Abstract
Although single-cell sequencing has provided a powerful tool to deconvolute cellular heterogeneity of diseases like cancer, extrapolating clinical significance or identifying clinically-relevant cells remains challenging. Here, we propose a novel computational method scAB, which integrates single-cell genomics data with clinically annotated bulk sequencing data via a knowledge- and graph-guided matrix factorization model. Once combined, scAB provides a coarse- and fine-grain multiresolution perspective of phenotype-associated cell states and prognostic signatures previously not visible by single-cell genomics. We use scAB to enhance live cancer single-cell RNA-seq data, identifying clinically-relevant previously unrecognized cancer and stromal cell subsets whose signatures show a stronger poor-survival association. The identified fine-grain cell subsets are associated with distinct cancer hallmarks and prognosis power. Furthermore, scAB demonstrates its utility as a biomarker identification tool, with the ability to predict immunotherapy, drug responses and survival when applied to melanoma single-cell RNA-seq datasets and glioma single-cell ATAC-seq datasets. Across multiple single-cell and bulk datasets from different cancer types, we also demonstrate the superior performance of scAB in generating prognosis signatures and survival predictions over existing models. Overall, scAB provides an efficient tool for prioritizing clinically-relevant cell subsets and predictive signatures, utilizing large publicly available databases to improve prognosis and treatments.
INTRODUCTION
Single-cell RNA sequencing, in particular single-cell RNA sequencing (scRNA-seq) and single-cell ATAC sequencing (scATAC-seq), has been widely adopted to dissect tissue heterogeneity of human diseases like cancer at the resolution of individual cells. While single-cell sequencing allows detailed cataloging of cancer subpopulations and surrounding niche cell subpopulations, its application to the clinic is hampered due to the high cost and labor associated with single-cell sequencing of large cohorts (1). The few cohorts with clinical information also make it difficult to ascertain the statistical associations between these subpopulations and clinical features such as survival and drug responses. Such limitation analysis hampers efforts for early prognosis and improved treatments of diseases. Therefore, it is highly important to extract more useful information for instructing precision diagnosis and treatment based on the available multidimensional data.
Fortunately, the rich bulk sequencing data provides clinical information of a large number of cohorts in publicly available databases such as The Cancer Genome Atlas (TCGA), International Cancer Genome Consortium (ICGC) (2), Genomics of Drug Sensitivity in Cancer (GDSC) (3) and Chinese Glioma Genome Atlas (CGGA) portal (4). To link the scRNA-seq derived subpopulations with clinical response, one main approach in existing studies was to identify marker genes of subpopulations and then assess their clinical significance using available bulk RNA-seq data of patients and machine learning models in survival analysis (5,6). Two recent methods scPrognosis (7) and scFeatures (8) infer features from scRNA-seq data and assess the clinical significance using the selected features. scPrognosis infers signature genes important in the Epithelial-to-Mesenchymal Transition (EMT) biological process, while scFeatures selects important features by presenting a very nice way for multi-view molecular representations of single-cell data. However, such approach lacks a systematical way to identify all possible cell subpopulations with prognostic significance and may depend on the pre-determined clustering results. Recently, Scissor (9) and DEGAS (10) were developed to identify cell subpopulations that were highly correlated with a given phenotype from scRNA-seq data via a sparse regression model and deep learning method, respectively. However, these two methods did not reveal the underlying patterns of phenotype-associated cells, which might be associated with distinct cancer hallmarks. More than ten cancer hallmarks have been recognized (11), implying the heterogeneity and plasticity of tumors and their associated microenvironments in clinical implications. In addition, methods designed for scATAC-seq data are lacking, preventing the identification of phenotype-associated chromatin accessible sites and transcription factors. Therefore, systematical identification of cell subsets and their patterns with clinical significance and distinct hallmarks in single-cell genomics remains challenging.
By integrating single-cell genomics And Bulk RNA sequencing data with phenotype information, here we propose a novel computational method called scAB. scAB links single-cell genomics and bulk data via pairwise correlations, and infers multiresolution cell states correlated with patients’ phenotypes via a knowledge- and graph-guided matrix factorization model. scAB can simultaneously dissect multiresolution phenotype-associated cell states and their predictive signatures. The multiresolution cell states include both coarse- and fine-grain cell states, which respectively correspond to all phenotype-associated cells and distinct subsets of phenotype-associated cells with distinct clinical significance. We apply scAB to several scRNA-seq and scATAC-seq datasets, layering key clinical information such as patient survival information and response to immunotherapeutic to efficiently resolve multiresolution cell states and molecular signatures for clinical prognosis from different modalities of single-cell genomics. In addition, we show that scAB provides an alternative survival model showing better performance over the traditional models such as CoxBoost (Cox model by likelihood-based boosting) and RSF (Random Survival Forests) when applying to bulk RNA-seq data only. Overall, scAB provides an efficient way to enhance the clinical power of single-cell genomics data.
MATERIALS AND METHODS
scAB model
To identify phenotype-associated cells from single-cell genomics, we first link scRNA-seq or scATAC-seq data with bulk RNA-seq data by computing Pearson's correlations based on the expression levels of shared genes. When taking scATAC-seq as input, we assume similar patterns between chromatin accessibility and gene expression, and transform the chromatin accessibility data into the gene activity data using ‘CreateGeneActivityMatrix’ function from Seurat package. Then we infer a set of phenotype-associated patterns by proposing a knowledge- and graph-guided matrix factorization model that incorporates available phenotype information from bulk samples. In detail, the optimization model of scAB is formulated as
 |
(1) |
where  is the calculated correlation matrix with rows representing n bulk samples and columns representing m individual cells. X is decomposed into a sample loading matrix
is the calculated correlation matrix with rows representing n bulk samples and columns representing m individual cells. X is decomposed into a sample loading matrix  and a cell loading matrix
and a cell loading matrix  . The ‘tr’ denotes the trace of a matrix, which is defined as the sum of its diagonal elements. k is the number of patterns, i.e. the number of rows of H. Each pattern, which typically corresponds to a known biological process/signal associated with a particular phenotype of interest, is obtained from each row of the resulted cell loading matrix H.
. The ‘tr’ denotes the trace of a matrix, which is defined as the sum of its diagonal elements. k is the number of patterns, i.e. the number of rows of H. Each pattern, which typically corresponds to a known biological process/signal associated with a particular phenotype of interest, is obtained from each row of the resulted cell loading matrix H. 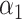 and
and  are regularization parameters to balance each term regarding the phenotype information and the graph regularization, respectively.
are regularization parameters to balance each term regarding the phenotype information and the graph regularization, respectively.
In the optimiation problem (Equation (1)), the second term is the constraint of the phenotype information of bulk samples ( )).
)). is a diagonal matrix in which small values represent the likelihood of bulk samples exhibiting the phenotype of interest (e.g, poor prognosis or strong response). Thus, minimizing this constraint allows the prioritization of phenotype-relevant bulk samples, which is reflected by large loading values of the bulk samples in the matrix W. The phenotype un-relevant bulk samples will be assigned with very small loading values (approximating to zeros) in W. In other words, matrix factorization in the first term will focus on the phenotype-relevant bulk samples. Then the high values in X lead to high loading values in both W and H, resulting in the identification of phenotype-relevant cells. In the scenario where the phenotype is binary group (e.g. response vs. non-response),
is a diagonal matrix in which small values represent the likelihood of bulk samples exhibiting the phenotype of interest (e.g, poor prognosis or strong response). Thus, minimizing this constraint allows the prioritization of phenotype-relevant bulk samples, which is reflected by large loading values of the bulk samples in the matrix W. The phenotype un-relevant bulk samples will be assigned with very small loading values (approximating to zeros) in W. In other words, matrix factorization in the first term will focus on the phenotype-relevant bulk samples. Then the high values in X lead to high loading values in both W and H, resulting in the identification of phenotype-relevant cells. In the scenario where the phenotype is binary group (e.g. response vs. non-response),  is assigned as 0 when the i-th sample is the phenotype of interest; otherwise,
is assigned as 0 when the i-th sample is the phenotype of interest; otherwise,  is taken as 1. In the scenario where the phenotype is survival information, we require the user to input a two-column table including the survival time and survival state of each bulk sample. To develop a unified computational framework, we transform the survival information to a single phenotype score of each patient, which is defined as follow,
is taken as 1. In the scenario where the phenotype is survival information, we require the user to input a two-column table including the survival time and survival state of each bulk sample. To develop a unified computational framework, we transform the survival information to a single phenotype score of each patient, which is defined as follow,
 |
where score(samplei) is the relative survival score of sample i that is computed using a generalized survival model (12). Compared to the Cox model, the relative survival score includes two additional survival cases, including early-censored–late-uncensored pairs and early-censored–late-censored pairs (Supplementary Table S1). In sum, such transformation allows us to develop a coherent mathematical model regardless of the type of phenotype information.
In the optimiation problem (Equation (1)), the third term is the graph regularization ( )). This term constrains the cell loading matrix H through the Laplacian matrix L of the cell-cell similarity matrix, which is helpful for identifying clinically relevant cells with high similarity. Specifically, L is a symmetric normalized Laplacian matrix, which is defined as
)). This term constrains the cell loading matrix H through the Laplacian matrix L of the cell-cell similarity matrix, which is helpful for identifying clinically relevant cells with high similarity. Specifically, L is a symmetric normalized Laplacian matrix, which is defined as
 |
where  is the cell–cell similarity matrix given by a shared nearest neighbor graph that is inferred by Seurat package (13) (see ‘Preprocessing of single-cell RNA-seq data’).
is the cell–cell similarity matrix given by a shared nearest neighbor graph that is inferred by Seurat package (13) (see ‘Preprocessing of single-cell RNA-seq data’).  is the degree matrix of the cell–cell similarity matrix, where
is the degree matrix of the cell–cell similarity matrix, where  and
and  for
for  .
.
Optimization algorithm for scAB
The optimization problem (Equation (1)) is solved by a multiplicative update rule (14), which updates variables W and H iteratively (Supplementary Text). The convergence of this algorithm can be theoretically proven based on the auxiliary function method (Supplementary Text).
Identification of coarse- and fine-grain phenotype-associated cell states
After solving the optimization problem (Equation (1)), we identify phenotype-associated cells based on the cell loading matrix H. The loading values, which are the entries of the matrix H, represent the contributions of each cell in each pattern (corresponding to each row of H), and cells with high loading values are defined as phenotype-associated cells. We examine the distribution of loading values in each pattern using Hartigan's Dip Test (the ‘dip.test’ function from the R package ‘diptest’). If the loading values exhibit a unimodal distribution, the cells with a z-score greater than 2 are defined as phenotype-associated cells. Specifically, we calculate the z-score for each element in each row of H by
 |
where  is the average value of the kth row of H and
is the average value of the kth row of H and  is the standard deviation of the kth row of H. If the loading values exhibit a bimodal distribution, the ‘locmodes’ function from the ‘multimode’ package is used to select an appropriate threshold to identify phenotype-associated cells.
is the standard deviation of the kth row of H. If the loading values exhibit a bimodal distribution, the ‘locmodes’ function from the ‘multimode’ package is used to select an appropriate threshold to identify phenotype-associated cells.
The fine-grain cell states represent the classification of all phenotype-relevant cells into certain subsets, which are identified based on each row of the matrix H (i.e. each inferred pattern). Specifically, for each row of the matrix H, we divide cells into two subsets by thresholding the z-score-scaled cell loadings (i.e. weights) and define the subset with high cell loadings as one fine-grain cell state. The coarse-grain cell state represents the classification of all single cells into phenotype-relevant cells and other cells, which are achieved by thresholding the z-score-scaled cell loadings in the whole matrix H. In other words, the phenotype-relevant cells in the coarse-grain cell state are the union of the phenotype-relevant cells across all fine-grain cell states. Therefore, the fine-grain and coarse-grain cell states can be simultaneously obtained based on the inferred cell loading matrix H in the scAB framework.
Identification of signature genes and loci for phenotype-associated cell states
For scRNA-seq data, after identifying cell subsets, we adopt a Wilcoxon ranksum test to identify differentially expressed genes between phenotype-associated cells and other cells. Genes are considered as the signature genes if (i) the adjusted P values are <0.05 and (ii) the absolute value of log fold-changes are >1. We use log-fold changes of 1 as the cutoff to identify the most differentially expressed signature genes, which is helpful for selecting potential biomarkers. The cutoff value may be dependent on the specific datasets and users can adjust this parameter value. Lower cutoff values lead to more candidate signature genes.
To account for the sparse nature of the nearly binary scATAC-seq data, aggregation is often used as an efficient strategy (15,16). Here we apply a previous method to aggregates the binary epigenomic profiles across similar cells (16). We then use the aggregated scATAC-seq data to perform differential accessibility analysis between phenotype-associated cells and other cells. Loci are considered as differentially expressed if (i) the adjusted P values are <0.05 and (ii) the absolute value of log fold-changes are >0.25. Moreover, we also performed motif enrichment analysis using chromVAR based on the aggregated scATAC-seq data. chromVAR (17) calculates the bias corrected deviations in accessibility. For each motif, there is a value for each cell, which measures how different the accessibility for loci with that motif is from the expected accessibility based on the average of all the cells.
Functional enrichment analysis
The function enrichment analysis for GO (gene oncology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) are performed using ‘enrichGO’ function and ‘enrichKEGG’ function in R package ‘clusterProfiler’ (18), respectively. The function ‘gseGO’ in R package ‘clusterProfiler’ is used for gene set enrichment analysis (GSEA). In addition, gene set variation analysis (GSVA) is done using the ‘gsva’ function with default settings in R package ‘GSVA’.
Survival analysis
Survival analysis is performed using R packages ‘survival’ and ‘glmnet’. In detail, patients were classified into a high-risk group and a low-risk group based on the median of computed prognostic scores. The survival curve of patients is then calculated using the Kaplan–Meier method in ‘survival’ package. Subsequently, the survival curve difference of patients between different groups is evaluated by log-rank test. In addition, survival models based on selected gene sets are established using the Cox-LASSO model via R package ‘glmnet’. The 5-fold cross-validation is performed on all methods that require cross-validation to determine parameters.
Implementation of the scAB model in the bulk RNA-seq data
The scAB model in (Equation (1)) can be naturally degenerated to the scenario when only bulk expression matrix and survival information are available. Then we can identify phenotype-associated gene signatures from bulk RNA-seq data with phenotype information (Supplementary Text).
Parameter selection
In scAB model, three parameters need to be determined, including the number of patterns k (i.e. the number of rows of the cell loading matrix H) and two regularization parameters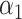
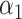 and
and  . First, the selection of k can be guided based on the errors between the original matrix X and the reconstructed matrix WH. Specifically, we define the rule for determining the k value as follows:
. First, the selection of k can be guided based on the errors between the original matrix X and the reconstructed matrix WH. Specifically, we define the rule for determining the k value as follows:
 |
where  is the average value of the objective function in ten repeated runs for
is the average value of the objective function in ten repeated runs for  . Therefore, the value of k is determined when the increase of k does not significantly reduce the reconstruction error. We then apply five-fold cross-validation to select the optimal
. Therefore, the value of k is determined when the increase of k does not significantly reduce the reconstruction error. We then apply five-fold cross-validation to select the optimal  and
and  with the maximum concordance index.
with the maximum concordance index.
Datasets
Liver cancer datasets
scRNA-seq dataset of liver cancer patients was downloaded from GEO (accession code: GSE125449), including seven cell types: B cell, cancer-associated fibroblasts (CAFs), cells with an unknown entity but express hepatic progenitor cell markers (HPC-like), malignant cells, T cells, tumor-associated macrophages (TAMs), and tumor-associated endothelial cells (TECs). Bulk RNA-seq datasets of liver cancer patients with survival information were downloaded from the Xena platform (19) (TCGA-LIHC cohort) and the ICGC Data Porta (LIRI cohort).
Melanoma datasets
scRNA-seq dataset of melanoma patients was downloaded from GEO (accession code: GSE115978), including nine cell types: B cell, cancer-associated fibroblasts (CAFs), endothelial cells, macrophages, malignant cells, Natural Killer (NK) cells, CD4+ T cells, CD8+ T cells, and T cells. The melanoma immunotherapy datasets PRJEB23709 and MGSP were downloaded from https://github.com/donghaixiong/Immune_cells_analysis, provided by Xiong et al. in a previous study (20). In addition, bulk RNA-seq datasets of immunotherapy of another melanoma cohort, thymic carcinoma cohort and non-small cell lung carcinoma cohort, were downloaded from GEO under accession codes GSE91061, GSE181815 and GSE135222, respectively. Besides, a bulk RNA-seq dataset of melanoma patients with survival information was downloaded from the Xena platform (TCGA-SKCM cohort). The Genomics of Drug Sensitivity in Cancer publicly available drug sensitivity data used in this study are available on the GDSC portal (https://www.cancerrxgene.org).
Glioma datasets
scATAC-seq dataset of glioma patients was downloaded from GEO (accession code: GSM4131779). Bulk RNA-seq datasets of glioma patients with survival information were downloaded from the Chinese Glioma Genome Atlas (CGGA) platform (training cohort: mRNAseq_693; the validation sets 2: mRNAseq_325; the validation set 3: mRNAseq_325) and the Xena platform (the validation set 1: TCGA-GBMLGG).
Bulk RNA-seq datasets for comparisons against existing survival models
We downloaded six cancer datasets from the Xena platform (https://xenabrowser.net/) collected by TCGA, including Breast Cancer (BRCA), Lung Adenocarcinoma (LUAD), Head and Neck Cancer (HNSC), Kidney Clear Cell Carcinoma (KIRC) and Bladder Cancer (BLCA), Lung Squamous Cell Carcinoma (LUSC).
Preprocessing of scRNA-seq and scATAC-seq data
The R package Seurat (v4.0.2) (13) was utilized to preprocess single-cell expression data in this study. First, genes were retained with detected expression in more than three cells. Cells were retained that with detected genes >200 features and the percentage of mitochondrial genes <5%. Subsequently, ‘NormalizeData’ function was utilized to normalize single-cell expression data with the default parameters. The highly variable genes were determined using ‘FindVariableFeatires’ function with the ‘vst’ method. Then we performed transformation on these data using ‘ScaleData’ function and principal component analysis on the scaled data using ‘RunPCA’ function. Next, we calculated cell-cell similarity using a shared nearest neighbor graph, which was given by ‘FindNeighbors’ function based on the first ten principal components. The cell-cell similarity matrix was then binarized and its diagonal elements were set to be zero. Finally, we used ‘RunUMAP’ function for dimensionality reduction to visualize cells in a two-dimensional space.
For scATAC-seq data, we performed the same preprocessing procedure by taking the gene activity data as an input.
Comparison of scAB against existing methods
We evaluate scAB against nine methods, including three methods designed for scRNA-seq data (Scissor (9), scPrognosis (7) and DEGAS (10)), one baseline method, and five traditional methods (tumor stage, patient sex, patient age, ImmuneCells.Sig (ImSig) (20) and PD- L1) on the studied datasets. Specifically, for the dataset with survival information, we compare scAB against Scissor, scPrognosis, DEGAS, the baseline method as well as clinical features such as tumor stage, patient sex and age. For the dataset with immunotherapy information, we compare scAB against Scissor, scPrognosis, DEGAS, the baseline method, the previously defined immunotherapy gene set ImSig and the expression levels of PD-L1. The detailed descriptions of how different methods are implemented are available in Supplementary Text.
The baseline method (or called ‘Markers_top100’ method) is an extension of standard single-cell data analysis. Specifically, we take the union of the top 100 marker genes of each cell cluster as candidate biomarkers, and run feature selection on this candidate single-cell derived biomarkers at the bulk RNA-seq training dataset with known clinical variables. We then evaluate the prediction accuracy of the selected biomarkers on the bulk RNA-seq testing dataset.
RESULTS
Overview of scAB
To identify phenotype-relevant cell subsets and predict genes, chromatin loci and transcription factors for clinical prognosis, we developed scAB by integrating scRNA-seq or scATAC-seq data with clinically annotated bulk RNA-seq data. scAB required three types of data as input, including single-cell expression data or chromatin accessibility data, bulk RNA-seq data, and the phenotype of patients in bulk samples (Figure 1A). To develop a unified mathematical model, we transformed the chromatin accessibility data into gene activity data (Materials and Methods). The phenotype annotations of bulk samples can be either survival data or binary groups such as response vs. non-response, wild type vs. mutation, and normal vs. disease (Materials and Methods).
Figure 1.
Overview of scAB. (A) scAB requires single-cell genomics data, bulk RNA-seq data, and the phenotype of patients in bulk samples as inputs. (B) scAB unveils a set of patterns for cell stratification via a knowledge- and graph- guided matrix decomposition of a similarity matrix between single cells and individual patients. (C) scAB enables simultaneous detection of coarse- and fine-grain phenotype-associated cell states from the inferred cell loading matrix H. Different fine-grain cell subsets may be associated with certain shared and specific cancer hallmarks. (D) scAB uncovers clinical-relevant gene signatures that are able to perform a variety of prognostic analyses, such as survival analysis, immunotherapy efficacy prediction, and synergistic treatment prediction.
Upon receiving the input data, scAB first calculated pairwise Pearson's correlations between individual cells and bulk samples using the single-cell and bulk expression data. Other similarity calculation strategies were also explored in this study (see Discussion). Second, scAB unveiled a set of patterns via a phenotype- and graph-guided non-negative matrix decomposition of the calculated correlation matrix (Figure 1B, Materials and Methods). Each pattern, which often corresponded to a known biological process/signal relating to a particular phenotype, was obtained from each row of the resulted cell loading matrix outputted by the model. The loading values (i.e. weights) represented the contribution of each cell in each pattern and cells with high loading values were defined as phenotype-associated cells. Third, we introduced the concept of multiresolution phenotype-associated cell states to represent both coarse- and fine-grain patterns of phenotype-associated cell landscape (Figure 1C). Each fine-grain cell state consisted of phenotype-associated cells in each learned pattern and the coarse-grain phenotype-associated cell state was defined by the union of phenotype-associated cells across all fine-grain cell states (see Materials and Methods). The fine-grain cell subsets may exhibit shared and specific cancer hallmarks, allowing to uncover latent patterns in phenotype-associated cells. Finally, we assessed the clinical significance of the identified phenotype-associated cell states and signatures using available bulk RNA-seq datasets. We demonstrated scAB’s utility as a biomarker identification tool, with the ability to predict survival risk, immunotherapy and drug responses (Figure 1D).
scAB identifies previously unrecognized cell subsets with high metabolism associated with metastasis and poor prognosis in liver cancer
To demonstrate the ability of our method, we first applied scAB to a scRNA-seq dataset for liver cancer to identify clinically-relevant cell states leading to poor survival. Cells in this scRNA-seq data were previously classified into seven cell types (21), including malignant cells, tumor-associated macrophages (TAMs), tumor-associated endothelial cells (TECs), HPC-like cells, and cancer-associated fibroblasts (CAFs), B cells, and T cells (Figure 2A). To identify survival-related cell states in the scRNA-seq data, we incorporated bulk RNA-seq data from 370 liver cancer patients from TCGA.
Figure 2.
scAB identifies multiresolution survival-relevant cell states and their enriched biological processes in liver cancer. (A, B) Uniform Manifold Approximation and Projection (UMAP) visualization of cells from the liver cancer microenvironment. (A) Cells are colored by the original cell annotations. (B) Cells are colored by the scAB’s prediction, where red and blue indicates scAB+ cells and other cells respectively. (C) The cell type compositions of scAB+ cells showing the percentage of each cell type over the total scAB+ cells. (D) The dot plot showing enriched cancer hallmarks in distinct cell subsets of scAB+ cells. The size and color of the dots indicate the calculated gene scores of each hallmark. (E) UMAP visualization of cells highlighting the two distinct cell subsets in scAB+ cells. (F) The volcano plot showing differential expressed genes in scAB+ cells versus other cells. (G) The dot plot showing the enriched biological processes (BP) of scAB+ cells versus other cells. (H, I) GSEA highlighting one upregulated and another downregulated biological processes in scAB+ cells compared to other cells.
By integrating the scRNA-seq and bulk RNA-seq data, scAB recognized around 11.2% cells (992 cells among the total 8853 cells) associated with poor survival of liver cancer and these cells were marked as scAB positive (scAB+) cells (Figure 2B). The scAB+ cells were the phenotype-associated cells defined in Materials and Methods. By assessing the cell type compositions of these identified scAB+ cells (n = 992 cells), we found that the majority of scAB+ cells (60.7%, n = 602 cells) were malignant cells (Figure 2C). Interestingly, scAB+ cells also included other type of cells, such as TECs (10.2%, n = 101 cells) and CAFs (7.3%, n = 72 cells), which was consistent with the increasing evidence of important roles of tumor microenvironment in affecting the survival of liver cancer (22–24). On the other hand, among each cell type, we found that 40.03% (602/1504) malignant cells, 9.95% (72/724) TAMs, 9.60% (101/1052) TECs, 8.09% (68/841) HPC-like cells, 7.60% (72/947) CAFs, 2.11% (21/993) B cells and 2.01% (56/2792) T cells were identified as scAB+ cells (Supplementary Figure S1), suggesting of the ability of scAB in prioritizing clinically-relevant cell subsets. Moreover, in addition to these coarse-grain survival-associated cell states, scAB also allowed the dissection of four fine-grain cell subsets. These four subsets could be categorized into two groups, defined by specific cancer hallmarks. The first group (cell subsets 1, 2 and 4) were associated with hallmarks such as autophagy, apoptosis, and proliferation while the second group (cell subset 3) was associated with metabolism, DNA repair, and response to stress (Figure 2D). These results indicated that different fine-grain cell subsets could represent distinct functional capabilities of cancer undergoing malignant transformation. Further examination showed that subset 3 (i.e. scAB+ cells in Pattern 3) dominantly represented malignant cells while subset 4 (i.e. scAB+ cells in Pattern 4) was enriched by various cell types in the tumor microenvironment, including a small subset of macrophage, T cells and fibroblasts (Figure 2E). Interestingly, fatty acid metabolism was highly enriched in subset 3 instead of subset 4 (Figure 2D and Supplementary Figure S2). Although previous studied showed that upregulated fatty acid metabolism contributed to cancer progression (25), our results suggested that cell subset 3 was likely the dominant driver.
To systemically gain insights into the biological significance of these predicted scAB+ cells, we performed differential gene expression analysis between the pooled scAB+ cells with other cells, leading to 26 downregulated and 108 upregulated genes in scAB+ cells (Figure 2F). Notably, these downregulated and upregulated genes can be also identified by performing differential gene expression analysis for scAB+ malignant cells versus all scAB– cells. Among these 108 up-regulated genes, a number of genes such as FGG and NUPR1 were reported to be associated with poor survival and metastatic potential in hepatocellular carcinoma (HCC, the most common type of primary liver cancer) in previous studies (26,27). Moreover, we performed functional enrichment analysis using positive genes from scAB+ cells and other cells and found that these up-regulated genes in scAB+ cells were dominantly enriched in several metabolism-related biological processes, such as ATP-, catabolic-, fatty acid- metabolic processes (Figure 2G). These findings were greatly consistent with the critical roles of metabolic process in contributing to the cellular processes linked to tumor progression (28). Cancer metabolism has been also identified as a key characteristic of tumor cell migration and invasion (29). In contrast, the down-regulated genes were related to T cell and leukocyte activation and ribosome biogenesis. Gene set enrichment analysis (GSEA) of the down-regulated and up-regulated genes further confirmed the significantly activation of lipid metabolic process (Padj = 7.2 × 10−11, Figure 2H) and inhibition of immune system (Padj = 8.3 × 10−07, Figure 2I), suggesting that scAB+ cells exhibited immune resistance and may escape immune surveillance (30). In summary, scAB identified a survival-related cell subset with high metabolism that was not recognized by the original study (21). Gene signature analysis suggested that the identified cell subset was linked to metastasis and poor prognosis of liver cancer. These results also demonstrate the superior performance of scAB in identifying survival-relevant cell subsets and their molecular signatures and biological processes. Our analysis shows that leveraging the available large amount of bulk data could increase the statistical power of single-cell transcriptomics.
Both coarse- and fine-grain survival-relevant gene signatures identified by scAB predict the prognosis of liver cancer
To further assess the clinical significance of these identified scAB+ cell states and their gene signatures in liver cancer, we utilized bulk RNA-seq data from liver cancer and performed survival prediction using a widely used LASSO-Cox model (31). We first used two bulk datasets from two large-scale clinical study cohorts, including the TCGA-LIHC (370 patients) and the ICGC-LIRI (231 patients) (32). These two datasets describe the survival information of patients with liver cancer.
Using the identified differential expressed genes (DEGs) of scAB+ cells versus other cells, we calculated the prognostic score of each patient in both bulk datasets using the LASSO-Cox model (Figure 3A). Patients were then classified into a high-risk group and a low-risk group based on the computed prognostic scores (Materials and Methods). We found that the high-risk group exhibited significantly worse survival time than the low-risk group in both datasets (TCGA-LIHC: hazard ratio (HR) = 2.38, P = 5.49 × 10−7; ICGC-LIRI: HR = 4.75, P = 3.79 × 10−6). Specifically, the median survival time of the high-risk group was 33.5 months in TCGA-LIHC and 48 months in ICGC-LIRI, while that of the low-risk group was 81.9 months in TCGA-LIHC. The median survival time of the low-risk group in ICGC-LIRI was not observed because more than half of patients were still alive. Furthermore, we assessed the clinical significance of the identified subsets of scAB+ cells (Figure 3B). Interestingly, patients in the high-risk group from both subset 3 and subset 4 (i.e. subset 3 high + subset 4 high) had the worst survival, while patients with low-risk from both subsets (i.e. subset 3 low + subset 4 low) had the highest survival rate in the early stage (about 0–40 months) compared to the case with a single high-risk group (i.e. subset 3 high + subset 4 low or subset 3 low + subset 4 high) (Figure 3B). These results indicated that both the scAB-identified coarse-grain and fine-grain survival-relevant cell states had clinical significance. Moreover, the fine-grain survival-relevant cell subsets indeed had distinct clinical significance and their signatures exhibited distinct ability in predicting patient prognosis of liver cancer.
Figure 3.
Assessment of clinical significance of the coarse- and fine-grain survival-relevant gene signatures identified by scAB in liver cancer. (A) Overall survival curves of patients with low or high risks according to DEGs in scAB+ cells versus other cells in LIHC and LIRI cohorts. (B) Overall survival curves of patients with the combinations of low- or high-risk groups according to gene signatures in two different subsets (subset 3 and subset 4) of scAB+ cells. S3H and S3L represent subset 3 high risk and subset 3 low risk, respectively. Similar meaning is for S4H and S4L. (C) Relapse free survival curves of patients with low or high risks according to DEGs in scAB+ cells in LIHC and GSE14520 cohorts. (D) Comparison of the prediction performance of scAB against different methods and clinical features. (E) Comparison of the tumor stage-only model with the model including tumor stage and scAB-derived signatures. The likelihood ratio test was used to compare the significance of the difference between the two models.
Due to the high metabolism of scAB+ cells that was linked to metastases and invasion (29) (Figure 2G), we further predicted patients’ recurrence risk using the DEGs identified based on the scAB+ cells in two datasets including TCGA-LIHC and GEO14520. Interestingly, a significant difference between the low- and high-risk groups was observed (Figure 3C; TCGA-LIHC: HR = 2.08, P = 9.02 × 10−6; GEO14520: HR = 1.84, P = 3.29 × 10−4). The median recurrence time of the high-risk group was 15.2 and 23.5 months, while that of the low-risk group was 41.0 and 57.7 months in TCGA-LIHC and GEO14520, respectively. This result indicated that the identified signatures using scAB could predict the recurrence for liver cancer.
Next we asked whether prognostic scores derived from scAB can better predict the patient survival over other methods designed for single-cell data, including Scissor, scPrognosis, DEGAS and Markers_top100 (a baseline method; see details in Materials and Methods), and the traditional clinical characteristics-based methods, including tumor pathological stage, patient sex and age. We thus examined the association of patient survival with derived prognostic scores from different methods and traditional clinical characteristics using the univariate Cox proportional hazard model and the concordance index (C-index). We found that scAB exhibited the highest C-index value compared to other methods survival in the testing bulk dataset (Figure 3D), suggesting the superior performance of scAB over other methods. Interestingly, compared to the traditional clinical characteristics-based methods, methods (except for DEGAS) designed for single-cell data consistently exhibited better performance, as reflected by higher C-index values. Moreover, the combination of scAB-derived prognostic score and pathological stage presented a higher survival prognosis efficiency than only the pathological stage in the testing bulk (Figure 3E), implying that the scAB derived prognostic score could improve the precision of prognosis in liver cancer patients.
scAB reveals macrophage-mediated immunotherapy response in melanoma microenvironment
In addition to the scenario where survival information is available, scAB can also identify cell states related to other phenotypes with binary information such as response vs. non-response and normal versus disease. Here, we took its application in immunotherapy response as an example. Immune checkpoint blockade (ICB) has been a hot spot in the field of cancer treatment in recent years, but only a few patients can benefit from it (33). To explore the mechanisms of ICB response and identify biomarkers that can predict response in melanoma, we utilized scAB to integrate gene expression in single-cell data and ICB response phenotype in bulk data. The single-cell melanoma data from GSE115978 were previously classified into nine cell types (34), including malignant cells, endothelial cells, NK cells, T cells, B cells, CAFs, CD8+ and CD4+ T cells, and macrophage (Figure 4A). The bulk data was from PRJEB23709 (35), including 73 melanoma samples treated with anti-PD-1 monotherapy or the combination of anti-PD-1 and anti-CTLA-413.
Figure 4.
scAB predicts cell subsets with better ICB response in melanoma microenvironment. (A, B) UMAP visualization of cells from the melanoma microenvironment. (A) Cells are colored by the original cell annotations. (B) Cells are colored by the scAB’ prediction, where red and blue indicates scAB_IR cells and other cells respectively. (C) The cell type compositions of scAB_IR cells showing the percentage of each cell type over the total scAB_IR cells. (D) The dot plot showing enriched cancer hallmarks in distinct cell subsets of scAB_IR cells. The size and color of the dots indicate the calculated gene scores of each hallmark. (E) UMAP visualization of cells highlighting the two distinct cell subsets in scAB_IR cells. (F) The volcano plot showing differential expression gene in scAB_IR cells versus other cells. (G) The dot plot showing the enriched biological processes (BP) of scAB_IR cells versus other cells. (H, I) GSEA highlighting one upregulated and another downregulated biological processes in scAB_IR cells compared to other cells.
scAB recognized around 80% (5465/6879) of cells associated with immunotherapy response. These responsive cells were indicated as scAB Immune Response (scAB_IR) cells (Figure 4B). By assessing the cell type compositions of these identified scAB_IR cells, we found that the vast majority of scAB_IR cells were T cells (CD8+ T cells: 31.34%, 1713/5465; CD4+ T cells: 15.32%, 837/5465; T cells: 12.04%, 658/5465) (Figure 4C). Interestingly, 6.48% (354/5465) scAB_IR cells were macrophages, which may offer a novel therapeutic strategy for cancer immunotherapy (36). In addition, scAB simultaneously identified the fine-grain cell subsets related to ICB. By examining the association of cancer hallmarks with these inferred five cell subsets, we found that these five cell subsets can be categorized into three groups: one group consisting of cell subsets 1, 2 and 5 was associated with senescence and angiogenesis, one group only consisting of cell subset 4 was associated with MYC_target_v2 (37) and G2M_checkpoint (38), and the other group only consisting of cell subset 3 was associated with PI3K/AKT/MTOR signaling (39) and KRAS signaling (40) (Figure 4D). Interestingly, the subset 3 was composed of immune cells, while the subset 5 was composed of a portion of malignant cells and a portion of macrophages (Figure 4E). Notably, macrophages in subset 5 exhibited higher expressions of CD274 (i.e. PD-L1) versus other macrophages (Supplementary Figure S3), which was consistent with the positive correlation between the PD-L1 expression in macrophages and the response to PD-1 blocked in previous reports (41,42).
To further gain insights into the biological significance of these predicted scAB_IR cells, we compared the gene expression differences between the pooled scAB_IR cells (i.e. coarse-grain cell state) with other cells, leading to 225 up-regulated and 102 down-regulated genes in scAB_IR cells compared to other cells (Figure 4F). Notably, among these 225 up-regulated genes, multiple genes, such as CCL5 (43–45), NKG7 (46–48) and CXCR3 (49), were reported to be associated with the efficacy of immunotherapy in previous studies. On the other hand, multiple down-regulated genes, such as CCND1 (50), S100B (51,52), SERPINA3 (53), were reported to have inhibitory effects in immunotherapy. We observed the up-regulated of ‘T cell activation’ and ‘lymphocyte differentiation’ in scAB_IR cells by performing the functional enrichment analysis (Figure 4G), suggesting that there is likely an immune active program in the identified scAB_IR cells. In addition, biological processes linked with ATP were down-regulated in scAB_IR cells. Further GSEA showed the over-activated adaptive immune response (Padj = 4.6х10−15) and suppressed oxidation-reduction process (Padj = 5.6 × 10−15) in scAB_IR cells compared to other cells (Figure 4H and I). In summary, scAB identified two distinct subsets of malignant cells activating different cancer hallmarks, and immune cell subsets dominant by T cells and macrophage associated with immune response.
scAB-derived biomarkers have strong predictive power for immunotherapy and drug response in melanoma
To further evaluate the clinical significance of the identified scAB_IR cell states in ICB response in melanoma, we calculated the gene set variation analysis (GSVA) score of each patient in bulk RNA-seq datasets, including ICB responders and non-responders. In the training dataset PRJEB23709, which was used to integrate with single-cell data using scAB, ICB responders showed higher scores than non-responders (P = 0.00035) (Figure 5A). By testing on another two independent bulk datasets from melanoma genome sequencing project (MGSP) and GSE91061, we observed consistently higher scores in ICB responders compared to non-responders (P = 0.028 and P = 0.0035) (Figure 5A). These results confirmed the association between scAB_IR and ICB response, and demonstrated that the scAB-derived signature genes can robustly distinguish ICB responders from non-responders.
Figure 5.
Evaluation of scAB’s performance in predicting immunotherapy response, survival rate and drug sensitivity. (A) Comparisons of GSVA scores calculated using scAB_IR cells’ gene signatures between response and non-response groups across three melanoma datasets and one Thymic carcinoma dataset. The Wilcoxon rank-sum test was used to assess the differences. (B) Progression-free survival curves of patients with low or high risks according to GSVA scores in non-small cell lung carcinoma patients. (C) Disease-specific survival curves of patients with low or high risks according to GSVA scores in the TCGA-SKCM melanoma dataset. (D) Comparison of the performance of biomarkers inferred from different methods in distinguishing responders and non-responders using the P-values of Wilcoxon tests. (E) Correlation analysis of GSVA scores and drug sensitivity IC50 scores across four melanoma datasets treated by different drugs. The Pearson's correlation coefficient and its associated p-values were shown. The line was the fitted using GSVA scores and IC50 scores via a linear model, and the shaded area was the 95% confidence interval for the line.
To assess whether the scAB-derived signature genes from ICB response in melanoma also represent prognostic biomarkers in other cancer types, we first calculated the GSVA scores of thymic carcinoma patients treated with pembrolizumab (GSE181815), and found that all responders had higher scores than all non-responders (Figure 5A). In another cohort of 27 advanced non-small cell lung carcinoma patients treated with anti-PD-1/PD-L1, there was a significant difference in the progression-free survival between the high-score and low-score groups based on the median of GSVA scores (HR = 2.37, P = 0.044) (Figure 5B). The observed distinguishing ability in these two non-melanoma immunotherapy cohorts indicated that the identified scAB_IR cells likely play an essential role in immunotherapy across different types of cancer. For another TCGA-SKCM dataset without immunotherapy information, we investigated the prognostic effect of the identified signature genes on patients with melanoma. Intriguingly, patients with higher scores exhibited a better survival prognosis than patients with lower scores (HR = 1.89, P = 1.29 × 10−5) (Figure 5C), suggesting the predictive power of the scAB-derived signature genes from ICB response.
To evaluate the superior performance of the transcriptomic characteristics of scAB_IR cells as an indicator of ICB treatment response, we calculated and compared GSVA scores for the gene sets identified by different methods, including the DEGs identified by scAB, the DEGs identified by another competitive methods (i.e. Scissor, scPrognosis, DEGAS and Markers_top100) and the previously discovered immunotherapy gene set ImmuneCells.Sig (ImSig) (20). On the three testing bulk RNA-seq datasets (MGSP, GSE91061, GSE181815), we used the expression values of PD-L1 as a basic measurement of ICB treatment response and evaluated the ability of different methods in distinguishing responders and non-responders using the P-values of Wilcoxon tests. Compared to other methods, scAB consistently exhibited the lowest P-values on MGSP and GSE91061 datasets, and comparable pvalues with Scissor, Markers_top100 and PD-L1 on GSE181815 (Figure 5D). Interestingly, Scissor, scPrognosis and Markers_top100 also showed better performance than ImSig and the sole expression of PD-L1, suggesting that integration of single-cell and bulk data can improve the prediction of ICB treatment response.
To further assess the prediction power of the scAB-derived signature genes in terms of drug sensitivity, we downloaded bulk datasets of melanoma cell lines from Genomics of Drug Sensitivity in Cancer (GDSC) (3). In these bulk datasets, melanoma was treated with different drugs. By calculating GSVA scores of cell lines using the scAB-derived signature genes from ICB response, we observed statistically positive correlations between the GSVA scores and the drug sensitivity (quantified by a conventionally metric IC50) of 54 drugs (Figure 5E, Supplementary Table S2). Note that a higher GSVA score also indicated a better ICB response, suggesting that the combination of these drugs with ICB may serve as a synergistic therapeutic effect in melanoma. In previous studies, Mitomycin C was confirmed to enhance the efficacy of PD-L1 blockade in non-small cell lung cancer (54), and Doxorubicin (55,56), Pyridostatin (57,58), and GSK650394 (59) were also reported to enhance ICB efficiency significantly. These results suggested that combining ICB with chemotherapy is a potential treatment strategy for melanoma patients.
Taken together, scAB identified immune-active cell subsets in the melanoma microenvironment that were associated with ICB response. These cell subsets were characterized by enhanced immune activity and low redox activity. More importantly, the signature genes derived from these cell subsets can robustly predict ICB response, survival rate and drug sensitivity in melanoma as well as other types of cancer.
scAB predicts prognosis signatures of glioma from scATAC-seq data
Finally, we show scAB’s capability in different modalities by utilizing a published human glioblastoma (GBM) scATAC-seq dataset (60) and a bulk RNA-seq dataset of 693 glioma patients with survival information from CGGA (4). To predict survival-relevant signatures including chromatin accessible loci and transcription factors (TFs), we first identified clinically-relevant cells using scAB (Figure 6A). To reveal the chromatin accessible sites associated with these scAB+ cells, we performed differential accessibility analysis (Materials and Methods). Encouragingly, we observed distinct chromatin accessibility patterns between scAB+ cells versus other cells (Figure 6B). By performing GO enrichment analysis of enriched chromatin accessible sites of scAB+ cells using GREAT tool (61), we revealed GBM- associated biological processes such as ‘maintenance of unfolded protein involved in ERAD pathway’ (Figure 6C), which contributed to the GBM development (62). In addition, scAB revealed four fine-grain cell subsets with distinct chromatin accessibility patterns (Figures 6D–F). GO analysis of the enriched chromatin accessible sites showed different biological processes across these four cell subsets. For example, subset 1 was enriched for ‘leukocyte activation’ (63) and subset 4 was enriched for ‘fibroblast growth factor receptor’ (64) (Figure 6G).
Figure 6.
scAB performs well in predicting prognosis signatures from scATAC-seq data. (A) UMAP visualization of cells from human glioblastoma scATAC-seq data. Cells are colored by the scAB’s prediction, where red and blue indicates scAB+ cells and other cells respectively. (B) Heatmap of the relative chromatin accessibility of the differential loci between scAB+ cells and other cells. (C) Enriched biological processes of the differential loci associated with scAB+ cells. (D) UMAP visualization of cells highlighting the distinct cell subsets in scAB+ cells. (E) Overlay the chromatin accessibility of two loci onto the UMAP space. Dark red and gray colors represent the high and zero expression, respectively. (F) Heatmap of the relative chromatin accessibility of the specific loci from fine-grained scAB+ cells and other cells. (G) Enriched biological processes of the differential loci associated with scAB+ subset 1 and subset 4 cells. (H) Heatmap of the chromVAR deviations for the top 10 differential TFs (columns) between scAB+ cells and other cells (rows). (I) Overall survival curves of patients with low or high risk according to the expression of the transcription factors ELK3 and TFTC in the three validation datasets. Log-rank test was used to compare the significance of the difference between high- and low-risk groups.
Moreover, we performed motif enrichment analysis using chromVAR and identified a number of motifs exhibiting differential patterns across scAB+ cells and other cells (Figure 6H). chromVAR (17) calculates the bias corrected deviations in accessibility. We then evaluated the clinical significance of TFs associated with these identified motifs using additional three bulk RNA-seq datasets. Survival analysis showed that overexpression of the transcription factor ELK3 was significantly associated with the poor prognosis of patients with gliomas (Figure 6I), which was in agreement with the in vitro experiment that ELK3 overexpression promoted proliferation and migration of glioma cells (65). In addition, we predicted a previous unrecognized transcription factor TFEC that was significantly linked to the poor prognosis of patients with gliomas, implying that TFEC could be a candidate target in the prognosis of patients with gliomas.
Together, these results showed scAB’s capability in predicting prognosis signatures of glioma from scATAC-seq data, suggesting that scAB was potentially applicable to different modalities of single-cell genomics.
scAB shows superior performance over existing prognosis models
Given the superior performance of scAB in predicting patient survival, we then asked whether it can serve as a better model over the classical survival models in bulk data analysis. To demonstrate this point, we first built a degradation model for bulk RNA-seq data only (Materials and Methods; For simplicity, we still refer it as scAB) and then applied it to bulk RNA-seq datasets from six types of cancer, including Breast Cancer (BRCA), Lung Adenocarcinoma (LUAD), Head and Neck Cancer (HNSC), Kidney Clear Cell Carcinoma (KIRC), Bladder Cancer (BLCA) and Lung Squamous Cell Carcinoma (LUSC).
We evaluated scAB against three widely used models, namely the Cox proportional hazard model with LASSO penalization (31), the iterative gradient boosting method CoxBoost, and the tree-based non-linear integration method RSF (66). For each dataset, samples were randomly divided into the training set (80%) and the test set (20%). After running each method ten times, we evaluated the performance on the test set using the C-index metric. scAB consistently exhibited higher C-index values than other three methods across these datasets except for the slightly lower values than RSF in the KIRC dataset (Figure 7A). On the contrast, the performance of other three methods varied across different datasets. These results indicated the superior performance of scAB over other classic methods in cancer prognosis.
Figure 7.
Applications of scAB to bulk RNA-seq data only and comparisons of its performance against existing survival models. (A) Comparison of C-indexes calculated using four survival models (i.e. scAB, Cox-LASSO model, RSF, CoxBoost model). Bulk datasets from six types of cancer including BRCA, LUAD, HNSC, BLCA, KIRC, LUSC are used. (B) The heatmap of the similarities of enriched GO terms. The word cloud annotation visualizes the summaries of biological functions in each GO cluster. Clustering and visualization of GO terms are performed with the R package ‘simplifyEnrichment’. (C) Overall survival curves of patients with low or high risks according to autophagy pathway in the training and the three validation datasets. Log-rank test was used to compare the significance of the difference between high- and low-risk groups.
Furthermore, we applied scAB in a CGGA (4) dataset and assessed the gene programs identified by scAB in multiple external independent glioma datasets. By clustering the enriched GO biological processes of signature genes, we observed that ‘telomere’ and ‘vesicle’ were associated with the survival of gliomas patients (Figure 7B), which was consistent with clinical results that glioma is a disease related to telomere imbalance (67) and that the changes in the vesicle transport system affect the growth of glioma (68). In addition, biological processes such as ‘ubiquitin-dependent’ and ‘mitochondrial’ were identified, which may serve as new prognostic factors in glioma survival (69,70).
To demonstrate the prognosis ability of scAB in glioma, we utilized genes from each of 24 enriched KEGG pathways and performed survival analysis in the training set and three external independent validation sets. Among the enriched pathways, the autophagy pathway plays an important role in providing energy for glioma progression (71). By computing prognostic scores using the autophagy pathway, we divided patients into high-risk and low-risk groups based on the median score. Regardless of the training set or the three validation sets, the survival rates of the high-risk and low-risk groups presented significant differences (P < 0.0001) (Figure 7C). Other 23 pathways also showed their ability in discriminating high-risk and low-risk groups in four data sets, suggesting the prognostic power of scAB (Supplementary Figure S4). Together, these results showed that the biological processes and pathways identified by scAB have the potential as therapeutic targets to predict prognosis in glioma, suggesting that scAB could be an effective method for prognosis in cancer.
DISCUSSION
While many computational methods have been developed to classify cell populations in single-cell data, including unsupervised clustering (72) and supervised cell type annotations (73), they have limited ability to identify cell states associated with phenotype of interest. The high cost of scRNA-seq limits the number of patients in available single-cell datasets, making it difficult to associate individual-level phenotype with single cells. To fill this critical gap, by integrating single-cell genomics and bulk RNA-seq data with phenotype information, we proposed a unified framework scAB to systematically dissect both coarse- and fine-grain phenotype-associated cell states and predictive signatures for early prognosis and treatments.
scAB is applicable to phenotype information including both survival information and binary group (e.g. response vs. non-response, normal vs. disease). By defining a phenotype score of each bulk sample, we were able to develop a coherent mathematical model regardless of the type of phenotype information. We demonstrated the capability of scAB in these two scenarios using scRNA-seq datasets from liver cancer with survival information, and melanoma with immune response information. Application to the prognosis of liver cancer survival, scAB identified a cell subset with high fatty acid metabolism and suppressed immune system. Using three bulk datasets, we showed that the signature genes of this cell subset can more accurately predict the survival and recurrence risk of liver cancer patients compared to the pathological stage only. Application to the immunotherapy of melanoma, scAB revealed a cell subset with immune-active program and low redox activity. Interestingly, the signature genes of this cell subset can better distinguish ICB responders from non-responders compared to the gene set from a previous study and the PD-L1 expression. Notably, this observation is not limited to patients with melanoma. Moreover, we identified 54 drugs that may have synergistic therapeutic effects with ICB using the drug sensitivity data, which could help to enhance the therapeutic effect of ICB. Together, these results demonstrate the strong potential of scAB in improving survival prognosis and treatments over traditional biomarkers.
The successful performance of scAB lies in utilizing a novel knowledge-guided matrix factorization model that incorporates phenotype information. Nonnegative matrix factorization (NMF) has been successfully applied to recover meaningful hidden patterns in bioinformatics and other fields (74,75). We treat the identification of phenotype-associated cells as a feature selection problem by assessing the contribution of each cell to the phenotype information of patients. As a part-based decomposition technique, NMF provides a natural way to reveal distinct patterns (76), that is the fine-grain phenotype-associated cell states in scAB. We showed that the identified distinct fine-grain cell subsets were characterized by shared or unique cancer and immunotherapy hallmarks in liver cancer and melanoma datasets. Moreover, these distinct cell subsets also exhibited distinct clinical significance. These results confirmed the known heterogeneity of tumors or other diseases in clinical implications. Combing all fine-grain cell subsets into one coarse-grain cell state allowed scAB to identify the conserved signatures and biological processes compared to other cells that are not highly correlated with phenotype of interest. Therefore, scAB provides a multiresolution dissection of phenotype-relevant cell subsets, which could offer new insights into the clinical significance of single cells compared to other methods like Scissor, scPrognosis and DEGAS.
In addition, scAB shows superior performance over the widely used survival models such as Cox regression model. There are two possible reasons. First, Cox regression model uses lasso penalty regularization for survival prognosis prediction, where lasso tends to select one gene or cell from a group of correlated genes or cells. However, genes or cells often coordinate together for biological functions and life activities. In contrast, the NMF utilized by scAB tend to recover biologically meaningful modules. When applying scAB to single-cell datasets, we also observed better performance of scAB in discriminating ICB responders from non-responders in melanoma and other cancer types, compared to the lasso-based method Scissor. Second, scAB adopts the penalty term of phenotype scores, that is the relative survival score in survival analysis. Instead of directly using the survival data of patients like in Scissor, we transformed the survival data to relative survival score of patients, which showed better performance than the Cox model in a previous study (12). Compared with the loss function of the Cox model, the relative survival score does not require the proportional hazard assumption and importantly includes two additional survival cases (i.e. early-censored–late-uncensored pairs and early-censored–late-censored pairs).
To establish a bridge between bulk data and single-cell data, scAB calculates a similarity matrix via Pearson correlation. We also explored the use of other similarity calculation strategies, including Spearman correlation, mutual information (MI), Euclidean similarity (ES1) and Euclidean similarity in the PCA-space (ES2). The comparisons of prediction performance among different similarity metrics indicate that Pearson correlation is a promising metric when integrating scRNA-seq data with bulk RNA-seq data (see details in Supplementary Text; Supplementary Figure S5). On the other hand, by randomly generating pseudo bulk RNA-seq datasets, we found that correlation-based metrics rather than Euclidean-based metrics can discriminate the real bulk data from the pseudo bulk data (see details in Supplementary Text; Supplementary Table S3), suggesting that correlation-based metrics can help to determine whether a bulk dataset was suitable to use for integrating single-cell genomics data. Taken together, while Pearson correlation was a simple measurement, these results indicated that it was suitable for the task of predicting clinically relevant cell subsets by linking single-cell genomics data with bulk data. This observation was consistent with the successful application of Pearson correlation or linear regression to linking single-cell and bulk data in many application scenarios from previous studies (9,77). However, instead of using these simple similarity metrics, more advanced methods such as transfer learning are likely better to link single-cell data with phenotype-annotated bulk data.
Given that clear positive correlations between the scRNA-seq data and the real bulk data rather than the pseudo bulk data were observed (Supplementary Table S3), one should select bulk data from closely related biological conditions and tissues, and most of bulk samples should be positively correlated with single cells. In addition, we explored the effect of the sample size of the bulk data on scAB’s performance. We thus constructed the bulk training set by randomly sampling n samples (n = 50,100,150,200,250,300) in the liver cancer bulk dataset (TCGA-LIHC), and then integrated each of them with single-cell data. We performed this procedure ten times and evaluated the predictive accuracy in the independent test set of bulk data (ICGC-LIRI). We found that the C-index values increased with the increase of the number of bulk samples, particularly when the number of bulk samples was greater than 200 (Supplementary Figure S6). These results showed that the increase of the sample size in the bulk training set can improve the prognostic performance of scAB model, and that few number of samples may reduce the prediction accuracy.
Although scAB exhibits good performance in identifying phenotype-related cell subsets and their signatures from single-cell genomics, further improvements could be done for linking single cells and bulk samples and for reducing the redundant fine-grain cell subsets with similar cancer hallmarks. To reduce the redundant cell subsets, orthogonality regularization may be helpful by adding it to the NMF model (78). In addition, scAB was tested on scRNA-seq and scATAC-seq data, but it is likely applicable to other modalities like single-cell proteomics. The scAB framework could be also coupled with our previous tool CellChat (79) to identify phenotype-associated cell-cell communication. Together, we anticipate that scAB will be widely used in the ever-growing single-cell studies and pave the way towards guiding the selection of effective therapeutic strategies targeting subsets of cells in precision medicine.
DATA AVAILABILITY
The datasets analyzed in this study are publicly available from the Gene Expression Omnibus (GEO) repository under the following accession numbers: GSE125449, GSE115978, GSE115978, GSE91061, GSE181815, GSE135222 and GSM4131779. Bulk RNA-seq datasets of liver cancer were downloaded from the Xena platform (TCGA-LIHC cohort) and the ICGC Data Porta (LIRI cohort). The melanoma immunotherapy datasets PRJEB23709 and MGSP were downloaded from https://github.com/donghaixiong/Immune_cells_analysis. The glioma bulk RNA-seq datasets were downloaded from the Xena platform (TCGA-GBMLGG cohort), and CGGA (http://www.cgga.org.cn) with accession codes mRNAseq_693, Rembrandt microarray, and mRNAseq_32, respectively. The Genomics of Drug Sensitivity in Cancer publicly available drug sensitivity data used in this study are available on the GDSC portal (https://www.cancerrxgene.org).
CODE AVAILABILITY
scAB is publicly available as an R package. Source codes, as well as tutorials have been deposited at the GitHub repository (https://github.com/jinworks/scAB).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Daniel William Haensel in Stanford University School of Medicine for his valuable suggestions on the manuscript. The numerical calculations in this paper have been done on the supercomputing system in the Supercomputing Center of Wuhan University.
Author contributions: S.J. and X.Z. conceived the project. Q.Z. conducted the research. Q.Z., S.J. and X.Z. wrote the paper. X.Z. and S.J. supervised the research. All authors approved the final manuscript.
Contributor Information
Qinran Zhang, School of Mathematics and Statistics, Wuhan University, Wuhan 430072, China; Hubei Key Laboratory of Computational Science, Wuhan University, Wuhan 430072, China.
Suoqin Jin, School of Mathematics and Statistics, Wuhan University, Wuhan 430072, China; Hubei Key Laboratory of Computational Science, Wuhan University, Wuhan 430072, China.
Xiufen Zou, School of Mathematics and Statistics, Wuhan University, Wuhan 430072, China; Hubei Key Laboratory of Computational Science, Wuhan University, Wuhan 430072, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [11831015]; Tian Yuan Mathematical Foundation [12126355]. Funding for open access charge: National Natural Science Foundation of China.
Conflict of interest statement. None declared.
REFERENCES
- 1. Suvà M.L., Tirosh I.. Single-Cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol. Cell. 2019; 75:7–12. [DOI] [PubMed] [Google Scholar]
- 2. Zhang J., Baran J., Cros A., Guberman J.M., Haider S., Hsu J., Liang Y., Rivkin E., Wang J., Whitty B.et al.. International cancer genome consortium data Portal–a one-stop shop for cancer genomics data. Database. 2011; 2011:bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., Bindal N., Beare D., Smith J.A., Thompson I.R.et al.. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic. Acids. Res. 2012; 41:D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao Z., Zhang K.-N., Wang Q., Li G., Zeng F., Zhang Y., Wu F., Chai R., Wang Z., Zhang C.et al.. Chinese glioma genome atlas (CGGA): a comprehensive resource with functional genomic data from chinese glioma patients. Genomics Proteomics Bioinformatics. 2021; 19:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim N., Eum H.H., Lee H.-O.. Clinical perspectives of single-cell RNA sequencing. Biomolecules. 2021; 11:1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan J.M., Quintanal-Villalonga Á., Gao V.R., Xie Y., Allaj V., Chaudhary O., Masilionis I., Egger J., Chow A., Walle T.et al.. Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell. 2021; 39:1479–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li X., Liu L., Goodall G.J., Schreiber A., Xu T., Li J., Le T.D.. A novel single-cell based method for breast cancer prognosis. PLOS Comput. Biol. 2020; 16:e1008133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao Y., Lin Y., Patrick E., Yang P., Yang J.Y.H.. scFeatures: Multi-view representations of single-cell and spatial data for disease outcome prediction. Bioinformatics. 2022; 38:4745–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sun D., Guan X., Moran A.E., Wu L.-Y., Qian D.Z., Schedin P., Dai M.-S., Danilov A.V., Alumkal J.J., Adey A.C.et al.. Identifying phenotype-associated subpopulations by integrating bulk and single-cell sequencing data. Nat. Biotechnol. 2021; 40:527–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson T.S., Yu C.Y., Huang Z., Xu S., Wang T., Dong C., Shao W., Zaid M.A., Huang X., Wang Y.et al.. Diagnostic evidence GAuge of single cells (DEGAS): a flexible deep transfer learning framework for prioritizing cells in relation to disease. Genome Med. 2022; 14:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li Y., Ma L., Wu D., Chen G.. Advances in bulk and single-cell multi-omics approaches for systems biology and precision medicine. Brief. Bioinform. 2021; 22:bbab024. [DOI] [PubMed] [Google Scholar]
- 12. Guan Y., Li H., Yi D., Zhang D., Yin C., Li K., Zhang P. A survival model generalized to regression learning algorithms. Nat. Comput. Sci. 2021; 1:433–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Butler A., Hoffman P., Smibert P., Papalexi E., Satija R.. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018; 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee D.D., Seung H.S.. Learning the parts of objects by non-negative matrix factorization. Nature. 1999; 401:788–791. [DOI] [PubMed] [Google Scholar]
- 15. Jin S., Zhang L., Nie Q.. scAI: an unsupervised approach for the integrative analysis of parallel single-cell transcriptomic and epigenomic profiles. Genome Biol. 2020; 21:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang L., Zhang J., Nie Q.. DIRECT-NET: an efficient method to discover cis-regulatory elements and construct regulatory networks from single-cell multiomics data. Sci. Adv. 2022; 8:eabl7393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schep A.N., Wu B., Buenrostro J.D., Greenleaf W.J.. chromVAR: inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods. 2017; 14:975–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu G., Wang L.-G., Han Y., He Q.-Y.. clusterProfiler: an r package for comparing biological themes among gene clusters. Omi. A J. Integr. Biol. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goldman M.J., Craft B., Hastie M., Repečka K., McDade F., Kamath A., Banerjee A., Luo Y., Rogers D., Brooks A.N.et al.. Visualizing and interpreting cancer genomics data via the xena platform. Nat. Biotechnol. 2020; 38:675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Xiong D., Wang Y., You M.. A gene expression signature of TREM2hi macrophages and γδ t cells predicts immunotherapy response. Nat. Commun. 2020; 11:5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma L., Hernandez M.O., Zhao Y., Mehta M., Tran B., Kelly M., Rae Z., Hernandez J.M., Davis J.L., Martin S.P.et al.. Tumor cell biodiversity drives microenvironmental reprogramming in liver cancer. Cancer Cell. 2019; 36:418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu J., Li P., Wang L., Li M., Ge Z., Noordam L., Lieshout R., Verstegen M.M.A., Ma B., Su J.et al.. Cancer-Associated fibroblasts provide a stromal niche for liver cancer organoids that confers trophic effects and therapy resistance. Cell. Mol. Gastroenterol. Hepatol. 2021; 11:407–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Q., Lou Y., Bai X.-L., Liang T.-B.. Immunometabolism: a novel perspective of liver cancer microenvironment and its influence on tumor progression. World J. Gastroenterol. 2018; 24:3500–3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang J.D., Nakamura I., Roberts L.R.. The tumor microenvironment in hepatocellular carcinoma: current status and therapeutic targets. Semin. Cancer Biol. 2011; 21:35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koundouros N., Poulogiannis G.. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer. 2020; 122:4–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee Y.-K., Jee B.A., Kwon S.M., Yoon Y.-S., Xu W.G., Wang H.-J., Wang X.W., Thorgeirsson S.S., Lee J.-S., Woo H.G.et al.. Identification of a mitochondrial defect gene signature reveals NUPR1 as a key regulator of liver cancer progression. Hepatology. 2015; 62:1174–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang X., Wang F., Huang Y., Ke K., Zhao B., Chen L., Liao N., Wang L., Li Q., Liu X.et al.. FGG promotes migration and invasion in hepatocellular carcinoma cells through activating epithelial to mesenchymal transition. Cancer Manag. Res. 2019; 11:1653–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Seo J., Jeong D.-W., Park J.-W., Lee K.-W., Fukuda J., Chun Y.-S.. Fatty-acid-induced FABP5/HIF-1 reprograms lipid metabolism and enhances the proliferation of liver cancer cells. Commun. Biol. 2020; 3:638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Röhrig F., Schulze A.. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer. 2016; 16:732–749. [DOI] [PubMed] [Google Scholar]
- 30. Makarova-Rusher O.V., Medina-Echeverz J., Duffy A.G., Greten T.F.. The yin and yang of evasion and immune activation in HCC. J. Hepatol. 2015; 62:1420–1429. [DOI] [PubMed] [Google Scholar]
- 31. Simon N., Friedman J., Hastie T., Tibshirani R.. Regularization paths for cox's proportional hazards model via coordinate descent. J. Stat. Softw. 2011; 39:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang J., Bajari R., Andric D., Gerthoffert F., Lepsa A., Nahal-Bose H., Stein L.D., Ferretti V.. The international cancer genome consortium data portal. Nat. Biotechnol. 2019; 37:367–369. [DOI] [PubMed] [Google Scholar]
- 33. Havel J.J., Chowell D., Chan T.A.. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer. 2019; 19:133–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jerby-Arnon L., Shah P., Cuoco M.S., Rodman C., Su M.-J., Melms J.C., Leeson R., Kanodia A., Mei S., Lin J.-R.et al.. A cancer cell program promotes t cell exclusion and resistance to checkpoint blockade. Cell. 2018; 175:984–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gide T.N., Quek C., Menzies A.M., Tasker A.T., Shang P., Holst J., Madore J., Lim S.Y., Velickovic R., Wongchenko M.et al.. Distinct immune cell populations define response to anti-pd-1 monotherapy and anti-pd-1/anti-ctla-4 combined therapy. Cancer Cell. 2019; 35:238–255. [DOI] [PubMed] [Google Scholar]
- 36. Long K.B., Beatty G.L.. Harnessing the antitumor potential of macrophages for cancer immunotherapy. Oncoimmunology. 2013; 2:e26860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qin Y., Liu H., Huang X., Huang L., Liao L., Li J., Zhang L., Li W., Yang J.. GIMAP7 as a potential predictive marker for pan-cancer prognosis and immunotherapy efficacy. J. Inflamm. Res. 2022; 15:1047–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sun L., Moore E., Berman R., Clavijo P.E., Saleh A., Chen Z., Van Waes C., Davies J., Friedman J., Allen C.T.. WEE1 kinase inhibition reverses G2/M cell cycle checkpoint activation to sensitize cancer cells to immunotherapy. Oncoimmunology. 2018; 7:e1488359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O’Donnell J.S., Massi D., Teng M.W.L., Mandala M.. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin. Cancer Biol. 2018; 48:91–103. [DOI] [PubMed] [Google Scholar]
- 40. Dong Z.-Y., Zhong W.-Z., Zhang X.-C., Su J., Xie Z., Liu S.-Y., Tu H.-Y., Chen H.-J., Sun Y.-L., Zhou Q.et al.. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin. Cancer Res. 2017; 23:3012–3024. [DOI] [PubMed] [Google Scholar]
- 41. Lin H., Wei S., Hurt E.M., Green M.D., Zhao L., Vatan L., Szeliga W., Herbst R., Harms P.W., Fecher L.A.et al.. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade–mediated tumor regression. J. Clin. Invest. 2018; 128:805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. DeNardo D.G., Ruffell B.. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 2019; 19:369–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tang Y., Hu Y., Niu Y., Sun L., Guo L.. CCL5 as a prognostic marker for survival and an indicator for immune checkpoint therapies in small cell lung cancer. Front. Med. 2022; 9:834725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mgrditchian T., Arakelian T., Paggetti J., Noman M.Z., Viry E., Moussay E., Van Moer K., Kreis S., Guerin C., Buart S.et al.. Targeting autophagy inhibits melanoma growth by enhancing NK cells infiltration in a CCL5-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E9271–E9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Huffman A.P., Lin J.H., Kim S.I., Byrne K.T., Vonderheide R.H.. CCL5 mediates CD40-driven CD4+ t cell tumor infiltration and immunity. JCI Insight. 2020; 5:e137263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Holt C. Study shows NKG7 mRNA improves tumor-killing ability of t cells. Oncol. Times. 2022; 44:18–18. [Google Scholar]
- 47. Wen T., Barham W., Li Y., Zhang H., Gicobi J.K., Hirdler J.B., Liu X., Ham H., Peterson Martinez K.E., Lucien F.et al.. NKG7 is a T-cell–Intrinsic therapeutic target for improving antitumor cytotoxicity and cancer immunotherapy. Cancer Immunol. Res. 2022; 10:162–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li X.-Y., Corvino D., Nowlan B., Aguilera A.R., Ng S.S., Braun M., Cillo A.R., Bald T., Smyth M.J., Engwerda C.R.. NKG7 is required for optimal antitumor T-cell immunity. Cancer Immunol. Res. 2022; 10:154–161. [DOI] [PubMed] [Google Scholar]
- 49. Han X., Wang Y., Sun J., Tan T., Cai X., Lin P., Tan Y., Zheng B., Wang B., Wang J.et al.. Role of CXCR3 signaling in response to anti-PD-1 therapy. EBioMedicine. 2019; 48:169–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chen Y., Huang Y., Gao X., Li Y., Lin J., Chen L., Chang L., Chen G., Guan Y., Pan L.K.et al.. CCND1 amplification contributes to immunosuppression and is associated with a poor prognosis to immune checkpoint inhibitors in solid tumors. Front. Immunol. 2020; 11:1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wagner N.B., Forschner A., Leiter U., Garbe C., Eigentler T.K.. S100B and LDH as early prognostic markers for response and overall survival in melanoma patients treated with anti-PD-1 or combined anti-PD-1 plus anti-CTLA-4 antibodies. Br. J. Cancer. 2018; 119:339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hauschild E., Brenner G., Mönig H.Christophers . Predictive value of serum S100B for monitoring patients with metastatic melanoma during chemotherapy and/or immunotherapy. Br. J. Dermatol. 1999; 140:1065–1071. [DOI] [PubMed] [Google Scholar]
- 53. Karlsson M.J., Costa Svedman F., Tebani A., Kotol D., Höiom V., Fagerberg L., Edfors F., Uhlén M., Egyhazi Brage S., Maddalo G.. Inflammation and apolipoproteins are potential biomarkers for stratification of cutaneous melanoma patients for immunotherapy and targeted therapy. Cancer Res. 2021; 81:2545–2555. [DOI] [PubMed] [Google Scholar]
- 54. Luo M., Wang F., Zhang H., To K.K.W., Wu S., Chen Z., Liang S., Fu L.. Mitomycin c enhanced the efficacy of PD-L1 blockade in non-small cell lung cancer. Signal Transduct. Target. Ther. 2020; 5:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Voorwerk L., Slagter M., Horlings H.M., Sikorska K., van de Vijver K.K., de Maaker M., Nederlof I., Kluin R.J.C., Warren S., Ong S.et al.. Immune induction strategies in metastatic triple-negative breast cancer to enhance the sensitivity to PD-1 blockade: the TONIC trial. Nat. Med. 2019; 25:920–928. [DOI] [PubMed] [Google Scholar]
- 56. Gao F., Zhang C., Qiu W.-X., Dong X., Zheng D.-W., Wu W., Zhang X.-Z.. PD-1 blockade for improving the antitumor efficiency of polymer-doxorubicin nanoprodrug. Small. 2018; 14:1802403. [DOI] [PubMed] [Google Scholar]
- 57. Miglietta G., Russo M., Duardo R.C., Capranico G.. G-quadruplex binders as cytostatic modulators of innate immune genes in cancer cells. Nucleic Acids Res. 2021; 49:6673–6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. De Magis A., Manzo S.G., Russo M., Marinello J., Morigi R., Sordet O., Capranico G.. DNA damage and genome instability by G-quadruplex ligands are mediated by r loops in human cancer cells. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:816–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Laino A.S., Betts B.C., Veerapathran A., Dolgalev I., Sarnaik A., Quayle S.N., Jones S.S., Weber J.S., Woods D.M.. HDAC6 selective inhibition of melanoma patient T-cells augments anti-tumor characteristics. J. Immunother. Cancer. 2019; 7:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Guilhamon P., Chesnelong C., Kushida M.M., Nikolic A., Singhal D., MacLeod G., Madani Tonekaboni S.A., Cavalli F.M., Arlidge C., Rajakulendran N.et al.. Single-cell chromatin accessibility profiling of glioblastoma identifies an invasive cancer stem cell population associated with lower survival. Elife. 2021; 10:e64090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. McLean C.Y., Bristor D., Hiller M., Clarke S.L., Schaar B.T., Lowe C.B., Wenger A.M., Bejerano G.. GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 2010; 28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fajardo Peñaranda, Meijer N.M., Kruyt F.A.E. The endoplasmic reticulum stress/unfolded protein response in gliomagenesis, tumor progression and as a therapeutic target in glioblastoma. Biochem. Pharmacol. 2016; 118:1–8. [DOI] [PubMed] [Google Scholar]
- 63. Desbaillets I., Diserens A.-C., Tribolet N.de, Hamou M.-F., Meir E.G.Van. Upregulation of interleukin 8 by Oxygen-deprived cells in glioblastoma suggests a role in leukocyte activation, chemotaxis, and angiogenesis. J. Exp. Med. 1997; 186:1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Auguste P., Gürsel D.B., Lemière S., Reimers D., Cuevas P., Carceller F., Di Santo J.P., Bikfalvi A.. Inhibition of fibroblast growth factor/fibroblast growth factor receptor activity in glioma cells impedes tumor growth by both Angiogenesis-dependent and -independent mechanisms1. Cancer Res. 2001; 61:1717–1726. [PubMed] [Google Scholar]
- 65. Liu Z., Ren Z., Zhang C., Qian R., Wang H., Wang J., Zhang W., Liu B., Lian X., Wang Y.et al.. ELK3: a new molecular marker for the diagnosis and prognosis of glioma. Front. Oncol. 2021; 11:608748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ishwaran H., Kogalur U.B., Blackstone E.H., Lauer M.S.. Random survival forests. Ann. Appl. Stat. 2008; 2:841–860. [Google Scholar]
- 67. Walsh K.M., Wiencke J.K., Lachance D.H., Wiemels J.L., Molinaro A.M., Eckel-Passow J.E., Jenkins R.B., Wrensch M.R.. Telomere maintenance and the etiology of adult glioma. Neuro. Oncol. 2015; 17:1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Portela M., Segura-Collar B., Argudo I., Sáiz A., Gargini R., Sánchez-Gómez P., Casas-Tintó S.. Oncogenic dependence of glioma cells on kish/TMEM167A regulation of vesicular trafficking. Glia. 2019; 67:404–417. [DOI] [PubMed] [Google Scholar]
- 69. Liang W., Fang J., Zhou S., Hu W., Yang Z., Li Z., Dai L., Tao Y., Fu X., Wang X.. The role of ubiquitin-specific peptidases in glioma progression. Biomed. Pharmacother. 2022; 146:112585. [DOI] [PubMed] [Google Scholar]
- 70. Hegazy A.M., Yamada D., Kobayashi M., Kohno S., Ueno M., Ali M.A.E., Ohta K., Tadokoro Y., Ino Y., Todo T.et al.. Therapeutic strategy for targeting aggressive malignant gliomas by disrupting their energy balance. J. Biol. Chem. 2016; 291:21496–21509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wang C., Haas M.A., Yeo S.K., Paul R., Yang F., Vallabhapurapu S., Qi X., Plas D.R., Guan J.-L.. Autophagy mediated lipid catabolism facilitates glioma progression to overcome bioenergetic crisis. Br. J. Cancer. 2021; 124:1711–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kiselev V.Y., Andrews T.S., Hemberg M.. Challenges in unsupervised clustering of single-cell RNA-seq data. Nat. Rev. Genet. 2019; 20:273–282. [DOI] [PubMed] [Google Scholar]
- 73. Abdelaal T., Michielsen L., Cats D., Hoogduin D., Mei H., Reinders M.J.T., Mahfouz A.. A comparison of automatic cell identification methods for single-cell RNA sequencing data. Genome Biol. 2019; 20:194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Pucher B.M., Zeleznik O.A., Thallinger G.G.. Comparison and evaluation of integrative methods for the analysis of multilevel omics data: a study based on simulated and experimental cancer data. Brief. Bioinform. 2019; 20:671–681. [DOI] [PubMed] [Google Scholar]
- 75. Baez-Ortega A., Gori K.. Computational approaches for discovery of mutational signatures in cancer. Brief. Bioinform. 2019; 20:77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Brunet J.-P., Tamayo P., Golub T.R., Mesirov J.P.. Metagenes and molecular pattern discovery using matrix factorization. Proc. Natl. Acad. Sci. U.S.A. 2004; 101:4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Jew B., Alvarez M., Rahmani E., Miao Z., Ko A., Garske K.M., Sul J.H., Pietiläinen K.H., Pajukanta P., Halperin E.. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 2020; 11:1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Pompili F., Gillis N., Absil P.-A., Glineur F.. Two algorithms for orthogonal nonnegative matrix factorization with application to clustering. Neurocomputing. 2014; 141:15–25. [Google Scholar]
- 79. Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.-H., Myung P., Plikus M.V., Nie Q.. Inference and analysis of cell-cell communication using cellchat. Nat. Commun. 2021; 12:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed in this study are publicly available from the Gene Expression Omnibus (GEO) repository under the following accession numbers: GSE125449, GSE115978, GSE115978, GSE91061, GSE181815, GSE135222 and GSM4131779. Bulk RNA-seq datasets of liver cancer were downloaded from the Xena platform (TCGA-LIHC cohort) and the ICGC Data Porta (LIRI cohort). The melanoma immunotherapy datasets PRJEB23709 and MGSP were downloaded from https://github.com/donghaixiong/Immune_cells_analysis. The glioma bulk RNA-seq datasets were downloaded from the Xena platform (TCGA-GBMLGG cohort), and CGGA (http://www.cgga.org.cn) with accession codes mRNAseq_693, Rembrandt microarray, and mRNAseq_32, respectively. The Genomics of Drug Sensitivity in Cancer publicly available drug sensitivity data used in this study are available on the GDSC portal (https://www.cancerrxgene.org).









