Highlights
-
•
NaB was a differential metabolite that affected the efficacy of OXA.
-
•
NaB can inhibit the proliferation, invasion and migration and induce the apoptosis of CRC cells.
-
•
Sodium butyrate combined with oxaliplatin has synergistic effect on CRC cells.
Keywords: Sodium butyrate, Oxaliplatin, Colorectal cancer, Synergistic effect, Clinical adjuvant
Abbreviations: OXA, Oxaliplatin; CRC, Colorectal cancer; NaB, Sodium butyrate; GC/MS, Gas chromatography–mass spectrometry; CCK-8, Cell counting kit-8; NCCN, National Comprehensive Cancer Network; DACH, Diaminocyclohexane; ICD, Immunogenic cell death; PD-L2, Programmed death ligand 2; CRT, Calreticulin; HMGB-1, High mobility group box 1; DC, Dendritic cells; DFS, Progression-free survival; OS, Overall survival; mCRC, metastatic colorectal cancer; SCFA, Short-chain-fatty acids
Abstract
Background
Oxaliplatin (OXA) is a chemotherapy agent commonly used in the treatment of colorectal cancer (CRC). Sodium butyrate (NaB) has an antitumor effect.
Methods
In total, 30 patients in stage III who completed 8 cycles of chemotherapy regimens were recruited for this study. The patients were divided into good and bad groups based on the chemotherapy efficacy. Gas chromatography–mass spectrometry (GC/MS) was used to detect microbial metabolites in stool samples from CRC patients. Cell counting kit-8 (CCK-8), Annexin-V APC/7-AAD double staining, Transwell assays, scratch-wound assays, and EdU assays were used to detect cell proliferation, apoptosis, invasion and migration, respectively. Fluoroelectron microscopy was used to observe the cell structures. To verify the inhibitory effect of NaB and OXA at animal level, a subcutaneous transplanted tumor model was established. Finally, 16S sequencing technology was used to detect intestinal bacteria. GC–MS was used to detect metabolites in mouse stools.
Results
NaB was a differential metabolite that affected the efficacy of OXA. NAB and oxaliplatin can synergically inhibit cell proliferation, migration and invasion, and induce cell apoptosis. Animal experiments confirmed the inhibitory effect of oxaliplatin and sodium butyrate on tumor in mice. In addition, the intestinal microbe detection and microbial metabolite detection in fecal samples from mice showed significant differences between butyrate-producing bacteria and NaB.
Conclusion
NaB and OXA can synergistically inhibit the proliferation, invasion and metastasis of CRC cells and promote the apoptosis of CRC cells. NaB, as an OXA synergist, has the potential to become a new clinical adjuvant in CRC chemotherapy.
Introduction
CRC is the most common gastrointestinal malignancy and the second leading cause of cancer-related death worldwide [1]. The incidence of CRC is increasing worldwide. It is expected that by 2030, the number of new cases will reach more than 2.2 million, and the number of deaths will reach 1.1 million [2], posing a serious threat to human life and health. Although therapeutic surgery is the initial treatment for CRC patients, most patients have advanced CRC secondary to unresectable and disseminated disease; therefore, systemic chemotherapy has become the main treatment method [3]. The National Comprehensive Cancer Network (NCCN) guidelines recommend that patients with advanced CRC receive a comprehensive treatment regimen with surgery as the mainstay and chemotherapy as the supplement. Oxaliplatin, as an alkylating agent, is the third generation of platinum drugs used in anti-tumor therapy. It has the molecular formula 1, 2-diaminocyclohexane (DACH), forms DACH-DNA adducts, and generates interstrand or intrastrand cross-links. With the same effect as other platinum drugs, they all target DNA, and platinum atoms form cross-blinding with DNA to antagonize its replication and transcription [4]. However, currently, more and more evidence has shown that the anti-tumor activity of OXA is more importantly related to its immune effects. (1) Regulation of immune microenvironment. In addition to inducing immunogenic cell death (ICD) and acting on STAT signaling pathway, oxaliplatin can regulate and change the tumor microenvironment and enhance the effect of tumor immune response [5,6]. In this process, the driving factors play an indispensable role. (2) Action on the STAT protein signaling pathway. The JAK-STAT signaling pathway, which is mainly composed of the JAK tyrosine kinase family and the STAT transcription factor family, is an important link in the regulation of immune response. The binding to the corresponding receptors can induce the phosphorylation of STAT6 and then activate the expression of programmed death ligand 2 (PD-L2), which has an inhibitory effect on T cells. Studies have shown that platinum can inhibit the phosphorylation of STAT6 and down-regulate the expression of PD-L2 in DC and tumor cells, thereby enhancing the activation of T cells and enhancing the lytic effect of killer T cells on tumor cells [7]. (3) Induction of ICD. After entering cells, oxaliplatin can interact with a variety of proteins, such as calreticulin (CRT) in the endoplasmic reticulum lumen and high mobility group box 1 (HMGB-1) in the nucleus, to regulate the key links of ICD. For example, after oxaliplatin enters tumor cells, it can cause ER stress, the translocation of CRT to the cell membrane, exposure to the tumor cell surface, and cause the phagocytosis of tumor cells by dendritic cells (DC) and macrophages [8,9]. FOLFOX and CapeOX with OXA were established as first-line chemotherapy in the 2017 NCCN guidelines. Approximately 50% of CRC patients benefit from OXA, and the median survival of CRC patients receiving oxaliplatin is significantly improved [10,11]. Therefore, OXA plays an important role in the treatment of CRC.
Among stage II and III CRC patients with high risk factors, the 3-year progression-free survival (DFS) and overall survival (OS) of patients who received first-line chemotherapy with OXA after surgery were significantly improved [12]. However, OXA is affected by various factors and can cause side effects, including neurotoxic reactions, gastrointestinal reactions and hematological toxicity, with increasing drug doses [13]. Among them, neurotoxicity is dose-limiting toxicity and is considered reversible peripheral paresthesia related to the cumulative dose that is triggered or aggravated by cold [14]. OXA is recommended as adjuvant CRC treatment (85 mg/m2 (iv drip), repeated every 2 weeks, with a maximum tolerated dose of 100 mg/m2 and a median cumulative dose of 810 mg/m2) and may increase the risk of early recurrence when the dose is reduced to reduce side effects [15,16]. In addition, chemotherapy resistance is a factor that cannot be ignored. The median tumor-free survival of patients with metastatic colorectal cancer (mCRC) is only 9.5 months [17]. The main factor affecting prognosis is whether tumor cells are resistant to chemotherapy, including congenital or acquired drug resistance. Approximately 40%-50% of stage II and III CRC patients using OXA develop chemotherapy resistance and relapse during treatment [18]. Therefore, the identification of a new adjuvant drug to improve the efficacy of OXA and reduce drug toxicity and side effects is an urgent issue in the treatment of CRC.
As the fecal storage organ, the gut is home to the largest microbial community in the human body, with a total of 1014 bacteria, accounting for 98.8% of the total bacterial community in the human body [19]. Interactions exist among intestinal microbial communities, such as competition, predation, symbiosis and cooperation [20,21]. The intestinal flora and its metabolites participate in the occurrence and development of CRC [22]. Butyrate-producing bacteria can stimulate the immune activity of the intestinal mucosa, inhibit the secretion of proinflammatory factors, promote the immune regulation of the intestinal microecology, and maintain the balance of the intestinal environment [23], [24], [25]. Butyric acid, a metabolite of intestinal flora, is a metabolite produced by butyric acid-producing bacteria, such as Ruminococcaceae, Butyricicoccus pullicaecorum and Clostridium leptum, during anaerobic fermentation of dietary fiber. Butyric acid can maintain the stability of the intestinal microecological environment [26]. Butyric acid exists stably in the intestinal tract in the form of NaB, and various foods, such as low-fat, low-protein, low-carbohydrate and high-cellulose foods, can promote butyric acid secretion and NaB production through microbial metabolism. NaB is a short-chain fatty acid produced by the intestinal flora during the anaerobic fermentation of dietary fiber. NaB can inhibit the occurrence and development of CRC through immune mechanisms, molecular signaling pathways and epigenetics [26]. In contrast to the low concentration of NaB in the colon, NaB has important biological activity in intestinal mucosal cells [27]. Multiple studies have shown that compared with healthy people, the content of NaB in stool samples from CRC patients is significantly reduced [28,29]. It has been proven that butyric acid can inhibit the proliferation, invasion and migration of CRC cells [30]. It has also been shown that NaB has antitumor properties to a certain extent and the potential to become an antitumor adjuvant new drug.
To further explore whether NaB can be used as an adjuvant drug to increase the chemotherapy efficacy of OXA and clarify the relationship between NaB and OXA, this study screened NaB as a differential metabolite affecting the chemotherapy efficacy of OXA through a clinical study. In addition, the relationship between NaB and OXA was further confirmed in cell function experiments and animal experiments. This study is expected to provide a theoretical basis for the clinical application of NaB as an adjuvant for OXA chemotherapy in CRC.
Material and methods
Population and disease progression assessment of the participants
CRC patients admitted to Huzhou Central Hospital from October 2019 to December 2020 were recruited for this study. This study was approved by the Ethics Committee of Huzhou Central Hospital (No. 20191104-01), and all subjects provided informed consent. The clinical protocols involving patients and the informed consent form were approved by the Chinese Clinical Trial Registry (http://www.chictr.org.cn, ChiCTR1900022432).
Inclusion criteria: The CRC subjects were diagnosed by a pathological examination. According to the American Joint Committee on Cancer (AJCC) staging criteria, all patients were in clinical stage III. The CRC subjects voluntarily acknowledged and finished 8 cycles of the FOLFOX chemotherapy regimen. The exclusion criteria were as follows: (1) subjects with diarrhea, constipation or other intestinal diseases before chemotherapy, such as ulcerative colitis and Crohn's disease; (2) subjects with a history of using oral antibiotics, hormones or intestinal flora preparations one month before chemotherapy; (3) patients who needed to interrupt the chemotherapy process due to disease progression; and (4) patients with other malignant tumors or serious cardiopulmonary diseases. In total, 30 patients with CRC were recruited for this study. The clinical characteristics of the patients are shown in Table 1.
Table 1.
Summary of clinical characteristic of CRC patients.
| Characteristics | Good effect group (n = 24) |
Bad effect group (n = 6) |
P | |
|---|---|---|---|---|
| Age (years) | 65.92±7.02 | 66.17±5.95 | 0.93 | |
| Gender(n) | male | 16 | 3 | 0.45 |
| female | 8 | 3 | ||
| Hypertension (n) | 5 | 1 | 0.82 | |
| Diabetes, (n) | 6 | 2 | 0.68 |
A total of 30 CRC patients who received and finished 8 cycles of chemotherapy were included in this study. We also recorded information on their characteristics, details were shown in the Table 1. There was no statistical difference in clinical data between the two groups (P>0.05).
Sample collection
Stool samples were collected one week before chemotherapy. The collection of the stool samples was completed within half an hour after the patient defecated without the use of laxatives. Approximately 5–10 g samples were collected and stored at -80°C.
GC–MS metabolite assay
A 1.5 mL centrifuge tube was prepared with 800 L precooled methanol solution (4:1) and stool samples; 160 L chloroform and 20 L internal standard (0.3 mg mL-1 2-chloro-L-phenylalanine prepared with methanol) were successively added. Then, the mixture was ground, and ultrasonic extraction was performed for 10 min at 4°C. After centrifugation at 4°C and 15,000 rpm for 10 min, the supernatant was obtained. A total of 500 L of supernatant was rapidly centrifuged and volatilized to dry. Then, 80 L methamine hydrochloride pyridine (15 mg mL-1) were added to the flask and swirled for 2 min. After a 90 min oxime reaction in a shaken incubator at 37°C, 80 L N,O-bis(trimethylsilyl) trifluoroacetamide (BSTFA) (containing 1% TMCS) derivatives and 20 L n-hexane were added to the mixture. Finally, the mixture was shaken by vortexing for 2 min, reacted at 70°C for 60 min, and placed at room temperature for 30 min for the GC/MS metabolomics analysis.
The analysis was performed using 7890B-5977A GC–MS (Agilent Technologies, Santa Clara, CA, USA). A 30 m 0.25 m DB-5MS fused silica capillary column (J&W Scientific, Folsom, CA) was used to perform the separation. The flow rate of high-purity helium was 1.0 mL min/-1. The inlet temperature was 260°C. The oven temperature increased from 80°C to 170°C at 8°C min-1, increased to 200°C at 5°C min-1, and finally increased to 305°C at 8°C min-1 for 4 min. The collection was started with a delay of 5 min, the collection speed was 36.966 spectra per second, and the mass-scanning range was 50–450 m/z. In total, 124 microbial metabolites were detected in the present study based on previous research [31].The results were shown in Table S1.
Intestinal microorganism detection
-
(1)
DNA extraction. The gene extraction from the stool samples was performed using a DNA extraction Kit (A E.Z.N.A.® Stool DNA Kit (Omega Bio-Tek, Norcross, GA, U.S.)). After the genomic DNA extraction, the extracted genomic DNA was detected by 1% agarose gel electrophoresis.
-
(2)
PCR amplification. Specific primers with barcodes were synthesized. The sequence of the 16S rDNA primer in the v1-V4 region was 357F: 5′-TacggGAGgCAGCAGG-3′; 1114 r: 5′- GCAACGAGCGCAACCC - 3′. A representative sample was randomly selected for a pre-experiment to ensure that most samples could be amplified at the appropriate concentration within the minimum number of cycles. Each sample was repeated 3 times. The PCR products of the same sample were mixed and detected by 2% agarose gel electrophoresis. An AxyPrepDNA Gel Recovery Kit (AXYGEN Company) was used to cut the gel and recover the PCR products, and Tris_HCl elution was performed. The PCR products were detected and quantified using a QuantiFluor™-ST blue fluorescence quantification system (Promega) based on the preliminary quantitative results of electrophoresis and then mixed in the appropriate proportion according to the sequencing volume requirements of each sample.
-
(3)
Miseq library construction. The "Y" shaped connector was connected. Magnetic beads were used to remove self-connecting segments. The library template was enriched by PCR amplification. Denatured sodium hydroxide was applied to produce single stranded DNA fragments.
-
(4)
Miseq sequencing. One end of the DNA fragment is complementary to the primer base and fixed on the chip. The other end is randomly complementary to another nearby primer that is also fixed to form a "bridge". PCR amplification was performed to produce DNA clusters. The DNA amplicon linearizes into a single strand. Modified DNA polymerase and dNTPs with four fluorescent markers were added, and only one base was synthesized in each cycle. The surface of the reaction plate was scanned by a laser, and the nucleotide species polymerized in the first reaction of each template sequence were read. The "fluorophorer" and "terminator" were chemically cleaved to restore the viscosity of the 3′ end and continue to polymerize the second nucleotide. The results of the fluorescence signal collected in each round were recorded, and the sequence of the template DNA fragment was obtained.
-
(5)
Bioinformatics analysis. The sequencing data processing and optimization were performed according to the criteria at http://en.wikipedia.org/wiki/Fastq. The Uparse (Version 7.1) method was used for the OTU clustering. The sequence similarity in the OTUs was set to 97%, and a representative sequence of the OTUs was obtained. Uchime (Version 4.2.40) was used to detect the chimeric sequences generated in the PCR amplification and remove them from the OTUs. The Usearch_global method was used to compare the optimized sequence map with the OTU representative sequence and obtain the sequence abundance statistics table of each OTU sample. The results were shown in Table S2. Microbial diversity studies were performed to investigate the ecology of the sample fecal microbiota, including a series of statistical analyses to estimate the species abundance and diversity of the environmental community. Mothur software was used to calculate the Chao index and Ace index to evaluate the abundance of bacteria. The results were shown in Table S3. The Shannon index and Simpson index were calculated to evaluate the diversity of the bacterial community. The RDP classifier Bayesian algorithm was used for the taxonomic analysis of the OTU representative sequences at 97% similar levels, and the community composition of each sample was calculated at each level (phylum, class, order, family, genus and species levels). Variance decomposition was used to reflect the differences in multiple sets of data using a two-dimensional coordinate graph, and a principal component analysis (PCA) was carried out by considering the two characteristic values that can best reflect the variance values on the coordinate axis. The positions of the samples in each dimension were recorded, the contribution of each OTU to each principal component was calculated, and a PCA statistical analysis was conducted using R language as a PCA sum diagram. The PCoA statistical analysis and mapping selection were carried out in R language.
Cell culture
The human CRC cell lines HCT-8, HCT116, SW480 and SW620 was obtained from the Cell Bank of the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI-1640 culture with 10% fetal bovine serum at 37°C in 5% CO2. The cell lines at the logarithmic growth stage were digested for 1 min with 1 mL 0.25% trypsin at 37°C. The cells were seeded in a 96-well plate with three duplicate wells at a density of 6 × 104 cells mL-1. The cells were cultured in various groups after 24 h incubation. 5-Fluorouracil (5-Fu), NaB and OXA were purchased from Selleck. 5-Fu and oxaliplatin were configured in 5% glucose solution as required.
CCK-8 assay
Cell proliferation was assayed using Cell Counting Kit-8 (CCK-8; Dojindo, Kumamoto, Japan). The cell lines at the logarithmic growth stage were digested for 1 min with 1 mL 0.25% trypsin at 37°C. The cells were seeded in a 96-well plate with three duplicate wells at a density of 6 × 104 cells mL-1. The cells were cultured in various groups after 48 h incubation. The concentrations of 5-Fu were set as 0.5, 1.0, 2.0, 4.0, 8.0, 16.0 and 32.0 μg/ml. The concentrations of NaB were set as 1, 2, 4, 8, 16 and 32 mmol/L. The concentrations of OXA were set as 1.25, 2.5, 5, 10, 20, 40, 80 and 160 µmol/L. After the treatment, 10% 5 mg mL-1 CCK-8 solution was added to each well. To determine the cell viability, a microplate reader (TECAN, Infinite M100 PRO, Switzerland) was used to obtain the OD value at a wavelength of 450 nm.
Apoptosis and cell cycle assay
An Annexin FITC/PI Apoptosis Detection Kit KGA101 (KeyGEN BioTECH, Jiangsu, China) was used to detect cell apoptosis and the cell cycle. CRC cells in 6-well plates (3.0 × 105 cells/well) with medium for 24 h were treated with different interventions. According to the IC50, 5 mmol/L and 100 μmol/L were selected as the NaB and OXA intervention concentrations, respectively. Then, NaB (5 mmol/L), OXA (100 μmol/L) and NaB (5 mmol/L)+OXA (100 μmol/L) were used to treat the cells for 48 h. Then, the cells were washed, collected, fixed using 70% ethanol and stored at 4°C overnight. Each sample was analyzed three times. Flow cytometry (FACSCalibur; Becton-Dickinson) was used to analyze the data.
Transwell assay
Cells were cultured in 6-well plates (3.0 × 105 cells/well) and treated with NaB (5 mmol/L), OXA (100 μmol/L) or NaB (5 mmol/L)+OXA (100 μmol/L) for 24 h. The cells were digested with 0.25% trypsin for 1 min, and the digestion was terminated by adding 3 mL medium. Then, the cells were centrifuged at 300 × g for 3 min, and a count plate was used for counting. The cell density was adjusted to 1106 cells/mL, 100 L/well were inoculated into a Matrigel-treated Transwell upper chamber, and the lower chamber was supplemented with 1640 complete medium. The chamber was incubated at 37°C with 5% CO2 for 24 h and then mixed with 4% paraformaldehyde for 30 min. Crystal violet was used to stain the cells for 20 min. Finally, PBS was used to wash the cells twice. After taking photos with a microscope, the cells in the back were counted.
Scratch-wound assay
Cells digested at the logarithmic growth stage were inoculated into 6-well plates. On the following day, when the cell aggregation reached approximately 80%, the six-well plate was evenly crossed with a sterile spear head. The floating cells were washed with PBS and then treated with NaB (5 mmol/L), OXA (100 μmol/L) and NaB (5 mmol/L)+OXA (100 μmol/L) for 24 h. The cells were removed and photographed under a biological inverted microscope (OLYMPUS IX51, Japan) (magnification: 100 ×) to measure the cell migration distance.
EdU assay
An EdU assay was performed to detect cell proliferation by using a KeyFluor488 Click-it EdU imaging kit (Jiangsu Kaiji Biotechnology Co., LTD., China, KGA331-100). The cells were digested, counted and prepared with a concentration of 1 × 104 cells/mL cell suspension. Two hundred microliters of cell suspension were added to each well of a 96-well cell culture plate (2 × 103 cells per well). A 96-well cell culture plate was placed in a 37°C, 5% CO2 incubator for 24 h. The medium was discarded, and the cells were washed twice with PBS. NaB (5 mmol/L), OXA (100 μmol/L) and NaB (5 mmol/L)+OXA (100 μmol/L) were added for 24 h, and then, the samples were washed with 100 μL PBS 1-3 times. A high-intensity cell imaging system (amplification 200 ×) was used for the detection.
Observation of the cell structure using fluoroelectron microscopy
First, the samples were collected, washed with PBS and placed in EP tubes. Then, the samples were fixed, dehydrated, embedded and solidified. The samples were sliced by an LKB-1 ultrathin slicing machine at 50-60 nm and dyed with 3% uranium acetate and lead citrate. Finally, the cell structures were observed using fluoroelectron microscopy (Japan, JEM-1400).
Animal model construction
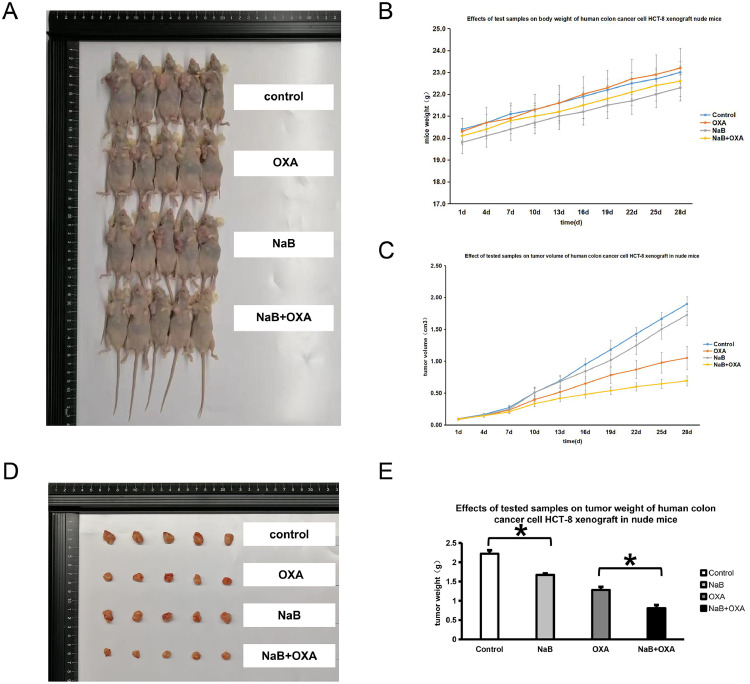
Cultured human colon cancer HCT-8 cell suspensions were collected at a concentration of 1 × 107/mL and inoculated subcutaneously in the right axilla of nude mice at 0.1 mL each. The drug plan was as follows: (1) in the control group, the mice were intraperitoneally injected with the same amount of normal saline once a day; (2) in the OXA group, the mice were intraperitoneally injected with 10 mg/kg or 200 µl/20 g once a week; (3) in the NaB group, the mice were intraperitoneally injected with 400 mg/kg or 200 µl/20 g NaB once a day; and (4) in the NaB+OXA group, the mice were intraperitoneally injected with 400 mg/kg+10 mg/kg or 200 µl/20 g once a day + once a week. All mice were observed for 28 days.
The diameter of the transplanted tumors in the nude mice was measured with Vernier calipers. After 16 days of inoculation, when the tumor grew to 80-100 mm3, the animals were randomly divided into 5 mice per group. Furthermore, the nude mice in each group were given drugs according to the drug plan. The tumor diameter was measured to dynamically observe the antitumor effect of the tested samples. After the intervention, the nude mice were immediately sacrificed, and the tumors were removed and weighed. The tumor volume, tumor weight and mouse weight were compared.
Hematoxylin eosin (HE) staining
The slices were dewaxed and hydrated following conventional methods. The PBS cleaning solution was obtained from China Jiangsu KGB5001 Biotechnology Co., Ltd. The operation followed the instructions of the hematoxylin-eosin dye kit (Jiangsu KGA224 Biotechnology Co., Ltd., China). The slices were dried, sealed and scanned with a digital pathology scanner. The morphology of tumor cells, the degree of necrosis, interstitial blood vessels, bleeding, and inflammatory cell infiltration were observed under a light microscope. The degree of tumor necrosis was rated as 0-4 (points). Tumor necrosis accounted for less than 1/4 of the tumor tissue was rated as 1 (1+, mild or small). Necrotic tumor accounted for about 1/4 to 2/4 of the tumor tissue and was rated as 2 points (2+, moderate or more). Tumor necrosis accounted for about 2/4-3/4 of the tumor tissue and was rated as 3+ (3+, severe or excessive). A score of 4 (4+, very severe or massive) was defined as tumor necrosis > 3/4 of the tumor tissue.
TUNEL assay
The sections were processed with a TUNEL assay kit (Jiangsu KGA702). The sections were treated with permeability, Proteinase K permeability, inactivation of enzyme, TUNEL reaction, addition of enzyme-labeled avidin, DAB color rendering, termination of color rendering, redyeing and sealing. Finally, the sections were observed under an optical microscope, and 3 regions with high expression were photographed and preserved.
Ki67 detection
The samples were treated according to the instructions of an EliVision Plus Kit (Fuzhou Mai Xin Biotechnology Co., LTD kit-9902, China), and Ki67 was obtained from Abcam AB92742, UK. After the sections were processed, the protein expression in tissue cells was observed under an optical microscope. Three expression regions were selected and photographed for preservation with a bioluminescence microscope (Olympus BX43, Japan).
Statistical analysis
The data are expressed as the mean standard deviation (SD). A chi-square test was used to analyze the counting data. A rank sum test was used to analyze the nonnormally distributed data. P<0.05 was considered indicative of a significant difference. The calculation and graph generation were performed using SPSS 16.0, Microsoft Excel 2007, and R studio.
Results
-
1.
Screening for differential microflora metabolites affecting the efficacy of OXA in human subjects
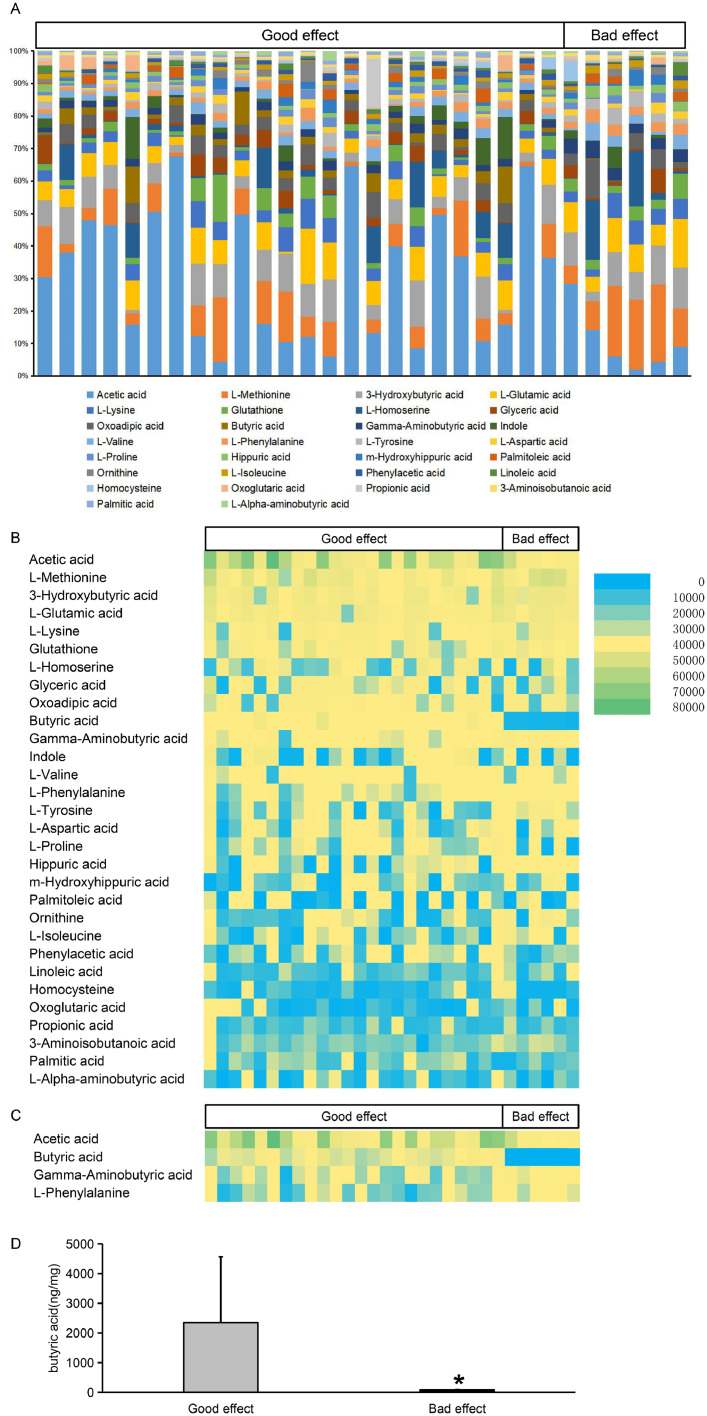
According to the efficacy of chemotherapy, we divided the patients into a good effect group and a bad effect group.The intestinal microflora metabolites of the patients in the two groups were analyzed. Fig. 1A shows that the top 30 concentrations of microbial metabolites in the same site were combined in two groups at the class level. Fig. 1B shows the content of the top 30 microbial metabolites in the two groups. The 30 types of intestinal microflora metabolites included acetic acid, L-methionine, 3-hydroxybutyric acid, L-glutamic acid, L-lysine, glutathione, L-homoserine, glyceric acid, oxoadipicic acid and butyric acid. Fig. 1C shows the content of the microbial metabolites with significant differences between the two groups. The results showed that the microbial metabolites with significant differences between the two groups included acetic acid, butyric acid, gamma-aminobutyric acid and L-phenylalanine. Fig. 1D shows that the butyric acid content in the good effect group was higher than that in the bad effect group (P<0.05).
-
2.
Verifying that NaB and OXA could synergistically inhibits the proliferation of CRC cells
Fig. 1.
Differential microflora metabolites affecting the efficacy of OXA
Fig. 1A shows that the top 30 concentrations of microbial metabolites in the same site were combined in two groups at the class level. Fig. 1B shows the content of the top 30 microbial metabolites in the two groups. Fig. 1C shows the content of the microbial metabolites with significant differences between the two groups. Fig. 1D shows a comparison of the butyric acid content between the two groups (P<0.05).
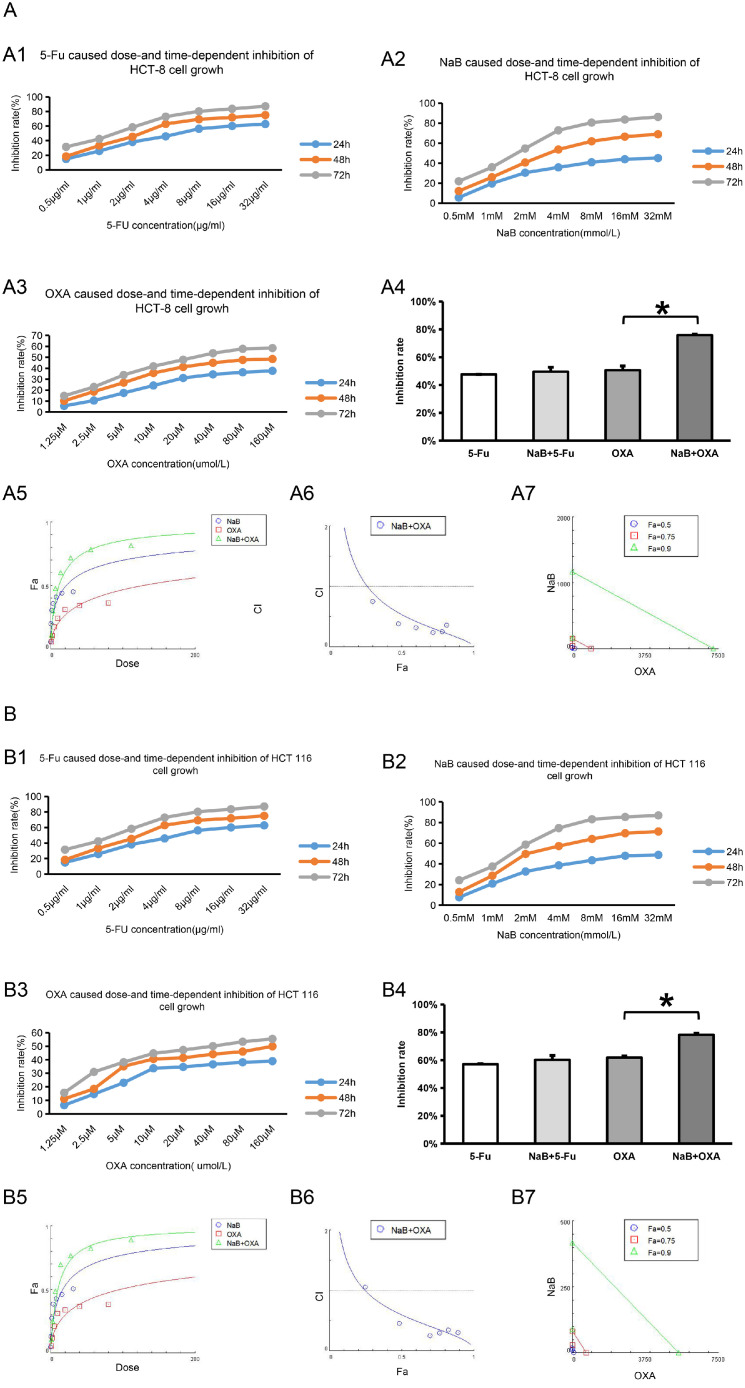
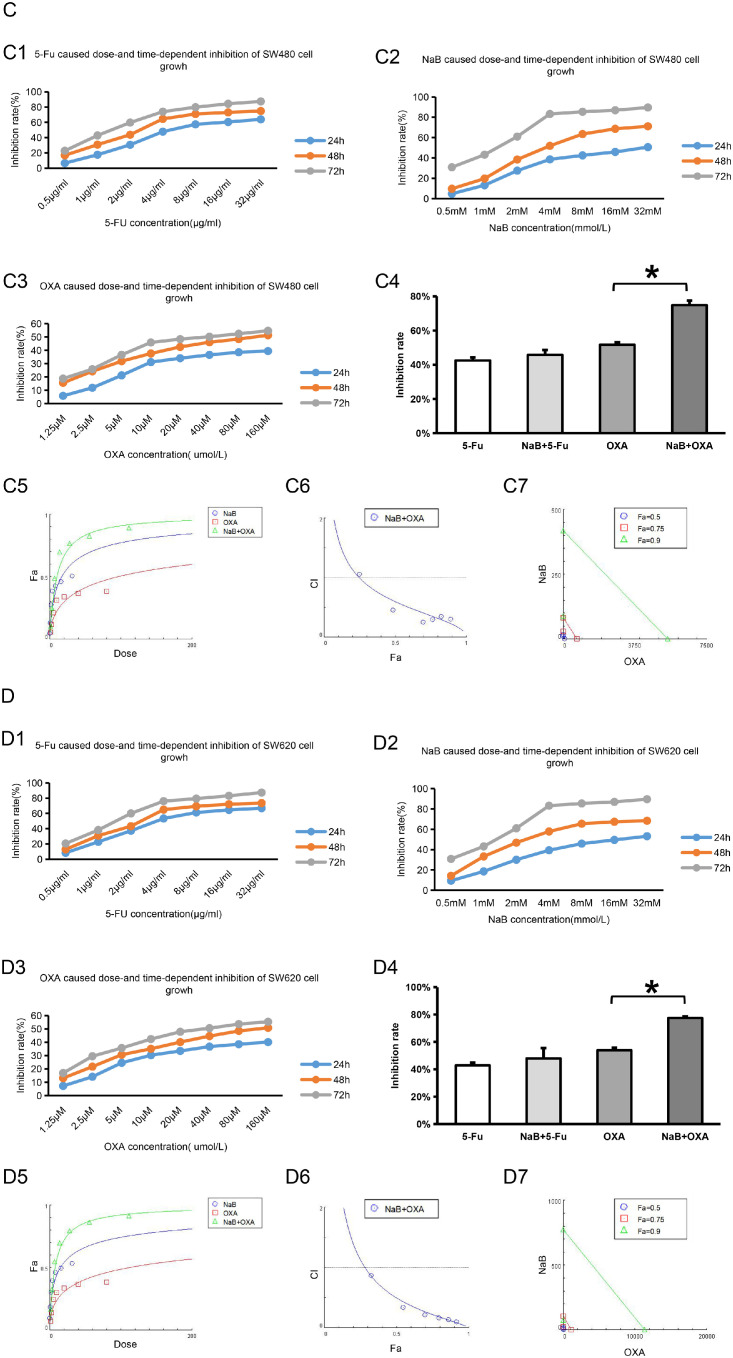
Cell proliferation was detected by CCK-8 assay. The effects of different interventions on the proliferation of CRC cells lines (HCT-8, HCT116, SW480 and HCT116) were evaluated. The values of IC50 are shown in Fig. 2 and Table 2.
Fig. 2.
NaB and OXA synergistically inhibits the proliferation of CRC cells
CCK-8 assay was used to detect the effects of different interventions on the proliferation of CRC cells lines (HCT-8, HCT116, SW480 and SW620). Chou-Talalay was used to analyze the synergistic relationship between NaB and OXA. CI>1 represents antagonistic effect, CI=1 represents additive effect and CI<1 represents synergistic effect. Lable Control represents the no intervention group, lable NaB represents the NaB intervention group, lable OXA represents the OXA intervention group, and OXA+NaB represents the OXA and NaB combined intervention group. * represents P<0.05.
Figures A1-A3, B1-B3, C1-C3, D1-D3, represent the effects of 5-Fu, NaB, and OXA intervention on the proliferation of CRC cell lines HCT-8, HCT116, SW480, and SW620, respectively. Figure A4, B4, C4, D4 represent the effects of 5-Fu and OXA intervention and combination with NaB on the inhibition ratio of CRC cell lines HCT-8, HCT116, SW480 and SW620 at 48h, respectively. Figure A5-A7, B5-B7, C5-C7, D5-D7 represent the synergistic relationship of NaB and OXA .
Table 2.
The half maximal inhibitory concentration (IC50) values on colorectal cancer cell lines with different interventions.
| 5-Fu | NaB | OXA | ||
|---|---|---|---|---|
| HCT8 | 24h | 11.103 | 26.081 | 300.067 |
| 48h | 3.047 | 5.035 | 91.426 | |
| 72h | 2.104 | 1.824 | 36.027 | |
| HCT116 | 24h | 6.654 | 19.213 | 156.498 |
| 48h | 2.781 | 3.860 | 87.844 | |
| 72h | 1.388 | 1.571 | 43.886 | |
| SW480 | 24h | 7.828 | 18.624 | 225.824 |
| 48h | 3.087 | 5.217 | 80.096 | |
| 72h | 1.518 | 1.168 | 44.435 | |
| SW620 | 24h | 5.797 | 17.275 | 250.923 |
| 48h | 3.360 | 3.841 | 87.636 | |
| 72h | 1.651 | 1.232 | 43.780 |
Cell proliferation was detected by CCK-8 assay. The effects of different interventions (5-FU, NaB and OXA) on the proliferation of CRC cells lines (HCT-8, HCT116, SW480 and HCT116) were evaluated.
Compared with the OXA intervention group, the cell inhibition rate in the OXA combined with NaB intervention group was higher (P<0.05) in HCT-8, HCT116, SW480 and HCT116, respectively (Fig. 2A4, B4, C4 and D4). Compared with the 5-Fu intervention group, the cell inhibition rate in the OXA combined with 5-Fu intervention group did not significantly difference (P>0.05). In order to further verify the synergistic effect of NaB and OXA, CompuSy, the third-generation drug combination dose-effect analysis software, developed by the mathematical model of Chou-Talalay was used to analyze. The results showed that NaB and OXA had synergistic effect in HCT-8 (Fig. 2 A5-A7), HCT116 (Fig. 2 B5-B7), SW480 (Fig. 2 C5-C7)and HCT116 (Fig. 2 D5-D7) (the values of CI are all less than 1). These results also indicated that 5-Fu combined with NaB could not synergistically inhibit cell proliferation, while NaB and OXA might have synergistic inhibitory effects on cell proliferation.
-
3.
Verifying that NaB and OXA can synergistically inhibit the invasion and metastasis of colorectal cancer cells and induce apoptosis
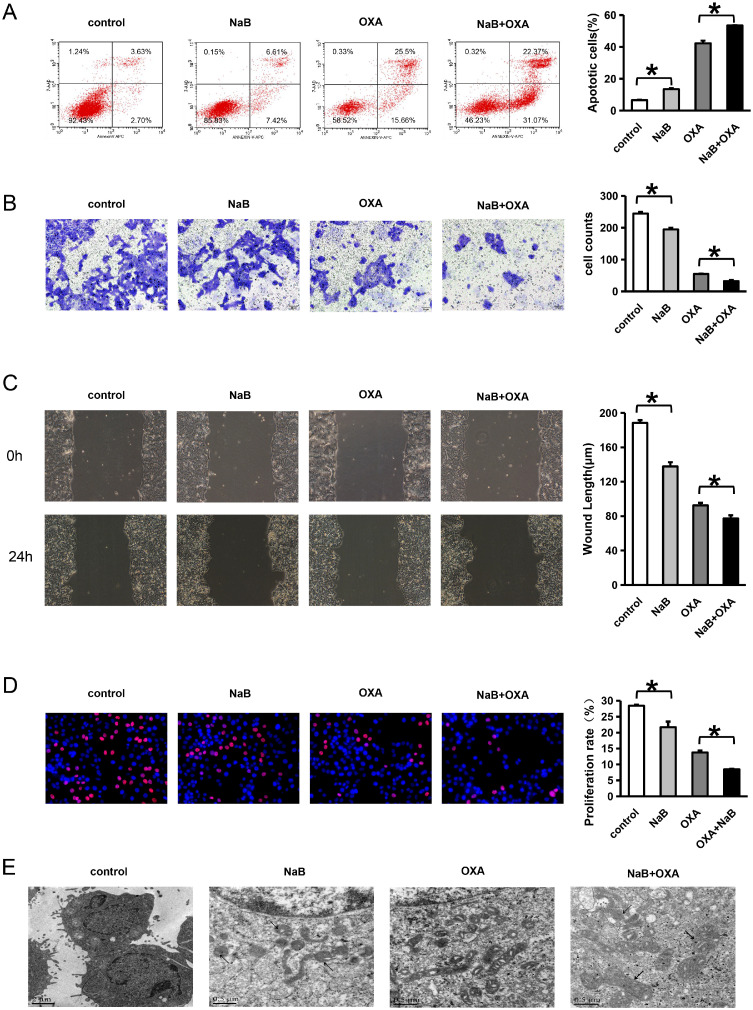
An Annexin FITC/PI Apoptosis double staining was used to detect cell apoptosis. Compared with the control group, the proportion of apoptosis increased after NaB intervention. In addition, compared with OXA alone, the proportion of apoptosis was highest in the NaB +OXA group (P<0.05) (Fig. 3A).
Fig. 3.
NaB and OXA can synergistically inhibit the invasion and metastasis of colorectal cancer cells and induce apoptosis
Fig. 3A shows a comparison of the cell apoptosis rate using different intervention methods. A Transwell assay was used to detect cell invasion. Panels A1, A2 and A3 represent the inhibition rates of 5-Fu, NaB and OXA on CRC cells, respectively. Panel A4 presents a comparison of the cell proliferation rate using different intervention methods. Annexin-v APC/7-AAD double staining was used to detect cell apoptosis. The number of cells in each quadrant in this figure is representative of (Q1) necrosis, (Q2) late apoptosis, (Q3) live cells, and (Q4) early apoptosis. Fig. 3B shows a comparison of cell invasion distance using different intervention methods. A scratch wound assay was used to detect cell migration. Fig. 3C shows a comparison of cell migration using different intervention methods. An EDU assay was used to detect cell proliferation. Red circles represent proliferative cells, and blue circles represent total cells. Fig. 3D represents a comparison of the cell proliferation rate using different intervention methods. Fluoroelectron microscopy was used to observe the cell structures of CRC cells. Fig. 2F represents the changes in the cell structures using different intervention methods.
Control represents the no intervention group, NaB represents the NaB intervention group, OXA represents the OXA intervention group, and OXA+NaB represents the OXA and NaB combined intervention group. * represents P<0.05.
Transwell assay was used to detect cell invasion. The results showed that compared with the control group, the number of invasive cells decreased after NaB intervention. Moreover, compared with oxaliplatin alone, the number of invasive cells was the least in NaB +OXA group (P<0.05) (Fig. 3B).
A scratch wound assay was used to detect cell migration. The results showed that compared with the control group, the number of migration distance decreased after NaB intervention. Moreover, compared with oxaliplatin alone, the migration distance was the least in NaB +OXA group (P<0.05) (Fig. 3C).
An EDU assay was used to detect cell proliferation. The results showed that compared with the control group, the cell proliferation ratio decreased after NaB intervention. Moreover, compared with oxaliplatin alone, cell proliferation ratio was the least in NaB +OXA group. (P<0.05) (Fig. 3D).
Fluoroelectron microscopy was used to observe the cell structures of CRC cells (Fig. 3E). In the control group, the nuclei were irregular, and the nucleoli tended to edge. The cytoplasm contained vesicles and mitochondria. In the NaB group, some cells died, and some mitochondrial pyknosis occurred. In the OXA group, the mitochondria were dense, and the space inside the mitochondrial crest was swollen. In the NaB combined with OXA group, some mitochondria shrank (thin arrow), the crest disappeared, and there were dense mitochondria.
-
4.
Verifying that NaB and OXA can synergistically regulate the function of CRC cells at the animal level
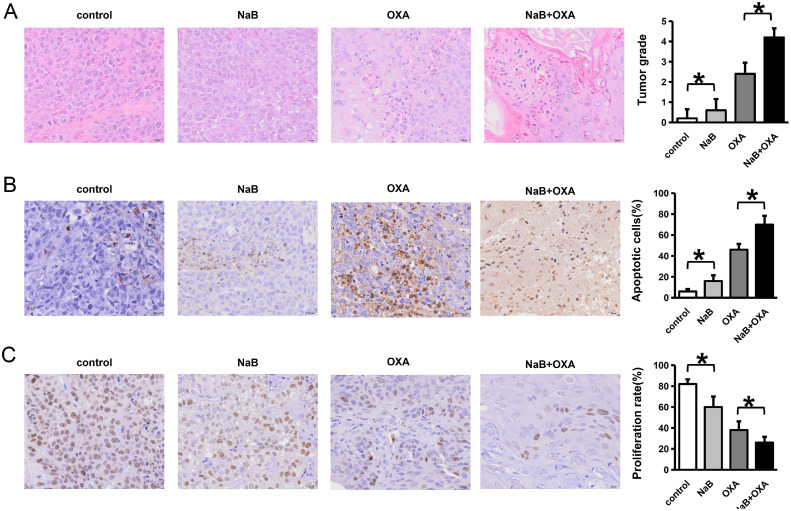
A mouse transplanted tumor model was established according to different intervention methods, and tumor tissues were collected from the mice in each group (Fig. 4A, D). The tumor tissues of the mice were removed, and the mouse weight, tumor volume and tumor weight were compared (Fig. 4B, C and E).The weight, tumor volume and weight of mice in the NaB group were lower than those in the control group, and the NaB + OXA group was the lowest. (P<0.05). Tumor tissue samples of the mice were obtained for HE staining, and the tumor cell morphology, necrosis degree, interstitial blood vessels, hemorrhage and inflammatory cell infiltration were observed by light microscopy. The scores were evaluated according to tumor necrosis degree (Fig. 5A). The results showed that the tumor tissue in the control group had a clear edge and compact texture. In the NaB group, there was no inflammatory cell infiltration or local hemorrhage in the tumor tissue and no neovascularization in the stroma. In the OXA group, the tumor tissue margins were unclear, and there was focal necrosis. There was a small amount of inflammatory cell infiltration and local hemorrhage in the tumor tissues. In the NaB+OXA group, the tumor tissue margins were unclear, and there was focal necrosis. A TUNEL assay was used to detect cell apoptosis. The results showed that the proportion of apoptosis increased after NaB intervention, and the proportion was the highest in NaB +OXA group (P<0.05) (Fig. 5B). The expression level of Ki67 was detected by IHC. The results showed that the proportion of cell proliferation decreased after NaB intervention. Moreover, compared with OXA alone, NaB combined with OXA showed the lowest cell proliferation ratio (Fig. 5C).
-
5.
Verifying the differential metabolites and butyrate-producing bacteria that affect the efficacy of OXA at the animal level
Fig. 4.
NaB and OXA can synergistically regulate the function of CRC cells at the animal level
Fig. 3A, D represents the transplanted tumor model established using different intervention methods and tumor tissues from mice in each group. Fig. 3C-E represents a comparison of the mouse weight, tumor weight and tumor tissues using different interventions.
Fig. 5.
Immunohistochemical changes in tumor tissues after different intervention methods
HE staining was used to evaluate the tumor cell morphology, necrosis degree, interstitial blood vessels, hemorrhage and inflammatory cell infiltration. Fig. 5A represents a comparison of the changes in the organizational form using different intervention methods. A TUNEL assay was used to detect cell apoptosis. Fig. 5B represents a comparison of cell apoptosis using different intervention methods. An IHC assay was used to detect Ki67. Fig. 5C represents a comparison of cell proliferation using different intervention methods.
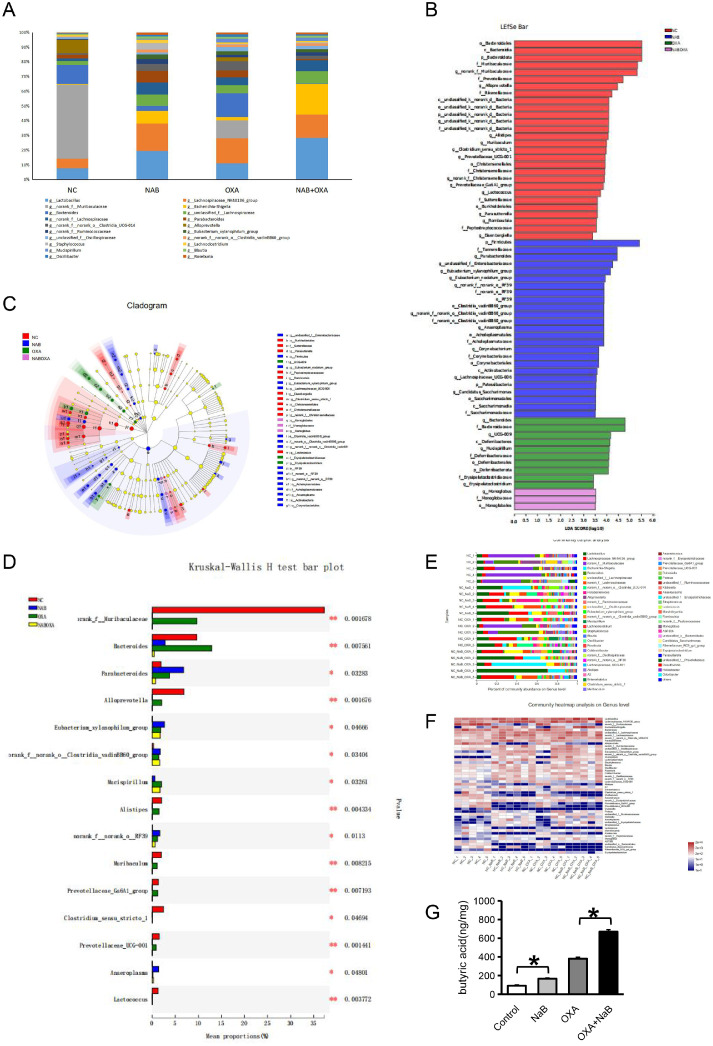
Fecal samples were obtained from each group of mice, and 16S sequencing technology was used to detect intestinal bacteria. Fig. 6A shows a percentile bar chart of the top 20 microbial communities. Five of these bacteria were butyric-producing bacteria, including Blautia, Lachnospiraceae, Eubacterium, Bacteroides and Ruminococcaceae. Fig. 6B shows the differences in the intestinal flora abundance among the four groups. In total, 58 intestinal bacteria showed significant differences, including the butyrate-producing bacteria Bacteroides. Fig. 6C shows four groups of bacteria with significant differences in abundance at the genus classification level. There were significant differences among 33 types of bacteria, including G__unclassifed fied_f__enterobacteriaceae, o__Burkholderiales, and f__Sutterellaceae. Fig. 6D shows a comparison and analysis of the differences among the 4 groups. A total, 15 intestinal bacteria showed significant differences, including the butyrate-producing bacteria Bacteroides. Fig. 6E shows the community composition and species of the 4 groups of samples at the bacterial genus level. The butyric-producing bacteria Blautia and Bacteroides accounted for a large proportion of bacteria. GC–MS was used to detect metabolites in mouse stools.The results were shown in Table S4. Fig. 6F shows a comparison of the butyric acid content. The results were as follows: NaB + OXA group> OXA group >NaB group > control group (P<0.05).
Fig. 6.
Differential metabolites and butyrate-producing bacteria that affect the efficacy of OXA at the animal level
16S sequencing technology was used to detect intestinal bacteria in mouse fecal samples. Fig. 6A represents a percentile bar chart of the top 20 microbial communities among the 4 groups. Fig. 6B represents the differences in intestinal flora abundance among the 4 groups. Fig. 6C represents the bacteria with significant differences in abundance at the genus classification level among the 4 groups. Fig. 6D represents a comparison and analysis of the differences among the 4 groups. Fig. 5E represents the community composition and species in the four groups of samples at the bacterial genus level. GC–MS was used to detect metabolites in mouse stools. Fig. 5F presents a comparison of the butyric acid content among the 4 groups.
Discussion
OXA is a chemotherapy drug commonly used in the treatment of CRC. Chemotherapy toxicity and drug resistance are the key factors that restrict the efficacy of OXA. Therefore, identifying new clinical adjuvant drugs is urgently needed to improve the side effects of chemotherapy and reduce the occurrence of drug resistance in the treatment of CRC. In this study, butyrate-producing bacteria and their metabolite NaB, which affect the efficacy of oxaliplatin chemotherapy, were screened. A series of cell and animal experiments was conducted to verify the synergistic inhibition of CRC cell function by NaB and OXA. This study can provide a theoretical basis for the use of NaB as a clinical adjunctive drug to OXA chemotherapy for CRC.
The occurrence and development of tumors are not only affected by the abundance and diversity of intestinal microorganisms but also regulated by microbial-related metabolites. Studies have shown that microbial primary metabolites, including amino acids, polysaccharides, lipids, and secondary metabolites, including SCFAs, secondary bile acids, and phenols, are related to the occurrence and development of CRC [23]. Some of these metabolites can promote the occurrence and development of tumors. For example, bile acids are metabolites produced by the liver as cholesterol is broken down and include primary and secondary bile acids [32]. When primary bile acids enter the large intestine, they are broken down and dehydroxylated by bacteria to produce secondary bile acids, deoxycholic acids and cholic acids [33]. Secondary bile acids have toxic biological effects, such as mutagenesis, cell lysis and DNA band rupture, and are risk factors for colorectal cancer and esophageal cancer [34]. In addition, some sulfate-metabolizing bacteria, such as Fusobacterium associated with CRC, produce some genotoxic hydrogen sulfide [35], which can induce the formation of CRC by inducing DNA damage, free radical release, colonic mucosal inflammation and hyperplasia, DNA methylation and other pathways [36]. In addition, as an important metabolite of the intestinal flora, trimethylamine oxide (TMAO) has been proven to be associated with cardiovascular disease, kidney disease, diabetes, and cancer and is an important marker of CRC risk [37]. In contrast, some metabolites, such as SCFAs, play a beneficial role in the development of tumors. As the main energy source of large intestine cells, SCFAs regulate the absorption of various nutrients and hormone production in the intestinal tract and are widely involved in energy metabolism. SCFAs can inhibit the occurrence and development of CRC through multiple pathways, such as immune regulation, epigenetics and molecular signaling pathways [23]. Some plant metabolites, such as phytophenols, have good antitumor and antioxidation properties. When plant polyphenols are consumed, intestinal bacteria can decompose them into small phenolic acids by enzymes to increase the bioavailability and activity of tea polyphenols, thus affecting the occurrence of cancer [38]. In addition, some dietary phytoestrogens can be converted into anticancer lignans by the intestinal flora [39].
Butyric acid is a short-chain fatty acid produced by anaerobic bacteria decomposition and the fermentation of dietary fiber in colon food that is not digested by the small intestine [40]. Butyric acid not only is the main energy source of colon epithelial cells but can also stimulate the release of colon peptides or growth factors to regulate the colon mucosal blood supply and promote the proliferation of epithelial cells [41]. NaB can inhibit the proliferation of various tumor cells and induce apoptosis in tumor cells, including CRC [30], liver cancer (LL) [42] and breast cancer (BC) [43]. To date, related studies have investigated NaB combined with other drugs against tumors. For example, Takuo et al. confirmed that NaB combined with cisplatin (DDP) has a synergistic antitumor effect [44]. In addition, Wang et al. not only found that NaB has a direct anticancer effect but also confirmed that NaB can enhance the sensitivity of anticancer drugs and has a synergistic antitumor effect when combined with cisplatin, mitomycin and adriamycin [45]. Shi et al. found that NaB can significantly improve the sensitivity of CRC and lung cancer (LL) cells to chloramphenicol and increase the resistance of LL cells to 5-Fu [46]. In addition, He et al. improved the infiltration and function of CD8+ T cells in the tumor microenvironment and saved the efficacy of the chemotherapy drug OXA by injecting NaB, a metabolite of the intestinal flora, into antibiotic-treated mice [47]. This finding also highlights the potential of NaB in the synergistic antitumor effect of OXA. However, relevant reports regarding the effect of NaB combined with OXA on CRC cells are lacking. To clarify the synergistic effect of NaB and OXA on CRC, this study was conducted at the cellular and animal levels. It was verified that NaB could not only directly inhibit the proliferation, invasion and metastasis of CRC cells but also induce cell apoptosis, which is consistent with our previous studies. Therefore, the synergistic antitumor effects of NaB and OXA were further verified. This study may provide new insight into the use of NaB as an adjunct to OXA in the treatment of CRC.
The intestinal microecology is complex and changeable, and there is mutual influence and interaction across intestinal microcommunities [47]. Although the antitumor effects of sodium butyrate have been well established, whether this beneficial effect is due to butyrate and/or a combination with other metabolites produced by these bacteria remains unclear. In the early stage, we summarized 17 types of butyric-producing bacteria reported in the literature [27], including Ruminococcus bromii, Blautia, Prevotell, Lactobacillus, Bifidobacterium longum, Lactobacillus Salivarius FP35, Butyrivibrio Fibrisolvens, bacteria, etc. In this study, intestinal microbes were detected by 16S sequencing technology, and 33 species of intestinal bacteria with significant differences were screened, including butyricoides-producing bacteria. However, whether the increase in NaB causes a change in the microecology or whether the antitumor effect of NaB combined with OXA causes a change in the microecology remains unclear. In our future studies, we aim to focus on this issue.
In order to overcome the toxic and side effects of OXA chemotherapy, OXA, a metabolite with differential efficacy, was found from environmental factors in the occurrence of CRC.There are still some limitations in this study. First, the study confirmed that NaB can have a synergistic antitumor effect with OXA, but its clinical application is limited due to the low titer intensity of NaB. Therefore, the focus of our next work is to develop high-potency NaB to promote its clinical application. Second, we only verified the synergistic anti–tumor effect of NaB and OXA at the cellular and animal levels, and the molecular mechanism of their synergistic regulation of CRC cell function remains to be further explored. In addition, the effect of the combination on the survival prognosis of the mice was not considered in the study. In order to promote the clinical application of the combination drugs, further studies on its impacts on survival benefits and other organs functions are needed to be conducted to improve the safety of combination of NaB and OXA in clinical application. Finally, the small number of included samples is a deficiency of this study. In future studies, we aim to expand the sample size and conduct further in vitro and in vivo experiments to further explore the role of NaB in CRC chemotherapy.
Conclusion
In order to overcome the toxic and side effects of OXA chemotherapy, NaB, a metabolite with differential efficacy, was found from environmental factors in the occurrence of CRC. A series of cell and animal experiments have confirmed that NaB and OXA had synergistic anti-tumor effects. NaB and OXA can synergistically inhibit the proliferation, invasion and metastasis of CRC cells and promote the apoptosis of CRC cells. The results not only suggest that the same antitumor effect may be achieved with the reduction of OXA dose, but also that side effects maybe reduced without reducing OXA dose. It is expected to provide new ideas for NaB as an adjuvant drug for chemotherapy.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Huzhou Central Hospital (No. 20191104-01), and all subjects provided informed consent. The clinical protocols involving patients and the informed consent form were approved by the Chinese Clinical Trial Registry (http://www.chictr.org.cn, ChiCTR1900022432).
Consent for publication
Not applicable.
Funding
This work was supported by the Zhejiang Provincial Natural Science Foundation (No. LQ20H160001), Medical and Health Technology Project of Zhejiang Province (No. 2022KY1218).
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
CRediT authorship contribution statement
Han Shuwen: Conceptualization, Methodology, Writing – review & editing. Wang Yangyanqiu: Visualization, Investigation. Chu Jian: Formal analysis, Data curation, Writing – original draft. Hu Boyang: Formal analysis, Data curation, Writing – original draft. Chen Gong: Validation. Zhuang Jing: Conceptualization, Methodology, Writing – review & editing.
Declaration of Competing Interest
The authors declare that no conflicts of interest exist.
Acknowledgements
Not applicable.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018 doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Arnold M, Sierra M S, Laversanne M, et al. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66(4):683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]
- 3.Haipeng H, Tingting L, Gengtai Y, et al. High expression of COL10A1 is associated with poor prognosis in colorectal cancer. OncoTargets Therapy. 2018;11:1571–1581. doi: 10.2147/OTT.S160196. Volume. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paola P, et al. Oxaliplatin in the era of personalized medicine: from mechanistic studies to clinical efficacy. Cancer Chemother. Pharmacol. 2016;77(1):5–18. doi: 10.1007/s00280-015-2901-x. [DOI] [PubMed] [Google Scholar]
- 5.He Y, Fu L, Li Y, et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8+ T cell immunity. Cell Metab. 2021;33(5):988–1000. doi: 10.1016/j.cmet.2021.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Yin Y, Yao S, Hu Y, et al. The immune-microenvironment confers chemoresistance of colorectal cancer through macrophage-derived IL6. Clin. Cancer Res. 2017;23(23):7375–7387. doi: 10.1158/1078-0432.CCR-17-1283. [DOI] [PubMed] [Google Scholar]
- 7.de Haas N, de Koning C, di Blasio S, et al. STAT family protein expression and phosphorylation state during moDC development is altered by platinum-based chemotherapeutics. J. Immunol. Res. 2019 doi: 10.1155/2019/7458238. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo SU, Min KJ, Woo SM, et al. Z-FL-COCHO, a cathepsin S inhibitor, enhances oxaliplatin-mediated apoptosis through the induction of endoplasmic reticulum stress. Exp. Mol. Med. 2018;50(8):1–11. doi: 10.1038/s12276-018-0138-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Limagne E, Thibaudin M, Nuttin L, et al. Trifluridine/tipiracil plus oxaliplatin improves PD-1 blockade in colorectal cancer by inducing immunogenic cell death and depleting macrophages. Cancer Immunol. Res. 2019;7(12):1958–1969. doi: 10.1158/2326-6066.CIR-19-0228. [DOI] [PubMed] [Google Scholar]
- 10.Bahrami A, Amerizadeh F, Hassanian S M, et al. Genetic variants as potential predictive biomarkers in advanced colorectal cancer patients treated with oxaliplatin-based chemotherapy. J. Cell. Physiol. 2017 doi: 10.1002/jcp.25966. [DOI] [PubMed] [Google Scholar]
- 11.Leung V, Huo Y R, Liauw W, et al. Oxaliplatin versus mitomycin C for HIPEC in colorectal cancer peritoneal carcinomatosis. Eur. J. Surg. Oncol. (EJSO) 2016 doi: 10.1016/j.ejso.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 12.Wolpin BM, Meyerhardt JA, Mamon HJ, et al. Adjuvant treatment of colorectal cancer. CA Cancer J. Clin. 2017;57:168–185. doi: 10.3322/canjclin.57.3.168. [DOI] [PubMed] [Google Scholar]
- 13.Saat K. Chemotherapy-induced peripheral neuropathy—part 2: focus on the prevention of oxaliplatin-induced neurotoxicity. Pharmacol. Rep.: PR, 2020;72:204–218. doi: 10.1007/s43440-020-00106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura, Masamichi, Maeda, et al. Hyperacute peripheral neuropathy is a predictor of oxaliplatin-induced persistent peripheral neuropathy. Support. Care Cancer Off. J. Multinational Assoc. Support. Care Cancer. 2017;25(5):1383–1389. doi: 10.1007/s00520-016-3514-6. [DOI] [PubMed] [Google Scholar]
- 15.Clingan P, Andre T, Hickish T, et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. 2013. [DOI] [PubMed]
- 16.Schmoll H-J, Tabernero J, Maroun J. Capecitabine plus oxaliplatin compared with fluorouracil/folinic acid as adjuvant therapy for stage III colon cancer: final results of the NO16968 randomized controlled phase III trial. J. Clin. Oncol. 2015 doi: 10.1200/JCO.2015.60.9107. [DOI] [PubMed] [Google Scholar]
- 17.Brodowicz T, Ciuleanu TE, Radosavljevic D, et al. FOLFOX 4 plus cetuximab administered weekly or every second week in the first-line treatment of patients with KRAs wild-type metastatic colorectal cancer: a randomized phase II CECOG study. Ann. Oncol. 2013;24:1769–1777. doi: 10.1093/annonc/mdt116. [DOI] [PubMed] [Google Scholar]
- 18.de Mestier L., et al. Primary tumor resection in colorectal cancer with unresectable synchronous metastases: a review. World. J. Gastrointest. Oncol. 2014;6(6):156–169. doi: 10.4251/wjgo.v6.i6.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ron S, Shai F, Ron M. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8) doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tilg H, Adolph T E, Gerner R R, et al. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33(6) doi: 10.1016/j.ccell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 21.Shen Z H, Zhu C X, Quan Y S, et al. Relationship between intestinal microbiota and ulcerative colitis: mechanisms and clinical application of probiotics and fecal microbiota transplantation. World J. Gastroenterol. 2018;24(1):5–14. doi: 10.3748/wjg.v24.i1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han S, Gao J, Zhou Q, et al. Role of intestinal flora in colorectal cancer from the metabolite perspective: a systematic review. Cancer Manag. Res. 2018;10 doi: 10.2147/CMAR.S153482. 199-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geirnaert A, Steyaert A, Eeckhaut V, et al. Butyricicoccus pullicaecorum, a butyrate producer with probiotic potential, is intrinsically tolerant to stomach and small intestine conditions. Anaerobe. 2014;30:70–74. doi: 10.1016/j.anaerobe.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 24.Eeckhaut V, Wang J, Van Parys A, et al. The probiotic Butyricicoccus pullicaecorum reduces feed conversion and protects from potentially harmful intestinal microorganisms and necrotic enteritis in broilers. Front. Microbiol. 2016;7:1416. doi: 10.3389/fmicb.2016.01416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kilner J, Corfe B M, Mcauley M T, et al. A deterministic oscillatory model of microtubule growth and shrinkage for differential actions of short chain fatty acids. Mol. Biosyst., 12. [DOI] [PubMed]
- 26.Han S, Da M, Qi Q, et al. Protective effect of the "food-microorganism-SCFAs" axis on colorectal cancer: from basic research to practical application. J. Cancer Res. Clin. Oncol. 2019;145(2) doi: 10.1007/s00432-019-02997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O"Keefe Stephen J D. Diet, microorganisms and their metabolites, and colon cancer. Nat. Rev. Gastroenterol. Hepatol. 2016;13(12):691–706. doi: 10.1038/nrgastro.2016.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H M, Yu Y N, Wang J L, et al. Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am. J. Clin. Nutr. 2013;97(5):1044–1052. doi: 10.3945/ajcn.112.046607. [DOI] [PubMed] [Google Scholar]
- 29.Hamer H. Review article: the role of butyrate on colonic function. Alimentary Pyarmacol. Therapeut. 2008;27(2):104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 30.Xi Y, Jing Z, Wei W, et al. Inhibitory effect of sodium butyrate on colorectal cancer cells and construction of the related molecular network. BMC Cancer. 2021;21(1) doi: 10.1186/s12885-021-07845-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang X, Pan Y, Wu W, et al. Analysis of prognosis, genome, microbiome, and microbial metabolome in different sites of colorectal cancer. J. Transl. Med. 2019;17(1):353. doi: 10.1186/s12967-019-2102-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Han S, Zhuang J, Wu Y, et al. Progress in research on colorectal cancer-related microorganisms and metabolites. Cancer Manag. Res. 2020;12:8703–8720. doi: 10.2147/CMAR.S268943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wenniger L, Beuers U. Bile salts and cholestasis. Digest. Liver Dis. Off. J. Italian Soc. Gastroenterol. Italian Assoc. Study Liver. 2010;42(6):409–418. doi: 10.1016/j.dld.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 34.Sayin S, Wahlstr?M A, Felin J, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17(2):225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Yazici C, Wolf P G, Kim H, et al. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut. 2017 doi: 10.1136/gutjnl-2016-313321. gutjnl-2016-313321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tjalsma H, Schller-Guinard M, Lasonder E, et al. Profiling the humoral immune response in colon cancer patients: diagnostic antigens from Streptococcus bovis. Int. J. Cancer. 2010;119(9):2127–2135. doi: 10.1002/ijc.22116. [DOI] [PubMed] [Google Scholar]
- 37.Xu R, Wang Q Q, Li L. A genome-wide systems analysis reveals strong link between colorectal cancer and trimethylamine N-oxide (TMAO), a gut microbial metabolite of dietary meat and fat. BMC Genomics. 2015;16(Suppl 7):S4. doi: 10.1186/1471-2164-16-S7-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brglez M E, Knez H M, Škerget M, et al. Polyphenols: extraction methods, antioxidative action, bioavailability and anticarcinogenic effects. Molecules. 2016;21(7):901. doi: 10.3390/molecules21070901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koltorov L, Lapk O, Strka L. Phytoestrogens and the intestinal microbiome. Physiol. Res. 2018;67(Suppl 3):S401–S408. doi: 10.33549/physiolres.934022. [DOI] [PubMed] [Google Scholar]
- 40.Leonel A J, Alvarezleite J I. Butyrate: implications for intestinal function. Curr. Opin. Clin. Nutr. Metab. Care. 2012;15(5):474. doi: 10.1097/MCO.0b013e32835665fa. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Li Y, Tian Y, et al. Interaction between microbes and host intestinal health: modulation by dietary nutrients and gut-brain-endocrine-immune axis. Curr. Protein Pept. Sci. 2015 doi: 10.2174/1389203716666150630135720. [DOI] [PubMed] [Google Scholar]
- 42.Wei Xu, et al. Histone deacetylase inhibitors upregulate Snail via Smad2/3 phosphorylation and stabilization of Snail to promote metastasis of hepatoma cells. Cancer Lett. 2018 doi: 10.1016/j.canlet.2018.01.068. [DOI] [PubMed] [Google Scholar]
- 43.Salimi V, Shahsavari Z, Safizadeh B, et al. Sodium butyrate promotes apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment. Lipids Health Dis. 2017;16(1):208. doi: 10.1186/s12944-017-0593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maruyama T, Yamamoto S, Qiu J, et al. Apoptosis of bladder cancer by sodium butyrate and cisplatin. J. Infect. Chemother. 2012;18(3):288–295. doi: 10.1007/s10156-011-0322-2. [DOI] [PubMed] [Google Scholar]
- 45.Wang D, Wang Z, Tian B, et al. Two hour exposure to sodium butyrate sensitizes bladder cancer to anticancer drugs. Int. J. Urol. 2010;15(5):435–441. doi: 10.1111/j.1442-2042.2008.02025.x. [DOI] [PubMed] [Google Scholar]
- 46.Shi Bin, et al. Effect of sodium butyrate on ABC transporters in lung cancer A549 and colorectal cancer HCT116 cells. Oncol. Lett. 2020 doi: 10.3892/ol.2020.12011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He Y, Fu L, Li Y, et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8+ T cell immunity. Cell Metab. 2021 doi: 10.1016/j.cmet.2021.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.