GDC-0077 was discovered to more potently inhibit PI3K signaling as well as reduce cell viability through p110α degradation, making it more effective at prolonging PI3K pathway suppression than other PI3K inhibitors.
Abstract
PIK3CA is one of the most frequently mutated oncogenes; the p110a protein it encodes plays a central role in tumor cell proliferation. Small-molecule inhibitors targeting the PI3K p110a catalytic subunit have entered clinical trials, with early-phase GDC-0077 studies showing antitumor activity and a manageable safety profile in patients with PIK3CA-mutant breast cancer. However, preclinical studies have shown that PI3K pathway inhibition releases negative feedback and activates receptor tyrosine kinase signaling, reengaging the pathway and attenuating drug activity. Here we discover that GDC-0077 and taselisib more potently inhibit mutant PI3K pathway signaling and cell viability through unique HER2-dependent mutant p110a degradation. Both are more effective than other PI3K inhibitors at maintaining prolonged pathway suppression. This study establishes a new strategy for identifying inhibitors that specifically target mutant tumors by selective degradation of the mutant oncoprotein and provide a strong rationale for pursuing PI3Kα degraders in patients with HER2-positive breast cancer.
Significance:
The PI3K inhibitors GDC-0077 and taselisib have a unique mechanism of action; both inhibitors lead to degradation of mutant p110a protein. The inhibitors that have the ability to trigger specific degradation of mutant p110a without significant change in wild-type p110a protein may result in improved therapeutic index in PIK3CA-mutant tumors.
See related commentary by Vanhaesebroeck et al., p. 20.
This article is highlighted in the In This Issue feature, p. 1
INTRODUCTION
Oncogenic mutations in the PIK3CA gene increase lipid kinase activity and transform cells (1–3). The α isoform of PI3K is a dimer composed of the p110a catalytic subunit and a p85 regulatory subunit that functions to stabilize p110a and reduce kinase activity (4). The binding of a phosphorylated receptor tyrosine kinase (RTK) activates p110a through the release of a subset of inhibitory contacts with p85 (5). Common hotspot mutations in PIK3CA helical (E542K, E545K) and kinase (H1047R) domains function by perturbing local interfaces between p85 and p110a (6, 7) and increasing dynamic events required for catalysis on membranes (8). Several inhibitors of PI3K have entered clinical trials, yet in patients with PIK3CA-mutant tumors, the efficacy has been modest, in part due to a limited therapeutic index (9–12). Hence, we reasoned that it might be possible to improve the therapeutic index by identifying compounds with increased specificity for mutant p110a.
RESULTS
PI3K Inhibitor Potency in PIK3CA-Mutant Cells
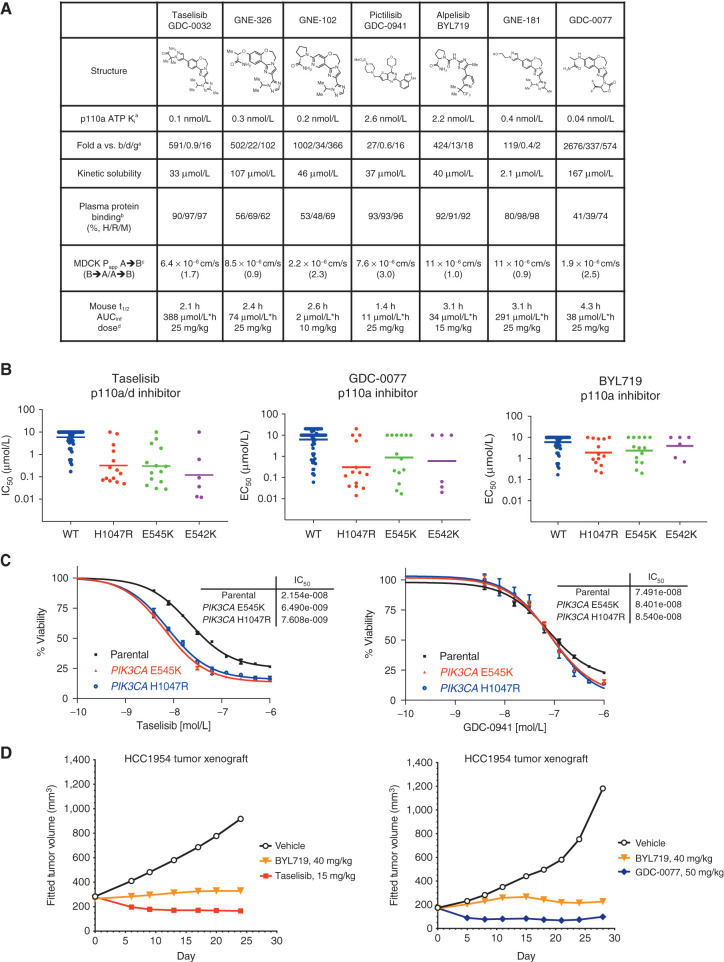
A selection of PI3K inhibitors was profiled for biochemical activity and pharmacokinetic properties, including inhibitors across several chemical classes (Fig. 1A). All PI3K inhibitors have varying isoform selectivity but showed no significant differences between inhibitor Ki for p110a wild-type (WT) and p110a-mutant enzyme complexed with either regulatory subunit p85a or p85b (Supplementary Fig. S1A). Taselisib, GDC-0077, pictilisib, and alpelisib showed no cytotoxic effects in primary human hepatocytes when tested up to 100 μmol/L. Therefore, it was surprising to discover that taselisib and GDC-0077 showed increased mutant potency in cell viability assays in a cancer cell line panel compared with other PI3K inhibitors (including the α isoform–selective inhibitor, BYL719; Fig. 1B). Furthermore, taselisib (and later, GDC-0077) was a stronger inducer of cell antiproliferation compared with other compounds, specifically in PIK3CA-mutant cancer cell lines (Supplementary Fig. S1B), suggesting that taselisib is more potent in PIK3CA-mutant cells compared with other PI3K inhibitors. We compared PI3K inhibitor potencies in parental isogenic SW48 colon cancer cells bearing WT PIK3CA and matched isogenic lines expressing H1047R or E545K hotspot mutants knocked into one allele of the PIK3CA locus. Taselisib potency [half maximal effective concentration (EC50)] increased 3-fold in PIK3CA-mutant cells versus parental WT SW48 cells, whereas GDC-0941 (13) displayed comparable EC50 in mutant and WT cells (Fig. 1C). To assess whether this potency shift was correlated with the taselisib chemical scaffold, structurally related analogues with increased α-isoform specificity, GNE-102 and GNE-326 (14), and a PI3Kα inhibitor from an unrelated chemical class (BYL719; ref. 15) were assessed and found to not have a potency differential in mutant versus WT isogenic cells (Supplementary Fig. S1C). We also confirmed that inhibition of multiple PI3K isoforms did not play a role in this increased potency. Taselisib binds equipotently to p110a and p110d isoforms but is selective against p110b and p110g isoforms (Fig. 1A). However, combination of a p110a inhibitor (GNE-102) with a p110d inhibitor (idelalisib; ref. 16) did not affect cell potency, nor did the combination of taselisib with a p110b inhibitor (TGX-221; ref. 17; Supplementary Fig. S1D and S1E). Neither cell permeability differences nor intracellular accumulation of inhibitors could explain the increased potency of taselisib in mutant cells (Supplementary Fig. S1F).
Figure 1.
GDC-0032 and GDC-0077 have increased potency in PIK3CA-mutant cancer cells. A, Chemical structures and physicochemical properties of PI3K inhibitors. aInhibition of ATP hydrolysis by PI3K isoforms in a biochemical assay, with ADP production measured by ADP-Glo. bPlasma protein binding determined by equilibrium dialysis. cPermeability measured using Madin-Darby canine kidney (MDCK) epithelial cells; A, apical; B, basolateral. B→A/A→B used to estimate efflux potential. dMouse oral administration dose as methylcellulose Tween (MCT) suspension. B, Cell viability IC50 values determined by quantifying ATP from all tumor lines at 5 days posttreatment. C, PI3K inhibitor potency in SW48 isogenic PIK3CA-mutant and PIK3CA-WT parental cells in a 4-day viability assay. Error bars are SD of quadruplicates. D,In vivo efficacy of taselisib, GDC-0077, and BYL719 in HCC1954 PIK3CA H1047R breast cancer xenograft model.
In Vivo Efficacy of GDC-0077
We next asked whether this greater potency and enhanced cell antiproliferation manifested in vivo in HCC1954-mutant PI3K tumor xenografts. Indeed, we observed greater efficacy for taselisib and GDC-0077 compared with a MTD of BYL719 (Fig. 1D; Supplementary Fig. S1G). In addition, GDC-0077 treatment at the MTD in vivo resulted in tumor regressions in multiple PIK3CA-mutant xenograft and patient-derived xenograft models (HCC1954, KPL-4, and HCI-003 PDX; Supplementary Fig. S1H).
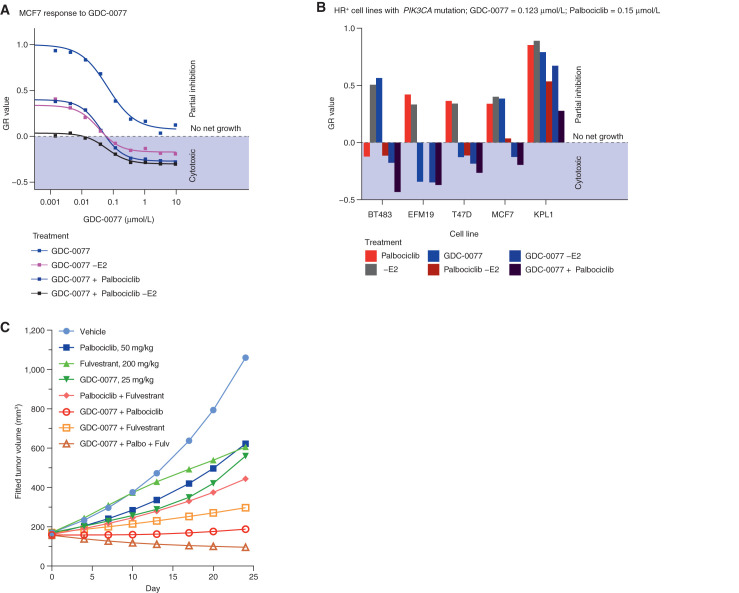
Given the potency of GDC-0077 and the significant improvement in PI3Kα isoform selectivity over both taselisib and BYL719 (Fig. 1A and B), we next evaluated the efficacy of GDC-0077 and the suitability for combination with standard of care in hormone receptor (HR)–positive/HER2-negative breast cancers. These included aromatase inhibitors and, more recently, CDK4/6 inhibitors such as palbociclib. We therefore assessed the effect of combining GDC-0077 with these drugs to evaluate efficacy and safety. First, we measured in vitro growth of five PI3KCA-mutant HR-positive lines across different concentrations of GDC-0077 with or without E2 [to mimic aromatase inhibitors (AI)] and with or without 0.15 μmol/L palbociclib. To assess growth, endpoint cell population, as measured by CyQuant assay, was normalized to the cell population at the time of treatment using the growth rate inhibition (GR) method (18). A GR value of 1 meant no inhibition, 0 meant no net growth, and negative values represented cell loss. Response in PIK3CAE545K-mutant MCF7 cells showed that addition of GDC-0077 induced a strong cytotoxic response in all combination treatments, as reflected by negative GR values for concentrations of 0.12 μmol/L and above (ref. 18; Fig. 2A). When assessing the efficacy of the combination treatments across all five cell lines, we found that the GR values measured in the condition with palbociclib and without E2 (equivalent to AI) decreased by a median of 0.25 when adding 0.123 μmol/L GDC-0077, showing a broad increase in efficacy by combining GDC-0077 with standard-of-care treatments (Fig. 2B; Supplementary Fig. S2A).
Figure 2.
Activity of PI3Kα inhibitors in combination with palbociclib in HR-positive breast cancer cells. A, Dose–response curve of MCF-7 cells treated with GDC-0077 either alone (blue), without E2 to mimic aromatase inhibitor (dark blue), with palbociclib at 0.15 μmol/L (purple), or with palbociclib at 0.15 μmol/L (purple) and without E2 (dark purple). Negative values indicate a cytotoxic response. The y-axis shows normalized growth inhibition (GR value). B, Growth response of five HR-positive breast cancer cells with PIK3CA mutations to treatment with palbociclib at 0.15 μmol/L (red), E2 withdrawal (gray), GDC-0077 at 0.123 μmol/L (blue), palbociclib at 0.15 μmol/L and E2 withdrawal (dark red), or the triple combination (dark purple). The y-axis shows normalized growth inhibition (GR value). C, Efficacy of palbociclib, fulvestrant, and GDC-0077 as single agent or in combination in MCF-7 xenografts. Each group contains 12 animals at the beginning of the experiment.
We then sought to validate this increased efficacy in vivo. Palbociclib (50 mg/kg) combined with fulvestrant (200 mg/kg) only conferred tumor growth inhibition (TGI) of 71%, whereas addition of GDC-0077 (25 mg/kg) further reduced tumor burden (mean TGI of 106%, n = 12, Fig. 2C; Supplementary Fig. S2B), consistent with the response observed in vitro (Fig. 2A). Weight loss was modest at 8.1% in the triple-combination cohort (Supplementary Fig. S2C), suggesting tolerability of adding GDC-0077 to a regimen with endocrine therapy plus CDK4/6 inhibitors. Similar efficacy results were obtained with taselisib combined with palbociclib in WHIM20 (Supplementary Fig. S2D), although weight loss was higher (Supplementary Fig. S2D) with taselisib as expected. Also, taselisib could only be combined with palbociclib and not the triple combination. Taken together, these data suggested the possibility of combining GDC-0077 with both palbociclib and fulvestrant for superior efficacy in PIK3CA-mutant, HER2-negative tumors and providing a therapeutic window not achievable with earlier PI3K inhibitors.
Taselisib and GDC-0077 Induce Mutant p110a Degradation
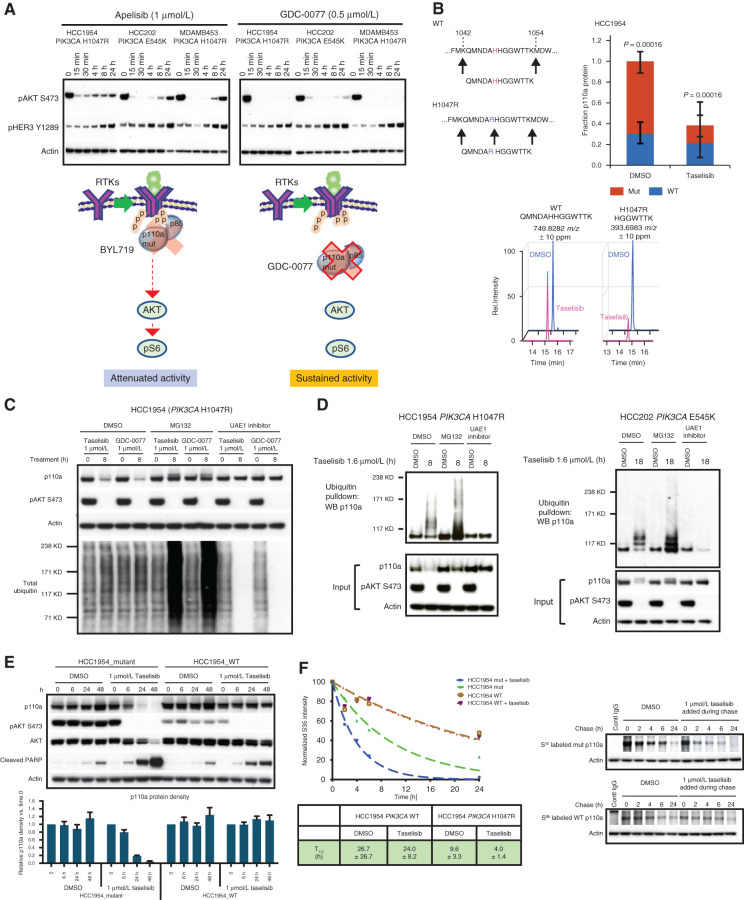
To determine the mechanistic basis for this efficacy differential in PIK3CA-mutant tumors, we compared the effects downstream (pAKT) and upstream (pHER3) of PI3K signaling for GDC-0077 versus BYL719 over a time course. In both cases, we observed robust acute inhibition of pAKT treatment; however, GDC-0077 demonstrated sustained inhibition of pAKT over the 24-hour treatment time despite inducing release of negative feedback, as measured by upregulation of pHER3 (as is also observed for BYL719; Fig. 3A).
Figure 3.
Taselisib depletes mutant p110a protein through ubiquitin and proteasome mechanism in a dose- and time-dependent manner. A, Western blots of the inhibitor response in PI3K signaling (pHER3 and pAKT) in PIK3CA-mutant (HCC1954, HCC202, and MDA-MB-453) cell lines treated with 1 μmol/L BYL719 or 0.5 μmol/L GDC-0077 for indicated time points. B, Mass spectrometry of HCC1954 cells treated for 24 hours with 500 nmol/L taselisib. A neotryptic peptide generated from PIK3CA H1047R was used to assess mutant protein levels compared with WT protein in the same lysate. C, Rescue of taselisib- or GDC-0077–mediated p110a degradation in HCC1954 cells by either a proteasome inhibitor (MG132) or a ubiquitin activating enzyme E1 (UAE1) inhibitor. D, HCC1954 PIK3CA H1047R cells were treated 8 hours or HCC202 PIK3CA E545K cells were treated for 18 hours with either DMSO or 1 μmol/L taselisib ± MG132 or ± UAE1 inhibitor. Protein lysates were run on Western blot and probed with antibodies to p110a, pAKT, and β-actin, or ubiquitinated proteins were pulled down with TUBE1 reagent and then blotted with anti-p110a antibody. E, HCC1954 cells engineered to be isogenic for PIK3CA-mutant or PIK3CA-WT were treated with 1 μmol/L taselisib for up to 48 hours followed by Western blotting. Experimental replicates n = 3 were used to quantify p110a. F, Pulse-chase of isogenic cell lines, HCC1954_mutant and HCC1954_WT. PI3K inhibitor taselisib at 1 μmol/L was added during the chase. Pulldown with p110a antibody was followed by autoradiography and data fit to a single exponential decay function.
We reasoned that one possible mechanism of enabling sustained inhibition of pAKT despite elevated pHER3 levels would be via drug-induced sequestration of PI3K away from the plasma membrane. To investigate this hypothesis, levels of p110a protein in subcellular fractions were evaluated by Western blot at various time points after taselisib treatment. Unexpectedly, we discovered time-dependent p110a protein depletion from the PIK3CA-mutant cells following taselisib treatment, regardless of the subcellular fraction evaluated (Supplementary Fig. S3A). BYL719 did not significantly affect p110a protein levels. Comparing whole-cell lysates from PIK3CA-mutant and PIK3CA-WT breast cancer cells by Western blot, taselisib treatment for 8 hours caused the dose-dependent depletion of p110a protein specifically in PIK3CAH1047R-mutant HCC1954 cells. No significant change was observed for p110a in PIK3CA-WT HDQ-P1 cells (Supplementary Fig. S3B). Treatment with BYL719 did not affect p110a levels in any cell lines tested. Transcription of PIK3CA alleles was not diminished following taselisib treatment (Supplementary Fig. S3C), implicating posttranscriptional regulation of mutant p110a protein.
We investigated p110a protein depletion in p110a-mutant xenograft breast cancer models and found that membrane-associated p110a was depleted 4 hours after a single oral dose of 15 mg/kg taselisib in individual tumors from a p110a-mutant HCC1954 xenograft model and that p110a depletion was not observed with 40 mg/kg BYL719 (Supplementary Fig. S3D). In the same xenograft model, a single oral dose of 50 mg/kg GDC-0077 depleted p110a protein expression for up to 8 hours, further confirming a similar mechanism of action for both taselisib and GDC-0077. In phase Ia clinical trials, taselisib and GDC-0077 had antitumor activity in PIK3CA-mutant tumors as assessed by response rates (19). The free drug exposures achieved in the clinic were evaluated in tissue culture experiments with PIK3CA-mutant breast cancer lines. Unlike pictilisib (GDC-0941), both taselisib (GDC-0032) and inavolisib (GDC-0077) treatment for 24 hours resulted in p110a degradation at clinically relevant concentrations (Supplementary Fig. S4A). Despite an understanding of their binding interactions with the p110a protein, it is still not clear why some inhibitors (taselisib, GDC-0077) result in p110a degradation, whereas others (BYL719, pictilisib) do not.
To generate direct evidence that taselisib or GDC-0077 treatment was preferentially depleting mutant p110a protein within a mixed WT and mutant allelic population, we took advantage of a neotryptic peptide encoded by the H1047R mutation to assess p110a protein levels by mass spectrometry. HCC1954 parental cells expressing both mutant and WT p110a were treated with taselisib or GDC-0077 for 24 hours, revealing loss of the mutant-specific peptide only in the taselisib- or GDC-0077–treated sample, P = 0.00016 (Fig. 3B; Supplementary Fig. S4B). There was no significant change in WT p110a, whereas the total p110a pool decreased commensurate with the initial abundance of the mutant protein (Supplementary Fig. S4B and S4C). Similarly, depletion of the E545K-mutant protein was observed in HCC202 heterozygous cells, as shown by assaying a second mutant-specific neotryptic peptide at this locus (P = 0.032; Supplementary Fig. S4D).
To further explore the molecular mechanisms underlying inhibitor-induced mutant p110a depletion, we examined whether reduction of p110a could be attributed to degradation. A proteasome inhibitor, MG132, and a ubiquitin-activating enzyme E1 inhibitor both rescued the p110a degradation induced by taselisib and GDC-0077 (Fig. 3C), whereas lysosomotropic agents failed to do so (Supplementary Fig. S4E). We next employed ubiquitin pulldown assays to confirm that mutant p110a is inducibly ubiquitinated on taselisib treatment and that this signal further accumulated when cells were cotreated with MG132 to prevent proteasomal degradation of ubiquitinated p110a (Fig. 3D). Accordingly, no ubiquitinated p110a was detected in cells treated with UAE1 inhibitor (Fig. 3D). Importantly, taselisib also induced ubiquitination and degradation of E545K p110a in the HCC202 cell model, with comparable rescue by MG132 and UAE1 inhibition (Fig. 3D).
Visualization of p110a depletion on Western blot was more easily discerned in HCC1954 cells, which have an increased copy number of the mutant p110a allele. Quantitative RT-PCR analysis confirmed higher expression of the mutant allele (Supplementary Fig. S4F). To generate a clean system to better compare the differential effect of inhibitors on WT and mutant p110a, we used CRISPR/Cas9 to generate isogenic HCC1954 cell lines bearing either the H1047 WT allele or the mutant H1047R allele, named HCC1954_mutant and HCC1954_WT (Supplementary Fig. S4F). Taselisib treatment resulted in ubiquitination and depletion of p110a protein only in the HCC1954_mutant cells and not in the matched HCC1954_WT cells (Fig. 3E; Supplementary Fig. S4G).
Because the basal level of ubiquitination was significantly higher for membrane-bound mutant p110a compared with WT p110a (Supplementary Fig. S4H), we hypothesized that the mutant p110a protein may be inherently less stable than the WT protein, as previously noted for mutant EGFR (20). Consistent with prior literature (4), pulse-chase experiments in HCC1954_WT isogenic cells indicated a protein half-life for WT p110a of approximately 26.7 hours. In contrast, the H1047R-mutant protein half-life was approximately 9.6 hours in the basal state and was further shortened to approximately 4 hours upon treatment with taselisib (Fig. 3F). Together, these data demonstrate that mutant p110a oncoprotein is inherently less stable and more vulnerable to inhibitor-mediated degradation in a ubiquitin- and proteasome-dependent manner.
p85b Potentiates p110a Degradation by Recruiting p110a to the Membrane
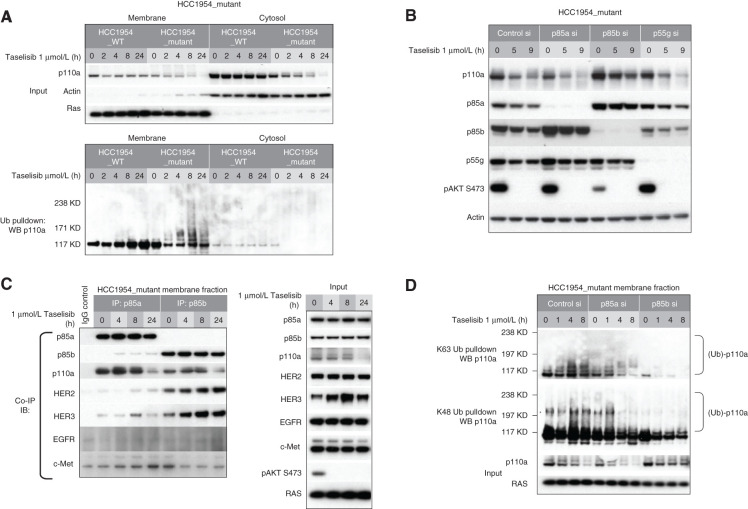
It has previously been shown that p110a/p85 dimers are activated by growth factor–stimulated RTK signaling (21). The mechanism of this activation involves release of inhibitory contacts between p110 and p85 once bound to phosphotyrosine residues of an RTK (5, 22). We next reasoned that the ability of our small-molecule degraders to accelerate the turnover of activated mutant protein might be through appropriation of the p110a activation process. In support of this hypothesis, cell fractionation studies showed that taselisib-inducible ubiquitination of p110a occurred preferentially in the membrane fraction (Fig. 4A).
Figure 4.
Taselisib-mediated degradation of mutant p110a occurs preferentially at the plasma membrane. A, Subcellular fractionation of isogenic HCC1954_mutant and HCC1954_WT cells. Pulldown of ubiquitinated protein was followed by Western blotting with anti-p110a. B, HCC1954_mutant cells were transfected with p85a, p85b, or p55g isoform-specific siRNA followed by 1 μmol/L taselisib treatment. C, HCC1954_mutant cell line was treated with 1 μmol/L taselisib for up to 24 hours. Cell lysates were precipitated with p85a or p85b antibody, followed by immunoblot with antibodies indicated to the left. IB, immunoblotting; IP, immunoprecipitation. D, HCC1954_mutant cells were transfected with p85a or p85b siRNA followed by taselisib treatment. Cells were harvested at various time points and fractionated. K63- or K48-linked ubiquitin conjugated protein was pulled down from membrane fraction and analyzed by immunoblotting with p110a antibody.
One major effector that recruits p110a to RTK is the p85 PI3K regulatory subunit. There are three p85 isoforms in class 1A PI3K: p85a, p85b, and p85g. We first confirmed comparable expression levels of all three isoforms in HCC1954 cells (Supplementary Fig. S5A). By coimmunoprecipitation, we also demonstrated that all three isoforms interact with mutant p110a (Supplementary Fig. S5A and S5C). We next evaluated the effect of p85 isoforms on inhibitor-induced p110a degradation. Knockdown (KD) of both p85a and p55g showed similar levels of taselisib-induced p110a degradation compared with control siRNA–treated cells. In contrast, we observed that p85b KD rescued taselisib-mediated p110a degradation (Fig. 4B). We further observed that p85b KD resulted in inhibition of pathway signaling, shown by reduced pAKT levels. These data suggest that p85 isoforms do not have redundant roles and that the p85b but not the p85a or p85g isoform is involved in mutant p110a degradation. Reduced pAKT levels with p85b KD is most likely a result of reduced p110a membrane localization.
This observation also suggested that p85 isoforms have differential affinity for RTK interaction. To address this question, we next performed a series of coimmunoprecipitation experiments. p85a or p85b was immunoprecipitated from the membrane fraction and immunoblotted with several antibodies, including four receptors that are highly activated in HCC1954 cells (Supplementary Fig. S5B). Both p85a and p85b interacted with p110a, consistent with previous results. We also observed a stronger association between p85b and HER2 and HER3 than observed for p85a or p85g protein (Fig. 4C; Supplementary Fig. S5C). We could not detect an interaction between p85 isoforms with EGFR or c-MET under these conditions.
To further confirm this observation, we next tested whether inhibition of HER2 phosphorylation blocked p85b binding to HER2 and rescue p110a degradation. Cells were treated with taselisib or lapatinib alone or in combination for various time points. These cell lysates were next immunoprecipitated using p85a or p85b antibody and immunoblotted with HER2 and HER3 antibodies. In taselisib-treated cells, a strong interaction between p85b and HER2/3 was observed. Treatment with lapatinib blocked this interaction, confirming that activated HER2 contributes to p85b membrane binding (Supplementary Fig. S5D). These results further demonstrated that p85b plays an important role in recruiting p110a to the membrane and that this is likely through binding to activated HER2 and HER3.
To further confirm the role of p85b in p110a degradation, we next tested the effects of p85 KD on K63 and K48 polyubiquitin chain formation on p110a. Upon knockdown of p85a or p85b and following inhibitor treatment, K63 or K48 ubiquitin–conjugated proteins were immunoprecipitated using linkage-specific antibodies and immunoblotted with p110a. In both control- and p85a-depleted cells, taselisib treatment induced both K63- and K48-linked ubiquitination on p110a. However, upon p85b depletion, ubiquitination of p110a was no longer detected (Fig. 4D). These data further confirm that the p85b regulatory subunit plays an important role in taselisib-induced mutant p110a degradation by recruiting p110a to the membrane where ubiquitination occurs.
Taselisib- and GDC-0077–Induced Mutant p110a Degradation Is Dependent on RTK Activity
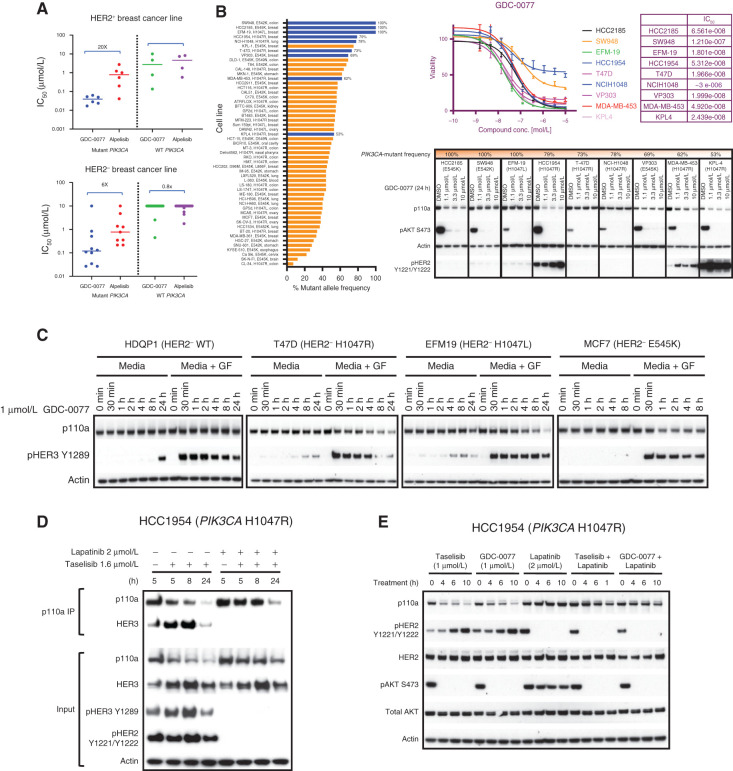
To further understand whether inhibitor-induced mutant p110a degradation is dependent on its recruitment to activated RTK, we next investigated the efficacy of the two clinically relevant molecules, GDC-0077 and BYL719 (alpelisib). Unexpectedly, in PIK3CA-mutant cells, we observed a significant difference in the sensitivity of HER2 amplified (∼20-fold difference between the mean IC50 values) versus HER2-negative cell lines (∼6-fold difference between two inhibitors) to GDC-0077 versus BYL719. The inhibitors were not differentiated in WT cell lines regardless of HER2 status (Fig. 5A). These data imply that RTK activity may define the sensitivity of a cell line to inducible degradation of mutant p110a. To confirm this finding, a panel of more than 50 cell lines harboring PIK3CA hotspot mutations was analyzed (Fig. 5B). Most of these were heterozygous, carrying both WT and mutant PIK3CA alleles at differing frequencies. For easier visualization of mutant p110a depletion, we analyzed the cell lines with higher copy numbers of mutant alleles that also represented each hotspot mutation across various tumors (Fig. 5B, colored in blue). In a cell proliferation assay, these cell lines have varying GDC-0077 sensitivity. All of the selected representative cell lines responded to GDC-0077, as measured by inhibition of pAKT. Not all cell lines showed visible p110a degradation; those that showed mutant p110a degradation were HCC2185, HCC1954, MDA-MB-453, and KPL-4 (Fig. 5B). It was particularly striking that all HER2-amplified cell lines showed p110a degradation. In contrast, the HER2-negative cell lines were resistant to degradation except HCC2185. To further understand this observation, we tested whether activating RTK by addition of growth factors would induce p110a degradation. A subset of HER2-negative cell lines, each harboring WT or one of three p110a hotspot mutations, was treated with GDC-0077 alone or in combination with growth factors. All responded to growth factors, as shown by induction of HER3 phosphorylation. Consistent with our data, p110a levels did not decrease in HER2-negative cell lines treated with GDC-0077 alone. However, the combination of growth factors and GDC-0077 induced p110a degradation in all three p110a-mutant lines. WT p110a protein expression was not affected by addition of growth factors (Fig. 5C). Immunoprecipitation and pulldown of p110a revealed that p110a degradation was markedly delayed when RTK interaction with the p110a/p85 complex was disrupted, with limited inducible degradation observed at the 8-hour time point (Fig. 5D, top). Conversely, inhibition of RTK phosphorylation by lapatinib partially rescued taselisib-mediated p110a degradation in HCC1954 cells (Fig. 5D, bottom). Similar to taselisib, GDC-0077–induced mutant p110a degradation was rescued by lapatinib treatment (Fig. 5E). In support of these results, we observed that in multiple patient-derived xenograft (PDX) models, high basal pHER2 and pHER3 expression level correlated with better taselisib-mediated p110a degradation, with weaker degradation observed in tumors with low levels of pRTK (Supplementary Fig. S6A). In addition, within this group of PDX models, the degree of degradation appeared to correlate with basal pRTK expression. Taken together, our data support a model where small molecule–induced p110a-mutant degradation depends on RTK activity in PIK3CA-mutant cancers.
Figure 5.
Taselisib- and GDC-0077–induced mutant p110a degradation is dependent on RTK activity. A, Cell viability IC50 values determined by quantifying ATP from breast tumor lines: HER2-positive PIK3CA-mutant (n = 6), HER2-positive PIK3CA-WT (n = 4), HER2-negative PIK3CA-mutant (n = 10), and HER2-negative PIK3CA-WT (n = 20) at 5 days posttreatment. B, Bar plot showing PIK3CA-mutant frequency among tumor lines harboring PIK3CA hotspot mutations. ATP-based cell viability assay in selected cell line (HCC2185, SW948, EFM-19, HCC1954, T-47D, NCIH1048, VP303, MDA-MB-453 and KPL-4). Western blot of the p110a protein levels and pAKT signaling in same cell lines treated with GDC-0077 for indicated concentrations for 24 hours. C, HER2-negative PIK3CA-WT or PIK3CA-mutant cells cultured in standard media with 10% FBS and treated with GDC-0077 alone or with addition of growth factors (EGF and neuregulin). D, HCC1954 PIK3CA H1047R cells treated with taselisib alone or combination of taselisib and lapatinib. Cell lysates were precipitated with p110a antibody, followed by Western blot with HER3 antibody. E, Cell lysates following treatment with taselisib or GDC-0077 alone or combination with lapatinib for indicated time points followed by Western blot analysis with indicated antibodies.
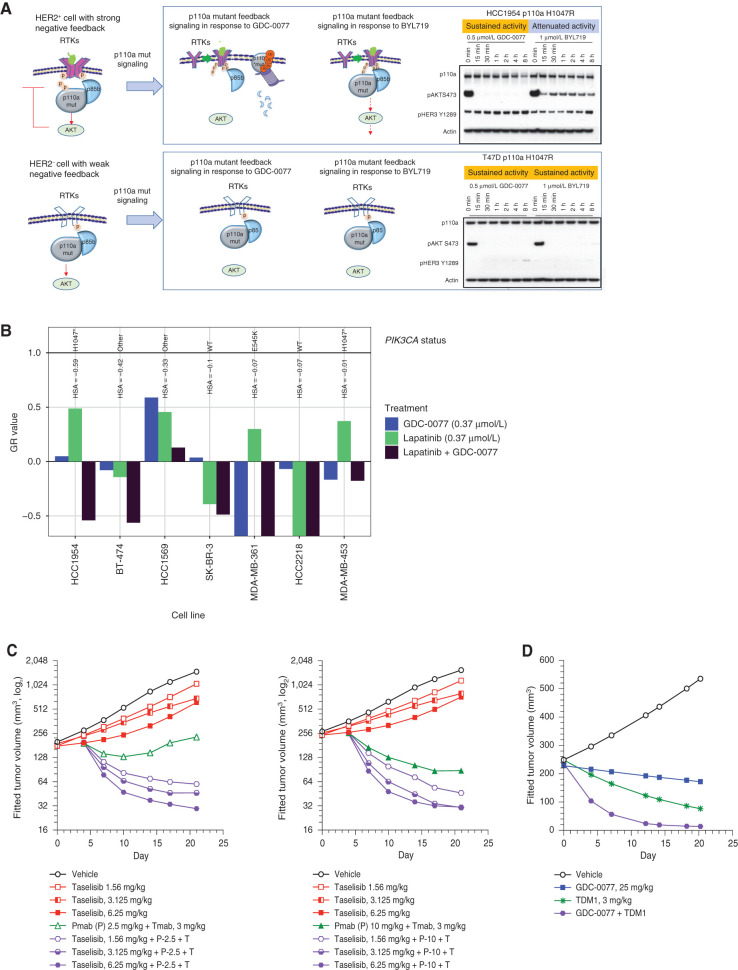
p110a Mutant–Degrading Inhibitors Provide More Sustained Benefit in HER2-Positive versus HER2-Negative p110a-Mutant Cancers
Given that RTK reactivation could potentially limit efficacy, we also aimed to understand activity in HER2-positive breast cancers. Degradation of mutant p110a blocked the feedback-induced pathway reactivation and resulted in enhanced potency of taselisib and GDC-0077 in cellular assays and an increase in antiproliferation in mutant cells. Our data thus far predict that drugs that induce degradation of mutant p110a may show more sustained benefit over nondegraders by preventing pathway reactivation. We speculated that the extent of negative feedback depends on the amount of RTK expression, implying that inhibitor-mediated feedback pathway reactivation would be weak in HER2-negative cells. Also, based on our findings, low RTK activity may result in inefficient mutant p110a degradation. Pathway reactivation was not observed in HER2-negative cells, in contrast to HER2-amplified cells, irrespective of the inhibitors used. This suggests that in HER2-negative mutant cells, the degrader mechanism of action may not provide additional benefit over drugs with a nondegrader mechanism (model in Fig. 6A). This was further confirmed in a panel of PIK3CA-mutant cell lines. In HER2-amplified cells, GDC-0077 resulted in sustained pathway inhibition, whereas BYL719 activity was attenuated, as evidenced by a rebound of pAKT levels (Supplementary Fig. S6B; 3A). In contrast, in a panel of HER2-negative lines, there was no difference in pathway inhibition by both inhibitors, GDC-0077 and BYL719.
Figure 6.
p110a-Mutant degrading inhibitors provide more benefit in HER2-positive versus HER2-negative p110a-mutant cancers. A, Mechanistic model of drug-induced p110a degradation in HER2-positive and HER2-negative PIK3CA-mutant cells. B, Growth response of seven HER2-amplified breast cancer cells with different PIK3CA status to treatment with GDC-0077 at 0.37 μmol/L (blue), lapatinib at 0.37 μmol/L (green), or the combination (dark purple). The y-axis shows normalized growth inhibition (GR value); HSA stands for excess over single agent. C, Tumor growth curve from KPL-4 (HER2+, PIK3CA H1047R) xenograft treated with vehicle, taselisib, trastuzumab plus pertuzumab, or the indicated drug combination. D, Tumor growth curve from KPL-4 (HER2+, PIK3CA H1047R) xenograft treated with vehicle, GDC-0077, TDM-1, or the indicated drug combination.
On the basis of these data, we posited that we could best leverage the degradation potential of GDC-0077 in HER2-amplified cancers. In HER2-driven breast cancers, HER2-targeted therapy is the standard of care. However, HER2-amplified tumors with p110a mutations are less responsive to HER2-targeted therapy. Therefore, combination treatment of the PI3K pathway inhibitors and HER2-targeted therapy should result in enhanced efficacy. Indeed, combination of the HER2 inhibitor lapatinib with GDC-0077 in vitro results in increased efficacy in six of seven HER2-amplified breast cancer cell lines (average excess over single agent of −0.22) with the strongest benefits observed in the p110a-mutant lines HCC1954, BT474, and HCC1569 (Fig. 6B). In vivo, the combination of HER2 inhibitors, trastuzumab and pertuzumab, with taselisib in a HER2-positive mutant p110a KPL-4 xenograft model showed better response compared with single drugs alone (Fig. 6C; Supplementary Fig. S6C). Furthermore, combining ado-trastuzumab emtansine (TDM-1) with GDC-0077 showed a synergistic effect in the KPL-4 xenograft model as well (Fig. 6D; Supplementary Fig. S6D).
Given that most p110a-mutant cells are heterozygous, selective mutant p110a degradation would imply a possible scenario where WT p110a protein is still present and can reactivate the same pathway and dampen inhibitor activity (Supplementary Fig. S6E). To test this possibility, we compared levels of feedback reactivation in an engineered homozygous versus heterozygous line. In a HCC1954 WT homozygous isogenic line, the pathway was reactivated by both GDC-0077 (degrader) and BYL719 (nondegrader), as expected (Supplementary Fig. S6F; Fig. 6A—panels from same study). In a HCC1954-mutant homozygous isogenic line, BYL719, but not GDC-0077 treatment, reactivated pathway signaling. Importantly, when compared, the phenotype of a heterozygous parental line behaved similar to that of a p110a-mutant homozygous line, suggesting that in heterozygous cells, mutant p110a functions as the main driver of cell signaling (Supplementary Fig. S6F). Furthermore, cell proliferation assays mirrored the inhibitor efficacy as well. GDC-0077 showed the same efficacy between homozygous and heterozygous mutant lines but had reduced efficacy in WT isogenic lines (Supplementary Fig. S6F), whereas no shift in IC50 was observed between all lines following BYL719 treatment. To further confirm these results, the same experiment was performed in HER2-negative lines. Consistent with our hypothesis for the role of HER2 in pathway reactivation, feedback pathway reactivation was not detected even in a homozygous WT line upon treatment with GDC-0077 or BYL719. In cell viability assays, neither inhibitor showed a shift in efficacy between isogenic lines (Supplementary Fig. S6G). Taken together, our data demonstrate that degradation potential of GDC-0077 would be most leveraged in HER2-positive breast cancers for more sustained pathway inhibition.
DISCUSSION
In summary, we have discovered that the mutant p110a, especially the H1047R mutation, has unique characteristics compared with WT p110a: a shorter half-life, ubiquitination in the membrane fraction, and proteasome-mediated turnover. Furthermore, the mutant oncoprotein is susceptible to increased proteasome-mediated degradation upon binding particular PI3K inhibitors such as taselisib and GDC-0077. Although we did not duplicate all experiments with GDC-0077, all the key mechanistic experiments suggested the same degradation mechanism of action between taselisib and GDC-0077. Therefore, based on all the data put together, we believe it is reasonable to assume the same results for both molecules. Our results suggest that RTK activity plays a key role in regulating p110a degradation by recruiting p110a to the membrane. The mutant oncoprotein may be particularly vulnerable to additional local conformational changes that affect membrane binding, and taselisib and GDC-0077 may be enhancing this effect to accelerate proteasome-mediated degradation. Although the underlying structural basis is still not clear, this discovery reveals a new mechanism of action to exploit in PIK3CA-mutant tumors, opening an exciting path to increasing drug efficacy for the predominant oncogene in cancer. With a combined mutant p110a degrader mechanism and exquisite p110a isoform selectivity, the PI3Kα-selective inhibitor and mutant PI3Kα degrader GDC-0077 may provide opportunities for previously inaccessible combination therapy with greater clinical responses. Moreover, this suggests that it may be possible to exploit endogenous mechanisms and the intrinsic instability of mutant oncoproteins more broadly to develop tumor-selective therapeutic agents. On the basis of these mechanistic discoveries, it may be possible to create efficacious compounds that specifically deplete the mutant p110a protein without blocking WT PI3K signaling, similar to engineered mouse models of cancer in which removal of the mutant PIK3CA oncogene induces regressions in the presence of WT PI3K (23, 24). For more than 2 million patients with cancer diagnosed annually with PIK3CA-mutant tumors, this discovery opens the possibility of a future therapeutic agent that solely targets tumor cells bearing mutant p110a without the systemic adverse effects of inhibiting WT signaling.
With respect to the clinical relevance of our findings, a first-in-human, open-label, phase I/IB dose escalation study of oral daily GDC-0077 alone and in combination with endocrine and targeted therapies for PIK3CA-mutant solid tumors is ongoing. The single-agent portion of the study showed that GDC-0077 had a manageable safety profile with a MTD of 9 mg once daily (supported by a linear pharmacokinetic profile), with promising antitumor activity (25). When combined at this dose with letrozole with and without palbociclib (26), or when combined with fulvestrant, the safety profile was also manageable with promising antitumor activity (27). No drug–drug interactions were observed in the letrozole ± palbociclib portion of the study (26). Phase III trials are now ongoing to assess efficacy and safety in a randomized, controlled manner in locally advanced or metastatic HR-positive/HER2-negative breast cancer in combination with palbociclib and fulvestrant (NCT04191499). The mechanism of T-DM1 is both through internalization to release DM1 and through trastuzumab inhibition of HER2–HER3 signaling, as well as trastuzumab-mediated antibody-dependent cell-mediated cytotoxicity. We showed that T-DM1 is active in a HER2-amplified mutant PIK3CA xenograft and caused enhanced activity by combining with GDC-0077 perhaps by overcoming resistance (from PIK3CA mut) to the trastuzumab component of T-DM1. To further leverage the degradation potential of GDC-0077 in HER2-positive breast cancers, this work has also provided the rationale for a first-in-human, open-label, phase I/IB dose escalation study of oral daily GDC-0077 in combination with standards of care (trastuzumab and pertuzumab) that is also being enabled (NCT03006172).
METHODS
Chemical Reagents
The proteasome inhibitor MG132 (474790) was obtained from Calbiochem EMD Millipore. Chloroquine (14774) was obtained from Cell Signaling Technology. Ammonium chloride (NH4Cl; 254134) was obtained from Sigma-Aldrich. PI3K inhibitors and ubiquitin-activating E1 inhibitor (28) were provided by the Chemistry Department at Genentech, Inc.
Antibody Reagents
Antibodies to p110a (4249), cleaved PARP (9541), pAKT Ser473 (4060), pS6 S235/236 (2111), antiubiquitin (3936), p110d (34050), HER3 (12708), HER2 (2242), pHER2 Y1221/Y1222 (2243), and pHER3 Y128 (4791) were obtained from Cell Signaling Technology. The antibody to β-actin (A5441) was from Sigma-Aldrich. Antibodies to RAS (ab52939), p85a (ab133595), and p85b (ab28356) were obtained from Abcam. Ubiquitin reagent TUBE1 (UM101) was obtained from LifeSensors, Inc. The p85g antibody (MAB6638) was obtained from R&D Systems. Antibodies to GAPDH (MAB374) and p110b (04-400) were obtained from EMD Millipore.
pPRAS40 ELISA
SW48 isogenic cells were plated in 384-well tissue culture–treated assay plates (catalog no. 781091; Greiner Bio-One) and incubated overnight at 37°C and 5% CO2. The three isogenic SW48 lines (WT, E545K, and H1047R) were plated and assayed in parallel. The next day, test compounds were serially diluted in DMSO and added to cells (final DMSO concentration of 0.5%). Cells were then incubated with drugs for 24 hours at 37°C and 5% CO2. After 24 hours, cells were lysed, and phosphorylated proline-rich AKT substrate of 40 kDa (pPRAS40) levels were measured using the Meso Scale Discovery (MSD) custom pPRAS40 384w Assay Kit (catalog no. L21CA-1). Cell lysates were added to assay plates precoated with antibodies against pPRAS40 and allowed to bind to the captured antibodies overnight at 4°C. The detection antibody (anti–total pPRAS40, labeled with an electrochemiluminescent SULFO-TAG) was added to the bound lysate and incubated for 1 hour at room temperature. The MSD Read Buffer was added such that when voltage was applied to the plate electrodes, the labels bound to the electrode surface emitted light. The MSD Sector Instrument measured the intensity of the light and quantitatively measured the amount of pPRAS40 in the sample. Percent inhibition of pPRAS40 per concentration of compounds was calculated relative to untreated controls. The EC50 values were calculated using the four-parameter logistic nonlinear regression dose–response model. Reported EC50 values indicate an average value from three independent experiments. SDs are reported as ± the reported EC50 values for each cell line. The EC50 in WT cells was divided by EC50 in mutants to derive the fold increase in potency in mutant cells.
Viability Assay CellTiter-Glo
Cells were seeded (1,000–2,000 cells/well) in 384-well plates for 16 hours. On day 2, nine serial 1:3 compound dilutions were made in DMSO in a 96-well plate. The compounds were then further diluted into growth media using a Rapidplate robot (Zymark Corp.). The diluted compounds were then added to quadruplicate wells in the 384-well cell plate and incubated at 37°C and 5% CO2. After 4 days, relative numbers of viable cells were measured by luminescence using CellTiter-Glo (Promega) according to the manufacturer's instructions and read on a Wallac Multilabel Reader (PerkinElmer). The EC50 calculations were carried out using Prism 6.0 software (GraphPad Software). The GR calculations and figures were performed using R scripts based on Hafner and colleagues (18).
Nucleosome ELISA
MDA-MB-453, HCC202, and Cal85-1 cells were plated in 96-well tissue culture–treated assay plates (catalog no. 3904; Corning) and incubated overnight at 37°C and 5% CO2. The following day, PI3K inhibitors were serially diluted in DMSO and added to cells (final DMSO concentration of 0.5%). Cells were then incubated with drugs for 72 hours at 37°C and 5% CO2. After 72 hours, cells were lysed and centrifuged at 200 × g for 10 minutes. Histone-associated DNA fragment levels were analyzed using the Cell Death Detection ELISAPLUS (catalog no. 11920685001; Roche). Then, 20 μL supernatant was added to each well of the streptavidin capture plate followed by 80 μL anti–histone-biotin and DNA-peroxidase immunoreagent. Plates were incubated at room temperature for 2 hours shaking (300 rpm). Contents of the plate were removed followed by three washes with incubation buffer. Then, 100 μL ABTS solution was added to each well, and plates were incubated for 10 to 20 minutes, after which 100 μL ABTS Stop Solution was added to each well. Absorbance was measured at 405 nm and 490 nm of each plate. Fold increases were generated by assessing the increased absorbance (A405–A490 nm) of wells from cells treated with compounds, normalized to those treated with DMSO alone.
Cell Lines and Cell Culture
Cell lines were obtained from the ATCC. All cell lines underwent authentication by short tandem repeat profiling, SNP fingerprinting, and Mycoplasma testing (29). The isogenic colon cancer cell lines SW48 human PIK3CA (H1047R/+; HD103–005) and SW48 human PIK3CA (E545K/+; HD103–001) and SW48 parental line were obtained from Horizon Discovery Ltd. Cell lines were grown under standard tissue culture conditions in RPMI media with 10% fetal bovine serum (Gibco, 10082–147), 100 U/mL penicillin-streptomycin (Gibco, 15140–122), and 2 mmol/L L-glutamine (Gibco, 15030–081). PIK3CA mutation status and frequencies of all cell lines are summarized in Fig. 5B. Cells were treated with compounds for the indicated periods of time. For rescue experiments, 1 μmol/L final concentration of proteasome inhibitor MG132 (cat. 474790–5MG; EMD Millipore), 2 μmol/L HER2 inhibitor lapatinib (cat. GW-572016; Selleckchem), or 2 μmol/L ubiquitin activating enzyme E1 inhibitor (synthesized at Genentech) was added before cell harvest.
CRISPR Engineering of HCC1954 Cells
HCC1954 breast cancer cells were engineered using CRISPR to knock out either the PIK3CA WT allele or the mutant alleles to create an isogenic pair of cell lines designated as HCC1954_mutant and HCC1954_WT. Guide RNAs (gRNAs) were designed using the CRISPRtool (http://crispr.mit.edu) to minimize potential off-target effects and cloned into pCas-Guide-EF1a-GFP vector (Blueheron Biotech). To generate the HCC1954_mutant line bearing p110a H1047R as a homozygous mutant, two CRISPR-Cas9 constructs were designed. One was designed to specifically target the WT allele in exon 21 (gRNA H1047R-2, ATGAATGATGCACATCATGG) and the second designed to target the intron of both WT and mutant alleles (gRNA H1047R-7, ACATTTGAGCAAAGACCTGA). To generate the HCC1954_WT line, two CRISPR-Cas9 constructs were designed with one gRNA targeting the mutant allele in exon 21 (H1047R-5, ATGAATGATGCACGTCATGG) and the second gRNA targeting the intron of both WT and mutant alleles (H1047R-8, TATTAAACTCCTGACATGCC). Plasmids for each targeting pair were cotransfected using Turbofectin (Thermofisher). After 48 hours, cells were put under selection with 1 μg/mL puromycin. Puromycin-resistant cells were further selected by collecting GFP-expressing cells by flow cytometry, and clones were expanded in standard cell culture conditions to create stable lines. Targeting efficiency of the CRISPR-induced allelic knockouts was assessed by PCR flanking the target sites (forward: TGCTGTGAAGGAAAATGGAA; reverse: TGCAGTGTGGAATCCAGAGTGAGC) and clones further validated with qRT-PCR and DNA sequencing.
siRNA Transfection
Transfection of siRNA was carried out using Lipofectamine RANiMAX reagent (Thermo Fisher Scientific) 72 hours in advance of drug treatment.
Western Blots
Protein concentration was determined using the Pierce BCA Protein Assay Kit. For immunoblots, equal protein amounts were loaded and then separated by electrophoresis through NuPAGE Novex Bis-Tris 4% to 12% gradient gels (Invitrogen). Proteins were transferred onto nitrocellulose membranes using the iBlot system and protocol from Invitrogen.
Subcellular Fractionation
Cells were washed once with phosphate-buffered saline before scraping into 0.8 mL/dish hypotonic lysis buffer (HLB: 25 mmol/L Tris-HCl pH 7.5, 10 mmol/L NaCl, 1 mmol/L EDTA, protease and phosphatase inhibitors). The cells were lysed by 30 strokes in a Dounce homogenizer, subjected to centrifugation at 1,500 × g (3,000 rpm) for 5 minutes to pellet nuclei and unbroken cells, followed by centrifugation of the supernatant at 100,000 × g (44,000 rpm) in a TLA55 rotor for 40 minutes. The supernatant (800 μL) was collected (S100 fraction) and the pellet resuspended in 200 μL HLB plus 1% NP40 (P100 fraction). The resuspended pellet was centrifuged 5 minutes at high speed in a microfuge and the supernatant collected.
Immunoprecipitation and Pulldown
Cells were lysed in 20 mmol/L Tris-HCL (pH 7.5), 137 mmol/L NaCl, 1 mmol/L EDTA, 1% NP40, and 10% glycerol plus protease and phosphatase inhibitors. For p110a immunoprecipitation, lysates were incubated with p110a antibody (Cell Signaling, 4249) overnight. Then, 50 μL protein A agarose beads was added to each sample and incubated additional 2 hours. For the ubiquitinated protein pulldown experiment, cells were lysed in lysis buffer containing 200 μg/mL TUBE1 (Lifesensors UM101). Lysates were isolated and added 50 μL glutathione agarose beads (Sigma, G4705). The samples were incubated overnight, and captured ubiquitinated protein was eluted in SDS reducing sample buffer.
RNA Isolation and PIK3CA Allele–Specific qRT-PCR
Total RNA was isolated from cells using the RNeasy Plus Mini Kit (Qiagen) following the protocol described in the kit. First-strand cDNA synthesis and qRT-PCR were carried out using One Step RT QPCR reagent (Roche). Resulting signal was detected on the Applied Biosystems Real Time PCR System. Primers and allele specific probes were as follows:
PIK3CA H1047R-forward: GGCTTTGGAGTATTTCATGAAACA
PIK3CA H1047R-reverse: GAAGATCCAATCCATTTTTGTTGTC
PIK3CA H1047R WT-probe: ATGATGCACATCATGGT
PIK3CA H1047R mut-probe: TGATGCACGTCATGGT
PIK3CA E545K-forward: GCAATTTCTACACGAGATCCTCTCT
PIK3CA E545K-reverse: CATTTTAGCACTTACCTGTGACTCCAT
PIK3CA E545K WT-probe: TGAAATCACTGAGCAGGAG
PIK3CA E545K mut-probe: TGAAATCACTAAGCAGGA
Intracellular Drug Concentration
Taselisib (1 μmol/L in RPMI medium) was added to PIK3CA-mutant and PIK3CA-WT cancer cell lines in triplicate and incubated under standard culture conditions (37°C, 5% CO2) for 18 hours. Medium containing taselisib was aspirated, and cells were washed twice with 1 mL ice-cold Hank's balanced salt solution (HBSS). Cells were lysed by sonication for 1 minutes in HBSS followed by the addition of an equal volume of acetonitrile containing analytical internal standard (propranolol, 100 nmol/L). The supernatant was analyzed in positive mode on a SCIEX API-4000 LC/MS/MS system with the transitions of 461/418 and 260/116 for taselisib and propranolol, respectively. The LC separations were achieved using a LUNA C18 column (4 μm, 50 × 2.1 mm) from Phenomenex, Inc. and Agilent 1100 series pumps consisting of mobile phase A (water containing 0.1% formic acid) and mobile phase B (acetonitrile containing 0.1% formic acid). The flow rate through the system was 1 mL/min. The initial condition was set at 5% B, which was ramped to 95% B over 40 seconds. After allowing the system to hold at 95% B for 20 seconds, the gradient was changed back to the initial condition of 5% B and allowed to equilibrate for 54 seconds before the next injection. A parallel replicate plate containing PIK3CA-mutant and PIK3CA-WT cancer cells was reserved for the determination of protein concentrations. Cells were washed with HBSS buffer and then lysed with 0.5 mL ProteaPrep cell lysis solution (Protea Biosciences). After 30 minutes, the solution was neutralized with 0.5 mL HBSS buffer, and 100 μL of the lysed cells was used to determine the protein concentration by the standard Bradford protein assay (Thermo Scientific), with bovine serum albumin as a standard.
PI3K ADP-Glo Assays for Ki Measurement
As described in WO 2017/001645 A1, the PI3K lipid kinase reaction was performed in the presence of PIP2:3PS lipid substrate (Promega #V1792) and ATP. Following the termination of the kinase reaction, turnover of ATP to ADP by the phosphorylation of the lipid substrate was detected using the Promega ADP-Glo (Promega #V1792) assay. Reactions were carried out using the following conditions for each PI3K isoform: PI3Kα (Millipore #14-602-K), kinase concentration 0.2 nmol/L, ATP 40 μmol/L, PIP2:3PS 50 μmol/L; PI3Kβ (Promega #V1751), kinase concentration 0.6 nmol/L, ATP 40 μmol/L, PIP2:3PS 50 μmol/L; PI3Kδ (Millipore #14–604-K), kinase concentration 0.25 nmol/L, ATP 40 μmol/L, PIP2:3PS 50 μmol/L; and PI3Kγ (Millipore #14-558-K), kinase concentration 0.4 nmol/L, ATP 25 μmol/L, PIP2:3PS 50 μmol/L in the presence of p85a. After 120 minutes of reaction time, the kinase reaction was terminated. After the reaction, any ATP remaining was depleted, leaving only ADP. Then the Kinase Detection Reagent was added to convert ADP to ATP, which was used in a coupled luciferin/luciferase reaction. The luminescent output was measured and correlated with kinase activity. All reactions were carried out at room temperature. For each PI3K isoform, a 3-μL mixture (1:1) of enzyme/lipid substrate solution was added to a 384-well white assay plate (Perkin Elmer #6007299) containing 50 μL test compound or DMSO only for untreated controls. The reaction was started by the addition of 2 μL ATP/MgCl2. The kinase reaction buffer contained 50 mmol/L HEPES, 50 mmol/L NaCl2, 3 mmol/L MgCl2, 0.01% BSA, 1% DMSO, and enzyme and substrate concentrations. The reaction was stopped by the addition of 10 μL ADP-Glo reagent. Plates were read in a Perkin Elmer Envision system using luminescence mode. A 10-point dose–response curve was generated for each test compound. Ki values for each compound were determined using the Morrison equation. For results shown in Supplementary Fig. S1G, same assays conditions were used as described above using p110a and p85 protein conditions shown as follows:
p110a/p85a (1.4 nmol/L, ATP 50 μmol/L,PIP2:3PS 50 μmol/L)
p110a/p85b (0.7 nmol/L, ATP 50 μmol/L,PIP2:3PS 50 μmol/L)
p110a/p85a E545K (0.7 nmol/L, ATP 50 μmol/L,PIP2:3PS 50 μmol/L)
p110a/p85b E545K (0.7 nmol/L, ATP 50 μmol/L,PIP2:3PS 50 μmol/L)
p110a/p85a H1047R (0.7 nmol/L, ATP 50 μmol/L,PIP2:3PS 50 μmol/L)
p110a/p85b H1047R (0.7 nmol/L, ATP 50 μmol/L,PIP2:3PS 50 μmol/L)
Kinetic Solubility
Compounds were dissolved in DMSO to a concentration of 10 mmol/L. These solutions were diluted into PBS buffer (pH 7.2, composed with NaCl, KCl, Na2HPO4, and KH2PO4) to a final compound concentration of 100 μmol/L, DMSO concentration of 2%, at pH 7.4. The samples were shaken for 24 hours at room temperature followed by filtration. LC/CLND (Chemiluminescent Nitrogen Detection) was used to determine compound concentration in the filtrate, with the concentration calculated by a caffeine calibration curve and the samples' nitrogen content. An internal standard compound was spiked into each sample for accurate quantification.
Plasma Protein Binding
As described in the literature (14), the extent of protein binding was determined in vitro in CD-1 mouse, Sprague−Dawley rat, and human plasma (Bioreclamation, Inc.) by equilibrium dialysis using the RED Device (Thermo Fisher Scientific). Compounds were added to pooled plasma (n ≥ 3) at a total concentration of 5 μmol/L. Plasma samples were equilibrated with phosphate-buffered saline (pH 7.4) at 37°C in 90% humidity and 5% CO2 for 4 hours. Following dialysis, compound concentration in plasma and buffer was measured by LC/MS-MS. The percentage of unbound compound in plasma was determined by dividing the concentration measured in the postdialysis buffer by that measured in the postdialysis plasma and multiplying by 100.
Madin-Darby Canine Kidney Cell Permeability
Experiments were carried out as previously described (30).
Drug Half-Life in Vivo
Experiments to determine half-life in mice were carried out as previously described (15, 31, 32).
Pulse-Chase
Cells were starved in methionine/cysteine-deficient media overnight. Cells were pulse labeled with 35S cysteine and methionine (Pro-mix l-[35S]; Amersham) in RPMI lacking cysteine and methionine for 6 hours, followed by three washes with RPMI containing no label. Cells were then incubated in normal media containing methionine and cysteine, either with or without compounds. Lysates were collected at various time points up to 48 hours. Radiolabeled p110a was immunoprecipitated and run on SDS-PAGE. Images were acquired on the Typhoon Scanner (GE Healthcare-Amersham) and signals quantified and normalized using ImageQuant TL software. Data were fit to a single exponential decay function using Prism (GraphPad Software) with Y = (Y0 – NS) * exp(–K * X) + NS and constraints Y0 = 100, NS = 0.
Animal Studies
The in vivo efficacy of PI3K inhibitors was tested in three breast cancer xenograft models that harbor PIK3CA mutations, HCC1954 (HER2 positive, PIK3CA H1047R), WHIM20 (ER positive/HER2 negative, PIK3CA H1047R), and HCI-003 (ER positive/HER2 negative, PIK3CA E545K). All in vivo studies were approved by Genentech and Institutional Animal Care and Use Committee and adhered to the NIH Guidelines for the Care and Use of Laboratory Animals. HCC1954 tumor cells (5 × 106) were inoculated in the 2/3 mammary fat pads of female NCR nude mice (Taconic Farms), whereas WHIM20 and HCI-003 tumors (50 mm3) were engrafted in 2/3 mammary fat pads in female NOD-SCID gamma mice (Jackson Laboratories). Tumor volumes were measured using Ultra Cal-IV calipers (Model 54–10–111; Fred V. Fowler Co.). The following formula was used in Excel, version 11.2 (Microsoft) to calculate tumor volume: Tumor Volume (mm3) = (Length × Width2) × 0.5. Mice for efficacy studies were distributed into 8 to 10 mice/group with a mean tumor volume of 200 to 250 mm3 at the initiation of dosing. A linear mixed effect modeling approach was used to analyze the repeated measurement of tumor volumes from the same animals over time (33). Cubic regression splines were used to fit a nonlinear profile to the time courses of log2 tumor volume at each dose level. These nonlinear profiles were then related to dose within the mixed model. Tumor growth inhibition as a percentage of vehicle control (%TGI) was calculated as the percentage of the AUC for the respective dose group per day in relation to the vehicle, using the following formula: %TGI = 100 × (1 – AUCdose/AUCvehicle). The PI3K inhibitors taselisib, GDC-0941, GDC-0077, and BYL719 were formulated in methylcellulose Tween vehicle consisting of 0.5% (w/v) methylcellulose, 0.2% (w/v) polysorbate 80 (Tween-80) and dosed orally by gavage daily. Tumor sizes and mouse body weights were recorded twice weekly, and mice with tumor volume exceeding 2,000 mm3 or body weight loss of 20% of starting weight were promptly euthanized. For pharmacodynamic and mechanistic studies, mice were dosed once by oral gavage and tumors harvested at 4 hours postdose. Following drug treatment, tumors were harvested and snap-frozen in liquid nitrogen and dissociated in lysis buffer containing 10 μmol/L Tris (pH 7.4), 100 μmol/L NaCl, 1 μmol/L EDTA, 1 μmol/L EGTA, 1 μmol/L NaF, 20 μmol/L Na4P2O7, 2 mmol/L Na3VO4, 1% Triton X-100, 10% glycerol, 0.1% SDS, and 0.5% deoxycholate supplemented with a phosphatase and protease inhibitor cocktail (Sigma). Protein concentrations were determined in whole-cell lysates using the BCA Protein Assay Kit (Pierce). Membrane fractions were isolated from xenograft tumors as described above and assessed by Western blot.
Mass Spectrometry
LC-MS/MS analysis was performed on p110a protein immunoprecipitated from three cell lines: HCC1954, HCC202, and HDQ-P1. Each cell line was treated for 24 hours with either DMSO (vehicle) or taselisib (500 nmol/L). Each experiment was performed beginning with 4 to 6 mg protein lysate per cell line/treatment (total of 6 samples/experiment) for n = 4 biological replicates. One gel region per sample, corresponding to the expected migration, was excised based on the migration of purified p110a protein in an adjacent lane. Gel pieces were diced into approximately 1-mm3 pieces and subjected to in-gel digestion as follows. Gel pieces were destained with 50 mmol/L ammonium bicarbonate/50% acetonitrile and dehydrated with 100% acetonitrile prior to reduction and alkylation using 50 mmol/L dithiothreitol (30 minutes, 50°C) and 50 mmol/L iodoacetamide (20 minutes, room temperature), respectively. Gel pieces were again dehydrated, allowed to reswell in 20 ng/μL trypsin in 50 mmol/L ammonium bicarbonate/5% acetonitrile digestion buffer on ice for 2 hours, and then transferred to a 37°C oven for overnight incubation. Digested peptides were transferred to microcentrifuge tubes and gel pieces were extracted twice, once with 50% acetonitrile/0.5% trifluoroacetic acid and a second round with 100% acetonitrile. Extracts were combined with digested peptides and speed-vac dried to completion. Samples were reconstituted in 5% formic acid/0.1% heptafluorobutyric acid/0.01% hydrogen peroxide 30 minutes prior to LC/MS-MS analysis. Samples were analyzed by LC/MS-MS with duplicate injection (with the exception of the first replicate in which samples were injected once) on a Thermo LTQ Orbitrap Elite coupled to a Waters nanoAcquity UPLC. Peptides were loaded onto a 0.1-mm × 100-mm Waters Symmetry C18 column packed with 1.7 μm BEH-130 resin and separated via a two-stage linear gradient in which solvent B (98% acetonitrile, 2% water) was ramped from 5% to 25% over 20 minutes and then from 25% to 50% over 2 minutes. Data were acquired in data-dependent mode with Orbitrap full MS scans collected at 60,000 resolution and the top 15 most intense precursors selected for Collision-induced dissociation MS/MS fragmentation in the ion trap. MS2 spectra were searched using Mascot, both against a concatenated target-decoy Uniprot database of human proteins as well as against a small database containing WT, E545K-mutant, and H1047R-mutant p110a sequences to identify mutant peptides. Peptide spectral matches for the Uniprot search were rough filtered using linear discriminant analysis to a 10% false discovery rate, then confirmed via manual inspection. Extracted ion chromatograms and peak area integration for p110a peptides were generated with 10-ppm mass tolerances using in-house software (MSPlorer). Peak area data for each of 14 peptides (see Fig. 3B; Supplementary Fig. S4C and S4D) were normalized on a per-replicate basis to the most abundant peak area among the six samples. In cases where duplicate injections (technical replicates) were available, normalized data for the two replicates were averaged to generate a single normalized peak area per peptide-condition-experiment for the protein sequence plots (Supplementary Fig. S4C). For statistical analysis, unnormalized peak areas across the four biological replicates were consolidated in R using linear mixed-effects modeling (lme4 package) to determine the relative ratio and measures of uncertainty (from which can be derived P values, confidence intervals, etc.) for the comparison of DMSO (vehicle) versus taselisib (500 nmol/L) treatments per cell line for each of total p110a, WT p110a, and mutant p110a (34). Total p110a was determined based on the data generated from the following peptides: EATLITIK (residues 39–46; 444.77481 m/z), DLNSPHSR (155–162; 463.22945 m/z), LCVLEYQGK (241–249), VCGCDEYFLEK (254–264; 710.30246 m/z), VPCSNPR (376–382), EAGFSYSHAGLSNR (503–516; 748.35281 m/z), YEQYLDNLLVR (641–651; 713.37540 m/z), FGLLLESYCR (684–693; 629.32042 m/z), and LINLTDILK (712–720; 521.83039 m/z). For cells bearing the H1047R mutation (i.e., HCC-1954), WT p110a was determined based on the QMNDAHHGGWTTK (1042–1054; 749.82824 m/z) peptide and mutant p110a based on QMNDAR (1042–1047; 375.66360 m/z) and HGGWTTK (1048–1054; 393.6983 m/z) peptides. For cells bearing the E545K mutation (i.e., HCC-202), WT p110a was determined based on the DPLSEITEQEK (538–548; 644.81917 m/z) peptide and mutant p110a based on DPLSEITK (538–545; 451.74627 m/z) peptides. Peptides from these two mutant loci were not considered when determining total p110a. Whenever applicable, cysteine residues within p110a peptides were analyzed in their carbamidomethylated form (+57.0215 Da) and methionine residues quantified based on their singly oxidized (Met630 sulfoxide) form (+15.9949 Da). Quantified peptide peak areas observed after treatment with taselisib or DMSO were analyzed to estimate differential abundances and accompanying measures of uncertainty for total, WT, and mutant, respectively, across the two treatments. These three fractional abundances were combined to estimate the relative proportions (and associated 95% confidence intervals) of WT versus mutant in each mutant-containing cell line in both the DMSO- and taselisib-treated conditions by applying the conservation of mass principle as follows. Suppose “p” and “1 – p” denote the fractions of WT and mutant p110a, respectively, in a DMSO-treated cell culture. Comparing peptides quantified after treatment with taselisib or DMSO from the total, WT-only, and mutant-only categories detailed above, define the three fractions: f_total = fraction of total p110a remaining after taselisib, relative to DMSO f_WT = fraction of WT p110a remaining after taselisib, relative to DMSO f_mut = fraction of mutant p110a remaining after taselisib, relative to DMSO. The conservation of mass principle requires that f_total = f_WT * p + f_mut * (1 – p). Solving for “p” (the fraction of WT p110a in DMSO-treated cells) yields an estimate of p = (f_total – f_mutant)/(f_WT – f_mutant), where log-scale estimates of the three fractions {f_total, f_WT, f_mutant} are obtained from the linear mixed-effects model. All other quantities of interest (with estimated SEs, confidence intervals, P values, etc.) likewise can be estimated as functions of these three fractions and their measures of uncertainty, which are also obtained from the linear mixed-effects model. Data are plotted as relative intensity values normalized to 1.0, and error bars represent the 95% confidence intervals for each measurement based on the linear effects model.
Authors' Disclosures
All Genentech authors are shareholders at Roche. K.W. Song reports a patent issued; and K.A. Edgar reports personal fees from Genentech outside the submitted work. E.J. Hanan reports other support from Genentech, Inc. outside the submitted work; in addition, E.J. Hanan has a patent for US 9,650,393 B2 issued and a patent for US 10,851,091 B2 issued. M. Hafner reports personal fees from Genentech during the conduct of the study. M. Merchant reports personal fees from Genentech/Roche outside the submitted work. G.D. Lewis Phillips reports other support from Genentech and other support from Roche outside the submitted work. L. Salphati is an employee of Genentech/Roche. T.P. Heffron is an employee of Genentech and a Roche shareholder. A.G. Olivero reports a patent for 20130079331 pending and issued to Genentech. S. Malek reports other support from Genentech during the conduct of the study; other support from Genentech outside the submitted work. D.S. Kirkpatrick reports other support from Genentech during the conduct of the study. L.S. Friedman reports other support from Genentech outside the submitted work; in addition, L.S. Friedman has a patent for Genentech pending. No disclosures were reported by the other authors.
Supplementary Material
Taselisib and GDC-0077 have increased potency in PIK3CA-mutant cancer cells
Activity of GDC-0077 in combination with Palbociclib and/ or Fulvestrant
Taselisib and GDC-0077 induce mutant p110a and not WT p110a degradation
Taselisib depletes mutant p110a protein through ubiquitin and proteasome mechanism in a dose and time dependent manner
Taselisib-mediated degradation of mutant p110a occurs preferentially at the plasma membrane.
RTK activity plays a key role in regulating p110a degradation
Acknowledgments
Support for third-party editing assistance for this manuscript, furnished by Daniel Clyde, PhD, of Health Interactions, was provided by F. Hoffmann-La Roche Ltd.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors' Contributions
K.W. Song: Investigation. K.A. Edgar: Investigation. E.J. Hanan: Investigation. M. Hafner: Investigation. J. Oeh: Investigation. M. Merchant: Investigation. D. Sampath: Investigation. M.A. Nannini: Investigation. R. Hong: Investigation. L. Phu: Investigation. W.F. Forrest: Investigation. E. Stawiski: Investigation. S. Schmidt: Investigation. N. Endres: Investigation. J. Guan: Investigation. J.J. Wallin: Investigation. J. Cheong: Investigation. E.G. Plise: Investigation. G.D. Lewis Phillips: Investigation. L. Salphati: Investigation. T.P. Heffron: Investigation. A.G. Olivero: Investigation. S. Malek: Writing–review and editing. S.T. Staben: Investigation. D.S. Kirkpatrick: Investigation. A. Dey: Conceptualization, supervision, writing–review and editing. L.S. Friedman: Conceptualization, supervision, writing–original draft.
References
- 1. Kang S, Bader AG, Vogt PK. Phosphatidylinositol 3-kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci U S A 2005;102:802–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Samuels Y, Diaz LA Jr, Schmidt-Kittler O, Cummins JM, Delong L, Cheong Iet al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 2005;7:561–73. [DOI] [PubMed] [Google Scholar]
- 3. Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RVet al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res 2005;65:10992–1000. [DOI] [PubMed] [Google Scholar]
- 4. Yu J, Zhang Y, McIlroy J, Rordorf-Nikolic T, Orr GA, Backer JM. Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol 1998;18:1379–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burke JE, Williams RL. Dynamic steps in receptor tyrosine kinase mediated activation of class IA phosphoinositide 3-kinases (PI3K) captured by H/D exchange (HDX-MS). Adv Biol Regul 2013;53:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Echeverria I, Liu Y, Gabelli SB, Amzel LM. Oncogenic mutations weaken the interactions that stabilize the p110alpha-p85alpha heterodimer in phosphatidylinositol 3-kinase alpha. FEBS J 2015;282:3528–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Miled N, Yan Y, Hon WC, Perisic O, Zvelebil M, Inbar Yet al. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science 2007;317:239–42. [DOI] [PubMed] [Google Scholar]
- 8. Burke JE, Perisic O, Masson GR, Vadas O, Williams RL. Oncogenic mutations mimic and enhance dynamic events in the natural activation of phosphoinositide 3-kinase p110alpha (PIK3CA). Proc Natl Acad Sci U S A 2012;109:15259–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol 2013;10:143–53. [DOI] [PubMed] [Google Scholar]
- 10. Krop IE, Mayer IA, Ganju V, Dickler M, Johnston S, Morales Set al. Pictilisib for oestrogen receptor-positive, aromatase inhibitor-resistant, advanced or metastatic breast cancer (FERGI): a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol 2016;17:811–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martín M, Chan A, Dirix L, O'Shaughnessy J, Hegg R, Manikhas Aet al. A randomized adaptive phase II/III study of buparlisib, a pan-class I PI3K inhibitor, combined with paclitaxel for the treatment of HER2- advanced breast cancer (BELLE-4). Ann Oncol 2017;28:313–20. [DOI] [PubMed] [Google Scholar]
- 12. Mayer IA, Abramson VG, Formisano L, Balko JM, Estrada MV, Sanders MEet al. A phase Ib study of alpelisib (BYL719), a PI3Kalpha-specific inhibitor, with letrozole in ER+/HER2− metastatic breast cancer. Clin Cancer Res 2017;23:26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Folkes AJ, Ahmadi K, Alderton WK, Alix S, Baker SJ, Box Get al. The identification of 2-(1H-indazol-4-yl)-6-(4-methanesulfonyl-piperazin-1-ylmethyl)-4-morpholin-4-yl-thieno[3,2-d]pyrimidine (GDC-0941) as a potent, selective, orally bioavailable inhibitor of class I PI3 kinase for the treatment of cancer. J Med Chem 2008;51:5522–32. [DOI] [PubMed] [Google Scholar]
- 14. Heffron TP, Heald RA, Ndubaku C, Wei B, Augistin M, Do Set al. The rational design of selective benzoxazepin inhibitors of the alpha-isoform of phosphoinositide 3-kinase culminating in the identification of (S)-2-((2-(1-Isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepin-9-yl)oxy)propanamide (GDC-0326). J Med Chem 2016;59:985–1002. [DOI] [PubMed] [Google Scholar]
- 15. Furet P, Guagnano V, Fairhurst RA, Imbach-Weese P, Bruce I, Knapp Met al. Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorg Med Chem Lett 2013;23:3741–8. [DOI] [PubMed] [Google Scholar]
- 16. Sadhu C, Masinovsky B, Dick K, Sowell CG, Staunton DE. Essential role of phosphoinositide 3-kinase delta in neutrophil directional movement. J Immunol 2003;170:2647–54. [DOI] [PubMed] [Google Scholar]
- 17. Jackson SP, Schoenwaelder SM, Goncalves I, Nesbitt WS, Yap CL, Wright CEet al. PI 3-kinase p110beta: a new target for antithrombotic therapy. Nat Med 2005;11:507–14. [DOI] [PubMed] [Google Scholar]
- 18. Hafner M, Niepel M, Chung M, Sorger PK. Growth rate inhibition metrics correct for confounders in measuring sensitivity to cancer drugs. Nat Methods 2016;13:521–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Juric D, Krop I, Ramanathan RK, Wilson TR, Ware JA, Sanabria Bohorquez SMet al. Phase I dose-escalation study of taselisib, an oral PI3K inhibitor, in patients with advanced solid tumors. Cancer Discov 2017;7:704–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Greig MJ, Niessen S, Weinrich SL, Feng JL, Shi M, Johnson TO. Effects of activating mutations on EGFR cellular protein turnover and amino acid recycling determined using SILAC mass spectrometry. Int J Cell Biol 2015;2015:798936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cantley LC, Auger KR, Carpenter C, Duckworth B, Graziani A, Kapeller Ret al. Oncogenes and signal transduction. Cell 1991;64:281–302. [DOI] [PubMed] [Google Scholar]
- 22. Backer JM. The regulation of class IA PI 3-kinases by inter-subunit interactions. Curr Top Microbiol Immunol 2010;346:87–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Engelman JA, Chen L, Tan X, Crosby K, Guimaraes AR, Upadhyay Ret al. Effective use of PI3K and MEK inhibitors to treat mutant Kras G12D and PIK3CA H1047R murine lung cancers. Nat Med 2008;14:1351–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cheng H, Liu P, Ohlson C, Xu E, Symonds L, Isabella Aet al. PIK3CA(H1047R)- and Her2-initiated mammary tumors escape PI3K dependency by compensatory activation of MEK-ERK signaling. Oncogene 2016;35:2961–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Juric D, Kalinsky K, Oliveira M, Cervantes A, Bedard P, Krop Iet al. A first-in-human phase Ia dose escalation study of GDC-0077, a p110a-selective and mutant-degrading PI3K inhibitor, in patients with PIK3CA-mutant solid tumors. Cancer Res 2019;80:Abstract OT1-08-4. [Google Scholar]
- 26. Jhaveri K, Kalinsky K, Bedard PL, Cervantes A, Saura C, Krop IEet al. Phase Ib dose escalation study evaluating the mutant selective PI3K-alpha inhibitor GDC-0077 in combination with letrozole with and without palbociclib in patients with PIK3CA-mutant HR+/HER2- breast cancer. Cancer Res 2019;80:Abstract P1-19-46. [Google Scholar]
- 27. Kalinsky K, Juric D, Bedard PL, Oliveira M, Cervantes A, Hamilton Eet al. A phase I/Ib study evaluating GDC-0077 plus fulvestrant in patients with PIK3CA-mutant, hormone receptor-positive/HER2-negative breast cancer. Cancer Res 2020;80: Abstract CT109. [Google Scholar]
- 28. Chen JJ, Tsu CA, Gavin JM, Milhollen MA, Bruzzese FJ, Mallender WDet al. Mechanistic studies of substrate-assisted inhibition of ubiquitin-activating enzyme by adenosine sulfamate analogues. J Biol Chem 2011;286:40867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu M, Selvaraj SK, Liang-Chu MM, Aghajani S, Busse M, Yuan Jet al. A resource for cell line authentication, annotation and quality control. Nature 2015;520:307–11. [DOI] [PubMed] [Google Scholar]
- 30. Irvine JD, Takahashi L, Lockhart K, Cheong J, Tolan JW, Selick HEet al. MDCK (Madin-Darby canine kidney) cells: a tool for membrane permeability screening. J Pharm Sci 1999;88:28–33. [DOI] [PubMed] [Google Scholar]
- 31. Pang J, Baumgardner M, Cheong J, Edgar K, Heffron TP, Le H. Preclinical evaluation of the β Isoform-sparing PI3K inhibitor GDC-0032 and prediction of its human pharmacokinetics. Drug Metab Rev 2014;45:1–286.24397791 [Google Scholar]
- 32. Salphati L, Pang J, Plise EG, Chou B, Halladay JS, Olivero AGet al. Preclinical pharmacokinetics of the novel PI3K inhibitor GDC-0941 and prediction of its pharmacokinetics and efficacy in human. Xenobiotica 2011;41:1088–99. [DOI] [PubMed] [Google Scholar]
- 33. Pinheiro J, Bates D, DebRoy S, Sarkar D; R Core Team. Linear and nonlinear mixed effects models. R package version 3.1–131.
- 34. Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw 2015;67:48. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Taselisib and GDC-0077 have increased potency in PIK3CA-mutant cancer cells
Activity of GDC-0077 in combination with Palbociclib and/ or Fulvestrant
Taselisib and GDC-0077 induce mutant p110a and not WT p110a degradation
Taselisib depletes mutant p110a protein through ubiquitin and proteasome mechanism in a dose and time dependent manner
Taselisib-mediated degradation of mutant p110a occurs preferentially at the plasma membrane.
RTK activity plays a key role in regulating p110a degradation