Summary
Background
HIV can infect multiple cells in the liver including hepatocytes, Kupffer cells and infiltrating T cells, but whether HIV can persist in the liver in people with HIV (PWH) on suppressive antiretroviral therapy (ART) remains unknown.
Methods
In a prospective longitudinal cohort of PWH and hepatitis B virus (HBV) co-infection living in Bangkok, Thailand, we collected blood and liver biopsies from 18 participants prior to and following ART and quantified HIV and HBV persistence using quantitative (q)PCR and RNA/DNAscope. Antiretroviral (ARV) drug levels were quantified using mass spectroscopy.
Findings
In liver biopsies taken prior to ART, HIV DNA and HIV RNA were detected by qPCR in 53% (9/17) and 47% (8/17) of participants respectively. Following a median ART duration of 3.4 years, HIV DNA was detected in liver in 61% (11/18) of participants by either qPCR, DNAscope or both, but only at very low and non-quantifiable levels. Using immunohistochemistry, HIV DNA was observed in both hepatocytes and liver infiltrating CD4+ T cells on ART. HIV RNA was not detected in liver biopsies collected on ART, by either qPCR or RNAscope. All ARVs were clearly detected in liver tissue.
Interpretation
Persistence of HIV DNA in liver in PWH on ART represents an additional reservoir that warrants further investigation.
Funding
National Health and Medical Research Council of Australia (Project Grant APP1101836, 1149990, and 1135851); This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. 75N91019D00024.
Keywords: HIV-HBV co-infection, Liver biopsy, Hepatocytes, HIV reservoir
Research in context.
Evidence before this study
People with HIV and HBV co-infection on antiretroviral therapy (ART) effective against both viruses have an increased risk of liver disease progression, liver-related mortality, and overall mortality compared to people with either HIV or HBV alone. The mechanisms behind this are not well understood. HIV can infect multiple cells in the liver including hepatocytes and infection of hepatocytes with HIV in vitro can directly impact HBV replication, leading to an increase in intracellular HBV surface antigen (HBsAg). In autopsied liver tissue and in studies of people living with HIV (PWH) on ART who underwent liver transplantation, HIV DNA or HIV RNA has been detected in low levels in the liver, however there have been no studies examining the liver as a reservoir prospectively prior to and following ART, nor has the location of viral persistence in the liver or impact on HBV replication been examined.
Added value of this study
In people living with HIV and HBV, we showed that in people off ART, HIV DNA and HIV RNA was easily detected in liver tissue. Following suppressive HBV-active ART, we found high levels of all antiretrovirals in liver tissue and only HIV DNA could be detected at very low levels. Using immunohistochemistry, we showed HIV DNA in both hepatocytes and infiltrating CD4+ T-cells. There was no relationship between detection of HIV DNA in the liver and liver function or markers of HBV persistence.
Implications of all the available evidence
Persistence of HIV DNA in liver in PWH on ART in both hepatocytes and CD4+ T-cells represents an additional HIV reservoir that warrants further investigation and should be considered in HIV cure strategies.
Introduction
There are approximately 38 million people globally living with human immunodeficiency virus (HIV), of which 5–20% are also living with hepatitis B virus (HBV).1 Antiretroviral therapy (ART) that can suppress both HIV and HBV replication, termed HBV-active ART, significantly reduces liver-related morbidity and mortality.2,3 However, people with HIV and HBV co-infection on suppressive ART still have an increased risk of liver disease progression, liver-related mortality, and overall mortality compared to people with either HIV or HBV alone.4, 5, 6 Given HIV can infect multiple cells in the liver7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18 including hepatocytes13,16,19, 20, 21, 22, 23 and infection of hepatocytes with HIV in vitro can have a direct impact on HBV replication leading to an increase in intracellular HBV surface antigen (HBsAg),20 we aimed to determine if the liver represents a significant reservoir in people with HIV and HBV co-infection on HBV-active ART, and whether persistence of HIV on ART was associated with HBV persistence or adverse liver outcomes.
The major barrier to a cure for HIV infection is long-lived and proliferating latenly infected CD4+ T cells.24 However, HIV can also persist in cells other than T cells, including monocytes and macrophages.25 In a humanised myeloid-only mouse model (MoM), HIV infection of tissue macrophages was rapidly suppressed by ART, and when ART was stopped, virus rebounded in a third of animals consistent with a long-lived reservoir of HIV in macrophages.26 In PWH on ART, HIV DNA or RNA has been detected in macrophages in spleen,27 brain28, 29, 30, 31, 32, 33 and other organs,34, 35, 36, 37 but whether this virus can result in viral rebound once ART is stopped remains unknown. Detection of HIV DNA in cerebrospinal fluid (CSF) in PWH on ART has been associated with impaired neurocognitive function, suggesting that identification of HIV DNA, even if not replication-competent, can also have a functional impact in specific tissue sites.38
In vitro, HIV can directly infect hepatocytes,13,16,19, 20, 21, 22, 23 Kupffer cells,7, 8, 9, 10, 11, 12, 13, 14, 15, 16 liver sinusoidal endothelial cells,16,17 hepatic stellate cells,18 and ex vivo, HIV DNA and/or RNA have been detected in liver biopsies from PWH off ART.31,39, 40, 41, 42 In studies of PWH on ART or non-human primates infected with either simian immunodeficiency virus (SIV) or a hybrid virus of SIV with an HIV envelope (SHIV) on ART who underwent autopsy, detection of HIV DNA or HIV RNA using quantitative (q)PCR or using in situ hybridisation has been reported in the liver at low but detectable levels in multiple studies,43, 44, 45, 46 however the location of infection within the liver has not been examined. Given that CD4+ T cells can be recruited to the liver during inflammation,47, 48, 49 and that the liver is rich with blood in liver sinusoids,50 detection of HIV DNA could potentially represent latently or productively infected CD4+ T cells and not true infection of liver cells.
Persistence of HIV in tissue sites in PWH on ART can be a consequence of multiple factors. First, there is enrichment of specific cells in tissue that favour HIV infection, such as resident memory T cells51 or Th17 cells.52, 53, 54 Second, there are anatomical barriers such as the blood brain barrier55 or blood testes barrier56 that inhibit entry of antiretrovirals (ARV) or anatomical compartments such as the B cell follicle that restrict entry of cytotoxic T cells.57 Finally, penetration of ARV in different tissues is variable.27,58,59 In one study, sub-optimal ARV concentrations in lymph node and the gastrointestinal tract in PWH on ART, correlated with increased detection of HIV RNA in these sites.60 The penetration of ARVs in the liver and the relationship of ARV concentration to HIV persistence has not previously been quantified.
We hypothesised that HIV infected cells persist in the liver in people with HIV-HBV co-infection on ART, possibly due to poor penetration of ARVs, and that ongoing production of HIV RNA could increase production of HBsAg, leading to persistent inflammation and adverse liver-related outcomes. In a prospective longitudinal study of people with HIV-HBV co-infection who initiated HBV-active ART, we found that HIV RNA and DNA were both detected in the liver prior to ART. There was no relationship between detection of HIV in the liver and markers of HBV or liver disease. Following an average of 3.4 years on ART, HIV DNA was detected in the liver at low levels in a subset of individuals using either qPCR or DNAscope. Using multi-parameter immunohistochemistry, we detected HIV DNA in both hepatocytes and CD4+ T cells. The implications of the liver as a persistent reservoir for HIV warrant further investigation.
Methods
Study recruitment
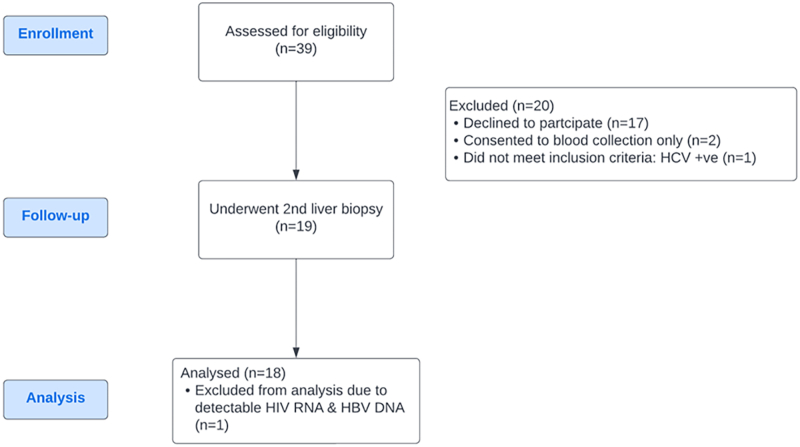
We previously reported on a cohort of 39 people living with HIV-HBV co-infection who were naïve to ART and underwent liver biopsy. They were recruited from the HIV-Netherlands-Australia-Thai Research Collaboration (HIV-NAT), Thai Red Cross AIDS Research Centre, Bangkok, Thailand.61 We enrolled 19 participants into this virological sub-study. Inclusion criteria for the sub-study was ART for a minimum of two years, a plasma HIV RNA <50 copies/mL over that time, HCV antibody negative and willingness to have a repeat liver biopsy. One individual did not meet enrolment criteria after the second liver biopsy as they were viremic and found to be non-adherent to ART and were therefore excluded from the analyses.
Ethics
This study was approved by the Institutional Review Board of the Faculty of Medicine, Chulalongkorn University, Bangkok, Thailand (Study number HIV-NAT178) and by the Alfred Hospital Human Research Ethics Committee, Melbourne, Australia (Study number 76/12). Written informed consent was obtained from all participants prior to enrolment.
HIV in peripheral blood CD4+ T cells
Total cellular DNA was extracted using the Qiagen AllPrep kit (Qiagen). HIV DNA was quantified in purified CD4+ T cells as described previously62 and normalised to cell number by quantification of the CCR5 gene.63 Unspliced (US) HIV-RNA was quantified using a semi-nested real-time quantitative PCR assay normalised to cell number by quantification of 18S as previously described64,65 but primers were modified to recognise HIV clade A/E which is the dominant clade in Thailand. The lower limit of detection for HIV RNA and HIV DNA was one copy per well. If there was no HIV PCR signal, this was recorded as zero and if there was a detectable signal but <1, this was recorded as 0.5 copies. For HIV RNA quantification, samples were run in quadruplicate with two no reverse transcriptase (RT) wells to control for DNA contamination. For HIV DNA quantification, samples were run in triplicate. For quantification of the CCR5 gene, samples were run in duplicate.
Liver biopsy processing
Liver biopsy samples were placed in RNAlater Stabilization Solution (Thermo Fisher Scientific) and stored at −80 °C until processing by scalpel disaggregation, lysis and homogenisation using Qiagen RLT buffer and a Qiagen Qiashredder column (Qiagen). Total cellular DNA and RNA was extracted from liver homogenates using a Qiagen AllPrep kit (Qiagen).
Quantification of HBV rcDNA and cccDNA in liver biopsy extracts by qPCR
Extracted DNA was also used to measure HBV covalently closed circular (ccc) DNA and relaxed circular (rc) DNA as previously described61,66 with the following modifications: extracted liver DNA was treated with Exonuclease I and III (New England Biolabs, Ipswich, MA, USA) to eliminate replicative intermediates. To each liver DNA sample, 1 μL (20 units) Exo I & 0.25 μL (25 units) Exo III, in 1X NEBuffer™ 1, and incubated at 37 °C for 3 h, followed by heat-treatment at 80 °C for 20 min, then cccDNA molecular analysis performed. Lower limit of quantification was 0.002 copies/genome equivalent (GEq). Samples that were undetectable were assigned a value of 0.002 for graphing on a log scale.
In situ hybridisation, RNAscope and DNAscope
In situ detection of HIV RNA and HIV DNA within liver biopsies was performed as previously described.67 The following probes were used to detect HIV clade A/E viral genome in paraformaldehyde (PFA) fixed tissues, HIV RNA: V-HIV1-CladeAE, targeting vif, vpr, tat, rev, vpu, env, nef and tar genes (#444031, ACD) and for HIV DNA: V-HIV1-CladeAE, targeting gag and pol (#444011, ACD). The probes were used individually, and the signal was detected using red chromogen. Slides mounted in Permount were scanned at high magnification (×400) using the ScanScope AT2 System (Aperio Technologies), yielding high-resolution data from the entire tissue section.
Phenotyping HIV+ cells
To phenotype the cells harbouring HIV DNA, fluorescence microscopy was performed using DNAscope with a sense probe targeting gag and pol of HIV clade A/E (#444011, ACD), and was then combined with immunofluorescence imaging using antibodies to CD4 (Rabbit; 1:100, Abcam), Hepatocytes (MA5-12417-OCH1E5, Thermo Fisher Scientific) and nuclear stain (DAPI). The detailed protocol was previously published.68 Up to 6 sections of 5 μm each were screened, and using a confocal microscope (Olympus, Fluoview FV10i) representative pictures were taken to evaluate the type of cells harbouring HIV DNA in all participants.
Quantitative assessment of ARVs in liver biopsies and plasma
Liver needle biopsies from 17/18 participants were analysed using infrared matrix-assisted laser desorption electrospray ionisation (IR-MALDESI) mass spectrometry imaging (MSI). Tissue concentrations of emtricitabine (FTC), tenofovir disoproxil fumarate (TFV), efavirenz (EFV), lamivudine (3TC) and rilpivirine (RPV) were measured. Tissue biopsies were evaluated on intact tissue without additional sample preparation or sectioning. Samples were thawed slightly to enable detachment from foil packaging and immediately mounted on a glass microscope slide. Calibration of IR-MALDESI antiretroviral response within a liver tissue matrix was performed using blank non-human primate liver tissue. This approach for quantitative MSI has been described in detail.69,70 Plasma ARV were measured by validated, multiplex LC-MS/MS assays as previously published.71,72 Briefly, all ARV were extracted using protein precipitation with the following isotopically labelled internal standards: TFV-13C5; FTC-13C, 15N2; 3TC-15N, d2; zidovudine (ZDV)-d4; EFV-d5; RPV-d6; Lopinavir (LPV)-d8; and Ritonavir (RTV)-d6. Using reverse phase chromatography, TFV, FTC, 3TC, ZDV, EFV, LPV and RTV were separated on a Waters Atlantis T3 (2.1 × 50 mm, 3 micron) analytical column and RPV was separated on a Waters XTERRA MS C18 (50 × 2.1 mm, 3.5 μm) analytical column. Analytes were then detected on an API-5000 Triple Quad mass spectrometer (AB SCIEX, Foster City, CA, USA). Calibrated assay ranges were 1.00–4000 ng/mL (TFV, FTC, 3TC, and ZDV); 50–20,000 ng/mL (EFV, LPV, and RTV); and 1–10,000 ng/mL (RPV). All calibration standards and quality control (QC) samples were prepared in blank human plasma with precision and accuracy acceptance criteria of ±15% (20% at the lower limit of quantification). To directly compare tissue ARV concentrations with plasma ARV concentrations, LC-MS/MS plasma concentration values were converted from units of ng/mL to μg/g based on an estimated liver tissue density of 1.06 g/cm3.
Plasma HBV DNA and HBV surface antigen (qHBsAg)
HBV DNA was quantified using the Abbott m2000sp/m2000rt Realtime HBV test (Abbott Laboratories) with a lower limit of quantification of 10 IU/mL. HBsAg was quantified using the Roche Diagnostics Elecsys® HBsAg II quant II as previously described.73
HIV DNA sequencing
PCR amplimers from low positive wells were directly sequenced with sanger sequencing. Alignments were performed using Clustal W in CLC Main Workbench (Qiagen) and neighbour joining trees were constructed using MEGAX (http://megasoftware.net).
Liver fibrosis
Liver fibrosis was evaluated in two ways as previously described.61 Liver biopsies were scored according to the Metavir classification.74 Liver stiffness was assessed by transient elastography (TE) using Fibroscan® (Echosens). We used TE cut-offs validated in HIV-HBV co-infection to stage fibrosis according to the Metavir system (F1–F4) such that F0/F1 < 5.9 kilopascals (kPa), F2 5.9 to <7.6 kPa, F3 7.6 to <9.4 kPa and F4 ≥ 9.4.kPa.75
Measure of inflammatory cytokines and chemokines in the plasma by ELISA or multiplex
We measured high mobility group box 1 (HMGB1, IBL International GMBH) and soluble CD14 (sCD14, R&D Systems) in the plasma by ELISA according to the manufacturer's instructions. We measured 10 inflammatory cytokines or chemokines (C-X-C motif chemokine ligand (CXCL)9, CXCL10, CXCL11, C–C motif chemokine ligand (CCL)2, CCL3, CCL4, regulated upon activation, normal T cell expressed and secreted (RANTES), interleukin (IL)-18, tumour necrosis factor (TNF)α, and IL-10) in plasma using a human custom multiplex kit (Human ProcartaPlex 10-plex, Invitrogen) according to the manufacturer's instructions. CCL2 and CXCL10 were also measured using a BioPlex kit (BioRad). The pre-ART BioPlex data had been analysed previously.61 However, to ensure consistency between pre-ART and on ART analyses, we re-ran the paired 18 pre-ART and on ART samples again on the same BioPlex kit using a fresh aliquot of plasma that had not been freeze-thawed. The repeat pre-ART analyses correlated with the previous results (unpublished).
Statistics
Data were presented descriptively, using n (%) for categorical data and median (25th to 75th percentile) for continuous data, and graphically using dot- and scatter-plots. Pre- and on ART data was compared using a two-tailed Wilcoxon signed rank test. Detectable versus undetectable HIV DNA in the liver while on ART were compared using a Wilcoxon rank-sum test. Comparisons between drug levels in plasma and liver were made using a Wilcoxon rank-sum test. Correlations were analysed using a Spearman correlation as HIV RNA and DNA in the liver were not normally distributed. All data were analysed using Prism v9.0. No adjustment of multiple testing was performed. p < 0.05 was considered statistically significant.
Role of funder
The study funder had no role in the study design, data collection, data analyses, interpretation of data; writing the report; nor in the decision to submit the paper for publication.
Results
Study participants & clinical features at enrolment (pre-ART) and at follow up (on ART)
Participants were recruited as part of a virology sub-study of people living with HIV and HBV in Bangkok, Thailand who underwent liver biopsy prior to initiating HBV-active ART (n = 39), previously reported in Singh et al.61 Clinical details of the original cohort and participants in this sub-study are shown in Table 1. After a minimum of two years of viral suppression (both HIV and HBV), participants could enrol in this sub-study and underwent a repeat liver biopsy and blood collection (n = 18, Supplementary Figure S1). Participants were predominantly young men (17/18) (sex data self-reported by study participants). Prior to initiation of HBV-active ART, the median CD4+ T cell count was 360 cells/mL and were HBsAg positive with a median qHBsAg of 3543 IU/mL and 55.6% were HB “e” antigen (HBeAg) positive (10/18). Following HBV-active ART, participants had a median CD4+ T cell count of 645 cells/uL and remained HBsAg positive with a median qHBsAg of 2878 IU/mL and 27.8% were HBeAg positive (5/18) (Table 1). The median time on HBV-active ART was 3.4 years.
Table 1.
Demographic and clinical characteristics (n = 18, paired pre- and on ART donors and n = 39 original pre-ART cohort).
| Clinical | Original cohorta | Substudy cohort |
|
|---|---|---|---|
| Pre-ART | On ART | ||
| Age, years (range) | 31.9 (25.3–35.8) | 32.5 (21.3–44.6) | 35.7 (24.8–47.5) |
| Sex, % male (n) | 89.7 (35) | 94.4 (17) | 94.4 (17) |
| Duration on ART (years) | NA | NA | 3.4 (2.9–3.7) |
| Alcohol intake, % (n) | |||
| Never or less than monthly | 43.6 (17) | 66.7 (12) | 72.2 (13) |
| Weekly-monthly | 56.4 (22) | 33.3 (6) | 26.3 (5) |
| Daily (or almost) | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| Number of participants on each ARV | |||
| Emtricitabine (FTC) | NA | NA | 16 |
| Tenofovir (TFV) | NA | NA | 17 |
| Efavirenz (EFV) | NA | NA | 16 |
| Lamivudine (3TC) | NA | NA | 2 |
| Zidovudine (AZT) | NA | NA | 1 |
| Laboratory | |||
| CD4, cells/μL | 360 (221–462) | 362 (262–425) | 645 (425–813) |
| CD8 total, cells/μL | 965 (662–1334) | 1070 (822–1411) | 844 (653–1074) |
| CD4/CD8 ratio | 0.32 (0.19–0.39) | 0.73 (0.64–1.07) | |
| HIV RNA, log10copies/mL (range) | 4.9 (4.5–5.5) | 4.8 (4.3–6.2) | 1.3 (1.3–1.6) |
| HBV DNA, log10IU/mL (range) | 7.4 (2.6–8.1) | 4.9 (1.0–8.6) | 1.0 (1.0–2.3) |
| HBV DNA, HBeAg+ (range) | 7.8 (7.3–8.4) | 7.5 (2.3–8.6) | 1.0 (1.0–2.3) |
| HBV DNA, HBeAg− (range) | 2.1 (1.5–3.5) | 2.2 (1.0–3.8) | 1.0 (1.0–1.2) |
| HBeAg positive, % (n) | 64.1% (25) | 55.6 (10) | 27.8 (5) |
| qHBsAg, IU/mL | 6403 (1327–18797) | 3543 (503–11,569) | 2878 (578–15,199) |
| Total bilirubin, mg/dL | 0.7 (0.5–0.8) | 0.6 (0.4–0.8) | 0.5 (0.4–0.6) |
| Alanine Aminotransferase, ALT, U/L | 38 (27–62) | 31 (27–50) | 39 (27–46) |
| Aspartate Aminotransaminase, AST, U/L | 33 (26–46) | 31 (26–40) | 27 (19–32) |
| Alkaline phosphatase, ALP, U/L | 70 (61–78) | 70 (63–78) | 80 (68–88) |
| γ-glutamyl transferase, GGT, U/L | 32 (19–61) | 31 (17–51) | 42 (21–65) |
| Fibrosis assessment | |||
| By transient elastography, TE, kPa | 6.3 (5.3–8.3) | 5.6 (5.3–7.9) | 5.2 (4.6–7.7) |
| TE, Fibrosis grade, % (n) | |||
| F0–F1 | 46.2 (18) | 55.5 (10) | 61.1 (11) |
| F2 | 35.9 (14) | 16.7 (3) | 16.7 (3) |
| F3 | 10.3 (4) | 16.7 (3) | 22.2 (4) |
| F4 | 7.7 (3) | 11.1 (2) | 0.0 (0) |
| Liver biopsy fibrosis grade (Metavir), % (n) | |||
| F0 | 61.5 (24) | 61.1 (11) | 83.3 (15) |
| F1 | 30.8 (12) | 33.3 (6) | 16.7 (3) |
| F2 | 0.0 (0) | 0.0 (0) | 0.0 (0) |
| F3 | 7.7 (3) | 5.6 (1) | 0.0 (0) |
| F4 | 0 (0) | 0.0 (0) | 0.0 (0) |
All values are presented as median (25th–75th percentiles) except where specified.
HBeAg = HB “e” antigen; HBsAg = HB surface antigen; qHBsAg = quantitative HBsAg; NA = not applicable.
Original cohort described in Singh et al.61 Demographic details of the current substudy are included at enrolment (prior to ART) and after at least 2 years on suppressive ART (on ART).
In all participants, plasma HIV RNA reduced following ART to below the limit of detection (20 copies/ml plasma) except for one participant who had low-level detection of HIV RNA at 40 copies/mL (Table 1). Plasma HBV DNA was significantly higher in HBeAg+ compared to HBeAg− participants at baseline as expected (Table 1). Following ART, most participants (67%, 12/18) had plasma HBV DNA below the limit of detection of the clinical assay used (limit of detection = 10 IU/mL plasma, Table 1). The remaining six participants had low-level detection of plasma HBV DNA between 11 and 204 IU/mL (Table 1). In contrast to HBV DNA, there was no significant decrease in qHBsAg following ART (p = 0.6705, Table 1).
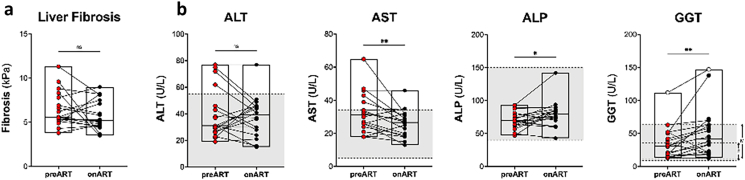
At baseline, the majority of participants on ART had mild fibrosis (F0–F2, measured by transient elastography or Metavir classification) and we did not find a significant change following ART (p = 0.3841, Supplementary Figure S2A). On ART, there was a significant decrease in AST (p = 0.0394) but no significant change in ALT. There was a significant increase in both ALP (p = 0.0086) and GGT (p = 0.0248) following ART but nearly all measurements were within the normal range as established for liver enzymes in uninfected Thai individuals at the Thai Red Cross (Supplementary Figure S2B). All other immunological and virological parameters measured in plasma and liver are summarised in Table 1.
Changes in HIV in the periphery and liver following ART
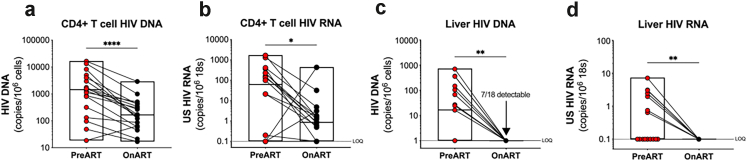
CD4+ T cells were isolated from peripheral blood mononuclear cells (PBMCs) and both cell-associated DNA and RNA were extracted. As expected, both prior to and on ART, cell-associated HIV DNA was detected in 100% (18/18) of participants, and the level of HIV DNA was significantly higher prior to ART compared to on ART (median = 1439 vs 166 copies/106 cells, respectively, p < 0.0001, Fig. 1A). Both prior to and on ART, cell-associated HIV RNA in circulating CD4+ T cells was detected in 67% (12/18) of participants. The median number of circulating CD4+ T-cells assayed was 120,203 (range = 76,545–156,750). Of the 12 participants, 8 participants had detectable HIV RNA pre- and on ART, two participants lost detectable HIV RNA on ART and two gained detectable HIV RNA on ART. However, the level of HIV RNA was significantly higher prior to ART compared to on ART (median = 62 vs 0.85 copies/106 18s, respectively, p = 0.0134, Fig. 1B).
Fig. 1.
Virological measures of HIV. Quantification of cell-associated HIV DNA (a) and RNA (b) in peripheral CD4+ T cells prior to initiation of ART and on ART. Quantification of cell-associated HIV DNA (c) and RNA (d) in liver biopsies prior to initiation of ART (red) and on ART (black). p values were determined by Wilcoxon signed-rank test. n = 18 except for the pre-ART data in panels c and d, which are n = 17, where one sample failed extraction. Boxes represent range with line at the median. LOQ = limit of quantification. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Cell-associated HIV DNA and RNA were also assessed in paired liver biopsies. Prior to ART, we found HIV DNA and RNA in the liver by quantitative PCR (qPCR) in 53% (9/17) and 47% (8/17) of participants, respectively (Fig. 1C and D). One sample failed extraction (this sample did not have detectable housekeeping genes) and could not be analysed by qPCR. On ART, we could not detect HIV RNA in the liver by qPCR in any participant, however, we detected HIV DNA by qPCR in 39% (7/18) of participants. In all participants where HIV DNA was detectable, the level was below the limit of quantification of 1 copy per well (Fig. 1C). The number of cell equivalents assayed for HIV DNA or RNA in the liver by qPCR was limited, with a median of 32,535 (range = 8667–47,895) cell equivalents per individual. Given that the limit of detection was 1 copy per well, this meant that the frequency of infection in the liver was <31 copies (range 20–115) copies per million cell equivalents.
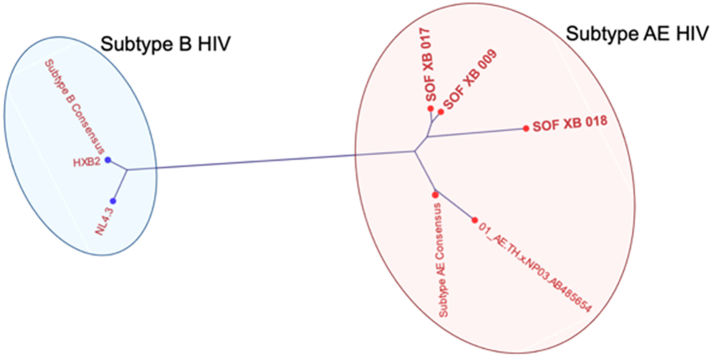
To confirm the low-level HIV DNA detected in liver biopsies by qPCR was not a PCR contaminant, we sequenced the amplicons from the three samples with the highest levels of HIV DNA. Sanger sequencing was performed using the PCR amplimers from 3 low positive wells. All resultant sequence reads had a single peak consistent with the low positive wells being at the limit of detection (1 copy HIV DNA per well). Phylogenetic analysis showed that sequences from these low positive wells clustered with subtype AE consensus sequences and away from subtype B consensus sequence and common lab contaminants (Supplementary Figure S3). Furthermore, each sequence from the three participants differed in sequence by at least one nucleotide.
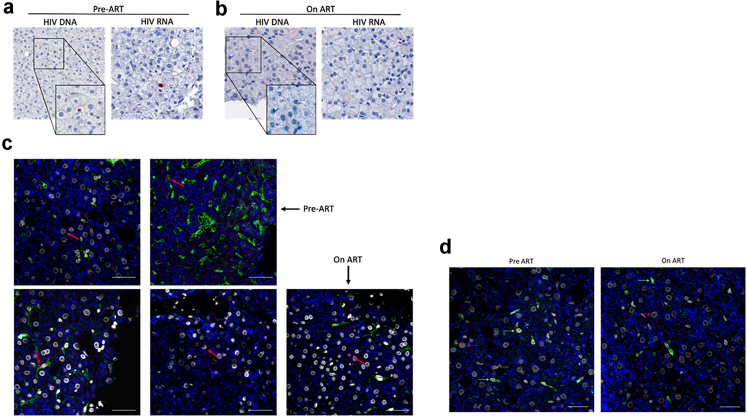
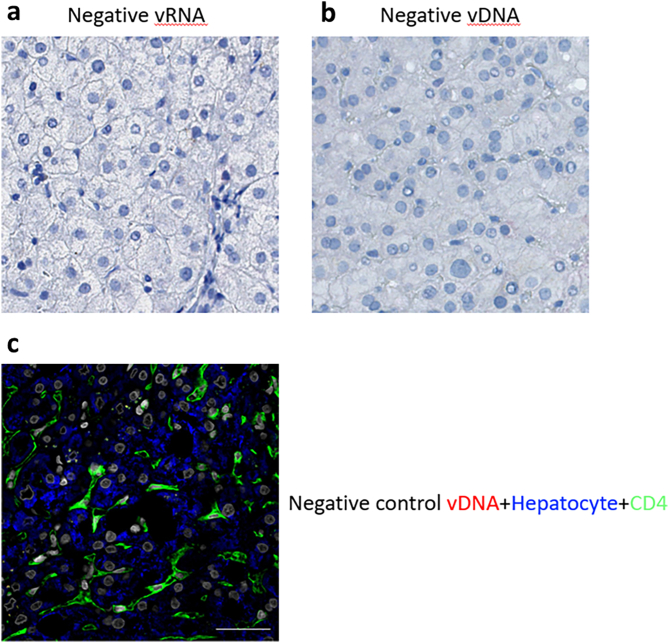
In addition to measuring HIV DNA and RNA in the liver by qPCR, we also performed DNA and RNAscope in liver biopsies from participants (Fig. 2A and B). For these studies, we had access to a larger number of liver samples pre-ART and therefore assessed 33 of 39 participants for HIV DNA and RNAscope. Prior to ART, HIV DNA and RNAscope was positive in 88% (29/33) and 67% (22/33) of participants, respectively (Fig. 2A). In participants on ART, we found low-level detection of HIV DNA by DNAscope in 33% (6/18) of individuals (Fig. 2B). In control samples from people without HIV or HBV, there was no detection of viral RNA or DNA (Supplementary Figure S4). Combining detection of HIV DNA in the liver using both methods, we found that on ART, HIV DNA was detected in 61% (11/18) of individuals by one or both methods. We could not detect any HIV RNA in the liver on ART by RNAscope, consistent with the qPCR results. Similar to our analysis with qPCR, the number of cells screened by DNAscope or RNAscope was limited, with a median of 10,165 cells (range = 210–23,480) per participant. In summary, we could detect HIV DNA and RNA in liver pre-ART. Following ART, in a subset of participants, HIV DNA could be detected at a low level, measured by either qPCR or DNAscope.
Fig. 2.
HIV DNA persists in CD4+ Tcells and hepatocytes on ART. HIV DNA or HIV RNA were measured by DNAscope and RNAscope and a chromogen stain in liver tissue sections prior to ART (a) and on ART (b). (c). Immunophenotyping of HIV DNA+ cells using DNAscope and anti-CD4 (green) in liver tissue collected pre-ART (top) and on ART (bottom). Representative images from two (pre-ART) and three (on-ART) participants are shown. (d). Immunophenotyping of HIV DNA+ cells using DNAscope and anti-CD3 (green) in liver tissue collected pre-ART (top) and on ART (bottom). The average total number of nuclei screened was 27,217.1 (range 3774–49,078). Red arrows point to HIV+ cells. red = HIV DNA, blue = hepatocytes, white = nuclei. Scale bars = 100 uM.
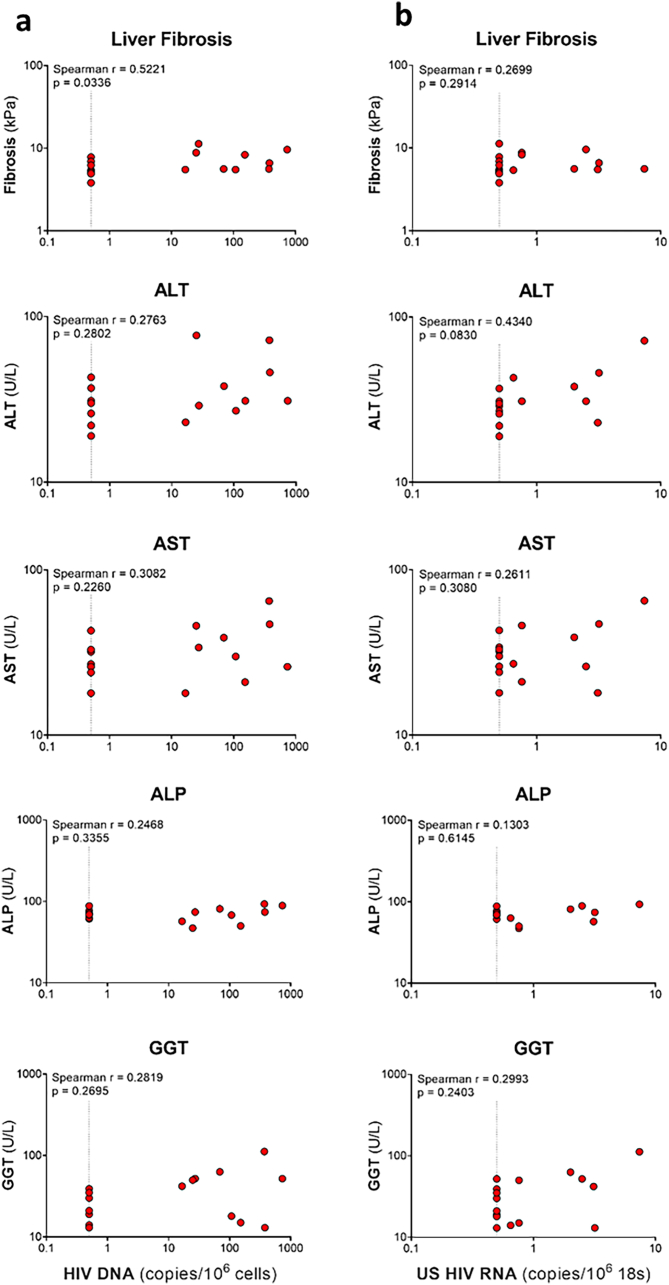
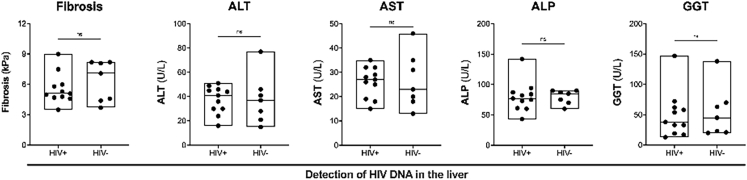
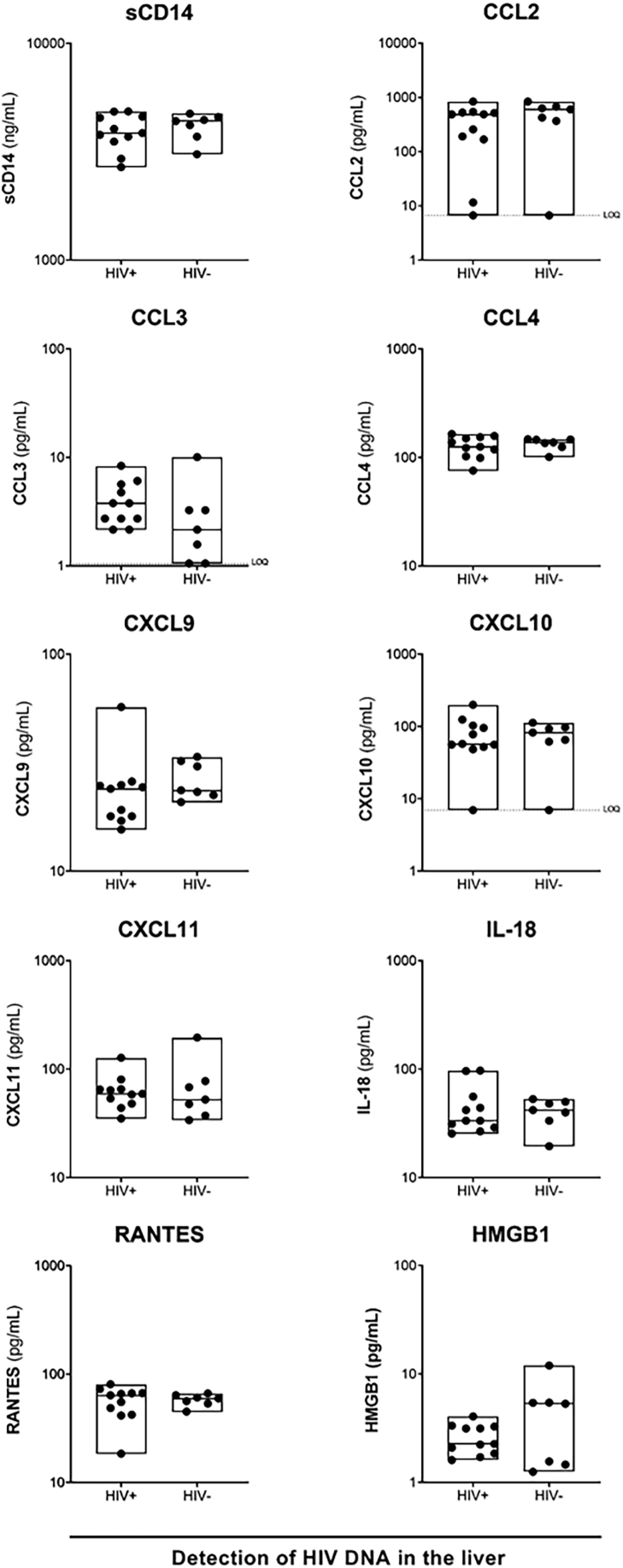
We next evaluated if there was a relationship between detection of HIV DNA and liver function. Prior to ART, there was a low but statistically significant correlation between HIV DNA in the liver and fibrosis (Spearman r = 0.5221, Supplementary Figure S5A). There were no significant correlations between HIV DNA in the liver and liver enzymes (Supplementary Figure S5A) or HIV RNA in the liver and fibrosis or liver enzymes (Supplementary Figure S5B). We were not able to quantitatively assess correlations between HIV DNA or RNA in the liver and fibrosis or liver enzymes on ART, given that HIV RNA in the liver was undetectable, and detection of HIV DNA was qualitative. We did, however, examine if there were differences in liver fibrosis and liver enzymes in those individuals who had detectable HIV DNA in the liver by either qPCR or DNAscope versus those who did not. We did not find any differences between individuals with detectable versus undetectable HIV DNA in the liver (Supplementary Figure S6). These findings were probably due to the fact that we were at the limit of detection for both assays, likely as a result of sampling bias, and therefore potentially underestimating participants with HIV DNA in the liver.
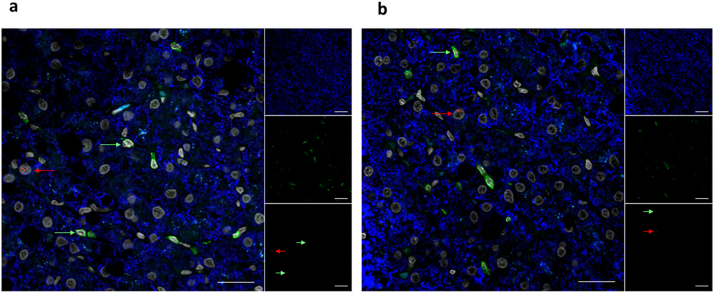
HIV DNA in liver is found in hepatocytes and CD4+ T cells
We wanted to determine which cells in the liver harboured HIV DNA on ART. Combined fluorescence microscopy with either DNA or RNAscope was used to phenotype cells that were HIV DNA+ or HIV RNA+. On ART, we found that HIV DNA persisted in both CD4+ cells (Fig. 2c, top right, bottom left panels) as well as in hepatocytes (Fig. 2c, top left, bottom middle and right panels). The CD4+ cells containing HIV DNA were morphologically consistent with T cells. We confirm that these cells were T cells as they were anti-CD3 positive (Fig. 2d). These data suggest that HIV DNA persists on ART in both CD4+ T cells and hepatocytes.
Antiretroviral concentrations in the liver are at or above plasma concentrations
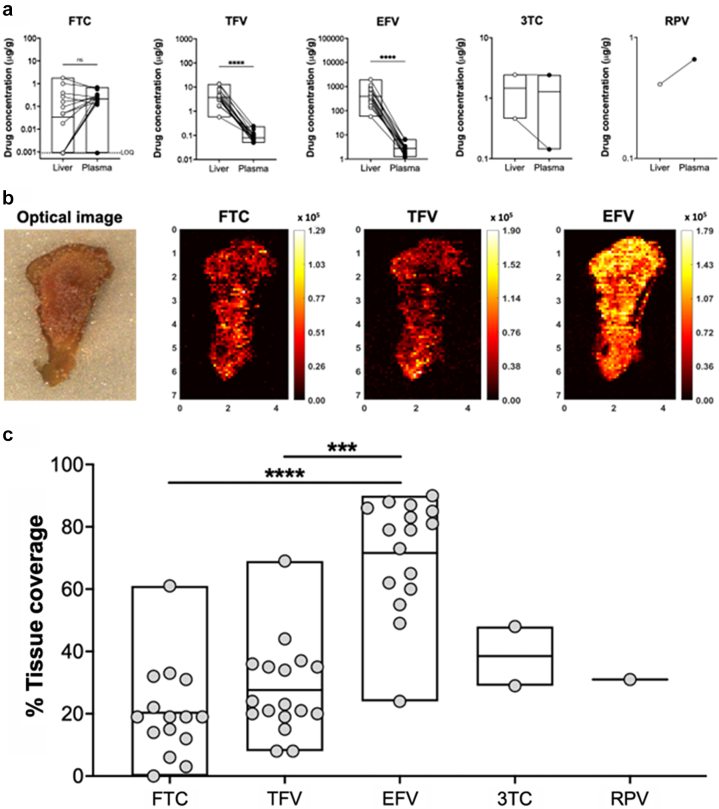
One of our main hypotheses was that HIV may persist in the liver due to poor ARV penetration. Therefore, we performed mass-spectrometry imaging on liver biopsy samples from 17/18 participants to measure intrahepatic ARV concentrations and compared this to ARV concentrations measured in the plasma. Five different ARVs were quantified (see Table 1). We found FTC concentrations to be similar between liver and plasma (p = 0.3180), whereas TFV and EFV were significantly higher in the liver than in the plasma (p < 0.0001 for both, Fig. 3a). Because 3TC and RPV were taken by few participants, we were not able to statistically compare liver and plasma concentrations. However, for both 3TC and RPV, the drug concentrations in the liver were similar to that seen in plasma (Fig. 3a). All ARVs showed a heterogeneous distribution pattern within liver tissue, as illustrated in representative ion maps of FTC, TFV, and EFV from a sample biopsy (Fig. 3b), where colour gradients ranging from black to yellow reflect localised changes in drug concentrations. The variability of ARV concentrations within the liver is consistent with previous findings of ARV heterogeneity throughout other tissues using mass-spectrometry imaging.59,70,76 To quantify heterogeneity within tissues, we evaluated the proportion of total tissue area with detectable drug (and the dynamic range of measurements across a sample). Coverage of tissue by detectable drug was consistent with the trend in cumulative ARV concentrations. We found the highest tissue coverage with EFV, reaching as high as 90% (median = 79%, range = 24–90%), which was significantly higher than both TFV (median = 22%, range = 8–69%) and FTC (median = 19%, range = below LOQ-61%) (p = 0.0005 and p < 0.0001, respectively, Fig. 3c). These findings suggest that there was good ARV penetration into the liver, yielding ARV concentrations that were similar to or higher than those found in the plasma.
Fig. 3.
Antiretroviral (ARV) drug levels in the liver and plasma. (a) Concentrations of ARVs in liver biopsies and plasma quantified by mass spec imaging are shown for liver (open circles) and plasma (filled circles). To directly compare liver and plasma ARV levels, plasma concentrations were converted from ng/mL to ug/g as described in the methods. (b) Representative images of ARV drug levels in liver biopsy sections. Lighter colours are associated with higher drug concentrations and darker colours are associated with lower drug concentrations. (c) Level of ARV coverage in the liver. FTC = emtricitabine, TFV = tenofovir, EFV = efavirenz, 3TC = lamivudine, RPV = rilpivirine. p values were determined by Wilcoxon signed-rank test. Boxes represent range with line at the median. ns = not significant. LOQ = limit of quantification. For panel c, statistics were not performed for 3TC and RPV due to low numbers. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
HIV DNA in the liver is not associated with any HBV parameter
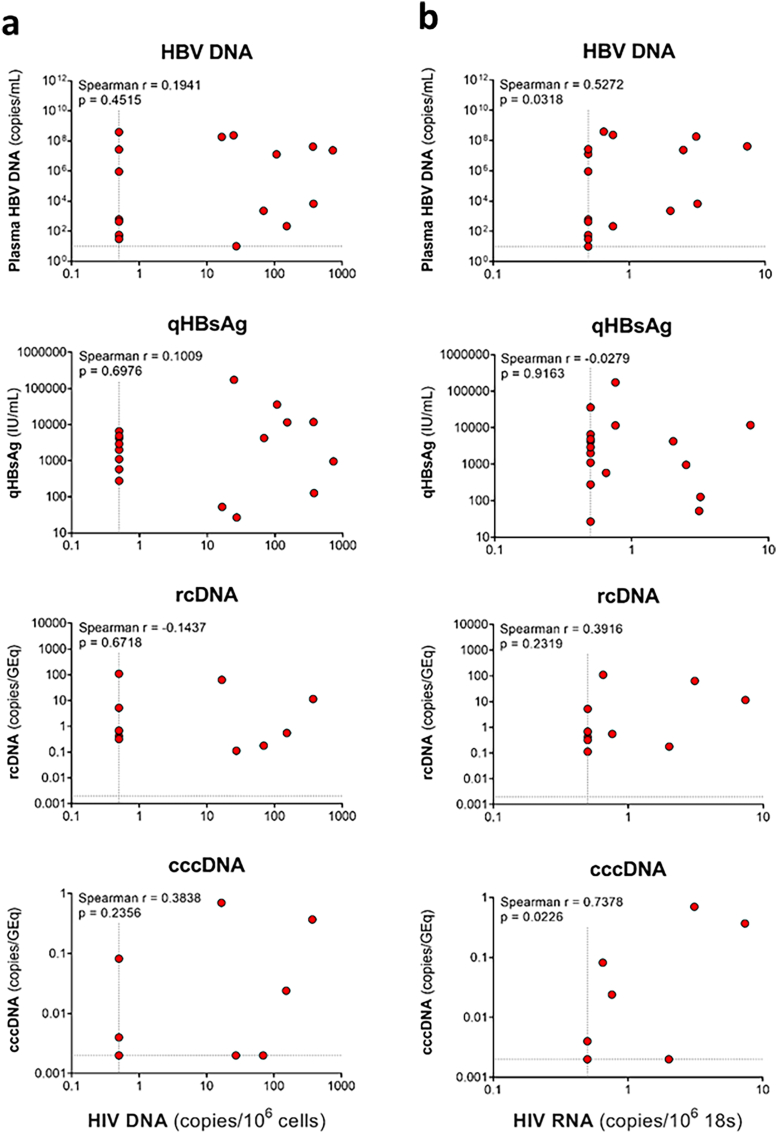
In addition to plasma HBV DNA and qHBsAg, we also measured both rc and ccc HBV DNA in 11 participants who had sufficient sample prior to ART. We did not find any statistically significant correlations between HIV DNA in the liver and markers of HBV replication, including plasma HBV DNA, qHBsAg, and liver rcDNA, and cccDNA (Supplementary Figure S7a). We found a statistically significant correlation between HIV RNA in the liver and plasma HBV DNA (Spearman r = 0.5272) but there were no correlations with qHBsAg, rcDNA, cccDNA (Supplementary Figure S7b).
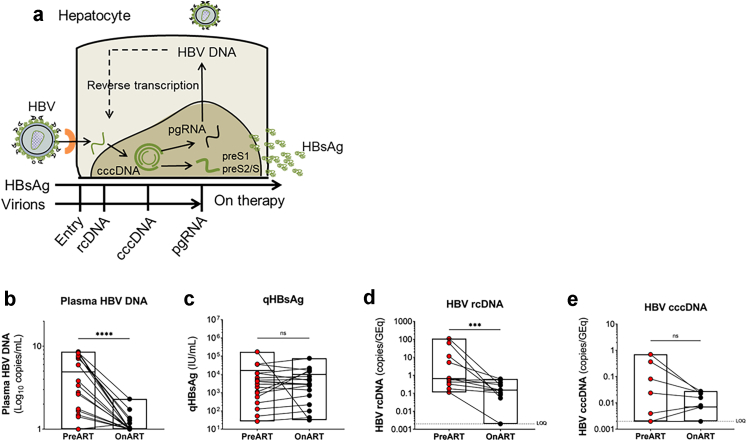
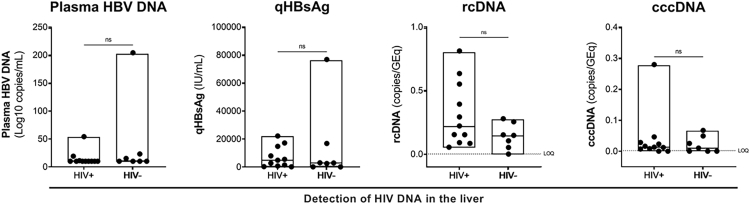
We next compared markers of HBV replication pre- versus on ART. The HBV replication cycle and different HBV intermediates are shown in Fig. 4a. As expected, there was a significant decrease in plasma HBV DNA on ART (p < 0.0001, Fig. 4b) but there was no decrease in qHBsAg (p = 0.6705, Fig. 4c). rcDNA was detectable in all individuals pre-ART, which significantly decreased on ART (p = 0.0010, Fig. 4d). cccDNA was detectable in 5/11 individuals pre-ART and in 6/11 individuals on ART with no significant decrease (p = 0.1562, Fig. 4e). Lastly, we compared markers of HBV replication in individuals with detectable versus undetectable HIV DNA in the liver and found no significant differences (Supplementary Figure S8).
Fig. 4.
Virological measures of HBV in plasma and liver. (a) Schematic representation of viral replication of HBV following entry of a hepatocyte after binding to its receptor and the formation of viral intermediates including rcDNA = relaxed circular DNA, cccDNA = covalently closed circular DNA and pgRNA = pre-genomic RNA. HBsAg = HBV surface antigen is produced from the preS1 and preS2 RNA transcripts. pgRNA undergoes reverse transcription to form HBV DNA which is then packaged with viral proteins and then released as a virion from the cell. (b–e) Quantification of HBV DNA (b) and quantitative (q)HBsAg (c) in plasma and HBV relaxed circular (rc)DNA (d) and covalently closed circular (ccc)DNA (e) in liver biopsies prior to initiation of ART (red) and on ART (black). p values were determined by Wilcoxon signed-rank test. n = 18 for panels b and c and n = 11 for panels d and e. Boxes represent range with line at the median. GEq = genome equivalents. ns = not significant. LOQ = limit of quantification. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.
Relationship of HIV persistence and inflammation
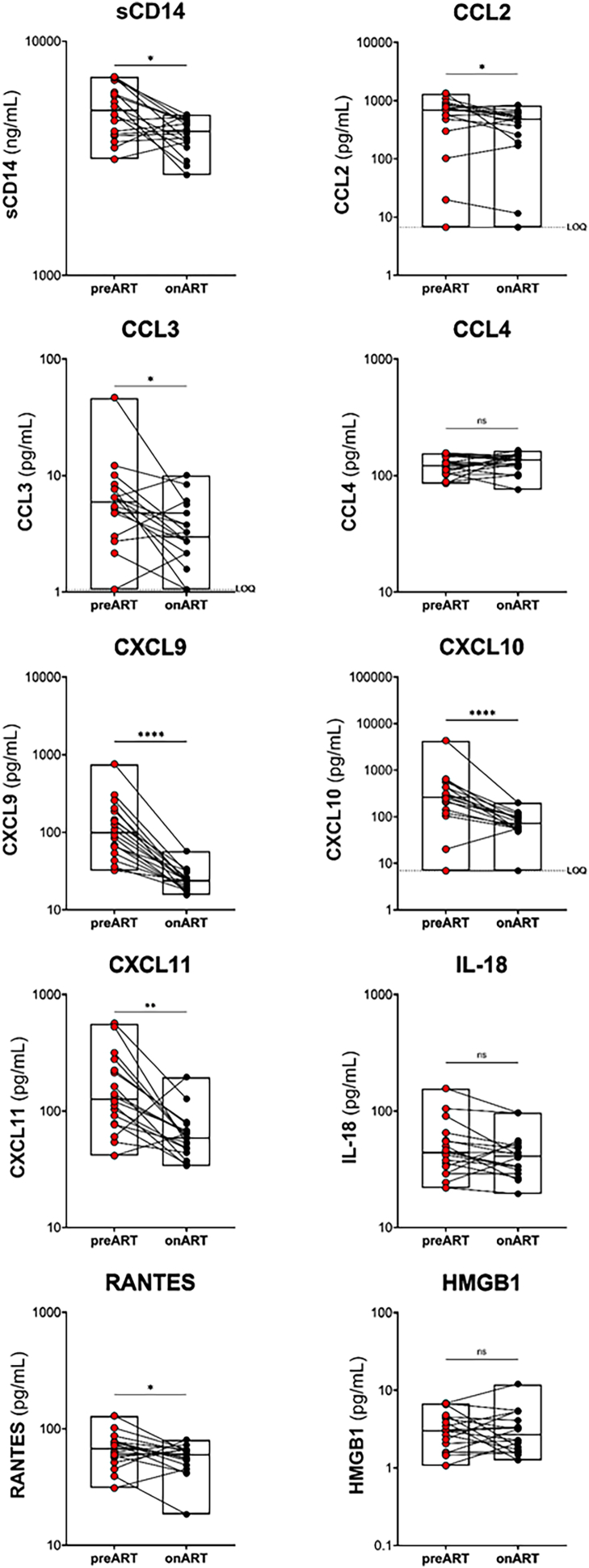
We hypothesised that HIV persistence in the liver could result in inflammation leading to adverse liver outcomes. Therefore, we measured a panel of 10 pro-inflammatory cytokines and chemokines by ELISA or Luminex both prior to and on ART (Supplementary Figure S9). We found that 7 out of 10 markers, including sCD14, CXCL9, CXCL10, CXCL11, RANTES, CCL2 and CCL3 all significantly decreased on ART compared to pre-ART levels (all p values < 0.05, Supplementary Figure S9). The chemokine CCL4, the cytokine interleukin (IL)-18, and high mobility group box 1 (HMGB1), a surrogate marker for tissue damage and inflammation,77 were not significantly changed on ART compared to baseline. These data suggest that global inflammation is decreased in people with HIV and HBV co-infection on HBV-active ART.
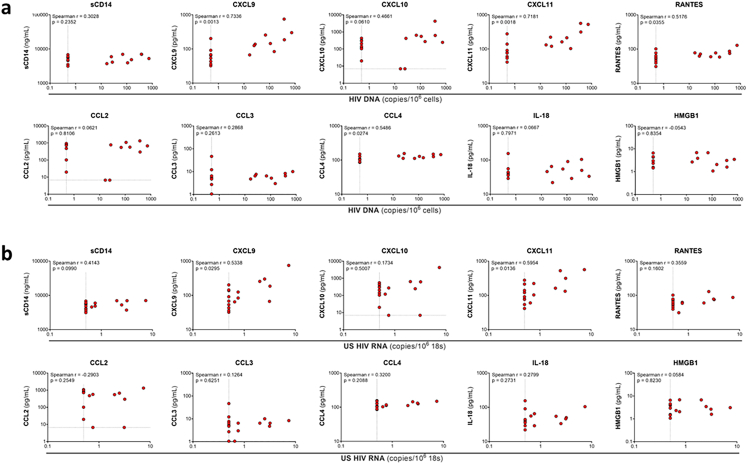
We assessed correlations between HIV DNA (Supplementary Figure S10a) and RNA (Supplementary Figure S10b) in the liver prior to ART with inflammatory cytokine and chemokine levels. We found that HIV DNA in the liver significantly correlated with CXCL9 (Spearman r = 0.7336), CXCL11 (Spearman r = 0.7181), RANTES (Spearman r = 0.5176), and CCL4 (Spearman r = 0.5486, Supplementary Figure S10a). Consistent with findings for HIV DNA, levels of HIV RNA in the liver were significantly correlated with CXCL9 (Spearman r = 0.5338) and CXCL11 (Spearman r = 0.5954, Supplementary Figure S10b). In participants on ART, we compared the levels of pro-inflammatory cytokines and chemokines in individuals with detectable versus undetectable HIV DNA in the liver and found no significant differences (Supplementary Figure S11). These data suggest that while there is an association between HIV DNA or RNA in the liver with circulating inflammation prior to ART, low-level detection of HIV DNA in the liver on ART was not associated with inflammation in people with HIV-HBV co-infection.
Discussion
In this study we examined the liver as a reservoir for HIV on ART in living individuals not undergoing liver transplantation due to decompensated liver disease or hepatocellular carcinoma. Prior to ART, both HIV DNA and RNA were detected in the liver in most participants. In contrast, following ART, HIV RNA was not detected while HIV DNA was detected at very low levels in 67% (12/18) of participants by either qPCR, DNAscope or both techniques. HIV DNA was found in both CD4+ T cells as well as hepatocytes. There was no relationship between detection of HIV DNA following ART and markers of HBV replication or inflammation. There was also no difference in markers of HBV replication or inflammation in participants on HBV-active ART with detectable versus undetectable HIV DNA in the liver.
Although we clearly found HIV DNA in the liver on ART, there are several important caveats to our findings. First, we do not know if the HIV DNA detected was intact or replication-competent. The relevant additional assays such as the intact proviral DNA assay78 or an assessment of inducible virus using either the Tat/Rev induced limiting dilution assay79 or quantitative viral outgrowth assay80 could not be performed using liver biopsy, given the very limited tissue available. We recently analysed blood samples for intact HIV virus from this same cohort, ie people with HIV-HBV co-infection,81 however sequencing virus from liver was not possible, given the very low level of HIV DNA and amount of tissue available. Second, it is possible that the low-level detection of HIV DNA was a laboratory contaminant. We do not believe this is likely given that we clearly showed in 3 participants that the viral sequence was AE, the most dominant subtype in Thailand,82 and a strain that we had not used in our lab when this study was being performed. Finally, although we detected HIV DNA in both CD4+ T cells and hepatocytes, one potential interpretation of our finding is that hepatocytes were not truly infected but had engulfed infected T cells, through a process termed enclysis.83
We recognise that the sample size is small, however, this study is the largest and most comprehensive study evaluating HIV in the liver in PWH on ART. Prior to the introduction of effective ART, there were many studies that looked at HIV in tissues, including liver, using various techniques, such as PCR, immunohistochemistry, and immunofluorescence.7,12,13,15,16,23,31,39, 40, 41, 42 The major strengths of these studies were that they were largely done on autopsy specimens, meaning large tissue samples could be obtained, however, the participants often had AIDS with high levels of HIV RNA in tissues.
Only a few studies have evaluated HIV in the liver in PWH on ART to date. Blackard et al. quantified HIV RNA in liver tissue from 16 participants using qPCR.40 Twelve of those participants were from liver biopsy while alive and four were from autopsy. Only one participant from each group was on suppressive ART while all other participants were viremic. While HIV RNA was detected in the liver in 9/16 participants in this study, HIV RNA was not detected in the liver of the two participants on ART with suppressed plasma HIV RNA.40 Lamers et al. quantified HIV DNA in liver tissue from 18 ART-suppressed individuals at autopsy by qPCR or droplet digital (dd)PCR.44 In this study, 14/18 individuals had detectable HIV DNA in the liver, ranging from 518 to 9722 copies/106 cells. The frequency of HIV DNA in liver tissue in this study, was much higher than what we found in our study, and could potentially be explained by the source of tissue ie liver biopsies versus autopsy specimens. Given the liver is a very vascular organ, a high level of blood contamination can occur when assessing autopsy tissue. Rose et al. measured HIV DNA in liver tissue from five ART-suppressed and two ART-naïve PWH at autopsy by ddPCR.28 Only one ART-naïve individual had detectable HIV in the liver. In a recent report of two individuals, who were on ART for 8 and 11 years and had samples taken at autopsy, the authors detected HIV DNA in the liver of both individuals, including detection of expanded clones.84
In our study, we were able to quantify the level of HIV DNA and RNA in the liver prior to and on ART prospectively and demonstrated that HIV DNA persisted in both CD4+ T cells as well as hepatocytes. Previous publications have clearly shown that macrophages can engulf HIV-infected CD4+ T cells.85,86 We believe our findings support persistent HIV infection in hepatocytes for the following reasons. First, using DNAscope and immunofluorescence imaging, we did not see multinucleated hepatocytes containing HIV DNA, which would suggest engulfment of an infected cell. Second, the HIV DNA stain was a distinct punctate dot in the nuclei of the hepatocytes, further suggesting specific infection of these cells. A future strategy that could be used to determine if HIV infection of hepatocytes in vivo was due to engulfment of an infected T cell or through true infection, is to use laser capture microscopy87 to assess individual cells for HIV together with quantification of rearranged T cell receptor-gamma/delta sequences as previously demonstrated.88 Unfortunately, this was not possible in the current study.
Although we could clearly detect HIV RNA in liver pre-ART, we were unable to detect HIV RNA in any participant on ART using either qPCR or RNAscope. There are several explanations for these findings. First, it is possible that all productively infected or transcriptionally active cells could have been cleared from the liver during ART, although this would be surprising, given that cell-associated HIV RNA was clearly detected in CD4+ T cells in blood on ART in this and many previous studies.46,89, 90, 91, 92, 93, 94, 95, 96 Second, the sensitivity of detection of HIV RNA in liver was greatly reduced compared to blood given the total cell numbers from liver biopsies were limited and we were also using whole liver biopsies, not sorted CD4+ T cells. Third, it is possible that the HIV DNA we detected in liver on ART was all defective and therefore transcription may not have been possible. Finally, it is possible that the liver micro-environment or specifically hepatocytes, in contrast to CD4+ T cells with variable activation states, could favour latency.
We found no relationships between HIV persistence in the liver and markers of HBV replication or liver disease in participants on ART. A possible explanation for our negative findings is that quantification of HIV DNA in liver in PWH on ART was at the very limit of detection and therefore may have not precisely classified participants with and without HIV DNA. Furthermore, we were unable to analyse HIV DNA in in liver biopsies collected on ART as a continuous variable, as we could do with samples collected prior to ART. Better strategies are urgently needed to quantify HIV persistence in tissue, including liver, in PWH on ART.97
There were some limitations in this study to examine the liver as a reservoir for HIV on ART in living people. First, the sensitivity for detection of HIV in liver was greatly reduced given the limited numbers of cells in a standard liver biopsy. A needle biopsy represents approximately 1/50,000th of the total mass of the liver98 and only a limited number of cells could be safely obtained. We were therefore unable to determine if virus was intact or replication-competent. Such studies could really only be done on autopsy tissue, but this approach can be complicated by poor tissue integrity and blood infiltration.99 Alternative approaches may be needed to better characterise HIV infection in the liver. One such approach would be in vivo imaging using radiolabelled antibodies that can detect HIV-infected cells, a technique that our group and others have recently developed.100,101 An alternative but still invasive approach would be to perform fluorescence in situ RNAsequencing, or FISSEQ. This would allow for spatio-temporal sequencing within biopsy or tissue samples, allowing for the identification of infected cells, transcriptionally active cells, gene expression changes within those cells, and the location of infected cells within the tissue itself.102 Second, we were unable to overlay the distribution of infected cells by RNAscope with the ARV spectroscopy as done previously in non-human primate studies after necropsy103 or in human biopsy samples.59 With advances in imaging, this would be an important avenue to pursue. Third, our participants all had HIV-HBV co-infection so it remains unclear if our findings can be translated to HIV mono-infection, however, previous studies of PWH on ART, have demonstrated HIV DNA in the liver at autopsy, consistent with our findings.43, 44, 45, 46 In addition, we recognise that the sample size is small, however, participants were recruited from a single site and therefore likely to harbor similar HBV genotype. Our findings should also be seen in context of the available literature, given there are no similar cohorts ever described for HIV-HBV co-infection and the challenges of obtaining liver biopsy specimens. Finally, given our small sample size, we could at most control for two parameters in a multivariate analysis and as such, multivariate analyses were not performed. Therefore, there may be some confounders that would only be revealed with a larger sample size.
In summary, we showed that HIV DNA can persist in the liver on ART in some individuals. HIV DNA was found in both CD4+ T cells and hepatocytes. Further work is needed to understand the liver as a reservoir for HIV infection in other PWH, including in HIV mono-infection, HIV-hepatitis C co-infection and participants from low- and middle-income countries where there are high rates of inflammation, co-infection and other liver related morbidities.
Contributors
Conceptualisation, MC, KPS, JA, AA, and SRL. Methodology, JMZ, AR, CD, MR, PR, ADMK, and SRL. Investigation, JMZ, KPS, WZ, AR, AD, ST, CT, MLC, ER, CD, HM, and MR. Resources, AA and SRL. Laboratory experimental analyses, JMZ, KPS, WZ, AR, MLC, ER, and CD; Formal analyses, JMZ, KPS, MLC, ER, CD, and JA; Statistical analyses, JMZ, KPS, DJP, SB, and SRL. Writing – original draft, JMZ and SRL. Writing – Review & Editing, all authors read and approved the final draft. Funding acquisition, MC, AA, and SRL. JMZ, KPS, MLC, DJP, SB, JA, and SRL verified the underlying data.
Data sharing statement
Files comprising the complete dataset are currently stored in the online repository for The University of Melbourne <melbourne.figshare.com>, https://doi.org/10.26188/21458415.v1 and https://doi.org/10.26188/21458130.
Declaration of interests
SRL has received consultancy fees from ViiV; and Honoraria from Gilead and Merck. There are no additional conflicts of interest to declare.
Acknowledgments
We thank the participants and the site study team. This work was supported by the National Health and Medical Research Council (NHMRC) of Australia including an NHMRC project grant (SRL, AA, MC, JA), NHMRC program grant (SRL), practitioner fellowship (SRL) and an NHMRC biomedical postgraduate scholarship (KPS). This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health (NIH), under Contract No. 75N91019D00024. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was also supported in part by the University of North Carolina at Chapel Hill Center for AIDS Research (CFAR), an NIH funded program P30 AI050410.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2022.104391.
Appendix A. Supplementary data
Supplementary Figure S1.
Supplementary Figure S2.
Supplementary Figure S3.
Supplementary Figure S4.
Supplementary Figure S5.
Supplementary Figure S6.
Supplementary Figure S7.
Supplementary Figure S8.
Supplementary Figure S9.
Supplementary Figure S10.
Supplementary Figure S11.
Supplementary Figure S12.
References
- 1.Singh K.P., Crane M., Audsley J., Avihingsanon A., Sasadeusz J., Lewin S.R. HIV-hepatitis B virus coinfection: epidemiology, pathogenesis, and treatment. AIDS (London, England) 2017;31(15):2035–2052. doi: 10.1097/QAD.0000000000001574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith C.J., Ryom L., Weber R., et al. Trends in underlying causes of death in people with HIV from 1999 to 2011 (D:A:D): a multicohort collaboration. Lancet. 2014;384(9939):241–248. doi: 10.1016/S0140-6736(14)60604-8. [DOI] [PubMed] [Google Scholar]
- 3.Boyd A., Gozlan J., Maylin S., et al. Persistent viremia in human immunodeficiency virus/hepatitis B coinfected patients undergoing long-term tenofovir: virological and clinical implications. Hepatology. 2014;60(2):497–507. doi: 10.1002/hep.27182. [DOI] [PubMed] [Google Scholar]
- 4.Rajbhandari R., Jun T., Khalili H., Chung R.T., Ananthakrishnan A.N. HBV/HIV coinfection is associated with poorer outcomes in hospitalized patients with HBV or HIV. J Viral Hepat. 2016;23(10):820–829. doi: 10.1111/jvh.12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weber R., Ruppik M., Rickenbach M., et al. Decreasing mortality and changing patterns of causes of death in the Swiss HIV Cohort Study. HIV Med. 2013;14(4):195–207. doi: 10.1111/j.1468-1293.2012.01051.x. [DOI] [PubMed] [Google Scholar]
- 6.Coffin C.S., Osiowy C., Myers R.P., Gill M.J. Virology and clinical sequelae of long-term antiviral therapy in a North American cohort of hepatitis B virus (HBV)/human immunodeficiency virus type 1 (HIV-1) co-infected patients. J Clin Virol. 2013;57(2):103–108. doi: 10.1016/j.jcv.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Hufert F.T., Schmitz J., Schreiber M., Schmitz H., Racz P., von Laer D.D. Human Kupffer cells infected with HIV-1 in vivo. J Acquir Immune Defic Syndr. 1993;6(7):772–777. [PubMed] [Google Scholar]
- 8.Mosoian A., Zhang L., Hong F., et al. Frontline science: HIV infection of Kupffer cells results in an amplified proinflammatory response to LPS. J Leukoc Biol. 2017;101(5):1083–1090. doi: 10.1189/jlb.3HI0516-242R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gendrault J.L., Steffan A.M., Schmitt M.P., Jaeck D., Aubertin A.M., Kirn A. Interaction of cultured human Kupffer cells with HIV-infected CEM cells: an electron microscopic study. Pathobiology. 1991;59(4):223–226. doi: 10.1159/000163650. [DOI] [PubMed] [Google Scholar]
- 10.Schmitt M.P., Gendrault J.L., Schweitzer C., et al. Permissivity of primary cultures of human Kupffer cells for HIV-1. AIDS Res Hum Retroviruses. 1990;6(8):987–991. doi: 10.1089/aid.1990.6.987. [DOI] [PubMed] [Google Scholar]
- 11.Schmitt M.P., Steffan A.M., Gendrault J.L., et al. Multiplication of human immunodeficiency virus in primary cultures of human Kupffer cells--possible role of liver macrophage infection in the physiopathology of AIDS. Res Virol. 1990;141(2):143–152. doi: 10.1016/0923-2516(90)90016-c. [DOI] [PubMed] [Google Scholar]
- 12.Housset C., Boucher O., Girard P.M., et al. Immunohistochemical evidence for human immunodeficiency virus-1 infection of liver Kupffer cells. Hum Pathol. 1990;21(4):404–408. doi: 10.1016/0046-8177(90)90202-g. [DOI] [PubMed] [Google Scholar]
- 13.Cao Y.Z., Dieterich D., Thomas P.A., Huang Y.X., Mirabile M., Ho D.D. Identification and quantitation of HIV-1 in the liver of patients with AIDS. AIDS (London, England) 1992;6(1):65–70. doi: 10.1097/00002030-199201000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Kandathil A.J., Sugawara S., Goyal A., et al. No recovery of replication-competent HIV-1 from human liver macrophages. J Clin Invest. 2018;128(10):4501–4509. doi: 10.1172/JCI121678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoda S.A., White J.E., Gerber M.A. Immunohistochemical studies of human immunodeficiency virus-1 in liver tissues of patients with AIDS. Mod Pathol. 1991;4(5):578–581. [PubMed] [Google Scholar]
- 16.Jiang T.J., Zhao M., Zhao J.M., et al. Immunohistochemical evidence for HIV-1 infection in the liver of HIV-infected patientsZhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi. 2005;19(2):152–154. [PubMed] [Google Scholar]
- 17.Steffan A.M., Lafon M.E., Gendrault J.L., et al. Primary cultures of endothelial cells from the human liver sinusoid are permissive for human immunodeficiency virus type 1. Proc Natl Acad Sci U S A. 1992;89(5):1582–1586. doi: 10.1073/pnas.89.5.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tuyama A.C., Hong F., Saiman Y., et al. Human immunodeficiency virus (HIV)-1 infects human hepatic stellate cells and promotes collagen I and monocyte chemoattractant protein-1 expression: implications for the pathogenesis of HIV/hepatitis C virus-induced liver fibrosis. Hepatology. 2010;52(2):612–622. doi: 10.1002/hep.23679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cao Y.Z., Friedman-Kien A.E., Huang Y.X., et al. CD4-independent, productive human immunodeficiency virus type 1 infection of hepatoma cell lines in vitro. J Virol. 1990;64(6):2553–2559. doi: 10.1128/jvi.64.6.2553-2559.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iser D.M., Warner N., Revill P.A., et al. Coinfection of hepatic cell lines with human immunodeficiency virus and hepatitis B virus leads to an increase in intracellular hepatitis B surface antigen. J Virol. 2010;84(12):5860–5867. doi: 10.1128/JVI.02594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kong L., Cardona Maya W., Moreno-Fernandez M.E., et al. Low-level HIV infection of hepatocytes. Virol J. 2012;9:157. doi: 10.1186/1743-422X-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao P., Usami O., Suzuki Y., et al. Characterization of a CD4-independent clinical HIV-1 that can efficiently infect human hepatocytes through chemokine (C-X-C motif) receptor 4. AIDS. 2008;22(14):1749–1757. doi: 10.1097/QAD.0b013e328308937c. [DOI] [PubMed] [Google Scholar]
- 23.Housset C., Lamas E., Courgnaud V., et al. Presence of HIV-1 in human parenchymal and non-parenchymal liver cells in vivo. J Hepatol. 1993;19(2):252–258. doi: 10.1016/s0168-8278(05)80579-3. [DOI] [PubMed] [Google Scholar]
- 24.Pitman M.C., Lau J.S.Y., McMahon J.H., Lewin S.R. Barriers and strategies to achieve a cure for HIV. Lancet HIV. 2018;5(6):e317–e328. doi: 10.1016/S2352-3018(18)30039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong M.E., Jaworowski A., Hearps A.C. The HIV reservoir in monocytes and macrophages. Front Immunol. 2019;10:1435. doi: 10.3389/fimmu.2019.01435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honeycutt J.B., Thayer W.O., Baker C.E., et al. HIV persistence in tissue macrophages of humanized myeloid-only mice during antiretroviral therapy. Nat Med. 2017;23(5):638–643. doi: 10.1038/nm.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nolan D.J., Rose R., Rodriguez P.H., et al. The spleen is an HIV-1 sanctuary during combined antiretroviral therapy. AIDS Res Hum Retroviruses. 2018;34(1):123–125. doi: 10.1089/aid.2017.0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brese R.L., Gonzalez-Perez M.P., Koch M., et al. Ultradeep single-molecule real-time sequencing of HIV envelope reveals complete compartmentalization of highly macrophage-tropic R5 proviral variants in brain and CXCR4-using variants in immune and peripheral tissues. J Neurovirol. 2018;24(4):439–453. doi: 10.1007/s13365-018-0633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ko A., Kang G., Hattler J.B., et al. Macrophages but not astrocytes harbor HIV DNA in the brains of HIV-1-infected aviremic individuals on suppressive antiretroviral therapy. J Neuroimmune Pharmacol. 2019;14(1):110–119. doi: 10.1007/s11481-018-9809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamers S.L., Rose R., Ndhlovu L.C., et al. The meningeal lymphatic system: a route for HIV brain migration? J Neurovirol. 2016;22(3):275–281. doi: 10.1007/s13365-015-0399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rose R., Lamers S.L., Nolan D.J., et al. HIV maintains an evolving and dispersed population in multiple tissues during suppressive combined antiretroviral therapy in individuals with cancer. J Virol. 2016;90(20):8984–8993. doi: 10.1128/JVI.00684-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tso F.Y., Kang G., Kwon E.H., et al. Brain is a potential sanctuary for subtype C HIV-1 irrespective of ART treatment outcome. PLoS One. 2018;13(7) doi: 10.1371/journal.pone.0201325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamers S.L., Salemi M., Galligan D.C., et al. Human immunodeficiency virus-1 evolutionary patterns associated with pathogenic processes in the brain. J Neurovirol. 2010;16(3):230–241. doi: 10.3109/13550281003735709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganor Y., Real F., Sennepin A., et al. HIV-1 reservoirs in urethral macrophages of patients under suppressive antiretroviral therapy. Nat Microbiol. 2019;4(4):633–644. doi: 10.1038/s41564-018-0335-z. [DOI] [PubMed] [Google Scholar]
- 35.Zalar A., Figueroa M.I., Ruibal-Ares B., et al. Macrophage HIV-1 infection in duodenal tissue of patients on long term HAART. Antiviral Res. 2010;87(2):269–271. doi: 10.1016/j.antiviral.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Deleage C., Moreau M., Rioux-Leclercq N., Ruffault A., Jegou B., Dejucq-Rainsford N. Human immunodeficiency virus infects human seminal vesicles in vitro and in vivo. Am J Pathol. 2011;179(5):2397–2408. doi: 10.1016/j.ajpath.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cribbs S.K., Lennox J., Caliendo A.M., Brown L.A., Guidot D.M. Healthy HIV-1-infected individuals on highly active antiretroviral therapy harbor HIV-1 in their alveolar macrophages. AIDS Res Hum Retroviruses. 2015;31(1):64–70. doi: 10.1089/aid.2014.0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spudich S., Robertson K.R., Bosch R.J., et al. Persistent HIV-infected cells in cerebrospinal fluid are associated with poorer neurocognitive performance. J Clin Invest. 2019;129(8):3339–3346. doi: 10.1172/JCI127413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van't Wout A.B., Ran L.J., Kuiken C.L., Kootstra N.A., Pals S.T., Schuitemaker H. Analysis of the temporal relationship between human immunodeficiency virus type 1 quasispecies in sequential blood samples and various organs obtained at autopsy. J Virol. 1998;72(1):488–496. doi: 10.1128/jvi.72.1.488-496.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blackard J.T., Ma G., Martin C.M., Rouster S.D., Shata M.T., Sherman K.E. HIV variability in the liver and evidence of possible compartmentalization. AIDS Res Hum Retroviruses. 2011;27(10):1117–1126. doi: 10.1089/aid.2010.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Donaldson Y.K., Bell J.E., Ironside J.W., et al. Redistribution of HIV outside the lymphoid system with onset of AIDS. Lancet. 1994;343(8894):383–385. doi: 10.1016/s0140-6736(94)91222-x. [DOI] [PubMed] [Google Scholar]
- 42.Lamers S.L., Salemi M., Galligan D.C., et al. Extensive HIV-1 intra-host recombination is common in tissues with abnormal histopathology. PLoS One. 2009;4(3) doi: 10.1371/journal.pone.0005065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chaillon A., Gianella S., Dellicour S., et al. HIV persists throughout deep tissues with repopulation from multiple anatomical sources. J Clin Invest. 2020;130(4):1699–1712. doi: 10.1172/JCI134815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamers S.L., Rose R., Maidji E., et al. HIV DNA is frequently present within pathologic tissues evaluated at autopsy from combined antiretroviral therapy-treated patients with undetectable viral loads. J Virol. 2016;90(20):8968–8983. doi: 10.1128/JVI.00674-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Estes J.D., Kityo C., Ssali F., et al. Defining total-body AIDS-virus burden with implications for curative strategies. Nat Med. 2017;23(11):1271–1276. doi: 10.1038/nm.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moron-Lopez S., Xie G., Kim P., et al. Tissue-specific differences in HIV DNA levels and mechanisms that govern HIV transcription in blood, gut, genital tract and liver in ART-treated women. J Int AIDS Soc. 2021;24(7) doi: 10.1002/jia2.25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crane M., Oliver B., Matthews G., et al. Immunopathogenesis of hepatic flare in HIV/hepatitis B virus (HBV)-coinfected individuals after the initiation of HBV-active antiretroviral therapy. J Infect Dis. 2009;199(7):974–981. doi: 10.1086/597276. [DOI] [PubMed] [Google Scholar]
- 48.Oo Y.H., Adams D.H. The role of chemokines in the recruitment of lymphocytes to the liver. J Autoimmun. 2010;34(1):45–54. doi: 10.1016/j.jaut.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 49.Pellicoro A., Ramachandran P., Iredale J.P., Fallowfield J.A. Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat Rev Immunol. 2014;14(3):181–194. doi: 10.1038/nri3623. [DOI] [PubMed] [Google Scholar]
- 50.Mak K.M., Shin D.W. Hepatic sinusoids versus central veins: structures, markers, angiocrines, and roles in liver regeneration and homeostasis. Anat Rec. 2021;304:1661–1691. doi: 10.1002/ar.24560. [DOI] [PubMed] [Google Scholar]
- 51.Cantero-Perez J., Grau-Exposito J., Serra-Peinado C., et al. Resident memory T cells are a cellular reservoir for HIV in the cervical mucosa. Nat Commun. 2019;10(1):4739. doi: 10.1038/s41467-019-12732-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson J.L., Khoury G., Fromentin R., et al. Human immunodeficiency virus (HIV)-infected CCR6+ rectal CD4+ T cells and HIV persistence on antiretroviral therapy. J Infect Dis. 2020;221(5):744–755. doi: 10.1093/infdis/jiz509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun H., Kim D., Li X., et al. Th1/17 polarization of CD4 T cells supports HIV-1 persistence during antiretroviral therapy. J Virol. 2015;89(22):11284–11293. doi: 10.1128/JVI.01595-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gosselin A., Monteiro P., Chomont N., et al. Peripheral blood CCR4+CCR6+ and CXCR3+CCR6+CD4+ T cells are highly permissive to HIV-1 infection. J Immunol. 2010;184(3):1604–1616. doi: 10.4049/jimmunol.0903058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Osborne O., Peyravian N., Nair M., Daunert S., Toborek M. The paradox of HIV blood-brain barrier penetrance and antiretroviral drug delivery deficiencies. Trends Neurosci. 2020;43(9):695–708. doi: 10.1016/j.tins.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang Y., Hoque M.T., Jenabian M.A., et al. Antiretroviral drug transporters and metabolic enzymes in human testicular tissue: potential contribution to HIV-1 sanctuary site. J Antimicrob Chemother. 2016;71(7):1954–1965. doi: 10.1093/jac/dkw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fukazawa Y., Lum R., Okoye A.A., et al. B cell follicle sanctuary permits persistent productive simian immunodeficiency virus infection in elite controllers. Nat Med. 2015;21(2):132–139. doi: 10.1038/nm.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fletcher C.V., Staskus K., Wietgrefe S.W., et al. Persistent HIV-1 replication is associated with lower antiretroviral drug concentrations in lymphatic tissues. Proc Natl Acad Sci U S A. 2014;111(6):2307–2312. doi: 10.1073/pnas.1318249111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thompson C.G., Rosen E.P., Prince H.M.A., et al. Heterogeneous antiretroviral drug distribution and HIV/SHIV detection in the gut of three species. Sci Transl Med. 2019;11(499):eaap8758. doi: 10.1126/scitranslmed.aap8758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee S.A., Telwatte S., Hatano H., et al. Antiretroviral therapy concentrations differ in gut vs. lymph node tissues and are associated with HIV viral transcription by a novel RT-ddPCR assay. J Acquir Immune Defic Syndr. 2020;83(5):530–537. doi: 10.1097/QAI.0000000000002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh K.P., Zerbato J.M., Zhao W., et al. Intrahepatic CXCL10 is strongly associated with liver fibrosis in HIV-hepatitis B co-infection. PLoS Pathog. 2020;16(9) doi: 10.1371/journal.ppat.1008744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Elliott J.H., Wightman F., Solomon A., et al. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 2014;10(10) doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hooker D.J., Mobarok M., Anderson J.L., et al. A new way of measuring apoptosis by absolute quantitation of inter-nucleosomally fragmented genomic DNA. Nucleic Acids Res. 2012;40(15):e113. doi: 10.1093/nar/gks334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lewin S.R., Vesanen M., Kostrikis L., et al. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J Virol. 1999;73(7):6099–6103. doi: 10.1128/jvi.73.7.6099-6103.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van der Sluis R.M., Zerbato J.M., Rhodes J.W., et al. Diverse effects of interferon alpha on the establishment and reversal of HIV latency. PLoS Pathog. 2020;16(2) doi: 10.1371/journal.ppat.1008151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bowden S., Jackson K., Littlejohn M., Locarnini S. Quantification of HBV covalently closed circular DNA from liver tissue by real-time PCR. Methods Mol Med. 2004;95:41–50. doi: 10.1385/1-59259-669-X:41. [DOI] [PubMed] [Google Scholar]
- 67.Deleage C., Wietgrefe S.W., Del Prete G., et al. Defining HIV and SIV reservoirs in lymphoid tissues. Pathog Immun. 2016;1(1):68–106. doi: 10.20411/pai.v1i1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brands C., Morcock D., Estes J., Deleage C. Next-generation viral RNA/DNA in situ hybridization applications in human immunodeficiency virus/simian immunodeficiency virus research. J Vis Exp. 2020;(160) doi: 10.3791/60318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bokhart M., Rosen E., Thompson C., Sykes C., Kashuba A.M., Muddiman D. Quantitative mass spectrometry imaging of emtricitabine in cervical tissue model using infrared matrix-assisted laser desorption electrospray ionization. Anal Bioanal Chem. 2015;407(8):2073–2084. doi: 10.1007/s00216-014-8220-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson C.G., Bokhart M.T., Sykes C., et al. Mass spectrometry imaging reveals heterogeneous efavirenz distribution within putative HIV reservoirs. Antimicrob Agents Chemother. 2015;59(5):2944–2948. doi: 10.1128/AAC.04952-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thurman A.R., Schwartz J.L., Cottrell M.L., et al. Safety and pharmacokinetics of a tenofovir alafenamide fumarate-emtricitabine based oral antiretroviral regimen for prevention of HIV acquisition in women: a randomized controlled trial. EClinicalMedicine. 2021;36 doi: 10.1016/j.eclinm.2021.100893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang J.H., Davis N.L., Corbett A.H., et al. Effect of efavirenz on levonorgestrel concentrations among Malawian levonorgestrel implant users for up to 30 months of concomitant use: a subanalysis of a randomized clinical trial. Contracept X. 2020;2 doi: 10.1016/j.conx.2020.100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sozzi V., Walsh R., Littlejohn M., et al. In vitro studies show that sequence variability contributes to marked variation in hepatitis B virus replication, protein expression, and function observed across genotypes. J Virol. 2016;90(22):10054–10064. doi: 10.1128/JVI.01293-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bedossa P., Poynard T., group M. An algorithm for the grading of activity in chronic hepatitis. Hepatology. 1996;24:289–293. doi: 10.1002/hep.510240201. [DOI] [PubMed] [Google Scholar]
- 75.Miailhes P., Pradat P., Chevallier M., et al. Proficiency of transient elastography compared to liver biopsy for the assessment of fibrosis in HIV/HBV-coinfected patients. J Viral Hepat. 2011;18(1):61–69. doi: 10.1111/j.1365-2893.2010.01275.x. [DOI] [PubMed] [Google Scholar]
- 76.Mavigner M., Habib J., Deleage C., et al. Simian immunodeficiency virus persistence in cellular and anatomic reservoirs in antiretroviral therapy-suppressed infant rhesus macaques. J Virol. 2018;92(18) doi: 10.1128/JVI.00562-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bianchi M.E., Crippa M.P., Manfredi A.A., Mezzapelle R., Rovere Querini P., Venereau E. High-mobility group box 1 protein orchestrates responses to tissue damage via inflammation, innate and adaptive immunity, and tissue repair. Immunol Rev. 2017;280(1):74–82. doi: 10.1111/imr.12601. [DOI] [PubMed] [Google Scholar]
- 78.Bruner K.M., Wang Z., Simonetti F.R., et al. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature. 2019;566(7742):120–125. doi: 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Procopio F.A., Fromentin R., Kulpa D.A., et al. A novel assay to measure the magnitude of the inducible viral reservoir in HIV-infected individuals. EBioMedicine. 2015;2(8):874–883. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Siliciano J.D., Siliciano R.F. Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol Biol. 2005;304:3–15. doi: 10.1385/1-59259-907-9:003. [DOI] [PubMed] [Google Scholar]
- 81.Wang X.Q., Zerbato J.M., Avihingsanon A., et al. Markers of immune activation and inflammation are associated with higher levels of genetically-intact HIV in HIV-HBV co-infected individuals. J Virol. 2022;96(16) doi: 10.1128/jvi.00588-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wirachsilp P., Kantakamalakul W., Foongladda S., et al. Surveillance of subtype and genetic variation of the circulating strains of HIV-1 in Thailand. Southeast Asian J Trop Med Public Health. 2007;38(5):814–827. [PubMed] [Google Scholar]
- 83.Davies S.P., Reynolds G.M., Wilkinson A.L., et al. Hepatocytes delete regulatory T cells by enclysis, a CD4(+) T cell engulfment process. Cell Rep. 2019;29(6):1610–1620.e4. doi: 10.1016/j.celrep.2019.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dufour C., Ruiz M., Pagliuzza A., et al. 2022. Expansion and extensive recirculation of HIV-infected cells in multiple organs, ABS 67. Conference on Retroviruses and Opportunistic Infections (CROI); Virtual. [Google Scholar]
- 85.Baxter A.E., Russell R.A., Duncan C.J., et al. Macrophage infection via selective capture of HIV-1-infected CD4+ T cells. Cell Host Microbe. 2014;16(6):711–721. doi: 10.1016/j.chom.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Calantone N., Wu F., Klase Z., et al. Tissue myeloid cells in SIV-infected primates acquire viral DNA through phagocytosis of infected T cells. Immunity. 2014;41(3):493–502. doi: 10.1016/j.immuni.2014.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Churchill M.J., Gorry P.R., Cowley D., et al. Use of laser capture microdissection to detect integrated HIV-1 DNA in macrophages and astrocytes from autopsy brain tissues. J Neurovirol. 2006;12(2):146–152. doi: 10.1080/13550280600748946. [DOI] [PubMed] [Google Scholar]
- 88.DiNapoli S.R., Ortiz A.M., Wu F., et al. Tissue-resident macrophages can contain replication-competent virus in antiretroviral-naive, SIV-infected Asian macaques. JCI Insight. 2017;2(4) doi: 10.1172/jci.insight.91214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zerbato J.M., Khoury G., Zhao W., et al. Multiply spliced HIV RNA is a predictive measure of virus production ex vivo and in vivo following reversal of HIV latency. EBioMedicine. 2021;65 doi: 10.1016/j.ebiom.2021.103241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yukl S.A., Kaiser P., Kim P., et al. HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med. 2018;10(430) doi: 10.1126/scitranslmed.aap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lassen K.G., Bailey J.R., Siliciano R.F. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. J Virol. 2004;78(17):9105–9114. doi: 10.1128/JVI.78.17.9105-9114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ishizaka A., Sato H., Nakamura H., et al. Short intracellular HIV-1 transcripts as biomarkers of residual immune activation in patients on antiretroviral therapy. J Virol. 2016;90(12):5665–5676. doi: 10.1128/JVI.03158-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pasternak A.O., de Bruin M., Jurriaans S., et al. Modest nonadherence to antiretroviral therapy promotes residual HIV-1 replication in the absence of virological rebound in plasma. J Infect Dis. 2012;206(9):1443–1452. doi: 10.1093/infdis/jis502. [DOI] [PubMed] [Google Scholar]
- 94.Pasternak A.O., Jurriaans S., Bakker M., Prins J.M., Berkhout B., Lukashov V.V. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS One. 2009;4(12) doi: 10.1371/journal.pone.0008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaiser P., Joos B., Niederost B., Weber R., Gunthard H.F., Fischer M. Productive human immunodeficiency virus type 1 infection in peripheral blood predominantly takes place in CD4/CD8 double-negative T lymphocytes. J Virol. 2007;81(18):9693–9706. doi: 10.1128/JVI.00492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kiselinova M., Pasternak A.O., De Spiegelaere W., Vogelaers D., Berkhout B., Vandekerckhove L. Comparison of droplet digital PCR and seminested real-time PCR for quantification of cell-associated HIV-1 RNA. PLoS One. 2014;9(1) doi: 10.1371/journal.pone.0085999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Deeks S.G., Archin N., Cannon P., et al. Research priorities for an HIV cure: International AIDS Society Global Scientific Strategy 2021. Nat Med. 2021;27(12):2085–2098. doi: 10.1038/s41591-021-01590-5. [DOI] [PubMed] [Google Scholar]
- 98.Bravo A.A., Sheth S.G., Chopra S. Liver biopsy. N Engl J Med. 2001;344(7):495–500. doi: 10.1056/NEJM200102153440706. [DOI] [PubMed] [Google Scholar]
- 99.Maldarelli F. The gift of a lifetime: analysis of HIV at autopsy. J Clin Invest. 2020;130(4):1611–1614. doi: 10.1172/JCI135905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McMahon J.H., Zerbato J.M., Lau J.S.Y., et al. A clinical trial of non-invasive imaging with an anti-HIV antibody labelled with copper-64 in people living with HIV and uninfected controls. EBioMedicine. 2021;65 doi: 10.1016/j.ebiom.2021.103252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Santangelo P.J., Rogers K.A., Zurla C., et al. Whole-body immunoPET reveals active SIV dynamics in viremic and antiretroviral therapy-treated macaques. Nat Methods. 2015;12(5):427–432. doi: 10.1038/nmeth.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee J.H., Daugharthy E.R., Scheiman J., et al. Fluorescent in situ sequencing (FISSEQ) of RNA for gene expression profiling in intact cells and tissues. Nat Protoc. 2015;10(3):442–458. doi: 10.1038/nprot.2014.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Scholz E.M.B., Mwangi J.N., De la Cruz G., et al. Quantitative imaging analysis of the spatial relationship between antiretrovirals, reverse transcriptase simian-human immunodeficiency virus RNA, and collagen in the mesenteric lymph nodes of nonhuman primates. Antimicrob Agents Chemother. 2021;65(6) doi: 10.1128/AAC.00019-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.