Abstract
Background
In recent years, the use of deep learning has become more commonplace in the biomedical field and its development will greatly assist clinical and imaging data interpretation. Most existing machine learning methods for coronary angiography analysis are limited to a single aspect.
Aims
We aimed to achieve an automatic and multimodal analysis to recognise and quantify coronary angiography, integrating multiple aspects, including the identification of coronary artery segments and the recognition of lesion morphology.
Methods
A data set of 20,612 angiograms was retrospectively collected, among which 13,373 angiograms were labelled with coronary artery segments, and 7,239 were labelled with special lesion morphology. Trained and optimised by these labelled data, one network recognised 20 different segments of coronary arteries, while the other detected lesion morphology, including measures of lesion diameter stenosis as well as calcification, thrombosis, total occlusion, and dissection detections in an input angiogram.
Results
For segment prediction, the recognition accuracy was 98.4%, and the recognition sensitivity was 85.2%. For detecting lesion morphologies including stenotic lesion, total occlusion, calcification, thrombosis, and dissection, the F1 scores were 0.829, 0.810, 0.802, 0.823, and 0.854, respectively. Only two seconds were needed for the automatic recognition.
Conclusions
Our deep learning architecture automatically provides a coronary diagnostic map by integrating multiple aspects. This helps cardiologists to flag and diagnose lesion severity and morphology during the intervention.
Introduction
Coronary artery disease (CAD) is the most common cardiovascular disease1, and the leading cause of death globally during the past two decades2. Therefore, the diagnosis and prevention of CAD is crucial for modern society. Coronary angiography (CAG), which provides assessments of luminal stenosis, plaque characteristics, and disease activity, is an important tool for CAD diagnosis and treatment guidance3,4. In recent years, the use of deep learning has become more commonplace in the biomedical field and its development will greatly assist clinical and imaging data interpretation5. Deep learning can simplify the procedure by directly learning predictive features, thereby strongly supporting the translation from artificial algorithms into clinical application6,7,8. However, much of the previous work to apply deep learning algorithms in the field of CAD has focused on single aspects of the analysis of the coronary artery, such as vessel segmentation9,10, coronary artery centreline extraction11, noise reduction12, coronary artery geometry synthesis13, coronary plaque characterisation14, and calcification detection. Thus, there remains a large gap between the results produced by the aforementioned algorithms and the actual diagnosis of CAD.
High diagnostic accuracy from a coronary angiogram requires correct recognition of lesion morphology and location. Herein, to tackle the above recognition tasks, two unique functional deep neural networks (DNN) were proposed, on which we created, trained, validated, and then tested a coronary angiography recognition system called DeepDiscern. DeepDiscern was evaluated on a test data set of consecutive angiograms collected from clinical cases.
Methods
STUDY POPULATION AND IMAGE ACQUISITION
To develop the DeepDiscern system, 20,612 angiograms from 10,073 patients were consecutively collected using the image acquisition data from a large single centre (Fu Wai Hospital, National Center for Cardiovascular Diseases, Beijing, China). For coronary segmentation DNN training, 13,373 angiograms were consecutively collected from 2,834 patients who underwent CAGs in July 2018. The remaining 7,239 angiograms, with at least one identifiable lesion morphology, such as stenotic lesion, total occlusion (TO), calcification, thrombus, and dissection, collected from 7,239 patients, were used for lesion morphology recognition. The collected angiogram information is listed in Table 1.
Table 1. Baseline patient and lesion characteristics.
| Coronary artery recognition | |
| Patients | (N=2,834) |
| Age, years | 61.6±17.5 |
| Female | 29.9% (848) |
| Left dominance | 24.1% (683) |
| Segmentation | (N=13,373)* |
| LM | 77.7% (10,390) |
| LAD | 64.2% (8,594) |
| DIA | 51.3% (6,854) |
| LCX | 36.6% (4,890) |
| OM | 36.6% (4,890) |
| L-PLA | 36.6% (4,890) |
| L-PDA | 36.6% (4,890) |
| RCA | 22.3% (2,983) |
| PDA | 22.3% (2,983) |
| PLA | 22.3% (2,983) |
| Lesion morphology detection | |
| Patients | (N=7,239) |
| Age, years | 65.5±16.0 |
| Female | 20.9% (1,513) |
| Left dominance | 22.8% (1,650) |
| Lesion morphology | (N=12,184)† |
| Stenosis (DS ≥50%) | 22.2% (2,700) |
| Total occlusion | 36.6% (4,458) |
| Lesion bending >45° | 44.2% (1,970) |
| Lesion length | 17.6±13.2 |
| ≥20 mm | 24.2% (1,079) |
| Blunt stump | 37.2% (1,658) |
| Moderate or heavy calcification | 19.5% (2,378) |
| Thrombus | 11.8% (1,439) |
| Dissection | 10.0% (1,209) |
| * The number of angiograms in the coronary artery recognition task. † The number of lesion samples in the lesion morphology detection task. DIA: diagonal; LAD: left anterior descending artery; LCX: left circumflex artery; LM: left main; L-PDA: left posterior descending; L-PLA: left posterolateral; OM: obtuse marginal; PDA: posterior descending; PLA: posterolateral; RCA: right coronary artery | |
Raw angiographic data for our work were acquired during interventional procedures of patients and saved in 512×512-pixel digital imaging and communications in medicine (DICOM) format with angiographic views and video information, without patient identifiers. Each patient’s angiographic DICOM included several angiographic sequences encompassing different angiographic views. The choice of angiographic views was left to the operator’s discretion, to delineate best the lesion severity and morphology. In general, the angiographic views for the left coronary artery included CRA (cranial view), CAU (caudal view), LAO_CRA (left anterior oblique-cranial view), LAO_CAU (left anterior oblique-caudal view), RAO_CRA (right anterior oblique-cranial view), and RAO_CAU (right anterior oblique-caudal view). For the right coronary artery the views included LAO, LAO_CAU, LAO_CRA and RAO. The data flow is presented in Figure 1.
Figure 1.
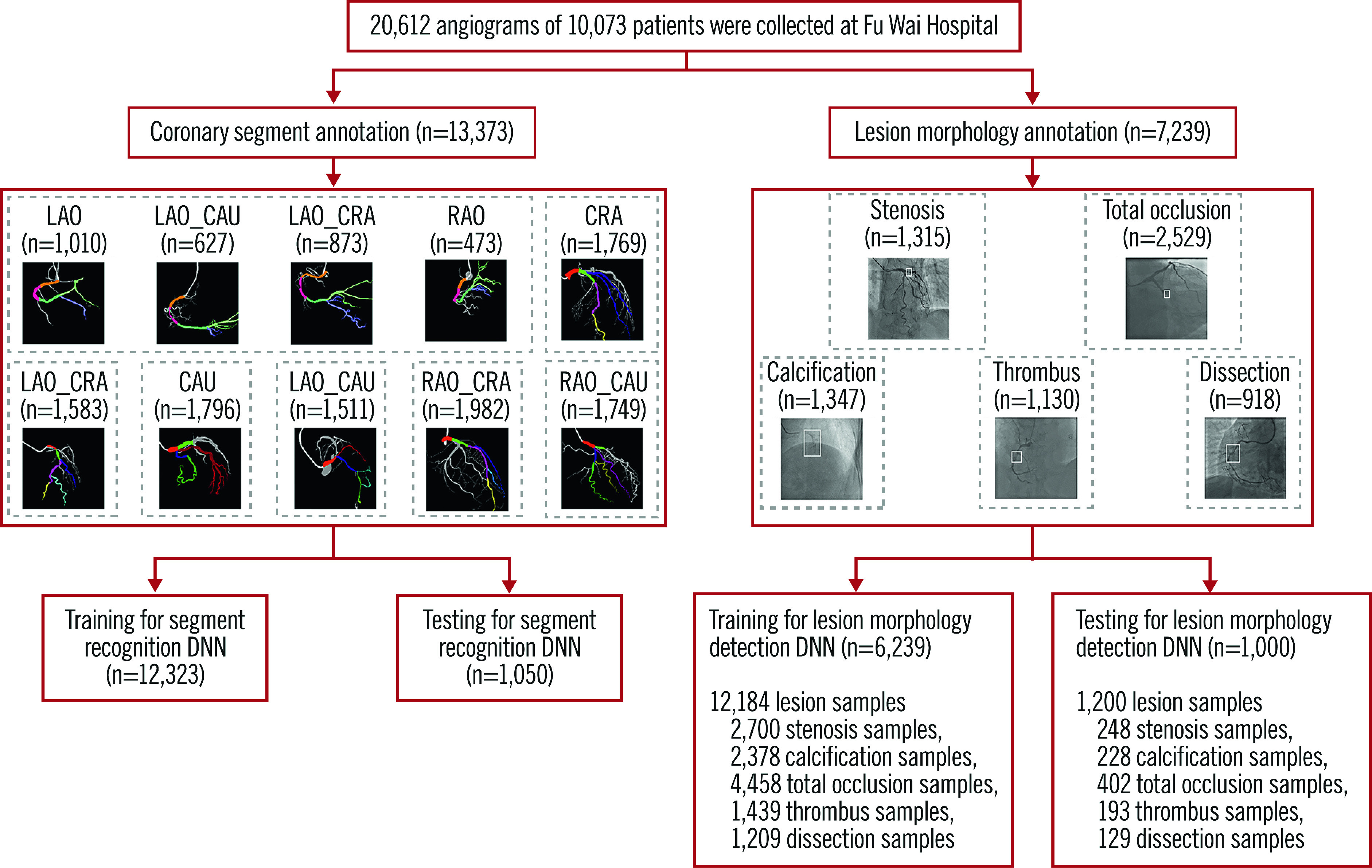
Data flow for the lesion morphology detection task and the coronary segment recognition task. Coronary segment recognition. In total, 13,373 angiograms were used, and divided into seven parts to train and test DeepDiscern DNN. Lesion morphology detection. In total, 7,239 angiograms with 1 to 3 lesion morphology were labelled for model training and testing. There were 12,184 lesion samples of five kinds of lesion morphology. CAU: caudal view; CRA: cranial view; LAO: left anterior oblique view; LAO_CAU: left anterior oblique-caudal view; LAO_CRA: left anterior oblique-cranial view: RAO: right anterior oblique view; RAO_CAU: right anterior oblique-caudal view; RAO_CRA: right anterior oblique-cranial view
REFERENCE STANDARD AND ANNOTATION PROCEDURES
DeepDiscern learns rules from the labelled images in the training phase. To this end, all angiograms collected over a period of 11 months for the training and testing data sets were reviewed by ten qualified analysts in the angiographic core lab at Fu Wai Hospital. Coronary segments were annotated based on pre-established diagnostic criteria and lesion morphology characterised.
For the coronary segment recognition data sets, each angiogram was labelled at a pixel-by-pixel level for coronary segmentation recognition. First, analysts annotated sketch labels of all coronary artery segments on the original angiograms, with different colours representing different arterial segments. Then a group of trained and certified technicians labelled fine ground-truth images pixel by pixel according to the sketch labels. Supplementary Figure 1 illustrates this process.
A total of 20 coronary artery segments were annotated (Supplementary Figure 2), including proximal right coronary artery (RCA prox), RCA mid, RCA distal, right posterior descending (PDA), right posterolateral, left main (LM), proximal left anterior descending (LAD prox), LAD mid, LAD distal, 1st diagonal, add. 1st diagonal, 2nd diagonal, add. 2nd diagonal, proximal circumflex (LCX prox), LCX distal, 1st obtuse marginal (OM), 2nd OM, left posterolateral, left posterior descending (L-PDA) and intermediate.
Although accurate coronary diagnosis requires coronary injections in multiple views to ensure that all coronary segments are seen clearly without foreshortening or overlap, it is not necessary to include all potential views in a given coronary segment. In clinical practice, several dominant projections are typically used to visualise a coronary segment and its morphology. Therefore, during the training and testing process of each coronary segment, DeepDiscern focused mainly on dominant projections provided. The corresponding relationship between the observed coronary segment and the angiographic views is shown in Supplementary Table 1.
For the lesion morphology detection data sets, expert analysts marked all lesion morphologies identified on the angiogram, including stenotic lesion, TO, calcification, thrombosis, and dissection. Stenotic lesion was defined as ≥50% diameter stenosis. TO was defined as angiographic evidence of TOs with Thrombolysis In Myocardial Infarction (TIMI) flow grade 0. Calcification was defined as readily apparent radiopacities noted within the apparent vascular wall (moderate: densities noted only with cardiac motion before contrast injection; severe: radiopacities noted without cardiac motion before contrast injection). Thrombus was defined as a discrete, intraluminal filling defect with defined borders and largely separated from the adjacent wall with or without contrast staining15. Dissection grade was diagnosed based on the National Heart, Lung and Blood Institute (NHLBI) coronary dissection criteria (Supplementary Table 2). The lesion type, location, and extent were labelled using a rectangular box. Supplementary Figure 1 illustrates this process. In total, 7,239 angiograms with one to three lesion morphologies were labelled for model training and testing. There were 12,184 positive samples in these angiograms. The lesion morphology classification data are shown in Table 1.
DEEP LEARNING MODEL
DeepDiscern was designed to use these two DNN to recognise coronary segments and detect lesion morphology (Figure 2). Features of the input angiogram carrying different semantic information were extracted including low-level features, such as vessel edges and background texture, and high-level features, such as the overall shape of the arteries.
Figure 2.
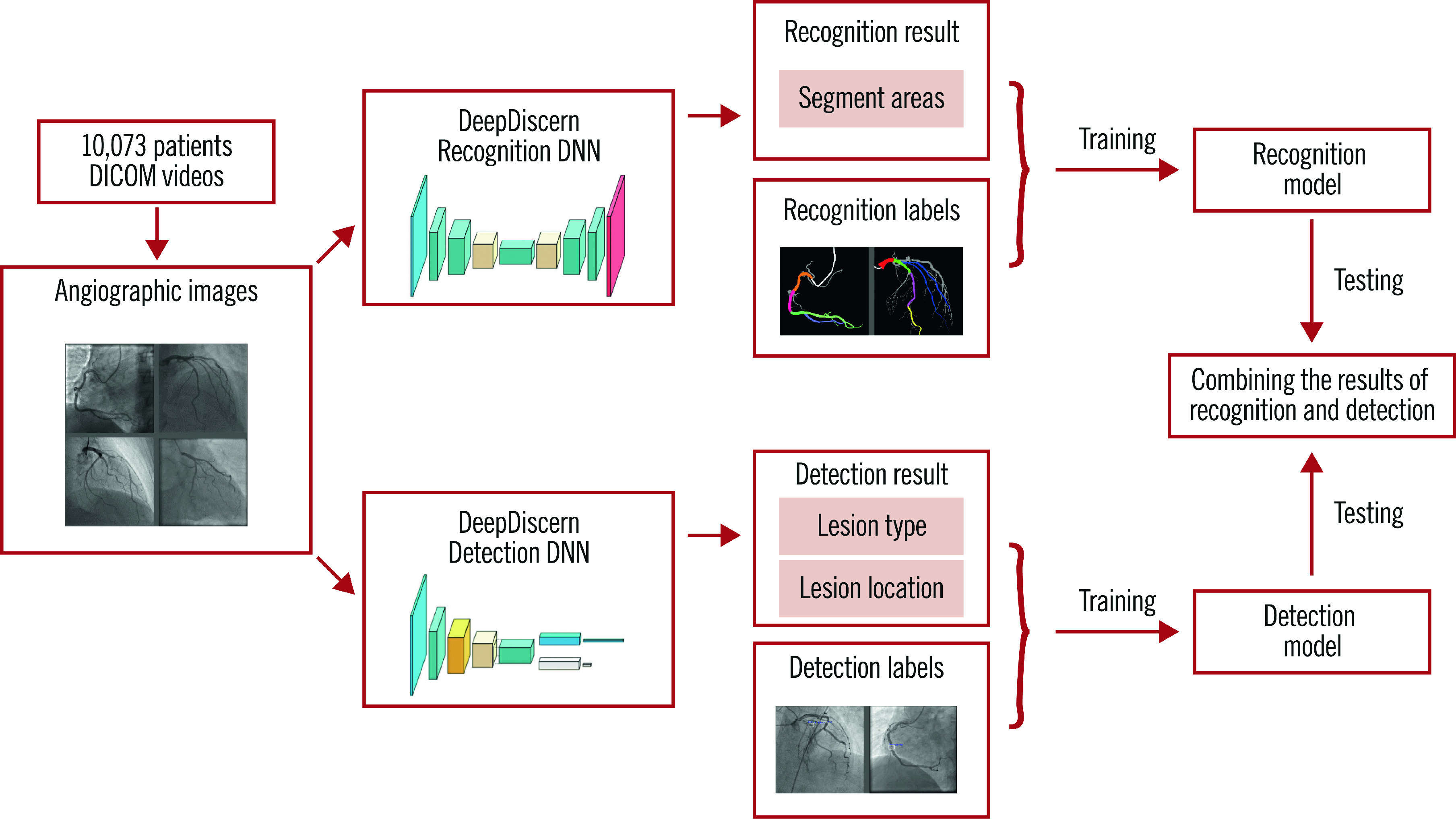
The workflow of DeepDiscern. In total, 20,612 angiographic images were collected from DICOM videos of 10,073 patients. Under the supervision of the labelled images, we trained the lesion morphology detection model and coronary artery recognition model of DeepDiscern. After training, the detection and recognition models generate the result images. These two results were combined to generate a high-level diagnosis.
For the coronary artery recognition task, we modified a special DNN - conditional generative adversarial network (cGAN) (Supplementary Figure 3) for image segmentation. For the lesion morphology detection task, we developed a convolutional DNN (Supplementary Figure 4), which outputs the location of all the lesion morphologies that appeared in the input angiogram. The network structure, implementation details, training process and testing process of segment recognition DNN are detailed in Supplementary Appendix 1, and the lesion detection DNN in Supplementary Appendix 2.
For each input angiogram, DeepDiscern combines the two output results from coronary artery recognition DNN and lesion morphology detection DNN to generate high-level diagnostic information, including identification of every coronary artery lesion and the coronary artery segment in which it is located.
MODEL EVALUATION AND STATISTICAL ANALYSIS
For arterial segment recognition, given an input angiogram, the DeepDiscern segment recognition DNN produced an output image with several identified areas that represent the different coronary segments (Supplementary Table 3). For each coronary segment, we calculated the predicted pixel number for true positive (TP), true negative (TN), false positive (FP) and false negative (FN). Based on these results, we evaluated the segment recognition model by several metrics including accuracy ([TP+TN]/[TP+TN+FP+FN]), sensitivity (TP/[TP+FN]), specificity (TN/[TN+FP]), positive predictive value (TP/[TP+FP]), and negative predictive value (TN/[TN+FN]). The recognition model was evaluated using 1,050 images including all the coronary segments.
In terms of lesion morphology detection, the DeepDiscern lesion detection DNN predicts several rectangular areas containing the lesions to describe their location and type. For the algorithmic analysis, lesion morphology is detected correctly if the overlap rate of a predicted rectangle and the ground-truth rectangle (labelled by cardiologists) exceeds a threshold λ_d= 0.5. We measured the performance of the lesion detection model using precision rate P, recall rate R, and F1 score. The precision rate is defined as the percentage of correctly detected lesion cases from all lesion cases detected by the models. The recall rate is defined as the percentage of correctly detected lesions from all ground-truth lesions labelled by cardiologists. The F1 score [F_1= 2×P×R⁄(P+R)], which combines the accuracy rate and recall rate, is a better measure of the overall performance of the detection DNN model. The evaluation process of the recognition model and of the detection model is illustrated in Supplementary Figure 5.
Results
Coronary segment recognition DNN was evaluated using 1,050 images that included all the coronary segments. For segment prediction, the average accuracy, sensitivity, specificity, positive predictive value, and negative predictive value of all coronary artery segments was 98.4%, 85.2%, 99.1%, 76.2%, and 99.5%, respectively. Higher accuracy and sensitivity rates were observed in the proximal segments of major epicardial vessels (99.9% and 91.8% for LM, 99.8% and 92.6% for LAD proximal, 99.8% and 87.9% for LCX proximal, and 99.8% and 87.9% for RCA proximal). Arterial segments that were identified incorrectly were mostly in the distal segments and side branches of the major epicardial vessels. The performance of coronary segment recognition DNN was improved as the amount of data increased (Supplementary Figure 6). Because the majority of pixels in the image were negative, DNN performance cannot be assessed only from specificity and negative predictive value. Table 2 provides detailed results including more metrics, and Figure 3A illustrates result images of the artery recognition task. The results under different angiographic views are shown in Supplementary Table 4.
Table 2. Performance of DeepDiscern segment recognition DNN for different coronary artery segments.
| Coronary artery segment | Accuracy % (95% CI) | Sensitivity % (95% CI) | Specificity % (95% CI) | PPV % (95% CI) | NPV % (95% CI) |
| All segments | 98.4 (98.3-98.4) | 85.2 (84.8-85.6) | 99.1 (99.1-99.1) | 76.2 (75.7-76.6) | 99.5 (99.5-99.5) |
| LM | 99.9 (99.9-99.9) | 91.8 (91.1-92.5) | 99.9 (99.9-99.9) | 80.7 (79.4-82.0) | 99.9 (99.9-99.9) |
| LAD proximal | 99.8 (99.8-99.8) | 92.6 (91.9-93.2) | 99.9 (99.8-99.9) | 80.9 (79.5-82.4) | 99.9 (99.9-99.9) |
| LAD mid | 99.8 (99.7-99.8) | 90.8 (90.1-91.4) | 99.8 (99.8-99.8) | 82.1 (81.0-83.2) | 99.9 (99.9-99.9) |
| LAD apical | 99.7 (99.7-99.7) | 84.5 (83.0-86.1) | 99.8 (99.8-99.8) | 67.8 (66.1-69.5) | 99.9 (99.9-99.9) |
| 1st DIA | 99.4 (99.4-99.5) | 78.1 (75.9-80.4) | 99.6 (99.6-99.6) | 60.0 (58.1-62.0) | 99.8 (99.8-99.9) |
| 2nd DIA | 99.7 (99.7-99.8) | 73.7 (68.0-79.3) | 99.8 (99.8-99.8) | 41.2 (36.5-45.9) | 99.9 (99.9-99.9) |
| LCX proximal | 99.8 (99.8-99.8) | 87.9 (86.4-89.4) | 99.9 (99.9-99.9) | 78.8 (77.1-80.5) | 99.9 (99.9-99.9) |
| LCX distal | 99.7 (99.6-99.7) | 81.3 (79.6-83.1) | 99.8 (99.8-99.8) | 78.3 (76.3-80.2) | 99.9 (99.8-99.9) |
| Intermediate | 99.6 (99.5-99.6) | 74.1 (69.8-78.4) | 99.7 (99.7-99.8) | 63.2 (58.1-68.4) | 99.9 (99.8-99.9) |
| OM | 99.7 (99.6-99.7) | 79.2 (75.9-82.5) | 99.8 (99.7-99.8) | 53.0 (48.8-57.2) | 99.9 (99.9-99.9) |
| L-PLA | 99.5 (99.5-99.5) | 80.6 (78.3-82.8) | 99.7 (99.6-99.7) | 69.1 (66.7-71.4) | 99.8 (99.8-99.9) |
| L-PDA | 99.6 (99.5-99.7) | 83.1 (79.6-86.6) | 99.7 (99.7-99.8) | 72.5 (69.1-75.9) | 99.9 (99.9-99.9) |
| RCA proximal | 99.8 (99.8-99.8) | 87.9 (87.0-88.8) | 99.9 (99.9-99.9) | 86.7 (85.9-87.5) | 99.9 (99.9-99.9) |
| RCA mid | 99.7 (99.7-99.8) | 85.6 (84.5-86.7) | 99.8 (99.8-99.9) | 76.6 (75.3-77.9) | 99.9 (99.9-99.9) |
| RCA distal | 99.8 (99.8-99.8) | 83.2 (82.0-84.4) | 99.9 (99.9-99.9) | 88.2 (87.1-89.3) | 99.9 (99.9-99.9) |
| PDA | 99.7 (99.7-99.7) | 75.4 (73.4-77.4) | 99.8 (99.8-99.9) | 70.6 (68.7-72.5) | 99.9 (99.9-99.9) |
| PLA | 99.5 (99.5-99.5) | 77.2 (75.6-78.7) | 99.7 (99.7-99.7) | 72.0 (70.3-73.7) | 99.8 (99.8-99.8) |
| CI: confidence interval; DIA: diagonal; LAD: left anterior descending artery; LCX: left circumflex artery; LM: left main; L-PDA: left posterior descending; L-PLA: left posterolateral; NPV: negative predictive value; OM: obtuse marginal; PDA: posterior descending; PLA: posterolateral; PPV: positive predictive value; RCA: right coronary artery | |||||
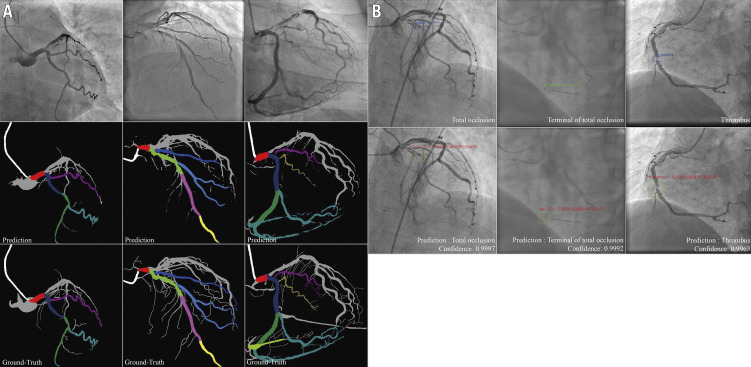
Figure 3.
Result imaging of the segment recognition model and the lesion morphology detection model. A) Segment recognition. First row: input angiograms. Second row: resulting images generated by DeepDiscern segment recognition DNN. Third row: ground-truth labelled images. Different identified areas represent the different coronary segments. B) Lesion morphology detection. First row: input angiograms and ground-truth bounding boxes. There is a TO morphology in the first and second angiograms, and a thrombus morphology in the third angiogram in this row. Second row: bounding boxes and lesion types generated by the DeepDiscern lesion morphology detection model.
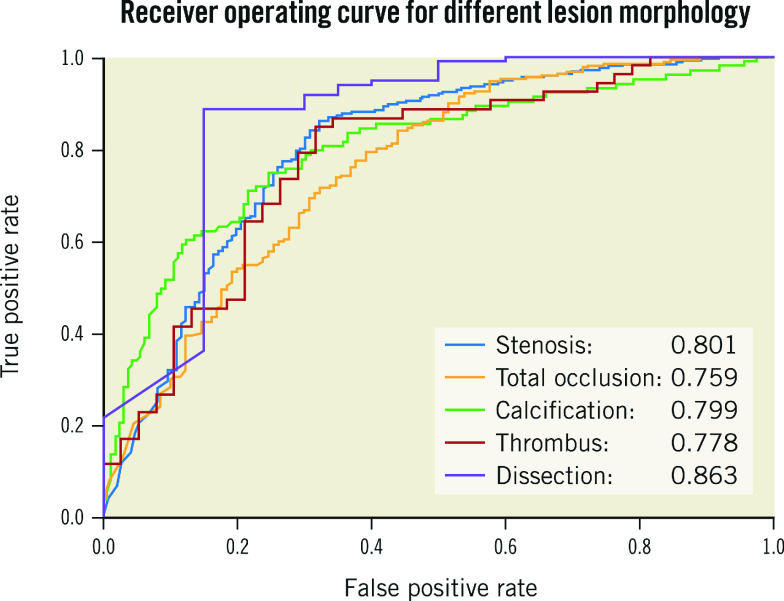
One thousand angiograms were used to test the lesion morphology detection DNN model. The test data set included 1,200 (248 stenotic, 228 calcification, 402 TO, 193 thrombus, and 129 dissection) lesion samples. The F1 score, which represents the overall performance of the DNN model, for stenotic lesion, TO, calcification, thrombus, and dissection was 0.829, 0.810, 0.802, 0.823 and 0.854, respectively. For all lesion morphologies, recall rates were higher than precision rates. Results are shown in Table 3 and examples of result images of the lesion morphology task are shown in Figure 3B. The receiver operating characteristic (ROC) curves for different lesion morphologies are shown in Figure 4. The area under the curve (AUC) of the lesion morphology detection DNN for stenotic lesion, TO, calcification, thrombus, and dissection was 0.801, 0.759, 0.799, 0.778, and 0.863, respectively.
Table 3. Diagnostic performance of DeepDiscern lesion detection DNN.
| Lesion type | Precision rate | Recall rate | F1 score |
| Stenosis | 0.769 | 0.901 | 0.829 |
| Total occlusion | 0.757 | 0.871 | 0.810 |
| Calcification | 0.751 | 0.862 | 0.802 |
| Thrombus | 0.742 | 0.925 | 0.823 |
| Dissection | 0.790 | 0.926 | 0.854 |
| DeepDiscern achieved an average recall of 89.7% for the five lesion types, namely, stenosis, total occlusion, calcification, thrombus and dissection (λd= 0.5). | |||
Figure 4.
ROC curves and AUC values of all lesions. DeepDiscern lesion detection DNN predict several bounding boxes, which may contain lesion morphologies. The bounding box with a correct location and a correct type is a positive sample. The bounding box with a wrong location or a wrong type is a negative sample.
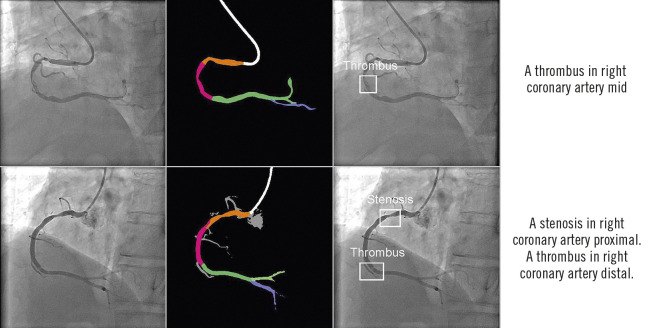
The DeepDiscern system provides an automatic and multimodal diagnosis in a two-step process. DeepDiscern first recognises all the arterial segments in the angiogram, and then it detects the lesions in the angiogram. Processing these two steps took less than two seconds on average for every angiogram (1.280 seconds for the segment recognition task, and 0.648 seconds for the lesion detection task). Combining these two results, DeepDiscern can analyse all lesions appearing in an angiogram (Figure 5).
Figure 5.
Combined results of DeepDiscern. In the first column are original angiograms. In the second column are resulting images of artery recognition DNN, where black areas represent background, white areas represent catheter, and other different colours represent different coronary artery segments. In the third column are resulting images of detection DNN. The location of lesion morphology on the angiogram is marked by several boundary boxes. The type of morphology is also predicted. In the fourth column are the combined results of recognition DNN and detection DNN.
Discussion
Many deep learning techniques that focus on a single aspect of the coronary angiogram and coronary computed tomography analysis, and therefore do not provide a high-level analysis, have been described previously9,10,11,12,13,14. Although a single neural network has been applied to medical imaging and other medical signals to generate high-level diagnosis16,17,18, these approaches are different from the DeepDiscern system, which provides a coronary diagnostic map by integrating multiple aspects, including the identification of different coronary artery segments, and the recognition of lesion location and lesion type. Because of the challenges of solving the complexities of coronary angiography using a single end-to-end DNN, DeepDiscern uses multiple DNN to solve multiple sub-problems and combines the results of multiple DNN to produce a high-level diagnosis.
With the current global population expansion, the number of patients with cardiovascular disease is increasing, resulting in a growing workload for cardiologists. DeepDiscern is capable of analysing a coronary angiogram in just a few seconds by learning and understanding medical knowledge from massive medical data. DeepDiscern can be used as an assistant to analyse lesion information quickly and help cardiologists to flag and diagnose lesion severity and morphology during the intervention before making a treatment decision. In addition, the amount of medical documentation routinely recorded has grown exponentially and now takes up to a quarter to a half of doctors’ time19. DeepDiscern can generate detailed angiographic reports automatically, saving cardiologists significant time for patient care. Thus, this approach has the potential to reduce workload and improve efficiency in coronary angiography diagnostics.
In clinical practice, visual interpretations of coronary angiograms by individuals are highly variable20. Inevitable subjective bias can have a great impact on diagnosis and treatment decisions21. Unlike the interpretation of cardiologists, the evaluation criteria of DeepDiscern are consistent for the same data set. In addition, deep learning has the ability to extract features automatically in digital angiographic images at a pixel scale, thereby impacting on angiographic interpretation by allowing analysis of angiographic images and identification of lesion features that are hard to discern by the human eye. Thus, the diagnosis of DeepDiscern is intended to be objective, accurate and reproducible.
DeepDiscern could also alleviate the growing problem of unequal distribution of medical resources and access to advanced health care. In 2017, at least half of the world’s population was unable to access essential health services22. In China, the difference between the highest value of healthcare access and quality (HAQ) and the lowest is 43.5 (the highest in Beijing is 91.5 and the lowest in Tibet is 48.0)23. The number of cardiovascular disease patients in China has reached 290 million. There are more than 2,000 primary hospitals in China providing coronary intervention treatment, with levels of diagnosis and treatment differing from place to place. The DeepDiscern technology can be extended easily and rapidly to major hospitals in the country and even the world. Implementing the DeepDiscern technology in primary hospitals countrywide could relieve the high demand for trained cardiologists, who are scarce, and provide consistency for improving angiographic diagnostic accuracy and treatment decisions, thereby achieving homogenisation of medical standards.
In the future, we will develop coronary artery lesion diagnostic systems that analyse more types of lesion morphology such as trifurcation, bifurcation, and severe tortuosity, among others. Thereafter, many decision-making tools based on recognition of lesion morphology and coronary artery segments can be automated without manual discrimination. For example, the SYNTAX score is a decision-making tool in interventional cardiology, which is determined simply by anatomical features in an angiogram24. The automation of SYNTAX score calculation is of great significance for the diagnosis of coronary angiography as it is an important tool for treatment selection (bypass surgery or percutaneous coronary intervention) in patients with more extensive CAD. The expected automatic SYNTAX score calculation system can generate a result in half a minute, detailing all the information about lesions that appear on a patient’s coronary arteries (Supplementary Figure 7).
Limitations
This study has several limitations. In this initial iteration, the input of DeepDiscern was a single frame angiogram obtained from an angiographic DICOM file, which provides limited information compared to a DICOM video. In actual use, after the procedure starts, the video stream of the contrast image is transmitted to our device. We used an automatic algorithm to extract a single frame with optimal contrast opacification and visualisation of the coronary artery tree and then used this single frame image as the input for DeepDiscern. The diagnosis of coronary lesions is based on the dynamic evaluation of lesion characteristics assessed in multiple coronary angiographic views. Additionally, DeepDiscern has a large requirement for training data volume. The lack of training data can seriously affect the recognition accuracy. In general, models trained with more data have better performance. Moreover, all the angiograms used for training and testing were collected from a large single centre; therefore, external validation by using data from other centres is warranted.
Conclusions
Deep learning technology can be used in the interpretation of diagnostic coronary angiography. In the future it may serve as a more powerful tool to standardise screening and risk stratification of patients with CAD.
Impact on daily practice
In clinical practice, inevitable subjective bias can have a great impact on diagnosis and treatment decisions. Unlike interpretation by cardiologists, the evaluation criteria of DeepDiscern are consistent for the same data set. In addition, DeepDiscern can analyse angiographic images and identify lesion features that are hard to discern by the human eye, which can also impact on angiographic interpretation. Thus, the diagnosis of DeepDiscern is intended to be objective, accurate and reproducible.
Supplementary data
Implementation detail about coronary artery recognition network.
Implementation detail about lesion morphology detection network.
The annotation procedure for coronary artery recognition and lesion morphology detection.
Annotated coronary artery segments.
The structure of the coronary artery recognition network.
The structure of the lesion morphology detection network.
Evaluation process of the coronary artery recognition model and the lesion morphology detection model.
Performance of vessel extraction.
Expected automatic calculation system for the SYNTAX score.
Coronary arteries labelled in different angiographic views.
National Heart, Lung and Blood Institute (NHLBI) coronary dissection criteria.
The mapping relationship between prediction label value and coronary artery segments.
Recognition performance of all segments under different angiographic views.
Acknowledgments
Funding
The study was supported by Beijing Municipal Science & Technology Commission - Pharmaceutical Collaborative Technology Innovation Research (Z18110700190000) and Chinese Academy of Medical Sciences - Medical and Health Science and Technology Innovation Project (2018-I2M-AI-007).
Conflict of interest statement
B. Xu reports grants from Beijing Municipal Science & Technology, and grants from the Chinese Academy of Medical Sciences during the conduct of the study. In addition, B. Xu has a patent for a method of coronary artery segmentation and recognition based on deep learning pending, and a patent for an automatic detection method system, and equipment of coronary artery disease based on deep learning pending. H. Zhang reports grants from Beijing Municipal Science & Technology during the conduct of the study. The other authors have no conflicts of interest to declare.
Abbreviations
- AI
artificial intelligence
- CVD
cardiovascular diseases
- DNN
deep neural networks
- LAD
left anterior descending
- LCX
left circumflex artery
- LM
left main
- OM
obtuse marginal
- RCA
right coronary artery
- TO
total occlusion
Contributor Information
Tianming Du, Beijing University of Posts and Telecommunications, Beijing, China.
Lihua Xie, Fu Wai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences, Beijing, China.
Honggang Zhang, Beijing University of Posts and Telecommunications, Beijing, China.
Xuqing Liu, Beijing University of Posts and Telecommunications, Beijing, China.
Xiaofei Wang, Beijing Redcdn Technology Co., Ltd, Beijing, China.
Donghao Chen, Beijing Redcdn Technology Co., Ltd, Beijing, China.
Yang Xu, Beijing Redcdn Technology Co., Ltd, Beijing, China.
Zhongwei Sun, Fu Wai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences, Beijing, China.
Wenhui Zhou, Beijing Redcdn Technology Co., Ltd, Beijing, China.
Lei Song, Fu Wai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences, Beijing, China.
Changdong Guan, Fu Wai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences, Beijing, China.
Alexandra J. Lansky, Yale University School of Medicine, New Haven, CT, USA.
Bo Xu, Fu Wai Hospital, National Center for Cardiovascular Diseases, Chinese Academy of Medical Sciences, Beijing, China.
References
- GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet. 2015;385:117–71. doi: 10.1016/S0140-6736(14)61682-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Global atlas on cardiovascular disease prevention and control. Geneva: World Health Organization; 2011. https://www.who.int/cardiovascular_diseases/publications/atlas_cvd/en/
- Dweck MR, Doris MK, Motwani M, Adamson PD, Slomka P, Dey D, Fayad ZA, Newby DE, Berman D. Imaging of coronary atherosclerosis - evolution towards new treatment strategies. Nat Rev Cardiol. 2016;13:533–48. doi: 10.1038/nrcardio.2016.79. [DOI] [PubMed] [Google Scholar]
- Authors/Task Force Members, Windecker S, Kolh P, Alfonso F, Collet JP, Cremer J, Falk V, Filippatos G, Hamm C, Head SJ, Jüni P, Kappetein AP, Kastrati A, Knuuti J, Landmesser U, Laufer G, Neumann FJ, Richter DJ, Schauerte P, Sousa Uva M, Stefanini GG, Taggart DP, Torracca L, Valgimigli M, Wijns W, Witkowski A. 2014 ESC/EACTS Guidelines on myocardial revascularization: The Task Force on Myocardial Revascularization of the European Society of Cardiology (ESC) and the European Association for Cardio-Thoracic Surgery (EACTS)Developed with the special contribution of the European Association of Percutaneous Cardiovascular Interventions (EAPCI). Eur Heart J. 2014;35:2541–619. doi: 10.1093/eurheartj/ehu283. [DOI] [PubMed] [Google Scholar]
- Rogers MA, Aikawa E. Cardiovascular calcification: artificial intelligence and big data accelerate mechanistic discovery. Nat Rev Cardiol. 2019;16:261–74. doi: 10.1038/s41569-018-0123-8. [DOI] [PubMed] [Google Scholar]
- Beam AL, Kohane IS. Translating Artificial Intelligence into Clinical Care. JAMA. 2016;316:2368–9. doi: 10.1001/jama.2016.17217. [DOI] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, Thrun S. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–8. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden JA. Deep Learning Algorithms for Detection of Lymph Node Metastases from Breast Cancer: Helping Artificial Intelligence Be Seen. JAMA. 2017;318:2184–6. doi: 10.1001/jama.2017.14580. [DOI] [PubMed] [Google Scholar]
- Nasr-Esfahani E, Karimi N, Jafari MH, Soroushmehr SMR, Samavi S, Nallamothu BK, Najarian K. Segmentation of vessels in angiograms using convolutional neural networks. Biomed Signal Proces. 2018;40:240–51. doi: 10.1016/j.bspc.2017.09.012. [DOI] [Google Scholar]
- Jun TJ, Kweon J, Kim YH, Kim D. T-Net: Nested encoder-decoder in encoder-decoder architecture for the main vessel segmentation in coronary angiography. Neural Netw. 2020;128:216–33. doi: 10.1016/j.neunet.2020.05.002. [DOI] [PubMed] [Google Scholar]
- Wolterink JM, van Hamersvelt RW, Viergever MA, Leiner T, Išgum I. Coronary artery centerline extraction in cardiac CT angiography using a CNN-based orientation classifier. Med Image Anal. 2019;51:46–60. doi: 10.1016/j.media.2018.10.005. [DOI] [PubMed] [Google Scholar]
- Wolterink JM, Leiner T, Viergever MA, Išgum I. Generative Adversarial Networks for Noise Reduction in Low-Dose CT. IEEE Trans Med Imaging. 2017;36:2536–45. doi: 10.1109/TMI.2017.2708987. [DOI] [PubMed] [Google Scholar]
- Wolterink JM, Leiner T, Isgum I. Blood Vessel Geometry Synthesis using Generative Adversarial Networks. 2018 [Google Scholar]
- Hwang YN, Lee JH, Kim GY, Shin ES, Kim SM. Characterization of coronary plaque regions in intravascular ultrasound images using a hybrid ensemble classifier. Comput Methods Programs Biomed. 2018;153:83–92. doi: 10.1016/j.cmpb.2017.10.009. [DOI] [PubMed] [Google Scholar]
- Pompa J, Almonacid A, Burke D. Qualitative and Quantitative Angiography. 2011 [Google Scholar]
- Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, Ng AY. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med. 2019;25:65–9. doi: 10.1038/s41591-018-0268-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh SLF, Calandriello L, Silva M, Sverzellati N. Deep learning for classifying fibrotic lung disease on high-resolution computed tomography: a case-cohort study. Lancet Respir Med. 2018;6:837–45. doi: 10.1016/S2213-2600(18)30286-8. [DOI] [PubMed] [Google Scholar]
- Gurovich Y, Hanani Y, Bar O, Nadav G, Fleischer N, Gelbman D, Basel-Salmon L, Krawitz PM, Kamphausen SB, Zenker M, Bird LM, Gripp KW. Identifying facial phenotypes of genetic disorders using deep learning. Nat Med. 2019;25:60–4. doi: 10.1038/s41591-018-0279-0. [DOI] [PubMed] [Google Scholar]
- Clynch N, Kellett J. Medical documentation: part of the solution, or part of the problem? A narrative review of the literature on the time spent on and value of medical documentation. Int J Med Inform. 2015;84:221–8. doi: 10.1016/j.ijmedinf.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Beauman GJ, Vogel RA. Accuracy of individual and panel visual interpretations of coronary arteriograms; implications for clinical decisions. J Am Coll Cardiol. 1990;16:108–13. doi: 10.1016/0735-1097(90)90465-2. [DOI] [PubMed] [Google Scholar]
- Généreux P, Palmerini T, Caixeta A, Cristea E, Mehran R, Sánchez R, Lazar D, Jankovic I, Corral MD, Dressler O, Fahy MP, Parise H, Lansky AJ, Stone GW. SYNTAX score reproducibility and variability between interventional cardiologists, core laboratory technicians, and quantitative coronary measurements. Circ Cardiovasc Interv. 2011;4:553–61. doi: 10.1161/CIRCINTERVENTIONS.111.961862. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Tracking universal health coverage: 2017 Global Monitoring Report. Geneva: World Health Organization; 2017. https://www.who.int/healthinfo/universal_health_coverage/report/2017/en/
- GBD 2016 Healthcare Access and Quality Collaborators. Measuring performance on the Healthcare Access and Quality Index for 195 countries and territories and selected subnational locations: a systematic analysis from the Global Burden of Disease Study 2016. Lancet. 2018;391:2236–71. doi: 10.1016/S0140-6736(18)30994-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sianos G, Morel MA, Kappetein AP, Morice MC, Colombo A, Dawkins K, van den Brand M, Van Dyck N, Russell ME, Mohr FW, Serruys PW. The SYNTAX Score: an angiographic tool grading the complexity of coronary artery disease. EuroIntervention. 2005;1:219–27. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Implementation detail about coronary artery recognition network.
Implementation detail about lesion morphology detection network.
The annotation procedure for coronary artery recognition and lesion morphology detection.
Annotated coronary artery segments.
The structure of the coronary artery recognition network.
The structure of the lesion morphology detection network.
Evaluation process of the coronary artery recognition model and the lesion morphology detection model.
Performance of vessel extraction.
Expected automatic calculation system for the SYNTAX score.
Coronary arteries labelled in different angiographic views.
National Heart, Lung and Blood Institute (NHLBI) coronary dissection criteria.
The mapping relationship between prediction label value and coronary artery segments.
Recognition performance of all segments under different angiographic views.