Abstract
The Mekong Delta River in Vietnam is facing salinity intrusion caused by climate change and sea-level rise that is severely affecting rice cultivation. Here, we evaluated salinity responses of 97 rice accessions (79 landraces and 18 improved accessions) from the Mekong Delta population by adding 100 mM NaCl to the nutrient solution for up to 20 days. We observed a wide distribution in salinity tolerance/sensitivity, with two major peaks across the 97 accessions when using the standard evaluation system (SES) developed by the International Rice Research Institute. SES scores revealed strong negative correlations (ranging from − 0.68 to − 0.83) with other phenotypic indices, such as shoot elongation length, root elongation length, shoot dry weight, and root dry weight. Mineral concentrations of Na+ in roots, stems, and leaves and Ca2+ in roots and stems were positively correlated with SES scores, suggesting that tolerant accessions lower their cation exchange capacity in the root cell wall. The salinity tolerance of Mekong Delta accessions was independent from the previously described salinity tolerance–related locus Saltol, which encodes an HKT1-type transporter in the salinity-tolerant cultivars Nona Bokra and Pokkali. Indeed, genome-wide association studies using SES scores and shoot dry weight ratios of the 79 accessions as traits identified a single common peak located on chromosome 1. This SNP did not form a linkage group with other nearby SNPs and mapped to the 3′ untranslated region of gene LOC_Os01g32830, over 6.5 Mb away from the Saltol locus. LOC_Os01g32830 encodes chloroplast glycolate/glycerate translocator 1 (OsPLGG1), which is responsible for photorespiration and growth. SES and shoot dry weight ratios differed significantly between the two possible haplotypes at the causal SNP. Through these analyses, we characterize Doc Phung, one of the most salinity-tolerant varieties in the Mekong Delta population and a promising new genetic resource.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12284-022-00613-0.
Keywords: Mekong Delta, Salinity tolerance, Rice, Doc Phung, GWAS (genome-wide association study), SES (standard evaluation system), Mineral concentrations, OsPLGG1
Background
Rice (Oryza sativa L.) is a main staple food source for about 90% of Asian people and is grown in areas with easy access to water. One of greatest threats to rice production is salinity stress (Maas and Grattan 1999). In coastal and/or acid and semi-arid areas, the effect of salinity conditions is a major issue for rice production. In particular, about 500,000 ha around coastal areas in the Mekong Delta/River are predicted to become affected by salinity intrusion caused by climate change and the rise of sea levels by 2050, with this number increasing tenfold when considering the worst case scenario of combined climate change, sea-level rise, and low river flows (Kontgis et al. 2019). Salinity intrusion from the sea can impose salinity stress during the dry season, when saltwater seeps into the ground freshwater (Nhung et al. 2019). The use of salinity-tolerant varieties is seen as a more efficient and effective mitigating measure than farming practices or salinity management facilities such as dikes, pumping stations, and sluices (Akbar et al. 1972).
Rice is generally sensitive to high-salinity conditions, especially at the seedling stage (Batayeva et al. 2018; De Leon et al. 2016; Kumari et al. 2009; Lutts et al. 1995; Mohammadi-Nejad et al. 2010; Puram et al. 2017; Tian et al. 2011). Much effort has been made to screen and select salinity-tolerant rice varieties (De Leon et al. 2017; Ganie et al. 2019; Hariadi et al. 2015; Lee et al. 2003), such as the high-salinity-tolerant ‘Nona Bokra’ and ‘Pokkali’, which have greatly contributed to salinity-tolerant rice breeding (Ren et al. 2005; Thomson et al. 2010). These indica strains are landraces from the Indian and Sri Lanka coastal regions, respectively. Their salinity-tolerance may have been achieved by adapting to high-salinity soils (Emon et al. 2015; Gregorio and Senadhira 1993). The Saltol locus defined a strong-effect quantitative trait locus (QTL) associated with this salinity-tolerance and was mapped to chromosome 1 (Bonilla et al. 2002; Xie et al. 2000). The Saltol locus was later shown to encode a HIGH-AFFINITY K+ TRANSPORTER1 (HKT1)-type sodium (Na+) transporter (Ren et al. 2005). However, the biological response to salt is extremely complex, and not all salinity-tolerance can be explained by the Saltol locus. In fact, other salinity-tolerant landrace varieties from Bangladesh and Saudi Arabia have a Saltol haplotype distinct from that of Nona Bokra or Pokkali (Bimpong et al. 2014; Rahman et al. 2017, 2019). Several QTL analyses have revealed various mechanisms involved in rice salinity-tolerance (Singh et al. 2021).
The mechanisms related to salinity-tolerance of rice can broadly be divided into three categories (Marschner 2012): Na+ exclusion, osmotic stress regulation, and tissue tolerance to Na+ accumulation (Munns and Tester 2008). Na+ absorbed by the roots is transported from the xylem to the shoots. Na+ accumulation in the shoots is negatively correlated with salinity-tolerance, as excess Na+ levels in cells have a detrimental effect on plant growth (Faiyue et al. 2012; Krishnamurthy et al. 2011; Platten et al. 2013). In the context of Na+ exclusion, the salinity-resistant Saltol haplotype, such as that harbored by Nona Bokra and Pokkali, rely on the activity of OsHKT1;5, a Na+ ion transporter that can avoid the accumulation of Na+ in shoots (Platten et al. 2013; Ren et al. 2005). Osmotic regulation modulates stomatal conductance before too much Na+ accumulates in shoots by reducing water evaporation and, thus, the flow of water from roots to shoots, resulting in salinity-tolerance (Rios et al. 2017; Termaat et al. 1985). Tissue tolerance refers to the mechanism that prevents toxic Na+ levels from accumulating in cells (Davenport et al. 2005). Several known mechanisms prevent Na+ from being released into the cytoplasm by sequestration of Na+ into vacuoles or by releasing Na+ into the soil (Anil et al. 2007; Bassil et al. 2012; Kader et al. 2007). Under saline conditions, concentrations for the cation Na+ and the anion Cl– increase in all plant organs, while the absorption of K+, Ca2+, Mg2+, and Mn2+ decrease (Grattan and Grieve 1999). Limiting the uptake of these essential ions negatively affects plant growth (Kumar et al. 2015).
In this study, we investigated the salinity-tolerance and physiological characteristics of a diverse panel of landrace rice accessions cultivated in the Mekong Delta region (Tam et al. 2019). We focused on the rice landrace variety ‘Doc Phung’, known to be particularly resistant to salt in the Mekong Delta (Ho et al. 2018; Le 1999; Tin et al. 2021). Based on the genomic structure of these Mekong Delta populations and GWAS results, we explored and resolved the relationship between the degree of salinity-tolerance, physiological characteristics, and genomic structure. Analysis of whole-genome sequences provided evidence that Doc Phung carries a Saltol allele different from that of Nona Bokra or Pokkali and that the causative region of salinity-tolerance might have enabled the adaptation to the coastal areas of the Mekong Delta.
Materials and Methods
Plant Materials
Ninety-seven rice accessions consisting of 79 landraces and 18 improved accessions were selected from the Genebank of Mekong Delta Development Research Institute (MDI), Can Tho University, Vietnam (Additional file 2: Table S1) (Tam et al. 2019). The group of accessions exhibited no strong population structure bias according to our previous analyses (Tam et al. 2019). ‘Nipponbare’ and ‘Pokkali’ were added as sensitive and tolerant cultivar controls, respectively, for salinity stress. However, as Pokkali supplied by the International Rice Research Institute (IRRI) did not show the expected high tolerance in our assays, we stopped using this material.
Screening for Salinity Tolerance
Seeds were treated with disinfectant (20% [v/v] solution of Tekurido C, Kumiai Chemical Industry Co., LTD) for 15 min for surface sterilization. Seeds were then washed with tap water three times and with distilled water two times. Surface-sterilized seeds were placed into petri dishes and germinated in dark conditions at 25 °C. Three days after germination, seedlings were transplanted to net sheets floating on distilled water in a 40-L container at 20–25 °C in the greenhouse (Additional file 1: Fig. S1). Four days after transfer (DAT), water was replaced with standard nutrient solution: 1.12 mM NH4NO3, 0.32 mM NaH2PO4, 0.19 mM K2SO4, 0.38 mM KCl, 1.25 mM CaCl2, 0.82 mM MgSO4, 35.8 µM Fe-EDTA, 9.1 µM Mn2SO4, 3.06 µM ZnSO4, 0.16 µM CuSO4, 46.2 µM H3BO3, and 0.052 µM (NH4)6Mo7O24. The nutrient solution was replaced every 3 days, and the pH was adjusted to 5.0 every day with 1 M NaOH or 0.5 M HCl, as needed. At 15 DAT, the salinity stress treatment was started by adding 100 mM NaCl to the nutrient solution. Immediately before treatment, the shoot and root lengths of all samples were measured. Five days under treatment (DUT), shoot and root elongation lengths were determined, as well as visible phenotypes in response to salinity stress, using the modified standard evaluation system (SES) score developed by IRRI (IRRI 2013) to assess salinity tolerance in each accession (Additional file 2: Table S2). At 20 DUT, all phenotypes were scored again with the SES, and shoot and root elongation lengths were measured, after which the samples were collected. All plants were washed in tap water (twice) and then in distilled water twice to remove salts on the plant surface. Plants were separated into two parts (shoot and root), dried at 70 °C for 80 h, and weighed as shoot dry weight and root dry weight. The shoot dry weight of each accession was used for the genome-wide association study (GWAS).
Mineral Analysis
To perform mineral analysis, 20 accessions with varying SES scores were selected from the initial 97 MDI accessions (Additional file 2: Supplementary Table S1). Plants grown in standard nutrient solution or with 100 mM NaCl for 20 days (20 DUT) were dried and divided into four parts: roots, upper stems (hereafter stems), lower stems, and leaves; lower stem samples were not analyzed due to contamination of the culture solution. Samples were ground to powder (30–40 mg) and digested with 2 mL of 60% (w/v) HNO3 in a tube at 107.5 °C in a DigiPREP apparatus (SCP Science, Quebec, Canada) for 4 h; 0.5 mL of 30% hydrogen peroxide was added, and the samples were heated to 107.5 °C. The tubes were allowed to cool to room temperature, and their volumes were adjusted to 10 mL with 2% (w/v) HNO3. The mineral ion composition for Na+, K+, Ca2+, and Mg2+ was determined by inductively coupled mass spectrometry (ICP-MS) (ELAN, DRC-e, Perkin-Elmer, Waltham, MA, USA) using a blank sample to account for chemical contaminations.
Mineral Data Analysis
The average value of three biological replications for each accession was calculated. The relative elongation rates of shoots and roots were calculated as follows:
where R is relative elongation, A is (shoot length/root length at 20 DUT—shoot length/root length at 0 DUT) of NaCl treatment, and B is (shoot length/root length at 20 DUT—shoot length/root length at 0 DUT) of control treatment.
Genome-Wide Association Study (GWAS)
A dataset of 578,704 single nucleotide polymorphisms (SNPs) distributed across the 12 rice chromosomes in the 97 accessions previously obtained by restriction site–associated DNA sequencing (RAD-seq) (Tam et al. 2019) was filtered for a minimum call rate of 60%, a heterozygous rate below 20%, and a minor allele frequency below 2%, resulting in 37,643 SNPs. Imputation of missing genotypes was performed using Beagle v5.0 (Browning et al. 2018). The set of 37,643 SNPs was also filtered for 79 landraces based on the same criteria above, yielding 29,881 SNPs. GWAS was performed using a general linear model (GLM) in TASSEL 5.2.50 (Bradbury et al. 2007). The population structure component was determined using principal component analysis (PCA) implemented in TASSEL. The significant threshold was set to p < 0.0001 (− Log10 p value > 4.0). Subsequently, SNPs with a q-value (false discovery rate [FDR]-adjusted p value) lower than 0.05 were selected as significant markers with R software (Storey et al. 2004).
Whole-Genome Resequencing
The whole-genome resequencing of the 20 MDI accessions selected from the 97 MDI accessions (Additional file 2: Table S1) was done using the Illumina Hiseq 150-bp Paired End platform, and 4.5 Gb data were obtained for each accession. After filtering and trimming low-quality reads, read mapping to reference genome Nipponbare and variant calling were performed using BWA-mem (Li and Durbin 2009) and the GATK pipeline (DePristo et al. 2011).
Haplotype Analysis
Candidate genes (near peak SNPs) were searched in QTL and gene databases in the rice SNP seek database (Mansueto et al. 2017). The position of the candidate SNP was identified by RAP-DP (https://rapdb.dna.affrc.go.jp/). Linkage disequilibrium (LD) analysis was performed using SNPs located within 200-kb windows upstream and downstream of the SNP peaks based on the Haploview method according to Barrett et al. (2005).
Results
Evaluation of 97 Rice Accessions from the Mekong Delta Region
We evaluated the salinity tolerance of 97 accessions from the Mekong Delta region to select highly tolerant accessions. In preliminary tests with 50 and 100 mM of NaCl solution, 100 mM NaCl clearly revealed differences across accessions (Additional file 2: Table S3). We scored the salinity responses according to SES developed by IRRI (Additional file 2: Table S2) (IRRI 2013). Distributions of SES scores for salinity-tolerance changed among different accessions at 5, 10, 15, and 20 days into salinity treatment (DUT). As the length of salinity exposure increased, so did the SES scores, with two peaks emerging with SES scores between 3–4 and 8–9 (Fig. 1). At 5 DUT, about 66% of all accessions presented a SES score below 3 (indicating high tolerance), while the remaining 34% of accessions had SES scores between 3 and 5 (tolerance). By contrast, only five accessions retained an SES score below 3 at 20 DUT, 47 accessions had an SES score of 3–5 (moderate tolerance), and 28 accessions had an SES score of 8–9 (sensitive) (Fig. 1). We concluded that 20 DUT is the most suitable screening condition to evaluate salinity responses of the accessions.
Fig. 1.
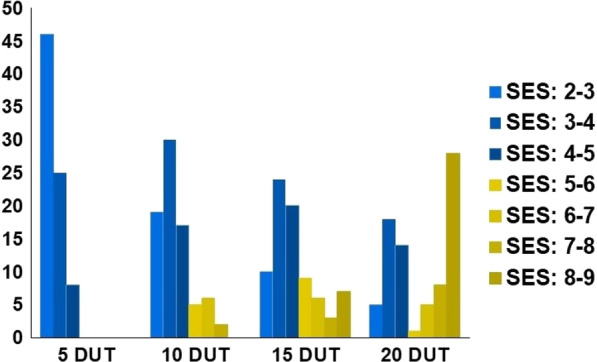
Changes in SES score distributions among 97 accessions from 5 to 20 days under treatment (DUT) with 100 mM NaCl. The responses of the 97 rice accessions to 100 mM NaCl treatment were assessed with three replications, evaluated by the SES score, and recorded at 5, 10, 15, and 20 DUT. As indicated in Additional file 2: Table S2, SES scores were grouped into seven classes: 2–3, 3–4, 4–5, 5–6, 6–7, 7–8, and 8–9, as indicated by different colors. SES scores of 2–5 defined salinity-tolerant accessions; SES score above 5 were considered salinity-sensitive accessions
Correlations Between Phenotypic Traits and SES Scores from 97 Accessions
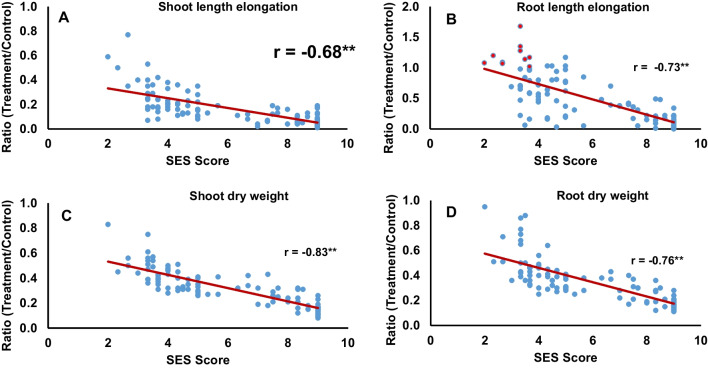
To explore possible correlations between SES scores and phenotypic traits, we examined shoot and root elongation lengths, as well as shoot and root dry weights after exposure to 100 mM NaCl for 20 days (Additional file 2: Table S4). Accordingly, we calculated the ratios of these four traits between NaCl-treated and control samples for all accessions. All ratios at 20 DUT were strongly and negatively correlated with SES scores (Fig. 2). Pearson’s correlation coefficients also supported these strong negative correlations, with values ranging from –0.68 to –0.83 (Fig. 2A–D). The shoot dry weight ratio showed the strongest correlation with SES score, with a Pearson’s correlation coefficient of –0.83 (Fig. 2C), while the shoot length ratio was least affected with a Pearson’s correlation coefficient of − 0.68 (Fig. 2A). When plotting the sum of the ratios for the four above traits as a function of SES score, we noticed that the top eight summed phenotypic values match eight of the nine lowest SES scores (Additional file 2: Table S4), indicating that SES scores are an acceptable index reflecting salinity tolerance. Among the examined accessions, MDI-21 showed the best performance in terms of salinity tolerance, as evidenced by a low SES score and high phenotypic ratios (Additional file 2: Table S4). Notably, MDI-21 was the variety Doc Phung, known to be highly salinity tolerant (Ho et al. 2018). Interestingly, according to our previous phylogenetic analyses of the MDI accessions (Tam et al. 2019), the closest accession to Doc Phung was MDI-13, called “Trang Phieu”, of which SES was 2.33, one of the most tolerant accessions (Additional file 2: Table S4).
Fig. 2.
Correlations between four phenotypic traits and SES scores across the 97 accessions at 20 DUT with 100 mM NaCl. A Shoot length elongation ratio. B Root length elongation ratio. C Shoot dry weight ratio. D Root dry weight ratio. All correlations were significant (p value ≤ 0.01). r, Pearson’s correlation coefficient
Mineral Concentrations in Saline Conditions
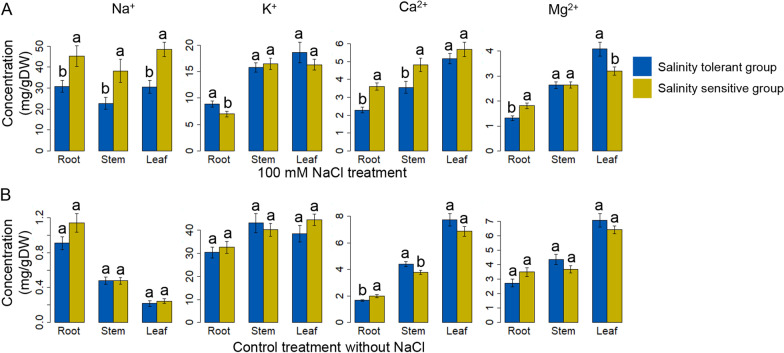
We selected 20 accessions with varying levels of salinity response from the initial collection of 97 MDI landraces to examine their mineral concentrations. These accessions comprised eight salinity-tolerant accessions (SES < 4) and 12 sensitive accessions (SES > 4) (Additional file 2: Table S5). We determined the concentrations of four elements (Na+, K+, Ca2+, and Mg2+) in roots, stems, and leaves (Fig. 3, Additional file 2: Table S5). Under high-salinity conditions (100 mM NaCl), Na+ concentrations were significantly lower at p-value < 0.05 in the tolerant accessions compared to sensitive accessions in all three organs, as were Ca2+ concentrations in roots and stems (Fig. 3A). By contrast, Mg2+ concentrations were significantly higher in leaves of tolerant accessions than in those of sensitive accessions (Fig. 3A). We observed no differences between tolerant or sensitive accessions for K+ in leaves, Ca2+ in leaves, and Mg2+ in stems (Fig. 3A). Under normal growth conditions, the concentrations for Na+, K+, Ca2+, and Mg2+ in roots, stems, and leaves were not different between salinity-sensitive and -tolerant accessions (Fig. 3B). We tested for correlations between mineral concentrations and tolerance/sensitivity, which revealed that Na+ and Ca2+ concentrations are positively correlated with SES scores in the 20 selected accessions, in particular Na+ concentrations in leaves (r = 0.66) and Ca concentrations in roots (r = 0.765) and stems (r = 0.64) (Additional file 1: Fig. S2). We obtained the lowest mineral concentrations in MDI-21 (Doc Phung), which also had the lowest SES at 20 DUT (Additional file 2: Table S5). We observed no clear correlations between K+ concentrations and SES scores among the 20 accessions.
Fig. 3.
Concentrations of four mineral ions (Na+, K+, Ca2+, and Mg2+) in roots, stems, and leaves for salinity-tolerant/sensitive groups. Twenty accessions, consisting of eight tolerant (green: SES scores 2–4) and 12 sensitive (red: SES scores 4–9) accessions, were selected from the initial 97 accessions. A Accessions treated with 100 mM NaCl for 20 days (20 DUT). B Accessions grown under control conditions
Comparison of the OsHKT1;5 Gene Among 20 MDI Accessions and Nona Bokra/Pokkali
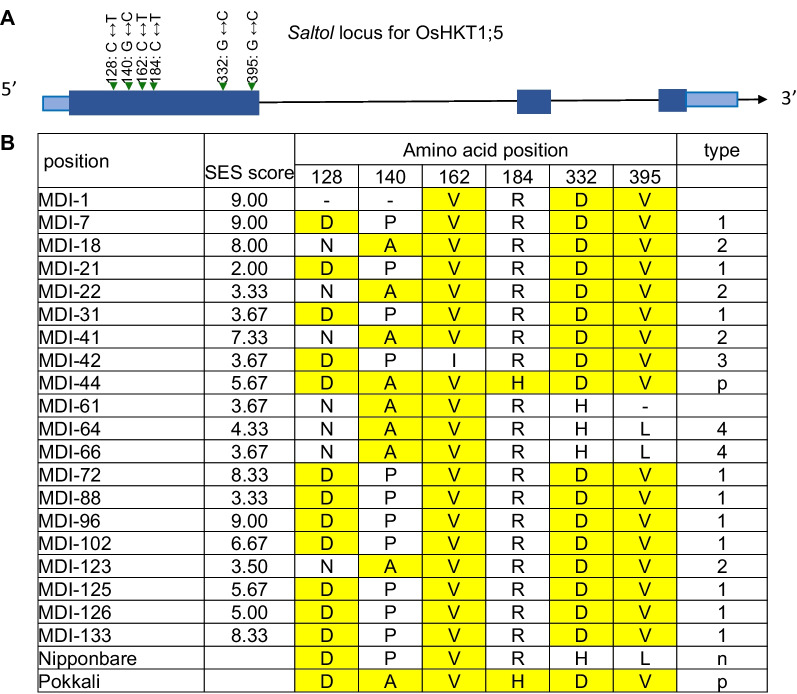
The OsHKT1;5 gene underlies the Saltol locus, whose alleles from the Nona Bokra and Pokkali varieties confer tolerance to high salinity. To assess whether Saltol contributes to salinity sensitivity/tolerance in the MDI accessions, we compared the OsHKT1;5 genomic sequences from the 20 MDI accessions to the Nona Bokra/Pokkali allele. To this end, we extracted the OsHKT1; 5 sequences and inferred the amino acid (aa) sequences from 20 MDI rice accessions representing 97 MDI rice accessions chosen for whole-genome resequencing. In Nona Bokra/Pokkali, the OsHKT1;5 open reading frame was 1,662 bp in length and encoded a protein of 554 aa (Fig. 4A). Six amino acid residues showed nonsynonymous polymorphisms for OsHKT1;5 among the 20 MDI accessions, Nipponbare and Nona Bokra/Pokkali (Fig. 4B), defining six haplotypes. The accession MDI-44 shared the same OsHKT1;5 haplotype with Nona Bokra/Pokkali, although it was not a highly tolerant accession, with an SES score of 4 (Fig. 4B). Haplotype 1 contained 10 accessions, including the highly tolerant MDI-21, and showed a broad range of SES scores (Fig. 4B). The four accessions belonging to haplotype 2 displayed varied SES scores, as with haplotype 1 (Fig. 4B). We concluded from these results that the genotype at OsHKT1;5 is not strongly correlated with salinity tolerance of the 20 MDI accessions tested here. These results thus suggested that the salinity tolerance in the MDI accessions is genetically distinct from the Saltol locus harbored by Nona Bokra/Pokkali.
Fig. 4.
Comparison of amino acid sequences between 20 MDI accessions and Nona Bokra/Pokkali at OsHKT1;5 encoded by the Saltol locus. A Schematic diagram of the OsHKT1;5 gene. Dark blue boxes, exons; light blue boxes, untranslated regions; lines, introns. The six positions of 20 MDI accessions indicate the polymorphisms leading to nonsynonymous amino acid (aa) changes relative to the Saltol allele of the Nona Bokra/Pokkali accessions. B Haplotype analysis of nonsynonymous amino acid substitutions at the six sites defined above in each of the 20 MDI accessions. Yellow and (−) represent shared amino acids with Nona Bokra/Pokkali and undetermined amino acids
Manhattan Plots for the SES Score of MDI Rice Accessions
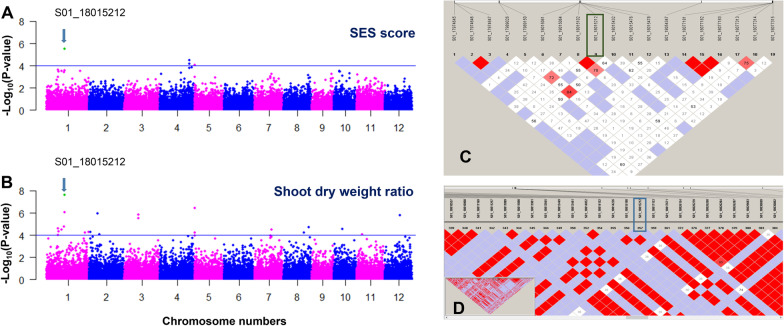
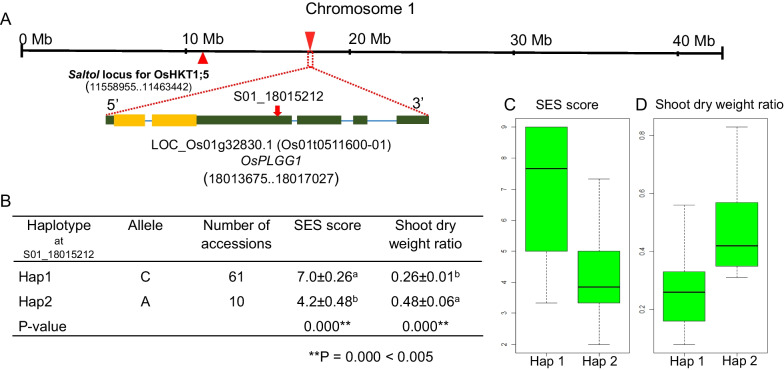
The above results showed that salinity tolerance in the MDI accessions may be regulated by loci different from the Saltol locus encoding the OsHKT1;5 transporter. To look for additional loci related to salinity tolerance, we performed GWASs using 37,643 imputed SNPs obtained from RAD-seq data from the 97 MDI accessions (Tam et al. 2019). We produced Manhattan plots using 29,881 SNPs from the 79 MDI landraces (97 MDI minus 18 improved varieties) using their SES scores at 20 DUT as the trait. We observed significant associations with three genomic regions for SES scores on chromosomes 1, 4, and 5. These three regions consisted of five SNP markers: one on chromosome 1, two on chromosome 4, and two on chromosome 5, with a significance threshold –Log (p-value) > 4.0 (Fig. 5A). When adjusting the threshold to account for a false discovery rate (q-value) < 0.1, only one SNP on chromosome 1 (S01_18015212) remained significant (Additional file 2: Table S6A).
Fig. 5.
GWAS of salinity responses with SES scores and shoot dry weights for the MDI accessions. A Manhattan plot for SES scores with 29,881 imputed SNPs (Tam et al. 2019) from the 79 accessions at 20 DUT with 100 mM NaCl. B Manhattan plot for shoot dry weight ratio. Both analyses identified a single and identical SNP peak, S01_18015212, on chromosome 1. C Linkage disequilibrium (LD) analysis showing that S01_18015212 forms a single block using 19 SNPs within 200 kb obtained by RAD-seq analysis for the 79 accessions. D LD analysis using 657 SNPs from the 20 accessions, showing a single LD block within the 200-kb window
We detected the same SNP peak when using shoot dry weight as the trait, which was the most highly correlated out of the four phenotypes measured, based on Pearson’s correlation coefficients, with SES scores (Fig. 5B). The GWAS returned 23 significant SNPs mapping to nine chromosomes for shoot dry weight with the threshold −Log (p value) ≥ 4.0, on chromosomes 1, 2, 3, 5, 7, 8, 10, 11, and 12 (Fig. 5B). We retained eight SNPs on five chromosomes with a q-value ≤ 0.05 (Fig. 5B). Importantly, SNP S01_18015212, which was associated with SES score, was the most significant for shoot dry weight (Additional file 2: Table S6B).
Linkage Disequilibrium (LD) and Haplotypes Around the SNP Peak Region
We analyzed the SNPs over a 200-kb window (100 kb upstream and 100 kb downstream of the peak SNP). After filtering, we retained 19 SNPs for LD analysis based on the Haploview method (Fig. 5C, D). Surprisingly, the peak SNP (S01_18015212) did not group with other SNPs within the neighboring 200-kb window (Fig. 5C), indicating that no other nearby SNP interacts with SNP (S01_18015212). Furthermore, we examined SNP (S01_18015212) using whole-genome resequencing data from the 20 accessions, resulting in 657 SNPs within the same 200-kb window. Again, the peak SNP S01_18015212 was the only site showing a peak in Manhattan plots and showed no LD with other linked SNPs (Fig. 5D).
SNP S01_18015212 mapped to the 3′ untranslated region (UTR) of LOC_Os01g32830 on chromosome 1, about 6.5 Mb away from the Saltol locus (Fig. 6A), clearly indicating that the Saltol allele does not play a role in salinity-tolerance in these accessions. LOC_Os01g32830 encodes a predicted chloroplast-localized transmembrane protein: chloroplast glycolate/glycerate translocator 1 (OsPLGG1) (Shim et al. 2019). PLGG1 transports glycolate/glycerate produced in chloroplasts to peroxisomes during photorespiration (Pick et al. 2013).
Fig. 6.
Location of the SNP peak on chromosome 1 and haplotype analysis. A Schematic representation of rice chromosome 1 and the position of SNP S01_18015212 within the 3′ UTR of LOC_Os01g3283 (OsPLGG1). This gene is 6.5 Mb away from the Saltol locus. According to RAP-DP, translated and untranslated regions in LOC_Os01g3283 were indicated by orange and green rectangles, respectively. ORF B S01_18015212 defines two haplotypes among 71 accessions: Hap 1 was found in 61 accessions with nucleotide C, and Hap 2 was found in 10 accessions with A. The remaining eight accessions had no exact haplotype (heterozygous alleles). C, D Significant differences in SES scores (C) and shoot dry weight ratio (D) values between Hap 1 and Hap 2 accessions
We identified 3 possible haplotypes among 79 rice accessions at SNP S01_18015212: 61 accessions harboring the C allele (Hap 1), 10 carrying the A allele (Hap 2), and 8 heterozygous accessions (not evaluated here for haplotype comparison) (Fig. 6B). The average SES score (4.2 ± 0.48) of accessions with the A allele was significantly lower than in accessions with the C allele (7.0 ± 0.26) (Fig. 6B, C). The shoot dry weight ratio showed the opposite pattern relative to SES scores, with Hap1 associated with a lower ratio than Hap 2 (Fig. 6B, D). We concluded that Hap 2 is a salinity-tolerant haplotype, while Hap 1 was sensitive to salinity. The variety Doc Phung belonged to Hap 2.
Discussion
Geographical Association with Salinity Stress Responses in MDI Strains
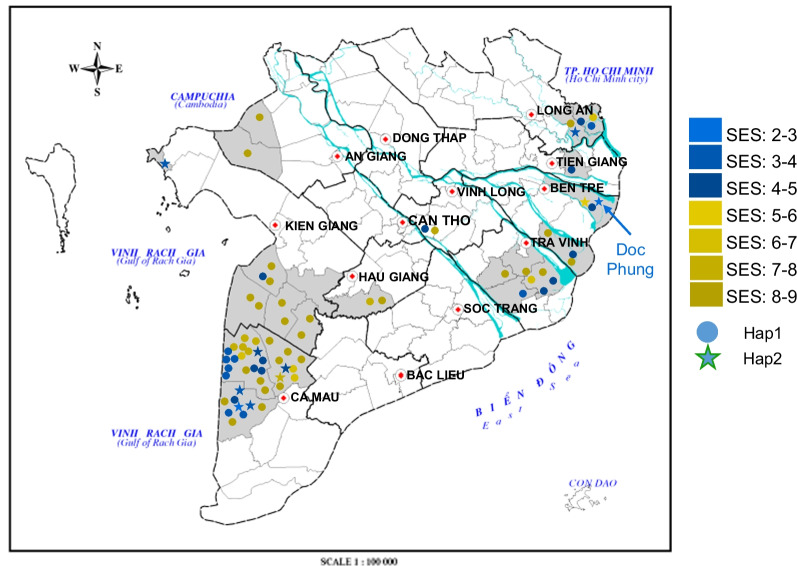
The evaluation of SES scores for 97 MDI accessions exposed to 100 mM NaCl for 20 days revealed two major peaks for salinity-tolerance: one peak from a group with high-salinity-tolerance (SES score between 3 and 5) and another peak from a group with low-salinity-tolerance (SES scores between 8 and 9) (Fig. 1). We observed regional differences between the coastal and inland areas of the Mekong Delta for the distribution of high- and low-salinity-tolerance groups, especially for high-salinity-tolerant accessions, which were mostly derived from the coastal Ca Mau region (Fig. 7, Additional file 2: Table S1). It is possible that adaptive selection for salinity-tolerance acted on these accessions originating from the Ca Mau coastal region, in an analogous manner to the salinity-tolerant varieties Nona Bokra and Pokkali, whose indigenous varieties originated from Bangladesh and coastal India, respectively (Emon et al. 2015; Gregorio and Senadhira 1993). On the other hand, Doc Phung does not originate from the Ca Mau region but from the Ben Tre region, and the salinity-tolerant allele of the Hap2 type is widely distributed in the coastal areas of the Mekong Delta (Fig. 7). In agreement, inland strains tended to be less salinity tolerant, may have grown with little exposure to salinity stress, and were likely to possess genomic structures similar to accessions from Cambodia and Laos (Tam et al. 2019). The three genomic regions identified by GWASs clearly illustrated the difference in genome structure between local coastal and inland accessions of the Mekong Delta (Fig. 7). The observed difference in salinity-tolerance, along with the differences in genomic structures, suggests that genetic exchange between the native lineages has been limited.
Fig. 7.
Geographical distribution of the 71 MDI landrace accessions in the Mekong Delta as a function of their S01_18015212 haplotype. The map shows the Mekong Delta region in Vietnam. Different colors indicate varying SES scores from Fig. 1. The circles and stars indicate Hap 1 (61 accessions) and Hap 2 (10 accessions), respectively
Genetic Association with Diversity of Salinity Stress Responses in MDI Strains
We used the SES score for salinity-tolerance developed by IRRI to evaluate our accessions at the seedling stage (IRRI 2013). SES scores for each accession were strongly and negatively correlated with their salinity response phenotypes and the ratios for shoot length, root length, shoot dry weight, and root dry weight, with Pearson’s correlation coefficients no higher than –0.67 (Fig. 2). These correlations demonstrated that the SES score is a good indicator of salinity-tolerance. Shoot dry weight ratios showed the strongest correlation with SES scores (Fig. 2). Using the shoot dry weight ratio and SES scores for GWAS, we identified a significant peak at a single SNP on chromosome 1 for both traits (Fig. 5A, B). We identified the same single SNP, S01_18015212, in a subsequent GWAS based on the resequencing data of 20 accessions with different SES scores (Fig. 5C, D). Although several salinity-response-related genes, including the Saltol locus (Ren et al. 2005; Thomson et al. 2010), are located on chromosome 1, the gene to which the peak SNP mapped is a previously uncharacterized gene, with the SNP located in the 3′ UTR (Fig. 6A).
Possible Functions of the SNP in the 3′ UTR of LOC_Os01g32830 (OsPLGG1)
The IRRI Rice SNP-Seek Database (Mansueto et al. 2017) and RAP DB (Sakai et al. 2013) revealed that SNP S01_18015212 maps to LOC_Os01g32830 (Os01t0511600-01). Os01t0511600-01 encodes OsPLGG1 responsible for the transport of glycolate/glycerate from chloroplasts to peroxisomes (Shim et al. 2019). Mutants in OsPLGG1 exhibit lower photosynthetic efficiency, starch accumulation, plant height, and crop productivity (Shim et al. 2019). Salinity stress is known to result in excess reactive oxygen species (ROS), which interferes with normal photosynthesis and negatively affect plant growth (Pawlowicz et al. 2018; Yang et al. 2020). We hypothesize that variation in OsPLGG1 activity in transporting glycolate/glycerate from chloroplasts to peroxisomes may contribute to the accumulation of ROS in response to salinity stress, which affects photosynthetic reactions and plant growth.
SNP S01_18015212 is located in the 3′ UTR of the gene, which typically participates in the regulation of mRNA stability, localization, or translation; this SNP may thus affect protein abundance (Sun et al. 2017). In addition, 3′ UTRs are known to control gene expression in plants, especially when exposed to environmental stress (Srivastava et al. 2018). To validate our prediction, the genome editing experiment will provide useful information on the relationships between salinity response and function of the candidate gene.
Physiological Characteristics of Distinct Salinity Tolerance in the MDR Landraces
In general, excess Na+ decreases Ca2+ uptake, while supplemental Ca2+ increases Ca2+ concentrations in plants, resulting in improved plant growth under salinity stress conditions (Cramer et al. 1991). To precisely evaluate the correlation between accessions tolerant or sensitive to salinity in this study, we analyzed the salinity response of 20 MDI accessions from the Vietnamese Mekong Delta (Fig. 3). The salinity response was highly variable among 20 MDI accessions, which can be divided into two distinct groups: a tolerant group of eight accessions (SES score < 4) and a sensitive group of 12 accessions (SES score > 4) (Additional file 2: Table S4). We also determined mineral concentrations for roots, leaves, and upper stems in these 20 accessions (Additional file 2: Table S5, Additional file 1: Fig. S2). Concentrations for Na+, Ca2+, the Na+/K+ ratio, and the Na+/Ca2+ ratio in the tolerant group were lower than those obtained for the sensitive group (Fig. 3A), which was consistent with the results reported by Kumar et al. (2008), except for Ca2+ concentrations, where Kumar et al. (2008) detected higher Ca2+ concentrations in the tolerant group relative to the sensitive group. Moreover, SNP S01_18015212 is located within the mapping interval of three QTLs for salinity tolerance (QTAROqtl-195, QTAROqtl-196, and QTAROqtl-197), which function in Na+ uptake, K+ concentrations, and the Na+/K+ ratio, respectively (Mansueto et al. 2017).
In roots, Ca2+ accumulates mainly in the cell wall (Demarty et al. 1984) by binding to the cation exchange sites of root cell wall pectin. Plants with low root Ca2+ concentrations thus tend to exhibit low cation exchange capacity (CEC) in their root cell walls (Marschner 2012). Similarly, Na+ binds to root cell wall pectin and causes cell wall damage (Byrt et al. 2018). Therefore, crops with low root CEC have been reported to be more tolerant to salinity than those with high root CEC (Bajwa and Bhumbla 1971). The positive correlations observed between SES scores and both Na+ and Ca2+ concentrations in roots in this study (Additional file 1: Fig. S2) suggest that root CEC is smaller in the tolerant accessions including Doc Phung. Compared to the salinity-sensitive accessions, the salinity-tolerant strains of the Mekong Delta population, represented by Doc Phung, may present lower cell wall damage due to lower Na+ and Ca2+ binding capacity to cell wall pectin (Byrt et al. 2018; Marschner 2012). Future exploration of the association between limited Na+ and Ca2+ binding to cell wall pectin and the SNP located in the 3′ UTR of OsPLGG1 will be useful to study the mechanism of salinity-tolerance in Doc Phung.
Genetic Specificity and Origin of Doc Phung
Several articles have reported that Doc Phung shows superior salinity-tolerance, and this study clearly validated the evidence for this promising salinity-tolerant accession (Ho et al. 2018; Le 1999; Tin et al. 2021). First, SES evaluation showed that Doc Phung has the highest salinity-tolerance and the highest values for shoot length ratio, shoot dry weight ratio, and root dry weight ratio, all highly correlated with salinity-tolerance (Fig. 2). GWASs based on these trait indices among the MDI accessions showed that a promising SNP associated with salinity-tolerance maps to chromosome 1 (Fig. 6), in a genomic location distinct from that of the Saltol locus encoding the HKT1;5 transporter, which is associated with salinity-tolerance in the varieties Nona Bokra and Pokkali (Fig. 6). The HKT1;5 genotype at the Saltol locus in Doc Phung was different from the allele harbored by Nona Bokra and Pokkali (Fig. 4). Therefore, the origin of salinity-tolerance in Doc Phung may have originated from accessions cultivated in the Mekong Delta or from the coastal periphery. SNP S01_1801521 in the 3′ UTR of OsPLGG1 provides a new option for salinity-tolerant alleles in rice breeding. It is necessary to analyze the relationship between this gene and salinity-tolerance. This SNP is useful for stacking multiple salinity tolerance alleles in rice breeding.
Conclusion
We analyzed the relationship between the degree of salinity-tolerance and physiological characteristics and performed GWASs for the Mekong Delta rice population. We focused on the landrace variety Doc Phung and confirmed it to be particularly resistant to salinity exposure (Ho et al. 2018; Le 1999; Tin et al. 2021). The Na+ and Ca2+ concentrations in the salinity-tolerant accessions allowed us to propose a salinity-tolerant mechanism caused by smaller root CEC. In addition, we identified a gene with a SNP specific to salinity-tolerant accessions obtained from GWASs. This SNP was located in the 3′ UTR of OsPLGG1. Overall, our results provide evidence that the salinity-tolerant accessions in the Mekong Delta population, such as Doc Phung, have a different Saltol allele from that of Nona Bokra and Pokkali. The causative genomic region of salinity-tolerance in Doc Phung, which might have been adapted to the Mekong Delta, is a new source for rice salinity tolerance. This study will provide a concrete countermeasure for rice cultivation against the salinity intrusion in the coastal areas of the Mekong Delta due to climate change.
Supplementary Information
Additional file 1: Figure S1. MDI accessions during the salinity tolerance test. Rice seedlings were grown hydroponically in a 40-L container with floating foam material in the greenhouse. Figure S2. Correlations between SES scores at 20 DUT and mineral concentrations across the 20 MDI accessions. Mineral concentrations and SES scores are plotted for four mineral ions (Na+, K+, Ca2+, and Mg2+) in roots, stems, and leaves. The 20 accessions were treated with 100 mM NaCl for 20 days (20 DUT). r, Pearson’s correlation coefficient.
Additional file 2: Table S1. List of Mekong Delta rice accessions examined in this study. Table S2. Phenotypic features for the standard evaluation system (SES) scores. Table S3. Preliminary test for salinity-tolerant/sensitive groups with 50 and 100 mM NaCl. Table S4. SES scores and phenotypic values in the 97 MDI accessions and their rankings. Table S5. Concentrations (mg/gDW) for mineral ions (Na+, K+, Ca2+, and Mg2+) and Na2+/K+ ratio in roots, stems, and leaves of 20 accessions exposed to 100 mM NaCl for 20 days (20 DUT) and control samples. Table S6A. SNPs above the threshold of p-value at 1.00E–04 in a Manhattan plot for SES score at 20 DUT (Fig. 5A). Table S6B. SNPs above the threshold of p-value at 1.00E–04 in a Manhattan plot for shoot dry weight ratio (Fig. 5B).
Acknowledgements
We gratefully acknowledge Dr. I. Takamure, Dr. N. Saito (Research Faculty of Agriculture, Hokkaido University) and Dr. H. Ky (College of Agriculture, Can Tho University) for their valuable suggestions concerning this study. We thank Mr. TH Phuc, Mr. HN Dien, and Mr. NV Chanh (Mekong Delta Development Research Institute, Can Tho University) for preparing and maintaining rice seeds for this study.
Abbreviations
- MDI
Mekong Delta Development Research Institute
- SES
The standard evaluation system
- CEC
Cation exchange capacity
- GWAS
Genome-wide association study
- OsPLGG1
Chloroplast glycolate/glycerate translocator 1
- HKT1
HIGH-AFFINITY K+ TRANSPORTER1
- IRRI
The International Rice Research Institute
- DAT
Days after transfer
- DUT
Days under treatment
- SNP
Single nucleotide polymorphism
- RAD-seq
Restriction site–associated DNA sequencing
- GLM
General linear model
- PCA
Principal component analysis
- LD
Linkage disequilibrium
- aa
The amino acid
Author contributions
TTN and YKi planned the research. TTN, TW, and YKi designed the research. TTN and YKi prepared rice seed samples. TTN, and TW carried out the experiments. TTN, MSD, SS and TW analyzed the data. DVL, and YKo supervised the materials preparation and data analysis. TTN, TW, MSD, and YKi wrote and improved the manuscript. MSD, TW, YKo, and YKi supervised TTN’s PhD study. All authors have read and approved the manuscript.
Funding
This study was funded in part by the Can Tho University Improvement Project VN14-P6, supported by a Japanese ODA loan.
Availability of data and materials
The RAD-seq data set of the 97-accession MDI has been deposited in the DDBJ database (accession number DRA008414). The data set of the whole-genome resequencing of the 20 MDI accessions has been registered as SUB10778038 into Sequence Read Archive (SRA). The request of availability of the accessions used in this study is asked to TTM (ngttam@ctu.edu.vn).
Declarations
Ethical approval and consent to participate
This study complied with the ethical standards of Japan, Vietnam, Hokkaido University, and Can Tho University, where this research was conducted.
Consent for publication
All authors have consented to the publication of this manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Tam Thanh Nguyen, Email: ngttam@ctu.edu.vn.
Toshihiro Watanabe, Email: nabe@chem.agr.hokudai.ac.jp.
Yuji Kishima, Email: kishima@abs.agr.hokudai.ac.jp.
References
- Akbar M, Yabuno T, Nakao S. Breeding for saline-resistant varieties of rice. I. Variability for salt tolerance among rice varieties. Jpn J Breed. 1972;22:8. doi: 10.1270/jsbbs1951.22.277. [DOI] [Google Scholar]
- Anil VS, Krishnamurthy H, Mathew MK. Limiting cytosolic Na+ confers salt tolerance to rice cells in culture: a two-photon microscopy study of SBFI-loaded cells. Physiol Plantarum. 2007;129:607–621. doi: 10.1111/j.1399-3054.2006.00854.x. [DOI] [Google Scholar]
- Bajwa MS, Bhumbla DR. Relationship between root cationexchange capacity and sodium tolerance of different crops. Plant Soil. 1971;34:7. doi: 10.1007/BF01372761. [DOI] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bassil E, Coku A, Blumwald E. Cellular ion homeostasis: emerging roles of intracellular NHX Na+/H+ antiporters in plant growth and development. J Exp Bot. 2012;63:5727–5740. doi: 10.1093/jxb/ers250. [DOI] [PubMed] [Google Scholar]
- Batayeva D, Labaco B, Ye C, Li X, Usenbekov B, Rysbekova A, Dyuskalieva G, Vergara G, Reinke R, Leung H. Genome-wide association study of seedling stage salinity tolerance in temperate japonica rice germplasm. BMC Genet. 2018;19:2. doi: 10.1186/s12863-017-0590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bimpong IK, Manneh B, Diop B, Ghislain K, Sow A, Amoah NKA, Gregorio G, Singh RK, Ortiz R, Wopereis M. New quantitative trait loci for enhancing adaptation to salinity in rice from Hasawi, a Saudi landrace into three African cultivars at the reproductive stage. Euphytica. 2014;200:45–60. doi: 10.1007/s10681-014-1134-0. [DOI] [Google Scholar]
- Bonilla P, Dvorak J, Mackill D, Deal K, Gregorio G. RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp Agric Sci. 2002;85:68–76. [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Browning BL, Zhou Y, Browning SR. A one-penny imputed genome from next-generation reference panels. Am J Hum Genet. 2018;103:338–348. doi: 10.1016/j.ajhg.2018.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrt CS, Munns R, Burton RA, Gilliham M, Wege S. Root cell wall solutions for crop plants in saline soils. Plant Sci. 2018;269:47–55. doi: 10.1016/j.plantsci.2017.12.012. [DOI] [PubMed] [Google Scholar]
- Cramer GR, Epstein E, Lauchli A. Effects of sodium, potassium and calcium on salt-stressed Barley. 2 Elemental Analysis. Physiol Plant. 1991;81:197–202. doi: 10.1111/j.1399-3054.1991.tb02129.x. [DOI] [Google Scholar]
- Davenport R, James RA, Zakrisson-Plogander A, Tester M, Munns R. Control of sodium transport in durum wheat. Plant Physiol. 2005;137:807–818. doi: 10.1104/pp.104.057307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon TB, Linscombe S, Subudhi PK. Molecular dissection of seedling salinity tolerance in rice (Oryza sativa L.) using a high-density GBS-based SNP linkage map. Rice (NY) 2016;9:52. doi: 10.1186/s12284-016-0125-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Leon TB, Linscombe S, Subudhi PK. Identification and validation of QTLs for seedling salinity tolerance in introgression lines of a salt tolerant rice landrace 'Pokkali'. PLoS ONE. 2017;12:e0175361. doi: 10.1371/journal.pone.0175361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demarty M, Morvan C, Thellier M. Calcium and the cell wall. Plant Cell Environ. 1984;7:8. doi: 10.1111/j.1365-3040.1984.tb01434.x. [DOI] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emon RM, Islam MM, Halder J, Fan YY. Genetic diversity and association mapping for salinity tolerance in Bangladeshi rice landraces. Crop J. 2015;3:440–444. doi: 10.1016/j.cj.2015.04.006. [DOI] [Google Scholar]
- Faiyue B, Al-Azzawi MJ, Flowers TJ. A new screening technique for salinity resistance in rice (Oryza sativa L.) seedlings using bypass flow. Plant Cell Environ. 2012;35:1099–1108. doi: 10.1111/j.1365-3040.2011.02475.x. [DOI] [PubMed] [Google Scholar]
- Ganie SA, Molla KA, Henry RJ, Bhat KV, Mondal TK. Advances in understanding salt tolerance in rice. Theor Appl Genet. 2019;132:851–870. doi: 10.1007/s00122-019-03301-8. [DOI] [PubMed] [Google Scholar]
- Grattan S, Grieve C. Mineral nutrient acquisition and response by plants grown in saline environments. Handb Plant Crop Stress. 1999;2:203–229. [Google Scholar]
- Gregorio GB, Senadhira D. Genetic-analysis of salinity tolerance in rice (Oryza-Sativa L.) Theor Appl Genet. 1993;86:333–338. doi: 10.1007/BF00222098. [DOI] [PubMed] [Google Scholar]
- Hariadi YC, Nurhayati AY, Soeparjono S, Arif I. Screening six varieties of rice (Oryza sativa) for salinity tolerance. Procedia Environ Sci. 2015;28:10. doi: 10.1016/j.proenv.2015.07.012. [DOI] [Google Scholar]
- Ho VT, NQ H, Pham BC. Evaluating salt tolerance of twenty traditional rice varieties from Vietnam. Tạp Chí Khoa Học Công Nghệ Và Thực Phẩm. 2018;14:9. [Google Scholar]
- IRRI (2013) SES Standard evaluation system for Rice, 5th edn, p 34
- Kader MA, Lindberg S, Seidel T, Golldack D, Yemelyanov V. Sodium sensing induces different changes in free cytosolic calcium concentration and pH in salt-tolerant and -sensitive rice (Oryza sativa) cultivars. Physiol Plantarum. 2007;130:99–111. doi: 10.1111/j.1399-3054.2007.00890.x. [DOI] [Google Scholar]
- Kontgis C, Schneider A, Ozdogan M, Kucharik C, Tri VD, Duc NH, Schatz J. Climate change impacts on rice productivity in the Mekong River Delta. Appl Geogr. 2019;102:71–83. doi: 10.1016/j.apgeog.2018.12.004. [DOI] [Google Scholar]
- Krishnamurthy P, Ranathunge K, Nayak S, Schreiber L, Mathew MK. Root apoplastic barriers block Na+ transport to shoots in rice (Oryza sativa L.) J Exp Bot. 2011;62:4215–4228. doi: 10.1093/jxb/err135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Shriram V, Nikam TD, Jawali N, Shitole MG. Sodium chloride-induced changes in mineral nutrients and proline accumulation in indica rice cultivars differing in salt tolerance. J Plant Nutr. 2008;31:1999–2017. doi: 10.1080/01904160802403466. [DOI] [Google Scholar]
- Kumar V, Singh A, Mithra SA, Krishnamurthy S, Parida SK, Jain S, Tiwari KK, Kumar P, Rao AR, Sharma S. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa) DNA Res. 2015;22:133–145. doi: 10.1093/dnares/dsu046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari S, Sabharwal VP, Kushwaha HR, Sopory SK, Singla-Pareek SL, Pareek A. Transcriptome map for seedling stage specific salinity stress response indicates a specific set of genes as candidate for saline tolerance in Oryza sativa L. Funct Integr Genom. 2009;9:109–123. doi: 10.1007/s10142-008-0088-5. [DOI] [PubMed] [Google Scholar]
- Le HN (1999) Biodiversity in the Mekong Delta. Community Biodi-versity Development and Conversation Project. Can Tho, Vietnam. Can Tho University
- Lee KS, Choi WY, Ko JC, Kim TS, Gregorio GB. Salinity tolerance of japonica and indica rice (Oryza sativa L.) at the seedling stage. Planta. 2003;216:1043–1046. doi: 10.1007/s00425-002-0958-3. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutts S, Kinet JM, Bouharmont J. Changes in plant response to NaCl during development of rice (Oryza sativa L.) varieties differing in salinity resistance. J Exp Bot. 1995;46:1843–1852. doi: 10.1093/jxb/46.12.1843. [DOI] [Google Scholar]
- Maas EV, Grattan SR. Crop yields as affected by salinity. Agric Drain. 1999;38:54. [Google Scholar]
- Mansueto L, Fuentes RR, Borja FN, Detras J, Abriol-Santos JM, Chebotarov D, Sanciangco M, Palis K, Copetti D, Poliakov A, Dubchak I, Solovyev V, Wing RA, Hamilton RS, Mauleon R, McNally KL, Alexandrov N. Rice SNP-seek database update: new SNPs, indels, and queries. Nucleic Acids Res. 2017;45:D1075–D1081. doi: 10.1093/nar/gkw1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H. Marschner's mineral nutrition of higher plants. 3. Amsterdam: Academic Press; 2012. [Google Scholar]
- Mohammadi-Nejad G, Singh RK, Arzani A, Rezaie AM, Sabouri H, Gregorio GB. Evaluation of salinity tolerance in rice genotypes. Int J Plant Prod. 2010;4:199–207. [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annu Rev Plant Biol. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Nhung TT, Vo PV, Nghi VV, Bang HQ. Salt intrusion adaptation measures for sustainable agricultural development under climate change effects: a case of Ca Mau Peninsula, Vietnam. Clim Risk Manag. 2019;23:88–100. doi: 10.1016/j.crm.2018.12.002. [DOI] [Google Scholar]
- Pawlowicz I, Waskiewicz A, Perlikowski D, Rapacz M, Ratajczak D, Kosmala A. Remodeling of chloroplast proteome under salinity affects salt tolerance of Festuca arundinacea. Photosynth Res. 2018;137:475–492. doi: 10.1007/s11120-018-0527-7. [DOI] [PubMed] [Google Scholar]
- Pick TR, Brautigam A, Schulz MA, Obata T, Fernie AR, Weber AP. PLGG1, a plastidic glycolate glycerate transporter, is required for photorespiration and defines a unique class of metabolite transporters. Proc Natl Acad Sci USA. 2013;110:3185–3190. doi: 10.1073/pnas.1215142110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platten JD, Egdane JA, Ismail AM (2013) Salinity tolerance, Na+ exclusion and allele mining of HKT1;5 in Oryza sativa and O-glaberrima: many sources, many genes, one mechanism? BMC Plant Biol 13:32 [DOI] [PMC free article] [PubMed]
- Puram VRR, Ontoy J, Linscombe S, Subudhi PK. Genetic dissection of seedling stage salinity tolerance in rice using introgression lines of a salt tolerant landrace Nona Bokra. J Hered. 2017;108:658–670. doi: 10.1093/jhered/esx067. [DOI] [PubMed] [Google Scholar]
- Rahman MA, Bimpong IK, Bizimana JB, Pascual ED, Arceta M, Swamy BPM, Diaw F, Rahman MS, Singh RK. Mapping QTLs using a novel source of salinity tolerance from Hasawi and their interaction with environments in rice. Rice (n y) 2017;10:47. doi: 10.1186/s12284-017-0186-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MA, Thomson MJ, De Ocampo M, Egdane JA, Salam MA, Shah EAM, Ismail AM. Assessing trait contribution and mapping novel QTL for salinity tolerance using the Bangladeshi rice landrace Capsule. Rice (n y) 2019;12:63. doi: 10.1186/s12284-019-0319-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, Zhu MZ, Wang ZY, Luan S, Lin HX. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet. 2005;37:1141–1146. doi: 10.1038/ng1643. [DOI] [PubMed] [Google Scholar]
- Rios JJ, Martinez-Ballesta MC, Ruiz JM, Blasco B, Carvajal M. Silicon-mediated improvement in plant salinity tolerance: the role of aquaporins. Front Plant Sci. 2017;8:948. doi: 10.3389/fpls.2017.00948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Lee SS, Tanaka T, Numa H, Kim J, Kawahara Y, Wakimoto H, Yang C, Iwamoto M, Abe T, Yamada Y, Muto A, Inokuchi H, Ikemura T, Matsumoto T, Sasaki T, Itoh T. Rice annotation project database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013;54:E6-+. doi: 10.1093/pcp/pcs183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim SH, Lee SK, Lee DW, Brilhaus D, Wu G, Ko S, Lee CH, Weber APM, Jeon JS. Loss of function of rice plastidic glycolate/glycerate translocator 1 impairs photorespiration and plant growth. Front Plant Sci. 2019;10:1726. doi: 10.3389/fpls.2019.01726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Kota S, Flowers TJ (2021) Salt tolerance in rice: seedling and reproductive stage QTL mapping come of age. Theor Appl Genet 134:3495–3533 [DOI] [PMC free article] [PubMed]
- Srivastava AK, Lu YM, Zinta G, Lang ZB, Zhu JK. UTR-Dependent Control of Gene Expression in Plants. Trends Plant Sci. 2018;23:248–259. doi: 10.1016/j.tplants.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Bass AJ, Dabney A, David Robinson D, Warnes G (2004) Q-value estimation for false discovery rate control R package version 2.10.0
- Sun HX, Li Y, Niu QW, Chua NH. Dehydration stress extends mRNA 3′ untranslated regions with noncoding RNA functions in Arabidopsis. Genome Res. 2017;27:1427–1436. doi: 10.1101/gr.218669.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam NT, Dwiyanti MS, Koide Y, Nagano AJ, Ky H, Tin HQ, Hien NL, Dung LV, Kishima Y. Profiling SNP and nucleotide diversity to characterize Mekong Delta rice landraces in southeast Asian populations. Plant Genome. 2019;12:1–11. doi: 10.3835/plantgenome2019.06.0042. [DOI] [PubMed] [Google Scholar]
- Termaat A, Passioura JB, Munns R. Shoot turgor does not limit shoot growth of nacl-affected wheat and barley. Plant Physiol. 1985;77:869–872. doi: 10.1104/pp.77.4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson MJ, de Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, Tumimbang-Raiz E, Blumwald E, Seraj ZI, Singh RK, Gregorio GB, Ismail AM. Characterizing the saltol quantitative trait locus for salinity tolerance in rice. Rice. 2010;3:148–160. doi: 10.1007/s12284-010-9053-8. [DOI] [Google Scholar]
- Tian L, Tan L, Liu F, Cai H, Sun C. Identification of quantitative trait loci associated with salt tolerance at seedling stage from Oryza rufipogon. J Genet Genom. 2011;38:593–601. doi: 10.1016/j.jgg.2011.11.005. [DOI] [PubMed] [Google Scholar]
- Tin HQ, Loi NH, Bjornstad Å, Kilian B. Participatory selection of CWR-derived salt-tolerant rice lines adapted to the coastal zone of the Mekong Delta. Crop Sci. 2021;61:277–288. doi: 10.1002/csc2.20405. [DOI] [Google Scholar]
- Xie JH, Zapata-Arias FJ, Shen M, Afza R. Salinity tolerant performance and genetic diversity of four rice varieties. Euphytica. 2000;116:105–110. doi: 10.1023/A:1004041900101. [DOI] [Google Scholar]
- Yang W, Wang F, Liu LN, Sui N. Responses of membranes and the photosynthetic apparatus to salt stress in cyanobacteria. Front Plant Sci. 2020;11:713. doi: 10.3389/fpls.2020.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. MDI accessions during the salinity tolerance test. Rice seedlings were grown hydroponically in a 40-L container with floating foam material in the greenhouse. Figure S2. Correlations between SES scores at 20 DUT and mineral concentrations across the 20 MDI accessions. Mineral concentrations and SES scores are plotted for four mineral ions (Na+, K+, Ca2+, and Mg2+) in roots, stems, and leaves. The 20 accessions were treated with 100 mM NaCl for 20 days (20 DUT). r, Pearson’s correlation coefficient.
Additional file 2: Table S1. List of Mekong Delta rice accessions examined in this study. Table S2. Phenotypic features for the standard evaluation system (SES) scores. Table S3. Preliminary test for salinity-tolerant/sensitive groups with 50 and 100 mM NaCl. Table S4. SES scores and phenotypic values in the 97 MDI accessions and their rankings. Table S5. Concentrations (mg/gDW) for mineral ions (Na+, K+, Ca2+, and Mg2+) and Na2+/K+ ratio in roots, stems, and leaves of 20 accessions exposed to 100 mM NaCl for 20 days (20 DUT) and control samples. Table S6A. SNPs above the threshold of p-value at 1.00E–04 in a Manhattan plot for SES score at 20 DUT (Fig. 5A). Table S6B. SNPs above the threshold of p-value at 1.00E–04 in a Manhattan plot for shoot dry weight ratio (Fig. 5B).
Data Availability Statement
The RAD-seq data set of the 97-accession MDI has been deposited in the DDBJ database (accession number DRA008414). The data set of the whole-genome resequencing of the 20 MDI accessions has been registered as SUB10778038 into Sequence Read Archive (SRA). The request of availability of the accessions used in this study is asked to TTM (ngttam@ctu.edu.vn).