Abstract
Alzheimer’s disease (AD) is the most common form of dementia, affecting approximately 6.5 million Americans age 65 or older. AD is characterized by increased cognitive impairment and treatment options available provide minimal disease attenuation. Additionally, diagnostic methods for AD are not conclusive with definitive diagnoses requiring postmortem brain evaluations. Therefore, miRNAs, a class of small, non-coding RNAs, have garnered attention for their ability to regulate a variety of mRNAs and their potential to serve as both therapeutic targets and biomarkers of AD. Several miRNAs have already been implicated with AD and have been found to directly target genes associated with AD pathology. The APP/PS1 mice is an AD model that expresses the human mutated form of the amyloid precursor protein (APP) and presenilin-1 (PS1) genes. In a previous study, it was identified that crossing long-living growth hormone (GH)-deficient Ames dwarf (df/df) mice with APP/PS1 mice provided protection from AD through a reduction in IGF-1, amyloid-β (Aβ) deposition, and gliosis. Hence, we hypothesized that changes in the expression of miRNAs associated with AD mediated such benefits. To test this hypothesis, we sequenced miRNAs in hippocampi of df/df, wild type (+ / +), df/ + /APP/PS1 (phenotypically normal APP/PS1), and df/df/APP/PS1 mice. Results of this study demonstrated significantly upregulated and downregulated miRNAs between df/df/APP/PS1 and df/ + /APP/PS1 mice that suggest the df/df mutation provides protection from AD progression. Additionally, changes in miRNA expression with age were identified in both df/df and wild-type mice as well as df/df/APP/PS1 and APP/PS1 mice, with predictive functional roles in the Pi3k-AKT/mTOR/FOXO pathways potentially contributing to disease pathogenesis.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-022-00633-0.
Keywords: Alzheimer’s disease (AD), Ames dwarf mice, MicroRNAs, Longevity
Introduction
Alzheimer’s disease (AD) is the most prevalent form of dementia, affecting an estimated 6.5 million Americans currently [1, 2]. The disease is commonly associated with aging and its course follows a progressive cognitive decline, with early symptoms involving memory loss and later symptoms including personality changes, functional and behavioral impairments that affect the ability to perform daily tasks, and deficits in language function [3]. Unfortunately, the average life expectancy following diagnosis averages 8–10 years, making AD the seventh leading cause of death worldwide [2]. AD pathology is caused by amyloid-β (Aβ) plaque accumulation and hypophosphorylated tau neurofibrillary tangles in the brain [4]. AD is also linked to increased reactive oxygen species (ROS) production with specific mutations in the amyloid precursor protein (APP) and presenilin-1 (PS1) genes ultimately contributing to APP processing and Aβ deposition [5].
APP/PS1 transgenic mice can be used to model oxidative stress-induced cerebral damage. These mice express the human mutations for APP and PS1 and have been found to exhibit similar outcomes such as increased Aβ and oxidative stress [5]. Meanwhile, Ames dwarf (df/df) mice have been extensively studied for their increased lifespan, which is widely attributed to their associated reduced inflammation, mitochondrial oxidative metabolism and enhanced cellular stress resistance [5, 6]. This increased stress resistance and enhanced longevity is ascribed to the absence of GH and combined loss of pituitary function as a result of the loss of function mutation in their prop-1 gene [7]. In two previous studies, df/df mice were also found to be particularly resistant to Aβ toxicity [5, 8].
To investigate the potential neuroprotective benefits of the hormone deficiencies experienced by df/df mice in AD progression, APP/PS1 transgenic mice were crossed with df/df mice [5]. This generated the following F2 generations: phenotypically normal mice carrying the dwarf gene (df/ +), dwarf mice (df/df), wild type mice (+ / +), df/ + /APP/PS1 mice, and df/df/APP/PS1 mice. In completing this study, Puig et al. developed a novel mouse model of AD that demonstrated a significant reduction in gliosis, Aβ levels, and IGF-1, suggesting the protective effects associated with GH deficiency in df/df mice can confer advantages for AD pathology as well.
MicroRNAs (miRNAs) have recently garnered attention for their ability to regulate a wide variety of pathways through targeted reduction of the translation of messenger RNAs (mRNAs) [9, 10]. Indeed, several miRNAs have been identified and correlated with AD pathology. For instance, miRNAs-200b, -135a, and -429, regulators of APP and BACE-1 (an enzyme involved in Aβ generation), were shown to be downregulated in the hippocampus of APP/PS1 mice [11]. Other potential miRNAs of interest have also been proposed and experimentally established to be involved in regulating processes crucial to AD outcome. Given the promising findings presented in the study by Puig et al., we were interested in identifying potential biomarkers or changes in expression profiles of miRNAs in the df/df x APP/PS1 crosses. Since AD can be difficult to diagnose, identifying potential biomarkers or factors that are altered with AD could provide added insight into future diagnostics and/or treatment methods. To investigate this, we sequenced miRNAs from hippocampal tissue of df/df, wild type, df/ + /APP/PS1, and df/df/APP/PS1 young and old mice.
Material and methods
Transgenic mice and tissue collection
For this study, C57BL6/APP/PS1 (APP/PS1; APPswe/PS1dE9; Mo/Hu APPswe PS1dE9; Tg(APPswe,PSEN1dE9)85Dbo) transgenic mice were bred with df/df mice to produce a heterozygous F1 generation that was then bred to produce F2 offspring by Puig et al. [5]. The F2 generation comprised dwarf (df/df), phenotypical normal heterozygous (df/ +), wild type (+ / +), APP/PS1, df/ + /APP/PS1, and df/df/APP/PS1 mice. Mice were genotyped and maintained under controlled light and temperature conditions with food and water provided ad libitum [5]. In this study, the following groups were selected: df/df, wild type, df/ + /APP/PS1, and df/df/APP/PS1 mice. The selected offspring were sacrificed at 3 months of age and 12 months of age for brain collection, hippocampus isolation, and downstream differential microRNA expression analyses (n = 5–6 per group). Harvested tissue was immediately frozen and stored at − 80 °C. All procedures involving animals were reviewed and approved by the UND Institutional Animal Care and Use Committee.
RNA isolation and library prep
Hippocampi collected was cut and weighed to obtain approximately 10 mg of tissue. Samples were lysed and homogenized with QIAzol lysis reagent and zirconium oxide beads (0.5 mm) in a bullet blender. Once homogenized, RNA extraction was achieved using the QIAGEN RNeasy mini kit (Hilden, Germany). All steps were performed in accordance with the provided protocol and total RNA concentrations were measured using the BioTek Epoch microplate spectrophotometer (BioTek, Agilent Technologies, Santa Clara, CA, USA). To prepare libraries for sequencing, 2 µg of total RNA was diluted in RNase-free water and combined with the appropriate NEXTFLEX Small RNA Seq. kit (V3) reagents used in accordance with the manufacturer’s protocol (Perkin-Elmer, Waltham, Massachusetts, USA). All of the samples were purified using the gel-free selection method. Following gel-free selection, libraries were pooled into two separate pools, precipitated using sodium acetate (3 M), ethanol (100%), and glycogen (20 mg/mL), centrifuged, washed with 70% ethanol, and then re-suspended in RNase-free water. The final, concentrated, library pools were then outsourced for QC and Illumina small RNA sequencing.
Statistical analysis
Fold change and relative expression
Alignment and quantification of miRNA libraries was performed using sRNAtoolbox as described before [12]. Statistical analyses of differentially expressed miRNAs was performed using EdgeR [13] on the R software (3.2.2) and miRNAs with a FDR < 0.05 and FC > 2.0 were considered as upregulated, and FDR < 0.05 and FC < 0.50 were considered as downregulated.
Prediction of miRNA target genes and their pathway interactions
DIANA Tool miRPath (v3) was used to generate lists of gene targets and pathways relevant to microRNAs of interest through the micro-T-CDS (V5.0). The DIANA-miRPath v3 was utilized for its ability to provide predicted and experimentally supported miRNA interactions and the pathways they regulate [14]. Alternatively, miRNA gene targets predicted to function in AD pathology were cross-referenced with the miRNA database (miRdb) [15, 16].
Results
Age impacts expression of miRNAs in hippocampi of df/df and wild type (+ / +) mice
To identify changes in miRNA expression with age, we evaluated differentially expressed miRNAs in df/df older mice (12 months of age) in comparison with df/df young mice (3 months of age) as well as in wild-type older mice (12 months of age) in comparison with wild-type young mice (3 months of age). Results of our analysis revealed downregulated expression of miR-17-5p, miR-19b-3p, miR-22-5p, miR-322-5p, miR-301a-3p, miR-19a-3p, miR-154-5p, miR-337-3p, miR-20a-5p, miR-34a-5p, miR-344b-3p, miR-467d-3p, miR-501-5p, and miR-296-5p in older df/df mice (Table 1). Pathway analysis of these down-regulated miRNAs revealed predicted gene targets involved in MAPK, FoxO, TGF-β, insulin, Pi3K-AKT, mTOR, Ras, and Hippo signaling (Supplemental Table 3). Conversely, miR-148b-5p, miR-1981-5p, miR-744-5p, miR-488-3p, and miR-873a-5p were found to be upregulated in df/df older mice (Table 1). These miRNAs are predicted to regulate genes involved in long-term depression, suggesting these middle-aged df/df mice may be less prone to long-term depression; however, experience increased insulin-signaling and associated pathways with age (Supplemental Tables 3 and 4). Similarly, wild-type older mice exhibited downregulated expression of a set of miRNAs that are also predicted to regulate MAPK, Pi3K-AKT, mTOR, and insulin signaling (Supplemental Table 6). These downregulated miRNAs include miR-296-5p, miR-138–2-3p, miR-669c-5p, miR-1264-5p, miR-204-5p, and miR-let-7c-5p. Both df/df and wild-type mice exhibited significant downregulation of miR-296-5p with age (Table 1). Conversely, miR-375-3p and miR-152-3p were significantly upregulated in older wild-type mice (Table 1). Pathway analysis revealed an association between these miRNAs and Hippo and FoxO signaling pathways, indicating that these two pathways are likely downregulated in these mice (Supplemental Table 5).
Table 1.
miRNA expression patterns significantly altered in df/df and wild-type (+ / +) older mice compared to young df/df and wild-type mice, respectively. p value and FDR < 0.05 were considered significant (refer to Supplemental Tables 1 and 2 for statistical values)
| MicroRNA ID | + / + | df/df |
|---|---|---|
| mmu-miR-17-5p | - | ↓ |
| mmu-miR-19b-3p | - | ↓ |
| mmu-miR-22-5p | - | ↓ |
| mmu-miR-322-5p | - | ↓ |
| mmu-miR-301a-3p | - | ↓ |
| mmu-miR-19a-3p | - | ↓ |
| mmu-miR-154-5p | - | ↓ |
| mmu-miR-337-3p | - | ↓ |
| mmu-miR-20a-5p | - | ↓ |
| mmu-miR-34a-5p | - | ↓ |
| mmu-miR-344b-3p | - | ↓ |
| mmu-miR-467d-3p | - | ↓ |
| mmu-miR-501-5p | - | ↓ |
| mmu-miR-296-5p | ↓ | ↓ |
| mmu-miR-148b-5p | - | ↑ |
| mmu-miR-1981-5p | - | ↑ |
| mmu-miR-744-5p | - | ↑ |
| mmu-miR-488-3p | - | ↑ |
| mmu-miR-837a-5p | - | ↑ |
| mmu-miR-138–2-3p | ↓ | - |
| mmu-miR-669c-5p | ↓ | - |
| mmu-miR-1264-5p | ↓ | - |
| mmu-miR-204-5p | ↓ | - |
| mmu-let-7c-5p | ↓ | - |
| mmu-miR-375-3p | ↑ | - |
| mmu-miR-152-3p | ↑ | - |
miRNAs predicted to regulate the mTOR and FoxO signaling pathways are differentially expressed in df/df/APP/PS1 older mice
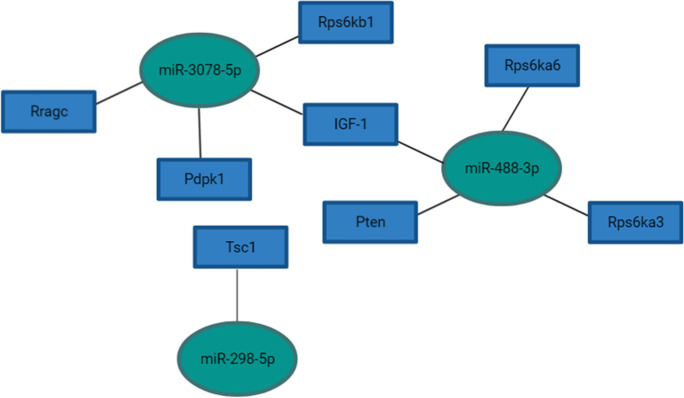
Puig et al. previously demonstrated reduced IGF-1 expression in the parietal cortex and hippocampi of df/df/APP/PS1 mice, which is in line with the df/df phenotype, suggesting the absence of GH and other pleiotropic hormones provide advantageous reductions in insulin signaling that might decelerate brain aging [5]. In our analysis of differentially expressed miRNAs in df/df/APP/PS1 mice compared to wild-type mice, we identified four miRNAs, miR-488a-5p, miR-488-3p, miR-3078-5p, and miR-298-5p, significantly upregulated in the hippocampi of df/df/APP/PS1 mice (Table 2). According to pathway analysis, these miRNAs target mTOR signaling and FoxO signaling (Figs. 1 and 2, Supplemental Table 10). Additionally, miR-488-3p may also play a crucial role in regulating onset of long-term depression (Supplemental Table 10). This miRNA was correspondingly upregulated in young df/df mice compared to wild-type young mice (Supplemental Table 11). miR-488-5p and miR-298-5p are also anticipated to regulate endocytosis (Supplemental Table 10) a process implicated in AD pathogenesis through APP and Aβ production [17].
Table 2.
miRNA expression patterns significantly altered in df/ + /APP/PS1 and df/df compared to wild-type (+ / +) middle-aged mice as well as df/df/APP/PS1 middle-aged mice compared to df/ + /APP/PS1 middle-aged mice. p value and FDR < 0.05 were considered significant (refer to Supplemental Tables 7–9 for statistical values)
| MicroRNA ID | df/ + /APP/PS1 | df/df | df/df/APP/PS1 |
|---|---|---|---|
| mmu-miR-1957a | ↓ | ↑ | - |
| mmu-miR-200b-3p | ↓ | ↓ | ↑ |
| mmu-miR-488-5p | - | ↑ | ↑ |
| mmu-miR-3078-5p | - | ↑ | ↑ |
| mmu-miR-412-5p | - | ↑ | ↑ |
| mmu-miR-298-5p | ↑ | ↑ | ↑ |
| mmu-miR-666-5p | - | ↑ | ↑ |
| mmu-miR-488-3p | - | ↑ | ↓ |
| mmu-miR-219b-3p | ↑ | ↓ | ↓ |
| mmu-miR-219a-5p | ↑ | ↓ | - |
| mmu-miR-29b-3p | ↑ | ↑ | ↓ |
| mmu-miR-342-5p | ↑ | ↑ | - |
| mmu-let-7b-3p | ↑ | - | ↓ |
| mmu-miR-451a | ↑ | - | ↓ |
| mmu-miR-1b-5p | ↑ | - | ↓ |
| mmu-miR-206-3p | ↑ | - | ↓ |
| mmu-miR-142a-3p | ↑ | - | ↓ |
| mmu-miR-669c-5p | ↑ | - | ↓ |
| mmu-miR-3065-5p | ↑ | - | ↓ |
| mmu-miR-1a-3p | ↑ | - | ↓ |
| mmu-miR-669a-5p | ↑ | - | ↓ |
| mmu-miR-669p-5p | ↑ | - | ↓ |
| mmu-miR-144-3p | ↑ | - | ↓ |
Fig. 1.
Predicted miRNA interactions with genes implicated in mTOR signaling. Pathway interactions were derived from DIANA Tools and predicted gene targets were determined using the microT-CDS target prediction algorithm (Vlachos et al., 2015). Refer to Supplemental Tables 7–9 for differential expression and statistical values for miRNAs selected. Figure was made with BioRender
Fig. 2.
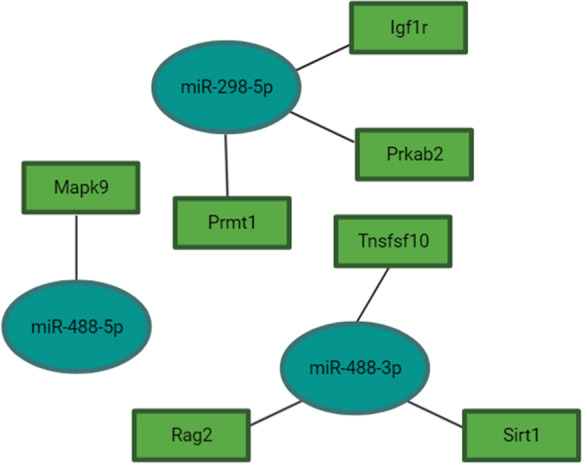
Predicted miRNA interactions with genes implicated in FoxO signaling. Pathway interactions were derived from DIANA Tools and predicted gene targets were determined using the microT-CDS target prediction algorithm (Vlachos et al., 2015). Refer to Supplemental Tables 7–9 for differential expression and statistical values for miRNAs selected. Figure was made with BioRender
miRNAs implicated with AD pathology are differentially expressed in older APP/PS1 mice compared to wild-type mice, as well as in older df/df/APP/PS1 compared to APP/PS1 mice
Several miRNAs have been identified as potential biomarkers or targets of AD. To investigate changes in expression of these miRNAs, we identified miRNAs differentially expressed in df/df, and df/ + /APP/PS1 older mice, compared to the wild-type older group, and in df/df/APP/PS1 older mice compared to the df/ + /APP/PS1 older mice. The results of this analysis demonstrated upregulated expression of miR-451a, miR-206-3p, miR-144-3p, and miR-142-3p in df/ + /APP/PS1 mice with downregulated expression of these miRNAs in df/df/APP/PS1 mice (Table 2). These miRNAs have previously been found to be differentially expressed in APP/PS1 mice and have been linked to increased APP and Aβ levels [18, 19], suggesting that the df/df phenotype may be conferring protection against AD progression through suppression of these miRNAs. The opposite expression profile was observed for miR-200b-3p and miR-219a-5p, which were found to be suppressed in df/df mice but upregulated in df/ + /APP/PS1 mice. Additionally, other miRNAs that may play a crucial role in facilitating AD pathology have also demonstrated similar expression profiles as miR-451a, miR-206-3p, and miR-144-3p. These miRNAs include miR-3065-5p, miR-1a-3p, miR-669a-5p, miR-669p-5p, and let-7b-3p (Table 2). Using the miRNA database (miRdb), we identified predicted gene targets that have been associated with preventing Aβ production through APP facilitation (Fig. 3). As such, these miRNAs upregulated in df/ + /APP/PS1 but downregulated in df/df/APP/PS1 mice may serve as novel miRNAs implicated with AD pathogenesis.
Fig. 3.
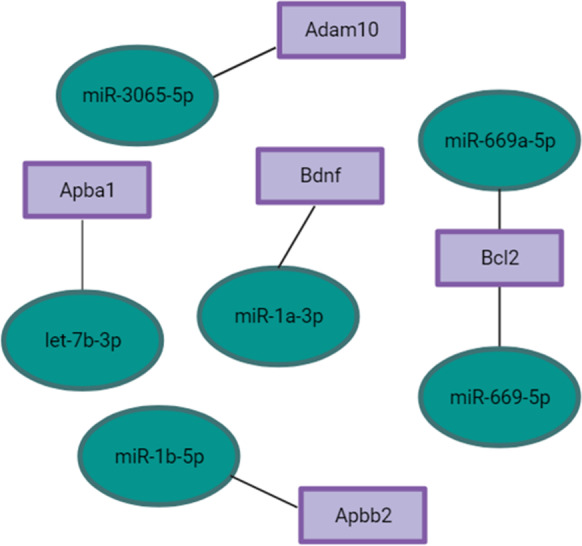
Predicted miRNA interactions with genes implicated in AD pathology. Genes were cross-referenced with the miRNA database (Chen et al., 2020 and Liu et al., 2019). Refer to Supplemental Tables 7–9 for differential expression and statistical values for miRNAs selected. Figure was made with BioRender
Discussion and conclusion
miRNA sequencing of hippocampal tissue in long-living GH-deficient df/df mice revealed 14 miRNAs downregulated and 5 miRNAs upregulated with age. Collectively, pathway analysis identified predicted gene targets involved in MAPK, FoxO, TGF-β, insulin, Pi3K-AKT, mTOR, Ras, and Hippo signaling pathways that correspond to the expected functional roles of the downregulated miRNAs. On the other hand, miRNAs upregulated with age in df/df mice demonstrated functional potential in regulating genes involved in long-term depression, suggesting df/df mice are less prone to long-term depression, however, demonstrate increased nutrient sensing and insulin-associated signaling in the brain with age. Previous studies have demonstrated a direct association between reduced IGF-1 with increased lifespan. Further, knockouts of insulin receptor substrate 1 (IRS-1), IRS-2, and IGF-1 have led to enhanced lifespan in mice. These findings specifically implicate the PI3k-AKT-mTOR and FoxO signaling pathways in modulating aging [20]. As such, our findings are in line with the literature, since miRNAs predicted to target the aforementioned signaling pathways are downregulated with age. Similarly, wild-type mice had decreased expression of miRNAs predicted to target MAPK, PI3K-AKT, mTOR, and insulin signaling. However, it has been widely established that df/df mice have notably enhanced insulin sensitivity with age when compared with their phenotypically normal littermates. These df/df mice exhibit hypersensitivity to insulin, have low fasting glucose and insulin levels [21], suggesting that GH deficiency and deficiencies in other hormones contribute to their overall improved insulin sensitivity with age and extended lifespan [22], which is also reflected in this study through the regulation of miRNAs in the hippocampi.
Additionally, miR-375-3p and miR-152-3p were significantly upregulated in wild-type mice with age. Pathway analysis revealed an association between these miRNAs and Hippo and FoxO signaling pathways. FoxO signaling plays a key role in insulin and IGF-1 signaling that is central to metabolic homeostasis [23]. Recent findings have demonstrated an association between dysregulation of FoxO signaling and type II diabetes, which interestingly, has been linked to increased risk of AD pathogenesis. This is likely due to the observed escalation in metabolic dysfunction in the AD brain [23]. Moreover, our findings demonstrated differentially expressed miRNAs that might be upregulated due to the absence of GH in df/df/APP/PS1 mice. These miRNAs include miR-3078-5p, miR-488-3p, miR-488-5p, and miR-298-5p. Pathway analysis demonstrated predicted functional roles for these miRNAs in regulating FoxO and mTOR signaling as well as endocytosis and long-term depression. FoxO and mTOR signaling pathways have been well established to be associated with AD pathogenesis, particularly due to the importance of both in maintaining metabolic homeostasis. Although suppression of FoxO signaling can bear negative impacts on the brain, such as through early depletion of neuronal stem cell pools [24], the genes regulated by these miRNAs are primarily associated with insulin-associated Pi3K-AKT-mTOR signaling as opposed to the broader function of FoxO signaling in maintaining cellular processes. Insulin-associated Pi3K-AKT signaling has widely been implicated with accelerating aging, a phenomenon that has deleterious effects on AD pathology [24, 25].
Furthermore, our findings revealed several mechanisms by which the reduction of Aβ plaque deposition and concentrations of Aβ1-40 and Aβ1-42 in df/df/APP/PS1 transgenic mice, demonstrated by Puig et al. [5], could be mediated. miR-451a, miR-206-3p, and miR-144-3p were found to be downregulated in df/df/APP/PS1 mice, while significantly upregulated in df/ + /APP/PS1 mice, suggesting the absence of GH and the benefits observed in df/df mice may confer advantages for the AD brain. miR-451a and miR-144-3p, miRNAs found to target ADAM10 and BCL2 as well as KH domain-containing RNA binding protein (QKI), modulate key regulators of Aβ and tau biosynthesis and transport, as well as regulate synaptic function and neuronal apoptosis. These miRNAs were found to be downregulated in APP/PS1 mice at different ages [19]. Correspondingly, our findings demonstrated a similar expression pattern in df/df/APP/PS1 mice; however, we found these miRNAs to be upregulated in df/ + /APP/PS1 mice compared to wild-type mice. Further, studies investigating the role of BCL2 in AD pathogenesis have reported a correlation between reduced BCL2 expression and Aβ1-40 levels, citing Aβ1-40 downregulates BCL2 expression [26]. BCL2 plays a critical role in regulating neuronal intracellular calcium signaling. Changes in calcium signaling is directly linked to neuronal loss in AD which can lead to attenuation of synapses, a phenomenon that is evident in early disease pathogenesis [27]. As such, downregulations in miR-451a and miR-144-3p may be advantageous for AD pathology. Similarly, miR-206, a miRNA associated with reduced brain-derived neurotrophic factor (BDNF) expression in APP/PS1 mice [18], was also significantly repressed in df/df/APP/PS1 mice, specifically through a reduction in the 3’ fragment (miR-206-3p). miR-206-3p is predicted to target BDNF, the most widely expressed neurotrophin in the brain [18]. BDNF functions primarily by regulating neurotransmitter release, neurite outgrowth, long-term potentiation, as well as gene transcription of genes involved in intracellular signaling pathways. In the context of AD, BDNF provides protection against Aβ toxicity [18]. As such, our findings indicate the df/df/APP/PS1 phenotype may be neuroprotective against Aβ toxicity through reduced miR-206-3p expression.
A previous study showed that APPtg and TAUtg mice have increased expression of miR-142a-5p while we demonstrated that df/df/APP/PS1 mice have reduced expression of miR-142a-3p. Human AD brain samples also demonstrate upregulated expression of miR-142a-5p, suggesting a potential role for this miRNA in AD progression [28]. On the other hand, miR-200b-3p, which belongs to the miR-200b family, has been implicated with regulation of APP and Aβ levels and is downregulated in APP/PS1 mice [11]. Our findings suggest a similar expression pattern in df/ + /APP/PS1 mice, while df/df/APP/PS1 mice exhibited upregulated expression of miR-200b-3p. Similarly, miR-219a-5p, a miRNA upregulated in both df/ + /APP/PS1 mice and in human AD brains [29] was also found to be suppressed in df/df mice. Taken together, it appears the GH deficiency in df/df mice provides protective advantages in AD pathology through the above-mentioned differentially expressed miRNAs. Other miRNAs that potentially play a crucial role in AD pathology were found to be repressed in df/df/APP/PS1 mice but increased in df/ + /APP/PS1 mice as well. These miRNAs include let-7b-3p, miR-3065-5p, miR-1a-3p, miR-669a-5p, and miR-669p-5p. Cross-analysis identified predicted gene targets and potential functional targets in regulating AD pathology [15, 16]. For instance, let-7b-3p is anticipated to target APBA1 (also known as X11α), a suppressor of the production of APP fragments (including Aβ peptides) [30]. Similarly, miR-3065-5p is predicted to target ADAM10, implicated with reducing Aβ production, tau pathology, as well as maintaining synaptic function, neurogenesis, and regulating neuronal networks in the hippocampus [31]. Further, miR-1a-3p is expected to target BDNF, which as described previously, is crucial in regulating AD. miR-669a-5p and miR-669p-5p, on the other hand, are anticipated to target BCL2 [15, 16], which plays a crucial role in regulating intracellular calcium signaling and thereby maintaining neuronal function [27]. With these miRNAs being effectively repressed in df/df/APP/PS1 in comparison to df/ + /APP/PS1, it is apparent that the GH deficiency provides protection from AD.
Overall, our findings provide a strong rationale and basis for the advantages conferred by the GH deficiency in AD progression in APP/PS1 transgenic mice. Our data support several potential mechanisms by which the df/df mutation provides protection against AD, as well as validates the current literature regarding the role of miRNAs in AD advancement. Future considerations include deriving the functional and mechanistic roles of the newly proposed miRNAs that could potentially serve as therapeutic targets in AD.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contribution
S, Noureddine—concept and design, analysis, interpretation of data, manuscript original draft, manuscript revision. T, Saccon—acquisition of data, manuscript revision. T, Rudeski-Rohr—acquisition of data, manuscript revision. A, Schneider—acquisition of data, analysis, manuscript revision. J, Dhabhi—analysis, manuscript revision. K, Puig—acquisition of data, analysis, manuscript revision. S, Rakoczy—acquisition of data, analysis, manuscript revision. H, Brown-Borg—acquisition of data, analysis, manuscript revision. M, Masternak—concept and design, funding acquisition, acquisition of data, analysis, interpretation of data, manuscript original draft, manuscript revision.
Funding
This work was supported by the National Institutes of Health/ National Institute on Aging grants R15 AG059190, R03 AG059846, R56 AG061414, and R21 AG062985 (to M.M.M.), 1R15AG061795-01A (to JBM), 1RO1 AG034206 (to HMBB), and the National Science Centre, Poland (2016/21/B/NZ4/03192) (grant no. 507/1–168-02/507–10-105 of the Medical University of Lodz, Poland (AG).
Declarations
Conflict of interest
Dr. Michal Masternak is an associate editor in the journal.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Organization, W.H. The top 10 causes of death 2020; Available from: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death.
- 2.Association, A.s., 2022 Alzheimer’s disease facts and figures. 2022;Alzheimers Dement. [DOI] [PubMed]
- 3.Nyul-Toth A, DelFavero J, Mukli P, Tarantini A, Ungvari A, Yabluchanskiy A, Csiszar A, Ungvari Z, Tarantini S. Early manifestation of gait alterations in the Tg2576 mouse model of Alzheimer's disease. GeroScience. 2021;43(4):1947–1957. doi: 10.1007/s11357-021-00401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weller J, Budson A. Current understanding of Alzheimer’s disease diagnosis and treatment. F1000 Research. 2018;7. [DOI] [PMC free article] [PubMed]
- 5.Puig KL, Kulas JA, Franklin W, Rakoczy SG, Taglialatela G, Brown-Borg HM, Combs CK. The Ames dwarf mutation attenuates Alzheimer’s disease phenotype of APP/PS1 mice. Neurobiol Aging. 2016;40:22–40. doi: 10.1016/j.neurobiolaging.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill CM, Fang Y, Miquet JG, Sun LY, Masternak MM, Bartke A. Long-lived hypopituitary Ames dwarf mice are resistant to the detrimental effects of high-fat diet on metabolic function and energy expenditure. Aging Cell. 2016;15(3):509–521. doi: 10.1111/acel.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gesing A, Al-Regaiey KA, Bartke A, Masternak MM. Growth hormone abolishes beneficial effects of calorie restriction in long-lived Ames dwarf mice. Exp Gerontol. 2014;219–229. [DOI] [PMC free article] [PubMed]
- 8.Schrag M, et al. Hippocampus of Ames dwarf mice is resistant to beta-amyloid-induced tau hyperphosphorylation and changes in apoptosis-regulatory protein levels. Hippocampus. 2008;18(3):239–244. doi: 10.1002/hipo.20387. [DOI] [PubMed] [Google Scholar]
- 9.Nunes ADC, Weigl M, Schneider A, Noureddine S, Yu L, Lahde C, Saccon TD, Mitra K, Beltran E, Grillari J, Kirkland JL, Tchkonia T, Robbins PD, Masternak MM. miR-146a-5p modulates cellular senescence and apoptosis in visceral adipose tissue of long-lived Ames dwarf mice and in cultured pre-adipocytes. GeroScience. 2022;44:503–215. doi: 10.1007/s11357-021-00490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pushpakumar S, et al. Exogenous hydrogen sulfide and miR-21 antagonism attenuates macrophage-mediated inflammation in ischemia reperfusion injury of the aged kidney. Geroscience. 2021;43(3):1349–1367. doi: 10.1007/s11357-020-00299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu CG, Wang JL, Li L, Xue LX, Zhang YQ, Wang PC. MicroRNA-135a and -200b, potential Biomarkers for Alzheimer׳s disease, regulate β secretase and amyloid precursor protein. Brain Res. 2014;1583:55–64. doi: 10.1016/j.brainres.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 12.Rueda A, et al. sRNAtoolbox: an integrated collection of small RNA research tools. Nucleic Acids Res. 2015;43(W1):W467–W473. doi: 10.1093/nar/gkv555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D, Vergoulis T, Dalamagas T, Hatzigeorgiou AG. DIANA-miRPath v30: deciphering microRNA function with experimental support. Nucleic Acids Res. 2015;43(W1):W460–W466. doi: 10.1093/nar/gkv403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen YWX. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020;(D1):D127-D131 [DOI] [PMC free article] [PubMed]
- 16.Liu W, Wang X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biology. 2019;20(1) [DOI] [PMC free article] [PubMed]
- 17.Ando K, Houben S, Homa M, de Fisenne MA, Potier MC, Erneux C, Brion JP, Leroy K. Alzheimer's disease: tau pathology and dysfunction of endocytosis. Front Mol Neurosci. 2020;13. [DOI] [PMC free article] [PubMed]
- 18.Tian N, Cao Z, Zhang Y. MiR-206 decreases brain-derived neurotrophic factor levels in a transgenic mouse model of Alzheimer’s disease. Neurosci Bull. 2014;30(2):191–197. doi: 10.1007/s12264-013-1419-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeng L, Jiang HL, Ashraf GM, Li ZR, Liu R. MicroRNA and mRNA profiling of cerebral cortex in a transgenic mouse model of Alzheimer’s disease by RNA sequencing. Neural Regen Res. 2021. [DOI] [PMC free article] [PubMed]
- 20.Bartke A. Impact of reduced insulin-like growth factor-1/insulin signaling on aging in mammals: novel findings. Aging Cell. 2008;7(3):285–290. doi: 10.1111/j.1474-9726.2008.00387.x. [DOI] [PubMed] [Google Scholar]
- 21.Masternak MM, Panici JA, Bonkowski MS, Hughes LF, Bartke A. Insulin sensitivity as a key mediator of growth hormone actions on longevity. J Gerontol A: Biol Sci Med Sci. 2009. [DOI] [PMC free article] [PubMed]
- 22.Wiesenborn DS, Ayala JE, King E, Masternak MM. Insulin sensitivity in long-living Ames dwarf mice. Age. 2014;36(5). [DOI] [PMC free article] [PubMed]
- 23.Zhang QS, Liu W, Lu GX. miR-200a-3p promotes b-amyloid-induced neuronal apoptosis through down-regulation of SIRT1 in Alzheimer's disease. J Biosci. 2017;42(3):397–404. doi: 10.1007/s12038-017-9698-1. [DOI] [PubMed] [Google Scholar]
- 24.Du S, Zheng H. Role of FoxO transcription factors in aging and age-related metabolic and neurodegenerative diseases. Cell Biosci. 2021;11. [DOI] [PMC free article] [PubMed]
- 25.Woodling NS, Rajasingam A, Minkley LJ, Rizzo A, Partridge L. Independent glial subtypes delay development and extend healthy lifespan upon reduced insulin-PI3K signalling. BMC Biol. 2020;18. [DOI] [PMC free article] [PubMed]
- 26.Paradis E, Douillard H, Koutroumanis M, Goodyer C, LeBlanc A. Amyloid β peptide of Alzheimer’s disease downregulates Bcl-2 and upregulates Bax expression in human neurons. J Neurosci. 1996;16(23):7533–7539. doi: 10.1523/JNEUROSCI.16-23-07533.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Callens M, Kraskovskaya N, Derevtsova K, Annaert W, Bultynck G, Bezprozvanny I, Vervliet T. The role of Bcl-2 proteins in modulating neuronal Ca2+ signaling in health and in Alzheimer's disease. Biochim Biophys Acta (BBA) - Mol Cell Res. 2021;1868(6). [DOI] [PMC free article] [PubMed]
- 28.Sierksma A, Lu A, Salta E, Eynden EV, Callaerts-Vegh Z, D'Hooge R, Blum D, Buee L, Fiers M, Strooper BD. Deregulation of neuronal miRNAs induced by amyloid-β or TAU pathology. Mol Neurodegeneration. 2018;13(54). [DOI] [PMC free article] [PubMed]
- 29.Cha DJ, Mengel D, Mustapic M, Liu W, Selkoe DJ, Kapogiannis D, Galasko D, Rissman R A, Bennet DA, Walsh DM. miR-212 and miR-132 are downregulated in neurally derived plasma exosomes of Alzheimer’s patients. Front Neurosci. 2019. [DOI] [PMC free article] [PubMed]
- 30.Lee J, Lau KF, Perkinton MS, Standen CL, Shemilt SJ, Mercken L, Cooper JD, McLoughlin DM, Miller CC. The neuronal adaptor protein X11alpha reduces Abeta levels in the brains of Alzheimer's APPswe Tg2576 transgenic mice. J Biol Chem. 2003;278(47). [DOI] [PubMed]
- 31.Yuan XZ, Sun S, Tan CC, Yu JT, Tan L. The role of ADAM10 in Alzheimer's disease. J Alzheimer's Dis. 2017;58(2) [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.