Abstract
The hybridization of complementary strands of DNA is the underlying principle of all microarray-based techniques for the analysis of DNA variation. In this paper, we study how probe immobilization at surfaces, specifically probe density, influences the kinetics of target capture using surface plasmon resonance (SPR) spectroscopy, an in situ label-free optical method. Probe density is controlled by varying immobilization conditions, including solution ionic strength, interfacial electrostatic potential and whether duplex or single stranded oligonucleotides are used. Independent of which probe immobilization strategy is used, we find that DNA films of equal probe density exhibit reproducible efficiencies and reproducible kinetics for probe/target hybridization. However, hybridization depends strongly on probe density in both the efficiency of duplex formation and the kinetics of target capture. We propose that probe density effects may account for the observed variation in target-capture rates, which have previously been attributed to thermodynamic effects.
INTRODUCTION
All microarray-based techniques for the analysis of DNA variation are based directly or indirectly on the hybridization of complementary strands of DNA (1–4). In principle, the fidelity of hybridization relies on differences in the thermodynamic equilibrium between bound and free oligomers. For surface-immobilized probes, however, thermodynamic equilibrium conditions may not be reached without excessively long incubation times and hybridization may be kinetically or sterically inaccessible for some probe sequences or for some surface probe densities.
Increasingly, researchers are exploiting surface-immobilized DNA oligomers for a variety of biotechnological, medical and nanoscience applications. Many studies have reported the detection of hybridization for immobilized DNA probes with solution-phase targets using techniques that include optical, electrochemical and mechanical methods (5–9). However, relatively few efforts have focused on understanding how probe immobilization at surfaces, specifically probe density, influences the kinetics of target capture. Measurements of the kinetics of target capture for surface-bound DNA can provide important insight into how hybridization at interfaces may be different from solution-phase duplex formation, and may also lead to the development of improved immobilization strategies for microarray applications.
In this paper, we use surface plasmon resonance (SPR) spectroscopy to monitor the kinetics of probe attachment as well as the process of target capture. This technique, which does not require fluorescence probes or other labels, has been used previously in our laboratory to study the kinetics and thermodynamics of DNA monolayer films (10–13). The attachment of a DNA probe to solid supports can be achieved by covalent or non-covalent attachment strategies. Here, we used covalent attachment based on gold/thiol bond formation a method shown previously to result in robust and reusable DNA probe films (6,10,11). In addition to the thiol modified 25mer DNA oligonucleotide component, the self-assembled monolayer film also contains mercaptohexanol. For the resulting films, the non-specific adsorption of DNA is negligible as reported previously (10–13).
Varying the amount of time that the solid support is exposed to the DNA–thiol solution can control the probe density of the film; however, other strategies can be used. In this paper, we use several additional approaches including varying the solution ionic strength and applying an attractive electrostatic field at the interface to assist in the immobilization of negatively charged single strand DNA-C6-SH. Also, we compare the immobilization of duplex DNA-C6-SH, which has a thiol linker on one of the oligonucleotide strands in contrast to ssDNA-C6-SH immobilization under the same solution conditions. While the immobilization of ssDNA is more commonly used, duplexes have been used by some researchers (14). One might expect that after denaturation of the duplex film, the probe density may be more optimum for hybridization compared with the immobilization of ssDNA, which could lead to films that are too tightly packed for efficient hybridization.
We find probe density to be a controlling factor for the efficiency of target capture as well as for the kinetics of the target/probe hybridization. In the lowest probe density regimes, essentially 100% of probes can be hybridized and the kinetics of binding follow Langmuir-like kinetics, whereas at high probe density the efficiencies drop to ∼10% and the kinetics are also slower. The high efficiencies achieved here are in contrast to the typically low efficiencies of hybridization found for many DNA microarrays, where sensitive fluorescence detection is used. For example, Forman et al. (15) report saturating adsorption densities at perfect match probe sites to be <10% of the probe density.
All quantitative measurements of DNA surface density for probe oligonucleotides and bound targets are obtained using SPR spectroscopy. This work is characterized by highly reproducible data: independent of which immobilization strategy is used, we find that DNA films of equal probe density exhibit reproducible efficiencies and reproducible kinetics for probe/target hybridization.
MATERIALS AND METHODS
Oligodeoxyribonucleotide sequences
All oligonucleotides (Table 1) were obtained from commercial vendors with purification by HPLC. Thiol functionalized DNA oligonucleotides were obtained from Integrated DNA Technologies; unfunctionalized DNA oligonucleotide targets were obtained from Alpha DNA.
Table 1. Oligodeoxyribonucleotide sequences.
Preparation of solutions
All solutions were prepared using purified water (18 M Ohm cm–1 resistance, Barnstead E-pure). All probe, target and duplex solutions (1 µM) were prepared at the specified salt concentration. Electrolyte solutions were KH2PO4 (1 M, Aldrich) and NaCl (1, 0.1 and 0.05 M, Fisher) with all NaCl solutions containing TE buffer (10 mM Tris buffer pH 7.2 and 1 mM EDTA) (Sigma). Mercaptohexanol (Aldrich) was prepared as a 1 mM solution in water.
Immobilization procedure
Before all immobilization experiments, the gold substrate was cleaned with piranha solution [7:3 mixture of H2SO4 (EM Science) and H2O2 (Mallinckrodt)]. For each DNA experiment, the gold SPR substrate was exposed to DNA solution for >10 h unless otherwise stated. The ssDNA-C6-SH probe film was treated with 1 mM mercaptohexanol solution for 1–2 h. For duplex immobilization, the double strand DNA-C6-SH film was immobilized as a pre-hybridized probe (ssDNA-C6-SH) and target (ssDNA) combination. Before the exposure of the target to the dsDNA-C6-SH film the duplex was denatured to create an ssDNA probe surface. No more than half the mass of the immobilized duplex film is lost during heating to 80°C, consistent with the removal of the target strand only; no thiol DNA is lost upon heating (10). For both types of prepared probe surfaces, a control experiment was done (using a non-complementary target solution) to ensure that no non-specific binding occurred.
Denaturation
For most experiments, repeated measurements were performed on the same DNA film requiring regeneration of the ssDNA probe film. This was achieved by the denaturation of the surface duplex by rinsing with hot water.
SPR measurements
The two-color SPR apparatus set-up and procedure for analysis of measurements have been described previously (16,17), as have details of how quantitative measurements of coverage are extracted from raw SPR reflectance data. Briefly, the SPR reflectance data were analyzed by fitting the data to a multilayer Fresnel model to extract the thickness and dielectric constant of the unknown DNA layer. The resulting best-fit parameters are converted to coverage of DNA (in molecules/cm2) as outlined previously (6,10,11). Hybridization efficiencies are calculated as the fraction of hybridized target coverage divided by the immobilized probe coverage. As in previous work, these calculations assumed an equivalent SPR response per unit coverage for ssDNA oligonucleotides at the surface of the SPR substrate regardless of whether the DNA consisted of surface-immobilized probe molecules or target DNA undergoing hybridization at the surface. Good agreement has been reported (5,9,18) between our SPR measurements and radiolabeling experiments and quantification by other methods such as electrochemistry.
RESULTS AND DISCUSSION
Probe fabrication strategies
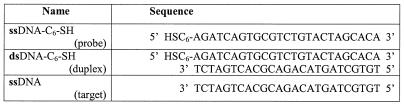
ssDNA versus dsDNA immobilization. Comparison of the immobilization of ssDNA-C6-SH on gold and dsDNA-C6-SH adsorption under the same solution conditions (1 M KH2PO4) is shown in Figure 1. The sequences used are shown in Table 1. For clarity and ease of comparison, the data in Figure 1 are reported in units of molecules/cm2.
Figure 1.
Representative data for immobilization kinetics of ssDNA-C6-SH (closed squares) and dsDNA-C6-SH (open circles) from 1 µM DNA solutions in 1 M KH2PO4.
Over repeated experimental runs, the final coverage for dsDNA-C6-SH, which is immobilized as a duplex, ∼3 × 1012 duplexes/cm2, is consistently lower than the density achieved for ssDNA-C6-SH. In addition, the kinetic behavior appears to follow a faster isotherm in contrast to the more complex kinetics for ssDNA-C6-SH immobilization (11). The observed difference in the shape of the kinetic isotherms is not yet fully understood but is likely to be due to a combination of effects, including differences in the conformation, flexibility and electrostatic interactions of duplex versus ssDNA under these identical solution conditions.
If we consider only the repulsive electrostatic interaction due to charge along the backbone of the DNA, we might expect duplex immobilization to result in a film with half the ssDNA-C6-SH probe density, simply because duplex DNA contains twice the anionic charge. The observation that the probe density for duplex immobilization is less than half of that obtained for ssDNA-C6-SH immobilization (Fig. 1) suggests that non-electrostatic effects, such as the conformation or flexibility of the DNA strands, may also be factors that control the ultimate probe density.
Benight and co-workers (19) report a 40% larger probe coverage for the immobilization of linear (ss) DNA probes compared with hairpin DNA probes. While different attachment chemistry is used, their results are consistent with our observations of achieving greater ssDNA probe density compared with duplex DNA under the same solution conditions. We assume here that hairpin oligonucleotides can be viewed as analogous to duplex DNA with regard to properties such as charge density and flexibility.
Over repeated experimental runs there is some variability in the resulting probe coverage. Interestingly, this variability is greatest for duplex immobilization. The average coverage for duplex immobilization is 2.8 × 1012 ± 0.6 × 1012 molecules/cm2. In contrast, for ssDNA-C6-SH immobilization, the average coverage shows much less variability between runs (11 × 1012 ± 0.2 × 1012 molecules/cm2). The average coverage for both cases is calculated after 10 h of exposure and the standard deviation is from six runs. For both cases, the variability in final probe coverage is not accompanied by a variation in the shape of the isotherm, which remains highly consistent between runs. The variability in the magnitude of the measured coverage is most likely caused by sample-to-sample variations in the gold surface roughness. Although there are no systematic studies of surface roughness on probe density, some data (20) suggest that higher probe densities are reached on rougher surfaces. This is due mainly to the greater available area for probe attachment on rough surfaces. At this time it is not understood why duplex DNA immobilization shows greater variability in probe density from run to run.
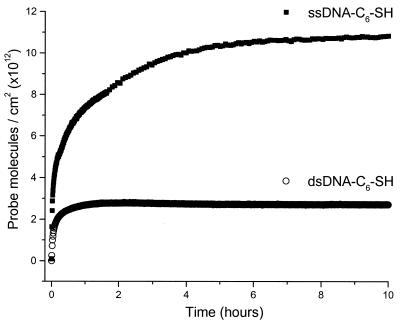
Ionic strength dependence of immobilization. As expected, the kinetics of immobilization depend strongly on solution ionic strength for both dsDNA-C6-SH (Fig. 2A) and ssDNA-C6-SH (Fig. 2B). In low ionic strength solutions, we observe less probe adsorption because of the larger electrostatic repulsion between probe strands. In high ionic strength solutions, the electrostatic repulsions between probe molecules are effectively screened, and higher probe coverage can be reached. These results are consistent with observations made by Herne and Tarlov (18) who report that maximum probe coverage is achieved when the KH2PO4 concentration is >0.4 M. We have found that immobilization in either 1 M KH2PO4 (pH 4) or 1 M NaCl (pH 7.2 w/TE buffer) show similar kinetics of film formation and similar final probe densities (not shown).
Figure 2.
Comparison of probe immobilization kinetics as a function of ionic strength formed from solutions containing (A) 1 µM dsDNA-C6-SH and (B) 1 µM ssDNA-C6-SH.
A more subtle effect, that has not been reported previously, arises in the shape of the adsorption isotherms. When the data in Figure 2 are scaled (not shown) it is clear that the rate of probe adsorption in the first few hours is consistently slower at a lower ionic strength compared with adsorption in a high ionic strength solution. Probe immobilization involves an interplay of forces including the short-range chemical interactions of the covalent gold/thiol attachment and the long-range electrostatic repulsion between DNA strands. In these experiments the electrostatics dominate: when the Debye length is large, the rate of probe attachment is low.
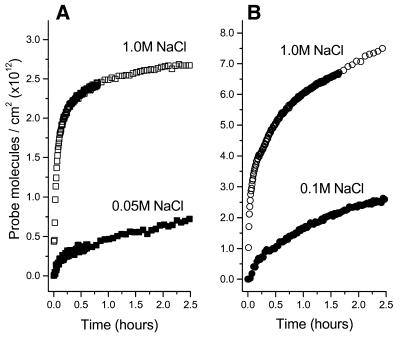
Electrostatic field control of probe immobilization. While it might be expected that an applied electrostatic field can control probe immobilization, this has not been investigated as a strategy for controlling DNA probe density. Interestingly, for n-alkanethiols on gold, Ma and Lennox (21) have shown that potential-assisted deposition leads to a faster and ultimately more complete self-assembled monolayer formation process. Here, we show that potential-assisted deposition can dramatically alter the process of attachment of thiol DNA to gold (Fig. 3). In related work from our laboratory, we have found that electrostatic fields can alter the rate of hybridization and denaturation (13).
Figure 3.
Immobilization kinetics from 1 µM ssDNA-C6-SH solutions in 1 M NaCl in the presence and absence of an applied potential. For potential-assisted immobilization, the potential is held at +0.3 V versus Ag/AgCl (triangles). The unassisted immobilization is at open circuit (open circles).
Figure 3 shows the effect of applying a potential of +0.3 V versus Ag/AgCl to the gold surface during the immobilization of ssDNA-C6-SH. Electrostatically controlled immobilization (closed triangles) has a larger initial rate of adsorption than ssDNA-C6-SH immobilization in the absence of an applied potential (open circles) under the same solution conditions. In the presence of the positive applied potential, the maximum attainable surface coverage remains similar to coverage attained at the open circuit but the rate is enhanced. Probe films formed by potential-assisted deposition are covalently attached; there is no significant probe loss when the potential is removed or when the film is rinsed. Target-capture rates for probe films formed with and without potential assistance are compared later (in ‘comparing probe films formed by different immobilization strategies’).
Probe density effects on probe/target hybridization
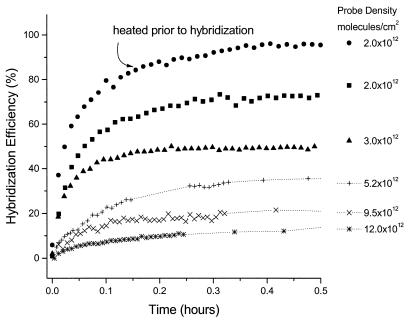
As expected, and in agreement with earlier reports (2,5,20), probe density strongly affects the target hybridization efficiency; that is, for high probe density films, we find that the efficiency of hybridization is low (Fig. 4). For the data, the probe density was controlled by varying the exposure time of the gold surface to ssDNA-C6-SH during the probe film fabrication process. For each hybridization isotherm shown in Figure 4, target/probe hybridization was performed under the same conditions of solution concentration and ionic strength. The efficiency of hybridization after 30 min varies from ∼15 to 95% depending on the probe density, with the highest density films exhibiting the lowest efficiencies. Longer exposure to target solutions for up to 14 h increases the efficiency of hybridization somewhat: the range increases from 30 to almost 100%.
Figure 4.
Target hybridization kinetics as a function of probe density. The probe density, determined by SPR, varies from 2 × 1012 to 12 × 1012 molecules/cm2. Heating of the probe film prior to hybridization increases the hybridization efficiency. All runs are 1 µM target in 1 M NaCl with TE buffer.
Our results are in general agreement with the amount of probe/target hybridization reported by other groups. For example, Demers et al. (7) used a fluorescence-based measurement to detect probe coverage and target hybridization on planar gold. For overnight immobilization, which is likely to lead to high-density probe coverage, the reported target hybridization efficiency for the resulting film was 33% even after 40 h, comparable with our observations. Steel et al. (5) used an electrochemical detection approach to observe a trend similar to our observations, that is, high-density probe films have reduced target hybridization efficiency.
Previous heating of the probe surface appears to have a significant effect on the hybridization efficiency. For example, a 22% increase in the hybridization efficiency was observed (solid circles in Fig. 4) when compared with the efficiency on a never heated surface. A similar increase was observed for a higher density film, as reported previously for a different 25mer sequence (10). This was not due to probe loss as there is no measurable decrease in probe coverage. At very high probe densities, prior probe heating does not improve the efficiency of hybridization.
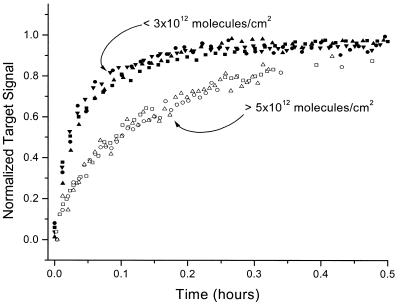
If we focus on the shape of the kinetic isotherms, we see that the kinetics of hybridization also have a distinct dependence on probe density. This effect, which is probably due to intermolecular interactions, has not been observed previously. Trends in the data are most clearly visualized when the kinetic isotherms are normalized, as in Figure 5. The data appear to fall into different regimes depending on the probe densities. For probe densities below ∼3 × 1012 molecules/cm2 the initial hybridization rate was faster reaching a maximum within ∼15 min. The distinct shape of the hybridization isotherms at high densities above ∼5 × 1012 molecules/cm2 (slower target-capture rate) is due most probably to repulsive electrostatic and steric interactions that increase with increasing probe density. Future work will focus on modeling these kinetic profiles.
Figure 5.
Target hybridization kinetics as a function of probe density. The same data as in Figure 4, however, the data are now normalized. The data show a distinct behavior that correlates with two regimes for probe density: low (<3 × 1012 molecules/cm2) and high (>5 × 1012 molecules/cm2).
The fact that probe density can alter target-capture rates is not unexpected but is not often considered, especially when differential measurements are made where the magnitudes of probe immobilization or hybridization are not recorded but only the ratio of matched to mismatched duplexes. It is interesting to note that the kinetics of hybridization for matched and mismatched DNA are distinct (12); however, it is not yet known whether probe density influences matched and mismatched duplex formation to the same degree. The influence of probe density on the formation perfectly matched versus partially mismatched hybrids is the subject of ongoing work in our laboratory.
In some cases, differences in probe density are measured; for example, in the work of Benight and co-workers (19) who propose that DNA hairpins could offer substantial advantages as nucleic acid capture moieties in solid support-based hybridization systems. They observe capture rates that depend on whether a linear or hairpin DNA probe is used and attribute the difference in kinetics to the increased stability of the hairpin probe/target duplex structure. However, the coupling density for linear probes is nearly twice that for hairpin probes. Based on our results, we would predict lower rates of target capture for higher density probe films. Thus, the higher density of the linear probe films could account, at least in part, for the different observed rates.
Comparing probe films formed by different immobilization strategies
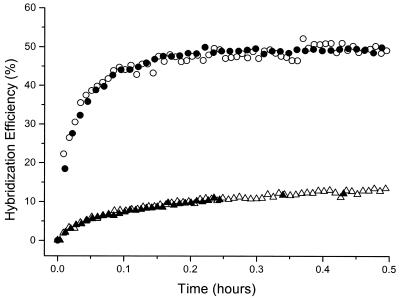
In this section, we compare the effectiveness of DNA probe films fabricated using different immobilization conditions by measuring (i) the rate of hybridization with targets and (ii) the overall efficiency of probe/target hybridization. In Figure 6, we compare the kinetics of hybridization on two sets of probe films (low-density and high-density probe). Low probe densities were obtained by immobilizing ssDNA-C6-SH for a short time period (<1.5 h) or via dsDNA-C6-SH immobilization followed by thermal dehybridization. High probe densities were obtained by immobilizing ssDNA-C6-SH for a longer time (>10 h) or via potential-assisted deposition of ssDNA-C6-SH. The hybridization conditions are identical for both sets of probe films. We observe no difference in the rate, magnitude or efficiency of hybridization for the films formed by a variety of methods provided that probe films of the same probe density are compared.
Figure 6.
Comparison of probe/target capture kinetics for probe films formed by different immobilization strategies. Hybridization kinetics are shown for two low probe density films, ∼3 × 1012 molecules/cm2 (circles) and two high probe density films, ∼1.2 × 1013 molecules/cm2 (triangles). Probe films at low density, formed either from ssDNA-C6-SH immobilization (filled circles) or via dsDNA-C6-SH immobilization (open circles) show comparable kinetics and overall efficiency of capture. Similarly, probe films formed via immobilization of ssDNA-C6-SH under electrostatic conditions of +0.3 V (open triangles) or via incubation overnight at open circuit (filled triangles) are indistinguishable. All hybridization kinetics obtained under the same conditions of 1 µM target in 1 M NaCl with TE buffer.
CONCLUSIONS
Surface density of the probe strand is a key factor that determines the extent to which immobilized probes are able to capture solution-phase targets. In low probe density regimes, essentially 100% of probes can be hybridized and the kinetics of binding follow comparatively faster kinetics, whereas at a high probe density the efficiencies drop and the kinetics are slower. In both cases, however, the hybridization isotherms remain complex and cannot be fit with simple kinetic models. Various strategies can be used to tailor immobilization and control the surface probe density. For all strategies used, the functionality of the film remains the same when compared at the same probe density. Most importantly, there is a strong dependence on probe density for both the efficiency of duplex formation and the kinetics of target capture. We propose that probe density effects may account for the observed variations in target capture which have previously been attributed (19) to thermodynamic effects.
Acknowledgments
ACKNOWLEDGEMENTS
We gratefully acknowledge support of the National Science Foundation through grants CHE-9709347 and DBI-0096731.
REFERENCES
- 1.Lockhart D.J., Dong,H.L., Byrne,M.C., Follettie,M.T., Gallo,M.V., Chee,M.S., Mittmann,M., Wang,C.W., Kobayashi,M., Horton,H. and Brown,E.L. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol., 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 2.Southern E., Mir,K. and Shchepinov,M. (1999) Molecular interactions on microarrays. Nature Genet., 21, 5–9. [DOI] [PubMed] [Google Scholar]
- 3.Mir K.U. and Southern,E.M. (1999) Determining the influence of structure on hybridization using oligonucleotide arrays. Nat. Biotechnol., 17, 788–792. [DOI] [PubMed] [Google Scholar]
- 4.Tillib S.V. and Mirzabekov,A. (2001) Advances in the analysis of DNA sequence variations using oligonucleotide microchip technology. Curr. Opin. Biotechnol., 12, 53–58. [DOI] [PubMed] [Google Scholar]
- 5.Steel A.B., Herne,T.M. and Tarlov,M.J. (1998) Electrochemical quantitation of DNA immobilized on gold. Anal. Chem., 70, 4670–4677. [DOI] [PubMed] [Google Scholar]
- 6.Levicky R., Herne,T.M., Tarlov,M.J. and Satija,S.K. (1998) Using self-assembly to control the structure of DNA monolayers on gold: a neutron reflectivity study. J. Am. Chem. Soc., 120, 9787–9792. [Google Scholar]
- 7.Demers L.M., Mirkin,C.A., Mucic,R.C., Reynolds,R.A., Letsinger,R.L., Elghanian,R. and Viswanadham,G. (2000) A fluorescence-based method for determining the surface coverage and hybridization efficiency of thiol-capped oligonucleotides bound to gold thin films and nanoparticles. Anal. Chem., 72, 5535–5541. [DOI] [PubMed] [Google Scholar]
- 8.Wu G.H., Ji,H.F., Hansen,K., Thundat,T., Datar,R., Cote,R., Hagan,M.F., Chakraborty,A.K. and Majumdar,A. (2001) Origin of nanomechanical cantilever motion generated from biomolecular interactions. Proc. Natl Acad. Sci. USA, 98, 1560–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mbindyo J.K.N., Reiss,B.D., Martin,B.R., Keating,C.D., Natan,M.J. and Mallouk,T.E. (2001) DNA-directed assembly of gold nanowires on complementary surfaces. Adv. Mater., 13, 249–254. [Google Scholar]
- 10.Peterlinz K.A. and Georgiadis,R.M. (1997) Observation of hybridization and dehybridization of thiol-tethered DNA using two-color surface plasmon resonance spectroscopy. J. Am. Chem. Soc., 119, 3401–3402. [Google Scholar]
- 11.Georgiadis R., Peterlinz,K.P. and Peterson,A.W. (2000) Quantitative measurements and modeling of kinetics in nucleic acid monolayer films using SPR spectroscopy. J. Am. Chem. Soc., 122, 3166–3173. [Google Scholar]
- 12.Peterson A.W., Heaton,R.J. and Georgiadis,R. (2000) Kinetic control of hybridization in surface immobilized DNA monolayer films. J. Am. Chem. Soc., 122, 7837–7838. [Google Scholar]
- 13.Heaton R.J., Peterson,A.W. and Georgiadis,R.M. (2001) Electrostatic surface plasmon resonance: direct electric field-induced hybridization and denaturation in monolayer nucleic acid films and label-free discrimination of base mismatches. Proc. Natl Acad. Sci. USA, 98, 3701–3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley S.O., Barton,J.K., Jackson,N.M., McPherson,L.D., Potter,A.B., Spain,E.M., Allen,M.J. and Hill,M.G. (1998) Orienting DNA helices on gold using applied electric fields. Langmuir, 14, 6781–6784. [Google Scholar]
- 15.Forman J.E., Walton,I.D., Stern,D., Rava,R.P. and Trulson,M.O. (eds) (1998) Chapter 13: Thermodynamics of duplex formation and mismatch discrimination on photolithographically synthesized oligonucleotide arrays. Molecular Modeling of Nucleic Acids. American Chemical Society, Washington, DC, Vol. 682, pp. 206–228.
- 16.Peterlinz K.A. and Georgiadis,R. (1996) Two-color approach for determination of thickness and dielectric constant of thin films using surface plasmon resonance spectroscopy. Opt. Commun., 130, 260–266. [Google Scholar]
- 17.Peterlinz K.A. and Georgiadis,R. (1996) In situ kinetics of self-assembly by surface plasmon resonance spectroscopy. Langmuir, 12, 4731–4740. [Google Scholar]
- 18.Herne T.M. and Tarlov,M.J. (1997) Characterization of DNA probes immobilized on gold surfaces. J. Am. Chem. Soc., 119, 8916–8920. [Google Scholar]
- 19.Riccelli P.V., Merante,F., Leung,K.T., Bortolin,S., Zastawny,R.L., Janeczko,R. and Benight,A.S. (2001) Hybridization of single-stranded DNA targets to immobilized complementary DNA probes: comparison of hairpin versus linear capture probes. Nucleic Acids Res., 29, 996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang E., Satjapipat,M., Han,S.B. and Zhou,F.M. (2001) Surface structure and coverage of an oligonucleotide probe tethered onto a gold substrate and its hybridization efficiency for a polynucleotide target. Langmuir, 17, 1215–1224. [Google Scholar]
- 21.Ma F.Y. and Lennox,R.B. (2000) Potential-assisted deposition of alkanethiols on Au: Controlled preparation of single- and mixed-component SAMs. Langmuir, 16, 6188–6190. [Google Scholar]