ABSTRACT
Salmonella enterica subsp. enterica serovar Infantis is the most prevalent serovar found in broilers and broiler meat and is among the top five serovars responsible for human infections in Europe. In 2008, a multidrug-resistant S. Infantis isolate emerged in Israel with a mosaic megaplasmid named pESI, associated with increased virulence, biofilm formation, and multidrug resistance. Since then, S. Infantis clones with pESI-like plasmids have been reported worldwide, replacing pESI-free clones. Here, we typed 161 S. Infantis isolates of poultry (n = 133) and human clinical (n = 28) origin using whole-genome sequencing. The isolates were collected between 2007 and 2021. In addition, we performed PacBio/Illumina sequencing for two representative pESI-like plasmids and compared them with publicly available sequences. All isolates belonged to sequence type 32 (ST32), except for one isolate that represented a novel single-locus variant of ST32. Core genome MLST (cgMLST) analysis revealed 14 clusters of genetically closely related isolates, of which four suggested broiler-to-human transmission of S. Infantis. pESI-like plasmids were present in 148/161 (91.9%) isolates; all were highly similar to the publicly available pESI-like sequences but lacked extended-spectrum beta-lactamase (ESBL) genes. PacBio/Illumina hybrid assembly allowed the reconstruction of two novel complete pESI variants. The present study revealed that the multidrug-resistant, pESI-positive S. Infantis clone became the predominant S. Infantis clone in Slovenian broilers and humans during the last decade. Continued surveillance of resistant S. Infantis clones along the food chain is needed to guide public health efforts.
IMPORTANCE Salmonella Infantis clones with pESI-like plasmids harboring several virulence and resistance genes have been reported worldwide. In the present study, we compared the population structure of 161 Salmonella Infantis isolates obtained from humans and broilers in Slovenia from 2007 to 2021. Whole-genome sequencing showed that most human isolates clustered apart from broiler isolates, suggesting an alternative source of infection. Most isolates were multidrug resistant due to the presence of pESI-like plasmids, of which two variants (pS89 and pS19) were fully reconstructed using long-read sequencing. Both exhibited high similarity with the original Israeli pESI plasmid and German p2747 plasmid. The prototype plasmid pS89 harbored the typical pESI-associated resistance genes aadA1, qacEΔ1, sul1, and tet(A), which were absent in the truncated plasmid pS19.
KEYWORDS: Salmonella Infantis, ST32, pESI, multidrug resistance, broilers, humans
INTRODUCTION
Salmonella enterica subsp. enterica serovar Infantis is the fourth most common Salmonella serovar causing human salmonellosis in Europe (1). It is the most prevalent serovar in broilers and can thus spread along the food chain (1). In 2014, multidrug-resistant (MDR) S. Infantis with a megaplasmid known as plasmid of emerging S. Infantis (pESI) was first reported for Israeli isolate 119944 from 2008 (2). Since then, S. Infantis clones with pESI-like plasmids have been reported worldwide (3–7).
These plasmids exhibit a mosaic structure and are associated with increased virulence and multidrug resistance, contributing to the success of pESI-positive S. Infantis clones observed over the past decade (2, 4, 8). The pESI-borne antimicrobial resistance genes (ARGs) are encoded in mobile genetic modules integrated into pESI resistance region 1 or 2 (4, 9). Recently, pESI-like plasmids with extended spectrum beta-lactamase (ESBL) genes have been reported worldwide (3, 4, 7, 9–13). Due to the large size of pESI-like plasmids, which can reach 323 kb (4), short-read next-generation sequencing needs to be complemented by long-read sequencing technologies. However, few studies have used the latter to reconstruct complete, gap-free sequences of pESI (4, 9, 10, 14, 15).
Here, we typed 161 S. Infantis isolates from poultry and humans using whole-genome sequencing (WGS). We performed a comprehensive genetic characterization of isolates to assess their epidemiology, population structure, and resistance in the Slovenian S. Infantis population. In addition, we used a state-of-the-art hybrid assembly to obtain complete sequences of two representative pESI-like plasmids and compared them with publicly available sequences.
RESULTS
WGS analysis.
Multilocus sequence typing (MLST) revealed that all isolates except one (n = 160) were of sequence type 32 (ST32); isolate S113 was a single locus variant of ST32 (thrA). In silico serotyping classified 158/161 isolates as S. Infantis (7:r:1,5), whereas three isolates (S20, S165, and S202) lacked the O antigen (−:r:1,5) but had been previously identified as S. Infantis by conventional slide agglutination (data not shown).
Presence/absence analysis of genes characteristic of pESI revealed that 148/161 (91.9%) isolates were pESI+ (see Tables S1 and S2 in the supplemental material). The first pESI+ isolates were observed in 2010 and have since completely replaced the pESI-negative isolates in broiler farms. The occurrence of pESI+ isolates was significantly higher in broiler isolates (128/132) than human isolates (20/28) (Fisher’s exact test, P = 0.0001). IncI1 plasmid MLST (pMLST) classified pESI-like plasmids into five types (Table S1); most pESI+ isolates (133/148; 89.9%) had the following allele profile: ardA_2, trbA_21, sogS_9, pilL_3.
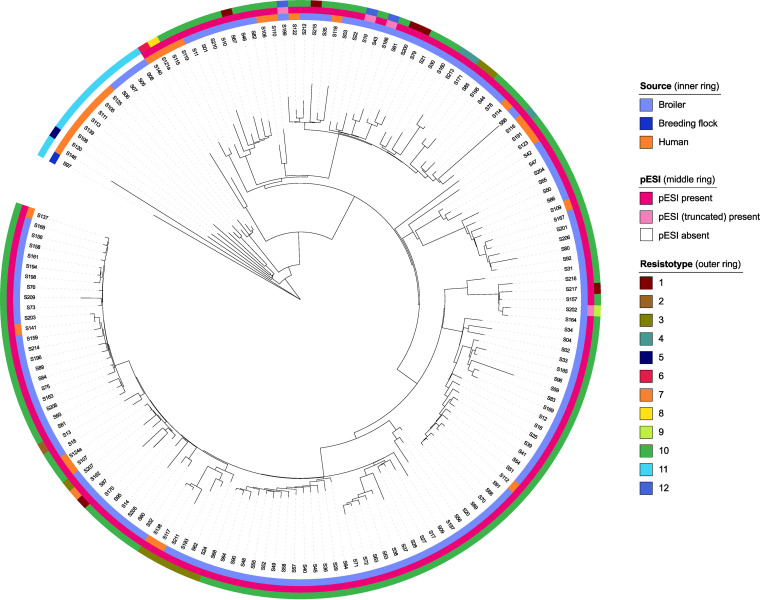
The core genome MLST (cgMLST) tree showed that the pESI− isolates clustered to the exclusion of pESI+ isolates (Fig. 1). The latter formed a single monophyletic clade and were genetically fairly homogeneous, with a median intracluster pairwise distance of 83 alleles (range, 2 to 184). pESI+ isolates differed from pESI− isolates in ≥102 alleles. With regard to pESI− isolates, the broiler isolates were more genetically homogenous and clustered apart from the human and breeding flock isolates. No evident clustering was observed with respect to isolation source or pESI variant (prototype versus truncated) (Fig. 1). On the contrary, general clustering of isolates according to the food business operator (FBO) was observed with few exceptions (see Fig. S1). Comparison of the isolates originating from the same farms revealed the presence of genetically closely related (≤30 allele differences) isolates in the farm; however, reintroduction of additional genetically distanced strains was also observed over time (data not shown).
FIG 1.
cgMLST tree of 161 Salmonella Infantis isolates. Isolation source, presence of pESI, and resistotype are provided. Resistotypes are numbered as follows: 1, tet(A), aac(6′)-Iaa, aadA1, sul1, parC (T57S), gyrA (S83Y), qnrS1, blaTEM-1B, nfsA (NS159); 2, tet(A), aac(6′)-Iaa, aadA1, sul1, parC (T57S), gyrA (S83Y), blaTEM-1B; 3, tet(A), aac(6′)-Iaa, aadA1, sul1, parC (T57S), gyrA (S83Y), blaTEM-1C, nfsA (NS159); 4, tet(A), aac(6′)-Iaa, aadA1, sul1, parC (T57S), gyrA (S83Y), blaTEM-1D, nfsA (NS159); 5, tet(A), aac(6′)-Iaa, ant(2′′)-Ia, sul1, dfrA1, cmlA1, parC (T57S), gyrA (D87Y), blaTEM-1B; 6, tet(A), aac(6′)-Iaa, aadA1, aph(3′′)-Ia, aph(3′′)-Ib, aph(6)-Id, sul1, sul2, dfrA14, floR, parC (T57S), gyrA (S83Y), blaTEM-1A, nfsA (NS159); 7, tet(A), aac(6′)-Iaa, aadA1, aadA2, sul1, sul3, dfrA8, cmlA1, parC (T57S), gyrA (S83Y), blaTEM-1B, nfsA (NS159); 8, tet(A), aac(6′)-Iaa, aadA1, sul1, dfrA14, parC (T57S), gyrA (S83Y), nfsA (NS159); 9, aac(6′)-Iaa, aadA1, sul2, dfrA1, parC (T57S), gyrA (S83Y), blaTEM-1B, nfsA (NS159); 10, tet(A), aac(6′)-Iaa, aadA1, sul1, parC (T57S), gyrA (S83Y), nfsA (NS159); 11, aac(6′)-Iaa, parC (T57S); 12, aac(6′)-Iaa, parC (T57S), gyrA (S83Y), nfsA (NS159).
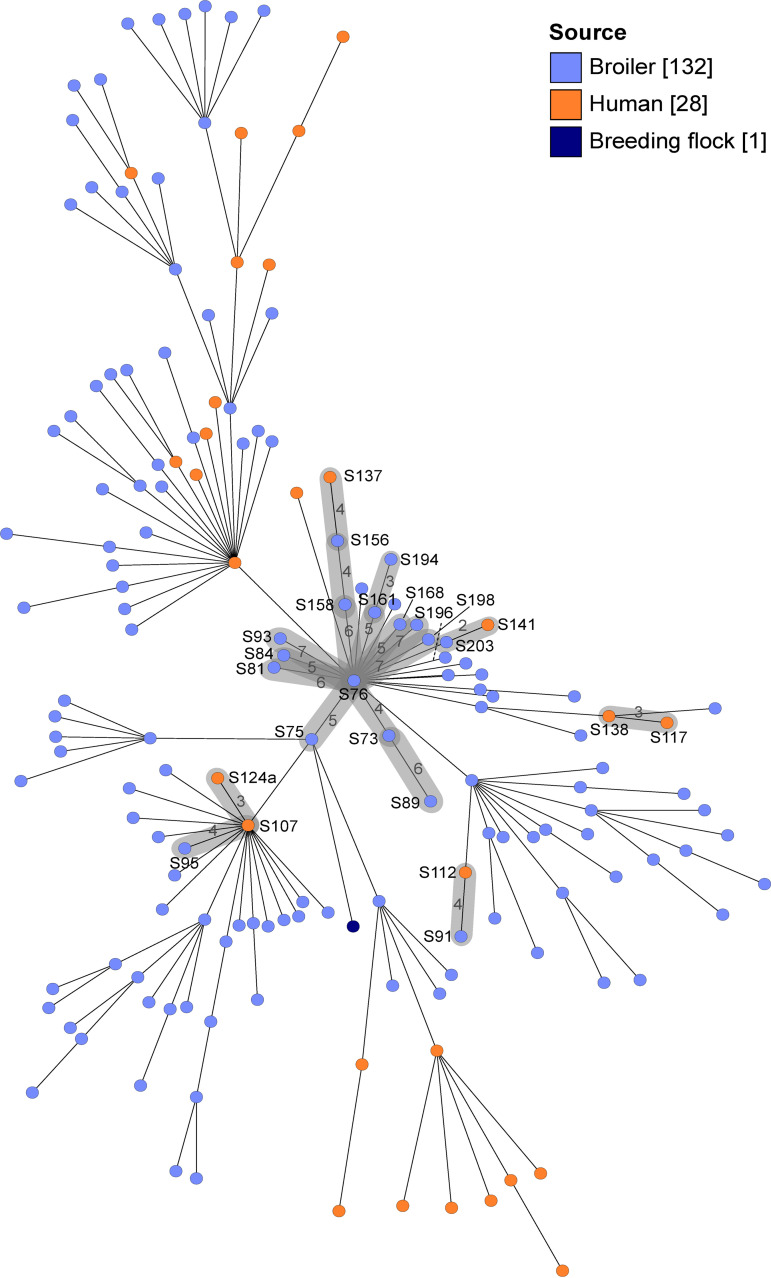
Four presumable broiler-to-human transmission clusters were detected, each comprising at least two isolates with ≤7 allele differences obtained over a limited time span (highlighted in Fig. 2). Additionally, one cluster included only two human isolates (S117 and S138), which originated from the same patient. Most (21/28) human isolates did not cluster with broiler isolates. Nine clusters containing only broiler isolates were observed but are not shown in Fig. 2 because of their de facto epidemiological linkage (Table S1). Six of nine broiler clusters were limited to a single farm and contained strains that presumably persisted up to 2 years. Isolates of cluster 1 (FBO 3) originated from eight farms and were isolated in 2017 to 2020 (Table S1), whereas isolates from FBOs 1 and 2 were more genetically diverse (Fig. S1).
FIG 2.
cgMLST minimum spanning tree of 161 Salmonella Infantis isolates. Isolates are colored according to the isolation source. Clusters of closely related (≤7 allele differences) isolates are highlighted in gray; isolates S117 and S138 originated from the same patient. Broiler-to-broiler transmission clusters are not shown. Numbers on the connecting lines indicate the number of allele differences. Other isolate names and distances are omitted for clarity.
The presence/absence profiles of ARGs and resistance-associated mutations are shown in Table S3; their genetic background is shown in Table S4 and summarized in Table 1. The isolates exhibited 12 different resistotypes, and 145/161 (90.1%) isolates were MDR (Fig. 1; Table S3). A total of 20 different acquired ARGs conferring resistance to six antimicrobial groups were observed, in addition to the nfsA mutation conferring nitrofurantoin resistance.
TABLE 1.
Resistance plasmids identified in silico in the studied Salmonella Infantis isolatesa
| Incompatibility group |
Best BlastN hit(s) | Mobility group | No. of isolates (% prevalence)b |
ARGsc |
|---|---|---|---|---|
| IncFIB (pESI) | CP047882.1 | Conjugative | 148 (91.9) | tet(A) (144/148), aadA1 (143/148), aph(3′)-Ia (1/148), sul1 (144/148), dfrA14 (2/148), floR (1/148), blaTEM-1C (8/148) |
| IncX1 | CP023660.1, CP088733.1, CP024467.1, KF362122.2, MH121702.1 | Conjugative | 8 (5.0) | qnrS1 (5/8), blaTEM-1D (2/8), blaTEM-1B (6/8) |
| NA | CP054381.1, CP035844.1, MN915013.1, CP038299.1 | Nonmobilizable | 3 (1.9) | tet(A) (1/3), aadA2b (1/3), ant(2′′)-Ia (1/3), sul3 (1/3), dfrA1 (1/3), dfrA8 (1/3), cmlA1 (2/3), blaTEM-1B (2/3) |
| IncIγ/K1 | MK070495.1 | Conjugative | 1 (0.6) | aadA1 (1/1), sul2 (1/1), dfrA1 (1/1), blaTEM-1B (1/1) |
| IncN | CP025233.1 | Conjugative | 1 (0.6) | aph(6)-Id (1/1), aph(3′′)-Ib (1/1), sul2 (1/1) |
| IncX4 | MW390518.1 | Conjugative | 1 (0.6) | blaTEM-1C (1/1) |
For a detailed description of the genetic background of each ARG, see Table S4. NA, not assigned.
n = 161.
Values in parentheses are numbers of ARG-positive isolates/plasmid-positive isolates.
Regarding pESI-encoded ARGs, 144/148 pESI+ isolates simultaneously harbored aadA1 (streptomycin resistance), tet(A) (tetracycline resistance), and sul1 (sulfonamide resistance) (Table 1; Tables S2 and S3). Seven different ARGs were encoded by pESI-like plasmids (Table 1; Table S4). Four pESI+ isolates had no ARGs in any of the resistance regions and thus harbored a presumed truncated pESI variant (Tables S1, S2, and S4). All pESI+ isolates harbored a chromosomal point mutation in gyrA (S83Y) leading to fluoroquinolone resistance.
WGS analysis revealed that most of the acquired ARGs were plasmid borne. In addition to tet(A), aadA1, and sul1, carried in resistance region 2, pESI-like plasmids also harbored blaTEM-1C (8/148 isolates) in resistance region 2 and aph(3′)-Ia (1/148 isolates), dfrA14 (2/148 isolates), and floR (1/148 isolates) in resistance region 1. In addition to pESI-like plasmids, nonmobilizable plasmids and conjugative plasmids of the incompatibility groups IncX1, IncN, IncIγ/K1, and IncX4 were also observed (Table 1).
All Slovenian pESI-like plasmids except the four truncated variants carried genes involved in resistance to quaternary ammonium compounds (qacEΔ1) and mercury (mer), both in resistance region 2, whereas none had ars genes, involved in arsenic tolerance. All pESI-like plasmids harbored the virulence- and fitness-associated traits located in the conserved pESI backbone, such as the yersiniabactin iron acquisition system (ybt) and two chaperone-usher fimbrial gene clusters (fae and ipf) (Table S2).
Comparative analysis of pESI-like plasmids.
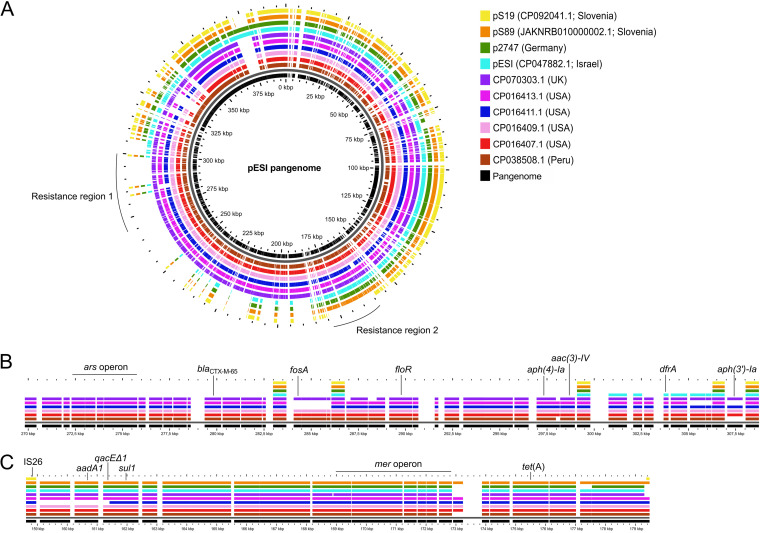
The complete, gap-free pESI-like sequence of the prototype Slovenian pESI-like plasmid pS89 confirmed the integration of the resistance genes aadA1, qacEΔ1, tet(A), and sul1 in resistance region 2 (Fig. 3). The complete, gap-free sequence of pS19 confirmed that it lacked all ARGs in resistance regions 1 and 2 (Fig. 3).
FIG 3.
Pangenome analysis of Slovenian pESI-like plasmids pS19 and pS89 compared with the original Israeli pESI and other pESI-like plasmids. (A) Pangenome analysis; (B) resistance region 1; (C) resistance region 2. Selected genes are highlighted in panels A and B. For isolate metadata, see Table S6.
The comparative analysis of pESI sequences revealed that pS19 and pS89 are genetically closely related (Fig. 3); this was confirmed by high average nucleotide identity based on the MUMmer algorithm (ANIm) (99.97% identity over a coverage of 89.97%) (Table S5). The Slovenian pESI-like plasmids pS89 and pS19 were most similar to the German p2747 and the original Israeli pESI (Fig. 3), with a minimum identity of 99.95% (Table S6). Plasmid pS89 harbored the same resistance genes as plasmid p2747, which both lacked dfrA14, in contrast to the original pESI (Fig. 3).
DISCUSSION
In the present study, we used WGS for the first time to elucidate the population structure of S. Infantis in Slovenia. Previously, we had observed an increasing rate of MDR S. Infantis in Slovenian broiler farms (16). We show here that the MDR pESI+ ST32 S. Infantis clone became the predominant clone in Slovenian broiler farms over the last decade. Most clinical isolates from humans were also of ST32 and harbored pESI. The success of S. Infantis ST32 can be explained, at least in part, by the presence of the virulence and resistance megaplasmid pESI (15).
We reconstructed two Slovenian pESI-like plasmids using state-of-the-art hybrid assembly, increasing the number of pESI sequences available in public databases. Their comparison with publicly available pESI-like plasmids from different geographical regions confirmed the highly mosaic structure with two variable resistance regions integrated into a conserved pESI backbone (4, 9, 10). Whereas the prototype variant pS89 had the resistance genes aadA1, tet(A), and sul1 (all in resistance region 2), the truncated variant pS19 lacked all ARGs, qacEΔ1, and the mer operon. Therefore, the isolates with truncated pESI were resistant only to fluoroquinolones and nitrofurantoin and were not MDR. Interestingly, the broiler isolate S202 harbored a truncated pESI but was MDR due to ARGs present on an additional IncIγ/K1 plasmid. A similar case was observed with the human isolate S120 (pESI−), which harbored ARGs on a nonmobilizable plasmid conferring resistance to five antimicrobial groups. With respect to the presence of additional resistance plasmids, no evident clustering of isolates was observed, suggesting their horizontal transfer between different S. Infantis clones.
In this study, seven pESI-encoded ARGs conferring resistance to five antimicrobial classes (tetracyclines, aminoglycosides, folate pathway antagonists, phenicols, and penicillins) were observed with variable presence; ESBL determinants and colistin resistance genes were not observed in any of the studied isolates. With regard to fluoroquinolone resistance, the S83Y mutation in gyrA was observed in all (n = 148) pESI+ isolates, the D87Y mutation in one pESI− human isolate (S120), and qnrS1 in 6/148 pESI+ isolates. Fluoroquinolones and third-generation cephalosporins are among the few treatment options for severe human salmonellosis cases (17). Therefore, treatment could be further complicated by integration of additional resistance genes into pESI or acquisition of additional resistance plasmids, both of which were observed here. High genome plasticity of pESI-like plasmids may facilitate evolutionary adaptation of S. Infantis to different environments and selection pressures, as previously suggested (2, 8).
Four isolates with the truncated pESI variant (including S19) were scattered throughout the pESI-positive clade in the cgMLST tree. Therefore, the truncated variant pS19 cannot be regarded as parental to the prototype plasmid pS89, but rather, it can be speculated that such variants arose independently multiple times. Truncated pESI variants without ARGs have also been reported previously (10, 18).
The predominant pMLST type of pESI observed in this study is associated with group 2 pESI, as defined by Bogomazova et al. (6), which has already been described as a carrier of the ESBL determinant blaCTX-M-1. In addition, the presence of certain ARGs in the resistance region 2 [aph(3′)-Ia, dfrA14, and floR] suggests that ESBL determinants could be easily integrated into the prototype Slovenian pESI-like plasmid.
pESI+ isolates predominated in both broilers and humans, although their prevalence was significantly higher in broilers. This suggests a stable integration of the pESI+ clone into both populations during the last decade. Here, four presumable broiler-to-human transmission clusters were detected, which has been rarely confirmed using WGS (4, 11). Nevertheless, most human isolates did not cluster with the broiler isolates, suggesting that Slovenian broilers cannot be regarded as an important source of infection for humans. However, future studies should also include WGS typing of isolates from imported meat. Another limitation of this study was the lack of data on human patient travel history, which would assess the impact of imported human cases. Due to the mosaic structure of pESI-like plasmids, continuous surveillance of antimicrobial resistance in S. Infantis is essential. Since conjugative transfer of pESI from S. Infantis to Escherichia coli has also been demonstrated both in vitro and in vivo (2, 8), detection of pESI in non-S. Infantis hosts should not be neglected.
We showed that the MDR pESI+ S. Infantis clone is predominant in broiler farms in Slovenia and has become increasingly common since 2010. The high genetic plasticity of pESI-like plasmids represents a threat for the introduction of ESBL genes into the Slovenian S. Infantis population. The presumable transmission clusters observed highlight the zoonotic potential of S. Infantis with limited treatment options.
MATERIALS AND METHODS
Isolate panel.
The analyzed S. Infantis panel (n = 161) consisted of isolates from broiler feces (n = 132), one isolate from a breeding flock, and 28 clinical isolates from humans (fecal, urine, and blood samples) from 2007 to 2021 (Table S1). The poultry isolates were selected from the isolate collection of the Institute of Microbiology and Parasitology, Veterinary Faculty. They originated from three large-scale FBOs in Slovenia, with farms distributed throughout the country, and eight independent farmers. The human isolates originated from sporadic human salmonellosis cases and represented the complete S. Infantis collection of the Institute of Microbiology and Immunology, Faculty of Medicine. For isolate metadata, see Table S1.
Illumina sequencing.
S. Infantis isolates were grown overnight on blood agar plates, and genomic DNA was extracted using a DNeasy blood and tissue kit (Qiagen, Hilden, Germany). All isolates underwent Illumina paired-end (2 × 150 bp) sequencing to a minimum coverage of 100×; details about the library preparation kits and sequencing platforms used in this study are provided in Table S1. The quality of the raw reads was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were assembled with Shovill v1.0.9 (https://github.com/tseemann/shovill) using SPAdes v3.13.1. The quality of the assembly was assessed using Quast (19); the following quality parameters were used: (i) N50 > 20,000, (ii) total number of contigs longer than 500 bp < 100, and (iii) total assembly size between 4.1 and 5.5 Mb.
PacBio sequencing.
Based on the preliminary gene content analysis of Illumina data, two strains (S19 and S89) were selected for PacBio long-read sequencing to reconstruct the complete sequences of pESI-like plasmids. Strain S89 harbored the Slovenian prototype pESI-like plasmid pS89 with the resistance genes sul1, aadA1, and tet(A). Strain S19 harbored its truncated variant pS19 with no ARGs. For this purpose, plasmid DNA was extracted from 500 mL of overnight culture grown in buffered peptone water using a NucleoBond Xtra midikit (Macherey-Nagel, Düren, Germany). SMRTbell libraries were sequenced on a PacBio Sequel platform. Hybrid assembly of Illumina and PacBio reads was performed using Unicycler v0.4.8 (20) run in normal mode.
WGS analysis.
In silico 7-gene MLST was performed using the Sequence query tool implemented in the PubMLST Salmonella database (21). pMLST of pESI-like plasmids was performed using pMLST v2.0 (22) according to the IncI1 pMLST scheme. In silico serotyping was performed using SeqSero2 v1.1.0 (23).
cgMLST allele calling was performed with chewBBACA (24) using the Prodigal training file for Salmonella enterica (https://github.com/B-UMMI/chewBBACA/tree/master/CHEWBBACA/prodigal_training_files). The initial ad hoc whole-genome MLST scheme consisted of 5,399 loci; after removal of 1,448 noncore loci, the final cgMLST scheme consisted of 3,951 loci. The circular cgMLST tree was created using the neighbor-joining algorithm implemented in Grapetree (25) and then annotated and visualized using iTOL v6.3 (26). The cgMLST minimum spanning tree was constructed using Grapetree v1.5.0 (25) and the MSTreeV2 algorithm. A single-linkage threshold of seven alleles (27) was used for cluster identification.
Detection of acquired ARGs and antimicrobial resistance-associated chromosomal mutations was performed using ResFinder v4.1 (28) using the cutoff thresholds of 60% coverage and 90% identity. In addition, the assemblies were screened for the presence of a nonsense mutation at position 159 of NfsA (nitrofurantoin resistance) in Geneious Prime v2021.1.1 (Biomatters, Auckland, New Zealand).
Comparative analysis of pESI-like plasmids.
BLASTn was used to detect the selected virulence- and fitness-associated genes in the conserved region of pESI (Table S2) by applying the cutoff values of 60% coverage and 90% identity. The isolates harboring the conserved pESI genes were defined as pESI+.
The reconstructed Slovenian pESI-like plasmids were compared with the publicly available sequences listed in Table S6. Pangenome analysis of pESI-like sequences was performed using GView Server v1.7 (20). MOB-recon integrated into MOB-suite tools v3.0.3 (29) was performed to reconstruct and type the plasmid sequences from draft genomes. The detected plasmid sequences were manually examined using BLASTn. ANIm values were calculated using JSpeciesWS (30).
Data availability.
All WGS read data (Table S1) and pESI-like plasmid sequences (Table S6) obtained in this study have been deposited in the NCBI Sequence Read Archive (SRA) and nucleotide database, respectively, under BioProject accession number PRJNA803004.
ACKNOWLEDGMENTS
This work was financially supported by the Administration for Food Safety, Veterinary Sector and Plant Protection (AFSVSPP), Slovenian Research Agency (“Management strategies and control of Salmonella Infantis infections in broiler flocks,” project no. V4-2004; “Animal health, environment and food safety,” research core funding no. P4-0092) and the Ministry of Agriculture, Forestry and Food of the Republic of Slovenia (project no. V4-2004).
AFSVSPP is acknowledged for providing the isolate metadata. We thank Nataša Peterka and Maja Kavalič for their major contribution to the management of isolate collection at the Institute of Microbiology and Parasitology, Veterinary Faculty, University of Ljubljana.
Footnotes
Supplemental material is available online only.
Contributor Information
Jana Avberšek, Email: jana.avbersek@vf.uni-lj.si.
Xianqin Yang, Agriculture and Agriculture-Food Canada.
REFERENCES
- 1.EFSA, ECDC. 2021. The European Union One Health 2020 zoonoses report. EFSA J 19:e6406. doi: 10.2903/j.efsa.2021.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aviv G, Tsyba K, Steck N, Salmon-Divon M, Cornelius A, Rahav G, Grassl GA, Gal-Mor O. 2014. A unique megaplasmid contributes to stress tolerance and pathogenicity of an emergent Salmonella enterica serovar Infantis strain. Environ Microbiol 16:977–994. doi: 10.1111/1462-2920.12351. [DOI] [PubMed] [Google Scholar]
- 3.Alba P, Leekitcharoenphon P, Carfora V, Amoruso R, Cordaro G, Di Matteo P, Ianzano A, Iurescia M, Diaconu EL, Pedersen SK, Guerra B, Hendriksen RS, Franco A, Battisti A, ENGAGE-EURL-AR Network Study Group . 2020. Molecular epidemiology of Salmonella Infantis in Europe: insights into the success of the bacterial host and its parasitic pESI-like megaplasmid. Microb Genom 6:e000365. doi: 10.1099/mgen.0.000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tate H, Folster JP, Hsu CH, Chen J, Hoffmann M, Li C, Morales C, Tyson GH, Mukherjee S, Brown AC, Green A, Wilson W, Dessai U, Abbott J, Joseph L, Haro J, Ayers S, McDermott PF, Zhao S. 2017. Comparative analysis of extended-spectrum-β-lactamase CTX-M-65-producing Salmonella enterica serovar Infantis isolates from humans, food animals, and retail chickens in the United States. Antimicrob Agents Chemother 61:e00488-17. doi: 10.1128/AAC.00488-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallejos-Sánchez K, Tataje-Lavanda L, Villanueva-Pérez D, Bendezú J, Montalván Á, Zimic-Peralta M, Fernández-Sánchez M, Fernández-Díaz M. 2019. Whole-genome sequencing of a Salmonella enterica subsp. enterica serovar Infantis strain isolated from broiler chicken in Peru. Microbiol Resour Announc 8:e00826-19. doi: 10.1128/MRA.00826-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bogomazova AN, Gordeeva VD, Krylova EV, Soltynskaya IV, Davydova EE, Ivanova OE, Komarov AA. 2020. Mega-plasmid found worldwide confers multiple antimicrobial resistance in Salmonella Infantis of broiler origin in Russia. Int J Food Microbiol 319:108497. doi: 10.1016/j.ijfoodmicro.2019.108497. [DOI] [PubMed] [Google Scholar]
- 7.Burnett E, Ishida M, de Janon S, Naushad S, Duceppe MO, Gao R, Jardim A, Chen JC, Tagg KA, Ogunremi D, Vinueza-Burgos C. 2021. Whole-genome sequencing reveals the presence of the blaCTX-M-65 gene in extended-spectrum β-lactamase-producing and multi-drug-resistant clones of Salmonella serovar Infantis isolated from broiler chicken environments in the Galapagos islands. Antibiotics 10:e267. doi: 10.3390/antibiotics10030267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aviv G, Rahav G, Gal-Mor O. 2016. Horizontal transfer of the Salmonella enterica serovar Infantis resistance and virulence plasmid pESI to the gut microbiota of warm-blooded hosts. mBio 7:e01395-16. doi: 10.1128/mBio.01395-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee WWY, Mattock J, Greig DR, Langridge GC, Baker D, Bloomfield S, Mather AE, Wain JR, Edwards AM, Hartman H, Dallman TJ, Chattaway MA, Nair S. 2021. Characterization of a pESI-like plasmid and analysis of multidrug-resistant Salmonella enterica Infantis isolates in England and Wales. Microb Genom 7:e000658. doi: 10.1099/mgen.0.000658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyson GH, Li C, Harrison LB, Martin G, Hsu CH, Tate H, Tran TT, Strain E, Zhao S. 2021. A multidrug-resistant Salmonella Infantis clone is spreading and recombining in the United States. Microb Drug Resist 27:792–799. doi: 10.1089/mdr.2020.0389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franco A, Leekitcharoenphon P, Feltrin F, Alba P, Cordaro G, Iurescia M, Tolli R, D’Incau M, Staffolani M, Di Giannatale E, Hendriksen RS, Battisti A. 2015. Emergence of a clonal lineage of multidrug-resistant ESBL-producing Salmonella Infantis transmitted from broilers and broiler meat to humans in Italy between 2011 and 2014. PLoS One 10:e0144802. doi: 10.1371/journal.pone.0144802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hindermann D, Gopinath G, Chase H, Negrete F, Althaus D, Zurfluh K, Tall BD, Stephan R, Nüesch-Inderbinen M. 2017. Salmonella enterica serovar Infantis from food and human infections, Switzerland, 2010–2015: poultry-related multidrug resistant clones and an emerging ESBL producing clonal lineage. Front Microbiol 8:e1322. doi: 10.3389/fmicb.2017.01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pietsch M, Simon S, Meinen A, Trost E, Banerji S, Pfeifer Y, Flieger A. 2021. Third generation cephalosporin resistance in clinical non-typhoidal Salmonella enterica in Germany and emergence of blaCTX-M-harbouring pESI plasmids. Microb Genom 7:e000698. doi: 10.1099/mgen.0.000698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen E, Rahav G, Gal-Mor O. 2020. Genome sequence of an emerging Salmonella enterica serovar Infantis and genomic comparison with other S. Infantis strains. Genome Biol Evol 12:151–159. doi: 10.1093/gbe/evaa048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Soto S, Abdel-Glil MY, Tomaso H, Linde J, Methner U. 2020. Emergence of multidrug-resistant Salmonella enterica subspecies enterica serovar Infantis of multilocus sequence type 2283 in German broiler farms. Front Microbiol 11:e1741. doi: 10.3389/fmicb.2020.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pate M, Mičunovič J, Golob M, Vestby LK, Ocepek M. 2019. Salmonella Infantis in broiler flocks in Slovenia: the prevalence of multidrug resistant strains with high genetic homogeneity and low biofilm-forming ability. Biomed Res Int 2019:4981463. doi: 10.1155/2019/4981463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO. Critically important antimicrobials for human use, 6th revision. https://apps.who.int/iris/bitstream/handle/10665/312266/9789241515528-eng.pdf. Accessed 22 June 2022.
- 18.Szmolka A, Szabó M, Kiss J, Pászti J, Adrián E, Olasz F, Nagy B. 2018. Molecular epidemiology of the endemic multiresistance plasmid pSI54/04 of Salmonella Infantis in broiler and human population in Hungary. Food Microbiol 71:25–31. doi: 10.1016/j.fm.2017.03.011. [DOI] [PubMed] [Google Scholar]
- 19.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jolley KA, Bray JE, Maiden MCJ. 2018. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:e124. doi: 10.12688/wellcomeopenres.14826.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. PlasmidFinder and pMLST: in silico detection and typing of plasmids. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S, den Bakker HC, Li S, Chen J, Dinsmore BA, Lane C, Lauer AC, Fields PI, Deng X. 2019. SeqSero2: rapid and improved Salmonella serotype determination using whole-genome sequencing data. Appl Environ Microbiol 85:e01746-19. doi: 10.1128/AEM.01746-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva M, Machado MP, Silva DN, Rossi M, Moran-Gilad J, Santos S, Ramirez M, Carriço JA. 2018. chewBBACA: a complete suite for gene-by-gene schema creation and strain identification. Microb Genom 4:e000166. doi: 10.1099/mgen.0.000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Z, Alikhan NF, Sergeant MJ, Luhmann N, Vaz C, Francisco AP, Carriço JA, Achtman M. 2018. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res 28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Di Marcantonio L, Janowicz A, Zilli K, Romantini R, Bilei S, Paganico D, Persiani T, Di Donato G, Di Giannatale E. 2020. Genomic comparison of Salmonella Enteritidis strains isolated from laying hens and humans in the Abruzzi region during 2018. Pathogens 9:349. doi: 10.3390/pathogens9050349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykäsenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM. 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. doi: 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robertson J, Nash JHE. 2018. MOB-suite: software tools for clustering, reconstruction and typing of plasmids from draft assemblies. Microb Genom 4:e000206. doi: 10.1099/mgen.0.000206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richter M, Rosselló-Móra R, Oliver Glöckner F, Peplies J. 2016. JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931. doi: 10.1093/bioinformatics/btv681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1-S6. Download spectrum.02481-22-s0001.xlsx, XLSX file, 0.08 MB (82.7KB, xlsx)
Fig. S1 and captions for Tables S1 to S6. Download spectrum.02481-22-s0002.pdf, PDF file, 0.2 MB (224.5KB, pdf)
Data Availability Statement
All WGS read data (Table S1) and pESI-like plasmid sequences (Table S6) obtained in this study have been deposited in the NCBI Sequence Read Archive (SRA) and nucleotide database, respectively, under BioProject accession number PRJNA803004.