Abstract
Epigenetic gene regulatory mechanisms play a central role in the biological control of cell and tissue structure, function, and phenotype. Identification of epigenetic dysregulation in cancer provides mechanistic into tumor initiation and progression and may prove valuable for a variety of clinical applications. We present an overview of epigenetically driven mechanisms that are obligatory for physiological regulation and parameters of epigenetic control that are modified in tumor cells. The interrelationship between nuclear structure and function are not mutually exclusive and are instead synergistic. We explore concepts influencing the maintenance of chromatin structures including phase separation, recognition signals, factors that mediate enhancer-promoter looping and insulation and how these are altered in during the cell cycle and in cancer. Understanding how these processes are altered in cancer provides potential for advancing capabilities for diagnosis and identification of novel therapeutic targets.
Keywords: Chromatin, Nuclear Structure, Epigenetic Control, Transcription, Nucleosomes, Spatial Transcriptomics, Cell Cycle Control, Histones, Tumor Suppression, Non-coding RNAs
An epigenetic perspective of biological control and pathology
A detailed understanding of the complex mechanisms that govern physiological gene expression states is rapidly evolving. Collectively, mechanisms that regulate chromatin structure, including the posttranslational modification of histone proteins (histone PTMs) DNA methylation, transcription factor (TF) binding, and chromatin accessibility, have been classified as so-called epigenetic gene regulatory mechanisms [1]. The plasticity of epigenetic mechanisms that intersect to regulate physiological responses to intrinsic and extrinsic cues (i.e. intracellular signaling, or interactions between specialized cells and the tissue microenvironment, respectively) have been demonstrated to play central roles in a variety of biological processes including, tissue remodeling, metabolism, cell growth, cell cycle, and cell survival as well as competency to establish and sustain the integrity of phenotype-specific requirements for cell and tissue structure and function [2-6].
Alterations in epigenetic mechanisms are increasingly recognized to drive the departure of cells from their specialized properties. Bidirectional epigenetic crosstalk between cancer cells and the tumor microenvironment are decisive for tumor onset and progression. Evidence is accruing for aberrant epigenetic control as key for tumor initiation, progression, and maintenance of the cancer-compromised phenotype [7, 8].
Nuclear structure-gene expression relationships are increasingly considered complementary dimensions to control. There is growing appreciation for the requirement to mechanistically understand the organization, assembly, and localization of regulatory complexes within the context of nuclear architecture [9, 10]. Intranuclear trafficking and phase condensates with requirements for regulatory determinants of transcription factors are being investigated to understand gene expression from a structure-function perspective [11-14]. Nuclear structure and function are no longer considered mutually exclusive and are instead synergistic. Nuclear structure mediates the physiological expression of genes, while the functional molecular mechanisms by which genes are expressed generate dynamic and phase-separated multi-dimensional mega-dalton protein/DNA/RNA complexes. Both processes continuously and synergistically redefine the components of nuclear architecture as a sophisticated factor that generates and responds to external demands for cell-based activities.
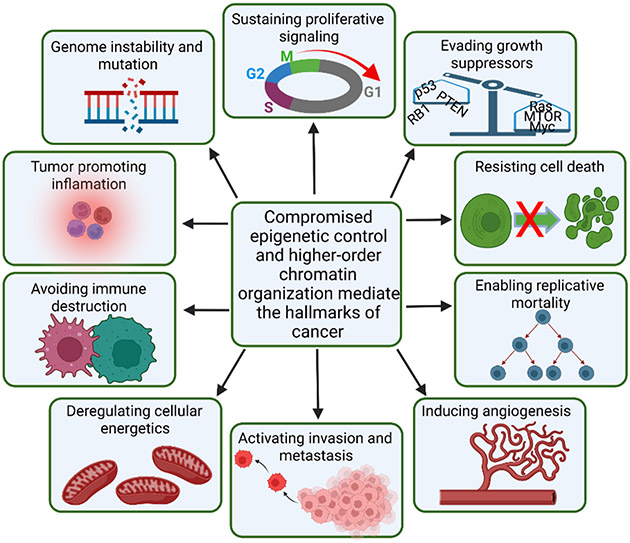
There is increasing recognition that perturbations in parameters of epigenetic control are functionally linked to the determinants of cancer that were identified by Hanahan and Weinberg [15] (Figure 1). A guiding concept is emerging that cancer is predominantly initiated by genetic modifications while predisposition and progression are epigenetically dominated. Clinically, epigenetic pathways provide therapeutic targets because the factors and enzymes that mediate epigenetics have active sites amenable to blockage by small molecules. Furthermore, epigenetic events are capable of priming and enhancing the responsiveness of treatment-refractory tumors to radiation, cytotoxic chemotherapy, and immunotherapy by lowering kinetic barriers to incoming cues emanating from signaling pathways [16-18].
Figure 1. Compromised epigenetic control and higher-order chromatin organization mediate the hallmarks of cancer.
Higher-order chromatin organization supports the gene expression required for cellular identity. Interconnected with the chromosome conformation are the epigenetic states of chromatin that are critical for this genomic regulation. Dysregulation of these chromatin states in the cancer-compromised genome drive all the hallmarks of cancer including sustained proliferation, evasion of growth suppressors, resistance to cell death, enabling of replicative immortality, induction of angiogenesis, deregulation of cellular energetics, avoidance of immune destruction, tumor promoting inflammation, and genome instability and mutation.
The overarching theme of this chapter is to highlight contributions of epigenetic mechanisms that alter gene expression leading to malignant transformation in normal cells. Understanding how these fundamental processes are altered during tumorigenesis will provide a potential for improved cancer diagnosis and identification of novel therapeutics. For the purposes of this chapter we will critically discuss: 1) DNA and histone post-translation modifications and their involvement in tumorigenesis; 2) maintenance of chromatin structures including chromatin fibers, topologically associating domains and compartments and their potential roles in disease states; 3) epigenetic and topological domains that are associated with distinct phases of the cell cycle that may be altered in cancer progression and 4) the requirement for mitotic bookmarking to maintain intranuclear localization of transcriptional regulatory machinery to reinforce cell identity throughout the cell cycle to prevent malignant transformation.
The human epigenome is organized in hierarchical, yet interdependent levels.
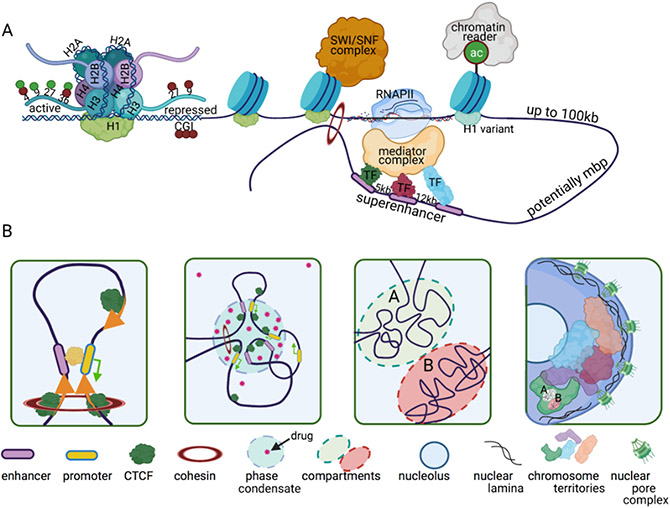
The structure and function of the human genome are integrated to support obligatory parameters of biological control including gene activation or suppression, DNA replication and repair, cell survival and fidelity of cell division[7, 9, 19-31]. Approximately two meters of DNA are contained and organized within the human cell nucleus with an average diameter of 10 micrometers. Proper organization of this length of DNA thus requires a scaling factor of up to five orders of magnitude to fit the genome into the confines of the nucleus. Scaling is primarily mediated by the sequestration of DNA into nucleosomes, but as such packaging of DNA affords gene regulation by altering steric access to genes with the nuclear space. Therefore, the linear DNA sequence of the genome conveys vital but only a limited component of the regulatory information required for physiological control. A hierarchy of interrelated structural and epigenetic information including, DNA methylation, nucleosomes to chromatin fibers, long-range chromosome interactions, topologically associating domains, chromosome territories-- is not encoded within the DNA sequence, but is superimposed on this basic blueprint thereby supporting biological activity (figure 2). Although nuclear compartments are not subdivided by membranes, the regulatory machinery for the various functions carried out by these regulatory compartments that include transcription, splicing, replication, and repair are architecturally organized in nuclear microenvironments that represent phase separated domains (discussed below). Many aspects of these levels of epigenetic and architectural control are compromised during cancer initiation and progression [2, 7, 9, 11, 22-24, 32-76]. While mutations at the level of the DNA sequence are often causative for disease pathology, epigenetic abnormalities are frequently involved in predisposition to cancer and tumor progression. While mutations are difficult to reverse, epigenetic alterations provide more tractable targets and in fact a number of pharmaceutical inhibitors of epigenetic processes have been examined in clinical trials and some have already become standards of care. A comprehensive understanding of the epigenetic alterations in cancer-compromised nuclei will inform enhanced capabilities for tumor diagnosis, prognosis, indications for recurrence, and options for targeted therapy and precision oncology [2, 7, 9, 53].
Figure 2. The human epigenome is organized in hierarchical, yet interdependent levels.
A hierarchy of interrelated structural and epigenetic information is not encoded within the DNA sequence, but is superimposed on this basic blueprint thereby supporting biological activity. A) PTMs of histones within active or repressed chromatin are depicted. While red circles indicate methylation, green circles indicate acetylation. DNA is methylated at CPG islands (CGI). A chromatin loop is depicted wherein a super-enhancer interacts with a promoter. B) A chromatin loop is depicted in which orange arrows indicate the directionality of CTCF motifs. At two convergent motifs a loop is formed. This loop is present in a topologically associating domain (TAD) within a phase condensate (blue circle). Anti-cancer drugs are concentrated within these phase condensates (red stars). These TADs coalesce into active A (green circle) or inactive B (red circle) compartments. At the highest-level compartments are contained within chromosome territories in the interphase nucleus. The chromatin interacts with the nuclear lamina at the nuclear periphery and at the around the nucleoli where heterochromatin in preferentially localized. Although nuclear compartments are not subdivided by membranes, the regulatory machinery for the various functions carried out by these regulatory compartments that include transcription, splicing, replication, and repair are architecturally organized in nuclear microenvironments.
Epigenetic modifications establishing lineage specification are crucial points of alterations during tumorigenesis.
DNA methylation in normal cells and dysregulation in the cancer-compromised genome.
DNA methylation of cytosine residue occurs at regions of chromatin containing CpG dinucleotides, and large repeat regions of CpGs are defined as “CpG islands” (CGI; Figure 2; reviewed extensively in [77-80]). In general, DNA methylation is associated with long term transcriptional silencing and wide-spread chromatin inactivation (e.g., X-chromosome inactivation[81], imprinting[82] and silencing of transposable genomic elements [79, 83]. The regulation of DNA methylation can be characterized by three distinct phases: establishment, maintenance, and demethylation. In humans, the methylation of DNA is established by two DNA methyltransferase enzymes, DNMT3A and DNMT3B, which contain two chromatin reading domains ATRX-DNMT3-DNMT3L (ADD) and PWWP and an MTase domain [79]. DNA methylation is maintained during replication by DNMT1 recruited by the E3 ubiquitin-protein ligase UHRF1 which binds to hemimethylated CpGs at replication forks through its SET and RING associated domains. Demethylation of DNA can occur passively, where non-maintained CpG methylation is diluted during successive rounds of DNA replication or can occur actively through the enzymatic activities of three ‘ten-eleven translocation’ (TET)-dioxygenases that mediate the Vitamin C dependent hydroxylation of methylcytosine: TET1, TET2 and TET3 [84, 85]. DNA methylation at gene promoter or enhancer regions has been demonstrated to affect the binding of transcriptional regulators and other DNA binding proteins through altered affinity to their consensus binding motifs [86]. In addition, methyl-CpG-binding domain (MBD) proteins interact directly with methylated CpGs and are bound by several zinc finger domains to contribute to DNA-methylation-based silencing [87]. Furthermore, 5-hydroxymethylation of cytosine (5hmC) is recognized as a highly relevant dynamic epigenetic mark on DNA [88]. Stringent control of DNA methylation and hydroxymethylation is therefore critical for development, to establish and sustain gene expression cell-type specific function, and required to support tissue remodeling.
Abnormal DNA methylation in cancer, mediated by compromised control of the methylation regulatory machinery, results in hypermethylation of CpG sites regulating several tumor suppressor genes (e.g. BRCA1, MLH1, MORT) [89]. The microsatellite landscape is another parameter of genome structure and function influenced by DNA methylation. For example, in Lynch Syndrome, it is known that mutations in the mismatch repair genes such as MLH1 result in microsatellite instability [89] and an increased predisposition to develop colorectal, endometrial, ovarian, or other cancers [89]. In tumor specimens from endometrial patients, methylation of the MLH1 promoter was found to be a common mechanism of gene silencing resulting in MLH1 deficiency [90, 91]. The clinical significance of MLH1 methylation was assessed and not linked to patient survival. However, a retrospective study determined that MLH1 methylation status could be used to predict patient response to adjuvant therapy [92]. Consequently, MLH1 methylation status may provide prognostic value. Other studies have assessed the methylation status of BRCA1 in mammary gland carcinomas that are predominantly associated with triple-negative breast cancer (TNBC). Sharma and colleagues reported methylation at the promoter of BRCA1 in 30% of patients (n=39) [93]. Methylation of the BRCA1 gene was also found to be associated with lower detectable levels of BRCA1 transcript. In a larger study (n=239), 57.3% of TNBC patients were found to have methylation at the promoter of BRCA1 [94]. Methylation of the BRCA1 promoter was found to be an independent predictor of overall survival and disease-free survival in TNBC cases. A recently identified long noncoding RNA, Mortal obligate RNA Transcript (MORT), is silenced by DNA methylation in the initial stages breast tumorigenesis.
Alterations in the enzymes involved in DNA methylation have been identified in cancer. For example, DNMT1 was found to be overexpressed in triple negative breast, pancreatic, gastric, lung, and thyroid cancers [95]. DNMT3A expression is increased in pituitary adenoma, and AML. Similarly, higher levels of DNMT3B are observed in the lung, hepatocellular carcinoma, ovarian and breast cancer. Consequently, targeting their DNA methyltransferase activity using 5-aza-2’deoxycytidine (decitabine, 5-azadmC) is an FDA-approved therapeutic strategy with proven effectiveness in TNBC and hematological malignancies [96]. Mutations or copy-number variations within methyl-CpG-binding domain (MBD) proteins occur in many cancers, leading to loss of MBD binding specificity to methylated sites and gene deregulation [87, 97].
Crosstalk between DNA methylation and post-translational histone modifications
The organization_of DNA into higher-order structures is critical for the regulation of biological functions such as gene transcription, replication, and DNA repair. This process is accomplished at the level of nucleosomes, in which ~146 base pairs of this DNA are wrapped around an octameric core of histone protein subunits (H2A, H2B, H3, H4) [98]. These histone proteins are assembled into nucleosomes, where their N-terminal domains can be post-translationally modified at specific residues by acetylation, methylation, phosphorylation, ubiquitination, and poly-ADP-ribosylation. Methylation of H3K4 has been extensively studied, and numerous studies have linked mono-methylation (H3K4me1) to active enhancers [99], whereas trimethylation (H3K4me3) is predominately associated with transcriptionally active or poised promoters [100]. The methylation of H3K4 is largely controlled through the actions of KMT2-family proteins (MLL1, MLL2, MLL3, MLL4, SETD1A, SETD1B) and demethylases of the KDM1 and KDM5 families (LSD1, LSD2, JARID1A, JARID1B, JARID1C, JARID1D) [101]. Acetylation of lysine residues by histone lysine acetyltransferases (KATs) is generally associated with loosening of chromatin for replication, or euchromatic transcriptional activation which is marked by histone H3 lysine 4 acetylation (H3K4ac) at active promoters and H3K27ac at enhancers. This epigenetic modification is reversed by histone deacetylases (HDACs) that remove acetyl groups from the histone tails. While acetylation is an active mark, methylation of histone tail residues denotes either heterochromatic inactivation or gene activation depending on which residue is modified. In contrast to H3K4 methylation which is associated with active enhancers or promoters, H3K27me3 is a facultative heterochromatic mark present over gene bodies that are repressed [102] and H3K9me3 is a constitutive heterochromatic modification present at silenced regions of the genome [103]. The H3K27me3 modification is written by the methyltransferase ‘Enhancer of Zeste Homolog’ (i.e., EZH1 and EZH2) that represents the enzymatic component of the Poly-comb Repressive Complex 2 (PRC2) multi-protein complex that also contains auxiliary proteins (e.g., EED, SUZ12). The H3K27me3 marks deposited by PRC2 are recognized by chromobox proteins (e.g., CBX2, CBX4, CBX6) and facilitate the recruitment of PRC1, that mediates H2AK119 ubiquitination, resulting in chromatin compaction and repression of transcription. The formation of heterochromatin propagates along chromatin by spreading these repressive post-translational modifications (PTMs), structural proteins, and the associated effector proteins outwards from nucleation sites. The exact extent of this heterochromatic spreading is heritable and has crucial consequences for establishing and maintaining cellular identity.
In pluripotent embryonic stem cells (ESCs), a great deal of functionally significant regulatory elements within the genome are simultaneously marked by both activating and suppressing PTMs. This phenomenon termed bivalency results in poised chromatin states which allows these ESCs cells to rapidly respond to differentiation stimuli to either activate or repress key genes involved in differentiation [104, 105]. While, both poised promoters and poised enhancers contain suppressive H3K27me3, bivalent promoters contain the activating H3K4me3 modification, while bivalent enhancers contain H3K4me1. As a result, this bivalency is resolved into active or suppressed chromatin states during differentiation [106, 107]. The histone code within chromatin is complex in that many residues on a single histone can be modified simultaneously, and different combinations have varying effects on chromatin states. The establishment and maintenance of these chromatin states through the epigenetic landscape of PTMs gives rise to cellular identity and tissue specificity. Recapitulation of bivalency within critical development related genes has been detected in early-stage breast cancer cells_and designated as oncofetal epigenetic control[108-110].
There is significant crosstalk between histone modifications to render genome domains poised for transcription, active or suppressed in a context-dependent manner [111]. While, histone phosphorylation is a critical component of the DNA damage response pathway in the case of H2AX phosphorylation, this epigenetic mark works in concert with acetylation to synergistically regulate gene expression. Moreover, there is significant crosstalk between histone modifications to control replication, transcriptional regulation to maintain cellular identity, and during DNA damage responses [112]. For example, H2BK120ub is associated with active transcription and signals for transcriptional elongation, H3 lysine methylation, and recruitment of the heterodimeric histone chaperone, FACT, including the SSRP1 and SPT16 proteins. During DNA damage and repair, histone tails at damaged chromatin are not only phosphorylated, but additionally acetylated and ubiquitinated [113]. Moreover, since many methyl PTMs on histone H3 are adjacent to residues that are phosphorylatable, a ‘phospho-methyl switch’ is a potential prevalent mechanism to control the association of reader proteins with chromatin in various biological processes [114, 115].
Not only do the epigenetic histone PTMs synergize and crosstalk with each other, but these histone modifications also interact with DNA methylation through the factors that write, erase, and read these modifications [116, 117]. For example, CpG DNA methylation is excluded from the promoters of actively transcribed genes since DNMT3A and 3B bind to H3K4, but are repelled by increased H3K4 methylation thereby activating auto-inhibition of these DNA methylases. Consequently, when H3K4 is methylated, de novo DNA methylation does not occur [79]. In contrast, during transcription RNA PolII recruits the histone methyltransferase, SETD2, to trimethylate H3K36. The PWWP of DNMT3A and B bind to this H3K36me3, thereby deactivating the auto-inhibition, resulting in subsequent DNA methylation of the gene body. Therefore, DNA methylation at the gene body is positively correlated with transcription [118]. H3K27me3 at CpG islands at promoters by Polycomb Repressive Complex 2, is more readily reversible than DNA methylation of these sites, DNA methylation is crucial for mechanisms that include gene imprinting, X chromosome inactivation, and suppression of transposable elements [119]. Maintenance of DNA methylation is also connected with histone H3K9 methylation in that UHRF1 binds to di and trimethylated H3K9 residues [120].
The complex interplay between activating and suppressive marks and DNA methylation at bivalent poised genes is highlighted by the fact that these genes only contain low levels of methylation in normal undifferentiated cells, but are hypermethylated in cancer cells. While DNA methylation is usually associated with decreased expression, in cancer cells is often indicative of upregulated expression [121]. Depletion of DNA methylation has been demonstrated to impact H3K27me3 distribution at bivalent promoters. Recently, bivalent promoters were subdivided into two groups, ones with a high HK27me3: HK4me3 ratio (hiBiv) and those with a low HK27me3: HK4me3 ratio (loBiv). LoBiv promoters are less enriched in canonical Polycomb components, had a lower level of intrachromosomal interactions, and were less sensitive to DNA hypomethylation. In cancer, hiBiv promoters lose HK27me3 and are more susceptible to DNA hypermethylation than loBiv promoters [122].
Alterations in histone modification enzymes in cancer.
Alterations due to dysregulated epigenetic machinery that writes, reads, and erases histone PTMs result in aberrant function of tumor suppressors and promoters in cancer. Disruption of the balance between active and repressive histone-modifying enzymes have been implicated in breast, lung, and colorectal cancer tumorigenesis through mis-regulated post-translational modification of both histone and non-histone substrates that are critical proteins involved in cancer progressions such as p53, MYC, and RB1 [123]. Mutations resulting in haploinsufficiency for these critical factors regulating the epigenetic landscape are among the most mutated genes in cancer [123, 124]. For example, aberrant SET1A activity results in the methylation of histones and YAP [125, 126]. In acute myeloid and lymphoid leukemia (AML and ALL), MLL1 (also known as KMT2A) is frequently translocated with other oncogenic partners resulting in the loss of its catalytic SET domain. These mutations are present in approximately 80% of childhood leukemia and 5–10% of adult leukemia [127]. ]. Moreover, MLL3 and MLL4 are often mutated in cancer as well [128-131]. Mutations within the PHD cluster of MLL3 interferes with its interaction with the tumor suppressor BAP1 which is the catalytic subunit within the Polycomb repressive deubiquitinase complex [129]. While, germline mutations in BAP1 have been demonstrated to predispose patients towards mesothelioma and melanocytic skin tumors, somatic mutations are prevalent in breast, lung, melanoma, clear cell renal cell carcinoma.
Altered KAT activity resulting from chromosomal translocations is involved in the initiation of some types of cancers, specifically lymphoid and hematopoietic tumors. Chromosomal translocations either result in the formation of oncogenic fusion proteins or activate oncogenes by generating new enhancers or promoters [132]. Such translocations have been identified in acute lymphoblastic leukemia (ALL) and acute myeloid leukemia (AML) in which (q23; p13) (11:16) translocations lead to the generation of the MLL-CBP fusion protein [133]. Other examples of hematopoietic malignancies resulting from oncogenic chromosomal translocations include MLL-P300 fusion protein resulting from t(11;22) (q23; p13) translocation and MOZ-CBP fusion protein because of the t(8;16) (p11;p13) translocation [132, 134]. The overall oncogenic or tumor-suppressive role of KATs is determined by the level of their activity i.e., enhanced activity is associated with oncogenesis or reduced expression results in reduced acetylation. This makes KATs a potential therapeutic target for cancer therapy.
Altered activity of HDACs (e.g., HDAC1, HDAC2) has been reported in many malignancies [135]. To date, eighteen genes encoding four major classes of HDACs (Class I, II, III, and IV) have been identified in the human genome. Class I, II, and IV share sequence and structure homology and require zinc ions for their catalytic activity, whereas class III HDACs or sir-2 like proteins (sirtuins e.g., SIRT1, SIRT2) depend on nicotinamide adenine dinucleotide (NAD) for their catalytic activity and share no sequence or structural homology with other classes of HDACs [17, 135, 136]. Changes in HDACs activity caused by chromosomal translocations can lead to aberrant deacetylation of tumor suppressor genes, and have been implicated in tumorigenesis. In acute promyelocytic leukemia (APL) the chromosomal translocations t(15;17) (11:17) leads to abnormal recruitment of HDACs which results in repression of RAR target genes [135, 137, 138]. Other examples where abnormal HDACs recruitment leads to gene silencing and contributes to tumorigenesis include AML-1-ETO t(8;21) and CBFβ-MYH11 (chromosome inversion 16(p13; q22) [137].
Another component of epigenetic machinery involved in regulation of gene expression are “reader” enzymes which selectively bind to individual or a combination of histone PTMs and read the molecular information defined by them [139]. These epigenetic readers recognize specific histone PTMs and have conserved domains, on basis of which they are classified into different families For instance bromodomains (BRDs) that are well known to recognize acetyled-lysine residues, while plant homeodomains (PHDs), chromatin organization modifier (chromodomains or CRDs), tudor domain, the proline-tryptophan-tryptophan-proline (PWWP) motif, and the malignant brain tumor (MBT) domains all bind to methyl-lysine or methyl-arginine residues [139-141]. Tumor cells are frequently associated with abnormalities due to mutations, insertions, deletions, or chromosomal translocations in histone reader enzymes. These aberrations can cause deregulated gene expression or expression of TFs that ultimately leads to oncogenesis and foster cancer progression. For instance, a loss-of-function mutation in the bromodomain BAF180 is associated with clear cell renal carcinoma. Moreover, mutations in PHD finger domains have been reported in breast cancer and melanoma and fusion of the PHD finger region of lysine demethylase 5A (KDM5A) with nucleoporin 98 (NUP98) results in abnormalities leading to leukemia [139]. Fusion of bromodomain proteins such as BRD-3 or BRD-4 to nuclear protein of the testis (NUT) transcriptional regulators results in formation of an oncoprotein that binds to acetylated histones which drives aberrant transcriptional machinery activity and ultimately leads to NUT carcinoma. Overexpression of another critical bromodomain reader protein ATAD2, recognizes the diacetylated histone H4K5ac and 4K12ac modification during replication, and is predictive of poor prognosis in breast cancer, lung cancer, and ovarian cancer [142-150]. Since these enzymes play a crucial role in the regulation of cell function, cancer initiation, and progression, they can serve as potential therapeutic targets in oncology. Current research suggests that small molecules targeting chromatin reader domains such as BET bromodomain inhibitors are efficacious in the treatment of cancer [151, 152].
Chromatin structures including chromatin fibers, topologically associating domains and compartments and histone modifications
Chromatin organization beyond the single nucleosome level is first characterized by their association into the chromatin fiber. These are arrangements of repeated nucleosomes that are periodically separated by 20 to 75bp of DNA [153, 154] that are heterogeneous in diameter ranging in size from 5 to 24nm [155]. In between these nucleosomes, the H1 linker histone protein binds at a ratio of approximately 1 per nucleosome [156]. Histone H1 variants are differentially localized in euchromatin or heterochromatin and perform various functions in chromatin structure and dynamics through differences in post-translational modifications (PTMs) [157]. Due to their capability to stabilize nucleosomal arrays and to promote compaction of the chromatin fiber, the H1 linker histones have been considered general transcriptional repressors. Despite this, knockouts of specific H1 variants do not result in global genomic activation, but differential patterns of down and upregulation of specific genes [158]. Knockout of three H1 variants in ES cells, also resulted in differences in DNA methylation , suggesting a further connection between the epigenetics of the H1 linker histones and DNA methylation [159]. Histone H1 PTMs and their impact on chromatin compaction, transcription, DNA repair, and differentiation are extensively reviewed by Andrés and colleagues [160]. Another dimension to H1 mediated control is suggested by the recent demonstration that ablation of the Histone Nuclear Factor P (HINFP) factor regulating H1 gene expression in drosophila results in altered chromatin organization, transposon activation, and genomic instability [161]. Mutations in genes encoding histones and their expression levels have been associated with oncogenesis. For example, downregulation of histone H1 mRNA expression 40% has been reported in ovarian adenocarcinoma. Moreover, different expression levels of histone H1 subtypes are associated with several malignancies such as upregulation of histone H1.5, which has been reported in metastatic prostate cancer [162-164].
A key component controlling chromatin remodeling is the activity of the SWI-SNF complex. SWItch - Sucrose Non-Fermentable (SWI-SNF) is a set of genes encoding enzyme complexes that are involved in transcriptional regulation through ATP-dependent chromatin remodeling. SWI-SNF complexes alter histone-DNA contacts thereby changing the position or completely disrupting the nucleosomal structure [165]. Consequently, the access of DNA regulatory factors to the chromatin is altered which in turn affects processes such as replication, transcription, recombination, and repair[166]. In addition to the composition of the SWI-SNF subunits, post-translational modifications including acetylation, sumoylation, methylation, and phosphorylation play a key role in modulating the activity of the SWI-SNF complex. For example, phosphorylation of SWI-SNF complex components by P38 kinases at the onset of myogenic differentiation modulates chromatin structure and facilitates activation of genes associated with myogenesis [167]. Mutations in genes encoding the components of SWI-SNF complex are emerging in tumor initiation and progression. It has been hypothesized that almost 20% of human tumor cases are associated with mutations in at least one component of SWI-SNF complex. ARID1A, a tumor suppressor gene of the SWI-SNF complex, is frequently mutated in cancer and is associated with a worse clinical prognosis [165, 166, 168]. Another example is with the SMRCB1 gene, which is also a component of SWI-SNF complex, where alterations are associated with multiple familial malignancies and familial schwannomatosis [166].
SWI/SNF is critical for paraspeckle (PS) assembly in the interchromatin domain of the nucleolus, distinct from nuclear speckles [169], by recruiting and facilitating essential Paraspeckle protein (PSP) interactions for proper assembly[170]. In fact, BRG1, the core ATPase subunit of the hSWI/SNF complex, physically interacts with PSPs and lncRNAs independent of ATPase activity [170, 171], suggesting a structural role of BRG1 in paraspeckles separate from its canonical chromatin remodeling function. While the exact function of these paraspeckles remains unclear, there is speculation that their purpose is associated with the presence and retention of specific RNAs involved in determining cell fate, such as pluripotency versus differentiation [171]. Current models suggest that coordinated assembly by long non-coding RNA (lncRNA) and DBHS proteins occurs in close proximity to chromosomal loci to create distinct sub-nuclear bodies. The initiating cellular signal or event that triggers paraspeckle assembly remains unclear. These unique nuclear bodies display a rich composition of RNA, especially the mammal-specific lncRNA, NEAT1, which is essential for paraspeckle assembly, architecture, and maintenance [171]. The remaining elements of paraspeckles consist of RNA-binding proteins (RBPs) involved in transcription and RNA processing [171]. At this point, more evidence is required to determine if the information for paraspeckle assembly and function is epigenetically regulated and conserved through subsequent cellular division cycles.
How the chromatin fiber folds into higher-order structures
The three-dimensional conformation of the chromatin fiber is a critical component regulating gene expression under a variety of physiological conditions, including cell cycle, differentiation, and disease states. Although enhancers can be megabases away from the gene(s) they regulate, they can support transcription by looping in 3D space [33, 34, 172, 173]. These enhancer-promoter contacts are re-organized in response to developmental and signaling cascades. Examples include IFNG [174], MHC Class II [175], the β-globin locus [176-178] and CFTR [179]. The binding of transcription factors is coincident with this conformation. For example, the transcription factor RUNX1 interacts with the CD34 gene promoter and its downstream regulatory elements in hematopoietic stem cells[35]. Similarly, the interaction between the beta-globin gene and its regulatory sites is dependent upon the transcription factors EKLF1 and GATA1 [180]. The osteocalcin gene is architecturally remodeled to support bone tissue-specific gene expression and promotor occupancy by the RUNX2 transcription factor [181-202]. The histone gene loci respond to transcriptional requirements at the G1/S phase cell cycle transition with modifications in chromatin structure that include histone modifications [187, 203-213], nucleosome organization [214], and higher-order chromatin confirmation [43, 187, 215, 216], to orchestrate competency for promotor occupancy by regulatory complexes in a cell cycle responsive manner [217-219] with modifications in tumor cells [43, 210, 220-230]. In addition, the transcription factor SATB1 is required for an enhancer to interact with the Rag1 and Rag2 genes during thermal cell development [231]. Although, many studies have demonstrated that transcription is simultaneous with enhancer-promoter contacts, it is still unclear as to whether these interactions are causative for gene expression. Despite this, evidence suggests that enhancer-promoter looping can influence transcription. For example, forced chromatin looping between the mouse beta globulin gene and its locus of control leads to a dramatic increase in transcription [172] More recently, with the advent of catalytically inactive dCAS9 variants that are dimerized after treatment with light [232] or small molecules [233], it has been demonstrated that such forced chromatin looping can reversibly activate transcription.
While there is considerable data demonstrating support for the requirement of enhancer-promoter interactions for the induction of transcription, several alterative lines of evidence suggest that they are not required. For example, the distance between the Sonic Hedgehog gene (Shh) and its enhancers was actually shown to increase during differentiation of neural progenitors [234]. Moreover, looping of enhancers to promoters that pre-exist before transcription have been described including TNF-responsive enhancers [235] and during thrombopoietin signaling in hematopoietic stem cells [236]. While studies have shown that these signaling cascades do not induce widespread alterations in enhancer-promoter interactions, other studies have attempted to interfere with transcription to determine if these interactions can be disrupted. For example, depletion of P53 resulted in decreased expression of target genes, but the persistence of long-range enhancer-promoter contacts [237]. Similarly, BET inhibition has been shown to inhibit transcription, but there were few changes in the configuration of chromatin [238]. While these are compelling findings suggesting that transcription and enhancer-promoter looping are independent, they are not mutually exclusive with the idea that these contacts are required in many cases. For example, the factors that are altered may not be driving specific contacts but are recruited to pre-existing loops.
Subnuclear trafficking and phase separation- mechanisms that contribute to the subnuclear organization assembly and activity of regulatory complexes in nuclear microenvironments.
One major question in the field of nuclear structure is how membrane-free compartments and microenvironments form and how the nuclear machinery is properly localized within these domains. Proper localization of nuclear machinery is critical for proper epigenetic regulation. Historically, one hypothesis has been that the nucleus contains a nuclear matrix wherein a 3-D filamentous network, analogous to the cytoskeleton, including the nuclear lamina, residual nucleolus, and ribonucleoprotein complexes, and nuclear matrix-associated DNA, collectively provides a structural framework for nuclear organization [24, 239]. The nuclear matrix is a composite organization of architecturally organized regulatory machinery within the nucleus. This architectural organization is dynamic and remodeled structurally and functionally in response to regulatory cues. Mechanisms that mediate the organization and assembly of the nuclear matrix are open-ended, however recent findings have demonstrated that many of these components are dynamically assembled. While an inclusive model for the steps that target proteins to subnuclear sites is unclear, there are many apparent intranuclear targeting signals within transcription factors. Alterations in the binding to aspects of this nuclear matrix affect the proper subnuclear localization of transcriptional machinery. For example, mutation of as little as one amino acid in the nuclear matrix targeting signal of RUNX factors results in their mis-localization. The addition of Matrix Attachment Regions (MARs) [240], which serve as chromatin attachment points within the DNA sequence, have been shown to increase transgene expression and transfection efficiency [241-246]. In contrast, while Lamina Associated Domains (as reviewed in [247, 248]) are also associated with organizing the nucleus, artificially tethering genes to the nuclear lamina by targeted recruitment of YY1 results in chromatin repression and re-localization to the nuclear periphery or nucleolus [249].
Another proposed mechanism driving the formation of membrane-free nuclear bodies is termed lipid-like phase separation (LLPS) in which liquid-like condensates or droplets form through weak multivalent interactions between macromolecules [250-252]. Physiological levels of cations have been shown to induce LLPS of reconstituted arrays of nucleosomes. The addition of the linker histone H1 to these reconstituted arrays promoted LLPS and increased the concentration of nucleosomes within condensates and while also decreasing the dynamics of droplets consistent with H1’s generally repressive function. While reconstituted chromatin microinjected into nuclei form condensates, nucleosomal arrays with acetylated histone tails did not readily form droplets [253]. The dissolution of droplet formation was restored upon the addition of BRD4 before microinjection [253]. The droplets with acetylated and BRD4-associated chromatin associate, but do not coalesce with condensates with chromatin that is not acetylated [254]. In addition, the presence of transcription factors such as HOXA13, HOXD13, RUNX2, and TBP, within these droplets is dependent upon the macromolecular interactions that occur with the transcriptional machinery [253]. Intrinsically Disordered Regions (IDRs) within these transcription factors drive their ability to form LLPS bodies. Lengthening of these IDRs interferes with their ability to form condensates and to co-condensate with transcriptional coactivators. These two mechanisms, subnuclear trafficking versus LLPS, are not mutually exclusive. Both models posit that the physical properties of macromolecular complexes drive principles of nuclear organization. LLPS of regulatory components that comprise transcriptional conjugates into nuclear sub-compartments such as the nuclear lamina, nucleolus, histone locus bodies, cajal bodies, splicing domains, and other subnuclear domains, contribute to the architectural organization of regulatory machinery in nuclear microenvironments. Recognition Signals within regulatory factors may drive their inclusion within nuclear microenvironments simultaneously with the physical properties in other domains such as IDRs.
Super-enhancers in cancer
Hotspots within the genome wherein multiple enhancers are generally clustered within 5-12.5kb of each other, are collectively termed super-enhancers (SEs). These sites are critical for supporting gene expression profiles to maintain cellular identity and regulate the expression of master transcription factors that control cell fate. As expected with the observation that nucleosomal arrays that are acetylated and associated with BRD4 form condensates that are separated from un-acetylated chromatin, LLPS has also been demonstrated to play a critical role in the formation of super-enhancers [252, 255-257]. Moreover, the LLPS of anti-cancer drugs into these critical genome regulatory elements is a key property to consider in that their inclusion or exclusion from condensates can affect efficacy. A comprehensive understanding of which SEs are active within disease states and the properties governing LLPS of SEs and drugs will provide insight into therapeutic strategies.
SEs have been linked to cancer development and progression. Li et al. performed ChIP-Seq of H3K27ac in tumors grown in the thoracic mammary gland of mice and identified a total of 661 SEs compared to control [258]. Elevated levels of histone H3K27ac and H4K8ac were found in clinical breast cancer patient samples (Luminal A, Luminal B, Basal-like, and Her-2) compared to native tissues when assessed by IHC arrays. Additionally, a subgroup of SEs marked with H3K27ac was identified (n=148) and associated with higher transcriptional activity and cancer-related pathways. Recently, Huang et al. performed ChIP-Seq for H3K27ac in TNBC and non-TNBC subtypes [259]. For example, the promoter of FOXC1 has a SE specific to TNBC and is more highly expressed in TNBC patients than in other subtypes (Her-2, Luminal A, Luminal B, and Normal-like) and correlates with an overall poorer survival. Depletion of FOXC1 by CRISPRi inhibits spheroid growth and invasiveness that is rescued by FOXC1 over-expression [259].
To study the role of SEs in colorectal cancer, 73 pairs of primary tumor tissues and corresponding adjacent tissues were collected from patients to generate 147 transcriptome profiles of H3K27ac using ChIP-Seq [260]. A total of 9896 and 10663 active promoters were identified in native and tumor tissues respectively. Comparison to other studies revealed that 32.4% of the enhancers identified in this study were novel enhancers. A total of 6690 variant enhancer loci were identified in colorectal cancer (5590 gain and 1100 lost, respectively). Of these, 455 variant super-enhancer loci (VSEL) were identified (334 gain and 121 lost, respectively). The VSEL in tumor tissues was located at well-known oncogenic targets such as MYC, VEGFA, and LIF. To verify the function of VSELs identified, 11 SEs were targeted by CRISPRi. Enhancers and super-enhancers are constrained within higher nuclear microenvironments described in the subsequent section.
Higher-order chromatin structures, TADs, and compartments
The chromatin fiber is folded into globular, self-interacting, insulated topologically associating domains (TADs) [261-264]. One of the major proteins involved in the insulation of TADs and mediating intra- and inter-chromosomal looping interactions is CTCF [265]. CTCF contributes to the interconnected nature of the various levels of nuclear organization through its interactions with chromatin remodeling proteins, histone-modifying enzymes, and transcription factors. Through these interactions it is implicated in a myriad of fundamental regulatory functions including imprinting [266], X chromosome inactivation [267], and organizing the major histone locus [43]. Although the mechanism by which TAD and chromatin loops are constructed is not fully understood, interaction between CTCF and the cohesin complex (SMC) for chromosomal structure maintenance are vital for the process known as the loop extrusion model [268, 269]. This model posits that two strands of DNA are held together by a cohesin ring and create loops by actively extruding the DNA. Once cohesin encounters a CTCF protein occupying a motif in a convergent orientation, a loop is formed [269]. Even though CTCF cannot be knocked out due to its essentiality [270], studies have depleted it using siRNAs or by tagging it with an auxin-inducible degron (AID). siRNA knockdown of CTCF decreased intra-TAD interactions slightly while increasing inter-TAD contacts [271]. Auxin-induced degradation of CTCF [272] resulted in a greater decrease in CTCF levels and led to a loss in TAD insulation, but did not alter intra-TAD contacts [273]. Less than 20% of TAD boundaries were unaffected by depletion of CTCF. Another study found that although CTCF depletion reduced genomic occupancy of cohesin, its knockdown slightly weakened TAD boundaries and most TADs remained unchanged [274]. More recently, it has been determined that some CTCF binding sites are more resistant to the removal of CTCF [275] . While CTCF sites that are maintained are unaltered, sites where CTCF is lost, are more altered in their TAD structure. Despite the fact that CTCF knockout is lethal, it was determined at a very early stage that the loss of CTCF results in disruption of TAD insulation [276].
TADs coalesce into two primary compartments that are either euchromatic, termed A, or heterochromatic, termed B [277, 278]. These compartments are further subdivided into six computationally distinct sub-compartments, two that are euchromatic and four that are heterochromatic [279]. More recently, these sub-compartments have been posited as a spectrum of compartments from those that are at the extremes and are either heterochromatic or euchromatic, while some TADs associate with both A and B compartments [280]. The euchromatic B compartments are noticeably less gene-rich, transcriptionally inactive, marked by inactive epigenetic signatures, and preferentially inaccessible to DNaseI than euchromatic A compartments [69, 281]. While, nucleosomal spacing is directly regulated by histone H1 PTMs, the triple knockout of three histone H1 variants in mESC cells resulted in more global alterations in higher-order nuclear organization. Although segregation between A and B compartments generally appears at TAD boundaries, depletion of cohesin and/or CTCF did not alter A and B compartmentalization [273, 282]. These results suggest that compartmentalization is independent of CTCF and the establishment of TADs.
Although CTCF is a major player in chromatin organization, other factors are likely are involved in these processes. Compelling evidence for other mechanisms driving TAD delineation include the fact that CTCF is absent at many TAD boundaries and that orthologues of CTCF do not exist in some species [283-287]. In these ancestral genomes, phase separation may be the predominant mechanism by which TAD structures form. As described above, high acetylation of histone tail leads to destabilization of chromatin domain [288]. This destabilization of the chromatin domain may explain the accumulation of epigenetic markings indicating genes actively transcribed at the TAD boundary (such as housekeeping genes) [263]. Indeed, expression data can predict the three-dimensional folding of the genome [289, 290]. Splitting the TAD boundary based on active expression independent of CTCF binding appears to be more common in Drosophila melanogaster [289, 291]. The Drosophila TAD boundary indicates that there are more transitions between open and closed compartments than in the nucleus of human cells [289]. The different properties of the active and inactive genes have been shown to predict Drosophila TAD boundaries based on polymer simulations [291]. Therefore, Drosophila TAD responds to transcriptional stimuli (e.g., recovery from heat shock [292] or activation or transcriptional inhibition of the conjugated genome [293]. The fact that higher-order chromatin organization within ancestral genomes are more specified by epigenetic states than that of the human genome suggests that human cells have greater control over these processes.
At the highest level of organization within the nucleus, chromosomes occupy discrete territories [24, 294]. These chromosome territories (CTs) are non-randomly distributed within the interphase nucleus. For example, the relative radial positioning of individual CTs has been demonstrated to be correlated with gene density, chromosome sequence length, and euchromatic state. Because transcriptionally inactive heterochromatin is preferentially localized around the nuclear periphery, the inactive X (Xi) CT is more peripheral than its active (Xa) counterpart. Chromosomes with longer sequence lengths have also been shown to be more peripheral than shorter ones. Several other factors have been implicated in the radial positioning of CTs such as interactions with nucleoli [295-297] or the nuclear lamina [298]. Nuclei from patients with Progeria express abnormally truncated lamin-A and as a result have a disordered radial arrangement of CTs [299].
Alteration of nuclear organization is implicated in hallmarks of cancer
The nucleus of cancer cells are generally larger, more irregularly shaped, and intranuclear sub-compartments, such as the nucleoli, are altered in their number and morphology [11, 59, 300]. Pathologists have long exploited these changes in nuclear architecture as useful diagnostic tools for recognizing malignancy [11, 59]. While it is well-known that epigenetics signals and nuclear morphology are disrupted in cancer onset and progression, emerging evidence demonstrates significant concomitant changes in higher-order chromatin organization.
Alterations in the epigenetic landscape in cancer nuclei affect chromatin structure. All of the functional properties of epigenetic regulation such as DNA methylation, histone PTM, nucleosome spacing, and chromatin remodeling are altered in cancer. Changes in these features influence the phase separation of condensates resulting in modifications to chromatin accessibility and/or long-range chromatin interactions at regulatory elements such as promoters, enhancers, insulators, and silencers. Moreover, alterations in these properties contribute to differences in topologically associating domains in cancer. Moreover, TAD boundaries can be shifted or disrupted in cancer. Since enhancer-promoter interactions are generally constrained within TADs, translocations, inversions or deletions that result in aberrant TADs can result in enhancers contacting inappropriate promoters. Even in unaltered TADs, mutations and/or dysregulated expression levels in transcription factors, such as YY1 and RUNX1, that facilitate enhancer-promoter interactions may perturb these contacts. Beyond the gene regulatory loops being altered, the fact that CTCF and its binding sites are mutated in many cancers, including breast cancer, suggests its functions in gene regulation and chromatin insulation are also perturbed upon malignant transformation [301-304].
Higher-order chromatin organization facilitates DNA repair and directly suppresses endogenous DNA damage. Chromatin with increased accessibility is more prone to exogenous and endogenous damage. Interestingly, heterochromatin is preferentially localized to the nuclear periphery where exogenous insults such as UV radiation are more prevalent [305]. Hence, heterochromatin could in principle absorb DNA damage insults. Indeed, it has recently been demonstrated that mutations occur more frequently in the chromatin at the nuclear periphery [305]. These regions of the genome are also less likely to contain critical regulatory genes. An open question is whether the nonrandom probabilistic radial positioning of chromosomes within nuclei as homologous recombination requiring physical interaction between homologs. Since, smaller chromosomes are confined to the nuclear interior and larger chromosomes are more peripheral the chance of self-interaction is likely higher. Alterations in the radial positioning of chromosome territories and interactions between small chromosomes has been identified in breast cancer [306].
Epigenetics and higher-order chromatin organization impact on all the hallmarks of cancer [15] including the resisting of induction of angiogenesis [307-309], activation of invasion and metastasis, cell death, replicative immortality [308, 310], evasion of growth suppressors, and sustained proliferative signaling [308, 311]. These hallmarks are all critical in the alterations that occur within the cell cycle of cancer cells.
Cell cycle
The cell cycle is controlled by complex and interdependent processes that require epigenetically mediated remodeling of the genome and the structural and functional regulatory machinery that governs fidelity of DNA replication and cell division. Crosstalk between mechanisms that coordinate proliferation and cell growth is obligatory, with unique requirements that are phenotype dependent. Many normal and tumor cells exhibit unrestricted proliferation. However, stringent control of the cell cycle is compromised in tumor cells, often reaching the balance between proliferation, cell survival, and responsiveness to physiological cues as well as acquiring resistance to therapy that targets cell cycle check points. We will provide an overview of the decisive regulatory parameters for cell cycle control that are mechanistically and clinically informative. Emphasis is on genome reconfiguration during the cell cycle that is required for epigenetically-mediated cell cycle and mitotic progression with competency to retain regulatory machinery during mitosis for transmission from parental to progeny cells.
Interplay between cell cycle progression, cellular transcriptional machinery, and epigenetics
Progression between distinct phases of cell cycle depends on correct timing of various events, which are coordinated by cellular transcriptional machinery and epigenetics controlling genome accessibility.
The cell cycle consists of interphase (G1, S, and G2 phases), mitotic phase (mitosis and cytokinesis, figure 3). During G1, cells nearly double their size, preparing for DNA replication. At the restriction point late in G1, the genes for deoxynucleotide biosynthesis are expressed, supporting competency for DNA synthesis. This preparation is carried out by assembling the pre-complex DNA replication at the replication origins. CDC6 is essential for the assembly of pre-replicative complex (pre-RCs), which are located on the origins of DNA replication [225, 228]. At S-phase, histone gene expression is initiated. The pre-replication complexes become active followed by the separation of the two DNA strands of the double helix, and by an elongation step catalyzed by DNA polymerase. Once DNA replication is completed, the chromosomes and Centrosomes are duplicated, and the amount of DNA is doubled in the cell. During G2 phase, the cells grow rapidly, and the checkpoint machinery ensures that the DNA is properly replicated and repaired when damage occurs. Centrosomes detach and undergo growth by the accumulation of pericentriolar material around the centrioles. This leads to centrosome maturation, which is a necessary step for assembly of the mitotic apparatus to enter M-phase.
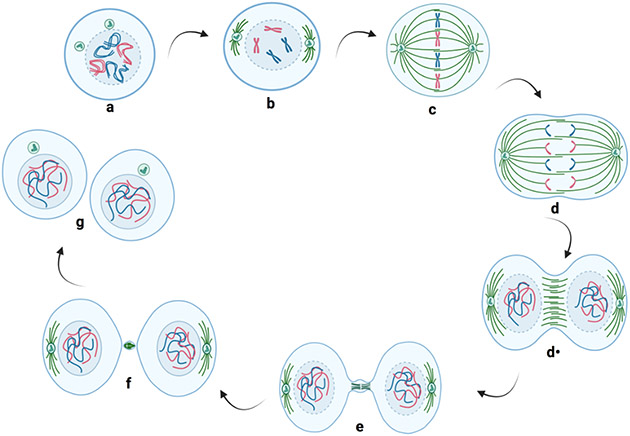
Figure 3. Chromosomes during interphase and mitotic M-phase.
a. The chromatin is present within chromosome territories during interphase. b. In prophase, the chromatin starts to condense into chromosomes and the nucleolus disappears. Centrosomes migrate to the opposite poles of the nucleus and initiate the formation of the mitotic spindle. c. During prometaphase-metaphase, the nuclear envelope breaks. Then, the chromosomes are oriented and aligned on the metaphase plate. d. In early anaphase, the sister chromatids separate, and the microtubules pull the chromatids apart toward the two opposite poles of the mitotic spindle. d· In late anaphase, the plasma membrane begins to invaginate to the equatorial plane. e. During telophase, the plasma membrane continues to invaginate, and the chromosomes decondense. f. During cytokinesis-abscission, the invagination of the plasma membrane appears through a contractile ring. The cleavage furrow progresses to create a midbody between the progeny cells, which disappears during the abscission process. g. This results in separation of the two progeny cells, which enter interphase and begin the process again.
Several studies investigated the structure, function, and regulation of the human histone genes, focusing especially on the cell cycle regulation of histone gene transcription and histone mRNA coupling with DNA replication. Genome-wide high-resolution chromosome conformation capture (HiC) technologies indicate that non-random higher-order organization of genes into structural domains supports regulatory activities involving non-contiguous sequences that influence transcription [212, 221, 312, 313]. The human histone locus bodies (HLBs) are nonmembrane bound, phase-separated domains that provide microenvironments for dynamic regulation of histone genes. The human histone gene transcription factor HINFP mediates coordinate expression of multiple histone H4 genes at the G1/S transition in normal and cancer cells, as well as the abbreviated human embryonic stem cell cycle [212, 221, 225, 228, 312]. Importantly, HINFP loss-of-function causes increased nucleosome spacing, induces replicative stress, and leads to genomic instability [313]. The overall increase in nucleosome spacing could be due to the insufficiency of newly synthesized histone H4 during DNA replication. In vivo mouse models, HINFP is essential for fidelity of histone H4 gene expression and required for early embryonic development [313, 314]. Furthermore, the role of HINFP in guarding normal chromatin organization is functionally conserved in flies [315]. NPAT is an essential and prototypical HLB resident protein that regulates expression of all replication-dependent histone gene classes (H4, H3, H2A, H2B, H1). HINFP is the critical anchor that tethers NPAT to histone H4 genes [316, 317]. In response to activation of the cyclin E/CDK2 cell cycle signaling cascade at the onset of S phase, NPAT is phosphorylated to initiate histone gene transcription [318-320]. Furthermore, deregulation of histone gene expression upon sustained loss of HINFP or NPAT causes disruption of HLBs and has drastic consequences for cell division and cell survival in both normal and cancer cells [313, 321, 322]. These findings establish that regulatory interactions involving HINFP and NPAT are essential for chromatin conformations at HLBs during the cell cycle to regulate histone gene transcription. Thus, understanding molecular mechanisms that control histone synthesis at the onset of S-phase would be a paradigm for transcriptional control that is obligatory for cell cycle progression.
GRO-seq, RNA-seq, and ChIP-seq analyzed the transcriptional and epigenetic dynamics during the cell cycle in breast cancer MCF-7 cells [323]. This study identified genes differentially transcribed at the G0/G1, G1/S, and M phases in MCF-7. There was no correlation between the transcription and steady-state mRNA abundance at the cell-cycle level. Active transcription occurred only during early mitosis. However, an overall increase in histone modification marks was reported at mitosis. Thousands of enhancer RNAs (eRNAs) and their associated transcription factors were identified, which were related to cell-cycle–regulated transcription but not to the steady-state expression.
Consequently, these results showed that the interplay between the transcriptional machinery and epigenetics are crucial for cell-cycle progression and that the precise understanding of cell cycle control is of great interest for understanding such pathologies as cancers.
Epigenetic control during mitosis
Mitosis plays a fundamental role to transmit stable inheritance of gene expression pattern from parental cells to their progeny [324]. During this phase, the nuclear envelop breaks down is disaggregated, triggered by the disassembly of nuclear pore complexes and lamina depolymerization [325, 326]. This process leads to chromatin becoming highly compacted into chromosomes [327]. During mitosis, many transcription factors dissociate from chromatin leading to significant downregulation of transcriptional activity [328]. However, there is little knowledge of how lineage-specific epigenetic information is faithfully maintained in progeny cells. Several studies suggest that the gene regulatory networks are controlled by epigenetic bookmarks that are maintained during mitosis [53-55, 328-331] (figure 4). In human embryonic stem cells (hESCs), bivalent genes with both the activation mark (H3K4me3) and repressive mark (H3K27me3) are important for maintaining pluripotency [332, 333]. However, during mitosis, the bivalent genes containing H3K4me3 are the most upregulated genes upon differentiation of hESC, showing a new aspect of epigenetic regulation crucial for the maintenance of pluripotency [332-334].
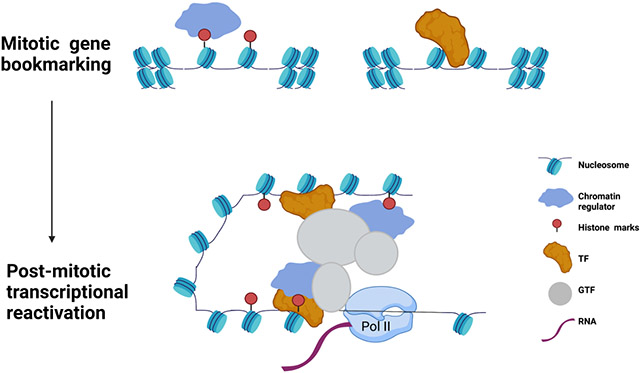
Figure 4. Mitotic bookmarking establishes post-mitotic reactivation of gene expression.
During mitosis, histone marks and chromatin regulators bind the open regions on the chromatin thus bookmarking specific loci for the memory program. Transcription factors can additionally associate to the chromatin targets. Consequently, these mechanisms result in a post-mitotic transcriptional activation after mitosis.
The entry of mitosis is controlled by CDK1/cyclin B kinase also known as MPF (M-phase Promoting Factors). CDK1/cyclin B is responsible for direct and indirect phosphorylation of substrates involved in mitosis. Histone H1 is a direct substrate of CDK1/cyclin B [335]. Other substrates include CDK1 regulators like CDC25 and Wee1, and cytoskeletal proteins like microtubules, nuclear lamina, and vimentin, are required for the proper progression of mitosis [336-339]. Most recently, it has demonstrated that the S-phase initiator CDC6 plays a crucial role in controlling mitotic M-phase entry. CDC6 regulates CDK1 activity and determines the timing of mitosis in Xenopus cell-free extract [340-344]. Inhibition of CDC6 results in earlier M-phase entry in Xenopus cell-free extract as well as in mouse zygotes. CDC6 modulates the activity of CDK1 via cyclin B and not cyclin A and acts through a bona fide CDK inhibitor Xic1 [340, 343]. Additionally, CDC6 regulates both G2/M transition and metaphase-to-anaphase transition during the first meiosis of mouse oocytes [345]. This suggests that the time of mitosis is not only under the control of cyclins as it has been described in the last 20 years [346, 347], but additionally under the control of CDC6 which can counterbalances the cyclins to control CDK1 activation, and subsequently to specify the timing of mitosis.
Taken together, these findings suggest that the epigenetic control and correct timing of mitosis play a coordinating role between gene regulatory networks, cell cycle machinery, and the developmental program, ensuring accurate inheritance of the genome during cell division.
Mechanisms of chromosome behavior during mitosis
Mitosis is composed of four basic phases: prophase, prometaphase-metaphase, anaphase, and telophase [348].
During prophase, the chromatin begins to condense into chromosomes and the nucleolus disappears. The chromosomes consist of two sister chromatids; each contains a DNA element called centromere that connects both chromatids [349]. The two sister chromatids are glued with cohesin. However, kinetochores are assembled at centromeres to ensure the interaction of chromosomes with spindle microtubules. In prophase, microtubules depolymerize, centrosomes migrate to opposite poles of the nucleus, and initiate the formation of the mitotic spindle.
During prometaphase-metaphase, the breakdown of the nuclear envelope (NEBD) occurs, and chromosome condensation begins. This is carried out by activation of the histone H1 kinase activity resulting in hyperphosphorylation of nuclear lamins, leads to NEBD, and shortens microtubules by phosphorylation of Microtubule-Associated Proteins (MAPs). Then, the chromosomes are captured by microtubules attached to kinetochores. At metaphase, the chromosomes are distributed at their kinetochores and assembled uniformly on the equatorial plate of the spindle. Three types of microtubules nucleated at the centrosomes can be distinguished i) kinetochore microtubules ii) polar microtubules pointing directly to the opposite spindle pole iii) astral microtubules radiating out of the spindle. The balance between ejection and pulling forces exerted on chromosomes permits the alignment of chromosomes on the metaphase plate. At this stage, the ubiquitin ligase Anaphase Promoting Complex/Cyclosome (APC/C), which is involved in the degradation of cyclins, becomes active [350]. This activation is carried out by a series of phosphorylation on APC/C complex induced by CDK1. Therefore, APC/C triggers degradation of the cyclins, allowing inactivation of CDK1 and subsequently separation of the sister-chromatid.
During anaphase, active segregation of sister chromatids takes place within the mitotic spindle. When APC/C becomes active, it catalyzes ubiquitination and degradation of securin relieving the inhibition from separase. The latter is responsible for the cleavage of cohesins allowing separation of sister chromatids [351]. In parallel, the microtubules pull the chromatids apart toward the two opposite poles of the mitotic spindle. In late anaphase, the plasma membrane starts to invaginate to the equatorial plane and the cell enters telophase.
In telophase, the nuclear envelope is reformed around the chromosomes entering the interphase state of de-condensation. The plasma membrane continues to invaginate, and the elongation of polar microtubules stops leading to the formation of the central spindle. In higher eukaryotes, this process is known as open mitosis, ensuring inheritance of chromosomes to the newly formed cells [352]. However, because the nuclear envelope does not break down during mitosis in lower eucaryotes, the mode of mitosis is referred to as closed mitosis [353].
During cytokinesis, the cytoplasm, organelles, cell membrane and the two formed nuclei split into two progeny cells. This is characterized by a cleavage furrow formed perpendicular to the mitotic spindle. The invagination of the plasma membrane appears through a contractile ring, where actin and myosin microfilaments play mechanical roles. The cleavage furrow progresses to create a midbody between the progeny cells. The midbody disappears during the abscission process leading to separation of the two daughter cells.
Taken together, these results showed how accurately chromosomes are distributed and organized at each stage of mitosis. Chromosome organization must be highly dynamic during mitosis to ensure inheritance to the progeny cells.
Recent reports have provided an update about the current knowledge on mechanisms of chromosome regulation across mitosis [354-356]. They examined how the pairs of homologous chromosomes are continuously separated at mitosis. It has been shown that mammalian cells disrupted homologous chromosome pairing by keeping the two haploid chromosome sets separately and by positioning them on either edge of the nuclear division axis, defined by centrosomes. Loss of mitotic anti-pairing causes loss of heterozygosity (LOH) and is correlated with gene mis-regulation in cancer cells. This suggests that anti-pairing plays a significant role in inhibition of genetic recombination or allelic mis-regulation across cell division. How mitotic recombination is prevented and how genomic stability is maintained during division are fundamental unanswered questions. For example, what are the regulatory mechanisms involved in haploid set chromosome sequestration? Is the cytoskeleton implicated in this process? In Drosophila, chromosome pairing can be actively inhibited, and several studies suggest that condensin II could play a role in regulating the anti-pairing activity [357-360]. Consequently, how does condensin II regulate mitotic anti-pairing in human cells?
Condensin-mediated remodeling of mitotic chromosomes
Nuclear DNA is highly condensed and wrapped around nucleosomes to fit inside the nucleus [361]. Each nucleosome is formed by a set of eight histone proteins — two copies each of the histone H2A, H2B, H3, H4, and 146-nucleotide pair DNA double helix. The latter is wrapped 1.65 times around the histone core to form a fiber structure of 10 nm width, described as ‘beads on a string’. Nucleosomes are stabilized by the histone H1 and compact the genomic DNA sevenfold. Additional compaction occurs during interphase to fully organize the DNA inside the nucleus. Upon mitotic entry, the chromatin highly condenses into mitotic chromosomes, positions at the metaphase plate, and avoids being disrupted during anaphase. The condensation of chromatin is controlled by condensins and post translational modifications [362]. The condensation events start before NEBD. The chromatin fibers fold and reach 700nm of diameter by the end of prophase approximately [363]. These events are in part due to the activation of CDK1/cyclin B and of condensins by phosphorylation [364]. Condensins depletion leads to a delay in mitotic compaction [365, 366].
Atomic force microscopy is an excellent technique to visualize condensins during mitosis [367]. Unlike other microscopes, it does not use photons nor electrons (like in electron microscopy) to produce images. Condensin is composed of two proteins of the family of SMC (Structural maintenance of chromosome) as well as three non-SMC subunits: a kleisin (NCAPH) and two HEAT domain proteins (NCAPG/D) [368]. There are two condensin types, condensin I and II, which differ by their non-SMC proteins. Condensin II binds the chromatin fiber throughout the cell cycle, while the condensin I complex binds the chromosomes only after NEBD. Both condensins I and II contribute to mitotic compaction [369]. However, it seems that condensin I act on the fibers that condensin II did not compact, thus promoting a lateral compaction of the fibers, and subsequently sustaining a stable condensed chromatin [370].
Cohesins interact with chromatin in G1 and attach the sister chromatids to each other during S-phase. A fraction of condensins II associates with duplicated regions during S-phase then initiates sister chromatid resolution. In prophase, the majority of cohesins are released from the chromosomal arms, condensins II associates with chromosomes inducing condensation. After NEBD, condensins I is directed to chromosomes and keep facilitating the condensation. In anaphase, the separase cleaves the kleisin subunit of cohesins, and allows separation of sister chromatids.
Several studies have shown that binding of condensin I to prometaphase requires the mitotic kinase Aurora B [371]. In the presence of topoisomerases I, Condensins fold the chromatin fiber into super loops to form the mitotic chromosome [372]. Later in mitosis, the axial compaction of chromosomal arms requires condensins activities in combination with sister chromatid resolution mediated by topoisomerase II and release of cohesins [373].
The condensation can take place in condensing-depleted cells; however, it is associated with chromosomal structural defects [370]. In S. pombe yeast, mutants of SMC proteins are not able to condense their chromosomes but remain capable of carrying out the elongation of the mitotic spindle and cell division [374]. In animals, condensation is affected but not completely abolished by the loss of condensins [370]. Moreover, Knock-out of SMC2 in chicken DT40 cells or Knock-down by RNAi (RNA interference) in C. elegans affects the architecture of the mitotic chromosome but has a limited impact on the condensation itself [365, 366]. Together these results suggest that several factors are important for the chromosome compaction during mitosis.
Chromatin decondensation upon the mitotic exit
In mitosis, Haspin and Aurora B are dissociated from the chromosomes and recruited to the centromeres [375]. Therefore, the histone H3S10PO4 and H3T3PO4 modification levels gradually decrease during metaphase/anaphase transition. Despite the reduction in H3 phosphorylation, the chromosomes remain condensed until telophase. During telophase, the mitotic chromosomes decondense and regain its interphase nuclear structure.
The exit from mitosis requires inactivation of kinases and reversion of mitotic dephosphorylations of mitotic substrates by phosphatases PP1 (protein phosphatase 1) and PP2A (protein phosphatase 2A) [341, 342, 344]. Depletion of PP1 and PP2A delays the exit from mitosis [376]. PP1 is responsible for the dephosphorylation of Thr23, Ser10 and Ser28 of histone H3 [377]. These dephosphorylations allow histone H4K16 reacetylation and chromosome decondensation. However, PP1 is directed to chromosomes by its Repo-man subunit. Depletion of Repo-Man alters the reformation of the nuclear envelope but has no visible consequence on the chromatin decondensation [378, 379]. PP1 is recruited at chromosomes undergoing decondensation by Ki-67 (MKI67) [380], which could compensate the loss of Repo-man. Because PP1 is recruited before the reformation of the nuclear envelope, this suggests a direct role of PP1 in mitotic decondensation; yet the mechanism is still unclear [381].
To study decondensation mechanisms upon mitosis exit, an interesting approach has been developed in vitro using Xenopus laevis cytoplasmic egg extract system which recapitulates mitotic decondensation [382]. These experiments showed that chromatin decondensation requires energy in the form of ATP and GTP, suggesting that decondensation is an active process and not just chromatin relaxation caused by the dissociation of condensation factors. This is consistent with the fact that decondensation is dependent on the presence of factors present in Xenopus eggs [382]. The dependency on ATP can be explained by the fact that the dissociation of Aurora B requires the ATPase protein p97/VCP [383]. Another ATPase complex has been involved in decondensation, the RuvB-like 1/2 complex [382]. In addition to its implications in many cellular processes, RuvB-like 1/2 is a component of several interphase ATP-dependent chromatin remodeling complexes [269], suggesting that it may act at the end of mitosis to regain an interphasic structure.
Involvement of TADs and CTCF in chromatin structure re-configuration after mitosis
As mentioned above, TADs play a key role in the regulation of gene expression along promoters, enhancers and silencers respectively activate or repress transcription [384]. For example, by folding DNA in space, enhancers and promoters interact and modulate transcription. It is then obvious that an enhancer must be in the same TAD as its target genes. This allows for several genes within the same TAD to be co-regulated by the same enhancer.
The formation of TADs was demonstrated by validating the Loop extrusion model [384]. This mechanism involves cohesin and CTCF proteins which recognize motifs around TADs and drag chromatin through rings [279, 285] . Both cohesin and CTCF are enriched at TAD boundaries [271]; however, their depletion makes TADs less prominent [385].
During mitosis TADs are absent; however, how TAD formation is controlled during the cell cycle remains unclear. Recent 5C analysis demonstrated that TADs and CTCF loops are present in interphase, but absent on mitotic chromatin [386], as stated in previous studies [387]. Additionally, ATAC-seq and CUT&RUN analysis showed reduced accessibility and loss of CTCF binding in prometaphase.
More recently, it has been demonstrated that depletion of CTCF during the M- to G1-phase progression alters the re-formation of chromatin domain boundary and structural loops, leading to inappropriate interactions between cis-regulatory elements (CREs) [388]. Thus, CTCF plays a crucial role in the formation of nuclear architecture during G1 entry, and along transcription, it contributes to chromatin structure re-configuration after mitosis.
Mitotic gene bookmarking: an epigenetic program for cell identity
After mitosis, cells remember their identity and faithfully reactivate their genetic programs [389]. This occurs through gene bookmarking, an epigenetic mechanism involved in the transmission of information during cell division [2, 53, 55]. It is defined as the retention of key regulatory proteins at gene loci on mitotic chromosomes (e.g., chromatin modifying factors, transcription factors, and components of RNA Pol I and II) [55]. This epigenetic mechanism ensures that genes important for the cellular phenotype will be expressed immediately after mitosis.
RUNX1 is a well characterized bookmark for defining mammary epithelial cell identity [329]. Disruption of RUNX1 and its heterodimeric partner CBFβ interaction and association with mitotic chromosomes leads to epithelial-to-mesenchymal transition (EMT) [329]. This shows the importance of bookmarking in stabilization of the mammary epithelial cell identity. Similarly, the HNF1B transcription factor is known to be involved in the conversion of fibroblast into hepatic stem cells and in renal tubular cells [390, 391]. When combined with three additional transcription factors involved in kidney functions, HNF1B induces conversation of mouse and human fibroblasts into induced renal tubular cells (iRECs) [391].
Nascent RNA sequencing technologies, such as precision run-on sequencing (PRO-seq) could be an excellent approach to study bookmarking mechanism [392-396]. PRO-seq directly measures nascent RNA transcription by creating high-resolution maps of all transcriptional engaged RNA polymerases, unlike the traditional RNA-seq or ChIP-seq analyses. PROseq can detect distinct steps of RNA transcription, including RNA polymerase recruitment, promoter-proximal pausing, and transcription elongation. It provides a direct measurement of enhancer activity and allows for quantifying transcription of enhancers and target genes simultaneously. Therefore, PROseq is extremely useful for studying bookmarking and enhancer-mediated regulation during mitosis.
A). Role of the histone post translational modifications (PTMs)
Histone post-translational modifications (PTMs) are responsible for the maintenance of the epigenetic memory, in this way the daughter cells decide which genes need to be reactivated or repressed after division. During mitosis, histones undergo many post-translational modifications, including phosphorylation, methylation, acetylation, and ubiquitylation [397]. These modifications can modify the overall chemical charge of histone proteins, which normally carry an acidic, negative charge, and they can also act on gene expression and chromatin condensation via recruitment or exclusion of proteins from chromatin. The question is whether these changes play a role in the organization of the structure of the mitotic chromosome.
Phosphorylation of histone H3 on serine 10 by Aurora B kinase is one of the most abundant PTM occurring in mitosis [398]. An additional four residues of histone H3 are phosphorylated in mitosis, including threonine 3 and 11 and serines 10 and 28.
Incomplete phosphorylation of histone H3 can lead to condensation and segregation of abnormal chromosomes because of the poor recruitment of condensin I [399, 400]. In early mitosis, the phosphorylation of threonine 3 on histone H3 (H3T3) by Haspin and phosphorylation of serine 10 on histone H3 (H3S10) by Aurora B recruits the deacetylase SIRT2 which in turn deacetylates lysine 16 on histone H4 (H4K16), leading to chromatin compaction [401, 402]. This mechanism has also been described in yeast [403]. In addition, phosphorylation modifications act on the attractive force between nucleosomes [403], which is generated by binding of H4 tails [404, 405]. It results in the displacement of regulatory complexes from mitotic chromatin [397, 406, 407]. For example, phosphorylation on Ser10 of H3K9me3 causes the displacement of the heterochromatin protein HP1 from H3K9me2 or H3K9me3 sites.
Methylation of histones is associated with both the active and repressive marks on the chromatin [403]. For example, mono-di and trimethylation at Lys4 (H3K4me1, H3K4me2 and H3K4me3) are responsible for gene activation and retained on chromatin during mitosis. However, trimethylation of histone H3 at Lys27 (H3K27me3) and Lys9 (H3K9me) are instead associated with repressed genes. So far, these methylations are maintained during mitosis, and are partly responsible for the epigenetic inheritance of cell identity [408, 409].
Acetylation of histones is generally associated with active transcription. Acetylation is decreased during mitosis, but some active promoters retain these marks, for example the acetylation at Lys27 (H3K27ac) on promoter and enhancers [323]. H3K27a seems to be involved in bookmarking enhancers and promoters’ genes critical for stem cell identity [323].
Ubiquitination to H2B is responsible for gene activation, which is decreased in mitosis, whereas ubiquitination H2A is responsible for gene repression [410].
In mitosis, the histone modifications are also able to recruit specific regulatory proteins on the chromatin. For example, phosphorylation on histone H3T3 act as a regulator together with Aurora B during mitosis, resulting in correct attachment of the kinetochore to the microtubules and chromosome separation [411-413].
B). Transcription factors (TFs), key regulators of memory program for resetting transcription
NEBD and DNA structural changes result in the dispersion of RNA polymerases, transcription factors and chromatin remodeling complexes from chromatin, and thus reduction of transcription. Many transcription factors are subjected to post-translational modifications, e.g., phosphorylation, subsequently resulting in changes in the protein structure. These changes alter the activity of the transcription factors, preventing them from recognizing their targets, and thus detaching from the DNA. For example, the transcription factor OCT1 once phosphorylated in mitosis is no longer able to bind DNA and after detaches [414]. Also, phosphorylation of transcription factors such as TFIIH, TFIID, Ikaros leads to reduction of transcription [414-418]. In addition, the transcription factor Sp1 which binds the human Hsp70 promoter shows reduced DNA-binding activity in mitotic cell extracts [419]. In contrast, the DNA-binding activities of three transcription factors bound to Hsp70 promoter (C/EBP, GBP, and HSF1) were normal. This suggests that retention or displacement of transcription factors during mitosis could provide another mechanism in the cell cycle for resetting transcription. Note that phosphorylation not only causes protein structural changes, but it can also cause degradation via ubiquitin degradation system, as in the case of EZH2. In mitosis, phosphorylation EZH2 by CDK1/Cyclin B leads to ubiquitination of EZH2, and subsequent degradation by the proteosome [420].
Other transcription factors, such as RUNX1/2, GATA1, FOXA1, TBP, HNF1B, bind to the chromosomes during mitosis to maintain the memory program [329, 421-426]. So far, it has been demonstrated that the degree of localization of TFs is correlated with some interphase TF features, such as the subnuclear localization, and the absolute charge of the DNA-binding domain [427]. These factors are of great importance to gene reactivation during early G1. For instance, the mitotic retention of Brd4 leads to accelerated decondensation and resumption of post-mitotic transcription [428]. Moreover, Brd4-deficient mouse embryonic fibroblasts exhibit delayed gene expression in early G1 [429].
C). Nucleosome re-positioning, an alternative bookmarking mechanism
The control of nucleosome positioning is another regulatory mechanism controlling the mitotic bookmarking. In mitosis, nucleosomes occupy the transcriptional start sites (TSSs), preventing the binding of the transcriptional machinery, and thus reducing the gene expression. When mitosis ends, nucleosomes recover their initial position to disclose the TSS [430].
D). Mitotic transmission of phenotype specific mRNAs in cancer cells
Beyond chromosome-related bookmarking mechanisms, cells may complement this genomic flagging of genes to maintain phenotype identity by the targeted transmission of mRNAs as a complementary level of translational control. For example, the osteogenic phenotype of osteosarcoma cells is directly linked to the expression of RUNX2. Osteosarcoma cells apply a non-genomic ‘mRNA stocking mechanism’ in which cells produce maximal levels of RUNX2 mRNA, but not protein, prior to mitotic cell division [431]. RUNX2 mRNA then partitions symmetrically between daughter cells in a non-chromosomal tubulin-containing compartment. Somatic transmission of RUNX2 mRNAs in these bone cancer cells ensures that RUNX2 levels can be rapidly replenished during early G1 by translational rather than transcriptional activation to sustain its mitotic bookmarking function during interphase in osteosarcoma cells.
Perturbation of chromosomal rearrangements in cancer
As mentioned above, both condensin I and condensin II are essential for chromosome condensation during mitosis. Equally, condensins regulate the spatial genome organization, replication, and transcription [432]. Therefore, it is well known that condensins are indispensable for maintaining genome stability. For example, depletion of condensin II provokes genome instability by damaging DNA throughout the genome and by impairing telomere function [433]. Condensin depletion can induce breakage–fusion–bridge (BFB) cycles and large chromosomal rearrangements [434]. Additionally, TCGA analysis shows that condensins are mutated in many types of cancers [435]. Most of these mutations are missense and are susceptible to drive sublethal effects on chromosome structure. Together, these results suggest that condensin inhibition may be a potential trigger involved in initiation and progression of tumorigenesis, because of perturbation of the mitotic chromosome condensation, genome instability and telomere dysfunction.
Transcription factor fusion proteins as therapeutic targets
Transcription factor fusions have been identified as a trigger of various subtypes of leukemias. Examples include AML1–ETO and core binding factor β (CBF-β) fused with SMMHC in acute myeloid leukemia. It has been shown that these protein fusions prevent differentiation, assuming a more stem cell-like state [436]. In addition, some of these protein fusions have been shown to cause significant changes in DNA repair genes [437], leading to more mutations and uncontrolled proliferation as well as clonal heterogeneity of leukemia. This finding suggests that transcription factor fusion proteins are a trigger for other genomic alterations, making these chimeric proteins desirable targets for drug development.
A recent approach called PROTACs (or PROteolysis Targeting Chimeras) has been employed to degrade transcription factors in cells with high accuracy [438-440]. PROTACs consist of two-headed small molecules that selectively join the target protein with an E3 ubiquitin ligase. This results in ubiquitylation and proteasome-mediated destruction of the target. Unlike traditional pharmacological approaches, this approach allows a single molecule to degrade multiple targets, and thus represents a key area for future therapy.
Conclusion
As our understanding of the regulatory dimensions of epigenetic control expand, insights into cancer-compromised gene expression are rapidly increasing. The rules governing cancer predisposition, initiation, and progression are, in part, responsive to mutations, but epigenetic signals that structurally and functionally support chromatin remodeling are emerging as a predominant mechanism to determine competency for gene expression or repression. Epigenetic mechanisms support crosstalk between cells and the tissue microenvironment that sustains physiologically responsive control of phenotype and proliferation status. They also contribute to the generation of regulatory states in chromatin necessary for stringently regulated growth control, tissue-specific gene expression, cell mobility, and cell motility. Epigenetic deregulation of these cellular processes directly compromises the essential mechanisms and checkpoints that are required for normal proliferation, survival, and aberrantly promotes the pathological expansion of cancer cells through synergy with the tumor microenvironment. Epigenetic regulation of normal biological processes contributes to several parameters of regulating chromatin organization to gene expression, and disruption of these signaling events promotes pathology. In addition, every cancer-compromised epigenetic mechanism has the potential to provide biomarkers for cancer risk assessment, diagnosis, and treatment responsiveness, and may provide unique opportunities to develop new, and highly specific therapeutics with minimal off-target consequences.
Acknowledgments
This work was supported by the following funding:
P01 CA240685 (GSS, JLS)
U54GM115516 (GSS)
R01GM129338
R01CA230618 (SF)
AR039588 (GSS, JBL)
DE029311 (JBL, GSS, JLS)
F32CA220935 (AJF)
Arthur Perelman Professorship (GSS)
Charlotte Perelman Research Fund (GSS)
References
- 1.Jaenisch R and Bird A, Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nature genetics, 2003. 33(3): p. 245–254. [DOI] [PubMed] [Google Scholar]
- 2.Zaidi SK, et al. , Mitotic gene bookmarking: An epigenetic program to maintain normal and cancer phenotypes, in Molecular Cancer Research. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skinner MK, Role of epigenetics in developmental biology and transgenerational inheritance. Birth Defects Research Part C: Embryo Today: Reviews, 2011. 93(1): p. 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osborne A, The role of epigenetics in human evolution. Bioscience Horizons: The International Journal of Student Research, 2017. 10. [Google Scholar]
- 5.Keyvani-Ghamsari S, et al. , Current understanding of epigenetics mechanism as a novel target in reducing cancer stem cells resistance. Clinical Epigenetics, 2021. 13(1): p. 1–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey A, et al. , Interplay between metabolites and the epigenome in regulating embryonic and adult stem cell potency and maintenance. Stem cell reports, 2019. 13(4): p. 573–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zaidi SK, et al. , Nuclear organization mediates cancer-compromised genetic and epigenetic control. Advances in biological regulation, 2018. 69: p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berdasco M and Esteller M, Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Developmental cell, 2010. 19(5): p. 698–711. [DOI] [PubMed] [Google Scholar]
- 9.Fritz A, et al. , Higher order genomic organization and epigenetic control maintain cellular identity and prevent breast cancer. Genes, Chromosomes and Cancer, 2019. 58(7): p. 484–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sexton T, et al. , Gene regulation through nuclear organization. Nature structural & molecular biology, 2007. 14(11): p. 1049–1055. [DOI] [PubMed] [Google Scholar]
- 11.Zaidi SK, et al. , Nuclear microenvironments in biological control and cancer. Nature Reviews Cancer, 2007. 7(6): p. 454–463. [DOI] [PubMed] [Google Scholar]
- 12.Boija A, Klein IA, and Young RA, Biomolecular condensates and cancer. Cancer cell, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sabari BR, Dall’Agnese A, and Young RA, Biomolecular condensates in the nucleus. Trends in biochemical sciences, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent S, et al. , Phase-separated transcriptional condensates accelerate target-search process revealed by live-cell single-molecule imaging. Cell reports, 2020. 33(2): p. 108248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan D and Weinberg RA, The hallmarks of cancer. cell, 2000. 100(1): p. 57–70. [DOI] [PubMed] [Google Scholar]
- 16.Quagliano A, Gopalakrishnapillai A, and Barwe SP, Understanding the mechanisms by which epigenetic modifiers avert therapy resistance in cancer. Frontiers in oncology, 2020. 10: p. 992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Y, et al. , Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal transduction and targeted therapy, 2019. 4(1): p. 1–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villanueva L, Álvarez-Errico D, and Esteller M, The contribution of epigenetics to cancer immunotherapy. Trends in immunology, 2020. [DOI] [PubMed] [Google Scholar]
- 19.Hildebrand EM and Dekker J, Mechanisms and functions of chromosome compartmentalization. Trends in biochemical sciences, 2020. 45(5): p. 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Misteli T, The self-organizing genome: principles of genome architecture and function. Cell, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghule PN, et al. , Higher order genomic organization and regulatory compartmentalization for cell cycle control at the G1/S-phase transition. Journal of cellular physiology, 2018. 233(10): p. 6406–6413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fritz AJ, et al. , Chromosomes at work: organization of chromosome territories in the interphase nucleus. Journal of cellular biochemistry, 2016. 117(1): p. 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barutcu AR, et al. , C-ing the genome: a compendium of chromosome conformation capture methods to study higher-order chromatin organization. Journal of cellular physiology, 2016. 231(1): p. 31–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fritz AJ, et al. , Chromosome territories and the global regulation of the genome. Genes, Chromosomes and Cancer, 2019. 58(7): p. 407–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kantidze OL and Razin SV, Weak interactions in higher-order chromatin organization. Nucleic acids research, 2020. 48(9): p. 4614–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cremer T, et al. , The 4D nucleome: Evidence for a dynamic nuclear landscape based on co-aligned active and inactive nuclear compartments. FEBS letters, 2015. 589(20): p. 2931–2943. [DOI] [PubMed] [Google Scholar]
- 27.Beagrie RA, et al. , Complex multi-enhancer contacts captured by genome architecture mapping. Nature, 2017. 543(7646): p. 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winick-Ng W, et al. , Cell-type specialization is encoded by specific chromatin topologies. Nature, 2021: p. 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pope BD, et al. , Topologically associating domains are stable units of replication-timing regulation. Nature, 2014. 515(7527): p. 402–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marchal C, Sima J, and Gilbert DM, Control of DNA replication timing in the 3D genome. Nature Reviews Molecular Cell Biology, 2019. 20(12): p. 721–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alberts B, et al. , Chromosomal DNA and its packaging in the chromatin fiber, in Molecular Biology of the Cell. 4th edition. 2002, Garland Science. [Google Scholar]
- 32.Zaidi SK, et al. , The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO reports, 2005. 6(2): p. 128–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kadauke S and Blobel GA, Chromatin loops in gene regulation. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 2009. 1789(1): p. 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krivega I and Dean A, Enhancer and promoter interactions—long distance calls. Current opinion in genetics & development, 2012. 22(2): p. 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levantini E, et al. , RUNX1 regulates the CD34 gene in haematopoietic stem cells by mediating interactions with a distal regulatory element. The EMBO journal, 2011. 30(19): p. 4059–4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stein GS and Berezney R, Nuclear structure and function. Journal of cellular biochemistry, 1996. 62(2): p. 147–148. [DOI] [PubMed] [Google Scholar]
- 37.Zeng C, et al. , Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-α transcription factors. Proceedings of the National Academy of Sciences, 1997. 94(13): p. 6746–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lamond AI and Earnshaw WC, Structure and function in the nucleus. Science, 1998. 280(5363): p. 547–553. [DOI] [PubMed] [Google Scholar]
- 39.McNeil S, et al. , Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situ: The C-terminus is a principal determinant for nuclear trafficking. Journal of cellular biochemistry, 1998. 68(4): p. 500–510. [PubMed] [Google Scholar]
- 40.Verschure PJ, et al. , Spatial relationship between transcription sites and chromosome territories. The Journal of cell biology, 1999. 147(1): p. 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matera AG, Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends in cell biology, 1999. 9(8): p. 302–309. [DOI] [PubMed] [Google Scholar]
- 42.Stein GS, et al. , Nuclear structure-gene expression interrelationships: implications for aberrant gene expression in cancer. Cancer research, 2000. 60(8): p. 2067–2076. [PubMed] [Google Scholar]
- 43.Fritz AJ, et al. , Intranuclear and higher-order chromatin organization of the major histone gene cluster in breast cancer. Journal of cellular physiology, 2018. 233(2): p. 1278–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stein GS, et al. , Temporal and spatial parameters of skeletal gene expression: targeting RUNX factors and their coregulatory proteins to subnuclear domains. Connective tissue research, 2003. 44(1): p. 149–153. [PubMed] [Google Scholar]
- 45.Stein GS, et al. , Intranuclear organization of RUNX transcriptional regulatory machinery in biological control of skeletogenesis and cancer. Blood Cells, Molecules, and Diseases, 2003. 30(2): p. 170–176. [DOI] [PubMed] [Google Scholar]
- 46.Stein GS, et al. , Nuclear microenvironments support assembly and organization of the transcriptional regulatory machinery for cell proliferation and differentiation. Journal of cellular biochemistry, 2004. 91(2): p. 287–302. [DOI] [PubMed] [Google Scholar]
- 47.Stein GS, et al. , Intranuclear trafficking of transcription factors: implications for biological control. Journal of cell science, 2000. 113(14): p. 2527–2533. [DOI] [PubMed] [Google Scholar]
- 48.Stein GS, et al. , Subnuclear organization and trafficking of regulatory proteins: implications for biological control and cancer. Journal of Cellular Biochemistry, 2000. 79(S35): p. 84–92. [PubMed] [Google Scholar]
- 49.Stein GS, et al. , An architectural perspective of cell-cycle control at the G1/S phase cell-cycle transition. Journal of cellular physiology, 2006. 209(3): p. 706–710. [DOI] [PubMed] [Google Scholar]
- 50.Stein GS, et al. , Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends in cell biology, 2003. 13(11): p. 584–592. [DOI] [PubMed] [Google Scholar]
- 51.Zaidi SK, et al. , A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. Journal of cell science, 2001. 114(17): p. 3093–3102. [DOI] [PubMed] [Google Scholar]
- 52.Zaidi SK, et al. , Alterations in intranuclear localization of Runx2 affect biological activity. Journal of cellular physiology, 2006. 209(3): p. 935–942. [DOI] [PubMed] [Google Scholar]
- 53.Zaidi SK, et al. , Mitotic gene bookmarking: An epigenetic mechanism for coordination of lineage commitment, cell identity and cell growth. In Advances in Experimental Medicine and Biology. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaidi SK, et al. , Bookmarking the genome: Maintenance of epigenetic information, in Journal of Biological Chemistry. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zaidi SK, et al. , Mitotic bookmarking of genes: A novel dimension to epigenetic control, in Nature Reviews Genetics. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spector DL, The dynamics of chromosome organization and gene regulation. Annual review of biochemistry, 2003. 72(1): p. 573–608. [DOI] [PubMed] [Google Scholar]
- 57.Barseguian K, et al. , Multiple subnuclear targeting signals of the leukemia-related AML1/ETO and ETO repressor proteins. Proceedings of the National Academy of Sciences, 2002. 99(24): p. 15434–15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harrington KS, et al. , Transcription factors RUNX1/AML1 and RUNX2/Cbfa1 dynamically associate with stationary subnuclear domains. Journal of cell science, 2002. 115(21): p. 4167–4176. [DOI] [PubMed] [Google Scholar]
- 59.Zink D, Fischer AH, and Nickerson JA, Nuclear structure in cancer cells. Nature reviews cancer, 2004. 4(9): p. 677–687. [DOI] [PubMed] [Google Scholar]
- 60.Handwerger KE and Gall JG, Subnuclear organelles: new insights into form and function. Trends in cell biology, 2006. 16(1): p. 19–26. [DOI] [PubMed] [Google Scholar]
- 61.Drobic B, et al. , Abnormalities of chromatin in tumor cells. Cancer: Cell Structures, Carcinogens and Genomic Instability, 2006: p. 25–47. [DOI] [PubMed] [Google Scholar]
- 62.Schneider R and Grosschedl R, Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes & development, 2007. 21(23): p. 3027–3043. [DOI] [PubMed] [Google Scholar]
- 63.Boisvert F-M, et al. , The multifunctional nucleolus. Nature reviews Molecular cell biology, 2007. 8(7): p. 574–585. [DOI] [PubMed] [Google Scholar]
- 64.Misteli T, Beyond the sequence: cellular organization of genome function. Cell, 2007. 128(4): p. 787–800. [DOI] [PubMed] [Google Scholar]
- 65.Dundr M and Misteli T, Biogenesis of nuclear bodies. Cold Spring Harbor perspectives in biology, 2010. 2(12): p. a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lever E and Sheer D, The role of nuclear organization in cancer. The Journal of Pathology: A Journal of the Pathological Society of Great Britain and Ireland, 2010. 220(2): p. 114–125. [DOI] [PubMed] [Google Scholar]
- 67.Rajapakse I and Groudine M, On emerging nuclear order. Journal of Cell Biology, 2011. 192(5): p. 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reddy KL and Feinberg AP. Higher order chromatin organization in cancer, in Seminars in cancer biology. 2013. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gibcus JH and Dekker J, The hierarchy of the 3D genome. Molecular cell, 2013. 49(5): p. 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sleeman JE and Trinkle-Mulcahy L, Nuclear bodies: new insights into assembly/dynamics and disease relevance. Current opinion in cell biology, 2014. 28: p. 76–83. [DOI] [PubMed] [Google Scholar]
- 71.Hancock R, The crowded nucleus. International review of cell and molecular biology, 2014. 307: p. 15–26. [DOI] [PubMed] [Google Scholar]
- 72.Pombo A and Dillon N, Three-dimensional genome architecture: players and mechanisms. Nature reviews Molecular cell biology, 2015. 16(4): p. 245–257. [DOI] [PubMed] [Google Scholar]
- 73.Fritz AJ, et al. , Cell type specific alterations in interchromosomal networks across the cell cycle. PLoS computational biology, 2014. 10(10): p. e1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fritz AJ, et al. , Wide-scale alterations in interchromosomal organization in breast cancer cells: defining a network of interacting chromosomes. Human molecular genetics, 2014. 23(19): p. 5133–5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sehgal N, et al. , Gene density and chromosome territory shape. Chromosoma, 2014. 123(5): p. 499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sehgal N, et al. , Large-scale probabilistic 3D organization of human chromosome territories. Human molecular genetics, 2016. 25(3): p. 419–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jones PA, Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature Reviews Genetics, 2012. 13(7): p. 484–492. [DOI] [PubMed] [Google Scholar]
- 78.Zhu H, Wang G, and Qian J, Transcription factors as readers and effectors of DNA methylation. Nature Reviews Genetics, 2016. 17(9): p. 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Greenberg MV and Bourc’his D, The diverse roles of DNA methylation in mammalian development and disease. Nature reviews Molecular cell biology, 2019. 20(10): p. 590–607. [DOI] [PubMed] [Google Scholar]
- 80.Petryk N, et al. , Staying true to yourself: mechanisms of DNA methylation maintenance in mammals. Nucleic Acids Research, 2021. 49(6): p. 3020–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sharp AJ, et al. , DNA methylation profiles of human active and inactive X chromosomes. Genome research, 2011. 21(10): p. 1592–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Elhamamsy AR, Role of DNA methylation in imprinting disorders: an updated review. Journal of Assisted Reproduction and Genetics, 2017. 34(5): p. 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choi JY and Lee YCG, Double-edged sword: The evolutionary consequences of the epigenetic silencing of transposable elements. PLoS genetics, 2020. 16(7): p. e1008872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rasmussen KD and Helin K, Role of TET enzymes in DNA methylation, development, and cancer. Genes & development, 2016. 30(7): p. 733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.An J, Rao A, and Ko M, TET family dioxygenases and DNA demethylation in stem cells and cancers. Experimental & molecular medicine, 2017. 49(4): p. e323–e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiong J, et al. , Cooperative action between SALL4A and TET proteins in stepwise oxidation of 5-methylcytosine. Molecular cell, 2016. 64(5): p. 913–925. [DOI] [PubMed] [Google Scholar]
- 87.Du Q, et al. , Methyl-CpG-binding domain proteins: readers of the epigenome. Epigenomics, 2015. 7(6): p. 1051–1073. [DOI] [PubMed] [Google Scholar]
- 88.Guibert S and Weber M, Functions of DNA methylation and hydroxymethylation in mammalian development. Current topics in developmental biology, 2013. 104: p. 47–83. [DOI] [PubMed] [Google Scholar]
- 89.Hegde M and Joshi MB, Comprehensive analysis of regulation of DNA methyltransferase isoforms in human breast tumors. Journal of Cancer Research and Clinical Oncology, 2021: p. 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zighelboim I, et al. , Microsatellite instability and epigenetic inactivation of MLH1 and outcome of patients with endometrial carcinomas of the endometrioid type. Journal of Clinical Oncology, 2007. 25(15): p. 2042–2048. [DOI] [PubMed] [Google Scholar]
- 91.Cosgrove CM, et al. , Epigenetic silencing of MLH1 in endometrial cancers is associated with larger tumor volume, increased rate of lymph node positivity and reduced recurrence-free survival. Gynecologic oncology, 2017. 146(3): p. 588–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Loukovaara M, Pasanen A, and Bützow R, Mismatch repair protein and MLH1 methylation status as predictors of response to adjuvant therapy in endometrial cancer. Cancer medicine, 2021. 10(3): p. 1034–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sharma P, et al. , The prognostic value of BRCA1 promoter methylation in early stage triple negative breast cancer. Journal of cancer therapeutics & research, 2014. 3(2): p. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhu X, et al. , Hypermethylation of BRCA1 gene: implication for prognostic biomarker and therapeutic target in sporadic primary triple-negative breast cancer. Breast cancer research and treatment, 2015. 150(3): p. 479–486. [DOI] [PubMed] [Google Scholar]
- 95.Wong KK DNMT1: A key drug target in triple-negative breast cancer. In Seminars in Cancer Biology. 2021. Elsevier. [DOI] [PubMed] [Google Scholar]
- 96.Christman JK, 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene, 2002. 21(35): p. 5483–5495. [DOI] [PubMed] [Google Scholar]
- 97.Jang J-S, et al. , Methyl-CpG binding domain 1 gene polymorphisms and risk of primary lung cancer. Cancer Epidemiology and Prevention Biomarkers, 2005. 14(11): p. 2474–2480. [DOI] [PubMed] [Google Scholar]
- 98.Mariño-Ramírez L, et al. , Histone structure and nucleosome stability. Expert review of proteomics, 2005. 2(5): p. 719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Calo E and Wysocka J, Modification of enhancer chromatin: what, how, and why? Molecular cell, 2013. 49(5): p. 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang Z, et al. , Combinatorial patterns of histone acetylations and methylations in the human genome. Nature genetics, 2008. 40(7): p. 897–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Collins BE, et al. , Histone H3 lysine K4 methylation and its role in learning and memory. Epigenetics & chromatin, 2019. 12(1): p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cao R, et al. , Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science, 2002. 298(5595): p. 1039–1043. [DOI] [PubMed] [Google Scholar]
- 103.Wagner EJ and Carpenter PB, Understanding the language of Lys36 methylation at histone H3. Nature reviews Molecular cell biology, 2012. 13(2): p. 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Grandy R, Tomaz RA, and Vallier L, Modeling disease with human inducible pluripotent stem cells. Annual Review of Pathology: Mechanisms of Disease, 2019. 14: p. 449–468. [DOI] [PubMed] [Google Scholar]
- 105.Grandy RA, et al. , Genome-wide studies reveal that H3K4me3 modification in bivalent genes is dynamically regulated during the pluripotent cell cycle and stabilized upon differentiation. Molecular and cellular biology, 2015. 36(4): p. 615–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Voigt P, Tee W-W, and Reinberg D, A double take on bivalent promoters. Genes & development, 2013. 27(12): p. 1318–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bernstein BE, et al. , A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell, 2006. 125(2): p. 315–326. [DOI] [PubMed] [Google Scholar]
- 108.Messier TL, et al. , Oncofetal epigenetic bivalency in breast cancer cells: H3K4 and H3K27 tri-methylation as a biomarker for phenotypic plasticity. Journal of cellular physiology, 2016. 231(11): p. 2474–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zaidi SK, et al. , Bivalent epigenetic control of oncofetal gene expression in cancer. Molecular and cellular biology, 2017. 37(23): p. e00352–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lu Y, et al. , Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Molecular cancer, 2020. 19(1): p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hosey AM, Chaturvedi C-P, and Brand M, Crosstalk between histone modifications maintains the developmental pattern of gene expression on a tissue-specific locus. Epigenetics, 2010. 5(4): p. 273–281. [DOI] [PubMed] [Google Scholar]
- 112.Kim JJ, Lee SY, and Miller KM, Preserving genome integrity and function: the DNA damage response and histone modifications. Critical reviews in biochemistry and molecular biology, 2019. 54(3): p. 208–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wojcik F, et al. , Functional crosstalk between histone H2B ubiquitylation and H2A modifications and variants. Nature communications, 2018. 9(1): p. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Varier RA, et al. , A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. The EMBO journal, 2010. 29(23): p. 3967–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Komar D and Juszczynski P, Rebelled epigenome: histone H3S10 phosphorylation and H3S10 kinases in cancer biology and therapy. Clinical Epigenetics, 2020. 12(1): p. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Du J, et al. , DNA methylation pathways and their crosstalk with histone methylation. Nature reviews Molecular cell biology, 2015. 16(9): p. 519–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kondo Y, Epigenetic cross-talk between DNA methylation and histone modifications in human cancers. Yonsei medical journal, 2009. 50(4): p. 455–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gujar H, Weisenberger DJ, and Liang G, The roles of human DNA methyltransferases and their isoforms in shaping the epigenome. Genes, 2019. 10(2): p. 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moore LD, Le T, and Fan G, DNA methylation and its basic function. Neuropsychopharmacology, 2013. 38(1): p. 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Rothbart SB, et al. , Association of UHRF1 with methylated H3K9 directs the maintenance of DNA methylation. Nature structural & molecular biology, 2012. 19(11): p. 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bernhart SH, et al. , Changes of bivalent chromatin coincide with increased expression of developmental genes in cancer. Scientific reports, 2016. 6(1): p. 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dunican DS, et al. , Bivalent promoter hypermethylation in cancer is linked to the H327me3/H3K4me3 ratio in embryonic stem cells. BMC biology, 2020. 18(1): p. 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Audia JE and Campbell RM, Histone modifications and cancer. Cold Spring Harbor perspectives in biology, 2016. 8(4): p. a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shen H and Laird PW, Interplay between the cancer genome and epigenome. Cell, 2013. 153(1): p. 38–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Fang L, et al. , SET1A-mediated mono-methylation at K342 regulates YAP activation by blocking its nuclear export and promotes tumorigenesis. Cancer Cell, 2018. 34(1): p. 103–118. e9. [DOI] [PubMed] [Google Scholar]
- 126.Salz T, et al. , Histone methyltransferase hSETD1A is a novel regulator of metastasis in breast cancer. Molecular Cancer Research, 2015. 13(3): p. 461–469. [DOI] [PubMed] [Google Scholar]
- 127.Smith E, Lin C, and Shilatifard A, The super elongation complex (SEC) and MLL in development and disease. Genes & development, 2011. 25(7): p. 661–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morgan MA and Shilatifard A, Chromatin signatures of cancer. Genes & development, 2015. 29(3): p. 238–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang L, et al. , Resetting the epigenetic balance of Polycomb and COMPASS function at enhancers for cancer therapy. Nature medicine, 2018. 24(6): p. 758–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sze CC and Shilatifard A, MLL3/MLL4/COMPASS family on epigenetic regulation of enhancer function and cancer. Cold Spring Harbor perspectives in medicine, 2016. 6(11): p. a026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fantini D, et al. , A Carcinogen-induced mouse model recapitulates the molecular alterations of human muscle invasive bladder cancer. Oncogene, 2018. 37(14): p. 1911–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zheng J, Oncogenic chromosomal translocations and human cancer. Oncology reports, 2013. 30(5): p. 2011–2019. [DOI] [PubMed] [Google Scholar]
- 133.Wang J, et al. , Conditional MLL-CBP targets GMP and models therapy-related myeloproliferative disease. The EMBO journal, 2005. 24(2): p. 368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ilencikova D and Kolenova A, MLL Gene Alterations in Acute Myeloid. Oncogene and Cancer: From Bench to Clinic, 2013: p. 225. [Google Scholar]
- 135.Li Y and Seto E, HDACs and HDAC inhibitors in cancer development and therapy. Cold Spring Harbor perspectives in medicine, 2016. 6(10): p. a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ramaiah MJ, Tangutur AD, and Manyam RR, Epigenetic modulation and understanding of HDAC inhibitors in cancer therapy. Life Sciences, 2021: p. 119504. [DOI] [PubMed] [Google Scholar]
- 137.Ellis L, Atadja PW, and Johnstone RW, Epigenetics in cancer: targeting chromatin modifications. Molecular cancer therapeutics, 2009. 8(6): p. 1409–1420. [DOI] [PubMed] [Google Scholar]
- 138.Ropero S and Esteller M, The role of histone deacetylases (HDACs) in human cancer. Molecular oncology, 2007. 1(1): p. 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Mio C, et al. , Reading cancer: Chromatin readers as druggable targets for cancer treatment. Cancers, 2019. 11(1): p. 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jain AK and Barton MC, Bromodomain histone readers and cancer. Journal of molecular biology, 2017. 429(13): p. 2003–2010. [DOI] [PubMed] [Google Scholar]
- 141.van Wijnen AJ, et al. , Biological functions of chromobox (CBX) proteins in stem cell self-renewal, lineage-commitment, cancer and development. Bone, 2021. 143: p. 115659. [DOI] [PubMed] [Google Scholar]
- 142.Perez-Salvia M and Esteller M, Bromodomain inhibitors and cancer therapy: From structures to applications. Epigenetics, 2017. 12(5): p. 323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zou JX, et al. , Kinesin family deregulation coordinated by bromodomain protein ANCCA and histone methyltransferase MLL for breast cancer cell growth, survival, and tamoxifen resistance. Molecular Cancer Research, 2014. 12(4): p. 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhang Y, et al. , ANCCA protein expression is a novel independent poor prognostic marker in surgically resected lung adenocarcinoma. Annals of surgical oncology, 2013. 20(3): p. 577–582. [DOI] [PubMed] [Google Scholar]
- 145.Kalashnikova EV, et al. , ANCCA/ATAD2 overexpression identifies breast cancer patients with poor prognosis, acting to drive proliferation and survival of triple-negative cells through control of B-Myb and EZH2. Cancer research, 2010. 70(22): p. 9402–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Caron C, et al. , Functional characterization of ATAD2 as a new cancer/testis factor and a predictor of poor prognosis in breast and lung cancers. Oncogene, 2010. 29(37): p. 5171–5181. [DOI] [PubMed] [Google Scholar]
- 147.Koo SJ, et al. , ATAD2 is an epigenetic reader of newly synthesized histone marks during DNA replication. Oncotarget, 2016. 7(43): p. 70323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Liu N, et al. , ATAD2 is associated with malignant characteristics of pancreatic cancer cells. Oncology letters, 2019. 17(3): p. 3489–3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Evans CM, et al. , Coordination of di-acetylated histone ligands by the ATAD2 bromodomain. International journal of molecular sciences, 2021. 22(17): p. 9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gay JC, et al. , Disulfide bridge formation influences ligand recognition by the ATAD2 bromodomain. Proteins: Structure, Function, and Bioinformatics, 2019. 87(2): p. 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ning B, et al. , Targeting epigenetic regulations in cancer. Acta biochimica et biophysica Sinica, 2016. 48(1): p. 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ramadoss M and Mahadevan V, Targeting the cancer epigenome: synergistic therapy with bromodomain inhibitors. Drug discovery today, 2018. 23(1): p. 76–89. [DOI] [PubMed] [Google Scholar]
- 153.Hübner MR, Eckersley-Maslin MA, and Spector DL, Chromatin organization and transcriptional regulation. Current opinion in genetics & development, 2013. 23(2): p. 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Maeshima K, et al. , Chromatin as dynamic 10-nm fibers. Chromosoma, 2014. 123(3): p. 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ou HD, et al. , ChromEMT: Visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science, 2017. 357(6349). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yue H, et al. , Single-molecule studies of the linker histone H1 binding to DNA and the nucleosome. Biochemistry, 2016. 55(14): p. 2069–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Prendergast L and Reinberg D, The missing linker: emerging trends for H1 variant-specific functions. Genes & Development, 2021. 35(1-2): p. 40–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hergeth SP and Schneider R, The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO reports, 2015. 16(11): p. 1439–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Yang S-M, et al. , H1 linker histone promotes epigenetic silencing by regulating both DNA methylation and histone H3 methylation. Proceedings of the National Academy of Sciences, 2013. 110(5): p. 1708–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Andrés M, et al. , Histone H1 post-translational modifications: update and future perspectives. International journal of molecular sciences, 2020. 21(16): p. 5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nirala NK, et al. , Hinfp is a guardian of the somatic genome by repressing transposable elements. Proceedings of the National Academy of Sciences, 2021. 118(41). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brockers K and Schneider R, Histone H1, the forgotten histone. 2019, Future Medicine. [DOI] [PubMed] [Google Scholar]
- 163.Scaffidi P, Histone H1 alterations in cancer. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 2016. 1859(3): p. 533–539. [DOI] [PubMed] [Google Scholar]
- 164.Soshnev AA, et al. , Histone H1 mutations in lymphoma: a link (er) between chromatin organization, developmental reprogramming, and cancer. Cancer Research, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Gill CM, et al. , SWI/SNF chromatin remodeling complex alterations in meningioma. Journal of Cancer Research and Clinical Oncology, 2021: p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Biegel JA, Busse TM, and Weissman BE. SWI/SNF chromatin remodeling complexes and cancer. in American Journal of Medical Genetics Part C: Seminars in Medical Genetics. 2014. Wiley Online Library. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Padilla-Benavides T, Reyes-Gutierrez P, and Imbalzano AN, Regulation of the Mammalian SWI/SNF Family of Chromatin Remodeling Enzymes by Phosphorylation during Myogenesis. Biology, 2020. 9(7): p. 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Masliah-Planchon J, et al. , SWI/SNF chromatin remodeling and human malignancies. Annual Review of Pathology: Mechanisms of Disease, 2015. 10: p. 145–171. [DOI] [PubMed] [Google Scholar]
- 169.Fox AH, et al. , Paraspeckles: a novel nuclear domain. Current Biology, 2002. 12(1): p. 13–25. [DOI] [PubMed] [Google Scholar]
- 170.Kawaguchi T, et al. , SWI/SNF chromatin-remodeling complexes function in noncoding RNA-dependent assembly of nuclear bodies. Proceedings of the National Academy of Sciences, 2015. 112(14): p. 4304–4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Fox AH and Lamond AI, Paraspeckles. Cold Spring Harbor perspectives in biology, 2010. 2(7): p. a000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Schoenfelder S and Fraser P, Long-range enhancer–promoter contacts in gene expression control. Nature Reviews Genetics, 2019. 20(8): p. 437–455. [DOI] [PubMed] [Google Scholar]
- 173.Kim J and Dean A, Enhancers navigate the three-dimensional genome to direct cell fate decisions. Current Opinion in Structural Biology, 2021. 71: p. 101–109. [DOI] [PubMed] [Google Scholar]
- 174.Eivazova ER and Aune TM, Dynamic alterations in the conformation of the Ifng gene region during T helper cell differentiation. Proceedings of the National Academy of Sciences, 2004. 101(1): p. 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Majumder P and Boss JM, CTCF controls expression and chromatin architecture of the human major histocompatibility complex class II locus. Molecular and cellular biology, 2010. 30(17): p. 4211–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Carter D, et al. , Long-range chromatin regulatory interactions in vivo. Nature genetics, 2002. 32(4): p. 623–626. [DOI] [PubMed] [Google Scholar]
- 177.Palstra R-J, et al. , The β-globin nuclear compartment in development and erythroid differentiation. Nature genetics, 2003. 35(2): p. 190–194. [DOI] [PubMed] [Google Scholar]
- 178.Tolhuis B, et al. , Looping and interaction between hypersensitive sites in the active β-globin locus. Molecular cell, 2002. 10(6): p. 1453–1465. [DOI] [PubMed] [Google Scholar]
- 179.Gheldof N, et al. , Cell-type-specific long-range looping interactions identify distant regulatory elements of the CFTR gene. Nucleic acids research, 2010. 38(13): p. 4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Drissen R, et al. , The active spatial organization of the β-globin locus requires the transcription factor EKLF. Genes & development, 2004. 18(20): p. 2485–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Owen TA, et al. , Postproliferative transcription of the rat osteocalcin gene is reflected by vitamin D-responsive developmental modifications in protein-DNA interactions at basal and enhancer promoter elements. Proceedings of the National Academy of Sciences, 1993. 90(4): p. 1503–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Montecino M, et al. , DNase I hypersensitive sites in promoter elements associated with basal and vitamin D dependent transcription of the bone-specific osteocalcin gene. Biochemistry, 1994. 33(1): p. 348–353. [DOI] [PubMed] [Google Scholar]
- 183.Banerjee C, et al. , An AML-1 consensus sequence binds an osteoblast-specific complex and transcriptionally activates the osteocalcin gene. Proceedings of the National Academy of Sciences, 1996. 93(10): p. 4968–4973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Montecino M, et al. , Changes in chromatin structure support constitutive and developmentally regulated transcription of the bone-specific osteocalcin gene in osteoblastic cells. Biochemistry, 1996. 35(15): p. 5093–5102. [DOI] [PubMed] [Google Scholar]
- 185.Montecino M, et al. , Requirement of distal and proximal promoter sequences for chromatin organization of the osteocalcin gene in bone-derived cells. Journal of cellular biochemistry, 1996. 63(2): p. 221–228. [DOI] [PubMed] [Google Scholar]
- 186.Guo B, et al. , YY1 regulates vitamin D receptor/retinoid X receptor mediated transactivation of the vitamin D responsive osteocalcin gene. Proceedings of the National Academy of Sciences, 1997. 94(1): p. 121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Guo B, et al. , ATF1 and CREB trans-activate a cell cycle regulated histone H4 gene at a distal nuclear matrix associated promoter element. Biochemistry, 1997. 36(47): p. 14447–14455. [DOI] [PubMed] [Google Scholar]
- 188.Montecino M, et al. , Chromatin hyperacetylation abrogates vitamin D-mediated transcriptional upregulation of the tissue-specific osteocalcin gene in vivo. Biochemistry, 1999. 38(4): p. 1338–1345. [DOI] [PubMed] [Google Scholar]
- 189.Montecino M, et al. , Phosphorylation-mediated control of chromatin organization and transcriptional activity of the tissue-specific osteocalcin gene. Journal of cellular biochemistry, 1999. 72(4): p. 586–594. [DOI] [PubMed] [Google Scholar]
- 190.Javed A, et al. , Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D-responsive transcription and contribute to chromatin organization. Molecular and cellular biology, 1999. 19(11): p. 7491–7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Gutiérrez J, et al. , Interaction of CBFα/AML/PEBP2α transcription factors with nucleosomes containing promoter sequences requires flexibility in the translational positioning of the histone octamer and exposure of the CBFα site. Biochemistry, 2000. 39(44): p. 13565–13574. [DOI] [PubMed] [Google Scholar]
- 192.Gutierrez S, et al. , CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. Journal of Biological Chemistry, 2002. 277(2): p. 1316–1323. [DOI] [PubMed] [Google Scholar]
- 193.Shen J, et al. , Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D-dependent, bone tissue-specific transcription. Journal of Biological Chemistry, 2002. 277(23): p. 20284–20292. [DOI] [PubMed] [Google Scholar]
- 194.Drissi H, et al. , Identification of novel protein/DNA interactions within the promoter of the bone-related transcription factor Runx2/Cbfa1. Journal of cellular biochemistry, 2002. 86(2): p. 403–412. [DOI] [PubMed] [Google Scholar]
- 195.Kundu M, et al. , Cbfβ interacts with Runx2 and has a critical role in bone development. Nature genetics, 2002. 32(4): p. 639–644. [DOI] [PubMed] [Google Scholar]
- 196.Paredes R, et al. , Interaction of the 1α, 25-dihydroxyvitamin D3 receptor at the distal promoter region of the bone-specific osteocalcin gene requires nucleosomal remodelling. Biochemical Journal, 2002. 363(3): p. 667–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shen J, et al. , Transcriptional induction of the osteocalcin gene during osteoblast differentiation involves acetylation of histones h3 and h4. Molecular Endocrinology, 2003. 17(4): p. 743–756. [DOI] [PubMed] [Google Scholar]
- 198.Sierra J, et al. , Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Molecular and cellular biology, 2003. 23(9): p. 3339–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Gutierrez S, et al. , The vitamin D response element in the distal osteocalcin promoter contributes to chromatin organization of the proximal regulatory domain. Journal of Biological Chemistry, 2004. 279(42): p. 43581–43588. [DOI] [PubMed] [Google Scholar]
- 200.Paredes R, et al. , Bone-specific transcription factor Runx2 interacts with the 1α, 25-dihydroxyvitamin D3 receptor to up-regulate rat osteocalcin gene expression in osteoblastic cells. Molecular and cellular biology, 2004. 24(20): p. 8847–8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Villagra A, et al. , Chromatin remodeling and transcriptional activity of the bone-specific osteocalcin gene require CCAAT/enhancer-binding protein β-dependent recruitment of SWI/SNF activity. Journal of Biological Chemistry, 2006. 281(32): p. 22695–22706. [DOI] [PubMed] [Google Scholar]
- 202.Cruzat F, et al. , SWI/SNF-independent nuclease hypersensitivity and an increased level of histone acetylation at the P1 promoter accompany active transcription of the bone master gene Runx2. Biochemistry, 2009. 48(30): p. 7287–7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Stein G, et al. , Regulation of cell cycle stage-specific transcription of histone genes from chromatin by non-histone chromosomal proteins. Nature, 1975. 257(5529): p. 764–767. [DOI] [PubMed] [Google Scholar]
- 204.Park WD, Stein JL, and Stein GS, Activation of in vitro histone gene transcription from Hela S3 chromatin by S-phase nonhistone chromosomal proteins. Biochemistry, 1976. 15(15): p. 3296–3300. [DOI] [PubMed] [Google Scholar]
- 205.Chrysogelos S, et al. , A human histone H4 gene exhibits cell cycle-dependent changes in chromatin structure that correlate with its expression. Proceedings of the National Academy of Sciences, 1985. 82(22): p. 7535–7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pauli U, et al. , Protein-DNA interactions in vivo upstream of a cell cycle-regulated human H4 histone gene. Science, 1987. 236(4806): p. 1308–1311. [DOI] [PubMed] [Google Scholar]
- 207.Kroeger P, et al. , Proximal and distal regulatory elements that influence in vivo expression of a cell cycle-dependent human H4 histone gene. Proceedings of the National Academy of Sciences, 1987. 84(12): p. 3982–3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Moreno M, et al. , Persistence of a micrococcal nuclease sensitive region spanning the promoter–coding region junction of a cell cycle regulated human H4 histone gene throughout the cell cycle. Biochemistry and Cell Biology, 1988. 66(2): p. 132–137. [DOI] [PubMed] [Google Scholar]
- 209.Chrysogelos S, et al. , Fine mapping of the chromatin structure of a cell cycle-regulated human H4 histone gene. Journal of Biological Chemistry, 1989. 264(2): p. 1232–1237. [PubMed] [Google Scholar]
- 210.Holthuis J, et al. , Tumor cells exhibit deregulation of the cell cycle histone gene promoter factor HiNF-D. Science, 1990. 247(4949): p. 1454–1457. [DOI] [PubMed] [Google Scholar]
- 211.Hovhannisyan H, et al. , Maintenance of open chromatin and selective genomic occupancy at the cell cycle-regulated histone H4 promoter during differentiation of HL-60 promyelocytic leukemia cells. Molecular and cellular biology, 2003. 23(4): p. 1460–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Mitra P, et al. , Identification of HiNF-P, a key activator of cell cycle-controlled histone H4 genes at the onset of S phase. Molecular and cellular biology, 2003. 23(22): p. 8110–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Miele A, et al. , HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Molecular and cellular biology, 2005. 25(14): p. 6140–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Moreno M, et al. , Reversible changes in the nucleosomal organization of a human H4 histone gene during the cell cycle. Biochemistry, 1986. 25(19): p. 5364–5370. [DOI] [PubMed] [Google Scholar]
- 215.Zeng C, et al. , Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proceedings of the National Academy of Sciences, 1998. 95(4): p. 1585–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Ghule PN, et al. , Maternal expression and early induction of histone gene transcription factor Hinfp sustains development in pre-implantation embryos. Developmental biology, 2016. 419(2): p. 311–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Van Wijnen AJ, et al. , Human H4 histone gene transcription requires the proliferation-specific nuclear factor HiNF-D: auxiliary roles for HiNF-C (Sp1-like) and HiNF-A (high mobility group-like). Journal of Biological Chemistry, 1989. 264(25): p. 15034–15042. [PubMed] [Google Scholar]
- 218.Van Wijnen AJ, et al. , Protein/DNA interactions involving ATF/AP1-, CCAAT-, and HiNF-D-related factors in the human H3-ST519 histone promoter: Cross-competition with transcription regulatory sites in cell cycle controlled H4 and H1 histone genes. Journal of cellular biochemistry, 1991. 47(4): p. 337–351. [DOI] [PubMed] [Google Scholar]
- 219.Ramsey-Ewing AL, et al. , Histone H4 proximal promoter mediates a complex transcriptional response during differentiation of 3T3L1 adipocytes. Journal of cellular physiology, 1995. 163(2): p. 312–320. [DOI] [PubMed] [Google Scholar]
- 220.Marashi F, et al. , Variations in the organization and expression of human histone genes in normal diploid and tumor cell lines. Anticancer research, 1984. 4(1-2): p. 69–74. [PubMed] [Google Scholar]
- 221.Holmes WF, et al. , Coordinate control and selective expression of the full complement of replication-dependent histone H4 genes in normal and cancer cells. Journal of Biological Chemistry, 2005. 280(45): p. 37400–37407. [DOI] [PubMed] [Google Scholar]
- 222.Ghule PN, et al. , The subnuclear organization of histone gene regulatory proteins and 3' end processing factors of normal somatic and embryonic stem cells is compromised in selected human cancer cell types. Journal of cellular physiology, 2009. 220(1): p. 129–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Vaughan PS, et al. , Activation of a cell-cycle-regulated histone gene by the oncogenic transcription factor IRF-2. Nature, 1995. 377(6547): p. 362–365. [DOI] [PubMed] [Google Scholar]
- 224.Becker KA, et al. , Self-renewal of human embryonic stem cells is supported by a shortened G1 cell cycle phase. Journal of cellular physiology, 2006. 209(3): p. 883–893. [DOI] [PubMed] [Google Scholar]
- 225.Becker KA, et al. , Establishment of histone gene regulation and cell cycle checkpoint control in human embryonic stem cells. Journal of cellular physiology, 2007. 210(2): p. 517–526. [DOI] [PubMed] [Google Scholar]
- 226.Xie R, et al. , The histone gene activator HINFP is a nonredundant cyclin E/CDK2 effector during early embryonic cell cycles. Proceedings of the National Academy of Sciences, 2009. 106(30): p. 12359–12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Becker KA, et al. , Cyclin D2 and the CDK substrate p220NPAT are required for self-renewal of human embryonic stem cells. Journal of cellular physiology, 2010. 222(2): p. 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Becker KA, et al. , Human embryonic stem cells are pre-mitotically committed to self-renewal and acquire a lengthened G1 phase upon lineage programming. Journal of cellular physiology, 2010. 222(1): p. 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Ghule PN, et al. , Reprogramming the pluripotent cell cycle: restoration of an abbreviated G1 phase in human induced pluripotent stem (iPS) cells. Journal of cellular physiology, 2011. 226(5): p. 1149–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Medina R, et al. , Epigenetic control of cell cycle-dependent histone gene expression is a principal component of the abbreviated pluripotent cell cycle. Molecular and cellular biology, 2012. 32(19): p. 3860–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Hao B, et al. , An anti-silencer–and SATB1-dependent chromatin hub regulates Rag1 and Rag2 gene expression during thymocyte development. Journal of Experimental Medicine, 2015. 212(5): p. 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Rege M, et al. , LADL: Light-activated dynamic looping for endogenous gene expression control. bioRxiv, 2018: p. 349340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Morgan SL, et al. , Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nature communications, 2017. 8(1): p. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Benabdallah NS, et al. , Decreased enhancer-promoter proximity accompanying enhancer activation. Molecular cell, 2019. 76(3): p. 473–484. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Jin F, et al. , A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature, 2013. 503(7475): p. 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Comoglio F, et al. , Thrombopoietin signaling to chromatin elicits rapid and pervasive epigenome remodeling within poised chromatin architectures. Genome research, 2018. 28(3): p. 295–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Melo CA, et al. , eRNAs are required for p53-dependent enhancer activity and gene transcription. Molecular cell, 2013. 49(3): p. 524–535. [DOI] [PubMed] [Google Scholar]
- 238.Crump NT, et al. , BET inhibition disrupts transcription but retains enhancer-promoter contact. Nature communications, 2021. 12(1): p. 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Razin S, Iarovaia O, and Vassetzky Y, A requiem to the nuclear matrix: from a controversial concept to 3D organization of the nucleus. Chromosoma, 2014. 123(3): p. 217–224. [DOI] [PubMed] [Google Scholar]
- 240.Mirkovitch J, Mirault M-E, and Laemmli UK, Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell, 1984. 39(1): p. 223–232. [DOI] [PubMed] [Google Scholar]
- 241.Wang TY, et al. , Enhanced expression of transgene in CHO cells using matrix attachment region. Cell biology international, 2008. 32(10): p. 1279–1283. [DOI] [PubMed] [Google Scholar]
- 242.Wang TY, et al. , Positional effects of the matrix attachment region on transgene expression in stably transfected CHO cells. Cell biology international, 2010. 34(2): p. 141–145. [DOI] [PubMed] [Google Scholar]
- 243.Girod PA, Zahn-Zabal M, and Mermod N, Use of the chicken lysozyme 5' matrix attachment region to generate high producer CHO cell lines. Biotechnology and bioengineering, 2005. 91(1): p. 1–11. [DOI] [PubMed] [Google Scholar]
- 244.Girod P-A, et al. , Genome-wide prediction of matrix attachment regions that increase gene expression in mammalian cells. Nature methods, 2007. 4(9): p. 747–753. [DOI] [PubMed] [Google Scholar]
- 245.Kim JD, et al. , Efficient selection of stable Chinese hamster ovary (CHO) cell lines for expression of recombinant proteins by using human interferon β SAR element. Biotechnology progress, 2005. 21(3): p. 933–937. [DOI] [PubMed] [Google Scholar]
- 246.Gorman C, et al. , Use of MAR elements to increase the production of recombinant proteins, in Cell Line Development. 2009, Springer. p. 1–32. [Google Scholar]
- 247.Briand N and Collas P, Lamina-associated domains: peripheral matters and internal affairs. Genome biology, 2020. 21(1): p. 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Van Steensel B and Belmont AS, Lamina-associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell, 2017. 169(5): p. 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Harr JC, et al. , Directed targeting of chromatin to the nuclear lamina is mediated by chromatin state and A-type lamins. Journal of Cell Biology, 2015. 208(1): p. 33–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Yoshizawa T, et al. , Biological phase separation: cell biology meets biophysics. Biophysical reviews, 2020. 12(2): p. 519–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Alberti S, Gladfelter A, and Mittag T, Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell, 2019. 176(3): p. 419–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Hnisz D, et al. , A phase separation model for transcriptional control. Cell, 2017. 169(1): p. 13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Boija A, et al. , Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell, 2018. 175(7): p. 1842–1855. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gibson BA, et al. , Organization of chromatin by intrinsic and regulated phase separation. Cell, 2019. 179(2): p. 470–484. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Sabari BR, et al. , Coactivator condensation at super-enhancers links phase separation and gene control. Science, 2018. 361(6400). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nair SJ, et al. , Phase separation of ligand-activated enhancers licenses cooperative chromosomal enhancer assembly. Nature structural & molecular biology, 2019. 26(3): p. 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Klein IA, et al. , Partitioning of cancer therapeutics in nuclear condensates. Science, 2020. 368(6497): p. 1386–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Li Q-L, et al. , The hyper-activation of transcriptional enhancers in breast cancer. Clinical epigenetics, 2019. 11(1): p. 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Huang H, et al. , Defining super-enhancer landscape in triple-negative breast cancer by multiomic profiling. Nature communications, 2021. 12(1): p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Li Q-L, et al. , Genome-wide profiling in colorectal cancer identifies PHF19 and TBC1D16 as oncogenic super enhancers. Nature communications, 2021. 12(1): p. 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Dixon JR, Gorkin DU, and Ren B, Chromatin domains: the unit of chromosome organization. Molecular cell, 2016. 62(5): p. 668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Dekker J and Heard E, Structural and functional diversity of Topologically Associating Domains. FEBS letters, 2015. 589(20): p. 2877–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Dixon JR, et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 2012. 485(7398): p. 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Sexton T, et al. , Three-dimensional folding and functional organization principles of the Drosophila genome. Cell, 2012. 148(3): p. 458–472. [DOI] [PubMed] [Google Scholar]
- 265.Phillips JE and Corces VG, CTCF: master weaver of the genome. Cell, 2009. 137(7): p. 1194–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Franco MM, Prickett AR, and Oakey RJ, The role of CCCTC-binding factor (CTCF) in genomic imprinting, development, and reproduction. Biology of reproduction, 2014. 91(5): p. 125, 1–9. [DOI] [PubMed] [Google Scholar]
- 267.Galupa R and Heard E, X-chromosome inactivation: new insights into cis and trans regulation. Current opinion in genetics & development, 2015. 31: p. 57–66. [DOI] [PubMed] [Google Scholar]
- 268.Sanborn AL, et al. , Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proceedings of the National Academy of Sciences, 2015. 112(47): p. E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Fudenberg G, et al. , Formation of chromosomal domains by loop extrusion. Cell reports, 2016. 15(9): p. 2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Moore JM, et al. , Loss of maternal CTCF is associated with peri-implantation lethality of Ctcf null embryos. PLoS One, 2012. 7(4): p. e34915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Zuin J, et al. , Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proceedings of the National Academy of Sciences, 2014. 111(3): p. 996–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Nishimura K, et al. , An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nature methods, 2009. 6(12): p. 917–922. [DOI] [PubMed] [Google Scholar]
- 273.Nora EP, et al. , Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell, 2017. 169(5): p. 930–944. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Kubo N, et al. , Preservation of chromatin organization after acute loss of CTCF in mouse embryonic stem cells. BioRxiv, 2017: p. 118737. [Google Scholar]
- 275.Khoury A, et al. , Constitutively bound CTCF sites maintain 3D chromatin architecture and long-range epigenetically regulated domains. Nature communications, 2020. 11(1): p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Franke M, et al. , CTCF knockout in zebrafish induces alterations in regulatory landscapes and developmental gene expression. Nature communications, 2021. 12(1): p. 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Lieberman-Aiden E, et al. , Comprehensive mapping of long-range interactions reveals folding principles of the human genome. science, 2009. 326(5950): p. 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Dixon JR, et al. , Chromatin architecture reorganization during stem cell differentiation. Nature, 2015. 518(7539): p. 331–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Rao SS, et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell, 2014. 159(7): p. 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Nagano T, et al. , Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature, 2017. 547(7661): p. 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hansen K, Reconstructing A/B compartments as revealed by Hi-C using long-range correlations in epigenetic data. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Schwarzer W, et al. , Two independent modes of chromatin organization revealed by cohesin removal. Nature, 2017. 551(7678): p. 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Crane E, et al. , Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature, 2015. 523(7559): p. 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Dong P, et al. , 3D chromatin architecture of large plant genomes determined by local A/B compartments. Molecular plant, 2017. 10(12): p. 1497–1509. [DOI] [PubMed] [Google Scholar]
- 285.Mizuguchi T, et al. , Cohesin-dependent globules and heterochromatin shape 3D genome architecture in S. pombe. Nature, 2014. 516(7531): p. 432–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Le TB and Laub MT, Transcription rate and transcript length drive formation of chromosomal interaction domain boundaries. The EMBO journal, 2016. 35(14): p. 1582–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Lioy VS, et al. , Multiscale structuring of the E. coli chromosome by nucleoid-associated and condensin proteins. Cell, 2018. 172(4): p. 771–783. e18. [DOI] [PubMed] [Google Scholar]
- 288.Pepenella S, Murphy KJ, and Hayes JJ, Intra-and inter-nucleosome interactions of the core histone tail domains in higher-order chromatin structure. Chromosoma, 2014. 123(1–2): p. 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Rowley MJ, et al. , Evolutionarily conserved principles predict 3D chromatin organization. Molecular cell, 2017. 67(5): p. 837–852. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Rennie S, et al. , Transcriptional decomposition reveals active chromatin architectures and cell specific regulatory interactions. Nature communications, 2018. 9(1): p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Ulianov SV, et al. , Active chromatin and transcription play a key role in chromosome partitioning into topologically associating domains. Genome research, 2016. 26(1): p. 70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Li L, et al. , Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Molecular cell, 2015. 58(2): p. 216–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Hug CB, et al. , Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell, 2017. 169(2): p. 216–228. e19. [DOI] [PubMed] [Google Scholar]
- 294.Cremer T and Cremer M, Chromosome territories. Cold Spring Harbor perspectives in biology, 2010. 2(3): p. a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Beatson GT, Meeting IX.—May 20, 1896: On the Treatment of Inoperable Cases of Carcinoma of the Mamma: Suggestions for a New Method of Treatment, with Illustrative Cases. Transactions. Medico-Chirurgical Society of Edinburgh, 1896. 15: p. 153. [PMC free article] [PubMed] [Google Scholar]
- 296.Ling Y, et al. , Loss of heterozygosity in thyroid hormone receptor beta in invasive breast cancer. Tumori Journal, 2015. 101(5): p. 572–577. [DOI] [PubMed] [Google Scholar]
- 297.Shi Y-B, Unliganded thyroid hormone receptor regulates metamorphic timing via the recruitment of histone deacetylase complexes. Current topics in developmental biology, 2013. 105: p. 275–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Shi Y-B, et al. , Thyroid hormone receptor actions on transcription in amphibia: The roles of histone modification and chromatin disruption. Cell & bioscience, 2012. 2(1): p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Heimeier RA, Hsia VS, and Shi Y-B, Participation of Brahma-related gene 1 (BRG1)-associated factor 57 and BRG1-containing chromatin remodeling complexes in thyroid hormone-dependent gene activation during vertebrate development. Molecular endocrinology, 2008. 22(5): p. 1065–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Derenzini M, et al. , Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. The Journal of pathology, 2000. 191(2): p. 181–186. [DOI] [PubMed] [Google Scholar]
- 301.Oh S, Oh C, and Yoo KH, Functional roles of CTCF in breast cancer. BMB reports, 2017. 50(9): p. 445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Katainen R, et al. , CTCF/cohesin-binding sites are frequently mutated in cancer. Nature genetics, 2015. 47(7): p. 818–821. [DOI] [PubMed] [Google Scholar]
- 303.Guo YA, et al. , Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nature communications, 2018. 9(1): p. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Dai J, et al. , Systematical analyses of variants in CTCF-binding sites identified a novel lung cancer susceptibility locus among Chinese population. Scientific reports, 2015. 5(1): p. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Smith KS, et al. , Nuclear topology modulates the mutational landscapes of cancer genomes. Nature structural & molecular biology, 2017. 24(11): p. 1000–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Barutcu AR, et al. , Chromatin interaction analysis reveals changes in small chromosome and telomere clustering between epithelial and breast cancer cells. Genome biology, 2015. 16(1): p. 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ciesielski O, et al. , The epigenetic profile of tumor endothelial cells. Effects of combined therapy with antiangiogenic and epigenetic drugs on cancer progression. International journal of molecular sciences, 2020. 21(7): p. 2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Flavahan WA, Gaskell E, and Bernstein BE, Epigenetic plasticity and the hallmarks of cancer. Science, 2017. 357(6348). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Cruickshank B, et al. , Dying to be noticed: epigenetic regulation of immunogenic cell death for cancer immunotherapy. Frontiers in immunology, 2018. 9: p. 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Yaswen P, et al. Therapeutic targeting of replicative immortality. in Seminars in cancer biology. 2015. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Darwiche N, Epigenetic mechanisms and the hallmarks of cancer: An intimate affair. American journal of cancer research, 2020. 10(7): p. 1954. [PMC free article] [PubMed] [Google Scholar]
- 312.Ghule PN, et al. , Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America, 2008. 105(44). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Ghule PN, et al. , Fidelity of Histone Gene Regulation Is Obligatory for Genome Replication and Stability. Molecular and Cellular Biology, 2014. 34(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Xie R, et al. , The histone gene activator HINFP is a nonredundant cyclin E/CDK2 effector during early embryonic cell cycles. Proceedings of the National Academy of Sciences of the United States of America, 2009. 106(30). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Nirala NK, et al. , Hinfp is a guardian of the somatic genome by repressing transposable elements. Proceedings of the National Academy of Sciences, 2021. 118(41). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Medina R, et al. , The histone gene transcription factor HiNF-P stabilizes its cell cycle regulatory co-activator p220NPAT. Biochemistry, 2006. 45(51). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Medina R, et al. , The HiNF-P/p220NPAT cell cycle signaling pathway controls nonhistone target genes. Cancer Research, 2007. 67(21). [DOI] [PubMed] [Google Scholar]
- 318.Bongiorno-Borbone L, et al. , FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle, 2008. 7(15). [DOI] [PubMed] [Google Scholar]
- 319.Ma T, et al. , Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in Cajal bodies promotes histone gene transcription. Genes and Development, 2000. 14(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Zhao J, et al. , NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes and Development, 2000. 14(18). [PMC free article] [PubMed] [Google Scholar]
- 321.Salzler HR, et al. , A Sequence in the Drosophila H3-H4 Promoter Triggers Histone Locus Body Assembly and Biosynthesis of Replication-Coupled Histone mRNAs. Developmental Cell, 2013. 24(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Yang XC, et al. , A conserved interaction that is essential for the biogenesis of histone locus bodies. Journal of Biological Chemistry, 2014. 289(49). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Liu Y, et al. , Transcriptional landscape of the human cell cycle. Proceedings of the National Academy of Sciences, 2017. 114(13): p. 3473–3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Alabert C and Groth A, Chromatin replication and epigenome maintenance, in Nature Reviews Molecular Cell Biology. 2012. [DOI] [PubMed] [Google Scholar]
- 325.Gerace L and Blobel G, The nuclear envelope lamina is reversibly depolymerized during mitosis. Cell, 1980. 19(1). [DOI] [PubMed] [Google Scholar]
- 326.Katsani KR, et al. , In vivo dynamics of Drosophila nuclear envelope components. Molecular Biology of the Cell, 2008. 19(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.de Castro IJ, Gokhan E, and Vagnarelli P, Resetting a functional G1 nucleus after mitosis, in Chromosoma. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Egli D, Birkhoff G, and Eggan K, Mediators of reprogramming: Transcription factors and transitions through mitosis, in Nature Reviews Molecular Cell Biology. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Rose JT, et al. , Inhibition of the RUNX1-CBFβ transcription factor complex compromises mammary epithelial cell identity: A phenotype potentially stabilized by mitotic gene bookmarking. Oncotarget, 2020. 11(26). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Lodhi N, Ji Y, and Tulin A, Mitotic Bookmarking: Maintaining Post-Mitotic Reprogramming of Transcription Reactivation. Current Molecular Biology Reports, 2016. 2(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Raccaud M and Suter DM, Transcription factor retention on mitotic chromosomes: regulatory mechanisms and impact on cell fate decisions, in FEBS Letters. 2018. [DOI] [PubMed] [Google Scholar]
- 332.Grandy RA, et al. , Genome-Wide Studies Reveal that H3K4me3 Modification in Bivalent Genes Is Dynamically Regulated during the Pluripotent Cell Cycle and Stabilized upon Differentiation. Molecular and Cellular Biology, 2016. 36(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Voigt P, Tee WW, and Reinberg D, A double take on bivalent promoters, in Genes and Development. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Bradbury EM, Inglis RJ, and Matthews HR, Control of cell division by very lysine rich histone (F1) phosphorylation. Nature, 1974. 247(5439). [DOI] [PubMed] [Google Scholar]
- 335.Slangy A, et al. , Phosphorylation by p34cdc2 regulates spindle association of human Eg5, a kinesin-related motor essential for bipolar spindle formation in vivo. Cell, 1995. 83(7). [DOI] [PubMed] [Google Scholar]
- 336.Hoffmann I, et al. , Phosphorylation and activation of human cdc25-C by cdc2-cyclin B and its involvement in the self-amplification of MPF at mitosis. EMBO Journal, 1993. 12(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Courvalin JC, et al. , The lamin B receptor of the inner nuclear membrane undergoes mitosis- specific phosphorylation and is a substrate for p34(cdc2)-type protein kinase. Journal of Biological Chemistry, 1992. 267(27). [PubMed] [Google Scholar]
- 338.Heald R, McLoughlin M, and McKeon F, Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated cdc2 kinase. Cell, 1993. 74(3). [DOI] [PubMed] [Google Scholar]
- 339.El Dika M, et al. , Mitotic timing is differentially controlled by A-and B-type cyclins and by CDC6 associated with a bona fide CDK inhibitor Xic1 in Xenopus laevis cell-free extract. International Journal of Developmental Biology, 2021. 65(7–8-9): p. 487–496. [DOI] [PubMed] [Google Scholar]
- 340.Kubiak JZ and El Dika M, Canonical and alternative pathways in cyclin-dependent kinase 1/cyclin B inactivation upon M-phase exit in Xenopus laevis cell-free extracts, in Enzyme Research. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Dika ME, et al. , Control of timing of embryonic M-phase entry and exit is differentially sensitive to CDK1 and PP2A balance. International Journal of Developmental Biology, 2014. 58(10–12): p. 767–774. [DOI] [PubMed] [Google Scholar]
- 342.Kubiak JZ, et al. , CDC6 regulates mitotic CDK1 via cyclin B and not cyclin A and acts through a bona fide CDK inhibitor Xic1. bioRxiv, 2020. [Google Scholar]
- 343.Debowski M, et al. , Flexibility vs. robustness in cell cycle regulation of timing of M-phase entry in Xenopus laevis embryo cell-free extract. International Journal of Developmental Biology, 2016. 60(7-9): p. 305–314. [DOI] [PubMed] [Google Scholar]
- 344.Yi ZY, et al. , CDC6 regulates both G2/M transition and metaphase-to-anaphase transition during the first meiosis of mouse oocytes. Journal of cellular physiology, 2020. 235(7–8): p. 5541–5554. [DOI] [PubMed] [Google Scholar]
- 345.Murray AW and Kirschner MW, Cyclin synthesis drives the early embryonic cell cycle. Nature, 1989. 339(6222): p. 275–280. [DOI] [PubMed] [Google Scholar]
- 346.Glotzer M, Murray AW, and Kirschner MW, Cyclin is degraded by the ubiquitin pathway. Nature, 1991. 349(6305): p. 132–138. [DOI] [PubMed] [Google Scholar]
- 347.Rieder CL and Khodjakov A, Mitosis and checkpoints that control progression through mitosis in vertebrate somatic cells, in Progress in cell cycle research. 1997. [DOI] [PubMed] [Google Scholar]
- 348.Bloom K and Costanzo V, Centromere Structure and Function, in Progress in molecular and subcellular biology. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Castro A, et al. , The anaphase-promoting complex: A key factor in the regulation of cell cycle, in Oncogene. 2005. [DOI] [PubMed] [Google Scholar]
- 350.Uhlmann F, Lottspelch F, and Nasmyth K, Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature, 1999. 400(6739). [DOI] [PubMed] [Google Scholar]
- 351.Güttinger S, Laurell E, and Kutay U, Orchestrating nuclear envelope disassembly and reassembly during mitosis, in Nature Reviews Molecular Cell Biology. 2009. [DOI] [PubMed] [Google Scholar]
- 352.De Souza CPC and Osmani SA, Mitosis, not just open or closed, in Eukaryotic Cell. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Zhou CY and Heald R, Emergent properties of mitotic chromosomes, in Current Opinion in Cell Biology. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Man T, et al. , The mechanics of mitotic chromosomes. Quarterly Reviews of Biophysics, 2021. [DOI] [PubMed] [Google Scholar]
- 355.Hua LL and Mikawa T, Mitotic antipairing of homologous and sex chromosomes via spatial restriction of two haploid sets. Proceedings of the National Academy of Sciences of the United States of America, 2018. 115(52). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Joyce EF, Erceg J, and C.t. Wu, Pairing and anti-pairing: A balancing act in the diploid genome, in Current Opinion in Genetics and Development. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Joyce EF, et al. , Identification of genes that promote or antagonize somatic homolog pairing using a high-throughput FISH-based screen. PLoS Genetics, 2012. 8(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Williams BR, et al. , Disruption of topoisomerase II perturbs pairing in Drosophila cell culture. Genetics, 2007. 177(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Hart TA, et al. , Condensin II resolves chromosomal associations to enable anaphase I segregation in Drosophila male meiosis. PLoS Genetics, 2008. 4(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Cutter AR and Hayes JJ, A brief review of nucleosome structure, in FEBS Letters. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Kschonsak M and Haering CH, Shaping mitotic chromosomes: From classical concepts to molecular mechanisms. BioEssays, 2015. 37(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Kireeva N, et al. , Visualization of early chromosome condensation: A hierarchical folding, axial glue model of chromosome structure. Journal of Cell Biology, 2004. 166(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Abe S, et al. , The initial phase of chromosome condensation requires Cdk1-mediated phosphorylation of the CAP-D3 subunit of condensin II. Genes and Development, 2011. 25(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Hagstrom KA, et al. , C. elegans condensin promotes mitotic chromosome architecture, centromere organization, and sister chromatid segregation during mitosis and meiosis. Genes and Development, 2002. 16(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Hudson DF, et al. , Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Developmental Cell, 2003. 5(2). [DOI] [PubMed] [Google Scholar]
- 366.Ryu J-K, et al. , The condensin holocomplex cycles dynamically between open and collapsed states. Nature Structural & Molecular Biology, 2020. 27(12): p. 1134–1141. [DOI] [PubMed] [Google Scholar]
- 367.Kalitsis P, et al. , Condensin, master organizer of the genome, in Chromosome Research. 2017. [DOI] [PubMed] [Google Scholar]
- 368.Ono T, et al. , Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell, 2003. 115(1). [DOI] [PubMed] [Google Scholar]
- 369.Green LC, et al. , Contrasting roles of condensin I and condensin II in mitotic chromosome formation. Journal of Cell Science, 2012. 125(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Lipp JJ, et al. , Aurora B controls the association of condensin I but not condensin II with mitotic chromosomes. Journal of Cell Science, 2007. 120(7). [DOI] [PubMed] [Google Scholar]
- 371.Hirano T, Condensins: Universal organizers of chromosomes with diverse functions, in Genes and Development. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Piskadlo E and Oliveira RA, Novel insights into mitotic chromosome condensation. F1000Research, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Petrova B, et al. , Quantitative Analysis of Chromosome Condensation in Fission Yeast. Molecular and Cellular Biology, 2013. 33(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Dai J, Sullivan BA, and Higgins JMG, Regulation of Mitotic Chromosome Cohesion by Haspin and Aurora B. Developmental Cell, 2006. 11(5). [DOI] [PubMed] [Google Scholar]
- 375.Hégarat N, Rata S, and Hochegger H, Bistability of mitotic entry and exit switches during open mitosis in mammalian cells, in BioEssays. 2016. [DOI] [PubMed] [Google Scholar]
- 376.Qi an J, et al. , PP1/repo-man dephosphorylates mitotic histone H3 at T3 and regulates chromosomal aurora B targeting. Current Biology, 2011. 21(9). [DOI] [PubMed] [Google Scholar]
- 377.Trinkle-Mulcahy L, et al. , Repo-Man recruits PP1γ to chromatin and is essential for cell viability. Journal of Cell Biology, 2006. 172(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Vagnarelli P, et al. , Repo-Man Coordinates Chromosomal Reorganization with Nuclear Envelope Reassembly during Mitotic Exit. Developmental Cell, 2011. 21(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Booth DG, et al. , Ki-67 is a PP1-interacting protein that organises the mitotic chromosome periphery. eLife, 2014. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Landsverk HB, et al. , PNUTS enhances in vitro chromosome decondensation in a PP1-dependent manner. Biochemical Journal, 2005. 390(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Magalska A, et al. , RuvB-like atpases function in chromatin decondensation at the end of mitosis. Developmental Cell, 2014. 31(3). [DOI] [PubMed] [Google Scholar]
- 382.Ramadan K, et al. , Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature, 2007. 450(7173). [DOI] [PubMed] [Google Scholar]
- 383.Jónsson ZO, et al. , Rvb1p and Rvb2p are Essential Components of a Chromatin Remodeling Complex that Regulates Transcription of over 5% of Yeast Genes. Journal of Biological Chemistry, 2001. 276(19). [DOI] [PubMed] [Google Scholar]
- 384.Sofueva S, et al. , Cohesin-mediated interactions organize chromosomal domain architecture. The EMBO journal, 2013. 32(24): p. 3119–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Oomen ME, et al. , CTCF sites display cell cycle–dependent dynamics in factor binding and nucleosome positioning. Genome research, 2019. 29(2): p. 236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Gibcus JH, et al. , A pathway for mitotic chromosome formation. Science, 2018. 359(6376). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Zhang H, et al. , CTCF and transcription influence chromatin structure re-configuration after mitosis. Nature communications, 2021. 12(1): p. 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Palozola KC, Lerner J, and Zaret KS, A changing paradigm of transcriptional memory propagation through mitosis, in Nature Reviews Molecular Cell Biology. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Yu B, et al. , Reprogramming fibroblasts into bipotential hepatic stem cells by defined factors. Cell Stem Cell, 2013. 13(3). [DOI] [PubMed] [Google Scholar]
- 390.Kaminski MM, et al. , Direct reprogramming of fibroblasts into renal tubular epithelial cells by defined transcription factors. Nature Cell Biology, 2016. 18(12). [DOI] [PubMed] [Google Scholar]
- 391.Mahat DB, et al. , Base-pair-resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nature protocols, 2016.11 (8): p. 1455–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Judd J, et al. , A rapid, sensitive, scalable method for precision run-on sequencing (PRO-seq). bioRxiv, 2020. [Google Scholar]
- 393.Kwak H, et al. , Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science, 2013. 339(6122): p. 950–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Duarte FM, et al. , Transcription factors GAF and HSF act at distinct regulatory steps to modulate stress-induced gene activation. Genes & development, 2016. 30(15): p. 1731–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Booth GT, et al. , Cdk9 regulates a promoter-proximal checkpoint to modulate RNA polymerase II elongation rate in fission yeast. Nature communications, 2018. 9(1): p. 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Kouzarides T, Chromatin Modifications and Their Function, in Cell. 2007. [DOI] [PubMed] [Google Scholar]
- 397.Cheung P, Allis CD, and Sassone-Corsi P, Signaling to chromatin through historic modifications, in Cell. 2000. [DOI] [PubMed] [Google Scholar]
- 398.Adams RR, et al. , Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. Journal of Cell Biology, 2001. 153(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Giet R and Glover DM, Drosophila aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. Journal of Cell Biology, 2001. 152(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Sawicka A and Seiser C, Sensing core histone phosphorylation - A matter of perfect timing, in Biochimica et Biophysica Acta - Gene Regulatory Mechanisms. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Vaquero A, et al. , SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes and Development, 2006. 20(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Wilkins BJ, et al. , A cascade of histone modifications induces chromatin condensation in mitosis. Science, 2014. 343(6166). [DOI] [PubMed] [Google Scholar]
- 403.Robinson PJJ, et al. , 30 nm Chromatin Fibre Decompaction Requires both H4-K16 Acetylation and Linker Histone Eviction. Journal of Molecular Biology, 2008. 381(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Sinha D and Shogren-Knaak MA, Role of direct interactions between the histone H4 tail and the H2A core in long range nucleosome contacts. Journal of Biological Chemistry, 2010. 285(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Varier RA and Timmers HTM, Histone lysine methylation and demethylation pathways in cancer, in Biochimica et Biophysica Acta - Reviews on Cancer. 2011. [DOI] [PubMed] [Google Scholar]
- 406.Varier RA, et al. , A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. EMBO Journal, 2010. 29(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.McManus KJ, et al. , Dynamic changes in histone H3 lysine 9 methylations: Identification of a mitosis-specific function for dynamic methylation in chromosome congression and segregation. Journal of Biological Chemistry, 2006. 281(13). [DOI] [PubMed] [Google Scholar]
- 408.Kelly T and Jones P, DAMD to epigenetic silence, in Proceedings of the National Academy of Sciences of the United States of America. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Wang F and Higgins JMG, Histone modications and mitosis: Countermarks, landmarks, and bookmarks, in Trends in Cell Biology. 2013. [DOI] [PubMed] [Google Scholar]
- 410.Wang F, et al. , Haspin inhibitors reveal centromeric functions of Aurora B in chromosome segregation. Journal of Cell Biology, 2012. 199(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Wang F, et al. , Histone H3 Thr-3 phosphorylation by haspin positions Aurora B at centromeres in mitosis. Science, 2010. 330(6001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.De Antoni A, et al. , A small-molecule inhibitor of Haspin alters the kinetochore functions of Aurora B. Journal of Cell Biology, 2012. 199(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Segil N, Roberts SB, and Heintz N. Cell-cycle-regulated phosphorylation of the transcription factor Oct-1. in Cold Spring Harbor Symposia on Quantitative Biology. 1991. [DOI] [PubMed] [Google Scholar]
- 414.Segil N, Roberts SB, and Heintz N, Mitotic phosphorylation of the Oct-1 homeodomain and regulation of Oct-1 DNA binding activity. Science, 1991. 254(5039). [DOI] [PubMed] [Google Scholar]
- 415.Bellier S, et al. , Phosphorylation of the RNA polymerase II largest subunit during Xenopus laevis oocyte maturation. Molecular and Cellular Biology, 1997. 17(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Akoulitchev S and Reinberg D, The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes and Development, 1998. 12(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Dovat S, et al. , A common mechanism for mitotic inactivation of C2H2 zinc finger DNA-binding domains. Genes and Development, 2002. 16(23). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Martínez-Balbás MA, et al. , Displacement of sequence-specific transcription factors from mitotic chromatin. Cell, 1995. 83(1). [DOI] [PubMed] [Google Scholar]
- 419.Wu SC and Zhang Y, Cyclin-dependent kinase 1 (CDK1)-mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. Journal of Biological Chemistry, 2011. 286(32). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Xing H, Vanderford NL, and Sarge KD, The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nature Cell Biology, 2008. 10(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Blobel GA, et al. , A Reconfigured Pattern of MLL Occupancy within Mitotic Chromatin Promotes Rapid Transcriptional Reactivation Following Mitotic Exit. Molecular Cell, 2009. 36(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Caravaca JM, et al. , Bookmarking by specific and nonspecific binding of FoxA1 pioneer factor to mitotic chromosomes. Genes and Development, 2013. 27(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 423.Verdeguer F, et al. , A mitotic transcriptional switch in polycystic kidney disease. Nature Medicine, 2010. 16(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Young DW, et al. , Mitotic retention of gene expression patterns by the cell fate-determining transcription factor Runx2. Proceedings of the National Academy of Sciences of the United States of America, 2007. 104(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Young DW, et al. , Mitotic occupancy and lineage-specific transcriptional control of rRNA genes by Runx2. Nature, 2007. 445(7126). [DOI] [PubMed] [Google Scholar]
- 426.Raccaud M, et al. , Mitotic chromosome binding predicts transcription factor properties in interphase. Nature Communications, 2019. 10(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Zhao R, et al. , Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nature Cell Biology, 2011. 13(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Yang Z, He N, and Zhou Q, Brd4 Recruits P-TEFb to Chromosomes at Late Mitosis To Promote G 1 Gene Expression and Cell Cycle Progression. Molecular and Cellular Biology, 2008. 28(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Ng RK and Gurdon JB, Epigenetic inheritance of cell differentiation status, in Cell Cycle. 2008. [DOI] [PubMed] [Google Scholar]
- 430.Sutani T, et al. , Condensin targets and reduces unwound DNA structures associated with transcription in mitotic chromosome condensation. Nature communications, 2015. 6(1): p. 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Varela N, et al. , Mitotic inheritance of mRNA facilitates translational activation of the osteogenic-Lineage commitment factor runx2 in progeny of osteoblastic cells. Journal of cellular physiology, 2016. 231(5): p. 1001–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Wallace HA, et al. , Condensin II subunit NCAPH2 associates with shelterin protein TRF1 and is required for telomere stability. Journal of cellular physiology, 2019. 234(11): p. 20755–20768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Umbreit NT, et al. , Mechanisms generating cancer genome complexity from a single cell division error. Science, 2020. 368(6488). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Leiserson MDM, et al. , Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nature genetics, 2015. 47(2): p. 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Martens JHA and Stunnenberg HG, The molecular signature of oncofusion proteins in acute myeloid leukemia. FEBS letters, 2010. 584(12): p. 2662–2669. [DOI] [PubMed] [Google Scholar]
- 436.Alcalay M, et al. , Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. The Journal of clinical investigation, 2003. 112(11): p. 1751–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Bondeson DP, et al. , Catalytic in vivo protein knockdown by small-molecule PROTACs. Nature chemical biology, 2015. 11(8): p. 611–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Scholes NS, Mayor-Ruiz C, and Winter GE, Identification and selectivity profiling of small-molecule degraders via multi-omics approaches. Cell Chemical Biology, 2021. [DOI] [PubMed] [Google Scholar]
- 439.Naito M, et al. , Targeted protein degradation by chimeric small molecules, PROTACs and SNIPERs. Frontiers in chemistry, 2019. 7: p. 849–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Neklesa TK, Winkler JD, and Crews CM, Targeted protein degradation by PROTACs. Pharmacology & therapeutics, 2017. 174: p. 138–144. [DOI] [PubMed] [Google Scholar]