Abstract
Cellular drug response (concentration required for obtaining 50% of a maximum cellular effect, EC50) can be predicted by the intracellular bioavailability (Fic) and biochemical activity (half-maximal inhibitory concentration, IC50) of drugs. In an ideal model, the cellular negative log of EC50 (pEC50) equals the sum of log Fic and the negative log of IC50 (pIC50). Here, we measured Fic’s of remdesivir, favipiravir, and hydroxychloroquine in various cells and calculated their anti-SARS-CoV-2 EC50’s. The predicted EC50’s are close to the observed EC50’s in vitro. When the lung concentrations of antiviral drugs are higher than the predicted EC50’s in alveolar type 2 cells, the antiviral drugs inhibit virus replication in vivo, and vice versa. Overall, our results indicate that both in vitro and in vivo antiviral activities of drugs can be predicted by their intracellular bioavailability and biochemical activity without using virus. This virus-free strategy can help medicinal chemists and pharmacologists to screen antivirals during early drug discovery, especially for researchers who are not able to work in the high-level biosafety lab.
Introduction
The coronavirus disease 2019 (COVID-19) vaccines are helping avoid people get infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). However, antiviral drugs are still useful to reduce morbidity and mortality associated with SARS-CoV-2 infection.1,2 It is imperative to develop antiviral drugs to end this pandemic. In early drug discovery, SARS-CoV-2 virus has been used in cell-based assay and animal study for evaluating the antiviral activity of drugs in vitro and in vivo.3,4 SARS-CoV-2 is classified as a Risk Group 3 (RG3) human pathogen, so antiviral experiments should be conducted in a validated biosafety level 3 (BSL-3) laboratory.5 Not all medicinal chemists and pharmacologists who work on antiviral research have BSL-3 laboratories, and this slows down the antiviral drug discovery progress around the world.6
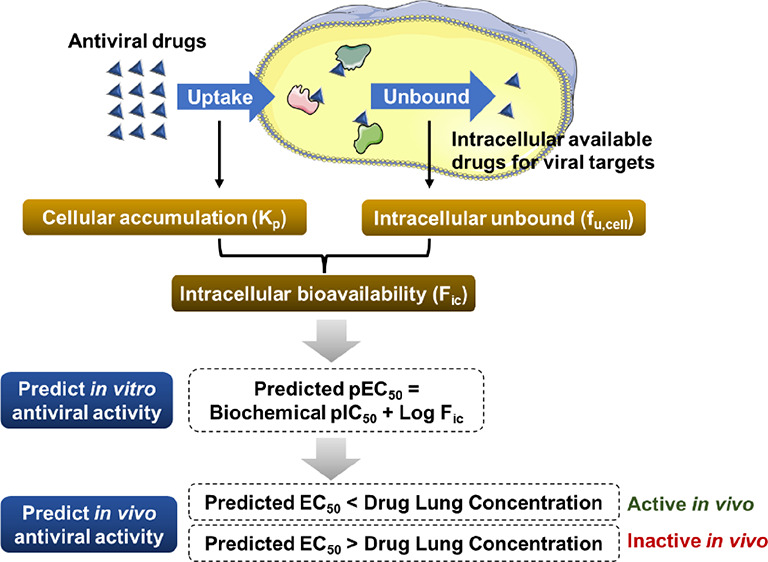
Most antiviral drugs target specific viral proteins. These antivirals, saquinavir and lopinavir as examples, are designed to directly bind to the viral protease and inhibit virus replication.7,8 The in vitro antiviral activity is proportional to the intracellular drug concentration;9,10 the more drug exposed to the targeting viral protein in the cytoplasm, the less viral replication in the cell.9,10 A rapid measurement of the intracellular bioavailability (Fic) of drugs in cultured cells has been developed.11 The intracellular unbound drug concentration can be measured as fu,cell, whereas the steady-state intracellular drug accumulation can be measured as Kp.11Fic is obtained by multiplying fu,cell and Kp.11 Moreover, the in vitro antiviral activity of drugs (pEC50) has been reported to be predicted by the formula pEC50 = pIC50 + Log Fic (Figure 1A).12 This formula successfully predicted the in vitro bioactivity of test compounds, including BACE-1, p38α, and PKIS inhibitors.12
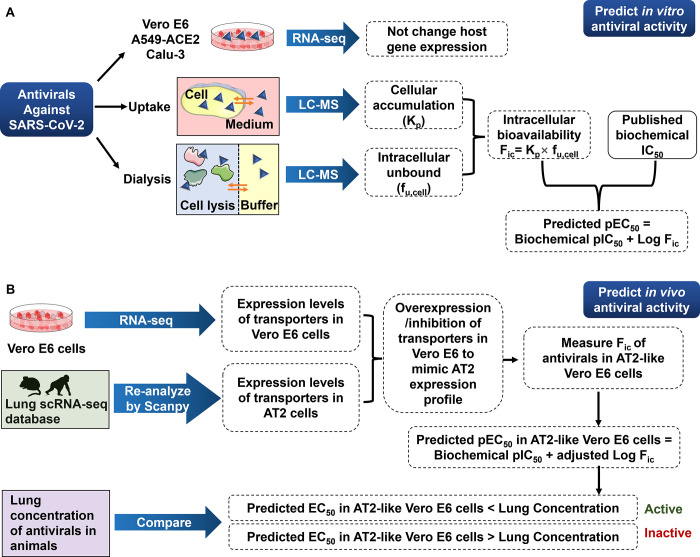
Figure 1.
Strategy of predicting the anti-SARS-CoV-2 activity of antivirals in this study. (A) Flowchart of prediction of in vitro antiviral activity by the intracellular bioavailability and biochemical activity. (B) Flowchart of prediction of in vivo antiviral activity by the predicted EC50 and lung concentration.
Drugs can directly bind to the target protein in the cytoplasm if the target protein is expressed in the host cells. This can cause less accurate fu,cell and therefore leads to an underestimated cellular activity of drugs. However, if the drug targets the viral protein, we have a good chance to obtain a more reliable prediction of fu,cell as well as cellular antiviral activity. In this study, we used remdesivir (RDV), GS-441524, favipiravir (FAV), chloroquine (CQ), and hydroxychloroquine (HCQ) as examples to test if their cellular antiviral activities can be predicted by their intracellular bioavailabilities and biochemical activities. This prediction could help scientists evaluate the in vitro antiviral activity of drugs without using virus (see the strategy in Figure 1A).
Additionally, the in vivo antiviral activity of drugs can be predicted by using the similar strategy. SARS-CoV-2 infects alveolar type 2 (AT2) cells in the lung.13 The antiviral drugs are supposed to enter AT2 cells and inhibit viral RNA-dependent RNA polymerase (RdRp) or main protease (Mpro) function in the cytoplasm.14,15 Like the in vitro model, the more drug exposed to the targeting viral protein in AT2 cells, the stronger antiviral activity in vivo should be observed.14,15 The intracellular bioavailability of antiviral drugs depends on drug transporters that are involved in the uptake or efflux of drugs.16 In our in vitro study, the key transporters that determine intracellular bioavailabilities of RDV, FAV, and HCQ were characterized. To predict the EC50 of drugs in AT2 cells, we used the Fic of drugs in cells with the similar expression profile of key transporters to AT2 cells. Here, we compared the predicted EC50 in AT2 cells with lung concentrations of RDV, FAV, and HCQ in their animal studies and predicted in vivo antiviral activities of these three drugs (see the strategy in Figure 1B). The lung concentration was identified as the maximum lung tissue concentration (Cmax), whereas the predicted EC50 was identified as the predicted concentration required to obtain a 50% antiviral effect in cell-based assay. If the lung Cmax is higher than the predicted EC50, it suggests that a 50% antiviral effect in the lung should be observed, therefore leading to a good antiviral activity in vivo.
Together, we developed a workflow (Figure 1) to predict both in vitro and in vivo anti-SARS-CoV-2 activities of RDV, FAV, and HCQ by their intracellular bioavailabilities and biochemical activities. The predicted EC50’s are close to the observed EC50’s in vitro. The prediction of in vivo antiviral activity is evidenced by animal studies. We believe that this virus-free strategy can help antiviral drug screenings, especially for scientists who are not able to work in BSL-3 labs.
Results
Predicting the Anti-SARS-CoV-2 Activity of Remdesivir
Remdesivir (RDV) is a prodrug that is metabolized to its active form GS-443902, a ribonucleotide analogue inhibitor of the RdRp of SARS-CoV-2 with IC50 at 32 nM (Figure 2A).17 RdRp catalyzes the replication of RNA from an RNA template.18 Thus, the inhibition of RdRp enzymatic activity terminates RNA genome replication and viral replication as well.19 RDV indeed has been reported to inhibit SARS-CoV-2 replication in cells with EC50 values of 0.07–1.65 μM (Figure 2A).20−22 If RDV inhibits SARS-CoV-2 replication in vitro mainly through its anti-RdRp activity, its cellular pharmacological response can be predicted by intracellular bioavailability (Fic) and biochemical activity (anti-RdRp IC50).
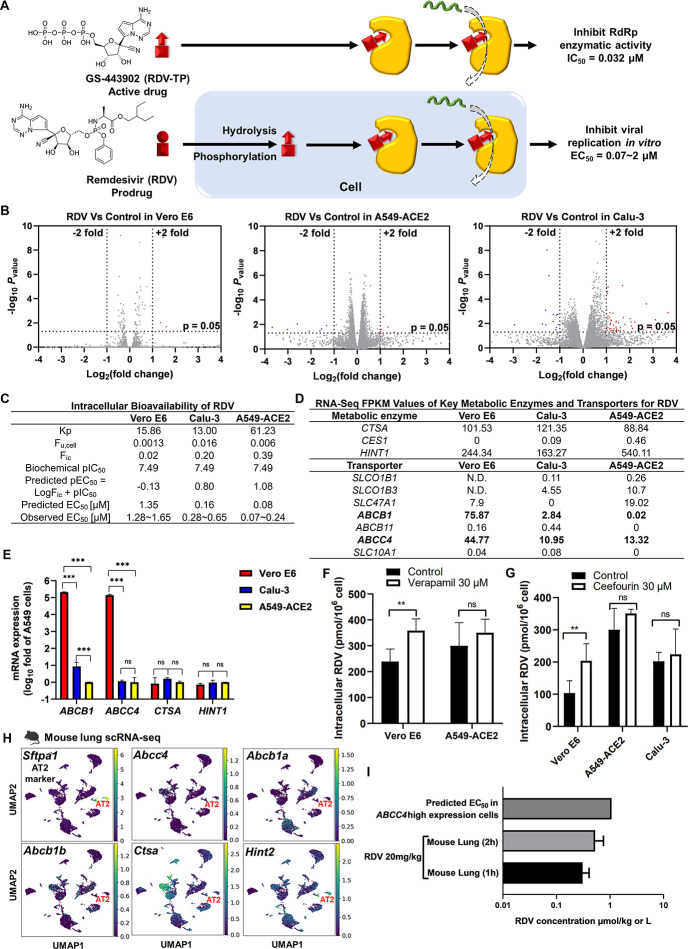
Figure 2.
Predicting the antiviral activity of RDV by the corresponding intracellular bioavailability and anti-RdRp activity. (A) The ideal bioactivation of RDV predominately occurs in vitro. (B) RDV did not significantly alter gene expression in Vero E6, Calu-3, and A549-ACE2 cells. Cells were treated with or without RDV for 48 h and then collected for bulk RNA sequencing. Volcano plot of gene expression changes (n = 3 biological replicates). Significant genes were called via DESeq2 (P < 0.05 and change of >2-fold). (C) Intracellular bioavailability of RDV and the corresponding predicted EC50. (D) FPKM of key metabolic enzymes and transporters of RDV. (E) qRT-PCR showed that ABCB1 and ABCC4 were highly expressed in Vero E6 cells but not in Calu-3 or A549-ACE2 cells. There was no significant difference in the expression of CTSA and HINT1 in the three cells. Data were processed and displayed as mean ± s.e.m. (n = 3 biological replicates) according to the 2–ΔΔCt method. (F) The P-gp inhibitor or (G) MRP4 inhibitor improved the cellular uptake of RDV in Vero E6 cells. (H) The scRNA-seq data of the mouse lung (GSE132901) were re-analyzed and visualized by Scanpy. Sftpa1 was used as an AT2 cell marker. (I) Levels of RDV in the lung of the mouse dosed with 20 mg/kg RDV and its comparison with predicted in vitro EC50.
In addition to directly inhibiting viral proteins, some antiviral drugs trigger the immune response to inhibit virus replication.23 For example, baicalin has been found to inhibit influenza through modulation of innate immune responses.23 We therefore treated Vero E6, A549-ACE2, and Calu-3 cells with RDV at their EC50 values for 48 h and collected the RNA for bulk RNA-seq analysis. We found that less than 1% host gene expression was significantly changed by RDV (Figure 2B). Interferon (IFN) activations are important for cellular defense against viruses.24 Increased transcript levels of IFN-activated genes were observed in SARS-CoV-2-infected cells compared to mock-infected cells (Figure S1A). However, RDV was not able to upregulate IFN-activated genes (Figure S1B), suggesting that the antiviral activity of RDV is independent of modulating the gene expression of infected host cells.
In an ideal model, cellular pEC50 equals the sum of log Fic and biochemical pIC50.12Fic’s of RDV in Vero E6, A549-ACE2, and Calu-3 cells are 0.02, 0.39, and 0.20, respectively. The corresponding predicted EC50’s are 1.35, 0.08, and 0.16 μM, respectively, which are close to the observed EC50’s (Figure 2C).20−22 Both Kp and Fic of RDV in A549-ACE2 are higher than Kp and Fic of RDV in Vero E6 and Calu-3, indicating that more RDV accumulates within the A549-ACE2 cells than others. These data suggest that RDV exhibits the strongest antiviral activity in the A549-ACE2 cells.
The intracellular unbound drug concentration depends on metabolic enzymes and transporters.16 The key metabolic enzymes of RDV are cathepsin A (CTSA) and histidine triad nucleotide-binding protein 1 (HINT1),25,26 which are both highly expressed in Vero E6, A549-ACE2, and Calu-3 cells (Figure 2D,E). RDV has been identified as a substrate of p-glycoprotein 1 (P-gp)27 and multidrug resistance-associated protein 4 (MRP4).28 P-gp is encoded by the ABCB1 gene, and MRP4 is encoded by ABCC4.29 According to RNA-seq analysis, the high expression of ABCB1 and ABCC4 was found in Vero E6 cells (Figure 2D), which was further evidenced by the qPCR analysis (Figure 2E). In our rescue assay, both verapamil (P-gp inhibitor) and ceefourin (MRP4 inhibitor) significantly increased RDV accumulation in Vero E6 cells (Figure 2F,G). Neither verapamil nor ceefourin changed RDV accumulation in A549-ACE2 cells (Figure 2F,G). We conclude here that the Fic of RDV depends on the expression of ABCB1 and ABCC4. The P-gp inhibitor verapamil showed a strong cytotoxicity in Calu-3 cells and led to the rescue assay technically unavailable.
We then applied scRNA-Seq to test whether these metabolic enzymes and transporters of RDV are highly expressed in AT2 cells. By curating public scRNA-seq data (GSE132901), we found that both Ctsa and Hint2 are highly expressed in AT2 cells of the mouse lung, indicating that there are sufficient metabolic enzymes for RDV biotransformation (Figure 2H). Meanwhile, Abcb1a and Abcb1b are barely expressed, whereas Abcc4 is highly expressed in mouse AT cells (Figure 2H). To better mimic the antiviral activity of RDV in AT2 cells, the cells used for in vitro assay should exhibit the high expression of ABCC4 but low expression of ABCB1. We therefore used verapamil to suppress ABCB1 and block the P-gp function. We took the Fic of verapamil-treated Vero E6 to predict the EC50 of RDV in mouse AT2 cells. The concentration of RDV in the mouse lung30 is lower than the predicted EC50 of RDV in mouse AT2 cells (Figure 2I and Table S1), indicating that RDV has little chance to inhibit SARS-CoV-2 replication in mice. However, RDV has been reported to successfully reduce virus titer in the lung in the SARS-CoV-2 mouse model.21 We speculated that most of RDV is transformed to its metabolite GS-441524 in the circulatory system before RDV gets into the lung.
Predicting the Anti-SARS-CoV-2 Activity of GS-441524
Since the in vivo antiviral activity of RDV may depend on the lung concentration of GS-441524, we further examined the activity of GS-441524. An intravenous (IV) injection of RDV in nonhuman primates leads to GS-441524 being present in the bloodstream at concentrations 1000-fold higher than RDV.26 It suggests that GS-441524 is the predominant metabolite in the plasma before RDV gets into the lung (Figure 3A). GS-441524 has been confirmed to inhibit SARS-CoV-2 replication with EC50 from 0.47 to 1.09 μM in Vero E6 cells (Figure 3A).21 Like RDV, GS-441524 significantly changed less than 1% host gene expression (Figure 3B), especially for IFN-activated genes (Figure S1B). These results suggest that the antiviral activity of GS-441524 is independent of modulating gene expression in Vero E6 cells. The Fic of GS-441524 in Vero E6 is 0.13, and the corresponding predicted EC50 is 0.27 μM, which is similar to the observed EC50 range from 0.47 to 1.09 μM (Figure 3C).21
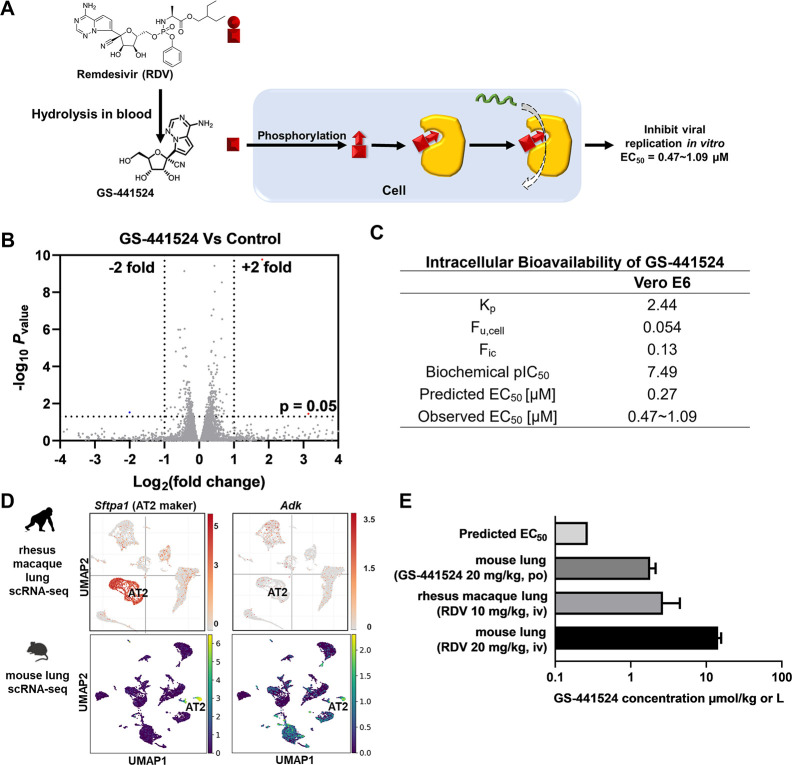
Figure 3.
Predicting the antiviral activity of GS-441524 by the corresponding intracellular bioavailability and anti-RdRp activity. (A) The ideal bioactivation of RDV predominately occurs in vivo. (B) GS-441524 did not significantly alter gene expression in Vero E6 cells. Vero E6 cells were treated with or without GS-441524 for 48 h and then collected for bulk RNA sequencing. Volcano plot of gene expression changes (n = 3 biological replicates). Significant genes were called via DESeq2 (P < 0.05 and change of >2-fold). (C) Intracellular bioavailability of GS-441524 and the corresponding predicted EC50. (D) The scRNA-seq data of the rhesus macaque lung were visualized by Single Cell Portal - Broad Institute, available from https://singlecell.broadinstitute.org. The scRNA-seq data of the mouse lung (GSE132901) were re-analyzed and visualized by Scanpy. Sftpa1 was selected as the AT2 cell marker. (E) Levels of GS-441524 in the lungs of mouse and rhesus macaque after RDV or GS-441524 administration and their comparison with the predicted in vitro EC50. iv: intravenous; po: oral administration.
Unlike RDV, GS-441524 is not a substrate of P-gp or MRP4 (Office of Clinical Pharmacology Review; U.S. Food and Drug Administration). GS-441524 is catalyzed by cellular adenylate kinase (ADK) for the conversion of its active drug GS-443902.26ADK is expressed in AT2 cells in both rhesus macaque lung and mouse lung (Figure 3D), indicating that GS-441524 can be successfully converted to its active drug. In studies of IV injection of RDV in rhesus macaques31 or mice,30 GS-441524 is present in the lungs at concentrations 10-fold higher than its predicted EC50 (Figure 3E and Table S2), which should lead to the reduction of virus titer in the lungs. This prediction is evidenced by the strong antiviral activities of RDV in both rhesus macaque and mouse.21,31 Furthermore, the lung concentration of GS-441524 is also higher than its predicted EC50 in mice via oral administration of GS-441524.32 This result indicates that the oral administration of GS-441524 should inhibit SARS-CoV-2 replication in vivo, which has been confirmed in a mouse model.20
Predicting the Anti-SARS-CoV-2 Activity of Favipiravir
Favipiravir (FAV) is another prodrug that is transformed into its active metabolite favipiravir-ribofuranosyl-5′-triphosphate (favipiravir-RTP), an RdRp inhibitor of SARS-CoV-2 with IC50 at ∼10 μM.33 FAV exhibits a mild inhibitory effect on SARS-CoV-2 replication in Vero E6 cells with EC50 at 65–400 μM (Figure 4A).34,35 In bulk RNA-seq analysis, FAV did not change the expression of IFN-activated genes (Figure S1B), suggesting that its antiviral activity is independent of modulating the immune response of the infected host cells. Interestingly, RNA-seq analysis showed that FAV reduced angiotensin-converting enzyme 2 (ACE2) expression in Vero E6 cells (Figure 4B,C), which was also evidenced by western blot (Figure 4D). SARS-CoV-2 spike protein directly binds to ACE2 and is primed by transmembrane serine protease 2 (TMPRSS2), therefore allowing the virus entry.36 The blockade of the interaction between ACE2 and spike protein or downregulation of ACE2 expression has been reported to inhibit SARS-CoV-2 replication.37 Recently, reduction of host ACE2 expression was proved to inhibit SARS-CoV-2 replication in the host cells.38 Thus, downregulation of ACE2 is likely to contribute the antiviral activity of FAV, but the mechanism remains unclear. In addition to acting as an entry inhibitor of SARS-CoV-2, FAV also exhibited a comparable inhibitory effect on virus replication in a post-infection treatment.35 All these data suggest that FAV has a good chance to inhibit SARS-CoV-2 replication through the RdRp inhibition.
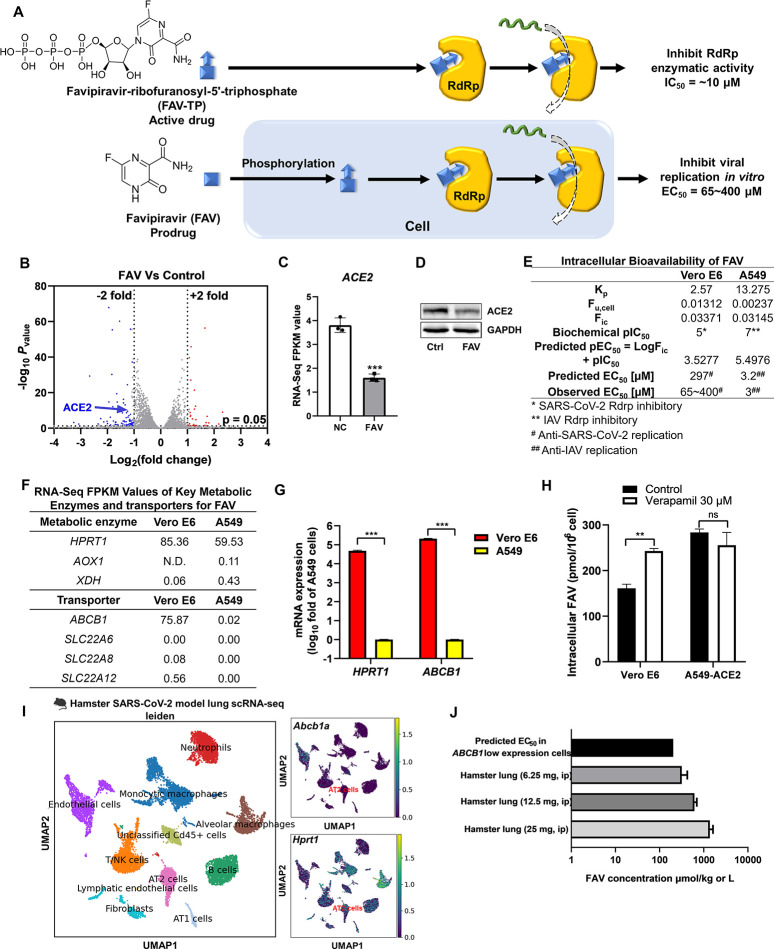
Figure 4.
Predicting the antiviral activity of FAV by the corresponding intracellular bioavailability and anti-RdRp activity. (A) The ideal bioactivation of FAV predominately occurs in vivo. (B) FAV significantly altered ACE2 expression in Vero E6 cells. Vero E6 cells were treated with or without FAV for 48 h, and mRNA was collected for bulk RNA sequencing. Volcano plot of gene expression changes (n = 3 biological replicates). Significant genes were called via DESeq2 (P < 0.05 and change of >2-fold). (C) FPKM values showed that FAV reduced ACE2 expression in Vero E6 cells. (D) The downregulation of ACE2 by FAV was confirmed by western blot. (E) Intracellular bioavailability of FAV and the corresponding predicted EC50. (F) FPKM values of key metabolic enzymes and transporters for FAV. (G) qRT-PCR showed that ABCB1 and HPRT1 were highly expressed in Vero E6 cells but not in A549 cells. Data were processed and displayed as mean ± s.e.m. (n = 3 biological replicates) according to the 2–ΔΔCt method. (H) The P-gp inhibitor improved the cellular uptake of FAV in Vero E6 cells. (I) The scRNA-seq data of the hamster lung (GSE162208) were re-analyzed and visualized by Scanpy. Sftpa1 was selected as the AT2 cell marker. (J) Levels of FAV in the lung of hamster dosed with 6.25, 12.5, and 25 mg of FAV and their comparison with the predicted in vitro EC50. ip: intraperitoneal injection.
We then tested whether the cellular antiviral EC50 of FAV can be predicted by Fic and anti-RdRp IC50. The Fic of FAV in Vero E6 cells is 0.03371, and the predicted anti-SARS-CoV-2 EC50 is 297 μM, which is comparable to the observed EC50 (65–400 μM). FAV also has been reported to inhibit influenza replication in A549 cells with EC50 at 3 μM.39 The Fic of FAV in A549 cells is 0.03145, and the predicted anti-influenza EC50 is 3.2 μM (Figure 4E). Together, we believe that the combination of Fic and biochemical IC50 can predict the antiviral activity of FAV in vitro. The key metabolic enzyme of FAV is hypoxanthine phosphoribosyltransferase 1 (HPRT1),40 which is highly expressed in Vero E6 but not in A549 cells (Figure 4F,G). Therefore, Vero E6 is more suitable than A549 cells for evaluating and predicting the antiviral effects of FAV. FAV also has been identified as a substrate of P-gp,41 and the P-gp inhibitor verapamil thus increased FAV accumulation in Vero E6 cells where ABCB1 is abundantly expressed (Figure 4G,H).
P-gp and HPRT1 have been identified as key proteins that determine the intracellular bioavailability of FAV, and we thus explored their expression profile in AT2 cells of the hamster lung. The scRNA-seq analysis shows that Abcb1a is barely expressed in AT2 cells, whereas Hprt1 is abundantly expressed in the AT2 cells of hamster (Figure 4I). We further took the Fic of verapamil-treated Vero E6 to predict the EC50 of FAV in hamster AT2 cells. The concentration of FAV in the lung34 is higher than the predicted EC50 of FAV in the AT2 cells of hamster (Figure 4J and Table S3), indicating that FAV has a good chance to inhibit SARS-CoV-2 replication in hamster. This prediction is consistent with the report that FAV leads to a reduction of infectious titers in the lung of hamster.34,42
Predicting the Anti-SARS-CoV-2 Activity of Hydroxychloroquine as an Inhibitor of Main Protease
Chloroquine (CQ) and its derivative hydroxychloroquine (HCQ) have been identified as entry inhibitors that inhibit SARS-CoV-2 replications in vitro with low micromolar EC50.43,44 However, their antiviral activities are abolished by overexpression of TMPRSS2.45,46 Notably, HCQ also has been identified as an inhibitor of Mpro of SARS-CoV-2.47 Mpro, known as 3C-like protease (3CLpro), cleaves the coronavirus polyproteins, giving nonstructural proteins (NSPs).48 The released NSPs are involved in replication and transcription of fresh virus in the host.48 Thus, the inhibition of Mpro suppresses the production of NSPs and then inhibits SARS-CoV-2 replication.49 Together, as an inhibitor of Mpro, we speculate that HCQ might have a chance to inhibit virus replication even in the TMPRSS2-overexpressed cells.
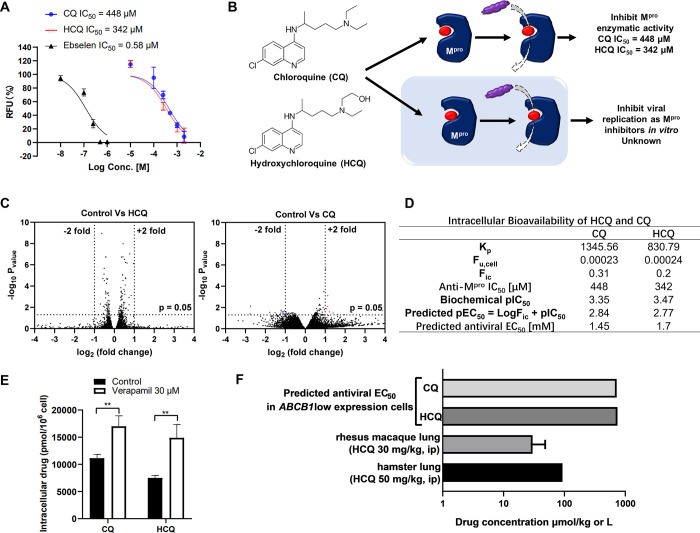
The inhibitory effect of HCQ on Mpro of SARS-CoV-2 is controversial. Li et al. have identified HCQ as a strong Mpro inhibitor with IC50 at 2.9 μM.47 However, Ma and Wang found that HCQ slightly inhibited the Mpro activities at 200 μM.50 In our assay, HCQ exhibited weak anti-Mpro activities with IC50 at 342 μM (Figure 5A), which is partially consistent with Ma and Wang’s result. CQ also inhibited Mpro activity with IC50 at 448 μM (Figure 5A). Therefore, the weak inhibitory effects of HCQ on viral Mpro might contribute to its antiviral activity in vitro (Figure 5B).
Figure 5.
Predicting the antiviral activity of HCQ and CQ by the corresponding intracellular bioavailability and anti-Mpro activity. (A) HCQ and CQ inhibited Mpro activity. The Mpro activity was evaluated by a SARS-CoV-2 Mpro inhibitor screening kit, and ebselen was used as a positive control. (B) HCQ and CQ were reported to inhibit SARS-CoV-2 replication in vitro through the inhibitory effects of Mpro. (C) Vero E6 cells were treated with or without HCQ/CQ for 48 h and then collected for bulk RNA sequencing. Volcano plot of gene expression changes (n = 3 biological replicates). Significant genes were called via DESeq2 (P < 0.05 and change of >2-fold). (D) Intracellular bioavailability of HCQ/CQ and the corresponding predicted EC50. (E) The P-gp inhibitor improved the cellular uptake of both HCQ and CQ in Vero E6 cells. (F) Levels of HCQ in the lung of hamster dosed with 50 mg/kg HCQ and in rhesus macaque dosed with 30 mg/kg HCQ and their comparison with the predicted in vitro EC50.
We next tested whether HCQ could trigger cellular antiviral responses. The bulk RNA-seq results showed that neither HCQ nor CQ significantly altered host gene expression compared to the control group (Figure 5C and Figure S1), indicating that HCQ or CQ did not inhibit virus replications through the regulation of host gene expression. To further explore whether HCQ inhibits virus replications through the inhibition of viral Mpro, we used the formula cellular pEC50 = log Fic + biochemical pIC50 to predict the antiviral EC50 of HCQ as an Mpro inhibitor. The Fic’s of CQ and HCQ in Vero E6 cells are 0.31 and 0.2, respectively, and the predicted EC50’s of CQ and HCQ are 1.45 and 1.70 mM, respectively (Figure 5D).
Since both CQ and HCQ have been identified as substrates of P-gp,51 it is not surprising that verapamil facilitated accumulation of both CQ and HCQ in Vero E6 cells (Figure 5E). We further selected the Fic of verapamil-treated Vero E6 to predict the EC50 of HCQ in the AT2 cells. If the lung concentration of HCQ can reach the predicted EC50, HCQ should have a capability to reduce virus titer in vivo even though TMPRSS2 is highly expressed in the AT2 cells. Unfortunately, HCQ lung concentrations in hamster42 or rhesus macaque52 are much lower than the predicted EC50 of HCQ in the AT2 cells (Figure 5F and Table S4), indicating that HCQ, as an Mpro inhibitor alone, cannot inhibit SARS-CoV-2 replication in vivo. This prediction is consistent with the results that HCQ failed to reduce infectious titers in lungs of hamster and rhesus macaque.42,52,53
Discussion
Insufficient target protein exposure makes drug less effective or even useless in cells, leading to failure in early drug discovery.9,10 To better evaluate the cellular activity of drugs, a quick assay has been developed to quantify the intracellular bioavailability of drugs. This assay allows to predict the drug cellular response by the formula pEC50 = pIC50 + Log Fic.12 In this study, we found that in vitro anti-SARS-CoV-2 activities of antiviral drugs can be successfully predicted by using this formula. The predicted EC50’s are close to the observed EC50’s. The workflow we developed in this study (Figure 1) enables scientists to quickly evaluate the antiviral activity of drugs without using SARS-CoV-2, which is required to be handled in a BSL-3 lab.
Additionally, we applied this strategy to predict the in vivo antiviral activity of drugs. Membrane transporters are important determinants of the intracellular bioavailability of drugs and play key roles in the drug uptake or efflux.16 In this study, we characterized the expression profiles of key transporters in both Vero E6 cells and AT2 cells. Some key transporters, P-gp for an example, are highly expressed in Vero E6 cells but barely expressed in AT2 cells. RDV, FAV, and HCQ have been identified as substrates of P-gp, suggesting that Vero E6 cells can use P-gp to pump out these antivirals. This makes antivirals accumulate more within AT2 cells than Vero E6 cells. To better mimic the transporter expression profiles of AT2 cells, we then selected specific inhibitors to block the function of these transporters of Vero E6 cells and measured the Fic of antivirals in these “AT2-like” Vero E6 cells. When the predicted EC50 of drugs in the “AT2-like” cells is lower than the lung concentrations of drugs, like FAV, the drugs are likely to reduce SARS-CoV-2 titer in vivo. Additionally, FAV was found to reduce ACE2 expression. Since ACE2 is required for coronavirus entry,37 the reduction of ACE2 expression might also contribute to the antiviral activity of FAV.
As an Mpro inhibitor, the lung concentration of HCQ is much lower than its predicted EC50. This is consistent with the report that HCQ failed to inhibit SARS-CoV-2 replication in vivo.42,52,53 SARS-CoV-2 infects cells through TMPRSS2-mediated membrane fusion or cysteine protease cathepsin L (CTSL)-mediated endocytosis.14 The spike protein of SARS-CoV-2 is activated by the endosomal pH-dependent CTSL.14 HCQ has the capability of increasing endosomal pH, therefore inhibiting CTSL-mediated endocytosis.54 Unlike CTSL, TMPRSS2-mediated cell entry is pH-independent, which cannot be blocked by HCQ.45,46 Therefore, it is not surprising that HCQ lost its anti-SARS-CoV-2 activity in vivo since TMPRSS2 is highly expressed in the AT2 cells in both hamster and rhesus macaque (Figure S2). HCQ has also been reported to inhibit virus replication through CTSL-independent pathways, including the inhibitory effect on Mpro.47 Our study suggests that as an inhibitor of Mpro, HCQ still loses its antiviral activity in vivo due to its low intracellular bioavailability in the AT2 cells.52,53,55
RDV has been reported to reduce virus titer in both mouse and rhesus macaque.21,31 However, in our study, the predicted EC50 of RDV in the AT2 cells is higher than the lung concentration of RDV in mice, suggesting that RDV is unlikely to inhibit virus replication in vivo. Nevertheless, RDV is hydrolyzed to GS-441524 in the circulatory system.26 The predicted EC50 of GS-441524 in the AT2 cells is much lower than the mouse lung concentration of GS-441524. Therefore, we speculate here that RDV reduces SARS-CoV-2 virus titer in vivo because of its metabolite GS-441512 rather than RDV itself. Together, to better evaluate the in vivo antiviral activity of prodrugs, we believe that the predominant metabolites in the plasma, instead of the prodrugs, should be tested in vitro in the preliminary screening.
Overall, our results indicate that both in vitro and in vivo antiviral activity drugs can be predicted by their intracellular bioavailability and biochemical activity without using virus. This virus-free strategy can help medicinal chemists and pharmacologists to screen antivirals during early drug discovery, regardless of whether they have high-level biosafety labs.
Materials and Methods
Compounds, Cell Culture, and Transfections
Vero E6 and A549-ACE2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin–streptomycin. In the case of Calu-3 cells, the medium was also supplemented with 1% non-essential amino acids and 1% sodium pyruvate. All cell lines were obtained from Otwo Biotech Corporation and incubated at 37 °C with 5% CO2. Lipo8000 (Beyotime, China) and pcDNA3.1-ACE2-Flag (Beyotime, China) were used for transfection. The expression of ACE2 was tested by western blot (Figure S3). Chloroquine diphosphate salt was purchased from Sigma-Aldrich (San Louis, MO, C6628; purity, 98.5–101.0%). Hydroxychloroquine sulfate was purchased from Sigma-Aldrich (San Louis, MO, 90527; purity, ≥95%). Favipiravir was purchased from Topscience (China, T6833; purity, ≥98%). Remdesivir was purchased from Cayman Chemical (Ann Arbor, MI, 30354; purity, ≥98%). GS-441524 was purchased from Selleck Chemicals (Houston, TX, S6814; purity, ≥99%). All chemicals were analyzed by HPLC (Figures S4–S8).
Western Blot Analysis
The cells were lysed in RIPA buffer (Thermo Fisher) on ice for 30 min, and the lysates were centrifuged at 12,000 rpm for 15 min. Proteins in the cell lysates were separated by 10% SDS polyacrylamide gel electrophoresis and electrotransferred to a polyvinylidene difluoride membrane. Then, the membrane was blocked in a solution of 5% nonfat milk and incubated overnight with primary antibodies against ACE2 (AF2335, Beyotime, China) or β-actin (sc-517582, Santa Cruz, US), followed by incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h. Specific bands were detected with Immobilon Chemiluminescent detection reagent (Millipore) and photographed with a Gel Doc imager (Bio-Rad, Hercules, CA).
RT-qPCR Analysis
A549-ACE2 cells were seeded in the 24-well plate (1 × 105 cell per well) and cultured for 48 h. Vero E6 and Calu-3 were seeded in the 24-well-plate (2 × 105 cell per well) and cultured for 48 h. The cells were then washed with phosphate-buffered saline (PBS) and lysed by the TRIzol reagent (Takara) according to the manufacturer’s instructions. Reverse transcription was performed by using PrimeScript RT Master Mix (TaKaRa). RT-qPCR was performed with TB Green (TaKaRa) using a LightCycler96. The sequences of primers are listed in Table S5. Data were processed and displayed according to the 2–ΔΔCT method.56
RNA-Seq and Data Analysis
A549-ACE2, Vero E6, and Calu-3 cells were seeded in the six-well plate and incubated for 48 h before the addition of drugs. Then, cells were incubated in DMEM containing antivirals for 48 h before being gathered and sent to BGI Genomics for RNA extraction and bulk mRNA sequencing. Libraries were constructed, and paired-end sequencing was performed according to the manufacturer’s instructions (Illumina, San Diego, CA, United States). The cDNA pools were loaded to an Illumina Hiseq2000 sequencer (Illumina, United States) for sequencing. Both library construction and RNA-seq were conducted at BGI.
Raw sequencing reads (fastq) were quality-checked using SOAPnuke (v1.5.2).57 Bowtie2 (v2.2.5) was applied to align the clean reads to the genomes.58 Also, expression levels of genes were calculated by RSEM (v1.2.12).59 Differential expression analysis was performed using DESeq2 (v1.4.5) with P < 0.05.60
Single-Cell Sequencing Analysis
The single-cell RNA-seq (scRNA-seq) data of the rhesus macaque lung are visualized by Single Cell Portal - Broad Institute, available from https://singlecell.broadinstitute.org.61 The scRNA-seq data of mice (GSE132901)62 and hamster (GSE162208)63 were downloaded from the public Gene Expression Omnibus (GEO) database and re-analyzed and visualized by Scanpy.64
Measurements of Intracellular Drug Accumulation
Cellular uptake was measured in A549, Vero E6, and Callu-3 cells seeded in 24-well plates. Compound solutions were diluted in HBSS from DMSO stock solutions to a final concentration according to drug’s EC50 against SARS-CoV-2 infected cells. Final concentrations of drugs are listed in Table S6. Before the experiment, cells were washed with prewarmed (37 °C) HBSS and 500 μL of compound solution was added followed by incubation at 37 °C on an orbital shaker (65 rpm). Uptake was stopped after 1 or 2 h by removing the incubated solutions of the compound. The cells were washed twice with HBSS and lysed by 500 μL of methanol, followed by 1 min ultrasonic procession. Samples were centrifuged for 15 min at 12,000 rpm. The supernatants were transferred to 1.5 mL Eppendorf tubes. Concentrations of CQ and HCQ in supernatants were quantified by measuring the absorbance in 328 nm; the concentration of FAV in supernatants was quantified by measuring the fluorescence intensity with excitation wavelength at 365 nm and emission wavelength at 430 nm. For RDV and GS-441524, concentrations were measured by a UPLC/MS/MS (Waters Xevo TQD) using an Agilent ZORBAX Eclipse Plus C18 column (100 mm × 3 mm, 1.8 μm). Chromatographic and MS conditions (Table S7) were previously described by Xie and Wang.32
The ratio between the total compound concentration in the cells and the compound concentration in the medium (Kp) at any time point or concentration was calculated according to:
where Acell is the amount of the compound in the cell lysates, and Vcell is the cell volume. By assuming cells to be spherical in shape, Vcell was calculated according to:
where n is the number of cells. A549, Vero E6, and Calu-3’s diameters were estimated to be 14, 17, and 20 μm, respectively. All experiments were performed with three to four replicates.65,66
Measurements of Intracellular Unbound Drug Fraction
Cells were harvested from T75 flasks using 0.25% trypsin–EDTA and then centrifuged at 1000 rpm for 5 min. Cell pellets were washed with HBSS and centrifuged at 1000 rpm for 5 min. After discarding the supernatant, the pellets were resuspended with HBSS to a specific concentration listed in Table S8 to achieve D = 100. These suspensions then underwent three freeze–thaw cycles to produce a cell homogenate. Dialysis was performed by using a Rapid Equilibrium Dialysis device (Thermo Fisher Scientific Inc.). One hundred microliters of cell homogenates spiked with drug was added into a homogenate chamber, and 350 μL of HBSS was added into a buffer chamber. The dialysis device was incubated at 200 rpm on an orbital shaker for 4 h at 37 °C. At the end of the incubation, 10 μL of sample from both the homogenate and buffer chamber was transferred to an Eppendorf tube containing 380 μL of methanol. Ten microliters of blank HBSS or cell homogenates was also added into this Eppendorf tube to eliminate the volume difference caused by sediments. Tubes were placed on ice for 30 min for proteins to be fully precipitated and then centrifuged at 12,000 rpm for 15 min. The concentration of drugs in the supernatant was quantified in aforementioned ways.
The unbound drug fraction in the cell homogenate (fu,hom) was calculated according to:
where Cbuffer is the compound concentration in the buffer chamber, and Chom is the compound concentration in the homogenate chamber. The unbound drug fraction in the cell (fu,cell) was calculated after correcting for homogenate dilution according to:
D was adjusted to be 100 by diluting different cells into suspensions with different concentrations. The unbound fraction of each compound was measured in triplicates.
Identification of Inhibitory Effects of CQ/HCQ against SARS-CoV-2 Mpro
SARS-CoV-2 Mpro inhibitor screening kit (P0312S, Beyotime, China) was used to measure inhibitory effects of CQ/HCQ against SARS-CoV-2 Mpro. To be specific, 592 nM enzyme, 400 μM substrate, and different concentrations of CQ/HCQ or ebselen (positive control) were added into different wells. The final concentrations of ebselen were 0.01, 0.1, 0.25, 0.5, and 1 μM. The final concentrations of HCQ/CQ were 10, 100, 250, 500, 1000, and 2000 μM. Compound solution (5 μL) was added into 47 μL of Mpro-containing buffer, and then the solutions were incubated at 37 °C for 10 min. The reaction was initiated by adding 48 μL of substrate-containing buffer. Fluorescence intensity was monitored every minute. The initial four measurements were used to calculate the slope through linear regression and converted to enzyme activity.
Cytotoxicity
Cell Counting Kit-8 (CCK8) (Dojindo, Kumamoto, Japan) was used for detection of cytotoxicity. A549, Vero E6, and Calu-3 cells were seeded in a 96-well plate and cultured for 24 h. Then, antiviral drugs then were added into a 96-well plate. After 48 h incubation, 10% CCK8 solution was added and incubated for 15 min. Absorbance was measured at 450 nm.
Statistical Analysis
The data are presented as mean ± SEM, and statistical comparisons were performed using two-sided Student’s t-test at ***P < 0.001, **P < 0.01, or *P < 0.05. RNA-seq-related statistical analysis was done with the embedded statistics in the DESeq2 package. P < 0.05 (95% confidence interval) was considered statistically significant.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.2c05376.
Antiviral drugs that do not change expression levels of IFN-activated genes; high expression of TMPRSS2 in the AT2 cells of the lung; overexpression of ACE2 in A549 cells; drug quality control by analytical HPLC; comparison of the predicted EC50 and the lung concentration of drugs; primers used in this study; final concentrations of different drugs in the accumulation assay; chromatographic and mass condition of RDV and GS-441524; specific concentration to achieve D = 100 (PDF)
Author Present Address
◆ Present address: Shanghai Institute of Materia Medica Chinese Academy of Sciences, 555 Zuchongzhi Road, Shanghai 201203, China (A.S.)
Author Present Address
○ Present address: Department of Surgery & Cancer, Faculty of Medicine, Imperial College London, South Kensington Campus, London SW7 2AZ, UK (J.Z.)
Author Contributions
∇ J.Z., M.H., and Q.X. contributed equally to this work.
Author Contributions
Y.W. and L.C. developed the concept. J.-W.Z., M.-F.H., Q.X., J.-H.L., Z.-Q.L., X.-H.H, C.H., Q.L., and B.S. conducted experimental work in cell lines and analyzed data. Y.W. and L.C. performed bioinformatic analysis. Y.W., L.C., and C.H. supervised research. Y.W. and L.C. wrote the manuscript.
This work was supported by the National Natural Science Foundation of China (81903875 and 82174033 to Y.W.), Guangdong Basic and Applied Basic Research Foundation (2020A1515011342 to Y.W.), the Shenzhen Science and Technology Innovation Fund (JCYJ20190807160601672 to Y.W.), Project for High Efficiency and Developed Hospital at Foshan City (202200087 to M.F.H.; 202000189, 202200089 to K.Y.Y.), Project of Traditional Chinese Medicine Bureau of Guangdong Province (20211367 to K.Y.Y.), and Foshan Science and Technology Innovation Project (2018AG100091 to K.Y.Y.).
The authors declare no competing financial interest.
Notes
The RNA-seq data of this study have been deposited in GEO (GSE194130).
Supplementary Material
References
- Pardi N.; Weissman D. Development of vaccines and antivirals for combating viral pandemics. Nat. Biomed. Eng. 2020, 4, 1128–1133. 10.1038/s41551-020-00658-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelly S.; Liotta D. Potent SARS-CoV-2 Direct-Acting Antivirals Provide an Important Complement to COVID-19 Vaccines. ACS Cent. Sci. 2021, 7, 396–399. 10.1021/acscentsci.1c00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist S. R.; Schäfer A.; Martinez D. R. Cell and animal models of SARS-CoV-2 pathogenesis and immunity. Dis. Model. Mech. 2020, 13, dmm046581. 10.1242/dmm.046581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz-Fontela C.; Dowling W. E.; Funnell S. G. P.; Gsell P. S.; Riveros-Balta A. X.; Albrecht R. A.; Andersen H.; Baric R. S.; Carroll M. W.; Cavaleri M.; Qin C.; Crozier I.; Dallmeier K.; De Waal L.; De Wit E.; Delang L.; Dohm E.; Duprex W. P.; Falzarano D.; Finch C. L.; Frieman M. B.; Graham B. S.; Gralinski L. E.; Guilfoyle K.; Haagmans B. L.; Hamilton G. A.; Hartman A. L.; Herfst S.; Kaptein S. J. F.; Klimstra W. B.; Knezevic I.; Krause P. R.; Kuhn J. H.; Le Grand R.; Lewis M. G.; Liu W. C.; Maisonnasse P.; McElroy A. K.; Munster V.; Oreshkova N.; Rasmussen A. L.; Rocha-Pereira J.; Rockx B.; Rodriguez E.; Rogers T. F.; Salguero F. J.; Schotsaert M.; Stittelaar K. J.; Thibaut H. J.; Tseng C. T.; Vergara-Alert J.; Beer M.; Brasel T.; Chan J. F. W.; Garcia-Sastre A.; Neyts J.; Perlman S.; Reed D. S.; Richt J. A.; Roy C. J.; Segales J.; Vasan S. S.; Henao-Restrepo A. M.; Barouch D. H. Animal models for COVID-19. Nature 2020, 586, 509–515. 10.1038/s41586-020-2787-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loibner M.; Langner C.; Regitnig P.; Gorkiewicz G.; Zatloukal K. Biosafety Requirements for Autopsies of Patients with COVID-19: Example of a BSL-3 Autopsy Facility Designed for Highly Pathogenic Agents. Pathobiology 2021, 88, 37–45. 10.1159/000513438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino B.; de Lamballerie X.; Charrel R. Biosafety and Biosecurity in European Containment Level 3 Laboratories: Focus on French Recent Progress and Essential Requirements. Front. Public Health 2017, 5, 121. 10.3389/fpubh.2017.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson C. S.; Chibale K.; Goss R. J. M.; Jaspars M.; Newman D. J.; Dorrington R. A. Antiviral drug discovery: preparing for the next pandemic. Chem. Soc. Rev. 2021, 50, 3647–3655. 10.1039/D0CS01118E. [DOI] [PubMed] [Google Scholar]
- Dolgin E. The race for antiviral drugs to beat COVID - and the next pandemic. Nature 2021, 592, 340–343. 10.1038/d41586-021-00958-4. [DOI] [PubMed] [Google Scholar]
- Hann M. M.; Simpson G. L. Intracellular drug concentration and disposition--the missing link?. Methods 2014, 68, 283–285. 10.1016/j.ymeth.2014.05.009. [DOI] [PubMed] [Google Scholar]
- Swinney D. C. Biochemical mechanisms of drug action: what does it take for success?. Nat. Rev. Drug Discovery 2004, 3, 801–808. 10.1038/nrd1500. [DOI] [PubMed] [Google Scholar]
- Mateus A.; Matsson P.; Artursson P. Rapid measurement of intracellular unbound drug concentrations. Mol. Pharmaceutics 2013, 10, 2467–2478. 10.1021/mp4000822. [DOI] [PubMed] [Google Scholar]
- Mateus A.; Gordon L. J.; Wayne G. J.; Almqvist H.; Axelsson H.; Seashore-Ludlow B.; Treyer A.; Matsson P.; Lundbäck T.; West A.; Hann M. M.; Artursson P. Prediction of intracellular exposure bridges the gap between target- and cell-based drug discovery. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E6231–E6239. 10.1073/pnas.1701848114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulay A.; Konda B.; Garcia G. Jr.; Yao C.; Beil S.; Villalba J. M.; Koziol C.; Sen C.; Purkayastha A.; Kolls J. K.; Pociask D. A.; Pessina P.; de Aja J. S.; Garcia-de-Alba C.; Kim C. F.; Gomperts B.; Arumugaswami V.; Stripp B. R. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 2021, 35, 109055 10.1016/j.celrep.2021.109055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Chen L. Tissue distributions of antiviral drugs affect their capabilities of reducing viral loads in COVID-19 treatment. Eur. J. Pharmacol. 2020, 889, 173634 10.1016/j.ejphar.2020.173634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Chen L. Lung tissue distribution of drugs as a key factor for COVID-19 treatment. Br. J. Pharmacol. 2020, 177, 4995–4996. 10.1111/bph.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateus A.; Treyer A.; Wegler C.; Karlgren M.; Matsson P.; Artursson P. Intracellular drug bioavailability: a new predictor of system dependent drug disposition. Sci. Rep. 2017, 7, 43047. 10.1038/srep43047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon C. J.; Tchesnokov E. P.; Woolner E.; Perry J. K.; Feng J. Y.; Porter D. P.; Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020, 295, 6785–6797. 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y.; Yan L.; Huang Y.; Liu F.; Zhao Y.; Cao L.; Wang T.; Sun Q.; Ming Z.; Zhang L.; Ge J.; Zheng L.; Zhang Y.; Wang H.; Zhu Y.; Zhu C.; Hu T.; Hua T.; Zhang B.; Yang X.; Li J.; Yang H.; Liu Z.; Xu W.; Guddat L. W.; Wang Q.; Lou Z.; Rao Z. Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 2020, 368, 779–782. 10.1126/science.abb7498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F.; Khan K. S.; Le T. K.; Paris C.; Demirbag S.; Barfuss P.; Rocchi P.; Ng W. L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell 2020, 80, 1136–1138. 10.1016/j.molcel.2020.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Cao L.; Li G.; Cong F.; Li Y.; Sun J.; Luo Y.; Chen G.; Li G.; Wang P.; Xing F.; Ji Y.; Zhao J.; Zhang Y.; Guo D.; Zhang X. Remdesivir Metabolite GS-441524 Effectively Inhibits SARS-CoV-2 Infection in Mouse Models. J. Med. Chem. 2022, 2785. 10.1021/acs.jmedchem.0c01929. [DOI] [PubMed] [Google Scholar]
- Pruijssers A. J.; George A. S.; Schafer A.; Leist S. R.; Gralinksi L. E.; Dinnon K. H. III; Yount B. L.; Agostini M. L.; Stevens L. J.; Chappell J. D.; Lu X.; Hughes T. M.; Gully K.; Martinez D. R.; Brown A. J.; Graham R. L.; Perry J. K.; Du Pont V.; Pitts J.; Ma B.; Babusis D.; Murakami E.; Feng J. Y.; Bilello J. P.; Porter D. P.; Cihlar T.; Baric R. S.; Denison M. R.; Sheahan T. P. Remdesivir Inhibits SARS-CoV-2 in Human Lung Cells and Chimeric SARS-CoV Expressing the SARS-CoV-2 RNA Polymerase in Mice. Cell Rep. 2020, 32, 107940 10.1016/j.celrep.2020.107940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihn S. J.; Merits A.; Bakshi S.; Turnbull M. L.; Wickenhagen A.; Alexander A. J. T.; Baillie C.; Brennan B.; Brown F.; Brunker K.; Bryden S. R.; Burness K. A.; Carmichael S.; Cole S. J.; Cowton V. M.; Davies P.; Davis C.; De Lorenzo G.; Donald C. L.; Dorward M.; Dunlop J. I.; Elliott M.; Fares M.; Da Silva Filipe A.; Freitas J. R.; Furnon W.; Gestuveo R. J.; Geyer A.; Giesel D.; Goldfarb D. M.; Goodman N.; Gunson R.; Hastie C. J.; Herder V.; Hughes J.; Johnson C.; Johnson N.; Kohl A.; Kerr K.; Leech H.; Lello L. S.; Li K.; Lieber G.; Liu X.; Lingala R.; Loney C.; Mair D.; McElwee M. J.; McFarlane S.; Nichols J.; Nomikou K.; Orr A.; Orton R. J.; Palmarini M.; Parr Y. A.; Pinto R. M.; Raggett S.; Reid E.; Robertson D. L.; Royle J.; Cameron-Ruiz N.; Shepherd J. G.; Smollett K.; Stewart D. G.; Stewart M.; Sugrue E.; Szemiel A. M.; Taggart A.; Thomson E. C.; Tong L.; Torrie L. S.; Toth R.; Varjak M.; Wang S.; Wilkinson S. G.; Wyatt P. G.; Zusinaite E.; Alessi D. R.; Patel A. H.; Zaid A.; Wilson S. J.; Mahalingam S. A plasmid DNA-launched SARS-CoV-2 reverse genetics system and coronavirus toolkit for COVID-19 research. PLoS Biol. 2021, 19, e3001091 10.1371/journal.pbio.3001091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayak M. K.; Agrawal A. S.; Bose S.; Naskar S.; Bhowmick R.; Chakrabarti S.; Sarkar S.; Chawla-Sarkar M. Antiviral activity of baicalin against influenza virus H1N1-pdm09 is due to modulation of NS1-mediated cellular innate immune responses. J. Antimicrob. Chemother. 2014, 69, 1298–1310. 10.1093/jac/dkt534. [DOI] [PubMed] [Google Scholar]
- McNab F.; Mayer-Barber K.; Sher A.; Wack A.; O’Garra A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. 10.1038/nri3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R.; Liclican A.; Xu Y.; Pitts J.; Niu C.; Zhang J.; Kim C.; Zhao X.; Soohoo D.; Babusis D.; Yue Q.; Ma B.; Murray B. P.; Subramanian R.; Xie X.; Zou J.; Bilello J. P.; Li L.; Schultz B. E.; Sakowicz R.; Smith B. J.; Shi P. Y.; Murakami E.; Feng J. Y. Key Metabolic Enzymes Involved in Remdesivir Activation in Human Lung Cells. Antimicrob. Agents Chemother. 2021, 65, e0060221 10.1128/AAC.00602-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan V. C.; Muller F. L. Advantages of the Parent Nucleoside GS-441524 over Remdesivir for Covid-19 Treatment. ACS Med. Chem. Lett. 2020, 11, 1361–1366. 10.1021/acsmedchemlett.0c00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegwater E.; Strik A.; Wilms E. B.; Bosma L. B. E.; Burger D. M.; Ottens T. H.; van Nieuwkoop C. Drug-induced Liver Injury in a Patient With Coronavirus Disease 2019: Potential Interaction of Remdesivir With P-Glycoprotein Inhibitors. Clin. Infect. Dis. 2021, 72, 1256–1258. 10.1093/cid/ciaa883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang K. What Do We Know About Remdesivir Drug Interactions?. Clin. Transl. Sci. 2020, 13, 842–844. 10.1111/cts.12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milosevic G.; Kotur N.; Krstovski N.; Lazic J.; Zukic B.; Stankovic B.; Janic D.; Katsila T.; Patrinos G. P.; Pavlovic S.; Dokmanovic L. Variants in TPMT, ITPA, ABCC4 and ABCB1 Genes As Predictors of 6-mercaptopurine Induced Toxicity in Children with Acute Lymphoblastic Leukemia. J. Med. Biochem. 2018, 37, 320–327. 10.1515/jomb-2017-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W. J.; Chang L.; Yang Y.; Wang X.; Xie Y. C.; Shen J. S.; Tan B.; Liu J. Pharmacokinetics and tissue distribution of remdesivir and its metabolites nucleotide monophosphate, nucleotide triphosphate, and nucleoside in mice. Acta Pharmacol. Sin. 2021, 42, 1195–1200. 10.1038/s41401-020-00537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson B. N.; Feldmann F.; Schwarz B.; Meade-White K.; Porter D. P.; Schulz J.; van Doremalen N.; Leighton I.; Yinda C. K.; Perez-Perez L.; Okumura A.; Lovaglio J.; Hanley P. W.; Saturday G.; Bosio C. M.; Anzick S.; Barbian K.; Cihlar T.; Martens C.; Scott D. P.; Munster V. J.; de Wit E. Clinical benefit of remdesivir in rhesus macaques infected with SARS-CoV-2. Nature 2020, 585, 273–276. 10.1038/s41586-020-2423-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J.; Wang Z. Can remdesivir and its parent nucleoside GS-441524 be potential oral drugs? An in vitro and in vivo DMPK assessment. Acta Pharm. Sin. B 2021, 11, 1607–1616. 10.1016/j.apsb.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naydenova K.; Muir K. W.; Wu L. F.; Zhang Z.; Coscia F.; Peet M. J.; Castro-Hartmann P.; Qian P.; Sader K.; Dent K.; Kimanius D.; Sutherland J. D.; Lowe J.; Barford D.; Russo C. J. Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2021946118 10.1073/pnas.2021946118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driouich J. S.; Cochin M.; Lingas G.; Moureau G.; Touret F.; Petit P. R.; Piorkowski G.; Barthelemy K.; Laprie C.; Coutard B.; Guedj J.; de Lamballerie X.; Solas C.; Nougairede A. Favipiravir antiviral efficacy against SARS-CoV-2 in a hamster model. Nat. Commun. 2021, 12, 1735. 10.1038/s41467-021-21992-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Cao R.; Zhang L.; Yang X.; Liu J.; Xu M.; Shi Z.; Hu Z.; Zhong W.; Xiao G. Remdesivir and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020, 30, 269–271. 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Kleine-Weber H.; Schroeder S.; Kruger N.; Herrler T.; Erichsen S.; Schiergens T. S.; Herrler G.; Wu N. H.; Nitsche A.; Muller M. A.; Drosten C.; Pohlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280. 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Xiang R.; Huo S.; Zhou Y.; Jiang S.; Wang Q.; Yu F. Molecular mechanism of interaction between SARS-CoV-2 and host cells and interventional therapy. Signal Transduction Targeted Ther. 2021, 6, 233. 10.1038/s41392-021-00653-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samelson A. J.; Tran Q. D.; Robinot R.; Carrau L.; Rezelj V. V.; Kain A. M.; Chen M.; Ramadoss G. N.; Guo X.; Lim S. A.; Lui I.; Nunez J. K.; Rockwood S. J.; Wang J.; Liu N.; Carlson-Stevermer J.; Oki J.; Maures T.; Holden K.; Weissman J. S.; Wells J. A.; Conklin B. R.; TenOever B. R.; Chakrabarti L. A.; Vignuzzi M.; Tian R.; Kampmann M. BRD2 inhibition blocks SARS-CoV-2 infection by reducing transcription of the host cell receptor ACE2. Nat. Cell Biol. 2022, 24, 24–34. 10.1038/s41556-021-00821-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta Y.; Takahashi K.; Fukuda Y.; Kuno M.; Kamiyama T.; Kozaki K.; Nomura N.; Egawa H.; Minami S.; Watanabe Y.; Narita H.; Shiraki K. In vitro and in vivo activities of anti-influenza virus compound T-705. Antimicrob. Agents Chemother. 2002, 46, 977–981. 10.1128/AAC.46.4.977-981.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naesens L.; Guddat L. W.; Keough D. T.; van Kuilenburg A. B.; Meijer J.; Vande Voorde J.; Balzarini J. Role of human hypoxanthine guanine phosphoribosyltransferase in activation of the antiviral agent T-705 (favipiravir). Mol. Pharmacol. 2013, 84, 615–629. 10.1124/mol.113.087247. [DOI] [PubMed] [Google Scholar]
- Ambrus C.; Bakos E.; Sarkadi B.; Ozvegy-Laczka C.; Telbisz A. Interactions of anti-COVID-19 drug candidates with hepatic transporters may cause liver toxicity and affect pharmacokinetics. Sci. Rep. 2021, 11, 17810. 10.1038/s41598-021-97160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaptein S. J. F.; Jacobs S.; Langendries L.; Seldeslachts L.; Ter Horst S.; Liesenborghs L.; Hens B.; Vergote V.; Heylen E.; Barthelemy K.; Maas E.; De Keyzer C.; Bervoets L.; Rymenants J.; Van Buyten T.; Zhang X.; Abdelnabi R.; Pang J.; Williams R.; Thibaut H. J.; Dallmeier K.; Boudewijns R.; Wouters J.; Augustijns P.; Verougstraete N.; Cawthorne C.; Breuer J.; Solas C.; Weynand B.; Annaert P.; Spriet I.; Vande Velde G.; Neyts J.; Rocha-Pereira J.; Delang L. Favipiravir at high doses has potent antiviral activity in SARS-CoV-2-infected hamsters, whereas hydroxychloroquine lacks activity. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 26955–26965. 10.1073/pnas.2014441117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Cao R.; Xu M.; Wang X.; Zhang H.; Hu H.; Li Y.; Hu Z.; Zhong W.; Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discovery 2020, 6, 16. 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.; Ye F.; Zhang M.; Cui C.; Huang B.; Niu P.; Liu X.; Zhao L.; Dong E.; Song C.; Zhan S.; Lu R.; Li H.; Tan W.; Liu D. In Vitro Antiviral Activity and Projection of Optimized Dosing Design of Hydroxychloroquine for the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Clin. Infect. Dis 2020, 71, 732–739. 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M.; Mosbauer K.; Hofmann-Winkler H.; Kaul A.; Kleine-Weber H.; Kruger N.; Gassen N. C.; Müller M. A.; Drosten C.; Pöhlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature 2020, 585, 588–590. 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- Ou T.; Mou H.; Zhang L.; Ojha A.; Choe H.; Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021, 17, e1009212 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z.; Li X.; Huang Y. Y.; Wu Y.; Liu R.; Zhou L.; Lin Y.; Wu D.; Zhang L.; Liu H.; Xu X.; Yu K.; Zhang Y.; Cui J.; Zhan C. G.; Wang X.; Luo H. B. Identify potent SARS-CoV-2 main protease inhibitors via accelerated free energy perturbation-based virtual screening of existing drugs. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 27381–27387. 10.1073/pnas.2010470117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- V’Kovski P.; Kratzel A.; Steiner S.; Stalder H.; Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol 2021, 19, 155–170. 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amporndanai K.; Meng X.; Shang W.; Jin Z.; Rogers M.; Zhao Y.; Rao Z.; Liu Z.-J.; Yang H.; Zhang L.; O’Neill P. M.; Samar Hasnain S. Inhibition mechanism of SARS-CoV-2 main protease by ebselen and its derivatives. Nat. Commun. 2021, 12, 3061. 10.1038/s41467-021-23313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.; Wang J. Dipyridamole, chloroquine, montelukast sodium, candesartan, oxytetracycline, and atazanavir are not SARS-CoV-2 main protease inhibitors. Proc. Natl. Acad. Sci. U. S. A. 2021, 118, e2024420118 10.1073/pnas.2024420118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J.; Bajraktari-Sylejmani G.; Haefeli W. E. Interaction of Hydroxychloroquine with Pharmacokinetically Important Drug Transporters. Pharmaceutics 2020, 12, 919. 10.3390/pharmaceutics12100919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonnasse P.; Guedj J.; Contreras V.; Behillil S.; Solas C.; Marlin R.; Naninck T.; Pizzorno A.; Lemaitre J.; Goncalves A.; Kahlaoui N.; Terrier O.; Fang R. H. T.; Enouf V.; Dereuddre-Bosquet N.; Brisebarre A.; Touret F.; Chapon C.; Hoen B.; Lina B.; Calatrava M. R.; van der Werf S.; de Lamballerie X.; Le Grand R. Hydroxychloroquine use against SARS-CoV-2 infection in non-human primates. Nature 2020, 585, 584–587. 10.1038/s41586-020-2558-4. [DOI] [PubMed] [Google Scholar]
- Rosenke K.; Jarvis M. A.; Feldmann F.; Schwarz B.; Okumura A.; Lovaglio J.; Saturday G.; Hanley P. W.; Meade-White K.; Williamson B. N.; Hansen F.; Perez-Perez L.; Leventhal S.; Tang-Huau T. L.; Callison J.; Haddock E.; Stromberg K. A.; Scott D.; Sewell G.; Bosio C. M.; Hawman D.; de Wit E.; Feldmann H., Hydroxychloroquine prophylaxis and treatment is ineffective in macaque and hamster SARS-CoV-2 disease models. JCI Insight 2020,5 (), 10.1172/jci.insight.143174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes C. P.; Fernandes D. E.; Casimiro F.; da Mata G. F.; Passos M. T.; Varela P.; Mastroianni-Kirsztajn G.; Pesquero J. B. Cathepsin L in COVID-19: From Pharmacological Evidences to Genetics. Front. Cell Infect. Microbiol. 2020, 10, 589505 10.3389/fcimb.2020.589505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A.; Duclos G.; Pastene B.; Bezulier K.; Guilhaumou R.; Solas C.; Zieleskiewicz L.; Leone M. Effects of Hydroxychloroquine on Covid-19 in Intensive Care Unit Patients: Preliminary Results. Int. J. Antimicrob. Agents 2020, 56, 106136 10.1016/j.ijantimicag.2020.106136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J.; Schmittgen T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Li R.; Li Y.; Kristiansen K.; Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics 2008, 24, 713–714. 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Langmead B.; Salzberg S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B.; Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011, 12, 323. 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I.; Huber W.; Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Han X. W.; Liu C.; Volpe G.; Wang Z.; Pan T.; Yuan Y.; Lei Y.; Lai Y.; Ward C.; Yu Y.; Wang M.; Shi Q.; Wu T.; Wu L.; Liu Y.; Wang C.; Zhang Y.; Sun H.; Yu H.; Zhuang Z.; Tang T.; Huang Y.; Lu H.; Xu L.; Xu J.; Cheng M.; Liu Y.; Wong C. W.; Tan T.; Ji W.; Maxwell P. H.; Yang H.; Wang J.; Zhu S.; Liu S.; Xu X.; Hou Y.; Esteban M. A.; Liu L.. South China Greater Bay Area-Single Cell Consortium, Single-cell atlas of a non-human primate reveals new pathogenic mechanisms of COVID-19. bioRxiv 2020. [Google Scholar]
- Kimmel J. C.; Penland L.; Rubinstein N. D.; Hendrickson D. G.; Kelley D. R.; Rosenthal A. Z. Murine single-cell RNA-seq reveals cell-identity- and tissue-specific trajectories of aging. Genome Res. 2019, 29, 2088–2103. 10.1101/gr.253880.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouailles G.; Wyler E.; Pennitz P.; Postmus D.; Vladimirova D.; Kazmierski J.; Pott F.; Dietert K.; Muelleder M.; Farztdinov V.; Obermayer B.; Wienhold S. M.; Andreotti S.; Hoefler T.; Sawitzki B.; Drosten C.; Sander L. E.; Suttorp N.; Ralser M.; Beule D.; Gruber A. D.; Goffinet C.; Landthaler M.; Trimpert J.; Witzenrath M. Temporal omics analysis in Syrian hamsters unravel cellular effector responses to moderate COVID-19. Nat. Commun. 2021, 12, 4869. 10.1038/s41467-021-25030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F. A.; Angerer P.; Theis F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018, 19, 15. 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R. D.; Shen H.; Piao Y. J. The morphometrical analysis on the ultrastructure of A549 cells. Rom. J. Morphol. Embryol. 2010, 51, 663–667. [PubMed] [Google Scholar]
- Kreft M. E.; Jerman U. D.; Lasic E.; Hevir-Kene N.; Rizner T. L.; Peternel L.; Kristan K. The characterization of the human cell line Calu-3 under different culture conditions and its use as an optimized in vitro model to investigate bronchial epithelial function. Eur. J. Pharm. Sci. 2015, 69, 1–9. 10.1016/j.ejps.2014.12.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.