Abstract
There are two alleles of the vacuolating cytotoxin gene from Helicobacter pylori, which code for toxins with different cell specificities. By analyzing the phenotypes of natural and artificial chimeras between the two forms of the protein, we have delimited a short stretch of amino acids which determine the cell specificity.
Helicobacter pylori produces a potent excreted cytotoxin, VacA, which causes a massive vacuolar degeneration in the target cells in vitro and gastric epithelial erosion in vivo (4, 13). VacA is found as high-molecular-weight oligomeric structures of an 87-kDa polypeptide with either six- or sevenfold radial symmetry (9, 10). Each monomer can be cleaved proteolytically at a specific site into two fragments of approximately 37 and 58 kDa that remain associated in the holotoxin (14). Biological and structural data suggest that VacA is similar to the AB family of dichain toxins, which consist of two distinct moieties, A and B, involved in toxic activity and membrane interaction, respectively (5, 11, 14).
VacA binds to the surfaces of target cells and then is translocated to the cytosol, where it is active (6, 7). Intracellular expression of a transfected truncated vacA gene from which most of the sequence coding for the 58-kDa subunit has been deleted results in cell vacuolation, indicating that 37-kDa toxin subunit is responsible for the vacuolating activity (6). More recently, Reyrat et al. (14) have shown that the 58-kDa subunit is involved in binding to the target cell. Toxicity has been associated with mosaicism in vacA genes (1, 5). Two variants of the middle region of the gene, m1 and m2, have been described (1). Most isolates with the m1 form are toxic to HeLa cells, whereas the m2 forms are essentially nontoxic to these cells. However, both forms are toxic to primary cultured human gastric cells and the rabbit kidney epithelial cell line RK-13 (11). Hence, the m2 form of VacA is fully toxic if the appropriate cells are used, which is consistent with the lack of statistical correlation between vacA allele or cytotoxicity to HeLa cells and disease (8) and the high incidence of peptic ulcer and gastric cancer found in the Chinese population, where the m2 allele is prevalent (12). We have previously shown that the absence of activity of the m2 cytotoxin in HeLa cells is due to a lack of interaction with the cell, indicating that the two forms of the toxin differ in their binding domains (11).
The major difference between the two types of protein is in an approximately 300-amino-acid region in the 58-kDa subunit (1, 5), and it is hence likely that it is this difference which determines the cell specificity of the toxin (11). In order to demonstrate this and to delimit the functional differences more precisely, we have analyzed the toxic activities of the products of natural and artificial chimeric vacA genes in which different parts of the midregion of an m1 gene have been replaced by the corresponding regions from an m2 gene.
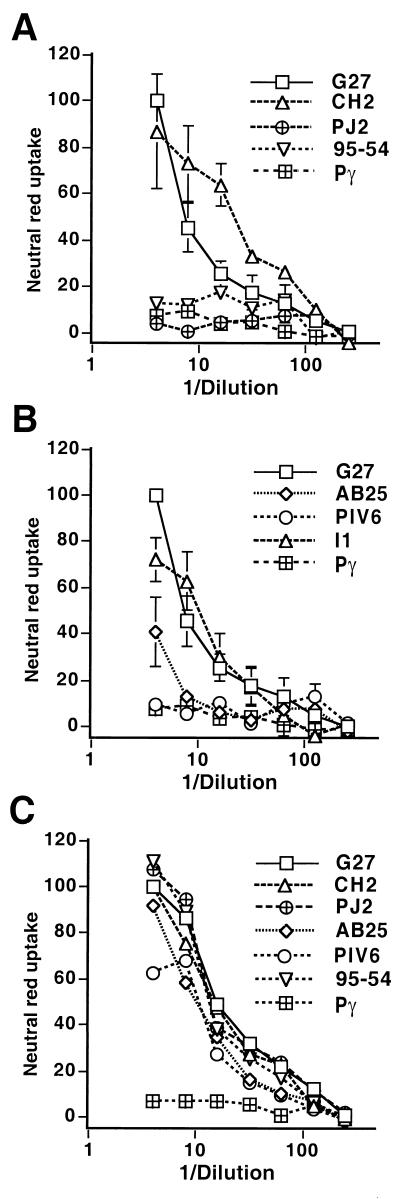
Recently, unusual vacA gene hybrids (m1-type proximal and m2-type distal) were identified (2, 12). We have sequenced the complete vacA gene from a naturally chimeric m1-m2 H. pylori strain, ch2 (GenBank accession no. AF191639) (2). The predicted product of this gene is highly similar to the m1 form of the protein up to amino acid 648 and is highly similar to the m2 form from amino acid 657 onwards (Fig. 1). Both water extracts and purified cytotoxin from ch2 induced vacuolation and neutral red uptake in HeLa cells to levels similar to those induced by a canonical m1 toxin from strain G27 (Fig. 2A). A canonical m2 toxin from strain 95-54 (11) failed to induce vacuolation in this assay but was fully toxic to RK13 cells (Fig. 2C). Hence, the chimeric ch2 toxin has the m1 toxic phenotype, indicating that the midregion from amino acid 657 onwards plays no role in the phenotypic difference between the m1 and m2 forms.
FIG. 1.
Box shade (http://www.ch.embnet.org/software/box_form.html) of predicted amino acid sequences of the 58-kDa subunit of ch2 VacA with the sequences of strain 11638 (m1) and 95-54 (m2) (GenBank accession no. AF191639, S72494, and U95971, respectively). Identical amino acid residues are shaded. The breakpoint between the m1-like and m2-like regions of ch2 is shown between amino acids 648 and 657. The bar above the sequence shows the region essential for the m1 phenotype. The white regions in the bar are above amino acids which are consistently different between the m1 and m2 forms.
FIG. 2.
Neutral red uptake assays of HeLa and RK13 cells treated with water extracts containing the chimeric VacA proteins. (A and B) Results of the vacuolation of HeLa cells (shown in two panels for clarity). (C) Results of vacuolation of RK13 cells. The data are the averages from three experiments, each in duplicate, and are normalized to the value of the lowest dilution of G27 in each experiment, which is taken as 100%. Error bars show standard deviations between experiments.
The above result indicates that the first 148 amino acids of the midregion between amino acids 501 and 647 contain the determinants of toxicity to HeLa cells. To demonstrate this, we have engineered the vacA gene in m1 strain G27 in order to replace this region with the corresponding region from the m2-type vacA gene from strain 95-54. In addition, we have dissected this region further by two chimeric vacA genes in which the sequences coding for amino acids 496 to 535 and 535 to 696 have been replaced by the corresponding m2 sequences.
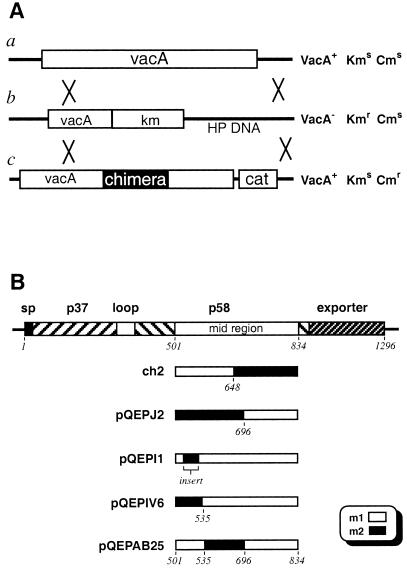
The strategy used to generate the chimeric vacA genes in G27 is shown in Fig. 3A. First, a G27 recipient strain was created by natural transformation with a plasmid construct containing the vacA gene sequences from m1 strain CCUG17874 (14) coding for the 37-kDa subunit, followed by a kanamycin resistance cassette, followed by about 1,000 bp of sequence immediately downstream of the stop codon (GenBank accession no. AF191640). Hence, in the resulting kanamycin-resistant strain (H. pylori G27::Pγ) the vacA gene sequences coding for the whole of the 58-kDa subunit and outer membrane exporter have been replaced by the kanamycin resistance cassette. Chimeric midregions were created in a delivery plasmid, PQE30Delivery (GenBank accession no. AF191638), which contains the complete CCUG17874 vacA gene, followed immediately by a chloramphenicol resistance gene, followed by vacA gene 3′ flanking sequences (3). The vacA gene sequences in this plasmid between the unique EcoNI and AflII sites were replaced by PCR products corresponding to the chimeric midregion. The PCR products were prepared in two steps. First, the parts of the m1 and m2 genes were amplified using primers based on their respective sequences. For each construct, the 3′ primer on the left part was complementary to the 5′ primer of the right part. The products of these PCRs were purified and mixed in a second reaction containing only the leftmost and rightmost primers such that a product could be obtained only if the complementary regions of the two fragments annealed through the overlapping complementary sequences. The resulting chimeric delivery constructs were transformed into the recipient strain G27::Pγ, and gene replacement strains PJ2 (GenBank accession no. AF191637), PIV6 (GenBank accession no. AF191636), and PAB25 (GenBank accession no. AF191635) were identified by resistance to chloramphenicol and sensitivity to kanamycin. The products of the constructs are shown schematically in Fig. 3B.
FIG. 3.
(A) Allelic exchange strategy. The vacA gene of wild-type strain G27 (a) is replaced by a double crossover event with that of construct Pr (b) to generate the recipitent strain G27::Pr, resulting in resistance to kanamycin. Allelic exchange between the Pr sequences and those of the delivery constructs (c) containing the chimeric genes results in resistance to chloramphenicol and sensitivity to kanamycin. The antibiotic resistance phenotypes of the resulting strains are shown at the right. (B) Schematic representation of the constructed chimeras. The shaded box shows replacement of m1 vacA sequence by m2 sequences. Amino acid positions starting from the initiation codon of the vacA gene (strain 11638) are shown.
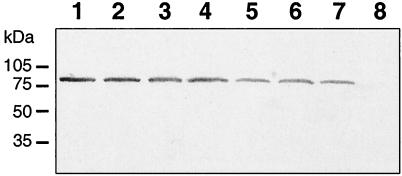
Water extracts from these strains were prepared, and the presence of VacA was determined by immunoblot analysis using rabbit sera raised against a recombinant m1 form of the protein (15). The results demonstrate that the mutant strains expressed mature 87-kDa VacA protein (Fig. 4). Laser densitometry of the filter revealed that the weakest band was 50% as intense as that of the parental G27 strain. However antibodies raised against the m1 form of the protein recognize the m2 form less well (11). The water extract of strain G27::PJ2 failed to induce vacuolation of HeLa cells (Fig. 2A). However, full toxic activity, equivalent to that of the parent G27 strain, was found when RK13 cells were used in the assay (Fig. 2C). Hence, the PJ2 chimera has the m2 toxic phenotype, thus confirming that the functional difference in toxic phenotype between the m1 and m2 forms resides in the first 160 amino acids of the midregion. A water extract from G27::PIV6 (Fig. 2B) also failed to induce vacuolation in HeLa cells but was fully toxic to RK13 cells (Fig. 2C). In contrast, a water extract from G27::AB25 induced vacuolation in HeLa cells but to a much lesser extent than the parental G27 strain. The reduction in toxicity was not due to lower expression or misfolding of this chimeric protein, since the same water extract was as toxic as that of the parental strain when RK13 cells were used in the assay.
FIG. 4.
Immunoblot of water extracts from H. pylori expressing chimeric VacA proteins. Lane 1, wild-type G27; lane 2, 95-54; lane 3, ch2; lane 4, G27::PJ2; lane 5, G27::PI1; lane 6, G27::PAB25; lane 7, G27::PIV6, lane 8, G27::Pr. Molecular mass standards are indicated on the left.
These data demonstrate that the amino acid differences between the m1 and m2 forms in a short segment of the toxin at the 5′ extremity of the midregion play a major role in defining cell specificity. m2 forms of the toxin have a 21-amino-acid insert in this region which is lacking in m1 forms. This insert, however, plays no role in determining cell specificity, since insertion of these sequences into the G27 vacA gene (G27::PI1) had no effect on the HeLa cell toxicity of the VacA product (Fig. 2B).
The data define a short region of 148 amino acids from the beginning of the midregion to the m1-m2 junction in strain ch2 which determines the phenotypic differences in target cell specificity between the m1 and m2 forms of the protein. Within this region, the first 35 amino acids must correspond to the m1 sequence for HeLa cell cytotoxicity. Excluding the 21-amino-acid insert, there are 13 amino acid differences between the m1 and m2 sequences used. However, an analysis of a number of genes from different geographic areas (X. Ji and J. L. Telford, unpublished data) reveals that only 10 of these positions consistently differ between the m1 and m2 forms (Fig. 1). The next 113 amino acids appear to influence the potency but do not completely eliminate toxicity to HeLa cells. In this region there are 49 amino acid differences between the genes used, of which 31 are consistently different between the m1 and m2 forms. These differences, however, do not affect the innate toxicity or functionality of the toxin, since all chimeric proteins tested were fully toxic to RK13 cells.
Pagliaccia et al. (11) have demonstrated that lack of toxicity of an m2 form of the protein correlates with lack of cell surface binding in HeLa cells and have postulated different receptors for the two forms. A key result from these studies was that the m2 form was fully toxic to HeLa cells if the lack of surface interaction could be overcome by intracellular expression of the protein. From these data, however, it is not possible to distinguish between the existence of two independent receptors, one for each form of the toxin, or a common receptor for both forms on RK13 cells and a different m1-specific receptor on HeLa cells. Hence, although it is clear that the region identified above is necessary for interaction with the HeLa cell receptor, it cannot be excluded that other regions of the protein are also involved. Furthermore, the results could also be explained if the receptor is polymorphic either in protein sequence or glycosylation such that the m2 toxin fails to recognize the HeLa cell isoform. Finally, the data raise the question of how or why the protein has evolved in two forms with such large differences over a 300-amino-acid segment if only a relatively short region determines cell specificity. These questions will require identification of the receptor(s) and an elucidation of the mechanism of interaction.
Acknowledgments
We thank A. Muzzi for synthesis of the oligonucleotides and S. Guidotti for automated sequencing of the vacA locus. We are grateful to Nathalie Norhais and Cristina Pagliaccia for their assistance in purification of the cytotoxin from the ch2 strain.
This work was supported by EU grants IC18CT95-0024 and National Natural Science Foundation of China grant 39670648.
REFERENCES
- 1.Atherton J C, Cao P, Reek R M, Tummuru M K R, Blaser M J. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 2.Atherton J C, Cover T L, Twells R J, Morales M R, Hawkey C J, Blaser M J. Simple and accurate PCR-based system for typing vacuolating cytotoxin alleles of Helicobacter pylori. J Clin Microbiol. 1999;37:2979–2982. doi: 10.1128/jcm.37.9.2979-2982.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burroni D, Lupetti P, Pagliaccia C, Reyrat J M, Dallai R, Rappuoli R, Telford J L. Deletion of the major proteolytic site of the Helicobacter pylori cytotoxin does not influence toxin activity but favors assembly of the toxin into hexameric structures. Infect Immun. 1998;66:5547–5550. doi: 10.1128/iai.66.11.5547-5550.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Covacci A, Telford J L, Del Giudice G, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 5.Cover T L, Tummuru M K R, Cao P, Thompson S A, Blaser M J. Divergence of genetic sequences for the vacuolating cytotoxin among Helicobacter pylori strains. J Biol Chem. 1994;269:10566–10573. [PubMed] [Google Scholar]
- 6.de Bernard M, Burroni D, Papini E, Rappuoli R, Telford J, Montecucco C. Identification of the Helicobacter pylori VacA toxin domain active in the cell cytosol. Infect Immun. 1998;66:6014–6016. doi: 10.1128/iai.66.12.6014-6016.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garner J A, Cover T L. Binding and internalization of the Helicobacter pylori vacuolating cytotoxin by epithelial cells. Infect Immun. 1996;64:4197–4203. doi: 10.1128/iai.64.10.4197-4203.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Go M F, Cissell L, Graham D Y. Failure to confirm association of vacA gene mosaicism with duodenal ulcer disease. Scand J Gastroenterol. 1998;33:132–136. doi: 10.1080/00365529850166842. [DOI] [PubMed] [Google Scholar]
- 9.Lanzavecchia S, Lupetti P, Bellon P L, Dallai R, Rappuoli R, Telford J L. Three dimensional reconstruction of metal replicas of the H. pylori vacuolating cytotoxin. J Struct Biol. 1998;121:9–18. doi: 10.1006/jsbi.1997.3941. [DOI] [PubMed] [Google Scholar]
- 10.Lupetti P, Heuser J E, Manetti R, Massari P, Lanzavecchia S, Bellon P L, Dallai R, Rappuoli R, Telford J L. Oligomeric and subunit structure of the Helicobacter pylori vacuolating cytotoxin. J Cell Biol. 1996;133:801–807. doi: 10.1083/jcb.133.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pagliaccia C, de Bernard M, Lupetti P, Ji X, Cover T L, Papini F, Rappuoli R, Telford J L, Reyrat J M. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc Natl Acad Sci USA. 1998;95:10212–10217. doi: 10.1073/pnas.95.17.10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Z J, Berg D E, van der Hulst R W M, Su W W, Raudonikiene A, Xiao S D, Dankert J, Tytgat G N J, van der Ende A. Prevalence of vacuolating cytotoxin production and distribution of distinct vacA alleles in Helicobacter pylori from China. J Infect Dis. 1998;178:220–226. doi: 10.1086/515601. [DOI] [PubMed] [Google Scholar]
- 13.Reyrat J M, Pelicic V, Papini E, Montecucco C, Rappuoli R, Telford J L. Towards deciphering the Helicobacter pylori cytotoxin. Mol Microbiol. 1999;34:197–204. doi: 10.1046/j.1365-2958.1999.01592.x. [DOI] [PubMed] [Google Scholar]
- 14.Reyrat J M, Lanzavecchia S, Lupetti P, de Bernard M, Pagliaccia C, Pelicic V, Charrel M, Ulivieri C, Norais N, Ji X, Cabiaux V, Papini E, Rappuoli R, Telford J. 3D imaging of the 58 kDa cell binding subunit of the Helicobacter pylori cytotoxin. J Mol Biol. 1999;290:459–470. doi: 10.1006/jmbi.1999.2877. [DOI] [PubMed] [Google Scholar]
- 15.Telford J L, Ghiara P, Dell Orco M, Burroni D, Bugnoli M, Tecce M, Censini S, Covacci A, Xiang Z, Pappini E, Rappuoli R. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. J Exp Med. 1994;179:420–460. doi: 10.1084/jem.179.5.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]