Abstract
Akkermansia muciniphila is a human intestinal tract bacterium that plays an important role in the mucus layer renewal. Several studies have demonstrated that it is a modulator for gut homeostasis and a probiotic for human health. The Akkermansia genus contains two species with standing in nomenclature but their genomic diversity remains unclear. In this study, eight new Akkermansia sp. strains were isolated from the human gut. Using the digital DNA-DNA hybridization (dDDH), average nucleotide identity (ANI) and core genome-based phylogenetic analysis applied to 104 A. muciniphila whole genomes sequences, strains were reclassified into three clusters. Cluster I groups A. muciniphila strains (including strain ATCC BAA-835T as type strain), whereas clusters II and III represent two new species. A member of cluster II, strain Marseille-P6666 differed from A. muciniphila strain ATCC BAA-835T and from A. glycaniphila strain PytT in its ability to grow in microaerophilic atmosphere up to 42 °C, to assimilate various carbon sources and to produce acids from a several compounds. The major fatty acids of strain Marseille-P6666 were 12-methyl-tetradecanoic and pentadecanoic acids. The DNA G + C content of strain Marseille-P6666 was 57.8%. On the basis of these properties, we propose the name A. massiliensis sp. nov. for members of cluster II, with strain Marseille-P6666T (= CSUR P6666 = CECT 30548) as type strain. We also propose the name “Candidatus Akkermansia timonensis” sp. nov. for the members of cluster III, which contains only uncultivated strains, strain Akk0196 being the type strain.
Subject terms: Microbiology, Bacterial evolution, Bacterial genomics
Introduction
The human gastro-intestinal tract is a complex ecosystem in which lives a dynamic microbial community. Most of this microbiota is represented by bacteria that play a crucial role in the body physiological balance, with an important impact on immunity, various metabolic processes and host health1. Akkermansia muciniphila, described in 2004 by Derien et al.2, is a bacterium able to use mucin as sole carbon source. It participates in the renewal of mucus and the integrity of the intestinal barrier3. It is a commensal of the intestinal flora (3–5%)4 and its abundance was suggested as a biomarker of an healthy gut microbiota5,6. In contrast, the number of studies depicting its decrease or depletion in inflammatory bowel diseases, such as Crohn's disease, colorectal cancer, ulcerative colitis7 and metabolic diseases including obesity8 and type 2 diabetes9, is constantly increasing. The reconstitution of its abundance by oral administration (murine model) has been correlated to several beneficial effects such as a reduction of hepatic injury, steatosis and neutrophil infiltration in animals with alcoholic liver disease10, to the decrease of atherosclerotic lesions and the metabolic endotoxemia-induced inflammation11, to an attenuation of dextran sulfate sodium (DSS)-induced acute colitis12, to the suppression of colonic tumorigenesis in ApcMin/+ mice, and to a reduced growth of implanted HCT116 or CT26 tumors in nude mice13.
Akkermansia muciniphila also possesses beneficial probiotic activities on the mucus layer3,14. Supplementation in A. muciniphila was demonstrated to significantly reduce the severity of nausea, vomiting, constipation during pregnancy, to play a key role in the regulation of metabolic functions to prevent obesity15,16, and to enhance glucose tolerance and attenuate adipose tissue inflammation17 and reduce diabetes incidence18.
The Akkermansia genus contains two validated species, i.e., A. muciniphila and A. glycaniphila2,19. In 2021, two putative new Akkermansia species, “Candidatus A. intestinavium” and “Candidatus A. intestinigallinarum” were described by Gilroy et al.20. Kim et al. observed a great genomic diversity within the species A. muciniphila21. Likewise, Orellana et al., described a potentially new genus within the family Akkermansiaceae, “Candidatus Mariakkermansia forsetii”, from a metagenomic analysis of surface seawater samples22. However, despite its role as a probiotic confirmed by numerous studies and its benefits on host health, very few Akkermansia species are described. In a project to search for new Akkermansia muciniphila strains and analyze their genomic diversity, we discovered two strains that may be members of a new Akkermansia species. Here, we report a taxonogenomic description of this new species and its phenotypic and biochemical characteristics. An analysis of complete A. muciniphila genomes available in public databases as well as those from eight isolates from our collection, allowed us to reclassify strains within this genus.
Results and discussion
Strain identification and phylogenetic analyses
All strains isolated in this study were first identified as Akkermansia muciniphila by MALDI-TOF–MS when we comparing their peptidic profiles to those available in the Bruker database. After sequencing the 8 strains, the 16S rRNA sequences of each isolate were extracted and compared to those of closely related species in the NCBI database (https://www.ncbi.nlm.nih.gov/). Strains Marseille-P6666, Marseille-P9185, Marseille-P5162, Marseille-P6566, Marseille-P7245, Marseille-P9184, Marseille-P9642 and Marseille-Q2586 showed highest 16S rRNA sequence similarities of 99.20, 98.95, 99.73, 99.79, 99.72, 99.96, 99.79 and 99.86%, respectively, with A. muciniphila strain ATCC BAA-835T (AY271254).
Phenotypic and biochemical analysis
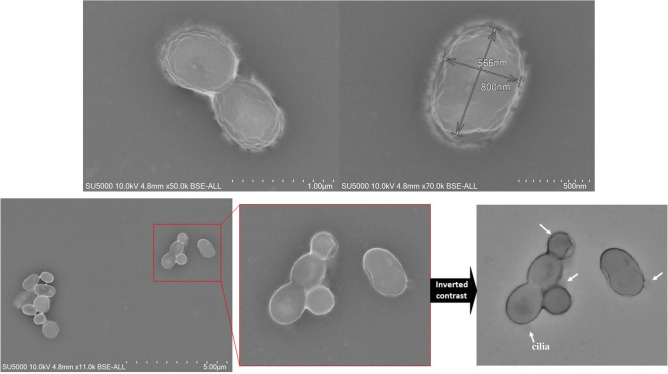
Cells from strain Marseille-P6666 were rod-shaped (0.5 × 0.8 μm), motile and Gram-negative (Fig. 1). In the presence of fluid, the cells turn on themselves and self-propel. Multiple cilia can be observed on the bacterial cell surface (Fig. 1). Colonies grown on Columbia agar plates appeared white, non-haemolytic, and circular with a diameter of 0.5 mm after 72 h of incubation. Optimal growth from strain Marseille-P6666 grew between from 37 to 42 °C, at a pH ranging from 6 to 7.5, and in the presence of 0 to 5 g/l NaCl. Strain Marseille-P6666 was able to grow in microaerophilic atmosphere, which enables it to survive in the mucus layer of the gastrointestinal tract23. The new isolate was able to use mucin as a solo carbon source.
Figure 1.
Transmission electron microscopy of Akkermansia massiliensis strain Marseille-P6666.
Cells were catalase-positive and oxidase-negative. Using an API ZYM strip (bioMérieux), production of alkaline phosphatase, esterase (C4), acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, β-glucuronidase and N-acetyl-β-glucosaminidase were positive. Using an API 20NE strips (bioMérieux), strain Marseille-P6666 was able to hydrolyze esculin and to produce β-galactosidase. Using an API 20A strip (bioMérieux), positive reactions were obtained for acidification of d-glucose, d-mannitol, d-lactose, d-maltose, esculin ferric citrate and d-mannose. Using an API 50CH strips (bioMérieux), strain Marseille-P6666 utilized l-arabinose, d-ribose, d-galactose, d-glucose, d-fructose, d-mannose, d-mannitol, d-sorbitol, N-acetylglucosamine, salicin, esculin ferric citrate, cellobiose, d-maltose, d-lactose, d-melibiose, amygdaline, d-saccharose, d-trehalose, arbutine, gentiobiose, d-turanose, d-tagalose and potassium gluconate as sole carbon sources. All negative properties obtained from the API ZYM, 50CH, 20A and 20NE strips were summarized in the description of the novel species. Furthermore, the physiological and biochemical characteristics of strain Marseille-P6666T were summarized and compared to those of other closely related species in Table 1. Strain Marseille-P6666 was found to be susceptible to trimethoprim-sulfamethoxazole, doxycycline, rifampicin, clindamycin, amoxicillin, oxacillin and benzylpenicillin but susceptible to vancomycin, amikacin, ciprofloxacin, tobramycin, ceftriaxone and ceftazidime. The major cellular fatty acids of strain Marseille-P6666 were saturated structures: 12-methyl-tetradecanoic acid (58.4%), pentadecanoic acid (15.8%) and 12-methyl-Tridecanoic acid (6%). The two major fatty acid namely anteiso-C15:0 and C15:0 are similar for strains Marseille-P6666, MucT and PytT (Table 2). Short fatty acids such as acetic acid (12 ± 7 mM) and propanoic acid (5 ± 2 mM) were produced. Strain Marseille-P6666 produced various polar lipids classes such as sphingomyelins, N-acyl ethanolamines, acyl carnitine, phosphatidylethanolamine, Lysophosphatidyléthanolamine, phosphatidylcholine, lysophosphatidylcholine, ceramides—glycero lipids, fatty acyls—glycero lipids, phosphatidic acid and several unknown structures and phospholipids (Supplementary Fig. 1 and Supplementary Table 1).
Table 1.
Physiological features of Akkermansia massiliensis strain Marseille-P6666 (a), Akkermansia muciniphila strain ATCC BAA-835T (b), Akkermansia glycaniphila strain PytT (c), Verrucomicrobium spinosum strain DSM4136T (d), Prosthecobacter debontii strain ATCC 700200T (e); (+): positive; (−): negative; (*): weak reaction; NA not available data.
| Properties | a | B | C | d | E |
|---|---|---|---|---|---|
| Size (μm) | 0.5 × 0.8 μm | 0.6 × 1.0 | 0.6 × 1.0 | 0.8–1.0 × 1.0–3.8 | 2–8 × 0.5 |
| Gram | − | − | − | − | − |
| Tolerance to oxygen | + | − | − | + | − |
| Motility | + | − | − | − | − |
| Endospore formation | − | − | − | NA | NA |
| Catalase | + | NA | + | + | NA |
| Nitrate reductase | − | NA | NA | − | NA |
| Urease | − | NA | − | + | NA |
| Indole production | − | NA | − | NA | NA |
| Aesculin hydrolysis | + | NA | + | + | NA |
| β-galactosidase | + | NA | NA | NA | NA |
| N-acetylglucosamine | + | + | + * | + | NA |
| Ribose | + | NA | NA | − | + |
| Arabinose | − | NA | − | − | − |
| Rhamnose | − | − | − | + | + |
| Raffinose | − | NA | − | + | − |
| Inulin | − | NA | NA | − | NA |
| Maltose | + | − | + | + | + |
| Cellobiose | + | − | − | + | + |
| Melezitose | − | NA | − | + | NA |
| Melibiose | − | NA | + | + | |
| Trehalose | + | NA | − | + | + |
| Galactose | + | − | + * | + | + |
| d-glucose | + | + | + * | + | + |
| d-fructose | + | − | − | + | − |
| Mannose | + | NA | − | + | + |
| Mannitol | + | NA | − | − | NA |
| Xylose | − | − | + | + | + |
| Lactose | + | − | + | + | + |
| Major faty acid product | Anteiso-C15 : 0 | Anteiso-C15 : 0 | Anteiso-C15 : 0 | NA | NA |
| DNA G + C content (mol%) | 57.8 | 55.6 | 58.2 | 57.9–59.3 | 57.1 |
Table 2.
Cellular fatty acid composition (%) of Akkermansia massiliensis strain Marseille-P6666, Akkermansia muciniphila strain ATCC BAA-835T and Akkermansia glycaniphila strain PytT.
| Fatty acids | Name | Mean relative %a | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| C 15:0 anteiso | 12-Methyl-tetradecanoic acid | 58.4 ± 6.1 | 53.6 | 42.3 |
| C 15:0 | Pentadecanoic acid | 15.8 ± 3.4 | 9.2 | 13.1 |
| C 14:0 iso | 12-Methyl-tridecanoic acid | 6.0 ± 1.5 | 3.1 | 1.3 |
| C 16:0 | Hexadecanoic acid | 6.0 ± 0.5 | 4 | 10.7 |
| C 17:0 | Heptadecanoic acid | 5.2 ± 1.2 | 6.3 | 0.5 |
| C 18:0 | Octadecanoic acid | 1.8 ± 0.2 | 1.1 | 1.1 |
| C 16:0 iso | 14-Methyl-pentadecanoic acid | 1.8 ± 0.2 | 0.5 | 0.6 |
| C 17:0 anteiso | 14-Methyl-hexadecanoic acid | 1.5 ± 0.2 | 4.5 | 2.5 |
| C 15:0 iso | 13-Methyl-tetradecanoic acid | TR | 2.6 | 1.2 |
| C 18:1n9 | 9-Octadecenoic acid | TR | - | - |
| C 16:1 iso | 14-Methyl-pentadecenoic acid | TR | - | - |
| C 14:0 | Tetradecanoic acid | TR | 0.7 | 3.9 |
| C 13:0 | Tridecanoic acid | TR | 0.2 | 0.2 |
| C 18:2n6 | 9,12-Octadecadienoic acid | TR | − | − |
| C 18:1n7 | 11-Octadecenoic acid | TR | − | − |
| C 13:0 anteiso | 10-Methyl-dodecanoic acid | TR | − | − |
| C 12:0 | Dodecanoic acid | TR | − | − |
Genome analysis and comparison
The genome properties of the eight strains sequenced in this study are summarized in Table 3. Genome sequences of these isolates had different sizes ranging from 2,740,501 to 3,280,190 bp.
Table 3.
Genomic characteristics of studied Akkermansia species and other closely-related species.
| Strain name | Genbank accession | Size, Mb | G + C mol% | Gene count |
|---|---|---|---|---|
| Marseille-P6666 | JAMGSI000000000 | 3,280,190 | 57.8% | 2,793 |
| Marseille-P9185 | JAMYIA000000000 | 3,193,520 | 57.9% | 2,672 |
| Marseille-P5162 | JAMZOC000000000 | 2,997,637 | 55.1% | 2,648 |
| Marseille-P6566 | JAMZOB000000000 | 2,784,480 | 55.2% | 2,402 |
| Marseille-P7245 | JAMGSH000000000 | 2,784,249 | 55.4% | 2,381 |
| Marseille-P9184 | JAMGSG000000000 | 2,814,301 | 55.6% | 2,471 |
| Marseille-P9642 | JAMGSF000000000 | 2,740,501 | 55.7% | 2,343 |
| Marseille-Q2586 | JAMGSE000000000 | 2,812,384 | 55.8% | 2,422 |
| ATCC BAA-835T | CP001071.1 | 2,664,051 | 55.6% | 2,183 |
| PytT | LIGX00000000 | 3,074,078 | 58.2% | 2,527 |
| ChiGjej6B6-8097 | DXEH00000000.1 | 2,313,406 | 65.09% | 1,836 |
| 14975 | DXFQ00000000.1 | 2,129,131 | 63.4% | 1,813 |
| Akk0196 | CP072051.1 | 3,212,887 | 56.7% | 2,705 |
| Akk7 | 20100303_Bin_52_1 | 2,025,900 | 51.1% | 1,838 |
The genome of Marseille-P6666 is 3,280,190 bp long with an average G + C content of 57.8%. It is composed of three contigs. Of the 2793 predicted genes, 2726 were protein-coding genes, 9 RNAs, 1 tmRNA and 57 tRNA genes. Circular maps of this strain are illustrated in Fig. 2. The genome of Marseille-P6666 is greater than that of A. muciniphila ATCC BAA-835T (= MucT), (2,664,051 bp) and A. glycaniphila pytT (3,074,078 bp). The G + C content of Marseille-P6666 is larger than that of A. muciniphila ATCC BAA-835T (55.6%) but smaller than that of A. glycaniphila PytT (58.2%). Distribution of genes into COGs functional categories between Marseille-P6666 and the other closely related species was presented in Fig. 3 and Table 4. The number of genes from each COG category was greater for strain Marseille-P6666 than for A. muciniphila strain ATCC BAA-835T, notably for genes encoding cell wall and membrane biogenesis, energy production and conversion, defense mechanisms, and transport and metabolism of carbohydrates, amino acids, nucleotides and inorganic ions. This is consistent with the fact that strain Marseille-P6666 has more coding genes (1983 genes) than A. muciniphila strain ATCC BAA-835T (1084 genes). The phylogenetic tree based on core genome identified tree clusters (I, II, and III), with strains considered previously belonging to the A. muciniphila species. Among the Akkermansia strains isolated in this study, six of them, clustered with A. muciniphila strain ATCC BAA-835T (Cluster I). Two strains formed a second cluster (Cluster II) with other strains previously classified as belonging to the A. muciniphila species. Comparison of the genomes from members of the clusters I, II and III with A. muciniphila strain ATCC BAA-835T, showed dDDH values higher than 70% with cluster I members (range 74.80% to 100%) but lower than 70% for cluster II and III members (range 33.8–34.2% and 17.1–24.9%, respectively, Supplementary Table 2). ANI values between cluster I, II and III isolates were 97–100%, 88% and 82% with A. muciniphila ATCC BAA-835T, respectively (Supplementary Fig. 2). ANI values between the three clusters (I, II and III) were significantly lower than the proposed cutoff value of 95% for defining a bacterial species24,25. A recent study even redefined the ANI threshold value to 96.5% for creating a new bacterial species26. In contrast, all ANI values within a given cluster were higher than 97%. Therefore, the distribution of strains previously considered as A. muciniphila into three distinct species was clearly supported by the genome-based phylogenetic analysis and the of DDH and ANI values.
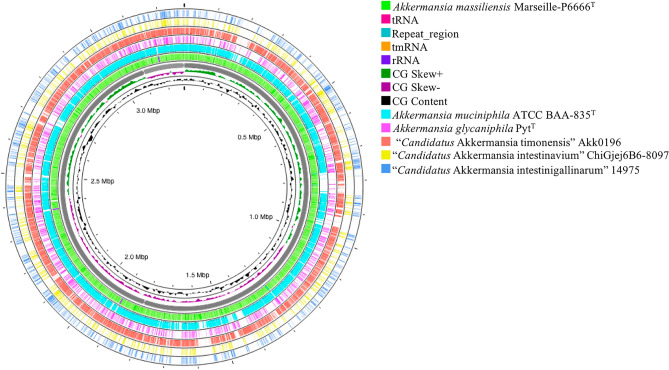
Figure 2.
Graphical circular map of A. massiliensis Marseille-P6666 uses as reference genome to compare the genomic organization of the closest species of published Akkermansia such as Akkermansia muciniphila strain ATCC BAA-835T, Akkermansia glycaniphila strain PytT, Candidatus Akkermansia intestinavium” strain ChiGjej6B6-8097T and Candidatus Akkermansia intestinigallinarum” strain 14975T, and the new species candidate “Candidatus Akkermansia timonensis” strain Akk0196. The different colors used in the circular map are indicated in the legend on the right part of the figure.
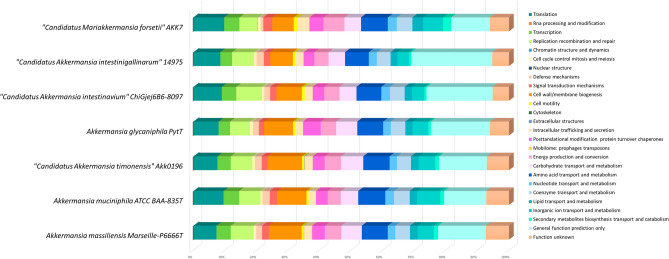
Figure 3.
Distribution of predicted protein-coding genes from Akkermansia muciniphila strain ATCC BAA-835T (Cluster I), Akkermansia massiliensis strain Marseille-P6666 (Cluster II), “Candidatus Akkermansia timonensis” strain Akk0196 (Cluster III), Akkermansia glycaniphila strain PytT, and other closely-related species, in COG categories.
Table 4.
Numbers of genes associated with the 25 functional categories in clusters of orthologous groups (COG).
| Description | COG | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|---|
| Translation | [J] | 144 | 103 | 147 | 151 | 140 | 130 | 133 |
| Rna processing and modification | [A] | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Transcription | [K] | 90 | 54 | 82 | 69 | 72 | 57 | 66 |
| Replication, recombination and repair | [L] | 144 | 73 | 133 | 120 | 127 | 107 | 79 |
| Chromatin structure and dynamics | [B] | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Cell cycle control, mitosis and meiosis | [D] | 16 | 7 | 16 | 19 | 12 | 12 | 13 |
| Nuclear structure | [Y] | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Defense mechanisms | [V] | 37 | 25 | 42 | 35 | 29 | 35 | 11 |
| Signal transduction mechanisms | [T] | 43 | 25 | 36 | 32 | 31 | 31 | 39 |
| Cell wall/membrane biogenesis | [M] | 206 | 101 | 212 | 174 | 126 | 110 | 93 |
| Cell motility | [N] | 14 | 7 | 13 | 15 | 14 | 13 | 18 |
| Cytoskeleton | [Z] | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Extracellular structures | [W] | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Intracellular trafficking and secretion | [U] | 51 | 26 | 51 | 46 | 40 | 39 | 50 |
| Posttanslational modification, protein turnover,chaperones | [O] | 79 | 40 | 73 | 106 | 57 | 52 | 61 |
| Mobilome: prophages, transposons | [X] | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Energy production and conversion | [C] | 104 | 45 | 101 | 93 | 76 | 69 | 91 |
| Carbohydrate transport and metabolism | [G] | 129 | 60 | 137 | 128 | 84 | 80 | 72 |
| Amino acid transport and metabolism | [E] | 163 | 93 | 162 | 156 | 123 | 115 | 116 |
| Nucleotide transport and metabolism | [F] | 47 | 30 | 46 | 46 | 43 | 39 | 39 |
| Coenzyme transport and metabolism | [H] | 93 | 54 | 86 | 88 | 73 | 67 | 68 |
| Lipid transport and metabolism | [I] | 48 | 25 | 47 | 42 | 36 | 33 | 46 |
| Inorganic ion transport and metabolism | [P] | 102 | 80 | 102 | 96 | 61 | 57 | 87 |
| Secondary metabolites biosynthesis, transport and catabolism | [Q] | 26 | 13 | 22 | 15 | 11 | 10 | 34 |
| General function prediction only | [R] | 301 | 145 | 296 | 357 | 328 | 393 | 169 |
| Function unknown | [S] | 145 | 78 | 136 | 115 | 81 | 81 | 83 |
Akkermansia massiliensis strain Marseille-P6666 (1), Akkermansia muciniphila strain ATCC BAA-835T (2), Candidatus Akkermansia timonensis strain Akk0196 (3), Akkermansia glycaniphila strain PytT (4), “Candidatus Akkermansia intestinavium” strain ChiGjej6B6-8097T (5), “Candidatus Akkermansia intestinigallinarum” strain 14975T (6) and “Candidatus Mariakkermansia forsetii” strain AKK7 (7).
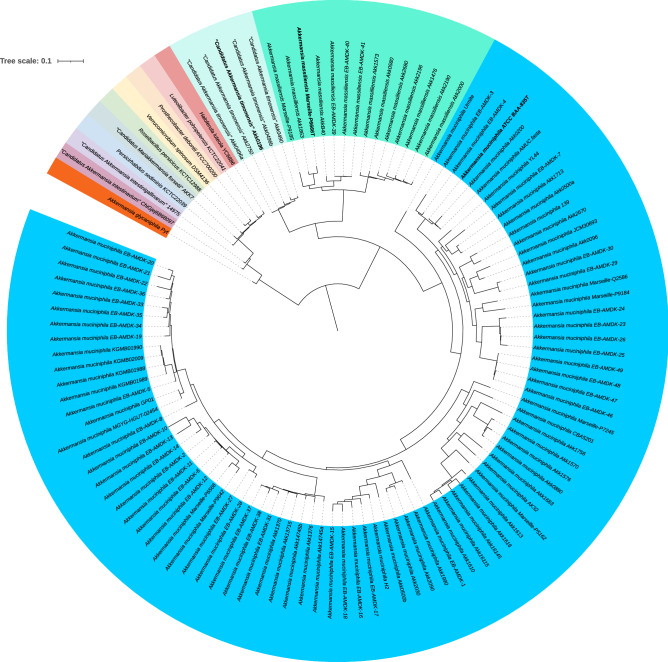
Hence, our genome analysis results strongly suggest that strains currently classified as A. muciniphila belong to three distinct species. However, the 98.7% 16SrRNA sequence similarity threshold defined to classify a bacterial species27 cannot discriminate between species in the Akkermansia genus. This highlights the limitation of 16S rRNA gene analysis for the correct species classification within some bacterial genera (Supplementary Fig. 3)28. Cluster I is formed by A. muciniphila strains, including the type strain ATCC BAA-835T (Fig. 4). Cluster II includes strains Marseille-P6666, Marseille-P9185 and 12 other strains described in previous studies (Fig. 4). Recently, Kumar et al.29 described a new Akkermansia strain, DSM 33459, that was phylogenetically close to strains EB-AMDK-39, EB-AMDK-40, and EB-AMDK-41, and proposed that this strain belongs to a new Akkermansia species. According to these authors, genomic comparison showed that strain Akkermansia sp. DSM 33459) exhibited OrthoANI values > 98% with strains EB-AMDK-39, EB-AMDK-40, and EB-AMDK-41, thus classifying it within the same species. This result also indicates that Akkermansia sp. strain DSM 33459 belongs to the same cluster, and probably to the same species, as strains Marseille-P6666 and Marseille-P9185. However, no accession number was provided by Kumar et al. for the genomic sequence of Akkermansia sp. strain DSM 33459, and therefore could not be included in our analysis. In addition, the authors deposited strain DSM 33459 in the DSMZ collection, but not in a second culture collection as requested by rule 30 from the international code of nomenclature of Prokaryotes for the description of a new species30. In addition, no name was proposed by the authors for this new species and Kumar and colleagues’ article does not contain any protolog to officially describe the properties of the new species. Cluster III includes the five strains Akk0196T, Akk0490, Akk0496a, Akk0496b and Akk2750 (Fig. 4).
Figure 4.
Core genome-based phylogenetic tree highlighting the position of Akkermansia muciniphila strain ATCC BAA-835T (Cluster I), Akkermansia massiliensis strain Marseille-P6666 (Cluster II), “Candidatus Akkermansia timonensis” strain Akk0196 (Cluster III), relative to other type strains within the genus Akkermansia and other members of the family Verrucomicrobiaceae. Sequence alignment was performed using MUGSY and a phylogenetic tree was obtained using the maximum-likelihood method. Numbers at the nodes are bootstrap values (≥ 70%) obtained by repeating the analysis 1,000 times. The scale bar indicates a 10% nucleotide sequence divergence.
We observed that A. glycaniphila strain PytT, initially described by Janneke et al., exhibited dDDH and ANI values of 22.5% and 73%, respectively, with A. muciniphila strain ATCC BAA-835T19. In addition, A. glycaniphila strain PytT also exhibited dDDH and ANI values ranging from 18.5 to 24% and from 73 to 74%, respectively, with all other members of the Akkermansia genus.
Pan-genome analysis of Akkermansia muciniphila, Akkermansia massiliensis sp. nov. and Candidatus Akkermansia timonensis sp. nov.
The pan- and core-genomes of A. muciniphila strains (85 strains) were composed of 6357 and 1193 genes, respectively. In addition, 1654 genes are accessory genes. A total of 1108 specific genes were found in only one A. muciniphila strain.
The pan- and core-genomes of A. massiliensis strains (14 strains) were composed of 3632 and 2138 genes, respectively. The accessory genome included 410 genes were strain-specific.
The pan- and core-genomes of A. timonensis strains (5 strains) were composed of 2767 and 2539 genes, respectively. The accessory genome sizes were 228 genes. A total of 125 specific genes were found in only one strain of A. timonensis.
The pan-genome of A. muciniphila ATCC BAA-835T (6357genes) is larger than that of A. massiliensis Marseille-P6666 (3632 genes). The percentage of the core-genome of A. muciniphila ATCC BAA-835T is smaller than that of A. massiliensis Marseille-P6666, 18.7% and 60.1%, respectively. However, due to the difference in the number of genomes used in the analysis of each pan-genome, the number of accessory genes of A. muciniphila ATCC BAA-835T (85 strains) is higher than that of A. massiliensis Marseille-P6666 (14 strains), 26% and 11.2% of the pan-genome, respectively.
Conclusion
From these results, we suggested the creation of two new species: Akkermansia massiliensis sp. nov. that includes strains Marseille-P6666 and Marseille-P9185, for which Marseille-P6666T is the type strain; and Candidatus Akkermansia timonensis sp. nov. that includes strains Akk0196, Akk0490, Akk0496a, Akk0496b and Akk2633.
Description of Akkermansia massiliensis sp. nov.
Akkermansia massiliensis (mas.si.li.en'sis L. fem. adj. massiliensis, of Massilia, the Latin name of Marseille where the strain was isolated).
Gram strain-negative, rod-shaped cells (0.5 × 0.8 μm). Bacteria are catalase-positive, oxidase-negative and motile. Non-spore forming. Colonies grown on Columbia agar are white, circular, convex and with entire margins and uniform. The optimal growth is observed in anaerobic atmosphere, at 37 °C, at pH 7 and in the presence of 5 g/l NaCl. Growth may also be obtained in microaerophilc atmosphere and at temperatures up to 42 °C. Nitrate reduction, indole production, gelatin hydrolysis and urease activities are absent. Strain Marseille-P6666T is positive for esculin hydrolysis and exhibits α-galactosidase, β-galactosidase, β-glucuronidase, N-acetyl-β-glucosaminidase, β-galactosidase, alkaline phosphatase, esterase (C4) Naphtol phosphohydrolase and acid phosphatase activities. Esterase lipase (C8), lipase (C14), leucine arylamidase, valine arylamidase, cystine arylamidase, trypsin α-chymotrypsin, α-glucosidase, β-glucosidase, α-mannosidase and α-fucosidase activities are not detected. The following substrates are used for growth and acid production: l-arabinose, d-ribose, d-galactose, d-glucose, d-fructose, d-mannose, d-mannitol, d-sorbitol, mucin, N-acetylglucosamine, salicin, esculin ferric citrate, cellobiose, d-maltose, d-lactose, d-melibiose, amygdaline, d-saccharose, d-trehalose, arbutine, gentiobiose, d-turanose, d-tagalose and potassium gluconate. Strain Marseille-P6666T does not utilize glycerol, erythritol, d-arabinose, d-xylose, l-xylose, d-adonitol, methyl-α d-mannopyranoside Methyl-α d-glucopyranoside, l-sorbose, l-rhamnose, dulcitol, inositol, Methyl-α d-Glucopyranoside, Methyl-α d-Mannopyranoside, inulin, d-melezitose, d-raffinose, glycogen, amidon, d-fucose, l-fucose, d-arabitol, l-arabitol, potassium 2-cetogluconate and potassium 5-cetogluconate. The major fatty acids are 12-methyl-tetradecanoic acid, pentadecanoic acid. Acetic acid and propanoic acid are produced. Various polar lipids classes are found: sphingomyelins, N-acyl ethanolamines, acyl carnitine, phosphatidylethanolamine, lysophosphatidyléthanolamine, phosphatidylcholine, lysophosphatidylcholine, ceramides—glycero lipids, fatty acyls—glycero lipids, phosphatidic acid and several unknown structures and phospholipids.
The DNA G + C content of the genomic DNA is 57.8%. The type strain Marseille-P6666T (= CSUR P6666 = CECT 30548), was isolated from human stool.
Description of Candidatus Akkermansia timonensis sp. nov.
Candidatus Akkermansia timonensis (ti.mo.nen’sis. L. fem. adj. timonensis, of Timone, the name of the hospital where this genome was analyzed).
Member of the Akkermansia genus, Candidatus Akkermansia timonensis is a bacterial species identified by metagenomic analyses. The genome length of the type genome is 3,212,887 bp and the G + C content is 56.7%.
The type material is Akk0196, a metagenome-assembled genome from human stool.
Materials and methods
Isolation and identification of strains by MALDI-TOF
Stools obtained from eight French patients as part of a culturomics project aiming at isolating as many distinct human-associated bacterial species from the gut, were included in the study from 2017 to 2020. All the methods used in this study were carried out in accordance with relevant guidelines and regulations conformed to the Declaration of Helsinki. Informed and oral consent was obtained from the stool donors. Approximately 1 g of each feces specimen was suspended in 2 ml of phosphate-buffered-saline (Life, Technologies, Carisbad, CA, USA). Then, 100 µl of each stool suspension was tenfold diluted up to 10–10. After that, 50 µl was inoculated on 5% sheep blood-enriched Columbia agar (BioMérieux, Marcy l’Etoile, France) and incubated at 37 °C in anaerobic atmosphere generated by AnaeroGen generator (bioMérieux). After 72 h of incubation, single colonies were selected and subcultured on the same medium in order to obtain pure isolates. Strains were identified using a Microflex MALDI-TOF MS spectrometer (Bruker, Daltonics, Leipzig, Germany) as previously described31.
Phenotypic and biochemical characterization
Cell morphology and characteristics of these isolates were observed using a TM4000 scanning electron microscope (Hitachi, Tokyo, Japan) from fresh colonies as previously described30. Colony morphology was described after observation of the strain grown after four days at 37 °C. Gram staining and spore formation were investigated and mobility was examined by microscopic observation32. Growth on Columbia agar at different temperatures (21 °C, 28 °C, 37 °C, 42 °C and 45 °C) and in microaerophilic conditions was tested using CampyGenTM (BioMérieux, ThermoFisher scientific) after 72 h of incubation. Growth in various NaCl concentrations (0, 5, 10 and 15 g/L) and at a pH range of 5 to 8.5 (at intervals of 0.5 pH unit) were assessed using Columbia agar plates33.
The ability to use mucin as sole carbon source was tested by using a modified basal media described by Derrien et al.2. Briefly, this modified medium contained 0.11 g CaCl2; 0.3 g NaCl; 0.53 g Na2HPO4; 0.4 g KH2PO4; 0.3 g NH4Cl; 0.1 g MgCl2.6H2O; 0.5 mg resazurin; 4 g NaHCO3; 0.25 g Na2S.7–9H2O and 1 ml of vitamin solution (0.10 mg Vitamin B12, 2.0 mg biotin, 5.0 mg riboflavin, 10.0 mg Pyridoxine–HCl, 5.0 mg Nicotinic acid, 5.0 mg p-Aminobenzoic acid, 2.0 mg Folic acid, 5.0 mg Lipoic acid, 5.0 mg D-Ca-pantothenate and 5.0 mg Thiamine-HClx 2H2O).
Catalase and oxidase activities were assessed by using a BBL™ DrySlide™ (Becton, Le Pont de Claix, France) according to the manufacturer’s instructions. Activities of other enzymes and metabolic characteristics were investigated by using the API ZYM, API 50CH, API 20A and API NE strips according to the manufacturer’s instructions (bioMérieux). Susceptibility to antibiotics was tested using the following E-test strip gradients: amoxicillin, benzylpenicillin, oxacillin, cefotaxine, ceftriaxone, amikacin, tobramicin, ciprofloxacin, clindamycin, doxycycline, rifampicin, vancomycin and trimethoprim-sulfamethoxazole. Plates with deposited strips were incubated at 37 °C for 48 h. Minimal inhibitory concentration (MIC) of each tested antibiotic was determined according to the manufacturer’s instructions34.
Chemotaxonomic characteristics
Cellular fatty acid methyl ester (FAME) analysis was performed by Gas Chromatography/ Mass Spectrometry (GC/MS) as previously described35. Approximately 65 mg of bacterial biomass collected from several Columbia agar plates cultured under anaerobic conditions for 3 days at 37 °C were distributed into each of two sterile tubes. FAMEs were extracted and prepared as described before by Sasser36. GC/MS analyses were done as previously described35.
Short chain fatty acids (SCFA) were extracted and analyzed from three independent culture bottles (both blank and samples). Strain Marseille-P6666 was cultured in anaerobic blood culture vial enriched with 5% sterilized sheep blood (Becton–Dickinson, Pont de Claix, France) for three days. SCFAs were measured with a Clarus 500 chromatography system connected to a SQ8s mass spectrometer (Perkin Elmer) as previously described by Diop et al.37.
Polar lipid analysis of strain Marseille-P6666 was performed by Hydrophilic Interaction Liquid Chromatography-Mass Spectrometry (HILIC-MS). Total lipids were extracted from cultures plates according to the Bligh and Dyer protocol38. Fifty percent chloroform/methanol was used to reconstitute the chloroformed extracts previously dried under a nitrogen stream, corresponding approximately to 0.5 mg of lipid content per 100 µL (v:v). Lipid extracts were injected (5 µL) into a HILIC column (BEH HILIC, 2.1 × 100 mm, 1.7 µm, Waters, Guyancourt, France). Elution of lipids from the column was performed according to their polarity using a gradient of the following solvent compositions: A = 5% water/95% acetonitrile, B = 50% H2O/50% acetonitrile, both at 10 mM ammonium acetate pH8. The HD-MS method (Vion ESI-IMS-Q-TOF mass spectrometer, Waters) with positive and negative modes was used for lipid control as previously described39. The assignment of lipid classes was done according to the retention times (RT) of an injected standard (Splash Lipidomix, Avanti Polar Lipids, Alabaster, AL, USA). A comparison of the corresponding masses with the COMP DB LipidMAPS database (tolerance of 0.0005 m/z; all chains are activated) was also performed to confirm the lipid classes.
Genome sequencing and assembly
Genomic DNAs (gDNAs) from all strains were extracted using an EZ1 biorobot and the EZ1 DNA Tissue kit (Qiagen, Hilden, Germany). The gDNAs were quantified by a Qubit assay with the high sensitivity kit (Thermofisher Scientific) to 0.2 ng/μl. gDNAs were sequenced using a MiSeq sequencer (Illumina, San Diego CA, USA) with the paired-end strategy. SPAdes was used to assemble the total reads of all genomes. Scaffolds smaller than 800 bp and those with depth values lower than 25% of the average depth (considered as possible contaminants) were deleted.
Genome annotation and comparison
Genome annotation was performed using the Prokka software40. Bacterial protein-coding sequences were predicted using BLASTP (E-value of 1e-03, coverage 0.7 and identity 30%) against the Clusters of Orthologous Groups (COG) database. Graphical circular maps of genomes was generated using CGView (Circular Genome Viewer) software41.
As of February 22, 2022, 191 complete A. muciniphila genomic sequences were available in the NCBI GenBank database and were downloaded. For genomic comparison, we eliminated duplicate sequences, retaining only 96 complete sequences from several studies (Supplementary Table 3). Overall, a total of 114 sequences were analyzed, including eight from this study, three from other Akkermansia species (A. glycaniphila, "Candidatus A. intestinigallinarum" and "Candidatus A. intestinavium"), and seven from closely related species from the Verrucomicrobiaceae family. Several genomic comparison approaches were used to delineate the species within the Akkermansia genus. The Genome-to Genome Distance Calculator (https://ggdc.dsmz.de/) and PyANI (a Python package and script that provides support for calculating ANI)42 were used to calculate the digital DNA-DNA hybridization (dDDH) and average nucleotide identity (ANI) between studied strains, retrospectively. The pan-genome of Akkermansia strains was analyzed using the Roary software43.
Nucleotide sequence accession numbers
The 16SrRNA gene/genome sequences are available in GenBank under accession numbers ON014381/JAMGSI000000000.1 (Akkermansia massiliensis strain Marseille-P6666), ON014382/JAMYIA000000000 (Akkermansia massiliensis strain Marseille-P9185), ON014383/JAMZOC000000000 (Akkermansia muciniphila strain Marseille-P5162), ON014384/JAMGSH000000000 (Akkermansia muciniphila strain Marseille-P7245), ON014385/JAMZOB000000000 (Akkermansia muciniphila strain Marseille-P6566), ON014386/JAMGSG000000000 (Akkermansia muciniphila strain Marseille-P9184), ON014387/JAMGSF000000000 (Akkermansia muciniphila strain Marseille-P9642) and ON014388/JAMGSE000000000 (Akkermansia muciniphila strain Marseille-Q2586). Genome sequence accession number of Candidatus Akkermansia timonensis AKK7 is 20100303_Bin_52_1.
Ethical statement
Oral informed consent was obtained from all patients. The study design was validated by the ethics committees of the IHU Méditerranée Infection under number 2016-011.
Supplementary Information
Abbreviations
- CSUR
Collection de Souches de l’Unité des Rickettsies
- DSM
Deutsche Sammlung von Mikroorganismen
- MALDI-TOF MS
Matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry
- FAME
Fatty acid methyl ester
- ORF
Open reading frame
- COGs
Clusters of orthologous groups
Author contributions
Conceptualization and supervision: P.-E.F. and D.R. Methodology, investigation and writing manuscript: S.N. Fatty acid and polar lipids analysis: N.A. Funding acquisition: D.R.
Funding
This work was supported by the Méditerranée Infection foundation (Marseille, France) and by the French Government under the «Investissements d’avenir» program managed by the Agence Nationale de la Recherche (ANR) under reference 10-IAHU-03. This work was also funded by Région Provence-Alpes-Côte d’Azur and European funding FEDER PRIMI.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Sokhna Ndongo, Email: sokhnandongo@gmail.com.
Pierre-Edouard Fournier, Email: pierre-edouard.fournier@univ-amu.fr.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-25873-0.
References
- 1.de Vos WM, Tilg H, Van Hul M, Cani PD. Gut microbiome and health: Mechanistic insights. Gut. 2022;71:1020–1032. doi: 10.1136/gutjnl-2021-326789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2004;54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 3.Liu X, et al. Transcriptomics and metabolomics reveal the adaption of Akkermansia muciniphila to high mucin by regulating energy homeostasis. Sci. Rep. 2021;11:9073. doi: 10.1038/s41598-021-88397-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belzer C, de Vos WM. Microbes inside—From diversity to function: The case of Akkermansia. ISME J. 2012;6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl. Environ. Microbiol. 2007;73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujio-Vejar S, et al. The gut microbiota of healthy Chilean subjects reveals a high abundance of the phylum Verrucomicrobia. Front. Microbiol. 2017;8:1221–1221. doi: 10.3389/fmicb.2017.01221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earley H, et al. A preliminary study examining the binding capacity of Akkermansia muciniphila and Desulfovibrio spp., to colonic mucin in health and ulcerative colitis. PLoS ONE. 2015;10:e0135280. doi: 10.1371/journal.pone.0135280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pascale A, Marchesi N, Govoni S, Coppola A, Gazzaruso C. The role of gut microbiota in obesity, diabetes mellitus, and effect of metformin: New insights into old diseases. Curr. Opin. Pharmacol. 2019;49:1–5. doi: 10.1016/j.coph.2019.03.011. [DOI] [PubMed] [Google Scholar]
- 9.Fassatoui M, et al. Gut microbiota imbalances in Tunisian participants with type 1 and type 2 diabetes mellitus. Biosci. Rep. 2019;39:BSR20182348. doi: 10.1042/BSR20182348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grander C, et al. Recovery of ethanol-induced Akkermansia muciniphila depletion ameliorates alcoholic liver disease. Gut. 2018;67:891–901. doi: 10.1136/gutjnl-2016-313432. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Lin S, Vanhoutte PM, Woo CW, Xu A. Akkermansia muciniphila protects against atherosclerosis by preventing metabolic endotoxemia-induced inflammation in Apoe−/− mice. Circulation. 2016;133:2434–2446. doi: 10.1161/CIRCULATIONAHA.115.019645. [DOI] [PubMed] [Google Scholar]
- 12.World Health Organization, World Food Programme, United Nations System Standing Committee on Nutrition, United Nations Children’s Fund, Community-Based Management of Severe Acute Malnutrition. Community-BasedManagementofSevereAcuteMalnutrition. www.who.int/nutrition/topics/statement_commbased_malnutrition/en/index.html (2007).
- 13.Fan L, et al. A. muciniphila suppresses colorectal tumorigenesis by inducing TLR2/NLRP3-mediated M1-like TAMs. Cancer Immunol. Res. 2021;9:1111–1124. doi: 10.1158/2326-6066.CIR-20-1019. [DOI] [PubMed] [Google Scholar]
- 14.Yaghoubfar R, et al. Effects of Akkermansia muciniphila and Faecalibacterium prausnitzii on serotonin transporter expression in intestinal epithelial cells. J. Diabetes Metab. Disord. 2021;20:1–5. doi: 10.1007/s40200-020-00539-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ashrafian F, et al. Extracellular vesicles and pasteurized cells derived from Akkermansia muciniphila protect against high-fat induced obesity in mice. Microb. Cell Factories. 2021;20:219. doi: 10.1186/s12934-021-01709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abuqwider JN, Mauriello G, Altamimi M. Akkermansia muciniphila, a new generation of beneficial microbiota in modulating obesity: A systematic review. Microorganisms. 2021;9:1098. doi: 10.3390/microorganisms9051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin N-R, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63:727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 18.Hänninen A, et al. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2018;67:1445–1453. doi: 10.1136/gutjnl-2017-314508. [DOI] [PubMed] [Google Scholar]
- 19.Ouwerkerk JP, Aalvink S, Belzer C, de Vos WM. Akkermansia glycaniphila sp. nov., an anaerobic mucin-degrading bacterium isolated from reticulated python faeces. Int. J. Syst. Evol. Microbiol. 2016;66:4614–4620. doi: 10.1099/ijsem.0.001399. [DOI] [PubMed] [Google Scholar]
- 20.Gilroy R, et al. Extensive microbial diversity within the chicken gut microbiome revealed by metagenomics and culture. PeerJ. 2021;9:e10941. doi: 10.7717/peerj.10941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim J-S, Kang SW, Lee JH, Park S-H, Lee J-S. The evolution and competitive strategies of Akkermansia muciniphila in gut. Gut Microbes. 2022;14:2025017. doi: 10.1080/19490976.2021.2025017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orellana LH, et al. Verrucomicrobiota are specialist consumers of sulfated methyl pentoses during diatom blooms. ISME J. 2022;16:630–641. doi: 10.1038/s41396-021-01105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouwerkerk JP, et al. Adaptation of Akkermansia muciniphila to the oxic-anoxic interface of the mucus layer. Appl. Environ. Microbiol. 2016;82:6983–6993. doi: 10.1128/AEM.01641-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Konstantinidis KT, Ramette A, Tiedje JM. Toward a more robust assessment of intraspecies diversity, using fewer genetic markers. Appl. Environ. Microbiol. 2006;72:7286–7293. doi: 10.1128/AEM.01398-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Varghese NJ, et al. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015;43:6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stackebrandt E, Ebers J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today. 2006;8:152–155. [Google Scholar]
- 28.Rossi-Tamisier M, Benamar S, Raoult D, Fournier P-E. Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int. J. Syst. Evol. Microbiol. 2015;65:1929–1934. doi: 10.1099/ijs.0.000161. [DOI] [PubMed] [Google Scholar]
- 29.Kumar R, et al. Identification and characterization of a novel species of genus Akkermansia with metabolic health effects in a diet-induced obesity mouse model. Cells. 2022;11:2084. doi: 10.3390/cells11132084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parker CT, Tindall BJ, Garrity GM. International code of nomenclature of prokaryotes. Prokaryotic code (2008 revision) Int. J. Syst. Evol. Microbiol. 2019;69(1A):S1–S111. doi: 10.1099/ijsem.0.000778. [DOI] [PubMed] [Google Scholar]
- 31.Seng P, et al. Ongoing revolution in bacteriology: Routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis. 2009;49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 32.Ndongo S, et al. Genome analysis and description of Xanthomonas massiliensis sp. nov., a new species isolated from human faeces. New Microbes New Infect. 2018;26:63–72. doi: 10.1016/j.nmni.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ndongo S, et al. Vitreoscilla massiliensis sp. nov., isolated from the stool of an Amazonian patient. Curr. Microbiol. 2021;78:3313–3320. doi: 10.1007/s00284-021-02577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Citron DM, Ostovari MI, Karlsson A, Goldstein EJ. Evaluation of the E test for susceptibility testing of anaerobic bacteria. J. Clin. Microbiol. 1991;29:2197–2203. doi: 10.1128/jcm.29.10.2197-2203.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dione N, et al. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microbes New Infect. 2016;10:66–76. doi: 10.1016/j.nmni.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sasser, M. Bacterial identification by gas chromatographic analysis of fatty acid methyl esters (GC-FAME). NewarkNYMicrob.ID (2006).
- 37.Diop K, et al. Vaginimicrobium propionicum gen. nov., sp. nov., a novel propionic acid bacterium derived from human vaginal discharge. Int. J. Syst. Evol. Microbiol. 2020;70:4091–4097. doi: 10.1099/ijsem.0.004106. [DOI] [PubMed] [Google Scholar]
- 38.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/y59-099. [DOI] [PubMed] [Google Scholar]
- 39.Hasni I, et al. Proteomics and lipidomics investigations to decipher the behavior of Willaertia magna C2c Maky according to different culture modes. Microorganisms. 2020;8:1791. doi: 10.3390/microorganisms8111791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinform. Oxf. Engl. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 41.Stothard P, Grant JR, Van Domselaar G. Visualizing and comparing circular genomes using the CGView family of tools. Brief. Bioinform. 2019;20:1576–1582. doi: 10.1093/bib/bbx081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods. 2015;8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 43.Page AJ, et al. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinform. Oxf. Engl. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.