Abstract
Background
Understanding the role and potential therapeutic targeting of tumor-associated macrophages (TAMs) is crucial to developing new biomarkers and therapeutic strategies for cancer immunotherapies. The epigenetic reader SP140 has emerged as a master regulator of macrophage transcriptional programs; however, its role in the signaling of TAMs and response to immunotherapy has not been investigated.
Methods
We evaluated the correlation between SP140 expression in head and neck squamous cell carcinoma (HNSCC) TAMs and clinical outcomes. We also used complementary bioinformatics and experimental approaches to study the association of SP140 expression with tumor mutation burden, patient survival, immunogenic signature of tumors, and signaling of TAMs. SP140 overexpression or knockdown was implemented to identify the role of SP140 in downstream signaling and production of inflammatory cytokine and chemokines. Chromatin immunoprecipitation and analysis of assay of transposase accessible chromatin sequencing data were used to demonstrate the direct binding of SP140 on the promoters of STAT1. Finally, correlation of SP140 with immune cell infiltrates and response to immune-checkpoint blockade in independent cohorts of HNSCC, metastatic melanoma, and melanoma was assessed.
Results
We found that SP140 is highly expressed in TAMs across many cancer types, including HNSCCs. Interestingly, higher expression of SP140 in the tumors was associated with higher tumor mutation burden, improved survival, and a favorable response to immunotherapy. Tumors with high SP140 expression showed enrichment of inflammatory response and interferon-gamma (IFN-γ) pathways in both pan-cancer analysis and HNSCC-specific analysis. Mechanistically, SP140 negatively regulates transcription and phosphorylation of STAT1 and induces IFN-γ signaling. Activating SP140 in macrophages and TAMs induced the proinflammatory macrophage phenotype, increased the antitumor activity of macrophages, and increased the production of IFN-γ and antitumor cytokines and chemokines including interleukin-12 and CXCL10. SP140 expression provided higher sensitivity and specificity to predict antiprogrammed cell death protein 1 immunotherapy response compared with programmed death-ligand 1 in HNSCCs and lung cancer. In metastatic melanoma, higher levels of SP140 were associated with a durable response to immunotherapy, higher immune score estimates, high infiltrations of CD8+ T cells, and inflammatory TAMs.
Conclusions
Our findings suggest that SP140 could serve as both a therapeutic target and a biomarker to identify immunotherapy responders.
Keywords: head and neck neoplasms; biomarkers, tumor; immunotherapy; macrophages
WHAT IS ALREADY KNOWN ON THIS TOPIC
Immune-checkpoint blockade is beneficial in a subset of patients. Combination of immune-checkpoint blockade with other therapies to improve clinical outcomes and targeting other immune cells such as tumor-associated macrophages (TAMs) are ongoing. There is an urgent need to investigate prognostic and predictive biomarkers for patient selection to improve immunotherapy response in different tumors.
WHAT THIS STUDY ADDS
In both pan-cancer and head and neck cancer analysis, high expression of SP140 in the tumors was associated with higher tumor mutation burden, improved survival, and a favorable response to immunotherapy. Mechanistically, SP140 negatively regulates transcription and phosphorylation of STAT1 and induces interferon gamma (IFN-γ) signaling. Activating SP140 in macrophages and TAMs induced the proinflammatory macrophage phenotype, increased the antitumor activity of macrophages, and increased the production of IFN-γ and antitumor cytokines and chemokines including interleukin-12 and CXCL10.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our findings prove for the first time that SP140 potentially serves as a prognostic marker for immunotherapy and a therapeutic target for sensitizing tumors to immunotherapy.
Introduction
Changes in the tumor microenvironment (TME) and especially changes in tumor-associated immune cells play an important role in both the development and progression of solid tumors including head and neck squamous cell carcinoma (HNSCC).1 2 In the presence of tumor-cell antigens, the immune system triggers a natural T-cell response via cytotoxic T lymphocytes. In contrast, tumor cells promote immunosuppression by various tumor immune evasion mechanisms including overexpression of programmed death-ligand 1 (PD-L1) in tumor cells, immune infiltration by programmed cell death protein 1 (PD-1) T lymphocytes, and recruitment and activation of macrophages.3
Macrophages associated with the TME are referred to as tumor-associated macrophages (TAMs). They usually reside in close proximity to tumor cells and may represent up to 50% of the tumor mass.4 Historically, macrophages were phenotypically classified into two types: proinflammatory phenotype (M1) and anti-inflammatory phenotype (M2). M1 macrophages demonstrate antitumor functions, produce proinflammatory cytokines, phagocytize microbes, and initiate an immune response. On the other hand, tumor cells activate or switch the macrophages to an M2-like phenotype via several pathways such as CCL-2, interleukin (IL)-6, CSF-1, PD-1/PD-L1, CD47/SIRPα, which support tumor growth.5 Recent reports suggest that a binary classification of M1 and M2 phenotypes cannot capture the complexity of macrophage signaling and phenotypes of the TME. Most TAMs demonstrate a mixture of both phenotypes with a dominance of anti-inflammatory and protumorigenic activity.6 7 A subset of TAMs in the TME express immune-checkpoint modulators including PD-L1, which directly suppress the T-cell activation and have been reported to be associated with a better response to immune-checkpoint blockade.8 However, the complete network of macrophage signaling responsible for the immunosuppressive tumor phenotype, potential molecules to target these pathways, and markers of tumor vulnerabilities in TAMs for patient selection have not been completely identified.
Speckled protein 140 (SP140) is a nuclear protein that belongs to the SP family, consisting of SP100, SP110, and SP140L; SP family proteins have high sequence homology with the autoimmune regulator and are highly expressed in innate and adaptive immune cells with SP140 being the immune restricted.9–11 SP140 is interferon gamma (IFN-γ) inducible, and variant forms or differential expression of SP140 have been associated with immune disorders such as Crohn’s disease,12 multiple sclerosis,13 14 inflammatory bowel disease,12 and biliary cirrhosis.15 Previous reports demonstrated that SP140 is a transcriptional repressor that plays a pivotal role in the transcriptional programming of macrophages by suppressing lineage-inappropriate genes and is essential for lipopolysaccharide (LPS)-induced macrophage transcriptional programs.12 However, SP140 roles in the pathogenesis of cancer, response to immunotherapy, as well as TAMs are not investigated.
In the present study, we investigated the role of SP140 and its downstream signaling in HNSCC and macrophages. High levels of SP140 augmented the antitumorigenic macrophage phenotype by inhibiting STAT1 at the transcription and post-transcription level and induction of IFN-γ-related genes. High levels of SP140 were associated with high immune infiltrate in tumors in HNSCC and pan-cancer studies. In patients who underwent immunotherapy with immune-checkpoint inhibitors, high levels of SP140 were associated with response to immunotherapy and increased overall survival in HNSCCs, lung cancer, and metastatic melanoma. SP140 expression in the tumor conferred higher diagnostic accuracy compared with PD-L1 for predicting response to anti-PD-1 immunotherapy.
Materials and methods
Clinical samples
HNSCC samples were collected at the time of surgical resection and reviewed by pathologists at Columbia University. After collection from surgical resection, all samples were embedded in formalin or snap-frozen. The HNSCC cohort used in this study included an untreated cohort of 48 human papillomavirus (HPV)-negative HNSCC and 20 age-matched and sex-matched controls. For the head and neck sample cohort, all samples were histologically confirmed with 80% tumor contact in each sample. The cohort demographics were the following: 65% men, 35% women; mean age of 68 (SD 3.2) years. Sixty-five percent of patients were stage I and II, and 35% were stage III and IV HNSCC.
For immunotherapy samples, pretreatment tumor tissue of 21 patients with HNSCC and lung cancer who received anti-PD-1 immunotherapy was obtained from Columbia Biobank. This cohort included 12 responders and 9 non-responders. The cohort demographics were 65% men, 35% women; mean age of 66 (SD 4.21) years. We used the Response Evaluation Criteria in Solid Tumors V.1.116 to retrospectively identify the treatment response to immune-checkpoint blockade. As previously described,17 clinical benefits (responders) were defined as having experienced partial response or stable disease lasting at least 6 months, whereas non-clinical benefits (non-responders) were defined as a primary progressive disease or stable disease lasting less than 6 months.
Data and bioinformatic analysis
Our analysis included the The Cancer Genome Atlas (TCGA) pan-cancer atlas dataset, which contains 10 522 tumors across 33 cancer types, including 528 HNSCC samples. The following data are available at gdc.cancer.gov/node/977: cancer type, mutation status, clinical data, HPV stats, reverse phase protein array, and mRNA gene expression. Immunogenomic studies of clinical datasets of metastatic melanoma treated with anti-PD-1 (n=38)18 and metastatic melanoma treated with CTLA-4 (n=110)19 were downloaded from cBioportal.20
Estimation of individual immune subtype fractions by xCell in TCGA samples was performed using xcell.ucsf.edu21 and TIMER.22 For the correlation analysis, Pearson’s correlation coefficient was calculated with a Bonferroni-corrected p value of ≤0.05, which was considered statistically significant. We generated survival curves of HNSCC cases in the TCGA cohort according to the expression status of the SP140 gene. A group cut-off of ‘median’ was identified, and the Kaplan-Meier curve was plotted. Immune Subtype Classifier in R was used for the classification of pan-cancer immune subtypes (five-gene signature)23 to identify the correlation of SP140 with pan-cancer immune subtypes. CIBERSORT and TIMER were used for the immune cell analysis of the immunogenomic studies.22 24
Gene set enrichment analysis (GSEA)
GSEA analysis was performed to identify pathways upregulated in SP140 high tumors compared with tumors with low expression of SP140. A total of 528 HNSCC TCGA samples were grouped into SP140 high tumors and SP140 low tumors based on the median expression value of SP140 in the entire cohort. Expression with a Bonferroni corrected p value of <0.01 was analyzed by the GSEA preranked algorithm.25 For each gene set, the normalized enrichment score, p value, and false discovery rate (FDR) q values were calculated based on the Cancer Hallmark pathway and Gene Ontology Pathways.
Chromatin immunoprecipitation (ChIP)
After being transfection with SP140 small interfering RNA (siRNA) for 48 hours, cells were fixed with 1% (v/v) formaldehyde for 7 min, and cross-linking was stopped with 0.125 M glycine. Chromatin was isolated by using the Zymo kit (D5209), according to the manufacturer’s instructions. Sheared chromatin was conjugated with ChIP grade SP140 and control IgG antibodies overnight at 4°C while rotating. ZymoMag Protein A beads (15 µL) were added to ChIP reactions and incubated for 1 hour at 4°C while rotating. Tubes were placed in a magnetic field. Protein A bead/antibody/chromatin complexes were washed with 1 mL of chromatin wash buffer, and the beads were diluted in 500 µL of elution buffer. After washing, elution, and de-cross-linking, the ChIP DNA was detected by PCR. The PCR primers used for this assay are listed in online supplemental table 1.
jitc-2022-005088supp001.pdf (562KB, pdf)
Cell culture
Human THP-1 monocytes were cultured in Roswell Park Memorial Institute (RPMI) medium with 10% FBS, 1% penicillin, and streptomycin. Cells were incubated at 37°C in a 5% v/v CO2 atmosphere. For monocyte–macrophage differentiation, cells were seeded at a density of 2.5×105 cells/mL, and macrophage differentiation was induced by exposing the cells to 100 nM/mL phorbol-12-myristate-13-acetate (Invivogen) for 24 hours and replaced with fresh media for 24 hours. Head and neck cancer cell lines (Cal27, FaDu) were grown in complete Dulbecco's Modified Eagle Medium (DMEM) media (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Corning), 1% penicillin, and streptomycin (Corning). SCC7, a murine head and neck carcinoma cell line, was cultured in DMEM media supplemented with 10% FBS, 2 mM L-glutamine, and 100 U/mL penicillin/streptomycin.
In vivo syngeneic tumor model and TAMs’ isolation
Eight-week C3H/HeOuJ female mice were obtained from The Jackson Laboratory (n=8). 1×105 SCC7 cells were inoculated subcutaneously and mice were sacrificed on day 21. Tumors were dissociated using a mouse tumor dissociation kit (Miltenyi Biotec) with the gentleMACS Dissociator according to the manufacturer’s protocol (Miltenyi Biotec). CD11b magnetic beads (Miltenyi) were used for isolation of TAMs after tumor dissociation as recommended by the manufacturer. TAMs were cultured in RPMI 1640 culture medium (Gibco) supplemented with 10% heat-inactivated FBS (Corning), 1% penicillin, and streptomycin (Corning) and were used for ex vivo experiments. This study was approved by the Columbia University IACUC committee (AC-AABS4603).
Flow cytometry
A flow cytometry panel consisting of human CD80 (PE, BioLegend), human CD86 (BV711, BioLegend), and human CD206 (PerCP-Cy5.5, BioLegend) was used for the characterization of macrophages. For intracellular staining of phospho-STAT1 (pSTAT1ty701), cells were prepared using the Fix and Perm kit as recommended by the manufacturer (Invitrogen). Antibody-capture beads (CompBeads, BD Biosciences) were used as single-color compensation controls. Cytometer calibration was performed daily by using rainbow fluorescent particles (BD Biosciences) after acquiring unstained and single-color control samples to calculate the compensation matrix. Data were analyzed using FCS Express software.
Cell viability assay
Cell viability was assessed using a Cell Viability Assay Kit (Fluorometric-Blue) (ab112120) following the manufacturer’s recommendation. The same amount of dye-loading solution (100 μL/well) was added to the wells. Cell were incubated at room temperature for 1 hour and the fluorescent intensity was quantified by excitation and emission of 405 and 460, respectively.
siRNA transfection
Human THP-1 and peripheral blood mononuclear cells (PBMC) naïve macrophages were transfected with SP140 siRNA for 6 hours in Opti-MEM using lipofectamine RNAimax (Thermo Fisher). The transfection procedure was followed as per manufacturer instructions. After transfection, Opti-MEM media were replaced with fresh RPMI complete media. Cells were collected for RNA and protein expression analysis at 24 and 48 hours, respectively.
RNA isolation and quantitative real time-PCR
RNA isolation from the formalin-fixed paraffin-embedded (FFPE) tissue section was performed using a quick DNA/RNA FFPE kit (Zymo Research) according to the manufacturer’s instructions. In cell culture experiments, cells were lysed with TRI reagent (Zymo Research), and RNA was extracted with Direct-zol RNA Miniprep plus (Zymo Research) according to the manufacturer’s instructions. The quantity and quality of the RNA were determined by NanoDrop 1000 (260:280 and 260:230 ratios). Typically, 500–1 μg of total RNA was reverse transcribed using iScript reverse transcription supermix (Bio-Rad) using a T100 PCR Thermal Cycler (Bio-Rad). The amplification protocol included incubations at 94°C for 15 s, 63°C for 30 s, and 72°C for 60 s. The cDNA generated was used for quantitative PCR (qPCR) on a CFX96 Real-Time thermocycler (Bio-Rad) as per the manufacturer’s instructions using iTaq Universal SYBR Green Supermix (Bio-Rad). Target gene expression was normalized to the expression of the housekeeping gene 18S. Relative gene expression was calculated using the standard 2-ΔΔCT method. The primers used for this study are presented in online supplemental table 1.
SP140 CRISPR/dCas9 lentiviral activation particle transduction
Cloning the SP140 overexpressed into macrophages (THP1-derived or TAMs isolated from syngeneic mouse tumors) was performed using SP140 CRISPR/dCas9 lentiviral activation transduction particles, a synergistic activation mediator transcription activation system for the transcriptional activation of endogenous genes, according to the manufacturer’s protocol (Santa Cruz Biotechnology). Briefly, cells were plated onto six-well plates 24 hours before transfection. The culture media was replaced with RPMI-1640 medium plus 10% fetal bovine serum (FBS) and 5 µg/mL polybrene (Thermo Fisher), then transduced with SP140 lentiviral activation transduction particles (Santa Cruz Biotechnology). Forty-eight hours after transduction, the cells were selected by the addition of 10 mg/mL puromycin dihydrochloride for at least 7 days. The mock-transfected cells were transduced with control lentiviral activation particles and went through the same selection protocol.
Assay of transposase accessible chromatin sequencing (ATAC-seq) data processing and analysis
ATAC-seq reads of macrophages treated with LPS (0 hour), macrophages treated with LPS (4 hours), SP140 KO macrophages (0 hour), and SP140 KO macrophages (4 hours) were obtained from the Gene Expression Omnibus (GSE89177).26 In addition, H3K27me3 ChIP-seq reads for macrophages treated with GM-CSF+IFN-γ (GSM1625982), along with ChIP-seq reads for baseline macrophages (GSM1625981), were obtained.26 Three sets of reads were available for each of the H3K27me3 conditions and, due to low read counts, the three reads in each condition were merged as BAM files after alignment. ATAC-seq data were aligned via Bowtie2 V.2.427 using alignment parameters—very-sensitive and −k 10 per Harvard FAS Informatics guidelines to an hg38 index provided by the Bowtie2 package (GRCh38 no-alt analysis set). H3K27me3 data were aligned using default package parameters. After coordinate sorting and indexing (and merging in the case of the H3Kme3 data) via Samtools V.1.11,28 BAM files were imported as GeneRegionTracks using Gviz V.1.34.129 in R V.4.0.2 and mapped against appropriate chromosomal regions (histogram plot, with a sliding window of an arbitrary size determined by package).
Immunofluorescence
HNSCC FFPE tissue array sections were deparaffinized and washed with xylene and graded alcohol, followed by antigen retrieval by using 10 mM citrate buffer (pH 6.0). Non-specific antibody binding was reduced by incubating the sections with 3% bovine serum albumin (BSA) for 30 min at room temperature. The sections were then incubated with anti-SP140 primary antibody (MyBioSource) at 4°C overnight. Samples were washed with phosphate-buffered saline (PBST) (Triton) three times and incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG (Invitrogen) secondary antibody (Green) and PE/Cyanine7 anti-human CD68 antibody (orange) for 2 hours under dark and subsequently washed with PBST. Slides were incubated with DAPI (1:1000) (Sigma) for 5 min at room temperature in the dark, protected from light. Tissue sections were washed with PBS three times and mounted with Prolong Diamond Antifade Mountant (Invitrogen). After 48 hours, tissue sections were digitalized under an automated bright-field and fluorescence scanner at 20× (Leica Aperio VERSA 8 Scanning System). The QuPath open-source software platform30 was used to identify and quantify TAMs (CD68+) and colocalization of SP140 with TAMs.
Cytokine analysis
The LegendPlex Human Proinflammatory Chemokine Panel and Cytokine Panels were used according to the manufacturer’s instructions. Briefly, the assay was carried out in V-bottom 96-well plates. Mixed beads (25 µL), streptavidin-phycoerythrin (PE), and detection antibodies were used. Cell culture supernatant was added to mixed beads and incubated for 2 hours; wells were washed twice with wash buffer. Samples and standards were incubated with detection antibodies for 1 hour. Streptavidin–PE was then added and the plate was incubated for 30 min. Finally, beads were washed and resuspended in 200 µL of wash buffer, and data were acquired on a four-laser BD LSR Fortessa X20. All incubation steps were carried out in the dark at room temperature, on an orbital shaker set at 600–800 RPM. Data were analyzed using the LegendPlex data analysis software as instructed by the manufacturer.
Immunoblotting
Whole-cell lysates were prepared on ice using RIPA lysis buffer solution (Thermo Fisher), 1% protease, and phosphatase inhibitor cocktail (Invitrogen). Protein concentrations were determined with the Protein BCA Assay (Thermo Fisher). Lysate protein was subjected to 10% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane. After blotting, membranes were probed with a primary antibody at 4°C overnight. The following primary antibodies were used: anti-SP140 (MyBioSource), anti-STAT5a (Invitrogen), anti-phospho-STAT5aY694 (Invitrogen), anti-STAT1 (Invitrogen), and anti-phospho-STAT1Y701 (p-STAT1Y701) (Cell Signaling Technology). After washing three times with Tris-buffered saline with 0.1% Tween 20 Detergent (TBST), membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary goat anti-rabbit (Bio-Rad) or anti-mouse (Bio-Rad) antibodies as indicated for 2 hours at room temperature. Blots were developed with the Clarity Max Western ECL detection system (Bio-Rad) according to the manufacturer’s instructions, and images were captured using an iBrightCL1000 (Invitrogen).
Statistical analysis
For all experiments, the number of technical and/or biological replicates is reported in the figure legends or text. To identify mRNA expression patterns that significantly correlate with SP140 expression, a Pearson’s correlation was performed, and p values were corrected using Bonferroni adjustment. For experimental data, parametric Student’s t-test or Mann-Whitney U test was performed to compare two groups based on the underlining distribution of data. Survival curves were calculated using the Kaplan-Meier method and compared using the log-rank test. Data are demonstrated as mean±SEM. Two-sided p values less than 0.05 were considered statistically significant. In GSEA, the p values were calculated by the permutation test. Data were analyzed using R V.4.0.3 and GraphPad Prism V.8 (GraphPad Software). In all cases, ****p<0.0001, ***p<0.001, **p<0.01, and *p<0.05.
Results
SP140 expression in HNSCC clinical samples
We first analyzed the expression levels of SP140 in different types of cancers. Overall, increased expression of SP140 was seen in many cancers in the TCGA, including HNSCC (online supplemental figure 1).
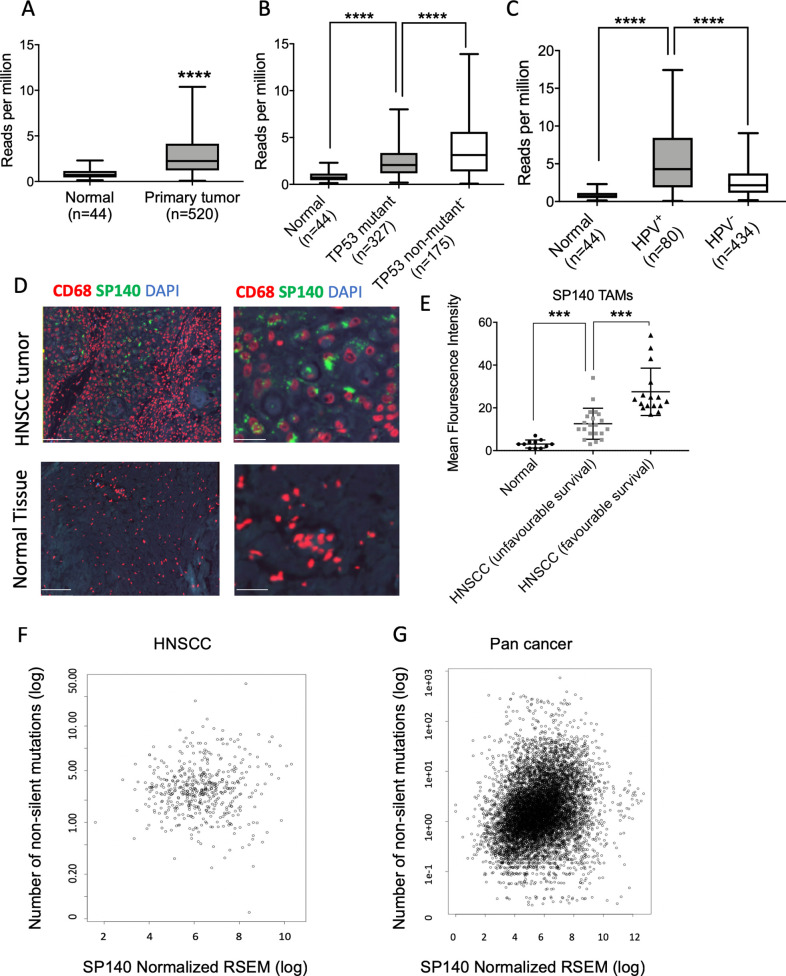
In the TCGA dataset, patients with HNSCC showed overall higher expression of SP140 in tumor tissue compared with normal adjacent tissue with some heterogeneity in the expression (n=44 normal, n=520 primary tumor, p<0.0001) (figure 1A). Since mutation in TP53 is the most common molecular driver of HNSCC, we assessed the SP140 expression levels in relation to TP53 mutation status. SP140 was upregulated in both TP53 mutant (n=327) and non-mutant (n=175) patients with HNSCC, with a significant increase in the TP53 non-mutant (p<0.0001) (figure 1B). Since a subset of HNSCCs is HPV+, we next compared SP140 expression in HPV+ and HPV− tumors. Although the level of SP140 was higher than in normal tissue for both HPV+ and HPV− HNSCC, the level of SP140 was significantly higher in HPV+ tumors (n=80) compared with HPV− tumors (n=434) (p<0.0001) (figure 1C).
Figure 1.
SP140 expressed heterogeneously in HNSCCs and its high expression in TAMs is associated with a favorable survival and high tumor mutation burden. (A) SP140 expression in primary HNSCC tumors and normal adjacent tissue in TCGA data (n=520 HNSCC, n=44 normal). (B) SP140 expression versus TP53 mutation status in patient with HNSCC tumors in the TCGA dataset (n=327, TP53 mutant, and n=175, TP53 non-mutant). (C) SP140 expression in primary HPV+ HNSCC tumors, HPV− HNSCC tumors, and normal adjacent tissue in the TCGA data (n=44, normal; n=80, HPV+ HNSC, and n=434, HPV− HNSCC). (D) Representative images of SP140 expression in HNSCC TAMs (CD68+) of normal tissues and HNSCC tumor tissues. (E) Mean fluorescent intensity was calculated in TAMs (CD68+) by quantification of SP140 expression using QuPath software and compared between controls, patients with HNSCC with unfavorable 5-year survival, and patients with HNSCC with favorable 5-year survival. (F) Correlation of SP140 expression and number of non-silent mutations in 497 HNSCCs in the TCGA dataset. (G) Correlation of SP140 expression and number of non-silent mutations in over 9766 cancers in the TCGA dataset. (A–C) Box and whisker plots, with whiskers representing minimum and maximum for the panel. (E) Scatter plot (median and 95% CI). P values (A–C) were calculated by Student’s t-test and corrected for multiple comparison by Bonferroni correction. The correlation between SP140 and mutation burden was quantified with Pearson’s correlation coefficient and corrected for multiple comparison. ***P< 0.001, ****P< 0.0001 after correction for multiple comparisons. The scale bars indicate 100 μm in the left images and 30 μm in the right images. HNSCC, head and neck squamous cell carcinoma; HPV, human papillomavirus; RSEM, RNA-Seq by Expectation-Maximization; SP140, speckled protein 140; TAM, tumor-associated macrophage; TCGA, The Cancer Genome Atlas.
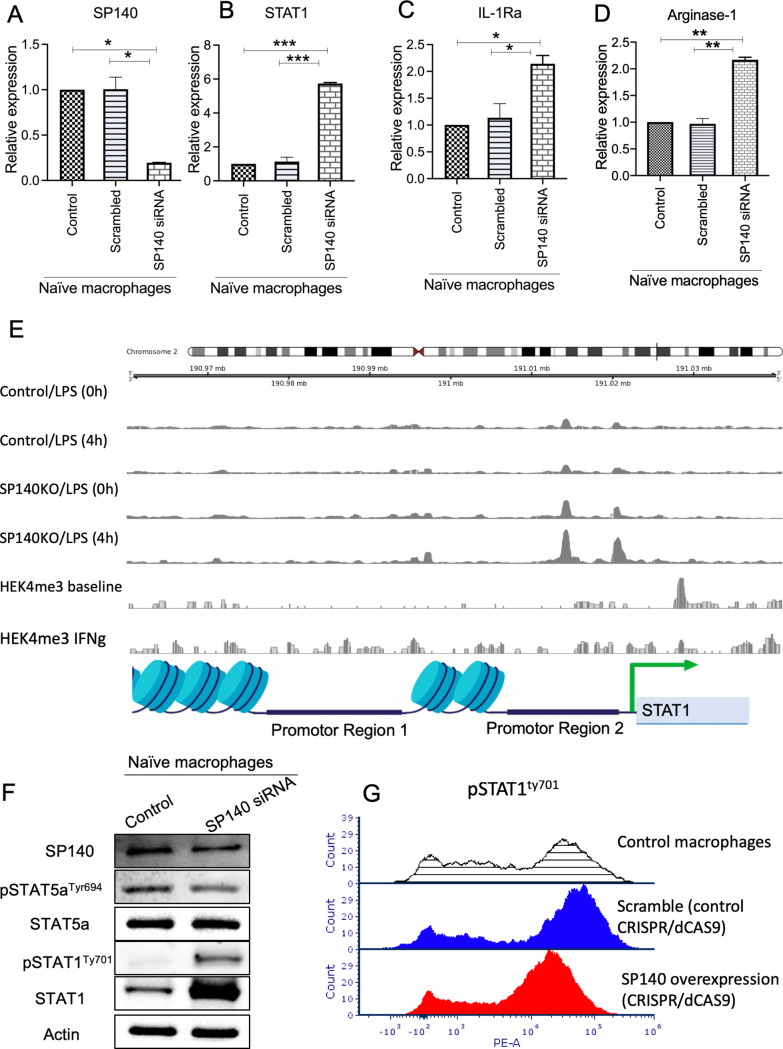
SP140 is described as an immune-restricted gene.26 Consistently, qPCR analysis of HNSCC lines (FaDu and Cal27) confirmed the low expression of SP140 in HNSCC cell lines and high expression in macrophages (online supplemental figure 2), consistent with SP140’s known role in immune cells. Multiplex immunofluorescence (IF) staining for SP140 showed high expression of SP140 localized in TAMs (CD68+) in HNSCC samples (figure 1D). Interestingly, levels of SP140 were higher in patients’ TAMs with favorable 5-year survival (figure 1E).
SP140 expression is positively associated with high tumor mutation burden, favorable survival outcome, and immune hot tumors
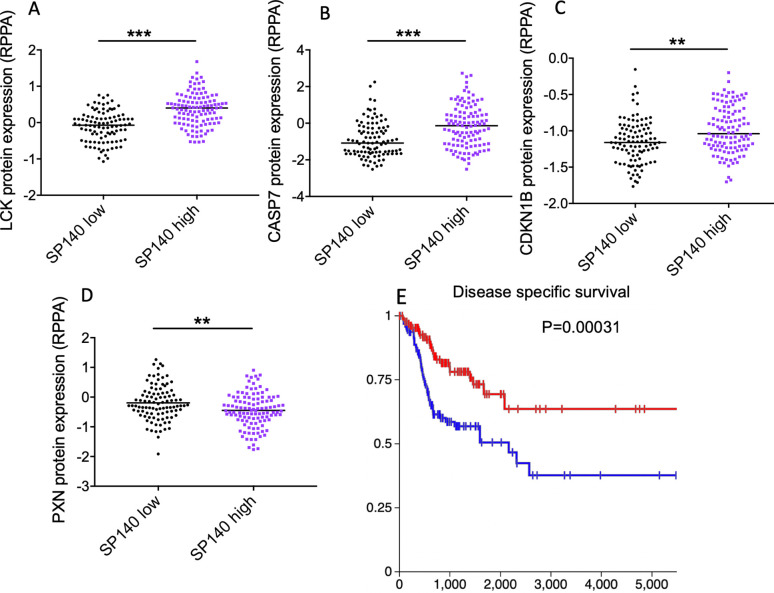
The SP140 expression level positively correlated with mutation load across cancers and specifically in HNSCC (p=9.604E-8 for pan-cancer and p=0.0170 for HNSCC after FDR correction, n=9766 for pan-cancer and n=497 for HNSCC) (figure 1F, G). High levels of SP140 were associated with an increase in lymphocyte-specific protein tyrosine kinase (LCK) protein expression in the TCGA HNSCC dataset (figure 2A). Higher expression of LCK is associated with expression of the immune response transcription module, infiltration of the tumor tissue by activated immune cells, and a favorable response to the immune blockade.31 High levels of SP140 were associated with high protein expression of proapoptotic proteins CASP7 (p<0.001) and CDKN1B (p<0.01) and low levels of PXN protein (p<0.01) in HNSCC (figure 2B–D). The high expression of PXN is associated with poor prognosis in HNSCC.32 High levels of SP140 were associated with better disease-specific survival (log-rank test p=0.01039) and overall survival (log-rank test, p=0.0205) in HNSCC (figure 2E and online supplemental figure 3).
Figure 2.
High levels of SP140 in HNSCC tumors are associated with higher expression of proapoptotic markers and favorable survival in HNSCC. (A–D) HNSCC TCGA tumors with available protein expression data (n=212) were divided to two groups of SP140 high and SP140 low based on the median of SP140 expression. The protein levels of LCK, CASP7, CDKN1B, and PXN were compared between the two groups by Student’s t-test, and the p value was calculated after Benjamini-Hochberg correction. (E) Disease-specific survival of patients with high levels of SP140 (n=261) and low levels of SP140 (n=261) in TCGA HNSCC. The p value was calculated based on the log-rank test. **P<0.01, ***P<0.001 after correction for multiple comparisons. HNSCC, head and neck squamous cell carcinoma; RPPA, reverse phase protein array; SP140, speckled protein 140; TCGA, The Cancer Genome Atlas.
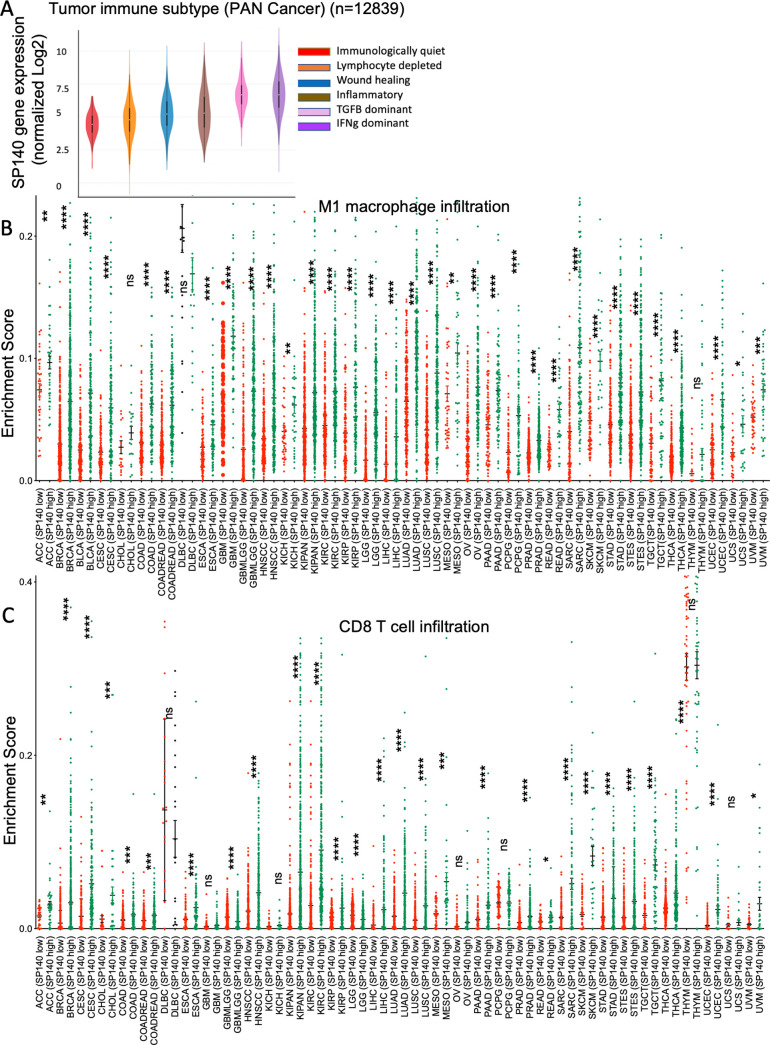
We next analyzed the correlation of SP140 expression levels in relation to tumor immune subtypes across 33 cancers in TCGA. This algorithm was developed based on immunogenomic analysis of more than 10 000 tumors and identified an immune response pattern impacting prognosis and classification of the TME to six immune subtypes, including wound healing, IFN-γ-dominant, inflammatory, lymphocyte depleted, immunologically quiet, and TGFβ-dominant.23 High SP140 expression was observed in the IFN-γ-dominant tumor type (p<0.0001 after Bonferroni correction) (figure 3A). Across the TCGA pan-cancer cohort, higher levels of SP140 were associated with significantly higher inflammatory macrophage infiltrates (M1) and CD8 T-cells infiltration in most TCGA cancer cohorts (figure 3B, C).
Figure 3.
High levels of SP140 in the pan-cancer TCGA tumors are associated with IFN-γ-dominant tumors and high infiltration of proinflammatory TAMs (M1) and CD8 T cells. (A) Association of SP140 gene expression with immune infiltrate subtypes in the TCGA pan-cancer cohort (n=12 839). (B, C) TCGA pan-cancer gene expression data were used to dichotomize SP140 expression based on the median. M1 macrophage and T-cell infiltrations were estimated for each cancer type in SP140 high and SP140 low tumors. Data are presented as scatter plots. The statistical significance is annotated by the number of stars. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. IFN-γ, interferon gamma; M1, proinflammatory phenotype; SP140, speckled protein 140; TAM, tumor-associated macrophage; TCGA, The Cancer Genome Atlas.
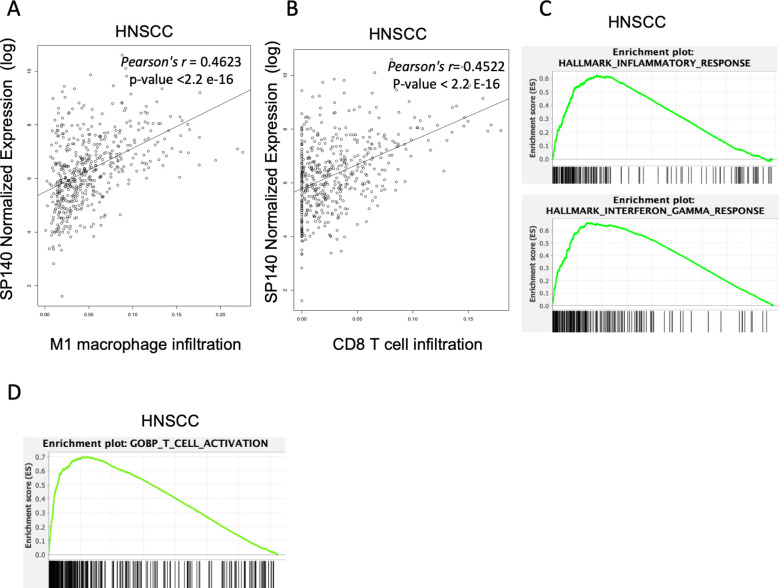
In the HNSCC cohort of TCGA samples, SP140 expression was positively associated with higher inflammatory macrophage infiltrates (M1) and CD8 T-cell infiltration (CD8 T cells: Pearson’s r=0.4522, p value <2.2 E-16; M1 macrophages, Pearson’s r=0.4623, p value <2.2 e-16) (figure 4A, B). We next investigated the expression levels of SP140 associated genes in HNSCC. High SP140 expression in HNSCC was associated with positive enrichment of IFN-γ, inflammatory response pathways, and T-cell activation based on GSEA33 (figure 4C, D, and online supplemental table 2). Expression of SP140 was positively associated with high infiltration of M1 TAMs and CD8 T cells in HNSCC in both HPV+ and HPV− HNSCC (online supplemental figure 4).
Figure 4.
(A, B) Correlation of SP140 expression with M1 macrophage and CD8 T-cell infiltration levels HNSCC (n=497). (C, D) Genes whose expression significantly correlated with SP140 expression were analyzed by GSEA in HNSCC samples (n = 522). The plot shows the top positively enriched pathways, hallmark inflammatory response, IFN-γ response, and T-cell activation. The Y-axis is the GSEA enrichment score. The X-axis is a list of genes ranked by differential expression correlation with SP140, with black bars representing genes in each gene set. GSEA, gene set enrichment analysis; HNSCC, head and neck squamous cell carcinoma; IFN-γ, interferon gamma; M1, proinflammatory phenotype; SP140, speckled protein 140.
SP140 inhibits STAT1 signaling in macrophages
As SP140 was shown to be IFN-γ inducible and STAT1 plays a role in the activation of IFN-γ signaling and TAMs, we designed mechanistic studies to study the dynamics of SP140-STAT1 interactions in macrophages. We knocked down SP140 using siRNAs in human THP1-derived unpolarized macrophages. We first analyzed the expression of SP140 mRNA and confirmed decreased expression in SP140 siRNA transfected cells (figure 5A). Interestingly, suppression of SP140 increased the expression of STAT1, IL-1Ra (IL-1 receptor antagonist), arginase 1 (ARG1), and IL-6 (figure 5B–D and online supplemental figure 5). However, the knockdown of SP140 did not affect STAT5a levels (online supplemental figure 5). Analysis of ATAC-seq data including macrophages treated with LPS (0 hour), macrophages treated with LPS (4 hours), SP140 KO macrophages (0 hour), and SP140 KO macrophages (4 hours) showed that SP140 acts as an epigenetic repressor at the promotor of STAT1 and controls its expression at the transcriptional level. Analysis of ChIP-seq data from baseline and IFN-γ stimulated macrophages suggests that this repression is correlated with an increase in repressive H3K27me3 modifications in the region of STAT1. (figure 5E). ChIP-qPCR analysis after silencing SP140 confirmed the interaction of SP140 with the STAT1 promotor (online supplemental figure 6). Protein analysis showed that STAT1 and pSTAT1Ty701 are significantly upregulated in SP140 knockdown cells, indicating inhibitory effects of SP140 on STAT1 activation (figure 5F). CRISPR/dCAS9 activation of SP140 confirmed the decrease in pSTAT1 Tyr701 in macrophages after overexpression of SP140 (figure 5G). Interestingly, SP140 knockdown in macrophages was associated with a significant increase in repressive H3K27me3 modifications upstream of the TSS of STAT1. These data indicate the repressor effects of SP140 on STAT1 activation. Mechanistic studies showed that an increase in STAT1 overexpression in macrophages contributes to the TAM immune suppressive activity in the TME and TAM-mediated T-cell deletion in the TME.34
Figure 5.
SP140 negatively regulates STAT1 transcription and phosphorylation in macrophages. (A–D) SP140 was downregulated using siRNA in naïve (undifferentiated) macrophages, and cells were collected for RNA expression analysis by qPCR for SP140, STAT1, IL-1Ra and arginase-1, after 24 hours. 18S was used as an endogenous normalizer. The delta–delta Ct method was used to identify the relative gene expression. Data are presented as mean and SD of fold change compared with the control. (E) ATAC-seq read coverage (average count over an arbitrary sliding window) of SP140 KO and non-KO macrophages treated with LPS for 4 hours (along with their respective 0-hour controls) and average H3K27me3 ChIP-seq coverage for IFN-γ stimulated and unstimulated macrophages in the neighborhood of the STAT1 gene. (F) SP140 was downregulated using siRNA in naïve (undifferentiated) macrophages, and cells were collected for protein isolation and western blot 48 hours post transfection. (I) Effect of SP140 overexpressed on Levels of pSTAT1Tyr701 was quantified by flow cytometry in controls (shaded line indicates control macrophages; blue line indicates scramble control (CRISPR/dCAS9 control), and red line indicates test sample (SP140 overexpression, CRISPR/dCAS9)) after 48 hours. *P<0.05, **P<0.01, ***P<0.001. ATAC-seq, assay of transposase accessible chromatin sequencing; CHiP, chromatin immunoprecipitation; IFN-γ, interferon gamma; IL, interleukin; qPCR, quantitative PCR; siRNA, small interfering RNA; SP140, speckled protein 140.
SP140 overexpression induces production of inflammatory cytokines and chemokines and reprograms macrophages to induce TAM-mediated tumor cytotoxicity
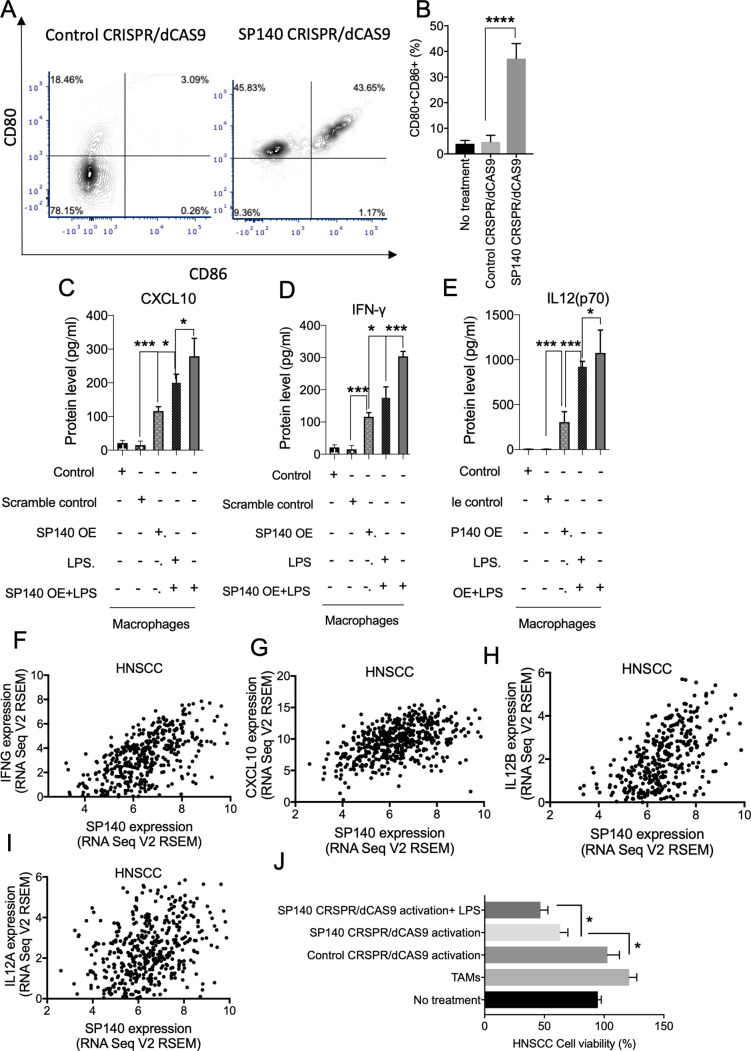
Overexpression of SP140 with a CRISPR/dCAS9 activation approach led to an increase in the percentage of M1 (CD80+CD86+) macrophages (figure 6A, B), indicating an enhancement in antigen presentation capacity of macrophages. Overexpression of SP140 induced increases in IFN-γ, CXCL10, and IL-12 in unstimulated and stimulated macrophages (figure 6C–E). Consistently, high levels of SP140 expression were associated with high levels of IFN-γ, IL-12, and CXCL10 in TCGA HNSCC tumors (p<0.0001 for all comparisons) (figure 6F–I). Interestingly, high levels of CXCL10 are reported to be required for an antitumor immune response following immune-checkpoint blockade.35 An increase in CRISPR/dCAS9 activation of SP140 in TAMs isolated from a syngeneic mouse model of HNSCC led to an increase in direct cytotoxicity of TAMs on the HNSCC SCC7 cell line (figure 6J).
Figure 6.
Overexpression of SP140 induces the production of inflammatory cytokines and chemokines and reprogram macrophages to induce TAM-mediated tumor cytotoxicity. (A, B) Control CRISPR/dCAS9 (control) or SP140 CRISPR/dCAS9 (SP140 overexpression) was transfected in naïve macrophages, and the expression of CD80 and CD86 was quantified by flow cytometry. (C–E) SP140 CRISPR/dCAS9 (SP140 OE) or scramble control (control CRISPR/dCAS9) were introduced to the naïve macrophages, and a multiplex fluorescence BioLegend assay was used to identify the levels of CXCL10, IFN-γ, and IL-12 (P70) in the supernatant after 48 hours. LPS (100 nM) was administered after 24 hours. (F–I) Correlation of SP140 expression and IFNG, CXCL10, IL-12B, and IL-12A levels in over 500 HNSCCs in the TCGA dataset. (J) Control CRISPR/dCAS9 or SP140 CRISPR/dCAS9 was introduced in TAMs isolated from syngeneic HNSCC tumor and cocultured with murine SCC7 cells. Cell viability after 48 hours of coculture was quantified with a quantitative viability assay kit. Data are presented based on the fold change of non-treated control. *P<0.05, ***P<0.001, ****P<0.0001. HNSCC, head and neck squamous cell carcinoma; IFN-γ, interferon gamma; IL, interleukin; SP140, speckled protein 140; TAM, tumor-associated macrophage; TCGA, The Cancer Genome Atlas.
Correlation of SP140 with immune cell infiltrates and response to anti-PD-1 immune-checkpoint blockade
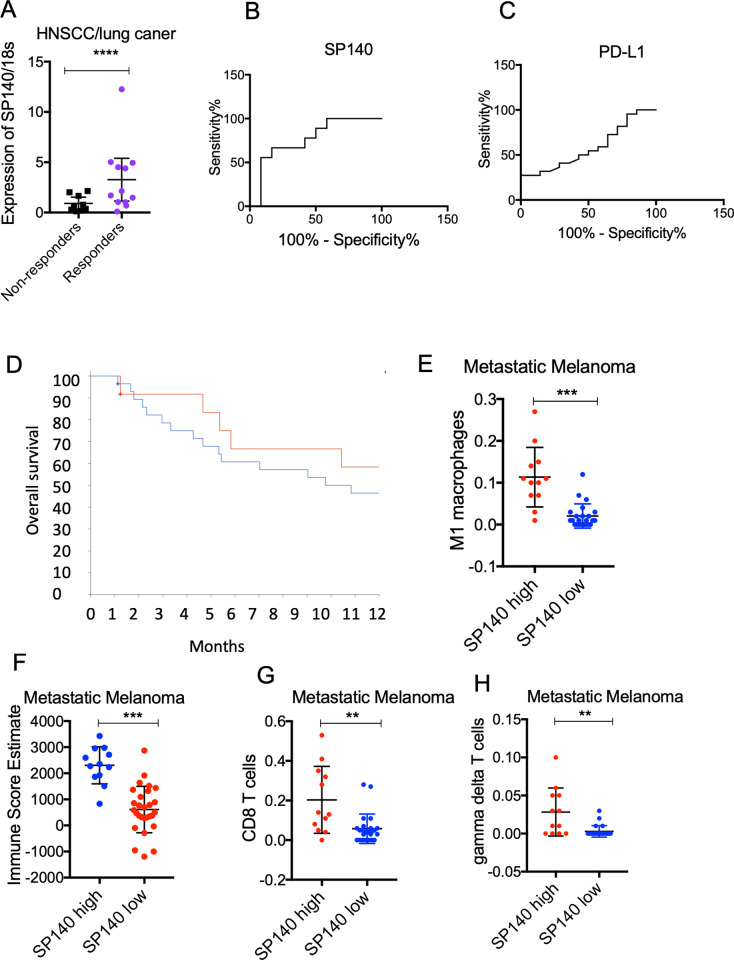
We next evaluated the expression of SP140 in patients who were treated with anti-PD-1 immune-checkpoint inhibitors in HNSCC and non-small cell lung cancer (n=21). Samples were collected for gene expression analysis before treatment. A significantly higher level of SP140 was associated with response to anti-PD-1 immunotherapy, defined as stable disease or remission after anti-PD-1 treatment over 6 months (p<0.05) (figure 7A). Receiver operating curve analysis showed better performance of SP140 compared with PD-L1 in diagnostic accuracy (area under the curve for SP140: 0.77±0.10 vs area under the curve for PD-L1: 0.55±0.13 (p>0.05) (figure 7B, C). Examining the dataset of metastatic melanoma patients who underwent CTLA-4 blockade immunotherapy,19 we found significantly more favorable survival in patients who had high SP140 levels (n=12) than in patients with low levels of SP140 (n=28) (log-rank test, p=4.98E-3) (figure 7D). High levels of SP140 were associated with a significantly higher immune score (p<0.001), higher CD8+ T cells (p<0.001), higher M1 macrophages, and higher gamma delta T cells (p<0.05) (figure 7E–H). Similar results were found in metastatic melanomas which were treated with anti-PD-1 immune blockade (online supplemental figure 7). Examining the datasets of patients with recurrent glioblastoma who underwent neoadjuvant anti-PD-1 therapy36 and patients with thymic carcinoma who underwent anti-PD-1 therapy,37 we found that higher levels of SP140 were associated with a favorable response to the neoadjuvant anti-PD-1 therapy in recurrent glioblastoma and a favorable response to anti-PD-1 in thymic carcinoma (online supplemental figure 8). These data indicate the vulnerability of SP140 high tumors to immunotherapy.
Figure 7.
High expression of SP140 in tumors is associated with a favorable response to immunotherapy. (A) RNA was isolated from pretreatment specimens of HNSCC and lung cancer responders and non-responders to anti-PD-1 immunotherapy. RNA was isolated and levels of SP140 were quantified by RT-qPCR (n=21). 18S was used for normalization. Student’s t-test was used for statistical analysis. (B, C) Receiver operating curves of SP140 and PD-L1 for discriminating responders or non-responder cases. Y-axis represents sensitivity (%) and X-axis represents 100% specificity (%). (D) Patients with metastatic melanoma were dichotomized to high expression and low expression groups based on expression of SP140 (the highest quartile vs the rest). Overall survival of patients with high levels of SP140 (n=12) and low levels of SP140 (n=28) was graphed and analyzed using the Kaplan-Meier estimate. (E, F) Tumors with high expression of SP140 (n=12) versus tumors with low levels of SP140 (n=28) showed higher infiltration of M1 macrophages, CD8 T cells, gamma delta T cells, and overall immune score. The Wilcoxon test was used for statistical analysis, and the p value was corrected for multiple comparisons. **P<0.01, ***P<0.001, ****P<0.0001. HNSCC, head and neck squamous cell carcinoma; M1, proinflammatory phenotype; PD-1, programmed cell death protein 1; PD-L1, programmed death-ligand 1; RT-qPCR, reverse transcription–quantitative PCR; SP140, speckled protein 140.
Discussion
In this study, our results showed that tumors with high levels of SP140 have more infiltrating M1 TAMs and an IFN-γ signature in the TME. Considering the outcome of immunotherapy using anti-PD-1 agents, patients who had a high level of SP140 in the tumor showed a favorable response to immunotherapy, defined as stable disease or remission after anti-PD-1 treatment over 6 months. SP140 mRNA conferred a higher diagnostic accuracy compared with PD-L1 for predicting response to immunotherapy in our cohort. Consistently, a higher level of SP140 in HNSCC, but not PD-L1, was associated with better survival outcomes in patients with HNSCC in the TCGA data. High SP140 level in tumors was associated with high CD8+ T cells in the tumor in both HPV+ and HPV− tumors. These data indicate the possible utility of using SP140 expression for patient selection in immunotherapy.
Recently, ChIP-seq data showed that the majority of endogenous SP140 in primary human macrophages is associated with promoters.38 It has been shown that SP140 preferentially occupied heterochromatic regions with low chromatin accessibility and low transcriptional activity, marked by high levels of histone H3 lysine 27 trimethylation (H3K27me3) and low histone H3 lysine 4 trimethylation (H3K4me3), in human macrophages and plays an important role in maintaining the macrophage transcriptional landscape.26 38 Interestingly, our bioinformatic analysis showed an association of SP140 with STAT1 promoter in macrophages. Given that the siRNA-mediated knockdown of SP140 in human macrophages resulted in an increased level of STAT1, IL-6, IL-1R, and ARG1. In HNSCC cells, high levels of SP140 were associated with high protein expression of proapoptotic and tumor suppressor proteins CASP7, CDKN1B, and BIM and low levels of PXN protein. A recent pan-cancer analysis showed that abnormal PXN expression is associated with a poor prognosis.32
In this study, we found an association between high levels of SP140 and high non-synonymous mutation burden. As previously described, tumors with a high mutation burden express a large number of abnormal proteins, neo-antigens, that could elicit T-cell immune response.39 In two independent immunotherapy cohorts of HNSCC/lung cancer and metastatic melanoma, tumors with a high mutation burden showed an improved clinical response to immune-checkpoint blockade and survival.40 Given the high accuracy of SP140 to predict response to anti-PD-1 observed in this study, SP140 levels could be potentially used for the prediction of mutation burden and response to immunotherapy. Furthermore, using the TME immune profile classification developed based on immunogenomic analysis of more than 10 000 tumors,23 we found an association of a high level of SP140 with IFN-γ-dominant tumor immune signature. These data are in accordance with the higher presence of M1 macrophages and CD8+ T cells in SP140 high tumors and their better response to immunotherapy.
In the present study, we showed that SP140 suppresses STAT1 at its transcriptional level and suppresses its phosphorylation at post-transcriptional levels, indicating that inhibition of SP140 could activate STAT1. The role of STAT1 in tumor progression and tumor immunity is not straightforward and is context dependent. Although the tumor-suppressive functions of STAT1 have already been demonstrated in several studies,41–43 in a mechanistic study performed by the Gabrilovich group,34 it was determined that STAT1, but not STAT3 or STAT6, is responsible for TAM immunosuppressive activity. These results are in accordance with the antitumor activity of macrophages high in SP140 and low in STAT1 as observed in this study. Interestingly, in a recent study, it has been shown that persistent activation of STAT1 is associated with the in vivo acquisition of transcriptomic features associated with relapse after radiation and anti-CTLA4 therapy.44 They also observed that prolonged STAT1 activation on interferon treatment is associated with resistance and clinical progression after anti-PD-1 therapy.44 Our observation regarding the inhibitory role of SP140 on STAT1 might be an explanation for the better response to immunotherapy in SP140 high tumors and inhibitory effects of SP140 on STAT1 activation.
As previously reported, SP140 expression is mostly associated with unpolarized (naive macrophages) and classically activated macrophages compared with the alternatively activated macrophages. Mechanistically, overexpression of SP140 increased CD80, the T-cell costimulatory molecule on macrophages and increased IFN-γ production by macrophages and their cytotoxicity of macrophages toward HNSCC cancer cell lines. Future preclinical and clinical studies are needed to investigate the potential use of SP140 activation as a therapeutic approach for cancer immunotherapy.
The results of our study showed that high levels of SP140 were associated with the presence of a higher inflammatory response, higher IFN-γ signature in the tumors, favorable survival in HNSCC, and favorable response to immunotherapy in HNSCC/lung squamous cancer, metastatic melanoma, thymic carcinoma, and recurrent glioblastoma who received neoadjuvant anti-PD-1 therapy. SP140 was expressed in higher abundance in undifferentiated (naïve) macrophages and classically activated macrophages. SP140 plays an inhibitory role in STAT1 activation. Overexpression of SP140 induced inflammatory and antitumor macrophage activity. SP140 could potentially serve as both a disease-associated biomarker to a priori identify immunotherapy responders and could be therapeutically upregulated as an adjunct to other immunotherapy modalities to elicit enhanced antitumor responses.
Acknowledgments
We thank the Herbert Irving Comprehensive Cancer Center Database Shared Resource and the Herbert Irving Comprehensive Cancer Center Molecular Pathology Core.
Footnotes
Twitter: @3kkranthi, @FatemehHeravi
Contributors: KKKT and FM-H planned and performed the majority of experiments and associated data analysis and wrote the paper; JM-B, MC, AT, and JN provided assistance in data analysis. SD, HN, RC, AM, JN, and RC provided technical assistance and data interpretation. AY and Y-SLC provided assistance with the characterization and interpretation of patient data and tissue specimens. FM-H provided reagents and resources for the project, supervised the whole project and performed data interpretation. All authors contributed to the editing of the manuscript and critical review. FM-H acts as the guarantor.
Funding: This work was supported by NIH/NIDCR (DE031112-01 and DE031112-01), the American Association of Cancer Research (AACR 20-60-51), and the Mark Foundation Grant, NIH/NCAT (KL2TR001874 and UL1TR001873) and NIH/NCI (P30CA013696).
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available in a public, open access repository. Data are available in a public, open-access repository from the following sources: https://gdc.cancer.gov/node/977; https://www.cbioportal.org/datasets; The Gene Expression Omnibus under accession number GSE121810, GSE181815, GSE89177, GSM1625982, GSM1625981.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human subjects and was approved by Columbia University Medical Center's institutional review board (IRB) (IRB number AAAS0169). We used a retrospective cohort of samples. The IRB waived the informed consent requirement. The samples were obtained based on IRB-approved protocols.
References
- 1.Peltanova B, Raudenska M, Masarik M. Effect of tumor microenvironment on pathogenesis of the head and neck squamous cell carcinoma: a systematic review. Mol Cancer 2019;18:63. 10.1186/s12943-019-0983-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Economopoulou P, Perisanidis C, Giotakis EI, et al. The emerging role of immunotherapy in head and neck squamous cell carcinoma (HNSCC): anti-tumor immunity and clinical applications. Ann Transl Med 2016;4:173. 10.21037/atm.2016.03.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang X, Wang J, Deng X, et al. Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol Cancer 2019;18:1–17. 10.1186/s12943-018-0928-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinogradov S, Warren G, Wei X. Macrophages associated with tumors as potential targets and therapeutic intermediates. Nanomedicine 2014;9:695–707. 10.2217/nnm.14.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evrard D, Szturz P, Tijeras-Raballand A, et al. Macrophages in the microenvironment of head and neck cancer: potential targets for cancer therapy. Oral Oncol 2019;88:29–38. 10.1016/j.oraloncology.2018.10.040 [DOI] [PubMed] [Google Scholar]
- 6.Vinogradov S, Warren G, Wei X. Macrophages associated with tumors as potential targets and therapeutic intermediates. Nanomedicine 2014;9:695–707. 10.2217/nnm.14.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solinas G, Germano G, Mantovani A, et al. Tumor-Associated macrophages (TAM) as major players of the cancer-related inflammation. J Leukoc Biol 2009;86:1065–73. 10.1189/jlb.0609385 [DOI] [PubMed] [Google Scholar]
- 8.Liu Y, Zugazagoitia J, Ahmed FS, et al. Immune cell PD-L1 colocalizes with macrophages and is associated with outcome in PD-1 pathway blockade therapy. Clin Cancer Res 2020;26:970–7. 10.1158/1078-0432.CCR-19-1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gibson TJ, Ramu C, Gemünd C, et al. The APECED polyglandular autoimmune syndrome protein, AIRE-1, contains the sand domain and is probably a transcription factor. Trends Biochem Sci 1998;23:242–4. 10.1016/S0968-0004(98)01231-6 [DOI] [PubMed] [Google Scholar]
- 10.Lallemand-Breitenbach V, de Thé H. PML nuclear bodies. Cold Spring Harb Perspect Biol 2010;2:a000661. 10.1101/cshperspect.a000661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bottomley MJ, Collard MW, Huggenvik JI, et al. The SAND domain structure defines a novel DNA-binding fold in transcriptional regulation. Nat Struct Biol 2001;8:626–33. 10.1038/89675 [DOI] [PubMed] [Google Scholar]
- 12.Mehta S, Cronkite DA, Basavappa M, et al. Maintenance of macrophage transcriptional programs and intestinal homeostasis by epigenetic reader SP140. Science Immunology 2017;2. 10.1126/sciimmunol.aag3160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matesanz F, Potenciano V, Fedetz M, et al. A functional variant that affects exon-skipping and protein expression of SP140 as genetic mechanism predisposing to multiple sclerosis. Hum Mol Genet 2015;24:5619–27. 10.1093/hmg/ddv256 [DOI] [PubMed] [Google Scholar]
- 14.Karaky M, Fedetz M, Potenciano V, et al. SP140 regulates the expression of immune-related genes associated with multiple sclerosis and other autoimmune diseases by NF-κB inhibition. Hum Mol Genet 2018;27:4012–23. 10.1093/hmg/ddy284 [DOI] [PubMed] [Google Scholar]
- 15.Granito A, Yang W-H, Muratori L, et al. PML nuclear body component Sp140 is a novel autoantigen in primary biliary cirrhosis. Am J Gastroenterol 2010;105:125–31. 10.1038/ajg.2009.596 [DOI] [PubMed] [Google Scholar]
- 16.Schwartz LH, Litière S, de Vries E, et al. RECIST 1.1-Update and clarification: from the RECIST Committee. Eur J Cancer 2016;62:132–7. 10.1016/j.ejca.2016.03.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zugazagoitia J, Liu Y, Toki M, et al. Quantitative assessment of CMTM6 in the tumor microenvironment and association with response to PD-1 pathway blockade in advanced-stage non-small cell lung cancer. J Thorac Oncol 2019;14:2084–96. 10.1016/j.jtho.2019.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugo W, Zaretsky JM, Sun L, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 2017;168:542. 10.1016/j.cell.2017.01.010 [DOI] [PubMed] [Google Scholar]
- 19.Van Allen EM, Miao D, Schilling B, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015;350:207–11. 10.1126/science.aad0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. 10.1186/s13059-017-1349-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res 2017;77:e108–10. 10.1158/0008-5472.CAN-17-0307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorsson V, Gibbs DL, Brown SD, et al. The immune landscape of cancer. Immunity 2018;48:812–30. 10.1016/j.immuni.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 2015;12:453–7. 10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian A, Kuehn H, Gould J, et al. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics 2007;23:3251–3. 10.1093/bioinformatics/btm369 [DOI] [PubMed] [Google Scholar]
- 26.Mehta S, Cronkite DA, Basavappa M, et al. Maintenance of macrophage transcriptional programs and intestinal homeostasis by epigenetic reader SP140. Sci Immunol 2017;2. doi: 10.1126/sciimmunol.aag3160. [Epub ahead of print: 03 Mar 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Danecek P, Bonfield JK, Liddle J, et al. Twelve years of SAMtools and BCFtools. Gigascience 2021;10. doi: 10.1093/gigascience/giab008. [Epub ahead of print: 16 02 2021]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hahne F, Ivanek R. Visualizing genomic data using Gviz and Bioconductor. Methods Mol Biol 2016;1418:335–51. 10.1007/978-1-4939-3578-9_16 [DOI] [PubMed] [Google Scholar]
- 30.Bankhead P, Loughrey MB, Fernández JA, et al. QuPath: open source software for digital pathology image analysis. Sci Rep 2017;7:16878. 10.1038/s41598-017-17204-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Silveira WA, Palma PVB, Sicchieri RD, et al. Transcription factor networks derived from breast cancer stem cells control the immune response in the basal subtype. Sci Rep 2017;7:2851. 10.1038/s41598-017-02761-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y, Zhao H, Xiao Y, et al. Pan-Cancer analysis reveals an immunological role and prognostic potential of PXN in human cancer. Aging 2021;13:16248–66. 10.18632/aging.203154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kusmartsev S, Gabrilovich DI. STAT1 signaling regulates tumor-associated macrophage-mediated T cell deletion. J Immunol 2005;174:4880–91. 10.4049/jimmunol.174.8.4880 [DOI] [PubMed] [Google Scholar]
- 35.House IG, Savas P, Lai J, et al. Macrophage-Derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res 2020;26:487–504. 10.1158/1078-0432.CCR-19-1868 [DOI] [PubMed] [Google Scholar]
- 36.Cloughesy TF, Mochizuki AY, Orpilla JR, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med 2019;25:477–86. 10.1038/s41591-018-0337-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He Y, Ramesh A, Gusev Y, et al. Molecular predictors of response to pembrolizumab in thymic carcinoma. Cell Rep Med 2021;2:100392. 10.1016/j.xcrm.2021.100392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fraschilla I, Jeffrey KL. The Speckled Protein (SP) family: immunity's chromatin readers. Trends Immunol 2020;41:572–85. 10.1016/j.it.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Braun DA, Burke KP, Van Allen EM. Genomic approaches to understanding response and resistance to immunotherapy. Clin Cancer Res 2016;22:5642–50. 10.1158/1078-0432.CCR-16-0066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rizvi NA, Hellmann MD, Snyder A, et al. Cancer immunology. mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348:124–8. 10.1126/science.aaa1348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koromilas AE, Sexl V. The tumor suppressor function of STAT1 in breast cancer. JAKSTAT 2013;2:e23353. 10.4161/jkst.23353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Molavi O, Su M, et al. The clinical and biological significance of STAT1 in esophageal squamous cell carcinoma. BMC Cancer 2014;14:791. 10.1186/1471-2407-14-791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turnquist C, Wang Y, Severson DT, et al. STAT1-induced ASPP2 transcription identifies a link between neuroinflammation, cell polarity, and tumor suppression. Proc Natl Acad Sci U S A 2014;111:9834–9. 10.1073/pnas.1407898111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Benci JL, Xu B, Qiu Y, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell 2016;167:1540–54. 10.1016/j.cell.2016.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2022-005088supp001.pdf (562KB, pdf)
Data Availability Statement
Data are available in a public, open access repository. Data are available in a public, open-access repository from the following sources: https://gdc.cancer.gov/node/977; https://www.cbioportal.org/datasets; The Gene Expression Omnibus under accession number GSE121810, GSE181815, GSE89177, GSM1625982, GSM1625981.