Abstract
Substance use disorder (SUD) is associated with severe health and social consequences. Continued drug use results in alterations of circuits within the mesolimbic dopamine system. It is critical to observe longitudinal impacts of SUD on neural activity in vivo to identify SUD predispositions, develop pharmaceuticals to prevent overdose, and help people suffering from SUD. However, implicated SUD associated areas are buried in deep brain which makes in vivo observation of neural activity challenging. The gradient index (GRIN) lens can probe these regions in mice and rats. In this short communications review, we will discuss how the GRIN lens can be coupled with other technologies such as miniaturized microscopes, fiberscopes, fMRI, and optogenetics to fully explore in vivo SUD research. Particularly, GRIN lens allows in vivo observation of deep brain regions implicated in SUD, differentiation of genetically distinct neurons, examination of individual cells longitudinally, correlation of neuronal patters with SUD behavior, and manipulation of neural circuits.
Keywords: GRIN lens, Addiction, Substance Use Disorder, miniscope
1.1. Introduction
Substance use disorder (SUD) involves chronic drug use [1] and can result in inabilities to meet responsibilities, brain damage, and overdose [2]. Researchers have used animal models to explore neural correlates of SUD and identify hallmarks such as “titrating” internal drug level [3], escalation [4], withdrawal [5], and drug-seeking [6]. Chronic SUD alters circuits within the mesolimbic system [7] such as nucleus accumbens (NAc) [8], amygdala [9], prefrontal cortex (PFC) [10], and hypothalamus [11]. Ideal technology for in vivo recording necessitates observation of deep brain regions, differentiation of genetically distinct cell types, longitudinal single-neuron tracking, and correlation of activity with behavior. In this short communication, we discuss how the gradient index (GRIN) lens can be implanted and used to achieve these criteria.
1.2. In Vivo Recording
Researchers have combined drug self-administration (SA) with in vivo recording technologies to correlate neural activity with behavior. Different techniques are useful for different circumstances and have pros and cons (Table 1). For instance, electrophysiology uses wires (steel, tungsten, platinum-iridium, etc.) to measure voltage changes in the extracellular environment. These act as a method to determine single-cell ‘spikes’ at high temporal resolution in deep brain regions [12, 13]. Neuronal subtypes are identified by distinct waveforms [14]. Drawbacks include data acquisition limited by channels, delicate microwires, and inability to differentiate between electrochemically similar neurons without optical manipulations [15]. Expression of immediate early gene C-fos labels large active neuronal populations and can be combined with transgenic animals [16, 17] at high spatial resolution but low temporal resolution [18] and activity does not always trigger fos expression [19]. Fast-scan cyclic voltammetry detects neurotransmission based on voltage oxidation at high temporal resolution in deep brain regions for specific neurotransmission studies [20, 21]. However, it cannot distinguish single neurons and can be clouded by high background current [22]. Fiber photometry quickly detects changes in population activity [23] using different types of sensors from deep brain regions [24] at axon terminals [25] and differentiates genetics at low cost [26] but lacks single-cell resolution [27]. The GRIN lens assists in vivo imaging in deep brain regions [28], using transgenic animals [29], and cost-effective open-source devices like miniaturized microscopes (miniscopes) [30–33] to record hundreds of individual neurons in vivo (Figure 1A) over months [34] which can be difficult to analyze. Neuron activity can be correlated with deep learning behavioral analyses [35], and optically manipulated with another LED without additional fibers [36]. However, care must be taken because physically damaging the lens or photobleaching neurons obscures activity.
Table 1:
In vivo recording techniques benefits/drawbacks
| In Vivo Techniques | Materials | Pros | Cons |
|---|---|---|---|
| Extracellular Electrophysiology |
|
|
|
| Protein c-Fos |
|
|
|
| Fast-Scan Cyclic Voltammetry |
|
|
|
| Fiber Photometry |
|
|
|
| GRIN Lens |
|
|
|
Figure 1A:
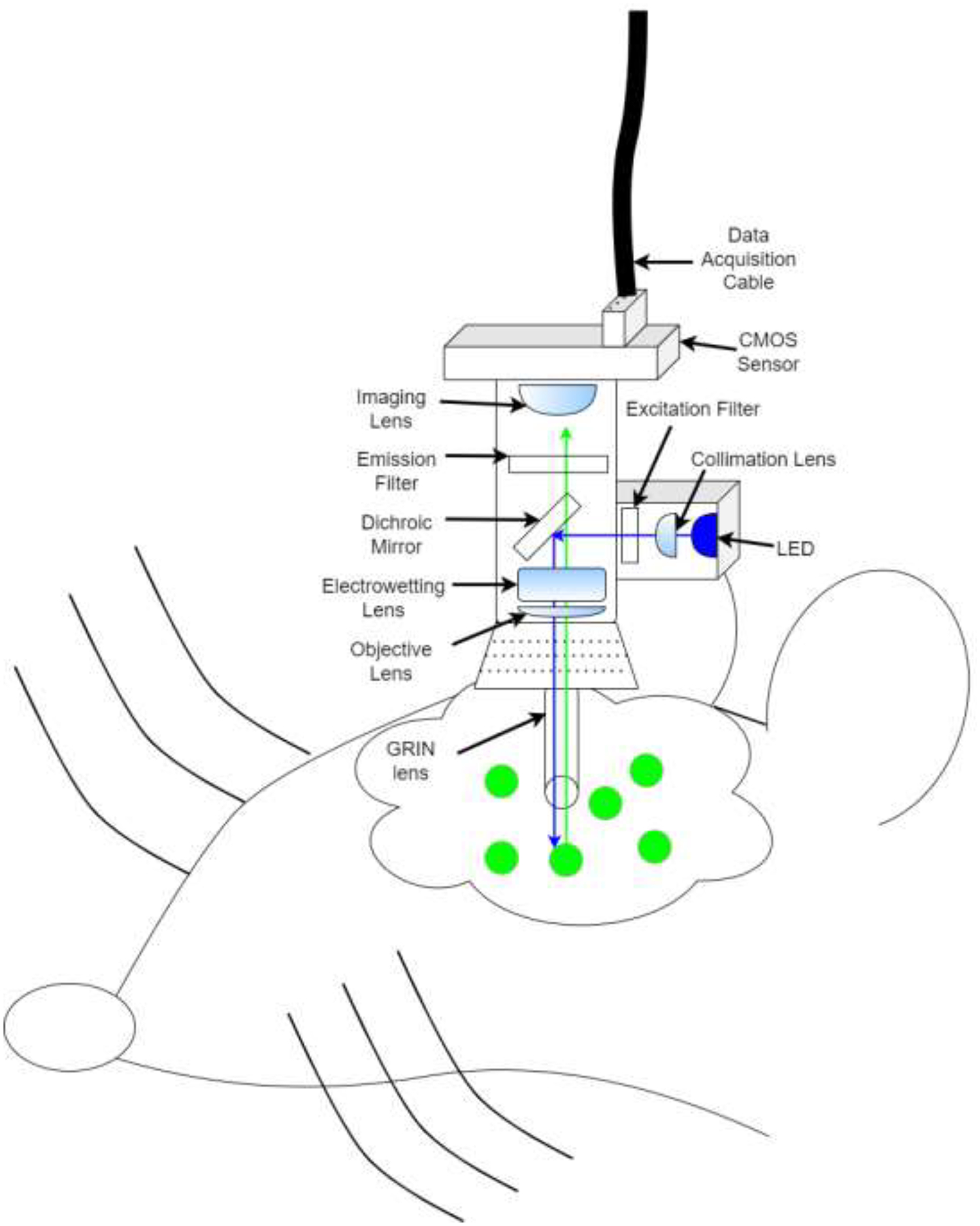
Depiction GRIN lens+ miniscope in vivo imaging. Miniscope is head mounted to a baseplate and allows for freely animal movement. A blue LED is triggered and light is reflected by a dichroic mirror into brain via the GRIN lens (see also, Figure 1b). Neurons infected with GCaMP emit green fluorescence while active in response to blue light. Green light (i.e., neural activity) is relayed back through the GRIN lens, past the dichroic mirror towards an imaging sensor and relayed offsite for analysis
1.3. GRIN lens overview
Light of a particular wavelength is transmitted from a source (e.g., LED) through a filter and dichroic mirror downward through the GRIN lens towards samples [37]). Active cells emit visible light in response to specific wavelengths and relay though the same GRIN lens towards a sensor/objective (Figure 1A) [38]. Traditional microscopy enhances signal visibility by refracting light through glass and transmits signals from glass to air by focusing light to a point [39]. The GRIN lens is crafted to refract light within one continuous glass tube to a single focal point outside of the skull (Figure 1B) [40, 41].
Figure 1B:
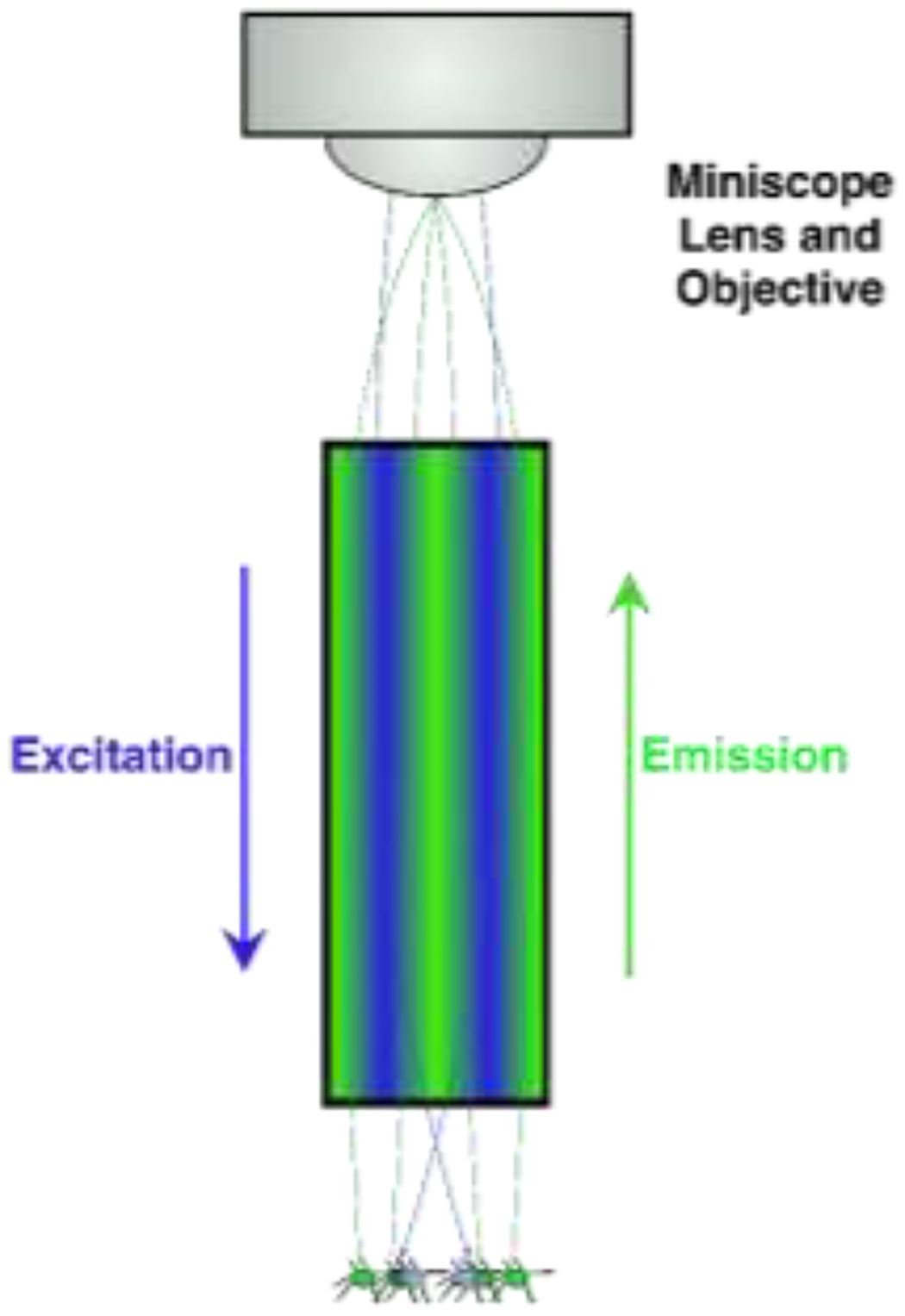
Schematic of an enlarged GRIN lens (seen in Figure 1A: Blue LED light is relayed through the GRIN lens. When neurons are active and simultaneously lit by the LED, these neurons fluoresce and produce light (green), some of which is gathered by the GRIN lens and relayed towards the objective for offsite analysis. Light is manipulated by the GRIN lens and meets at a specific focal point where the image is clear. Out-of-focus (i.e., focal length adjustment) can be via physical placement of the objectives or with an electrowetting lens (see Figure 1A) to resolve the image.
Without the GRIN lens light cannot pass deep tissue, and visualization > ~1mm [42] is restricted to ex vivo. Combining GRIN lens with 1-photon [34, 35, 43, 44] and 2-photon [45] excitation techniques helps reach target areas. Additionally, commercial GRIN lenses made from thallium-containing glass and salt melts can leach toxins but bio-compatible coatings reduce toxic-effects without dropping image quality [41, 46].
1.4. Samples of GRIN lens use in the field
1.4.1. GRIN Lens Combined with Miniscope Technology
Miniscope development revolutionized in vivo neuronal detection [34, 47, 48]. Originally, miniscopes used fibers to transmit light away from the organism [49] but have compartmentalized [50]. This decreased cost for behavioral microscopy and coincided with open-source miniscopes [31, 33, 50]. Miniscope and GRIN lens combination allows for single-cell visualization in deep regions such as NAc [34], and ventral tegmental area [43]. Potential GRIN lens limitations include out-of-focus fluorescence and poor optical sectioning (i.e., the ability to resolve samples embedded in tissue from noise [51], but are attenuated using an electrowetting lens to adjust the focal point [52, 53]. Motion blur can be adjusted in post-collection processes [34, 47, 48]. Major GRIN lens benefits involve targeting populations while preserving individualistic neural data. To target specific cell types transgenic animals can be bred to express cre-recombinase [54] in a multitude of cell types [55] and receptors such as μ-opioid [56], DRD1-[57], DRD2-[58], and other receptors [59].
An example experiment using cre-dependent viruses (e.g., GCaMP) injected into NAc (Figure 1A). In a DRD1-iCre rat only D1- expressing cells emit fluorophores when active and illuminated with a specific frequency [34]. Multiple genetic experiments using dual-color imaging through one GRIN lens [60] inject two spectrally distinct viruses: green-emitting cre-dependent GCaMP labels D1 cells and non-cre dependent red-emitting TdTomato [61] labels all cells. Intermittently shining distinct lights onto cells induces distinct fluorescence. Post-processing distinguishes D1 cells from remaining population. Deep learning analyses correlate cell-type specific activity associated with behavioral sequences [35] and can be theoretically applied to drug use during miniscope imaging [44, 62].
Optogenetic tools for excitation/inhibition can be integrated with the GRIN lens [36, 63]. Optogenetics use viruses containing proteins for excitation/inhibition of cells in response to specific wavelengths [64, 65]. Light optically manipulates neurons and resulting activity changes pass back to visualize activity in vivo. GRIN lens-coupled techniques can be applied in future studies to visualize neuronal progression throughout drug SA, abstinence, and relapse which have been pioneered using other in vivo techniques but can be adapted using GRIN lenses to record more neurons over longer periods of time.
1.4.2. GRIN lens in tandem with fMRI and fiberscope
An alternative GRIN lens approach is in conjunction with functional magnetic resonance imaging (fMRI) and a fiberscope. fMRI measures whole brain activity based on changes in cerebral blood flow [66]. A fiberscope uses calcium signaling to visualize neurons through an implanted GRIN lens, but relays towards CMOS-sensor [67, 68]. Researchers can use fiberscopes as a control for fMRI signal to understand effects of drug-associated cues across brain regions. For example, NAc and PrL are interconnected [69] and implicated in SUD [70, 71]. Following lens implants, animals would be trained for drug SA paired with a specific cue (e.g., a tone or odor). Animals would be placed in an fMRI (whole-brain) while asleep or head-fixed but awake as the animal is re-exposed to drug cues and fiberscope imaging (single-cell) would simultaneously carry signals outside of the fMRI [72]. Variations in drug-associated cue signals between the imaging techniques, PrL vs. NAc, awake vs. asleep, and correlations with relevant limbic brain regions are also possible.
1.5. Future Directions
The GRIN lens enables a wide range of possibilities for future deep-brain microscopy. New techniques and tools can extract precise neurophysiological information such as sensors which detect changes in endocannabinoid [73], serotonin [74], dopamine [75], or voltage [76]. A novel “clear optically matched panoramic access channel technique (COMPACT) works by inserting a GRIN lens into an implanted tube which can be adjusted dorsoventrally to capture refracted light [77]. This method reduces GRIN lens scar tissue and allows for multiple within-subject mesolimbic targets implicated in SUD (i.e., PrL and NAc [78]). Overall, the GRIN lens provides powerful research and technological developments to ultimately help people suffering from SUD.
Acknowledgments
This research was supported by NIH NIDA IRP. NJB, and YZ were supported by post-doctoral Fellowship from the Center on Compulsive Behaviors (CCB), National Institutes of Health. KAW was supported by NIDA IRP Scientific Director’s Fellowship for Diversity in Research (SDFDR). YL was supported by National Institute of Health (NIH) grants 5P20GM121310, R61NS115161, and UG3NS115608.
Abbreviations:
- SA
Self-Administration
- SUD
Substance Use Disorder
- NAc
Nucleus Accumbens
- PFC
Prefrontal Cortex
- PrL
Prelimbic Cortex
- GRIN
Gradient Index
- fMRI
functional magnetic resonance imaging
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].A. P. Association, Diagnostic and statistical manual of mental disorders (5th ed., text rev.). 2022. [Google Scholar]
- [2].De Marchi S, Cecchin E, Basile A, Bertotti A, Nardini R, and Bartoli E, “Renal tubular dysfunction in chronic alcohol abuse--effects of abstinence,” New England Journal of Medicine, vol. 329, no. 26, pp. 1927–1934, 1993. [DOI] [PubMed] [Google Scholar]
- [3].Yokel RA and Pickens R, “Drug level of d-and l-amphetamine during intravenous self-administration,” Psychopharmacologia, vol. 34, no. 3, pp. 255–264, 1974. [DOI] [PubMed] [Google Scholar]
- [4].Ahmed SH and Koob GF, “Transition from moderate to excessive drug intake: change in hedonic set point,” Science, vol. 282, no. 5387, pp. 298–300, Oct 09 1998. [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/9765157. [DOI] [PubMed] [Google Scholar]
- [5].Weiss F, Markou A, Lorang MT, and Koob GF, “Basal extracellular dopamine levels in the nucleus accumbens are decreased during cocaine withdrawal after unlimited-access self-administration,” Brain research, vol. 593, no. 2, pp. 314–318, 1992. [DOI] [PubMed] [Google Scholar]
- [6].Shaham Y, Shalev U, Lu L, De Wit H, and Stewart J, “The reinstatement model of drug relapse: history, methodology and major findings,” Psychopharmacology, vol. 168, no. 1, pp. 3–20, 2003. [DOI] [PubMed] [Google Scholar]
- [7].Koob GF and Volkow ND, “Neurocircuitry of addiction,” Neuropsychopharmacology, vol. 35, no. 1, pp. 217–238, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Carelli RM and Wightman RM, “Functional microcircuitry in the accumbens underlying drug addiction: insights from real-time signaling during behavior,” Current opinion in neurobiology, vol. 14, no. 6, pp. 763–768, 2004. [DOI] [PubMed] [Google Scholar]
- [9].See RE, Fuchs RA, Ledford CC, and McLaughlin J, “Drug addiction, relapse, and the amygdala,” Annals of the New York Academy of Sciences, vol. 985, no. 1, pp. 294–307, 2003. [DOI] [PubMed] [Google Scholar]
- [10].McFarland K and Kalivas PW, “The circuitry mediating cocaine-induced reinstatement of drug-seeking behavior,” Journal of Neuroscience, vol. 21, no. 21, pp. 8655–8663, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].DiLeone RJ, Georgescu D, and Nestler EJ, “Lateral hypothalamic neuropeptides in reward and drug addiction,” Life sciences, vol. 73, no. 6, pp. 759–768, 2003. [DOI] [PubMed] [Google Scholar]
- [12].Coffey KR et al. , “Electrophysiological evidence of alterations to the nucleus accumbens and dorsolateral striatum during chronic cocaine self-administration,” European Journal of Neuroscience, vol. 41, no. 12, pp. 1538–1552, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pachitariu M, Steinmetz N, Kadir S, and Carandini M, “Kilosort: realtime spike-sorting for extracellular electrophysiology with hundreds of channels,” BioRxiv, p. 061481, 2016. [Google Scholar]
- [14].Bakhurin KI, Mac V, Golshani P, and Masmanidis SC, “Temporal correlations among functionally specialized striatal neural ensembles in reward-conditioned mice,” Journal of neurophysiology, vol. 115, no. 3, pp. 1521–1532, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Coffey KR, Nader M, Bawa J, and West MO, “Homogeneous processing in the striatal direct and indirect pathways: single body part sensitive type II b neurons may express either dopamine receptor D1 or D2,” European Journal of Neuroscience, vol. 46, no. 8, pp. 2380–2391, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Barth AL, Gerkin RC, and Dean KL, “Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse,” Journal of Neuroscience, vol. 24, no. 29, pp. 6466–6475, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Karcz-Kubicha M et al. , “Enabling role of adenosine A1 receptors in adenosine A2A receptor-mediated striatal expression of c-fos,” European Journal of Neuroscience, vol. 18, no. 2, pp. 296–302, 2003. [DOI] [PubMed] [Google Scholar]
- [18].Morgan JI, Cohen DR, Hempstead JL, and Curran T, “Mapping patterns of c-fos expression in the central nervous system after seizure,” Science, vol. 237, no. 4811, pp. 192–197, 1987. [DOI] [PubMed] [Google Scholar]
- [19].Labiner DM, Butler LS, Cao Z, Hosford D, Shin C, and McNamara J, “Induction of c-fos mRNA by kindled seizures: complex relationship with neuronal burst firing,” Journal of Neuroscience, vol. 13, no. 2, pp. 744–751, 1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].John CE and Jones SR, “Fast scan cyclic voltammetry of dopamine and serotonin in mouse brain slices,” Electrochemical methods for neuroscience, 2007. [PubMed] [Google Scholar]
- [21].Robinson DL, Venton BJ, Heien ML, and Wightman RM, “Detecting subsecond dopamine release with fast-scan cyclic voltammetry in vivo,” Clinical chemistry, vol. 49, no. 10, pp. 1763–1773, 2003. [DOI] [PubMed] [Google Scholar]
- [22].Castagnola E et al. , “Real-time fast scan cyclic voltammetry detection and quantification of exogenously administered melatonin in mice brain,” Frontiers in bioengineering and biotechnology, vol. 8, p. 602216, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kim CK et al. , “Simultaneous fast measurement of circuit dynamics at multiple sites across the mammalian brain,” Nature methods, vol. 13, no. 4, pp. 325–328, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cui G et al. , “Deep brain optical measurements of cell type–specific neural activity in behaving mice,” Nature protocols, vol. 9, no. 6, p. 1213, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Bruno CA et al. , “pMAT: An open-source software suite for the analysis of fiber photometry data,” Pharmacology Biochemistry and Behavior, vol. 201, p. 173093, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Simone K, Füzesi T, Rosenegger D, Bains JS, and Murari K, “Open-source, cost-effective system for low-light in vivo fiber photometry,” Neurophotonics, vol. 5, no. 2, p. 025006, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lütcke H et al. , “Optical recording of neuronal activity with a genetically-encoded calcium indicator in anesthetized and freely moving mice,” Frontiers in neural circuits, p. 9, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Murray TA and Levene MJ, “Singlet gradient index lens for deep in vivo multiphoton microscopy,” Journal of biomedical optics, vol. 17, no. 2, p. 021106, 2012. [DOI] [PubMed] [Google Scholar]
- [29].Parkitna JR, Engblom D, and Schütz G, “Generation of Cre recombinase-expressing transgenic mice using bacterial artificial chromosomes,” in Gene Knockout Protocols: Springer, 2009, pp. 325–342. [DOI] [PubMed] [Google Scholar]
- [30].Aharoni D, Khakh BS, Silva AJ, and Golshani P, “All the light that we can see: a new era in miniaturized microscopy,” Nature methods, vol. 16, no. 1, pp. 11–13, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Barbera G et al. , “Spatially Compact Neural Clusters in the Dorsal Striatum Encode Locomotion Relevant Information,” Neuron, vol. 92, no. 1, pp. 202–213, Oct 05 2016, doi: 10.1016/j.neuron.2016.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Barbera G, Liang B, Zhang L, Li Y, and Lin DT, “A wireless miniScope for deep brain imaging in freely moving mice,” J Neurosci Methods, vol. 323, pp. 56–60, May 19 2019, doi: 10.1016/j.jneumeth.2019.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].de Groot A et al. , “NINscope, a versatile miniscope for multi-region circuit investigations,” Elife, vol. 9, p. e49987, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang L et al. , “Miniscope GRIN Lens System for Calcium Imaging of Neuronal Activity from Deep Brain Structures in Behaving Animals,” Curr Protoc Neurosci, vol. 86, no. 1, p. e56, Jan 2019, doi: 10.1002/cpns.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zhang Y et al. , “Detailed mapping of behavior reveals the formation of prelimbic neural ensembles across operant learning,” Neuron, vol. 110, no. 4, pp. 674–685. e6, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Stamatakis AM et al. , “Simultaneous optogenetics and cellular resolution calcium imaging during active behavior using a miniaturized microscope,” Frontiers in neuroscience, p. 496, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Akerboom J et al. , “Optimization of a GCaMP calcium indicator for neural activity imaging,” Journal of neuroscience, vol. 32, no. 40, pp. 13819–13840, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Levene MJ, Dombeck DA, Kasischke KA, Molloy RP, and Webb WW, “In vivo multiphoton microscopy of deep brain tissue,” Journal of neurophysiology, vol. 91, no. 4, pp. 1908–1912, 2004. [DOI] [PubMed] [Google Scholar]
- [39].Abramowitz M, Spring KR, Keller HE, and Davidson MW, “Basic principles of microscope objectives,” Biotechniques, vol. 33, no. 4, pp. 772–781, 2002. [DOI] [PubMed] [Google Scholar]
- [40].Leiner DC and Prescott R, “Correction of chromatic aberrations in GRIN endoscopes,” Applied Optics, vol. 22, no. 3, pp. 383–386, 1983. [DOI] [PubMed] [Google Scholar]
- [41].Yang Y et al. , “A Two-Step GRIN Lens Coating for In Vivo Brain Imaging,” Neurosci Bull, Mar 9 2019, doi: 10.1007/s12264-019-00356-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Benninger RK and Piston DW, “Two-photon excitation microscopy for the study of living cells and tissues,” Current protocols in cell biology, vol. 59, no. 1, pp. 4.11. 1–4.11. 24, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Engelhard B et al. , “Specialized coding of sensory, motor and cognitive variables in VTA dopamine neurons,” Nature, vol. 570, no. 7762, pp. 509–513, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Liang B et al. , “Distinct and Dynamic ON and OFF Neural Ensembles in the Prefrontal Cortex Code Social Exploration,” Neuron, Sep 13 2018, doi: 10.1016/j.neuron.2018.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Piyawattanametha W et al. , “In vivo brain imaging using a portable 2.9 g two-photon microscope based on a microelectromechanical systems scanning mirror,” Optics letters, vol. 34, no. 15, pp. 2309–2311, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kyogoku T, Suzuki T, and Mino M, “Ion beam assisted deposition of a thin film coating on a gradient-index lens array,” Applied optics, vol. 29, no. 28, pp. 4071–4076, 1990. [DOI] [PubMed] [Google Scholar]
- [47].Aharoni D and Hoogland TM, “Circuit investigations with open-source miniaturized microscopes: past, present and future,” Frontiers in cellular neuroscience, vol. 13, p. 141, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Jacob AD et al. , “A Compact Head-Mounted Endoscope for In Vivo Calcium Imaging in Freely Behaving Mice,” Current Protocols in Neuroscience, vol. 84, no. 1, p. e51, 2018. [DOI] [PubMed] [Google Scholar]
- [49].Helmchen F, “Miniaturization of fluorescence microscopes using fibre optics,” Experimental Physiology, vol. 87, no. 6, pp. 737–745, 2002. [DOI] [PubMed] [Google Scholar]
- [50].Ghosh KK et al. , “Miniaturized integration of a fluorescence microscope,” Nat Methods, vol. 8, no. 10, pp. 871–8, Sep 11 2011, doi: 10.1038/nmeth.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Agard DA, “Optical sectioning microscopy: cellular architecture in three dimensions,” Annual review of biophysics and bioengineering, vol. 13, no. 1, pp. 191–219, 1984. [DOI] [PubMed] [Google Scholar]
- [52].Acosta E, Gomez-Reino C, and Liñares J, “Effective radius and numerical aperture of GRIN lenses with revolution symmetry,” Applied optics, vol. 26, no. 15, pp. 2952–2955, 1987. [DOI] [PubMed] [Google Scholar]
- [53].Hayashi Y, Kobayakawa K, and Kobayakawa R, “Large-scale calcium imaging with a head-mount axial scanning 3D fluorescence microscope,” bioRxiv, 2021. [Google Scholar]
- [54].Sato Y et al. , “Establishment of Cre/LoxP recombination system in transgenic rats,” Biochemical and biophysical research communications, vol. 319, no. 4, pp. 1197–1202, 2004. [DOI] [PubMed] [Google Scholar]
- [55].Bäck S et al. , “Neuron-specific genome modification in the adult rat brain using CRISPR-Cas9 transgenic rats,” Neuron, vol. 102, no. 1, pp. 105–119. e8, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Bossert JM et al. , “Effect of selective lesions of nucleus accumbens μ-opioid receptor-expressing cells on heroin self-administration in male and female rats: a study with novel Oprm1-Cre knock-in rats,” bioRxiv, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Golden SA et al. , “Compulsive Addiction-like Aggressive Behavior in Mice,” Biol Psychiatry, vol. 82, no. 4, pp. 239–248, Aug 15 2017, doi: 10.1016/j.biopsych.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Yu Q, Liu Y-Z, Zhu Y-B, Wang Y-Y, Li Q, and Yin D-M, “Genetic labeling reveals temporal and spatial expression pattern of D2 dopamine receptor in rat forebrain,” Brain Structure and Function, vol. 224, no. 3, pp. 1035–1049, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Blanco-Centurion C, Bendell E, Zou B, Sun Y, Shiromani PJ, and Liu M, “VGAT and VGLUT2 expression in MCH and orexin neurons in double transgenic reporter mice,” IBRO reports, vol. 4, pp. 44–49, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chien Y-F et al. , “Dual GRIN lens two-photon endoscopy for high-speed volumetric and deep brain imaging,” Biomedical Optics Express, vol. 12, no. 1, pp. 162–172, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shaner NC, Steinbach PA, and Tsien RY, “A guide to choosing fluorescent proteins,” Nature methods, vol. 2, no. 12, pp. 905–909, 2005. [DOI] [PubMed] [Google Scholar]
- [62].Kahan A et al. , “Light-guided sectioning for precise in situ localization and tissue interface analysis for brain-implanted optical fibers and GRIN lenses,” Cell reports, vol. 36, no. 13, p. 109744, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Stamatakis AM et al. , “Miniature microscopes for manipulating and recording in vivo brain activity,” Microscopy, vol. 70, no. 5, pp. 399–414, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Deisseroth K, “Optogenetics,” Nature methods, vol. 8, no. 1, pp. 26–29, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gradinaru V, Thompson KR, and Deisseroth K, “eNpHR: a Natronomonas halorhodopsin enhanced for optogenetic applications,” Brain cell biology, vol. 36, no. 1, pp. 129–139, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].DeYoe EA, Bandettini P, Neitz J, Miller D, and Winans P, “Functional magnetic resonance imaging (FMRI) of the human brain,” Journal of neuroscience methods, vol. 54, no. 2, pp. 171–187, 1994. [DOI] [PubMed] [Google Scholar]
- [67].Abramov A, Minai L, and Yelin D, “Multiple-channel spectrally encoded imaging,” Optics express, vol. 18, no. 14, pp. 14745–14751, 2010. [DOI] [PubMed] [Google Scholar]
- [68].Mekhail SP, Arbuthnott G, and Chormaic SN, “Advances in fibre microendoscopy for neuronal imaging,” Optical Data Processing and Storage, vol. 2, no. 1, 2016. [Google Scholar]
- [69].Ma Y-Y et al. , “Bidirectional modulation of incubation of cocaine craving by silent synapse-based remodeling of prefrontal cortex to accumbens projections,” Neuron, vol. 83, no. 6, pp. 1453–1467, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Peoples LL and West MO, “Phasic firing of single neurons in the rat nucleus accumbens correlated with the timing of intravenous cocaine self-administration,” Journal of Neuroscience, vol. 16, no. 10, pp. 3459–3473, 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].West EA, Saddoris MP, Kerfoot EC, and Carelli RM, “Prelimbic and infralimbic cortical regions differentially encode cocaine-associated stimuli and cocaine-seeking before and following abstinence,” European Journal of Neuroscience, vol. 39, no. 11, pp. 1891–1902, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Schlegel F et al. , “Fiber-optic implant for simultaneous fluorescence-based calcium recordings and BOLD fMRI in mice,” Nature protocols, vol. 13, no. 5, pp. 840–855, 2018. [DOI] [PubMed] [Google Scholar]
- [73].Dong A et al. , “A fluorescent sensor for spatiotemporally resolved imaging of endocannabinoid dynamics in vivo,” Nature biotechnology, vol. 40, no. 5, pp. 787–798, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Wan J et al. , “A genetically encoded sensor for measuring serotonin dynamics,” Nature neuroscience, vol. 24, no. 5, pp. 746–752, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Sun F et al. , “Next-generation GRAB sensors for monitoring dopaminergic activity in vivo,” Nature methods, vol. 17, no. 11, pp. 1156–1166, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Nakajima R, Jung A, Yoon B-J, and Baker BJ, “Optogenetic monitoring of synaptic activity with genetically encoded voltage indicators,” Frontiers in Synaptic Neuroscience, vol. 8, p. 22, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wei B, Wang C, Cheng Z, Lai B, Gan W-B, and Cui M, “Clear optically matched panoramic access channel technique (COMPACT) for large-volume deep brain imaging,” Nature Methods, vol. 18, no. 8, pp. 959–964, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Paxinos G, Watson CR, and Emson PC, “AChE-stained horizontal sections of the rat brain in stereotaxic coordinates,” Journal of neuroscience methods, vol. 3, no. 2, pp. 129–149, 1980. [DOI] [PubMed] [Google Scholar]


