Abstract
Previous studies indicated that the Plasmodium yoelii circumsporozoite protein (PyCSP) 57–70 region elicits T cells capable of eliminating infected hepatocytes in vitro. Herein, we report that the PyCSP58–67 sequence contains an H-2d binding motif, which binds purified Kd molecules in vitro with low affinity (3,267 nM) and encodes an H-2d-restricted cytotoxic T lymphocyte (CTL) epitope. Immunization of BALB/c mice with three doses of a multiple antigen peptide (MAP) construct containing four branches of amino acids 57 to 70 linked to a lysine-glycine core [MAP4(PyCSP57–70)] and Lipofectin as the adjuvant induced both T-cell proliferation and a peptide-specific CTL response that was PyCSP59–67 specific, H-2d restricted, and CD8+ T cell dependent. Immunization with either DNA encoding the PyCSP or irradiated sporozoites demonstrated that this CTL epitope is subdominant since it is not recognized in the context of whole CSP immunization. The biological relevance of this CTL response was underlined by the demonstration that it could mediate genetically restricted, CD8+- and nitric-oxide-dependent elimination of infected hepatocytes in vitro, as well as partial protection of BALB/c mice against sporozoite challenge. These findings indicate that subdominant epitopes with low major histocompatibility complex affinity can be used to engineer epitope-based vaccines and have implications for the selection of epitopes for subunit-based vaccines.
In general, immune responses are not directed against all possible cytotoxic T lymphocyte (CTL) epitopes but are rather remarkably restricted and focused on one or a few immunodominant epitopes. According to the nomenclature originally derived from Sercarz and collaborators, dominant epitopes are the ones recognized by T-cell responses induced by immunization with whole unprocessed antigen (31).
These epitopes correspond to the epitopes recognized by T cells elicited by immunization with whole intact antigens or generated during the course of natural infection. Subdominant epitopes, by contrast, are epitopes that are not normally recognized by responses generated by whole antigens or natural infection but nevertheless are immunogenic and are generated by natural antigen processing. As a result, T cells elicited by deliberate immunization with subdominant peptides can recognize antigens naturally processed by infected cells or other antigen-presenting cells (31). Finally, cryptic epitopes are defined as epitopes that elicit T cells capable of recognizing the immunizing peptide only but not antigens naturally processed by infected cells or antigen-processing cells.
In the last few years, epitope-based vaccines have received considerable attention as a possible means to develop novel prophylactic vaccines and immunotherapeutic strategies. Selection of an appropriate mixture of dominant and subdominant T- and B-cell epitopes from the pathogen of interest should, in principle, allow one to focus the immune systems toward the desired type of response. Examples of this type of situation include focusing the immune response toward conserved epitopes of pathogens which are characterized by high sequence variability (such as human immunodeficiency virus, hepatitis C virus, and Plasmodium spp.).
Epitope-based vaccines may also allow one to focus the immune response toward protective subdominant determinants. This feature could be particularly valuable in the case of various chronic viral diseases and cancers, where T cells directed against the immunodominant epitopes might have been inactivated while T cells specific for subdominant epitopes might have escaped T-cell tolerance (3, 15). The use of epitope-based vaccines may also allow one to avoid suppressive or inappropriate determinants such as T-cell epitopes which, either because of their major histocompatibility complex (MHC) binding capacity or T-cell activation features, induce TH2 responses in conditions where a TH1 response is desirable, or vice versa.
Once appropriate epitope determinants have been defined, they can be combined and delivered by various means, including lipopeptides, viral delivery vectors, particles of viral or synthetic origin, naked or particle-absorbed cDNA, and addition or covalent attachment of helper peptides (6, 17, 21, 29, 30, 37, 41). However, before appropriate epitopes can be defined, one major obstacle has to be overcome, namely, the very high degree of polymorphism of the MHC molecules expressed in the human population. More than 200 different types of HLA class I and class II molecules have already been identified (1). Our own group has provided evidence to demonstrate that, in the case of HLA class I molecules, peptides capable of binding several different HLA class I molecules can be identified. Over 60% of the known HLA class I molecules can, in fact, be grouped into four broad HLA supertypes, characterized by similar peptide binding specificities (HLA supermotifs) (33).
Previous studies have shown that a peptide corresponding to amino acids 59 to 79 (YNRNIVNRLLGDALNGKPEEK) of the Plasmodium yoelii circumsporozoite protein (PyCSP) primes for specific T-cell proliferation and CD8+-dependent elimination of hepatic-stage parasites from culture (5, 26). However, the exact nature of the epitope recognized was not determined. In the present study, we have mapped an H-2Kd-restricted CTL epitope to residues 58 to 67 of the CSP of P. yoelii. This epitope bound purified Kd only weakly but was demonstrated to be immunogenic in the context of a multiple antigen peptide (MAP) construct.
PyCSP58–67-specific cytolytic responses were not detected in spleen cells from mice immunized with irradiated sporozoites or DNA encoding the PyCSP; however, T cells elicited by peptide immunization recognized naturally processed antigen as produced by infected hepatocytes in vitro, demonstrating that the PyCSP58–67 is a classical subdominant epitope. Furthermore, immunization with the MAP construct containing PyCSP58–67 also afforded partial in vivo protection from P. yoelii sporozoite challenge.
MATERIALS AND METHODS
Mice.
Four- to eight-week-old female BALB/cByJ mice were purchased from The Jackson Laboratory (Bar Harbor, Maine).
Peptides.
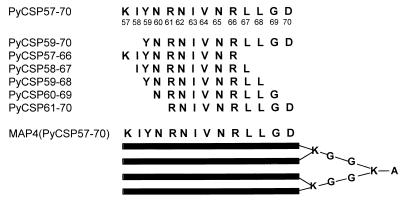
A MAP composed of four branches of amino acids 57 to 70 was colinearly synthesized with a glycine-lysine core [MAP4(PyCSP57–70)] (Fig. 1). A series of overlapping 10-amino-acid peptides spanning the 57–70 region were used in the CTL assay and for determining MHC class I binding affinities (Fig. 1). The linear peptides, PyCSP57–70 and PyCSP59–70, were also used in CTL assays for stimulating effectors and coating target cells. Control peptides used in the in vitro assays were PfSSP2-15 (PNPPNPPNPPNPPNP-amide) and PfSSP2-9 (RRHNWVNHA). In some experiments, mice were immunized with a combination of MAP4(PyCSP57–70) and a linear 20-amino-acid peptide (PyCSP280–299; SYVPSAEQILEFVKOI) containing the immunodominant PyCSP CD8+ CTL epitope (280 to 288; SYVPSAEQI). In challenge experiments, control mice were immunized with MAPp2p30, which is a MAP containing four branches of two colinearly synthesized universal T-helper epitopes from tetanus toxin (p2 = QYIKANSKFIGITE; p30 = FNNFTVSFWLRVPKVSASHLE) (23).
FIG. 1.
Peptides from the PyCSP that were used as immunogens and in the in vitro assays.
Cell lines.
P815 (H-2d) cells and EL-4 (H-2b) cells were obtained from the American Type Culture Collection (Manassas, Va.) and maintained in RPMI 1640 (Gibco-BRL, Gaithersburg, Md.) supplemented with 10% heat-inactivated fetal bovine serum (Gibco-BRL).
Affinity purification of H-2Kd molecules.
P815 cells were lysed at a concentration of 108 cells/ml in phosphate-buffered saline (PBS) containing 1% NP-40 and 1 mM phenylmethylsulfonyl fluoride. Lysates were cleared of debris and nuclei by centrifugation at 10,000 × g for 20 min. MHC molecules were then purified by affinity chromatography as previously described (32). Briefly, columns of inactivated Sepharose CL4B and protein A-Sepharose were used as precolumns. Lysates were filtered through 0.8- and 0.4-μm-pore-size filters and then H-2Kd purified by passage over a Y3 monoclonal antibody column. Antibody columns were washed with 15 column volumes of 10 mM Tris in 1.0% NP-40–PBS and 2 column volumes of PBS containing 0.4% n-octylglucoside. Finally, the class I molecules were eluted with 50 mM diethylamine in 0.15 M NaCl containing 0.4% n-octylglucoside, pH 11.5. A 1/25 volume of 2.0 M Tris, pH 6.8, was added to the eluate to reduce the pH to ∼8.0 and then concentrated by centrifugation in Centriprep 30 concentrators (Amicon, Beverly, Mass.) at 2,000 rpm. Protein purity, concentration, and effectiveness of depletion steps were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Class I peptide binding assays.
Purified H-2d molecules (5 to 500 nM) were incubated with 1 to 10 nM 125I-radiolabeled probe peptide (KFNPMKTYI, a Kd consensus peptide iodinated by the chloramine T method) (2) for 48 h at room temperature in the presence of 1 μM human β2-microglobulin (Scripps Laboratories, San Diego, Calif.) and a cocktail of protease inhibitors. The final concentrations of protease inhibitors were 1 mM phenylmethylsulfonyl fluoride, 1.3 nM 1,10-phenanthroline, 73 μM pepstatin A, 8 mM EDTA, and 200 μM Nα-p-tosyl-l-lysine chloromethyl ketone (TLCK).
Class I peptide complexes were separated from free peptide by gel filtration on TSK2000 columns, and the fraction of bound peptide was calculated as previously described (32). In preliminary experiments, the titers of the HLA class I preparation were determined in the presence of fixed amounts of radiolabeled peptides to determine the concentration of class I molecules necessary to bind 10 to 20% of the total radioactivity. All subsequent inhibition and direct binding assays were then performed using these class I concentrations.
In the inhibition assays, peptide inhibitors were typically tested at concentrations ranging from 120 μg/ml to 1.2 ng/ml. The data were then plotted, and the dose yielding 50% inhibition was measured (50% inhibitory concentration [IC50]). Peptides were tested in two to four completely independent experiments. Since under these conditions the label concentration is less than the MHC concentration and the IC50 is equal to or more than the MHC concentration, the measured IC50s are reasonable approximations of the true values. To allow comparison of the data obtained from different experiments, a relative binding figure was calculated for each peptide by dividing the IC50 of a positive control for inhibition by the IC50 for each test peptide. The positive control is represented by the unlabeled version of the radiolabeled probe.
Adjuvant.
The cationic lipid Lipofectin (Gibco-BRL) was mixed with the peptides in PBS to give a dose of 15 μg of Lipofectin/mouse. Lipofectin was selected as the adjuvant because immunization of mice with a PyCSP peptide (amino acids 280 to 299) and Lipofectin induces CTLs and protection against sporozoite challenge (9a). Cationic lipids have been used as adjuvants in immunization with recombinant proteins (40, 42) in order to facilitate induction of CD8+ CTLs by directing proteins into the class I MHC presentation pathway.
Immunizations.
Mice were immunized by subcutaneous injection at the base of the tail of the peptide or a mixture of peptides with Lipofectin in PBS in a volume of 50 μl using a 26-gauge needle. Two or three doses were given with a 3-week interval between doses. Spleens were removed 13 to 36 days after the last immunization, and splenocytes were used in the lymphocyte proliferation assay and as effectors in the CTL assay. Splenocytes from two to three mice in each group were pooled and used in the assays; experiments were repeated at least twice.
For irradiated sporozoite immunizations, sporozoites were isolated by the discontinuous gradient technique (22) from infected Anopheles stephensi mosquitoes that had been irradiated at 10,000 rads (137Ce). Mice were immunized via the tail vein at 0 (50,000 sporozoites), 2 (30,000 sporozoites), and 6 (20,000 sporozoites) weeks.
A plasmid DNA encoding the PyCSP (PyCSP DNA) was obtained from Vical Corporation (San Diego, Calif.) (30). Mice were injected intramuscularly in the right and left tibialis anterior muscles with a total of 40 μg of PyCSP DNA in 50 μl of PBS (25 μl in each leg).
Lymphocyte proliferation assay.
Splenocytes were plated at a concentration of 2.5 × 106 cells/ml in the presence of 0.38 to 20 μg of the test or control peptides per ml in 96-flat-bottom-well plates. Tritiated thymidine (1 μCi/well) was added to wells on day 4, and plates were harvested 18 h later with a 96-well plate harvester (Skatron, Sterling, Va.).
Chromium release CTL assay.
Effector cells were stimulated in vitro for 6 days with 1.0 to 2.5 μM peptide in 24-well plates. Target cells were labeled overnight with 0.1 mCi 51Cr (sodium chromate; Dupont NEN, Boston, Mass.) and 0.025 to 2.5 μM peptide. Effectors and targets were incubated for 6 h in the presence of 0.013 to 1.3 μM peptide. Supernatants were harvested with a Supernatant Collection System (Skatron). Maximum release was determined by lysing target cells with 5% Triton X-100. Minimum or spontaneous release was less than 18% of maximum. Percent specific lysis was calculated as the experimental release minus spontaneous release divided by the maximum release minus spontaneous release multiplied by 100. Data are presented as the percent peptide specific lysis, which is the percent specific lysis of target cells with peptide minus percent specific lysis without peptide. In experiments where CD4+ and CD8+ cells were depleted, effector cells were incubated with monoclonal antibodies against CD4+ (GK 1.5; American Type Culture Collection) and CD8+ (2.43; American Type Culture Collection) and 10% rabbit complement (Low-Tox M; Cedarlane Laboratories, Hornby, Ontario, Canada) for 30 min at 37°C.
Inhibition of liver stage development assay.
Mouse hepatocytes were obtained by in situ collagenase perfusion of mouse liver as previously described (9, 19). Briefly, livers were perfused in situ, sequentially, with Hanks balanced salt solution and a collagenase solution. The cell suspension generated was then centrifuged over a Percoll gradient to remove dead cells. The hepatocytes were then seeded onto eight-well Lab-Tek chamber slides (Miles Research, Elkhardt, Ind.) in complete medium (minimal essential medium with Earle's balanced salts supplemented with 0.2% bovine serum albumin [fraction 5], 10% fetal bovine serum, 2% penicillin-streptomycin solution, insulin, 1% l-glutamine solution [100×], and 1% nonessential amino acid solution [100×]) at a concentration of 105 cells/well. The slides were incubated overnight at 37°C in a 5% CO2–95% air environment. The medium was changed on the following day, and fresh medium containing dexamethasone (7 × 10−5 M) was added to the cultures.
Hepatocyte cultures, which had been seeded onto eight-well chamber slides 24 h previously, were incubated with 7.5 × 104 P. yoelii sporozoites for 3 h. The cultures were then washed with medium and incubated for 24 h. Effector cells (1 × 105, 2.5 × 105, 5 × 105, and 1 × 106 lymphocytes) from mice that had been immunized with MAP4(PyCSP57–70) or Lipofectin and stimulated in vitro for 6 days with 2.5 μM MAP4(PyCSP57–70) were then added and allowed to remain in culture with the infected hepatocytes for an additional 24 h. In some experiments, restimulated spleen cells were depleted of CD8+ T cells using Dynabeads (Dynal, Lake Success, N.Y.) according to the manufacturer's instructions. Other cultures were incubated with 5 μM [N-(3-aminomethyl)benzyl] acetamidine (also designated 1400W), an inhibitor of inducible nitric oxide synthase (iNOS) (10). The chamber slides were then fixed for 10 min in ice-cold absolute methanol and stained with NYLS3 (4) and fluorescein isothiocyanate-labeled goat anti-mouse immunoglobulin G. The stained slides were then viewed, and hepatocytes were counted using an epifluorescence microscope. Percent inhibition was calculated according to the formula (control − test)/control × 100.
Protection against challenge with sporozoites.
Mice received an intravenous injection in the tail of 50 to 100 sporozoites of P. yoelii (nonlethal; strain 17XNL). Mice were considered protected if no parasites were found on blood smears prepared on days 5, 7, 9, 11, and 14 postchallenge. Mice have never become positive after day 14 postchallenge.
The experiments reported herein were conducted according to the principles set forth in reference 13a.
RESULTS
PyCSP58–67 binds purified Kd with low affinity.
In order to map potential CD8+-restricted epitopes on the PyCSP, we scanned the 59–79 region for the presence of H-2d class I binding motifs. An H-2Kd binding motif was found between amino acids 58 and 67 (IYNRNIVNRL), carrying the anchor residues tyrosine at position 2 and leucine at position 10 (27). Using purified Kd molecules, we determined the in vitro binding affinities to H-2Kd of five overlapping 10-amino-acid peptides between amino acids 57 and 70. The peptide PyCSP58–67 (IYNRNVNRL) had low (3,267 nM) albeit detectable affinity. No significant affinity (up to the 10 μM level) was detected for any of the other 10-amino-acid peptides (PyCSP57–66, PyCSP59–68, PyCSP60–69, and PyCSP61–70). In conclusion, these data identified PyCSP58–67 as a low binder to H-2Kd.
Definition of the PyCSP58–67 CTL epitope.
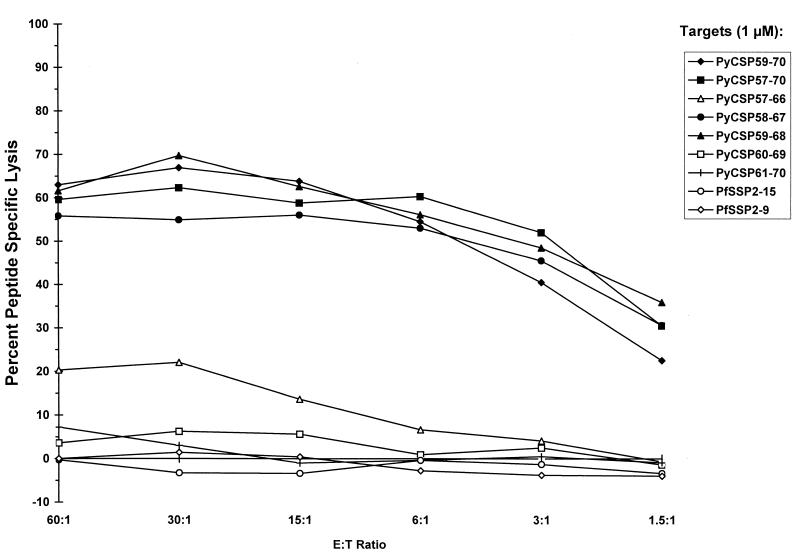
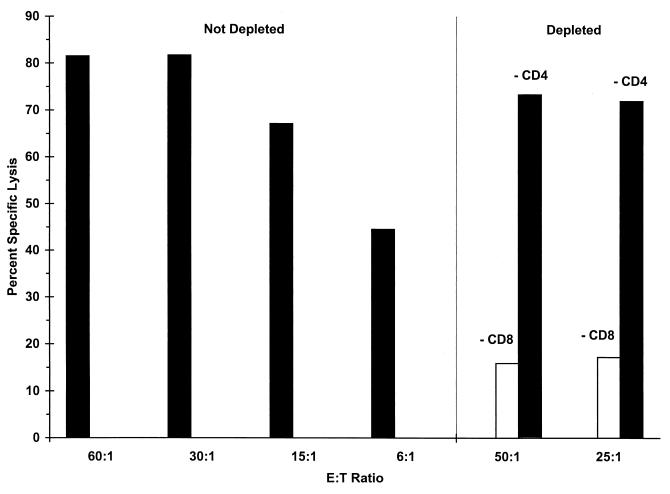
The next experiments were designed to determine if immunization of H-2d mice with MAP4(PyCSP57–70) induced CTLs against peptides containing the H-2Kd motif. Splenocytes from BALB/c mice immunized with three doses of 40 μg of MAP4(PyCSP57–70) and 15 μg of Lipofectin were stimulated in vitro with 2.5 μM PyCSP57–70. Five overlapping 10-amino-acid peptides included in the region between amino acids 57 and 70, as well as PyCSP59–70 and PyCSP57–70, were used to coat P815 target cells in a standard CTL assay. Target cells coated with 1 μM PyCSP59–70, PyCSP57–70, PyCSP58–67, and PyCSP59–68 were all lysed by effector cells (Fig. 2). The amino acids 59 to 67 (YNRNIVNRL) were common to the four peptides that sensitized target cells for lysis and correspond to the peptide indicated by the MHC binding assay as the likely CTL epitope. Furthermore, it was also found that the CTL response induced by immunization with MAP4(PyCSP57–70) was genetically restricted and CD8+ T cell dependent. More specifically, lysis of target cells by splenocytes derived from mice immunized with MAP4(PyCSP57–70) was shown to be solely dependent on the presence of CD8+ T cells, since in vitro depletion of CD8+ T cells significantly reduced lysis whereas depletion of CD4+ T cells had no effect on lysis of target cells coated with PyCSP59–70 (Fig. 3). Genetic restriction of the response was demonstrated by the inability of effector cells to lyse EL-4 (H-2b) cells coated with PyCSP59–70 (data not shown).
FIG. 2.
Definition of the CTL epitope. Mice were immunized two times with 40 μg of MAP4(PyCSP57–70) plus 15 μg of Lipofectin. Results shown are those of the CTL assay done with splenocytes removed 14 days after the second immunization. Target P815 cells were coated with 1 μM PyCSP59–70, PyCSP57–70, PyCSP57–66, PyCSP58–67, PyCSP59–68, PyCSP60–69, or PyCSP61–70 or the irrelevant peptides, PfSSP2-15 and PfSSP2-9. E:T Ratio, effector/target cell ratio.
FIG. 3.
Depletion of CD8+ (−CD8) or CD4+ (−CD4) T cells from splenocyte cultures of mice immunized three times with 40 μg of MAP4(PyCSP57–70) and 15 μg of Lipofectin. Effectors were stimulated with 2.5 μM PyCSP59–70. Lysis of target P815 cells coated with 2.5 μM PyCSP59–70 is shown. E:T Ratio, effector/target cell ratio.
In conclusion, the data shown in this section demonstrate that immunization with the MAP4(PyCSP57–70) construct can induce a specific CTL response. Furthermore, these results also demonstrate the existence of a CD8+ H-2d-restricted CTL epitope corresponding to PyCSP58–67.
PyCSP58–67 is a subdominant or cryptic CTL epitope.
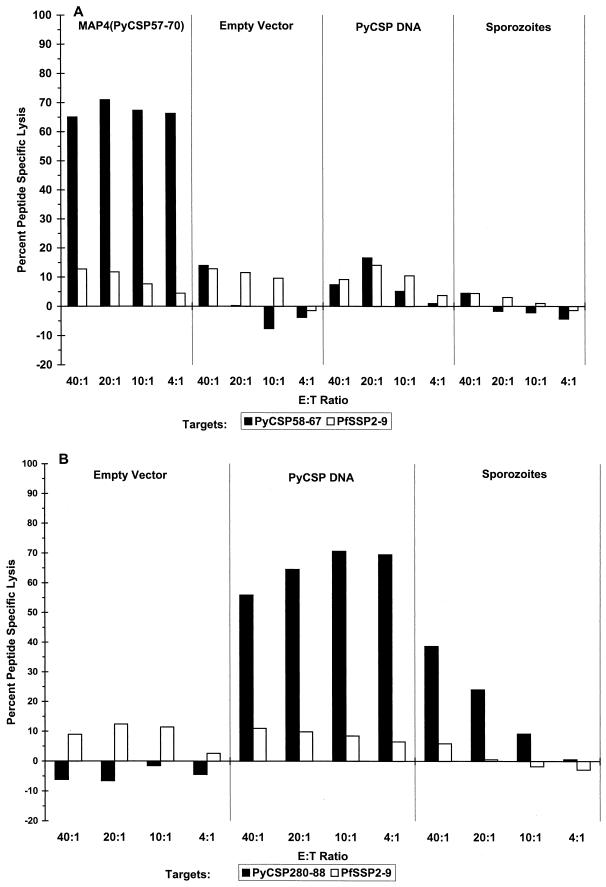
In order to determine whether the PyCSP58–67 epitope was recognized in the context of the responses elicited by the whole PyCSP antigen, we examined the pattern of T-cell responses from BALB/c mice immunized with irradiated P. yoelii sporozoites or PyCSP plasmid DNA. It has been shown previously that more than 50% of BALB/c mice immunized with plasmid DNA coding for PyCSP are protected against challenge with sporozoites (7, 30), and we have recently described how cDNA immunization can be utilized as a tool for definition of dominant and subdominant CTL epitopes (38). Splenocytes from mice immunized with irradiated sporozoites or PyCSP plasmid DNA were tested for their capacity to lyse target cells. As shown in Fig. 4A, splenocytes from mice immunized with three doses of irradiated P. yoelii sporozoites or PyCSP plasmid DNA and stimulated in vitro for 6 days with 1 μM PyCSP58–67 did not lyse target cells coated with 1 μM PyCSP58–67. In the same series of experiments, it was also shown that the control dominant CD8+ CTL epitope (PyCSP280–288) was indeed recognized by sporozoite- and PyCSP DNA-immunized mice (Fig. 4B).
FIG. 4.
(A) Immunization with PyCSP DNA or irradiated P. yoelii sporozoites did not induce CTLs specific for PyCSP58–67; however, immunization with MAP4(PyCSP57–70) induced high levels of CTLs specific PyCSP58–67. Effector cells were stimulated for 6 days with 1 μM PyCSP58–67; target cells were coated with 1 μM PyCSP58–67. (B) In the same experiment, immunization with PyCSP DNA or irradiated P. yoelii sporozoites induced CTLs against PyCSP280–288; the dominant CD8+ CTL epitope. Effector cells were stimulated for 6 days with 2.5 μM PyCSP280–296; target cells were coated with 0.025 μM PyCSP280–288. E:T Ratio, effector/target cell ratio.
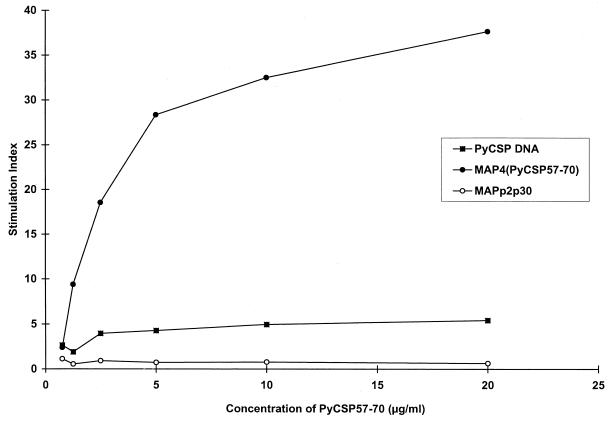
In addition, we wanted to determine if immunization with PyCSP DNA induces proliferation in the presence of PyCSP57–70. Splenocytes from these mice proliferated in vitro in the presence of PyCSP57–70; however, stimulation indices were lower than those in mice immunized with MAP4(PyCSP57–70) (Fig. 5). Splenocytes from mice immunized with irradiated sporozoites did not proliferate in the presence of PyCSP57–70 (data not shown). In conclusion, these data demonstrate that PyCSP58–67 is either a subdominant or a cryptic epitope not commonly recognized by immune responses elicited by the whole PyCSP antigen.
FIG. 5.
Immunization with PyCSP DNA induces low levels of proliferation against PyCSP57–70. Mice were immunized three times with either 40 μg of MAP4(PyCSP57–70) and Lipofectin, 40 μg of MAPp2p30 and Lipofectin, or 40 μg of PyCSP DNA and PBS. Results shown are those of the lymphocyte proliferation assay done with splenocytes removed 14 days after the last immunization.
MAP4(PyCSP57–70) immunization mediates genetically restricted, CD8+-dependent, and nitric oxide-dependent elimination of infected hepatocytes from culture.
To demonstrate the biological relevance of the responses induced by MAP4(PyCSP57–70), we initially tested spleen cells from mice immunized with two doses of MAP4(PyCSP57–70) and Lipofectin or Lipofectin alone, and stimulated in vitro for 6 days with MAP4(PyCSP57–70), for their capacity to eliminate infected hepatocytes in vitro. Compared to spleen cells from mice immunized with Lipofectin alone and stimulated in vitro with MAP4(PyCSP57–70), spleen cells from mice immunized with MAP4(PyCSP57–70) in Lipofectin and stimulated with MAP4(PyCSP57–70) eliminated 60 to 79% of infected hepatocytes from cultures (Table 1) (106 spleen cells). Spleen cells stimulated with the control peptide (PfSSP2-15) eliminated only 15% of infected hepatocytes. Because the T cells elicited by peptide immunization recognized naturally processed antigens, as produced by infected cells, these results demonstrate that the epitope recognized is not a cryptic one but rather a subdominant one according to the definition of Sercarz and colleagues (31).
TABLE 1.
Specific elimination of P. yoelii-infected hepatocytes by restimulated spleen cells from mice immunized with MAP4(PyCSP57–70) in Lipofectin
| No. of expt and condition | Immunogen | No. of spleen cells | Spleen cell stimulant | No. of schizonts/well (mean ± SD) | % Inhibition |
|---|---|---|---|---|---|
| Expt 1 | |||||
| BALB/c hepatocytes with BALB/c effector cells | Lipofectin | 1 × 106 | MAP4(PyCSP57–70) | 96, 108, 90 (98 ± 9) | |
| Lipofectin | 1 × 106 | PfSSP2-15 | 113, 117, 91 (107 ± 14) | ||
| MAP4(PyCSP57–70) | 1 × 106 | MAP4(PyCSP57–70) | 24, 21, 17 (21 ± 4) | 79 | |
| MAP4(PyCSP57–70) | 1 × 106 | PfSSP2-15 | 87, 99, 88 (91 ± 7) | 15 | |
| Expt 2 | |||||
| BALB/c hepatocytes with BALB/c effector cells | Lipofectin | 1 × 106 | MAP4(PyCSP57–70) | 66, 71, 76 (71 ± 4) | |
| Lipofectin | 5 × 105 | MAP4(PyCSP57–70) | 59, 57, 73 (63 ± 7) | ||
| MAP4(PyCSP57–70) | 1 × 106 | MAP4(PyCSP57–70) | 29, 21, 36 (28 ± 6) | 60 | |
| MAP4(PyCSP57–70) | 5 × 105 | MAP4(PyCSP57–70) | 42, 29, 27 (32 ± 6) | 49 | |
| MAP4(PyCSP57–70) | 2.5 × 105 | MAP4(PyCSP57–70) | 61, 62, 60 (61 ± 0.8) | 3 | |
| MAP4(PyCSP57–70) | 1 × 105 | MAP4(PyCSP57–70) | 74, 70, 73 (72 ± 1.6) | ||
| CD8+ depletion | MAP4(PyCSP57–70) | 1 × 106 | MAP4(PyCSP57–70) | 60, 59, 70 (63 ± 5) | 11 |
| MAP4(PyCSP57–70) | 5 × 105 | MAP4(PyCSP57–70) | 63, 56, 66 (61 ± 4) | 3 | |
| 5 μM 1400W (iNOS inhibitor) | MAP4(PyCSP57–70) | 1 × 106 | MAP4(PyCSP57–70) | 63, 83, 66 (70 ± 9) | 1 |
| C57BL/6 hepatocytes with BALB/c effector cells | Lipofectin | 1 × 106 | MAP4(PyCSP57–70) | 156, 152, 137 (148 ± 8) | |
| Lipofectin | 5 × 105 | MAP4(PyCSP57–70) | 200, 170, 183 (177 ± 6) | ||
| MAP4(PyCSP57–70) | 1 × 106 | MAP4(PyCSP57–70) | 120, 176, 145 (147 ± 23) | 1 | |
| MAP4(PyCSP57–70) | 5 × 105 | MAP4(PyCSP57–70) | 147, 169, 198 (171 ± 20) | 3 | |
| MAP4(PyCSP57–70) | 2.5 × 105 | MAP4(PyCSP57–70) | 135, 154, 159 (150 ± 3) | 15 | |
| MAP4(PyCSP57–70) | 1 × 105 | MAP4(PyCSP57–70) | 160, 159, 142 (156 ± 4) | 11 | |
| CD8+ depletion | MAP4(PyCSP57–70) | 1 × 106 | MAP4(PyCSP57–70) | 165, 149, 161 (158 ± 7) | 0 |
| MAP4(PyCSP57–70) | 5 × 105 | MAP4(PyCSP57–70) | 174, 148, 156 × (161 ± 13) | 12 |
We next wanted to determine whether the elimination of infected hepatocytes in vitro is genetically restricted and mediated by CD8+ T cells and nitric oxide. Spleen cells from BALB/c mice immunized with MAP4(PyCSP57–70) in Lipofectin eliminated infected BALB/c hepatocytes but not infected C57BL/6 hepatocytes from culture (Table 1), indicating that inhibition of liver stage development is MHC restricted. Depletion of CD8+ T cells reduced inhibition from 60 to 11% (Table 1). The inhibition of liver stage development by spleen cells from mice immunized with MAP4(PyCSP57–70) and stimulated with MAP4(PyCSP57–70) is reversed by an iNOS inhibitor (1400W) (Table 1), indicating that infected hepatocytes produced nitric oxide, which killed developing parasites.
Encouraged by these findings, we next tested whether MAP4(PyCSP57–70) induced protection against in vivo sporozoite challenge. Immunization of mice with three doses of 40 μg of MAP4(PyCSP57–70) in Lipofectin protected 18% (4 of 22) of the mice challenged with sporozoites in three experiments. None of the control mice (0 of 22) immunized with three doses of 16 μg of MAPp2p30 and Lipofectin were protected. The low-level protection was indeed statistically significant (exact Fisher's P value = 0.048).
DISCUSSION
In the present study, we demonstrated that immunization of mice with a MAP construct containing four branches of amino acids 57 to 70 from the PyCSP linked to a lysine-glycine core [MAP4(PyCSP57–70)] induced a peptide-specific, CD8+-dependent, genetically restricted CTL response. An H-2Kd binding motif is located between amino acids 58 and 67 (IYNRNIVNRL) with anchor sequences at positions 2 (tyrosine) and 10 (leucine) (27). We found that PyCSP58–67 bound to purified Kd in vitro, although the binding affinity was low (3,267 nM), and was also the epitope recognized by a CD8+ H-2d-restricted CTL response. In contrast, the epitope was not recognized in the context of immunization with whole PyCSP (in the form of either sporozoites or PyCSP DNA). However, the T cells resulting from peptide immunization recognized naturally processed antigen as produced from infected cells, as demonstrated by their ability to eliminate infected hepatocytes in vitro, and therefore, the epitope was classified as a subdominant epitope. The biological relevance of the responses induced by the MAP construct was demonstrated by the fact that clearance of infected hepatocytes was genetically restricted and CD8+ and nitric oxide dependent. Partial protection in vivo against sporozoite challenge was also demonstrated. To our knowledge, this represents the first study in which the exact molecular nature of a Plasmodium-derived, subdominant CTL epitope has been defined. Previous reports have described cryptic T-cell epitopes in the CSP of Plasmodium falciparum (11), Plasmodium berghei (16), and P. yoelii (34).
These results can be discussed in the context of their implications for the design of subunit- and epitope-based vaccines. The present results demonstrate that, by the combined use of motif analysis, quantitative molecular assays, and biological and immunological in vitro assays, subdominant Plasmodium-derived T-cell epitopes can be identified. Identification of subdominant CTL epitopes restricted by common HLA types and encoded in conserved regions of Plasmodium species of human pathogenic significance could represent an important aspect of the design of a prophylactic malaria vaccine. Focusing the immune response toward conserved pathogen regions may be crucial for obtaining broad efficacy in the case of infectious agents characterized by high mutation rates, which may use hypervariable peptides as a decoy system to derail the effectiveness of immune responses.
It is of interest to discuss the current results in the context of previous studies which reported T-cell reactivities directed against this CSP region derived from either P. yoelii or P. berghei. In this study, we showed that CD8+ T cells are required for CTL activity against target cells coated with the peptide PyCSP59–70 and for elimination of infected hepatocytes from culture. However, we cannot rule out the possibility that CD4+ T cells recognizing an epitope located between amino acids 57 and 70 of the PyCSP are also required for the in vitro inhibition of liver stage development reported here. Indeed, a large panel of CD4+ T-cell clones belonging to the TH1 and TH2 subsets were derived from BALB/c mice immunized with a peptide (PyCSP59–79) from this region (12, 13, 25, 34). It has previously been shown, unequivocally, that these CD4+ T-cell clones inhibit parasite development in vitro (5, 18, 26) and in vivo (34). However, none of the clones were shown to lyse target cells coated with the peptide (PyCSP59–79) in a CTL assay (25). Proliferation assays using truncated peptides showed that the epitope recognized by the CD4+ T cells was contained within the 13-amino-acid sequence from 59 to 71 (5), which overlaps with the amino acid sequence of the peptide (PyCSP57–70) used in our experiments. Immunization with amino acids 59 to 79 conjugated to the PyCSP repeat sequence (QGPGAP) provides help for the production of antibodies to peptides containing the repeat region (5, 26). In fact, the peptide containing amino acids 57 to 70, as opposed to 58 to 67, was originally selected for use in our studies because it differs in only two amino acids from the analogous T-cell epitope on the P. berghei CSP (28) and because mice immunized with a synthetic peptide containing this sequence (PbCSP57–70; KIYNRNTVNRLLAD) were indeed protected against P. berghei sporozoite challenge (20). In the case of PyCSP57–70, similar immunization studies with linear peptide were ineffective in inducing either T-cell reactivity or protection from sporozoite challenge (data not shown). It is thus apparent that the mechanisms and nature of T cells elicited by the PyCSP58–67 and PbCSP59–70 epitopes are quite distinct. The results presented herein demonstrate that the PyCSP58–67 reactivity is mediated by a classical subdominant CTL epitope. The observation that this CTL epitope binds Kd with low affinity (545-fold lower than the dominant Kd-restricted PyCSP280–288 epitope, which binds with an affinity of 6 nM) suggests that suboptimal MHC binding capacity might, in all or in part, explain the subdominance of this particular epitope, which would, therefore, according to a recently proposed classification scheme (39), be categorized as a type I subdominant CTL epitope. Similar examples of type I subdominant epitopes have recently been reported for the lymphocytic choriomeningitis virus system (36).
We would also like to comment on the fact that we are aware that the construct utilized to induce protective responses is not yet fully optimized. We plan in the near future to test the immunogenicity of constructs incorporating optimal helper T-cell epitopes, as well as the minimal PyCSP58–67 CTL epitope. We also plan to utilize lipopeptide constructs which do not require the use of adjuvants and may therefore be a closer model for human prophylactic use. One type of modification which we plan to evaluate is introducing specific changes within the CTL epitope in order to increase its binding affinity and immunogenicity, according to the strategy exemplified by Kast and colleagues (14), Parkhurst and colleagues (24), and Topalian and colleagues (35).
Eberl and colleagues (8) described how in certain cases immunodominance may apparently not correlate with the affinity of peptides for MHC molecules but rather may be explained at the T-cell level by the existence of a limited peptide-specific T-cell repertoire. This repertoire may have a low chance of encountering peptide-MHC complexes, expands slowly due to limited availability of T-cell growth factors in the local environment, and is immunodominated by a bigger, faster-expanding T-cell repertoire. In this case, the use of exogenous growth factors, such as interleukin 12, in the immunization protocol could overcome the immunodominance effect (8).
In conclusion, we have shown that Plasmodium subdominant epitopes can be identified and utilized to induce elimination of infected hepatocytes in culture and partial protective immunity effective against sporozoite challenge. Future studies may identify similar subdominant epitopes derived from conserved regions of the P. falciparum genome and evaluate their use in development of malaria vaccines destined for human use.
ACKNOWLEDGMENTS
This study was supported by the Naval Medical Research and Development Command work units 612787A.870.00101.EFX.1432 and 612787A.870.00101.EVX.1435.
REFERENCES
- 1.Bodmer J G, Marsh S G E, Albert E D, Bodmer W F, Dupont B, Erlich H A, Mach B, Mayr W R, Parham P, Sasazuki T, et al. Nomenclature for factors of the HLA system, 1994. Tissue Antigens. 1994;44:1–18. doi: 10.1111/j.1399-0039.1994.tb02351.x. [DOI] [PubMed] [Google Scholar]
- 2.Buus S, Sette A, Colon S M, Miles C, Grey H M. The relation between major histocompatibility complex (MHC) restriction and the capacity of Ia to bind immunogenic peptides. Science. 1987;235:1353–1358. doi: 10.1126/science.2435001. [DOI] [PubMed] [Google Scholar]
- 3.Celis E, Sette A, Grey H M. Epitope selection and development of peptide based vaccines to treat cancer. Semin Cancer Biol. 1995;6:329–336. doi: 10.1016/1044-579x(95)90002-0. [DOI] [PubMed] [Google Scholar]
- 4.Charoenvit Y, Mellouk S, Sedegah M, Toyoshima T, Leef M F, De la Vega P, Beaudoin R L, Aikawa M, Fallarme V, Hoffman S L. Plasmodium yoelii: 17-kDa hepatic and erythrocytic stage protein is the target of an inhibitory monoclonal antibody. Exp Parasitol. 1995;80:419–429. doi: 10.1006/expr.1995.1054. [DOI] [PubMed] [Google Scholar]
- 5.Del Giudice G, Grillot D, Renia L, Muller I, Corradin G, Louis J A, Mazier D, Lambert P H. Peptide-primed CD4+ cells and malaria sporozoites. Immunol Lett. 1990;25:59–64. doi: 10.1016/0165-2478(90)90092-5. [DOI] [PubMed] [Google Scholar]
- 6.Deres K, Schild H, Wiesmuller K H, Jung G, Rammensee H G. In vivo priming of virus-specific cytotoxic T lymphocytes with synthetic lipopeptide vaccine. Nature. 1989;342:561–564. doi: 10.1038/342561a0. [DOI] [PubMed] [Google Scholar]
- 7.Doolan D L, Sedegah M, Hedstrom R C, Hobart P, Charoenvit Y, Hoffman S L. Circumventing genetic restriction of protection against malaria with multigene DNA immunization: CD8+ cell-, interferon gamma-, and nitric oxide-dependent immunity. J Exp Med. 1996;183:1739–1746. doi: 10.1084/jem.183.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberl G, Kessler B, Eberl L P, Brunda M J, Valmori D, Corradin G. Immunodominance of cytotoxic T lymphocyte epitopes co-injected in vivo and modulation by interleukin-12. Eur J Immunol. 1996;26:2709–2716. doi: 10.1002/eji.1830261124. [DOI] [PubMed] [Google Scholar]
- 9.Franke E D, Hoffman S L, Sacci J B, Jr, Wang R, Charoenvit Y, Appella E, Chesnut R, Alexander J, Del Guercio M F, Sette A. Pan DR binding sequence provides T-cell help for induction of protective antibodies against Plasmodium yoelii sporozoites. Vaccine. 1999;17:1201–1205. doi: 10.1016/s0264-410x(98)00341-7. [DOI] [PubMed] [Google Scholar]
- 9a.Franke E D, Corradin G, Hoffman S L. Induction of protective CTL responses against the Plasmodium yoelii circumsporozoite protein by immunization with peptides. J Immunol. 1997;159:3424–3433. [PubMed] [Google Scholar]
- 10.Garvey E P, Oplinger J A, Furfine E S, Kiff R J, Laszlo F, Whittle B J, Knowles R G. 1400W is a slow, tight binding, and highly selective inhibitor of inducible nitric-oxide synthase in vitro and in vivo. J Biol Chem. 1997;272:4959–4963. doi: 10.1074/jbc.272.8.4959. [DOI] [PubMed] [Google Scholar]
- 11.Good M F, Branigan J, Smith G, Houghten R A. Peptide immunization can elicit malaria protein-specific memory helper but not proliferative T cells. Pep Res. 1990;3:110–115. [PubMed] [Google Scholar]
- 12.Grillot D, Michel M, Muller I, Tougne C, Renia L, Mazier D, Corradin G, Lambert P H, Louis J A, Del Guidice G. Immune responses to defined epitopes of the circumsporozoite protein of the murine malaria parasite, Plasmodium yoelii. Eur J Immunol. 1990;20:1215–1222. doi: 10.1002/eji.1830200604. [DOI] [PubMed] [Google Scholar]
- 13.Grillot D, Valmori D, Lambert P H, Corradin G, Del Giudice G. Presentation of T-cell epitopes assembled as multiple-antigen peptides to murine and human T lymphocytes. Infect Immun. 1993;61:3064–3067. doi: 10.1128/iai.61.7.3064-3067.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Institute of Laboratory Animal Resources. Guide for the care and use of laboratory animals. Washington, D.C.: National Academy Press; 1996. [Google Scholar]
- 14.Kast W M, Brandt R M, Sidney J, Drijfhout J W, Kubo R T, Grey H M, Melief C J, Sette A. Role of HLA-A motifs in identification of potential CTL epitopes in human papillomavirus type 16 E6 and E7 proteins. J Immunol. 1994;152:3904–3912. [PubMed] [Google Scholar]
- 15.Kawakami Y, Eliyahu S, Jennings C, Sakaguchi K, Kang X, Southwood S, Robbins P F, Sette A, Appella E, Rosenberg S A. Recognition of multiple epitopes in the human melanoma antigen gp100 by tumor-infiltrating T lymphocytes associated with in vivo tumor regression. J Immunol. 1995;154:3961–3968. [PubMed] [Google Scholar]
- 16.Krzych U, Jereed T, Link H T, Loomis L D, Ballou W R. Distinct T cell specificities are induced with the authentic versus recombinant Plasmodium berghei circumsporozoite protein. J Immunol. 1992;148:2530–2538. [PubMed] [Google Scholar]
- 17.Li S, Rodrigues M, Rodriguez D, Rodriguez J R, Esteban M, Palese P, Nussenzweig R S, Zavala F. Priming with recombinant influenza virus followed by administration of recombinant vaccinia virus induces CD8+ T-cell-mediated protective immunity against malaria. Proc Natl Acad Sci USA. 1993;90:5214–5218. doi: 10.1073/pnas.90.11.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marussig M, et al. Linear and multiple antigen peptides containing defined T and B epitopes of the Plasmodium yoelii circumsporozoite protein: antibody-mediated protection and boosting by sporozoite infection. Int Immunol. 1997;9:1817–1824. doi: 10.1093/intimm/9.12.1817. [DOI] [PubMed] [Google Scholar]
- 19.Mellouk S, Berbiguier N, Druilhe P, Sedegah M, Galey B, Yuan L, Leef M, Charoenvit Y, Paul C, Hoffman S, et al. Evaluation of an in vitro assay aimed at measuring protective antibodies against sporozoites. Bull W H O. 1990;68(Suppl.):52–59. [PMC free article] [PubMed] [Google Scholar]
- 20.Migliorini P, Betschart B, Corrradin G. Malaria vaccine: immunization of mice with a synthetic T cell helper epitope alone leads to protective immunity. Eur J Immunol. 1993;23:582–585. doi: 10.1002/eji.1830230245. [DOI] [PubMed] [Google Scholar]
- 21.Nardelli B, Tam J P. Cellular immune responses induced by in vivo priming with a lipid-conjugated multimeric antigen peptide. Immunology. 1993;79:355–361. [PMC free article] [PubMed] [Google Scholar]
- 22.Pacheco N D, Strome C P, Mitchell F, Bawden M P, Beaudoin R L. Rapid, large-scale isolation of Plasmodium berghei sporozoites from infected mosquitoes. J Parasitol. 1979;65:414–417. [PubMed] [Google Scholar]
- 23.Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol. 1989;19:2237–2242. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- 24.Parkhurst M R, Salgaller M L, Southwood S, Robbins P F, Sette A, Rosenberg S A, Kawakami Y. Improved induction of melanoma-reactive CTL with peptides from the melanoma antigen gp100 modified at HLA-A*0201-binding residues. J Immunol. 1996;157:2539–2548. [PubMed] [Google Scholar]
- 25.Renia L, Grillot D, Marussig M, Corradin G, Miltgen F, Lambert P H, Mazier D, Del Giudice G. Effector functions of circumsporozoite peptide-primed CD4+ T cell clones against Plasmodium yoelii liver stages. J Immunol. 1993;150:1471–1478. [PubMed] [Google Scholar]
- 26.Renia L, Marussig M S, Grillot D, Pied S, Corradin G, Miltgen F, Del Giudice G, Mazier D. In vitro activity of CD4+ and CD8+ T lymphocytes from mice immunized with a synthetic malaria peptide. Proc Natl Acad Sci USA. 1991;88:7963–7967. doi: 10.1073/pnas.88.18.7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romero P, Corradin G, Luescher I F, Maryanski J L. H-2Kd-restricted antigenic peptides share a simple binding motif. J Exp Med. 1991;174:603–612. doi: 10.1084/jem.174.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero P J, Tam J P, Schlesinger D, Calvijo P, Gibson H, Barr P J, Nussenzweig R S, Nussenzweig V, Zavala F. Multiple T helper cell epitopes of the circumsporozoite protein of Plasmodium berghei. Eur J Immunol. 1988;18:1951–1957. doi: 10.1002/eji.1830181213. [DOI] [PubMed] [Google Scholar]
- 29.Schild H, Deres K, Wiesmuller K H, Jung G, Rammensee H G. Efficiency of peptides and lipopeptides for in vivo priming of virus-specific cytotoxic T cells. Eur J Immunol. 1991;21:2649–2654. doi: 10.1002/eji.1830211102. [DOI] [PubMed] [Google Scholar]
- 30.Sedegah M, Hedstrom R, Hobart P, Hoffman S L. Protection against malaria by immunization with plasmid DNA encoding circumsporozoite protein. Proc Natl Acad Sci USA. 1994;91:9866–9870. doi: 10.1073/pnas.91.21.9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sercarz E E, Lehmann P V, Ametani A, Benichou G, Miller A, Moudgil K. Dominance and crypticity of T cell determinants. Annu Rev Immunol. 1993;11:729–766. doi: 10.1146/annurev.iy.11.040193.003501. [DOI] [PubMed] [Google Scholar]
- 32.Sette A, Buus S, Colon S, Miles C, Grey H M. Structural analysis of peptides capable of binding to more than one Ia antigen. J Immunol. 1989;142:35–40. [PubMed] [Google Scholar]
- 33.Sidney J, Grey H M, Kubo R T, Sette A. Practical, biochemical and evolutionary implications of the discovery of HLA class I supermotifs. Immunol Today. 1996;17:261–266. doi: 10.1016/0167-5699(96)80542-1. [DOI] [PubMed] [Google Scholar]
- 34.Takita-Sonoda Y, Tsuji M, Kamboj K, Nussenzweig R S, Clavijo P, Zavala F. Plasmodium yoelii: peptide immunization induces protective CD4+ T cells against a previously unrecognized cryptic epitope of the circumsporozoite protein. Exp Parasitol. 1996;84:223–230. doi: 10.1006/expr.1996.0108. [DOI] [PubMed] [Google Scholar]
- 35.Topalian S L, Gonzales M I, Parkhurst M, Li Y F, Southwood S, Sette A, Rosenberg S A, Robbins P F. Melanoma-specific CD4+ T cells recognize nonmutated HLA-DR-restricted tyrosinase epitopes. J Exp Med. 1996;183:1965–1971. doi: 10.1084/jem.183.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Most R G, Sette A, Oseroff C, Alexander J, Murali-Krishna K, Lau L L, Southwood S, Sidney J, Chesnut R W, Matloubian M, Ahmed R. Analysis of cytotoxic T cell responses to dominant and subdominant epitopes during acute and chronic lymphocytic choriomeningitis virus infection. J Immunol. 1996;157:5543–5554. [PubMed] [Google Scholar]
- 37.Vitiello A, Ishioka G, Grey H M, Rose R, Farness P, LaFond R, Yuan L, Chisari F V, Furze J, Bartholomeuz R, Chesnut R W. Development of a lipopeptide-based therapeutic vaccine to treat chronic HBV infection. I. Induction of a primary cytotoxic T lymphocyte response in humans. J Clin Investig. 1995;95:341–349. doi: 10.1172/JCI117662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vitiello A, Sette A, Yuan L, Farness P, Southwood S, Sidney J, Chesnut R W, Grey H M, Livingston B. Comparison of cytotoxic T lymphocyte responses induced by peptide or DNA immunization: implications on immunogenicity and immunodominance. Eur J Immunol. 1997;27:671–678. doi: 10.1002/eji.1830270315. [DOI] [PubMed] [Google Scholar]
- 39.Vitiello A, Yuan L, Chesnut R W, Sidney J, Southwood S, Farness P, Jackson M R, Peterson P A, Sette A. Immunodominance analysis of CTL responses to influenza PR8 virus reveals two new dominant and subdominant Kb-restricted epitopes. J Immunol. 1996;157:5555–5562. [PubMed] [Google Scholar]
- 40.Walker C, Selby M, Erickson A, Cataldo D, Valensi J P, Van Nest G. Cationic lipids direct a viral glycoprotein into the class I major histocompatibility complex antigen-presentation pathway. Proc Natl Acad Sci USA. 1992;89:7915–7918. doi: 10.1073/pnas.89.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Widmann C, Romero P, Maryanski J L, Corradin G, Valmori D. T helper epitopes enhance the cytotoxic response of mice immunized with MHC class I-restricted malaria peptides. J Immunol Methods. 1992;155:95–99. doi: 10.1016/0022-1759(92)90275-x. [DOI] [PubMed] [Google Scholar]
- 42.Wizel B, Rogers W O, Houghten R A, Lanar D E, Tine J A, Hoffman S L. Induction of murine cytotoxic T lymphocytes against Plasmodium falciparum sporozoite surface protein 2. Eur J Immunol. 1994;24:1487–1495. doi: 10.1002/eji.1830240705. [DOI] [PubMed] [Google Scholar]