ABSTRACT
Streptococcus pneumoniae (Spn) strains cause pneumonia that kills millions every year worldwide. Spn produces Ply, a hemolysin that lyses erythrocytes releasing hemoglobin, and also produces the pro-oxidant hydrogen peroxide (Spn-H2O2) during growth. The hallmark of the pathophysiology of hemolytic diseases is the oxidation of hemoglobin, but oxidative reactions catalyzed by Spn-H2O2 have been poorly studied. We characterized the oxidation of hemoglobin by Spn-H2O2. We prepared a series of single-mutant (ΔspxB or ΔlctO), double-mutant (ΔspxB ΔlctO), and complemented strains in TIGR4, D39, and EF3030. We then utilized an in vitro model with oxyhemoglobin to demonstrate that oxyhemoglobin was oxidized rapidly, within 30 min of incubation, by Spn-H2O2 to methemoglobin and that the main source of Spn-H2O2 was pyruvate oxidase (SpxB). Moreover, extended incubation caused the release and the degradation of heme. We then assessed oxidation of hemoglobin and heme degradation by other bacterial inhabitants of the respiratory tract. All hydrogen peroxide-producing streptococci tested caused the oxidation of hemoglobin and heme degradation, whereas bacterial species that produce <1 μM H2O2 neither oxidized hemoglobin nor degraded heme. An ex vivo bacteremia model confirmed that oxidation of hemoglobin and heme degradation occurred concurrently with hemoglobin that was released from erythrocytes by Ply. Finally, gene expression studies demonstrated that heme, but not red blood cells or hemoglobin, induced upregulated transcription of the spxB gene. Oxidation of hemoglobin may be important for pathogenesis and for the symbiosis of hydrogen peroxide-producing bacteria with other species by providing nutrients such as iron.
KEYWORDS: hydrogen peroxide, Streptococcus pneumoniae, heme, hemoglobin, iron
INTRODUCTION
Streptococcus pneumoniae (Spn) colonizes the lower respiratory epithelium, causing pneumonia that can progress to invasive pneumococcal disease (IPD) (1–4). Spn mainly affects the most vulnerable populations: young children, immunocompromised patients, and the elderly (1, 4, 5). Globally, Spn causes ~15 million cases of illness each year, leading to more than a million deaths due to pneumonia (6–8). Pneumonia is an acute infection of the pulmonary parenchyma that presents with pulmonary hemorrhage, inflammatory congestion, hepatization, suppurative infiltration, and lung parenchymal injury (4, 9–13). If left untreated, Spn invades the bloodstream, producing septic shock, multiorgan failure, cardiotoxicity, and death within days from onset (4, 9–12, 14, 15).
Spn strains produce the toxin pneumolysin (Ply), which has cytotoxic activity against lung epithelial and endothelial cells (16–18). Ply is a 53-kDa surface-located protein (19, 20) that can also be released through autolysis (21, 22). Besides its toxicity against cells, Ply has hemolytic activity against erythrocytes of several mammalian species, and hemolysis is observed whether Ply is released into the supernatant or located on the bacterial membrane (20). Under standard bacterial culture conditions, i.e., 37°C in a 5% CO2 atmosphere, Ply is responsible for almost all hemolytic activity in cultures of Spn strains (11), causing the release of a large amount of hemoglobin into the supernatant (20, 21, 23). Transcription of the gene encoding this pneumolysin, ply, is upregulated when Spn is cultured under conditions mimicking lung infection (24), and therefore, Ply-induced release of hemoglobin is expected to occur during pneumococcal lung infection.
Hemoglobin is a tetrameric protein (64 kDa) made of four subunits, two alpha and two beta chains. Each subunit contains a heme center that reversibly binds oxygen through a penta-coordinate heme molecule containing ferrous iron (Fe2+), known as oxyhemoglobin (25, 26). When hemoglobin is released from erythrocytes, heme-hemoglobin can be observed by spectroscopy at ~415 nm (25, 27, 28). This region is known as the Soret region peak and represents heme-hemoglobin, while the alpha and beta chains are characterized by two absorption peaks of ~540 and ~570 nm (27, 28). Cell-free heme is toxic to cells because of its potential to generate toxic radicals (29).
Release of hemoglobin during hemolytic disease, such as sickle cell disease and other hemorrhagic conditions, causes oxidation of hemoglobin (30). Oxidized hemoglobin releases heme, which in turn induces adverse effects, including leukocyte activation, cytokine upregulation, and production of oxidants, leading to lung epithelial injury and damage to the vascular system (29–31). Reactive oxygen species (ROS), such as nitric oxide (NO) (32), hypochlorous acid conjugate base (OCl−) (24), and H2O2 (33, 34), drive a catalytic cycle including the oxidation of oxyhemoglobin to ferryl hemoglobin (Hb-Fe+4) and autoreduction of the ferryl intermediate to methemoglobin (Hb-Fe3+), which is detected by spectroscopy at 405 nm (35, 36). Additional pro-oxidant molecules, such as H2O2, produce more ferryl hemoglobin and cause heme degradation (28, 36). Thus, the oxidation of hemoglobin results in highly oxidizing ferryl hemoglobin (Hb-Fe4+) and methemoglobin (Hb-Fe3+), and the release of toxic heme, which disrupts the mitochondrial membrane potential of lung epithelial cells and endothelial cells and causes lipid peroxidation (28, 30, 36–38).
Because Spn causes hemorrhage in the lung and produces a large amount of highly reactive H2O2 (39, 40), oxidation of cell-free hemoglobin might occur during pneumococcal disease. H2O2 is a by-product of the oxidation of pyruvate, which is the end product of the glycolysis produced by the enzyme pyruvate oxidase (SpxB) (41, 42). This reaction consumes oxygen and produces H2O2. In the absence of oxygen, pyruvate is converted to l-lactate, while in the presence of molecular O2, l-lactate feeds back to pyruvate, via lactate oxidase (LctO), in a reaction that also produces H2O2 (41, 42). It has been calculated that ~85% of H2O2 released in cultures of Spn occurs through the reaction catalyzed by SpxB, whereas the remaining ~15% is attributed to LctO (43, 44). Experiments using animal models have demonstrated that Spn-H2O2 plays a role in lung colonization and in the translocation of Spn from the lungs to the bloodstream (45–48). Details of the mechanism(s) are largely unknown.
We recently discovered that Spn-H2O2 oxidizes hemoglobin to methemoglobin, a non-oxygen-binding form of hemoglobin (23). We also demonstrated that the oxidation of hemoglobin causes the characteristic greenish halo around pneumococcal colonies grown on blood agar plates, known as alpha-hemolysis (23). Thus, the phenotype known as alpha-hemolysis is not hemolysis but the oxidation of hemoglobin to methemoglobin (23). Another alpha-hemolytic streptococcus, Streptococcus gordonii, oxidizes hemoglobin to methemoglobin through catalytic reactions driven by H2O2 (49).
Oxidation of cell-free hemoglobin induces lung injury in patients with acute respiratory distress syndrome (31), and in children with sickle cell disease (SCD), oxidation of hemoglobin S (HbS) to Hb-Fe4+ and then its reduction to methemoglobin result in an acute hemolytic vascular inflammatory process causing acute lung injury (30, 50). We hypothesize that the oxidation of hemoglobin during pneumococcal disease can exacerbate cytotoxicity in the lungs, but details of these oxidative reactions in cultures of Spn strains are not available. Therefore, in this study, we comprehensively investigated the oxidation of hemoglobin by Spn cultures. We discovered that the oxidative capacity of Spn cultures, under the culture conditions utilized here, was attributed to H2O2 produced through catalytic reactions of SpxB but not of LctO. We also found that Spn-H2O2 caused the degradation of heme and that a number of H2O2-producing, alpha-hemolytic streptococci oxidized hemoglobin and degraded heme from hemoglobin. An ex vivo model of pneumococcal bacteremia further demonstrated that oxidation of hemoglobin and heme degradation occurred rapidly and that heme, but not hemoglobin or red blood cells, caused an upregulated transcription of spxB. Oxidative reactions driven by Spn-H2O2 might contribute to the pathogenesis of pneumococcal disease and can also be important for the symbiosis with other non-hydrogen peroxide-producing bacteria by providing nutrients such as heme iron.
RESULTS
Streptococcus pneumoniae-produced hydrogen peroxide oxidizes heme-hemoglobin, causing the loss of the Soret peak.
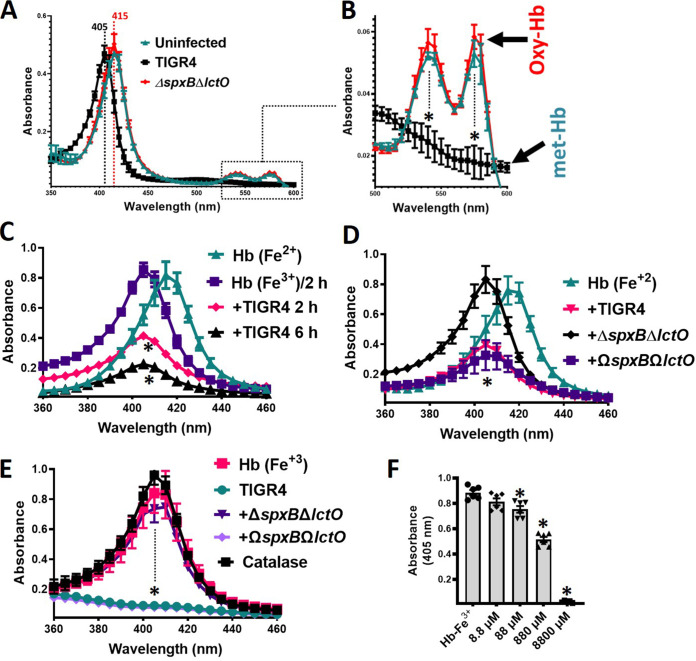
We previously demonstrated that the alpha and beta chains of oxyhemoglobin are deoxygenated by S. pneumoniae strains, but further oxidation of heme-Fe2+ was not investigated (23). To assess this, Todd-Hewitt broth (THY) supplemented with sheep oxyhemoglobin (oxyHb), which contains heme-Fe2+, was inoculated with strain TIGR4 and incubated at 37°C in a 5% CO2 and ~20% O2 atmosphere. As a control, uninoculated THY-oxyHb was incubated under the same conditions. After 30 min of incubation, TIGR4 cultures caused the shift of the Soret curve from heme-Fe2+, which peaks at 415 nm, to a Soret curve of heme-Fe3+ at 405 nm (Fig. 1A). As expected, this hemoglobin preparation was deoxygenated, producing methemoglobin (Fig. 1B). Similar oxidation of heme-Fe2+ to heme-Fe3+ was observed when oxyhemoglobin was purified from horse erythrocytes and incubated with Spn strain TIGR4 or D39 (data not shown). Cultures of THY-oxyHb inoculated with hydrogen peroxide-deficient strain TIGR4ΔspxBΔlctO or D39ΔspxBΔlctO did not cause the shift of the Soret curve (Fig. 1A and data not shown). Hemoglobin in uninfected cultures was not auto-oxidized after this 30-min incubation period (Fig. 1A).
FIG 1.
Hydrogen peroxide produced in S. pneumoniae cultures oxidizes human hemoglobin. TIGR4, TIGR4ΔspxBΔlctO, or TIGR4ΩspxBΩlctO was inoculated in THY containing oxyhemoglobin [Hb (Fe2+)] or methemoglobin [Hb (Fe3+)] or left uninfected, and cultures were incubated for 30 min (A and B), 2 or 6 h (C), or 6 h (D to F) at 37°C in a 5% CO2 atmosphere. Supernatants were collected, and the spectra were obtained using an Omega spectrophotometer (BMG LabTech). (B) Boxed area in panel A, corresponding to the absorbance peaks of the alpha chain and the beta chain of oxyhemoglobin, flattened in methemoglobin. Dotted lines (A, B, and E) indicate the Soret peak for oxyhemoglobin (415 nm) or methemoglobin (405 nm). (F) THY supplemented with methemoglobin (Hb-Fe3+) was treated with the indicated concentration of hydrogen peroxide or left untreated. These cultures were incubated for 6 h at 37°C in a 5% CO2 atmosphere, after which the supernatants were purified and the spectra were obtained as above. The absorbance at 405 nm obtained for each assessed concentration was utilized to construct the graphic. Error bars in all panels represent the standard errors of the means calculated using data from at least three independent experiments performed with two technical replicates. (B to F) One-way ANOVA with Dunnett’s test for multiple comparison was performed. *, P < 0.05 compared to the Soret peak of uninfected THY-Hb-Fe2+ (B and D), uninfected THY-Hb-Fe2+ incubated for 2 h (C), or uninfected THY-Hb-Fe3+ (E and F).
Extended incubation of TIGR4 in THY-oxyHb revealed that the Soret curve was lost in a time-dependent manner (Fig. 1C). To assess if Spn-H2O2 had caused the loss of the Soret peak, we inoculated THY-oxyHb cultures with TIGR4ΔspxBΔlctO, and the culture was incubated for 6 h. A Soret curve was observed in this culture, but the heme-Fe2+ peak (415 nm), observed after 30 min postinoculation (Fig. 1A), had shifted to 405 nm, representing heme-Fe3+ (Fig. 1D). However, the deoxygenation of hemoglobin in THY-oxyHb cultures of TIGR4ΔspxBΔlctO was due to autoxidation, because uninfected cultures incubated under the same conditions, i.e., at 37°C with a 5% CO2 and ~20% O2 atmosphere, had shifted to heme-Fe3+ as well (Fig. 1C).
To further confirm that the loss of the Soret peak was due to Spn-H2O2, we prepared a complemented strain by cloning back spxB and lctO (ΩspxB ΩlctO) between SP_1113 and SP_1114 (Fig. 2). We also complemented TIGR4ΔspxB with a single copy of spxB to be utilized later (Fig. 2). This chromosomal location is unaffected by exogenous regulatory elements, such that complementing genes were controlled by their own promoters and insertion of genes within SP_1113 and SP_1114 did not affect virulence (51–54). Compared to the wild type (wt) and TIGR4ΔspxBΔlctO, complemented strains showed a slight but not significant growth delay (Fig. 2B); however, H2O2 production was restored to wt levels, as assessed with the Prussian blue-forming reaction and the Amplex Red kit (Fig. 2C and D) (55). In THY-oxyHb cultures of the ΩspxB ΩlctO strain incubated for 6 h, the Soret curve had flattened (Fig. 1C). These results suggest that Spn-H2O2 causes heme degradation.
FIG 2.
Preparation of TIGR4ΔspxBΔlctO-derivative complemented strains carrying spxB and lctO or only spxB. (A) Schematic diagram showing the chromosomal region where spxB and lctO (top) or spxB (bottom) was used to complement TIGR4ΔspxBΔlctO, creating TIGR4ΩspxBΩlctO or TIGR4ΩspxB. Promotor regions of each gene (P) are indicated, as well as terminators (red symbols); gene IDs were included using TIGR4 nomenclature. (B to D) TIGR4, TIGR4ΔspxBΔlctO, TIGR4ΩspxBΩlctO, or TIGR4ΩspxB was inoculated in THY and incubated at 37°C in a 5% CO2 atmosphere. (B) The OD600s of the cultures were obtained at the indicated times. Error bars represent the standard errors of the means calculated using data from at least three independent experiments performed with two technical replicates. (C) Supernatants from 4-h cultures were sterilized, and an aliquot was inoculated onto Prussian blue agar plates (55). The plates were incubated for 2 h at room temperature and photographed. This experiment was repeated at least three times. (D) Hydrogen peroxide was quantified from the supernatants using the Amplex Red assay. Error bars represent the standard errors of the means calculated using data from two independent experiments performed with two technical replicates. The limit of detection was 0.56 μM.
Given that heme-Fe2+ in THY-oxyHb cultures was oxidized to heme-Fe2+ (Fig. 1A) prior to losing the Soret curve (Fig. 1C), we next assessed if the oxidation state of heme was important for the subsequent loss of the Soret curve. To investigate this, we supplemented THY with deoxyhemoglobin (i.e., methemoglobin) containing heme-Fe3+ (THY-Hb), and the culture was incubated with Spn. A similar loss of the Soret curve was observed with wt and ΩspxB ΩlctO TIGR4 (Fig. 1E), but an intact curve was obtained in cultures of TIGR4ΔspxBΔlctO (Fig. 1E), indicating that heme-Fe3+ was further degraded.
Exogenous hydrogen peroxide supplementation at levels produced in S. pneumoniae cultures recapitulates hemoglobin oxidation.
We then assessed whether a concentration of hydrogen peroxide similar to that observed in wt cultures of Spn (<1 mM) (Fig. 2) (39, 40) can alter the heme-Fe3+ curve. For easy comparison of the multiple concentrations utilized in this experiment, we plotted the maximum absorbance obtained by spectroscopy for THY-Hb (405 nm) and compared it against that of THY-Hb treated with an increasing concentration of hydrogen peroxide. As shown in Fig. 1F, 10 μM hemoglobin in THY-Hb peaked at an absorbance of ~0.8. After 4 h of incubation with hydrogen peroxide, the absorbance was significantly reduced with a concentration of 88 μM and above, including the concentration of hydrogen peroxide observed in Spn cultures (~880 μM) (Fig. 1F). Altogether, these results indicate that the heme moiety in oxidized hemoglobin is degraded by Spn-H2O2.
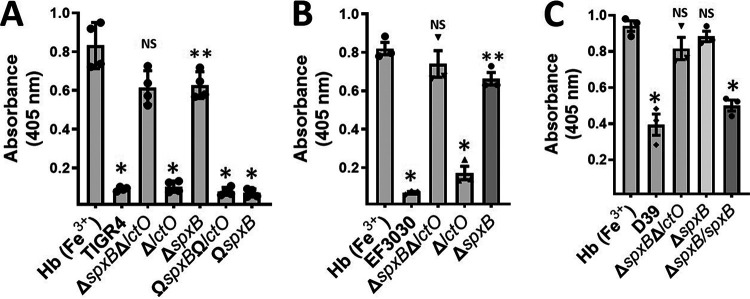
Oxidation of hemoglobin in S. pneumoniae cultures is mainly caused by hydrogen peroxide catalyzed through pyruvate oxidase SpxB.
Under aerobic conditions, hydrogen peroxide in pneumococcal cultures is produced through catalytic reactions of pyruvate oxidase (SpxB) and lactate oxidase (LctO) (43, 44). To assess the contribution of each enzyme, single ΔspxB or ΔlctO mutants prepared in the TIGR4 or EF3030 background were assessed in THY-Hb cultures. In cultures of wt TIGR4 and EF3030, the isogenic mutant TIGR4ΔlctO, and the EF3030ΔlctO, ΩspxBΩlctO and TIGR4ΔspxBΩspxB complemented strains, the Soret Heme-Hb3+ curve was flattened, causing a statistically significant reduction of the 405-nm peak (Fig. 3A). In contrast, in THY-Hb cultures of TIGR4ΔspxB and EF3030ΔspxB, the Soret curve remained intact, similar to THY-Hb cultures inoculated with the corresponding ΔspxB ΔlctO mutant (Fig. 3A and B). Additional D39ΔspxBΔlctO or D39ΔspxB (P878) hydrogen peroxide-deficient mutants did not alter the Soret curve, while the D39ΔspxB complemented strain, P1221, caused the loss of the Soret curve (Fig. 3C). These experiments demonstrated that oxidation of hemoglobin in cultures of Spn is mainly caused by hydrogen peroxide sourced from reactions catalyzed by SpxB.
FIG 3.
Oxidation of hemoglobin occurs through hydrogen peroxide produced by SpxB activity but not LctO. (A) Strains TIGR4, TIGR4ΔspxBΔlctO, TIGR4ΔspxB, TIGR4ΔlctO, TIGR4ΩspxBΩlctO, and TIGR4ΩspxB, (B) strains EF3030, EF3030ΔspxBΔlctO, EF3030ΔspxB, and EF3030ΔlctO, and (C) strains D39 and D39ΔspxBΔlctO were inoculated into THY containing methemoglobin [Hb (Fe3+)], and cultures were incubated for 4 h at 37°C in a 5% CO2 atmosphere. Supernatants were collected, and the spectra were obtained using an Omega spectrophotometer (BMG LabTech). The absorbance at 405 nm obtained from each strain was utilized to construct the graphics. Error bars in all panels represent the standard errors of the means calculated using data from at least three independent experiments performed with two technical replicates each. One-way ANOVA with Dunnett’s test for multiple comparison was performed. NS, not significant compared to THY-Hb (Fe3+); *, P < 0.0001 compared with untreated THY-Hb (Fe3+) or the ΔspxB ΔlctO or ΔspxB strain; **, P < 0.003 compared with untreated THY-Hb (Fe3+).
Spn-produced H2O2 degrades heme-Fe3+.
Heme degradation occurs through enzymatic reactions or by nonselective destruction of heme double bonds caused by reactive oxygen species, such as H2O2 (35, 56). To investigate whether Spn-H2O2 caused degradation of heme, THY-Hb medium was inoculated with TIGR4, EF3030, their hydrogen peroxide mutant derivatives, or complemented strains, and heme was assessed in supernatants using an in-gel heme assay (57, 58). The signals of heme and heme bound to hemoglobin monomers or dimers were detected in uninfected THY-Hb cultures incubated for 6 h (Fig. 4A). Heme signal in the supernatant of THY-Hb cultures incubated with TIGR4 for 2 h was similar to that of the uninoculated control, but it decreased after 4 h postinoculation and was not detected 6 h postinoculation (Fig. 4A). In THY-Hb cultures of TIGR4, TIGR4ΔlctO, TIGR4ΩspxBΩlctO, TIGR4ΔspxBΩspxB, EF3030, or EF3030ΔlctO, the signal of heme had already disappeared after 6 h of incubation (Fig. 4B and C). In contrast, in THY-Hb cultures of TIGR4ΔspxBΔlctO, TIGR4ΔspxB, EF3030ΔspxBΔlctO, and EF3030ΔspxB, signals of free heme and heme bound to hemoglobin were detected, thus confirming that degradation of heme had occurred in these cultures through hydrogen peroxide produced by SpxB.
FIG 4.
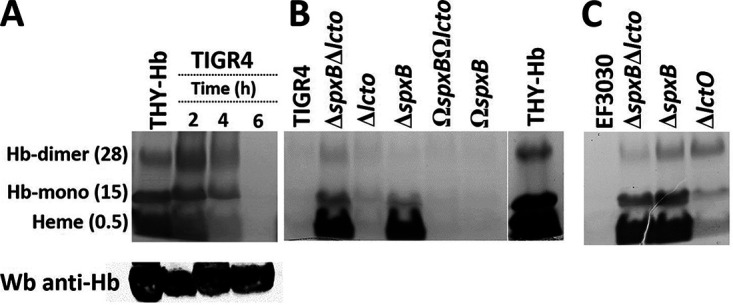
Oxidation of human hemoglobin by Spn-H2O2 leads to heme degradation. (A) TIGR4 wt was inoculated in THY supplemented with methemoglobin (THY-Hb) and incubated for 2, 4, or 6 h at 37°C in a 5% CO2 atmosphere. Supernatant was harvested and run in a 12% polyacrylamide gel under nondenaturing conditions, and heme was stained by an in-gel heme staining procedure. Free heme and heme bound to hemoglobin monomer (Hb-monomer) or to hemoglobin dimer (Hb-dimer), are indicated, with the observed molecular size (in kilodaltons). (B) TIGR4, TIGR4ΔspxBΔlctO, TIGR4ΔlctO, TIGR4ΔspxB, TIGR4ΩspxB, and TIGR4ΩspxBΩlctO and (C) EF3030 wt strains and EF3030ΔspxBΔlctO, EF3030ΔspxB, and EF3030ΔlctO were inoculated into THY-Hb and incubated for 6 h. Supernatants were harvested and analyzed as for panel A.
As a loading control and to additionally investigate if hemoglobin had been degraded, supernatants were probed by Western blotting with a polyclonal antihemoglobin antibody. Our results revealed a similar concentration of hemoglobin in those supernatants, confirming that the apoprotein was not degraded after 2, 4, or 6 h of incubation with Spn (Fig. 4A, bottom). As expected since heme was degraded by Spn-H2O2, the intracellular concentrations of iron were 79.19, 18.21, and 108.79 μM Fe/μg of protein in TIGR4, TIGR4ΔspxBΔlctO, and TIGR4ΩspxBΩlctO, respectively, confirming that iron was released and taken up by wt and complemented strains. Altogether, these experiments demonstrated that Spn-H2O2 degrades free heme and heme bound to hemoglobin.
Streptococci that produce hydrogen peroxide, but not other respiratory bacteria, cause degradation of heme.
Nonencapsulated Spn (NESpn) and other alpha-hemolytic streptococci produce hydrogen peroxide (59, 60). We therefore assessed a collection of NESpn strains and oral streptococci for heme degradation. We included Streptococcus mutans as an additional control, since it does not produce H2O2 (61, 62). As shown in Fig. 5, degradation of heme-Fe3+ was observed in THY-Hb cultures of NESpn strains MNZ41 and MNZ67, S. salivarius, S. mitis, S. pseudopneumoniae, and S. oralis but not in cultures of S. mutans (Fig. 5A). We also tested cultures of other bacterial species that cohabit the upper airways along with oral streptococci, such as Haemophilus influenzae and Staphylococcus aureus, and assessed cultures of Klebsiella pneumoniae. The incubation of THY-Hb with any of these bacterial species did not cause heme degradation (Fig. 5A). Streptococcal species that caused heme degradation produced hydrogen peroxide, while those unable to affect the heme-hemoglobin Soret curve did not (Fig. 5B). Altogether, these experiments demonstrated that Spn and other alpha-hemolytic streptococci that produce abundant hydrogen peroxide oxidize hemoglobin, causing the degradation of heme.
FIG 5.
Oxidation of hemoglobin occurs in cultures of oral streptococci that produce hydrogen peroxide. (A) THY supplemented with methemoglobin (THY-Hb) was left uninoculated or inoculated with the indicated strain and incubated for 4 h at 37°C in a 5% CO2 atmosphere. Supernatants were collected, and the spectra were obtained using an Omega spectrophotometer (BMG LabTech). The absorbance obtained at 405 nm for each assessed strain was utilized to construct the graphic. Error bars represent the standard errors of the means calculated using data from at least three independent experiments performed with two technical replicates. One-way ANOVA with Dunnett’s test for multiple comparisons was performed. *, P < 0.05, and NS, not significant, compared with untreated THY-Hb. (B) Hydrogen peroxide was quantified from 4-h-culture supernatants of each strain using the Amplex Red assay. Error bars represent the standard errors of the means calculated using data from two independent experiments performed with two technical replicates. The limit of detection was 0.56 μM.
Heme is oxidized and degraded in an ex vivo model of pneumococcal bacteremia.
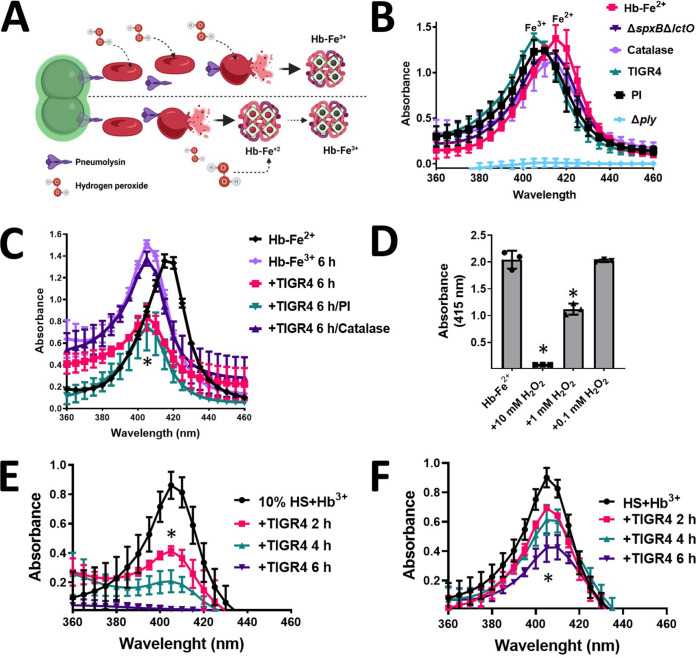
During the course of pneumococcal bacteremia, erythrocytes are lysed by pneumolysin (Ply), which is released through autolysis (63). Ply is also located on the bacterial membrane, where it lyses red blood cells through contact (19, 20). Abundant Spn-H2O2 is produced as early as 2 h postinoculation (Fig. 2D); therefore, we sought to assess whether oxidation of hemoglobin and heme degradation occur in an ex vivo model of pneumococcal bacteremia (Fig. 6A). To assess this, we supplemented THY broth with a suspension of red blood cells (THY-RBC), and this medium was infected with TIGR4, TIGR4Δply, or TIGR4ΔspxBΔlctO. The oxidation and concentration of hemoglobin as well as heme degradation were investigated.
FIG 6.
Hemoglobin is oxidized by Spn-H2O2 in an ex vivo bacteremia model with erythrocytes. (A) Model of ex vivo pneumococcal bacteremia showing pneumococci with pneumolysin (Ply) located in the membrane; Ply is also released through autolysis. Membrane-permeative hydrogen peroxide (H2O2) is produced as a by-product of the oxidation of pyruvate. (Bottom) Oxyhemoglobin (Hb-Fe2+) is then released from erythrocytes by Ply-induced erythrocyte lysis, and once released, oxyhemoglobin is oxidized by Spn-H2O2 to methemoglobin (Hb-Fe3+). (Top) Alternatively, Spn-H2O2 permeates inside erythrocytes causing the oxidation of hemoglobin, which is then released from lysed erythrocytes as methemoglobin (Hb-Fe3+). (B and C) A suspension (2%) of sheep erythrocytes in THY was inoculated with TIGR4, TIGR4 supplemented with catalase, TIGR4 supplemented with protease inhibitors (PI), TIGR4ΔspxBΔlctO, or TIGR4Δply, and the inoculated cultures were incubated for 30 min (B) or 6 h (C) at 37°C in a 5% CO2 atmosphere. As a control, erythrocytes were lysed with saponin, and the oxyhemoglobin (THY-Hb2+) that was released into the supernatant was considered to represent the maximum hemoglobin release. (D and E) Human serum was supplemented with 10 μM methemoglobin (HS+Hb3+), or serum was used to supplement THY to a final concentration of 10% (vol/vol), and 10 μM methemoglobin (10% HS+Hb3+) was added. These media were inoculated with TIGR4 and incubated for 2, 4, or 6 h. (F) Oxyhemoglobin was released from a 2% suspension of erythrocytes and left untreated (Hb-Fe2+) or supplemented with hydrogen peroxide (H2O2) and incubated for 4 h. Supernatants from experiments in panels B to F were collected, and the spectra were obtained using an Omega spectrophotometer (BMG LabTech). (B to F) Error bars represent the standard errors of the means calculated using data from at least three independent experiments. One-way ANOVA with Dunnett’s test for multiple comparisons was performed. *, P < 0.05, and NS, not significant, compared with (C) TIGR4/catalase, (D) untreated HS+Hb3+, (E) untreated 10% HS+Hb3+, or (F) untreated Hb-Fe2+.
As expected, the Soret curve was not observed in supernatants from the nonlysed THY-RBC control, and it was also absent in supernatants of THY-RBC infected with TIGR4Δply, because this strain does not lyse erythrocytes (Fig. 6B). Because RBC contains oxyhemoglobin (Hb-Fe2+), the Soret curve of the positive control, where RBC had been lysed with saponin, peaked at 415 nm with an average absorbance of 1.38 (Fig. 6B). The concentration of cell-free hemoglobin in this control supernatant was 17.35 μM, as measured by the Quantichrom assay (Table 1). The Soret curve from THY-RBC cultures infected with TIGR4 peaked at 405 nm with an average absorbance of 1.39 (Fig. 6B) and hemoglobin concentration of 12.8 μM (Table 1). These results indicate that maximum TIGR4-induced release of hemoglobin is achieved within 30 min of incubation, compared with the saponin-lysed control, and that the oxidation of heme-Fe2+ to heme-Fe3+ had already occurred (Fig. 6B). Similar to the wt and because TIGR4ΔspxBΔlctO produces Ply (21, 23), a Soret curve was observed in THY-RBC cultures infected with this hydrogen peroxide-deficient mutant as early as 30 min postinoculation (Fig. 6B). This Soret curve peaked at 415 nm with an average absorbance of 1.21 and a concentration of hemoglobin of 16.4 μM (Table 1), indicating that most hemoglobin had been released but, unlike in the wt strain, oxidation of heme-Fe2+ did not occur, because of the lack of hydrogen peroxide. Accordingly, oxidation of hemoglobin did not occur in TIGR4 cultures treated with catalase, but it was oxidized to methemoglobin in TIGR4 cultures treated with protease inhibitors (Fig. 6B).
TABLE 1.
Quantification of cell-free hemoglobin and the heme-hemoglobin Soret curve
| Strain | Cell-free hemoglobin (μM) | Absorbance at 415 nm |
|---|---|---|
| Uninfected | 17.35 | 1.38 |
| TIGR4 | 12.8 | 1.39 |
| TIGR4ΔspxBΔlctO | 16.4 | 1.21 |
After 6 h of incubation, autoxidation of uninoculated, saponin-lysed THY-RBC cultures (Fig. 6C, Hb-Fe3+) and those infected with TIGR4 treated with catalase (Fig. 6C) was seen. THY-RBC infected with TIGR4 had already exhibited a flattened heme-Fe3+ curve, and therefore, heme had been degraded, but this was not seen with TIGR4 treated with catalase (Fig. 6C). Degradation of heme was not inhibited by supplementing THY-RBC with protease inhibitors before infection (Fig. 6C). We next treated saponin-lysed oxyhemoglobin with increasing amounts of H2O2, and oxidation to methemoglobin and then degradation of heme were recapitulated (Fig. 6D). Altogether, our results indicate that oxidation of hemoglobin and degradation of heme by Spn-H2O2 occur during pneumococcal bacteremia.
To further mimic bacteremia, we added THY with 10% of human serum and supplemented this broth culture with 10 μM methemoglobin. In addition, nondiluted human serum was supplemented with 10 μM methemoglobin. These hemoglobin-containing serum cultures were infected with TIGR4 and incubated at different times. A time-dependent degradation of heme from hemoglobin was observed in both culture conditions (Fig. 6E and F), confirming that hemoglobin is oxidized and heme is degraded when Spn grows in human serum.
Because the release of hemoglobin and its oxidation were observed early during the simulated bacteremia (i.e., 30 min), to gain insights into the mechanism leading to hemoglobin oxidation and heme degradation, we investigated expression of ply, spxB, and lctO during incubation of Spn in THY-RBC, THY-Hb, and THY supplemented with heme. Compared to THY control cultures, the expression of ply, spxB, and lctO did not significantly change when TIGR4 was grown in broth cultures with RBC or hemoglobin. However, transcription of spxB was upregulated when TIGR4 was incubated in THY containing heme, whereas the transcription of lctO and ply was downregulated (Fig. 7). Together, these experiments indicate that under these culture conditions, heme was the inducer for Spn-H2O2 production.
FIG 7.

Regulation of ply, spxB and lctO genes by incubation with erythrocytes, hemoglobin, or heme. TIGR4 wt was inoculated into THY broth alone, THY broth containing 1% sheep erythrocytes, THY broth with 10 μM hemoglobin (Hb), or THY broth with 5 μM heme. Cultures were incubated for 2 h at 37°C in a 5% CO2 atmosphere; bacteria were then harvested, and RNA was extracted from these cultures and utilized as the template in qRT-PCRs with primers that amplified ply, spxB, or lctO. Average CT values were normalized to the 16S rRNA gene, and the fold differences relative to the THY control were calculated using the comparative CT method (2−ΔΔCT) (78).
DISCUSSION
We have demonstrated in this study that H2O2 produced by the pneumococcus, Spn-H2O2, and other alpha-hemolytic streptococci caused the oxidation of hemoglobin to methemoglobin with a subsequent degradation of heme. The current study also demonstrated that the oxidation of hemoglobin and heme degradation occurred in a simulated bacteremia infection, suggesting that these oxidative reactions may occur during pneumococcal disease. It has been found that nearly 3% of oxyhemoglobin is auto-oxidized inside erythrocytes, and this autoxidation is caused by 2 × 10−7 mM hydrogen peroxide generated in the cytosol of red blood cells (64). Although autoxidation by CO2 occurred in our in vitro system 4 h postinoculation (i.e., autoxidation was not observed upon incubation at 37°C with no CO2), Spn cultures produced 107-fold more (~1 mM) hydrogen peroxide within a few hours of incubation and caused the rapid oxidation of hemoglobin leading to the release of heme, a toxic molecule, with its subsequent degradation (65). We demonstrated that heme degradation caused an increased of free iron in the medium, which was taken up by pneumococci. These novel observations might have important implications for pathogenesis of hydrogen peroxide-producing streptococci but also for the symbiosis observed in bacterial communities, where some species can benefit from heme and iron release by hydrogen peroxide-producing bacteria.
In vitro oxidation of hemoglobin with hydrogen peroxide produces ferryl hemoglobin (Hb-Fe4+), a highly reactive form of hemoglobin that is reduced to methemoglobin (66). With an increased presence of hydrogen peroxide molecules, methemoglobin is oxidized back to Hb-Fe4+, which in turn causes heme degradation (65). Iron stored in heme-hemoglobin is poorly reactive, but once freed up as a consequence of heme degradation, free iron can react with hydrogen peroxide to produce another highly reactive hydroxyl radical (67, 68). Indeed, we have demonstrated that free iron present in THY medium is enough to generate hydroxyl radicals (·OH) (40), killing S. aureus strains rapidly (39, 69, 70). The increase of iron in the culture medium due to heme degradation should result in an increased generation of ·OH. The potential presence of two highly reactive radicals, Hb-Fe4+ and ·OH, that can contribute to the cytotoxicity seen during pneumococcal disease is being investigated in our laboratories.
In children with SCD, hemoglobin (Hb) S is oxidized to Hb-Fe4+ and then reduced to methemoglobin, resulting in an acute hemolytic vascular inflammatory process that causes acute lung injury (30, 50). Both Hb-Fe4+ and methemoglobin induce a drop in mitochondrial oxygen consumption and mitochondrial membrane potential in epithelial lung cells (36). Acute chest syndrome is an important cause of hospitalization and mortality in children with SCD, and Spn is a leading etiologic agent of acute chest syndrome. While the precise role of Spn is still unclear, children with SCD are ~100-fold more susceptible to pneumococcal infection (71). Studies of the oxidation of hemoglobin by Spn-H2O2, however, have been neglected. We recently demonstrated that the growth of planktonic Spn, as well as the formation of pneumococcal biofilms, is enhanced by supplementing the culture medium with human hemoglobin (72, 73). Besides iron, other nutrients may become accessible to pneumococcus, since the oxidation of hemoglobin by hydrogen peroxide releases globin-derived peptides (29), but also, a striking transcriptome remodeling was identified that included upregulated transcription of genes encoding transporters of glyco-conjugated molecules (72, 73).
Besides making nutrients available, oxidative reactions may lead to unintended consequences. During pneumococcal pneumonia, the pulmonary parenchyma presents with hemorrhage, inflammatory congestion, hepatization, suppurative infiltration, and lung parenchymal injury (4, 9–13). Whereas there are a number of virulence factors implicated in the pathophysiology of lung infection (63), the oxidation of hemoglobin through Spn-H2O2 may also be an important contributor to pneumococcal disease. For example, mice intranasally inoculated with Spn develop lung hemorrhage, at necropsy, and histological analysis reveals lung consolidation associated with alveolar septal edema and pleomorphic alveolar, interstitial, and perivascular cellular inflammation (74). Spn strains with mutations in the spxB gene, whether with a pleotropic capsule defect or not, were attenuated for virulence in mouse models of pneumococcal disease (46, 47, 75).
There are other important implications as a result of the oxidation of hemoglobin by hydrogen peroxide produced by streptococci. Heme release and/or heme degradation through hydrogen peroxide may be beneficial to other bacteria which do not synthetize large amounts of this pro-oxidant but require iron for their metabolism and pathogenesis. For instance, it was recently demonstrated that dental plaque bacteria such as S. gordonii, which produces hydrogen peroxide, oxidize hemoglobin to release heme, and this free heme facilitates the colonization of the dental plaque by the Gram-negative bacterium Porphyromonas gingivalis (49). P. gingivalis does not produce siderophores, but it has a sophisticated mechanism for heme iron acquisition (76). Conversion of oxyhemoglobin to methemoglobin by hydrogen peroxide-producing bacteria allows the release of heme, which can then be taken up by these microorganisms.
In summary, we demonstrated that Spn and other hydrogen peroxide-producing streptococci oxidize hemoglobin from human and other species, releasing and degrading heme. These oxidative reactions occurred early during the lag phase of growth, indicating that metabolic adaptation for growth in the presence of hemoglobin stimulated production of Spn-H2O2. This upregulated metabolic adaptation was the result in part of the release of heme, since gene expression studies demonstrated an upregulation of spxB transcription, but not that of lctO or the pneumolysin gene ply, under the culture conditions tested. It is noteworthy that incubation with erythrocytes or hemoglobin did not induce regulation of spxB in the current study. We observed production of a potential toxic radical during the oxidation of hemoglobin, spanning the incubation time utilized in gene expression studies, which may explain the observed downregulation of spxB (A. Scasny, P. Smith, & J. E. Vidal., unpublished observations). Efforts are in place to assess the specific role for these oxidative reactions for pathogenesis and bacterial symbiosis.
MATERIALS AND METHODS
Bacterial strains, material, and growth conditions.
All wild-type Spn strains and mutant derivatives used in this study are listed in Table 2. Bacterial stocks were prepared in medium containing skim milk-tryptone-glucose-glycerin (STGG) and stored at −80°C (77). Pneumococci were cultivated at 37°C in a 5% CO2 and ~20% O2 atmosphere. Experiments were performed using Todd-Hewitt broth with 0.5% yeast extract (THY), THY with 10 μM human methemoglobin (Sigma-Aldrich), THY with 5 μM heme (Sigma-Aldrich), THY with sheep erythrocytes (Quad Five), and THY with horse erythrocytes (Lampire). Brain heart infusion (BHI) broth, Bacto agar, Bacto tryptic soy broth, Bacto yeast extract, and Bacto Todd-Hewitt broth were purchased from Becton, Dickinson and Company. Antibiotics utilized were gentamicin, erythromycin, spectinomycin, and chloramphenicol, and were all sourced from Sigma-Aldrich. Other materials used include hydrogen peroxide (Fisher Scientific), catalase (Sigma-Aldrich), cOmplete mini-protease inhibitor cocktail (Roche), FeCl3·6H2O and potassium hexacyanoferrate(III) (Sigma-Aldrich), o-dianisidine (Alfa Aesar)/sodium acetate (Sigma-Aldrich), and ethanol and methanol (Fisher Scientific). Human serum was purchased from MP Biomedicals.
TABLE 2.
Strains utilized in this study
| Strain | Characteristics | Reference or source |
|---|---|---|
| TIGR4 | Reference strain, whole genome sequenced, capsular serotype 4 | 83 |
| SPJV41 (TIGR4ΔspxBΔlctO) | TIGR4 with a deleted spxB and lctO gene by insertion-deletion with an erythromycin and spectinomycin cassette, respectively | 40 |
| SPJV29 (TIGR4ΔspxB) | TIGR4 with an insertionally inactivated spxB::ermB (Eryr) | 23 |
| SPJV42 (TIGR4ΔlctO) | TIGR4 with an insertionally inactivated lctO:: aad9 (Spcr) | 40 |
| SPJV43 (TIGR4ΔspxBΩspxB) | SPJV29 complemented with the gene spxB and its promoter | This study |
| SPJV44 (TIGR4ΔspxBΔlctOΩspxB-lctO) | SPJV41 complemented with the gene spxB and lctO | This study |
| D39 | Avery strain, clinical isolate, capsular serotype 2 | 84 |
| P878 | D39 with a deletion in spxB | 39 |
| P1221 | P878 complemented with pMU1328::spxB | 43 |
| SPJV45 (D39ΔspxBΔlctO) | D39 with a deleted spxB and lctO gene by insertion-deletion with an erythromycin and spectinomycin cassette, respectively | 23 |
| EF3030 | Clinical isolate, caccine serotype 19F strain | 85, 86 |
| SPJV49 (EF3030ΔspxBΔlctO) | EF3030 with a deleted spxB and lctO gene by insertion-deletion with an erythromycin and spectinomycin cassette, respectively | 23 |
| SPJV50 (EF3030ΔspxB) | EF3030 with an insertionally inactivated spxB::ermB (Eryr) | This study |
| SPJV51 (EF3030ΔlctO) | EF3030 with an insertionally inactivated lctO:: aad9 (Spcr) | This study |
| MNZ41 | Non-encapsulated S. pneumoniae strain | 87 |
| MNZ67 | Non-encapsulated S. pneumoniae strain | 87 |
| S. pseudopneumoniae | ATCC 960 | Laboratory stock |
| S. salivarius | ATCC 7073 | Laboratory stock |
| S. oralis | ATCC 35037 | Laboratory stock |
| K. pneumoniae | ATCC 700603 | 88 |
| Staphylococcus aureus strain Newman | NCTC 8178, ATCC 13420 | 89 |
| Non-typeable Haemophilus influenzae (NTHI) | M5029 | Laboratory stock |
| S. mitis | ATCC 49456 | Laboratory stock |
| S. mutans | ATCC 25175 | Laboratory stock |
Preparation of inoculum for experiments.
The inoculum was prepared as previously described (40). Briefly, bacteria were inoculated on blood agar plates (BAP) and incubated overnight at 37°C in a 5% CO2 atmosphere. Bacteria were then harvested from plates by phosphate-buffered saline (PBS) washes, and this suspension was used to inoculate THY with or without additives, which was brought to a final optical density at 600 nm (OD600) of ~0.1. This suspension contained ~5.15 × 108 CFU/mL, as confirmed by dilution and plating of aliquots of the suspension.
Bacterial broth culture medium containing oxyhemoglobin.
Sheep or horse erythrocytes were washed three times with sterile PBS (pH 7.4) at 300 × g for 5 min in a refrigerated centrifuge (Eppendorf). The washed erythrocyte suspension was lysed with 0.1% saponin, and the concentration of oxyhemoglobin in this preparation was determined using the QuantiChrom hemoglobin assay kit (BioAssay Systems); oxyhemoglobin was then added to THY broth to a final concentration of ~10 μM. The concentration and the redox state of oxyhemoglobin were verified by spectroscopy before each experiment. In some experiments, THY containing oxyhemoglobin was supplemented with an excess of catalase to a final concentration of 1,000 U/mL or with 1× cOmplete protease inhibitor cocktail (Roche).
Studies of the oxidation of oxyhemoglobin and methemoglobin and heme degradation by spectroscopy.
Experiments were conducted using six-well microplates (Genesee Scientific) containing THY and a 1% suspension of washed sheep erythrocytes, 10 μM oxyhemoglobin, or 10 μM methemoglobin. In some experiments, human serum was supplemented with 10 μM methemoglobin, or THY was supplemented with human serum to a final 10% (vol/vol) concentration, and 10 μM methemoglobin was added. These media were inoculated with Spn or the other bacterial species listed for each experiment and incubated for the indicated time at 37°C in a 5% CO2 and ~20% O2 atmosphere. In another set of experiments, THY containing 10 μM human methemoglobin was treated with different concentrations of hydrogen peroxide for 4 h. At the of the incubation time, supernatants containing planktonic bacteria were removed from cultures, and bacteria were pelleted by centrifugation at 13,000 rpm for 5 min in a refrigerated microcentrifuge (Eppendorf). The oxidation of oxyhemoglobin to methemoglobin and further oxidation and degradation of methemoglobin were determined by analyzing the spectra from 200 nm to 1,000 nm in bacterium-free supernatants using a spectrophotometer Omega BMG LabTech (Thermo Fisher).
Quantification of hydrogen peroxide production by S. pneumoniae strains.
Hydrogen peroxide production was quantified from Spn cultures inoculated in six-well microplates (Genesee Scientific) containing THY. These cultures were incubated at 37°C in a 5% CO2 and ~20% O2 atmosphere for the indicated time. At the end of the incubation, the culture supernatant was collected and centrifuged at 4°C for 5 min at 15,000 rpm, and then the supernatant was transferred to a new tube and kept on ice for no more than 30 min. Some supernatants were filter sterilized using a 0.4-μm syringe filter (Fisher Scientific). Collected supernatants were diluted with 1× reaction buffer from the Amplex Red H2O2 assay kit (Molecular Probes), and hydrogen peroxide levels were quantified according to the manufacturer’s instructions using a spectrophotometer Omega BMG LabTech (Thermo Fisher).
Qualitative detection of hydrogen peroxide production by S. pneumoniae strains.
Qualitative detection of H2O2 production by Spn was performed using the Prussian blue agar assay, whose limit of detection is 2.5 nm H2O2 (55). Prussian blue agar plates were made with 1.0 g/L of FeCl3·6H2O, 1.0 g/L of potassium hexacyanoferrate(III) {K3[Fe(CN)6]; also known as Prussian blue}, 37 g/L of dehydrated BHI broth, and 15 g/L of agar. Prior to experiments, detection of H2O2 was assessed by dropwise addition of 10 μL of a 1 mM H2O2 solution that reduced K3[Fe(CN)6] and causing its precipitation and then a blue halo within minutes. PBS was used as a negative control. Spn strains or culture supernatants (10 μL), obtained as described above, were inoculated or spotted, respectively, onto Prussian blue agar plates. These plates were incubated at 37°C in a 5% CO2 and ~20% O2 atmosphere overnight (Spn) or for 4 h (supernatants), after which plates were photographed with a Canon Rebel EOS T5 camera system, and digital pictures were analyzed.
Western blotting.
Bacteria were grown in THY with 10 μM hemoglobin at 37°C for 2 h, 4 h, and 6 h. After incubation, the cells were harvested by centrifuge at 15,000 rpm for 5 min, and the supernatant were transferred to new tubes. The collected samples were combined with 4× reducing sample buffer and boiled for 5 min. The mixture was loaded into 10-well 4 to 12% Mini-Protean TGX precast gels (Bio-Rad), which were run for 2 h at 90 V in running buffer. Gels were transferred to nitrocellulose membranes in transfer buffer supplemented with 20% ethanol with the Trans-Blot Turbo transfer system. Membranes were blocked 1 h at room temperature in 5% nonfat dry milk in Tris-buffered saline supplemented with 0.1% Tween 20 (TBST). Then, a membrane was incubated with the Hb polyclonal antibody (Invitrogen) at a 1:1,000 dilution in 5% nonfat dry milk in TBST overnight at 4°C. The next day, the blot was washed three times for 5 min each with TBST and incubated with donkey anti-goat IgG conjugated to horseradish peroxidase (HRP) (Santa Cruz) as the secondary antibody at 1:5,000 in 5% nonfat dry milk in TBST 1 h at room temperature. Blots were washed three times for 5 min each in TBST, and 2 mL of each SuperSignal substrate (Thermo Fisher Scientific) reagent was added to the blots, which were incubated for 5 min at room temperature. Blots were imaged on a ChemiDoc MP imager (Bio-Rad) using Image Lab 5.0 software, with automatic exposure settings for chemiluminescence and high specificity, optimizing for bright bands.
In-gel heme staining.
Bacteria were grown in THY with 10 μM hemoglobin at 37°C for 2, 4, or 6 h, after which supernatants were collected, spun down, and combined with nonreducing loading buffer. The mixtures were loaded into a nondenaturing 12% Mini-Protean TGX precast gel (Bio-Rad), which was run for 2 h at 90 V in running buffer lacking SDS. The gel was then stained using a published protocol (35). Briefly, after electrophoresis, gels were immersed in a methanol-sodium acetate (pH 5) solution and incubated at room temperature on a rocking platform (VWR) at 1 speed for 2 min with constant shaking. After incubation, a solution of o-dianisidine–sodium acetate (pH 5) was added to the gel, which was incubated for 20 min in the dark. To visualize heme in the gel, 3% hydrogen peroxide was added, and the reaction was stopped by washing with distilled water. The gel was then photographed with a Canon Rebel EOS T5 camera system, and digital pictures were analyzed.
RNA extraction and qRT-PCR analysis.
Bacteria were inoculated into THY or into THY containing 5 μM heme, 5 μM Hb, or a 1% suspension of RBC and incubated at 37°C for 2 h. After incubation, the bacterial suspension was collected, and mixed with RNAprotect reagent (Qiagen). Then cells were harvested and the total RNA was extracted using the RNeasy minikit (Qiagen) following the manufacturer’s instructions. DNA was removed using the Turbo DNase-free kit (Life Technologies). The RNA concentration was obtained using a NanoDrop spectrophotometer (Thermo Fisher Scientific), and 200 ng of RNA was cDNA transcribed using the iScript cDNA synthesis kit (Bio-Rad). Gene expression analysis was carried out using PerfeCTa SYBR green supermix (Quantabio) and a CFX96 Touch real-time PCR system (Bio-Rad). Primers used for the qRT-PCR analysis are listed in Table 3. The following conditions were utilized: 1 cycle at 95°C for 3 min and 40 cycles of 95°C for 15 s, 60°C for 30 s and 72°C for 30 s. Melting curves were generated to confirm the absence of primer dimers. The relative quantitation of mRNA expression was normalized to the constitutive expression of the housekeeping 16S rRNA gene and calculated by the comparative cycle threshold (2−ΔΔCT) method (78).
TABLE 3.
Oligonucleotides designed and used in this study
| Primer | Sequence (5′→3′) |
|---|---|
| hlpA-up1 | TCAGCAGGTTCATGAGGGAA |
| hlpA-up2 | TCATTTCTGTTTTATAACAAAGTCCGGATCCTTTAACAGCGT |
| hlpA-down1 | CTCGCCGAAAATCAAATATGATCACTCACGGCATGGATGA |
| hlpA-down2 | CAAAACAACATTGCCCGACG |
| spxB1 | CTTTGTTATAAAACAGAAATGA |
| spxB2 | CATATTTGATTTTCGGCGAG |
| hlpA-up3 | TTTCCCCAATTATTCACCCCACATATTTGATTTTCGGCGAG |
| lcto1 | TGGGGTGAATAATTGGGGAAA |
| lcto2 | CATTCAGTGGAGGCAATCTGT |
| lcto-spec1 | TTTCTGAGTAGGCGGAGTGG |
| lcto-spec2 | GCAATTTTCAGAGCAGCTTGG |
| JVS109L | TCTACTCAATCGACGTAGGTAACA |
| JVS110R | TATTGAACGTTTGTGATAACGTCT |
| JVS59L | TGAGACTAAGGTTACAGCTTACAG |
| JVS60R | CTAATTTTGACAGAGAGATTACGA |
| JVS105L | TTTGCAATGTAGAAAATCCAAGTA |
| JVS106R | AAATCTCTGGAAGGTCAACAGTAG |
| JVS49L | CTGATGACTGTCAATCGAGATACC |
| JVS50R | AATGGACGAATCAACTCCATAA |
| JVS35L | AACCAAGTAACTTTGAAAGAAGAC |
| JVS36R | AAATTTAGAATCGTGGAATTTTT |
Preparation of lctO mutation in Spn strains EF3030 and D39.
To prepare an isogenic lctO mutant, we PCR amplified, using the primers lcto-spec1 and lcto-spec2 (Table 3), an insertionally inactivated lctO-spectinomycin fragment from strain TIGR4ΔlctO (40). This PCR product was purified using the QIAquick PCR purification kit (Qiagen) and then transformed into EF3030 or D39 using a standard transformation procedure (79). Transformants were harvested in BAP with spectinomycin (100 μg/mL), and the insertional inactivation of lctO was confirmed by PCR with the primers lcto-spec1 and lcto-spec2 and sequencing.
Construction of complemented strains.
To complement the gene spxB or the genes spxB and lctO in TIGR4ΔspxB or TIGR4ΔspxBΔlctO, respectively, we chose a “neutral” chromosomal location within the hlpA gene (SP1113) in which insertions do not affect virulence (52). A spxB-complemented fragment was prepared by amplifying an upstream region carrying a fragment of the hlpA gene, using DNA purified from TIGR4 as a template and primers hlpA-up1 and hlpA-up2 (Table 3). We then generated a PCR fragment, using DNA purified from TIGR4 as a template, containing the spxB gene and its promoter with the primers spxB1 and spxB2 (Table 3). A downstream fragment containing the catP gene, encoding chloramphenicol resistance, and a fragment of open reading frame (ORF) SP1114 was amplified using DNA from strain JWV500 as a template and primers hlpA-down1 and hlpA-down2. PCR products were purified using a QIAquick PCR purification kit (Qiagen) and ligated by splicing overlap extension PCR with the primers hlpA-up1 and hlpA-down2. The PCR-ligated product was verified in a 0.8% DNA gel, purified from the gel using a QIAquick gel extraction kit (Qiagen), and reamplified by PCR with the primers hlpA-up1 and hlpA-down2. This PCR product was purified as described above and used to transform TIGR4ΔspxB using a standard procedure (79). Transformants were harvested in BAP containing chloramphenicol (5 μg/mL) and screened by PCR using the primers spxB1 and spxB2 to confirm the complementation of spxB.
To construct a spxB- and lctO-complemented strain, an upstream DNA fragment containing hlpA and spxB was amplified using DNA purified from TIGR4ΔspxBΩspxB and the primers hlpA1 and spxB2. Then, a DNA fragment was prepared by PCR amplification of the lctO gene and its putative promoter region using TIGR4 DNA as a template and the primers lctO1 and lctO2. The downstream fragment containing the catP gene and a fragment of ORF SP1114, prepared as described above, was used. After purification with a QIAquick PCR purification kit (Qiagen), these three DNA fragments were ligated by splicing overlap extension PCR with the primers hlpA-up1 and hlpA-down2 and purified. This construct was reamplified with the primers hlpA-up1 and hlpA-down2, purified, and transformed into TIGR4ΔspxBΔlctO. Transformants were screened by PCR using the primers hlpA-up1 and hlpA-down2, and selected completed clones were further confirmed by H2O2 production.
Metal content analysis.
Pneumococcal biofilms cultured in THY-Hb for 4 h were washed three times with ice-cold PBS, resuspended in 100 μL PBS, and sonicated at high intensity for 5 min in cycles of 30 s on and 30 s off using a Bioruptor (Diagenode, Denville, NJ, USA). Protein in all samples was measured by the Bradford method, and a known mass of sample was mineralized in concentrated HNO3, trace metal grade (Thermo Fisher Scientific), as previously described (80). Iron in each sample was measured by atomic absorbance spectroscopy (AAS) using a 55B AA flame atomic absorption spectrometer (Agilent, Santa Clara, CA, USA). Analytical grade standards for iron (Thermo Fischer Scientific) were diluted in ultrapure water (18 Mohm). The iron content of each sample was normalized to the initial mass of protein as previously reported for other metals (81, 82).
Statistical analysis.
We performed one-way analysis of variance (ANOVA) followed by Dunnett’s multiple-comparison test when more than two groups were compared or Student’s t test to compare two groups, as indicated. All statistical analysis was performed using the software GraphPad Prism (version 8.3.1).
ACKNOWLEDGMENTS
This study was supported in part by a grant from the National Institutes of Health (NIH; 5R21AI144571-03) to J.E.V. and Wesleyan University institutional funds to T.P.-B. B.A. was supported by a Fulbright scholarship awarded by the U.S. Department of State. F.M.F. is supported by the Ronald E. McNair Postbaccalaureate Fellowship program. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH or the U.S. Department of State.
We thank Jaime Carrazco-Carrillo from Wesleyan University for his assistance in some laboratory assays.
Footnotes
This article is a direct contribution from Jorge E. Vidal, a member of the Infection and Immunity Editorial Board, who arranged for and secured reviews by Elsa Bou Ghanem, University at Buffalo, State University of New York; Karthik Subramanian, Rajiv Gandhi Centre for Biotechnology Bio-Innovation Centre; and Cheryl Okumura, Occidental College.
Contributor Information
Jorge E. Vidal, Email: jvidal@umc.edu.
Andreas J. Bäumler, University of California, Davis
REFERENCES
- 1.Jedrzejas MJ. 2001. Pneumococcal virulence factors: structure and function. Microbiol Mol Biol Rev 65:187–207. 10.1128/MMBR.65.2.187-207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kadioglu A, Weiser JN, Paton JC, Andrew PW. 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 6:288–301. 10.1038/nrmicro1871. [DOI] [PubMed] [Google Scholar]
- 3.Tomasz A. 2000. Streptococcus pneumoniae: molecular biology & mechanisms of disease. Mary Ann Liebert, Inc., Larchmont, NY. [Google Scholar]
- 4.van der Poll T, Opal SM. 2009. Pathogenesis, treatment, and prevention of pneumococcal pneumonia. Lancet 374:1543–1556. 10.1016/S0140-6736(09)61114-4. [DOI] [PubMed] [Google Scholar]
- 5.Klugman KP, Madhi SA, Albrich WC. 2008. Novel approaches to the identification of Streptococcus pneumoniae as the cause of community-acquired pneumonia. Clin Infect Dis 47(Suppl 3):S202–S206. 10.1086/591405. [DOI] [PubMed] [Google Scholar]
- 6.O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, Lee E, Mulholland K, Levine OS, Cherian T, Hib and Pneumococcal Global Burden of Disease Study Team . 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893–902. 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 7.Walker CL, Rudan I, Liu L, Nair H, Theodoratou E, Bhutta ZA, O’Brien KL, Campbell H, Black RE. 2013. Global burden of childhood pneumonia and diarrhoea. Lancet 381:1405–1416. 10.1016/S0140-6736(13)60222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holyoake LV, Hunt S, Sanguinetti G, Cook GM, Howard MJ, Rowe ML, Poole RK, Shepherd M. 2016. CydDC-mediated reductant export in Escherichia coli controls the transcriptional wiring of energy metabolism and combats nitrosative stress. Biochem J 473:693–701. 10.1042/BJ20150536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chao Y, Marks LR, Pettigrew MM, Hakansson AP. 2014. Streptococcus pneumoniae biofilm formation and dispersion during colonization and disease. Front Cell Infect Microbiol 4:194. 10.3389/fcimb.2014.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simell B, Auranen K, Kayhty H, Goldblatt D, Dagan R, O’Brien KL, Pneumococcal Carriage Group . 2012. The fundamental link between pneumococcal carriage and disease. Expert Rev Vaccines 11:841–855. 10.1586/erv.12.53. [DOI] [PubMed] [Google Scholar]
- 11.Eurich DT, Marrie TJ, Minhas-Sandhu JK, Majumdar SR. 2017. Risk of heart failure after community acquired pneumonia: prospective controlled study with 10 years of follow-up. BMJ 356:j413. 10.1136/bmj.j413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Musher DM, Rueda AM, Kaka AS, Mapara SM. 2007. The association between pneumococcal pneumonia and acute cardiac events. Clin Infect Dis 45:158–165. 10.1086/518849. [DOI] [PubMed] [Google Scholar]
- 13.Masters IB, Isles AF, Grimwood K. 2017. Necrotizing pneumonia: an emerging problem in children? Pneumonia (Nathan) 9:11. 10.1186/s41479-017-0035-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown AO, Millett ER, Quint JK, Orihuela CJ. 2015. Cardiotoxicity during invasive pneumococcal disease. Am J Respir Crit Care Med 191:739–745. 10.1164/rccm.201411-1951PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brissac T, Shenoy AT, Patterson LA, Orihuela CJ. 2018. Cell invasion and pyruvate oxidase-derived H2O2 are critical for Streptococcus pneumoniae-mediated cardiomyocyte killing. Infect Immun 86:e00569-17. 10.1128/IAI.00569-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimoto AT, Rosch JW, Tuomanen EI. 2020. Pneumolysin: pathogenesis and therapeutic target. Front Microbiol 11:1543. 10.3389/fmicb.2020.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubins JB, Duane PG, Charboneau D, Janoff EN. 1992. Toxicity of pneumolysin to pulmonary endothelial cells in vitro. Infect Immun 60:1740–1746. 10.1128/iai.60.5.1740-1746.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rubins JB, Duane PG, Clawson D, Charboneau D, Young J, Niewoehner DE. 1993. Toxicity of pneumolysin to pulmonary alveolar epithelial cells. Infect Immun 61:1352–1358. 10.1128/iai.61.4.1352-1358.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Price KE, Greene NG, Camilli A. 2012. Export requirements of pneumolysin in Streptococcus pneumoniae. J Bacteriol 194:3651–3660. 10.1128/JB.00114-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shak JR, Ludewick HP, Howery KE, Sakai F, Yi H, Harvey RM, Paton JC, Klugman KP, Vidal JE. 2013. Novel role for the Streptococcus pneumoniae toxin pneumolysin in the assembly of biofilms. mBio 4:e00655-13. 10.1128/mBio.00655-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bryant JC, Dabbs RC, Oswalt KL, Brown LR, Rosch JW, Seo KS, Donaldson JR, McDaniel LS, Thornton JA. 2016. Pyruvate oxidase of Streptococcus pneumoniae contributes to pneumolysin release. BMC Microbiol 16:271. 10.1186/s12866-016-0881-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacques LC, Panagiotou S, Baltazar M, Senghore M, Khandaker S, Xu R, Bricio-Moreno L, Yang M, Dowson CG, Everett DB, Neill DR, Kadioglu A. 2020. Increased pathogenicity of pneumococcal serotype 1 is driven by rapid autolysis and release of pneumolysin. Nat Commun 11:1892. 10.1038/s41467-020-15751-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDevitt E, Khan F, Scasny A, Thompson CD, Eichenbaum Z, McDaniel LS, Vidal JE. 2020. Hydrogen peroxide production by Streptococcus pneumoniae results in alpha-hemolysis by oxidation of Oxyhemoglobin to Methemoglobin. mSphere 5:e01117-20. 10.1128/mSphere.01117-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aprianto R, Slager J, Holsappel S, Veening JW. 2018. High-resolution analysis of the pneumococcal transcriptome under a wide range of infection-relevant conditions. Nucleic Acids Res 46:9990–10006. 10.1093/nar/gky750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kakar S, Hoffman FG, Storz JF, Fabian M, Hargrove MS. 2010. Structure and reactivity of hexacoordinate hemoglobins. Biophys Chem 152:1–14. 10.1016/j.bpc.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perutz MF. 1990. Mechanisms regulating the reactions of human hemoglobin with oxygen and carbon monoxide. Annu Rev Physiol 52:1–25. 10.1146/annurev.ph.52.030190.000245. [DOI] [PubMed] [Google Scholar]
- 27.Ludlow JT, Wilkerson RG, Sahu KK, Nappe TM. 2020. Methemoglobinemia. StatPearls, Treasure Island, FL. [PubMed] [Google Scholar]
- 28.Balla J, Jacob HS, Balla G, Nath K, Eaton JW, Vercellotti GM. 1993. Endothelial-cell heme uptake from heme proteins: induction of sensitization and desensitization to oxidant damage. Proc Natl Acad Sci USA 90:9285–9289. 10.1073/pnas.90.20.9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Posta N, Csősz É, Oros M, Pethő D, Potor L, Kalló G, Hendrik Z, Sikura KÉ, Méhes G, Tóth C, Posta J, Balla G, Balla J. 2020. Hemoglobin oxidation generates globin-derived peptides in atherosclerotic lesions and intraventricular hemorrhage of the brain, provoking endothelial dysfunction. Lab Invest 100:986–1002. 10.1038/s41374-020-0403-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato GJ, Steinberg MH, Gladwin MT. 2017. Intravascular hemolysis and the pathophysiology of sickle cell disease. J Clin Invest 127:750–760. 10.1172/JCI89741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shaver CM, Upchurch CP, Janz DR, Grove BS, Putz ND, Wickersham NE, Dikalov SI, Ware LB, Bastarache JA. 2016. Cell-free hemoglobin: a novel mediator of acute lung injury. Am J Physiol Lung Cell Mol Physiol 310:L532–L541. 10.1152/ajplung.00155.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almeida CB, Souza LE, Leonardo FC, Costa FT, Werneck CC, Covas DT, Costa FF, Conran N. 2015. Acute hemolytic vascular inflammatory processes are prevented by nitric oxide replacement or a single dose of hydroxyurea. Blood 126:711–720. 10.1182/blood-2014-12-616250. [DOI] [PubMed] [Google Scholar]
- 33.Rice RH, Lee YM, Brown WD. 1983. Interactions of heme proteins with hydrogen peroxide: protein crosslinking and covalent binding of benzo[a]pyrene and 17 beta-estradiol. Arch Biochem Biophys 221:417–427. 10.1016/0003-9861(83)90160-1. [DOI] [PubMed] [Google Scholar]
- 34.Nagababu E, Rifkind JM. 1998. Formation of fluorescent heme degradation products during the oxidation of hemoglobin by hydrogen peroxide. Biochem Biophys Res Commun 247:592–596. 10.1006/bbrc.1998.8846. [DOI] [PubMed] [Google Scholar]
- 35.Maitra D, Byun J, Andreana PR, Abdulhamid I, Diamond MP, Saed GM, Pennathur S, Abu-Soud HM. 2011. Reaction of hemoglobin with HOCl: mechanism of heme destruction and free iron release. Free Radic Biol Med 51:374–386. 10.1016/j.freeradbiomed.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kassa T, Jana S, Strader MB, Meng F, Jia Y, Wilson MT, Alayash AI. 2015. Sickle cell hemoglobin in the ferryl state promotes betaCys-93 oxidation and mitochondrial dysfunction in epithelial lung cells (E10). J Biol Chem 290:27939–27958. 10.1074/jbc.M115.651257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nader E, Romana M, Connes P. 2020. The red blood cell-inflammation vicious circle in sickle cell disease. Front Immunol 11:454. 10.3389/fimmu.2020.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buehler PW, Humar R, Schaer DJ. 2020. Haptoglobin therapeutics and compartmentalization of cell-free hemoglobin toxicity. Trends Mol Med 26:683–697. 10.1016/j.molmed.2020.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Pericone CD, Overweg K, Hermans PW, Weiser JN. 2000. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun 68:3990–3997. 10.1128/IAI.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu X, Gordon O, Jiang W, Antezana BS, Angulo-Zamudio UA, Del Rio C, Moller A, Brissac T, Tierney ARP, Warncke K, Orihuela CJ, Read TD, Vidal JE. 2019. Interaction between Streptococcus pneumoniae and Staphylococcus aureus generates .OH radicals that rapidly kill Staphylococcus aureus strains. J Bacteriol 201:e00474-19. 10.1128/JB.00474-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lisher JP, Tsui HT, Ramos-Montanez S, Hentchel KL, Martin JE, Trinidad JC, Winkler ME, Giedroc DP. 2017. Biological and chemical adaptation to endogenous hydrogen peroxide production in Streptococcus pneumoniae D39. mSphere 2:e00291-16. 10.1128/mSphere.00291-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taniai H, Iida K, Seki M, Saito M, Shiota S, Nakayama H, Yoshida S. 2008. Concerted action of lactate oxidase and pyruvate oxidase in aerobic growth of Streptococcus pneumoniae: role of lactate as an energy source. J Bacteriol 190:3572–3579. 10.1128/JB.01882-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pericone CD, Park S, Imlay JA, Weiser JN. 2003. Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction. J Bacteriol 185:6815–6825. 10.1128/JB.185.23.6815-6825.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramos-Montanez S, Kazmierczak KM, Hentchel KL, Winkler ME. 2010. Instability of ackA (acetate kinase) mutations and their effects on acetyl phosphate and ATP amounts in Streptococcus pneumoniae D39. J Bacteriol 192:6390–6400. 10.1128/JB.00995-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Echlin H, Frank M, Rock C, Rosch JW. 2020. Role of the pyruvate metabolic network on carbohydrate metabolism and virulence in Streptococcus pneumoniae. Mol Microbiol 10.1111/mmi.14557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Echlin H, Frank MW, Iverson A, Chang TC, Johnson MD, Rock CO, Rosch JW. 2016. Pyruvate oxidase as a critical link between metabolism and capsule biosynthesis in Streptococcus pneumoniae. PLoS Pathog 12:e1005951. 10.1371/journal.ppat.1005951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramos-Montanez S, Tsui HC, Wayne KJ, Morris JL, Peters LE, Zhang F, Kazmierczak KM, Sham LT, Winkler ME. 2008. Polymorphism and regulation of the spxB (pyruvate oxidase) virulence factor gene by a CBS-HotDog domain protein (SpxR) in serotype 2 Streptococcus pneumoniae. Mol Microbiol 67:729–746. 10.1111/j.1365-2958.2007.06082.x. [DOI] [PubMed] [Google Scholar]
- 48.Rai P, Parrish M, Tay IJ, Li N, Ackerman S, He F, Kwang J, Chow VT, Engelward BP. 2015. Streptococcus pneumoniae secretes hydrogen peroxide leading to DNA damage and apoptosis in lung cells. Proc Natl Acad Sci USA 112:E3421–E3430. 10.1073/pnas.1424144112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown JL, Yates EA, Bielecki M, Olczak T, Smalley JW. 2018. Potential role for Streptococcus gordonii-derived hydrogen peroxide in heme acquisition by Porphyromonas gingivalis. Mol Oral Microbiol 33:322–335. 10.1111/omi.12229. [DOI] [PubMed] [Google Scholar]
- 50.van Beers EJ, Samsel L, Mendelsohn L, Saiyed R, Fertrin KY, Brantner CA, Daniels MP, Nichols J, McCoy JP, Kato GJ. 2014. Imaging flow cytometry for automated detection of hypoxia-induced erythrocyte shape change in sickle cell disease. Am J Hematol 89:598–603. 10.1002/ajh.23699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guiral S, Henard V, Laaberki MH, Granadel C, Prudhomme M, Martin B, Claverys JP. 2006. Construction and evaluation of a chromosomal expression platform (CEP) for ectopic, maltose-driven gene expression in Streptococcus pneumoniae. Microbiology (Reading) 152:343–349. 10.1099/mic.0.28433-0. [DOI] [PubMed] [Google Scholar]
- 52.Kjos M, Aprianto R, Fernandes VE, Andrew PW, van Strijp JA, Nijland R, Veening JW. 2015. Bright fluorescent Streptococcus pneumoniae for live-cell imaging of host-pathogen interactions. J Bacteriol 197:807–818. 10.1128/JB.02221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gallay C, Sanselicio S, Anderson ME, Soh YM, Liu X, Stamsas GA, Pelliciari S, van Raaphorst R, Denereaz J, Kjos M, Murray H, Gruber S, Grossman AD, Veening JW. 2021. CcrZ is a pneumococcal spatiotemporal cell cycle regulator that interacts with FtsZ and controls DNA replication by modulating the activity of DnaA. Nat Microbiol 6:1175–1187. 10.1038/s41564-021-00949-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mercy C, Ducret A, Slager J, Lavergne JP, Freton C, Nagarajan SN, Garcia PS, Noirot-Gros MF, Dubarry N, Nourikyan J, Veening JW, Grangeasse C. 2019. RocS drives chromosome segregation and nucleoid protection in Streptococcus pneumoniae. Nat Microbiol 4:1661–1670. 10.1038/s41564-019-0472-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saito M, Seki M, Iida K, Nakayama H, Yoshida S. 2007. A novel agar medium to detect hydrogen peroxide-producing bacteria based on the prussian blue-forming reaction. Microbiol Immunol 51:889–892. 10.1111/j.1348-0421.2007.tb03971.x. [DOI] [PubMed] [Google Scholar]
- 56.Nagababu E, Rifkind JM. 2004. Heme degradation by reactive oxygen species. Antioxid Redox Signal 6:967–978. 10.1089/ars.2004.6.967. [DOI] [PubMed] [Google Scholar]
- 57.Pamplona A, Ferreira A, Balla J, Jeney V, Balla G, Epiphanio S, Chora A, Rodrigues CD, Gregoire IP, Cunha-Rodrigues M, Portugal S, Soares MP, Mota MM. 2007. Heme oxygenase-1 and carbon monoxide suppress the pathogenesis of experimental cerebral malaria. Nat Med 13:703–710. 10.1038/nm1586. [DOI] [PubMed] [Google Scholar]
- 58.Mingone CJ, Gupte SA, Chow JL, Ahmad M, Abraham NG, Wolin MS. 2006. Protoporphyrin IX generation from delta-aminolevulinic acid elicits pulmonary artery relaxation and soluble guanylate cyclase activation. Am J Physiol Lung Cell Mol Physiol 291:L337–L344. 10.1152/ajplung.00482.2005. [DOI] [PubMed] [Google Scholar]
- 59.Keller LE, Robinson DA, McDaniel LS. 2016. Nonencapsulated Streptococcus pneumoniae: emergence and pathogenesis. mBio 7:e01792-15. 10.1128/mBio.01792-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Facklam R. 2002. What happened to the streptococci: overview of taxonomic and nomenclature changes. Clin Microbiol Rev 15:613–630. 10.1128/CMR.15.4.613-630.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kajfasz JK, Rivera-Ramos I, Abranches J, Martinez AR, Rosalen PL, Derr AM, Quivey RG, Lemos JA. 2010. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J Bacteriol 192:2546–2556. 10.1128/JB.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Hoeven JS, Hoogendoorn H. 1990. Uptake of oxygen, release and degradation of hydrogen peroxide by Streptococcus mutans NCTC 10449. Antonie Van Leeuwenhoek 57:91–95. 10.1007/BF00403160. [DOI] [PubMed] [Google Scholar]
- 63.Weiser JN, Ferreira DM, Paton JC. 2018. Streptococcus pneumoniae: transmission, colonization and invasion. Nat Rev Microbiol 16:355–367. 10.1038/s41579-018-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giulivi C, Hochstein P, Davies KJ. 1994. Hydrogen peroxide production by red blood cells. Free Radic Biol Med 16:123–129. 10.1016/0891-5849(94)90249-6. [DOI] [PubMed] [Google Scholar]
- 65.Nagababu E, Rifkind JM. 2000. Heme degradation during autoxidation of oxyhemoglobin. Biochem Biophys Res Commun 273:839–845. 10.1006/bbrc.2000.3025. [DOI] [PubMed] [Google Scholar]
- 66.De Jesus-Bonilla W, Ramirez-Melendez E, Cerda J, Lopez-Garriga J. 2002. Evidence for nonhydrogen bonded compound II in cyclic reaction of hemoglobin I from Lucina pectinata with hydrogen peroxide. Biopolymers 67:178–185. 10.1002/bip.10082. [DOI] [PubMed] [Google Scholar]
- 67.Mao GD, Thomas PD, Poznansky MJ. 1994. Oxidation of spin trap 5,5-dimethyl-1-pyrroline-1-oxide in an electron paramagnetic resonance study of the reaction of methemoglobin with hydrogen peroxide. Free Radic Biol Med 16:493–500. 10.1016/0891-5849(94)90127-9. [DOI] [PubMed] [Google Scholar]
- 68.Iuliano L, Violi F, Pedersen JZ, Pratico D, Rotilio G, Balsano F. 1992. Free radical-mediated platelet activation by hemoglobin released from red blood cells. Arch Biochem Biophys 299:220–224. 10.1016/0003-9861(92)90267-z. [DOI] [PubMed] [Google Scholar]
- 69.Khan F, Wu X, Matzkin GL, Khan MA, Sakai F, Vidal JE. 2016. Streptococcus pneumoniae eradicates preformed Staphylococcus aureus biofilms through a mechanism requiring physical contact. Front Cell Infect Microbiol 6:104. 10.3389/fcimb.2016.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Regev-Yochay G, Trzcinski K, Thompson CM, Malley R, Lipsitch M. 2006. Interference between Streptococcus pneumoniae and Staphylococcus aureus: in vitro hydrogen peroxide-mediated killing by Streptococcus pneumoniae. J Bacteriol 188:4996–5001. 10.1128/JB.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Assad Z, Michel M, Valtuille Z, Lazzati A, Boizeau P, Madhi F, Gaschignard J, Pham LL, Caseris M, Cohen R, Kaguelidou F, Varon E, Alberti C, Faye A, Angoulvant F, Koehl B, Ouldali N. 2022. Incidence of acute chest syndrome in children with sickle cell disease following implementation of the 13-valent pneumococcal conjugate vaccine in France. JAMA Netw Open 5:e2225141. 10.1001/jamanetworkopen.2022.25141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Akhter F, Womack E, Vidal JE, Le Breton Y, McIver KS, Pawar S, Eichenbaum Z. 2021. Hemoglobin induces early and robust biofilm development in Streptococcus pneumoniae by a pathway that involves comC but not the cognate comDE two-component system. Infect Immun 89:e00779-20. 10.1128/IAI.00779-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Akhter F, Womack E, Vidal JE, Le Breton Y, McIver KS, Pawar S, Eichenbaum Z. 2020. Hemoglobin stimulates vigorous growth of Streptococcus pneumoniae and shapes the pathogen’s global transcriptome. Sci Rep 10:15202. 10.1038/s41598-020-71910-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gotts JE, Bernard O, Chun L, Croze RH, Ross JT, Nesseler N, Wu X, Abbott J, Fang X, Calfee CS, Matthay MA. 2019. Clinically relevant model of pneumococcal pneumonia, ARDS, and nonpulmonary organ dysfunction in mice. Am J Physiol Lung Cell Mol Physiol 317:L717–L736. 10.1152/ajplung.00132.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Spellerberg B, Cundell DR, Sandros J, Pearce BJ, Idanpaan-Heikkila I, Rosenow C, Masure HR. 1996. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol Microbiol 19:803–813. 10.1046/j.1365-2958.1996.425954.x. [DOI] [PubMed] [Google Scholar]
- 76.Smalley JW, Olczak T. 2017. Heme acquisition mechanisms of Porphyromonas gingivalis - strategies used in a polymicrobial community in a heme-limited host environment. Mol Oral Microbiol 32:1–23. 10.1111/omi.12149. [DOI] [PubMed] [Google Scholar]
- 77.O’Brien KL, Bronsdon MA, Dagan R, Yagupsky P, Janco J, Elliott J, Whitney CG, Yang YH, Robinson LG, Schwartz B, Carlone GM. 2001. Evaluation of a medium (STGG) for transport and optimal recovery of Streptococcus pneumoniae from nasopharyngeal secretions collected during field studies. J Clin Microbiol 39:1021–1024. 10.1128/JCM.39.3.1021-1024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 79.Vidal JE, Ludewick HP, Kunkel RM, Zahner D, Klugman KP. 2011. The LuxS-dependent quorum-sensing system regulates early biofilm formation by Streptococcus pneumoniae strain D39. Infect Immun 79:4050–4060. 10.1128/IAI.05186-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raimunda D, Padilla-Benavides T, Vogt S, Boutigny S, Tomkinson KN, Finney LA, Arguello JM. 2013. Periplasmic response upon disruption of transmembrane Cu transport in Pseudomonas aeruginosa. Metallomics 5:144–151. 10.1039/c2mt20191g. [DOI] [PubMed] [Google Scholar]
- 81.Tavera-Montanez C, Hainer SJ, Cangussu D, Gordon SJV, Xiao Y, Reyes-Gutierrez P, Imbalzano AN, Navea JG, Fazzio TG, Padilla-Benavides T. 2019. The classic metal-sensing transcription factor MTF1 promotes myogenesis in response to copper. FASEB J 33:14556–14574. 10.1096/fj.201901606R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gordon SJV, Xiao Y, Paskavitz AL, Navarro-Tito N, Navea JG, Padilla-Benavides T. 2019. Atomic absorbance spectroscopy to measure intracellular zinc pools in mammalian cells. J Vis Exp 2019:59519. 10.3791/59519. [DOI] [PubMed] [Google Scholar]
- 83.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 84.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery’s virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keller LE, Luo X, Thornton JA, Seo KS, Moon BY, Robinson DA, McDaniel LS. 2015. Immunization with pneumococcal surface protein K of nonencapsulated Streptococcus pneumoniae provides protection in a mouse model of colonization. Clin Vaccine Immunol 22:1146–1153. 10.1128/CVI.00456-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Junges R, Maienschein-Cline M, Morrison DA, Petersen FC. 2019. Complete genome sequence of Streptococcus pneumoniae serotype 19F strain EF3030. Microbiol Resour Announc 8:e00198-19. 10.1128/MRA.00198-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Keller LE, Thomas JC, Luo X, Nahm MH, McDaniel LS, Robinson DA. 2013. Draft genome sequences of five multilocus sequence types of nonencapsulated Streptococcus pneumoniae. Genome Announc 1:e00520-13. 10.1128/genomeA.00520-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ostria-Hernandez ML, Juarez-de la Rosa KC, Arzate-Barbosa P, Lara-Hernandez A, Sakai F, Ibarra JA, Castro-Escarpulli G, Vidal JE. 2018. Nosocomial, multidrug-resistant Klebsiella pneumoniae strains isolated from Mexico City produce robust biofilms on abiotic surfaces but not on human lung cells. Microb Drug Resist 24:422–433. 10.1089/mdr.2017.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boake WC. 1956. Antistaphylocoagulase in experimental staphylococcal infections. J Immunol 76:89–96. [PubMed] [Google Scholar]