Abstract
The initial stages of the pandemic caused by SARS-CoV-2 showed that early detection of the virus in a simple way is the best tool until the development of vaccines. Many different tests are invasive or need the patient to cough up or even drag a sample of mucus from the throat area. Besides, the manufacturing time has proven insufficient in pandemic conditions since they were out of stock in many countries. Here we show a new method of manufacturing virus sensors and a proof of concept with SARS-CoV-2. We found that a fluorogenic peptide substrate of the main protease of the virus (Mpro) can be covalently immobilized in a polymer, with which a cellulose-based material can be coated. These sensory labels fluoresce with a single saliva sample of a positive COVID-19 patient. The results matched with that of the antigen tests in 22 of 26 studied cases (85% success rate).
Keywords: Peptide, Substrate, Fluorimetry, Immobilization, COVID-19, Paper-supported, Coating, Pandemics
Graphical Abstract
1. Introduction
At the beginning of 2022, the SARS-CoV-2 virus caused one of the most severe pandemics in the history of humanity and claimed the lives of more than 6 million people [1], [2], [3]. After two years of the pandemic, we now know that in the initial stages, where the virus advances rapidly, early detection methods for those infected and the isolation of these people are the most useful tools against the virus's progression [4], [5].
Since the pandemic, polymerase chain reaction (PCR) and antigen detection tests have been the most widely used methods for detecting infected people. However, these methods belong to entirely different families, and each of them has advantages and disadvantages concerning the other. The choice of detection method is usually based on the rush for the result (antigens) or the desired precision and sensitivity (PCR).
PCRs belong to the group of nuclear acid-based detection methods, and antigen tests to the group of protein-based detection methods [6]. On the one hand, nuclear acid-based detection methods, such as PCR or Loop-Mediated Isothermal Amplification (LAMP), require an amplification process in which a few DNA strands can be replicated to produce a much larger sample. This amplification process makes the sensitivity of these methods very high. Within this group, we could include aptamer-based detection methods [7]. Unlike DNA, aptamers are single-stranded chains made of 20–80 nucleotides. With them, receptors can be synthesized for specific targets of the coronavirus, such as protein N and protein S [8].
On the other hand, the most relevant protein-based detection methods are called antigen and antibody tests [9]. These systems are composed of a rapid immunoassay kit (Enzyme-Linked ImmunoSorbent, ELISA, or lateral flow kit) in which a protein is detected by antigen-antibody interaction. However, most methods start with taking a sample using a swab, an invasive process that is uncomfortable and unpleasant. Some commercially available kits are non-invasive and work with a saliva sample, but they require a large saliva sample and an additional experimental procedure based on a lateral flow test.
One of the most promising techniques for detecting these types of viruses is a subgroup of the protein-based detection methods called protein function-based detection methods [6], [10], [11], [12], [13]. They are based on using a peptide substrate to detect an enzyme closely related to the coronavirus, in this case, the Mpro protein [6]. Mpro (also called 3CLpro), is a cysteine hydrolase-like protease essential in viral replication that cleavages polyproteins and generates the corresponding functional proteins necessary for virion formation [14]. This protease has been well characterised for its possible role as a therapeutic target against the SARS-Cov2 virus [15], [16], [17]. For example, the peptide substrates designed for Mpro can be chemically modified with fluorophores and quenchers (FRET pairs), so that the interaction with the target enzyme generates a visual response, i.e., an OFF-ON fluorescence change [18]. Furthermore, this response is specific for the Mpro of SARS-CoV-2 and other highly similar Mpro (such as SARS-CoV) [15], [16].
Considering all ordinary citizens and children, we believe that developing a non-invasive, visual, and simple detection method for respiratory viruses is worthy, not only for coronavirus but also for other present and future viruses. In this work, we have applied our knowledge in detecting species of interest to develop a polymer that becomes fluorescent in the presence of the Mpro protein, indicative of the presence of SARS-CoV-2. Unlike the published works concerning the detection of this enzyme [10], [11], [12], [13], our proposal remains on the synthesis of a linear polymer that can be applied as a coating on a cellulose-based support. The user must only apply a single saliva sample above the smart label and check the fluorescence after the established time with the naked eye or, more precisely, with a smartphone.
2. Experimental
2.1. Materials
All materials and solvents were commercially available and used as received unless otherwise indicated. The following materials and solvents were used: fluorogenic peptide substrate Dabcyl-KTSAVLQSGFRKME-Edans (GenScript, N-Terminus: DABCYL, ≥95.0%, additional information in SI-Section S1), methylmethacrylate (MMA) (Merck, 99%), 1-vinyl-2-pyrrolidone (VP) (Acros Organic, 99%), 4-aminostyrene (SNH2) (TCI, 98%), glutaraldehyde aqueous solution (VWR, 25%), ethanol (VWR-Prolabo, 99.9%), methanol (VWR-Prolabo, 99.9%), diethyl ether (VWR-Prolabo, 99.9%), dimethylsulfoxide (VWR, 99%), acetonitrile (VWR, 99.9%), dimethylformamide (Supelco, 99.9%) distilled water, acetonitrile (VWR, 99.9%), dimethylsulfoxide-d 6 (VWR, 99.8%), 5-aminonaphthalene-1-sulfonic acid (Alfa Aesar, 90%), Mpro 3CL protease from coronavirus SARS-CoV-2 (Sigma), recombinant MERS-CoV 3CL protease, CF (R&D Systems), Trizma (Aldrich, 99.5%), NaCl (VWR, 98%), Ethylenediaminetetraacetic acid (EDTA) (VWR, 99.8%), dithiothreitol (DTT) (TCI, 98%), filter paper in reams (Filter Lab, 73 g/m2). Azo-bis-isobutyronitrile (AIBN, Aldrich, 98%) was recrystallized twice from methanol.
The tissues and the reagents employed in the In vitro EpiDerm™ skin irritation test (EPI-200-SIT) were provided by MatTek In Vitro Life Science Laboratories: EpiDerm™ tissues (EPI-200-SIT), DMEM medium (EPI-100), 5% sodium dodecyl sulfate (SDS) solution (TC-SDS-5%), MTT-100 assay kit (MTT-100), which include the following components: MTT concentrate (MTT-100-CON), MTT diluent (MTT-100-DIL) and extractant solution (MTT-100-EXT). DPBS (Dulbecco’s Phosphate-Buffered Saline) was provided by Corning.
2.2. Instrumentation and methods
1H and 13C{1H} NMR spectra (Avance III HD spectrometer, Bruker Corporation, Billerica, Massachusetts, USA) were recorded at 300 MHz for 1H and 75 MHz for 13C using deuterated dimethyl sulfoxide (DMSO-d 6) at 25 °C as solvent.
The powder X-ray diffraction (PXRD) patterns were obtained using a diffractometer (D8 Discover Davinci design, Bruker Corporation, Billerica, Massachusetts, USA) operating at 40 kV, using Cu(Kα) as the radiation source, a scan step size of 0.02°, and a scan step time of 2 s
The polymers thermal characterization was performed by thermogravimetric analysis (Q50 TGA analyzer, TA Instruments, New Castle, DE, USA) with 10–15 mg of sample under synthetic air and nitrogen atmosphere at 10 °C·min−1; and differential scanning calorimetry, with 10–15 mg of the sample under a nitrogen atmosphere at a heating rate of 10 °C min−1 (Q200 DSC analyzer, TA Instruments, New Castle, DE, USA).
Infrared spectra (FTIR) were recorded with an infrared spectrometer (FT/IR-4200, Jasco, Tokyo, Japan) with an ATR-PRO410-S single reflection accessory.
Enzymatic activity assays were performed using a Synergy HT microplate reader (BioTek®, Winooski, Vermont, USA), measuring fluorescence with 360/40–460/40 nm excitation/emission filters. Digital photographs were taken with a smartphone (Mi 9, Xiaomi, Pekín, China).
Solution fluorescence spectra were recorded using a F-7000 Hitachi Fluorescence spectrophotometer (Hitachi, Tokyo, Japan). A rectangular 10 mm cuvette was used for the fluorescence measurements, measuring all data at 25ºC ± 0.1 ºC.
2.3. Design of sensory polymers
The design of the polymers was carried out thinking about the application as solid sensory material. Our hypothesis relies on the fact that the virus synthesised the protease Mpro when it starts replicating inside infected cells, so, as a respiratory virus, it could be detected in a saliva sample.
First, we believe that a water-soluble polymer does not fit our work for one main reason: the user could inadvertently suck on the sensor tag, ingesting small amounts of polymer. Therefore, the polymer must contain a high enough amount of a hydrophobic monomer, such as methyl methacrylate, so the resulting copolymer is insoluble in water. Second, certain hydrophilicity of the material may favour the interaction between the Mpro enzyme and the substrate, which is why a specific mol% of the hydrophilic monomer 1-vinyl-2-pyrrolidone was included in the formulations. Therefore, two polymers were designed with different hydrophilicity.
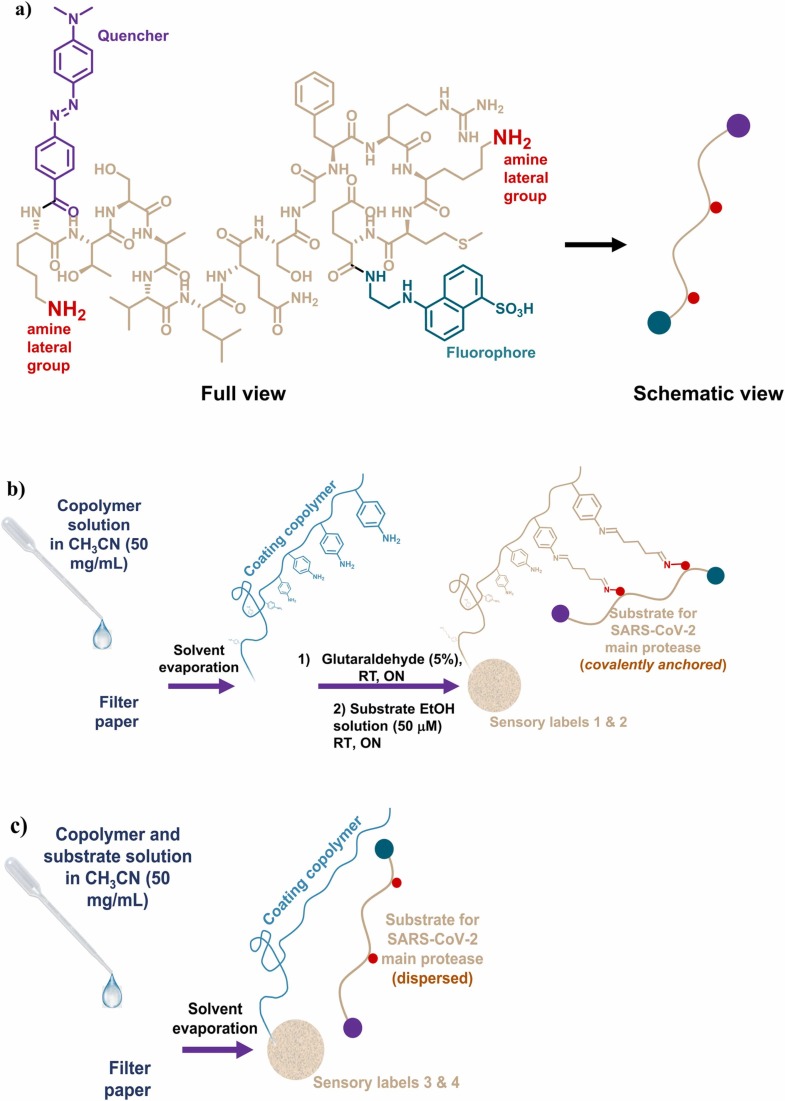
Finally, the polymers must have functional groups that serve as anchors for the immobilization of the peptide substrate ( Fig. 1a, characterization available in SI-Section S1). In our case, this immobilization occurs through the aniline side groups provided by the monomer 4-aminostyrene. All the monomers provide polarity to the final copolymers, which we believe is necessary to favour the affinity with the chosen support, i.e., cellulose paper.
Fig. 1.
Preparation of sensory labels: a) chemical structure and schematic view of the fluorogenic peptide substrate for SARS-CoV-2 main protease; b) graphical abstract of the sensory labels preparation procedure containing covalently anchored substrates; and c) graphical abstract of the sensory labels preparation procedure containing non-covalently anchored substrates.
2.4. Synthesis of polymers
Linear polymers were prepared by radical co-polymerization of the commercially available monomers 1-vinyl-2-pyrrolidone (VP), methyl methacrylate (MMA) and 4-aminostyrene (SNH2) in different molar ratios, following the experimental procedure described below.
The amounts of each monomer, specified in Table 1, were dissolved in DMF and the solution was added to a round-bottom pressure flask. Subsequently, radical thermal initiator AIBN was added, and the solution was sonicated for 10 min and heated overnight at 60 °C, under a nitrogen atmosphere, and without stirring. The solution was then dropwise added to diethyl ether (100 mL) with magnetic stirring, yielding the desired copolymers as whitish precipitates. Finally, polymers were purified in a Soxhlet apparatus with diethyl ether as the washing solvent to eliminate DMF traces. The characterization of polymers can be found in the electronic Supporting information (SI-Section S2).
Table 1.
Copolymers´ formulations indicating monomers´ mol%. The table shows the amounts of monomers, solvent (DMF), and radical thermal initiator (AIBN) used in different co-polymerizations.
 |
Copolymers 3 and 4 were prepared as blanks of copolymers 1 and 2, respectively. These blanks are highly relevant for the storage stability study depicted in Section 3.2, and the understanding of the importance of the peptide to be covalently anchored to the copolymers.
The followed methodology gives rise to polymers with relatively low molecular weight so that the viscosity of coating solutions does not reach high values. These low viscosities can be obtained by carrying out the polymerization with high concentrations of the thermal radical initiator (0.1 M). The resulting copolymers are compatible with coating techniques such as drop-, spray- or dip-coating but incompatible with bar coating.
2.5. Preparation of sensory labels
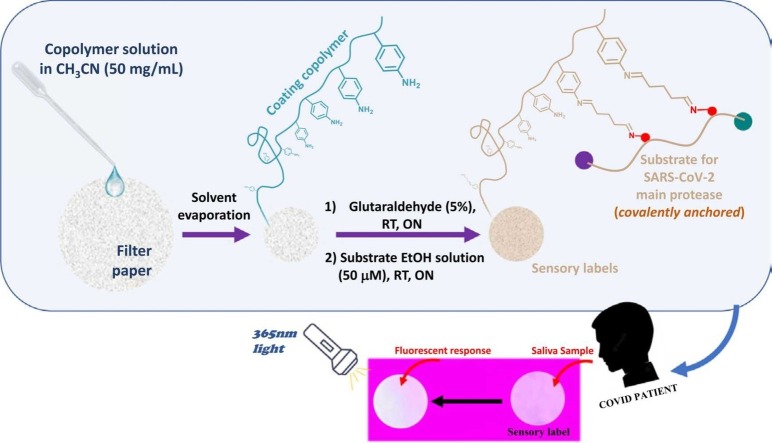
The preparation of the sensory labels by drop coating is schematically shown in Fig. 1b. 50 mg of copolymers 1 or 2 were dissolved in acetonitrile (1 mL). Then, the resulting solution was deposited twice (2 × 10 μL) on the surface of a filter paper disc (6 mm diameter, 28 mm2), and the solvent was evaporated at 60 °C for 10 min. Analogously, 8 mm disks were prepared specifically for cytotoxicity assays.
After that, the substrate for Mpro was immobilized following two steps. Firstly, the coated paper discs were dipped in an aqueous solution of glutaraldehyde (5%), and left overnight at 25 °C. Then, the discs were washed by dipping them in distilled water for 10 min, and the process was repeated 5 times until the glutaraldehyde odour finally disappeared [19]. Secondly, the discs were dipped in a substrate solution (50 μM in ethanol), and left overnight at 25 °C. Finally, several dip washes were carried out with absolute ethanol (3 × 10 min), 75% ethanol (1 × 10 min), 50% ethanol (1 × 10 min), 25% ethanol (1 × 10 min), distilled water (2 × 10 min), and 20 mM Tris-HCl pH 7.3, 100 mM NaCl, 1 mM EDTA, 1 mM DTT buffer (2 × 10 min). Sensory labels 1 and 2 were air-dried and stored in zip bags. The video included as Supporting information (SI-Video) explains the complete preparation procedure of the sensory labels.
Supplementary material related to this article can be found online at doi:10.1016/j.snb.2022.133165.
The following is the Supplementary material related to this article Video 1..
Additionally, sensory labels (3 and 4) without covalently anchored substrates were also prepared, as depicted in Fig. 1c. In this case, the coating solutions contain 0.042 mg of the substrate (50 μM) and 2.21 mg of copolymer 3 or 4.
2.6. Preliminary enzyme tests and storage stability study
Measurements were carried out in a microplate reader with 96-well plates, including a 6 mm diameter sensory label at the bottom of each well and 20 μL of a 0.5 μM solution of Mpro in buffer (negative controls were performed with 20 μL of 20 mM Tris-HCl pH 7.3, 100 mM NaCl, 1 mM EDTA, 1 mM DTT buffer). The assay was carried out at the optimum enzyme temperature (30ºC), and the fluorescence emission at 460 nm was recorded over time (15, 30, 60, 120, 180, and 240 min). Measuring conditions: excitation slit = 40 nm; emission slit = 40 nm; excitation wavelength = 360 nm.
The stability of sensory labels with and without covalently anchored substrates was studied over time. All labels were room stored in zip bags without more care, and enzyme tests were performed at 1, 7, 14, 28 and 60 days.
The limit of detection (LOD) was estimated in vitro for the sensory label 1 and 2 using different concentrations of Mpro enzyme (from 0 to 1 µM) after 1 h of incubation with the sensory labels. We estimated the limit of detection (LOD) by the following equation: LOD= 3.3xSD/s, where SD is the standard deviation of blank sample and s is the slope of the calibration curve in the region of low Mpro content, respectively.
2.7. In vitro skin irritation test
The skin irritation test was performed according to the in vitro EpiDerm™ skin irritation test (EPI-200-SIT, MatTek In Vitro Life Science Laboratories, 2020) after confirming the inability of the sensors to interfere with and/or to reduce the MTT following the guideline recommendations.
Upon receipt, the tissues were inspected for damage according to the manufacturer´s instructions, transferred to 6-well plates prefilled with 0.9 mL of assay medium (EPI-100-NMM). and incubated at optimal conditions (37 ºC ± 1 ºC, 5 ± 1% CO2, 90% ± 10% RH) for 1 h. Then, the tissues were transferred to a freshly prepared medium and incubated overnight (18 ± 3 h) at optimal conditions to release transport-stress, after which the tissues were exposed to the sensors for 1 h (37 ºC ± 1 ºC, 5 ± 1% CO2, 90% ± 10% RH). As negative and positive controls, the tissues were exposed to DPBS or 5% SDS, respectively. Three tissues were used per test material and controls. After the exposure, tissues were washed 15 times with DPBS, blotted in a sterile blotting paper, dried with a sterile cotton-tipped swab, transferred to a 6-well plate with 0.9 mL culture medium and incubated at optimal conditions for 24 ± 2 h. Finally, the culture medium was removed, fresh medium was added, and the tissues were subsequently incubated again for 18 ± 2 h at optimal conditions.
Tissue viability after exposure to the sensors was determined using the MTT viability assay, following procedures described in the OECD guideline Test Nº 439. At the end of the 18 ± 2 h incubation, the tissues were transferred to a 24-well plate containing 0.3 mL of a MTT solution at 1 mg mL−1, and incubated for 3 h at optimal conditions. After this step, tissues were rinsed twice with DPBS, and formazan crystals were solubilized by adding 2 mL of isopropanol (MTT-100-EXT) for 2 h at RT with agitation. At the end of the extraction period, tissues were pierced with an injection needle, and the extract ran into the well from which the insert was taken. Afterwards, the tissues were discarded, and the extraction solutions were homogenized and transferred to a 96–well plate. Tissue viability is reported as % of negative control, measuring the OD of each isopropanol extract in duplicate at 570 nm by using a plate reader (BioTek Synergy HT). Isopropanol alone was used as a blank. The viability % of each tissue was calculated relative to negative control using the following equation:
| % Viability tissue = [ODtissue / Mean ODNC] x 100% | (1) |
2.8. Tests with subjects. Proof of concept
The tests with COVID patients and controls were authorized by the bioethics committee of the University of Burgos on April 22, 2022 (Ref. IO 05/2022). All the participants in this study were informed of the entire procedure and signed an informed consent document. Due to the restrictions, and the responsibility we had with a virus as harmful as SARS-CoV-2, we decided the patients themselves would pick up the kit, and carry out the tests at home.
This study was conducted with 26 subjects (14 men and 12 women). Among the participants, there were 15 patients (mild symptoms or asymptomatic patients) and 11 controls confirmed with antigen tests. The age range was from 23 to 69 years, and the tests were performed between 1 and 7 days after the first symptoms, or first positive result with PCR/antigen test.
All tests were performed between 8:00 p.m. and 9:00 p.m., and for security reasons, they were not collected and analyzed until 12 h later. The participants did not eat, drink or smoke during the 10 min before the test, and they first performed an antigen test on themselves (Boson Biotech, Hotgen, or Deepblue). They then discarded a saliva sample and placed four separate saliva samples over four sensory labels (2 made with copolymer 1, and another 2 with copolymer 2) contained in 2 Petri dishes, so that the discs looked completely covered by saliva. This step was done without forcing a cough, or a tear in the throat, just spitting up naturally. The Petri dishes remained open overnight, and finally, saliva excess was removed with a tissue, and the Petri dishes were sealed with parafilm.
Since the patients kept the tests during the first 12 h, in this case, we could not follow the response of the material with a fluorimeter. However, as it is a visual sensor, photographs of all the sensory labels were taken once received under 365 nm light illumination, always in the same dark room, and always replicating the same lighting conditions, since the photographs were taken on different days. For that, two lamps were placed at 23 cm from the discs and at a 45-degree angle. The smartphone was placed at 20 cm from the disks without tilting. However, from the final user's point of view (1 single photo), it is unnecessary to be so careful with the lighting conditions. It is only required to see the difference in fluorescence between the positive and negative control with the naked eye and take the photo under these conditions.
Each photo contained 3 discs, a positive control, a negative control, and the test disc, and the measurements were carried out by duplicate. G parameter (form RGB digital colour space) were extracted from discs in photographs using the smartphone app “Colorimetric Titration”, and the G parameters of the negative and positive control were assigned as 0 and 100 G%, respectively. Thus, Gtest% values were obtained with the following equation:
| Gtest% = (Gtest−Gneg) / (Gpos−Gneg) × 100 | (2) |
In this way, values above 55 were considered positive tests for COVID, while values below 45 were considered negative. The range between 45 and 55 G% was considered borderline and would require a repeat test.
2.9. Selectivity/specificity study
To study the substrate specificity for Mpro SARS-CoV-2 of our sensory labels two analyses were performed. Similarity matrix percentage and multiple alignments of amino acid sequences from 3CL proteases of different human coronaviruses were performed using MUSCLE program [20], and displayed by ESPript 3.0 web server [21]. In addition, a phylogenetic tree was obtained from Phylogeny.fr software [22]. Amino acid sequences of Mpro or 3CL protease from human coronaviruses were obtained from NCBI database (SARS-CoV: pdb|3V3M|A; SARS-CoV-2: YP_009742612.1; MERS-CoV: pdb|7D3C|A; HCoV-OC43: YP_009924323.1; HCoV-HKU1: YP_009944273.1; HCoV-NL63: pdb|7E6R|A and HCoV-229E: AGT21366.1).
Then, in vitro analyses were developed to determine the kinetic parameters of the Mpro 3CL proteases from SARS-CoV-2 and MERS-CoV. These parameters were determined using different substrate concentrations ranging from 2.5 to 40 μM and 100 nM of the respective enzyme in a final volume of 100 μL. Initial velocities were determined from the linear part of the curve and converted to the amount of hydrolysed substrate per unit of time (μM/min). Kinetic parameters were obtained using the Michaelis-Menten equation in OriginPro Program software. Edans standard curve was performed using known amounts (0–40 µM) in reaction buffer 20 mM Tris-HCl pH 7.3, 100 mM NaCl, 1 mM EDTA, 1 mM DTT and the fluorescence was measured at 30 °C using an excitation filter of 360/40 nm and an emission filter of 460/40 nm.
To study the in vitro functionality and selectivity, a 6 mm diameter of the sensory labels 1 and 2 discs were placed at the bottom of 96-well plates and 20 μL of 0.5 µM solution of Mpro from SARS-CoV-2 and MERS-CoV in 20 mM Tris-HCl pH 7.3, 100 mM NaCl, 1 mM EDTA, 1 mM DTT buffer were added. Fluorescence was recorded at 30 and 60 min after enzyme addition.
3. Results and discussion
3.1. Characterization of copolymers
The four synthesized copolymers were characterized by infrared spectroscopy, 1H and 13C nuclear magnetic resonance, thermogravimetric analysis, differential scanning calorimetry, and powder X-ray diffraction. As shown in SI-Section S2, copolymers 1 and 2 show aromatic proton signals between 6.2 and 6.8 ppm in the 1H NMR spectra, confirming the presence of aniline side groups in the polymer structure. On the other hand, 13C NMR and FT-IR spectra show typical signals of random copolymers prepared with 1-vinyl-2-pyrrolidone and methyl methacrylate. FT-IR spectra of copolymers 2 and 4 (MMA 95 mol%) contain typical peaks of PMMA at 1718 and 1138 cm−1, assigned to C O stretching and -O-CH3 stretching vibrations, respectively [23]. Signals related to PVP are only appreciable at FT-IR spectra of copolymer 1 and 3, in which the broad band assigned to the O–H stretching vibration can be shown between 3060 and 3703 cm−1, probably related to water molecules associated with the copolymer´s hydrophilicity [24]. Other characteristic peaks of PVP can also be seen at 1665 cm−1 (carbonyl group), and at 1021 cm−1 (C–N stretching) [25].
The most relevant information came from the PXRD and the polymers' thermal analysis. Thus, the PXRD spectra indicate that the separation between chains is not affected by the introduction of the aniline side groups in the polymer structure since the values of 2θ MAX do not vary significantly. In the same way, concerning thermal analysis, copolymers 1 and 3 do not present significant differences in the values of T 5, T 10 and Tg. However, copolymer 2 has T 5 and T 10 values 16 and 12 °C higher than copolymer 4, respectively. Therefore, our interpretation is that adding aniline groups improves the interaction between chains through H bonds with the carbonyl groups of the rest of the comonomers, which improves the material's thermal properties. This can also be seen in the behaviour of the Tg, which increases 5 °C when only 0.5% mol of vinylaniline is introduced in the copolymer formulation.
3.2. Response time and storage stability of sensory labels
Fluorescence emission was measured at a fixed wavelength, and three replicates were performed for each measurement in 96-well plates to obtain statistically robust data, and the fluorescence data were transformed into enzymatic activity data through the following equation:
| (3) |
where “RFUs” are the relative fluorescence units, and “F” is the emitted fluorescence.
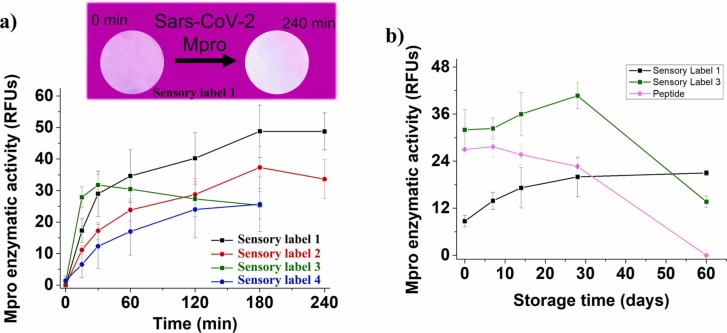
The enzyme tests that were carried out with the sensory labels showed that an OFF-ON fluorescence process occurs when exposed to the Mpro enzyme, as shown in Fig. 2a. The system reached equilibrium 180 min after the enzyme addition for all sensory labels except the Nº3, in which the response time was 30 min. However, after half an hour, the response is intense enough to be detected with the naked eye. These experiments were performed with an enzyme concentration of 0.5 µM, but the LODs of the sensory labels 1 and 2 are 0.177 µM (6.018 µg/mL) and 0.396 µM (13.4 µg/mL), respectively.
Fig. 2.
Enzyme tests carried out in 96-well plates, including a 6 mm diameter sensory label at the bottom of each well and 20 μL of a 0.5 μM solution of Mpro in 20 mM Tris-HCl pH 7.3, 100 mM NaCl, 1 mM EDTA, 1 mM DTT buffer. Experimental conditions: temperature = 30ºC, λex = 360 nm, λem = 460 nm, excitation slit = 40 nm, emission slit = 40 nm. (a) Response time study of sensory labels 1–4, by monitoring the fluorescence at 15, 30, 60, 120, 180, and 240 min; image of a sensory label 1 before and after interaction with Mpro. (b) Storage stability study of sensory labels 1, 3 and free peptide by measuring the fluorescence response at 60 min after 1, 7, 14, 28 and 60 days of storage using zip bags. Data are means ± standard error of 3 independent replicates.
The stability study (Fig. 2b) showed that sensory labels with covalently anchored substrates, such as sensory label 1, have a longer shelf-life and remain suitable for at least two months after preparation (black line). However, as we have shown in previous works [26], this stability is not exhibited when using labels with substrate dispersed (not covalently anchored, as sensory label 3), showing the same behaviour as the free peptide substrate (green and pink lines, respectively). Additionally, results for sensory labels 2 and 4 were equivalent. Therefore, our interpretation is that the polymeric chains exert a protective effect only on the covalently anchored substrates. This fact is one of the novel keys of this study since this kind of substrates are usually unstable, so they are not used in the preparation of sensory materials.
3.3. Cytotoxicity assays
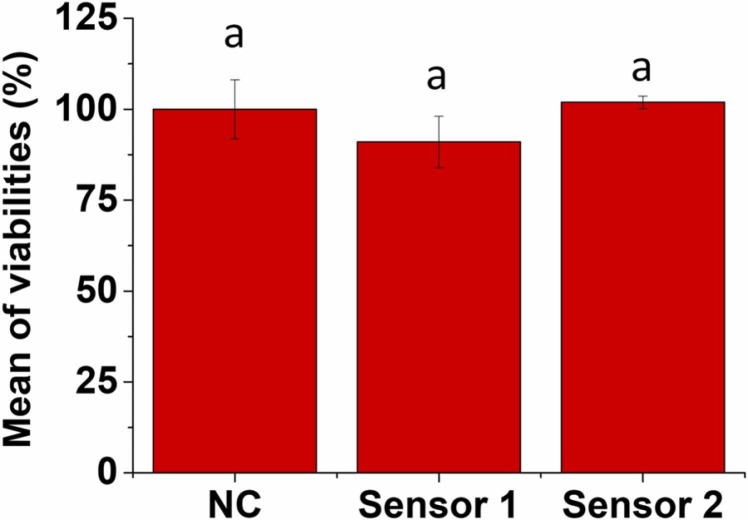
In vitro EpiDerm™ skin irritation test (MatTek) is a test compliant with the OECD Test Guideline (TG) No. 439 to evaluate the skin irritation potential of the test chemicals in the context of identification and classification of skin irritation hazards according to the EU and Classification Harmonized System of Classification and Labelling Chemicals, GHS, (R38 / Category 2 or no label). Thus, an irritant is predicted if the mean relative tissue viability of three individual tissues exposed to the test substance is reduced below 50% of the mean viability of the negative controls.
As shown in Fig. 3, sensory labels 1 and 2 did not reduce the viability by over 50% when compared with the controls. Therefore, according to the EU and Globally Harmonized System of Classification and Labelling Chemicals, GHS, (R38/ Category 2 or no label), none of them could be considered irritants in the conditions tested.
Fig. 3.
EpiDermTM tissues were exposed to sensory labels 1 and 2 for 1 h. The viability was analysed by MTT assay, and it is expressed as a percentage of negative control. Data represented the mean ± standard error of 3 independent replicates. Differences were established using a one-way ANOVA followed by a multiple comparisons test (Tukey test) and considered significant when p ≤ 0.05. The same letter indicates no significant differences between treatments.
3.4. Selectivity/Specificity of the sensory labels
The most common human respiratory viruses are influenza A and B, syncytial, rhinoviruses, adenoviruses, and coronaviruses. Among them, only rhinoviruses and coronaviruses use their 3 C-like proteases to replicate [27]. However, the 3 C-like proteases of rhinoviruses only have a 20% homology with the protease studied in this work, they recognize a substantially different peptide sequence (LEVLFQ/GP) than that of the SARS-CoV-2, and their active site is made up of three amino acids instead of two as is the case of coronaviruses [28], [29], [30]. Therefore, the specificity and selectivity studies were carried out with viruses from the coronaviruses group.
Seven human coronaviruses (HCoV) have been described until now, the endemic viruses HCoV-OC43, HCoV-229E, HCoV-NL63, HCoV-HKU1 and the epidemic viruses MERS-CoV, SARS-CoV, and SARS-CoV-2. HCoV-OC43 and HCoV-229E are included in the alphacoroviruses genera, while HCoV-NL63, HCoV-HKU1, MERS-CoV, SARS-CoV, and SARS-CoV-2 are members of the betacoronaviruses genera [31], [32].
To study the specificity of our labels, the encoding amino acid sequences of the 3 C-like proteases of these seven coronaviruses were obtained and aligned. Fig. S8 (SI-Section S3) summarizes the multiple alignment results, showing the highly conserved residues of the catalytic site of the proteases (blue), the strictly equal residues in all the sequences (red), and the residues belonging to the same group of amino acids (yellow). The residues of catalytic site (blue) and those around them (red) are conserved, although their positions are slightly displaced. For instance, while in SARS-CoV and SARS-CoV-2 this catalytic dyad is located at residues Hys41 and Cys145, in MERS-CoV it is located at residues Hys41 and Cys148 [33].
The results of the identity matrix (Table S1, SI-Section S3) show that the alpha genera viruses are those with the lowest degree of conservation and, therefore, the lowest similarity with the rest of the coronaviruses. Consequently, on the one hand, we can conclude that the amino acid sequences of the 3 C-like protease of SARS-CoV-1 and SARS-CoV-2 are practically identical (96.08% of similarity). On the other hand, the similarity between SARS-CoV-2 and MERS-CoV is quite lower (∼50%), and even lower when comparing SARS-CoV-2 with the rest of the coronaviruses as previously reported [12], [33]. The phylogenetic tree analysis also supports these results, in which coronaviruses are sorted by genera (Fig. S9, SI-Section S3). SARS coronaviruses 1 and 2 have evolved separately from the rest of the betacoronaviruses. In fact, the MERS-CoV, HCoV-HKU1 and HCoV-OC43 viruses seem to have the same evolutionary origin, highly influenced by their zoonotic origin (from animals to humans), and definitively they have evolved differently and have adapted to their new niche [32], [33].
Accordingly, we performed the enzymatic analyses summarized in Table S2 (SI-Section S4), showing the kinetic parameters obtained for Mpro 3CL proteases from SARS-CoV-2 and MERS-CoV using the substrate Dabcyl-KTSAVLQSGFRKME-Edans. As expected, KM of the Mpro from SARS-CoV-2 is lower than KM of the Mpro from MERS-CoV, i.e., 43.02 ± 3.23 µM and 117.06 ± 10.61 µM values, respectively. This parameter indicates that the Mpro-SARS-CoV-2 presents a higher affinity for the substrate than Mpro-MERS-CoV. In addition, the Mpro-MERS-CoV's enzymatic efficiency (Kcat/KM) is remarkably lower than the Mpro-SARS-CoV-2, confirming our in silico studies and previous reports from other authors [12], [33], [34].
Regarding the tests with sensory labels, the Mpro-SARS-CoV-2 has higher activity than the Mpro-MERS-CoV, which means a higher fluorescence emission when performing fluorescence analysis in the microplate reader, and also higher visual response with the naked eye, as shown in Fig. S10.
3.5. Proof of concept. Saliva test with 26 subjects (15 patients and 11 controls)
This study was carried out with 26 subjects, 15 COVID patients, and 11 controls. It is a relatively small number to be considered a medical study, but large enough for a work in which the main claim and novelty is the strategy of preparing these new virus sensors straightforwardly, only anchoring a fluorogenic peptide substrate to a polymer and then using this sensory polymer as a coating for paper.
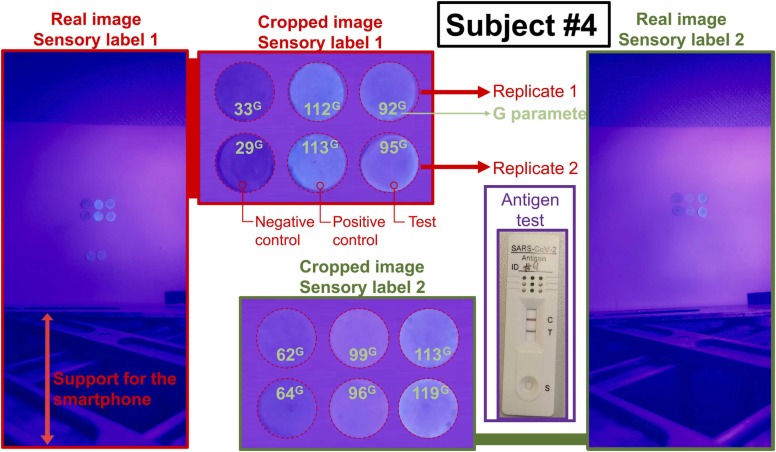
For example, Fig. 4 shows the results of Subject #4 (asymptomatic), who tested positive for COVID with the antigen test and our sensory labels 1 and 2. The figures for the rest of the subjects are included in the SI-Section S5.
Fig. 4.
Results for Subject #4 testing with sensory label coated with copolymer 1 (sensory label 1), sensory label coated with copolymer 2 (sensory label 2), and antigen test. The image shows the real photograph and the cropped image. Each photo contains 2 negative controls, 2 positive controls, and 2 replicates.
The COVID patients generated fluorescence in the sensory labels to the naked eye, while the negative cases did not. To assign a numerical value to that response, the digital colour of each label was analysed in two different digital colour spaces (RGB and HSV), and we found G parameter from the RGB colour space was the one that best distinguishes between patients and controls.
Considering the G parameter of the negative control as 0 G%, and the G parameter of the positive control as 100 G%, Subject #4 gave a G% result greater than 55 (75 G% for sensory label 1 and 157 G% for sensory label 2), so the result was positive for COVID in line with the antigen test. Table 2 shows the results for all subjects.
Table 2.
Results of the study with 26 subjects, testing COVID with sensory label 1, sensory label 2, and antigen test. The table shows the mean ± standard error of 2 replicates of the G% extracted from photographs, and calculated with the equation Gtest% = (Gtest-Gneg) / (Gpos-Gneg) × 100 (Eq. 3).
 |
 |
The risk of making a mistake in the experimental procedure was high and difficult to control since all the responsibility lies within the participants. Despite this, our sensory label 1 and the used antigen tests provided the same result in 21 of the 26 cases (81%), which suggests that our proposed idea can be a leading methodology for the industrial production of this type of sensors in the short term. Furthermore, from a comparative point of view, the success rate of self-testing antigen tests is 82.5% [35], very similar to the result obtained with our material.
Sensory label 1 has better results than sensory label 2 (85% match with antigen test versus 65%). Our interpretation is that the greater hydrophilicity of copolymer 1 makes the environment generated for the reaction with the Mpro enzyme more appropriate. Regarding the non-satisfactory results of sensory label 1, we must underline that result for subject #17 is on the borderline, that is, the subject should have repeated the test. The other four unsuccessful cases are one false positive (subject#8) and three false negatives (subjects #13, #14 and #20). In the case of the false positive, he/she presented symptoms compatible with COVID, and therefore there may be interference with some other rhinovirus. This interference problem can be addressed by increasing the length of the substrate peptide, i.e., making it much more specific for Mpro. Regarding the false negatives, we believe some other substance in the saliva may be quenching the fluorescent signal through deactivation processes. We think that this could be improved by including a tooth and tongue brushing 10 min before the test, but in our case could not be carried out since it is contraindicated in many of the antigen tests.
4. Conclusions
Early detection methods are essential tools against the spread of infectious diseases, as seen in the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) caused pandemic with the width spread use of rapid and reliable immunoassay to qualitatively detect antibodies against the virus. Herein, we report on a strategy to prepare an easy-to-use protein function-based detection method using saliva of potentially infected people, based on peptide substrate containing a FRET pair (fluorophore and quencher) to detect the Mpro protein. Specifically, we have chemically anchored the Mpro substrate to linear polymers with different hydrophilicity, and coated cellulose supports with it to prepare the sensory labels. Results show that the higher hydrophilicity of the copolymer, the higher performance of the sensory labels. Upon contacting the sensor with the saliva (Mpro is present in infected people, as demonstrated), the OFF-ON fluorescence response allows for the qualitative visual detection of the infection (LOD = 0.177 µM for sensory label 1). Moreover, a picture of the sensors provides a quantitative and statistical infection result. The advantages of these sensors are that they can be easily prepared, inexpensively, quickly, and in high quantities. Even more relevant is the proposed methodology that can be applied to detecting saliva or other body fluids and expired air for virus and bacterial infections.
Open data
Open Data is available at https://riubu.ubu.es/handle/10259/5684 (Dataset of the work " Lab-on-a-chip for the easy and visual detection of SARS-CoV-2 based on a sensory polymer").
Supporting Information
Characterization of the fluorogenic peptide substrate for Mpro; characterization of polymers; amino acid sequence analyses of Mpro 3 C-like main proteases of human coronaviruses; in vitro comparative analysis of Mpro-SARS-CoV-2 and Mpro-MERS-CoV; proof of concept with 26 participants; explanatory video.
CRediT authorship contribution statement
Ana Arnaiz: Methodology, Validation, Formal analysis, Investigation, Writing – original draft. Jose Carlos Guirado-Moreno: Methodology, Conceptualization, Validation, Investigation, Resources. Marta Guembe-García: Validation, Investigation, Writing – original draft. Rocio Barros: Methodology, Validation, Formal analysis. Juan A. Tamayo-Ramos: Validation, Formal analysis, Investigation, Supervision. Natalia Fernández-Pampín: Methodology, Validation, Formal analysis. José M. García: Conceptualization, Methodology, Writing – review & editing. Supervision, Funding acquisition. Saul Vallejos: Conceptualization, Funding acquisition, Project administration, Methodology, Investigation, Writing – original draft, Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We gratefully acknowledge the financial support provided by all funders. Author Saul Vallejos coordinates the project leading to these results, which has received funding from "La Caixa" Foundation, under agreement LCF/PR/PR18/51130007. This work was supported by the Regional Government of Castilla y León (Junta de Castilla y León) and by the Ministry of Science and Innovation MICIN and the European Union NextGenerationEU PRTR. Author Jose Miguel García received grant PID2020–113264RB-I00 funded by MCIN/AEI/ 10.13039/501100011033 and by “ERDF A way of making Europe”. Ana Arnaiz received funding from Ministerio de Universidades-European Union in the frame of NextGenerationEU RD 289/2021 (Universidad Politécnica de Madrid). Finally, all the authors want to thank the support provided by City Hall of Villadiego “Ayuntamiento de Villadiego” when looking for participants for the proof of concept.
Biographies
Dr. Ana Arnaiz (F) completed her Ph.D. in Biotechnology and Genetic Resources in Plants and Associated Microorganisms at the Universidad Politécnica de Madrid in 2019. In addition, she has a postdoctoral Margarita Salas Grant at the University of Burgos to expand her research in the development of sensory polymeric materials oriented to detect biological targets.
José Carlos Guirado-Moreno (M) received her Master's degrees in drug discovery and advanced chemistry at the University of Alcalá de Henares (Spain, 2018) and the University of Burgos (Spain, 2019), respectively. He started his Ph.D. in Advanced Chemistry in 2019, and his research interests include smart polymers with applications in biomedicine and food safety.
Dr. Marta Guembe-García (F) received her Ph.D. in Advanced Chemistry at the University of Burgos, Spain, in 2021. In addition, she has a postdoctoral Margarita Salas Grant at the University of Pavia (Italy) to expand her research in the development of smart materials with different applications in analytical chemistry.
Dr. Rocío Barros (F) has a degree in Environmental Biology and a PhD in Environmental Science. She is Head of the ICCRAM – Environmental, Sustainability and Toxicology Research Group and collaborates in several H2020 / HE Projects, coordinating GREENER Project. She has been involved in more than 25 European projects mainly related to developing different environmental technologies for soil, water and air treatment.
Dr. Juan Antonio (M) Tamayo has Ph.D. in Molecular Microbiology and is the scientific leader of the Toxicology Research line. He has achieved 24 publications in top sciences journals (h-index=12, i-10 index=13, total citations=744), 2 patents, and participation in more than 20 R&D projects at national and international level. He is the Coordinator of the on-going DIAGONAL Project, H2020-MSCA-RISE NANOGENTOOLS.
Natalia Fernández-Pampín (F) is a Ph.D. student that currently works as a researcher at the International Research Center in Critical Raw Materials (ICCRAM), University of Burgos. Her project thesis is focused on the study of the antitumoral and antimicrobial properties of new metal Pt(II),Ir(III),Ru(II) and Rh(III) complexes. Since 2021,she has been part of the Environmental,Sustainability and Toxicological Research Group at ICCRAM,where she is involved in studying the toxicity of nanomaterials employing human cell lines,3D Reconstructed Human Epidermis (RhE) and different prokaryotic and eukaryotic microorganisms as in vitro models.
Prof. José Miguel García (M) is Full Professor at the Department of Chemistry at the University of Burgos, Spain. He carried out his doctoral studies at the Institute of Polymer Science & Technology, Spanish National Research Council (CSIC), receiving his Ph.D. in Chemistry at the Complutense University of Madrid, Spain in 1995. Prof. Garcia is a co-author of 120 + peer-reviewed scientific publications and has a number of patents, along with co-author books and book chapters. His principal research areas are high-performance materials, functional polymers, and sensory polymers as sensing materials for food, biomedical, environmental, and civil security applications.
Dr. Saúl Vallejos (M) received his Ph.D. in Chemistry in 2014 from the University of Burgos, Spain. Now he is a postdoc researcher at the Department of Chemistry, University of Burgos. Dr. Vallejos is the author/co-author of 45 + peer-reviewed scientific publications and has 22 patents, along with co-author books and book chapters. His research interests are functional polymers with receptor motifs as sensory materials for anions, cations, and neutral molecules.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.snb.2022.133165.
Appendix A. Supplementary material
Supplementary material
.
Data Availability
No data was used for the research described in the article.
References
- 1.Arroyo-Marioli F., Bullano F., Kucinskas S., Rondón-Moreno C. Tracking R of COVID-19: a new real-time estimation using the Kalman filter. PLoS One. 2021;16 doi: 10.1371/JOURNAL.PONE.0244474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.University of Oxford, COVID-19 Data Explorer, (2022). 〈https://ourworldindata.org/explorers/coronavirus-data-explorer〉 (Accessed March 7, 2022).
- 3.Hale T., Angrist N., Goldszmidt R., Kira B., Petherick A., Phillips T., Webster S., Cameron-Blake E., Hallas L., Majumdar S., Tatlow H. A global panel database of pandemic policies (Oxford COVID-19 Government Response Tracker) Nat. Hum. Behav. 2021;5:529–538. doi: 10.1038/S41562-021-01079-8. [DOI] [PubMed] [Google Scholar]
- 4.Artik Y., Kurtulmus M.S., Cesur N.P., Mart Komurcu S.Z., Kazezoglu C., Kocatas A. Clinic evaluation of the destrovir spray effectiveness in SARS-CoV-2 disease. Electron. J. Gen. Med. 2022;19:em357. doi: 10.29333/EJGM/11578. [DOI] [Google Scholar]
- 5.Fernandes Q., Inchakalody V.P., Merhi M., Mestiri S., Taib N., Moustafa Abo El-Ella D., Bedhiafi T., Raza A., Al-Zaidan L., Mohsen M.O., Yousuf Al-Nesf M.A., Hssain A.A., Yassine H.M., Bachmann M.F., Uddin S., Dermime S. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann. Med. 2022;54:524–540. doi: 10.1080/07853890.2022.2031274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diao M., Lang L., Feng J., Li R. Molecular detections of coronavirus: current and emerging methodologies. Expert Rev. Anti. Infect. Ther. 2022;20:199–210. doi: 10.1080/14787210.2021.1949986. [DOI] [PubMed] [Google Scholar]
- 7.Chen H., Park S.G., Choi N., Kwon H.J., Kang T., Lee M.K., Choo J. Sensitive detection of SARS-CoV-2 using a SERS-based aptasensor. ACS Sens. 2021;6:2378–2385. doi: 10.1021/ACSSENSORS.1C00596. [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y., Hu M., Liu A.A., Lin Y., Liu L., Yu B., Zhou X., Pang D.W. Detection of SARS-CoV-2 by CRISPR/Cas12a-enhanced colorimetry. ACS Sens. 2021;6:1086–1093. doi: 10.1021/ACSSENSORS.0C02365. [DOI] [PubMed] [Google Scholar]
- 9.Baker A.N., Richards S.J., Pandey S., Guy C.S., Ahmad A., Hasan M., Biggs C.I., Georgiou P.G., Zwetsloot A.J., Straube A., Dedola S., Field R.A., Anderson N.R., Walker M., Grammatopoulos D., Gibson M.I. Glycan-based flow-through device for the detection of SARS-COV-2. ACS Sens. 2021;6:3696–3705. doi: 10.1021/ACSSENSORS.1C01470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuo C.J., Chi Y.H., Hsu J.T.A., Liang P.H. Characterization of SARS main protease and inhibitor assay using a fluorogenic substrate. Biochem. Biophys. Res. Commun. 2004;318:862–867. doi: 10.1016/J.BBRC.2004.04.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu L., Ye F., Feng Y., Yu F., Wang Q., Wu Y., Zhao C., Sun H., Huang B., Niu P., Song H., Shi Y., Li X., Tan W., Qi J., Gao G.F. Both Boceprevir and GC376 efficaciously inhibit SARS-CoV-2 by targeting its main protease. Nat. Commun. 2020;11:1–8. doi: 10.1038/s41467-020-18233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van de Plassche M.A.T., Barniol-Xicota M., Verhelst S.H.L. Peptidyl acyloxymethyl ketones as activity-based probes for the main protease of SARS-CoV-2. ChemBioChem. 2020;21:3383–3388. doi: 10.1002/CBIC.202000371. [DOI] [PubMed] [Google Scholar]
- 14.Hu Q., Xiong Y., Zhu G.-H., Zhang Y.-N., Zhang Y.-W., Huang P., Ge G.-B., Xiong Y. The SARS-CoV-2 main protease (Mpro): structure, function, and emerging therapies for COVID-19. MedComm. 2022;3 doi: 10.1002/MCO2.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sacco M.D., Ma C., Lagarias P., Gao A., Townsend J.A., Meng X., Dube P., Zhang X., Hu Y., Kitamura N., Hurst B., Tarbet B., Marty M.T., Kolocouris A., Xiang Y., Chen Y., Wang J. Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against Mpro and cathepsin L. Sci. Adv. 2020;6 doi: 10.1126/SCIADV.ABE0751/SUPPL_FILE/ABE0751_SM.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a-ketoamide inhibitors. Sci. (80-. ) 2020;368:409–412. doi: 10.1126/SCIENCE.ABB3405/SUPPL_FILE/PAPV2.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cully M. A tale of two antiviral targets - and the COVID-19 drugs that bind them. Nat. Rev. Drug Discov. 2022;21:3–5. doi: 10.1038/d41573-021-00202-8. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu N., Aoki K., Yamada M., Yukinaga H., Fujita Y., Kamioka Y., Matsuda M. Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol. Biol. Cell. 2011;22:4647–4656. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dekker R.F.H. Immobilization of a lactase onto a magnetic support by covalent attachment to polyethyleneimine-glutaraldehyde-activated magnetite. Appl. Biochem. Biotechnol. 1989;22:289–310. doi: 10.1007/BF02921763. [DOI] [PubMed] [Google Scholar]
- 20.Edgar R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinforma. 2004;5:1–19. doi: 10.1186/1471-2105-5-113/FIGURES/16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/NAR/GKU316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard J.F., Guindon S., Lefort V., Lescot M., Claverie J.M., Gascuel O. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/NAR/GKN180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramesh S., Leen K.H., Kumutha K., Arof A.K. FTIR studies of PVC/PMMA blend based polymer electrolytes. Spectrochim. Acta - Part A Mol. Biomol. Spectrosc. 2007;66:1237–1242. doi: 10.1016/j.saa.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Sreekanth K., Siddaiah T., Gopal N.O., Jyothi N.K., Kumar K.V., Ramu C. Thermal, structural, optical and electrical conductivity studies of pure and Mn2+ doped PVP films. South Afr. J. Chem. Eng. 2021;36:8–16. doi: 10.1016/j.sajce.2020.09.003. [DOI] [Google Scholar]
- 25.Alver E., Metin A., Çiftçi H. Synthesis and characterization of chitosan/polyvinylpyrrolidone/zeolite composite by solution blending method. J. Inorg. Organomet. Polym. Mater. 2014;24:1048–1054. doi: 10.1007/s10904-014-0087-z. [DOI] [Google Scholar]
- 26.Arnaiz A., Guembe-García M., Delgado-Pinar E., Valente A.J.M., Ibeas S., García J.M., Vallejos S. The role of polymeric chains as a protective environment for improving the stability and efficiency of fluorogenic peptide substrates. Sci. Rep. 2022;12:8818. doi: 10.1038/s41598-022-12848-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weston S., Frieman M.B. Respiratory viruses. Encycl. Microbiol. 2019:85. doi: 10.1016/B978-0-12-801238-3.66161-5. [DOI] [Google Scholar]
- 28.May Wang Q., Chen S.-H. Human rhinovirus 3C protease as a potential target for the development of antiviral agents. Curr. Protein Pept. Sci. 2007;8:19–27. doi: 10.2174/138920307779941523. [DOI] [PubMed] [Google Scholar]
- 29.Matthews D.A., Smith W.W., Ferre R.A., Condon B., Budahazi G., Slsson W., Villafranca J.E., Janson C.A., McElroy H.E., Gribskov C.L., Worland S. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 30.Cordingley M.G., Callahan P.L., Sardana V.V., Garsky V.M., Colonno R.J. Substrate requirements of human rhinovirus 3C protease for peptide cleavage in vitro. J. Biol. Chem. 1990;265:9062–9065. doi: 10.1016/S0021-9258(19)38811-8. [DOI] [PubMed] [Google Scholar]
- 31.Zhou P., Lou Yang X., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Di Jiang R., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corman V.M., Muth D., Niemeyer D., Drosten C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 2018;100:163–188. doi: 10.1016/BS.AIVIR.2018.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tomar S., Johnston M.L., St. John S.E., Osswald H.L., Nyalapatla P.R., Paul L.N., Ghosh A.K., Denison M.R., Mesecar A.D. Ligand-induced dimerization of middle east respiratory syndrome (MERS) coronavirus nsp5 protease (3CLpro) J. Biol. Chem. 2015;290:19403–19422. doi: 10.1074/jbc.m115.651463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X.Y., Zhang Y.Y., Zhang X.G., Liu X., Chen M., Liu F., Zhang D.Z., Ling P.X. Research progress of new coronavirus SARS-CoV-2 detection technology. Prog. Biochem. Biophys. 2020;47:275–285. doi: 10.16476/J.PIBB.2020.0060. [DOI] [Google Scholar]
- 35.Lindner A.K., Nikolai O., Rohardt C., Kausch F., Wintel M., Gertler M., Burock S., Hörig M., Bernhard J., Tobian F., Gaeddert M., Lainati F., Corman V.M., Jones T.C., Sacks J.A., Seybold J., Denkinger C.M., Mockenhaupt F.P. Diagnostic accuracy and feasibility of patient self-testing with a SARS-CoV-2 antigen-detecting rapid test. J. Clin. Virol. 2021;141 doi: 10.1016/J.JCV.2021.104874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
No data was used for the research described in the article.