Abstract
The oral bacterium Fusobacterium nucleatum is often found in colorectal cancer (CRC). In the 21 July 2020 issue of Science Signaling, Casasanta et al. show that CRC cell-resident F. nucleatum promote cytokine secretion that may potentiate tumor growth and metastatic progression in patients.
The gut is filled with bacteria. Current estimates suggest that there are roughly 40 trillion bacterial cells in the average adult human colon (1), with over 1000 different species that have been identified (2). It is therefore natural to wonder whether any of these abundant bacterial populations might promote the growth of intestinal cancers. High-throughput genome and transcriptome sequencing of the tissue microbiome provided an initial clue. Almost a decade ago, two independent laboratories found through the comparison of RNA (4) and DNA (5) from tumor and normal colon tissues that the bacterial species Fusobacterium nucleatum is highly abundant in specimens of colorectal tumors surgically removed from patients.
There are several questions raised by the association of F. nucleatum with colorectal cancer (CRC). First, F. nucleatum is known to be able to invade colonic epithelial cells (6,7). What is the connection between cellular invasion and carcinogenesis? Second, F. nucleatum abundance is associated with higher levels of expression of inflammatory genes, including many interleukin genes, such as IL1B and IL6 (8). Could the induction of inflammation-associated transcripts be a factor by which F. nucleatum promotes colorectal carcinogenesis?
Casasanta et al. (3) tackle these questions. Until recently, there was no systematic method to perform targeted gene deletions in Fusobacterium. Based on a recently reported method for gene deletion in Fusobacterium (9), the authors further developed an elegant way to generate gene deletions and replacements in F. nucleatum genomes. In brief, wild-type fusobacteria cannot grow in the presence of 2-deoxy-D-galactose. Deletion of the galKT operon means that the bacteria cannot generate 2-deoxy-galactose-1-phosphate, the toxic metabolic product, so the fusobacteria now survive on medium containing 2-deoxy-galactose. Finally, the use of a plasmid targeting a specific locus by homologous recombination, positive selection for thiamphenicol resistance to get initial selection, and finally negative selection or a galK gene that has been introduced on deoxygalactose, leads to markerless deletion of the targeted gene.
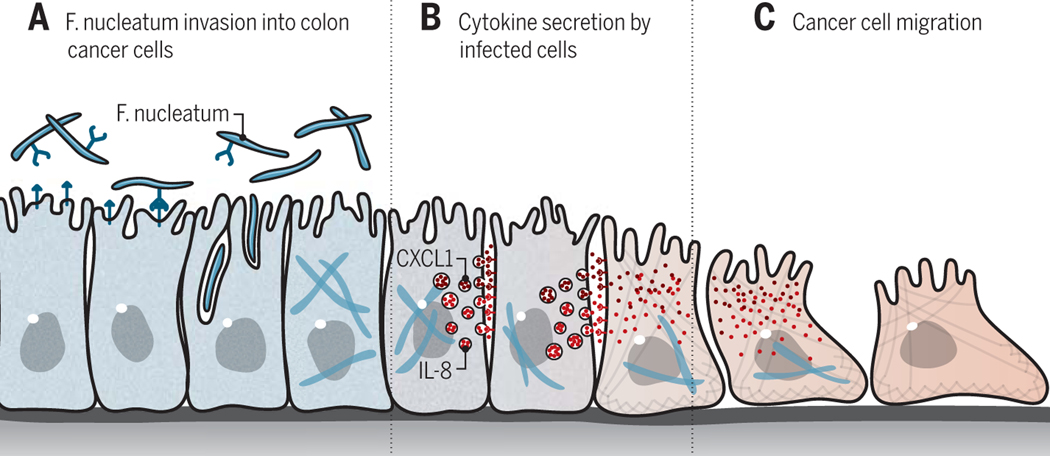
Casasanta et al. used their method to evaluate which bacterial genes were required for the bacteria to invade CRC cells. The results revealed that the fap2 gene is required for F. nucleatum to invade HCT116 CRC cells in culture (3). Given that fap2 reportedly modulates the immune response to colorectal cancers (10), the authors went on to ask whether F. nucleatum could induce cytokine secretion. They demonstrated that not only does F. nucleatum induce the secretion of IL-8 and CXCL1 from CRC cells, but they also showed that this secretion is dependent on a functional fap2 gene (3). They further showed that F. nucleatum induces the migration of HCT116 cells in a manner that is dependent on the secretion of IL-8 and CXCL1 (Figure 1), and that depletion of these proteins reduces migration. Thus, the findings that Casasanta et al. indicate that the presence of F. nucleatum in colorectal tumors might induce metastatic progression of the cancer in patients by promoting gut inflammation (3). If this concept holds, this could prove an important mechanism by which the microbiome leads to causation of intestinal cancers.
Figure 1: A model in which Fusobacterium nucleatum invasion into colorectal cancer cells induces cytokine secretion and invasion.
(A to C) A conceptual model of the findings of Casasanta et al. using a cultured CRC cell line, not actual CRC tissues. (A) F. nucleatum can invade CRC cells in a manner dependent on the bacterial adhesin protein FAP2. (B) Invasion of F. nucleatum induces secretion of IL-8 and CXCL1 by the host cell. (C) These cytokines, in turn, promote migration in both infected and uninfected cancer cells.
As the role of the microbiome in human cancer is increasingly understood, the methods and findings of the Casasanta study provide a rigorous and effective way to assess and dissect many of the claims that are now being made in the literature. Through their rigorous analysis, the authors confirmed some previous hypotheses in the literature and have refined our understanding of others. Similar studies will be needed to further explore and refine these findings in CRC and the broader context of the cancer-associated microbiome.
Funding:
Work in the Meyerson laboratory is supported by Bayer, Janssen, Novo Ventures, and Ono.
Footnotes
Competing interests: M.M. is chair of the Scientific Advisory Board and a consultant for OrigiMed, holds several patents on various diagnostic and therapeutic inventions (licensed to Bayer and LabCorp), and is an inventor on several patent applications related to Fusobacterium and colon cancer.
References
- 1.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016. Aug 19;14(8):e1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev. 2014. Sep;38(5):996–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casasanta MA, Yoo CC, Udayasuryan B, Sanders BE, Umana A, Zhang Y, Peng H, Duncan AJ, Wang Y, Li L, Verbridge SS, Slade DJ. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci. Signal. 13, eaba9157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, Holt RA. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012. Feb;22(2):299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, Baselga J, Liu C, Shivdasani RA, Ogino S, Birren BW, Huttenhower C, Garrett WS, Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012. Feb;22(2):292–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011. Sep;17(9):1971–8. [DOI] [PubMed] [Google Scholar]
- 7.Bullman S, Pedamallu CS, Sicinska E, Clancy TE, Zhang X, Cai D, Neuberg D, Huang K, Guevara F, Nelson T, Chipashvili O, Hagan T, Walker M, Ramachandran A, Diosdado B, Serna G, Mulet N, Landolfi S, Ramon Y Cajal S, Fasani R, Aguirre AJ, Ng K, Élez E, Ogino S, Tabernero J, Fuchs CS, Hahn WC, Nuciforo P, Meyerson M. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science. 2017. Dec 15;358(6369):1443–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, El-Omar EM, Brenner D, Fuchs CS, Meyerson M, Garrett WS. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013. Aug 14;14(2):207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu C, Al Mamun AAM, Luong TT, Hu B, Gu J, Lee JH, D’Amore M, Das A, Ton-That H. Forward genetic dissection of biofilm development by Fusobacterium nucleatum: novel functions of cell division proteins FtsX and EnvC. mBio. 2018. Apr 24;9(2):e00360-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015. Feb 17;42(2):344–355. [DOI] [PMC free article] [PubMed] [Google Scholar]