Abstract
The ability of Campylobacter jejuni to penetrate normally nonphagocytic host cells is believed to be a key virulence determinant. Recently, kinetics of C. jejuni intracellular survival have been described and indicate that the bacterium can persist and multiply within epithelial cells and macrophages in vitro. Studies conducted by Pesci et al. indicate that superoxide dismutase contributes to intraepithelial cell survival, as isogenic sod mutants are 12-fold more sensitive to intracellular killing than wild-type strains. These findings suggest that bacterial factors that combat reactive oxygen species enable the organism to persist inside host cells. Experiments were conducted to determine the contribution of catalase to C. jejuni intracellular survival. Zymographic analysis indicated that C. jejuni expresses a single catalase enzyme. The gene encoding catalase (katA) was cloned via functional complementation, and an isogenic katA mutant strain was constructed. Kinetic studies indicate that catalase provides resistance to hydrogen peroxide in vitro but does not play a role in intraepithelial cell survival. Catalase does however contribute to intramacrophage survival. Kinetic studies of C. jejuni growth in murine and porcine peritoneal macrophages demonstrated extensive killing of both wild-type and katA mutant strains shortly following internalization. Long-term cultures (72 h postinfection) of infected phagocytes permitted recovery of viable wild-type C. jejuni; in contrast, no viable katA mutant bacteria were recovered. Accordingly, inhibition of macrophage nitric oxide synthase or NADPH oxidase permitted recovery of katA mutant C. jejuni. These observations indicate that catalase is essential for C. jejuni intramacrophage persistence and growth and suggest a novel mechanism of intracellular survival.
Campylobacter jejuni is a microaerobic, highly motile, gram-negative bacterium and the primary agent of the most frequent form of human bacillary gastroenteritis, campylobacteriosis (6, 32). Campylobacteriosis is an acute illness, the signs and symptoms of which vary with socioeconomic conditions. In underdeveloped areas, the disease, which affects mainly infants and young children (<2 years), is endemic and is observed as a watery diarrhea suggestive of infection with a toxigenic organism (15). In developed areas, where the disease predominantly affects young adults, campylobacteriosis is most often observed as a dysentery suggestive of infection with an invasive organism (15). Though C. jejuni is the leading cause of human gastroenteritis worldwide, little is known of its virulence determinants. A number of studies in vivo have established that C. jejuni is a facultatively intracellular bacterium (1, 5, 17, 19). Electron microscopy studies have demonstrated the organism residing within epithelial cells lining the gut lumen as well as granulocytes and parenchymal cells located within the lamina propria (1, 5). This intracellular existence provides the fastidious, asaccharolytic, slow-growing organism an unoccupied niche where microbial competition is relaxed or nonexistent. Additionally, an intracellular lifestyle is thought to shelter organisms from immune surveillance; however, there is little evidence to support this idea regarding campylobacteriosis.
Once inside, C. jejuni may be exposed to a variety of host killing mechanisms, including various reactive oxygen species generated by the respiratory burst oxidase as the bacterium remains bound within an endosome (9, 17). These products include superoxide, hydrogen peroxide, and halogenated oxygen molecules. Hydrogen peroxide, which is generated during aerobic metabolism, has bacteriocidal activity, as suggested by the ubiquitous presence of at least two catalase enzymes expressed in organisms which use oxygen as final electron acceptors. Furthermore, interaction of hydrogen peroxide with myeloperoxidase, reduced iron, or products of nitric oxide synthase may lead to formation of more toxic intermediates, such as hypochlorous anion, hydroxyl radicals, hydroxide anions, nitrogen dioxide, and peroxynitrite (11). Therefore, it has been postulated that bacterial factors that inactivate hydrogen peroxide, such as catalase, may interrupt production of these toxic species and aid persistence and survival within host cells and tissues. Experiments have been conducted to examine the effect of oxygen radicals on the survival of bacteria (10). It was found that the Salmonella enterica serovar Typhimurium sodC mutant was more susceptible to killing in the presence of both the respiratory burst and nitric oxide synthase. This killing was reduced with the addition of NG-l-monomethyl arginine or acetovanillone, and the attenuated virulence of the sodC mutant was restored completely with the elimination of the respiratory burst. These results suggest there are synergistic antimicrobial properties resulting from the combination of the phagocytic respiratory burst and NO synthase and that the Cu-Zn Sod protects S. enterica serovar Typhimurium from the products of both these phagocytic properties. Still other studies have indicated that production of catalase and not nitric oxide correlates to virulence of Staphylococcus aureus, Neisseria meningitidis, Legionella pneumophila, Nocardia asteroides, and Mycobacterium tuberculosis (7).
These observations suggest that the ability to inactivate hydrogen peroxide contributes to survival in the host (2) and led us to examine the role of catalase in C. jejuni hydrogen peroxide resistance and intracellular survival. In this report, we demonstrate that catalase is required for C. jejuni hydrogen peroxide resistance in vitro as well as persistence within macrophages.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and plasmids used in these studies are described in Table 1. All strains of Escherichia coli were routinely cultured at 37°C using Luria-Bertani (LB) agar media or LB broth in an aerobic atmosphere. Broth cultures were incubated at 37°C with agitation (250 rpm) in an orbital shaking water bath. All strains of C. jejuni were routinely cultured at 37°C on Mueller-Hinton (MH) agar media containing 5% citrated bovine blood in an atmosphere of 10% CO2–10% H2–80% N2. The following antibiotics were used where appropriate: ampicillin (Amp) (100 μg/ml), cephalothin (Cef) (100 μg/ml), chloramphenicol (Cm) (15 μg/ml), kanamycin sulfate (Kan) (40 μg/ml), nalidixic acid (Nal) (20 μg/ml), streptomycin sulfate (Sm) (25 μg/ml), and tetracycline hydrochloride (Tet) (12.5 μg/ml).
TABLE 1.
Bacterial strains and plasmids used in these studies
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strain | ||
| C. jejuni | ||
| M129 | Wild type; isolated from individual presenting frank dysentery | 18 |
| M129N | Spontaneous nalidixic acid-resistant strain, derivative of M129 | This study |
| JD900 | M129N derivative, katA::kan, Kanr Nalr | This study |
| JD901 | JD900 derivative, JD900::pJD107, Kanr Cmr Nalr | This study |
| E. coli | ||
| DH5α | F− φ80d lacZΔM15 Δ(lacZYA-argF) U169 endA1 recA1 hsdR17 deoR thi-1 supE44 gyrA96 relA1, Nalr | Bethesda Research Laboratories |
| UM255 | pro leu rpsL hsdM hsdR endI lacY recA katG2 katE12::Tn10, Tetr | 27 |
| S17-1 | thi thr leu tonA lacY supE recA::RP4-Z-Tc::(Mu kanr::smr) Smr | 20 |
| Plasmid | ||
| pUOA20 | E. coli-C. jejuni shuttle vector; source of cat gene (Cmr) | 36 |
| pRY107 | E. coli-C. jejuni shuttle vector; RP4 oriT aphA-3 (Kanr) Campylobacter ori | 36 |
| pRY107S | pRY107 derivative lacking a 1.8-kb HindIII fragment of the Campylobacter ori, Kanr | This study |
| pJD100 | pRY107 derivative containing C. jejuni katA gene at BamHI site | This study |
| pJD107 | pJD100 derivative containing the pUOA20 cat gene at the XbaI site, Cmr | This study |
| pJD108 | pRY107S derivative containing the 1.1-kb internal katA PCR product | This study |
Molecular methods and strain construction.
All restriction endonucleases, T4 DNA ligase, calf intestinal alkaline phosphatase, and Klenow fragment of DNA polymerase I were purchased from Promega (Madison, Wis.) and used as directed by the manufacturer. C. jejuni chromosomal DNA was extracted using sodium dodecyl sulfate–proteinase K lysis and chloroform extractions as described by Meade et al. (25). Plasmid DNA was extracted via alkaline lysis extraction as described by Birnboim and Doly (4). Transformation of all E. coli strains used in these studies was accomplished via heat shock of hexamminecobalt-treated cells as described by Hanahan (14). Automated DNA sequencing using an ABI 377 cycle sequencer was accomplished at the Arizona Macro-molecular Structural Facility.
Construction of a katA mutant strain of C. jejuni was accomplished by insertion mutagenesis. A katA internal sequence that excludes N- and C-terminal residues and domains critical to catalase function amplified using PCR and primers internal to the C. jejuni katA open reading frame. The 1.1-kb product was cloned into a T-tailed EcoRV site of pBluescript KSII(+). Construction of a katA mutagenic plasmid carrying the internal katA sequence required the generation of a suicide vector encoding a Campylobacter selectable marker and oriT for conjugative delivery of the construct to C. jejuni that would replicate in E. coli. The E. coli-C. jejuni shuttle vector pRY107 encodes these factors as well as lacZα complementarity via the pBluescript multiple cloning site (MCS); however, this vector contains an E. coli ori and will exist as a plasmid in C. jejuni (36). Labigne-Rousel et al. have demonstrated that deletion of any HindIII fragment from the Campylobacter ori eliminates function (20). To generate a C. jejuni suicide vector, pRY107 was completely digested with restriction endonuclease HindIII. The 4.0- and 1.4-kb fragments were gel purified and ligated, and the products were introduced into E. coli DH5α. Transformants were selected on LB agar plates containing kanamycin and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), permitting identification of clones harboring a C. jejuni suicide vector, designated pRY107S, which retains the MCS and lacZα complementarity. To generate the katA mutagenic vector, the internal katA sequence was excised on an EcoRI-SalI fragment and ligated to identically processed pRY107S-generating plasmid pJD108, which was transformed into E. coli strain S17-1 to direct conjugative delivery of the mutagenic construct to C. jejuni by following methods described by Labigne-Roussel et al. (20). Recombinant C. jejuni M129N exconjugants were harvested in MH broth and spread on MH blood agar plates containing nalidixic acid, cephalothin, and additional antibiotics whose resistance was encoded on the mobilized plasmid. Verification of the katA locus as site of pJD108 integration was accomplished by Southern analysis of kanamycin-resistant clones of C. jejuni.
Catalase activity gels.
Overnight cultures of C. jejuni and E. coli strains were established on two MH agar plates containing no bovine blood and two LB agar plates, respectively. Bacteria were harvested from the cultures by using phosphate-buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 1.5 mM KH2PO4, and 8 mM Na2HPO4, pH 7.4) containing phenylmethylsulfonyl fluoride (1 mM) (Sigma, St. Louis, Mo.) and mixed with approximately one-third the volume of 0.1- to 0.15-mm-diameter glass beads. To produce a lysate, the mixture was vortexed for 10 min and then centrifuged for 5 min (10,000 × g). Amounts of proteins present in each clarified supernatant were determined using the bicinchoninic acid protein assay (Pierce, Rockford, Ill.). One hundred micrograms of each lysate was electrophoresed through an 8% nondenaturing polyacrylamide gel overnight and stained for catalase activity as described by Woodbury et al. (33).
Quantification of catalase activity.
Kinetics studies of catalase activity made use of C. jejuni lysates prepared for zymographic analysis (see above). At time zero, 1.8 ml of each lysate (250 μg/ml) was mixed with 0.2 ml of a phosphate buffer containing 10 mM hydrogen peroxide. One milliliter of the mixture was immediately added to a disposable cuvette (0.1 cm) and placed into a spectrophotometer (Beckman, Palo Alto, Calif.). Catalase activity was observed via degradation of hydrogen peroxide as determined by a decrease in UV light (240 nm) absorbance over time. Measurements of absorbance were taken at 15, 30, 60, and 120 s after addition of the lysate to the hydrogen peroxide buffer. Units of catalase activity present in 1 ml of lysate were calculated as described by C. C. Worthington (34). Experiments were completed in triplicate.
Determination of hydrogen peroxide sensitivity.
Strains of C. jejuni examined for hydrogen peroxide sensitivity were cultured overnight on MH agar plates without bovine blood containing appropriate antibiotics at 37°C under the modified atmosphere (see above). Cells were harvested in MH broth and diluted to an optical density of 0.3 (600 nm) in 7 ml. Precise numbers of viable bacteria in each preparation were determined via dilutional plating on MH blood agar plates. Hydrogen peroxide (Sigma) was added to the bacteria to a final concentration of 1 mM. One-milliliter aliquots of the suspensions were dispersed into 17- by-100-mm culture tubes (VWR Scientific, West Chester, Pa.) and incubated 5, 15, 30, or 60 min under the modified atmosphere at 37°C. Viable numbers of bacteria present in each aliquot were determined via dilutional plating on MH blood agar plates containing appropriate antibiotics. Experiments were completed in triplicate.
Kinetics of intracellular survival. (i) Epithelial cell culture.
Human epidermoid tissue cells (HEp-2) were obtained from the American Type Culture Collection (Manassas, Va.). The cells were cultured in modified Eagle's medium (MEM) supplemented with 10% fetal bovine serum (FBS) containing no antibiotics and were incubated at 37°C in a humidified, 5% CO2 incubator. For survival kinetic studies, each well of a 24-well tissue culture plate (Falcon) was seeded with 5 × 104 cells and incubated 18 h as described above. Immediately prior to use, the semiconfluent monolayers were washed once in MEM containing 1% FBS and no antibiotics.
(ii) Macrophage cell culture.
Eight- to 12-week-old BALB/c mice (Jackson Laboratories, Bar Harbor, Maine) were euthanatized using a Halothane chamber. Peritoneal macrophages were harvested by peritoneal lavage with ice-cold sterile 0.34 M sucrose. The peritoneal cells were washed three times in RPMI tissue culture media containing 15% FBS, diluted to 104 cells/ml in media containing gentamicin (50 μg/ml), seeded into each well of a 24-well tissue culture plate, and incubated overnight at 37°C in a humidified, 5% CO2 incubator. The next day, nonadherent cells were removed by washing each well three times in 15% FBS–RPMI without antibiotics. This procedure generated cultures of >95% macrophages, as judged by differential staining (Difquik). Immediately prior to use, the wells were washed once in 15% FBS–RPMI without antibiotics.
(iii) Intraepithelial cell survival assays.
Strains of C. jejuni examined were cultured at 37°C overnight on MH blood agar plates without antibiotics under the modified atmosphere. Bacteria were harvested in MEM containing 1% FBS and no antibiotics, washed once, and resuspended in the media to an optical density of 0.2 (600 nm). Precise numbers of bacteria in each preparation were determined by dilutional plating on MH blood agar plates containing the appropriate antibiotics. Each suspension was overlaid onto three wells containing semiconfluent HEp-2 monolayers and incubated 3 h at 37°C in the tissue culture incubator to allow bacterial adherence and internalization as described by Konkel and Joens (18). After incubation, the infected monolayers were washed three times with 1% FBS–MEM and then cultured in washing media containing gentamicin sulfate (250 μg/ml) and incubated an additional 3 h to kill extracellular bacteria. Following incubation in extracellular killing medium, the monolayers were either washed three times with PBS and lysed with 0.5% deoxycholate to recover intracellular bacteria (6-h time point) or washed three times with 1% FBS–MEM and cultured an additional 18, 42, 66, or 90 h in washing media containing no antibiotics. Following prolonged incubation, the cultures were washed three times with 1% FBS–MEM and PBS and then cultured 3 h in washing media containing gentamicin sulfate (250 μg/ml). Following the final incubation, the monolayers were washed three times with PBS and lysed with 0.5% deoxycholate to recover intracellular bacteria. Numbers of viable intracellular bacteria were determined by dilutional plate counts on MH blood agar medium containing the appropriate antibiotics. Experiments were completed in triplicate.
(iv) Intramacrophage survival assays.
C. jejuni strains examined were prepared and enumerated as described above with the following modifications. Bacteria were harvested in 15% FBS–RPMI without antibiotics and then were diluted to an optical density of 0.05 (600 nm), corresponding to roughly 5 × 106 bacteria/ml. Precise numbers of bacteria in the suspension were determined by dilutional plate counts on MH blood agar plates containing appropriate antibiotics. The suspension was applied to wells with macrophages to establish a multiplicity of infection of roughly 50 bacteria per macrophage. The cocultures were incubated 3 h at 37°C in the humidified incubator to allow phagocytosis of ∼50% of the inoculated bacteria as reported by Kiehlbauch et al. (17). Following incubation, the infected cells were either washed three times with PBS and lysed with 0.5% deoxycholate (3-h time point) or washed three times with 15% FBS–RPMI without antibiotics and incubated an additional 9, 18, 42, or 66. Following prolonged incubation, the cells were washed three times with PBS and lysed in 0.5% deoxycholate to recover intracellular bacteria. Viable intracellular bacteria were enumerated using dilutional plate counts on MH blood agar medium containing the appropriate antibiotics. Experiments were completed in triplicate.
(v) Porcine peritoneal macrophages.
Colostrum-deprived newborn piglets were obtained from sows at farrowing, rinsed with Betadine, and transported to the laboratory. The piglets were sacrificed with Beuthanasia-D, cleaned, and rinsed in alcohol. An area of the lower abdomen was scrubbed with Betadine, the skin was separated by incision, and the incision was bathed in alcohol. A 0.34 M sucrose solution was injected into the peritoneum through the exposed facia, the abdomen was massaged, and peritoneal macrophages were aspirated from the peritoneum by using a 50-ml syringe and a 16-gauge needle. The harvested cells were counted and plated at 104 per well in a 24-well plate containing RPMI medium supplemented with 15% fetal bovine serum, 50 μg of gentamicin/ml, 1 mg of vancomycin/ml, and 250 U of polymyxin B/ml. Following a 24-h incubation period, the peritoneal macrophages were washed to remove the antibiotics and used in the endocytic experiments. Macrophages were infected with either M129 (3 × 105 bacteria/well) or JD900 (4 × 105 bacteria/well), resulting in a multiplicity of infection of approximately 10 bacteria per macrophage. After incubation for 24, 48, or 72 h in a 37°C humidified 5% CO2 incubator, the infected cells were washed three times with PBS and lysed with 0.5% sodium deoxycholate to recover intracellular bacteria. The viable intracellular bacteria were quantified using plate counts on MH agar supplemented with 5% FBS. Experiments were completed in triplicate.
(vi) J774A.1 cell line.
J774A.1 cells, a murine macrophage-like cell line, were obtained from the American Type Culture Collection. These macrophage-like cells have retained the ability to display a respiratory burst. The ability of the katA mutant to survive intracellularly in macrophages was determined with the use of C. jejuni strain JD900 (katA mutant) and parent strain M129. J774A.1 cells were cultured in RPMI tissue culture media containing 10% FBS and incubated at 37°C in a humidified 5% CO2 incubator. For survival studies, each well of a 24-well tissue culture plate was seeded with 107 cells and incubated overnight as previously described. Following incubation and immediately prior to use, semiconfluent monolayers were washed three times with 10% FBS–RPMI. Bacterial cells were grown as described above, harvested, and inoculated into wells at a concentration of 109 cells/well. For inhibitor experiments, NG-l-monomethyl arginine, which competes for nitric oxide synthase, was added to wells as described above at a concentration of 400 μm to inhibit nitric oxide production. Apocynin, which inhibits NADPH oxidase, was added to wells at a concentration of 400 μm to inhibit the respiratory burst. Control wells were maintained without the use of inhibitors. The plates were then incubated as mentioned above for 24, 48, and 72 h. At each respective time point, cells were washed three times and lysed by 0.5% deoxycholate. Viable plate counts were done to determine the number of surviving bacteria. The assay was repeated five times.
RESULTS
Zymographic analysis of C. jejuni catalase activity.
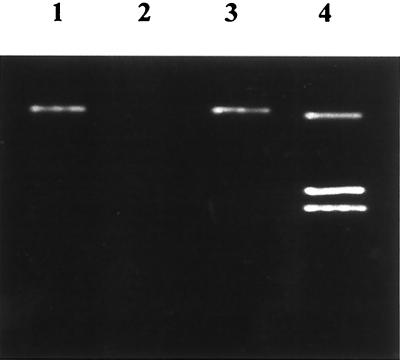
Our strategy to determine the role of catalase in C. jejuni intracellular survival involved the production of isogenic strains of C. jejuni which differ only in their ability to produce catalase. Therefore, the first step was determination of the number of catalases expressed by this bacterium, as it is known that numerous enteric bacteria, as well as others, produce two catalases which would confound efforts to generate isogenic C. jejuni catalase mutants deficient in catalase production. Examination of the number of catalases produced by C. jejuni employed zymographic analysis via catalase activity gels. To this end, a lysate was prepared from C. jejuni cells cultured on MH agar plates without blood under microaerophilic conditions. Lysate produced from E. coli strain DH5α cultured on LB agar plates grown overnight in an aerobic atmosphere served as control for the experiment. Constituents in the lysates were electrophoretically separated through a nondenaturing polyacrylamide gel and stained for catalase activity. Clear bands in the stained gel correspond to areas of catalase activity (Fig. 1). Three bands, indicative of the activity of KatE and KatG catalases (top and bottom bands, respectively) expressed by E. coli are consistent with previous zymographic reports (Fig. 1, lane 4) (27). In contrast, a single clear band, resultant of catalase activity, was identified from the C. jejuni lysate, indicating the expression of one catalase protein (Fig. 1, lane 1). No additional clear bands were apparent from C. jejuni treated with 1 to 10 mM hydrogen peroxide, indicating that no other catalases are induced upon exposure to peroxide (data not shown). Consequently, we believe that C. jejuni expresses a single catalase enzyme.
FIG. 1.
Zymographic analysis of C. jejuni catalase activity. Lane 1, C. jejuni strain M129; lane 2, E. coli UM255; lane 3, E. coli UM255 containing the plasmid pJD100; lane 4, E. coli DH5α.
Cloning and sequence analysis of the C. jejuni catalase gene.
To clone the C. jejuni catalase gene, genomic DNA was extracted from strain M129, partially digested with restriction endonuclease Sau3A1, size fractionated (4 to 7 kb), and used to construct a genomic library in the C. jejuni-E. coli shuttle vector pRY107. A library of 6,000 recombinant clones was generated in E. coli DH5α, ensuring 99.9% probability of complete representation of the 1.712-Mb C. jejuni chromosome (8, 28). To isolate recombinant plasmids containing the C. jejuni catalase gene, the library was pooled and the plasmids were extracted via alkaline lysis and transformed into E. coli strain UM255 (catalase negative). Clones harboring plasmids encoding catalase were identified using the bubble assay. A solution of 0.3% hydrogen peroxide in PBS was poured onto the colonies. Clones that produced bubbles, the result of catalase-mediated breakdown of hydrogen peroxide to water and oxygen gas, were immediately subcultured on fresh LB agar plates. Six clones with catalase activity were identified. Zymographic analysis of lysates derived from these clones demonstrated the production of a single catalase enzyme that comigrates with the C. jejuni catalase (Fig. 1, lane 3). Verification of C. jejuni as the origin of the catalase gene contained within plasmid pJD100 was accomplished using Southern analysis (data not shown). Sequence analysis of the pJD100 insert identified a single complete open reading frame of 1,524 bp 99% homologous to the reported C. jejuni katA gene (GenBank accession no. X85130). A putative ribosomal binding site containing six of seven bases of the E. coli consensus sequence and that is complementary to the 3′ end of the C. jejuni 16S rRNA gene product is present six bases upstream of the open reading frame start codon ATG. A putative promoter containing 23 of 27 bases defining the −10, −16, and −35 elements of the reported C. jejuni ς70 promoter consensus sequence was identified 26 bp upstream of the ribosomal binding site (35). Alignment analysis of the 58-kDa protein by using the BLASTp algorithmic search program revealed high homology to numerous eubacterial and eukaryotic heme-containing catalases (13).
Generation and characterization of an isogenic katA mutant strain.
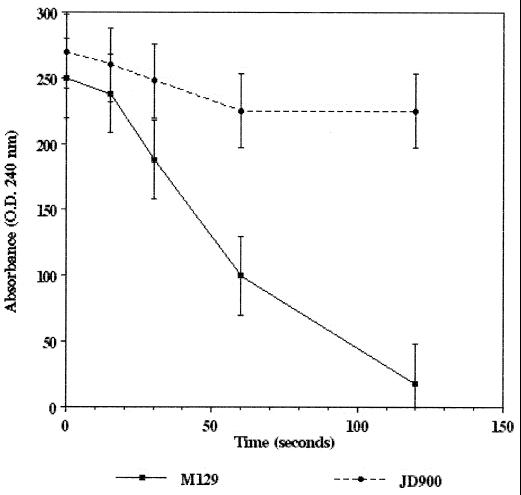
To evaluate the role of catalase in C. jejuni intracellular survival, an M129N isogenic strain containing an insertion mutation in the katA gene was constructed and designated JD900. The inability of JD900 to produce a functional catalase was immediately obvious, as colonies did not produce gas upon addition of hydrogen peroxide. The inability of strain JD900 to produce a functional catalase was further demonstrated using spectrophotometric analysis as described by Beers and Sizer (3). This assay exploits the absorption spectra of hydrogen peroxide to provide an indirect method of observation and quantification of catalase activity. Absorption of UV light (240 nm) corresponds to levels of hydrogen peroxide present in the sample. Lysates derived from the wild-type katA strain M129N produced catalase activity corresponding to 190 U/mg degrading the hydrogen peroxide in the cuvette in a time-dependent manner (Fig. 2). Lysates derived from the katA mutant strain JD900 demonstrated no catalase activity over the period examined (Fig. 2). Low-level degradation of hydrogen peroxide observed in the presence of this lysate likely reflects spontaneous breakdown or interaction with cellular targets, such as nucleic acids and proteins. This absence of catalase activity was statistically significant relative to strain M129N at 60 and 120 s (P < 0.05).
FIG. 2.
Catalase activity of C. jejuni strains M129N (wild-type katA) and JD900 (katA::kan). Decreased absorbance corresponds to breakdown of hydrogen peroxide due to catalase activity. Error bars indicate confidence level (95%); P values were determined using the Student t test (see text).
Catalase is essential for C. jejuni hydrogen peroxide resistance in vitro.
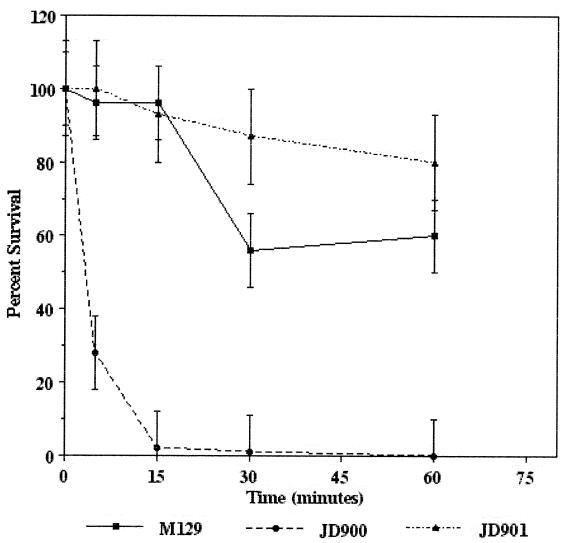
To determine the role of KatA in C. jejuni hydrogen peroxide sensitivity in vitro, survival curves were determined for strains M129N and JD900 cultured in 1 mM hydrogen peroxide (Fig. 3). Viability of strain M129N remained over 50% throughout the assay period. In contrast, strain JD900 was sensitive to the 1 mM hydrogen peroxide, as more than 98% of the bacteria were nonculturable within 15 min. The viability of the katA mutant strain was significantly decreased relative to that of the wild-type strain and the complemented strain (JD901) at all times examined (P < 0.05). This sensitivity was not due to sensitivity of strain JD900 to the culture conditions, as bacteria cultured in MH broth without hydrogen peroxide remained viable (>99%) over the assay period (data not shown). To verify that the effects associated with the katA insertion mutation were due to changes in katA alone and not the result of polar effects on adjacent gene expression, plasmid pJD107, encoding the wild-type katA gene, was introduced into strain JD900 via conjugation to generate strain JD901. This strain was less sensitive to 1 mM hydrogen peroxide than the wild-type strain M129N (P < 0.05 at 30 min), as more than 80% of the bacteria remained viable throughout the assay period. This phenomenon may result from high KatA levels in strain JD901 being a consequence of the high copy number of plasmids harboring the Campylobacter replicon (20). Whatever the case, these results demonstrate that hydrogen peroxide sensitivity of strain JD900 is due to mutation of katA alone and indicate that catalase plays a central role in C. jejuni hydrogen peroxide resistance in vitro.
FIG. 3.
Hydrogen peroxide sensitivity of C. jejuni strains M129N (wild-type katA), JD900 (katA::kan), and JD901 (katA::kan::pJD106). Bacteria were incubated in 1 mM hydrogen peroxide; at times indicated, numbers of surviving bacteria were determined using dilutional plate counts. Numbers of surviving bacteria are represented as the percentage of the original inoculum. Error bars indicate confidence level (95%); P values were determined using the Student t test (see text).
Catalase role in C. jejuni intracellular survival.
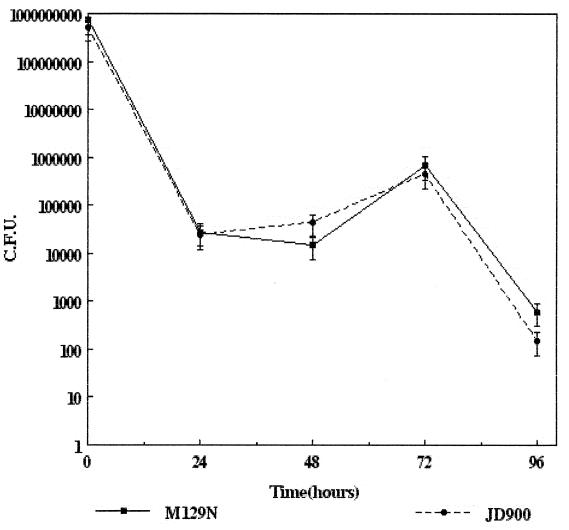
A recent report by Pesci et al. described the role of superoxide dismutase in intraepithelial cell survival in vitro (29). Their findings, such as sodA mutants being 12-fold less viable than an isogenic wild-type sodA strain, suggest that reactive oxygen molecules, such as superoxide anion, hydrogen peroxide, and products of myeloperoxidase, have a critical role in killing intracellular C. jejuni. To determine the role of catalase in C. jejuni intraepithelial cell survival in vitro, HEp-2 cells were infected with strains M129N and JD900 and survival kinetics were analyzed over a 96-h period. Both strains exhibited similar survival characteristics undergoing an initial death phase (6- to 48-h postinfection), as approximately 90% of the intracellular bacteria were killed (Fig. 4). Seventy-two hours postinfection, both strains were able to multiply intracellularly (Fig. 4). Neither strain was present in high numbers at 96-h postinfection (Fig. 4). This lack of viability is likely due to cytopathic effects imposed by the growing bacteria upon the host cell rather than killing of intracellular C. jejuni, as microscopic analysis of the infected monolayers revealed rounded refractory vacuolated and dead cells. These results suggest that catalase plays a minor role, if any, in intraepithelial cell survival. Of note, the growth kinetics and observed changes in infected host cell morphology are consistent with those described for wild-type C. jejuni by Konkel et al. (19).
FIG. 4.
Intraepithelial cell survival of C. jejuni strains M129N (wild-type katA) and JD900 (katA::kan) within cultured HEp-2 epithelial cells was determined using dilutional plate counts. Numbers of surviving bacteria are represented as CFU. Error bars indicate confidence level (95%).
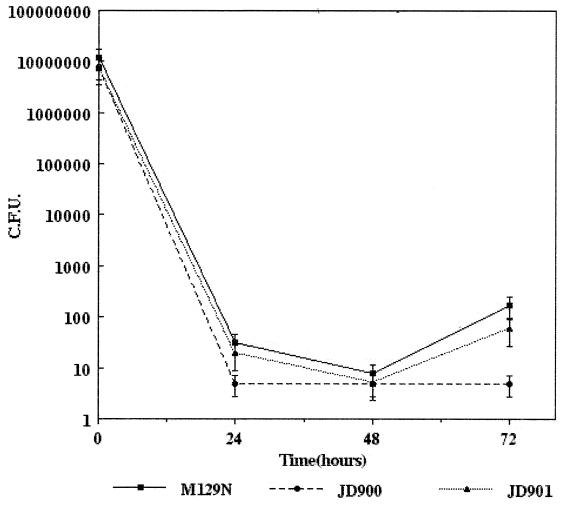
To determine the role of catalase in C. jejuni survival within professional phagocytes in vitro, cultured murine peritoneal macrophages were infected with strains M129N, JD900, and JD901 and survival kinetics were analyzed over a 72-h period. Within 30 min postinfection, the macrophages appeared lacy and highly vacuolated. These observations, which have been correlated to production of a respiratory burst and killing of intracellular C. jejuni, are consistent with those described by Kiehlbauch et al. (17). Twenty-four hours postinfection, few catalase mutant C. jejuni JD900 cells were recovered (<99.9% killed), indicating extensive killing by the macrophages. Viable wild-type and complemented strains were recovered in higher numbers over the same period; however, these results are not significantly different relative to those of the katA mutant strain. Forty-eight hours postinfection, no viable katA mutant JD900 bacteria were recovered (detectable limit, 5 bacteria) (Fig. 5). Comparable results were obtained for the wild-type and complemented strains, as an average of 10 and 5 bacteria were recovered, respectively (Fig. 5). Seventy-two hours postinfection, strain JD900 remained unculturable (detection limit, 5 bacteria) (Fig. 5). In contrast, both catalase-producing strains, M129N and JD901, were recovered in significantly higher numbers (P < 0.05), indicating that each strain was able to persist and eventually multiply within the peritoneal macrophages.
FIG. 5.
Intramacrophage survival of C. jejuni strains M129N, JD900, and JD901. Survival of C. jejuni strains M129N (wild-type katA), JD900 (katA::kan), and JD901 (katA::kan::pJD107) in cultured peritoneal macrophages was determined using dilutional plate counts. Numbers of surviving bacteria are represented as CFU. Error bars indicate confidence level (95%); P values were determined using the Student t test (see text).
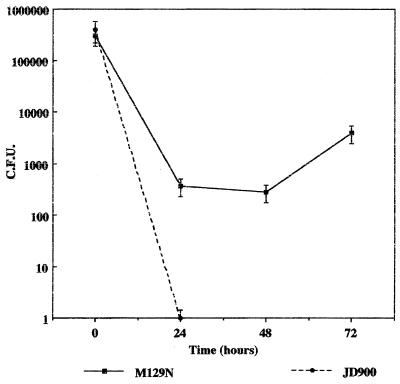
Survival studies with porcine peritoneal macrophages yielded results similar to those of the murine macrophage studies. Porcine peritoneal macrophages were infected with M129 and JD900, and survival kinetics were analyzed over a 72-h period. C. jejuni JD900 (katA mutant) was not detected 24 h postinfection (detection limit, 5 bacteria). Conversely, the catalase-producing wild-type strain M129 was cultured in significantly higher numbers, indicating that this strain was able to multiply and persist within porcine peritoneal macrophages (Fig. 6). Similarly, after 24 h postinfection in J774A.1 cells, C. jejuni JD900 bacteria were not recovered, while the wild-type M129 persisted (Fig. 7). The survival kinetics plotted for wild-type C. jejuni are consistent, albeit at significantly reduced numbers, compared to those reported by Kiehlbauch et al. (17). The disparity of intramacrophage survival exhibited by katA mutant and wild-type bacteria is not due to differences in numbers of infecting bacteria, as inocula for all strains examined were approximately equal (data not shown) or equivalent to cell numbers, as in the case of the J774A.1 experiments. These results suggest that catalase plays a significant role in C. jejuni intramacrophage survival, allowing the bacteria to persist and multiply within these cells in vitro.
FIG. 6.
Survival of C. jejuni strains M129 (wild type) and JD900 (katA mutant) in porcine peritoneal macrophages. Survival of C. jejuni was determined using dilutional plate counts. Numbers of surviving bacteria are represented as CFU.
FIG. 7.
Survival of C. jejuni strains M129 (wild-type) and JD900 (katA mutant) in J774A.1 cells treated with and without a nitric oxide inhibitor. Survival of C. jejuni was determined using dilutional plate counts. Numbers of surviving bacteria are represented as CFU.
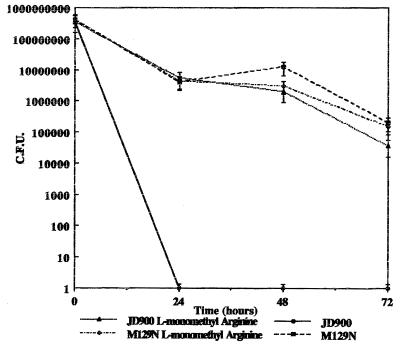
Effects of nitric oxide inhibitor and respiratory burst inhibitor on survival of the katA mutant. (i) Nitric oxide inhibitor.
To look at the role of nitric oxide in the killing of the C. jejuni katA mutant cells by macrophages, NG-l-monomethyl arginine was added to J774A.1 cells during survival assays in order to prevent the formation of nitric oxide synthase. After 72 h, JD900 harvested from the treated cells and the wild-type M129 strain appeared to have similar survival kinetics, while in untreated J774A.1 cells, the katA mutant was not detected past 48 h. Control M129 cells treated with NG-l-monomethyl arginine were unaffected (Fig. 7).
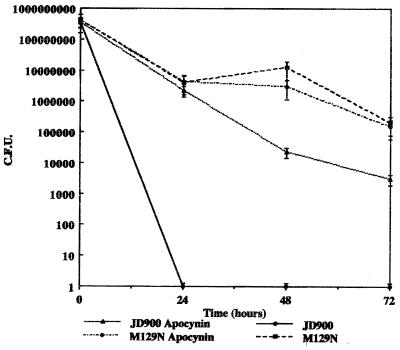
(ii) Respiratory burst inhibitor.
High levels of hydrogen peroxide formed during the respiratory burst in macrophages are lethal to bacteria. In order to determine if hydrogen peroxide produced during the respiratory burst has a role in killing of the katA mutant, apocynin, a respiratory burst inhibitor, was added to J774A.1 cells during intracellular survival assays. Survival kinetics for the C. jejuni katA mutant were similar to those of the wild-type strain M129 over the course of the 72-h incubation period while the mutant failed to survive past 48 h when incubated with untreated J774A.1 cells. M129 cells treated with apocynin were unaffected (Fig. 8).
FIG. 8.
Survival of C. jejuni strains M129 (wild type) and JD900 (katA mutant) in J774A.1 cells treated with and without a respiratory burst inhibitor. Survival of C. jejuni was determined using dilutional plate counts. Numbers of surviving bacteria are represented as CFU.
DISCUSSION
Catalase, which catalyzes the breakdown of hydrogen peroxide to water and oxygen, is produced by most, if not all, aerobic organisms, including humans and bacteria, as well as many microaerophilic and aerotolerant species. This broad distribution reflects the toxic nature of hydrogen peroxide, which can induce single- and double-stranded breaks in DNA and oxidize biological membranes and proteins (31). Our studies indicate that C. jejuni expresses a single heme-containing catalase which is highly homologous to other heme catalases produced by many eukaryotic and prokaryotic organisms. Data indicating the presence of a single catalase were provided using established biochemical assays. Genetic evidence of the existence of a single catalase was provided using BLASTp searches of the nearly completed C. jejuni chromosome employing primary sequences of a number of eubacterial and eukaryotic proteins found no significant homologies other than the KatA reported herein and elsewhere (data not shown) (13).
To examine the role that catalase plays in C. jejuni hydrogen peroxide resistance and intracellular survival, mutants were constructed using insertional mutagenesis. Consistent with findings in other organisms, disruption of catalase production generates bacteria which are hypersensitive to hydrogen peroxide relative to wild-type strains. These findings, which indicate that KatA is central to C. jejuni hydrogen peroxide resistance in vitro, do not address or dispel the existence of other factors which may also contribute to peroxide resistance, such as glutathione and peroxidases (24, 31).
Our findings suggest that catalase does not contribute to survival within human epithelial cells in vitro, as katA mutant strain JD900 exhibited intracellular growth kinetics nearly identical to those of the wild-type strain. These results do not rule out the possibility that reactive oxygen species play roles in epithelial cell killing of intracellular C. jejuni. Recent work by Pesci et al. determined that superoxide dismutase contributes to C. jejuni intraepithelial survival, as isogenic sod mutants were 12 times more sensitive to host killing than the isogenic wild-type strain (29). The apparent disparity between the two studies findings may be explained by the potential interaction of superoxide with nitric oxide. Numerous investigators have indicated that nitric oxide generated by the inducible nitric oxide synthase isozyme, which is activated in the presence of proinflammatory cytokines, is a key mechanism employed by human epithelial cells to combat bacterial infections (16, 21, 22, 23, 26). Interaction of superoxide and nitric oxide may produce highly toxic products, including nitrogen dioxide and peroxynitrite (11). Evidence of this synergy was recently provided by studies using S. enterica serovar Typhimurium sodC mutants (10). Elimination of copper-zinc Sod production yielded strains which were sensitive to peritoneal macrophage killing. Bacteriocidal activity was exacerbated with agents that block superoxide or nitric oxide production (10). Although these studies were conducted in a different cell type and a different sod allele, the potential for similar synergistic actions provide a mechanism by which the iron-containing C. jejuni Sod may function to eliminate half of the cogeners involved.
We next sought to determine the role of catalase in C. jejuni survival within cells which exhibit a respiratory burst. Peritoneal macrophages from the murine and porcine species were chosen as host cells for these studies, as previous reports have demonstrated that C. jejuni survives long-term (>72 h) within these cells (17). In addition, immunohistological studies of gut tissue infected with C. jejuni suggest that intramacrophage survival occurs in vivo (1, 5). Our studies demonstrate that catalase contributes to intramacrophage survival. The data indicate that the katA mutant was more sensitive to killing than the wild-type strain, as JD900 was uniformly recovered in lower numbers than the wild-type strain. We speculate that the detection base of the assay (>5 bacteria) limited its sensitivity and may not allow accurate statistical representation of viable bacteria present at 24 and 48 h postinfection. Nevertheless, strain JD900 remained nonculturable 72 h postinfection while wild-type and complemented strain JD901 survived and multiplied to significantly higher numbers. Kinetics of growth within the macrophages suggest that the absence of catalase produced bacteria which were hypersensitive to killing by reactive oxygen molecules produced in the respiratory burst which was observed shortly after infection. Alternatively, due to restrictions inherent in the assay's sensitivity, we cannot rule out possibility that both JD900 and the catalase-producing strains were present 24 and 48 h postinfection and that catalase contributes to conditions permitting bacterial growth in the phagocyte. Whichever is the case, these results support the hypothesis that catalase contributes to C. jejuni intramacrophage survival. As reported by Fang (11), the reactive species derived from NO synthesis are important in the antimicrobial action of host cells, especially against intracellular pathogens. NO is thought to work in conjunction with reactive oxygen species to damage microbial DNA, proteins, and lipids. The microbicidal activity of phagocytic cells resulting from NO synthase is l-arginine dependent and can be inhibited by NG-l-monomethyl arginine. NO is thought to protect mammalian cells against oxidative damage while enhancing the antimicrobial activity of the respiratory burst. Additionally, inducible NO synthase and NADPH oxidase are thought to be costimulated by inflammatory stimuli resulting in the formation of reactive nitrogen and oxygen intermediates which may in turn lead to the formation of distinct antimicrobial species. An example of this would be the interaction of NO with superoxide, forming peroxinitrate. NO2 may also be formed by the auto-oxidation of NO or the oxidation of NO2− by myeloperoxidase and hydrogen peroxide. With the use of nitric oxide synthase and nitric oxide production inhibitor NG-l-monomethyl arginine, the katA mutant was able to survive within J774A.1 cells at a rate analogous to that of the wild-type strain M129. In addition, use of apocynin, an NADPH oxidase-respiratory burst inhibitor, also resulted in the increased survival of the katA mutant within macrophages. This data supports the hypothesis that catalase is an important factor in C. jejuni intramacrophage survival by counteracting the effects of nitric oxide synthesis as well as the respiratory burst.
These results contrast to those observed in other facultative intracellular pathogens. Disruption of catalase production in S. enterica serovar Typhimurium does not affect virulence in macrophages cultured in vitro or in infected mice (7). Similar results have been described for Shigella flexneri and Listeria monocytogenes in similar models (12, 22). Recent insights into the biology of these organisms may provide an explanation for the disparate observations. Work by Rathman has demonstrated that Salmonella, like many other intracellular pathogens, modifies its vacuole inside host cells and may inhibit entry of bacteriocidal factors, including products of the respiratory burst oxidase (30). Several investigators have observed that both Shigella species and L. monocytogenes escape the phagosome gaining entry to the host cell cytosol, where hydrogen peroxide concentrations are low (2, 31). These activities suggest that catalase activity may not be required for intracellular survival, as these organisms are not subjected to hydrogen peroxide and its derivatives. To date, studies of C. jejuni interaction with its endosomal compartment have not been described.
Kinetics of intracellular growth in both cell types examined are consistent with previous reports by Konkel et al. and Kiehlbauch et al. which describe C. jejuni growth in cultured epithelial cells and peritoneal macrophages, respectively, in vitro (17, 19). Interestingly, striking similarities are observed in growth kinetics for organisms cultured within each cell type. This phenomenon suggests that following a signal transmitted in both cell types, the bacterium may express factors which promote growth. These studies as well as those reported herein indicate that many important issues await examination, including intracellular trafficking and constitution of the C. jejuni-containing vacuole and role of macrophages in development of campylobacteriosis.
REFERENCES
- 1.Babakhani F K, Bradley G A, Joens L A. Newborn piglet model for campylobacteriosis. Infect Immun. 1993;61:3466–3475. doi: 10.1128/iai.61.8.3466-3475.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beaman L, Beaman B L. The role of oxygen and its derivatives in microbial pathogenesis and host defense. Annu Rev Microbiol. 1984;38:27–48. doi: 10.1146/annurev.mi.38.100184.000331. [DOI] [PubMed] [Google Scholar]
- 3.Beers R F, Jr, Sizer I W. A spectrophotometric method for measuring the breakdown of hydrogen peroxide by catalase. J Biol Chem. 1952;195:133–140. [PubMed] [Google Scholar]
- 4.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black R E, Levine M M, Clements M L, Hughes T P, Blaser M J. Experimental Campylobacter jejuni infection in humans. J Infect Dis. 1988;157:472–479. doi: 10.1093/infdis/157.3.472. [DOI] [PubMed] [Google Scholar]
- 6.Blaser M J, Taylor D N, Feldman R A. Epidemiology of Campylobacter jejuni infections. Epidemiol Rev. 1983;5:157–176. doi: 10.1093/oxfordjournals.epirev.a036256. [DOI] [PubMed] [Google Scholar]
- 7.Buchmeirer N A, Libby S J, Xu Y, Loewen P C, Switala J, Guiney D G, Fang F C. DNA repair is more important than catalase for Salmonella virulence in mice. J Clin Investig. 1995;95:1047–1053. doi: 10.1172/JCI117750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clark L, Carbon J. A colony bank containing synthetic ColE1 hybrids representative of the entire E. coli genome. Cell. 1976;9:91–99. doi: 10.1016/0092-8674(76)90055-6. [DOI] [PubMed] [Google Scholar]
- 9.De Melo M A, Gabbiani G, Pechere J C. Cellular events and intracellular survival of Campylobacter jejuni during infection of HEp-2 cells. Infect Immun. 1989;57:2214–2222. doi: 10.1128/iai.57.7.2214-2222.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeGroote M A, Ochsner U A, Shiloh M A, Nathan C, McCord J M, Dinauer M C, Libby S J, Vazquez-Torres A, Xu Y, Fang F. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc Natl Acad Sci USA. 1997;94:13997–14001. doi: 10.1073/pnas.94.25.13997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang F C. Mechanisms of nitric oxide-related antimicrobial activity. J Clin Investig. 1997;99:2818–2825. doi: 10.1172/JCI119473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franzon V L, Arondel J, Sansonetti P J. Contribution of superoxide dismutase and catalase activities to Shigella flexneri pathogenesis. Infect Immun. 1990;58:529–535. doi: 10.1128/iai.58.2.529-535.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant K A, Park S F. Molecular characterization of katA from Campylobacter jejuni and generation of a catalase-deficient mutant of Campylobacter coli by interspecific allelic exchange. Microbiology. 1995;141:1369–1376. doi: 10.1099/13500872-141-6-1369. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 15.Healing T D, Greenwood M H, Pearson A D. Campylobacters and enteritis. Rev Med Microbiol. 1992;3:159–167. [Google Scholar]
- 16.Igietseme J U, Perry L L, Ananaba G A, Uriri I M, Ojior O O, Kumar S N, Caldwell H D. Chlamydia infection in inducible nitric oxide synthase knockout mice. Infect Immun. 1998;66:1282–1286. doi: 10.1128/iai.66.4.1282-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kiehbauch J A, Albach R A, Baum L L, Chang K P. Phagocytosis of Campylobacter jejuni and its intracellular survival in mononuclear phagocytes. Infect Immun. 1985;48:446–451. doi: 10.1128/iai.48.2.446-451.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konkel M E, Joens L A. Adhesion to and invasion of HEp-2 cells by Campylobacter spp. Infect Immun. 1989;57:2984–2990. doi: 10.1128/iai.57.10.2984-2990.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Konkel M E, Hayes S F, Joens L A, Cieplak W., Jr Characteristics of the internalization and intracellular survival of Campylobacter jejuni in human epithelial cell cultures. Microb Pathog. 1992;13:357–370. doi: 10.1016/0882-4010(92)90079-4. [DOI] [PubMed] [Google Scholar]
- 20.Labigne-Roussel A, Harel J, Tompkins L. Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J Bacteriol. 1987;169:5320–5323. doi: 10.1128/jb.169.11.5320-5323.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamarque D, Kiss J, Tankovic J, Flejou J F, Delchier J C, Whittle B J. Induction of nitric oxide synthase in vivo and cell injury in rat duodenal epithelium by a water soluble extract of Helicobacter pylori. Br J Pharmacol. 1998;123:1073–1078. doi: 10.1038/sj.bjp.0701706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leblond-Francillard M, Gaillard J-L, Berche P. Loss of catalase activity in Tn1545-induced mutants does not reduce growth of Listeria monocytogenes in vivo. Infect Immun. 1989;57:2569–2573. doi: 10.1128/iai.57.8.2569-2573.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li C K, Seth R, Gray T, Bayston R, Mahida Y R, Wakelin D. Production of proinflammatory cytokines and inflammatory mediators in human intestinal epithelial cells after invasion by Trichinella spiralis. Infect Immun. 1998;66:2200–2206. doi: 10.1128/iai.66.5.2200-2206.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma M, Eaton J W. Multicellular oxidant defense in unicellular organisms. Proc Natl Acad Sci USA. 1992;89:7924–7928. doi: 10.1073/pnas.89.17.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meade H M, Long S R, Ruvkun C B, Brown S E, Ausubel F M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meng Q H, Springall D R, Bishop A E, Morgan K, Evans T J, Habib S, Gruenert D C, Gyi K M, Hodson M E, Yacoub M H, Polak J M. Lack of inducible nitric oxide synthase in bronchial epithelium: a possible mechanism of susceptibility to infection in cystic fibrosis. J Pathol. 1998;184:323–331. doi: 10.1002/(SICI)1096-9896(199803)184:3<323::AID-PATH2>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 27.Mulvey M R, Sorby P A, Triggs-Raine B L, Loewen P C. Cloning and physical characterization of katE and katF required for catalase HPII expression in Escherichia coli. Gene. 1988;73:337–345. doi: 10.1016/0378-1119(88)90498-2. [DOI] [PubMed] [Google Scholar]
- 28.Nuitjten P J, Bartels C, Bleumink N M, Gaastra W, van der Zeijst B A. Size and physical map of the Campylobacter chromosome. Nucleic Acids Res. 1990;18:6211–6214. doi: 10.1093/nar/18.21.6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pesci E C, Cottie D L, Pickett C L. Genetic, enzymatic, and pathogenic studies of the iron superoxide dismutase of Campylobacter jejuni. Infect Immun. 1994;62:2687–2694. doi: 10.1128/iai.62.7.2687-2694.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rathman M, Baker L P, Falkow S. The unique trafficking pattern of Salmonella typhimurium-containing phagosomes in murine macrophages is independent of the mechanism of entry. Infect Immun. 1997;65:1475–1485. doi: 10.1128/iai.65.4.1475-1485.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storz G, Tartaglia L A, Farr S B, Ames B N. Bacterial defenses against oxidative stress. Trends Genet. 1990;6:363–368. doi: 10.1016/0168-9525(90)90278-e. [DOI] [PubMed] [Google Scholar]
- 32.Tauxe R V. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In: Nachamkin I, Blaser M J, Tompkins L S, editors. Campylobacter jejuni: current status and future trends. Washington, D.C.: American Society for Microbiology; 1992. pp. 9–19. [Google Scholar]
- 33.Woodbury W, Spencer A K, Stahmann M A. An improved procedure using ferric cyanide for detecting catalase isozymes. Annals Biochem. 1971;44:301–305. doi: 10.1016/0003-2697(71)90375-7. [DOI] [PubMed] [Google Scholar]
- 34.Worthington C C, editor. Worthington enzyme manual. Freehold, N.J: Worthington Biochemical Corporation; 1988. pp. 72–74. [Google Scholar]
- 35.Wosten M M S M, Boeve M, Koot M G A, van Nuenen A C, van der Zeijst A M. Identification of Campylobacter jejuni promoter sequences. J Bacteriol. 1998;180:594–599. doi: 10.1128/jb.180.3.594-599.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yao R, Alm R A, Trust T J, Guerry P. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene. 1993;130:127–130. doi: 10.1016/0378-1119(93)90355-7. [DOI] [PubMed] [Google Scholar]