Abstract
This paper presents a consensus-based approach that incorporates three microarray and three RNA-Seq methods for unbiased and integrative identification of differentially expressed genes (DEGs) as potential biomarkers for critical disease(s). The proposed method performs satisfactorily on two microarray datasets (GSE20347 and GSE23400) and one RNA-Seq dataset (GSE130078) for esophageal squamous cell carcinoma (ESCC). Based on the input dataset, our framework employs specific DE methods to detect DEGs independently. A consensus based function that first considers DEGs common to all three methods for further downstream analysis has been introduced. The consensus function employs other parameters to overcome information loss. Differential co-expression (DCE) and preservation analysis of DEGs facilitates the study of behavioral changes in interactions among DEGs under normal and diseased circumstances. Considering hub genes in biologically relevant modules and most GO and pathway enriched DEGs as candidates for potential biomarkers of ESCC, we perform further validation through biological analysis as well as literature evidence. We have identified 25 DEGs that have strong biological relevance to their respective datasets and have previous literature establishing them as potential biomarkers for ESCC. We have further identified 8 additional DEGs as probable potential biomarkers for ESCC, but recommend further in-depth analysis.
Keywords: Differential expression analysis, Biomarker identification, Esophageal squamous cell carcinoma, Differentially expressed gene, RNA-sequencing, Microarray
Introduction
For a critical disease of interest, the knowledge of differentially expressed genes (DEG) are a crucial step toward biomarker identification. This is achieved through Differential Expression Analysis (DEA) which monitors the behavior of each gene in isolation over normal and disease conditions and streamlines the search for biomarkers by providing a candidate list of these discriminative candidate genes. DNA microarray and RNA Sequencing (RNA-Seq) are indispensable methods for DEA. Previously, microarray technology was the most widely used approach. However, there are inherent limitations such as the pre-requisite prior knowledge of the sequence for the array design or the fact that cross-hybridization makes it difficult to analyze highly correlated sequences. Furthermore, the lack of sensitivity to highly or lowly expressed genes as well as the lack of reproducibility across laboratories and platforms pose major challenges. These limitations are overcome by RNA-Seq technology.
Numerous DEA methods have been developed to serve both microarray and RNA-Seq technologies. Furthermore, a large number of datasets related to critical diseases that are compatible with both technologies are widely available. Keeping in mind the fact that most methods developed for these technologies are not effective for all cases, we propose a consensus-based integrative approach that ensembles a selected few of these methods with the aim to achieve improved performance. In other words, in this paper, we present a consensus-based approach that employs a few chosen microarray DEA methods (Limma [1], SAM [2] and EBAM [3]) and RNA-Seq (Limma+Voom [4], edgeR [5], DESeq2 [6]) to present an unbiased list of DEGs as candidates for potential biomarkers. As the primary focus of our work is on the critical disease ESCC, we validate our results on two ESCC microarray datasets (GSE20347 and GSE23400) datasets and one ESCC RNA-Seq (GSE130078) dataset.
The rest of the paper is organized as follows. Section “Related Work” provides a brief overview of related work. Section “Proposed DE Framework” describes our proposed framework for biomarker identification of critical disease(s) employing three microarray and three RNA-Seq methods. We also introduce our consensus function for unbiased integration of DEGs individually detected by previously mentioned methods. Section “Analysis” reports a detailed experimental analysis and validation of our method on two benchmark microarray gene expression datasets and one RNA Sequencing (RNA-Seq) dataset. Section “Discussion” presents a detailed analysis and discussion on candidate genes that have been identified as potential biomarkers for ESCC. In this section, we also present a comparison of our algorithm with two recent works with similar approaches. Finally, the concluding remarks are given in Section “Conclusion”.
Related Work
A number of statistical approaches are used by Limma [1] to achieve effectiveness in large-scale expression studies. Limma takes advantage of the flexibility of linear models and fits one to each row (gene) of data in a gene expression matrix where columns correspond to samples. Limma has the inherent ability to model correlations between samples through analysis of the entire dataset as one entity. Significance Analysis of Microarrays (SAM) [2] assimilates a set of gene-specific t-tests to identify genes that exhibit statistically significant changes in expression. For each gene, based on the changes in gene expression in terms of standard deviation for repeated measurements, SAM assigns a score. Potentially significant genes are identified using a threshold for these scores. Empirical Bayes Analysis of Microarrays (EBAM) [3] employs the removal Bayes rule to obtain the posterior probability that a gene was affected or unaffected under the various conditions. EBAM makes multiple testing a possibility by establishing a connection between prior probabilities and local false discovery rate (local fdr) in turn handling the issues that arise from simultaneous tests.
Voom [4] works on the idea that commonly used microarray-based statistical methods can be applied to read counts of corresponding RNA-Seq through robust and non-parametric estimation of mean-variance. In other words, Voom incorporates the mean-variance trend into the empirical Bayes procedure of Limma. edgeR [5] and DESeq2 [6] are estimations of gene-wise dispersion by conditional maximum likelihood, conditioning on the total count for the gene. edgeR effectively borrows information within genes to shrink the dispersion towards a consensus value through the use of empirical Bayes. It adapts for overdispersed data and incorporates an exact test to assess each gene. DESeq2 accurately estimates the expected dispersion value for genes of a given expression strength, which is then used to conform the gene-wise dispersion towards the predicted values . It also accounts for gene-specific variations and makes it possible to estimate fitted curves and testing even in settings with less information.
Proposed DE Framework
The proposed framework aims to work with microarray and RNA Sequencing data. We have chosen three methods that work on micro-array (Limma, SAM and EBAM) and three on RNA-Seq (Limma+voom, DESeq and EdgeR). First, both types of data require pre-processing. For microarray, pre-processing consists of the removal of unwanted and redundant information, normalization of the dataset, missing value estimation while for RNA-Seq data, we perform removal of low read counts, normalization, and transformation.
Pre-processing is followed by DE analysis that results in differentially expressed genes (DEG). For each data type, we employ a consensus function that filters out all common DEGs for each dataset. In other words, depending on the type of the input dataset, the DE analysis unit detects DEGs using three corresponding methods, followed by a consensus function that filters the DEGs common to all three methods as well as identify other relevant DEGs. Our consensus function is given by
| 1 |
where
and
Here, and are q-value and local.fdr significance values that are chosen according to their relevance to the experiment. Through multiple iterations of implementation, we observed that consideration of only genes common to all three methods leads to information loss. Thus, to overcome this we introduced q-value into the consensus function. The main idea behind this is that while a p-value of 0.05 gives the implication that 5% of the tests will be false positive (FP), q-value, which is an FDR adjusted p-value, implies that 5% of the test found to be significant will be FP. q-value requires a very important adjustment for multiple tests on the same data sample. Our consensus function considers all genes common to all three methods with as detected DEGs. Furthermore, all genes that are not among the common genes but have a , i.e. (Eq. 1), are added to the list of DEGs. However, in the microarray datasets, we implement the proposed consensus function given by Eq. 1 to start off by taking the DEGs common to all three methods. Unlike RNA-Seq, instead of q-value, we incorporate its useful counterpart local fdr ( in Eq. 1). Local fdr is a measure of the posterior probability that the null hypothesis is true. We use local fdr since Limma and SAM calculate p-value. EBAM, on the other hand, estimates the posterior probability and local fdr. It is worth mentioning that posterior probability and p-value are not interchangeable. However, local fdr can be estimated from p-values. Our consensus function (Eq. 1) considers all genes common to all three methods with as detected DEGs. Furthermore, all genes that are not among the common genes but have a () are added to the list of DEGs.
The relevant DEGs are then taken as input to the DCE analysis unit. The idea behind performing DCE analysis is that it leads to the creation of biologically relevant modules which are easier for further analysis and validation. The DCE unit identifies differentially co-expressed modules and performs preservation analysis on these modules to identify biologically relevant modules. This is followed by the identification of hub genes in these modules using WGCNA [7] intramodular connectivity.
We validate our results using several approaches. First, we consider all relevant DEGs detected by the DE analysis unit in isolation and perform Gene Ontology (GO) and KEGG pathway enrichment analysis to validate biological relevance. We consider all the hub genes in the biologically relevant modules identified by the DCE unit as biomarker candidates. Furthermore, all DEGs that are annotated with the most enriched GO term in all three databases as well as the most enriched KEGG pathway are also considered as candidates for biomarkers. We term such genes as Top Enriched DEGs (TEDs). Secondly, we further analyze the biomarker candidates through observation of their interactions and behavioral changes. Finally, we trace literature evidence for the relevant genes in other scientific sources and works for further validation of these identified genes as possible biomarkers.
Analysis
Our focus is on ESCC, a cancer very common in developing countries, especially in North-East India, and is highly attributed to tobacco and betelnut chewing, alcohol consumption as well as poor diet. Two microarray datasets GSE20347 and GSE23400 and an RNA-Sequencing dataset GSE130078 were chosen to validate our proposed DE framework (see Fig. 1). Details of each dataset is provided in Table 1. All three datasets examined gene expression in tumor and matched normal adjacent tissue. The test platform is a DELL workstation with Intel(R) Xeon(R) W-2145 with 3.70GHz processor, 64 GB RAM running Windows 10 Pro for workstations.
Fig. 1.
Proposed framework for differential expression analysis
Table 1.
Datasets
Preprocessing
RNA-Seq dataset GSE130078 has 57,783 genes and 46 samples. Large datasets tend to add complications to the analysis and as such, we filter out genes with low read counts. We achieve this by calculating the counts per million (CPM) for each sample for each gene and keep only those genes that have CPM for at least two samples. This reduces the dataset size from 57,783 to 22,270. We then follow up by normalization of the dataset. We also consider two microarray datasets GSE20347 and GSE23400 for analysis. The inputs to these datasets are expression values of genes across samples. First, we pre-process the data through the removal of unwanted and redundant genes, missing value estimation, and normalization. However, for both GSE20347 and GSE23400, there are no missing values and as such we proceed further down the pipeline.
DE Analysis
For the microarray datasets, Limma takes the pre-processed dataset as input and outputs the equivalent DEGs with a significance of 5% () and False Discovery Rate (FDR) of 0.05. On the other hand, for the other two methods SAM and EBAM, we employ findDelta with FDR fiving us an estimate of the delta values at which FDR is closest to 0.05 and chose accordingly. In SAM, delta is the distance between the observed and the expected test scores, whereas in EBAM, delta is the probability that a gene with a specific test score is differentially expressed. Table 2 summarizes the DEGs detected by all three methods on all three datasets.
Table 2.
Summary of detected DEGs by the three RNA-Seq methods and the three microarray methods
In the case of the RNA-Seq dataset, the pre-processed data are the input to all three methods, i.e., Limma+Voom, edgeR and DESeq2. However, it is to be noted that while DESeq2 directly takes the count data as input, the other two methods require the count data to be transformed into a DGEList (Digital Gene Expression Data) object. All the methods perform multiple tests on all the 22,270 genes in the dataset across 46 samples. We consider a significance of 5%, i.e., and the corresponding DEGs detected by the 3 these methods are summarized in Table 2.
We implement the proposed consensus Eq. 1 to identify the common genes detected by these three methods. First, we consider the DEGs detected by all three methods, i.e. common genes. In GS20347, there are such 7706 DEGs. So as not to bypass crucial information, we use in Eq. 1, i.e., the consensus function. With () another 662 genes are considered DEGs resulting in a list of 8,368 DEGs. Similarly, in GSE23400, Limma, SAM, and EBAM find 3,431 common DEGs. With (), another 4,066 genes are considered as DEGs, resulting in a list of 7,497 DEGs. In the case of GSE130078, the three methods Limma+Voom, edgeR, and DESeq2 discover 2,765 common DEGs and a q-value () adds another 9,945 genes resulting in a list of 12,710 DEGs.
DCE Analysis
To analyze the interactions among the DEGs as well as the variations in behavior under normal and disease circumstances, we construct co-expression networks (CEN) using WGCNA [7]. The pipeline for DCE analysis is to detect DEGs by our method is as follows.
Divide the dataset into separate datasets: All genes with only normal samples and all genes with only disease samples
Choose the soft thresholding power to which co-expression similarity is raised to calculate an adjacency matrix. Soft thresholding power is based on the criterion of approximate scale-free topology.
Construct two separate CENs in the form of an adjacency matrix: normal and disease.
Transform adjacency matrices into topological overlap matrices (TOM [8]) to minimize the effects of noise and spurious associations
Extract all connections that correspond to the subset of DEGs from both CENs.
Extract normal and disease modules using hierarchical clustering.
Merge modules through eigenvector module selection and MEDissThres threshold merging.
Identify modules extracted in the normal dataset that are non-preserved in disease dataset and vice versa through preservation analysis [9]. We consider such modules as modules of interest for further downstream analysis.
Identify the top 20 hub genes using intramodular connectivity [7] in all modules of interest.
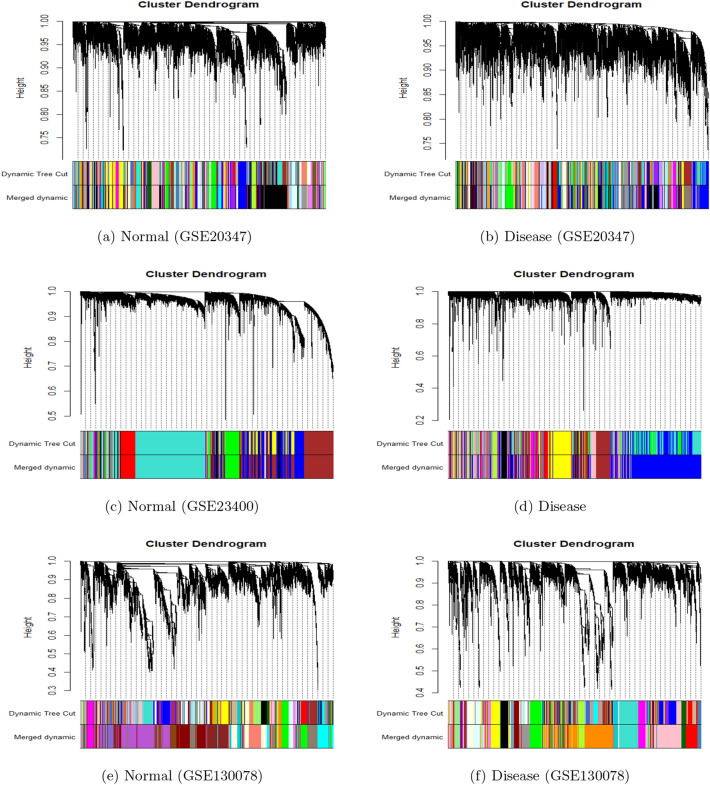
We start DCE analysis by clustering the samples using the hierarchical approach to detect outlier samples. We remove the outlier samples with the aim of creating a more robust CEN. For GSE23047, as seen in Fig. 2a, b, we find a single outlier in the case of normal samples with a tree cut at height h = 70 (Blue). However, in disease samples, there are two outliers with a cut at h = 130 (Red). Similarly, in the case of GSE23400, as seen in Fig. 2c, d, a tree cut at height h = 105 (Blue) and at h = 95 (Red) removes one and two outliers from normal and disease samples, respectively. In the case of GSE130078, a cut at h = 1,500,000 (Blue) and h = 2,000,000 (Red) removes one normal (Fig. 2e) and one disease Fig. 2f sample.
Fig. 2.
Outlier hierarchical trees for normal and disease samples for all three datasets
In GSE20347, hierarchical Clustering and tree cut results in 50 and 75 normal and disease modules, respectively. Figure 3a shows the dendrogram while the first strip of colors below represents the corresponding module colors for the normal dataset. Similarly, Fig. 3b shows the dendrogram for the disease dataset. To merge modules, we choose a height cut of 0.25, corresponding to a correlation of 0.75. Merging of the modules with tree cut at h = 0.25 further reduces the number of modules to 38 and 61 for normal and disease datasets, respectively. The second color strip in Fig. 3a, b shows the colors for the merged normal and disease modules respectively. In GSE23400, hierarchical clustering results in 9 normal (the first color strip in Fig. 3c) and 13 (the first color strip in Fig. 3d) disease modules, which are then reduced to 8 normal (the second color strip in Fig. 3c) and 11 disease (second color strip in Fig. 3d) modules after merging. Finally in GSE130078, hierarchical clustering results in 65 normal (the first color strip in Fig. 3e) and 40 disease (the first color strip in Fig. 3f) modules, which are then reduced to 21 normal (the second color strip in Fig. 3e) and 24 disease (the second color strip in Fig. 3f) modules after merging.
Fig. 3.
Dendrograms where the first strip of colors represents the colors initially assigned to corresponding modules while the second strip of colors represents the colors assigned to corresponding merged modules for the ESCC dataset: GSE20347 (a, b); GSE23400 (c, d); GSE130078 (e, f)
We follow module extraction by module preservation analysis with the aim of analyzing the distinction between preserved and non-preserved modules. According to Langfelder et al. [9], while the preserved modules retain a majority of their co-expressed connections (or edges between two genes), the same cannot be perceived from non-preserved modules. According to Langfelder et al. [9], a module with is considered non-preserved [9]. It is noteworthy that, GSE23400 due to its inherent nature, extracts a smaller number of modules with significantly larger sizes and higher densities (Fig. 4). There are no non-preserved modules with and most modules are either moderately preserved or highly preserved. We take into consideration moderately preserved modules with [9].
Fig. 4.
on ESCC dataset GSE20347 (a, b); on ESCC dataset GSE23400 (c, d); on ESCC dataset GSE130078 (e, f). All modules below the red line () are non-preserved, all modules between the red and blue lines () are weak to moderately preserved and all modules above the blue line () have strong evidence of being preserved
Table 3 summarizes the preservation analysis for non-preserved modules in all three datasets. The second column highlights the module preservation reference and test networks. For example, the table reading for module pink in Normal/Disease subset of dataset GSE20347 can be interpreted module pink of size 278 detected in the normal network that is non-preserved in disease network with a value of – 0.118842161.
Table 3.
Preservation analysis of modules in all three datasets
| Ref/Test | Module | Size | ||
|---|---|---|---|---|
| GSE20347 | Normal/Disease | pink | 276 | – 0.118842161 |
| bisque4 | 62 | 0.84895892 | ||
| orangered4 | 82 | 1.10844007 | ||
| grey | 17 | 1.38809779 | ||
| Disease/Normal | grey | 3 | – 0.11691952 | |
| greenyellow | 149 | 0.21296495 | ||
| brown2 | 39 | 0.40355693 | ||
| darkgreen | 201 | 0.57638054 | ||
| lightpink4 | 61 | 0.58347786 | ||
| white | 99 | 0.72904092 | ||
| lightyellow | 122 | 0.88046344 | ||
| darkolivegreen4 | 40 | 1.13783059 | ||
| antiquewhite4 | 58 | 1.19766058 | ||
| lightsteelblue1 | 143 | 1.42400619 | ||
| mediumpurple3 | 77 | 1.74769518 | ||
| black | 775 | 1.79866799 | ||
| skyblue1 | 51 | 1.79952166 | ||
| lavenderblush3 | 60 | 1.83004964 | ||
| lightgreen | 123 | 1.88961457 | ||
| GSE23400 | Normal/Disease | magenta | 45 | 5.63609823 |
| Disease/Normal | magenta | 231 | 5.59355067 | |
| salmon | 44 | 5.64755802 | ||
| greenyellow | 172 | 6.47180995 | ||
| purple | 225 | 9.42312232 | ||
| GSE130078 | Normal/Disease | magenta | 248 | – 1.80627625 |
| skyblue2 | 37 | – 0.62266325 | ||
| bisque4 | 1000 | 0.01985689 | ||
| maroon | 82 | 0.58940588 | ||
| grey | 70 | 1.90096828 | ||
| Disease/Normal | lightyellow | 240 | – 1.18378716 | |
| red | 759 | – 0.18883575 | ||
| lightcyan | 321 | 0.68868388 | ||
| steelblue | 145 | 0.92648735 | ||
| skyblue3 | 104 | 1.01960350 | ||
| violet | 142 | 1.38497020 |
We only consider non-preserved modules of substantial size () as modules of interest for further downstream analysis and validation. To find the hub genes for each module of interest extracted previously we employ WGCNA intramodular-connectivity proposed by Langfelder et al. [7]. Intramodular-connectivity calculates the connectivity of a node to other nodes in the same module.
Validation
Enrichment Analysis of Modules
For a module of interest to be regarded as Gene Ontology (GO) or pathway enriched, at least one gene in the module must be assigned to an enriched GO term or pathway, respectively with a significance of 5% (i.e., ). To perform functional enrichment analysis, we use the easily available online tool DAVID [10, 11]. Table 5 summarizes the percentages of genes in the modules of interest annotated to enriched GO terms as well as enriched KEGG pathways. We observe that all modules of interest are GO and pathway enriched.
Table 5.
Percentages of genes in each module that are annotated to the Gene Ontology (GO) databases (BP: Biological Processes, CC: Cellular components or MF: Molecular function) and KEGG pathways
| Dataset | Module | Size | GO_BP (%) | GO_MF (%) | GO_CC (%) | KEGG (%) |
|---|---|---|---|---|---|---|
| GSE130078 | lightyellow | 240 | 78.9 | 80.5 | 78.9 | 38.3 |
| red | 759 | 83.3 | 87.6 | 84.8 | 38.9 | |
| skyblue3 | 104 | 82.6 | 87.2 | 85.3 | 46.8 | |
| steelblue | 145 | 73.1 | 76.6 | 71.9 | 26.9 | |
| violet | 142 | 82.0 | 84.7 | 80.0 | 37.3 | |
| lightcyan | 321 | 69.1 | 70.7 | 69.4 | 27.7 | |
| bisque4 | 1000 | 87.5 | 91.4 | 89.1 | 40.5 | |
| magenta | 249 | 84.4 | 89.9 | 84.9 | 31.0 | |
| GSE20347 | pink | 276 | 90.1 | 96.0 | 95.2 | 47.2 |
| greenyellow | 149 | 98.4 | 98.4 | 97.7 | 49.2 | |
| darkgreen | 201 | 95.6 | 96.7 | 95.6 | 55.0 | |
| lightyellow | 122 | 95.0 | 96.0 | 97.0 | 53.5 | |
| lightsteelblue1 | 143 | 95.2 | 94.4 | 94.4 | 49.2 | |
| black | 775 | 94.9 | 97.0 | 96.1 | 52.9 | |
| lightgreen | 123 | 90.7 | 98.1 | 92.6 | 56.5 | |
| GSE23400 | greenyellow | 172 | 97.7 | 98.3 | 96.5 | 63.4 |
| magenta | 231 | 94.8 | 96.5 | 93.4 | 44.5 | |
| purple | 225 | 96.6 | 98.1 | 96.3 | 58.2 |
Candidate Genes
As mentioned earlier, we select DEGs as candidates for potential biomarkers based on the following two criteria:
All hub genes detected by the DCE analysis unit of our framework in all modules of interest are candidate genes.
DEGs that have been annotated to the most enriched GO terms in all three GO databases (BP, CC and MF) and are also annotated to the most enriched pathway after GO and Pathway enrichment analysis on the entire dataset are also considered as potential biomarkers. We rename these DEGs as TEDs (Top Enriched DEGs)
Thus, alongside all DEGs that are among Top 20 hub genes in modules of interest (as summarized in Table 4), our second criterion adds 22, 18 and 11 TEDs to the list of candidate genes in GSE20347, GSE23400 and GSE130078, respectively. We summarize these DEGs (TEDs) in Table 6. The numbers of candidate genes for GSE20347,GSE23400 and GSE130078 increase from 140, 60 and 160 to 162, 78 and 171, respectively. We perform the enrichment analysis on the entire dataset or in more specific terms the list of all genes in the dataset. This leads to the observation that as the lists of genes in GSE20347 (22,278 genes) and GSE23400 (22,283 genes) are almost the same, the list of top enriched genes (35 genes) extracted are the same. However, the differences in TEDs are seen (Table 6) due to the fact that there are DEGs identified in one dataset that might not be detected in the other.
Table 4.
Top 20 hub genes for each extracted module of interest in all three datasets using WGCNA [7] intramodular connectivity
Hub genes with strong literature evidence of association to esophageal squamous cell carcinoma (ESCC) are marked in Red while hub genes with evidence of association with five other SCCs namely, Oral, Tongue, Head and Neck, Tongue or Laryngeal squamous cell carcinoma are marked in Blue
Table 6.
DEGs that are annotated to most enriched GO term in all three GO databases (BP, CC and MF) as well as the most enriched pathway

DEGs with strong literature evidence of association to Esophageal squamous cell carcinoma (ESCC) are marked in Red while hub genes with evidence of association to five other SCC namely, Oral, Tongue, Head and Neck, Tongue or Laryngeal Squamous Cell Carcinoma are marked in Blue
Biological Analysis
To establish the biological relevance of the candidate genes detected by our method, we use functional enrichment analysis and the construction of a gene regulatory network (GRN). Transcription Factors (TF) have remarkable diversity as well potency as drivers of cell transformation. Bhagwat et al. [12] justify the continued pursuit of TFs as potential biomarkers across many forms cancer by the prevalent deregulation of the same. We observe that 26 (hub genes:21, TEDs:5), 11 (hub genes:6, TEDs:5) and 23 (hub genes:23, TEDs:0) candidate genes detected by our method in GSE20347, GSE23400 and GSE130078, respectively are TFs. These TFs exhibit regulatory behavior in their respective modules, establishing their biological relevance. For easy visualization, we extract a manageable subset of hub genes from the non-preserved modules detected by our method (Figs. 5a–f and 6a–f). We construct a Gene Regulatory Network (GRN) with these hub genes and associated Transcription Factors (TFs) so as to observe the regulatory behavior of the corresponding genes. The resulting GRN is in the form of an adjacency list with weighted directed edges from TFs to other target genes (TGs).
Fig. 5.
Gene Regulatory Network (GRN) on the subset of hub genes detected by our method for modules pink, greenyellow, darkgreen, lightsteelblue1, black and magenta. Regulatory behavior is represented by a weighted directed edge from a Transcription Factor (TF) to a Target Gene (TG)
Fig. 6.
Gene Regulatory Network (GRN) on the subset of hub genes detected by our method for modules purple, greenyellow, blue, lightyellow, violet and steelblue. Regulatory behavior is represented by a weighted directed edge from a Transcription Factor (TF) to a Target Gene (TG)
As in the the case of validation of modules, we employ DAVID [10, 11] to perform functional enrichment analysis of all candidate genes detected by our method. A candidate gene can be regarded as GO enriched considering a GO database (GO_BP, GO_CC, GO_MF) if it is annotated to at least one GO term in that database with significance of 5% (). Tables 7, 8 and 9 summarize the candidate genes annotated to the top 3 GO terms in each GO database in GSE20347, GSE23400 and GSE130078, respectively. Similarly, a candidate gene is KEGG pathway enriched if it is annotated to at least one KEGG pathway term with significance of 5%. Table 10 summarizes the candidate genes annotated to top 3 enriched KEGG pathways in GSE20347, GSE23400 and GSE130078.
Table 7.
Summary of candidate genes detected by our method in the microarray dataset, GS20347 that are annotated to top 3 GO terms in the three GO databases
| GO Term | Annotated Candidate Genes | |
|---|---|---|
| GO_BP | GO:0007165 signal transduction | PRKACB, BCR, AR, LYN, APPL1, STAT1, SHC1, NFKB2, PIK3CD, PIK3CB, PIK3R2, PIK3R1, EXT2, RAC2, PPFIA1, RAF1, GNB5, GSK3B, MAPK10, KRAS, RARA, MAP2K1, TXNRD1, FLT3LG, FADD, RANBP1, FAS, MAPK1 |
| GO:0045944 positive regulation of transcription from RNA polymerase II promoter | AR, HOXC11, STAT1, PITX1, NFKB2, TP63, PIK3R2, PIK3R1, PTMA, RAF1, MED1, ZNF148, RARA, FADD, CXCR3 | |
| GO:0016032 viral process | ITSN2, POM121, POM121C, LYN, BRD2, PSMB5, STAT1, ABI2, SHC1, PIK3R1, FADD, RANBP1, MAPK1 | |
| GO_CC | GO:0005829 cytosol | ITSN2, PRKACB, BCR, CASP10, MSRA, RAC2, PTMA, RAF1, UBA7, HOMER3, RARA, MAP2K1, MAP3K20, MCL1, MCM7, UBASH3A, AR, LYN, APPL1, STAT1, KPNA2, PIK3CD, PIK3CB, PIK3R2, PIK3R1, TRIO, RPL22, NEDD4L, PNO1, ABI2, NEB, SARS1, TXNRD1, FADD, CLIC4, PRMT1, FAS, SENP5, EPB41L1, HOXC11, SHC1, ODF2, CEP290, EEF1B2, NABP1, GNB5, KRAS, HAUS7, PSAT1, FLT3LG, MAPK1, PSMD4, RUFY3, PSMB5, TCOF1, PSMC2, NFKB2, JPT2, EXOSC4, CCT4, PPFIA1, SBF1, KIF4A, GSK3B, MAPK10, HPRT1, ITPKC, ENOX2, SPAG5, MAPRE1, RANBP1 |
| GO:0005654 nucleoplasm | PRKACB, CIAPIN1, TP63, MSRA, PTMA, UBA7, SLC3A2, RRP7A, RARA, ORC3, MCL1, MCM7, TIMELESS, NONO, UBASH3A, AR, STAT1, KPNA2, PIK3CB, DBF4, MED1, NEDD4L, PNO1, ABI2, DNMT3B, TXNRD1, PRMT1, SENP5, POP7, MAGOHB, HOXC11, NABP1, TERF1, NSD2, RFC2, NPIPB3, RAB8B, MAPK1, PSMD4, UQCRC2, PSMB5, TCOF1, PSMC2, NFKB2, IMP4, EXOSC4, CCT4, KIF4A, GSK3B, MAPK10, TRRAP, POM121, ZNF148, BRD2, ESF1, NMD3 | |
| GO:0016020 membrane | TFRC, BCR, ITGB7, CEP290, NEU1, EXT2, RAC2, GNAQ, SLC3A2, MAN1C1, KRAS, MCL1, MCM7, FLT3LG, NONO, LSS, RUFY3, APPL1, PSMC2, KPNA2, PIK3CD, PIK3CB, ENTPD7, PIK3R1, CD52, CD48, SBF1, KIF4A, DDX24, MED1, LST1, NMD3, CLIC4, FAS | |
| GO_MF | GO:0005515 protein binding | TFRC, PRKACB, BCR, ZKSCAN5, NEU1, MSRA, RAC2, RAF1, RALY, CD2, HOMER3, RRP7A, APOOL, RARA, MAP2K1, MCL1, MCM7, TIMELESS, UBASH3A, AR, APPL1, STAT1, PITX1, PIK3CD, PIK3CB, HSD17B10, EPHA2, PIK3R2, PIK3R1, RCN1, MED1, RPL22, NEDD4L, PNO1, ABI2, DNMT3B, TXNRD1, GMCL2, FADD, ITM2A, CLIC4, PRMT1, FAS, TGFB2, POP7, EPB41L1, MAGOHB, HOXC11, CNPY2, SHC1, CEP290, MAN1C1, KRAS, TERF1, RFC2, PSAT1, FLT3LG, RAB8B, VGLL4, MAPK1, UQCRC2, CKS2, TCOF1, NFKB2, IMP4, EXOSC4, KIF4A, GSK3B, MAPK10, TNS1, RANBP1 |
| GO:0042802 identical protein binding | TFRC, PSMD4, APPL1, STAT1, CEP290, TP63, CD3D, RAF1, RALY, CD2, KRAS, HOMER3, RPL22, HPRT1, TERF1, ABI2, PSAT1, FADD, PRMT1, MAPRE1, FAS, TIMELESS, NONO, MAPK1 | |
| GO:0003723 RNA binding | POP7, TFRC, PSMD4, MAGOHB, TCOF1, KPNA2, HSD17B10, CCT4, NABP1, DDX24, SLC3A2, RALY, RPL22, RRP7A, PNO1, EBNA1BP2, MAP3K20, SARS1, ENOX2, ESF1, TNS1, NMD3, PRMT1, MAPRE1, PUS7, NONO |
Table 8.
Summary of candidate genes detected by our method in the microarray dataset GSE23400 that have been annotated to top 3 GO terms in the three GO databases
| Data base | GO Term | Annotated Candidate Genes |
|---|---|---|
| GO_BP | GO:0007165 signal transduction | BCR, AR, APPL1, STAT1, STAT2, CXCL10, CXCL11, PIK3CD, PIK3R1, RAC2, CD53, IL15, TYROBP, RAF1, GSK3B, PRKCB, HIF1A, RARA, MAP2K1, VRK1, TXNRD1, FADD |
| GO:0045944 positive regulation of transcription from RNA polymerase II promoter | AR, STAT1, STAT2, CXCL10, PIK3R1, RAF1, HIF1A, RARA, FADD, TFEC | |
| GO:0045893 positive regulation of transcription | PRKCB, AR, HIF1A, RARA, STAT1, MAP2K1, CD86 | |
| GO_CC | GO:0005829 cytosol | BCR, MTHFD1, SCO2. SRM, GZMB, RAC2, RAF1, PRKCB, IFIH1, RARA, MAP2K1, IFIT1, IFIT3, OAS3, AR, APPL1, STAT1, STAT2, HNMT, PIK3CD, UBE2L6, PIK3R1, LAPTM5, CD163, TXNRD1, MNDA, FADD, DLGAP5, CDKN3, IL15, VRK1, EIF2S1, PSMA3, PSMB8, PSMB9, PSMC1, IFI35, ISG15, GSK3B, HIF1A, TUBG1 |
| GO:0005654 nucleoplasm | MNAT1, PRKCB, RARA, OAS3, AR, STAT1, STAT2, HNMT, UBE2L6, TXNRD1, MNDA, IL15, NASP, VRK1, PSMA3, PSMB8, PSMB9, PSMC1, ISG15, GSK3B, HIF1A, TFEC | |
| GO:0016020 membrane | BCR, MTHFD1, ITGB2, GZMB, RAC2, PLXNC1, HLA-C, HLA-F, HLA-G, EIF2S1, OAS3, APPL1, PSMC1, PIK3CD, IFI35, ENTPD1, PIK3R1, TAP1, CD163 | |
| GO_MF | GO:0005515 protein binding | BCR, SCO2, MNAT1, SRM, RAC2, FCER1G, RAF1, PRKCB, RARA, MAP2K1, AR, APPL1, STAT1, STAT2, PIK3CD, UBE2L6, ENTPD1, PIK3R1, GLRX5, CD163, TXNRD1, FADD, IL15, TIMM9, PLXNC1, VRK1, EIF2S1, FCGR2A, IFI35, GSK3B, HIF1A, TUBG1 |
| GO:0042802 identical protein binding | APPL1, STAT1, STAT2, SRM, IFI35, CD53, FCER1G, TYROBP, RAF1, IFIH1, IFIT3, HLA-G, FADD, TUBG1, C1QB | |
| GO:0003723 RNA binding | PSMC1, IFIH1, EBNA1BP2, IFIT1, IFIT3, EIF2S1 |
Table 9.
Summary of candidate genes detected by our method in GS130078 that have been annotated to top 3 GO terms in the three GO databases
| Data base | GO Term | Hub Genes |
|---|---|---|
| GO_BP | GO:0007165 signal transduction | PIP5K1A, RHBDL1, RRAS2, CXCL16, HPGDS, PIK3CD, PIK3CB, VEGFC, RIT1, RALB, CDC42SE1, HIF1A, GPI, PGF, CDK4, PDE1B, PDE2A, PDE3A, TENM1, TYMP, PDE9A, CSF2RB, PI4KA, PI4KB, PDE3B, PDE4A, PDE4D, PPP2R5A, PDE5A, GNA13, LIMK1 |
| GO:0000122 negative regulation of transcription from RNA polymerase II promoter | MEF2A, H1-2, ZNF239, ZNF85, OSR2, CAV1, NFIB, ZNF568, PDE2A | |
| GO:0045944 positive regulation of transcription from RNA polymerase II promoter | TBK1, MEF2A, LRP6, OSR2, ZNF91, HIF1A, NFIB, ESRRG, LMO4, SENP1 | |
| GO_CC | GO:0005886 plasma membrane | FEZ1, KCNH3, ARL4A, SDE2, RIT1, RALB, PI4K2A, COA6, TENM1, CDH22, CDH23, RRAS2, CXCL16, PIK3CD, PIK3CB, LRP6, EPHA3, CAVIN1, CARMIL1, PDE2A, PDE9A, PDE4A, PDE4D, PIP5K1A, ADGRL3, EXO1, SYT15, CACNA1B, CACNA1E, CAV1, GPI, CSF2, HRH1, PI4KA, GNA13, ZAN, PCDH10, ANO4, CDC42SE1, CDH4, MUCL1 |
| GO:0005829 cytosol | ARFGAP1, ARL4A, SDE2, PI4K2A, ELP6, TYMP, NEXMIF, SAMD4A, HPGDS, PIK3CD, PIK3CB, DTL, EPHA3, CAVIN1, CHPF, PPP6C, PDE1B,CARMIL1, PDE2A, PDE3A, PDE9A, PDE3B, PDE4A, PDE4D, PDE5A, TBK1, PIP5K1A, CRLF1, GMNN, IGF2BP2, MON2, GPI, POLR2D, PANK2, HRH1, PI4KA, PI4KB, PPP2R5A, RBMS3, GNA13, LIMK1, PPP4R4, MEF2A, THUMPD3, KIF15, CCT2 | |
| GO:0005654 nucleoplasm | MSH2, ARL4A, SDE2, ZNF85, ERCC3, NFIB, COA6, NEXMIF, HPGDS, PIK3CB, SUMO3, DTL, EPHA3, CAVIN1, APPBP2, RNF112, ESRRG, PPP6C, RBBP5, CARMIL1, PDE9A, CTDSPL2, PDE4A, SENP1, TBK1, PIP5K1A, LIN52, ZNF470, GMNN, EXO1, UBE2T, GPI, ZNF367, DHRS2, RFC5, POLR2D, DNA2, MEF2A, ZNF232, CLUAP1, HIF1A, CDK4 | |
| GO_MF | GO:0005515 protein binding | ORAI2, KCNH3, MSH2, PIWIL1, SDE2, RALB, XIRP1,GSE1, TYMP, RRAS2, HPGDS, MFHAS1, PIK3CD, PIK3CB, DTL, LRP6, EPHA3, CRELD2, APPBP2, RNF112, ESRRG, RBBP5, PDE1B, PDE2A, PDE3A, IGFN1, PDE9A, SOHLH1, PDE3B, PDE4A, PDE4D, PDE5A, ACAN, PIP5K1A, LIN52, CRLF1, TMBIM4, VEGFC, CACNA1B, UBE2T, GNS, GPI, RFC5, DNA2, PI4KA, PI4KB, PPP2R5A |
| GO:0042802 identical protein binding | TBK1, CAV1, PGF, EMILIN3, LRP6, PCSK1, CAVIN1, ESRRG, PDE2A, PDE9A | |
| GO:0004712 protein serine/threonine/tyrosine kinase activity | TBK1, CDK18, CDK4, EPHA3, LIMK1 |
Table 10.
Summary of candidate genes detected by our method in all three databases that have been annotated to the top 5 KEGG enriched pathways in these two microarray datasets
| Data set | KEGG Pathways | Annotated Candidate Genes |
|---|---|---|
| GSE20347 | hsa05200:Pathways in cancer | BCR, AR, APPL1, CKS2, RARA, STAT1, MAP2K1, NFKB2, PIK3CD, PIK3CB, TXNRD1, FLT3LG, PIK3R2, PIK3R1, FADD, RAC2, FAS, RAF1, TGFB2, GNAQ, GNB5, GSK3B, MAPK1, MAPK10, KRAS |
| hsa04010:MAPK signaling pathway | PRKACB, FLT3LG, EPHA2, RAC2, FAS, MAP2K1, RAF1, MAP3K20, TGFB2, NFKB2, MAPK1, MAPK10, KRAS | |
| hsa05169:Epstein-Barr virus infection | PSMD4, PIK3R2, PIK3R1, FADD, LYN, CD3D, STAT1, PSMC2, FAS, NFKB2, PIK3CD, PIK3CB, MAPK10 | |
| hsa04151:PI3K-Akt signaling pathway | FLT3LG, EPHA2, PIK3R2, PIK3R1, MAP2K1, RAF1, MCL1, GNB5, GSK3B, PIK3CD, PIK3CB, ITGB7, MAPK1, KRAS | |
| hsa05171:Coronavirus disease - COVID-19 | RPL22, PIK3R2, PIK3R1, STAT1, PIK3CD, PIK3CB, MAPK1, MAPK10 | |
| GSE23400 | hsa05200:Pathways in cancer | PRKCB, BCR, AR, HIF1A, APPL1, RARA, STAT1, STAT2, MAP2K1, PIK3CD, TXNRD1, PIK3R1, FADD, RAC2, IL15, RAF1, GSK3B |
| hsa04010:MAPK signaling pathway | PRKCB, CD14, RAC2, MAP2K1, RAF1 | |
| hsa04151:PI3K-Akt signaling pathway | PIK3R1, MAP2K1, RAF1, GSK3B, PIK3CD | |
| hsa05169:Epstein-Barr virus infection | ENTPD1, HLA-C, HLA-F, HLA-G, PIK3R1, FADD, STAT1, STAT2, PSMC1, TAP1, ISG15, CXCL10, PIK3CD, OAS3 | |
| hsa05171:Coronavirus disease - COVID-19 | C3AR1, PRKCB, PIK3R1, IFIH1, C1QB, C1QA, STAT1, STAT2, ISG15, CXCL10, PIK3CD, OAS3, FCGR2A | |
| GSE130078 | hsa01100:Metabolic pathways | PIP5K1A, PIGT, HPGDS, PIK3CD, PIK3CB, ST6GALNAC5, ST6GAL1, MOGS, ARSB, NAT8B, PI4K2A, GNS, GPI, CHPF, PANK2, PDE1B, PDE2A, PDE3A, TYMP, PDE9A, PI4KA, PI4KB, PDE3B, PDE4A, PDE4D, PDE5A, SGPP1 |
| hsa05200:Pathways in cancer | HIF1A, PGF, MSH2, CDK4, PIK3CD, PIK3CB, LRP6, VEGFC, CSF2RB, RALB, GNA13 | |
| hsa04144:Endocytosis | PIP5K1A, CAV1, ARFGAP1, FOLR3 | |
| hsa04010:MAPK signaling pathway | RRAS2, VEGFC, PGF, CACNA1B, CACNA1E | |
| hsa05165:Human papillomavirus infection | TBK1, CDK4, PIK3CD, PIK3CB, PPP2R5A |
Literature Trace
Zhu et al. [13] highlight that prothymosin alpha (PTMA) expression was up-regulated in ESCC tissues, thus presenting PTMA as a potential candidate for ESCC. Tang et al. [14] indicated that the expression of PTPRF interacting protein alpha 1 is significantly increased and is related to some malignant clinical features and poor outcomes in ESCC patients, thus establishing it as a valuable biomarker for early detection, treatment formulation and prognosis evaluation for ESCC. Jiang et al. [15] suggested that downregulation of VGLL4 was very important in the progression of ESCC, and restoring the function of VGLL4 might be a promising therapeutic strategy for ESCC. In Shen et al. [16], homer scaffolding protein 3 (HOMER3) is one of the three genes presented as candidate cancer-associated genes and may play a tumorigenic role in ESCC. Ma et al. [17] summarized that upregulation of Proteasome 26S subunit non-ATPase 4 (PSMD4) promotes the progression of ESCC through the reduction of ERS-induced cell apoptosis. Chen et al. [18] found that overexpression of DNA methyltransferase 3b (DNMT3b) is responsible for more aggressive tumor growth and resistance to treatment in ESCC and is linked to activated STAT3 signaling. Liu et al. [19] concluded that Phosphoserine Aminotransferase 1 (PSAT1) expression was elevated in ESCC tissues compared to normal esophageal tissues and increase in the same is significantly associated with stage of disease, lymph node metastasis, distant metastasis and poor prognosis. Findings by Cheng et al. [20] suggested that through activation of the Akt and Erk1/2 signaling pathways, Non-POU Domain Containing Octamer Binding (NONO) plays a potent role in multiple biological aspects of ESCC. Wada et al. [21] highlighted the clinically important implications associated with Transferrin Receptor (TFRC) and concluded that it offers an independent prognostic factor. By employing Cox regression He et al. [22] demonstrated the prognostic value of Canopy FGF Signaling Regulator 2 (CNPY2) for ESCC. Yu et al. [23] demonstrated that Myeloid cell leukemia 1 (MCL-1) contributes to the development of ESCC. Yang et al. [24] concluded that a lower expression of Processing Of Precursor 7 (POP7) predicts a worse prognosis in esophageal cancer. Qiu et al. [25] suggested that through activation of AKT1/mTOR signaling pathway, maintenance complex component 7 (MCM7) promotes tumor cell proliferation, colony formation and migration of ESCC cells. Choy et al. [26] and [27] further suggested MCM7 as a more sensitive proliferation markers for evaluation and for predicting various clinical outcomes of ESCC respectively. Miyazaki et al. [28] concluded that ephrin receptor A2 (EphA2) overexpression appears to be related to poor degree of tumor differentiation and lymph node metastasis in ESCC. Ma et al. [29] suggested that Karyopherin 2 (KPNA2) protein levels were high in ESCC tumors, and siRNA against KPNA2 could inhibit the growth of ESCC cells, suggesting it may be a new potent marker and therapeutic target for ESCC. Sakai et al. [30] further concluded that KPNA2 expression is associated with poor differentiation, tumor invasiveness, and tumor proliferation in ESCC. Wang et al. [31] identified kinesin family member 4A (KIF4A) as a facilitator of proliferation, cell cycle, migration, and invasion of ESCC in vivo and in vitro. Similarly, Sun et al. [32] stated that through the Hippo signaling pathway, KIF4A regulates the biological function of ESCC cells thus promoting ESCC cell proliferation and migration. Kita et al. [33] demonstrated that the expression of cyclin-dependent kinase subunit 2 (CKS2) in ESCC was elevated relative to levels in normal tissue, and that CKS2 overexpression is associated with the depth of tumor invasion, lymphatic invasion, clinical stage, distant metastasis and poor prognosis. Zheng et al. [34] found that the expression of cytokine induced apoptosis inhibitor 1 (CIAPIN1) was statistically correlated with the degree of differentiation, depth of invasion, and lymph node metastasis of ESCC and thus has been considered as a valuable prognostic indicator in ESCC. Zhao et al. [35] highlighted that Protein arginine methyltransferase 1 (PRMT1) activates and maintains esophageal TICs by mediating transcription alteration through histone H4 arginine methylation. Zhou et al. [36] highlighted that PRMT1 activates Hedgehog signaling and up-regulated the expression of target genes downstream of Hedgehog signaling thus taking an oncogenic role of PRMT1 in the progression of ESCC.
Zhang et al. [37] provided evidence that Human Leukocyte Antigen-F (HLA-F) antigen expression was associated with survival in patients with ESCC. Yie et al. [38] established that Human Leukocyte Antigen-G (HLA-G) expression has a strong and independent prognostic value in human ESCC. According to Sato et al. [39], high chemokine (CXC motif) ligand 10 (CXCL10) expression is an independent prognostic factor and has the potential to serve as a clinically useful marker of the need for adjuvant chemotherapy after surgery in patients with advanced thoracic ESCC. Yuan et al. [40] suggested the tumor promotion role of Interferon-stimulated gene 15 (ISG15) in ESCC via c-MET/Fyn/-catenin pathway. Yu et al. [41] and Wang et al. [42] identified that Cyclin-dependent kinase inhibitor 3 (CDKN3) regulates tumor progression through activation of AKT signaling pathway in ESCC. Liu et al. [43] further suggested that CDKN3 acted as an oncogene in human ESCC and may accelerate the G1/S transition by affecting CyclinD-CDK4 complex via regulating pAKT-p53-p21 axis and p27 independent of AKT. Preliminary studies by Hu et al. [44] suggested that disks large-associated protein 5 (DLGAP5) promotes cell proliferation in ESCC. According to Liu et al. [45], vaccinia-related kinase (VRK) serine/threonine kinase 1 promotes CDDP resistance through c-MYC by activating c-Jun and potentiating a malignant phenotype in ESCC. Liu et al. [46] provided a potential target for the immuno-oncology effect of roteasome alpha-subunit 3 (PSMA3) in ESCC therapy. Wang et al. [47] detected the major role of Pleckstrin-2 (PLEK2) in driving metastasis and chemoresistance in ESCC by regulating LCN2. Qu et al. [48] found that Component 3a Receptor 1 (C3AR1) might be the cause of an immunosuppressive microenvironment by affecting the polarization of macrophages to M2 phenotype and lead to the progression of ESCC. Zhang et al. [49] suggested that Signal Transducer and Activator of Transcription-1 (STAT1) is a tumor suppressor in ESCC. According to Shao et al. [50], Hypoxia-inducible factor 1 (HIF-1), p53, and vascular endothelial growth factor (VEGF) are important factors that facilitate tumor progression. The results from the study conducted by Hu et al. [51] indicated that HIF-1 promotes metastasis of ESCC by targeting SP1 in a hypoxic microenvironment. Bolidong et al. [52] suggested that via cyclin D1/CDK4-mediated cell cycle progression Glycogen synthase kinase 3 (GSK3) has a tumor promoting role in ESCC. According to Gao et al. [53], GSK3 expression promotes ESCC progression through STAT3 in vitro and in vivo, and GSK3-STAT3 signaling could be a potential therapeutic target for ESCC treatment.
According to Kato et al. [54] and [55] caveolin-1 (CAV1) is a biomarker for ESCC. Lu et al. [56] and Shu et al. [57] indicated that insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) serves a major carcinogenic role in ESCC. According to [58], the expression of vascular endothelial growth factor C (VEGF-C) correlates with lymph node metastasis and poor prognosis. Similarly, [59] suggests that VEGF-C expression in ESCC may play a great key role in lymphatic spread. Feng et al. [60] indicated that ras-like without CAAX1 (RIT1) displays tumor-suppressing functions in ESCC, and these functions were carried out by inhibiting MAPK and PI3K/AKT signaling pathway, inhibiting EMT, and downregulating cancer stemness of ESCC cells. Zhou et al. [61] demonstrated that Dehydrogenase/reductase member 2 (DHRS2) had an important part in ESCC development and progression. Li et al. [62] suggested a tumor suppression function for RNA Binding Motif Single Stranded Interacting Protein 3 (RBMS3) gene in ESCC. According to Wang et al. [63], UBE2T is involved in the development of ESCC, and gene signatures derived from UBE2T-associated genes are predictive of prognosis in ESCC. Gao et al. [64] demonstrated that High Mobility Group Box 3 (HMBG3) may be a potential molecular marker for predicting the prognosis of ESCC patients. According to Huang et al. [65], cyclin-dependent kinase 4 (CDK4) amplification was identified as an independent prognostic factor for survival, which could be incorporated into the tumor-node-metastasis staging system to refine risk stratification of patients with esophageal squamous cell carcinoma. Ling et al. [66] suggested that MutS Homolog 2 (MSH2) methylation in the plasma would be a good predictor of DFS for these ESCC patients before oesophagectomy. Xu et al. [67] identified Estrogen-related receptor gamma (ESRRG) as one of four molecular markers that may be helpful in the diagnosis and treatment of ESCC. Chen et al. [68] demonstrated that silencing EphA3 in KYSE410 cells triggers epithelial–mesenchymal transition, and promoted cell migration and invasion in ESCC. Luo et al. [69] found that knockdown of Insulin-like growth factor binding protein-3 (IGFBP-3) confers resistance to the cell killing effects of IR on ESCC in vitro and in vivo. Zhao et al. [70] indicated that the increased ESCC chemosensitivity might be dependent on IGFBP-3 upregulation through EGFR-dependent pathway. Furthermore, according to Luo et al. [71], high level of IGFBP-3 expression in ESCC associates with early clinical stages and are predictive for favorable survival of the patients treated with radiotherapy. According to Wang et al. [72], carbohydrate sulfotransferase 15 (CHST15) promotes the proliferation of TE-1 cells via multiple pathways in ESCC.
Discussion
In Table 11, we give a detailed summary of all DEGs that have been identified by our method as candidates for potential biomarkers for ESCC. In our method, we consider strong literature evidence for association with ESCC and five other SCCs related to ESCC as the necessary criterion for a candidate gene to be a potential biomarker, and the findings from literature are summarized in Table 11. In the table, we also highlight the enriched GO terms and pathways to which the candidate genes has been annotated. Furthermore, it also details whether the same is a hub gene, a transcription factor (TF) or whether it is upregulated or down-regulated. A DEG is upregulated if and downregulated when . We take into consideration logFC values calculated by limma for the microarray datasets, and edgeR in the RNA-Seq dataset.
Table 11.
Summary of potential biomarkers identified by our proposed framework (Fig. 1)
| Detected hub gene | Enriched GO Term | Enriched cancer pathway(s) | HG or TED | TF? | +/– | Literature Evidence | |
|---|---|---|---|---|---|---|---|
| GSE20347 | PTMA | BP, CC, MF | Nil | Yes | Yes (Fig 5a) | + | ESCC [13], HNSCC [73] |
| PPFIA1 | BP, CC | Nil | HG | No | ESCC [14], HNSCC [74] | ||
| PIK3CB | BP, CC, MF | hsa05200,hsa05169,hsa04151,hsa05171,hsa05165, hsa05417,hsa05161,hsa04510,hsa04015,hsa05162, hsa05164,hsa05205,hsa05166,hsa04210,hsa05418, hsa05167,hsa05163,hsa04380,hsa05146,hsa05222, hsa04933,hsa05135,hsa05215,hsa05017, hsa04722, hsa04722,hsa05170,hsa04810,hsa04066,hsa01100, hsa05131,hsa05010,hsa04024,hsa04014,hsa05210, hsa05221,hsa05160,hsa05212,hsa04218,hsa04630, hsa05203 | HG | No | OSCC [75] | ||
| VGLL4 | BP, CC, MF | Nil | HG | Yes (Fig. 5a) | + | ESCC [15] | |
| HOMER3 | BP, CC, MF | hsa04068,hsa04724 | HG | No | + | ESCC [16] | |
| SHC1 | BP, CC, MF | hsa04510,hsa04722,hsa04014,hsa04072,hsa04935, hsa04910,hsa01521,hsa04062,hsa05220,hsa05225, hsa05226,hsa04917,hsa05224,hsa04915,hsa04012, hsa05214,hsa04926,hsa01522,hsa04650,hsa05100 | HG | No | + | LSCC [76] | |
| PSMD4 | BP, CC, MF | hsa05169,hsa05017,hsa05022,hsa05010,hsa05020, hsa05016,hsa05014,hsa05012,hsa03050 | HG | No | + | ESCC [17] | |
| CLIC4 | BP, CC, MF | Nil | HG | No | + | HNSCC [77] | |
| DNMT3B | BP, CC, MF | hsa01100 | HG | Yes (Fig. 5b) | + | ESCC [78], HNSCC [79], OSCC | |
| MYO1B | BP, CC, MF | hsa05130 | HG | No | + | HNSCC [80] | |
| PSAT1 | CC, MF | hsa01100,hsa01200 | HG | No | + | ESCC [19] | |
| SLC3A2 | BP, CC, MF | hsa04150,hsa04216 | HG | NO | + | OSCC [81] | |
| NONO | BP, CC, MF | Nil | HG | Yes (Fig. 5c) | ESCC [20] | ||
| TFRC | BP, CC, MF | hsa04066,hsa04145,hsa04640,hsa04144,hsa04216 | HG | No | + | ESCC [21] | |
| SENP5 | BP, CC | Nil | HG | No | + | OSCC [82] | |
| RCN1 | BP, CC, MF | Nil | HG | No | + | OSCC [83] | |
| CNPY2 | BP, CC, MF | Nil | HG | No | + | ESCC [22] | |
| SBF1 | BP, CC, MF | Nil | HG | No | – | HNSCC [84] | |
| MCL1 | BP, CC, MF | hsa04151,hsa04210,hsa04630 | HG | No | + | ESCC [23], OSCC [85] | |
| POP7 | BP, CC, MF | Nil | HG | No | + | ESCC [24] | |
| MCM7 | BP, CC, MF | hsa04110,hsa03030 | HG | Yes (Fig. 5d) | + | ESCC [25–27], OSCC [86] | |
| POM121 | BP, CC, MF | hsa03013,hsa05014 | HG | No | OSCC [87] | ||
| PSMC2 | BP, CC, MF | hsa05169,hsa05017,hsa05022,hsa05010,hsa05020, hsa05016,hsa05014,hsa05012,hsa03050 | HG | Yes (Fig 5d) | + | OSCC [88] | |
| EPHA2 | BP, CC, MF | hsa04010,hsa04151,hsa04015,hsa04014,hsa04360 | HG | No | + | ESCC [28] ,LSCC [89], HNSCC [90, 91] | |
| KPNA2 | BP, CC, MF | hsa05164,hsa03013,hsa05207 | HG | No | + | ESCC [29, 30], OSCC [92] | |
| KIF4A | BP, CC, MF | Nil | HG | No | + | ESCC [31, 32], OSCC [93] | |
| CCT4 | BP, CC, MF | Nil | HG | Yes (Fig. 5e) | + | HNSCC [94] | |
| GSE20347 | CKS2 | BP, CC, MF | hsa05200,hsa05222 | HG | No | + | ESCC [33], TSCC [95] |
| CIAPIN1 | BP, CC, MF | Nil | HG | No | + | ESCC [34] | |
| PRMT1 | BP, CC, MF | hsa04068,hsa04922 | HG | No | + | ESCC [35, 36], HNSCC [96] | |
| HPRT1 | BP, CC, MF | hsa01100 | HG | No | + | OSCC [97], HNSCC [98] | |
| NDD4L | Nil | Nil | HG | No | + | OSCC [99] | |
| KRAS | BP, CC, MF | hsa05200,hsa04010,hsa04151,hsa05165,hsa05417, hsa05161,hsa04015,hsa05205,hsa05166,hsa04210, hsa05167,hsa05163,hsa04933,hsa05215,hsa05022, hsa04722,hsa05170,hsa04810,hsa04066,hsa05010, hsa04014,hsa05210,hsa05221,hsa05160,hsa05212, hsa04218,hsa05203,hsa04072,hsa04919,hsa04935, hsa04660,hsa04068,hsa04910,hsa01521,hsa04062, hsa05211,hsa05220,hsa05225,hsa04625,hsa05226, hsa05223,hsa05230,hsa04917,hsa05224,hsa04540, hsa04915,hsa04550,hsa05218,hsa04012,hsa05214, hsa04071,hsa04720,hsa04926,hsa05213,hsa05219, hsa05208,hsa05235,hsa04140,hsa04662,hsa04912, hsa05231,hsa01522,hsa04650,hsa04929,hsa04664, hsa04360, hsa04730,hsa04916,hsa04371,hsa04921, hsa05216,hsa04725,hsa04213,hsa04211,hsa04137, hsa04960,hsa04370,hsa04150,hsa05207,hsa04726 | TED | No | – | LSCC [100] | |
| PIK3R2 | BP, CC, MF | Nil | TED | No | – | LSCC [101] | |
| GSE23400 | TAP1 | BP, CC, MF | hsa05169,hsa05163,hsa05170,hsa04145,hsa04612 | HG | No | + | TSCC [102] |
| HLA-F | BP, CC, MF | hsa05169,hsa05165,hsa05166,hsa05163,hsa05167, hsa05170,hsa05203,hsa04218,hsa04145,hsa04144, hsa04612,hsa05416,hsa05320,hsa04940,hsa05332, hsa04514,hsa05330 | HG | No | + | ESCC [37] | |
| IFIT3 | BP, CC, MF | Nil | HG | No | + | OSCC [103, 104] | |
| HLA-G | BP, CC, MF | hsa05163,hsa05167,hsa05170,hsa05203,hsa04218, hsa04145,hsa04144,hsa04612,hsa05416,hsa05320, hsa04940,hsa05332,hsa04514,hsa05330 | HG | No | + | ESCC[38] , HNSCC [105, 106], OSCC [107] | |
| IFI44L | BP, CC, MF | Nil | HG | No | + | OSCC [108] | |
| CXCL10 | BP, CC, MF | hsa05169,hsa05171,hsa05164,hsa05160,hsa04060, hsa04062,hsa04668,hsa04620,hsa04622,hsa04657, hsa04061,hsa04623 | HG | No | + | ESCC [39], TSCC [109], HNSCC [110] | |
| IFIT1 | BP, CC, MF | hsa05160 | HG | No | + | OSCC [103], HNSCC [111] | |
| ISG15 | BP, CC, MF | hsa05169,hsa05171,hsa05165, hsa04622 | HG | No | + | ESCC [40] , OSCC [112, 113] | |
| CDKN3 | BP, CC, MF | Nil | HG | No | + | ESCC [41–43] | |
| DLGAP5 | BP, CC, MF | Nil | HG | No | + | ESCC [44] | |
| VRK1 | BP, CC, MF | Nil | HG | No | + | ESCC [45], HNSCC [114] | |
| PSMA3 | BP, CC, MF | hsa05017,hsa05022,hsa05010,hsa05020,hsa05016, hsa05012,hsa05014,hsa03050 | HG | No | + | ESCC [46], OSCC [115] | |
| PLEK2 | BP, CC | Nil | HG | Yes (Fig. 5f) | + | ESCC [47], HNSCC [116] | |
| CD163 | BP, CC, MF | Nil | HG | No | + | HNSCC [117], OSCC [118, 119] | |
| ITGB2 | BP, CC, MF | hsa04015,hsa05166,hsa05152,hsa05146,hsa04810, hsa04145,hsa04610,hsa05134,hsa05140,hsa04613, hsa05416,hsa04650,hsa04670,hsa04390,hsa05133, hsa05323,hsa05144,hsa04514 | HG | No | + | OSCC [120] | |
| FCGR2A | BP, CC, MF | hsa05171,hsa05152,hsa04380,hsa05135,hsa04145, hsa05130,hsa04611,hsa04666,hsa05140,hsa04613 | HG | No | + | ESCC [121] , HNSCC [122, 123] | |
| C3AR1 | BP, CC, MF | hsa05171,hsa04936,hsa04080,hsa04610 | HG | No | + | ESCC [48] | |
| HIF1A | BP, CC, MF | hsa05200,hsa05205 | TED | Yes 6a | + | ESCC[50, 51], OSCC [124–126], TSCC [127] | |
| CAV1 | BP, CC, MF | hsa04144,hsa04510,hsa05205 | HG | No | + | ESCC[54, 55], OSCC[128], HNSCC[129], TSCC[130] | |
| GSE130078 | IGF2BP2 | BP, CC, MF | Nil | HG | No | + | ESCC[56, 57], OSCC[131, 132] |
| VEGFC | BP, CC, MF | hsa05200,hsa04010,hsa04151,hsa04015,hsa04510, hsa04020,hsa04926 | HG | No | + | ESCC[58, 59], OSCC[133, 134] | |
| HIF1A | BP, CC, MF | hsa05200,hsa05205 | HG | Yes 6d | + | ESCC[50, 51], OSCC [124–126], TSCC [127] | |
| GNA13 | BP, CC, MF | hsa05200,hsa04270 | HG | No | + | LSCC [135] | |
| RIT1 | BP, CC | Nil | HG | No | - | ESCC [60] | |
| DHRS2 | BP, CC | Nil | HG | No | + | ESCC [61] | |
| RBMS3 | BP, CC, MF | Nil | HG | No | + | ESCC [62], LSCC [136] | |
| RFC5 | BP, CC, MF | Nil | HG | No | + | LSCC [137] | |
| UBE2T | BP, CC, MF | Nil | HG | No | + | ESCC [63] | |
| HMGB3 | BP, CC, MF | Nil | HG | Yes 6f | + | ESCC [64] | |
| LMO4 | BP, CC, MF | Nil | HG | Yes 6f | + | HNSCC [138], TSCC[139] | |
| CDK4 | BP, CC, MF | hsa05200,hsa05165,hsa04151,hsa04530,hsa05224, hsa04110 | HG | No | + | ESCC [65], HNSCC[140, 141] | |
| EXO1 | BP, C C, MF | Nil | HG | No | + | HNSCC[142] | |
| MSH2 | BP, CC, MF | hsa05200 | HG | No | + | ESCC[66], HNSCC [143] | |
| TBK1 | BP, CC, MF | hsa05165 | HG | No | + | HNSCC [144] | |
| HPDL | CC, MF | Nil | HG | No | + | TSCC [145] | |
| LRP6 | BP, CC, MF | hsa05200,hsa05224 | HG | No | + | OSCC [146] | |
| ESRRG | BP, CC, MF | Nil | HG | Yes 6e | – | ESCC [67], LaSCC [147] | |
| EPHA3 | BP, CC, MF | Nil | HG | Yes | + | ESCC [68] | |
| TRAM2 | CC | Nil | HG | Yes | + | OSCC [148] | |
| IGFBP3 | BP, CC, MF | Nil | HG | Yes | + | ESCC [69–71], OSCC[149, 150] | |
| CLPTM1L | BP, CC | Nil | HG | No | + | OSCC[151, 152] | |
| CHST15 | CC | Nil | HG | No | + | ESCC[72] | |
| GSE20347 & GSE23400 | TXNRD1 | BP, CC, MF | hsa05200,hsa05225 | TED | No | + | OSCC [153], HNSCC [154] |
| FADD | BP, CC, MF | hsa05200,hsa05169,hsa05165,hsa05161,hsa05162, hsa05164,hsa04210,hsa05167,hsa05152,hsa05163, hsa05022,hsa05170,hsa04936,hsa05010,hsa05160, hsa05132,hsa05142,hsa04668,hsa04620,hsa05130, hsa01524,hsa04622,hsa04657,hsa04217,hsa04621 | TED | No | + | OSCC [155], HNSCC [156, 157] | |
| STAT1 | BP, CC, MF | hsa05200,hsa05169,hsa05171,hsa05165,hsa05161, hsa05162,hsa05164,hsa05167,hsa05152,hsa04380, hsa04933,hsa05145,hsa05160,hsa05212,hsa04630, hsa04919,hsa04935,hsa04062,hsa04625,hsa04620, hsa04659,hsa04917,hsa04658,hsa05140,hsa05235, hsa04217,hsa05321,hsa04621 | TED | Yes 6b | + | ESCC [49], HNSCC [158, 159] | |
| AR | BP, CC, MF | hsa05200,hsa05215,hsa05207 | TED | Yes 6c | - | OSCC [160, 161] | |
| RAF1 | BP, CC, MF | hsa05200,hsa04010,hsa04151,hsa05165,hsa05161, hsa04510,hsa04015,hsa05164,hsa05205,hsa04210, hsa05167,hsa05152,hsa05163,hsa05215,hsa05022, hsa04722,hsa05170,hsa04810,hsa05010,hsa04024, hsa04014,hsa05210,hsa05221,hsa05160,hsa05212, hsa04218,hsa04630, hsa04072,hsa04919,hsa05132, hsa04935,hsa04660,hsa04068,hsa04910,hsa01521, hsa04062,hsa04022,hsa05211,hsa04928,hsa05220, hsa05225,hsa04625,hsa05226,hsa05223,hsa05230, hsa04917,hsa05224,hsa04540,hsa04915,hsa04550, hsa05218,hsa04012,hsa05214,hsa04071,hsa04720, hsa04926,hsa05213,hsa04666,hsa05219,hsa05208, hsa05235,hsa04140,hsa04662,hsa04270,hsa04912, hsa05231,hsa01522,hsa04650,hsa04613,hsa04929, hsa04664,hsa04360,hsa04730,hsa04916,hsa04371, hsa04921,hsa04370,hsa04150,hsa05207,hsa04726 | TED | No | – | OSCC [162] | |
| GSE20347 & GSE23400 | GSK3B | BP, CC, MF | hsa05200,hsa04151,hsa05165,hsa05417,hsa04510, hsa05162,hsa05167,hsa05163,hsa05135,hsa05215, hsa05022,hsa04722,hsa05131,hsa05010,hsa05210, hsa05160,hsa04919,hsa04935,hsa04660,hsa04910, hsa01521,hsa04062,hsa05225,hsa05226,hsa04110, hsa04917,hsa05224,hsa04932,hsa04550,hsa04012, hsa04728,hsa04934,hsa05020,hsa05415,hsa05213, hsa04931,hsa04662,hsa04657,hsa04360,hsa04916, hsa04390,hsa04310,hsa04150 | TED | No | + | ESCC [52, 53], OSCC [163, 164] |
| MAP2K1 | BP, CC, MF | hsa05200,hsa04010,hsa04151,hsa05165,hsa05161, hsa04510,hsa04015,hsa05164,hsa05205,hsa05166, hsa04210,hsa05167,hsa05163,hsa04380, hsa05135,hsa05215,hsa05022,hsa04722,hsa05170, hsa04810,hsa04066,hsa05010,hsa04024,hsa04014, hsa05210,hsa05221,hsa05160,hsa05212,hsa04218, hsa04072,hsa04919,hsa05132,hsa04935,hsa04660, hsa04068,hsa04910, hsa01521,hsa04062,hsa04022, hsa05211,hsa04928,hsa05220,hsa04668,hsa05225, hsa04620, hsa05226,hsa05223,hsa05230,hsa04917, hsa05224,hsa04540,hsa04915,hsa04550,hsa05218, hsa04012,hsa04934,hsa05214,hsa04071,hsa04720, hsa04926,hsa05213,hsa04666,hsa05219,hsa05208, hsa05235,hsa04140,hsa04662,hsa04270,hsa04912, hsa05231,hsa01522,hsa04650,hsa04613,hsa04929, hsa04664,hsa04360,hsa04730,hsa04916,hsa04371, hsa04921,hsa04370,hsa05216,hsa04725,hsa04150, hsa05207,hsa04726 | TED | No | – | HNSCC [165] |
HG hub gene, TED Top Enriched DEG, DEG annotated to most enriched GO term in all three databases as well as to the most enriched pathway, TF transcription factor, + Upregulated DEG and – Downregulated DEG
The biological relevance of a candidate to its respective dataset is considered based on three criteria:
Annotated to at least one GO term in 2 of 3 GO databases with ,
Annotated to at least one KEGG pathway with , and
It’s a TF and thus exhibits regulatory behavior towards other DEGs in the network.
For a candidate gene to be considered a potential biomarker, we consider following four cases.
Case 1: Strong literature evidence of association with ESCC and biologically relevant to its dataset based on all three criteria a,b and c,
Case 2: Strong literature evidence of association with ESCC and biologically relevant to its dataset based on criteria a and b,
Case 3: Strong literature evidence of association with ESCC and biologically relevant to its dataset based on criteria a or b,
Case 4: Biologically relevant to its dataset using all three criteria a,b and c, and has literature evidence of association with previously mentioned 5 SCCs related to ESCC, namely, oral SCC, Lung SCC, Tongue SCC, Head and Neck SCC and Laryngeal SCC.
All candidate genes that fall under Case 1 and Case 2 are considered potential biomarkers for ESCC because of existing evidence of association with ESCC in the form of other works while our biological validation of these genes establishes their relevance to their respective datasets. For candidate genes that fall under Case 3, although there is strong literature evidence of association with ESCC, we have weak evidence of their biological relevance to their datasets. On the other hand, for candidate genes that fall under Case 4, although we strongly validate their biological relevance to their datasets, there is only literature evidence of association with other SCCs related to ESCC. For both these cases, the candidates can be considered probable potential biomarkers, but need further in-depth analysis.
Top Enriched DEGs (TEDs), STAT1 and HIF1A detected in both microarray datasets (GSE20347 and GSE23400) and GSE130078, respectively, belong to Case 1. In GSE20347, two candidates DNMT3B and MCM7 also belong to Case 1. Thus, STAT1, HIF1A, DNMT3B and MCM7 are potential biomarkers for ESCC. GSK3B is a TED detected in both microarrays, and belongs to Case 2. In dataset GSE20347, 9 candidate genes HOMER3, PSMD4, PSTAT1, TFRC, MCL1, EPHA2, KPNA2, CKS2 and PRMT1 belong to Case 2, and thus are potential biomarkers for ESCC. Similarly, 7 candidate genes HLA-F, HLA-G, CXCL10, ISG15, PSMA3, FCGR2A and C3AR1 are potential biomarkers for ESCC as they fall under Case 2. Four candidate genes in the RNASeq dataset GSE130078, CAV1, VEGFC, CDK4 and MSH2 fall under Case 2 and are potential biomarkers for ESCC.
Three candidates genes in GSE20347, PTMA, VGLL4 and NONO fall in Case 3. In other words, although there are other works that establish their role as potential biomarkers for ESCC, the biological relevance to their respective datasets is not that strong. However, they can still be regarded as probable potential biomarkers for ESCC, but need further in-depth validation. Similarly in GSEE23400 and GSE130078, one (PLEK2) and 2 (HMGB3 and ESRRG) genes fall under Case 3. PSMC2 detected in GSE20347, on the other hand falls under Case 4. We validate its strong association with the dataset as this candidate gene has been annotated to GO terms in all three GO databases as well as several enriched pathways. They further exhibit regulatory behavior in a GRN, but there are no previous works that relate the same to ESCC. However, its worth mentioning that there is literature evidence that identify PSMC2 as potential biomarker for OSCC. Similarly, the TED identified in the two microarray datasets, AR, also falls under Case 4. Both, PSMC2 and AR are probable potential biomarkers for ESCC, but need further in-depth analysis.
In Table 12, we put forward a comparison between our work and two recent works presented by Patowary et al. [166] and Hu. et al. [44] that perform DE analysis by employing approaches and methods similar to our work.
Table 12.
Comparison of our method with two recent works that employ DE analysis on ESCC datasets
| Parameter | Our Method | Patowary et al. [166], 2020 | Hu. et al. [44], 2020 |
|---|---|---|---|
| Datasets | Microarray: GSE20347 and GSE23400, RNA-Seq: GSE130078 | Microarray: GSE20347 and GSE23400, RNA-Seq: SRP064894 | Microarray: GSE20347 and GSE26886 |
| Identification of DEGs | DEA methods: Limma, SAM, EBAM, limma-voomm, edgeR and DESeq2 with | DEA methods: Limma, SAM, EBAM, limma-voomm, edgeR and DESeq2 with | Limma; and |
| Consensus function | (a) Common genes given by the methods with (b) For each method, DEGs not in common genes with (RNA-Seq) or (Microarray) | (a) Common genes given by the methods with and (b) top ranked DEGs (other than common) given by each method with | Up- and downregulated DEGs common to both microarray datasets |
| Module Extraction | Heiarchical Tree Classification; Eigenmodule selection and MEDissThres threshold merging | Hierarchical Tree Classification; Eigenmodule selection and MEDissThres threshold merging | (a) PPI network; Nodes with edge of as hub genes; (b) WGCNA and Heiarchical Tree Classification |
| Preservation Analysis | Yes | Yes | No |
| Modules of Interest | All non-preserved modules of size larger that 100 nodes | Least Preserved module | Relevance between each module and hub genes identified in PPI network |
| hub gene | Intramodular Connectivity | Intramodular Connectivity; Degree and confidence in PPI Network | Nodes with edge of in PPI network |
| Candidate Identification | Top 20 hub genes in all modules of interest; DEGs that are annotated to most enriched GO term in all three GO databases as well as the most enriched pathway | DEG with highest intramodular connectivity in each module of interest | Top hub gene in module of interest |
| Enrichment Analyses | DAVID; GO and KEGG pathway | DAVID; GO and pathway | DAVID; GO and pathway and GSEA |
| Diseases considered for literature trace | ESCC and five other SCCs, namely oral SCC, Lung SCC, Tongue SCC, Head and Neck SCC, Laryngeal SCC | ESCC and all other cancers | ESCC and all other cancers |
| Identified potential biomarkers | STAT1,HIF1A,DNMT3B, MCM7,GSK3B,HOMER3, PSMD4,PSTAT1,TFRC, MCL1,EPHA2,KPNA2, CKS2,PRMT1,HLA-F, HLA-G,CXCL10,ISG15, PSMA3,FCGR2A,C3AR1, CAV1,VEGFC,CDK4 and MSH2 | COL27A1,SOX11,BAG6, TOP3,CDC6,EZH2, COL7A1,G6PD and AKR1C2 | DLGAP5 |
Conclusion
All six methods, three microarray and three RNA-sequencing, employed by the proposed integrative approach based differential expression (DE) Analysis framework were found effective in extracting differentially expressed genes (DEGs) with a p-value of 0.05. We further proposed a consensus function that takes into account the information loss due to the DEGs common to all three respective methods and further employs local fdr (for microarray) and q-value (for RNA-Seq). Through differential co-expression (DCE) and preservation analysis, we studied the behavioral changes among the DEGs under normal and disease circumstances. All non-preserved modules of reasonable sizes are considered modules of interest and analyzed further down the pipeline. DEGs are considered candidates for potential biomarkers for ESCC when they are either: (a) hub genes in the modules of interest, or (b) Top Enriched DEG (TED), i.e., a DEG annotated to the most enriched GO term in all three GO databases as well as the most enriched KEGG pathway in their respective datasets. Our proposed framework was validated on two microarray datasets (GSE20347 and GSE23400) and one RNA-Sequencing dataset (GSE130078). From, 7, 3 and 8 modules of interest in GSE20347, GSE23400 and GSE130078 respectively, 124, 59 and 160 hub genes were identified. The consideration of 22, 18 and 16 TEDs detected by GSE20347, GSE23400 and GSE130078 respectively results in 146, 77 and 176 as candidates for potential biomarkers of ESCC. Biological relevance for each candidate to their respective datasets is analysed based on (a) annotation to enriched GO terms in the GO databases, (b) annotation to enriched KEGG pathways, and (c) if the candidate gene is a transcription factor in a gene regulatory network. Another very important criterion that we considered for a candidate gene to be a potential biomarker is previous literature that has either (a) established them as potential biomarkers for ESCC itself, or (b) established them as potential biomarkers for 5 other SCCs related to ESCC, namely, Oral SCC, Tongue SCC, Lung SCC, Head and Neck SCC and Laryngeal SCC.
Our method identified 4 candidate genes, STAT1, HIF1A , DNMT3B and MCM7, that are transcription factors (TFs), have strong biological relevance to their respective datasets and have previous literature works that establish their role as potential biomarkers in ESCC. Our method identified GSK3B , detected as DEG by both microarray datasets (GSE20347 and GSE23400), as a TED and has both strong literature evidence as potential biomarker of ESCC and we established its strong biological relevance to both microarray datasets. Similarly, nine (HOMER3 , PSMD4, PSTAT1, TFRC, MCL1, EPHA2, KPNA2, CKS2 and PRMT1), seven (HLA-F, HLA-G, CXCL10, ISG15, PSMA3, FCGR2A and C3AR1) and four (CAV1, VEGFC, CDK4 and MSH2) candidates genes in GSE20347, GSE23400 and GSE130078 are established as potential biomarkers for ESCC. We further identified 3 (PTMA, VGLL4 and NONO), 1 (PLEK2) and 2 (HMGB3 and ESRRG) TFs in GSE20347, GSE23400 and GSE130078 respectively, that have strong literature evidence as potential biomarkers of ESCC, but have moderate evidence for biological relevance to their respective datasets, and thus can be regarded as probable potential biomarkers for ESCC. On the other side of the spectrum, the transcription factor AR, a TED that is identified as a DEG in both microarray datasets and PSMC2 have strong biological relevance to their datasets, but have been identified as potential biomarkers for other SCC related to ESCC. They can also be considered probable potential biomarkers for ESCC, but need further in-depth analysis. For future work, there is scope for improvement in the framework in general and consensus function specifically, towards detection of a smaller number of DEGs with minimum loss of relevant information.
Declarations
Conflict of interest
On the behalf of all authors, the corresponding authors states that there is no conflict of interest.
Footnotes
This article is part of the topical collection “Pattern Recognition and Machine Learning” guest edited by Ashish Ghosh, Monidipa Das and Anwesha Law.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dhruba K. Bhattacharyya has contributed to this work.
Contributor Information
Manaswita Saikia, Email: saikiamanaswita@gmail.com.
Dhruba K. Bhattacharyya, Email: dkb@tezu.ernet.in
Jugal K. Kalita, Email: jkalita@uccs.edu
References
- 1.Smyth GK. Linear Models forMicroarray Data. In: Gentleman R, CareyVJ Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY: Springer; 2005. pp. 397–420. [Google Scholar]
- 2.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Efron B, Tibshirani R, Storey JD, Tusher V. Empirical Bayes analysis of a microarray experiment. Journal of the American statistical association. 2001;96(456):1151–60. doi: 10.1198/016214501753382129. [DOI] [Google Scholar]
- 4.Law CW, Chen Y, Shi W, Smyth GK. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15(2):R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabási AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297(5586):1551–1555. doi: 10.1126/science.1073374. [DOI] [PubMed] [Google Scholar]
- 9.Langfelder P, Luo R, Oldham MC, Horvath S. Is My Network Module Preserved and Reproducible? PLoS Comput Biol. 2011;7(1):e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherman BT, Lempicki RA, et al. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 11.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhagwat AS, Vakoc CR. Targeting Transcription Factors in Cancer. Trends Cancer. 2015;1(1):53–65. doi: 10.1016/j.trecan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y, Qi X, Yu C, Yu S, Zhang C, Zhang Y, et al. Identification of prothymosin alpha (PTMA) as a biomarker for esophageal squamous cell carcinoma (ESCC) by label-free quantitative proteomics and Quantitative Dot Blot (QDB) Clinical Proteomics. 2019;16(1):12. doi: 10.1186/s12014-019-9232-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang P, Jia R, Gong L, Sui Z, Xiao W, Yang Y, Gong L, Sui Z, Xiao W, Yang Y, Wu X and Yu Z, Zhang, H. High expression of PPFIA1 in human esophageal squamous cell carcinoma correlates with tumor progression and poor prognosis. Res Square; 2022. 10.21203/rs.3.rs-554718/v2.
- 15.Jiang W, Yao F, He J, Lv B, Fang W, Zhu W, et al. Downregulation of VGLL4 in the progression of esophageal squamous cell carcinoma. Tumor Biology. 2015;36(2):1289–1297. doi: 10.1007/s13277-014-2701-7. [DOI] [PubMed] [Google Scholar]
- 16.Shen TY, Mei LL, Qiu YT, Shi ZZ. Identification of candidate target genes of genomic aberrations in esophageal squamous cell carcinoma. Oncol Lett. 2016;12(4):2956–2961. doi: 10.3892/ol.2016.4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma AG, Yu LM, Zhao H, Qin CW, Tian XY, Wang Q. PSMD4 regulates the malignancy of esophageal cancer cells by suppressing endoplasmic reticulum stress. Kaohsiung J Med Sci. 2019;35(10):591–597. doi: 10.1002/kjm2.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen WC, Chen MF, Lin PY. Significance of DNMT3b in oral cancer. PLoS One. 2014;9(3):e89956. doi: 10.1371/journal.pone.0089956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Jia Y, Cao Y, Wu S, Jiang H, Sun X, et al. Overexpression of phosphoserine aminotransferase 1 (PSAT1) predicts poor prognosis and associates with tumor progression in human esophageal squamous cell carcinoma. Cell Physiol Biochem. 2016;39(1):395–406. doi: 10.1159/000445633. [DOI] [PubMed] [Google Scholar]
- 20.Cheng R, Zhu S, Guo S, Min L, Xing J, Guo Q, et al. Downregulation of NONO induces apoptosis, suppressing growth and invasion in esophageal squamous cell carcinoma. Oncol Rep. 2018;39(6):2575–2583. doi: 10.3892/or.2018.6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wada S, Noguchi T, Takeno S, Kawahara K. PIK3CA and TFRC located in 3q are new prognostic factors in esophageal squamous cell carcinoma. Ann Surg Oncol. 2006;13(7):961–966. doi: 10.1245/ASO.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 22.He JZ, Wu ZY, Wang SH, Ji X, Yang CX, Xu XE, et al. A decision tree-based combination of ezrin-interacting proteins to estimate the prognostic risk of patients with esophageal squamous cell carcinoma. Hum Pathol. 2017;66:115–125. doi: 10.1016/j.humpath.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Yu X, Li W, Xia Z, Xie L, Ma X, Liang Q, et al. Targeting MCL-1 sensitizes human esophageal squamous cell carcinoma cells to cisplatin-induced apoptosis. BMC Cancer. 2017;17(1):1–13. doi: 10.1186/s12885-017-3442-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Han B, He Z, Zhang Y, Lin K, Su H, Hosseini DK, Sun H, Yang M, Chen X. RNA-Binding Proteins CLK1 and POP7 as Biomarkers for Diagnosis and Prognosis of Esophageal Squamous Cell Carcinoma. Front Cell Dev Biol. 2021;9:715027. doi: 10.3389/fcell.2021.715027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu YT, Wang WJ, Zhang B, Mei LL, Shi ZZ. MCM7 amplification and overexpression promote cell proliferation, colony formation and migration in esophageal squamous cell carcinoma by activating the AKT1/mTOR signaling pathway. Oncol Rep. 2017;37(6):3590–3596. doi: 10.3892/or.2017.5614. [DOI] [PubMed] [Google Scholar]
- 26.Choy B, LaLonde A, Que J, Wu T, Zhou Z. MCM4 and MCM7, potential novel proliferation markers, significantly correlated with Ki-67, Bmi1, and cyclin E expression in esophageal adenocarcinoma, squamous cell carcinoma, and precancerous lesions. Hum Pathol. 2016;57:126–135. doi: 10.1016/j.humpath.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong X, Chen X, Guan X, Zhang H, Ma Y, Zhang S, et al. Overexpression of G9a and MCM7 in oesophageal squamous cell carcinoma is associated with poor prognosis. Histopathology. 2015;66(2):192–200. doi: 10.1111/his.12456. [DOI] [PubMed] [Google Scholar]
- 28.Miyazaki T, Kato H, Fukuchi M, Nakajima M, Kuwano H. EphA2 overexpression correlates with poor prognosis in esophageal squamous cell carcinoma. Int J Cancer. 2003;103(5):657–663. doi: 10.1002/ijc.10860. [DOI] [PubMed] [Google Scholar]
- 29.Ma S, Zhao X. KPNA2 is a promising biomarker candidate for esophageal squamous cell carcinoma and correlates with cell proliferation. Oncol Rep. 2014;32(4):1631–1637. doi: 10.3892/or.2014.3381. [DOI] [PubMed] [Google Scholar]
- 30.Sakai M, Sohda M, Miyazaki T, Suzuki S, Sano A, Tanaka N, et al. Significance of karyopherin- 2 (KPNA2) expression in esophageal squamous cell carcinoma. Anticancer Res. 2010;30(3):851–856. [PubMed] [Google Scholar]
- 31.Wang L, Liu G, Bolor-Erdene E, Li Q, Mei Y, Zhou L. Identification of KIF4A as a prognostic biomarker for esophageal squamous cell carcinoma. Aging (Albany NY) 2021;13(21):24050. doi: 10.18632/aging.203585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun X, Chen P, Chen X, Yang W, Chen X, Zhou W, et al. KIF4A enhanced cell proliferation and migration via Hippo signaling and predicted a poor prognosis in esophageal squamous cell carcinoma. Thorac Cancer. 2021;12(4):512–524. doi: 10.1111/1759-7714.13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kita Y, Nishizono Y, Okumura H, Uchikado Y, Sasaki K, Matsumoto M, et al. Clinical and biological impact of cyclin-dependent kinase subunit 2 in esophageal squamous cell carcinoma. Oncol Rep. 2014;31(5):1986–1992. doi: 10.3892/or.2014.3062. [DOI] [PubMed] [Google Scholar]
- 34.Zheng X, Zhao Y, Wang X, Li Y, Wang R, Jiang Y, et al. Decreased expression of CIAPIN1 is correlated with poor prognosis in patients with esophageal squamous cell carcinoma. Dig Dis Sci. 2010;55(12):3408–3414. doi: 10.1007/s10620-010-1212-7. [DOI] [PubMed] [Google Scholar]
- 35.Zhao Y, Lu Q, Li C, Wang X, Jiang L, Huang L, et al. PRMT1 regulates the tumour-initiating properties of esophageal squamous cell carcinoma through histone H4 arginine methylation coupled with transcriptional activation. Cell Death Dis. 2019;10(5):1–17. doi: 10.1038/s41419-019-1595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou W, Yue H, Li C, Chen H, Yuan Y. Protein arginine methyltransferase 1 promoted the growth and migration of cancer cells in esophageal squamous cell carcinoma. Tumor Biol. 2016;37(2):2613–2619. doi: 10.1007/s13277-015-4098-3. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Lin A, Zhang JG, Bao WG, Xu DP, Ruan YY, et al. Alteration of HLA-F and HLA I antigen expression in the tumor is associated with survival in patients with esophageal squamous cell carcinoma. Int J Cancer. 2013;132(1):82–89. doi: 10.1002/ijc.27621. [DOI] [PubMed] [Google Scholar]
- 38.Yie Sm, Yang H, Ye Sr, Li K, Dong DD, Lin Xm. Expression of HLA-G is associated with prognosis in esophageal squamous cell carcinoma. Am J Clin Pathol. 2007;128(6):1002–1009. doi: 10.1309/JNCW1QLDFB6AM9WE. [DOI] [PubMed] [Google Scholar]
- 39.Sato Y, Motoyama S, Nanjo H, Wakita A, Yoshino K, Sasaki T, et al. CXCL10 expression status is prognostic in patients with advanced thoracic esophageal squamous cell carcinoma. Ann Surg Oncol. 2016;23(3):936–942. doi: 10.1245/s10434-015-4909-1. [DOI] [PubMed] [Google Scholar]
- 40.Yuan H, Zhou W, Yang Y, Xue L, Liu L, Song Y. ISG15 promotes esophageal squamous cell carcinoma tumorigenesis via c-MET/Fyn/ -catenin signaling pathway. Exp Cell Res. 2018;367(1):47–55. doi: 10.1016/j.yexcr.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 41.Yu H, Yao J, Du M, Ye J, He X, Yin L. CDKN3 promotes cell proliferation, invasion and migration by activating the AKT signaling pathway in esophageal squamous cell carcinoma. Oncol Lett. 2020;19(1):542–548. doi: 10.3892/ol.2019.11077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W, Liao K, Guo HC, Zhou S, Yu R, Liu Y, et al. Integrated transcriptomics explored the cancer-promoting genes CDKN3 in esophageal squamous cell cancer. J Cardiothorac Surg. 2021;16(1):1–7. doi: 10.1186/s13019-021-01534-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Min L, Zhu S, Guo Q, Li H, Zhang Z, et al. Cyclin-dependent kinase inhibitor 3 promoted cell proliferation by driving cell cycle from G1 to S phase in esophageal squamous cell carcinoma. J Cancer. 2019;10(8):1915. doi: 10.7150/jca.27053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu J, Li R, Miao H, Wen Z. Identification of key genes for esophageal squamous cell carcinoma via integrated bioinformatics analysis and experimental confirmation. J Thorac Dis. 2020;12(6):3188. doi: 10.21037/jtd.2020.01.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu ZC, Cao K, Xiao ZH, Qiao L, Wang XQ, Shang B, et al. VRK1 promotes cisplatin resistance by up-regulating c-MYC via c-Jun activation and serves as a therapeutic target in esophageal squamous cell carcinoma. Oncotarget. 2017;8(39):65642. doi: 10.18632/oncotarget.20020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J, Shao J, Zhang C, Qin G, Liu J, Li M, et al. Immuno-oncological role of 20S proteasome alpha-subunit 3 in aggravating the progression of esophageal squamous cell carcinoma. Eur J Immunol. 2022;52(2):338–351. doi: 10.1002/eji.202149441. [DOI] [PubMed] [Google Scholar]
- 47.Wang F, Zhang C, Cheng H, Liu C, Lu Z, Zheng S, et al. TGF- -induced PLEK2 promotes metastasis and chemoresistance in oesophageal squamous cell carcinoma by regulating LCN2. Cell Death Dis. 2021;12(10):1–12. doi: 10.1038/s41419-021-04155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qu J, Zhao Q, Yang L, Ping Y, Zhang K, Lei Q, et al. Identification and characterization of prognosis-related genes in the tumor microenvironment of esophageal squamous cell carcinoma. Int Immunopharmacol. 2021;96:107616. doi: 10.1016/j.intimp.2021.107616. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Molavi O, Su M, Lai R. The clinical and biological significance of STAT1 in esophageal squamous cell carcinoma. BMC Cancer. 2014;14(1):1–14. doi: 10.1186/1471-2407-14-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shao N, Han Y, Song L, Song W, et al. Clinical significance of hypoxia-inducible factor 1, and its correlation with p53 and vascular endothelial growth factor expression in resectable esophageal squamous cell carcinoma. J Cancer Res Ther. 2020;16(2):269. doi: 10.4103/jcrt.JCRT_781_19. [DOI] [PubMed] [Google Scholar]
- 51.Hu X, Lin J, Jiang M, He X, Wang K, Wang W, et al. HIF-1 promotes the metastasis of esophageal squamous cell carcinoma by targeting SP1. J Cancer. 2020;11(1):229. doi: 10.7150/jca.35537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bolidong D, Domoto T, Uehara M, Sabit H, Okumura T, Endo Y, et al. Potential therapeutic effect of targeting glycogen synthase kinase 3in esophageal squamous cell carcinoma. Sci Rep. 2020;10(1):1–13. doi: 10.1038/s41598-020-68713-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao S, Li S, Duan X, Gu Z, Ma Z, Yuan X, et al. Inhibition of glycogen synthase kinase 3 beta (GSK3) suppresses the progression of esophageal squamous cell carcinoma by modifying STAT3 activity. Mol Carcinog. 2017;56(10):2301–2316. doi: 10.1002/mc.22685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kato K, Hida Y, Miyamoto M, Hashida H, Shinohara T, Itoh T, et al. Overexpression of caveolin-1 in esophageal squamous cell carcinoma correlates with lymph node metastasis and pathologic stage. Cancer. 2002;94(4):929–933. doi: 10.1002/cncr.10329. [DOI] [PubMed] [Google Scholar]
- 55.Ando T, Ishiguro H, Kimura M, Mitsui A, Mori Y, Sugito N, et al. The overexpression of caveolin-1 and caveolin-2 correlates with a poor prognosis and tumor progression in esophageal squamous cell carcinoma. Oncol Rep. 2007;18(3):601–609. [PubMed] [Google Scholar]
- 56.Lu F, Chen W, Jiang T, Cheng C, Wang B, Lu Z, et al. Expression profile, clinical significance and biological functions of IGF2BP2 in esophageal squamous cell carcinoma. Exp Ther Med. 2022;23(4):1–11. doi: 10.3892/etm.2022.11177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shu W, Lin Y, Yan Y, Sun Y, Wu X, Cao Q. IGF2BP2 Promotes the Proliferation, Invasion and Migration of Esophageal Carcinoma Cells via Activation of the PI3K/AKT/EMT Signaling Pathway. Research Square; 2021. 10.21203/rs.3.rs-711778/v1.
- 58.Tanaka T, Ishiguro H, Kuwabara Y, Kimura M, Mitsui A, Katada T, et al. Vascular endothelial growth factor C (VEGF-C) in esophageal cancer correlates with lymph node metastasis and poor patient prognosis. J Exp Clin Cancer Res. 2010;29(1):1–7. doi: 10.1186/1756-9966-29-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kimura Y, Watanabe M, Ohga T, Saeki H, Kakeji Y, Baba H, et al. Vascular endothelial growth factor C expression correlates with lymphatic involvement and poor prognosis in patients with esophageal squamous cell carcinoma. Oncol Rep. 2003;10(6):1747–1751. doi: 10.3892/or.10.6.1747. [DOI] [PubMed] [Google Scholar]
- 60.Feng YF, Lei YY, Lu JB, Xi SY, Zhang Y, Huang QT, et al. RIT1 suppresses esophageal squamous cell carcinoma growth and metastasis and predicts good prognosis. Cell Death Dis. 2018;9(11):1–13. doi: 10.1038/s41419-018-0979-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhou Y, Wang L, Ban X, Zeng T, Zhu Y, Li M, et al. DHRS2 inhibits cell growth and motility in esophageal squamous cell carcinoma. Oncogene. 2018;37(8):1086–1094. doi: 10.1038/onc.2017.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li Y, Chen L, Nie Cj, Zeng Tt, Liu H, Mao X, et al. Downregulation of RBMS3 is associated with poor prognosis in esophageal squamous cell carcinoma. Cancer Res. 2011;71(19):6106–6115. doi: 10.1158/0008-5472.CAN-10-4291. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Liu Y, Leng X, Cao K, Sun W, Zhu J, Ma J. UBE2T Contributes to the Prognosis of Esophageal Squamous Cell Carcinoma. Pathol. Oncol. Res. 2021;27:632531. doi: 10.3389/pore.2021.632531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao J, Zou Z, Gao J, Zhang H, Lin Z, Zhang Y, et al. Increased expression of HMGB3: a novel independent prognostic marker of worse outcome in patients with esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2015;8(1):345. [PMC free article] [PubMed] [Google Scholar]
- 65.Huang J, Wang X, Zhang X, Chen W, Luan L, Song Q, et al. CDK4 amplification in esophageal squamous cell carcinoma associated with better patient outcome. Front Genet. 2021;12:375. doi: 10.3389/fgene.2021.616110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ling Z, Zhao Q, Zhou S, Mao W. MSH2 promoter hypermethylation in circulating tumor DNA is a valuable predictor of disease-free survival for patients with esophageal squamous cell carcinoma. Eur J Surg Oncol (EJSO) 2012;38(4):326–332. doi: 10.1016/j.ejso.2012.01.008. [DOI] [PubMed] [Google Scholar]
- 67.Xu Y, Wang N, Liu R, Lv H, Li Z, Zhang F, et al. Epigenetic study of esophageal carcinoma based on methylation, gene integration and weighted correlation network analysis. Onco Targets Ther. 2021;14:3133. doi: 10.2147/OTT.S298620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen X, Lu B, Ma Q, Ji CD, Li JZ. EphA3 inhibits migration and invasion of esophageal cancer cells by activating the mesenchymal-epithelial transition process. Int J Oncol. 2019;54(2):722–732. doi: 10.3892/ijo.2018.4639. [DOI] [PubMed] [Google Scholar]
- 69.Luo LL, Zhao L, Wang YX, Tian XP, Xi M, Shen JX, et al. Insulin-like growth factor binding protein-3 is a new predictor of radiosensitivity on esophageal squamous cell carcinoma. Sci Rep. 2015;5(1):1–12. doi: 10.1038/srep17336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao L, Li QQ, Zhang R, Xi M, Liao YJ, Qian D, et al. The overexpression of IGFBP-3 is involved in the chemosensitivity of esophageal squamous cell carcinoma cells to nimotuzumab combined with cisplatin. Tumor Biol. 2012;33(4):1115–1123. doi: 10.1007/s13277-012-0352-0. [DOI] [PubMed] [Google Scholar]
- 71.Luo LL, Zhao L, Xi M, He LR, Shen JX, Li QQ, et al. Association of insulin-like growth factor-binding protein-3 with radiotherapy response and prognosis of esophageal squamous cell carcinoma. Chin J Cancer. 2015;34(3):1–8. doi: 10.1186/s40880-015-0046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Cheng G, Zhang T, Deng L, Xu K, Xu X, et al. CHST15 promotes the proliferation of TE-1 cells via multiple pathways in esophageal cancer. Oncol Rep. 2020;43(1):75–86. doi: 10.3892/or.2019.7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tripathi SC, Matta A, Kaur J, Grigull J, Chauhan SS, Thakar A, et al. Overexpression of prothymosin alpha predicts poor disease outcome in head and neck cancer. PLoS One. 2011;6(5):e19213. doi: 10.1371/journal.pone.0019213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tan KD, Zhu Y, Tan HK, Rajasegaran V, Aggarwal A, Wu J, et al. Amplification and overexpression of PPFIA1, a putative 11q13 invasion suppressor gene, in head and neck squamous cell carcinoma. Genes Chromosom Cancer. 2008;47(4):353–362. doi: 10.1002/gcc.20539. [DOI] [PubMed] [Google Scholar]
- 75.Al-Rawi NH, Merza MS, Ghazi AM, et al. PIK3CB and K-ras in oral squamous Cell carcinoma. A possible cross-talk! J Orofac Sci. 2014;6(2):99. doi: 10.4103/0975-8844.143049. [DOI] [Google Scholar]
- 76.Liang Y, Lei Y, Du M, Liang M, Liu Z, Li X, et al. The increased expression and aberrant methylation of SHC1 in non-small cell lung cancer: Integrative analysis of clinical and bioinformatics databases. J Cell Mol Med. 2021;25(14):7039–7051. doi: 10.1111/jcmm.16717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xue H, Lu J, Yuan R, Liu J, Liu Y, Wu K, et al. Knockdown of CLIC4 enhances ATP-induced HN4 cell apoptosis through mitochondrial and endoplasmic reticulum pathways. Cell Biosci. 2016;6(1):1–9. doi: 10.1186/s13578-016-0070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen MF, Lu MS, Lin PY, Chen PT, Chen WC, Lee KD. The role of DNA methyltransferase 3b in esophageal squamous cell carcinoma. Cancer. 2012;118(16):4074–4089. doi: 10.1002/cncr.26736. [DOI] [PubMed] [Google Scholar]
- 79.Liu Z, Wang L, Wang LE, Sturgis EM, Wei Q. Polymorphisms of the DNMT3B gene and risk of squamous cell carcinoma of the head and neck: a case-control study. Cancer Lett. 2008;268(1):158–165. doi: 10.1016/j.canlet.2008.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ohmura G, Tsujikawa T, Yaguchi T, Kawamura N, Mikami S, Sugiyama J, et al. Aberrant myosin 1b expression promotes cell migration and lymph node metastasis of HNSCC. Mol Cancer Res. 2015;13(4):721–731. doi: 10.1158/1541-7786.MCR-14-0410. [DOI] [PubMed] [Google Scholar]
- 81.Liang J, Sun Z. Overexpression of membranal SLC3A2 regulates the proliferation of oral squamous cancer cells and affects the prognosis of oral cancer patients. J Oral Pathol Med. 2021;50(4):371–377. doi: 10.1111/jop.13132. [DOI] [PubMed] [Google Scholar]
- 82.Ding X, Sun J, Wang L, Li G, Shen Y, Zhou X, et al. Overexpression of SENP5 in oral squamous cell carcinoma and its association with differentiation. Oncol Rep. 2008;20(5):1041–1045. doi: 10.3892/or_00000107. [DOI] [PubMed] [Google Scholar]
- 83.Ueda S, Hashimoto K, Miyabe S, Hasegawa S, Goto M, Shimizu D, et al. Salivary NUS1 and RCN1 levels as biomarkers for oral squamous cell carcinoma diagnosis. In Vivo. 2020;34(5):2353–2361. doi: 10.21873/invivo.12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhu MH, Ji SL, Zhang CY, Cui L, Xiong L, Zheng HL. DNA microarray reveals ZNF195 and SBF1 are potential biomarkers for gemcitabine sensitivity in head and neck squamous cell carcinoma cell lines. Int J Clin Exp Pathol. 2014;7(4):1514. [PMC free article] [PubMed] [Google Scholar]
- 85.Maji S, Samal SK, Pattanaik L, Panda S, Quinn BA, Das SK, et al. Mcl-1 is an important therapeutic target for oral squamous cell carcinomas. Oncotarget. 2015;6(18):16623. doi: 10.18632/oncotarget.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Feng CJ, Li HJ, Li JN, Lu YJ, Liao GQ. Expression of MCM7 and CDC6 in oral squamous cell carcinoma and precancerous lesions. Anticancer Res. 2008;28(6A):3763–3769. [PubMed] [Google Scholar]
- 87.Ma H, Li L, Jia L, Gong A, Wang A, Zhang L, et al. POM121 is identified as a novel prognostic marker of oral squamous cell carcinoma. J Cancer. 2019;10(19):4473. doi: 10.7150/jca.33368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang Z, Xiong H, Zuo Y, Hu S, Zhu C, Min A. PSMC2 knockdown inhibits the progression of oral squamous cell carcinoma by promoting apoptosis via PI3K/Akt pathway. Cell Cycle. 2022;21(5):477–488. doi: 10.1080/15384101.2021.2021722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Faoro L, Singleton PA, Cervantes GM, Lennon FE, Choong NW, Kanteti R, et al. EphA2 mutation in lung squamous cell carcinoma promotes increased cell survival, cell invasion, focal adhesions, and mammalian target of rapamycin activation. J Biol Chem. 2010;285(24):18575–18585. doi: 10.1074/jbc.M109.075085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu Y, Zhang X, Qiu Y, Huang D, Zhang S, Xie L, et al. Clinical significance of EphA2 expression in squamous-cell carcinoma of the head and neck. J Cancer Res Clin Oncol. 2011;137(5):761–769. doi: 10.1007/s00432-010-0936-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rivera RS, Gunduz M, Nagatsuka H, Gunduz E, Cengiz B, Fukushima K, et al. Involvement of EphA2 in head and neck squamous cell carcinoma: mRNA expression, loss of heterozygosity and immunohistochemical studies. Oncol Rep. 2008;19(5):1079–1084. doi: 10.3892/or.19.5.1079. [DOI] [PubMed] [Google Scholar]
- 92.Lin F, Gao L, Su Z, Cao X, Zhan Y, Li Y, et al. Knockdown of KPNA2 inhibits autophagy in oral squamous cell carcinoma cell lines by blocking p53 nuclear translocation. Oncol Rep. 2018;40(1):179–194. doi: 10.3892/or.2018.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Minakawa Y, Kasamatsu A, Koike H, Higo M, Nakashima D, Kouzu Y, et al. Kinesin family member 4A: a potential predictor for progression of human oral cancer. PLoS One. 2013;8(12):e85951. doi: 10.1371/journal.pone.0085951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dong Y, Lu S, Wang Z, Liu L. CCTs as new biomarkers for the prognosis of head and neck squamous cancer. Open Med. 2020;15(1):672–688. doi: 10.1515/med-2020-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gao F, Li C, Zhao X, Xie J, Fang G, Li Y. CKS2 modulates cell-cycle progression of tongue squamous cell carcinoma cells partly via modulating the cellular distribution of DUTPase. J Oral Pathol Med. 2021;50(2):175–182. doi: 10.1111/jop.13116. [DOI] [PubMed] [Google Scholar]
- 96.Chuang CY, Chang CP, Lee YJ, Lin WL, Chang WW, Wu JS, et al. PRMT1 expression is elevated in head and neck cancer and inhibition of protein arginine methylation by adenosine dialdehyde or PRMT1 knockdown downregulates proliferation and migration of oral cancer cells. Oncol Rep. 2017;38(2):1115–1123. doi: 10.3892/or.2017.5737. [DOI] [PubMed] [Google Scholar]
- 97.Wu T, Jiao Z, Li Y, Su X, Yao F, Peng J, et al. HPRT1 promotes chemoresistance in oral squamous cell carcinoma via activating MMP1/PI3K/Akt signaling pathway. Cancers. 2022;14(4):855. doi: 10.3390/cancers14040855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ahmadi M, Eftekhari Kenzerki M, Akrami SM, Pashangzadeh S, Hajiesmaeili F, Rahnavard S, et al. Overexpression of HPRT1 is associated with poor prognosis in head and neck squamous cell carcinoma. FEBS Open Bio. 2021;11(9):2525–2540. doi: 10.1002/2211-5463.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang G, Zhao X, Liu W. NEDD4L inhibits glycolysis and proliferation of cancer cells in oral squamous cell carcinoma by inducing ENO1 ubiquitination and degradation. Cancer Biol Ther. 2022;23(1):243–253. doi: 10.1080/15384047.2022.2054244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Acker F, Stratmann J, Aspacher L, Nguyen NTT, Wagner S, Serve H, Wild PJ, Sebastian M. KRAS Mutations in Squamous Cell Carcinomas of the Lung. Front Oncol. 2021;11:788084. doi: 10.3389/fonc.2021.788084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vallejo-Díaz J, Olazabal-Morán M, Cariaga-Martínez AE, Pajares MJ, Flores JM, Pio R, et al. Targeted depletion of PIK3R2 induces regression of lung squamous cell carcinoma. Oncotarget. 2016;7(51):85063. doi: 10.18632/oncotarget.13195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Attaran N, Gu X, Coates PJ, Fåhraeus R, Boldrup L, Wilms T, et al. Downregulation of TAP1 in tumor-free tongue contralateral to squamous cell carcinoma of the oral tongue, an indicator of better survival. Int J Mol Sci. 2020;21(17):6220. doi: 10.3390/ijms21176220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pidugu VK, Wu MM, Yen AH, Pidugu HB, Chang KW, Liu CJ, et al. IFIT1 and IFIT3 promote oral squamous cell carcinoma metastasis and contribute to the anti-tumor effect of gefitinib via enhancing p-EGFR recycling. Oncogene. 2019;38(17):3232–3247. doi: 10.1038/s41388-018-0662-9. [DOI] [PubMed] [Google Scholar]
- 104.Lee TC, Pidugu VK, Wu MM, Liu CJ. IFIT1 and IFIT3 modulate the drug response via enhancing EGFR signaling and in human oral squamous cell carcinoma cells. In: Proceedings for Annual Meeting of The Japanese Pharmacological Society WCP2018 (The 18th World Congress of Basic and Clinical Pharmacology), pp. PO4–6, 2018, Japanese Pharmacological Society.
- 105.Sarmah N, Baruah MN, Baruah S. Immune modulation in HLA-G expressing head and neck squamous cell carcinoma in relation to human papilloma virus positivity: a study from Northeast India. Front Oncol. 2019;9:58. doi: 10.3389/fonc.2019.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bora M, Sarmah N, Das B, Baruah MN, Deka G, Hazarika SG, Baruah S. A comparative study on regulation of HLA-G expression in bad obstetric history and in head and neck squamous cell carcinoma from Northeast India. Human Immunol. 2022;83(5):453–457. doi: 10.1016/j.humimm.2022.02.006. [DOI] [PubMed] [Google Scholar]
- 107.Imani R, Seyedmajidi M, Ghasemi N, Moslemi D, Shafaee S, Bijani A. HLA-G Expression is Associated with an Unfavorable Prognosis of Oral Squamous Cell Carcinoma. Asian Pac J Cancer Prev. 2018;19(9):2527–2533. doi: 10.22034/APJCP.2018.19.9.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ou D, Wu Y. The prognostic and clinical significance of IFI44L aberrant downregulation in patients with oral squamous cell carcinoma. BMC Cancer. 2021;21(1):1327. doi: 10.1186/s12885-021-09058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rentoft M, Coates PJ, Loljung L, Wilms T, Laurell G, Nylander K. Expression of CXCL10 is associated with response to radiotherapy and overall survival in squamous cell carcinoma of the tongue. Tumour Biol. 2014;35(5):4191–8. doi: 10.1007/s13277-013-1549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li Y, Wu T, Gong S, Zhou H, Yu L, Liang M, Shi R, Wu Z, Zhang J, Li S. Analysis of the Prognosis and Therapeutic Value of the CXC Chemokine Family in Head and Neck Squamous Cell Carcinoma. Front Oncol. 2021;10:570736. doi: 10.3389/fonc.2020.570736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Li H, Yang LL, Wu CC, Xiao Y, Mao L, Chen L, Zhang WF, Sun ZJ. Expression and Prognostic Value of IFIT1 and IFITM3 in Head and Neck Squamous Cell Carcinoma. Am J Clin Pathol. 2020;153(5):618–629. doi: 10.1093/ajcp/aqz205. [DOI] [PubMed] [Google Scholar]
- 112.Laljee RP, Muddaiah S, Salagundi B, Cariappa PM, Indra AS, Sanjay V, Ramanathan A. Interferon stimulated gene-ISG15 is a potential diagnostic biomarker in oral squamous cell carcinomas. Asian Pac J Cancer Prev. 2013;14(2):1147–50. doi: 10.7314/apjcp.2013.14.2.1147. [DOI] [PubMed] [Google Scholar]
- 113.Zhang Q, He Y, Nie M, Cai W. Roles of miR-138 and ISG15 in oral squamous cell carcinoma. Exp Ther Med. 2017;14(3):2329–2334. doi: 10.3892/etm.2017.4720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Santos CR, Rodríguez-Pinilla M, Vega FM, Rodríguez-Peralto JL, Blanco S, Sevilla A, Valbuena A, Hernández T, van Wijnen AJ, Li F, de Alava E, Sánchez-Céspedes M, Lazo PA. VRK1 signaling pathway in the context of the proliferation phenotype in head and neck squamous cell carcinoma. Mol Cancer Res. 2006;4(3):177–85. doi: 10.1158/1541-7786.MCR-05-0212. [DOI] [PubMed] [Google Scholar]
- 115.Cao X, Luan K, Yang J, Huang Y. Targeting lncRNA PSMA3-AS1, a Prognostic Marker, Suppresses Malignant Progression of Oral Squamous Cell Carcinoma. Dis Markers. 2021;2021:3138046. doi: 10.1155/2021/3138046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang J, Sun Z, Wang J, Tian Q, Huang R, Wang H, Wang X, Han F. Expression and prognostic potential of PLEK2 in head and neck squamous cell carcinoma based on bioinformatics analysis. Cancer Med. 2021;10(18):6515–6533. doi: 10.1002/cam4.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Troiano G, Caponio VCA, Adipietro I, Tepedino M, Santoro R, Laino L, Lo Russo L, Cirillo N, Lo Muzio L. Prognostic significance of CD68+ and CD163+ tumor associated macrophages in head and neck squamous cell carcinoma: A systematic review and meta-analysis. Oral Oncol. 2019;93:66–75. doi: 10.1016/j.oraloncology.2019.04.019. [DOI] [PubMed] [Google Scholar]
- 118.Kubota K, Moriyama M, Furukawa S, Rafiul HASM, Maruse Y, Jinno T, Tanaka A, Ohta M, Ishiguro N, Yamauchi M, Sakamoto M, Maehara T, Hayashida JN, Kawano S, Kiyoshima T, Nakamura S. CD163+CD204+ tumor-associated macrophages contribute to T cell regulation via interleukin-10 and PD-L1 production in oral squamous cell carcinoma. Sci Rep. 2017;7(1):1755. 10.1038/s41598-017-01661-z. [DOI] [PMC free article] [PubMed]
- 119.He KF, Zhang L, Huang CF, Ma SR, Wang YF, Wang WM, Zhao ZL, Liu B, Zhao YF, Zhang WF, Sun ZJ. CD163+ tumor-associated macrophages correlated with poor prognosis and cancer stem cells in oral squamous cell carcinoma. Biomed Res Int. 2014;2014:838632. doi: 10.1155/2014/838632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang X, Dong Y, Zhao M, Ding L, Yang X, Jing Y, Song Y, Chen S, Hu Q, Ni Y. ITGB2-mediated metabolic switch in CAFs promotes OSCC proliferation by oxidation of NADH in mitochondrial oxidative phosphorylation system. Theranostics. 2020;10(26):12044–12059. doi: 10.7150/thno.47901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Lu S, Li N, Peng Z, Lu Z, Tu X, Zhang W, Kang Y. Fc fragment of immunoglobulin G receptor IIa (FCGR2A) as a new potential prognostic biomarker of esophageal squamous cell carcinoma. Chin Med J (Engl). 2021;135(4):482–484. doi: 10.1097/CM9.0000000000001776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dai Y, Chen W, Huang J, Cui T. FCGR2A Could Function as a Prognostic Marker and Correlate with Immune Infiltration in Head and Neck Squamous Cell Carcinoma. Biomed Res Int. 2021;2021:8874578. doi: 10.1155/2021/8874578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Magnes T, Melchardt T, Hufnagl C, Weiss L, Mittermair C, Neureiter D, Klieser E, Rinnerthaler G, Roesch S, Gaggl A, Greil R, Egle A. The influence of FCGR2A and FCGR3A polymorphisms on the survival of patients with recurrent or metastatic squamous cell head and neck cancer treated with cetuximab. Pharmacogenomics J. 2018; 18(3):474-479. 10.1038/tpj.2017.37. [DOI] [PubMed]
- 124.Santos Md, Mercante AMdC, Louro ID, Gonçalves AJ, Carvalho MBd, da Silva EHT, et al. HIF1-Alpha Expression Predicts Survival of Patients with Squamous Cell Carcinoma of the Oral Cavity. PLoS ONE. 2012;7(9):e45228. doi: 10.1371/journal.pone.0045228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhou J, Huang S, Wang L, Yuan X, Dong Q, Zhang D, et al. Clinical and prognostic significance of HIF-1 overexpression in oral squamous cell carcinoma: a meta-analysis. World J Surg Oncol. 2017;15(1):1–8. doi: 10.1186/s12957-017-1163-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Fillies T, Werkmeister R, van Diest PJ, Brandt B, Joos U, Buerger H. HIF1-alpha overexpression indicates a good prognosis in early stage squamous cell carcinomas of the oral floor. BMC Cancer. 2005;5:84. doi: 10.1186/1471-2407-5-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liang J, Zhang Z, Liang L, Shen Y, Ouyang K. HIF-1α regulated tongue squamous cell carcinoma cell growth via regulating VEGF expression in a xenograft model. Ann Transl Med. 2014;2(9):92. doi: 10.3978/j.issn.2305-5839.2014.08.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Routray S. Caveolin-1 in oral squamous cell carcinoma microenvironment: an overview. Tumour Biol. 2014;35(10):9487–95. doi: 10.1007/s13277-014-2482-z. [DOI] [PubMed] [Google Scholar]
- 129.Jung AC, Ray AM, Ramolu L, Macabre C, Simon F, Noulet F, Blandin AF, Renner G, Lehmann M, Choulier L, Kessler H, Abecassis J, Dontenwill M, Martin S. Caveolin-1-negative head and neck squamous cell carcinoma primary tumors display increased epithelial to mesenchymal transition and prometastatic properties. Oncotarget. 2015;6(39):41884–901. doi: 10.18632/oncotarget.6099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Xue J, Chen H, Diao L, Chen X, Xia D. Expression of Caveolin-1 in tongue squamous cell carcinoma by quantum dots. Eur J Histochem. 2010;54(2):e20. doi: 10.4081/ejh.2010.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhou L, Li H, Cai H, Liu W, Pan E, Yu D, He S. Upregulation of IGF2BP2 Promotes Oral Squamous Cell Carcinoma Progression That Is Related to Cell Proliferation, Metastasis and Tumor-Infiltrating Immune Cells. Front Oncol. 2022;12:809589. doi: 10.3389/fonc.2022.809589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wang X, Xu H, Zhou Z, Guo S, Chen R. IGF2BP2 maybe a novel prognostic biomarker in oral squamous cell carcinoma. Biosci Rep. 2022;42(2):BSR20212119. doi: 10.1042/BSR20212119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Rapone B, Ferrara E. Vascular Endothelial Growth Factor Expression in the Pathological Angiogenesis in Oral Squamous Cell Carcinoma. In: Sridharan G, Sukumaran A, Al-Ostwani AEO, editors. Oral Diseases. Rijeka: IntechOpen; 2020
- 134.Almeida ADS, Oliveira DT, Pereira MC, Faustino SES, Nonogaki S, Carvalho AL, et al. Podoplanin and VEGF-C immunoexpression in oral squamous cell carcinomas: prognostic significance. Anticancer Res. 2013;33(9):3969–3976. [PubMed] [Google Scholar]
- 135.Na J, Zhou W, Yin M, Hu Y, Ma X. GNA13 promotes the proliferation and migration of lung squamous cell carcinoma cells through regulating the PI3K/AKT signaling pathway. Tissue Cell. 2022;76:101795. doi: 10.1016/j.tice.2022.101795. [DOI] [PubMed] [Google Scholar]
- 136.Liang Yn, Liu Y, Meng Q, Li X, Wang F, Yao G, et al. RBMS3 is a tumor suppressor gene that acts as a favorable prognostic marker in lung squamous cell carcinoma. Med Oncol. 2015;32(2):1–9. doi: 10.1007/s12032-014-0459-9. [DOI] [PubMed] [Google Scholar]
- 137.Wang M, Xie T, Wu Y, Yin Q, Xie S, Yao Q, et al. Identification of RFC5 as a novel potential prognostic biomarker in lung cancer through bioinformatics analysis. Oncol Lett. 2018;16(4):4201–4210. doi: 10.3892/ol.2018.9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Simonik EA, Cai Y, Kimmelshue KN, Brantley-Sieders DM, Loomans HA, Andl CD, et al. LIM-only protein 4 (LMO4) and LIM domain binding protein 1 (LDB1) promote growth and metastasis of human head and neck cancer (LMO4 and LDB1 in head and neck cancer) PLoS One. 2016;11(10):e0164804. doi: 10.1371/journal.pone.0164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kwong RA, Scarlett CJ, Kalish LH, Cole IE, Kench JG, Sum EY, Musgrove EA, Henshall SM, Lindeman GJ, Biankin AV, Visvader JE, Sutherland RL. LMO4 expression in squamous cell carcinoma of the anterior tongue. Histopathology. 2011;58(3):477–80. doi: 10.1111/j.1365-2559.2011.03765.x. [DOI] [PubMed] [Google Scholar]
- 140.van Caloen G, Machiels JP. Potential role of cyclin-dependent kinase 4/6 inhibitors in the treatment of squamous cell carcinoma of the head and neck. Curr Opin Oncol. 2019;31(3):122–130. doi: 10.1097/CCO.0000000000000513. [DOI] [PubMed] [Google Scholar]
- 141.Ku BM, Yi SY, Koh J, Bae YH, Sun JM, Lee SH, Ahn JS, Park K, Ahn MJ. The CDK4/6 inhibitor LY2835219 has potent activity in combination with mTOR inhibitor in head and neck squamous cell carcinoma. Oncotarget. 2016;7(12):14803–13. doi: 10.18632/oncotarget.7543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lourenco GJ, Nogueira GAS, Oliveira CBM, Marson FAL, Lopes-Aguiar L, Costa EFD, Lima TRP, Liutti VT, Leal F, Santos VA, Rinck JA, Lima CSP. MSH3 and EXO1 polymorphisms and head and neck squamous cell carcinoma risk and prognosis. Journal of Clinical Oncology. 2015;33(15_suppl):6063–6063. doi: 10.1200/jco.2015.33.15_suppl.6063. [DOI] [Google Scholar]
- 143.Pereira CS, Oliveira MV, Barros LO, Bandeira GA, Santos SH, Basile JR, Guimarães AL, De Paula AM. Low expression of MSH2 DNA repair protein is associated with poor prognosis in head and neck squamous cell carcinoma. J Appl Oral Sci. 2013;21(5):416–21. doi: 10.1590/1679-775720130206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang Y, Lu Z, Hu Z, Zheng A, Wang F, Xu Y, et al. The up-regulation of TANK-binding kinase 1 in head and neck squamous cell carcinoma. Differentiation. 2017;1(14):46–7. [Google Scholar]
- 145.Zhang H, Liu J, Fu X, Yang A. Identification of Key Genes and Pathways in Tongue Squamous Cell Carcinoma Using Bioinformatics Analysis. Med Sci Monit. 2017;23:5924–5932. doi: 10.12659/msm.905035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yuan Y, Xie X, Jiang Y, Wei Z, Wang P, Chen F, et al. LRP6 is identified as a potential prognostic marker for oral squamous cell carcinoma via MALDI-IMS. Cell Death Dis. 2017;8(9):e3035–e3035. doi: 10.1038/cddis.2017.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shen Z, Hu Y, Zhou C, Yuan J, Xu J, Hao W, et al. ESRRG promoter hypermethylation as a diagnostic and prognostic biomarker in laryngeal squamous cell carcinoma. J Clin Lab Anal. 2019;33(6):e22899. doi: 10.1002/jcla.22899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fukushima R, Kasamatsu A, Nakashima D, Higo M, Fushimi K, Kasama H, et al. Overexpression of translocation associated membrane protein 2 leading to cancer-associated matrix metalloproteinase activation as a putative metastatic factor for human oral cancer. J Cancer. 2018;9(18):3326. doi: 10.7150/jca.25666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang SH, Chen YL, Hsiao JR, Tsai FY, Jiang SS, Lee AYL, et al. Insulin-like growth factor binding protein 3 promotes radiosensitivity of oral squamous cell carcinoma cells via positive feedback on NF-B/IL-6/ROS signaling. J Exp Clin Cancer Res. 2021;40(1):1–18. doi: 10.1186/s13046-021-01898-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sakata J, Hirosue A, Yoshida R, Matsuoka Y, Kawahara K, Arita H, et al. Enhanced expression of IGFBP-3 reduces radiosensitivity and is associated with poor prognosis in oral squamous cell carcinoma. Cancers. 2020;12(2):494. doi: 10.3390/cancers12020494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hano K, Hatano K, Saigo C, Kito Y, Shibata T, Takeuchi T. Combination of Clptm1L and TMEM207 expression as a robust prognostic marker in oral squamous cell carcinoma. Front Oral Health. 2021;2:12. doi: 10.3389/froh.2021.638213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hou Y, Xue F, Fu Y, Feng G, Wang R, Yuan H. CLPTM1L is a novel putative oncogene promoting tumorigenesis in oral squamous cell carcinoma. Cell Transplant. 2021;30:09636897211045970. doi: 10.1177/09636897211045970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Iwasawa S, Yamano Y, Takiguchi Y, Tanzawa H, Tatsumi K, Uzawa K. Upregulation of thioredoxin reductase 1 in human oral squamous cell carcinoma. Oncol Rep. 2011;25(3):637–644. doi: 10.3892/or.2010.1131. [DOI] [PubMed] [Google Scholar]
- 154.Feng J, Han B, Yu C, Shen C, Wen Z. Co-expression Network Identification and Clinical Prognostic Evaluation of Hub Genes in Head and Neck Squamous Cell Carcinoma. Research Square; 2020. 10.21203/rs.3.rs-77378/v1.
- 155.Chien HT, Cheng SD, Chuang WY, Liao CT, Wang HM, Huang SF. Clinical implications of FADD gene amplification and protein overexpression in Taiwanese oral cavity squamous cell carcinomas. PLoS One. 2016;11(10):e0164870. doi: 10.1371/journal.pone.0164870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.González-Moles MÁ, Ayén Á, González-Ruiz I, de Porras-Carrique T, González-Ruiz L, Ruiz-Ávila I, et al. Prognostic and clinicopathological significance of FADD upregulation in head and neck squamous cell carcinoma: a systematic review and meta-analysis. Cancers. 2020;12(9):2393. doi: 10.3390/cancers12092393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rasamny JJ, Allak A, Krook KA, Jo VY, Policarpio-Nicolas ML, Sumner HM, et al. Cyclin D1 and FADD as biomarkers in head and neck squamous cell carcinoma. Otolaryngol-Head Neck Surg. 2012;146(6):923–931. doi: 10.1177/0194599811435052. [DOI] [PubMed] [Google Scholar]
- 158.Knitz MW, Darragh LB, Bickett TE, Bhatia S, Bukkapatnam S, Gadwa J, et al. Loss of cancer cell STAT1 improves response to radiation therapy and promotes T cell activation in head and neck squamous cell carcinoma. Cancer Immunol Immunother. 2022;71(5):1049–1061. doi: 10.1007/s00262-021-03059-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xi S, Dyer KF, Kimak M, Zhang Q, Gooding WE, Chaillet JR, et al. Decreased STAT1 expression by promoter methylation in squamous cell carcinogenesis. J Natl Cancer Inst. 2006;98(3):181–189. doi: 10.1093/jnci/djj020. [DOI] [PubMed] [Google Scholar]
- 160.Tomasovic-Loncaric C, Fucic A, Andabak A, Andabak M, Ceppi M, Bruzzone M, et al. Androgen receptor as a biomarker of oral squamous cell carcinoma progression risk. Anticancer Res. 2019;39(8):4285–4289. doi: 10.21873/anticanres.13593. [DOI] [PubMed] [Google Scholar]
- 161.Liu X, Qing S, Che K, Li L, Liao X. Androgen receptor promotes oral squamous cell carcinoma cell migration by increasing EGFR phosphorylation. Onco Targets Ther. 2019;12:4245. doi: 10.2147/OTT.S200718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kordi-Tamandani D, Sabers E, Jamali S, Rigi Ladiz M. ERK and RAF1 genes: analysis of methylation and expression profiles in patients with oral squamous cell carcinoma. Br J Biomed Sci. 2014;71(3):100–103. doi: 10.1080/09674845.2014.11669972. [DOI] [PubMed] [Google Scholar]
- 163.Mishra R, Nagini S, Rana A. Expression and inactivation of glycogen synthase kinase 3 alpha/beta and their association with the expression of cyclin D1 and p53 in oral squamous cell carcinoma progression. Mol Cancer. 2015;14(1):1–16. doi: 10.1186/s12943-015-0300-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Matsuo FS, Andrade MF, Loyola AM, da Silva SJ, Silva MJB, Cardoso SV, et al. Pathologic significance of AKT, mTOR, and GSK3 proteins in oral squamous cell carcinoma-affected patients. Virchows Arch. 2018;472(6):983–997. doi: 10.1007/s00428-018-2318-0. [DOI] [PubMed] [Google Scholar]
- 165.Jain AP, Patel K, Pinto S, Radhakrishnan A, Nanjappa V, Kumar M, et al. MAP2K1 is a potential therapeutic target in erlotinib resistant head and neck squamous cell carcinoma. Sci Rep. 2019;9(1):1–12. doi: 10.1038/s41598-019-55208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Patowary P, Bhattacharyya DK, Barah P. Identifying critical genes in esophageal squamous cell carcinoma using an ensemble approach. Inf Med Unlock. 2020;18:100277. doi: 10.1016/j.imu.2019.100277. [DOI] [Google Scholar]