Abstract
Mutational analysis has shown that the integrity of the region in domain III of 25S rRNA that is involved in binding of ribosomal protein L25 is essential for the production of mature 25S rRNA in the yeast Saccharomyces cerevisiae. However, even structural alterations that do not noticeably affect recognition by L25, as measured by an in vitro assay, strongly reduced 25S rRNA formation by inhibiting the removal of ITS2 from the 27SB precursor. In order to analyze the role of L25 in yeast pre-rRNA processing further we studied the effect of genetic depletion of the protein or mutation of each of its three previously identified functional domains, involved in nuclear import (N-terminal), RNA binding (central) and 60S subunit assembly (C-terminal), respectively. Depletion of L25 or mutating its (pre-)rRNA-binding domain blocked conversion of the 27SB precursor to 5.8S/25S rRNA, confirming that assembly of L25 is essential for ITS2 processing. However, mutations in either the N- or the C-terminal domain of L25, which only marginally affect its ability to bind to (pre-)rRNA, also resulted in defective ITS2 processing. Furthermore, in all cases there was a notable reduction in the efficiency of processing at the early cleavage sites A0, A1 and A2. We conclude that the assembly of L25 is necessary but not sufficient for removal of ITS2, as well as for fully efficient cleavage at the early sites. Additional elements located in the N- as well as C-terminal domains of L25 are required for both aspects of pre-rRNA processing.
INTRODUCTION
Ribosomes are complex ribonucleoprotein particles, present in every cell, that are responsible for production of the entire complement of cellular proteins and, thus, are critical for the vitality and survival of the cell. It is, therefore, important to fully understand the many interconnected steps required for production of mature ribosomes.
Eukaryotic ribosomes contain one copy each of four different rRNA species as well as approximately 80 different ribosomal proteins (r-proteins). The assembly of this large number of constituents into a functional ribosome is an intricate process that takes place largely in a specialized nuclear compartment, the nucleolus. Apart from the actual rRNA and r-protein components, this process also requires the participation of a multitude of non-ribosomal factors, which are necessary both for directing correct assembly of r-proteins and for concomitant conversion of the primary pre-rRNA transcript into the mature rRNA species (reviewed in 1,2).
Three of the four eukaryotic rRNA species (18S, 5.8S and 25–28S rRNA) are transcribed by RNA polymerase I as a single large precursor molecule that subsequently undergoes a number of structural alterations. These consist of nucleotide modifications, predominantly 2′-O-methylation and pseudouridylation, as well as removal (processing) of the transcribed spacer sequences present at both ends [5′- and 3′-external transcribed spacers (ETS)] and in between the mature sequences [internal transcribed spacers (ITS) 1 and 2] (reviewed in 3–5; cf. Fig. 1A).
Figure 1.
The S.cerevisiae rDNA unit and formation of the mature rRNA species. (A) Genetic organization of the rDNA unit. Black bars correspond to the mature sequences, lines to the external and internal spacer sequences. The processing sites are indicated, as are the positions of the various probes used for the northern and primer extension analyses. (B) The rRNA processing pathway in S.cerevisiae. Vertical arrows correspond to endonucleolytic cleavages, kinked arrows to exonucleolytic processing. (C) Sequences of the probes depicted in (A).
Processing of pre-rRNA has been studied most extensively in the yeast Saccharomyces cerevisiae. As a result of these studies the yeast pre-rRNA processing pathway is now almost fully known (3,6,7) (Fig. 1B). The primary transcript first undergoes rapid endonucleolytic cleavage at its 3′-end resulting in the 35S pre-rRNA, which carries only a small portion of the original 3′-ETS (8). This precursor is then cleaved at sites A0, A1 and A2 to give the 20S and 27SA2 pre-rRNAs. The former is exported to the cytoplasm, where it is converted into 18S rRNA by endonucleolytic cleavage at site D (9–11). The 27SA2 precursor is further processed in the nucleolus by two alternative pathways. The majority (∼90%) is cleaved by RNase MRP at site A3 to produce 27SA3 pre-rRNA (12–16), which is then processed to 27SBS pre-rRNA by 5′→3′ exonucleolytic trimming (17). The remaining 10% is converted into 27SBL pre-rRNA, which contains several extra nucleotides at the 5′-end compared to its 27SBS counterpart, by an as yet unknown mechanism. The two types of 27SB precursor are further processed in the same manner. Endonucleolytic cleavage at site C2/C2′ within ITS2 produces the readily detectable 7S pre-rRNA, which is converted into 5.8S rRNA by multistep 3′→5′ trimming up to site E (18), and the very short-lived 25.5S pre-rRNA, from which 25S rRNA is produced by 5′→3′ exonucleolytic degradation to site C1 (7).
Both biochemical analysis (19) and kinetic labeling studies (20) have provided evidence that the yeast 35S pre-rRNA is already associated with a number of r-proteins in a 90S pre-ribosomal particle and that further r-proteins are added in an ordered manner at subsequent stages of the processing pathway. The large subunit species L4, L12, L17, L20, L25 and L30 (the nomenclature for yeast r-proteins used in this paper is that of Mager et al.; 21) have been found to bind specifically to 35S pre-rRNA and/or 25S rRNA in vitro (22–25). Thus, these r-proteins may be among the first to be assembled into the pre-ribosome.
The concurrence of pre-rRNA processing and r-protein assembly suggests that the two processes may be interdependent. However, there is as yet only limited information on the role of individual r-proteins in pre-rRNA processing. So far four large subunit r-proteins have been identified in yeast whose absence or functional inactivation affects processing. These include the 5S rRNA-binding protein L5 (26) and the large subunit proteins L11 (27), L28 (28) and L30 (29). Interestingly, in all cases a very similar processing phenotype resulted from either genetic depletion or mutation of the r-protein, namely accumulation of the 35S as well as the 27S pre-rRNA species, indicating a delay in processing at the early cleavage sites A0, A1 and A2, as well as the late cleavage at C2. Furthermore, two small subunit r-proteins, S31 (30) and S0 (31), have been implicated in conversion of the 20S pre-rRNA into mature 18S rRNA.
We have previously described a mutational analysis of the L25-binding region located within domain III of 25S rRNA, which showed that most of the structural alterations introduced into this region lead to a disturbance, in fact in many cases a complete block, in ITS2 processing, as concluded from the loss of 7S pre-rRNA and accumulation of the 27SB precursors (32). These alterations included structural changes that did, as well as changes that did not, interfere with binding of L25, according to their analysis by an in vitro binding assay using synthetic rRNA and r-protein (33). In order to obtain a better understanding of the role of the L25–(pre-)rRNA interaction in pre-rRNA processing we have now studied the effect of genetic depletion of L25 and of mutations in each of the three previously identified functional domains of the protein, involved in nuclear import (34), RNA binding (33,35) and large subunit assembly (36), respectively, on levels of the various pre-rRNAs. The results clearly show that all three domains of L25 contain elements that are crucial not only for processing steps specific to formation of the mature large subunit rRNAs, i.e. removal of ITS2 from the 27SB precursor, but also for fully efficient cleavage of the 35S pre-rRNA at the early processing sites A0, A1 and A2.
MATERIALS AND METHODS
Strains and mutants
Saccharomyces cerevisiae strain YCR61 (MATa, leu2-3, leu2-112, ade2, his4-519, ura3-52) containing a wild-type L25 gene under control of the GAL1-10 promoter has been described elsewhere (35,36). YCR61 cells were transformed with the YCplac111 shuttle vector carrying one of a set of mutant alleles of the L25 gene (33,34,36) using the LiAc method (37). In order to avoid differences in expression at the transcriptional and translational levels all mutant genes were placed under control of the authentic L25 promoter and carried the same untranslated leader and trailer sequences. Transformants were selected at 30°C on Sgal medium supplemented with adenine and histidine.
Northern and primer extension analysis
YCR61 transformants were grown on liquid YNB-Gal to an OD660 of 0.5, transferred to YNB-Glu medium at an OD660 of 0.1 and further cultured. Samples were taken at different times after the shift, from which cells were collected by centrifugation. After washing the cells in 1 ml of buffer containing 0.1 M Tris–HCl pH 7.4, 0.1 M LiCl and 0.1 M EDTA, total RNA was isolated and analyzed by northern hybridization as described previously (38,39). An amount of RNA corresponding to 0.2 OD660 units of cells was separated on either a 1.2% agarose gel or an 8% polyacrylamide gel, blotted and hybridized sequentially with the various oligonucleotide probes shown in Figure 1A. Primer extension analysis was performed according to Beltrame and Tollervey (40). The sequences of the probes are shown in Figure 1C.
RESULTS
L25 is an essential ribosomal protein in yeast (35) that possesses at least three distinct functional domains, as revealed by mutational analysis both in vitro and in vivo. The N-terminal 61 amino acids contain two nuclear localization signals (NLS), both of which are required for full functionality of the protein (34). The information necessary and sufficient for specific binding of L25 to a region within domain III of 25S rRNA is located within the region encompassing amino acids 62–126 (33). Finally, the C-terminal 15 amino acids of the protein starting at residue 127 contain another element that is indispensable for its function, supposedly because it plays a crucial role in a step in ribosome biogenesis subsequent to the association of L25 (36).
To study the involvement of yeast r-protein L25 in pre-rRNA processing we used yeast strain YCR61, which contains a genomic L25 gene under control of the GAL1-10 promoter (35). Thus, shifting the cells from a galactose- to a glucose-based medium shuts off production of wild-type L25, allowing it to be depleted or replaced by plasmid-encoded mutant forms.
L25 is required for both pre-rRNA processing pathways
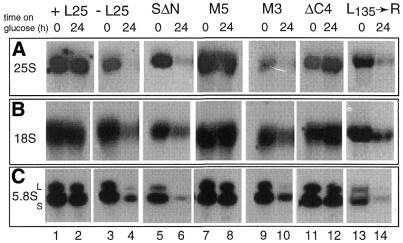
In a first series of experiments we analyzed the effect of depletion of the protein on pre-rRNA processing. Total RNA was prepared from YCR61 cells carrying an ‘empty’ YCplac111 plasmid immediately before and 24 h after the shift to glucose-based medium and the levels of the various mature and precursor RNAs were assayed by northern hybridization. RNA prepared from cells transformed with the same plasmid carrying a wild-type L25 gene was used as a control. As shown in Figure 2 (lanes 1 and 2), in the latter cells the levels of the mature rRNAs did not change. Depletion of L25 leads to a severe reduction in the levels of the large subunit rRNAs (Fig. 2A and C, lanes 3 and 4), in agreement with our earlier finding that the protein is necessary for 60S subunit formation (36). Surprisingly, however, there was also a considerable reduction in the level of 18S rRNA (Fig. 2B), suggesting that lack of L25 affects 40S subunit formation as well.
Figure 2.
Effect of depletion or mutation of L25 on accumulation of the mature rRNA species. YCR61 cells were transformed with the YCplac111 plasmid carrying the wild-type L25 gene (lanes 1 and 2), an ‘empty’ vector (lanes 3 and 4) or a vector containing one of several mutant forms of the L25 gene (lanes 5–14). Tranformants were grown on galactose-based medium and shifted to glucose-based medium. Total RNA was isolated immediately before and 24 h after the shift and analyzed by northern hybridization using oligonucleotides complementary to 25S rRNA (probe G, A), 18S rRNA (probe F, B) and 5.8S rRNA (probe H, C). See Figure 1 for location of the probes. SΔN, replacement of the 41 N-terminal amino acids with the NLS of the SV40 large T antigen (34); M3 and M5, mutations in the conserved motif at the C-terminus of the RNA-binding domain (M3, L126→K; M5, K120K121→RR; 33,36); ΔC4, deletion mutant lacking the four C-terminal amino acids; L135→R, point mutation in the C-terminal functional element of L25 (36).
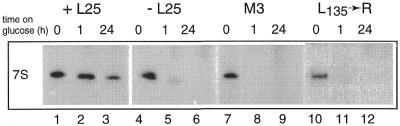
To ascertain whether the effect of L25 depletion on accumulation of mature rRNAs is due to defective pre-rRNA processing, the same blot was probed sequentially with a set of oligonucleotides that detect the various known processing intermediates (cf. Fig. 1). As shown in Figures 3B (lanes 3 and 4) and 4 (lanes 4–6), blocking L25 synthesis causes an increase in the level of the 27SB precursor species accompanied by a drastic reduction in 7S pre-rRNA, which already becomes apparent shortly after the shift to glucose (Fig. 4), indicating a strong requirement for L25 in ITS2 processing. These results demonstrate that the presence of L25 is crucial for initiating the removal of ITS2 from the 27SB pre-rRNA by cleavage at site C2/C2′ (7).
Figure 3.
Effect of depletion or mutation of L25 on pre-rRNA processing. YCR61 cells were transformed with the YCplac111 plasmid carrying the wild-type L25 gene (lanes 1 and 2), an ‘empty’ plasmid (lanes 3 and 4) or a vector containing one of several mutant forms of the gene (lanes 5–14; see legend to Fig. 2 for details). Transformants were grown on galactose-based medium and shifted to glucose-based medium. Total RNA was isolated immediately before and 24 h after the shift and analyzed by northern hybridization with oligonucleotides complementary to different spacer regions to determine the levels of the various pre-rRNA intermediates using the same blot as shown in Figure 2. (A) Probe A; (B) probe D; (C) probe C; (D) probe C; (E) probe E. The pre-rRNA species detected is indicated in each of the different panels. See Figure 1 for location and sequence of the probes.
Figure 4.
Effect of depletion or mutation of L25 on the accumulation of 7S pre-rRNA. YCR61 cells transformed with YCplac111 carrying the wild-type L25 gene (lanes 1–3), an ‘empty’ plasmid (lanes 4–6) or a plasmid carrying the M3 (lanes 7–9) or L135→R mutant forms of L25 (lanes 10–12), respectively, were shifted from galactose- to glucose-based medium (see legend to Fig. 2 for details). Total RNA was isolated immediately before and at different times after the shift, separated on an 8% polyacrylamide gel and subjected to northern analysis using probe D complementary to the upstream region of ITS2 (see Fig. 1).
A further effect of L25 depletion on pre-rRNA processing is apparent from Figure 3 (lanes 3 and 4), which shows a notable reduction in the level of 27SA2 (Fig. 3C) as well as the 20S (Fig. 3E) pre-rRNA. At the same time there was an increase in the aberrant 23S precursor species (Fig. 3D), which extends from the 5′-end of the primary transcript to site A3. This pattern is diagnostic for inhibition of the early processing cleavages at sites A0, A1 and A2 (2,41). Further support for this conclusion comes from the experiment shown in Figure 5, which demonstrates a strong decrease in the amount of the A0–A1 spacer fragment upon depletion of L25 (lanes 3 and 4).
Figure 5.
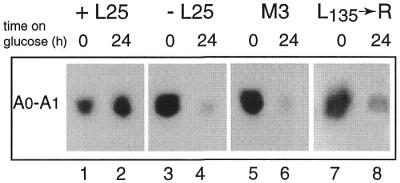
Effect of depletion or mutation of L25 on accumulation of the A0–A1 spacer fragment. YCR61 cells transformed with YCplac111 containing the wild-type L25 gene (lanes 1 and 2), the ‘empty’ plasmid (lanes 3 and 4) or a plasmid carrying the M3 (lanes 5 and 6) or L135→R (lanes 7 and 8) mutant forms of the L25 gene, respectively, were shifted from galactose- to glucose-based medium (see legend to Fig. 2 for details). Total RNA was isolated immediately before and 24 h after he shift, separated on an 8% polyacrylamide gel and subjected to northern analysis using probe B complementary to the 5′-ETS downstream from site A0 (see Fig. 1).
To determine whether the defect in pre-rRNA processing at sites A0–A2 is a direct consequence of the absence of L25 rather than a secondary effect due to the insufficiency of large ribosomal subunits, we performed northern hybridization on RNA isolated at shorter intervals after shutting off L25 expression. We have previously found that YCR61 cells start to show a significant reduction in growth rate ∼6 h after having been shifted to glucose-based medium (36). The results depicted in Figure 6 demonstrate, however, that even 3 h after the shift the level of 20S pre-rRNA, the immediate precursor of the mature 18S rRNA, is already notably reduced. At the same time there is an increase in the level of 35S and a reduction in 32S pre-rRNA (cf. lanes 2 and 1), indicating partial inhibition of the early processing cleavages. This supports the conclusion that lack of L25 not only causes a severe defect in ITS2 processing but also has a significant negative influence on the efficiency with which the initial cleavages at sites A0–A2 are carried out. This phenotype is similar to that caused by depletion or mutation of several other 60S subunit r-proteins in yeast (26,28,29) and, in fact, is also seen upon inactivation or depletion of a number of non-ribosomal proteins specifically involved in maturation of the large subunit rRNAs (3).
Figure 6.

Effect of depletion of L25 on pre-rRNA processing. YCR61 cells transformed with the empty YCplac111 plasmid were shifted from galactose- to glucose-based medium. Total RNA was isolated immediately before (lane 1) and 3 h after (lane 2) the shift. The RNA was subjected to northern analysis using probe E complementary to the region of ITS1 upstream from site A2.
We also assessed the effect of L25 depletion on the extent of cleavage at individual processing sites by primer extension experiments. As shown in Figure 7 (lanes 3 and 4), the data are consistent with the conclusions drawn from the northern hybridization experiments. The intensity of the primer extension stops corresponding to sites A1 and A2 decreases substantially upon depletion of the r-protein (Fig. 7B and C), whereas the intensity of that corresponding to site A3 remains constant (Fig. 7D). The stop corresponding to site B1S, on the other hand, increases in intensity (Fig. 7E), confirming accumulation of the 27SB precursor species. The only discrepancy between the northern and primer extension data is the increase in the signal corresponding to A0 in the L25-depleted cells. However, the reduced efficiency of cleavage at A0 in L25-depleted cells is amply substantiated by the increased level of 35S pre-rRNA, accumulation of the 23S species and a reduction in the level of the A0–A1 fragment detected by northern hybridization (Figs 3, 5 and 6).
Figure 7.
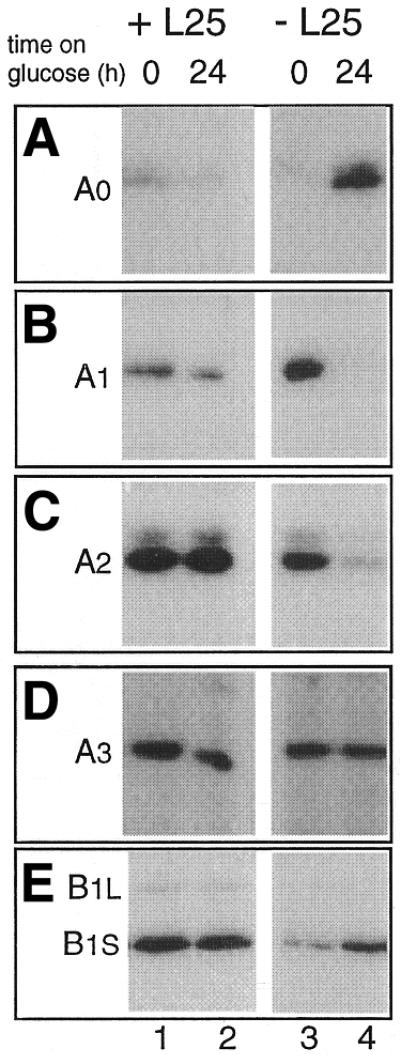
Effect of depletion of L25 on cleavage at different processing sites. YCR61 cells transformed with the YCplac111 plasmid either with the wild-type L25 gene (lanes 1 and 2) or without an insert (lanes 3 and 4) were shifted from galactose- to glucose-based medium. Total RNA was isolated immediately before (lanes 1 and 3) and 24 h after (lanes 2 and 4) the shift. The RNA was subjected to primer extension analysis. (A and B) Probe F; (C–E) probe H. See Figure 1 for location of the probes. The site detected in each of the different panels is indicated.
Inactivation of any of the three functional domains of L25 affects pre-rRNA processing in the same manner
As outlined above, in addition to a central, (pre-)rRNA-binding domain L25 possesses two further distinct functional domains. One of these, located N-terminally, contains the signals for nucle(ol)ar import of the protein (34), while the role of the other, located C-terminally, has not been precisely determined (36). We have previously characterized various mutations in each of these three functional domains with respect to their effect on cell growth and the rRNA-binding capacity of L25 (33,34,36)
We first addressed the role of the rRNA-binding domain using mutants M3 and M5, each of which carries amino acid substitution(s) in the highly conserved motif at the C-terminus of this domain. In mutant M5 the two Lys residues at positions 120 and 121 have been replaced by Arg, which reduces the in vitro rRNA-binding capacity of the protein to 20% of that shown by its wild-type counterpart, but has no discernable effect on growth (36). Mutant M3, on the other hand, in which the Leu residue at position 126 has been replaced by Lys, has completely lost the ability to bind to 25S rRNA in vitro and does not support growth (33). The northern hybridization data obtained with the two mutants are shown in Figures 2 and 3 (lanes 7–10). In mutant M5 we observed normal levels of the mature and precursor species in RNA isolated after the shift to glucose (lanes 7 and 8), which is in accordance with the normal growth rate of this mutant. RNA prepared from mutant M3, on the other hand, showed a pattern of decreased and elevated levels of the various mature and precursor species very similar to that seen in cells depleted of L25 (lanes 9 and 10). M3 mutant cells also suffer the same rapid loss of 7S pre-rRNA (Fig. 4, lanes 7–9) and severely reduced levels of the A0–A1 spacer fragment (Fig. 5, lanes 5 and 6) as L25-depleted cells. These results clearly indicate that assembly of L25 with the pre-rRNA is essential to allow pre-rRNA processing, both at the early sites and within ITS2, to proceed normally.
The N-terminal 61 amino acids of L25 are completely dispensable for its interaction with the (pre-)rRNA (33). However, this domain contains the information for nuclear import of the r-protein, which is located within the first 41 residues (34). In order to ascertain the possible involvement of this N-terminal domain in pre-rRNA processing we used mutant SΔN lacking these 41 N-terminal amino acids. In order to ensure that the truncated protein can still be actively translocated to the nucleus, however, it was fused to the heterologous NLS from the SV40 large T antigen, which is known to act efficiently in yeast (34,42,43).
The SΔN mutant displays a processing phenotype very similar to that of M3 mutant cells (Figs 2 and 3, lanes 5 and 6), indicating that although its capacity to associate with the (pre-)rRNA is not disturbed, it has nevertheless lost the ability to support efficient pre-rRNA processing at sites A0–A2 and C2/C2′. This observation explains why the mutant protein, although it does support slow growth of YCR61 cells on glucose, fails to restore the growth rate to its wild-type value (34).
Finally, we analyzed mutations in the C-terminal domain of L25 downstream from its rRNA-binding domain. Previous experiments demonstrated the presence of an important functional element in this region encompassing all or some of residues 134–138 (36). Introduction of a non-conservative point mutation (L→R) at position 135 reduced the growth rate ∼3-fold and the rRNA-binding capacity to 70% of the wild-type value. Removal of the four C-terminal amino acids 139–142 (mutant ΔC4) caused the same small reduction in (pre-)rRNA-binding capacity but had no effect on growth (36).
Mutant ΔC4 shows the expected wild-type pattern of mature and precursor RNAs (Figs 2 and 3, lanes 11 and 12), while the processing phenotype of mutant L135→R is once more highly similar to that seen for cells depleted of L25 or expressing either the M3 or SΔN mutant forms of the r-protein (Figs 2 and 3, lanes 13 and 14). This includes the rapid loss of 7S pre-rRNA (Fig. 4, lanes 10–12) as well as disappearance of the A0–A1 fragment (Fig. 5, lanes 7 and 8). Thus, the previously identified functional element within the C-terminal domain is a further feature of L25 required to allow normally efficient pre-rRNA processing at sites A0–A2 and C2/C2′.
DISCUSSION
Over the past decade a host of non-ribosomal factors that are required for pre-rRNA processing in eukaryotic cells have been identified and the manner in which the processing pathway is disturbed by their absence or mutation has been established. In stark contrast, very little is known about the role of individual ribosomal proteins in formation of the mature rRNA species, although the available data, all from yeast, do make it clear that the absence of at least some r-proteins has a negative effect on pre-rRNA processing (26–31).
In this paper we describe an investigation of the role in pre-rRNA processing of the yeast large subunit r-protein L25, probably the most extensively studied eukaryotic r-protein to date. Kinetic labeling experiments indicate that L25 is one of the first r-proteins to be assembled with the pre-rRNA (20). Further analysis has shown that L25 specifically and independently recognizes a region located in domain III of the 25S rRNA, which we have characterized in detail (22,32,44), and that the protein is able to bind directly to the 35S pre-rRNA (24). Finally, in vitro and in vivo mutational analysis of L25 (33,34,36) has provided detailed structure–function information unavailable for other eukaryotic r-proteins, except the exchangeable, acidic species (45)
Using a yeast strain in which the wild-type L25 gene is conditionally expressed we find that genetic depletion of L25 has a drastic effect on the removal of ITS2. Upon shutting off transcription of the L25 gene there is a rapid drop in the steady-state level of the 7S rRNA precursor (Fig. 4), while the 27SB pre-rRNA accumulates (Figs 3 and 7), indicating strong inhibition of processing at site C2/C2′. Furthermore, the northern analysis (Figs 3–6) and primer extension (Fig. 7) experiments clearly indicate that the lack of L25 causes a decrease in the efficiency of processing at the early cleavage sites A1 and A2. The latter indeed appears to be a direct consequence of the failing supply of L25 as it becomes noticeable even before growth of the L25-depleted cells is affected (Fig. 6).
With respect to the effect of L25 depletion on the processing cleavage at site A0, the primer extension and northern hybridization data (Figs 3 and 6) are in apparent contradiction. The latter demonstrates accumulation of the 35S pre-rRNA but not of the 33S precursor having its 5′-end at A0 (Figs 3 and 6), indicating that cleavage at A0 is diminished, whereas primer extension shows an unmistakable increase in the stop corresponding to this cleavage site (Fig. 7A). A similar discrepancy has been previously observed upon depletion of several snoRNP components as well as Rrp5p, a non-ribosomal protein that is required for the early cleavages at A0–A2 (3). A satisfactory explanation for this phenomenon, however, is still lacking. Possibly, it reflects changes in the relative efficiency of cleavage at A0 and A1 leading to an increase in the steady-state level of the A0–A1 fragment. Alternatively, depletion of these trans-acting factors, including L25, causes accumulation of a product extending from A0 to an aberrant cleavage site located upstream of the position of probe E, which would not have been detected by the probes used in the various northern hybridization experiments. It should be noted that potential downstream complements of such an aberrant processing intermediate have been observed in mutants defective in cleavage at A2 and/or A3 (39,46).
The M3 point mutation (L126→K), which abolishes the interaction of L25 with its binding site in 25S rRNA (33), results in a processing phenotype indistinguishable from that seen for L25-depleted cells. This demonstrates that the processing defects observed upon depletion are due to the failure of L25 to associate with the pre-ribosomal particle and not to some other possible role of the r-protein.
The results for the M3 RNA-binding mutant corroborate our previous finding that mutations in 25S rRNA that severely compromise L25 binding inhibit cleavage of ITS2 at site C2/C2′ (32). Although in these previous experiments no detailed analysis of the early processing steps was performed, the mutations also appear to cause a reduction in the level of 18S rRNA, indicating that they do lower the efficiency of cleavage at sites A0–A2 (see fig. 3 in Van Beekvelt et al.; 32). Interestingly, structural alterations in the same region of 25S rRNA that inhibit ITS2 processing but have no effect on L25 binding do not show a drop in 18S rRNA levels, which further supports the argument that assembly of L25 is required for fully efficient cleavage at the early processing sites.
Some mutations in the other two functional domains of L25 that do not or only marginally affect RNA binding (33,36) also result in severe inhibition of ITS2 processing and decrease efficiency of cleavage at the early sites A0–A2. One possible explanation for these observations would be that the expression level of the mutant proteins is very low. We have taken precautions to avoid differences in expression at the level of transcription and translation (see Materials and Methods), leaving metabolic instability of the mutant proteins as the only reasonable cause of such reduced expression levels. As yeast, like other eukaryotes, do not contain significant pools of free r-proteins the cellular levels of these proteins are directly linked to their incorporation in stable ribosomes. Thus, it is impossible to directly assess the effect of a particular mutation on the in vivo half-life of the r-protein. However, we deem it very unlikely that the deleterious mutations in the N- and C-terminal domains have a significant influence on the stability of L25 since other, similar mutations in the same regions have no effect on cell growth and ribosome biogenesis (34,36; this paper). We therefore interpret the negative effect of the SΔN and L135→R mutations on pre-rRNA processing to indicate that both the N- and C-terminal domains contain elements important for both the early and late stages of processing. In the case of the N-terminal domain these element(s) appear to be located in the same region (amino acids 1–41) that contains the information for nuclear import of L25 (34). In the case of the C-terminal domain the necessary element is located within the previously identified, but not functionally characterized, sequence spanning amino acids 134–138 (36).
The precise role of the N- and C-terminal domains of L25 in pre-rRNA processing remains to be determined. A plausible assumption, however, is that they are required for the association of further proteins, either later assembling ribosomal proteins or non-ribosomal proteins involved in subsequent steps in ribosome formation. This is supported by the fact that depletion or mutation of several other large subunit r-proteins, all of which assemble subsequent to L25 according to the labeling data (20), also leads to kinetic delay of the early processing steps as well as inhibition of ITS2 processing (1,3,26–29; and references therein). Furthermore, a number of trans-acting factors primarily involved in removal of ITS2 from the 27SB pre-rRNA also appear to be required for fully efficient cleavage at sites A0–A2 (1,47–50). Unfortunately, there is as yet no information that allows us to deduce whether any of these proteins depend on direct interaction with L25 for their association with the pre-ribosome. Moreover, the involvement of L25 might also be indirect in that it effects a conformational change in the pre-rRNA required for subsequent protein association, as has been observed for a number of bacterial r-proteins (51–56). This would of course imply that the regions of L25 flanking its rRNA-binding domain interact with the pre-rRNA in some manner after the r-protein has been assembled.
The reason for the partial interdependence of 40S and 60S subunit formation in cells in which the latter process is disturbed is still unknown, but two explanations have been proposed (46,50,57–59). On the one hand, the pre-rRNA processing machinery might form a single large complex, whose structural integrity is essential for efficient progression of all steps. On the other hand, the early processing machinery might check for the presence of functional components needed in later steps as part of a quality control mechanism. The latter explanation seems the most plausible one in view of the observation that 18S and 5.8S/25S rRNA can be efficiently produced from physically separated transcription units (60). The negative effect of depletion or mutation of the various large subunit r-proteins tested so far on processing at A0–A2 is a clear indication that this whole set of r-proteins is already present in the 90S pre-ribosome. The same conclusion holds for the various trans-acting factors involved in 60S subunit biogenesis, even though they may not be required until a later stage of the processing pathway.
In summary, the results presented in this paper demonstrate that assembly of L25 with the pre-ribosome is necessary but not sufficient to allow processing at both early (A0–A2) and late (C2/C2′) cleavage sites to proceed normally. Elements in both the N- and C-terminal domains of the protein not involved in efficient recognition of its binding site on the pre-rRNA are also required, underscoring the central role of L25 in 60S subunit formation.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr J. C. Vos for critical reading of the manuscript. This work was supported in part by the Council for Chemical Sciences (CW) with financial aid from the Netherlands Foundation for Scientific Research (NWO).
REFERENCES
- 1.Kressler D., Linder,P. and de la Cruz,J. (1999) Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 19, 7897–7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tollervey D. (1996) Trans-acting factors in ribosome synthesis. Exp. Cell Res., 229, 226–232. [DOI] [PubMed] [Google Scholar]
- 3.Venema J. and Tollervey,D. (1999) Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet., 33, 216–311. [DOI] [PubMed] [Google Scholar]
- 4.Bachellerie J.-P., Cavaillé,J. and Qu,L.-H. (2000) Nucleotide modifications of eukaryotic rRNAs: the world of small nucleolar RNAs guides revisited. In Garrett,R.A., Douthwaite,S.A., Liljas,A., Matheson,A.T., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. American Society for Microbiology, Washington, DC, pp. 191–204.
- 5.Kiss T. (2001) Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J., 20, 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raué H.A. and Planta,R.J. (1995) The pathway to maturity: processing of ribosomal RNA in Saccharomyces cerevisiae. Gene Expr., 5, 71–77. [PMC free article] [PubMed] [Google Scholar]
- 7.Geerlings T.H., Vos,J.C. and Raué,H.A. (2000) The final step in the formation of 25S rRNA in Saccharomyces cerevsiae is performed by 5′→3′ exonucleases. RNA, 6, 1698–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kufel J., Dichtl,B. and Tollervey,D. (1999) Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA, 5, 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stevens A., Hsu,C.L., Isham,K.R. and Larimer,F.W. (1991) Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′→3′ exoribonuclease-1. J. Bacteriol., 173, 7024–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moy T.I. and Silver,P.A. (1999) Nuclear export of the small ribosomal subunit requires the Ran-GTPase cycle and certain nucleoporins. Genes Dev., 13, 2118–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanrobays E., Gleizes,P.-E., Bousquet-Antonelli,C., Noaillac-Depeyre,J., Caizergues-Ferrer,M. and Gélugne,J.-P. (2001) Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essentail non-ribosomal cytoplasmic protein. EMBO J., 20, 4204–4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shuai K. and Warner,J.R. (1991) A temperature-sensitive mutant of Saccharomyces cerevisiae defective in pre-rRNA processing. Nucleic Acids Res., 19, 5059–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindahl L., Archer,R.H. and Zengel,J.M. (1992) A new rRNA processing mutant of Saccharomyces cerevisiae. Nucleic Acids Res., 20, 295–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmitt M.E. and Clayton,D.A. (1993) Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 7935–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindahl L. and Zengel,J.M. (1996) RNase MRP and rRNA processing. Mol. Biol. Rep., 22, 69–73. [DOI] [PubMed] [Google Scholar]
- 16.Lygerou Z., Allmang,C., Tollervey,D. and Séraphin,B. (1996) Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science, 272, 268–270. [DOI] [PubMed] [Google Scholar]
- 17.Henry Y., Wood,H., Morrissey,J.P., Petfalski,E., Kearsey,S. and Tollervey,D. (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J., 13, 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell P., Petfalski,E. and Tollervey,D. (1996) The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev., 10, 502–513. [DOI] [PubMed] [Google Scholar]
- 19.Trapman J., Retèl,J. and Planta,R.J. (1975) Ribosomal precursor particles from yeast. Exp. Cell Res., 90, 95–104. [DOI] [PubMed] [Google Scholar]
- 20.Kruiswijk T., Planta,R.J. and Krop,J.M. (1978) The course of the assembly of ribosomal subunits in yeast. Biochim. Biophys. Acta, 517, 378–389. [DOI] [PubMed] [Google Scholar]
- 21.Mager W.H., Planta,R.J., Ballesta,J.-P.G., Lee,J.C., Mizuta,K., Suzuki,K., Warner,J.R. and Woolford,J. (1997) A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae. Nucleic Acids Res., 25, 4872–4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.El-Baradi T.T.A.L., Raué,H.A., De Regt,V.C.H.F., Verbree,E.C. and Planta,R.J. (1985) Yeast ribosomal protein L25 binds to an evolutionary conserved site on yeast 26S and E.coli 23S rRNA. EMBO J., 4, 2101–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Baradi T.T.A.L., de Regt,V.C.H.F., Einerhand,S.W., Teixido,J., Planta,R.J., Ballesta,J.P.G. and Raué,H.A. (1987) Ribosomal proteins EL11 from Escherichia coli and L15 from Saccharomyces cerevisiae bind to the same site in both yeast 26S and mouse 28S rRNA. J. Mol. Biol., 195, 909–917. [DOI] [PubMed] [Google Scholar]
- 24.Yeh L.C. and Lee,J.C. (1998) Yeast ribosomal proteins L4, L17, L20 and L25 exhibit different binding characteristics for the yeast 35S precursor rRNA. Biochim. Biophys. Acta, 1443, 139–148. [DOI] [PubMed] [Google Scholar]
- 25.Vilardell J., Yu,S.J. and Warner,J.R. (2000) Multiple functions of an evolutionarily conserved RNA binding domain. Mol. Cell, 5, 761–766. [DOI] [PubMed] [Google Scholar]
- 26.Deshmukh M., Tsay,Y.F., Paulovich,A.G. and Woolford,J.L. (1993) Yeast ribosomal protein L1 is required for the stability of newly synthesized 5S rRNA and the assembly of 60S ribosomal subunits. Mol. Cell. Biol., 13, 2835–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moritz M., Paulovich,A.G., Tsay,G.Y.-F. and Woolford,J.L. (1990) Depletion of yeast ribosomal proteins L16 or rp59 disrupts ribosome assembly. J. Cell Biol., 111, 2261–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Underwood M.R. and Fried,H.M. (1990) Characterization of nuclear localizing sequences derived from yeast ribosomal protein L29. EMBO J., 9, 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vilardell J. and Warner,J.R. (1997) Ribosomal protein L32 of Saccharomyces cerevisiae influences both the splicing of its own transcript and the processing of rRNA. Mol. Cell. Biol., 17, 1959–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finley D., Bartel,B. and Varshavsky,A. (1989) The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature, 338, 394–401. [DOI] [PubMed] [Google Scholar]
- 31.Ford C.L., Randal-Whitis,L. and Ellis,S.R. (1999) Yeast proteins related to the p40/laminin receptor precursor are required for 20S ribosomal RNA processing and the maturation of 40S ribosomal subunits. Cancer Res., 59, 704–710. [PubMed] [Google Scholar]
- 32. Van Beekvelt C.A., Kooi,E.A., De Graaff-Vincent,M., Van ’t Riet,J., Venema,J. and Raué,H.A. (2000) Domain III of Saccharomyces cerevisiae 25S ribosomal RNA: its role in binding of ribosomal protein L25 and 60S subunit formation. J. Mol. Biol., 296, 7–17. [DOI] [PubMed] [Google Scholar]
- 33.Rutgers C.A., Rientjes,J.M.J., Van ’t Riet,J. and Raué,H.A. (1991) rRNA binding domain of yeast ribosomal protein L25. Identification of its borders and a key leucine residue. J. Mol. Biol., 218, 375–385. [DOI] [PubMed] [Google Scholar]
- 34.Schaap P.J., Van ’t Riet,J., Woldringh,C.L. and Raué,H.A. (1991) Identification and functional analysis of the nuclear localization signals of ribosomal protein L25 from Saccharomyces cerevisiae. J. Mol. Biol., 221, 225–237. [DOI] [PubMed] [Google Scholar]
- 35.Rutgers C.A., Schaap,P.J., van ’t Riet,J., Woldringh,C.R. and Raué,H.A. (1990) In vivo and in vitro analysis of structure-function relationships in ribosomal protein L25 from Saccharomyces cerevisiae. Biochim. Biophys. Acta, 1050, 74–79. [DOI] [PubMed] [Google Scholar]
- 36.Kooi E.A., Rutgers,C.A., Kleijmeer,M.J., Van ’t Riet,J., Venema,J. and Raué,H.A. (1994) Mutational analysis of the C-terminal region of Saccharomyces cerevisiae ribosomal protein L25 in vitro and in vivo demonstrates the presence of two distinct functional elements. J. Mol. Biol., 240, 243–255. [DOI] [PubMed] [Google Scholar]
- 37.Gietz D., Jean,A.S., Woods,R.A. and Schiestl,R.H. (1992) Improved methods for high efficiency transformation of intact yeast cells. Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venema J., Planta,R.J. and Raué,H.A. (1998) In vivo mutational analysis of ribosomal RNA in Saccharomyces cerevisiae. In Martin,R. (ed.), Protein Synthesis: Methods and Protocols. Humana Press, Totowa, NJ, Vol. 77, pp. 257–270. [DOI] [PubMed]
- 39.Venema J. and Tollervey,D. (1996) RRP5 is required for formation of both 18S and 5.8S rRNA in yeast. EMBO J., 15, 5701–5714. [PMC free article] [PubMed] [Google Scholar]
- 40.Beltrame M. and Tollervey,D. (1992) Identification and functional analysis of two U3 binding sites on yeast preribosomal RNA. EMBO J., 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Venema J. and Tollervey,D. (1995) Processing of pre-ribosomal RNA in Saccharomyces cerevisiae. Yeast, 11, 1629–1650. [DOI] [PubMed] [Google Scholar]
- 42.Kalinich J.F. and Douglas,M.G. (1989) In vitro translocation through the yeast nuclear envelope—signal-dependent transport requires ATP and calcium. J. Biol. Chem., 264, 17979–17989. [PubMed] [Google Scholar]
- 43.Nelson M. and Silver,P. (1989) Context affects nuclear protein localization in Saccharomyces cerevisiae. Mol. Cell. Biol., 9, 384–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kooi E.A., Rutgers,C.A., Mulder,A., van ’t Riet,J., Venema,J. and Raué,H.A. (1993) The phylogenetically conserved doublet tertiary interaction in domain III of the large subunit rRNA is crucial for binding of ribosomal protein. Proc. Natl Acad. Sci. USA, 90, 213–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ballesta J.P.G., Guarinos,E., Zurdo,J., Parada,P., Nusspaumer,G., Lalioti,V.S., Perez-Fernandez,J. and Remacha,M. (2000) Structure of the yeast ribosomal stalk. In Garrett,R.A., Douthwaite,S.A., Matheson,A., Liljas,A., Moore,P.B. and Noller,H.F. (eds), The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. American Society for Microbiology, Washiongton, DC, pp. 115–125.
- 46.Allmang C., Henry,Y., Morrissey,J.P., Wood,H., Petfalski,E. and Tollervey,D. (1996) Processing of the yeast pre-rRNA at sites A2 and A3 is linked. RNA, 2, 60–73. [PMC free article] [PubMed] [Google Scholar]
- 47. de la Cruz J., Kressler,D., Rojo,M., Tollervey,D. and Linder,P. (1988) Sbp4p, an essential putative RNA helicase, is required for a late step in the assembly of 60S ribosomal subunits in Saccharomyces cerevisiae. RNA, 4, 1268–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gautier T., Berges,T., Tollervey,D. and Hurt,E. (1997) Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol., 17, 7088–7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong B., Brockenbrough,J.C., Wu,P. and Aris,J.P. (1997) Nop2p is required for pre-rRNA processing and 60S ribosome subunit synthesis in yeast. Mol. Cell. Biol., 17, 378–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zanchin N.I.T., Roberts,P., Desilva,A., Sherman,F. and Goldfarb,D.S. (1997) Saccharomyces cerevisiae Nip7p is required for efficient 60S ribosome subunit biogenesis. Mol. Cell. Biol., 17, 5001–5015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Svensson P., Changchien,L.-M., Craven,G.R. and Noller,H. (1988) Interaction of ribosomal proteins, S6, S8, S15 and S18 with the central domain of 16S rRNA. J. Mol. Biol., 200, 301–308. [DOI] [PubMed] [Google Scholar]
- 52.Stern S., Wilson,R.C. and Noller,H.F. (1986) Localization of the binding site for protein S4 on 16S ribosomal RNA by chemical and enzymatic probing and primer-extension. J. Mol. Biol., 192, 101–110. [DOI] [PubMed] [Google Scholar]
- 53.Powers T., Changchien,L.-M., Craven,G.R. and Noller,H.F. (1988) Probing the assembly of the 3′ major domain of 16S ribosomal RNA. Quaternary interactions involving ribosomal proteins S7, S9 and S19. J. Mol. Biol., 200, 309–319. [DOI] [PubMed] [Google Scholar]
- 54.Mandiyan V., Tumminia,S., Wall,J.S., Hainfeld,J.F. and Boublik,M. (1989) Protein-induced conformational changes in 16S ribosomal RNA during the initial assembly steps of the Escherichia coli 30S ribosomal subunit. J. Mol. Biol., 210, 323–336. [DOI] [PubMed] [Google Scholar]
- 55.Leffers H., Egebjerg,J., Andersen,A., Christensen,A. and Garrett,R.A. (1988) Domain VI of Escherichia coli 23S ribosomal RNA. Structure, assembly and function. J. Mol. Biol., 204, 507–522. [DOI] [PubMed] [Google Scholar]
- 56.Egebjerg J., Leffers,H., Christensen,A., Andersen,H. and Garrett,R.A. (1987) Structure and accessibility of domain I of Escherichia coli 23S RNA in free RNA, in the L24-RNA complex and in 50S subunits. J. Mol. Biol., 196, 125–136. [DOI] [PubMed] [Google Scholar]
- 57.Allmang C. and Tollervey,D. (1988) The role of the 3′ external transcribed spacer in yeast pre-rRNA processing. J. Mol. Biol., 78, 67–78. [DOI] [PubMed] [Google Scholar]
- 58.Kressler D., de la Cruz,J., Rojo,M. and Linder,P. (1998) Dbp6p is an essential putative ATP-dependent RNA helicase required for 60S ribosomal subunit assembly in Saccharomyces cerevisiae. Mol. Cell. Biol., 18, 1855–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Daugeron M.-C. and Linder,P. (1998) Dbp7p, a putative ATP-dependent RNA helicase from Saccharomyces cerevisiae, is required for 60S ribosomal subunit assembly. RNA, 4, 566–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liang W.-Q. and Fournier,M.J. (1997) Synthesis of functional eukaryotic ribosomal RNAs in trans: development of a novel in vitro rDNA system for dissecting ribosome biogenesis. Proc. Natl Acad. Sci. USA, 94, 2864–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]