGaP: phs000424.v8.p2. TCGA data are publicly available through National Cancer Institute’s Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). Access to the custom code: https://github.com/pingjie/EURASN_GWAS/.