Abstract
Vietnam is a low- and middle-income country (LMIC), a primary food producer, and an antimicrobial resistance (AMR) hotspot. AMR is recognized as a One Health challenge since it may transfer between humans, animals and the environment. This study aimed to apply systematic review and meta-analysis to investigate the phenotypic profiles and correlations of antimicrobial-resistant Enterobacteriaceae across three compartments: humans, animals and the environment in Vietnam. A total of 89 articles found in PubMed, Science Direct, and Google Scholar databases were retrieved for qualitative synthesis. E. coli and non-typhoidal Salmonella (NTS) were the most common bacterial species in studies of all compartments (60/89 studies). Among antimicrobials classified as critically important, the resistance levels were observed to be highest to quinolones, 3rd generation of cephalosporins, penicillins, and aminoglycosides. Of 89 studies, 55 articles reported the resistance prevalence of E. coli and NTS in healthy humans, animals and the environment against ciprofloxacin, ceftazidime, ampicillin, gentamicin, sulfamethoxazole-trimethoprim, chloramphenicol was used for meta-analysis. The pooled prevalence was found highest in E. coli against ampicillin 84.0% (95% CI 73.0–91.0%) and sulfamethoxazole-trimethoprim 66.0% (95% CI 56.0–75.0%) while in NTS they were 34.0% (95% CI 24.0–46.0%), 33.0% (95% CI 25.0–42.0%), respectively. There were no significant differences in the pooled prevalence of E. coli and NTS to these antimicrobials across healthy humans, animals and the environment, except for ceftazidime-resistant E. coli (χ2 = 8.29, p = 0.02), chloramphenicol-resistant E.coli (χ2 = 9.65, p < 0.01) and chloramphenicol-resistant NTS (χ2 = 7.51, p = 0.02). Findings from the multiple meta-regression models indicated that the AMR levels in E. coli (β = 1.887, p < 0.001) and the North (β = 0.798, p = 0.047) had a higher fraction of AMR than NTS and other regions of Vietnam. The outcomes of this study play an important role as the baseline information for further investigation and follow-up intervention strategies to tackle AMR in Vietnam, and more generally, can be adapted to other LMICs.
Keywords: Antimicrobial resistance, Enterobacteriaceae, One Health, Systematic review, Meta-analysis, Vietnam
Highlights
-
•
This review reports the high AMR levels of E. coli and NTS in Vietnam, especially to critically important antimicrobials.
-
•
There was a marginal difference in the AMR levels in the three compartments of humans, animals and the environment.
-
•
The North of Vietnam and E. coli had higher levels of AMR compared with other regions of Vietnam and NTS.
-
•
The results highlight the gaps in research for further investigation and future strategies to mitigate AMR in Vietnam.
-
•
Implementation of AMR surveillance- under the One Health approach is required to tackle the AMR problems in Vietnam.
1. Introduction
Antimicrobial resistance (AMR) has been declared by the World Health Organization (WHO) as a top ten global public health threat [1]. AMR leads to ineffective treatment of infectious diseases, resulting in increased mortality rates, economic losses and compromised food security [2]. Annually, it is estimated that around 5 million persons die due to AMR bacterial infections, with LMICs bearing the heaviest burden [3].
Bacterial species belonging to the Enterobacteriaceae family are included in AMR surveillance programmes worldwide. In food animal production systems, both commensal E. coli and non-typhoidal Salmonella (NTS) are commonly included in monitoring programs [4,5]. In addition to NTS and E. coli (both commensal and pathogenic), other Enterobacteriaceae including Klebsiella spp., and Shigella spp., have been monitored for their AMR profiles in human populations [6]. Resistance to the last-resort antimicrobial drugs (such as carbapenem and colistin) among Enterobacteriaceae has increased globally, especially in LMICs [[7], [8], [9]]. Increased resistance among Enterobacteriaceae is partly associated with the overuse of antimicrobials in animal production systems [[10], [11], [12], [13]].
Vietnam is currently classified by the World Bank as a low- and middle-income country. The agricultural sector (including animal production) represents 15.0% of the national gross domestic product [14]. Vietnam is also a hotspot of AMR [15,16]. A significant driver of AMR in Vietnam and elsewhere is the indiscriminate use of antimicrobials in animal production, which is estimated to account for ∼70.0% of the total consumption of antimicrobials in the country [17]. In the last decade, annual poultry production in the country has increased from 2010 to 2020 (from 300.5 to 512.7 million chickens, a 70.6% increase). Pig populations also increased after the incursion of African Swine Fever in January 2019; the total population resumed by 13.3% in 2020, reaching 22.0 million pigs from 19.6 million pigs in 2019 [18]. The increased antimicrobial consumption is linked to the intensification of animal production, especially for prevention and growth promotion purposes [19]. The most commonly identified antimicrobial-resistant bacterial species belong to the Enterobacteriaceae family, mainly E. coli, followed by non-typhoidal Salmonella and Klebsiella spp. [20].
Since AMR bacteria and AMR-encoding genes can flow between humans, animals and the environment, applying a One Health approach is crucial to tackling this problem [21]. The present study aims to describe and analyze the phenotypic profiles and correlations of antimicrobial-resistant Enterobacteriaceae in Vietnam's three One Health compartments (humans, animals and the environment). Findings from this review highlight the gaps in research for further investigation and future strategies to mitigate AMR in Vietnam. These findings may potentially be used as a model for other LMICs in the region.
2. Materials and methods
2.1. Study protocol
A systematic review was performed according to the PRISMA guideline [22], and all 27 checklist items were addressed in the study (Supplementary File 1).
2.2. Search strategy
Research articles were identified through (1) PubMed, (2) ScienceDirect, (3) Google Scholar, and (4) Vietnamese journal websites. Boolean Logic tools with the connectors ‘AND’, ‘OR’ were applied to search the articles. The keywords for advanced search used in PubMed were: (antimicrobial resistance OR antibiotic resistance) AND (human OR animal OR environment) AND (Enterobacteriaceae OR E. coli OR Escherichia coli OR Salmonella OR Klebsiella OR Proteus OR Shigella OR Enterobacter OR Citrobacter OR Cronobacter) AND (livestock OR poultry OR cattle OR swine OR pig OR chicken OR fish OR shrimp OR shellfish) AND (meat OR pork OR beef OR milk) AND (environment OR soil OR water OR wastewater OR sludge OR vegetable OR drainage) AND (Vietnam OR Vietnamese); and in ScienceDirect: (antimicrobial OR antibiotic) AND (resistance OR susceptibility OR sensitivity) AND (Vietnam OR Vietnamese). Furthermore, hand-searching in Google Scholar, Vietnamese journal websites, and reference lists of selected articles were also applied to have a higher range of articles that satisfy eligibility criteria.
2.3. Selection of publications
The selection of articles for review was completed based on three stages: title, abstract and full text. Articles included must describe the prevalence of AMR in Enterobacteriaceae, and sample collection was from (1) humans (healthy/ diseased humans in hospitals); (2) animals (both terrestrial and aquatic) healthy and diseased; animal food products (meat, egg, and milk); (3) the environment (water, drainage, soil, sludge, vegetables, commercial feed, and wildlife animals). Additionally, all studies must apply antimicrobial susceptibility testing methods according to the Clinical and Laboratory Standards Institute (CLSI) guidelines. Articles that did not contain sufficient information on bacterial species, phenotypic resistance prevalence data or disaggregated data by bacterial species were excluded. Moreover, review articles, book chapters, conference abstracts, letters, and articles written in languages other than English and duplicated among databases were also excluded.
In order to minimize selection bias, the study quality assessment of included studies was appraised using the Joanna Briggs Institute (JBI) Critical Appraisal Checklist for studies reporting prevalence data [23]. A total number of 9 questions in the checklist were accessed by two independent reviewers based on the JBI guideline with the following answers: ‘yes’, ‘no’, ‘unclear’, and ‘not applicable’. Only studies appraised with ‘yes’ for all questions were included. If there were discrepant results between reviewers for any question, a third independent reviewer participated in the assessment process; studies were included when there was a consensus of ‘yes’ from two out of three reviewers.
2.4. Data extraction
Data from each retrieved publication were reviewed and the following information were extracted and saved in a Microsoft Excel 2019 spreadsheet: (1) region of Vietnam (North/ Center/ South); (2) publication year; (3) host (humans/ animals/ environment/ integrated studies between animals, humans and environment); (4) type of sample (feces, stool, rectal swab/ wound/ nasal swabs/ blood/ pus/ urine/ vaginal/ meat/ milk/ eggs/ water/ soil/ vegetables, etc.); (5) bacterial species (E.coli, Samonella spp., Klebsiella spp., Proteus spp., Shigella spp., Enterobacter spp., Citrobacter spp., Cronobacter spp., etc.); (6) number of isolates; (7) animal species (chicken or chicken meat/ pig or pork/ cattle or beef/ fish/ shrimp/ rodent/ shellfish, etc.); (8) laboratory methods for antimicrobial susceptibility testing (minimum inhibitory concentration or disk diffusion); (9) prevalence (%) of resistant bacteria to antimicrobials, antimicrobial agents and antimicrobial classes were classified by WHO (highest priority critically important/ high priority critically important/ highly important/important) [24]; (10) prevalence of multi-drug resistance (MDR) isolates, which were resistant to at least one antimicrobial agent of more than three antimicrobial classes [25]. The data from included studies were extracted by two independent reviewers to reduce the information bias. The results were rechecked by a third reviewer. The data were ready for analysis after reaching a consensus among all reviewers.
2.5. Statistical analyses
Only studies that included data on AMR in bacterial species covering at least two articles of three compartments (humans, animals and environment) were chosen for meta-analysis. Heterogeneity assessment of selected studies for meta-analysis was applied using inverse variance index (I2), an index >75.0% and a p-value <0.05 is considered as significant heterogeneity [26]. One recognized problem in meta-analysis with prevalence data derives from studies reporting very small or large prevalence; such estimates have minimal variance and, therefore, a greater weight (pooling effect size) [27]. Therefore, the logit-transformed proportions were applied using a generalised linear mixed-effect model (GLMM), which includes a random-effect model to identify the within-studies variance (VYi) and estimate of between-studies variance (τ2) [28]. Moreover, subgroup analysis was applied in each selected antimicrobial for meta-analysis of AMR prevalence to identify the significant difference among study compartments of animals, humans, and the environment with AMR prevalence. Besides, multiple meta-regression models were used to identify the risk factors associated with the MDR prevalence with bacterial species, study compartments, study area, and year of publication as explanatory variables. Multiple meta-regression was built using a stepwise approach to select the final model. Variables of a multivariable model with p-value <0.05 were considered as a significant association with the MDR prevalence. The results of the meta-analysis were displayed by forest plots. Publication bias in the meta-analysis was applied using both methods, including contour-enhanced funnel plots and Egger's regression test [29]. All statistical analyses were applied by R software with the packages ‘meta’ and “metafor” used for meta-analysis, subgroup analyses, and multiple meta-regression; package ‘tidyverse’ used for testing publication bias; and “ggplot2” for visualization of the study results.
3. Results
3.1. Study selection
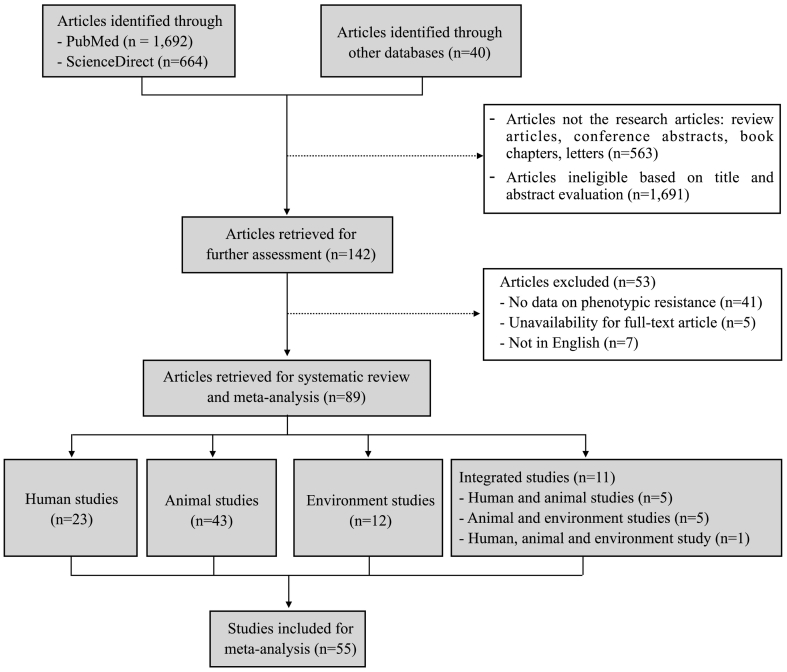
A total of 2396 articles were initially identified. Of those, 142 were further selected after excluding non-research articles (n = 563) and ineligible articles (n = 1691) based on the evaluation of the title and abstract. Thereafter, 53 articles were further excluded because they contained no phenotypic AMR data (n = 41), the full-text was not available (n = 5), or they were published in languages other than English (n = 7). Finally, 89 articles were selected for the systematic review, including 23 human studies, 43 studies of animals and food, 12 environmental studies, and 11 studies on AMR in isolates from more than one compartment (animals, humans or the environment). Furthermore, out of 89 articles, we selected the articles reporting data on the AMR prevalence of each antimicrobial agent with at least two studies for each bacterial species in all three compartments of humans, animals and environments. Finally, there was a total of 55 articles used for meta-analysis. All 89 selected studies are presented in Supplementary File 2. The process of study selection followed the PRISMA flow diagram and is shown in Fig. 1.
Fig. 1.
PRISMA flow diagram of the study selection.
3.2. Qualitative syntheses of selected studies
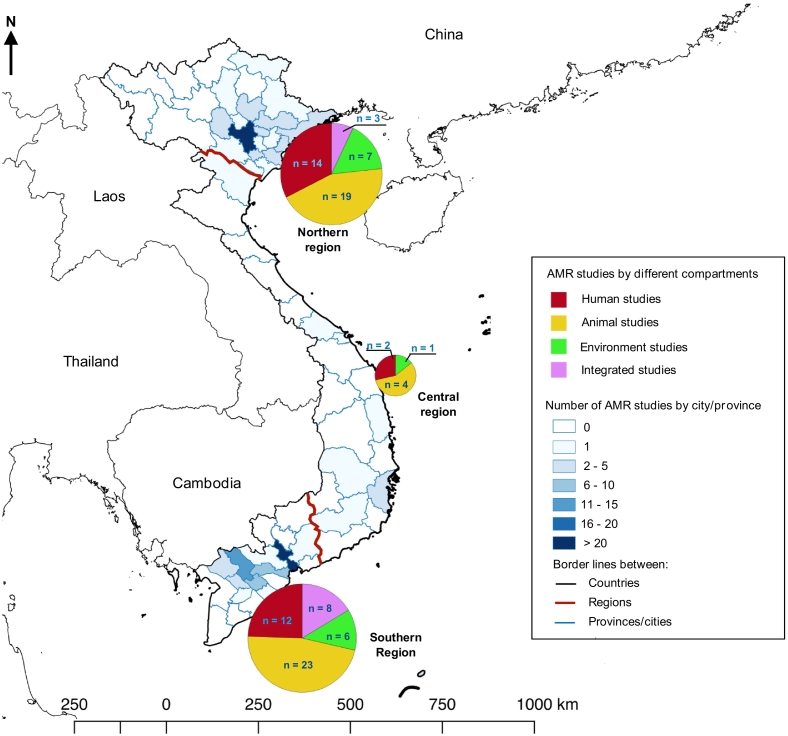
The description of selected studies for qualitative syntheses is presented in Table 1. All 89 selected studies were published between 2002 and 2022. Most selected studies were published in last recent years over the 2016–2022 period (54/89, 60.7%) and mostly conducted in the southern (43/89, 48.3%) and the northern regions of Vietnam (37/89, 41.6%). The study sites and the number of articles published in three different regions are illustrated in Fig. 2.
Table 1.
Number of studies in each category.
| Description of selected studies | Human studies (n = 23) (25.8) | Animal studies (n = 43) (48.3) | Environment studies (n = 12) (13.5) | Integrated studies (n = 11) (12.4) |
Total (n = 89) (%) | ||
|---|---|---|---|---|---|---|---|
| Human and animal (n = 5) (5.6) | Animal and environment (n = 5) (5.6) | Human, animal and environment (n = 1) (1.1) | |||||
| Bacterial species | |||||||
| E. coli | 7 (7.9) | 26 (29.2) | 6 (6.7) | 2 (2.2) | 1 (1.1) | _ | 42 (47.2) |
| Salmonella spp. | 3 (3.4) | 14 (15.7) | 5 (5.6) | 3 (3.4) | 3 (3.4) | 1 (1.1) | 29 (32.6) |
| Klebsiella spp. | 2 (2.2) | _ | _ | _ | _ | _ | 2 (2.2) |
| Shigella spp. | 1 (1.1) | _ | _ | _ | _ | _ | 1 (1.1) |
| E. coli, Salmonella spp. | _ | 3 (3.4) | _ | _ | 1 (1.1) | _ | 4 (4.5) |
| E. coli, Klebsiella spp. | 3 (3.4) | _ | _ | _ | _ | _ | 3 (3.4) |
| E. coli, Shigella spp. | 3 (3.4) | _ | _ | _ | _ | _ | 3 (3.4) |
| E. coli, Salmonella spp., Shigella spp. | 1 (1.1) | _ | _ | _ | _ | _ | 1 (1.1) |
| E. coli, Klebsiella spp., Enterobacter spp. | 1 (1.1) | _ | _ | _ | _ | _ | 1 (1.1) |
| E. coli, Klebsiella spp., Proteus spp. | 1 (1.1) | _ | _ | _ | _ | _ | 1 (1.1) |
| E. coli, Klebsiella spp., Enterobacter spp., Proteus spp. | 1 (1.1) | _ | _ | _ | _ | _ | 1 (1.1) |
| E. coli, Salmonella spp., Klebsiella spp., Enterobacter spp., Cronobacter spp. | _ | _ | 1 (1.1) | _ | _ | _ | 1 (1.1) |
| Year of publication | |||||||
| 2002–2005 | 3 (3.4) | _ | _ | _ | _ | _ | 3 (3.4) |
| 2006–2010 | 5 (5.6) | 7 (7.9) | 2 (2.2) | 2 (2.2) | _ | _ | 16 (18.0) |
| 2011–2015 | 4 (4.5) | 9 (11.2) | 1 (1.1) | _ | 2 (2.2) | _ | 16 (18.0) |
| 2016–2020 | 8 (9.0) | 21 (23.6) | 7 (7.9) | 3 (3.4) | 2 (2.2) | 1 (1.1) | 42 (47.2) |
| 2021–2022 | 3 (3.4) | 6 (6.7) | 2 (2.2) | _ | 1 (1.1) | _ | 12 (13.5) |
| Study area | |||||||
| Northern region | 11 (12.4) | 17 (19.1) | 6 (4.5) | 1 (1.1) | 2 (2.2) | _ | 37 (41.6) |
| Central region | _ | 3 (3.4) | _ | _ | _ | _ | 3 (3.4) |
| Southern region | 9 (10.1) | 21 (23.6) | 5 (5.6) | 4 (4.5) | 3 (3.4) | 1 (1.1) | 43 (48.3) |
| Northern and Southern regions | 1 (1.1) | 1 (1.1) | _ | _ | _ | _ | 2 (2.2) |
| Northern, Central and Southern regions | 2 (2.2) | 1 (1.1) | 1 (1.1) | _ | _ | _ | 4 (4.5) |
| Sample type | |||||||
| Stool/ faecal/ rectal swab | 17 (19.1) | 22 (24.7) | 1 (1.1) | 3 (3.4) | _ | 43 (48.3) | |
| Blood | 4 (4.5) | _ | _ | _ | _ | 4 (4.5) | |
| Carcass rinse/ meat (animals) | _ | 19 (21.3) | _ | _ | 1 (1.1) | 20 (22.5) | |
| Sludge/wastewater | _ | _ | 3 (3.4) | _ | _ | 3 (3.4) | |
| Vegetables/ fruit/ edible ice | _ | _ | 4 (4.5) | _ | _ | 4 (4.5) | |
| Mixed sample collection | 2 (2.2) | 2 (2.2) | 4 (4.5) | 2 (2.2) | 4 (4.5) | 1 (1.1) | 15 (16.9) |
| Antimicrobial susceptibility testing method | |||||||
| Disc diffusion (DD) | 9 (10.1) | 29 (32.6) | 7 (7.9) | 4 (4.5) | 4 (4.5) | 1 (1.1) | 54 (60.7) |
| Minimum Inhibitory Concentration (MIC) | 11 (12.4) | 12 (13.5) | 4 (4.5) | 1 (1.1) | _ | _ | 28 (31.5) |
| Both DD and MIC | 3 (3.4) | 2 (2.2) | 1 (1.1) | _ | 1 (1.1) | _ | 7 (7.9) |
Fig. 2.
Study sites and the number of articles in each compartment in the review.
E. coli was the most common species in the selected studies (42/89, 47.2%), followed by Salmonella spp. (29/89, 32.6%), Klebsiella spp. (2/89, 2.2%) and Shigella spp. (1/89, 1.1%). There were 15 studies (16.9%) covering more than one bacterial species: E. coli and Salmonella spp. (4/89, 4.5%), E. coli and Klebsiella spp. (3/89, 3.4%), E. coli and Shigella spp. (3/89, 3.4%), and E. coli, Salmonella spp., Klebsiella spp., Enterobacter spp., and Cronobacter spp. (1/89, 1.1%). Most (54/89, 60.7%) selected studies applied the agar disc diffusion method for antimicrobial susceptibility testing, and other studies (28/89, 31.5%) used agar/broth methods to determine the minimum inhibitory concentration (MIC) of antimicrobial substances.
In the compartments of humans and animals, the new subsets were split into healthy/ diseased groups. Regarding 29 human studies (23 human studies only, 6 integrated studies related to humans), we found that E. coli were the common bacteria in humans, 5 and 10 studies in the healthy and diseased group, while NTS was found in 2 studies and 4 studies in healthy and diseased human studies. Most other bacterial species (Klebsiella spp., Enterobacter spp., Shigella spp., Proteus spp., and ESBL-producing E. coli) were conducted in diseased humans. In animal studies, there were 21 and 25 studies performed in E.coli and NTS, 4 studies worked on the ESBL-producing E. coli in healthy animals. In the group of diseased animal studies, only 3 studies conducted E. coli in diarrheal pigs. Similarly, in the environment compartment, E. coli (9 studies) and NTS (9 studies) were primarily found in environmental studies. The details of studies by bacterial species in each compartment of human, animal, and environment are shown in Table 2.
Table 2.
Total of studies by bacterial species in each compartment of human, animal, and environment.
| Bacteria | Human studies |
Animal studies |
Environment studies | ||
|---|---|---|---|---|---|
| Healthy | Diseased (in hospitals) | Healthy animals/animal food products | Diseased | ||
| E. coli | 5 [[30], [31], [32], [33], [34]] | 10 [[35], [36], [37], [38], [39], [40], [41], [42], [43], [44]] | 21 [12,13,[45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63]] | 3 [[64], [65], [66]] | 9 [52,58,[67], [68], [69], [70], [71], [72], [73]] |
| ESBL-producing E. coli | _ | 2 [74,75] | 4 [[76], [77], [78], [79]] | _ | _ |
| ESBL/pAmpC-producing E. coli | _ | _ | 2 [80,81] | _ | _ |
| Cephalosporin-resistance E. coli | 1 [82] | _ | 1 [82] | _ | _ |
| mcr1_positive E. coli | 1 [83] | _ | 1 [84] | _ | _ |
| Non-typhoidal Salmonella (NTS) | 2 [85,86] | 4 [37,85,87,88] | 25 [53,[57], [58],60,85,86,[88], [89], [90], [91], [92], [93], [94], [95], [96], [97], [98], [99], [100], [101], [102], [103], [104], [105], [106]] | _ | 9 [58,73,97,104,[107], [108], [109], [110], [111]] |
| Typhoidal Salmonella | _ | 3 [87,112,113] | _ | _ | _ |
| Enterobacter spp. | _ | 2 [35,36] | _ | _ | 1 [73] |
| Cronobacter spp. | _ | _ | _ | _ | 1 [73] |
| Klebsiella spp. | 1 [33] | 5 [35,36,38,114,115] | _ | _ | 1 [73] |
| ESBL-producing Klebsiella | _ | 2 [74,75] | _ | _ | _ |
| Proteus spp. | _ | 1 [36] | _ | _ | _ |
| ESBL-producing Proteus | _ | 1 [75] | _ | _ | _ |
| Shigella spp. | _ | 5 [37,[40], [41], [42],116] | _ | _ | _ |
ESBL-producing E. coli/ Klebsiella/ Proteus: Extended-spectrum β-lactamase- (ESBL-) producing Escherichia coli/ Klebsiella/ Proteus; ESBL/pAmpC-producing E. coli: Extended-spectrum β-lactamase- (ESBL-) and plasmid-mediated AmpC β-lactamase- (pAmpC-) producing Escherichia coli; mcr1_positive E. coli: Colistin-resistant Escherichia coli harboring mcr-1 gene.
3.3. Quantitative syntheses of selected studies
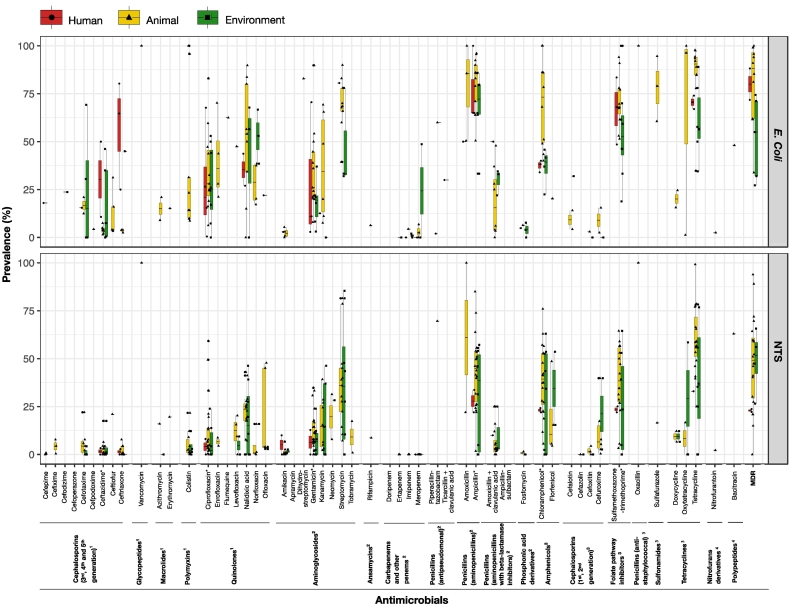
A total of 89 studies included AMR phenotypic susceptibility for 56 different antimicrobial agents (21 antimicrobial classes) among 7 Enterobacteriaceae species (Supplementary File 3). E. coli and NTS were by far the most common bacterial species (73/89 studies). We selected only studies that reported the AMR prevalence of E. coli and NTS in healthy humans, healthy animals/animal food products and environment studies (60/89 studies) for quantitative syntheses (Fig. 3). The detailed description is represented in Supplementary Table 1.
Fig. 3.
The AMR prevalence of E. coli and NTS in healthy human, healthy animal/animal food products and environment studies. 1highest priority critically important antimicrobials, 2high priority critically important antimicrobials, 3highly important antimicrobials, 4important antimicrobials, *antimicrobials of studies used for meta-analysis.
Regarding the highest priority critically important antimicrobials in the three compartments, the highest levels of resistance were observed against 3rd generation of cephalosporins and quinolones. E. coli, resistance to ceftriaxone in humans was 56.7% (SE ± 16.5), followed by ceftazidime 30.4% (SE ± 19.7), and ciprofloxacin 27.6% (SE ± 14.4). In animal isolates, resistance levels were higher in quinolones than other drugs in animals, ranging from 28.4–62.5% for E. coli and 4.4–22.0% for NTS. For environmental isolates, the prevalence of E. coli ranged from 17.8–45.0% to 3rd generation of cephalosporins and 22.7–41.4% to quinolones, while in NTS the resistance prevalence was found highest in quinolones, 7.0–17.9%. Colistin-resistant E. coli was found at 34.1% in healthy animals. Neither E. coli nor Salmonella isolates were resistant to colistin in human studies, and the resistance level to colistin was observed in only NTS in the environment at 4.1% (SE ± 2.8).
With regards to high priority critically important antimicrobials, levels of resistance to ampicillin of E. coli and NST were at 67.9–80.2% and 27.9–43.6%; and for gentamicin 18.2–35.0% and 6.4–12.3%, respectively. In carbapenem antimicrobials, the resistance level at 24.4% (SE ± 24.4) was observed in only E. coli to meropenem in the environment, and there were no observations of resistance of Salmonella to carbapenem class.
For antimicrobials classified as highly important, the resistance prevalence of E. coli and NTS to tetracycline in all sectors ranged from 60.9–84.6% and 33.0–60.1%, with the highest level being in animals, followed by humans and the environment. For other drugs, sulfamethoxazole-trimethoprim and chloramphenicol were also found high in all groups, ranging from 54.4–70.8% and 37.4–70.2% in E. coli, 23.6–38.9% and 23.1–38.8% in NTS, respectively.
The highest MDR prevalence of E. coli was found at 80.4% and 80.0% in animals and humans, respectively. The MDR prevalence of E. coli was observed in the environment studies at 52.0%. In contrast, in NTS the highest MDR was observed in the environment (48.9%), followed by animals (47.9%) and humans (23.0%).
3.4. Meta-analysis of selected studies
According to the results from the systematic review, only 55 studies representing the resistance levels of E. coli and NTS to 6 antimicrobials of ciprofloxacin, ceftazidime, ampicillin, gentamicin, sulfamethoxazole-trimethoprim, chloramphenicol and MDR in the groups of healthy human, healthy animal and environment were used for meta-analysis (Supplementary File 4). The forest plots of pooled prevalence of E.coli and NTS to these antimicrobials are shown in Supplementary Fig. 1. A summary of the subgroup meta-analysis of the pooled AMR prevalence across the human, animal and environment studies is represented in Table 3.
Table 3.
Subgroup meta-analysis of pooled AMR prevalence of E. coli and NTS across the human, animal and environment studies.
| Antimicrobials |
E. coli |
NTS |
||||||
|---|---|---|---|---|---|---|---|---|
| Human | Animal | Environment | Total | Human | Animal | Environment | Total | |
| Ceftazidime | ||||||||
| Pooled prevalence (%) | 25.0 | 4.0 | 8.0 | 6.0⁎ | 3.0 | 1.0 | 2.0 | 1.0 |
| 95% CI | [0−100] | [2.0–6.0] | [0–58.0] | [3.0–15.0] | [0–81.0] | [0–3.0] | [0–16.0] | [0–3.0] |
| Number of studies (n) | 2 | 7 | 5 | 14 | 2 | 16 | 6 | 24 |
| Ciprofloxacin | ||||||||
| Pooled prevalence (%) | 14.0 | 32.0 | 24.0 | 26.0 | 8.0 | 6.0 | 6.0 | 5.0 |
| 95% CI | [0–85.0] | [19.0–49.0] | [12.0–42.0] | [16.0–39.0] | [0–67.0] | [3.0–12.0] | [1.0–31.0] | [3.0–10.0] |
| Number of studies (n) | 4 | 11 | 8 | 23 | 2 | 21 | 6 | 29 |
| Gentamicin | ||||||||
| Pooled prevalence (%) | 17.0 | 33.0 | 16.0 | 25.0 | 12.0 | 8.0 | 8.0 | 8.0 |
| 95% CI | [0–96.0] | [21.0–48.0] | [6.0–34.0] | [16.0 0–37.0] | [1.0–66.0] | [5.0–13.0] | [4.0–16.0] | [5.0–12.0] |
| Number of studies (n) | 3 | 13 | 6 | 22 | 2 | 23 | 9 | 34 |
| Ampicillin | ||||||||
| Pooled prevalence (%) | 88.0 | 88.0 | 71.0 | 84.0 | 30.0 | 40.0 | 17.0 | 34.0 |
| 95% CI | [5.0–100] | [73.0–96.0] | [50.0–86.0] | [73.0–91.0] | [6.0–73.0] | [28.0–54.0] | [3.0–58.0] | [24.0–46.0] |
| Number of studies (n) | 3 | 13 | 6 | 22 | 2 | 22 | 10 | 34 |
| Chloramphenicol | ||||||||
| Pooled prevalence (%) | 39.0 | 82.0 | 34.0 | 65.0⁎⁎ | 24.0 | 37.0 | 18.0 | 33.0⁎ |
| 95% CI | [33.0–46.0] | [51.0–95.0] | [20.0–52.0] | [43.0–82.0] | [4.0–70.0] | [29.0–46.0] | [4.0–56.0] | [25.0–42.0] |
| Number of studies (n) | 3 | 10 | 4 | 17 | 2 | 20 | 9 | 31 |
| Sulfamethoxazole-trimethoprim | ||||||||
| Pooled prevalence (%) | 66.0 | 73.0 | 53.0 | 66.0 | 25.0 | 37.0 | 23.0 | 33.0 |
| 95% CI | [30.0–90.0] | [62.0–82.0] | [34.0–71.0] | [56.0–75.0] | [4.0–71.0] | [27.0–47.0] | [6.0–57.0] | [25.0–42.0] |
| Number of studies (n) | 3 | 11 | 8 | 22 | 2 | 18 | 7 | 27 |
p < 0.05.
p < 0.01.
In meta-analyses of all studies reported data on 6 selected drugs, the inverse variance indexes (I2) were found high in selected studies of each antimicrobial, ranging from 80.0%–98.0% (all p < 0.01), implying that there is significant heterogeneity among studies' results.
The pooled prevalence of E. coli and NTS to ciprofloxacin was 26.0% (95% CI 16.0–39.0%) and 5.0% (95% CI 3.0–10.0%), and for ceftazidime they were at 6.0% (95% CI 3.0–15.0%) and 1% (95% CI 0–3.0%). There was a significant difference observed in only ceftazidime-resistant E. coli, with the pooled prevalence in humans at 25.0% higher than in animals at 4.0% and environment at 8% (χ2 = 8.29, p = 0.02). For ampicillin, the pooled prevalence of E. coli was high at 84.0% (95% CI 73.0–91.0%) and 34.0% (95% CI 24.0–46.0%) for NTS; and for gentamicin, the pooled prevalence was 25% (95% CI 16.0–37.0%) and 8% (95% CI 5.0–12.0%) in E. coli and NTS, respectively. The pooled prevalence of E. coli to chloramphenicol and sulfamethoxazole-trimethoprim was high at 65.0% (95% CI 43.0–82.0%) and 66.0% (95% CI 56.0–75.0%) respectively, and for NTS the prevalence was the similar between chloramphenicol and sulfamethoxazole-trimethoprim at 33.0% (95% CI 25.0–42.0%). The significant differences were found with the pooled prevalence in humans, animals and environment of E. coli 39.0%, 82.0% and 34.0% (χ2 = 9.65, p < 0.01) and in NTS 24.0%, 37.0% and 18.0% (χ2 = 7.51, p = 0.02).
The statistical models investigating the factors associated with the pooled MDR prevalence using meta-regression models are presented in Table 4. In univariable models, all four independent variables (1) bacterial, (2) sectors, (3) study regions and (4) year of publication were selected as potential variables used in multivariable meta-regression models. Findings from the final model of multivariable found that the AMR level in E. coli (β = 1.887, p < 0.001) and the northern region (β = 0.798, p = 0.047) had a higher fraction of AMR than NTS and other regions of Vietnam.
Table 4.
Multiple meta-regression analyses of studies with data on the pooled prevalence of MDR.
| Variables | Univariable models |
Multivariable model⁎ |
||||
|---|---|---|---|---|---|---|
| β | 95% CI | p-value | β | 95% CI | p-value | |
| Bacterial species (baseline = NTS) | ||||||
| E. coli | 1.952 | 1.19–2.71 | <0.001 | 1.887 | 1.16–2.61 | <0.001 |
| Sector (baseline = environment) | ||||||
| Human | 0.139 | −1.82-2.10 | 0.887 | 0.533 | −0.92-1.99 | 0.462 |
| Animal | 0.706 | −0.50-1.91 | 0.147 | 1.027 | 1.36–1.92 | 0.225 |
| Study regions (baseline = Southern region) | ||||||
| Northern region | 1.276 | 0.24–2.31 | 0.017 | 0.798 | −0.04-1.64 | 0.047 |
| Central region | 2.140 | −0.12-4.40 | 0.063 | 0.660 | −1.23-2.55 | 0.485 |
| Multi-region | 1.652 | −0.20-3.50 | 0.079 | 1.370 | −0.177-2.916 | 0.081 |
| Year of publication (baseline = 2006–2010) | ||||||
| 2002–2005 | 1.77 | −1.63-5.18 | 0.299 | −0.253 | −2.98-2.48 | 0.853 |
| 2011–2015 | 0.923 | −0.45-2.30 | 0.184 | 0.291 | −0.76-1.34 | 0.579 |
| 2016–2020 | 0.628 | −0.53-1.79 | 0.283 | 0.060 | −0.83-0.95 | 0.892 |
| 2021–2022 | 1.170 | −0.27-2.61 | 0.108 | 0.876 | −0.25-2.00 | 0.123 |
Intercept: -0.904; SE: 0.541.
Analyses for publication bias found the asymmetry among selected articles for meta-analysis. In the contour-enhanced funnel plots, the scatters illustrating selected articles are unevenly distributed far away from the pooled effect size (vertical line) and mostly plotted on the shaded regions of p < 0.05 and p < 0.01. The findings are confirmed by the results of Egger's regression tests. The selected studies showing the intercept (βo) of selected studies for meta-analyses of ciprofloxacin, ceftazidime and gentamicin, sulfamethoxazole-trimethoprim, chloramphenicol and MDR were different from zero value (all βo either <−0.72 or > 1.09), whereas in studies reported data on ampicillin, although the intercept of Egger's regression tests was equal to zero, no significance was detected (p = 0.998). This indicates that there is the existence of publication bias in the selected studies for meta-analysis. All contour-enhanced funnel plots illustrating the asymmetry among selected studies and the results of Egger's regression tests are shown in Supplementary Fig. 2.
4. Discussion
This is the first systematic review and meta-analysis of integrated studies on AMR under the One Health perspective in Vietnam. A total of 89 studies on Enterobacteriaceae isolates over three compartments of humans, animals and environment were selected in the review. The sub-groups of healthy and diseased humans and animals were classified. Furthermore, the separation of bacterial species into various subsets and selection of only 55 studies on commensal E. coli and NTS in healthy humans and animals for meta-analysis helps to minimize the information bias and accurate appraisal of the associations of antimicrobial phenotypic resistance of E. coli and NTS across three compartments.
There were marginal differences in the AMR prevalence of E. coli and NTS across the three compartments. The high AMR levels could be stemmed from the transfer of AMR bacteria and AMR-encoding genes due to close contact between humans, animals and the environment [117]. A recent One Health study in Vietnam reported a high similarity in AMR profiles of E. coli from humans and chickens from the same farms higher than those from different farms implying the potential transmission of cross-species [118]. Furthermore, the presence of bacterial isolates resistant to antimicrobials commonly administered to humans and animals from wastewater has been demonstrated in Vietnam [71,97]. Of 18 articles regarding environment studies in our review, eight studies included samples collected from wastewater from hospitals and slaughterhouses. This might suggest that AMR bacteria from human and animal sewage are discharged into the environment. There have been demonstrated that bacteria resistant to antimicrobials commonly administered to humans (penicillins, aminoglycosides, quinolones) were present in the hospital wastewater, and seriously there was no difference in AMR levels of isolates that samples collected before and after the treatment of water [69]
The generation of antimicrobial resistance stems from selection pressure caused by the widespread usage of antimicrobials in communities, healthcare settings, and animal production [119]. In terms of antimicrobial use (AMU) of antimicrobial active ingredients per kg of human and animal biomass, the use of antimicrobials in humans and animals in Vietnam was estimated to be 261.7 mg and 247.3 mg in comparison to 122.0 mg and 151.5 mg in the European countries; particularly in the chicken production system the levels of AMU was about 6 times higher with 84.0% usage was for the prophylactic purpose [[17], [19]]. A recent study measured the antimicrobials usage in humans and animals in Vietnam found that the antimicrobials mostly consumed in humans were cephalosporins (49.4%), followed by penicillins (40.0%) and quinolones (9.3%), whereas in animals penicillins (35.5%), tetracyclines (30.7%) and macrolides (19.1%) were commonly given to pigs and chickens [120]. Results about AMR levels in our studies found high AMR levels of E. coli and NTS in these drugs showing that the high levels of AMR might be associated with the usage of antimicrobials.
We found similar results as in a review on AMR from Cameroon with levels of E. coli resistant to tetracycline (85.5%), trimethoprim-sulfamethoxazole (83.3%), and ampicillin (65.6%) [121]. Another systematic review in Ethiopia conducted in three compartments of humans, animals and environment also observed the highest levels of E. coli to ciprofloxacin (50.0%) and ceftriaxone (28.0%) among reviewed drugs [122]. However, this review found a higher prevalence in humans compared with animals and the environment.
In our review, levels of AMR were considerably higher for E. coli than for Salmonella for almost all antimicrobials. The findings were in agreement with a previous study in chickens and pigs that found that the AMR of E. coli was higher than Salmonella in all tested antimicrobials [121]. The difference in AMR development among bacterial species is influenced by the ability of selective pressures of a bacterial species under the presence of antimicrobials to be survival than other species. Compared with other bacterial species, E. coli has a greater capacity to integrate ARGs from other bacteria and as well as transfer the genes to others [123].
We found that the northern region of Vietnam has higher AMR levels than the other two regions. Most studies in the north were conducted in the Red River Delta, known to have some of the highest densities of human and animal populations in Vietnam [18]. It has been proposed that higher population and animal production densities are correlated with AMU and AMR [124]. Recent studies demonstrated to a high-level detail amounts of antimicrobials used in human and chicken production systems in the south of Vietnam [[7], [125],118]. However, there is an existing gap in AMU studies in the northern region. Thus, studies about AMU quantification in different regions in the country are suggested [53].
Following the Global Action Plan on AMR [126], Vietnam also has launched its National Action Plan for the management of AMU and control of AMR, including key activities to support surveillance, improve the awareness, governance and good practices of AMU in humans and animals [127]. Antimicrobials used for growth promotion purposes have been banned in Vietnam since January 2018 [128]. The new legislation on AMU to gradually ban prophylactic use of antimicrobials in animal production systems was also issued in 2020: the critically important antimicrobials were banned from 2021, the highly important from 2022, and the important antimicrobials from 2023 and any other antimicrobial classes from 2026 [129,130]. Since almost selected studies in our reviews were conducted before the time the legislation on the prohibition of prophylactic AMU was released, we suggested that follow-up studies should be conducted in Vietnam to see how well the new regulation policy works in practice.
There were a number of limitations in our study. Given that our review mainly evaluated phenotypic data of AMR in all three compartments of published studies in humans, animals and the environment, the suggestions for the follow-up review on genotypic resistance to have a comprehensive approach to AMR situation in Vietnam. Our review assessed the articles conducted in various geographical locations, mostly the North and the South, and different sample types; the heterogeneity in the study results was therefore found significant in the meta-analysis. Moreover, the high heterogeneity could stem from the publication bias by the effects of small studies in our review. This may result in difficulty in assessing the representativeness of AMR situation in the whole country.
5. Conclusion
This review confirms that Vietnam is a hotspot of AMR. Our study revealed the high and similar AMR prevalence of E. coli and NTS over the three compartments of humans, animals and the environment. Importantly, bacteria were resistant to several antimicrobials categorized as critically important. Our analyses have yielded some findings on the burden of AMR that should be helpful to baseline information for further investigation to understand the evolution and transmission mechanisms of antimicrobial-resistant bacteria in all three compartments of humans, animals and the environment. Given the observed high prevalence of resistance against the highest critically important antimicrobials, we recommend that their use be restricted, particularly in animal production. Since only a single compartment does not have adequate ability to tackle this global health problem, we strongly recommend implementing AMR surveillance through an integrated collaboration, including multiple scales and levels under the One Health approach to tackle the AMR problem in Vietnam.
The following are the supplementary data related to this article.
Forest plot of pooled prevalence of E.coli and NTS to six antimicrobials in meta-analysis.
The contour-enhanced funnel plots show the asymmetry among selected studies. The dashed vertical lines show the average effect sizes, each scatter presents each selected article, and the shaded regions illustrate the statistical significance of the asymmetry of the publications. CIP - Ciprofloxacin, CAZ - Ceftazidime, AMP - Ampicillin, GEN- Gentamicin, SXT - Sulfamethoxazole-trimethoprim, CHL-Chloramphenicol, MDR - Multi-drug Resistance.
The PRISMA item checklist.
A total number of 89 selected studies were used for systematic review and meta-analysis.
The data on AMR extracted from 89 selected studies for systematic review
The data on AMR extracted from 55 selected studies for meta-analysis
The average AMR prevalence of E. coli and Non-typhoidal Salmonella (NTS) in healthy humans, healthy animals/ animal food products and the environment.
Author contributions
DHP, TW, TT, JCM designed the study and developed the protocol. DHP, DBT, TW selected the articles and extracted the data. DHP, DBT contributed to data analyses. DHP, TW, DBT, NVC, TT and JCM contributed to writing up the manuscript. All authors have approved the submitted version of the manuscript.
Funding
This work has been supported by Walailak University Graduate Studies Research Fund (No. CGS-RF 2022/09), awarded to Hoang Phu Doan (Doan Hoang Phu).
Conflicts of interest
All authors declare no conflict of interest.
Acknowledgments
The author would like to express the deepest gratitude to Walailak University Ph.D. Scholarships for High Potential Candidates to Enroll in Doctoral Programs (Contract No. HP009/2021) awarded to Hoang Phu Doan (Doan Hoang Phu).
Data availability
The research data are available in supplementary materials.
References
- 1.WHO WHO - Antimicrobial Resistance 2021. 2021. https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed Nov. 28, 2021)
- 2.Rather I.A., Kim B.-C., Bajpai V.K., Park Y.-H. Self-medication and antibiotic resistance: crisis, current challenges, and prevention. Saudi J. Biol. Sci. May 2017;24(4):808–812. doi: 10.1016/j.sjbs.2017.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murray C.J., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. Feb. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anjum M.F., et al. The potential of using E. coli as an indicator for the surveillance of antimicrobial resistance (AMR) in the environment. Curr. Opin. Microbiol. Dec. 2021;64:152–158. doi: 10.1016/j.mib.2021.09.011. [DOI] [PubMed] [Google Scholar]
- 5.McDermott P.F., Zhao S., Tate H. Antimicrobial resistance in nontyphoidal Salmonella. Microbiol. Spectr. Jul. 2018;6(4):6.4.16. doi: 10.1128/microbiolspec.ARBA-0014-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization . World Health Organization; Geneva: 2020. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2020.https://apps.who.int/iris/handle/10665/332081 Accessed: Aug. 11, 2022. [Online]. Available: [Google Scholar]
- 7.Cuong N.V., et al. High-resolution monitoring of antimicrobial consumption in Vietnamese small-scale chicken farms highlights discrepancies between study metrics. Front. Vet. Sci. Jun. 2019;6:174. doi: 10.3389/fvets.2019.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao W., Ivanovska V., Schweickert B., Muller A. Proxy indicators for antibiotic consumption; surveillance needed to control antimicrobial resistance. Bull. World Health Organ. Jan. 2019;97(1) doi: 10.2471/BLT.18.227348. pp. 3-3A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yen N.T.P., et al. Prevalence of carbapenem resistance and its potential association with antimicrobial use in humans and animals in rural communities in Vietnam. JAC-Antimicrob. Resist. Mar. 2022;4(2):dlac038. doi: 10.1093/jacamr/dlac038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aeksiri N., et al. First detection and genomic insight into mcr-1 encoding plasmid-mediated colistin-resistance gene in Escherichia coli ST101 isolated from the migratory bird species Hirundo rustica in Thailand. Microb. Drug Resist. Dec. 2019;25(10):1437–1442. doi: 10.1089/mdr.2019.0020. [DOI] [PubMed] [Google Scholar]
- 11.Eiamphungporn W., Yainoy S., Jumderm C., Tan-arsuwongkul R., Tiengrim S., Thamlikitkul V. Prevalence of the colistin resistance gene mcr-1 in colistin-resistant Escherichia coli and Klebsiella pneumoniae isolated from humans in Thailand. J. Glob. Antimicrob. Resist. Dec. 2018;15:32–35. doi: 10.1016/j.jgar.2018.06.007. [DOI] [PubMed] [Google Scholar]
- 12.Hoa T.T.T., et al. Extended-spectrum beta-lactamase-producing Escherichia coli harbouring sul and mcr - 1 genes isolates from fish gut contents in the Mekong Delta, Vietnam. Lett. Appl. Microbiol. Jul. 2020;71(1):78–85. doi: 10.1111/lam.13222. [DOI] [PubMed] [Google Scholar]
- 13.Kawahara R., et al. Most domestic livestock possess colistin-resistant commensal Escherichia coli harboring mcr in a rural community in Vietnam. Antimicrob. Agents Chemother. Jun. 2019;63(6) doi: 10.1128/AAC.00594-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shira D. “Why the Agtech Industry Will Aid Vietnam's Hi-Tech Growth,” Why the Agtech Industry Will Aid Vietnam's Hi-Tech Growth. 2021. https://www.vietnam-briefing.com/news/why-agtech-industry-will-aid-vietnams-hi-tech-growth.html/
- 15.Rousham E.K., Unicomb L., Islam M.A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and One Health approaches. Proc. R. Soc. B Biol. Sci. Apr. 2018;285(1876):20180332. doi: 10.1098/rspb.2018.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitchell M.E.V., Alders R., Unger F., Nguyen-Viet H., Le T.T.H., Toribio J.-A. The challenges of investigating antimicrobial resistance in Vietnam - what benefits does a One Health approach offer the animal and human health sectors? BMC Public Health. Dec. 2020;20(1):213. doi: 10.1186/s12889-020-8319-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrique-Mas J.J., Choisy M., Van Cuong N., Thwaites G., Baker S. An estimation of total antimicrobial usage in humans and animals in Vietnam. Antimicrob. Resist. Infect. Control. Dec. 2020;9(1):16. doi: 10.1186/s13756-019-0671-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.General Statistics Office . Statistical Publishing House; Vietnam: 2021. Statistical Year Book of Vietnam 2020.https://www.gso.gov.vn/wp-content/uploads/2021/07/Sach-NGTK-2020Ban-quyen.pdf [Online]. Available: [Google Scholar]
- 19.Carrique-Mas J.J., et al. Antimicrobial usage in chicken production in the Mekong Delta of Vietnam. Zoonoses Public Health. Apr. 2015;62:70–78. doi: 10.1111/zph.12165. [DOI] [PubMed] [Google Scholar]
- 20.Nhung N., Cuong N., Thwaites G., Carrique-Mas J. Antimicrobial usage and antimicrobial resistance in animal production in Southeast Asia: a review. Antibiotics. Nov. 2016;5(4):37. doi: 10.3390/antibiotics5040037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McEwen S.A., Collignon P.J. Antimicrobial resistance: a One Health perspective. Microbiol. Spectr. Apr. 2018;6(2):6.2.10. doi: 10.1128/microbiolspec.ARBA-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Page M.J., et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. Mar. 2021:n71. doi: 10.1136/bmj.n71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munn Z., Moola S., Lisy K., Riitano D., Tufanaru C. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and incidence data. Int. J. Evid. Based Heal. 2015;13(3):147–153. doi: 10.1097/XEB.0000000000000054. [DOI] [PubMed] [Google Scholar]
- 24.WHO . 6th rev. World Health Organization; Geneva: 2019. Critically Important Antimicrobials for Human Medicine.https://apps.who.int/iris/handle/10665/312266 Accessed: Mar. 29, 2022. [Online]. Available: [Google Scholar]
- 25.Magiorakos A.-P., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. Mar. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 26.Wada Y., et al. Prevalence of vancomycin-resistant enterococcus (VRE) in companion animals: the first meta-analysis and systematic review. Antibiotics. Jan. 2021;10(2):138. doi: 10.3390/antibiotics10020138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barendregt J.J., Doi S.A., Lee Y.Y., Norman R.E., Vos T. Meta-analysis of prevalence. J. Epidemiol. Community Health. Nov. 2013;67(11):974–978. doi: 10.1136/jech-2013-203104. [DOI] [PubMed] [Google Scholar]
- 28.Lin L., Xu C. Arcsine-based transformations for meta-analysis of proportions: pros, cons, and alternatives. Health Sci. Rep. Sep. 2020;3(3) doi: 10.1002/hsr2.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harrer M., Cuijpers P., Furukawa T.A., Ebert D.D. 1st ed. Chapman and Hall/CRC; Boca Raton: 2021. Doing Meta-Analysis with R: A Hands-on Guide. [DOI] [Google Scholar]
- 30.Dyar O.J., et al. High prevalence of antibiotic resistance in commensal Escherichia coli among children in rural Vietnam. BMC Infect. Dis. Dec. 2012;12(1):92. doi: 10.1186/1471-2334-12-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoang P.H., et al. Antimicrobial resistance profiles and molecular characterization of Escherichia coli strains isolated from healthy adults in Ho Chi Minh City, Vietnam. J. Vet. Med. Sci. 2017;79(3):479–485. doi: 10.1292/jvms.16-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nhi L. Thi Quynh, et al. Excess body weight and age associated with the carriage of fluoroquinolone and third-generation cephalosporin resistance genes in commensal Escherichia coli from a cohort of urban Vietnamese children. J. Med. Microbiol. Oct. 2018;67(10):1457–1466. doi: 10.1099/jmm.0.000820. [DOI] [PubMed] [Google Scholar]
- 33.Duong B.T., et al. Antibiotic-resistant gram-negative bacteria carriage in healthcare workers working in an intensive care unit. Infect. Chemother. 2021;53(3):546. doi: 10.3947/ic.2021.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trung N.V., et al. Colonization of enteroaggregative Escherichia coli and Shiga toxin-producing Escherichia coli in chickens and humans in southern Vietnam. BMC Microbiol. Dec. 2016;16(1):208. doi: 10.1186/s12866-016-0827-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vu T.V.D., et al. Antimicrobial susceptibility testing and antibiotic consumption results from 16 hospitals in Viet Nam: the VINARES project 2012–2013. J. Glob. Antimicrob. Resist. Sep. 2019;18:269–278. doi: 10.1016/j.jgar.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Biedenbach D.J., et al. Antimicrobial susceptibility and extended-spectrum beta-lactamase rates in aerobic gram-negative bacteria causing intra-abdominal infections in Vietnam: report from the study for monitoring antimicrobial resistance trends (SMART 2009–2011) Diagn. Microbiol. Infect. Dis. Aug. 2014;79(4):463–467. doi: 10.1016/j.diagmicrobio.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 37.Isenbarger D.W., et al. Comparative antibiotic resistance of diarrheal pathogens from Vietnam and Thailand, 1996-1999. Emerg. Infect. Dis. Feb. 2002;8(2):175–180. doi: 10.3201/eid0802.010145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garpvall K., et al. Admission screening and cohort care decrease carbapenem resistant enterobacteriaceae in Vietnamese pediatric ICU’s. Antimicrob. Resist. Infect. Control. Dec. 2021;10(1):128. doi: 10.1186/s13756-021-00994-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Son T.V., et al. Molecular detection of blaCTX-M gene to predict phenotypic cephalosporin resistance and clinical outcome of Escherichia coli bloodstream infections in Vietnam. Ann. Clin. Microbiol. Antimicrob. Dec. 2021;20(1):60. doi: 10.1186/s12941-021-00466-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nguyen T.V., Le P.V., Le C.H., Weintraub A. Antibiotic resistance in Diarrheagenic Escherichia coli and Shigella strains isolated from children in Hanoi, Vietnam. Antimicrob. Agents Chemother. Feb. 2005;49(2):816–819. doi: 10.1128/AAC.49.2.816-819.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hien B.T.T., et al. Diarrheagenic Escherichia coli and Shigella strains isolated from children in a hospital case-control study in Hanoi, Vietnam. J. Clin. Microbiol. Mar. 2008;46(3):996–1004. doi: 10.1128/JCM.01219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hien B.T.T., Trang D.T., Scheutz F., Cam P.D., Mølbak K., Dalsgaard A. Diarrhoeagenic Escherichia coli and other causes of childhood diarrhoea: a case–control study in children living in a wastewater-use area in Hanoi, Vietnam. J. Med. Microbiol. Aug. 2007;56(8):1086–1096. doi: 10.1099/jmm.0.47093-0. [DOI] [PubMed] [Google Scholar]
- 43.Hung P.N., et al. Antibiotic resistance profile and diversity of subtypes genes in Escherichia coli causing bloodstream infection in Northern Vietnam. Open Access Maced. J. Med. Sci. Dec. 2019;7(24):4393–4398. doi: 10.3889/oamjms.2019.842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tuan-Anh T., et al. Pathogenic Escherichia coli possess elevated growth rates under exposure to sub-inhibitory concentrations of azithromycin. Antibiotics. Oct. 2020;9(11):735. doi: 10.3390/antibiotics9110735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vounba P., Rhouma M., Arsenault J., Bada Alambédji R., Fravalo P., Fairbrother J.M. Prevalence of colistin resistance and mcr-1/mcr-2 genes in extended-spectrum β-lactamase/AmpC-producing Escherichia coli isolated from chickens in Canada, Senegal and Vietnam. J. Glob. Antimicrob. Resist. Dec. 2019;19:222–227. doi: 10.1016/j.jgar.2019.05.002. [DOI] [PubMed] [Google Scholar]
- 46.Trung N.V., et al. Prevalence and risk factors for carriage of antimicrobial-resistant Escherichia coli on household and small-scale chicken farms in the Mekong Delta of Vietnam. J. Antimicrob. Chemother. Mar. 2015:dkv053. doi: 10.1093/jac/dkv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sary K., Fairbrother J.M., Arsenault J., de Lagarde M., Boulianne M. Antimicrobial resistance and virulence gene profiles among Escherichia coli isolates from retail chicken carcasses in Vietnam. Foodborne Pathog. Dis. Apr. 2019;16(4):298–306. doi: 10.1089/fpd.2018.2555. [DOI] [PubMed] [Google Scholar]
- 48.Ngoc T.T.B., Oanh D.T., Pineda L., Ayudhya S., de Groot N., Han Y. The effects of synergistic blend of organic acid or antibiotic growth promoter on performance and antimicrobial resistance of bacteria in grow–finish pigs. Transl. Anim. Sci. Oct. 2020;4(4):txaa211. doi: 10.1093/tas/txaa211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nguyen N.T., et al. A novel method for measuring phenotypic colistin resistance in Escherichia coli populations from chicken flocks. Microbiology. Oct. 2020 doi: 10.1101/2020.10.22.351577. preprint. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van T.T.H., Chin J., Chapman T., Tran L.T., Coloe P.J. Safety of raw meat and shellfish in Vietnam: an analysis of Escherichia coli isolations for antibiotic resistance and virulence genes. Int. J. Food Microbiol. Jun. 2008;124(3):217–223. doi: 10.1016/j.ijfoodmicro.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 51.Nguyen N.T., et al. Use of Colistin and other critical antimicrobials on pig and chicken farms in southern Vietnam and its association with resistance in commensal Escherichia coli bacteria. Appl. Environ. Microbiol. Jul. 2016;82(13):3727–3735. doi: 10.1128/AEM.00337-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nhung N.T., et al. High levels of antimicrobial resistance among Escherichia coli isolates from livestock farms and synanthropic rats and shrews in the Mekong Delta of Vietnam. Appl. Environ. Microbiol. Feb. 2015;81(3):812–820. doi: 10.1128/AEM.03366-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tuat C.V., et al. Antimicrobial resistance pilot surveillance of pigs and chickens in Vietnam, 2017–2019. Front. Vet. Sci. Jul. 2021;8:618497. doi: 10.3389/fvets.2021.618497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Usui M., et al. Antimicrobial susceptibility of indicator bacteria isolated from chickens in southeast Asian countries (Vietnam, Indonesia and Thailand) J. Vet. Med. Sci. 2014;76(5):685–692. doi: 10.1292/jvms.13-0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.K L.T.L., L L.T.C. Antimicrobial resistance of Escherichia coli causing edema disease in post-weaning pigs in Vinh Long province. Can Tho Univ. J. Sci. 2019;11(2):1. doi: 10.22144/ctu.jen.2019.017. [DOI] [Google Scholar]
- 56.T H.T.V., A D.T.L., D L.V. Escherichia coli infection in ducks in the Mekong Delta: bacterial isolation, serogroup distribution and antibiotic resistance. Can Tho Univ. J. Sci. 2019;11(1):24. doi: 10.22144/ctu.jen.2019.003. [DOI] [Google Scholar]
- 57.Van T.T.H., Moutafis G., Tran L.T., Coloe P.J. Antibiotic resistance in food-borne bacterial contaminants in Vietnam. Appl. Environ. Microbiol. Dec. 2007;73(24):7906–7911. doi: 10.1128/AEM.00973-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thai T.H., Ngoc P.T., Lan N.T., Huong C.T.T. 2018. High prevalence of Antibiotic Resistance in Salmonella and Escherichia coli isolated from Pig Farms and Slaughterhouses in North Vietnam; p. 7. [Google Scholar]
- 59.Kha N.H.N., Sarter S., Legarve T., Montet D. 2006. Characterization Of Bacteria Isolated From Farmed Catfish Coming From Vietnam And Study Of Their Antibiotic Resistance; p. 7. [Google Scholar]
- 60.Xuan Binh D., Ngoc Minh N., Thi Nguyet D. Prevalence of Listeria monocytogenes, E. coli, Salmonella Spp. and Staphylococcus aureus bacteria contamination on meat at public market in the North of Vietnam. SOJ Microbiol. Infect. Dis. Dec. 2017;5(5):1–22. doi: 10.15226/sojmid/5/5/00184. [DOI] [Google Scholar]
- 61.Toan Nguyen Tat. Identification of genotype and phenotype of antimicrobial resistance of Escherichia coliisolates from pigs in southern Vietnam. Thai J. Vet. Med. 2021;51:125. doi: 10.14456/TJVM.2021.17. [DOI] [Google Scholar]
- 62.Vounba P., Arsenault J., Bada-Alambédji R., Fairbrother J.M. Pathogenic potential and the role of clones and plasmids in beta-lactamase-producing E. coli from chicken faeces in Vietnam. BMC Vet. Res. Dec. 2019;15(1):106. doi: 10.1186/s12917-019-1849-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nhung N.T., et al. Method for measuring phenotypic colistin resistance in Escherichia coli populations from chicken flocks. Appl. Environ. Microbiol. Feb. 2021;87(5) doi: 10.1128/AEM.02597-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oanh T.K.N., Nguyen V.K., Do T.N., Goddeeris B.M., De Greve H. Escherichia coli strains causing edema disease in northern Vietnam share an identical verotoxin 2e. Trop. Anim. Health Prod. Dec. 2010;42(8):1797–1804. doi: 10.1007/s11250-010-9639-6. [DOI] [PubMed] [Google Scholar]
- 65.Do N.T., et al. Antimicrobial resistance phenotypes of ETEC isolates from piglets with Diarrhea in North Vietnam. Ann. N. Y. Acad. Sci. Oct. 2006;1081(1):543–545. doi: 10.1196/annals.1373.082. [DOI] [PubMed] [Google Scholar]
- 66.Smith M., Do T.N., Gibson J.S., Jordan D., Cobbold R.N., Trott D.J. Comparison of antimicrobial resistance phenotypes and genotypes in enterotoxigenic Escherichia coli isolated from Australian and Vietnamese pigs. J. Glob. Antimicrob. Resist. Sep. 2014;2(3):162–167. doi: 10.1016/j.jgar.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 67.Le Huy H., et al. Antibiotic-resistant Escherichia coli isolated from urban rodents in Hanoi, Vietnam. J. Vet. Med. Sci. 2020;82(5):653–660. doi: 10.1292/jvms.19-0697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schutzius G., Nguyen M., Navab-Daneshmand T. Antibiotic resistance in fecal sludge and soil in Ho Chi Minh City, Vietnam. Environ. Sci. Pollut. Res. Nov. 2019;26(33):34521–34530. doi: 10.1007/s11356-019-06537-5. [DOI] [PubMed] [Google Scholar]
- 69.Lien L., et al. Antibiotic resistance and antibiotic resistance genes in Escherichia coli isolates from hospital wastewater in Vietnam. Int. J. Environ. Res. Public Health. Jun. 2017;14(7):699. doi: 10.3390/ijerph14070699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thai P.K., et al. Occurrence of antibiotic residues and antibiotic-resistant bacteria in effluents of pharmaceutical manufacturers and other sources around Hanoi, Vietnam. Sci. Total Environ. Dec. 2018;645:393–400. doi: 10.1016/j.scitotenv.2018.07.126. [DOI] [PubMed] [Google Scholar]
- 71.Duong H.A., et al. Occurrence, fate and antibiotic resistance of fluoroquinolone antibacterials in hospital wastewaters in Hanoi, Vietnam. Chemosphere. Jun. 2008;72(6):968–973. doi: 10.1016/j.chemosphere.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 72.Tong N.T.A. Assessment of antibiotic resistance and bacterial contamination of ice sold in Cantho City, Vietnam. Vietnam J. Sci. Technol. Nov. 2019;57(3B):49. doi: 10.15625/2525-2518/57/3B/14205. [DOI] [Google Scholar]
- 73.Harada T., et al. Prevalence and antimicrobial susceptibility of Enterobacteriaceae isolated from retail pepper in Vietnam. J. Food Prot. May 2016;80(5):716–724. doi: 10.4315/0362-028X.JFP-16-501. [DOI] [PubMed] [Google Scholar]
- 74.Trang N.H.T., et al. The characterization of ESBL genes in Escherichia coli and Klebsiella pneumoniae causing nosocomial infections in Vietnam. J. Infect. Dev. Ctries. Dec. 2013;7(12):922–928. doi: 10.3855/jidc.2938. [DOI] [PubMed] [Google Scholar]
- 75.Cao V., et al. Distribution of extended-Spectrum -lactamases in clinical isolates of Enterobacteriaceae in Vietnam. Antimicrob. Agents Chemother. 2002;46:5. doi: 10.1128/AAC.46.12.3739-3743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Q.P., et al. Characteristics of extended-Spectrum β-lactamase–producing Escherichia coli in retail meats and shrimp at a local market in Vietnam. Foodborne Pathog. Dis. Aug. 2015;12(8):719–725. doi: 10.1089/fpd.2015.1954. [DOI] [PubMed] [Google Scholar]
- 77.Hang B.P.T., Wredle E., Börjesson S., Sjaunja K.S., Dicksved J., Duse A. High level of multidrug-resistant Escherichia coli in young dairy calves in southern Vietnam. Trop. Anim. Health Prod. Jul. 2019;51(6):1405–1411. doi: 10.1007/s11250-019-01820-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.L T.H., H T.T.T., H H.M. Antibiotic resistance and molecular characteristics of extended-spectrum beta-lactamase-producing Escherichia coli isolated from fish pond. Can Tho Univ. J. Sci. 2018;54(8):114. doi: 10.22144/ctu.jen.2018.045. [DOI] [Google Scholar]
- 79.Hon N.T.N., et al. Spread of antibiotic and antimicrobial susceptibility of ESBL-producing Escherichia coli isolated from wild and cultured fish in the Mekong Delta, Vietnam. Fish Pathol. 2016;51(Special-issue):S75–S82. doi: 10.3147/jsfp.51.S75. [DOI] [Google Scholar]
- 80.Nguyen D.P., et al. Dissemination of extended-spectrum β -lactamase- and AmpC β -lactamase-producing Escherichia coli within the food distribution system of Ho Chi Minh City, Vietnam. Biomed. Res. Int. 2016;2016:1–9. doi: 10.1155/2016/8182096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nakayama T., et al. Frequent use of colistin-based drug treatment to eliminate extended-spectrum beta-lactamase-producing Escherichia coli in backyard chicken farms in Thai Binh Province, Vietnam. Trop. Anim. Health Prod. Jan. 2017;49(1):31–37. doi: 10.1007/s11250-016-1154-y. [DOI] [PubMed] [Google Scholar]
- 82.Dang S.T.T., Bortolaia V., Tran N.T., Le H.Q., Dalsgaard A. Cephalosporin-resistant Escherichia coli isolated from farm workers and pigs in northern Vietnam. Tropical Med. Int. Health. Apr. 2018;23(4):415–424. doi: 10.1111/tmi.13054. [DOI] [PubMed] [Google Scholar]
- 83.Kawahara R., et al. Prevalence of mcr-1 among cefotaxime-resistant commensal Escherichia coli in residents of Vietnam. Infect. Drug Resist. Oct. 2019;12:3317–3325. doi: 10.2147/IDR.S224545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Le P.Q., et al. Prevalence of mobile colistin resistance (mcr) genes in extended-spectrum β-lactamase-producing Escherichia coli isolated from retail raw foods in Nha Trang, Vietnam. Int. J. Food Microbiol. May 2021;346:109164. doi: 10.1016/j.ijfoodmicro.2021.109164. [DOI] [PubMed] [Google Scholar]
- 85.Parisi A., et al. The role of animals as a source of antimicrobial resistant nontyphoidal salmonella causing invasive and non-invasive human disease in Vietnam. Infect. Genet. Evol. Nov. 2020;85:104534. doi: 10.1016/j.meegid.2020.104534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Trung N.V., et al. Non-Typhoidal salmonella colonization in chickens and humans in the Mekong Delta of Vietnam. Zoonoses Public Health. Mar. 2017;64(2):94–99. doi: 10.1111/zph.12270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ly T.L.-K., et al. Prevalence of salmonella and Escherichia coli O157 from acute diarrheic children in the Mekong Delta, Vietnam. J. Vet. Epidemiol. 2010;14(1):55–61. doi: 10.2743/jve.14.55. [DOI] [Google Scholar]
- 88.Vo A.T.T., van Duijkeren E., Gaastra W., Fluit A.C. Antimicrobial resistance, class 1 integrons, and genomic island 1 in salmonella isolates from Vietnam. PLoS One. Feb. 2010;5(2):e9440. doi: 10.1371/journal.pone.0009440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yen N.T.P., et al. Antimicrobial residues, non-typhoidal Salmonella, Vibrio spp. and associated microbiological hazards in retail shrimps purchased in Ho Chi Minh city (Vietnam) Food Control. Jan. 2020;107:106756. doi: 10.1016/j.foodcont.2019.106756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lettini A.A., et al. Distribution of salmonella Serovars and antimicrobial susceptibility from poultry and swine farms in Central Vietnam. Zoonoses Public Health. Nov. 2016;63(7):569–576. doi: 10.1111/zph.12265. [DOI] [PubMed] [Google Scholar]
- 91.Ta Y.T., et al. Quantification, Serovars, and antibiotic resistance of salmonella isolated from retail raw chicken meat in Vietnam. J. Food Prot. Jan. 2014;77(1):57–66. doi: 10.4315/0362-028X.JFP-13-221. [DOI] [PubMed] [Google Scholar]
- 92.Nhung N.T., et al. Antimicrobial residues and resistance against critically important antimicrobials in non-typhoidal salmonella from meat sold at wet markets and supermarkets in Vietnam. Int. J. Food Microbiol. Feb. 2018;266:301–309. doi: 10.1016/j.ijfoodmicro.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tu L.T.P., et al. High levels of contamination and antimicrobial-resistant non-typhoidal salmonella serovars on pig and poultry farms in the Mekong Delta of Vietnam. Epidemiol. Infect. Oct. 2015;143(14):3074–3086. doi: 10.1017/S0950268815000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nguyen D.T.A., et al. Prevalence, antibiotic resistance, and extended-spectrum and AmpC β-lactamase productivity of salmonella isolates from raw meat and seafood samples in Ho Chi Minh City, Vietnam. Int. J. Food Microbiol. Nov. 2016;236:115–122. doi: 10.1016/j.ijfoodmicro.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 95.Thai T.H., Hirai T., Lan N.T., Yamaguchi R. Antibiotic resistance profiles of salmonella serovars isolated from retail pork and chicken meat in North Vietnam. Int. J. Food Microbiol. May 2012;156(2):147–151. doi: 10.1016/j.ijfoodmicro.2012.03.016. [DOI] [PubMed] [Google Scholar]
- 96.Van T.T.H., Moutafis G., Istivan T., Tran L.T., Coloe P.J. Detection of salmonella spp. in retail raw food samples from Vietnam and characterization of their antibiotic resistance. Appl. Environ. Microbiol. Nov. 2007;73(21):6885–6890. doi: 10.1128/AEM.00972-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.A H.T.T., K L.T.L. The prevalence and antibiotic resistance of salmonella spp. isolated from pigs and farm environments in Vinh Long province. Can Tho Univ. J. Sci. 2018;54(Agriculture):26. doi: 10.22144/ctu.jsi.2018.091. [DOI] [Google Scholar]
- 98.Ellerbroek L., et al. Antibiotic resistance in salmonella isolates from imported chicken carcasses in Bhutan and from pig carcasses in Vietnam. J. Food Prot. Feb. 2010;73(2):376–379. doi: 10.4315/0362-028X-73.2.376. [DOI] [PubMed] [Google Scholar]
- 99.Ngoc Nghiem M., Thanh Nguyen V., Hoai Nguyen T.T., Dang Nguyen T., Bich Vo T.T. Antimicrobial resistance gene expression associated with multidrug resistant salmonella spp. isolated from retail meat in Hanoi, Vietnam. Int. Microbiol. 2017;no. 20:85–93. doi: 10.2436/20.1501.01.288. [DOI] [PubMed] [Google Scholar]
- 100.Thai T.H., Hirai T., Lan N.T., Shimada A., Ngoc P.T., Yamaguchi R. Antimicrobial resistance of salmonella Serovars isolated from beef at retail Markets in the North Vietnam. J. Vet. Med. Sci. 2012;74(9):1163–1169. doi: 10.1292/jvms.12-0053. [DOI] [PubMed] [Google Scholar]
- 101.Thai T.H., Lan N.T., Hirai T., Yamaguchi R. Antimicrobial resistance in salmonella Serovars isolated from meat shops at the Markets in North Vietnam. Foodborne Pathog. Dis. Nov. 2012;9(11):986–991. doi: 10.1089/fpd.2011.1121. [DOI] [PubMed] [Google Scholar]
- 102.González-Santamarina B., et al. Genomic characterization of multidrug-resistant salmonella Serovars Derby and Rissen from the pig value chain in Vietnam. Front. Vet. Sci. Aug. 2021;8:705044. doi: 10.3389/fvets.2021.705044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ogasawara N., et al. Antimicrobial susceptibilities of salmonella from domestic animals, food and human in the Mekong Delta, Vietnam. J. Vet. Med. Sci. 2008;70(11):1159–1164. doi: 10.1292/jvms.70.1159. [DOI] [PubMed] [Google Scholar]
- 104.Nguyen T.K., et al. Prevalence and antibiotic resistance of salmonella isolated from poultry and its environment in the Mekong Delta, Vietnam. Vet. World. Dec. 2021:3216–3223. doi: 10.14202/vetworld.2021.3216-3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Noor Uddin G.Md., Larsen M.H., Barco L., Minh Phu T., Dalsgaard A. Clonal occurrence of salmonella Weltevreden in cultured shrimp in the Mekong Delta, Vietnam. PLoS One. Jul. 2015;10(7) doi: 10.1371/journal.pone.0134252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Holohan N., et al. Analysis of antimicrobial resistance in non-typhoidal salmonella collected from pork retail outlets and slaughterhouses in Vietnam using whole genome sequencing. Front. Vet. Sci. Mar. 2022;9 doi: 10.3389/fvets.2022.816279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nguyen D.T.A., et al. Prevalence, Serovar, and antimicrobial resistance of Nontyphoidal salmonella in vegetable, fruit, and water samples in Ho Chi Minh City, Vietnam. Foodborne Pathog. Dis. May 2021;18(5):354–363. doi: 10.1089/fpd.2020.2891. [DOI] [PubMed] [Google Scholar]
- 108.Nguyen T.K., et al. Retail fresh vegetables as a potential source of salmonella infection in the Mekong Delta, Vietnam. Int. J. Food Microbiol. Mar. 2021;341 doi: 10.1016/j.ijfoodmicro.2021.109049. [DOI] [PubMed] [Google Scholar]
- 109.Minh D.K., Hounmanou Y.M.G., Mai H.B.T., Olsen J.E., Dalsgaard A. Prevalence and genomic characterization of salmonella Weltevreden in commercial pig feed. Vet. Microbiol. Jul. 2020;246 doi: 10.1016/j.vetmic.2020.108725. [DOI] [PubMed] [Google Scholar]
- 110.Huong L.Q., Forslund A., Madsen H., Dalsgaard A. Survival of salmonella spp. and fecal indicator bacteria in Vietnamese biogas digesters receiving pig slurry. Int. J. Hyg. Environ. Health. Sep. 2014;217(7):785–795. doi: 10.1016/j.ijheh.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ly T.L.-K., et al. Isolation of salmonella from flies in the Mekong Delta, Vietnam. J. Vet. Epidemiol. 2010;14(1):41–46. doi: 10.2743/jve.14.41. [DOI] [Google Scholar]
- 112.Chiou C.-S., et al. Antimicrobial resistance in salmonella enterica Serovar Typhi isolates from Bangladesh, Indonesia, Taiwan, and Vietnam. Antimicrob. Agents Chemother. Nov. 2014;58(11):6501–6507. doi: 10.1128/AAC.03608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Trung N.D., Suthisarnsuntorn U., Kalambaheti T., Wonglumsom W., Tunyong W. Antimicrobial susceptibility patterns and phage types of salmonella Typhi from Vietnam. Southeast Asian J. Trop. Med. Publ. Health. 2007;38(3):6. [PubMed] [Google Scholar]
- 114.Tran G.M., et al. Patterns of antimicrobial resistance in intensive care unit patients: a study in Vietnam. BMC Infect. Dis. Dec. 2017;17(1):429. doi: 10.1186/s12879-017-2529-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Berglund B., et al. Molecular and phenotypic characterization of clinical isolates belonging to a KPC-2-producing strain of ST15 Klebsiella pneumoniae from a Vietnamese pediatric hospital. Antimicrob. Resist. Infect. Control. Dec. 2019;8(1):156. doi: 10.1186/s13756-019-0613-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Vinh H., et al. A changing picture of shigellosis in southern Vietnam: shifting species dominance, antimicrobial susceptibility and clinical presentation. BMC Infect. Dis. Dec. 2009;9(1):204. doi: 10.1186/1471-2334-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vale F.F., Lehours P., Yamaoka Y. Editorial: the role of Mobile genetic elements in bacterial evolution and their adaptability. Front. Microbiol. Feb. 2022;13 doi: 10.3389/fmicb.2022.849667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Nhung N.T., et al. Antimicrobial resistance in commensal Escherichia coli from humans and chickens in the Mekong Delta of Vietnam is driven by antimicrobial usage and potential cross-species transmission. JAC-Antimicrob. Resist. May 2022;4(3):dlac054. doi: 10.1093/jacamr/dlac054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Melnyk A.H., Wong A., Kassen R. The fitness costs of antibiotic resistance mutations. Evol. Appl. Mar. 2015;8(3):273–283. doi: 10.1111/eva.12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cuong N.V., et al. Feasibility study of a field survey to measure antimicrobial usage in humans and animals in the Mekong Delta region of Vietnam. JAC-Antimicrob. Resist. Jul. 2021;3(3):dlab107. doi: 10.1093/jacamr/dlab107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mouiche M.M.M., et al. Antimicrobial resistance from a one health perspective in Cameroon: a systematic review and meta-analysis. BMC Public Health. Dec. 2019;19(1):1135. doi: 10.1186/s12889-019-7450-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fujita A.W., et al. Antimicrobial resistance through the lens of One Health in Ethiopia: a review of the literature among humans, animals, and the environment. Int. J. Infect. Dis. Jun. 2022;119:120–129. doi: 10.1016/j.ijid.2022.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Varga C., et al. 2008. Comparison of Antimicrobial Resistance in Generic Escherichia coli and Salmonella spp. Cultured from Identical Fecal Samples in Finishing Swine; p. 7. [PMC free article] [PubMed] [Google Scholar]
- 124.Carrique-Mas J., et al. Mortality, disease and associated antimicrobial use in commercial small-scale chicken flocks in the Mekong Delta of Vietnam. Prev. Vet. Med. Apr. 2019;165:15–22. doi: 10.1016/j.prevetmed.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Phu D.H. Reducing antimicrobial usage in small-scale chicken farms in Vietnam: a 3-year intervention study. Front. Vet. Sci. 2021;7:14. doi: 10.3389/fvets.2020.612993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.WHO Implementation of Global Action Plan on Antimicrobial Resistance. 2017. https://www.who.int/antimicrobial-resistance/news/WHO-GAP-AMR-Newsletter-No-32-Nov-2017.pdf [Online]. Available:
- 127.FAO . Food and Agriculture Organization of the United Nations; Rome: 2016. Livestock-Related Interventions during Emergencies: The how-to-Do-it Manual. [Google Scholar]
- 128.Anon “Law on animal husbandry.” National Assembly of the Socialist Republic of Vietnam, 2018. 2018. https://www.economica.vn/Content/files/LAW%20%26%20REG/Law%20on%20Animal%20Husbandry%202018.pdf [Online]. Available.
- 129.The Government of the Socialist and Republic of Vietnam Decree No. 13/2020/ND-CP guiding the Law on Animal Husbandry. 2020. https://english.luatvietnam.vn/decree-no-13-2020-nd-cp-dated-january-21-2020-of-the-government-on-detailing-a-number-of-articles-of-the-law-on-animal-husbandry-180147-Doc1.html (accessed Nov. 28, 2021)
- 130.Ha L.T.T., et al. Antimicrobial usage surveillance through sales at veterinary drug shops intended for livestock in Vietnam. Front. Sustain. Food Syst. Dec. 2021;5:784500. doi: 10.3389/fsufs.2021.784500. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forest plot of pooled prevalence of E.coli and NTS to six antimicrobials in meta-analysis.
The contour-enhanced funnel plots show the asymmetry among selected studies. The dashed vertical lines show the average effect sizes, each scatter presents each selected article, and the shaded regions illustrate the statistical significance of the asymmetry of the publications. CIP - Ciprofloxacin, CAZ - Ceftazidime, AMP - Ampicillin, GEN- Gentamicin, SXT - Sulfamethoxazole-trimethoprim, CHL-Chloramphenicol, MDR - Multi-drug Resistance.
The PRISMA item checklist.
A total number of 89 selected studies were used for systematic review and meta-analysis.
The data on AMR extracted from 89 selected studies for systematic review
The data on AMR extracted from 55 selected studies for meta-analysis
The average AMR prevalence of E. coli and Non-typhoidal Salmonella (NTS) in healthy humans, healthy animals/ animal food products and the environment.
Data Availability Statement
The research data are available in supplementary materials.