Abstract
During prolonged mitotic arrest induced by antimicrotubule drugs, cell fate decision is determined by two alternative pathways, one leading to cell death and the other inducing premature escape from mitosis by mitotic slippage. FBWX7, a member of the F-box family of proteins and substrate-targeting subunit of the SKP1–CUL1–F-Box E3 ubiquitin ligase complex, promotes mitotic cell death and prevents mitotic slippage, but molecular details underlying these roles for FBWX7 are unclear. In this study, we report that WDR5 (WD-repeat containing protein 5), a component of the mixed lineage leukemia complex of histone 3 lysine 4 methyltransferases, is a substrate of FBXW7. We determined by coimmunoprecipitation experiments and in vitro binding assays that WDR5 interacts with FBXW7 in vivo and in vitro. SKP1–CUL1–F-Box-FBXW7 mediates ubiquitination of WDR5 and targets it for proteasomal degradation. Furthermore, we find that WDR5 depletion counteracts FBXW7 loss of function by reducing mitotic slippage and polyploidization. In conclusion, our data elucidate a new mechanism in mitotic cell fate regulation, which might contribute to prevent chemotherapy resistance in patients after antimicrotubule drug treatment.
Keywords: FBXW7, WDR5, ubiquitylation (ubiquitination), mitotic slippage, chemoresistance
Abbreviations: cDNA, complementary DNA; CHX, cycloheximide; SCF, Skp1–Cullin–F-box; GSK3β, glycogen synthase kinase-3β; GST, glutathione-S-transferase; HEK293T, human embryonic kidney 293T cell line; MLL, mixed lineage leukemia; MS, mass spectrometry; SAC, spindle assembly checkpoint; WDR5, WD-repeat containing protein 5
FBXW7/hCdc4 is a substrate specificity component of the Skp1–Cullin–F-box (SCF) E3 ubiquitin ligase and a member of the F-box protein family, which is responsible for recognizing and binding phosphorylated substrates to regulate their turnover (1). Most of the FBXW7 substrates are widely studied protooncogenes, including cyclin E, c-Myc, Notch, and c-Jun (2). FBXW7 functions as tumor suppressor, and mutations in the FBXW7 gene have been observed in a variety of human cancers (2, 3). FBXW7 phosphorylation by Plk2 leads to its ubiquitin-mediated degradation and stabilization of cyclin E (4).
Several lines of evidence have implicated loss or mutation of FBXW7 in chemotherapy resistance against antimicrotubule drugs (5, 6). For example, taxanes are microtubule-stabilizing drugs that inhibit the dynamic instability of mitotic spindles, and taxane-based chemotherapy agents are used for treatment of a wide range of cancers. Antimicrotubule drugs cause a prolonged mitotic delay and mitotic cell death. Perturbation of microtubule dynamics induces activation of the spindle assembly checkpoint (SAC) and mitotic arrest. However, cells can escape from the SAC-induced mitotic arrest through a process called mitotic slippage (7). Upon mitotic arrest caused by antimicrotubule drugs, FBXW7 promotes cell death and prevents mitotic slippage (8) and is counteracted by the E3 ligase FBXO45/MYCBP2, a regulator of FBXW7 protein abundance in mitosis (9). Two competing cellular networks, cyclin B1 degradation and activation of proapoptotic caspases, were proposed to regulate cell fate following mitotic arrest (10). Mcl-1, a prosurvival member of the Bcl-2 family central to the intrinsic apoptosis pathway, is degraded during a prolonged mitotic arrest and may therefore act as a mitotic death timer (6). It is being debated whether FBXW7 has a direct role in regulating Mcl-1 protein levels (6, 11, 12), and identification of FBXW7 substrates in mitotic cell fate regulation is a major task.
WDR5 (WD-repeat containing protein 5) is a highly conserved core scaffolding subunit of the KMT2 (mixed lineage leukemia [MLL]/SET) enzymes that deposit histone H3 lysine 4 methylation (13, 14, 15, 16). Apart from regulating gene expression by chromatin remodeling in interphase, WDR5 also functions in cell division (17, 18). In particular, WDR5 localizes to the midbody and regulates abscission (19). Moreover, WDR5 also regulates localization of the kinesin motor protein Kif2A during mitosis to facilitate chromosome congression and proper spindle assembly (20). In this study, we identified WDR5 as a new substrate of SCF–FBXW7. FBXW7 ubiquitinates WDR5, and its depletion leads to increased WDR5 protein levels. Furthermore, we find that WDR5 promotes mitotic slippage. Our study also has translational implication to provide a rationale for the FBXW7–WDR5 axis as a possible target for chemotherapy resistance treatment in combination with antimicrotubule drug therapies.
Results
Proteomics-based approach to identify putative substrates of FBXW7
We have previously shown that the SCF–FBXW7 complex is targeted for degradation by the FBXO45-MYCBP2 E3 ubiquitin ligase specifically during mitotic arrest induced by antimicrotubule drugs. This in turn leads to induction of mitotic slippage and prevention of mitotic cell death (9). Now we aimed at identifying ubiquitination substrates of FBXW7 in mitotic slippage induction. We performed a screen with the aim to search for FBXW7 substrates that function in mitotic cell fate regulation (Fig. 1A). FLAG-tagged FBXW7α was immunopurified from human embryonic kidney 293T (HEK293T) cell and analyzed by mass spectrometry (MS). As a negative control, we used a FLAG-tagged FBXW7α-WD40 mutant that lacks the ability to bind substrates but not Skp1 and Cul1 (21). Among the proteins that we identified by MS were c-Myc and members of the MLL complex, including KMT2D, WDR5, and ASH2L (Figs. 1A, S1, Tables S1 and S2). To verify the findings of our screen, we transfected FLAG-tagged FBXW7 in HEK293T cells. Immunoprecipitation using anti-FLAG affinity beads identified WDR5 and KMT2D (MLL4) as putative FBXW7-binding proteins. We find that the interaction between FBXW7 is likely specific for KMT2D as no interaction with two other MLL family members SetD1A and MLL1 was detected (Fig. 1B). The interaction between FBXW7 and KMT2D, as well as WDR5, was abolished when the WD40 domain of FBXW7 that includes the conserved amino acid residues R465, R479, and R505, required for high-affinity substrate binding, was mutated (FBXW7ARG) (22). An interaction between FBXW7 and KMT2D was previously also identified (23). As our major aim was to identify FBXW7 substrates involved in mitotic slippage, we first checked whether KMT2D could act as a FBXW7 substrate in mitotic slippage. We analyzed this by depleting KMT2D in U2OS cells and performed live-cell imaging in the presence of nocodazole. Our results shown in Figure 1C reveal that KMT2D does not function in mitotic slippage.
Figure 1.
Proteomics-based approach to identify putative FBXW7 substrates.A, volcano plot showing enrichment of FBXW7-WD40 interactors identified by IP–MS. FLAG-FBXW7 or FLAG-FBXW7 (T439I, S462A, T463A, and R465A) were overexpressed in HEK293T for 48 h and interactors identified by FLAG-IP, TMT10-plex labeling, and MS (Table S1). Significance was determined via t-statistics using LIMMA R-package; proteins were considered significant if they showed a fold change of at least 50% and an adjusted p value after Benjamini and Hochberg below 5%, n = 2 (Table S2). B, FLAG-FBXW7 or FLAG-FBXW7ARG (R465H, R479Q, and R505C mutations) were overexpressed in HEK293T cells for 48 h with or without inhibition of cullin-RING E3 ligases by 5 μM MLN4924 for 5 h prior to harvest, as indicated. FLAG-FBXW7 constructs were used for an IP against the FLAG tag. + marks nonspecific bands. C, U2OS were transfected twice with 30 nM siRNA targeting GL2, KMT2D, or FBXW7, as indicated. About 830 nM nocodazole were added 48 h after the second siRNA transfection, and mitotic cell fates of n = 50 cells were determined by live-cell imaging. ∗p < 0.05, ns p > 0.05, one-way ANOVA with Tukey post hoc test, n = 3. MS, mass spectrometry; ns, not significant; HEK293T, human embryonic kidney 293T cell line; IP, immunoprecipitation.
FBXW7 regulates WDR5 protein levels by ubiquitination and proteasomal degradation
It was previously shown that MLL regulates M-phase progression in association with the WRAD complex and a conserved subunit of WRAD, WDR5, localizes to the mitotic spindle and to the midbody in dividing human cells (20).
Interestingly, WDR5 was identified as a major interacting protein of FBXW7 in our screen, and protein levels of WDR5 were decreased in response to FBXW7 overexpression (Fig. 1B). We therefore checked whether WDR5 protein levels would be affected by FBXW7 ablation. We found that in two FBXW7 KO cell lines HCT116 and DLD1, as well as in HeLa cells transfected with siFBXW7, WDR5 protein levels were significantly increased (Fig. 2A), which might suggest that FBXW7 could also regulate WDR5 protein stability. Quantitative PCR experiments indicated that WDR mRNA levels remain unchanged and that WDR5 is upregulated at the post-translational level (Fig. B). WDR5 protein levels were also upregulated in FBXW7 KO cells, whether or not KMT2D was present, showing that WDR5 upregulation is independent of KMT2D (Fig. 2C). A phenylalanine residue within the WIN site of WDR5 (F133A) is required for the interaction with KMT2 methyltransferases (24). We asked whether FBXW7 could still bind to the WDR5-F133A mutant and therefore independently of KMT2D. Our results shown in Figure 2D showed that this mutant can still coprecipitate FBXW7 but not KMT2D. As seen in Figure 2E, endogenous WDR5 was identified in FBXW7 immunoprecipitates, and vice versa, endogenous FBXW7 was detected in immunoprecipitation experiments using antibodies against WDR5. To show that the interaction between FBXW7 and WDR5 is direct, we performed in vitro binding assays where we incubated insect cell–derived His-FBXW7/Skp1 with bacterially expressed glutathione-S-transferase (GST)-WDR5 and performed GST pull-down assays. Our results presented in Figure 2F show that FBXW7 directly binds to WDR5.
Figure 2.
WDR5 is upregulated by FBXW7 depletion and interacts with FBXW7 independently of KMT2D.A, HeLa were transfected twice with siRNA against Gl2 or FBXW7 and harvested 72 h after the first transfection. ∗p < 0.05, unpaired two-way Student’s t test with Welch’s correction, n = 3. B, HeLa were transfected twice with siRNA against Gl2 or FBXW7 and harvested 72 h after the first transfection. ns p > 0.05, unpaired two-way Student’s t test with Welch’s correction, n = 3. C, HCT116 WT and FBXW7 KO cell lines were transfected twice with siRNA against Gl2 or KMT2D and harvested 72 h after the first transfection. ∗∗∗p < 0.001, ns p > 0.05, one-way ANOVA with Tukey post hoc test, n = 3. D, GFP-WDR5 WT or a F133A mutant were overexpressed in HEK293T cells for 24 h and immunoprecipitated using GFP-trap beads. ns p > 0.05, unpaired two-way Student’s t test with Welch’s correction, n =3. E, endogenous WDR5 or FBXW7 were immunoprecipitated from HEK293T lysates using specific antibodies and protein A or protein G Sepharose. F, purified recombinant GST-WDR5 and His-FBXW7 were combined, and GST pulldowns were performed using reduced glutathione on CL-4B beads. GST, glutathione-S-transferase; HEK293T, human embryonic kidney 293T cell line; mIgG, mouse control immunoglobulin G; ns, not significant; rIgG, rabbit control immunoglobulin G.
In order to assess the effect of FBXW7 on WDR5 protein stability, we used FBXW7 WT and KO DLD1 cell lines and added cycloheximide (CHX) to block translation. Cells were harvested at different time points after CHX addition. In FBXW7-depleted cells, overexpressed FLAG-WDR5 (Fig. 3A) and endogenous WDR5 (Fig. 3B) were stabilized compared with FBXW7-WT cells (Fig. 3A), indicating that FBXW7 promotes the degradation of WDR5. To address whether WDR5 protein levels are regulated by proteasomal degradation, we treated cells with the proteasomal inhibitor MG132. In the presence of MG132, the decay of WDR5 was prevented (Fig. 3C). Expression of FBXW7 but not FBXW7 ΔF-box mutant, which is deficient of SCF-E3 ligase activity, promoted the degradation of endogenous WDR5 in CHX-treated HEK293T cells (Fig. 3D). To analyze whether the regulation of WDR5 by FBXW7 is mediated by ubiquitination, we performed in vivo ubiquitination assays. We found that overexpression of FBXW7 but not FBXW7 ΔF-box or FBXW7ARG mutants leads to an increase of WDR5 polyubiquitination (Fig. 3E). Taken together, our findings suggest that WDR5 is a new substrate of FBXW7, which regulates its ubiquitin-dependent proteasomal degradation.
Figure 3.
WDR5 protein stability is regulated by FBXW7-mediated ubiquitination.A, FLAG-WDR5 was overexpressed in DLD1 WT and FBXW7 KO cell lines for 24 h, followed by blockage of protein synthesis with 300 μg/ml cycloheximide (CHX) for the indicated durations prior to harvest. Protein half-lives were determined by fitting to a one-phase decay model, n = 3. + marks a nonspecific band. B, DLD1 WT and FBXW7 KO cell lines were treated with 300 μg/ml CHX for the indicated durations prior to harvest. C, FLAG-WDR5 was overexpressed in DLD1 WT for 24 h, followed by inhibition of protein synthesis with 300 μg/ml CHX in the presence of 10 μM MG132 or DMSO for the indicated durations prior to harvest. Normalized protein levels were fit to a one-phase decay model to determine protein half-lives, n = 3. D, FLAG-FBXW7 or FLAG-FBXW7 ΔF-box (amino acids 284–321 deletion) were overexpressed in HEK293T for 48 h, followed by inhibition of protein synthesis with 100 μg/ml CHX for the indicated durations prior to harvest. Normalized protein levels were fit to a one-phase decay model to determine protein half-lives, n = 3. E, the indicated proteins were overexpressed in HEK293T for 24 h, and the 26S proteasome was blocked by 10 μM MG132 for 5 h prior to harvest. FLAG-WDR5 was immunoprecipitated using α-FLAG M2 beads in the presence of 20 mM N-ethylmaleimide. DMSO, dimethyl sulfoxide; HEK293T, human embryonic kidney 293T cell line; l.e., long exposure; s.e., short exposure.
The interaction between FBXW7 and WDR5 is partially regulated by glycogen synthase kinase-3β–dependent phosphorylation
Degradation of FBXW7 substrates depends on phosphorylation of a phosphodegron (2). To find out whether the interaction between FBXW7 and WDR5 is regulated by phosphorylation, we treated FLAG-FBXW7 immunoprecipitates with λ-phosphatase and observed that the interaction with WDR5 was impaired (Fig. 4A). In a variety of substrates including cyclin E, c-Myc, and c-Jun, glycogen synthase kinase-3β (GSK3β) phosphorylation plays a pivotal role (25, 26, 27). We therefore analyzed whether GSK3β-dependent phosphorylation of WDR5 is also required for the interaction between FBXW7 and WDR5. We made use of a small-molecule inhibitor, CHIR-99021, that blocks the enzymatic activity of GSK3β. Treatment of WDR5 overexpressing HEK293T cells with the GSK3β inhibitor reduced but did not completely block the binding between FBXW7 and WDR5 (Fig. 4B). Moreover, we found that siRNA-mediated ablation of GSK3β leads to upregulation of WDR5 protein levels (Fig. 4C). Coexpression of WDR5 and GSK3β followed by immunoprecipitation of WDR5 revealed an interaction of WDR5 with both ectopically expressed and endogenous GSK3β (Fig. 4D). We then analyzed whether treatment of HEK293T cells with CHIR-99021 would affect WDR5 ubiquitination. While WDR5 was ubiquitinated by FBXW7 in the control, this increase in ubiquitination was markedly reduced in the presence of the GSK3β inhibitor as well as by inhibition of cullin-RING E3 ubiquitin ligases using the small-molecule neddylation inhibitor, MLN4924 (Fig. 4E). Furthermore, in vitro ubiquitination of WDR5 was observed in the presence of Uba1 (E1), CDC34 and UbcH5b (E2), GSK3β, and SCF–FBXW7 but not SCF–FBXW7ARG (Fig. 4F). Together, these results propose that GSK3β cooperates with FBXW7 to regulate WDR5 protein abundance.
Figure 4.
The regulation of WDR5 by FBXW7 depends in part on GSK3β.A, FLAG-FBXW7 was overexpressed in HEK293T for 24 h, and all cultures were treated with 5 μM MLN4924 for 5 h prior to harvest. FLAG-FBXW7 was immunoprecipitated, and the indicated sample was dephosphorylated with λ-phosphatase at 30 °C for 3 h. B, GFP-WDR5 was overexpressed in HEK293T for 24 h, and cultures were treated with 5 μM CHIR-99021 or DMSO for 5 h prior to cell harvest, as indicated. GFP-WDR5 was immunoprecipitated using GFP-trap beads. ∗∗p < 0.01, unpaired two-way Student’s t test with Welch’s correction, n = 3. C, U2OS cells were transfected twice with 30 nM against GL2 or GSK3β and harvested 72 h after the first transfection. ∗p < 0.05, ∗∗p < 0.01, one-way ANOVA, n = 3. D, GFP-WDR5 and Myc-GSK3β were overexpressed in HEK293T cells for 24 h. GFP-WDR5 was immunoprecipitated using GFP-trap beads. E, the indicated proteins were overexpressed in HEK293T for 24 h. 5 μM CHIR-99021 or 5 μM MLN4924 were added for 9 h and 10 μM MG132 for 5 h prior to harvest. FLAG-WDR5 was immunoprecipitated using α-FLAG M2 beads in the presence of 20 mM N-ethylmaleimide. F, FLAG-FBXW7 or FLAG-FBXW7ARG (R465H, R479Q, and R505C mutations) were coexpressed with Myc-Cullin1 in HEK293T for 48 h and immobilized on α-FLAG M2 beads. In vitro ubiquitination assays of 200 nM GST-WDR5 or GST-c-Myc were performed with 170 nM Uba1, 250 nM of UbcH5b and CDC34, 30 μM ubiquitin, 200 nM His-GSK3β, 5 mM ATP, and immobilized FLAG-FBXW7 constructs at 37 °C for 90 min. DMSO, dimethyl sulfoxide; GSK3β, glycogen synthase kinase-3β; GST, glutathione-S-transferase; HEK293T, human embryonic kidney 293T cell line; l.e., long exposure; s.e., short exposure.
WDR5 and cyclin E1 promote mitotic slippage
Previous reports indicate that FBXW7 prevents mitotic slippage by promoting mitotic cell death in cells that were arrested in mitosis upon treatment with antimicrotubule drugs (5, 6). In addition (20, 28), it was shown that MLL and WDR5 regulate M-phase progression. Given the role of WDR5 in mitosis and the fact FBXW7 depletion leads to an upregulation of WDR5 protein levels (23) (Fig. 2), we asked whether overexpression of WDR5 would influence mitotic slippage. We generated an U2OS cell line where WDR5 could be ectopically induced by doxycycline addition. We analyzed mitotic cell fate by live-cell imaging (Fig. 5A, Videos S1 and S2) in the presence of spindle poisons at concentrations that prevent cell division and caused either mitotic cell death or mitotic slippage (29). Our results show that upregulation of WDR5 leads to an increase in mitotic slippage in cells that were arrested in mitosis with nocodazole (Fig. 5B).
Figure 5.
WDR5 and cyclin E1 promote mitotic slippage.A, still images from live-cell imaging with U2OS T-Rex WDR5 showing different mitotic cell fates in response to 830 nM nocodazole. Arrows indicate cells undergoing mitotic arrest and slippage or apoptosis. B, U2OS T-Rex WDR5 were cultured in medium containing 2 μg/ml doxycycline for 4 days before the addition of 830 nM nocodazole and live-cell imaging. Mitotic cell fate of n = 50 cells was determined for each experiment. ∗p < 0.05, two-way unpaired Student's t test, n = 3. C, U2OS Tet-Off cyclin E1 were cultured with or without 2 μg/ml doxycycline for 4 days. At days 2 and 3, cells were transfected with 30 nM siRNA targeting Gl2 of FBXW7, as indicated. About 48 h after the second transfection, 830 nM nocodazole were added and mitotic cell fates of n = 50 cells determined by live-cell imaging. ∗∗p < 0.01, ∗p < 0.05, ns p > 0.05, one-way ANOVA with Tukey post hoc test, n = 3. D, U2OS were transfected twice with 30 nM siRNA targeting Gl2, FBXW7, cyclin E1, or WDR5, as indicated. About 830 nM nocodazole were added 48 h after the second transfection, and mitotic cell fates of n = 50 cells were determined by live-cell imaging. ∗∗∗p < 0.001, ∗∗p < 0.01, p < 0.05, ns p > 0.05, one-way ANOVA with Tukey post hoc test, n = 3. ns, not significant.
Another key substrate of FBXW7 is cyclin E1. Previous work has shown that overexpression of cyclin E1 leads to accumulation of cells in early mitosis and chromosomal instability (30, 31). In addition, overexpression of cyclin E1 has been suggested to weaken nocodazole-dependent mitotic arrest (32). Therefore, we asked whether overexpression of cyclin E1 leads to increased mitotic slippage and less cell death in response to nocodazole. We used an U2OS cell line where cyclin E1 can be ectopically expressed after removal of doxycycline and performed live-cell imaging experiments (33). We observed that overexpression of cyclin E1 led to an increase in mitotic slippage to a similar extent as FBXW7 depletion. Simultaneous overexpression of cyclin E1 and FBXW7 depletion did not further increase slippage (Fig. 5C). These data suggest that mitotic slippage could be promoted by ectopic expression of single FBXW7 substrates.
Next, we asked whether downregulation of cyclin E1 and WDR5 could rescue the mitotic slippage phenotype induced upon depletion of FBXW7. As shown in Figure 5D, we find that the effect of FBXW7 depletion was partially rescued by knockdown of cyclin E1 but more pronounced by the depletion of WDR5 or the combined depletion of both proteins. Interestingly, knockdown of cyclin E1 or WDR5 did not reduce mitotic slippage in comparison to the siRNA control. We also showed that KMT2D depletion did not affect mitotic cell fate (Fig. 1C), suggesting that WDR5 exerts this function in regulation of mitotic cell fate independent of KMT2D. Taken together, our results support that WDR5 and cyclin E1 are involved in the FBXW7-dependent regulation of mitotic cell fate.
WDR5 and cyclin E1 are equally required for the formation of polyploidy in response to antimicrotubule drugs
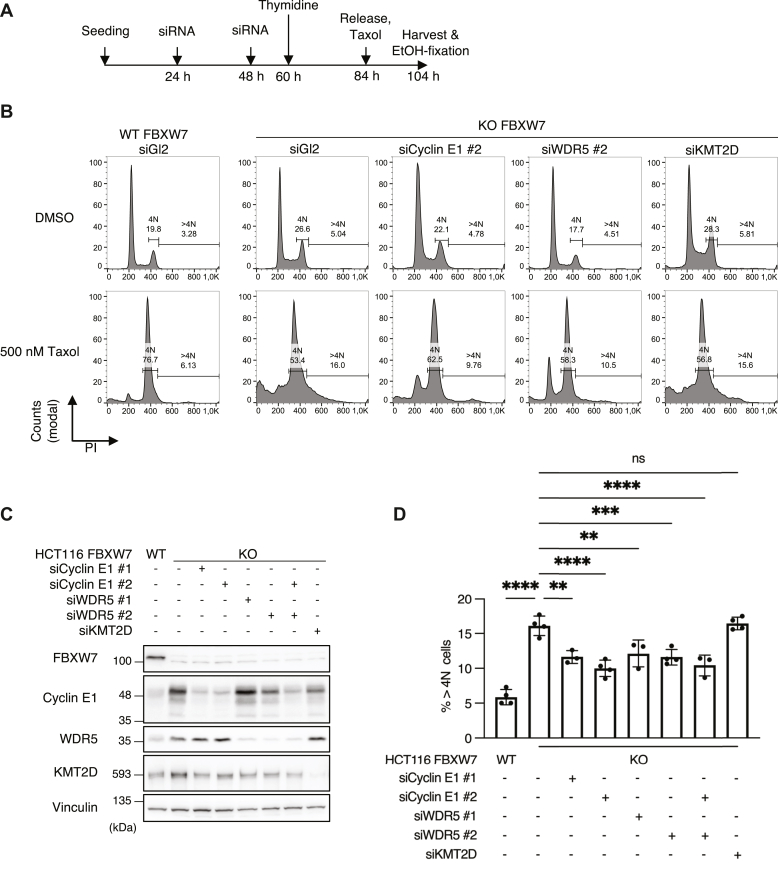
Mitotic slippage leads to the formation of tetraploid cells and further to polyploidy, which is reflected by an increase in DNA content. There is evidence for an involvement of cyclin E1 and aurora A in the formation of polyploidy after treating FBXW7-depleted cells with antimicrotubule drugs (5). Therefore, we asked whether WDR5 also plays a role in drug-induced polyploidy. We performed a fluorescence-activated cell sorting analysis of the DNA content in cells arrested in mitosis by addition of the microtubule-stabilizing drug taxol. HCT116 FBXW7 KO cells were treated with control, cyclin E1, WDR5, or KMT2D-specific siRNAs, respectively and further treated following the scheme shown in Figure 6A. Verification of increased WDR5 and cyclin E1 levels in response to FBXW7 KO and reduction of target proteins by siRNA treatment is demonstrated in Figure 6B. As previously shown (5), downregulation of FBXW7 leads to an increase of DNA content >4N in the presence of antimicrotubule drugs. Upon downregulation of WDR5 or cyclin E1 in FBXW7 KO HCT116, respectively, the increase in DNA content could be markedly reduced (Fig. 6C), suggesting that WDR5 and cyclin E1 function are required for formation of polyploid cells. Codepletion of WDR5 and cyclin E1 did not further decrease the DNA content (Fig. 6D). This could hint that both proteins support polyploidization through similar mechanisms. Depletion of KMT2D in HCT116 FBXW7 KO cells did not lead to a decrease in DNA content. Similar results were obtained when vincristine, a microtubule-destabilizing drug, was used instead of taxol to arrest cells in mitosis (Fig. S2). Taken together, these data provide evidence that WDR5 and cyclin E1 are equally required for the polyploidization of FBXW7-depleted cells in response to different antimicrotubule drugs.
Figure 6.
WDR5 and cyclin E1 are equally required for drug-induced polyploidy in response to antimicrotubule drugs.A, schematic depicting the protocol followed to determine drug-induced polyploidy of HCT116. HCT116 WT and FBXW7 KO were transfected twice with 30 nM siRNA and blocked in S-phase with 2 mM thymidine for 24 h. Cells were released into medium containing 500 nM taxol and harvested and fixed after 20 h. B, exemplary histograms showing DNA contents of HCT116 determined by FACS-Scan analysis. HCT116 were treated following the protocol in A, and DNA was stained with PI for 15 min at RT. Histograms are shown as modal counts and fraction of G2/M (4N) and polyploid (>4N) cells. C, HCT116 were transfected following the protocol from A but without synchronization and taxol treatment. Asynchronous cells were harvested 48 h after the second siRNA transfection. D, statistical analysis of B. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01, ns p > 0.05, one-way ANOVA with Tukey post hoc test, n = 3 to 4. FACS, fluorescence-activated cell sorting; ns, not significant; PI, propidium iodide; RT, room temperature.
Discussion
WDR5 localizes to the spindle microtubules during mitosis, and its depletion leads to chromosome misalignment and spindle assembly defects (20). Here, we report a new function of WDR5 in mitotic cell fate. We found not only that WDR5 plays a role in mitotic slippage caused by prolonged mitotic arrest in response to treatment with antimicrotubule drugs but also that WDR5 is a substrate and binding partner of the tumor suppressor FBXW7, which is involved in mitotic cell fate decision (5, 6, 12). High levels of WDR5 induced by the absence of FBXW7 promote mitotic slippage. In the future, it would be interesting to find out whether FBXW7-mediated degradation of WDR5 is also involved in canonical functions of WDR5, for instance, histone methylation (histone H3 lysine 4).
Antimicrotubule drugs arrest or delay cells in mitosis because of the activation of the mitotic or SAC (6). If the SAC cannot be satisfied anymore, mitosis remains uncompleted as cells either undergo cell death or perform mitotic slippage (12). MCL-1, a member of the antiapoptotic Bcl-2 family of proteins that suppress the activation of caspases, is degraded by ubiquitin-mediated degradation (6, 10, 34).
The tumor suppressor FBXW7 has been also implicated in cell fate decision, but it is not yet clear whether it plays a role in regulating MCL-1 protein levels (6, 11, 12). Therefore, it is important to identify and characterize substrates of FBXW7 involved in mitotic cell fate.
Similar to the regulation of KMT2D by FBXW7 (23), we show that WDR5 protein levels are regulated by FBXW7 through ubiquitin-mediated degradation suggesting that two proteins of the MLL4 complex are substrates of FBXW7. However, while WDR5 upregulation promotes mitotic slippage, KMT2D upregulation does not affect exit from drug-induced mitotic arrest suggesting that the deregulation of both proteins affects cancer cells differently. In addition, binding of FBXW7 to WDR5 does not require KMT2D (Fig. 2D). Together, our data implicate that FBXW7 targets WDR5 for degradation and that WDR5 upregulation supports chemoresistance of FBXW7-depleted cancer cells by mitotic slippage, whereas the FBXW7–KMT2D axis was previously shown to function in oxidative phosphorylation in B-cell malignancies (23). As the overexpression of each of the two FBXW7 substrates, WDR5 and cyclin E1, induced mitotic slippage, we suggest that both substrates work downstream of FBXW7 in mitotic slippage induction, but whether they act in the same pathway remains unclear. In addition, we cannot exclude that other FBXW7 substrates could also function in mitotic slippage. For example, c-Myc, another key substrate of FBXW7, promotes cell death following prolonged mitotic arrest (35).
In summary, our results open the venue for development of drugs to prevent mitotic slippage in response to treatment with antimicrotubule drugs. WDR5 has been characterized as a target for the development of small-molecule inhibitors (36, 37). Future work could involve testing of WDR5 inhibitors in FBXW7-deficient or mutated cancer types.
Experimental procedures
Cell lines and cell culture
HEK293T (catalog no.: ACC 635; DSMZ), HeLa (American Type Culture Collection CCL-2), HCT116 and HCT116 KO FBXW7 (B. Vogelstein, Johns Hopkins University), U2OS (American Type Culture Collection HTB-96), U2OS Flp-In-T-rex (J.D. Pravin, Ohio State University), and U2OS Tet-Off cyclin E1 (J. Bartek, Karolinska Institute) were maintained in Dulbecco’s modified Eagle’s medium containing 4.5 g/l glucose (Gibco; catalog no.: 41965-039). For U2OS Tet-Off cyclin E1, 2 μg/ml doxycycline (Sigma; catalog no.: D-9891) were added to the cell culture medium. DLD1 and DLD1 KO FBXW7 (B. Vogelstein) were cultivated in RPMI1640 (Sigma; catalog no.: R8758). All media were supplemented with 10% FBS (Gibco; catalog no.: 10270-106) and 1% penicillin/streptomycin (Sigma; catalog no.: P0781). Cell lines were grown in a humidified CO2 incubator at 37 °C and 5% CO2. U2OS Flp-In-T-Rex WDR5 and WDR5 F133A stable cell lines were generated following the manufacturer’s instructions (Life Technologies). Protein expression was induced by addition of 2 μg/ml doxycycline for at least 72 h. GSK3β activity was blocked with 5 μM CHIR-99021 (MedChemExpress; catalog no.: HY-10182) and cullin-RING E3 ubiquitin ligases with 5 μM MLN4924 (Cell Signaling Technology; catalog no.: 85923) for 5 h prior to cell harvest.
Plasmids, cloning, and mutagenesis
Human FBXW7α, WDR5, and c-Myc complementary DNAs (cDNAs) were obtained from the Genomics and Proteomics Core Facility at the DKFZ. Full-length FBXW7α was cloned into pCMV-3Tag1C and pEGFP-C1 via EcoRI and HindIII sites. WDR5 cDNA was subcloned to pCMV-3TagA using HindIII and XhoI to pEGFP-C2 with ApaI and EcoRI and to pGEX-4T1 with EcoRI and XhoI. c-Myc was subcloned to pGEX-4T1 using BamHI and XhoI. pCDNA3-FLAG-FBXW7α and pCDNA3-FLAG-FBXW7α T439I, S642A, T463A, R465A mutants were a gift from Michele Pagano (NYU Grossmann School of Medicine). pCVM-Tag5A-Myc-GSK3β was a gift from Mien-Chie-Hung (38) (Addgene; no.: 16260) and pcDNA3-myc3-Cullin1 was from Yue Xiong (39) (Addgene; no.: 19896). Mutants of cDNA were generated by site-directed mutagenesis. For the generation of stable WDR5-expressing cell lines using the T-Rex system, WDR5 constructs were subcloned to pcDNA5-FRT-TO using HindIII and XhoI.
Transient plasmid and siRNA transfections
HEK293T cells were transiently transfected with plasmid DNA using polyethylenimine (Polysciences) at a ratio of DNA 1:3.4 polyethylenimine. Proteins were overexpressed for 24 to 48 h. Lipofectamine 2000 (Invitrogen) was used to transfect DLD1 cell lines with plasmid DNA following the manufacturer’s instructions. Transfection mixes were removed 6 h after transfection, and fresh growth medium was added.
Lipofectamine 2000 was used for transfection of HeLa or U2OS, and Lipofectamine RNAiMAX (Invitrogen) was used for transfection of HCT116 with siRNA. Cells were transfected with 30 nM of siRNA 24 and 48 h after seeding and further cultivated for at least 48 h after the second siRNA transfection.
The following siRNA sequences were used:
Gl2 (firefly luciferase), 5′-CGUACGCGGAAUACUUCGAdTdT-3‘
pan-FBXW7, 5’-ACAGGACAGUGUUUACAAAdTdT-3‘ (40)
WDR5 1, 5’-UUAGCAGUCACUCUUCCACUUdTdT-3‘ (20)
WDR5 2, 5’- GCUCAGAGGAUAACCUUGUdTdT-3‘ (41)
Cyclin E1 1, 5‘-CCUCCAAAGUUGCACCAGUUUdTdT-3‘ (42)
Cyclin E1 2, 5‘-UGACUUACAUGAAGUGCUAdTdT-3‘
siGSK3β 1, 5’-GAAGUCAGCUAUACAGACAdTdT-3‘
siGSK3β 2, 5’-GGUCACGUUUGGAAAGAAUdTdT-3‘ (43)
siKMT2D, 5’-CCCACCUGAAUCAUCACCUdTdT-3’ (44)
Cell lysis, coimmunoprecipitation, and Western blot analysis
For Western blot analysis, cells were harvested and washed with ice-cold PBS. Cell pellets were lysed in three to five volumes of NP-40 or radioimmunoprecipitation assay buffer. NP-40 buffer (40 mM Tris–HCl [pH 7.5], 150 mM NaCl, 5 mM EDTA, 10 mM β-glycerophosphate, 5 mM NaF, 0.5% NP-40, 1 mM DTT, 10 μg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone, 5 μg/ml tosyl lysyl chloromethyl ketone, 0.1 mM NA3VO4, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 10 μg/ml trypsin inhibitor from soybean) was used for all immunoprecipitations. For all other samples, radioimmunoprecipitation assay buffer (50 mM Tris–HCl [pH 7.4], 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 150 mM NaCl, 2 mM EDTA, 50 mM NaF, 1 mM DTT, 10 μg/ml l-1-tosylamido-2-phenylethyl chloromethyl ketone, 5 μg/ml tosyl lysyl chloromethyl ketone, 0.1 mM NA3VO4, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 10 μg/ml trypsin inhibitor from soybean) was used. Cell lysates were incubated on ice for 30 min and cleared by centrifugation at 16,100g for 20 min. For SDS-PAGE, cell extracts were mixed with 2× Laemmli buffer or 2× LDS sample buffer (samples for KMT2D analysis) and incubated at 95 °C for 5 min or 72 °C for 10 min, respectively. Immunoprecipitations were performed as described previously (9). Immunoprecipitation of endogenous WDR5 and FBXW7 was performed using 12 mg lysate from HEK293T and 4 μg of mouse anti-WDR5 (G9; Santa Cruz Biotechnology) or rabbit anti-FBXW7 (catalog no.: A301-720; Bethyl) antibodies overnight. Precipitates were bound to protein G (mouse) or protein A (rabbit) Sepharose beads at 4 °C for 2 h and washed three times with NP-40 buffer adjusted to 600 mM NaCl. Proteins were eluted by incubation with 2× Laemmli buffer at 95 °C for 5 min.
Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membranes. For the analysis of KMT2D protein levels, a Tris-acetate gel and buffer system was used (45). Proteins were detected using following antibodies: α-FBXW7α (catalog no.: A301-720), α-Rbbp5 (catalog no.: A300-109A), α-KMT2D (catalog no.: A300-BL1185), α-MLL1 (catalog no.: A300-374A), and α-SETD1A (catalog no.: A300-289A) antibodies were obtained from Bethyl. α-WDR5 (G9), α-cyclin E1 (HE12), and α-Myc (9E10) antibodies were purchased from Santa Cruz Biotechnology, Inc; α-FLAG-tag (F3165), α-vinculin (V9131), and α-tubulin (T6074) antibodies were purchased from Sigma. α-c-Myc (9402) antibody was purchased from Cell Signaling Technology; α-HA-tag (16B12) antibody was purchased from Babco; and the α-GFP antibody was produced in-house.
GST pull-down assay
Recombinant GST-WDR5 was expressed in Escherichia coli Rosetta (DE3) and purified using glutathione CL-4B Sepharose (Sigma; catalog no.: 49739). His-FBXW7/Skp1 was a gift from Frauke Melchior. For in vitro GST pull-down assays, 10 μg of purified GST-WDR5 and 10 μg of His-FBXW7/Skp1 were mixed in 200 μl NP-40 buffer and incubated on a rotating wheel at 4 °C for 1 h. About 10 μl glutathione Sepharose beads were resuspended in 200 μl NP-40 buffer, added to the mixture, and incubated at 4 °C for 2 h. Beads were washed four times with NP-40 buffer, and proteins were eluted by incubation with 25 μl 2× Laemmli buffer at 95 °C for 5 min.
CHX chase
For the analysis of WDR5 protein stability, DLD1 WT and DLD1 KO FBXW7 were transfected with pCMV-3Tag1A-WDR5 using Lipofectamine 2000, and HEK293T were transfected with pCMV-3Tag1C-FBXW7 or FBXW7 ΔF-box (AA284–321 deletion) using polyethyleneimine. About 24 h after transfection, 300 μg/ml CHX (ChemCruz, Santa Cruz Biotechnology; catalog no.: SC-3508) were added to inhibit protein synthesis in DLD1 cells. HEK293T cells were treated with 100 μg/ml CHX 48 h after transfection. Samples were harvested at different time points for up to 24 h. Protein degradation via the 26S proteasome was blocked by adding 10 μM MG132 (Sigma–Aldrich; catalog no.: C2211) 3 h before the addition of CHX. About 10 μM MG132 were then also added to the medium with CHX.
Ubiquitination assays
HEK293T cells were transiently cotransfected with the indicated plasmids. About 24 h after transfection, cells were treated with 10 μM MG132 for 5 h. GSK3β or cullin-RING E3 ligase activity was blocked by addition of 5 μM CHIR-99021 or 5 μM MLN4924, respectively, for 4 h prior to addition of MG132. Cells were collected and lysed in complete NP-40 buffer with 20 mM N-ethylmaleimide (Sigma; catalog no.: E3876), and FLAG-immunoprecipitations were performed as described previously. Samples of whole cell extracts and eluates were denatured with LDS sample buffer for 10 min at 72 °C. Proteins were resolved by SDS-PAGE and detected after Western blotting.
In vitro ubiquitination assay
Recombinant GST-WDR5 and GST-c-Myc were expressed in E. coli Rosetta (DE3) and purified using glutathione CL-4B Sepharose. Uba1, CDC34, UbcH5b, and ubiquitin were kind gifts from Frauke Melchior (University of Heidelberg) (46). His-GSK3β was purchased from MRC PPU, University of Dundee. HEK293T cells were transfected with pCMV-3Tag1C-FBXW7 or FBXW7ARG and pCDNA3-3myc-Cullin1 and harvested after overexpression for 48 h. FLAG-immunoprecipitations were performed as described previously, but proteins were not eluted after washing. Ubiquitination assays were performed in 50 mM Tris–HCl (pH 7.50), 100 mM NaCl, 10 mM MgCl2, 0.05% CHAPS, 1 mM DTT, 0.1 mM NA3VO4, 1 μg/ml aprotinin, 1 μg/ml leupeptin, and 10 μg/ml trypsin inhibitor from soybean in 20 μl total volume. About 200 nM GST-WDR5 or GST-c-Myc were combined with 170 nM Uba1, 250 nM UbcH5b and 250 nM CDC34, 200 nM His-GSK3β, 30 μM ubiquitin, and α-FLAG M2 beads with immobilized FBXW7. Reactions were started by addition of 5 mM ATP, incubated at 37 °C, shaking for 90 min, and terminated by incubation with 2× LDS at 72 °C for 10 min. Proteins were resolved by SDS-PAGE and detected after Western blotting.
MS-based SCF–FBXW7 substrate screen
Eluates from α-FLAG-FBXW7 and FLAG-FBXW7 WD40 mutant (T439I, S642A, T463A, and R465A) immunoprecipitations were analyzed by SDS-PAGE and stained by Colloidal Coomassie. MS and data processing were performed by the EMBL Proteomics Core Facility Heidelberg. Briefly, whole gel lanes were cut, and proteins were reduced with 10 mM DTT in 100 mM NH4HCO3 for 30 min at 56 °C. About 180 μl acetonitrile were added at room temperature for 15 min, followed by replacement with 200 μl of 55 mM chloroacetamide in 100 mM NH4HCO3 and alkylation for 20 min in the dark. Gel pieces were washed twice with acetonitrile at room temperature for 15. Proteins were in-gel digested by addition of 200 μl of 2 ng/μl trypsin (Promega; catalog no.: V511A) in resuspension buffer (Promega; catalog no.: V542A) and incubation on ice for 30 min and then at 37 °C overnight. Peptides were extracted with 50% acetonitrile and 1% formic acid, dried, and reconstituted in 0.1% (v/v) formic acid. After a reverse-phase cleanup step, peptides were reconstituted in 10 μl of 100 mM Hepes/NaOH, pH 8.5, and reacted with 80 μg of TMT10plex (Thermo Scientific; catalog no.: 90111) dissolved in 4 μl acetonitrile for 1 h at room temperature. Peptides were mixed 1:1, subjected to reverse-phase clean-up step, and analyzed by LC–MS/MS on a Q Exactive Plus (Thermo Scientific) as previously described (47). Acquired data were analyzed using isobarQuant and Mascot V2.4 (Matrix Science). Peptide mass error tolerance was 10 ppm for full scan MS spectra and 0.02 Da for MS/MS spectra, and a false discovery rate below 0.01 was required on peptide and protein levels. Two replicates were performed for each condition, and results were compared using the LIMMA package for R.
Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Potential genomic DNA contaminants were digested with DNAse (Sigma; AMPD1), and first strand cDNA was synthesized with RevertAid cDNA Synthesis Kit (Thermo Scientific; K1621). Quantitative real-time PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems; catalog no.: 4368577), and PCRs were performed in a QuantStudio 5 cycler (Applied Biosystems). Relative mRNA expression was calculated using the ΔΔCT method.
WDR5: forward 5′-ATGCGACAGAGACCATCATAG-3‘
WDR5: reverse 5’-CGTGAGGATATGGGATGTGAA-3‘
GAPDH: forward 5’-CAAGGCTGAGAACGGGAAG-3‘
GAPDH: reverse 5’-TGAAGACGCCAGTGGACTC-3′
Live-cell imaging
Mitotic cell fates were analyzed by live-cell imaging. U2OS were transfected with 30 nM siRNA 24 and 48 h after seeding. About 24 h after second transfection, cells were seeded to 8-well imaging dishes (Sarstedt; catalog no.: 94.6170.802) at 50% confluency. For U2OS T-Rex WDR5 clones and U2OS Tet-Off cyclin E1, subcultures were cultivated in medium with or without 2 μg/ml doxycycline for 72 h before seeding to imaging dishes. In addition, U2OS Tet-Off cyclin E1 were transfected twice with 30 nM siRNA. About 24 h after seeding, 830 nM nocodazole were added to all wells, and the dish was placed in a microscopy incubation chamber at 5% CO2 and 37 °C. Cells undergoing mitotic arrest were monitored using a Zeiss Cell Observer Z1 inverted microscope and a 10×/0.3 EC PlnN Ph1 DICI objective. Phase-contrast images of multiple positions were taken every 10 min for up to 60 h using the Zeiss ZEN blue software. Cell fates during mitotic arrest were analyzed with ImageJ Fiji (NIH).
Flow cytometry analysis
DNA content of HCT116 and HCT116 KO FBXW7 was determined by DNA staining of EtOH-fixed cells with 30 μg/ml propidium iodide and flow cytometric analysis. DNA content was determined on a FACSCalibur (BD), and raw data were analyzed using FlowJo, version 10 (BD).
Statistical analysis
All statistical analyses, if not stated otherwise, were performed with GraphPad Prism, version 9 (GraphPad Software, Inc). Quantitative data were collected from at least three independent experiments and represented as mean ± SD. Statistical significance was analyzed by unpaired, two-tailed, Student’s t test with Welch’s correction or one-way ANOVA with Tukey post hoc test, as indicated in each figure legend. p Values of less than 0.05 were considered statistically significant (not significant, p > 0.05, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, and ∗∗∗∗p < 0.0001).
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD035501.
All other data are included in this article. Further information and raw data are available upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank J. Bartek, F. Melchior, M. Pagano, and B. Vogelstein for providing reagents and cell lines. We acknowledge the Microscopy Core Facility for providing equipment and technical assistance. We thank A. Turi da Fonte Dias for comments on the article. This work was supported by a grant from the Wilhelm Sander-Stiftung (grant no.: 2019.049.1.).
Author contributions
I. H. conceptualization; S. H.-K. and K. T. R. validation; S. H.-K. and K. T. R. data curation; S. H.-K., K. T. R., and I. H. writing–review & editing; I. H. supervision; I. H. project administration.
Edited by George DeMartino
Supporting information
2
References
- 1.Harper J.W., Schulman B.A. Cullin-RING ubiquitin ligase regulatory circuits: a quarter century beyond the F-box hypothesis. Annu. Rev. Biochem. 2021;90:403–429. doi: 10.1146/annurev-biochem-090120-013613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis R.J., Welcker M., Clurman B.E. Tumor suppression by the Fbw7 ubiquitin ligase: Mechanisms and opportunities. Cancer Cell. 2014;26:455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yumimoto K., Nakayama K.I. Recent insight into the role of FBXW7 as a tumor suppressor. Semin. Cancer Biol. 2020;67:1–15. doi: 10.1016/j.semcancer.2020.02.017. [DOI] [PubMed] [Google Scholar]
- 4.Cizmecioglu O., Krause A., Bahtz R., Ehret L., Malek N., Hoffmann I. Plk2 regulates centriole duplication through phosphorylation-mediated degradation of Fbxw7 (human Cdc4) J. Cell Sci. 2012;125:981–992. doi: 10.1242/jcs.095075. [DOI] [PubMed] [Google Scholar]
- 5.Finkin S., Aylon Y., Anzi S., Oren M., Shaulian E. Fbw7 regulates the activity of endoreduplication mediators and the p53 pathway to prevent drug-induced polyploidy. Oncogene. 2008;27:4411–4421. doi: 10.1038/onc.2008.77. [DOI] [PubMed] [Google Scholar]
- 6.Wertz I.E., Kusam S., Lam C., Okamoto T., Sandoval W., Anderson D.J., et al. Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature. 2011;471:110–114. doi: 10.1038/nature09779. [DOI] [PubMed] [Google Scholar]
- 7.Rieder C.L., Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell. 2004;7:637–651. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Cheng B., Crasta K. Consequences of mitotic slippage for antimicrotubule drug therapy. Endocr. Relat. Cancer. 2017;24:T97–T106. doi: 10.1530/ERC-17-0147. [DOI] [PubMed] [Google Scholar]
- 9.Richter K.T., Kschonsak Y.T., Vodicska B., Hoffmann I. FBXO45-MYCBP2 regulates mitotic cell fate by targeting FBXW7 for degradation. Cell Death Differ. 2020;27:758–772. doi: 10.1038/s41418-019-0385-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gascoigne K.E., Taylor S.S. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 2008;14:111–122. doi: 10.1016/j.ccr.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Sloss O., Topham C., Diez M., Taylor S. Mcl-1 dynamics influence mitotic slippage and death in mitosis. Oncotarget. 2016;7:5176–5192. doi: 10.18632/oncotarget.6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allan L.A., Skowyra A., Rogers K.I., Zeller D., Clarke P.R. Atypical APC/C-dependent degradation of Mcl-1 provides an apoptotic timer during mitotic arrest. EMBO J. 2018;37 doi: 10.15252/embj.201796831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., et al. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 14.Miller T., Krogan N.J., Dover J., Erdjument-Bromage H., Tempst P., Johnston M., et al. Compass: a complex of proteins associated with a trithorax-related SET domain protein. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuettengruber B., Martinez A.M., Iovino N., Cavalli G. Trithorax group proteins: Switching genes on and keeping them active. Nat. Rev. Mol. Cell Biol. 2011;12:799–814. doi: 10.1038/nrm3230. [DOI] [PubMed] [Google Scholar]
- 16.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 17.Orpinell M., Fournier M., Riss A., Nagy Z., Krebs A.R., Frontini M., et al. The ATAC acetyl transferase complex controls mitotic progression by targeting non-histone substrates. EMBO J. 2010;29:2381–2394. doi: 10.1038/emboj.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ali A., Veeranki S.N., Tyagi S. A SET-domain-independent role of WRAD complex in cell-cycle regulatory function of mixed lineage leukemia. Nucl. Acids Res. 2014;42:7611–7624. doi: 10.1093/nar/gku458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey J.K., Fields A.T., Cheng K., Lee A., Wagenaar E., Lagrois R., et al. WD repeat-containing protein 5 (WDR5) localizes to the midbody and regulates abscission. J. Biol. Chem. 2015;290:8987–9001. doi: 10.1074/jbc.M114.623611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ali A., Veeranki S.N., Chinchole A., Tyagi S. MLL/WDR5 complex regulates Kif2A localization to ensure chromosome congression and proper spindle assembly during mitosis. Dev. Cell. 2017;41:605–622.e607. doi: 10.1016/j.devcel.2017.05.023. [DOI] [PubMed] [Google Scholar]
- 21.Hao B., Oehlmann S., Sowa M.E., Harper J.W., Pavletich N.P. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 22.Welcker M., Clurman B.E. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 23.Saffie R., Zhou N., Rolland D., Onder O., Basrur V., Campbell S., et al. FBXW7 triggers degradation of KMT2D to favor growth of diffuse large B-cell lymphoma cells. Cancer Res. 2020;80:2498–2511. doi: 10.1158/0008-5472.CAN-19-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel A., Dharmarajan V., Cosgrove M.S. Structure of WDR5 bound to mixed lineage leukemia protein-1 peptide. J. Biol. Chem. 2008;283:32158–32161. doi: 10.1074/jbc.C800164200. [DOI] [PubMed] [Google Scholar]
- 25.Welcker M., Singer J., Loeb K.R., Grim J., Bloecher A., Gurien-West M., et al. Multisite phosphorylation by Cdk2 and GSK3 controls cyclin E degradation. Mol. Cell. 2003;12:381–392. doi: 10.1016/s1097-2765(03)00287-9. [DOI] [PubMed] [Google Scholar]
- 26.Gregory M.A., Qi Y., Hann S.R. Phosphorylation by glycogen synthase kinase-3 controls c-myc proteolysis and subnuclear localization. J. Biol. Chem. 2003;278:51606–51612. doi: 10.1074/jbc.M310722200. [DOI] [PubMed] [Google Scholar]
- 27.Wei W., Jin J., Schlisio S., Harper J.W., Kaelin W.G., Jr. The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer cell. 2005;8:25–33. doi: 10.1016/j.ccr.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Ali M., Hom R.A., Blakeslee W., Ikenouye L., Kutateladze T.G. Diverse functions of PHD fingers of the MLL/KMT2 subfamily. Biochim. Biophys. Acta. 2014;1843:366–371. doi: 10.1016/j.bbamcr.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brito D.A., Yang Z., Rieder C.L. Microtubules do not promote mitotic slippage when the spindle assembly checkpoint cannot be satisfied. J. Cell Biol. 2008;182:623–629. doi: 10.1083/jcb.200805072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spruck C.H., Won K.A., Reed S.I. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 31.Keck J.M., Summers M.K., Tedesco D., Ekholm-Reed S., Chuang L.C., Jackson P.K., et al. Cyclin E overexpression impairs progression through mitosis by inhibiting APC(Cdh1) J. Cell Biol. 2007;178:371–385. doi: 10.1083/jcb.200703202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bagheri-Yarmand R., Nanos-Webb A., Biernacka A., Bui T., Keyomarsi K. Cyclin E deregulation impairs mitotic progression through premature activation of Cdc25C. Cancer Res. 2010;70:5085–5095. doi: 10.1158/0008-5472.CAN-09-4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartkova J., Horejsi Z., Koed K., Kramer A., Tort F., Zieger K., et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 34.Harley M.E., Allan L.A., Sanderson H.S., Clarke P.R. Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its Cdc20-dependent destruction during mitotic arrest. EMBO J. 2010;29:2407–2420. doi: 10.1038/emboj.2010.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topham C., Tighe A., Ly P., Bennett A., Sloss O., Nelson L., et al. MYC is a major determinant of mitotic cell fate. Cancer Cell. 2015;28:129–140. doi: 10.1016/j.ccell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grebien F., Vedadi M., Getlik M., Giambruno R., Grover A., Avellino R., et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPalpha N-terminal leukemia. Nat. Chem. Biol. 2015;11:571–578. doi: 10.1038/nchembio.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu X., Li D., Kottur J., Shen Y., Kim H.S., Park K.S., et al. A selective WDR5 degrader inhibits acute myeloid leukemia in patient-derived mouse models. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abj1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou B.P., Deng J., Xia W., Xu J., Li Y.M., Gunduz M., et al. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 2004;6:931–940. doi: 10.1038/ncb1173. [DOI] [PubMed] [Google Scholar]
- 39.Ohta T., Michel J.J., Schottelius A.J., Xiong Y. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol. Cell. 1999;3:535–541. doi: 10.1016/s1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 40.Busino L., Millman S.E., Scotto L., Kyratsous C.A., Basrur V., O'Connor O., et al. Fbxw7alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat. Cell Biol. 2012;14:375–385. doi: 10.1038/ncb2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen X., Xie W., Gu P., Cai Q., Wang B., Xie Y., et al. Upregulated WDR5 promotes proliferation, self-renewal and chemoresistance in bladder cancer via mediating H3K4 trimethylation. Sci. Rep. 2015;5:8293. doi: 10.1038/srep08293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takada M., Zhang W., Suzuki A., Kuroda T.S., Yu Z., Inuzuka H., et al. FBW7 loss promotes chromosomal instability and tumorigenesis via cyclin E1/CDK2-mediated phosphorylation of CENP-A. Cancer Res. 2017;77:4881–4893. doi: 10.1158/0008-5472.CAN-17-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weikel K.A., Cacicedo J.M., Ruderman N.B., Ido Y. Knockdown of GSK3beta increases basal autophagy and AMPK signalling in nutrient-laden human aortic endothelial cells. Biosci. Rep. 2016;36 doi: 10.1042/BSR20160174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang L., Jin M., Jung N., Jeong K.W. MLL2 regulates glucocorticoid receptor-mediated transcription of ENACalpha in human retinal pigment epithelial cells. Biochem. Biophys. Res. Commun. 2020;525:675–680. doi: 10.1016/j.bbrc.2020.02.046. [DOI] [PubMed] [Google Scholar]
- 45.Cubillos-Rojas M., Amair-Pinedo F., Tato I., Bartrons R., Ventura F., Rosa J.L. Simultaneous electrophoretic analysis of proteins of very high and low molecular mass using Tris-acetate polyacrylamide gels. Electrophoresis. 2010;31:1318–1321. doi: 10.1002/elps.200900657. [DOI] [PubMed] [Google Scholar]
- 46.Schweiggert J., Habeck G., Hess S., Mikus F., Beloshistov R., Meese K., et al. SCF(Fbxw5) targets kinesin-13 proteins to facilitate ciliogenesis. EMBO J. 2021;40 doi: 10.15252/embj.2021107735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Becher I., Andres-Pons A., Romanov N., Stein F., Schramm M., Baudin F., et al. Pervasive protein thermal stability variation during the cell cycle. Cell. 2018;173:1495–1507.e1418. doi: 10.1016/j.cell.2018.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
2
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD035501.
All other data are included in this article. Further information and raw data are available upon request.