ABSTRACT
Inorganic phosphate (Pi) is a central nutrient and signal molecule for bacteria. Pi limitation was shown to increase the virulence of several phylogenetically diverse pathogenic bacteria with different lifestyles. Hypophosphatemia enhances the risk of death in patients due to general bacteremia and was observed after surgical injury in humans. Phosphate therapy, or the reduction of bacterial virulence by the administration of Pi or phosphate-containing compounds, is a promising anti-infective therapy approach that will not cause cytotoxicity or the emergence of antibiotic-resistant strains. The proof of concept of phosphate therapy has been obtained using primarily Pseudomonas aeruginosa (PA). However, a detailed understanding of Pi-induced changes at protein levels is missing. Using pyocyanin production as proxy, we show that the Pi-mediated induction of virulence is a highly cooperative process that occurs between 0.2 to 0.6 mM Pi. We present a proteomics study of PA grown in minimal medium supplemented with either 0.2 mM or 1 mM Pi and rich medium. About half of the predicted PA proteins could be quantified. Among the 1,471 dysregulated proteins comparing growth in 0.2 mM to 1 mM Pi, 1,100 were depleted under Pi-deficient conditions. Most of these proteins are involved in general and energy metabolism, different biosynthetic and catabolic routes, or transport. Pi depletion caused accumulation of proteins that belong to all major families of virulence factors, including pyocyanin synthesis, secretion systems, quorum sensing, chemosensory signaling, and the secretion of proteases, phospholipases, and phosphatases, which correlated with an increase in exoenzyme production and antibacterial activity.
IMPORTANCE Antibiotics are our main weapons to fight pathogenic bacteria, but the increase in antibiotic-resistant strains and their consequences represents a major global health challenge, revealing the necessity to develop alternative antimicrobial strategies that do not involve the bacterial killing or growth inhibition. P. aeruginosa has been placed second on the global priority list to guide research on the development of new antibiotics. One of the most promising alternative strategies is the phosphate therapy for which the proof of concept has been obtained for P. aeruginosa. This article reports the detailed changes at the protein levels comparing P. aeruginosa grown under Pi-abundant and Pi-depleted conditions. These data describe in detail the molecular mechanisms underlying phosphate therapy. Apart from Pi, several other phosphate-containing compounds have been used for phosphate therapy and this study will serve as a reference for comparative studies aimed at evaluating the effect of alternative compounds.
KEYWORDS: Pseudomonas aeruginosa, phosphate starvation, proteomics, anti-infective therapy, pyocyanin, quorum sensing, virulence
INTRODUCTION
Pseudomonas aeruginosa (PA) is among the most feared human pathogens. It is an opportunistic pathogen that frequently causes nosocomial disease by infecting debilitated patients. PA is omnipresent in the environment and has been detected in different soil, water, human and animal-derived samples, different foods like salads, vegetables, and milk, as well as in plumbing systems or hospital settings (1–3). PA is a highly versatile pathogen, able to infect almost all human tissues, including the respiratory tract, ear, eye, brain, heart, and urinary tract, and causes a general bacteremia (4). PA is of important clinical relevance since: (i) infections are associated with significant death rates; (ii) it is among the most frequent causes of nosocomial infections; and (iii) multidrug-resistant strains are rapidly emerging (5–8). In addition to humans, PA infects animals and plants (9, 10).
Phosphorous is an essential element for all living cells and required for the synthesis of ATP, nucleic acids, phospholipids, and other biomolecules. In addition to its metabolic importance, inorganic phosphate (Pi) is an important signal molecule that modulates virulence in many different pathogens, including human-pathogenic bacteria like Vibrio cholerae (11), Bacillus anthracis (12), Mycobacterium tuberculosis (13), Staphylococcus aureus (14), the bacterial phytopathogens Agrobacterium tumefaciens (15) and Xanthomonas oryzae (16), or the human fungal pathogen Candida albicans (17). In the context of PA, Pi starvation was found to elicit important transcriptional changes (18) that were shown to shift this bacterium toward virulent phenotypes (19–21). The role of Pi as a signal molecule that controls virulence is likely to be related to the low phosphate availability in a healthy person (1.25 mM) that then becomes limited in patients following chemotherapy or who have undergone a recent surgical intervention (<0.03 mM) (20–22).
Pi is sensed by a sophisticated signaling complex comprised by the PstABCS phosphate transporter that is linked through the coupling protein PhoU to the PhoRB two-component system. Under Pi-limiting conditions, the PhoR kinase phosphorylates its response regulator PhoB to regulate gene expression (23). Several studies show that Pi starvation causes a PhoRB-mediated activation of the Rhl and PQS quorum sensing (QS) systems, whereas the Las QS system is dispensable (24–26).
PA was found to be present in the intestine of 20% of healthy individuals and up to 50% of hospitalized patients (27). Furthermore, the intestinal tract was identified as the primary site from which PA spreads, causing sepsis (28). Surgical interventions were shown to deplete intestinal Pi, in turn causing an activation of bacterial virulence (20, 21). A mouse model was established in which surgical injury was induced followed by intestinal PA inoculation (21). Several studies showed that the oral administration of Pi prior to the surgical injury of mice protected the animals against infections caused by PA (21, 29) and C. albicans (30). This approach was termed phosphate therapy and subsequently optimized using different phosphate-containing polyethylene glycol (PEG) polymers combining the favorable properties of Pi and PEG (31–34) or the use of nanomaterials that release Pi (35). The administration of Pi-PEG polymers not only suppressed virulence in PA but also in Serratia marcescens, Klebsiella oxytoca, Enterococcus faecalis, Klebsiella oxytoca, C. albicans (31), and in multispecies communities isolated form the intestine (33). In addition, the application of polyphosphate also caused a reduction of S. marcescens and PA virulence treats (36). These data suggest a universal nature of phosphate therapy to fight different classes of pathogens.
The increase in antibiotic-resistant microorganisms and their consequences represents a major global health challenge. Antimicrobial-resistant infections kill 700,000 patients per year (37, 38) and estimations indicate that this toll will rise to 10 million deaths annually by 2050 (38). Due to several different reasons, the pipeline to develop novel antibiotics is drying out (39). The current situation is referred to as antibiotics resistance crisis (40) and the World Health Organization (WHO) warns that a postantibiotic era, in which common infection and minor injuries kill, is far from being an apocalyptic fantasy, but a real possibility (41). As a result, the WHO has rated the development of new antimicrobial agents as critical and has placed PA second on global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics (42). However, fighting antibiotic-resistant bacteria with novel antibiotics will cause again the selection of resistant strains, resulting in a vicious cycle that may not permit to tackle antimicrobial resistance over the long term. Although there is a desperate need for the discovery of novel antibiotics, approaches like the phosphate therapy are promising alternatives to fight pathogenic microorganisms (43–46).
Comprehensive information on the changes at the protein level caused by Pi scarceness in PA is lacking. However, this information is essential to comprehend the molecular basis underlying phosphate therapy and it is the primary objective of this article to fill this gap in knowledge. We report here a proteomics study of PA grown in rich medium, or minimal medium supplemented with 0.2 mM or 1 mM Pi. About half of the predicted PA proteins could be quantified and vast changes at the protein level were detected comparing samples containing 0.2 mM and 1 mM Pi.
RESULTS AND DISCUSSION
Pyocyanin production is stimulated over a very narrow Pi window in a highly cooperative process.
To determine the effect of different Pi concentrations on the regulation of virulence in PA, initial experiments were conducted to study the production of the virulence factor pyocyanin as a proxy. The canonical reference strain in PA research, PAO1, was used as a model. We conducted growth experiments in minimal medium (MM) supplemented with different Pi concentrations. Growth in the presence of 50 and 1 mM could be considered identical, whereas a slight reduction in the growth rate was observed in the presence of 0.2 mM Pi (Fig. 1A). This deviation was significantly larger at 0.1 mM Pi (Fig. 1A). The visual inspection of the Erlenmeyer flasks at the end of the growth experiment (Fig. 1B) revealed an intense blue-green color at 0.2 mM and 0.1 mM, indicative of an elevated pyocyanin production, whereas the cultures with 1 and 50 mM were pale. To precisely determine the Pi concentration range at which pyocyanin production was altered, we repeated growth experiments incrementing the Pi concentration in 0.1 mM steps and extracted pyocyanin (Fig. 1C) for its spectrometric quantification (Fig. 1D). Experiments showed that the transition occurs between 0.6 mM and 0.2 mM Pi. This 3-fold reduction in the Pi concentration caused about a 75-fold increase in the pyocyanin production (Fig. 1D), indicative of a regulatory mechanism characterized by very strong positive cooperativity.
FIG 1.
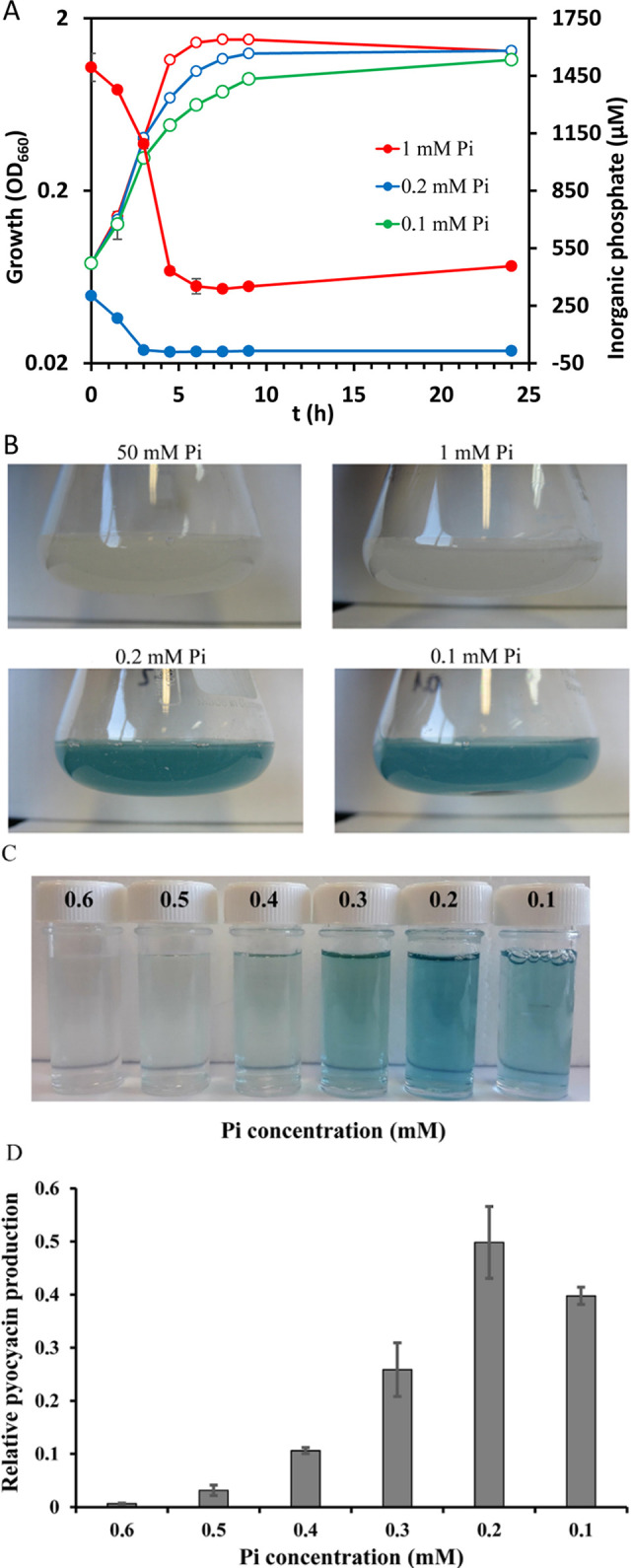
Growth of P. aeruginosa in the presence of different inorganic phosphate (Pi) concentrations. (A) Growth curves in minimal medium supplemented with different concentrations of Pi. The growth kinetics of cultures grown in 50 mM and 1 mM Pi are almost identical; hence the curve for 50 mM Pi has been omitted. The following growth rates were derived (in number of generations per h): 50 mM Pi, 1.01 ± 0.03; 1 mM Pi, 1.06 ± 0.03; 0.2 mM Pi, 0.87 ± 0.02; 0.1 mM Pi, 0.71 ± 0.02. The Pi concentration in supernatants is also shown. Growth (open symbols), Pi consumption (filled symbols). (B) Erlenmeyer flasks containing PA cultures after culture for 24 h at 37°C. (C) Cell-free supernatants from PA cultures grown in the presence of different Pi concentrations. (D) Quantification of pyocyanin from supernatants shown in C. Data were normalized using the cell density.
Phosphate starvation stimulates exoenzyme production and antibacterial activity.
Among the hallmarks of PA virulence is the increased production of extracellular invasive enzymes such as elastases and proteases (47). As shown in Fig. 2A, the elastase activity of cell-free supernatants of PA grown in the presence of 0.2 mM Pi was about 5-fold higher than supernatants derived from cultures in 1 mM Pi. Fig. 2B illustrates the proteolytic capacity of these cell-free supernatants on skim milk agar, which is illustrated by the appearance of a clearance area around the bacterial colony. In analogy to the elastase activity, the protease activity under Pi starvation was significantly increased. Dramatic changes were observed in the measurements of the antibacterial activity of these supernatants, as monitored by a bioassay to kill Chromobacterium violaceum cells forming a lawn on agar plates. The placement of supernatants of PA grown with 0.2 mM Pi into holes punched into the plate caused a very significant antibacterial halo, whereas no antibacterial activity was detected for supernatants derived from cultures grown with 1 mM Pi (Fig. 2C).
FIG 2.
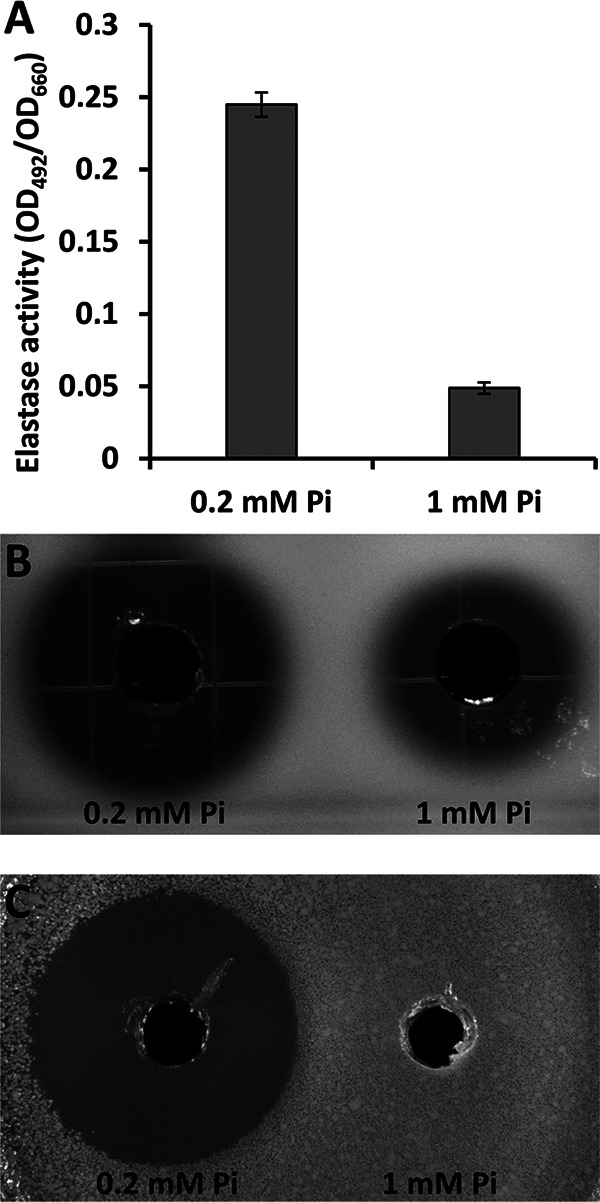
Exoenzyme production and antibacterial activity of P. aeruginosa PAO1 under Pi-deficient (0.2 mM) and -sufficient (1 mM) conditions. (A) Elastase activity in P. aeruginosa PAO1 supernatants. Data were normalized for culture density (OD660). Data are the means and standard deviations of three biological replicates. (B) Protease activity in P. aeruginosa PAO1 supernatants in skim milk agar plates. (C) Halos of antibiosis against Chromobacterium violaceum CV026 of filter-sterilized supernatants of P. aeruginosa PAO1. In B and C, the bioassays were conducted in triplicate and representative images are shown.
Reduction of Pi from 1 to 0.2 mM Pi causes large proteome changes.
To get insight into the changes in the proteome caused by altered Pi levels, we prepared protein extracts from cultures grown in MM supplemented with 1 mM and 0.2 mM Pi as well as in LB rich medium. Samples were taken at an optical density at 660 nm (OD660) of 0.6, which implies that the supernatant Pi concentration at the moment of sample taking for cultures grown in 1 mM and 0.2 mM Pi was of approximately 700 and 13 μM, respectively (Fig. 1A). An SDS-PAGE gel of the three replicate samples used for proteomics is shown in Fig. 3. Whereas, visually the proteomes of PA grown in LB and MM + 1 mM Pi are similar, there are very important changes comparing samples grown in MM + 1 mM Pi and MM + 0.2 mM Pi (Fig. 3).
FIG 3.
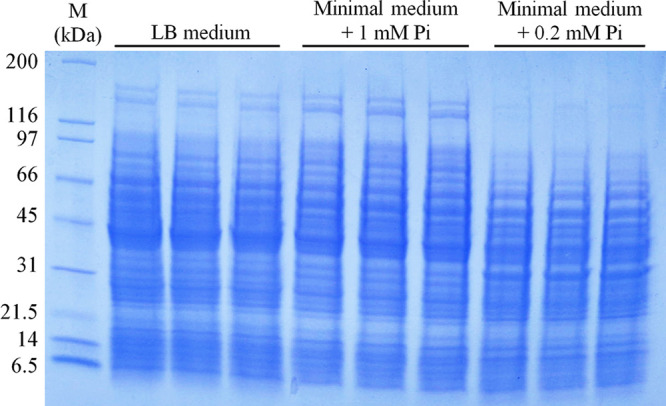
SDS-PAGE gel of protein extracts of P. aeruginosa cultures grown in LB rich medium or minimal medium supplemented with 1 mM or 0.2 mM Pi. Shown are the samples for three biological replicates that were used for the proteomics analysis. M, protein marker.
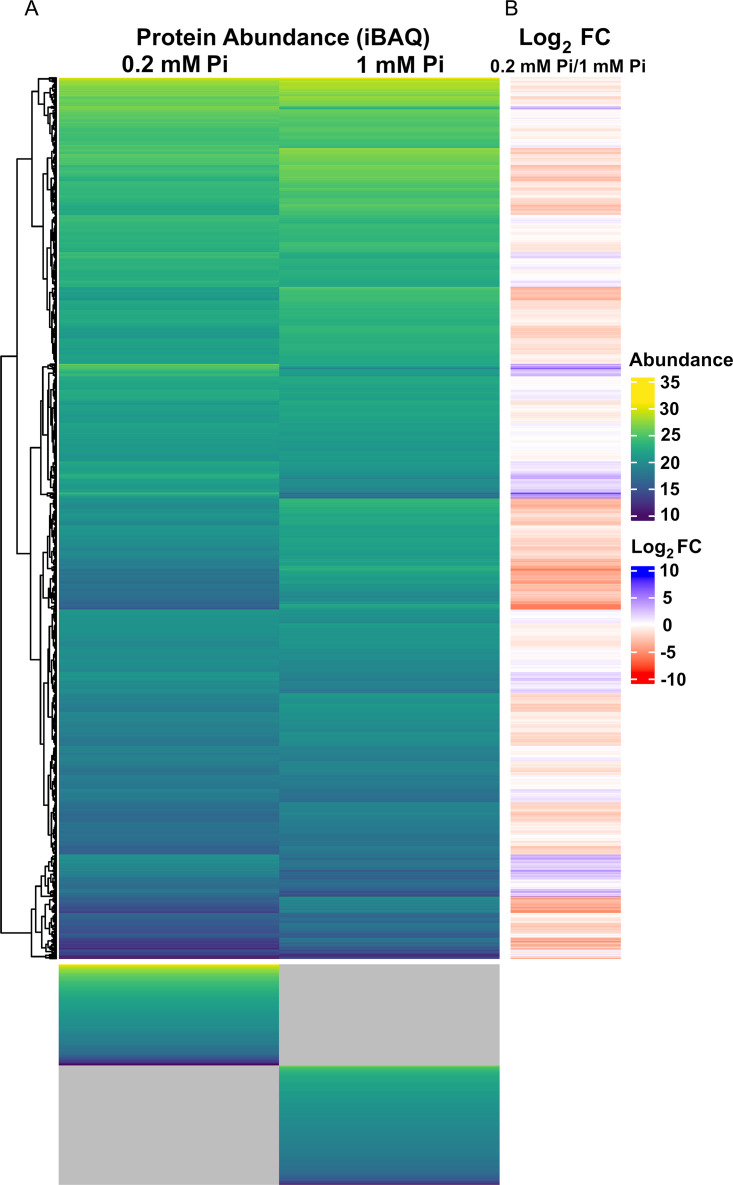
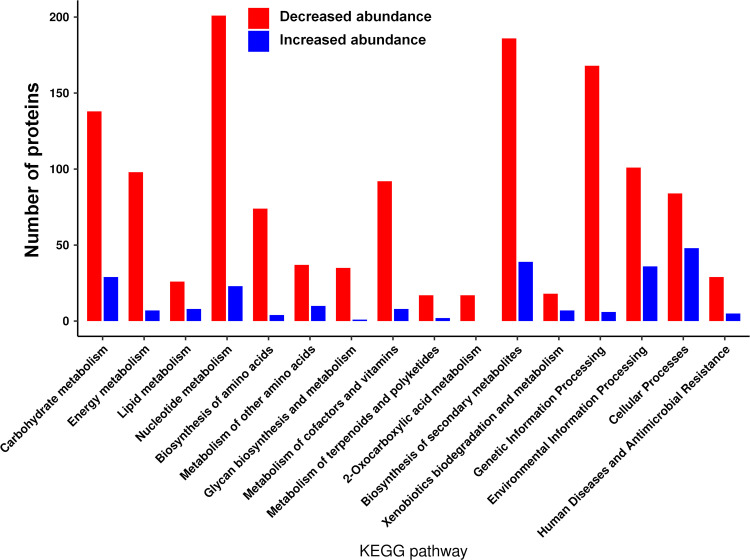
The proteomics analyses of these three conditions resulted in the quantification of a similar number of proteins, namely 2,716; 2,747; and 2,733 for the MM + 0.2 mM Pi, MM + 1 mM Pi, and LB samples, respectively (see Data Set S1 in the supplemental material). Considering that there are 5,587 annotated protein coding genes in the PA PAO1 genome (48), this corresponds to a coverage of about 49%. In this article, we analyze the changes in the proteome that occurred in cultures grown in the presence of 0.2 mM Pi compared to 1 mM Pi. Differentially abundant proteins are defined by a magnitude of the log2 fold change superior to 0.8 as well as a P value inferior to 0.01 in a Student's t test comparing the two growth conditions. In total, 295 and 1,022 proteins were found to be enriched or depleted, respectively, comparing growth in 0.2 mM Pi to 1 mM Pi. In addition, there were several cases where a given protein was detected in the three replicates of one growth condition but was not identified in the three replicates from the other growth conditions. It is very likely that the failure to detect a given protein in one growth condition is due to its concentration being below the dynamic detection range of the mass spectrometer. These proteins were thus included into the lists of depleted (Data Set S2) and accumulated proteins (Data Set S3). Growth in 0.2 mM Pi compared to 1 mM Pi resulted primarily in the downregulation of protein abundance (1,100 proteins [Data Set S2]), and in an accumulation of 371 proteins (Data Set S3) (Table 1, Fig. 4).
TABLE 1.
Proteins with altered abundance in P. aeruginosa grown in minimal medium plus 0.2 mM Pi compared to minimal medium plus 1 mM Pi
| Sense of change | Detected in both conditions and statistically different | Not detectable in the 0.2 mM Pi samples, but detected in the 1 mM Pi samples | Detected in the 0.2 mM Pi samples, but not detected in the 1 mM Pi samples | Total |
|---|---|---|---|---|
| Enriched proteins | 295 | 76 | 371 | |
| Depleted proteins | 1,022 | 78 | 1,100 |
FIG 4.
Heatmaps derived from P. aeruginosa growth in minimal medium supplemented with 0.2 mM or 1 mM Pi. (A) Protein abundance (intensity-based absolute quantification [iBAQ] values) in both samples. The gray shading indicates that the protein could not be detected in any of the three replicates of one condition, but was quantified in the three replicates of the other condition (referred to as “ON” or “OFF” at “low Pi” or “high Pi” in Data Set S2 and S3). (B) Log2 fold changes (FC) for growth in 0.2 mM compared to 1 mM Pi.
Lower abundance of proteins involved in metabolism and biosynthesis during Pi limitation.
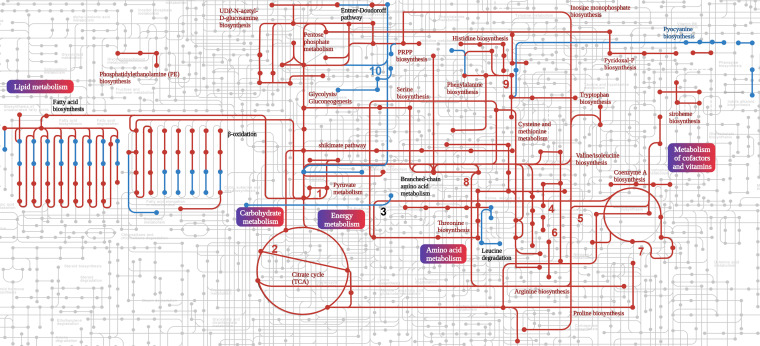
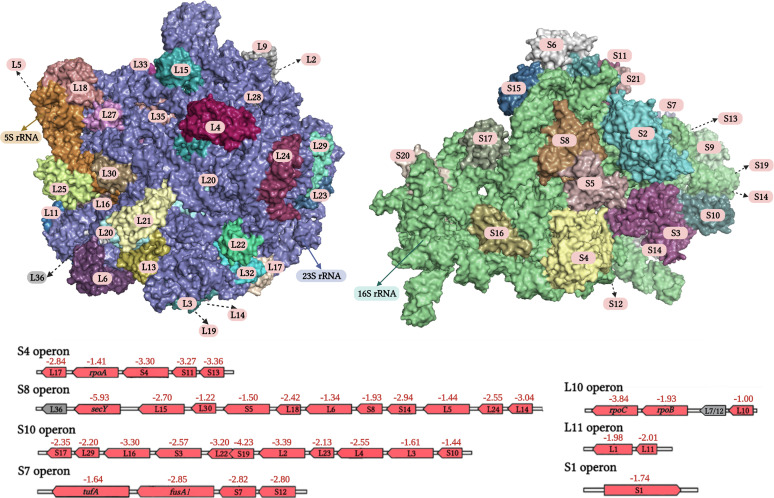
The reduction in the Pi concentration resulted in lowered amounts of about 40% of the detected proteins that correspond to ~20% of the proteins encoded in the genome of PAO1 (Data Set S2). The 10 proteins with the strongest depletion are listed in Table 2 and include proteins involved in the general metabolism, like the maintenance of redox homeostasis or respiratory electron transfer chain, transport, biosynthesis, and catabolism. Fig. 5 shows a map of PA metabolic processes in which the downregulated processes are highlighted in red and the classification of differentially abundant proteins according to the KEGG Orthology (KO terms) is shown in Fig. 6. Major downregulated processes include the glycolysis/gluconeogenesis and Krebs cycle, the glyoxylate cycle, the pentose phosphate metabolism, and the pathway for dissimilatory nitrate reduction. Importantly, biosynthetic pathways for many amino acids, phosphatidylethanolamine, inosine 5’-monophosphate, pyridoxal phosphate, Coenzyme A, UDP-N-acetyl-d-glucosamine, siroheme, polyamines, and glutathione were also downregulated (Fig. 5 and 6; Data Set S2). Furthermore, protein levels for several metabolic pathways were reduced, like those for different amino biosynthesis, and phosphonate and phosphinate metabolism. Several enzymes of the fatty acid biosynthesis and degradation as well as the Entner-Doudoroff pathways were depleted whereas others showed increased levels (Fig. 5 and 6). The reduction in the Pi level caused significantly lowered amounts of enzymes in the biosynthesis and metabolism of all major groups of biological molecules, like carbohydrates, lipids, amino acids, cofactors, and vitamins as well as secondary metabolites (Fig. 6). Next to the reduction in the abundance of proteins involved in the biosynthesis and metabolism of biomolecules, there were numerous proteins with decreased amounts that participate in energy metabolism as well as signal processing like transcriptional regulation. The fact that the primary consequence of the reduction in Pi from 1 mM to 0.2 mM is a reduction in abundance of 1,100 proteins is also reflected in a general depletion of ribosomal proteins. In total, 36 ribosomal proteins were detected at lower levels with an average magnitude of 2.51 ± 0.98 log2 fold change that corresponds to about a 6-fold mean reduction of ribosomal proteins under Pi-limiting conditions (Fig. 7).
TABLE 2.
Proteins with largest changes in abundance after growth in minimal medium plus 0.2 mM Pi compared to minimal medium plus 1 mM Pi
| Locus tag | Gene | Protein name | Protein function/comment | Log2 FC | Ref. |
|---|---|---|---|---|---|
| Depleted | |||||
| PA0196 | pntB | Pyridine nucleotide transhydrogenase, beta subunit | Regulation of NAD(P)+/NAD(P)H redox homeostasis | −7.27 | 111 |
| PA1680 | Predicted 3-oxoadipate enol-lactonase activity | Predicted role in benzoate catabolism | −6.46 | 112 | |
| PA1555 | ccoP2 | Cytochrome c oxidase, cbb3-type, CcoP subunit | Respiratory electron transfer chain component | −6.36 | |
| PA2840 | ATP-dependent RNA helicase | Regulation of type 3 secretion system | −6.30 | 113 | |
| PA1297 | aitP | Metal transporter | Fe2+ and Co2+ export | −6.18 | 114 |
| PA3321 | Transcriptional regulator | −6.10 | |||
| PA0141 | Polyphosphate kinase | GTP synthesis | −6.06 | 115 | |
| PA5118 | thiI | Thiazole biosynthesis protein | Thiazole synthesis | −6.02 | |
| PA1964 | ATP-binding component of ABC transporter | Transport | −5.98 | ||
| PA4243 | secY | Secretion protein SecY | Protein export | −5.93 | |
| Enriched | |||||
| PA3330 | Probable short-chain dehydrogenase | Regulated by quorum sensing | 8.62 | 116 | |
| PA4214a PA1903a |
phzE1
phzE2 |
Phenazine biosynthesis protein PhzE Phenazine biosynthesis protein PhzE |
Phenazine (pyocyanin) biosynthesis, host interaction Phenazine (pyocyanin) biosynthesis, host interaction |
7.90 7.90 |
117 |
| PA4216a PA1905a |
phzG1
phzG2 |
Probable pyrridoxamine 5′-phosphate oxidase Probable pyrridoxamine 59-phosphate oxidase |
7.33 7.33 |
||
| PA3280 | oprO | Pyrophosphate-specific outer membrane porin OprO | Phosphate and diphosphate transport | 6.45 | 51 |
| PA3279 | oprP | Phosphate-specific outer membrane porin OprP | 6.36 | ||
| PA5360 | phoB | Two-component response regulator PhoB | Transcriptional regulation in response to Pi starvation | 6.33 | 118 |
| PA4211 | phzB1 | Phenazine biosynthesis protein | Phenazine biosynthesis | 6.13 | 117 |
| PA0690 | pdtA | Phosphate depletion regulated two-partner secretion system partner A | Secretion system involved in virulence | 5.97 | 119 |
| PA0696 | Hypothetical protein | Unknown, part of the σVreI regulon, upregulated by Pi | 5.92 | 120, 121 | |
| PA0698 | Hypothetical protein | 5.80 | |||
Proteins of the same sequence.
FIG 5.
Metabolic map summarizing the principal differences in the proteomes of P. aeruginosa grown in minimal medium in the presence of 0.2 mM and 1 mM Pi. Up- and downregulated routes are shown in blue and red, respectively. Routes that contain both depleted and enriched proteins are annotated in black. 1, Phosphate acetyltransferase-acetate kinase pathway; 2, glyoxylate cycle; 3, lysine degradation; 4, isoleucine biosynthesis; 5, arginine and proline metabolism; 6, dissimilatory nitrate reduction; 7, polyamine biosynthesis; 8, glutathione biosynthesis; 9, tyrosine biosynthesis; 10, phosphonate and phosphinate metabolism.
FIG 6.
Analysis of differences in the proteomes of P. aeruginosa grown in the presence of 0.2 mM Pi compared to 1 mM. Classification of observed changes according to KEGG Orthology (KO) terms.
FIG 7.
Changes in the abundance of ribosomal proteins of P. aeruginosa grown in the presence of 0.2 mM Pi compared to 1 mM. Red, decrease; gray, no significant difference. The numbers indicate the log2 fold change of iBAQ values. The fact that the fold change values are associated with genes does not imply that regulation is at the transcriptional level. The upper part shows the three-dimensional structures of the large and small ribosomal subunits of the PA ribosome (pdb: 6SPD).
Many accumulated proteins under Pi-limiting conditions participate in virulence processes.
Among the proteins with the highest enrichment under Pi limitation (Table 2) were 5 proteins required for the synthesis of phenazine virulence factors (e.g., pyocyanin), 3 proteins involved in phosphate transport (OprO, OprP) and regulation (PhoB), a protein of a secretion system (PdtA), and two proteins of unknown function that belong to the regulon of the VreRI extracytoplasmic function (ECF) sigma factor system that is active during infection, promoting transcription of potential virulence determinants (49). An analysis of the totality of accumulated proteins permits their classification into several categories (Table 3) that are mainly related to phosphate acquisition or virulence processes.
TABLE 3.
A selection of proteins with increased abundance comparing growth in minimal medium plus 0.2 mM Pi with minimal medium plus 1 mM Pi
| Locus tag | Gene | Protein name/function | Log2 FCa |
|---|---|---|---|
| Proteins related to phosphate and phosphonate transport and regulation | |||
| PA3279 | oprP | Phosphate-specific outer membrane porin OprP precursor | 6.36 |
| PA3280 | oprO | Pyrophosphate-specific outer membrane porin OprO precursor | 6.45 |
| PA3383 | phnD | Binding protein component of ABC phosphonate transporter | 3.23 |
| PA3384 | phnC | ATP-binding component of ABC phosphonate transporter | ON/OFF |
| PA5360 | phoB | Two-component response regulator PhoB | 6.33 |
| PA5365 | phoU | Phosphate uptake regulatory protein PhoU | 3.56 |
| PA5366 | pstB | ATP-binding component of ABC phosphate transporter | 3.44 |
| PA5367 | pstA | Membrane protein component of ABC phosphate transporter | 3.73 |
| PA5368 | pstC | Membrane protein component of ABC phosphate transporter | 2.68 |
| PA5369 | pstS | Phosphate ABC transporter, periplasmic phosphate-binding protein | 2.66 |
| Secretion systems | |||
| Type II secretion systems | |||
| PA0681 | hxcT | Hxc type II secretion system | ON/OFF |
| PA0684 | hxcZ | Hxc type II secretion system | ON/OFF |
| PA0685 | hxcQ | Hxc type II secretion system | ON/OFF |
| PA0688 | lapA | Alkaline phosphatase A (secreted by Hxc secretion system) | ON/OFF |
| PA0689 | lapB | Alkaline phosphatase B (secreted by Hxc secretion system) | ON/OFF |
| PA0690 | pdtA | Phosphate depletion regulated TPS partner A, type II secretion system | ON/OFF |
| PA3105 | xcpQ | Xcp type II secretion system | 2.67 |
| PA3102 | xcpS | Xcp type II secretion system | ON/OFF |
| PA0026 | plcB | Phospholipase C (secreted by Xcp secretion system) | ON/OFF |
| PA0852 | cbpD | Chitin-binding protein CbpD (secreted by Xcp secretion system) | 4.46 |
| PA3296 | phoA | Alkaline phosphatase (secreted by Xcp secretion system) | 3.03 |
| PA3319 | plcN | Nonhemolytic phospholipase C (secreted by Xcp secretion system) | ON/OFF |
| PA2939 | paAP | Aminopeptidase (secreted by Xcp secretion system) | ON/OFF |
| PA0572 | pmpA | Putative protease (secreted by Xcp secretion system) | 5.10 |
| PA1871 | lasA | Elastase A (secreted by Xcp secretion system) | ON/OFF |
| PA0347 | glpQ | Glycerophosphoryl diester phosphodiesterase (secreted by Xcp secretion system) | 5.31 |
| PA3910 | eddA | Extracelullar DNA degradation protein (secreted by Xcp secretion system) | ON/OFF |
| Type VI secretion systems | |||
| PA0082 | tssA1 | Type VI secretion system H1 | 1.22 |
| PA0083 | tssB1 | Type VI secretion system H1 | 0.85 |
| PA0086 | tagJ1 | Type VI secretion system H1 | 0.82 |
| PA2363 | hsiJ3 | Type VI secretion system H3 | ON/OFF |
| PA2365 | hsiB3 | Type VI secretion system H3 | ON/OFF |
| PA2366 | hsiC3 | Type VI secretion system H3 | ON/OFF |
| PA3905 | tecT | Type VI effector chaperone for Tox-Release | 1.46 |
| PA0263b PA5267b PA1512b |
hcpC
hcpB hcpA |
Type VI secretion system (protein of the spear) Type VI secretion system (protein of the spear) Type VI secretion system (protein of the spear) |
2.35 2.35 2.35 |
| PA1844 | tse1 | Type VI secretion system (secreted protein, peptidoglycan amidase) | 1.30 |
| Other enzymes involved in Pi metabolism (phosphatases, phospholipases, and diesterases) | |||
| PA3885 | tpbA | Protein tyrosine phosphatase | 4.02 |
| PA5292 | pchP | Phosphorylcholine phosphatase | 1.82 |
| PA0843 | plcR | Phospholipase accessory protein PlcR precursor | ON/OFF |
| PA2856 | tesA | Lysophospholipase A | 1.1 |
| PA2990 | Probable phosphodiesterase | 0.95 | |
| Pyocyanin synthesis and activity | |||
| PA4209 | phzM | Phenazine-specific methyltransferase | 2.99 |
| PA4210 | phzA1 | Phenazine biosynthesis protein | ON/OFF |
| PA4211 PA1900 |
phzB1
phzB2 |
Phenazine biosynthesis protein Phenazine synthesis protein |
6.13 4.16 |
| PA4213b PA1902b |
phzD1
phzD2 |
Phenazine biosynthesis proteins Phenazine biosynthesis proteins |
ON/OFF ON/OFF |
| PA4214b PA1903b |
phzE1
phzE2 |
Phenazine biosynthesis proteins Phenazine biosynthesis proteins |
7.9 7.9 |
| PA4215b PA1904b |
phzF1
phzF2 |
Phenazine biosynthesis proteins Phenazine biosynthesis proteins |
4.46 4.46 |
| PA4216b PA1905b |
phzG1
phzG2 |
Pyridoxamine 5′-phosphate oxidase Pyridoxamine 59-phosphate oxidase |
7.33 7.33 |
| PA4217 | phzS | Flavin-containing monooxygenase | 2.92 |
| Che2 chemosensory pathway | |||
| PA0176 | aer2 | Chemoreceptor Aer2/McpB | 1.62 |
| PA0177 | cheW2 | Coupling protein | 3.33 |
| PA0179 | cheY2 | Response regulator | 2.53 |
| Quorum sensing | |||
| PA0997 | pqsB | HHQ/PQS synthesis | 3.74 |
| PA0998 | pqsC | HHQ/PQS synthesis | 2.59 |
| PA0999 | pqsD | 3-oxoacyl-[acyl-carrier-protein] synthase III | 4.43 |
| PA1001 | phnA | Anthranilate synthase component I | ON/OFF |
| PA1002 | phnB | Anthranilate synthase component II | ON/OFF |
| PA2303 | ambD | Synthesis of the IQS signal | 3.63 |
| PA2304 | ambC | Synthesis of the IQS signal | 3.37 |
| PA3476 | rhlI | C4-HSL autoinducer synthesis protein | 2.22 |
| PA3477 | rhlR | Transcriptional regulator RhlR | 3.88 |
| Signal transduction | |||
| PA0675 | vreI | ECF sigma factor | ON/OFF |
| PA0676 | vreR | Sigma factor regulator | ON/OFF |
| PA0696 | Unknown function, part of the σVreI regulon | 5.92 | |
| PA0698 | Unknown function, part of the σVreI regulon | 5.80 | |
| PA2047 | cmrA | Transcriptional regulator of multidrug efflux pump | 1.66 |
| PA2895 | sbrR | Anti-sigma factor, inhibits SbrI | 1.43 |
| PA2899 | atvR | Atypical virulence-related response regulator AtvR | 1.52 |
| PA3161 | himD | Integration host factor beta subunit | 1.67 |
| PA3347 | hsbA | HptB-dependent secretion and biofilm anti-sigma factor | 1.41 |
| PA3385 | amrZ | Alginate and motility regulator Z | 1.81 |
| PA3622 | rpoS | Sigma factor RpoS | 2.33 |
| PA3678 | mexL | Transcriptional repressor of the mexJK multidrug efflux operon | 0.81 |
| PA4101 | bfmR | Response regulator involved in biofilm formation | 2.17 |
| PA4381 | colR | Response regulator involved in polymyxin resistance | 1.36 |
| PA4608 | mapZ | C-di-GMP-binding adaptor protein, interacts with a chemotaxis methyltransferase to control flagellar motor switching | 1.52 |
| PA4778 | cueR | Transcriptional regulator activated by LasR | 0.81 |
| PA4878 | brlR | Pyocyanin binding transcriptional regulator that activates multidrug efflux pump | 3.14 |
| PA5261 | algR | Alginate biosynthesis regulator | 1.12 |
| PA5360 | phoB | Two-component response regulator PhoB | 6.33 |
| Other virulence related genes | |||
| PA0122 | rahU | Rhamnolipid-interacting protein that modulates innate immunity and inflammation in host cells. Involved in biofilm formation | 0.91 |
| PA0355 | pfpi | Protease involved in antibiotic resistance, swarming and biofilm formation | 4.05 |
| PA1249 | aprA | Alkaline protease, degrades transferrin, complement proteins, cytokines, and components of the extracellular matrix | ON/OFF |
| PA3479 | rhlA | Rhamnosyltransferase chain A | ON/OFF |
| PA3550 | algF | Alginate o-acetylltransferase | 2.53 |
| PA3552 | arnB | Uridine 5′-(beta-1-threo-pentapyranosyl-4-ulose diphosphate) aminotransferase with critical role in colistin resistance | ON/OFF |
| PA3692 | lptF | Lipotoxin F, outer membrane protein, necessary for adhesion to human cells | 1.34 |
| PA4379 | warA | Methyltransfrase involved in LPS biosynthesis | ON/OFF |
| PA4667 | lbcA | Lipoprotein binding partner of CtpA, required for type III secretion system function and virulence | 0.91 |
ON/OFF corresponds to proteins that were detected in the three replicates of samples derived from growth in 0.2 mM Pi, but that were not detected any of the three replicates of samples derived from growth at 1 mM Pi.
Proteins of the same sequence.
(i) Proteins related to phosphate and phosphonate transport and regulation. As mentioned above, the induction of pyocyanin production is a highly cooperative process (Fig. 1B–D). Pi is sensed by a complex comprising the PstABCS high-affinity phosphate transporter, the PhoU coupling protein and the PhoRB two-component system (50). The reduction in Pi in the growth medium resulted in a higher abundance of these proteins involved in Pi transport and sensing (Table 3; Data Set S3). OprP and OprO are outer membrane channels that permit the specific uptake of phosphate or pyrophosphate (51, 52) and under Pi starvation both proteins were found to be enriched at similar levels, namely, by factors of 82- and 87-fold (Table 3), indicating that these vicinal genes are subject to similar regulatory processes. Transcript levels of oprO and oprP were found to be upregulated when PA makes contact with lung epithelial cells (53). Next to the higher abundance of the PstABCS phosphate transporter, two of the three subunits of the PhnCDE phosphonate transporter were also detected in larger amounts. Phosphonates are phosphorus-containing organic molecules in which the phosphorous atom is linked directly to a carbon atom in a stable chemical bond. However, studies of E. coli PhnCDE revealed that it also transports Pi and the authors conclude that PhnCDE has to be considered a genuine phosphate transporter (54).
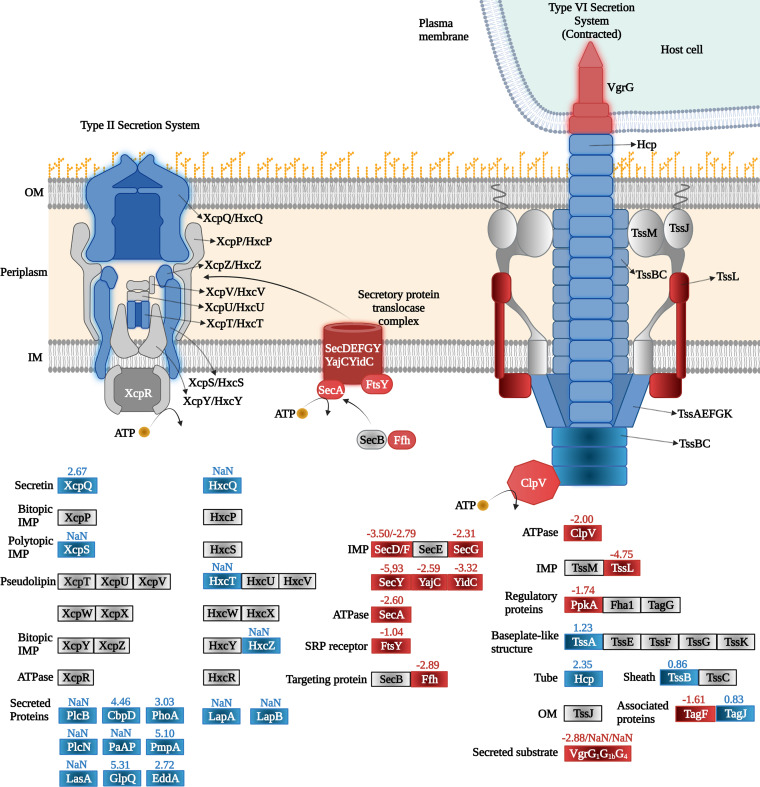
(ii) Secretion systems. Protein secretion systems are multiprotein complexes that are used to release proteins, toxins, or enzymes, for example, enzymes that hydrolyze complex carbon sources or proteins that capture essential elements such as iron (55). These systems are also used to colonize and survive within eukaryotic hosts, causing acute or chronic infections, subverting the host cell response, and escaping the immune system (56). Pi limitation modulated protein levels of both type II and type VI secretion systems (Fig. 8, Table 3).
FIG 8.
Changes in the abundance of secretion system proteins of P. aeruginosa grown in the presence of 0.2 mM Pi compared to 1 mM. Blue, increase; red, decrease; gray, no significant difference; NaN, not a number (not detected in the three replicates of the 1 mM Pi samples but identified in the three replicates of the 0.2 mM Pi sample). The numbers indicate the log2 fold changes of iBAQ values.
PA has two homologous type II secretion systems, Hxc and Xcp, and several proteins of both systems were among the enriched proteins under Pi limitation (Fig. 8, Table 3). This accumulation was accompanied by an increase in the levels of numerous proteins that were shown to be secreted by these systems (Fig. 8). Some of these proteins, namely, different phosphatases and phosphodiesterases, are involved in the Pi metabolism, whereas others were proteases and a chitin-binding protein (Table 3). Among the accumulated proteins that are secreted by the type II system is the protease LasA that is among the most relevant PA virulence factors. LasA acts synergistically with elastase LasB to degrade elastin (57). Elastin fibers are abundant in different human tissues like lung, skin, or auricular cartilage (58). The importance of LasA and LasB as virulence factors is demonstrated by a study reporting an about 70% reduction in in vitro invasion of epithelia cells for mutants deficient in one of the enzymes, whereas loss of both enzymes caused a further decrease in the invasive capacity of the cell (59). As indicated above, we observed an increase of about 5-fold in elastase levels in vitro at 0.2 mM Pi with respect to 1 mM Pi (Fig. 2A).
Two other exported proteins, the LapA and LapB phosphatases, were both not detected in the 1 mM Pi condition, whereas significant levels were detected under Pi scarceness (Data Set S1). In a study monitoring biofilm formation using an ex vivo chronic wound model, the lapA gene was the most upregulated (log2 FC = 11.32) compared to planktonic growth, indicating that this phosphatase plays a key role in wound infection, making it a potential drug target (60). LapA/LapB and PhoA, another phosphatase with increased abundance, are thought to be independently exported by the Hxc and Xcp type II secretion systems, respectively (61, 62). The secretion of LapA via the Hxc system is considered a complementary means to acquire Pi in case PhoA secretion by the Hcp system has become limited (61).
The phospholipases PlcB and PlcN are secreted by the Xcp type II secretion system and a hyperbiofilm phenotype was reported in mutants defective in plcB or plcN (63). Both proteins were not detected in 1 mM Pi but were present at significant amounts under Pi limitation. Another enriched protein, EddA, has alkaline phosphatase and phosphodiesterase activities and was found to be essential for the defense against extracellular traps that are produced by neutrophil immune cells as a mechanism to fight microbial pathogens (64).
In PA, there are three gene clusters that encode three homologous type VI secretion systems (H1- to H3-T6SS) (65). These systems are molecular nanomachines that inject effectors into target eukaryotic and prokaryotic cells and promote survival under harmful conditions, challenging survival of competitor organisms and helping PA to prevail in specific environments (66). Pi starvation had a differential impact on the levels of type VI secretion systems (Table 3, Fig. 8). Several proteins encoded in the H3 cluster were accumulated (Table 3), whereas the level of proteins encoded in cluster H2 remained unchanged. Pi starvation had a differential impact on proteins of the H1 cluster for which abundances were either increased or decreased (Table 3, Fig. 8; Data Set S2). One of the toxins secreted by the H1 system, Tse1, is among the accumulated proteins under Pi limitation.
(iii) Other enzymes involved in Pi metabolism. Next to the enriched phosphatases and phospholipases that are substrates for secretion systems, several other enzymes involved in Pi metabolism showed increased levels in Pi-limited conditions (Table 3). Among them is the tyrosine phosphatase TpbA that plays a key role in PA physiology. TpbA specifically controls the phosphorylation state of the transmembrane diguanylate cyclase TpbB (PA1120) by dephosphorylating Tyr and Ser/Thr residues. This control of the phosphorylation state regulates TpbB activity which in turn determines central features in PA physiology like the rugose colony morphology, biofilm production, and swarming motility (67, 68). Pi starvation also increased levels of the phosphorylcholine phosphatase PchP. Among the mechanisms required for the colonization of different tissues is the breakdown of host membrane phospholipids, a reaction catalyzed by PchP resulting in choline and Pi (69). A mutant in plcR was 200-fold less virulent in a mouse infection model (70). PlcR is an accessory protein that was found to be essential for the secretion of the PlcH phospholipase. In our study, PlcR was enriched (Table 3) whereas PlcH was depleted (log2 FC = 3; Data Set S2) (71). It may be possible that PlcR is also required for the secretion of other phospholipases.
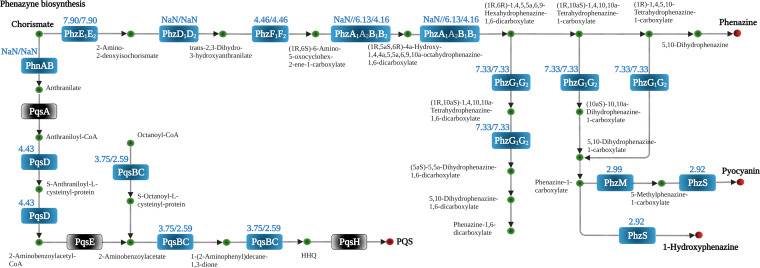
(iv) Pyocyanin synthesis and activity. Chorismate is the end product of the shikimate pathway that was downregulated under Pi-limiting conditions (Fig. 5). However, the routes that convert chorismate into the QS signal 4-hydroxy-2-heptylquinoline (HHQ) and the phenazines pyocyanin, phenazine, and 1-hydroxyphenazine were strongly upregulated (Fig. 9). Several proteins involved in phenazine biosynthesis were among those with the largest increase, as detailed in Table 2, and pyocyanin production was dramatically enhanced under Pi starvation (Fig. 1B–D). Pyocyanin is a redox-active compound that is secreted into the medium. It is one of PA’s primary virulence factors and is required to establish infection (72). Due to its redox activity, pyocyanin toxicity has been associated with the inhibition of aerobic respiration and the production of reactive oxygen species (73), and promoting virulence by interfering with numerous cellular functions in host cells (74). The different phenazines are considered biomarkers of PA infections (72).
FIG 9.
Changes in the abundance of proteins involved in the synthesis of quorum sensing signals and phenazines of P. aeruginosa grown in the presence of 0.2 mM Pi compared to 1 mM. Blue, increase; gray, no significant difference; NaN, not a number (not detected in the three replicates of the 1 mM Pi sample but identified in the three replicates of the 0.2 mM Pi sample). The numbers indicate the log2 fold changes of iBAQ values. Values separated by a slash are those of the individual subunits of an enzyme.
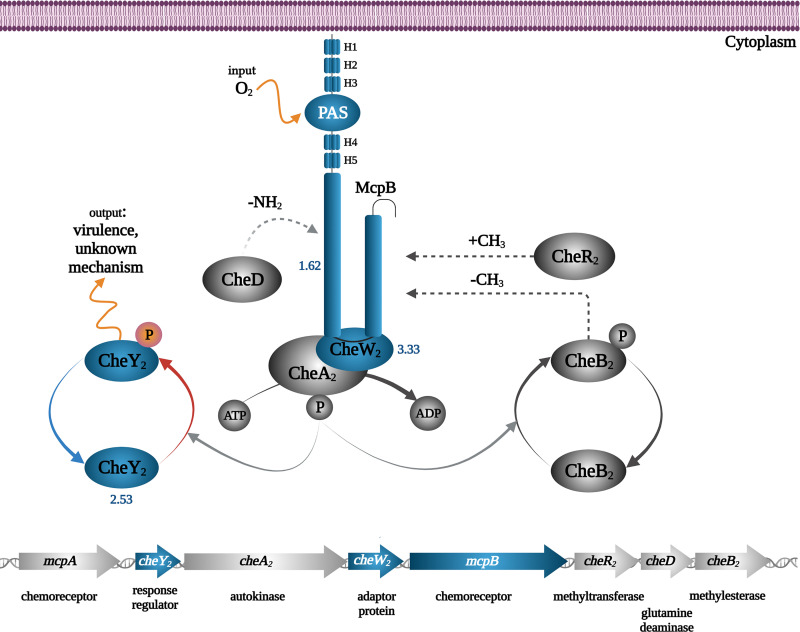
(v) Che2 chemosensory pathway. PA has four chemosensory pathways. Of these, the Che pathway mediates chemotaxis and the Wsp pathway controls c-di-GMP levels, whereas the Chp pathway is associated with mechanosensing and type IV pili-based motility (75). Although the function of the fourth pathway, Che2, is unknown, there are multiple pieces of evidence indicating that it plays an important role in virulence. It has been shown that the deletion of the genes encoding the sole chemoreceptor that stimulates this pathway, Aer2/McpB, as well as the CheB2 methylesterase caused a reduction in virulence (76, 77). Under Pi scarceness, three proteins of the Che2 pathway were accumulated, namely, the McpB chemoreceptor, the CheW2 adaptor protein, and the CheA2 autokinase (Fig. 10). In accordance with our results, different genes of the Che2 pathway were upregulated in a transcriptomic study at low Pi levels (18).
FIG 10.
Changes in the abundance of signaling proteins of the Che2 pathway of P. aeruginosa grown in the presence of 0.2 mM Pi compared to 1 mM. Blue, increase; gray, no significant difference or not detected; the numbers indicate the log2 fold changes of iBAQ values. H1 to H5: HAMP (found in histidine kinases, adenylate cyclases, methyl accepting proteins) domains.
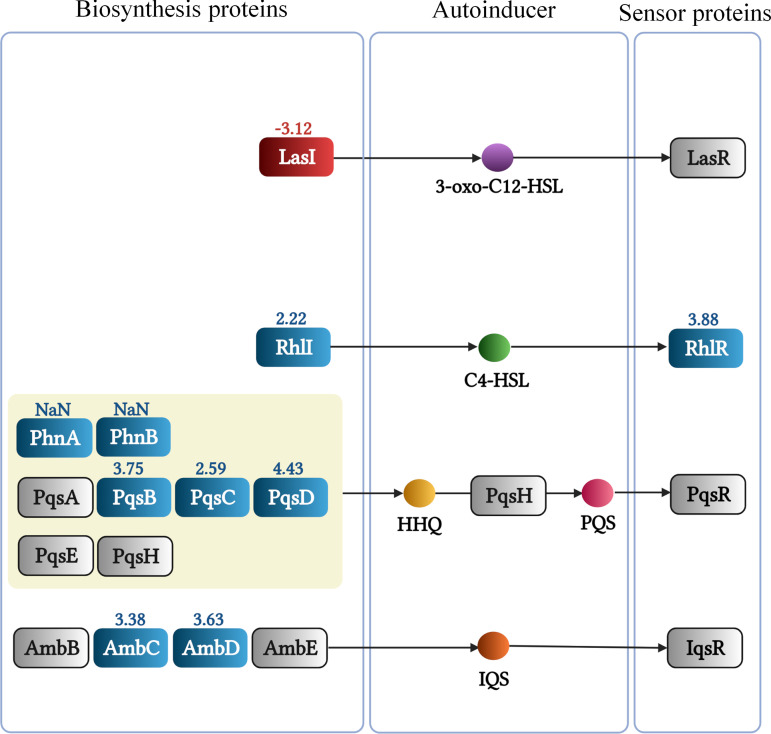
(vi) Quorum sensing. So far four QS systems have been reported in PA, namely, the Las, Rhl, Pqs, and Iqs systems. The Las and Rhl systems involve the synthesis and detection of the acylhomoserine lactone QS signals N-3-oxododecanoylhomoserine lactone (3-oxo-C12-HSL) and N-butanoylhomoserine lactone (C4-HSL), respectively (Fig. 11). Alternatively, the Pqs system uses HHQ and 2-heptyl-3-hydroxy-4-quinolone (PQS) as signals, whereas the Iqs system is based on the production of 2-(2-hydroxyphenyl)-thiazole-4-carbaldehyde (IQS) (Fig. 11) (78). Under Pi-sufficient conditions, the function of these four systems is closely interwoven with the Las system at the top of the regulatory hierarchy controlling the expression of different parts of the remaining three systems (78). The concerted action of these QS systems controls the expression of 10% of PA genes, including many virulence factor genes (73, 78). Many of the proteins known to be controlled by QS were detected at increased amounts in our study, including enzymes involved in the pyocyanin synthesis, the LasA protease, the PlcB phospholipase, Che2 pathway proteins, components of type II and type VI secretion system, or other proteins involved in virulence like the PfpI protease, lipotoxin F (LptF) or the RahU aegerolysin (Table 3) (78, 79). These data suggest a central role of the QS systems in mediating Pi responses. Pi starvation caused significant increases in the protein levels of the RhlI/RhlR pair and those of several proteins that are required for the synthesis of quinolone and IQS QS signals (Fig. 11). In contrast, an important depletion of the LasI synthase was noted, suggesting that the accumulation of the Rhl, Pqs, and Iqs proteins is not primarily mediated by the Las system under Pi-limited conditions, in accordance with transcriptomics studies (24, 80).
FIG 11.
Changes in the abundance of proteins involved in quorum sensing of P. aeruginosa grown in the presence of 0.2 mM Pi compared to 1 mM. Blue, increase; red, decrease; gray, no significant difference or not detected; the numbers indicate the log2 fold changes of iBAQ values. NaN, not a number (not detected in the three replicates of the 1 mM Pi sample but identified in the three replicates of the 0.2 mM Pi sample). HSL, homoserine lactone; HHQ, 4-hydroxy-2-heptylquinoline; PQS, 2-heptyl-3-hydroxy-4-quinolone.
(vii) Signal transduction proteins. Pi scarceness also altered the levels of a significant number of signal transduction proteins of which the most relevant examples are listed in Table 3. Next to the very significant increase observed for proteins of the VreIR ECF system and the PhoB response regulator, notable increases were also observed for other response regulators, ColR, AtvR, and BfmR, as well as the transcription factors CmrA, MexL, and AmrZ. ColR is involved in conferring polymyxin resistance (81) and AtvR was associated with PA adaptation to anaerobic conditions during infection (82). BfmR was shown to be essential for biofilm maturation (83) and its activation caused a shift from acute to chronic infection (84). MexL, BrlR, and CmrA regulate the expression of multidrug efflux pumps (85, 86). Interestingly, pyocyanin was shown to bind to BrlR, in turn enhancing BrlR-DNA affinity (87). AmrZ regulates twitching motility and alginate biosynthesis, two processes required for biofilm development (88, 89). Furthermore, this regulator orchestrates the assembly of all three type VI secretion systems (90). Further proteins with increased levels include the RpoS sigma factor that plays a central role in QS-mediated regulation (91), and MapZ, that controls the methylation state of chemoreceptors and consequently chemotaxis (92), another important virulence treat (78).
(viii) Other proteins involved in virulence. Several additional proteins with diverse roles in virulence were among the accumulated proteins (Table 3). An example is the rhamnolipid-interacting protein RahU, that is under the control of the Rhl QS system (93). RahU is involved in biofilm formation (94) and was shown to interfere with host innate immunity by inhibiting the accumulation of nitric oxide and chemotaxis of different immune cells (95). The protease PfpI was upregulated more than 16-fold (Table 3) and mutants defective in pfpI showed an increased resistance to ciprofloxacin, reduced swarming motility, and biofilm formation (96). Higher protein amounts were also observed for the RhlA rhamnosyltransferase and the ArnB aminotransferase, two enzymes essential for the synthesis of rhamnolipids (97) and involved in the production of an arabinose derivative that forms part of lipopolysaccharides (LPSs) (98), respectively. Colistin is a last-resort antibiotic to fight PA and colistin resistant mutants showed increased amounts of ArnB (99). Furthermore, the WarA methyltransferase interacts with the LPS biogenesis machinery and contributes to the ability of PA to evade detection by the host (100), most likely due to the role of LPSs as host immune response modulator. LbcA is a lipoprotein that forms a complex with the CtpA protease. LbcA is essential for the proteolytic activity of CtpA that in turn is required for the function of a type III secretion system and virulence in a mouse model of acute pneumonia (101, 102). Lastly, it is worth mentioning that the outer membrane protein, LptF, which has been associated with antibiotic resistance (103), was also enriched (Table 3).
Conclusions and future perspectives.
An increase in bacterial virulence caused by Pi limitation appears to be a general regulatory mechanism since it has been observed for several bacterial pathogens that differ in phylogeny, lifestyle, and virulence mechanisms (11–17). Phosphate therapy, or the reduction of bacterial virulence by the administration of Pi (or phosphate-containing compounds), is a promising approach to fight microbial pathogens that will not cause cytotoxicity nor the development of antibiotic resistance. The proof of concept for phosphate therapy has been obtained using primarily PA as a model organism (21, 29, 31, 32, 35, 36). In this study, we have determined the changes in the PA proteome caused by alterations in the Pi concentration.
Using the synthesis of pyocyanin as a proxy for the regulatory action of Pi, we show that changes occur in a very small window of Pi concentrations. Whereas almost no pyocyanin is synthesized in the presence of 0.6 mM Pi, a 3-fold decrease in the Pi concentration caused about a 75-fold increase in the pyocyanin levels, indicative of a highly cooperative process. Our proteomics analyses identified at least part of the molecular basis for the cooperativity observed. Pi is sensed by the PstABCS/PhoRBU complex (50), and Pi scarceness caused large increases in the protein levels of this sensory complex (Table 2 and 3) resulting in a feed-forward regulatory circuit and positive cooperativity.
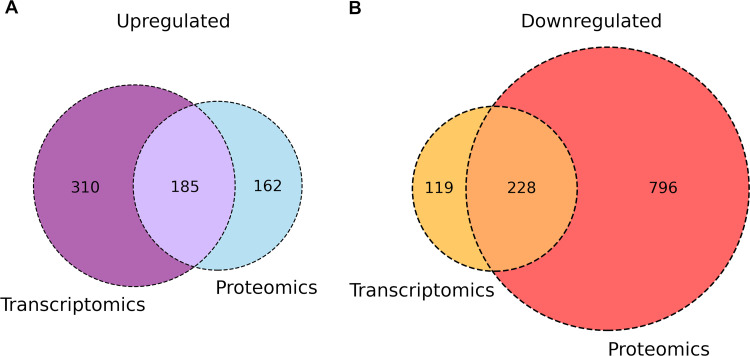
As mentioned above, Pi scarceness increases the virulence of many different pathogens by an overexpression of different virulence factors. However, our study shows that of the 1,471 dysregulated proteins, the large majority, 1,100, were detected with lower abundance. Most of the depleted proteins are involved in general and energy metabolism, transport, and many different biosynthetic and catabolic routes (Fig. 5) or form part of the ribosome (Fig. 7). This issue is also illustrated by comparing the transcriptomics study of Bains et al. (18) with our proteomics data. In both studies, PAO1 was cultured under the same growth conditions and exposed to either 0.2 mM or 1 mM Pi. Fig. 12 is a Venn diagram of the up- and downregulated genes/proteins in both studies. Whereas a balance in the number of upregulated genes/proteins is observed between both studies, there is a large excess of downregulated proteins compared to the genes that were downregulated in the transcriptomics study (Fig. 12) (18). These data suggest the existence of strong posttranscriptional regulatory mechanisms that are responsible for the vast majority of depleted proteins.
FIG 12.
Venn diagram comparing genes/proteins regulated by an exposure of PA PAO1 to 0.2 mM and 1 mM Pi in a transcriptomics study (18) and our proteomics analysis. The culture conditions in both studies were identical. Since in the transcriptomics study, a cutoff 2-fold change was applied, the dysregulated proteins in the proteomics study were truncated at the same level.
Many of the accumulated proteins in our study are known to be regulated by quorum sensing, suggesting that QS is a key mechanism by which Pi induces the corresponding changes. In general, the four QS systems in PA are arranged in a hierarchical order with the Las system on top, controlling the activity of the Rhl, Pqs, and Iqs systems (78). However, this general scheme does not appear to apply to QS-mediated changes under Pi-scarce conditions. Indeed, a recent study (25) has shown that: (i) the Las system is globally dispensable for mediating responses to low Pi; (ii) LasR expression is not affected by phosphate limitation; (iii) PhoB directly regulates rhlR transcription; and (iv) mutation of pqsR reduced pyocyanin production as a whole, suggesting that under phosphate limitation, the Rhl system is on top of the QS regulatory hierarchy. However, contrary to these findings, an alternative study revealed that PhoB activated lasI expression, the gene encoding the homoserine lactone (HSL) synthesizing enzyme of the Las system, under phosphate depletion conditions (26). Our proteomics data largely support the notion that QS-mediated regulation in response to Pi limitation is Las independent and governed by the Rhl system. Pi scarceness significantly reduced LasI levels, did not alter LasR levels, but increased components of the Rhl (e.g., RhlI, RhlR, RhlA), Pqs (e.g., PhnA, PhnB, PqsB, PqsC, PqsD) and Iqs (e.g., AmbC, AmbD) systems (Fig. 9 and 11).
A recent review has classified the different virulence hallmarks of PA according to their function into the following 6 groups: factors for host colonization and bacterial motility, biofilm formation, production of destructive enzymes, toxic secondary metabolites, iron-chelating siderophores, and toxins (47). Representatives of all six categories were among the proteins that were upregulated by Pi limitation. Abundances of several key virulence factors identified for each of these six categories were increased in our study, including PstS, LasA, AprA, PlcB, PlcN, PhzABDEFG, RhlAIR, PqsBCD, and AlgFR (47). Pi limitation has been observed following surgical injury, leading to an increased bacterial virulence (20, 21). This enhanced virulence is due to an induction of proteins that belong to all six major categories of virulence hallmarks. However, the administration of Pi to patients to increase its concentration above 0.5 mM will result in a reversal of virulence protein induction. Phosphate therapy thus holds a strong promise as an alternative strategy to tackle microbial pathogens.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
PA PAO1 was grown at 37°C with orbital shaking (200 rpm) in LB (5 g/L yeast extract, 10 g/L bactotryptone, 5 g/L NaCl) or MM (0.1 M HEPES, 7 mM [NH4]2SO4, 20 mM succinate, 1 mM MgSO4, 6 mg/L Fe-citrate, 300 mg/L HBO3, 50 mg/L ZnCl2, 30 mg/L MnCl2 × 4 H2O, 200 mg/L CoCl2, 10 mg/L CuCl2 × 2 H2O, 20 mg/L NiCl2 × 6 H2O, 30 mg/L NaMoO4 × 2 H2O, pH 7.0) containing 0.1 to 50 mM potassium phosphate (K2HPO4/KH2PO4, pH 7.0).
Quantification of the supernatant Pi concentration during bacterial growth.
Determination of Pi concentration along the growth curve was carried out using the malachite green phosphate assay kit (Sigma-Aldrich; Catalog number MAK307) following the manufacturer’s instructions. Reaction assays were incubated for 30 min at room temperature for color development, followed by an absorbance measurement at 660 nm using a Sunrise plate reader from Tecan (Männedorf, Switzerland).
Phenotypic assays.
Assays for pyocyanin quantification, antibacterial activity, and extracellular enzyme production (e.g., proteases and elastase) were carried out on culture supernatants from PA PAO1 cells grown in MM with different Pi concentrations. Overnight cultures of PAO1 grown in MM containing 1 mM inorganic phosphate (K2HPO4/KH2PO4) were washed twice with phosphate-free MM and then used to inoculate flasks containing 15 mL MM with 0.1 to 1 mM Pi at an initial OD660 of 0.075 and grown for 24 h at 37°C with orbital shaking (200 rpm). After overnight growth, bacterial cultures were centrifuged at 9,000 × g for 10 min and the supernatants were filtered (0.2 μm cutoff). The resulting filter-sterilized supernatants were used to conduct the following bioassays.
(i) Pyocyanin quantification. Pyocyanin was extracted into chloroform by mixing 7.5 mL of each of the supernatants with 4.5 mL of chloroform. Pyocyanin was further extracted from the solvent phase into 1.5 mL of 0.2 M HCl. The absorbance of the resulting solution was measured at 520 nm and data normalized against cell density at 660 nm.
(ii) Antibacterial assays. Antibiotic activity was tested by the agar lawn assay using 0.8% LB agar plates containing a top lawn of 200 μL of an overnight culture of Chromobacterium violaceum CV026 (104). Next, 300 μL of filter-sterilized PAO1 supernatants were added to wells punched into the plates and incubated at 30°C for 48 h.
(iii) Elastase activity determination. One milliliter supernatant was added to tubes containing 10 mg elastin-Congo red (ECR; Merck) and 1 mL of 0.1 M Tris/HCl, 1 mM CaCl2 (pH 7.0). Tubes were incubated at 37°C with shaking (200 rpm) for 24 h and the reactions stopped by the addition of 1 mL 0.7 M sodium phosphate buffer pH 6.0. Residual, solid ECR was removed by centrifugation and the OD492 of the reaction was measured. These measurements were normalized against the absorbance at OD492 of the cell-free supernatants prior to the addition of ECR and against cell density at 660 nm.
Protease activity. Qualitative proteolysis assays were conducted using the skim milk assay. Briefly, 300 μL supernatants were added to wells punched into the skim milk plates (1% skim milk – quarter-strength LB) and incubated at 30°C for 48 h. Skim milk proteolysis was evaluated as the zone of clearance from the well edge.
Sample preparation for mass spectrometry (MS).
Overnight cultures of PA PAO1 grown in MM containing 1 mM Pi (K2HPO4/KH2PO4, pH 7.0) were washed twice with Pi-free MM. Washed cultures were then used to inoculate flasks with 50 mL MM containing high (1 mM) and low (0.2 mM) Pi concentrations at an OD660 of 0.075 and grown at 37°C with orbital shaking (200 rpm). At an OD660 of 0.6, 40 mL aliquots were centrifuged at 9,000 × g for 10 min. The resulting pellets were resuspended in 2 mL extraction buffer (6 M urea, 2 M thiourea, 50 mM Tris/CL, pH 7.5) and cells were broken by sonication. After centrifugation at 9,200 × g for 10 min, supernatants were collected, and protein concentration was determined by the Bradford assay (Bio-Rad, reference no. 500-0006). Supernatant aliquots were subsequently mixed with 5× Laemmli sample buffer containing 1 mM β-mercaptoethanol. After 1 h of incubation at room temperature, aliquots corresponding to 25 μg of protein were run on a 4 to 20% precast polyacrylamide gel (Bio-Rad, reference no. 4561094) at 95 V. The polyacrylamide gel was fixed with solution A (40% [vol/vol] methanol, 7% [vol/vol] acetic acid) for 10 min and stained with solution B (0.1% [wt/vol] Coomassie brilliant blue R [Sigma-Aldrich, Ref. B7920], 40% [vol/vol] methanol, 10% [vol/vol] acetic acid). The gel was finally destained with solution A. Proteins were prepared for tandem mass spectrometry analyses, as previously described (105). Briefly, gel lanes were fractionated into 10 gel pieces, cut into smaller blocks, and transferred into low binding tubes. Samples were washed until gel blocks were destained and dried in a vacuum centrifuge before they were covered with a trypsin solution. Samples were digested overnight at 37°C prior to peptide elution into water by ultrasonication treatment. The peptide-containing supernatants were transferred into fresh tubes, desiccated in a vacuum centrifuge and peptides were resolubilized in 0.1% (vol/vol) acetic acid for mass spectrometric analysis.
MS/MS analysis.
LC-MS/MS analyses were performed on a LTQ Orbitrap VelosPro instrument (ThermoFisher Scientific, Waltham, MA, USA) using an EASY-nLC II liquid chromatography system. Tryptic peptides were subjected to liquid chromatography separation and electrospray ionization-based MS applying the same injected volumes in order to allow for label-free relative protein quantification between and within samples. Therefore, peptides were loaded on a self-packed analytical column (OD 360 μm, ID 100 μm, length 20 cm) filled with 3-μm diameter C18 particles (Maisch, Ammerbuch-Entringen, Germany) and eluted by a binary nonlinear gradient of 5 to 99% acetonitrile in 0.1% (vol/vol) acetic acid over 87 min with a flow rate of 300 nl/min. For MS analysis, a full scan in the Orbitrap with a resolution of 30,000 was followed by collision-induced dissociation (CID) of the 20 most abundant precursor ions. MS/MS experiments were acquired in the linear ion trap.
MS data analysis.
Database searches against a database of PA PAO1 downloaded from the Pseudomonas Genome DB (48) on 6 October 2021 (5,587 entries) as well as Intensity-Based Absolute Quantification (iBAQ) were performed using MaxQuant (version 1.6.17.0) (106). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies, and proteome-wide protein quantification (107). Common laboratory contaminants and reversed sequences were included by MaxQuant. Search parameters were set as follows: trypsin/P-specific digestion with up to two missed cleavages, methionine oxidation and N-terminal acetylation as variable modification, match between runs with default parameters enabled. The false discovery rates (FDRs) of protein and peptide spectrum match (PSM) levels were set to 0.01. Two identified unique peptides were required for protein identification. Results were filtered for proteins quantified in at least two out of three biological replicates before statistical analysis. Here, protein from PAO1 grown in MM supplemented with 0.2 mM and 1 mM Pi were compared by a Student's t test applying a threshold P value of 0.01, which was based on all possible permutations. Proteins were considered differentially abundant if the log2 fold change was greater than 0.8. “ON/OFF proteins” were defined as being identified in all bioreplicates of one condition, whereas the protein was not identified in any replicate of the other condition.
Bioinformatics.
Heatmap and hierarchical clustering analyses of the detected and differentially expressed proteins were made using the ComplexHeatmap version 2.9.3 package in R (108). Biological pathway enrichment analysis of differentially expressed proteins was performed using the BlastKOALA tool from the Kyoto Encyclopedia of Genes and Genomes (KEGG) (109). Fig. 4 to 10 were created using BioRender.com.
Data availability.
The MS data reported in this publication have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (110) with data set identifier PXD033250.
ACKNOWLEDGMENTS
This work was supported by grants PID2019-103972GA-I00 (to M.A.M.) and PID2020-112612GB-I00 (to T.K.) from the Spanish Ministry for Science and Innovation/Agencia Estatal de Investigación 10.13039/501100011033 and grant P18-FR-1621 (to T.K.) from the Junta de Andalucía.
The authors do not declare a conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Miguel A. Matilla, Email: miguel.matilla@eez.csic.es.
Tino Krell, Email: tino.krell@eez.csic.es.
Giordano Rampioni, University Roma Tre.
REFERENCES
- 1.Crone S, Vives-Flórez M, Kvich L, Saunders AM, Malone M, Nicolaisen MH, Martínez-García E, Rojas-Acosta C, Catalina Gomez-Puerto M, Calum H, Whiteley M, Kolter R, Bjarnsholt T. 2020. The environmental occurrence of Pseudomonas aeruginosa. APMIS 128:220–231. doi: 10.1111/apm.13010. [DOI] [PubMed] [Google Scholar]
- 2.Bel Hadj Ahmed A, Salah Abbassi M, Rojo-Bezares B, Ruiz-Roldán L, Dhahri R, Mehri I, Sáenz Y, Hassen A. 2020. Characterization of Pseudomonas aeruginosa isolated from various environmental niches: new STs and occurrence of antibiotic susceptible “high-risk clones. Int J Environ Health Res 30:643–652. doi: 10.1080/09603123.2019.1616080. [DOI] [PubMed] [Google Scholar]
- 3.Bédard E, Prévost M, Déziel E. 2016. Pseudomonas aeruginosa in premise plumbing of large buildings. Microbiologyopen 5:937–956. doi: 10.1002/mbo3.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iglewski BH. 1996. Chapter 27 Pseudomonas. In Baron S (ed), Medical Microbiology, 4th edition. University of Texas Medical Branch, Galveston, Galveston (TX). [Google Scholar]
- 5.Morata L, Cobos-Trigueros N, Martínez JA, Soriano A, Almela M, Marco F, Sterzik H, Núñez R, Hernández C, Mensa J. 2012. Influence of multidrug resistance and appropriate empirical therapy on the 30-day mortality rate of Pseudomonas aeruginosa bacteremia. Antimicrob Agents Chemother 56:4833–4837. doi: 10.1128/AAC.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sindeldecker D, Stoodley P. 2021. The many antibiotic resistance and tolerance strategies of Pseudomonas aeruginosa. Biofilm 3:100056. doi: 10.1016/j.bioflm.2021.100056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jean SS, Chang YC, Lin WC, Lee WS, Hsueh PR, Hsu CW. 2020. Epidemiology, Treatment, and Prevention of Nosocomial Bacterial Pneumonia. JCM 9:275. doi: 10.3390/jcm9010275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang CI, Kim SH, Kim HB, Park SW, Choe YJ, Oh MD, Kim EC, Choe KW. 2003. Pseudomonas aeruginosa bacteremia: risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin Infect Dis 37:745–751. doi: 10.1086/377200. [DOI] [PubMed] [Google Scholar]
- 9.Walker TS, Bais HP, Deziel E, Schweizer HP, Rahme LG, Fall R, Vivanco JM. 2004. Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol 134:320–331. doi: 10.1104/pp.103.027888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rahme LG, Ausubel FM, Cao H, Drenkard E, Goumnerov BC, Lau GW, Mahajan-Miklos S, Plotnikova J, Tan MW, Tsongalis J, Walendziewicz CL, Tompkins RG. 2000. Plants and animals share functionally common bacterial virulence factors. Proc Natl Acad Sci USA 97:8815–8821. doi: 10.1073/pnas.97.16.8815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Kruger WM, Lery LM, Soares MR, de Neves-Manta FS, Batista e Silva CM, Neves-Ferreira AG, Perales J, Bisch PM. 2006. The phosphate-starvation response in Vibrio cholerae O1 and phoB mutant under proteomic analysis: disclosing functions involved in adaptation, survival and virulence. Proteomics 6:1495–1511. doi: 10.1002/pmic.200500238. [DOI] [PubMed] [Google Scholar]
- 12.Aggarwal S, Somani VK, Bhatnagar R. 2015. Phosphate starvation enhances the pathogenesis of Bacillus anthracis. Int J Med Microbiol 305:523–531. doi: 10.1016/j.ijmm.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Elliott SR, White DW, Tischler AD. 2019. Mycobacterium tuberculosis requires regulation of ESX-5 secretion for virulence in Irgm1-deficient mice. Infect Immun 87:e00660-18. doi: 10.1128/IAI.00660-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelliher JL, Radin JN, Kehl-Fie TE. 2018. PhoPR contributes to Staphylococcus aureus growth during phosphate starvation and pathogenesis in an environment-specific manner. Infect Immun 86:e00371-18. doi: 10.1128/IAI.00371-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aoyama T, Takanami M, Makino K, Oka A. 1991. Cross-talk between the virulence and phosphate regulons of Agrobacterium tumefaciens caused by an unusual interaction of the transcriptional activator with a regulatory DNA element. Mol Gen Genet 227:385–390. doi: 10.1007/BF00273927. [DOI] [PubMed] [Google Scholar]
- 16.Zheng D, Xue B, Shao Y, Yu H, Yao X, Ruan L. 2018. Activation of PhoBR under phosphate-rich conditions reduces the virulence of Xanthomonas oryzae pv. oryzae. Mol Plant Pathol 19:2066–2076. doi: 10.1111/mpp.12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Köhler JR, Acosta-Zaldívar M, Qi W. 2020. Phosphate in virulence of Candida albicans and Candida glabrata. JoF 6:40. doi: 10.3390/jof6020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bains M, Fernandez L, Hancock RE. 2012. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl Environ Microbiol 78:6762–6768. doi: 10.1128/AEM.01015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zaborin A, Romanowski K, Gerdes S, Holbrook C, Lepine F, Long J, Poroyko V, Diggle SP, Wilke A, Righetti K, Morozova I, Babrowski T, Liu DC, Zaborina O, Alverdy JC. 2009. Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc Natl Acad Sci USA 106:6327–6332. doi: 10.1073/pnas.0813199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shor R, Halabe A, Rishver S, Tilis Y, Matas Z, Fux A, Boaz M, Weinstein J. 2006. Severe hypophosphatemia in sepsis as a mortality predictor. Ann Clin Lab Sci 36:67–72. [PubMed] [Google Scholar]
- 21.Long J, Zaborina O, Holbrook C, Zaborin A, Alverdy J. 2008. Depletion of intestinal phosphate after operative injury activates the virulence of P aeruginosa causing lethal gut-derived sepsis. Surgery 144:189–197. doi: 10.1016/j.surg.2008.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salem RR, Tray K. 2005. Hepatic resection-related hypophosphatemia is of renal origin as manifested by isolated hyperphosphaturia. Ann Surg 241:343–348. doi: 10.1097/01.sla.0000152093.43468.c0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamarche MG, Wanner BL, Crepin S, Harel J. 2008. The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol Rev 32:461–473. doi: 10.1111/j.1574-6976.2008.00101.x. [DOI] [PubMed] [Google Scholar]
- 24.Lee J, Wu J, Deng Y, Wang J, Wang C, Wang J, Chang C, Dong Y, Williams P, Zhang LH. 2013. A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol 9:339–343. doi: 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- 25.Soto-Aceves MP, Cocotl-Yañez M, Servín-González L, Soberón-Chávez G. 2021. The Rhl Quorum-Sensing System Is at the Top of the Regulatory Hierarchy under Phosphate-Limiting Conditions in Pseudomonas aeruginosa PAO1. J Bacteriol 203:e00475-20. doi: 10.1128/JB.00475-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng X, Ahator SD, Zhang LH. 2020. Molecular mechanisms of phosphate stress activation of Pseudomonas aeruginosa quorum sensing systems. mSphere 5:e00119-20. doi: 10.1128/mSphere.00119-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thuong M, Arvaniti K, Ruimy R, de la Salmoniere P, Scanvic-Hameg A, Lucet JC, Regnier B. 2003. Epidemiology of Pseudomonas aeruginosa and risk factors for carriage acquisition in an intensive care unit. J Hosp Infect 53:274–282. doi: 10.1053/jhin.2002.1370. [DOI] [PubMed] [Google Scholar]
- 28.Murono K, Hirano Y, Koyano S, Ito K, Fujieda K. 2003. Molecular comparison of bacterial isolates from blood with strains colonizing pharynx and intestine in immunocompromised patients with sepsis. J Med Microbiol 52:527–530. doi: 10.1099/jmm.0.05076-0. [DOI] [PubMed] [Google Scholar]
- 29.Romanowski K, Zaborin A, Fernandez H, Poroyko V, Valuckaite V, Gerdes S, Liu DC, Zaborina OY, Alverdy JC. 2011. Prevention of siderophore- mediated gut-derived sepsis due to P aeruginosa can be achieved without iron provision by maintaining local phosphate abundance: role of pH. BMC Microbiol 11:212–212. doi: 10.1186/1471-2180-11-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanowski K, Zaborin A, Valuckaite V, Rolfes RJ, Babrowski T, Bethel C, Olivas A, Zaborina O, Alverdy JC. 2012. Candida albicans isolates from the gut of critically ill patients respond to phosphate limitation by expressing filaments and a lethal phenotype. PLoS One 7:e30119. doi: 10.1371/journal.pone.0030119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zaborin A, Defazio JR, Kade M, Kaiser BL, Belogortseva N, Camp DG, 2nd, Smith RD, Adkins JN, Kim SM, Alverdy A, Goldfeld D, Firestone MA, Collier JH, Jabri B, Tirrell M, Zaborina O, Alverdy JC. 2014. Phosphate-containing polyethylene glycol polymers prevent lethal sepsis by multidrug-resistant pathogens. Antimicrob Agents Chemother 58:966–977. doi: 10.1128/AAC.02183-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mao J, Zaborin A, Poroyko V, Goldfeld D, Lynd NA, Chen W, Tirrell MV, Zaborina O, Alverdy JC. 2017. De novo synthesis of phosphorylated triblock copolymers with pathogen virulence-suppressing properties that prevent infection-related mortality. ACS Biomater Sci Eng 3:2076–2085. doi: 10.1021/acsbiomaterials.7b00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaborin A, Smith D, Garfield K, Quensen J, Shakhsheer B, Kade M, Tirrell M, Tiedje J, Gilbert JA, Zaborina O, Alverdy JC. 2014. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. mBio 5:e01361-14–e01314. doi: 10.1128/mBio.01361-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiegerinck M, Hyoju SK, Mao J, Zaborin A, Adriaansens C, Salzman E, Hyman NH, Zaborina O, van Goor H, Alverdy JC. 2018. Novel de novo synthesized phosphate carrier compound ABA-PEG20k-Pi20 suppresses collagenase production in Enterococcus faecalis and prevents colonic anastomotic leak in an experimental model. Br J Surg 105:1368–1376. doi: 10.1002/bjs.10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nichols D, Pimentel MB, Borges FTP, Hyoju SK, Teymour F, Hong SH, Zaborina OY, Alverdy JC, Papavasiliou G. 2019. Sustained release of phosphates from hydrogel nanoparticles suppresses bacterial collagenase and biofilm formation in vitro. Front Bioeng Biotechnol 7:153. doi: 10.3389/fbioe.2019.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyoju SK, Klabbers RE, Aaron M, Krezalek MA, Zaborin A, Wiegerinck M, Hyman NH, Zaborina O, Van Goor H, Alverdy JC. 2018. Oral polyphosphate suppresses bacterial collagenase production and prevents anastomotic leak due to Serratia marcescens and Pseudomonas aeruginosa. Ann Surg 267:1112–1118. doi: 10.1097/SLA.0000000000002167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Neil J. 2016. Tackling drug-resistant infections globally: final report and recommendations. Review on Antimicrobial Resistance. https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf. [Google Scholar]
- 38.Taylor J, Hafner M, Yerushalmi E, Smith R, Bellasio J, Vardavas R, Bienkowska T, Rubin J. 2018. Estimating the economic costs of antimicrobial resistance. Rand Corporation. https://www.rand.org/pubs/research_reports/RR911html. [Google Scholar]
- 39.Ardal C, Balasegaram M, Laxminarayan R, McAdams D, Outterson K, Rex JH, Sumpradit N. 2020. Antibiotic development - economic, regulatory and societal challenges. Nat Rev Microbiol 18:267–274. doi: 10.1038/s41579-019-0293-3. [DOI] [PubMed] [Google Scholar]
- 40.Ventola CL. 2015. The antibiotic resistance crisis: part 1: causes and threats. P T 40:277–283. [PMC free article] [PubMed] [Google Scholar]
- 41.WHO. 2014. Antimicrobial Resistance: Global report on Surveillance. https://appswhoint/iris/bitstream/handle/10665/112642/9789241564748_engpdf;jsessionid=D6567BD288AB09C8AE4C5D5F85608685?sequence=1.
- 42.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, Pulcini C, Kahlmeter G, Kluytmans J, Carmeli Y, Ouellette M, Outterson K, Patel J, Cavaleri M, Cox EM, Houchens CR, Grayson ML, Hansen P, Singh N, Theuretzbacher U, Magrini N, WHO Pathogens Priority List Working Group. 2018. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis 18:318–327. https://www.sciencedirect.com/science/article/pii/S1473309917307533?via%3Dihub. [DOI] [PubMed] [Google Scholar]
- 43.Cegelski L, Marshall GR, Eldridge GR, Hultgren SJ. 2008. The biology and future prospects of antivirulence therapies. Nat Rev Microbiol 6:17–27. doi: 10.1038/nrmicro1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bjarnsholt T, Ciofu O, Molin S, Givskov M, Hoiby N. 2013. Applying insights from biofilm biology to drug development - can a new approach be developed? Nat Rev Drug Discov 12:791–808. doi: 10.1038/nrd4000. [DOI] [PubMed] [Google Scholar]
- 45.Allen RC, Popat R, Diggle SP, Brown SP. 2014. Targeting virulence: can we make evolution-proof drugs? Nat Rev Microbiol 12:300–308. doi: 10.1038/nrmicro3232. [DOI] [PubMed] [Google Scholar]
- 46.Krell T, Matilla MA. 2022. Antimicrobial resistance: progress and challenges in antibiotic discovery and anti-infective therapy. Microb Biotechnol 15:70–78. doi: 10.1111/1751-7915.13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chadha J, Harjai K, Chhibber S. 2022. Revisiting the virulence hallmarks of Pseudomonas aeruginosa: a chronicle through the perspective of quorum sensing. Environ Microbiol 24:2630–2656. doi: 10.1111/1462-2920.15784. [DOI] [PubMed] [Google Scholar]
- 48.Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA, Brinkman FS. 2016. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res 44:D646–53. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Otero-Asman JR, Quesada JM, Jim KK, Ocampo-Sosa A, Civantos C, Bitter W, Llamas MA. 2020. The extracytoplasmic function sigma factor σ(VreI) is active during infection and contributes to phosphate starvation-induced virulence of Pseudomonas aeruginosa. Sci Rep 10:3139. doi: 10.1038/s41598-020-60197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peng YC, Lu C, Li G, Eichenbaum Z, Lu CD. 2017. Induction of the pho regulon and polyphosphate synthesis against spermine stress in Pseudomonas aeruginosa. Mol Microbiol 104:1037–1051. doi: 10.1111/mmi.13678. [DOI] [PubMed] [Google Scholar]
- 51.Modi N, Ganguly S, Bárcena-Uribarri I, Benz R, van den Berg B, Kleinekathöfer U. 2015. Structure, dynamics, and substrate specificity of the OprO porin from Pseudomonas aeruginosa. Biophys J 109:1429–1438. doi: 10.1016/j.bpj.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chevalier S, Bouffartigues E, Bodilis J, Maillot O, Lesouhaitier O, Feuilloley MGJ, Orange N, Dufour A, Cornelis P. 2017. Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol Rev 41:698–722. doi: 10.1093/femsre/fux020. [DOI] [PubMed] [Google Scholar]
- 53.Chugani S, Greenberg EP. 2007. The influence of human respiratory epithelia on Pseudomonas aeruginosa gene expression. Microb Pathog 42:29–35. doi: 10.1016/j.micpath.2006.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stasi R, Neves HI, Spira B. 2019. Phosphate uptake by the phosphonate transport system PhnCDE. BMC Microbiol 19:79. doi: 10.1186/s12866-019-1445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rapisarda C, Tassinari M, Gubellini F, Fronzes R. 2018. Using cryo-EM to investigate bacterial secretion systems. Annu Rev Microbiol 72:231–254. doi: 10.1146/annurev-micro-090817-062702. [DOI] [PubMed] [Google Scholar]
- 56.Filloux A. 2011. Protein secretion systems in Pseudomonas aeruginosa: an essay on diversity, evolution, and function. Front Microbiol 2:155. doi: 10.3389/fmicb.2011.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peters JE, Park SJ, Darzins A, Freck LC, Saulnier JM, Wallach JM, Galloway DR. 1992. Further studies on Pseudomonas aeruginosa LasA: analysis of specificity. Mol Microbiol 6:1155–1162. doi: 10.1111/j.1365-2958.1992.tb01554.x. [DOI] [PubMed] [Google Scholar]
- 58.Kielty CM, Sherratt MJ, Shuttleworth CA. 2002. Elastic fibres. J Cell Sci 115:2817–2828. doi: 10.1242/jcs.115.14.2817. [DOI] [PubMed] [Google Scholar]
- 59.Cowell BA, Twining SS, Hobden JA, Kwong MSF, Fleiszig SMJ. 2003. Mutation of lasA and lasB reduces Pseudomonas aeruginosa invasion of epithelial cells. Microbiology (Reading) 149:2291–2299. doi: 10.1099/mic.0.26280-0. [DOI] [PubMed] [Google Scholar]
- 60.Tan X, Cheng X, Hu M, Zhang Y, Jia A, Zhou J, Zhu G. 2021. Transcriptional analysis and target genes discovery of Pseudomonas aeruginosa biofilm developed ex vivo chronic wound model. AMB Express 11:157–157. doi: 10.1186/s13568-021-01317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ball G, Durand E, Lazdunski A, Filloux A. 2002. A novel type II secretion system in Pseudomonas aeruginosa. Mol Microbiol 43:475–485. doi: 10.1046/j.1365-2958.2002.02759.x. [DOI] [PubMed] [Google Scholar]
- 62.Filloux A, Michel G, Bally M. 1998. GSP-dependent protein secretion in gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol Rev 22:177–198. doi: 10.1111/j.1574-6976.1998.tb00366.x. [DOI] [PubMed] [Google Scholar]
- 63.Lewenza S, Charron-Mazenod L, Afroj S, van Tilburg Bernardes E. 2017. Hyperbiofilm phenotype of Pseudomonas aeruginosa defective for the PlcB and PlcN secreted phospholipases. Can J Microbiol 63:780–787. doi: 10.1139/cjm-2017-0244. [DOI] [PubMed] [Google Scholar]
- 64.Wilton M, Halverson TWR, Charron-Mazenod L, Parkins MD, Lewenza S. 2018. Secreted phosphatase and deoxyribonuclease are required by Pseudomonas aeruginosa to defend against neutrophil extracellular traps. Infect Immun 86:e00403-18. doi: 10.1128/IAI.00403-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Filloux A, Hachani A, Bleves S. 2008. The bacterial type VI secretion machine: yet another player for protein transport across membranes. Microbiology (Reading) 154:1570–1583. doi: 10.1099/mic.0.2008/016840-0. [DOI] [PubMed] [Google Scholar]
- 66.Sana TG, Berni B, Bleves S. 2016. The T6SSs of Pseudomonas aeruginosa Strain PAO1 and Their Effectors: beyond Bacterial-Cell Targeting. Front Cell Infect Microbiol 6:61. doi: 10.3389/fcimb.2016.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ueda A, Wood TK. 2009. Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog 5:e1000483. doi: 10.1371/journal.ppat.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pu M, Wood TK. 2010. Tyrosine phosphatase TpbA controls rugose colony formation in Pseudomonas aeruginosa by dephosphorylating diguanylate cyclase TpbB. Biochem Biophys Res Commun 402:351–355. doi: 10.1016/j.bbrc.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Domenech CE, Otero LH, Beassoni PR, Lisa AT. 2011. Phosphorylcholine Phosphatase: a Peculiar Enzyme of Pseudomonas aeruginosa. Enzyme Res 2011:561841. doi: 10.4061/2011/561841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ostroff RM, Wretlind B, Vasil ML. 1989. Mutations in the hemolytic-phospholipase C operon result in decreased virulence of Pseudomonas aeruginosa PAO1 grown under phosphate-limiting conditions. Infect Immun 57:1369–1373. doi: 10.1128/iai.57.5.1369-1373.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cota-Gomez A, Vasil AI, Kadurugamuwa J, Beveridge TJ, Schweizer HP, Vasil ML. 1997. PlcR1 and PlcR2 are putative calcium-binding proteins required for secretion of the hemolytic phospholipase C of Pseudomonas aeruginosa. Infect Immun 65:2904–2913. doi: 10.1128/iai.65.7.2904-2913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vilaplana L, Marco MP. 2020. Phenazines as potential biomarkers of Pseudomonas aeruginosa infections: synthesis regulation, pathogenesis and analytical methods for their detection. Anal Bioanal Chem 412:5897–5912. doi: 10.1007/s00216-020-02696-4. [DOI] [PubMed] [Google Scholar]
- 73.Biswas L, Götz F. 2021. Molecular Mechanisms of Staphylococcus and Pseudomonas Interactions in Cystic Fibrosis. Front Cell Infect Microbiol 11:824042. doi: 10.3389/fcimb.2021.824042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hall S, McDermott C, Anoopkumar-Dukie S, McFarland AJ, Forbes A, Perkins AV, Davey AK, Chess-Williams R, Kiefel MJ, Arora D, Grant GD. 2016. Cellular Effects of Pyocyanin, a Secreted Virulence Factor of Pseudomonas aeruginosa. Toxins 8:236. doi: 10.3390/toxins8080236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matilla MA, Martin-Mora D, Gavira JA, Krell T. 2021. Pseudomonas aeruginosa as a model to study chemosensory pathway signaling. Microbiol Mol Biol Rev 85:e00151-20. doi: 10.1128/MMBR.00151-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garcia-Fontana C, Vilchez JI, Gonzalez-Requena M, Gonzalez-Lopez J, Krell T, Matilla MA, Manzanera M. 2019. The involvement of McpB chemoreceptor from Pseudomonas aeruginosa PAO1 in virulence. Sci Rep 9:13166. doi: 10.1038/s41598-019-49697-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garvis S, Munder A, Ball G, de Bentzmann S, Wiehlmann L, Ewbank JJ, Tummler B, Filloux A. 2009. Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathog 5:e1000540. doi: 10.1371/journal.ppat.1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee J, Zhang L. 2015. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6:26–41. doi: 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Schuster M, Lostroh CP, Ogi T, Greenberg EP. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jensen V, Löns D, Zaoui C, Bredenbruch F, Meissner A, Dieterich G, Münch R, Häussler S. 2006. RhlR expression in Pseudomonas aeruginosa is modulated by the Pseudomonas quinolone signal via PhoB-dependent and -independent pathways. J Bacteriol 188:8601–8606. doi: 10.1128/JB.01378-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gutu AD, Sgambati N, Strasbourger P, Brannon MK, Jacobs MA, Haugen E, Kaul RK, Johansen HK, Høiby N, Moskowitz SM. 2013. Polymyxin resistance of Pseudomonas aeruginosa phoQ mutants is dependent on additional two-component regulatory systems. Antimicrob Agents Chemother 57:2204–2215. doi: 10.1128/AAC.02353-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kaihami GH, Breda LCD, de Almeida JRF, de Oliveira Pereira T, Nicastro GG, Boechat AL, de Almeida SR, Baldini RL. 2017. The atypical response regulator atvr is a new player in Pseudomonas aeruginosa response to hypoxia and virulence. Infect Immun 85:e00207-17. doi: 10.1128/IAI.00207-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Petrova OE, Sauer K. 2009. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog 5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cao Q, Yang N, Wang Y, Xu C, Zhang X, Fan K, Chen F, Liang H, Zhang Y, Deng X, Feng Y, Yang CG, Wu M, Bae T, Lan L. 2020. Mutation-induced remodeling of the BfmRS two-component system in Pseudomonas aeruginosa clinical isolates. Sci Signal 13:eaaz1529. doi: 10.1126/scisignal.aaz1529. [DOI] [PubMed] [Google Scholar]
- 85.Chuanchuen R, Gaynor JB, Karkhoff-Schweizer R, Schweizer HP. 2005. Molecular characterization of MexL, the transcriptional repressor of the mexJK multidrug efflux operon in Pseudomonas aeruginosa. Antimicrob Agents Chemother 49:1844–1851. doi: 10.1128/AAC.49.5.1844-1851.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Juarez P, Jeannot K, Plésiat P, Llanes C. 2017. Toxic electrophiles induce expression of the multidrug efflux Pump MexEF-OprN in Pseudomonas aeruginosa through a novel transcriptional regulator, CmrA. Antimicrob Agents Chemother 61:e00585-17. doi: 10.1128/AAC.00585-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang F, He Q, Yin J, Xu S, Hu W, Gu L. 2018. BrlR from Pseudomonas aeruginosa is a receptor for both cyclic di-GMP and pyocyanin. Nat Commun 9:2563. doi: 10.1038/s41467-018-05004-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jones CJ, Ryder CR, Mann EE, Wozniak DJ. 2013. AmrZ modulates Pseudomonas aeruginosa biofilm architecture by directly repressing transcription of the psl operon. J Bacteriol 195:1637–1644. doi: 10.1128/JB.02190-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Xu A, Zhang M, Du W, Wang D, Ma LZ. 2021. A molecular mechanism for how sigma factor AlgT and transcriptional regulator AmrZ inhibit twitching motility in Pseudomonas aeruginosa. Environ Microbiol 23:572–587. doi: 10.1111/1462-2920.14985. [DOI] [PubMed] [Google Scholar]
- 90.Allsopp LP, Wood TE, Howard SA, Maggiorelli F, Nolan LM, Wettstadt S, Filloux A. 2017. RsmA and AmrZ orchestrate the assembly of all three type VI secretion systems in Pseudomonas aeruginosa. Proc Natl Acad Sci USA 114:7707–7712. doi: 10.1073/pnas.1700286114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Potvin E, Sanschagrin F, Levesque RC. 2008. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev 32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- 92.Xu L, Xin L, Zeng Y, Yam JK, Ding Y, Venkataramani P, Cheang QW, Yang X, Tang X, Zhang LH, Chiam KH, Yang L, Liang ZX. 2016. A cyclic di-GMP-binding adaptor protein interacts with a chemotaxis methyltransferase to control flagellar motor switching. Sci Signal 9:ra102. doi: 10.1126/scisignal.aaf7584. [DOI] [PubMed] [Google Scholar]
- 93.Miklavič Š, Kogovšek P, Hodnik V, Korošec J, Kladnik A, Anderluh G, Gutierrez-Aguirre I, Maček P, Butala M. 2015. The Pseudomonas aeruginosa RhlR-controlled aegerolysin RahU is a low-affinity rhamnolipid-binding protein. FEMS Microbiol Lett 362:fnv069. [DOI] [PubMed] [Google Scholar]
- 94.Rao J, DiGiandomenico A, Artamonov M, Leitinger N, Amin AR, Goldberg JB. 2011. Host derived inflammatory phospholipids regulate rahU (PA0122) gene, protein, and biofilm formation in Pseudomonas aeruginosa. Cell Immunol 270:95–102. doi: 10.1016/j.cellimm.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rao J, Elliott MR, Leitinger N, Jensen RV, Goldberg JB, Amin AR. 2011. RahU: an inducible and functionally pleiotropic protein in Pseudomonas aeruginosa modulates innate immunity and inflammation in host cells. Cell Immunol 270:103–113. doi: 10.1016/j.cellimm.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fernández L, Breidenstein EB, Song D, Hancock RE. 2012. Role of intracellular proteases in the antibiotic resistance, motility, and biofilm formation of Pseudomonas aeruginosa. Antimicrob Agents Chemother 56:1128–1132. doi: 10.1128/AAC.05336-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Abdel-Mawgoud AM, Lépine F, Déziel E. 2010. Rhamnolipids: diversity of structures, microbial origins and roles. Appl Microbiol Biotechnol 86:1323–1336. doi: 10.1007/s00253-010-2498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Moskowitz SM, Ernst RK, Miller SI. 2004. PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol 186:575–579. doi: 10.1128/JB.186.2.575-579.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee JY, Chung ES, Na IY, Kim H, Shin D, Ko KS. 2014. Development of colistin resistance in pmrA-, phoP-, parR- and cprR-inactivated mutants of Pseudomonas aeruginosa. J Antimicrob Chemother 69:2966–2971. doi: 10.1093/jac/dku238. [DOI] [PubMed] [Google Scholar]
- 100.McCarthy RR, Mazon-Moya MJ, Moscoso JA, Hao Y, Lam JS, Bordi C, Mostowy S, Filloux A. 2017. Cyclic-di-GMP regulates lipopolysaccharide modification and contributes to Pseudomonas aeruginosa immune evasion. Nat Microbiol 2:17027. doi: 10.1038/nmicrobiol.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Srivastava D, Seo J, Rimal B, Kim SJ, Zhen S, Darwin AJ. 2018. A Proteolytic Complex Targets Multiple Cell Wall Hydrolases in Pseudomonas aeruginosa. mBio 9:e00972-18. doi: 10.1128/mBio.00972-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Seo J, Darwin AJ. 2013. The Pseudomonas aeruginosa periplasmic protease CtpA can affect systems that impact its ability to mount both acute and chronic infections. Infect Immun 81:4561–4570. doi: 10.1128/IAI.01035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Harrison LB, Fowler RC, Abdalhamid B, Selmecki A, Hanson ND. 2019. lptG contributes to changes in membrane permeability and the emergence of multidrug hypersusceptibility in a cystic fibrosis isolate of Pseudomonas aeruginosa. Microbiologyopen 8:e844. doi: 10.1002/mbo3.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart G, Williams P. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 105.Bonn F, Bartel J, Buttner K, Hecker M, Otto A, Becher D. 2014. Picking vanished proteins from the void: how to collect and ship/share extremely dilute proteins in a reproducible and highly efficient manner. Anal Chem 86:7421–7427. doi: 10.1021/ac501189j. [DOI] [PubMed] [Google Scholar]
- 106.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. 2011. Global quantification of mammalian gene expression control. Nature 473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 107.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 108.Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 109.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–62. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vizcaino JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44:11033. doi: 10.1093/nar/gkw880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Clarke DM, Loo TW, Gillam S, Bragg PD. 1986. Nucleotide sequence of the pntA and pntB genes encoding the pyridine nucleotide transhydrogenase of Escherichia coli. Eur J Biochem 158:647–653. doi: 10.1111/j.1432-1033.1986.tb09802.x. [DOI] [PubMed] [Google Scholar]
- 112.Jones CJ, Newsom D, Kelly B, Irie Y, Jennings LK, Xu B, Limoli DH, Harrison JJ, Parsek MR, White P, Wozniak DJ. 2014. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog 10:e1003984. doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Intile PJ, Balzer GJ, Wolfgang MC, Yahr TL. 2015. The RNA helicase DeaD stimulates ExsA translation to promote expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol 197:2664–2674. doi: 10.1128/JB.00231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Salusso A, Raimunda D. 2017. Defining the roles of the cation diffusion facilitators in Fe(2+)/Zn(2+) homeostasis and establishment of their participation in virulence in Pseudomonas aeruginosa. Front Cell Infect Microbiol 7:84. doi: 10.3389/fcimb.2017.00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ishige K, Zhang H, Kornberg A. 2002. Polyphosphate kinase (PPK2), a potent, polyphosphate-driven generator of GTP. Proc Natl Acad Sci USA 99:16684–16688. doi: 10.1073/pnas.262655299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol 185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. 2001. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol 183:6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Siehnel RJ, Worobec EA, Hancock RE. 1988. Regulation of components of the Pseudomonas aeruginosa phosphate-starvation-inducible regulon in Escherichia coli. Mol Microbiol 2:347–352. doi: 10.1111/j.1365-2958.1988.tb00038.x. [DOI] [PubMed] [Google Scholar]
- 119.Faure LM, Garvis S, de Bentzmann S, Bigot S. 2014. Characterization of a novel two-partner secretion system implicated in the virulence of Pseudomonas aeruginosa. Microbiology 160:1940–1952. doi: 10.1099/mic.0.079616-0. [DOI] [PubMed] [Google Scholar]
- 120.Faure LM, Llamas MA, Bastiaansen KC, de Bentzmann S, Bigot S. 2013. Phosphate starvation relayed by PhoB activates the expression of the Pseudomonas aeruginosa σvreI ECF factor and its target genes. Microbiology 159:1315–1327. doi: 10.1099/mic.0.067645-0. [DOI] [PubMed] [Google Scholar]
- 121.Quesada JM, Otero-Asman JR, Bastiaansen KC, Civantos C, Llamas MA. 2016. The activity of the Pseudomonas aeruginosa virulence regulator σ(VreI) is modulated by the anti-σ factor VreR and the transcription Factor PhoB. Front Microbiol 7:1159. doi: 10.3389/fmicb.2016.01159. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download spectrum.02590-22-s0001.xlsx, XLSX file, 0.7 MB (750.5KB, xlsx)
Data Set S2. Download spectrum.02590-22-s0002.xlsx, XLSX file, 0.3 MB (334.6KB, xlsx)
Data Set S3. Download spectrum.02590-22-s0003.xlsx, XLSX file, 0.1 MB (121.2KB, xlsx)
Data Availability Statement
The MS data reported in this publication have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository (110) with data set identifier PXD033250.