Abstract
The Lyra SARS-CoV-2 assay was the primary method for molecular testing performed at Barnes-Jewish Healthcare System in St. Louis, Missouri during the initial COVID-19 surge from mid-March to late-April 2020. We performed a retrospective analysis of 1,043 positive Lyra SARS-CoV-2 results during these 36 days to investigate associations between cycle threshold (CT) value and patient characteristics. Total RNA were extracted from NP or OP swabs using either the EasyMag or KingFisher automated extraction systems and quantified with RotorGene Q (Qiagen) or Applied Biosystems 7500 Fast Dx thermocyclers respectively. Notably, we found lower a significant median lower CT for samples tested on the KingFisher-ABI 7500 fastDX (KF/ABI) system compared to the EasyMag/RotorGene (EM/RGQ) platform. Since 77.5% of our tests were ran on the EM/RGQ pipeline we then perform additional analysis on these values and found that C T values in outpatient care settings compared to samples obtained in the emergency department or inpatient had significantly lower C T values. These collective findings suggests a difference in viral load amongst various patient populations.
Keywords: Clinical virology, SARS-CoV-2, Molecular assays
1. Introduction
Molecular detection of SARS-CoV-2 RNA is the primary method used to identify viral presence in patients with suspected COVID-19 (Ransom et al., 2020). Upper respiratory specimens, most commonly nasopharyngeal (NP) and oropharyngeal swabs (OP), are utilized for testing (Han et al., 2020; Ransom et al., 2020). Currently, molecular platforms range from sample-to-answer assays that require minimal hands-on time or technical expertise to systems requiring separate instruments for RNA extraction and real time RT-PCR systems (Han et al., 2020; Ransom et al., 2020). The Lyra SARS-CoV-2 Assay (Quidel) is a RT-PCR assay for detection of SARS-CoV-2 presence using fluorescent probes that bind to non-structural polyprotein (pp1ab)/Orf1ab RNA within the SARS-CoV-2 genome. The analysis is preceded by total RNA extraction and the assay uses a separate extraction system and amplification/detection system (Ransom et al., 2020).
The Barnes-Jewish Healthcare System, a network of hospitals in the St. Louis metropolitan area (Missouri, USA) implemented the Lyra SARS-CoV-2 Assay as one of our initial tests for molecular identification of SARS-CoV-2. Here we report on the experience with the assay during March and April 2020, using two different extraction and quantification platforms. Samples were sent to either the EasyMag (bioMerieux)/RotorGene Q (Qiagen) or KingFisher (ThermoFisher)/Applied Biosystems 7500 Fast Dx (ThermoFisher) extraction/amplification systems. We have recently demonstrated that the KingFisher/7500 Fast Dx system results in a statistically significant -1 cycle threshold (CT) difference compared to the EasyMag/RotorGeneQ system but was not likely to be biologically meaningful (Ransom et al., 2020). We performed a retrospective analysis of the CT values from 1,043 valid positive clinical results (from a total of 7,411 samples t during this time frame) to identify differences between platform and patient demographic information.
2. Materials & methods
2.1. Molecular detection of SARS-CoV-2
Patients symptomatic for COVID-19 or with known sick contacts had mucosal sites sampled using NP or OP swabs (including BD, Copan, Puritan, and Hardy) as part of routine clinical management. The encounter types occurred primarily in outpatient (i.e. symptomatic healthcare worker testing), inpatient (e.g. intensive care unit,) or emergency department (ED) settings at various locations in the Barnes-Jewish Healthcare System in the St. Louis region. All samples were submitted to the Barnes-Jewish Hospital Molecular Infectious Disease (BJH-MID) Lab for SARS-CoV-2 testing. Due to potential challenges with procurement of reagents, the BJH-MID Laboratory established two different testing pipelines for detection of SARS-CoV-2 RNA (Ransom et al., 2020). Total RNA was extracted from samples by trained clinical laboratory staff using either the NUCLISENS® EasyMag (bioMerieux) or KingFisher (Thermo Fisher) instruments. For extraction, 20 μL of the Lyra Processing Control was added to 180 μL aliquots of the VTM or UTM. The KingFisher system elutes in 50 μL while the EasyMag system elutes in 100 μL. To perform the PCR reaction, 135 μL of the reagent solvent is added to lyophilized master mix and 15 μL of reconstituted solution is used with 5 μL of input RNA. This method is in accordance with the manufacturer's instructions for use required for compliance with the emergency use authorization granted by the FDA. Samples extracted on the EasyMag were paired with amplification/detection on the RotorGene Q (Qiagen) thermocycler and samples from KingFisher were detected on the Applied Biosystems 7500 Fast Dx (ThermoFisher). Samples were deemed valid if the Lyra processing control (added to every specimen during the extraction phase) had a detectable CT value and runs were deemed valid if both the Lyra positive and negative SARS-CoV-2 control samples, provided in the test kit, had detectable processing control and the SARS-CoV-2 target was respectively detected or not detected. CT values were manually recorded.
2.2. Patient demographic information
Health information pertaining to each patient sampled for SARS-CoV-2 presence was recorded and retrieved from the Epic electronic medical record from mid-March to late-April 2020 after approval from the Washington University in St. Louis Medical Center Human Research Protection Office (IRB #202005002). Encounter type was clustered into three categories, inpatient, outpatient, and ED. Other non-identifying medical information procured for the purpose of our study was sample type, gender, and age.
2.3. Statistics
Pairwise comparison of CT values for the different conditions was analyzed in Prism v8.4.3 using an unpaired parametric t-test. One-way ANOVA was used to determine significance in CT values for comparisons with >2 groups.
3. Results
3.1. Overall trends in test positivity
Using the first positive sample per patient, we found a total percent positivity of 14.3% (1043/7257). The highest proportion of positive tests came from the ED (20.9%, 497/2369) compared to outpatient (13.2%, 472/3579) and inpatient (5.6%, 74/1309) facilities. When examining positivity by age we observed a stepwise increase between larger age bins. The 20-29 and 30-39 groups had the lowest positivity rate (12.8% and 12.1%) which increased in the 40-49 and 50-59 groups (16.6% and 17.6%) and again in the 60-69 and 70-79 (20.0% and 20.62%) before rising in the 80-89 and 90+ groups (30.3% and 37.2%). The major trend observed for test order setting was a higher count of outpatient tests compared to ED for 20-29 (73 vs 35 tests), 30-39 (75 vs 46 tests), and 40-49 (76 vs 57 tests) groups but an inverse of this for 50-59 (80 vs 96 tests), 60-69 (71 vs 108 tests), 70-79 (44 vs 73 tests), and 80-89 (37 vs 52 tests). Contingency analysis within the inpatient group was underpowered for further statistical evaluation.
3.2. CT values differ between testing platform and are not normally distributed
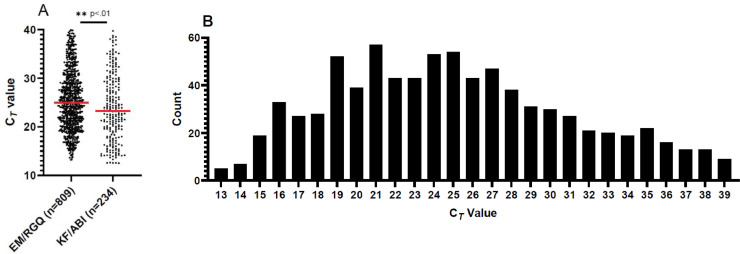
We retrieved the CT values for 1043 positive Lyra SARS-CoV-2 assays (from 7,411 samples tested) from mid-March to late-April 2020 at Barnes-Jewish Healthcare system. Since the Lyra assay does not include the first 10 cycles in the report out of CT from the instrument, we added 10 to each value for more consistent comparison with other molecular assays (Ransom et al., 2020). Given supply procurement challenges and difficulty obtaining reagents necessary for SARS-CoV-2 molecular testing, we used two separate extraction and detection platforms. Both methods were used interchangeably through the course of testing, with decisions for use based upon the reagent availability or test batch size. Initially we split the 1,043 positive samples by their respective test platform and found a significant (p=.0012) difference between the CT values produced by EM/RGQ versus KF/ABI (Fig. 1 .A). The median for EM/RGQ was 24.96 while the median for KF/ABI was 23.25. Despite having fewer samples, the KF/ABI values had a slightly larger standard deviation (6.8) compared to the EM/RGQ (6.1) values. Given that the EM/RGQ represents 77.5% (809/1043) of positive tests and because of the difference in CT value by platform we chose to analyze the 809 EM/RGQ samples. The 25%-75% interquartile range of for the EM/RGQ data was 20.76 – 29.58. The minimum and maximum CT values obtained for this assay was 13.20 and 39.94, respectively (Fig. 1B). Interestingly, the data was not normally distributed as the Shapiro-Wilk test (p value <.0001) did not pass normality. Only 1.12% (9/809) of samples had CT values of 39, near the limit of detection in this assay.
Fig. 1.
Comparison of CT values for both testing platforms and distribution of EM/RGQ Lyra SARS-CoV-2 test CT values from March-April, 2020. (A) Scatter dots depict each positive CT value as a point. Red line depicts the median CT value. We find differences in CT values obtained between platform (unpaired parametric t-test p value .0012). (B) Histogram depicting number of samples binned to their lowest whole number for EM/RGQ samples. Distribution of CT values indicates our data is not normally distributed as the Shapiro-Wilk test (p value <.0001) did not pass normality.
3.3. Differences in CT value between test order setting and age
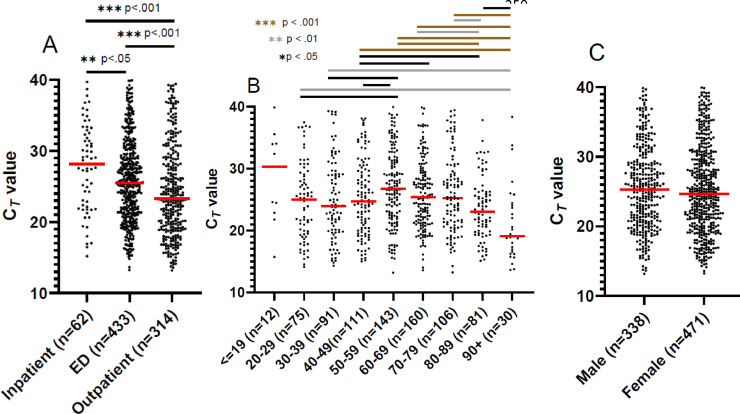
Given that our samples originated from a variety of patient care settings, we stratified positive results into inpatient, emergency department (ED), and outpatient bins to examine CT value trends. Interestingly we found significant separation amongst all groups using ANOVA (p < .001) and amongst all possible pairwise comparisons (Fig. 2A). Outpatient tests had the lowest median CT value (23.28) compared to ED (25.5) and inpatient (28.16) positive tests.
Fig. 2.
CT values by age, setting, and gender. Dotplot for EM/RGQ CT values depicting depict each positive CT value as a point for test order setting (A),age (B), and gender (C). Red line depicts the median CT value. Overall difference (ANOVA p <0.0001) in CT for age (A) and test order setting (B) but not gender (C). Individual pairwise comparisons significant by unpaired parametric t-test denoted in figure.
Since age has been indicated as a major risk factor for development of severe respiratory problems during SARS-CoV-2 infection, we performed an analysis of our institutional data by stratifying patients into 10-year age brackets. ANOVA found significant (p <0.0001) separation amongst all groups (Fig. 2B). We excluded positive values from patients <= 19 years of age given the smaller sample size from that population. We observed that the highest median CT values were detected in patients 50-59, 60-69, and 70-79 (26.77, 25.3, and 25.24 respectively). The 20-29 age bracket had significant differences in median CT values compared to the 50-59 and 90+ groups. While the middle ages had the highest median CT values, the 30-39, 80-89, and 90+ age ranges have the lowest median CT values (23.93, 23.00, and 19.07 respectively).We did not find a significant difference for other patient demographic factors such as gender (p=.23) (Fig. 2C).
3.4. Consecutive repeat positive CT values
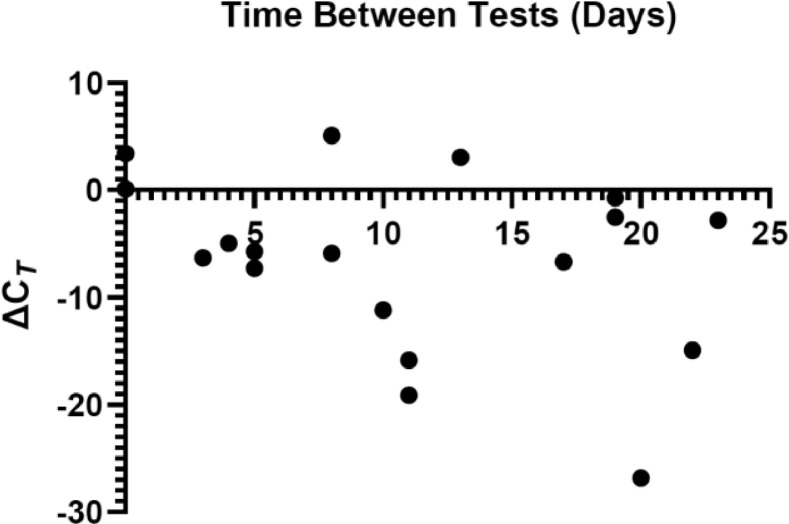
During the period evaluated, 527 patients had multiple tests performed. Of these tests, we had 18 occurrences of two consecutive positive Lyra SARS-CoV-2 assay results using the EM/RGQ system for 18 patients (Fig. 3 ). These additional 18 CT values were excluded from all previously mentioned analysis. We were interested in comparing the ΔCT value (CT1- CT2) as a function of time between tests that were consecutively positive. We found that the median 10.5 days between positive tests with 2/18 (11.1%) actually retested on the same day and a median of -5.7 ΔCT value. 3/18 (16.67%) instances had a positive ΔCT indicating that the level of viral burden increased between testing. In our study cohort, covering a 36-day time frame, the greatest time difference between two positive tests was 23 days. 5 individuals were both tested outpatient, 6 were initially tested in the ED then were admitted and tested as an inpatient. 5 were tested consecutively while inpatient and 2 were tested twice in the ED.
Fig. 3.
Association of repeat CT values in unique patients and the time between results. Repeat XY plot showing the EM/RGQ ΔCT (CT1-CT2) as a function of days elapsed between the two tests for patients who consecutively tested positive for SARS-CoV-2. A negative ΔCT value indicates a decrease in viral load between tests.
4. Discussion
While molecular RT-PCR tests have become the primary means for reliable detection of SARS-CoV-2 RNA for diagnostic determination of COVID-19, there exists a relative dearth of knowledge on the relationship between CT values, patient information, and outcome. True quantification of viral levels for kinetics relies on a standard curve, but further investigation of CT values could yield clinical insight (Han et al., 2020; Tom and Mina, 2020). This highlights the need for integration of CT values as a proxy for relative viral load with studies that include patient demographic and clinical factors. Overall, our study found that testing platform, age, and patient setting are factors that can result in different CT values. Our result on the difference in CT values between the two extraction/quantification system used fits with a separate investigation by our group comparing extraction platforms which found around a 1 CT lower CT in KingFisher compared to EasyMag extraction, likely due to elution of RNA in a smaller volume (Ransom et al., 2020). Both methods showed exceptional sensitivity when known positive samples were diluted and had RNA extracted (Ransom et al., 2020).
One recent investigation has found associations between death, intubation and high viral loads (CT < 25) compared to medium (CT 25-30) and low viral loads (CT >30) (Magleby et al., 2020). This study used the Cobas 6800 RT-PCR system (Roche) which detects the Orf1ab and the E gene within the SARS-CoV-2 genome (Magleby et al., 2020). Given that the Lyra SARS-CoV-2 Assay also analyses Orf1ab levels, there is opportunity to compare our findings with the findings of this group. Using their criteria, our EM/RGQ results had 50.2% (406/809) in the high viral load, 30.0% (243/809) with medium viral load, and 19.8% (160/809) with low viral loads. As a laboratory serving a large healthcare system, we were able to categorize tests as being ordered from an inpatient, ED, or outpatient setting and found significant differences in the CT values between all pairwise groups. The median for inpatient and ED settings were both in the medium viral load range but the median for our outpatient tests was in the high viral load category. The observation that outpatient setting tests had lower CT values compared to patients presenting at the ED or tested from an inpatient hospital floor was unexpected but could possibly be due to greater levels of aerosol exposure as these patients are predominantly healthcare workers. One investigation of influenza burden in children came to a similar conclusion to our own observation that CT values in outpatients tend to be lower than inpatients (Fuller et al., 2013). One explanation given by these authors is that outpatients present earlier for testing than inpatients when viral loads are likely to be higher (Fuller et al., 2013). It has been demonstrated that CT values can be similar between asymptomatic patients and those with COVID-19 disease (Zou et al., 2020). When stratified for age (excluding the low n <19 group) we found that the age intervals with the highest median CT value (20-29, 50-59, 60-69, and 70-79) were in the medium viral load but all other groups had medians in the high viral load category. Further work is therefore warranted to investigate if indicators of severe disease were also more prevalent in these groups. This trend of lower CT values amongst the youngest population and oldest adults but higher CT values amongst the middle age has also been documented for norovirus (Shioda et al., 2017). Shioda et al hypothesized that this is likely due to age related senescence in the elderly and lack of prior norovirus exposure to the youngest. A group from Germany found no meaningful differences amongst age range and viral load, however they did not use CT values but rather a viral load estimation using standard curve for their two different platforms (Jones et al., 2020). Additionally, these authors included lower respiratory samples although they make up a small (∼3%) of the total volume. Differences between molecular target, master mix efficiency, and threshold cutoffs may make interpretation of CT values between assays difficult. For instance, the US CDC assay detects the nucleocapsid gene and not the Orf1ab gene (the target of the Lyra assay). This could explain why an analysis of the first 12 US COVID-19 patients found a higher mean CT value than our analysis (Anonymous, 2020).
Importantly, the Lyra SARS-CoV-2 Assay and other molecular tests detect presence of viral RNA and not infectious virions. A recent study investigated the relationship between CT value and growth in Vero cells, finding that no growth occurred in NP samples with CT value >24 cycles (Bullard et al., 2020). CT values were obtained using real time RT-PCR for a 122 bp region within the E gene. In line with this, another group found that patients with CT values >33 cycles are noninfectious and according to their model can be discharged (La Scola et al., 2020). Considerable interest exists for the use of repeat PCR testing to diagnose patients with suspected COVID-19 but are PCR negative, as well as monitor viral levels in patients with resolving symptoms (Fisher et al., 2020). Previous investigations have shown that CT values generally spike 5-6 days following symptom onset and gradually decline overtime, in some cases taking 6 weeks before becoming negative (Xiao et al., 2020). We therefore were interested in investigating instances where patients tested positive consecutively using the Lyra SARS-CoV-2 assay and found that most have higher CT values in the second test indicating a decreased viral burden.
In this study we retrospectively analyzed 809 positive Lyra SARS-CoV-2 Assay tests to investigate CT value overall distribution and differences in CT values and positive tests as it relates to test ordering setting and patient demographics. One of the limitations of our study is that given the short timeframe of our study, patients could test persistently positive longer than the time range we investigated. Another limitation is that we are unable to ascertain an accurate day of symptom onset for our patients, if they were symptom-free at the time of testing, or whether they developed symptoms at a later date. This makes it difficult to use our data to track natural progression of SARS-CoV-2 levels within patients. Additionally, we have not integrated clinical factors with our laboratory data to ascertain which patients had severe COVID-19 disease. These correlative findings could be valid only during the initial surge of cases and may not reflect the CT distribution in the timeframe following the initial surge, therefore continued monitoring of CT values is ongoing. Variability in purity or quality between samples of extracted nucleic acid may be partially responsible for different CT values. Finally there could be a difference between CT values from NP and OP swab samples but we are underpowered given the low number of OP swabs analyzed to address this.
Our investigation has many strengths. We were able to evaluate a large number of samples from a variety of patient encounter-types. We used the Lyra Assay as our testing platform and thus we were able to obtain a CT value for quantification which is not possible on all testing platforms used clinically. An advantage of our investigation is that as the central laboratory for a multi-hospital system we were able to achieve a high number of positive samples quickly for analysis and could stratify those tests into relevant bins. Additionally, our testing population includes healthcare workers and local residents. We found that within these groups there were significant differences between CT values by test setting and age bins. We have also demonstrated that patients can remain consecutively positive for greater than 10 days but further work is warranted to investigate the infectivity of patients with high CT values. In conclusion, Lyra Assay CT values can be used within our system to identify differences in viral load between patients within different age brackets and from testing sites.
Author’ contributions
RFP and BA were responsible for data curation, writing, revising, and editing. CSE, CDB, NWA, BAP, were involved in writing, revising, and editing.
Declaration of competing interest
There are no conflicts of interest.
Acknowledgments
Acknowledgments
We thank the Molecular Infectious Disease Lab of Barnes-Jewish Hospital for their tireless efforts in running SARS-CoV-2 tests and their help in compiling the data.
Funding
No external funding was obtained to support this study.
References
- Anonymous. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med. 2020;26:861–868. doi: 10.1038/s41591-020-0877-5. [DOI] [PubMed] [Google Scholar]
- Bullard J, Dust K, Funk D, Strong JE, Alexander D, Garnett L, et al. Predicting infectious SARS-CoV-2 from diagnostic samples. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher B, Seese L, Sultan I, Kilic A. The importance of repeat testing in detecting coronavirus disease 2019 (COVID-19) in a coronary artery bypass grafting patient. J Card Surg. 2020;35:1342–1344. doi: 10.1111/jocs.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller JA, Njenga MK, Bigogo G, Aura B, Ope MO, Nderitu L, et al. Association of the CT values of real-time PCR of viral upper respiratory tract infection with clinical severity, Kenya. J Med Virol. 2013;85:924–932. doi: 10.1002/jmv.23455. [DOI] [PubMed] [Google Scholar]
- Han MS, Byun JH, Cho Y, Rim JH. RT-PCR for SARS-CoV-2: quantitative versus qualitative. Lancet Infect Dis. 2020 doi: 10.1016/s1473-3099(20)30424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TC, Mühlemann B, Veith T, Biele G, Zuchowski M, Hoffmann J, et al. An analysis of SARS-CoV-2 viral load by patient age. medRxiv. 2020 doi: 10.1101/2020.06.08.20125484:2020.06.08.20125484. [DOI] [Google Scholar]
- La Scola B, Le Bideau M, Andreani J, Hoang VT, Grimaldier C, Colson P, et al. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39:1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magleby R, Westblade LF, Trzebucki A, Simon MS, Rajan M, Park J, et al. Impact of SARS-CoV-2 viral load on risk of intubation and mortality among hospitalized patients with coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom EM, Potter RF, Wallace MA, Mitchell KF, Yarbrough ML, Burnham CA, et al. Comparison of extraction methods and thermocyclers for SARS-CoV-2 molecular detection using clinical specimens. J Clin Microbiol. 2020 doi: 10.1128/jcm.01622-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioda K, Barclay L, Becker-Dreps S, Bucardo-Rivera F, Cooper PJ, Payne DC, et al. Can use of viral load improve norovirus clinical diagnosis and disease attribution? Open Forum Infect Dis. 2017;4:ofx131. doi: 10.1093/ofid/ofx131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tom MR, Mina MJ. To interpret the SARS-CoV-2 test, consider the cycle threshold value. Clin Infect Dis. 2020;71:2252–2254. doi: 10.1093/cid/ciaa619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao AT, Tong YX, Zhang S. Profile of RT-PCR for SARS-CoV-2: a preliminary study from 56 COVID-19 patients. Clin Infect Dis. 2020;71:2249–2251. doi: 10.1093/cid/ciaa460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]