Summary
Pathologists diagnose prostate cancer by core needle biopsy. In low-grade and low-volume cases, they look for a few malignant glands out of hundreds within a core. They may miss a few malignant glands, resulting in repeat biopsies or missed therapeutic opportunities. This study developed a multi-resolution deep-learning pipeline to assist pathologists in detecting malignant glands in core needle biopsies of low-grade and low-volume cases. Analyzing a gland at multiple resolutions, our model exploited morphology and neighborhood information, which were crucial in prostate gland classification. We developed and tested our pipeline on the slides of a local cohort of 99 patients in Singapore. Besides, we made the images publicly available, becoming the first digital histopathology dataset of patients of Asian ancestry with prostatic carcinoma. Our multi-resolution classification model achieved an area under the receiver operating characteristic curve (AUROC) value of 0.992 (95% confidence interval [CI]: 0.985–0.997) in the external validation study, showing the generalizability of our multi-resolution approach.
Keywords: prostate cancer, histopathology, digital pathology, computational pathology, whole slide image analysis, artificial intelligence, machine learning, deep learning
Highlights
-
•
Multi-resolution models outperform single-resolution models in gland classification
-
•
Both morphology and neighborhood information are vital in gland classification
-
•
Multi-resolution models generalize across institutes and patients’ ancestries
-
•
Multi-resolution models focus on similar features used in the clinic
The bigger picture
Deep-learning-based assistive tools are becoming an integral part of pathology clinics. They promise to reduce pathologists’ workloads and improve patients’ outcomes. This study focuses on a challenging task rather than an easy one to enhance these promises. It develops a deep-learning-based pipeline detecting low-grade and low-volume prostate cancer that can be easily overlooked, potentially resulting in missed therapeutic opportunities. The pipeline detects the few low-grade cancerous components within a low volume of prostate tissue and highlights high-risk regions for detailed analysis by pathologists. It can help early diagnosis of prostate cancer and effective use of other therapeutic tools at early stages, like active surveillance, rather than aggressive treatments eligible at later stages. Besides, this study conducts an external validation on data of patients from different ancestries, which can be a critical factor in the success of deep-learning-based assistive tools.
Diagnosis of prostate cancer in low-grade and low-volume cases is a challenging task for pathologists. They may miss a few malignant components within the tissue, resulting in repeat biopsies or missed therapeutic opportunities. This study developed a multi-resolution pipeline to assist pathologists in such cases. An external validation study demonstrated the generalizability of the multi-resolution approach across institutes and patients’ ancestries. Besides, the analysis of models revealed their focus in their predictions.
Introduction
Prostate cancer is the second most common cancer diagnosed in men worldwide.3 It is diagnosed by core needle biopsy analysis, involving a collection of about 12 cores from different parts of the prostate. Pathologists analyze individual prostate glands on the slides of the collected cores for malignancy. For low-grade and low-volume cases, pathologists have to carefully examine hundreds of glands in each core to avoid missing any malignant glands. This is a tedious and time-consuming process that is prone to errors and inter-observer variability. Besides, increasing incident rates and decreasing number of actively working pathologists escalate the workload per pathologist.4
Prostatic carcinoma is graded based on Gleason patterns (GPs) from 1 to 5 (although GP1 and GP2 are not routinely reported in the clinic). The sum of the two most common patterns inside the slide (for biopsy slides, the highest grade is reported as the second pattern if it is >5%) is the Gleason score (GS) and is used as the grade of prostatic carcinoma. Recently, the 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma updated the definitions of GPs and introduced a new grading system.5 There are five prognostically different grade groups from GG1 to GG5 (from the most favorable to the least favorable), which are based on the modified GS groups: GG1 (GS ≤ 6), GG2 (GS 3 + 4 = 7), GG3 (GS 4 + 3 = 7), GG4 (GS 8), and GG5 (GS 9–10). It is vital to diagnose cancer at low grades for a better prognosis and patient life quality.
Recently, it has been shown that the assistance of machine-learning systems significantly improves the diagnosis and grading of prostate cancer by pathologists.6,7 A few studies developed successful machine-learning systems for prostate cancer diagnosis and grading to assist pathologists.8,9,10,11,12,13,14,15,16,17 They covered a broad spectrum of grade groups. Nevertheless, it is easier for an expert pathologist to diagnose high-grade cancers such as GG4 or GG5. On the other hand, it becomes a real challenge to discriminate rare malignant glands among numerous benign glands in low-grade cancers such as GG1 and GG2.18 Besides, undetected malignant glands may result in repeat biopsies and missed therapeutic opportunities. Therefore, this study concentrates on low-grade prostatic carcinoma of GG1 and GG2. It develops a deep-learning pipeline detecting rare malignant glands in core needle biopsy slides to help pathologists quickly and accurately diagnose prostate cancer in low-grade and low-volume cases. Given a core needle biopsy slide, the pipeline produces a vivid heatmap highlighting the malignant glands inside the slide to pathologists. Moreover, contrary to previous studies’ single resolution and patch-based methods, this study uses a multi-resolution and gland-based classification approach, providing pathologists with individual gland labels.
Our pipeline consists of two stages: gland segmentation using a Mask R-CNN19 model and a multi-resolution gland classification model (see experimental procedures and Figure 1). While glands in biopsy cores were detected by the gland segmentation model, each detected gland was classified into benign versus malignant by the gland classification model. Our multi-resolution gland classification model jointly analyzed a gland’s high-resolution (40× and 20×) and low-resolution (10× and 5×) patches to exploit morphology information (of nuclei and glands) and neighborhood information (for architectural patterns), respectively (see experimental procedures). The multi-resolution models imitate pathologists’ comprehensive workflow of analyzing both macro and micro structures inside the slides,20,21 and even naive multi-resolution models obtained as ensembles perform better than single-resolution models.8 The code is publicly available.2
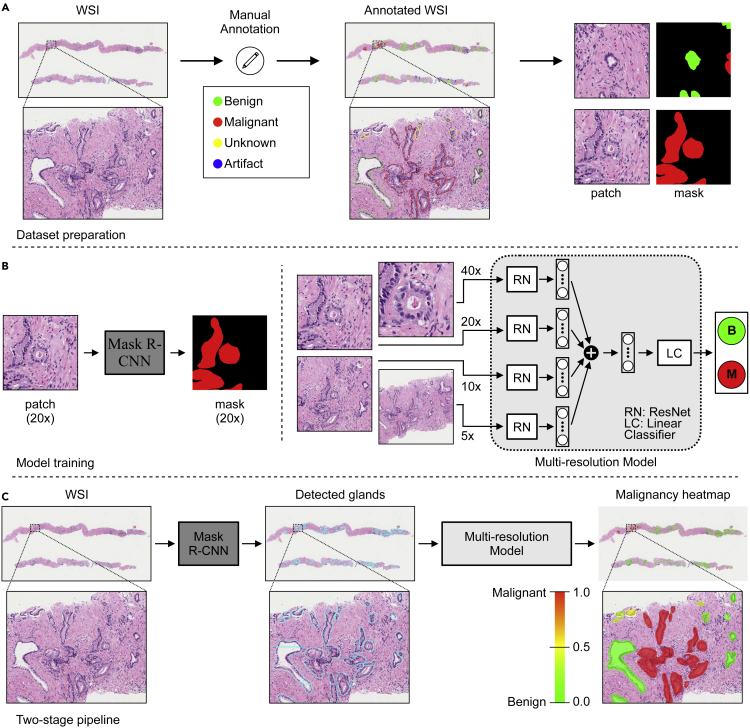
Figure 1.
Malignant gland detection pipeline in prostate core needle biopsies
(A) Glands in collected WSIs were annotated by drawing contours around each gland and assigning a malignant, benign, unknown, or artifact label. Then, patches centered around a particular gland were cropped at different resolutions for each gland while preparing datasets for training machine-learning models. See also Figure S1.
(B) A Mask R-CNN19 model was trained for gland segmentation on patch and mask pairs prepared at 20× resolution. A multi-resolution (four-resolution) deep-learning model was trained for gland classification on patches (cropped at 5×, 10×, 20×, and 40× resolutions) and label pairs.
(C) After gland segmentation and classification models were successfully trained, a two-stage pipeline was constructed. The trained Mask R-CNN model detected the glands in a WSI, and the trained multi-resolution model obtained the malignancy probability for each detected gland. Finally, a malignancy heatmap was generated to support pathologists in prostate cancer diagnosis from core needle biopsy WSIs.
This study was conducted on a local cohort of 99 patients collected in Singapore (see datasets and Table 1). Data collected from each patient contained hundreds of glands for machine learning. The data are publicly available,1 and this is the first digital histopathology dataset of patients of Asian ancestry with prostatic carcinoma. The global research community can benefit from this valuable dataset to develop and test machine-learning models and ultimately improve patients’ outcomes.
Table 1.
Singapore data: The number of slides and patches in training, validation, and test sets for gland segmentation and classification tasks
| Gland segmentation | ||||||||
|---|---|---|---|---|---|---|---|---|
| # slides |
# patches |
|||||||
| Train | Valid | Test | Total | Train | Valid | Test | Total | |
| Prostatectomy | 17 | 8 | 15 | 40 | 7,795 | 3,753 | 7,224 | 18,772 |
| Biopsy | 26 | 13 | 20 | 59 | 5,559 | 4,028 | 5,981 | 15,568 |
| Total | 43 | 21 | 35 | 99 | 13,354 | 7,781 | 13,205 | 34,340 |
| Gland classification | ||||||||
| # slides (3 + 3:3 + 4:4 + 3) | # patches (benign:malignant) | |||||||
| Train | Valid | Test | Total | Train | Valid | Test | Total | |
| Biopsy | 10:9:1 | 3:7:0 | 6:10:0 | 19:26:1 | 1,557:2,277 | 1,216:1,341 | 1,543:2,718 | 4,316:6,336 |
There is one H&E-stained WSI for each prostatectomy or core needle biopsy specimen. The gland classification datasets are subsets of the gland segmentation datasets. See also Table S1.
The pipeline’s performance was evaluated on the data of held-out patients in the test set. The performance metric was the area under the receiver operating characteristic curve (AUROC) value with a 95% confidence interval (CI) constructed using the percentile bootstrap method.22 We obtained an AUROC value of 0.997 (95% CI: 0.987–1.000) on benign versus malignant classification of core needle biopsy parts (81 parts: 50 benign and 31 malignant) in 16 slides of 16 patients in the test set. Furthermore, we produced spatial malignancy maps with a gland-level resolution to assist pathologists in reading prostate core needle biopsy slides (Figure 2D). Our pipeline can help pathologists detect prostate cancer in core needle biopsy slides at early stages and shorten turnaround times by presenting high-risk regions via malignancy maps to pathologists.
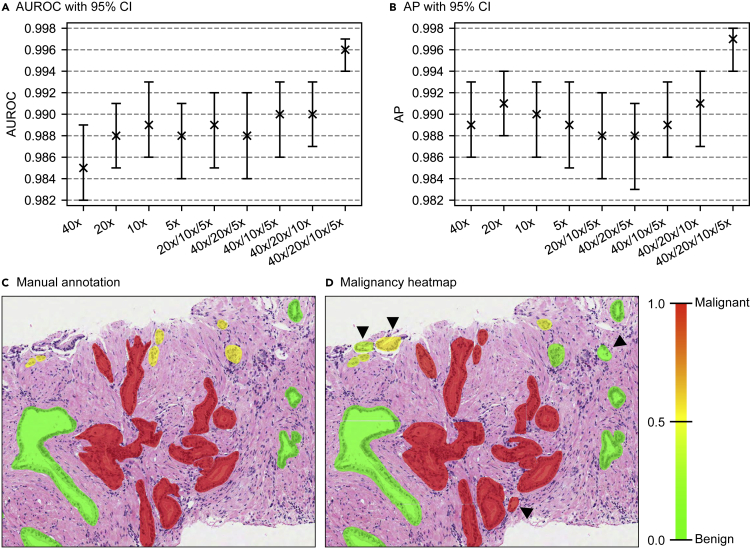
Figure 2.
Performance evaluation on the test set of gland classification dataset and spatial malignancy map
(A and B) Area under receiver operating characteristics curve (AUROC) (A) and average precision (AP) (B) calculated over precision versus recall curve together with 95% confidence intervals (obtained using the percentile bootstrap method22) are presented for the single-resolution models, three-resolution models, and a four-resolution model.
(C and D) Example manual annotations and spatial malignancy map produced using a deep-learning-based pipeline, respectively. Arrow heads show tissue components detected by the pipeline but not manually annotated.
An external validation study also showed that our multi-resolution classification model generalized across institutions that had patients from different ancestries. A three-resolution classification model was trained on the publicly available PANDA challenge dataset consisting of thousands of prostate core needle biopsy slides of patients with European ancestry.18 Then, the model was tested on our gland classification dataset, which consisted of slides of patients with Asian ancestry. An AUROC value of 0.992 (95% CI: 0.985–0.997) was obtained on benign versus malignant classification of core needle biopsy parts (280 parts: 179 benign and 81 malignant) (Table 3).
Table 3.
External validation set performance of algorithms in prostate cancer detection
| Study | External dataset (B: benign, M: malignant) | AUROC (95% CI) |
|---|---|---|
| Campanella et al.8 | 12,727 core needle biopsy slides (B = 314, M = 12,413) | 0.932 (0.911–0.952)a |
| Pantanowitz et al.13 | 355 parts with multiple slides (B = 225, M = 130) | 0.991 (0.979–1.000)b |
| Ström et al.14 | 330 core needle biopsy slides (B = 108, M = 222) | 0.986 (0.972–0.996) |
| Bulten et al.15 | 245 tissue microarray cores (B = 10, M = 235) | 0.988 (0.984–1.000) |
| Current study | 280 core needle biopsy parts (B = 179, M = 81) | 0.992 (0.985–0.997) |
Results
We trained our models on the training set and chose the best set of model weights based on validation set performance (see datasets and Tables 1 and S1). Finally, we evaluated the performance of our trained models on the data of completely unseen patients in the hold-out test set. Each patient in the test set was like a new patient walking into the clinic.23
Mask R-CNN model successfully segmented prostate glands
The Mask R-CNN model’s performance was evaluated on the test set of gland segmentation dataset. Using an intersection over union (IoU) threshold of 0.5, a recall of 0.945 and a precision of 0.830 were obtained at gland level. The low precision (compared with recall) was due to glands appearing inside dataset patches but which were not annotated since they were partial glands at the edges of the biopsy cores (Figure 2D).
Moreover, we compared the Mask R-CNN model’s performance with other literature methods on a publicly available gland segmentation dataset.24 We trained the model on the provided training set and checked the trained model’s performance on the hold-out test set. The Mask R-CNN model slightly outperformed deep-learning-based segmentation methods (Table 2). Besides, all the deep-learning-based methods vastly outperformed traditional image processing or machine-learning-based methods.
Table 2.
Performances of different methods in prostate gland segmentation in terms of pixel-based metrics
| Method | Accuracy | Precision | Recall | Dice |
|---|---|---|---|---|
| Farjam et al.25,a | 0.6378 ± 0.1586 | 0.7183 ± 0.3034 | 0.4372 ± 0.1736 | 0.5070 ± 0.2059 |
| Naik et al.26,a | 0.7402 ± 0.1151 | 0.7958 ± 0.2021 | 0.5819 ± 0.2275 | 0.6357 ± 0.2105 |
| Peng et al.27,a | 0.7957 ± 0.1535 | 0.6508 ± 0.2568 | 0.9305 ± 0.1124 | 0.7334 ± 0.2198 |
| Nguyen et al.28,a | 0.7703 ± 0.1632 | 0.8260 ± 0.1588 | 0.7041 ± 0.2998 | 0.7145 ± 0.2556 |
| Singh et al.29,a | 0.6734 ± 0.1247 | 0.9001 ± 0.1743 | 0.3869 ± 0.2493 | 0.4931 ± 0.2557 |
| Ren et al.30,b | 0.8576 ± 0.1139 | 0.8199 ± 0.1638 | 0.8861 ± 0.1673 | 0.8308 ± 0.1495 |
| Xu et al.31,b | 0.8250 ± 0.1106 | 0.7407 ± 0.1597 | 0.9273 ± 0.1079 | 0.8079 ± 0.1264 |
| Salvi et al.24,b | 0.9325 ± 0.0684 | 0.8897 ± 0.1359 | 0.9356 ± 0.0964 | 0.9016 ± 0.1087 |
| Mask R-CNN19,b,c | 0.9410 ± 0.0010 | 0.9002 ± 0.0026 | 0.9468 ± 0.0011 | 0.9229 ± 0.0015 |
The performances were on the hold-out test set of Salvi et al.24 Note that accuracy values were the balanced accuracy values as in Salvi et al.,24 and all performance values except the one for the Mask R-CNN model were collected from Salvi et al.24
Traditional image processing or machine-learning-based methods.
Deep-learning-based methods.
Standard deviations were calculated using bootstrapping.22
The four-resolution model outperformed single-resolution models in gland classification
The four-resolution deep neural network model incorporated information from different levels in gland classification task. While 40× and 20× patches provided detailed morphology of the gland under consideration, 10× and 5× patches provided spatial neighboring information. We also trained single-resolution models for comparison. The models were evaluated using AUROC and average precision (AP) calculated over precision versus recall curve. 95% CIs were constructed using the percentile bootstrap method.22
The four-resolution model achieved an AUROC of 0.996 (95% CI: 0.994–0.997) and an AP of 0.997 (95% CI: 0.994–0.998). While the single-resolution models also produced satisfactory results, the four-resolution model outperformed them (Figures 2A and 2B).
Gland morphology and neighborhood information were important in prostate gland classification
To assess the contribution of each resolution to the gland classification performance, we trained three-resolution models by dropping a different resolution each time from the four-resolution model. Then, the performance drop on the test set was used as a metric for that particular resolution. The highest and second-highest performance drops were observed in both AUROC and AP when the resolutions of 10× and 40×, respectively, were excluded (Figures 2A and 2B). Our analysis showed that 10× and 40× patches provided valuable information for the four-resolution model. This also validated that both morphology (from 40×) and neighborhood (from 10×) information were important in prostate gland classification.
The pathologist’s annotations served as landmarks and guided the researcher in creating viable annotations for machine learning
Having a pathologist annotate every single gland in a slide is expensive and not feasible. Therefore, we followed a different strategy (see Figure S1 for details). A senior pathologist annotated only 10% of the glands in each slide. Based on these annotations, a researcher annotated the rest of the glands.
We checked the effectiveness of our annotation strategy. In the training set of gland classification dataset, we trained two four-resolution models: the first model using glands annotated by the pathologist (modelP), and the second model using glands annotated by the researcher (modelR). Then, in the test set of gland classification dataset, each model’s performance was calculated on only the glands annotated by the pathologist (glandsP) and only the glands annotated by the researcher (glandsR).
On the glandsP, while the modelP achieved an AUROC of 0.969 (95% CI: 0.950–0.985) and an AP of 0.950 (95% CI: 0.918–0.976), the modelR achieved an AUROC of 0.975 (95% CI: 0.958–0.989) and an AP of 0.962 (95% CI: 0.933–0.986). Similarly, on the glandsR, while the modelP achieved an AUROC of 0.987 (95% CI: 0.983–0.991) and an AP of 0.988 (95% CI: 0.983–0.992), the modelR achieved an AUROC of 0.990 (95% CI: 0.987–0.994) and an AP of 0.991 (95% CI: 0.987–0.994). Obtaining a similar performance for both models on each subset of the test set (see Figure S2 for details), we concluded that the pathologist’s annotations served as landmarks and guided the researcher in creating viable annotations for machine learning.
Deep-learning-based pipeline successfully classified biopsy parts into negative and positive
There were multiple needle biopsy cores in a whole-slide image (WSI), and these cores could be broken into parts during slide preparation. Each core needle biopsy part within WSIs in the test set of gland classification dataset was classified into positive versus negative based on the manual annotations. A part was assigned a positive label if it contained at least one malignant gland and a negative label otherwise. Then, the pipeline was tested end to end on the core needle biopsy part classification task (81 parts in 16 slides of 16 patients in the test set: 50 benign and 31 malignant). The glands in each part were detected by the trained Mask R-CNN model. For each detected gland, a malignancy probability was obtained from the trained four-resolution model (see experimental procedures). The maximum of the predicted malignancy probabilities in a part was used as the part’s malignancy probability. An AUROC value of 0.997 (95% CI: 0.987–1.000) was obtained.
Furthermore, to assist pathologists in reading prostate core needle biopsy slides, a spatial malignancy map for each slide was constructed using malignancy probabilities of detected glands obtained from the trained four-resolution model (Figure 2D).
External validation demonstrated the generalizability of our multi-resolution approach
To check the generalizability of our multi-resolution approach, we trained a three-resolution benign versus malignant classification model on the publicly available development set of the PANDA challenge.18 The PANDA dataset consisted of 10,512 prostate biopsy slides of different grade groups collected from two institutes in Europe (see PANDA dataset for details). We randomly segregated 10,512 slides into training (6,279 - benign [B]: 1,719 versus malignant [M]: 4,560), validation (2,093 - B: 573 versus M: 1,520), and test (2,140 - B: 580 versus M: 1,560) sets (see Table S1 for details). The model was trained on the training set for 327 epochs with early stopping criteria on the validation set performance. Then, besides checking the model’s slide classification performance on the unseen test set of the PANDA dataset (see Table S2), an external validation study was performed on the Singapore (SG) gland classification dataset.
We obtained malignancy probability scores of patches within a core needle biopsy part from the trained model and used the maximum of the scores as the part’s malignancy score. Then, we performed a receiver operating characteristics curve analysis for benign versus malignant classification on the SG gland classification dataset (280 cores from 46 patients - B: 179 versus M: 81). An AUROC value of 0.992 (95% CI: 0.985–0.997) was obtained (Table 3), showing the generalizability of our multi-resolution approach across institutions with patients of different ancestries. Moreover, the PANDA model achieved an AUROC value of 0.980 (95% CI: 0.953–0.997) on the test set of the SG gland classification dataset, which was similar to our pipeline’s performance of 0.997 (95% CI: 0.987–1.000) in the previous section (see Table S2).
Post-hoc analysis revealed the focuses of the models in their predictions
To obtain deeper insights into the four-resolution model trained on the SG dataset (SG model) and the three-resolution model trained on the PANDA dataset (PANDA model), we conducted a post-hoc analysis on images in the test set of the SG gland classification dataset. We used integrated gradients attribution method32 to obtain the contribution of each element inside the images of different resolutions (Figures 3A, S3, and S4). The post-hoc analysis revealed that elements of images from different resolutions contributed to the models’ predictions. While the SG model exploited information from both high- and low-resolution images, the PANDA model focused mostly on low-resolution images.
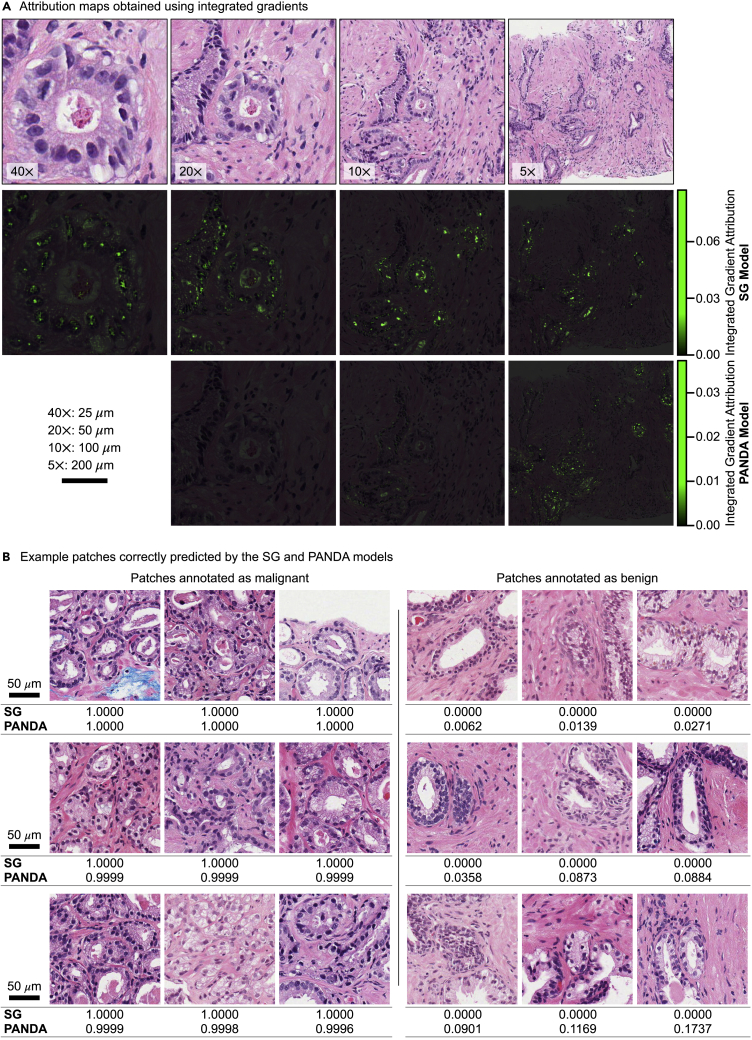
Figure 3.
Post-hoc analysis using the trained SG and PANDA models
(A) Attribution maps obtained using integrated gradients32 with blurred images as baselines are presented for a malignant sample in the test set of SG gland classification dataset. This sample is predicted correctly by the SG and PANDA models. See also Figures S3 and S4.
(B) One malignant patch and one benign patch with the highest correct class probability scores (by the SG model) from nine different slides in the test set of the SG gland classification dataset. Predicted malignancy scores by the trained SG and PANDA models are presented under each image. See also Figure S5. Scale bars are shown in black.
We observed in SG model’s predictions that prominent nucleoli in malignant glands (Figure 3A and S3) had high attribution scores, consistent with prostate cancer histology in the clinic. Similarly, nuclei regions and lumen contours were highlighted in attribution maps of low-resolution images for both models (Figures 3A and S3), showing the contribution of nuclear morphology and arrangement in the models’ predictions. Another interesting observation was that, beside nuclear areas, cytoplasm of columnar epithelial cells in benign glands had high attributions (Figure S4). This might be thought of as a criterion of the machine-learning model similar to nucleus-cytoplasm ratio used in the clinic.
To have a better understanding of the SG and PANDA models, we inspected some patches annotated by the pathologist in the test set of the SG gland classification dataset. Among patches correctly classified by the models with high confidence (Figure 3B), there were malignant patches with both GP3 and GP4, indicating that models learned to identify these patterns. Similarly, there were benign glands with both cross-sectional and tangential cuts. Besides, we observed that the PANDA model made better predictions on the patches with infiltrating malignant glands (Figure S5), which might be due to better generalization from having more patient data. On the other hand, the SG model’s performance was better on benign glands. One reason could be that many of these benign glands were annotated by the pathologist upon the researcher’s request since they were hard cases. Moreover, it is to be noted that the test set of the SG gland classification dataset was an internal test set for the SG model. However, it was an external test set for the PANDA model. This could be another reason of the SG model’s slightly better performance.
Grade group prediction using a multi-resolution GP classifier was promising
To determine the multi-resolution GP classifier’s viability in grade group prediction, our multi-resolution benign versus malignant gland classification model was modified to classify a patch into benign, pattern3, pattern4, or pattern5 (see supplemental experimental procedures for details). We conducted our experiments with this model on the Radboud dataset, a partition of the PANDA dataset (Table S1). Of note, this was the only dataset with pixel-level GP annotations. The annotations were generated semi-automatically using a trained deep-learning model and contained label noise.18
In the GP classification task, the model achieved a patch-level accuracy of 0.864 on the test set of the Radboud dataset. As seen in the confusion matrix (Figure S6), the model performed well on benign patches. However, it had difficulty in discriminating malignant patches. Many pattern5 patches, for example, were classified as pattern4. There were also malignant patches classified as benign. To assess the severity of misclassification, we conducted a benign versus malignant slide classification experiment on slide malignancy scores obtained by aggregating malignant classes (see supplemental experimental procedures for details). An AUROC value of 0.960 (95% CI: 0.949–0.970) was obtained (Figure S7), showing that these errors were not severe.
After obtaining pattern predictions for all patches in a slide, we obtained the slide’s grade group based on the pattern percentages within the slide (see supplemental experimental procedures for details). To check the agreement between predicted grade groups and reference grade groups in the dataset, we used quadratically weighted Cohen’s κ.33 A κ value of 0.707 (95% CI: 0.665–0.748) was obtained. Besides, many of the wrong predictions were within one grade group (Figure S8). Although the model’s performance was not as high as the performance of the PANDA challenge models,18 it was promising for further exploration. One of the main reasons for this performance gap could be the label noise in pixel-level annotations. All the reported models were multiple-instance-learning-based models trained directly on slide-level reference grade groups and avoided noisy pixel-level annotations.18 Hence, we concluded that grade group prediction based on our multi-resolution GP predictor was promising. It could help us interpret the prediction by providing pattern percentages and their distribution over the slide (Figures S9 and S10). However, to improve the performance, we needed more reliable pixel-level annotations, which was kept as future work.
Discussion
Manual reading of core needle biopsy slides by pathologists is the gold standard in the prostate cancer diagnosis in the clinic. However, it requires the analysis of around 12 (6–18) biopsy cores, including hundreds of glands. Especially for low-grade and low-volume prostate cancer (GS 3 + 3 and 3 + 4), identifying the few malignant glands among vastly benign glands is a tedious and challenging task. These few malignant glands can be easily overlooked, potentially resulting in missed therapeutic opportunities. This study developed a deep-learning-based pipeline to detect malignant glands in core needle biopsy slides of patients with low-grade prostate cancer. The pipeline can help early diagnosis of prostate cancer and effective use of other therapeutic tools at early stages, like active surveillance, rather than aggressive prostatectomy eligible for later stages. Moreover, the pipeline can reduce pathologists’ workload as an assistive tool.
Deep-learning-based pipeline can assist pathologists
Our pipeline successfully classified biopsy cores as negative or positive. It can be deployed as a pre-analysis stratification tool and help pathologists effectively manage their time on each biopsy core. For instance, they can spend less time validating negative cores while devoting more time to positive cores. Besides, spatial malignancy maps can help them concentrate on high-risk regions and decide if further cuts are required (for example, in the regions with a malignancy probability of 0.5) to make a diagnosis.
Furthermore, our pipeline can be deployed as a second-read system. The system can generate a flag for a second opinion in case of a contradiction between the pathologist’s diagnosis and the system’s classification. This can help reduce false negatives and false positives, which potentially result in missed therapeutic opportunities and aggressive treatment, respectively.
Challenges of gland-level annotation
Gland-level manual annotation is a challenging task, especially in core needle biopsies. A tissue core starts drying out from the surface after excision. If the core is not put into formalin buffer immediately, the morphology of the glands at the edges becomes distorted, making benign versus malignant classification more challenging. Besides, the glands at the edges are usually partial, and partial glands are not used for diagnosis in the clinical routine. If all glands within the core are partial, pathologists usually make a diagnosis based on other viable cores. Moreover, most glandular structures inside the cores are tangential cuts, which are hard to annotate. They are considered secondary for diagnosis and mostly require deeper cuts to reveal the glandular structure for diagnosis.
Furthermore, artifacts occur during the sample preparation, such as detached glands, folded tissue, uneven cuts, and poor preservation. These also make the annotation of each gland challenging. Another challenge appears in identifying the boundaries of the glands. It can be difficult to draw the boundaries of branching glands and fused glands during manual annotation.
Despite these challenges, gland-level annotation provides a fine-level resolution to identify individual glands as benign or malignant. This helps us train machine-learning models with fewer slides than we would need with slide-level annotations.
Limitations and future work
Gland-level annotation enabled us to train highly accurate machine-learning models. However, we had a limited number of annotated slides since manual annotation was tedious and time consuming. It would have been better if we had more slides to consolidate our model’s performance. Our external cohort study showed the robustness of our model against inter-institution differences. Yet, the coverage of our external cohort (our gland classification dataset) was limited to 3 + 3 and 3 + 4 slides of 46 patients.
In the future, we wish to deploy our pipeline as a second-read system in Singapore and check its performance in the real-world clinical flow. Moreover, extending our Asian cohort to cover all Gleason grade groups and adapting our multi-resolution approach to predict grade group directly are kept as future work.
Experimental procedures
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Mustafa Umit Oner (mustafaumit.oner@eng.bau.edu.tr).
Materials availability
This study did not generate new unique reagents.
Datasets
SG dataset
Digitized hematoxylin and eosin (H&E)-stained WSIs of 40 prostatectomy and 59 core needle biopsy specimens were collected from 99 patients with prostate cancer at Tan Tock Seng Hospital, Singapore. There were 99 WSIs in total such that each specimen had one WSI. H&E-stained slides were scanned at 40× magnification (specimen-level pixel size 0.25 × 0.25 μm) using Aperio AT2 Slide Scanner (Leica Biosystems).
We developed models for the gland segmentation and gland classification tasks using the 99 WSIs. The 99 WSIs were randomly segregated into training (43), validation (21), and test sets (35) at the patient level to avoid data leakage while training the models.23 While all the slides were utilized in the gland segmentation task, only a subset of the slides in each set (training: 20, validation: 10, and test: 16) was used in the gland classification task (Table 1). The models were trained on the training sets. The best sets of model weights were chosen on the validation sets using early stopping to avoid overfitting, and the best models were evaluated on the test sets.
Prostate glandular structures in core needle biopsy slides were manually annotated and classified into four classes, benign, malignant, unknown, and artifact (Figure 1A), using the ASAP annotation tool (https://computationalpathologygroup.github.io/ASAP/). A senior pathologist reviewed 10% of the annotations in each slide, ensuring that some reference annotations were provided to the researcher at different regions of the core (see Figure S1 for details). It is to be noted that partial glands appearing at the edges of the biopsy cores were not annotated.
PANDA dataset
Publicly available development set of the PANDA challenge consisted of 10,616 prostate biopsy slides from two institutes (Radboud University Medical Center, the Netherlands, and Karolinska Institutet, Sweden) in Europe.18 The slides covered all range of GSs (see Table S1 for details). They were scanned using different scanners, and the highest available resolution inside the slides was 20× (≈0.5 μm/pixel).
We dropped 104 slides because they were empty or did not have pixel-level annotation masks. Then, we randomly segregated 10,512 slides into training (6,279), validation (2,093), and test (2,140) sets to train a three-resolution benign versus malignant classification model (see Table S1 for details). It is to be noted that there were multiple slides for a patient in the PANDA challenge development set.18 However, the mapping between slides and patients was not provided. Therefore, our segregation might suffer from data leakage.23 To avoid spurious results, we used European data only for training of our model and conducted an external validation on Singapore data.
Ethics statement
This study complies with the ethical principles of the Declaration of Helsinki. Institutional review board approval was obtained for this study (National Healthcare Group, Domain Specific Review Board 2009/00144, Singapore). Besides, we were granted a waiver of informed consent for this study.
All the data were de-identified. The slides were scanned by excluding the ID tags. Then, digital slides were labeled with arbitrary file names in the form of “patient_RRRRRR_slide_01,” where “R” stands for a random digit (e.g., patient_040551_slide_01). There is no record of mapping between original patient IDs and arbitrary file names.
Prostate gland segmentation and classification pipeline
This study developed a deep-learning-based pipeline detecting malignant glands in core needle biopsy slides of prostate tumors. The ultimate aim was twofold: to improve patients’ outcomes by helping pathologists diagnose prostate cancer in low-grade and low-volume cases and to reduce pathologists’ workload by providing them with an assistive tool during diagnosis. The pipeline consisted of two stages: gland segmentation and gland classification models.
Gland segmentation using a mask R-CNN model
The first stage used a Mask R-CNN19 model to segment glands. The Mask R-CNN had a ResNet5034 as its region proposal network. The box predictor and mask predictor had two classes (gland versus background). The model was trained end to end from scratch.
The dataset used in this stage consisted of cropped patches of size 512 × 512 pixels at 20× magnification from WSIs such that an annotated gland was centered at each patch (Figure 1B). The patch size and resolution were selected such that both nuclei morphology and gland structure information were available to be exploited by the Mask R-CNN model. For each patch, binary masks of all glands present in the patch, including incomplete glands at the edges, were created as labels.
Data augmentation techniques, namely random horizontal and vertical flip, color augmentation (contrast, brightness), and rotation, were applied to the patches and binary masks at training. After augmentation, the patches were cropped to 362 × 362 pixels around the center and passed as input to Mask R-CNN.
Gland classification using a four-resolution model
The second stage used a four-resolution deep-learning model that emulates pathologists’ workflow to perform gland classification. Patches of size 512 × 512 pixels were cropped from WSIs at resolutions 5×, 10×, 20×, and 40× with an annotated gland centered at each patch. To predict whether the center gland was benign or malignant, patches of these resolutions from the same tissue region (around a particular gland) were passed into the multi-resolution model simultaneously (Figure 1B).
Specifically, each patch of a different resolution was passed to a different ResNet-1834 feature extractor. Extracted features from patches of all resolutions were then summed and passed to a linear classifier to predict whether the center gland was benign or malignant. The same data augmentation techniques used in the first stage were applied during the training of the multi-resolution model. The model was trained end to end.
Training of deep-learning models
Models trained on SG datasets
The Mask R-CNN model was trained using the Adam optimizer with a batch size of 4 for 142 epochs. The learning rate was initially set to 3e−4. After the training loss plateaued at the end of epochs 60 and 110, it was reduced to 3e−5 and 3e−6, respectively. Similarly, the multi-resolution model was trained using the Adam optimizer with a learning rate of 5e−4 and a batch size of 32 for 76 epochs.
Mask-RCNN model trained on RINGS algorithm dataset
The Mask-RCNN model had the same architecture with the one used on SG dataset. The model was trained end to end from scratch on the training set of the RINGS algorithm dataset24 using the Adam optimizer with a learning rate of 3e−5 for 50 epochs. Batch size was 2.
Three-resolution model trained on PANDA dataset
The three-resolution model was trained on the training set of the PANDA dataset using the Adam optimizer with a learning rate of 5e−4 for 111 iterations and then 5e−5 for 216 iterations. Batch size was 16.
Inference using trained pipeline
The trained pipeline accepted a WSI of prostate core needle biopsy as input. It detected the glands within the slide and predicted whether each detected gland was malignant or benign (Figure 1C).
Firstly, overlapping patches of size 512 × 512 pixels at 20× magnification were cropped from the tissue regions inside the slide in a sliding window fashion with stride 256 pixels. These patches were passed into the trained Mask R-CNN model to segment glands present. Secondly, predicted masks from the Mask R-CNN model were grayscale and converted to binary using thresholding. Then, binary masks were post-processed to merge partial predictions and eliminate redundant predictions arising from overlapping patch cropping (see supplemental experimental procedures for details). Finally, for each detected gland (instance) in a slide, patches at multiple resolutions were cropped from the slide. The trained multi-resolution model classified each detected gland as benign or malignant.
Post-hoc analysis
A post-hoc analysis was conducted on images in the test set of the SG gland classification dataset to gain deeper insights into the four-resolution model trained on SG dataset (SG model) and the three-resolution model trained on PANDA dataset (PANDA model). Integrated gradients attribution method32 implemented in Captum package35 was used for the analysis. Blurred images were used as baselines, and global attribution scores obtained by factoring the model inputs’ multiplier were used while obtaining attribution maps.
Acknowledgments
This study was funded by the Biomedical Research Council of the Agency for Science, Technology, and Research, Singapore.
Author contributions
M.U.O., M.Y.N., C.E.C.X., and L.A.Y.X. selected the patients and collected the data. M.Y.N. and D.M.G. annotated the data. M.U.O. and M.Y.N. verified the underlying data, conducted the experiments, and analyzed the results with the help of M.S.. M.U.O. and M.Y.N. wrote the manuscript. M.S., W.Y., W.-K.S., C.F.W., and H.K.L. contributed to the manuscript preparation. C.F.W. and H.K.L. supervised the study. All authors reviewed the manuscript and agreed with its contents.
Declaration of interests
The authors declare no competing interests.
Published: November 29, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.patter.2022.100642.
Supplemental information
Data and code availability
-
•
All images have been deposited at Zenodo under https://doi.org/10.5281/zenodo.5971763 and are publicly available.1
-
•
All original code has been deposited at Zenodo under https://doi.org/10.5281/zenodo.5982397 and is publicly available.2 The repository provides a detailed step-by-step explanation, from training of gland segmentation and classification models to inference with the trained models.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Oner M.U., Ng M.Y., Giron D.M., Xi C.E.C., Xiang L.A.Y., Singh M., Yu W., Sung W.-K., Wong C.F., Lee H.K. Zenodo; 2022. Digital Pathology Dataset for Prostate Cancer Diagnosis. [DOI] [Google Scholar]
- 2.Oner M.U., Ng M.Y., Singh M. Zenodo; 2022. Multi-lens Neural Machine (Mlnm) [DOI] [Google Scholar]
- 3.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: Globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Metter D.M., Colgan T.J., Leung S.T., Timmons C.F., Park J.Y. Trends in the US and Canadian pathologist workforces from 2007 to 2017. JAMA Netw. Open. 2019;2:e194337. doi: 10.1001/jamanetworkopen.2019.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epstein J.I., Egevad L., Amin M.B., Delahunt B., Srigley J.R., Humphrey P.A., Grading Committee The 2014 international society of urological pathology (isup) consensus conference on gleason grading of prostatic carcinoma. Am. J. Surg. Pathol. 2016;40:244–252. doi: 10.1097/PAS.0000000000000530. [DOI] [PubMed] [Google Scholar]
- 6.Raciti P., Sue J., Ceballos R., Godrich R., Kunz J.D., Kapur S., Reuter V., Grady L., Kanan C., Klimstra D.S., Fuchs T.J. Novel artificial intelligence system increases the detection of prostate cancer in whole slide images of core needle biopsies. Mod. Pathol. 2020;33:2058–2066. doi: 10.1038/s41379-020-0551-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulten W., Balkenhol M., Belinga J.-J.A., Brilhante A., Çakır A., Egevad L., Eklund M., Farré X., Geronatsiou K., Molinié V., et al. Artificial intelligence assistance significantly improves gleason grading of prostate biopsies by pathologists. Mod. Pathol. 2021;34:660–671. doi: 10.1038/s41379-020-0640-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campanella G., Hanna M.G., Geneslaw L., Miraflor A., Werneck Krauss Silva V., Busam K.J., Brogi E., Reuter V.E., Klimstra D.S., Fuchs T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019;25:1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagpal K., Foote D., Liu Y., Chen P.-H.C., Wulczyn E., Tan F., Olson N., Smith J.L., Mohtashamian A., Wren J.H., et al. Development and validation of a deep learning algorithm for improving gleason scoring of prostate cancer. NPJ Digit. Med. 2019;2:113–210. doi: 10.1038/s41746-019-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucas M., Jansen I., Savci-Heijink C.D., Meijer S.L., de Boer O.J., van Leeuwen T.G., de Bruin D.M., Marquering H.A. Deep learning for automatic gleason pattern classification for grade group determination of prostate biopsies. Virchows Arch. 2019;475:77–83. doi: 10.1007/s00428-019-02577-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh M., Kalaw E.M., Jie W., Al-Shabi M., Wong C.F., Giron D.M., Chong K.-T., Tan M., Zeng Z., Lee H.K. Cribriform pattern detection in prostate histopathological images using deep learning models. arXiv. 2019 doi: 10.48550/arXiv.1910.04030. Preprint at. [DOI] [Google Scholar]
- 12.Nagpal K., Foote D., Tan F., Liu Y., Chen P.-H.C., Steiner D.F., Manoj N., Olson N., Smith J.L., Mohtashamian A., et al. Development and validation of a deep learning algorithm for gleason grading of prostate cancer from biopsy specimens. JAMA Oncol. 2020;6:1372–1380. doi: 10.1001/jamaoncol.2020.2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pantanowitz L., Quiroga-Garza G.M., Bien L., Heled R., Laifenfeld D., Linhart C., Sandbank J., Albrecht Shach A., Shalev V., Vecsler M., et al. An artificial intelligence algorithm for prostate cancer diagnosis in whole slide images of core needle biopsies: a blinded clinical validation and deployment study. Lancet. Digit. Health. 2020;2:e407–e416. doi: 10.1016/S2589-7500(20)30159-X. [DOI] [PubMed] [Google Scholar]
- 14.Ström P., Kartasalo K., Olsson H., Solorzano L., Delahunt B., Berney D.M., Bostwick D.G., Evans A.J., Grignon D.J., Humphrey P.A., et al. Artificial intelligence for diagnosis and grading of prostate cancer in biopsies: a population-based, diagnostic study. Lancet Oncol. 2020;21:222–232. doi: 10.1016/S1470-2045(19)30738-7. [DOI] [PubMed] [Google Scholar]
- 15.Bulten W., Pinckaers H., van Boven H., Vink R., de Bel T., van Ginneken B., van der Laak J., Hulsbergen-van de Kaa C., Litjens G. Automated deep- learning system for gleason grading of prostate cancer using biopsies: a diagnostic study. Lancet Oncol. 2020;21:233–241. doi: 10.1016/S1470-2045(19)30739-9. [DOI] [PubMed] [Google Scholar]
- 16.Pinckaers H., Bulten W., van der Laak J., Litjens G. Detection of prostate cancer in whole-slide images through end-to-end training with image-level labels. IEEE Trans. Med. Imaging. 2021;40:1817–1826. doi: 10.1109/TMI.2021.3066295. [DOI] [PubMed] [Google Scholar]
- 17.Mun Y., Paik I., Shin S.-J., Kwak T.-Y., Chang H. Yet another automated gleason grading system (yaaggs) by weakly supervised deep learning. NPJ Digit. Med. 2021;4:1–9. doi: 10.1038/s41746-021-00469-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bulten W., Kartasalo K., Chen P.-H.C., Ström P., Pinckaers H., Nagpal K., Cai Y., Steiner D.F., van Boven H., Vink R., et al. Artificial intelligence for diagnosis and gleason grading of prostate cancer: the panda challenge. Nat. Med. 2022;28:154–163. doi: 10.1038/s41591-021-01620-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He K., Gkioxari G., Dollár P., Girshick R. Proceedings of the IEEE international conference on computer vision. IEEE; 2017. Mask r-cnn; pp. 2961–2969. [Google Scholar]
- 20.D’Alfonso T.M., Ho D.J., Hanna M.G., Grabenstetter A., Yarlagadda D.V.K., Geneslaw L., Ntiamoah P., Fuchs T.J., Tan L.K. Multi-magnification- based machine learning as an ancillary tool for the pathologic assessment of shaved margins for breast carcinoma lumpectomy specimens. Mod. Pathol. 2021;34:1487–1494. doi: 10.1038/s41379-021-00807-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hatami N., Bilal M., Rajpoot N. Deep multi-resolution dictionary learning for histopathology image analysis. arXiv. 2021 doi: 10.48550/arXiv.2104.00669. Preprint at. [DOI] [Google Scholar]
- 22.Efron B. Breakthroughs in statistics. Springer; 1992. Bootstrap methods: another look at the jackknife; pp. 569–593. [Google Scholar]
- 23.Oner M.U., Cheng Y.-C., Lee H.K., Sung W.-K. Training machine learning models on patient level data segregation is crucial in practical clinical applications. medRxiv. 2020 doi: 10.1101/2020.04.23.20076406. Preprint at. [DOI] [Google Scholar]
- 24.Salvi M., Bosco M., Molinaro L., Gambella A., Papotti M., Acharya U.R., Molinari F. A hybrid deep learning approach for gland segmentation in prostate histopathological images. Artif. Intell. Med. 2021;115:102076. doi: 10.1016/j.artmed.2021.102076. [DOI] [PubMed] [Google Scholar]
- 25.Farjam R., Soltanian-Zadeh H., Jafari-Khouzani K., Zoroofi R.A. An image analysis approach for automatic malignancy determination of prostate pathological images. Cytometry B Clin. Cytom. 2007;72:227–240. doi: 10.1002/cyto.b.20162. [DOI] [PubMed] [Google Scholar]
- 26.Naik S., Doyle S., Agner S., Madabhushi A., Feldman M., Tomaszewski J. 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro. IEEE; 2008. Automated gland and nuclei segmentation for grading of prostate and breast cancer histopathology; pp. 284–287. [Google Scholar]
- 27.Peng Y., Jiang Y., Eisengart L., Healy M.A., Straus F.H., Yang X.J. Computer-aided identification of prostatic adenocarcinoma: segmentation of glandular structures. J. Pathol. Inform. 2011;2:33. doi: 10.4103/2153-3539.83193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nguyen K., Sabata B., Jain A.K. Prostate cancer grading: gland segmentation and structural features. Pattern Recogn. Lett. 2012;33:951–961. [Google Scholar]
- 29.Singh M., Kalaw E.M., Giron D.M., Chong K.-T., Tan C.L., Lee H.K. Gland segmentation in prostate histopathological images. J. Med. Imaging. 2017;4:027501. doi: 10.1117/1.JMI.4.2.027501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren J., Sadimin E., Foran D.J., Qi X. Vol. 10133. International Society for Optics and Photonics; 2017. Computer aided analysis of prostate histopathology images to support a refined gleason grading system; p. 101331V. (Medical Imaging 2017: Image Processing). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu Y., Li Y., Wang Y., Liu M., Fan Y., Lai M., Chang E.I.C., Chang C. Gland instance segmentation using deep multichannel neural networks. IEEE Trans. Biomed. Eng. 2017;64:2901–2912. doi: 10.1109/TBME.2017.2686418. [DOI] [PubMed] [Google Scholar]
- 32.Sundararajan M., Taly A., Yan Q. International conference on machine learning. PMLR; 2017. Axiomatic attribution for deep networks; pp. 3319–3328. [Google Scholar]
- 33.Cohen J. A coefficient of agreement for nominal scales. Educ. Psychol. Meas. 1960;20:37–46. [Google Scholar]
- 34.He K., Zhang X., Ren S., Sun J. Proceedings of the IEEE conference on computer vision and pattern recognition. IEEE; 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 35.Kokhlikyan N., Miglani V., Martin M., Wang E., Alsallakh B., Reynolds J., Melnikov A., Kliushkina N., Araya C., Yan S., et al. Captum: a unified and generic model interpretability library for pytorch. arXiv. 2020 doi: 10.48550/arXiv.2009.07896. Preprint at. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All images have been deposited at Zenodo under https://doi.org/10.5281/zenodo.5971763 and are publicly available.1
-
•
All original code has been deposited at Zenodo under https://doi.org/10.5281/zenodo.5982397 and is publicly available.2 The repository provides a detailed step-by-step explanation, from training of gland segmentation and classification models to inference with the trained models.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.