Abstract
Objective:
Higher optimism is associated with reduced mortality and a lower risk of age-related chronic diseases. DNA methylation (DNAm) may provide insight into mechanisms underlying these relationships. We hypothesized DNAm would differ among older individuals who are more versus less optimistic.
Methods:
Using cross-sectional data from two population-based cohorts of women with diverse races/ethnicities (N=3,816) and men (only white, N=667), we investigated the associations of optimism with epigenome-wide leukocyte DNAm. Random-effects meta-analyses were subsequently used to pool the individual results. Significantly differentially methylated cytosine-phosphate-guanines (CpGs) were identified by the number of independent degrees of freedom approach: effective degrees of freedom correction using the number of principal components (PCs), explaining>95% of the variation of the DNAm data (PC-correction). We performed regional analyses using comb-p and pathways analyses using the Ingenuity Pathway Analysis software.
Results:
We found essentially all CpGs (total probe N= 359,862) were homogeneous across sex and race/ethnicity in the DNAm~optimism association. In the single CpG site analyses based on homogeneous CpGs, we identified 13 significantly differentially methylated probes using PC-correction. We found four significantly differentially methylated regions and two significantly differentially methylated pathways. The annotated genes from the single CpG site and regional analyses are involved in psychiatric disorders, cardiovascular disease, cognitive impairment, and cancer. Identified pathways were related to cancer, neurodevelopmental and neurodegenerative disorders.
Conclusion:
Our findings provide new insights into possible mechanisms underlying optimism and health.
Keywords: optimism, DNA methylation, epigenome-wide association study, mechanism
Introduction
By 2035, the number of people in the U.S. aged 65 years and older is estimated to reach 78 million (1). Promoting the health and well-being of older populations is therefore critical. Optimism, defined either as a generalized expectation that good things will happen (i.e., dispositional optimism) (2) or as an explanatory style regarding how individuals explain the causes of good and bad events (e.g., bad events are explained as being due to external, transient, and specific causes; i.e., exploratory optimism) (3), has been identified as a positive health asset. Growing evidence suggests associations of higher optimism with lower risk of aging-related chronic cardiopulmonary diseases, neurodegenerative diseases, all-cause mortality, and higher likelihood of healthy aging (4–6), and these findings appear to be independent of effects of psychological distress (4). However, limited empirical work has considered the molecular mechanisms underlying these associations.
Epigenetic modifications are reversible processes that can turn on or off the activity of select genes without changing the DNA sequence. DNA methylation (DNAm) is an epigenetic process that attaches small chemical groups called methyl groups predominantly to cytosine-phosphate-guanine (CpG) sites (7). DNAm is thought to be susceptible to environmental (e.g., pollution) and psychosocial (e.g., perceived stress, socioeconomic status) influences (8–10) and may be a mechanism by which psychosocial factors affect health. DNAm has been linked to adverse health outcomes, such as accelerated biological aging (11), cardiovascular disease (12), and cancer (13). Meanwhile, DNAm has also been associated with psychological disorders (e.g., depression) (14–16), but limited work has examined DNAm in relation to positive psychological factors (17). In the only study to date, Baselmans et al. conducted an epigenome-wide association study (EWAS) of psychological wellbeing as assessed by life satisfaction and found associations for DNAm with six CpG sites (18). In previous work (17) conducted by our group, we examined the associations between optimism and either of two well-established epigenetic clocks, which are based on DNAm and summary measures of aging. However, the associations were null (17). These epigenetic clocks provide only a limited assessment of epigenetic processes due to the use of a small number of CpG sites (19). Further, optimism is partly heritable (23–32%) (20), and in fact is quite stable in adulthood absent deliberate efforts to modify it (although it can shift in major life transitions either transiently or more persistently) (21). However, experimental research has also demonstrated optimism is modifiable with accessible methods such as writing exercises and cognitive-behavioral strategies (22), and thus may be a novel target for intervention to improve health. To date, no study has examined the relationship of optimism with DNAm, especially in an epigenome-wide study.
Prior studies have generally found higher optimism is associated with better health in both men and women and in diverse populations (4), although biological underpinnings of these associations may differ by sex or by race/ethnicity. For example, populations with different races/ethnicities experience different levels of social stressors such as racism and poverty, and may therefore experience different levels of or have a different response to optimism (23). To identify whether DNAm could serve as one biological mechanism linking higher optimism to better health, we conducted EWAS obtained from whole-blood samples drawn from participants in the Women’s Health Initiative (WHI, women only) and the Normative Aging Study (NAS, men only). We examined whether optimism was associated with DNAm in both men and women and across racial/ethnic groups.
Materials and Methods
Study Population
This study used cross-sectional data from both WHI and NAS. The study in WHI was approved by Institutional Review Boards at each clinical center and at the coordinating center. The study in NAS was approved by the Institutional Review Boards by the Harvard T.H. Chan School of Public Health and the Department of Veterans Affairs. All participants provided their written informed consent.
WHI
WHI is a nationwide prospective cohort of 161,808 women, who were postmenopausal and aged 50–79 years at recruitment (24). Participants were enrolled in this study between 1993 and 1998, when they completed an initial visit at baseline to provide physical measures and blood specimens, at which time they also completed psychosocial questionnaires. Extensive description on recruitment methods is available elsewhere (25). The present study includes white (non-Hispanic), Black/African-American (AA), and Hispanic/Latina women who participated in two sub-studies in which epigenome-wide methylation was measured. Because the sub-study samples were derived differently, we considered them in separate analyses. One sub-study (Epigenetic Mechanisms of PM-Mediated Cardiovascular Disease; WHI_AS315) comprised a stratified random sample representative of a clinical trial (17). The second sub-study (Integrative Genomics and Risk of Coronary Heart Disease (CHD) and Related Phenotypes; WHI_BAA23) comprised women from both a clinical trial and an observational study who were included in a nested case-control study of CHD (26). Of the 118 women who were included in both sub-studies, we included their data only in analyses with the WHI_AS315 sub-study.
We restricted each sample to participants with data on optimism and DNAm at the baseline visit. In total, 1,914 subjects were included in WHI_AS315, of whom 56.3% were white (not of Hispanic origin), 27.9% were Black/AA and 15.8% were Hispanic/Latina; 1,902 subjects were included in WHI_BAA23, of whom 49.2% were white (not of Hispanic origin), 31.2% were Black/AA and 19.6% were Hispanic/Latina.
NAS
NAS is an on-going longitudinal cohort established in 1963 at the Veteran Affairs Boston Outpatient Clinic. This study initially enrolled 2,280 community-dwelling men from the Greater Boston Area who were free of known chronic medical conditions at baseline. Optimism was assessed using a mail survey to all active cohort members in 1986. Every 3 to 5 years, participants attended in-person examinations, which involved collecting information on demographic factors, medical history, lifestyle factors, and a blood draw. The DNA samples obtained from blood draw were collected starting in 1999. We restricted study participants to those with data on both optimism and its closest DNAm assessment. We further excluded non-white men to reduce study heterogeneity since they accounted for approximately 3% of the participants (27). In total, 667 white men were included in this study.
Measures
Optimism
Optimism is defined either as having generalized expectations for positive outcomes (i.e., dispositional optimism) or based on an individual’s explanation of causes for past events (i.e., exploratory optimism). We used different optimism measures in the two cohorts: dispositional optimism in the WHI and explanatory optimism in the NAS.
WHI.
Dispositional optimism was assessed at baseline using the 6-item Life Orientation Test-Revised (LOT-R), which has good predictive and discriminant validity (2). Optimism scores were calculated as the sum of six items, coded from 1=strongly disagree to 5=strongly agree. Three negatively worded items were reverse coded before summing and overall higher scores indicate higher optimism. Following general practice in WHI, this score was considered as missing if any of the six items was missing (17). The internal consistency reliability of the measure was high in the analytic sample (Cronbach α = 0.75) (17).
NAS.
We obtained explanatory optimism using the Revised Optimism-Pessimism Scale (PSM-R) assessed in 1986 (28). The bipolar scale was developed by applying the Content Analysis of Verbatim Explanations technique to Minnesota Multiphasic Personality Inventory-II items (3). The scale measures explanatory style on a continuum from optimistic to pessimistic using 263 dichotomous items weighted according to their levels on three explanatory style domains including internality, stability, and globality. Following the scoring algorithm, items were combined into a composite score (3) with a lower PSM-R score indicating a more optimistic explanatory style. The PSM-R score has good internal consistency in NAS as well (Cronbach alpha=0.78) (3) and has demonstrated high test-retest reliability in other studies (28).
These two measures of optimism have been shown to be moderately correlated (r=−0.49, p-value<0.01) (29) and have similar associations with health outcomes (30). To improve comparability of findings between the two cohorts whereby a higher score on the optimism measure indicates higher optimism, we reverse coded the PSM-R score in NAS and also derived an optimism score in both WHI and NAS by converting an individual’s score on the optimism measure to the percent of the maximum possible score that could be achieved on that measure (31). Also, for comparability, we expressed effect estimates as difference in DNAm (% 5-mC) per 1% increase in the percent of the maximum possible optimism score obtained.
DNAm
Blood specimens were collected in EDTA tubes after on overnight fast and stored at −70 °C. DNA was extracted from the peripheral blood leukocytes and DNAm quantification was performed using the Illumina Infinium Human Methylation450K BeadChip (450K) (Illumina Inc.; San Diego, CA, USA), which uses probes to measure DNAm levels at 485,577 unique CpG sites. DNAm occurs at the 5 position of the pyrimidine ring of the cytosine residues within CpG sites to form 5-methylcytosine (i.e., 5-mC). DNAm level was expressed as the signal intensity for methylated cytosines over the sum of the signal intensities for methylated and unmethylated cytosines at the position, and then multiplied by 100 (%5-mC). Thus, the DNAm level ranged from 0 % to 100 % 5-mC. We conducted quality control and normalization of DNAm data in both WHI and NAS. See the Supplemental Digital Content for details of these protocols.
WHI.
Samples in WHI_AS315 were processed at the Northwestern University Genomics Core, and samples in WHI_BAA23 were processed at the HudsonAlpha Institute of Biotechnology. In WHI, we included 359,944 probes in AS315 and 360,307 probes in BAA23.
NAS.
Samples in NAS were measured at the Northwestern University Genomics Core. The ewastools package was used to exclude low-quality samples (32). In NAS, we included 360,272 probes in our working set, with 359,921 common probes in the two sub-studies in WHI working set. The common probes (N=359,921) across three groups (i.e., WHI_AS315, WHI_BAA23, and NAS) were used in our analysis.
Covariate Assessment
Covariates were selected a priori based on the incorporation of prior findings in relevant literature evaluating linkages of optimism with health outcomes, such as DNAm-related biomarkers of aging, CHD, stress reactivity, and cause-specific mortality (17, 29, 33, 34).
WHI.
Covariates were obtained from the baseline questionnaire. Demographic variables included age (years), race/ethnicity (white, Black/AA, Hispanic/Latina), education (less than high school, high school graduate, some college or associate degree, college or more), smoking status (never/past/current smoker), pack-years (only for current/past smoker), body mass index (BMI, derived from self-reported weight and height, kg/m2).
NAS.
Covariates were assessed with questionnaires completed closest in time to collection of the blood samples from each participant. Demographic variables included age at blood draw (years), and education (years), smoking status (never/past/current smoker), pack-years (only for current/past smoker), and BMI (derived from self-reported weight and height, kg/m2).
Statistical Analyses
To fully elucidate the genes and pathways potentially related to optimism, we assessed the association between DNAm and optimism at three levels: single CpG site, regional, and pathway analysis. In the single CpG site analysis, there were three steps: 1) we conducted analyses in each of the three sub-studies (WHI_AS315, WHI_BAA23, NAS) separately; 2) we tested whether there were heterogeneous CpG sites that modified the DNAm-optimism associations across sex or race/ethnicity; 3) based on homogeneous CpGs across sex and race/ethnicity, we performed random-effects meta-analyses to pool the results from the three sub-studies. Regional and pathway analyses were also performed based on homogeneous CpGs.
Single CpG site analyses
We considered DNAm the dependent variable and optimism the independent variable. We examined whether optimism was associated with DNAm, and assessed if associations varied by sex and race/ethnicity in three steps.
In Step 1, we used linear mixed-effect (LME) models to estimate the associations of optimism with DNAm for each of common probes (N=359,921) (26). All analyses were adjusted for the covariates described above (Covariate Assessment section). Since WHI_BAA23 is a case-control study (case status is defined by presence/absence of coronary heart disease), we additionally adjusted for the case/control status and used inverse probability weighting (35) to control for the potential selection bias. For more details, see the supplement.
To account for multiple testing in the context of the high correlation among CpG sites, we used the “number of independent degrees of freedom” (NIDF) approach to generate the effective number of independent tests (34). More specifically, we used principal component analysis to calculate the number of principal components that could explain > 95% variation of the DNAm data (i.e., 95% NIDF). Figure S1 (Supplemental Digital Content) shows the NIDF (scree plot) and 95%NIDF (cumulative scree plot) that explained more than 95% of the variation of the DNAm data, in each sub-study. Thus, we set the number of independent degrees of freedom to be 95%NIDF in each sub-study. We then obtained the threshold for statistical significance of each estimate by dividing 0.05 by the 95%NIDF (i.e., PC-correction) (36); the thresholds are detailed in Table S1, Supplemental Digital Content. In a separate analysis, we defined the threshold for statistical significance of each CpG using the traditional approach (i.e., probe-correction; 0.05/359,921) (18), which is overly conservative in the presence of correlated data (36). We presented these results in the supplemental material (see Table S2).
In Step 2, we first conducted random-effects meta-regression combining the results from the two sub-studies in WHI among white women and NAS (white men) to test whether sex modified DNAm~optimism associations. We defined the CpG site as having heterogeneity in sex if the associated p-value was less than the smallest threshold based on PC-correction from the two sub-studies in WHI among white women and NAS (white men) (i.e., 5.160×10−05 in AS315 whites). We also conducted random-effects meta-regressions combining the results from the WHI sub-studies to test whether race/ethnicity modified DNAm-optimism associations. We considered the CpG as having heterogeneity in race/ethnicity if the p-value of moderator was less than the smallest threshold based on PC-correction among the six WHI sub-studies (i.e., 5.160×10−05 in WHI_AS315 whites).
In Step 3, we extracted the homogeneous CpGs across sex and race/ethnicity and performed random-effects meta-analyses to examine DNAm~optimism associations with these probes. Significance thresholds based on PC- and probe-correction depended on the number of the homogeneous CpGs we identified in Step 2. We then used the I-squared (I2) test to assess between-study heterogeneity potentially caused by differences in study design, DNAm measures, etc. When I2 < 50% and p-value > 0.05 for the Cochran’s Q test, we considered this as evidence of no heterogeneity across sub-studies (37).
Regional and pathway analyses
In addition to examining the associations of optimism and DNAm at the level of single CpG site, we also investigated differently methylated regions (DMRs) in relation to optimism. We used the comb-p tool because it has the best sensitivity and highest control of false-positive rate compared to other DMR tools, including DMRcate, probe lasso and bump hunting (38). The input contained individual CpG sites with their p-values from single CpG site analyses as well as information on chromosomal locations (39). We defined a significant DMR as one with ≥3 CpGs and Sidak p-value <0.05. The combination of single CpG site and regional analysis may not fully capture the mechanisms underlying optimism and health because CpGs that are not located in the same region could function together. Hence, we used the Ingenuity Pathway Analysis (IPA) database (QIAGEN Inc.) to identify canonical pathways significantly enriched with genes located around the top 100 homogeneous CpGs associated with optimism (40). We defined significant pathways if permutation p-value <0.05 and gene set contained ≥4 genes.
Sensitivity Analyses
To test the robustness of the results, we conducted two sensitivity analyses for the single CpG site analyses based on the homogeneous CpGs across sex and race/ethnicity. First, we dropped all cases from the WHI case-control study (WHI_BAA23) to minimize potential confounding by significant health issues, and then we performed random-effects meta-regression on the revised analytic sample. Second, we re-ran models using fixed-effects meta-regressions which have more commonly been used in previous EWAS (41). Both sensitivity analyses adjusted for the same covariates as in the main analyses except we dropped case status and inverse probability weighting in the first sensitivity analyses as it was no longer appropriate.
Replication Analyses
We conducted the replication analyses in two ways: First, based on the significantly differentially methylated probes (DMPs) from the single CpG site analyses, we assessed the replication of results derived from the main meta-analyses, in each of the three groups separately (given their slightly different compositions regarding race/ethnicity or sex): WHI_AS315, WHI_BAA23, and NAS. Second, we examined the replication across the three groups (without including the main meta-analyses), WHI_AS315, WHI_BAA23, and NAS. More specifically, we extracted the significant DMPs (based on PC-correction) in one of the three groups and checked whether these DMPs were also significant (based on nominal significance) in the other two groups.
Statistical analyses were performed using R (Version 3.5.1, Vienna, Austria) with stats (principal component analysis), lme4 (LME models), metafor (meta-analyses), and ENmix (regional analyses) packages.
Results
Population description
Characteristics of the study populations are presented in Table 1. In WHI_AS315, the mean age was 62 years [(standard deviation) SD=7]. Almost half the women never smoked and one-third had a college education or more. Summary statistics in WHI_BAA23 were similar to those in WHI_AS315. The NAS consisted of 667 white men with mean age of 73 years (SD=7), with a mean of 15 years (SD=3) of education. The mean (SD) of optimism were 23.1 (SD=3.4), 23.0 (SD=3.3), and 44.6 (SD=10.7) in WHI_AS315, WHI_BAA23, and NAS, respectively.
Table 1.
Characteristics of study participants in the WHI_AS315, WHI_BAA23, and the NAS.
| Characteristics | WHI_AS315 (N=1,914) | WHI_BAA23 (N=1,902) | NAS (N=667) |
|---|---|---|---|
|
| |||
| Female (%) | 100 | 100 | 0 |
| Optimism (mean ± SD) | 23.1 ± 3.4 | 23.0 ± 3.3 | 44.6 ± 10.7 |
| Age (mean ± SD) | 62 ± 7 | 65 ± 7 | 73 ± 7 |
| Pack-years (mean ± SD) | 9.1 ± 17.4 | 9.4 ± 18.7 | 21.2 ± 25.5 |
| BMI (mean ± SD) | 29.62 ± 5.94 | 29.84 ± 6.07 | 28.09 ± 4.10 |
| Race/ethnicity (n (%)) | |||
| white (not of Hispanic origin) | 1,078 (56.3%) | 935 (49.2%) | 667 (100%) |
| Black/African-American | 534 (27.9%) | 594 (31.2%) | 0 (0%) |
| Hispanic/Latin | 302 (15.8%) | 373 (19.6%) | 0 (0%) |
| Smoking status | |||
| Never Smoked | 988 (51.6%) | 1,004 (52.8%) | 207 (31.0%) |
| Past Smoker | 737 (38.5%) | 687 (36.1%) | 434 (65.1%) |
| Current Smoker | 163 (8.5%) | 187 (9.8%) | 26 (3.90%) |
| Missing (%) | 26 (1.4%) | 24 (1.3%) | 0 (0%) |
| Education (n, (%)) | |||
| WHI | |||
| Less than high school | 406 (21.2%) | 493 (25.9%) | - |
| High school graduate | 357 (18.7%) | 368 (19.3%) | - |
| Some college or associate | 522 (27.3%) | 478 (25.1%) | - |
| College or more | 614 (32.1%) | 548 (28.8%) | - |
| Missing (%) | 15 (0.8%) | 15 (0.8%) | - |
| NAS | |||
| Years of education (mean ± SD) | - | - | 15.0 ± 3.0 |
| Missing (n (%)) | - | - | 0 (0%) |
Abbreviations: WHI, Women’s Health Initiative; NAS, Normative Aging Study; SD, standard deviation; BMI, body mass index.
Optimism and DNAm associations
Single CpG site analyses
In Step 1, we found a different number of significant DMPs in each of the seven sub-groups (see Table S2, Supplemental Digital Content). In the supplement, we also present the Manhattan plots and the quantile-quantile (Q-Q) plots with the estimated genomic inflation factor for each sub-study (see Figure S2, Supplemental Digital Content). In Step 2, we identified a total of 359,862 homogeneous CpGs across both sex and race/ethnicity (see Figure S3). In Step 3, we performed random-effects meta-analyses for all homogeneous probes. We set the significance threshold of probe-correction as 1.389×10−07 (0.05/359,862). Because the number of the homogeneous CpGs (N=359,862) was very close to the number of the common probes (N=359,921), the difference in the 95%NIDF between the two beta matrices in each sub-group were negligible. Hence, we set the significance threshold of PC-correction as 5.160×10−05 (0.05/969, the smallest PC-correction threshold among the individual studies in Table S1). Figure 1a shows the Manhattan plot for these homogeneous CpGs (N=359,862). Figure 1b shows the Q-Q plot for each of these CpGs with the estimated genomic inflation factor (0.70).
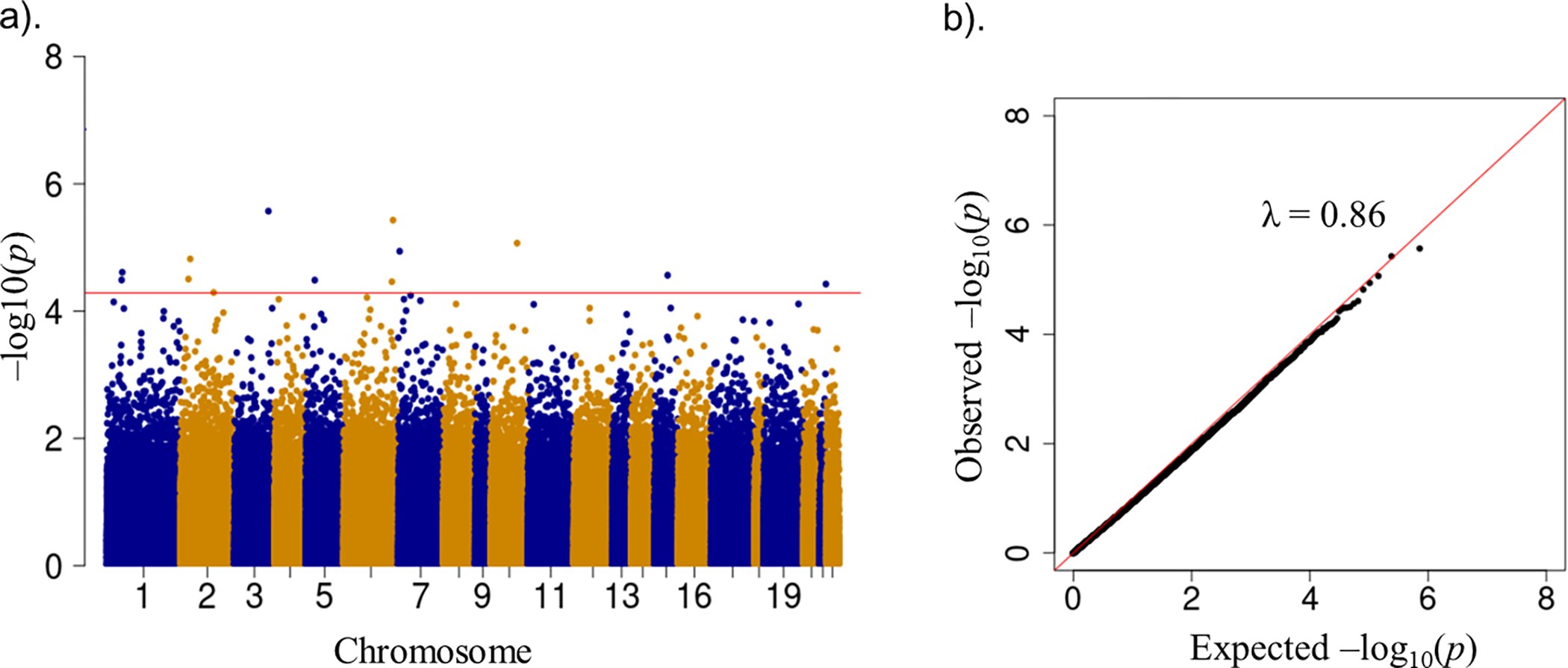
Figure 1a. Manhattan plot in random-effects meta-analyses for homogeneous CpGs (N=359,862). The red horizontal line represents the PC-correction threshold (5.160×10−05).
Figure 1b. Quantile-quantile plot of the observed log10-transformed p-value for each CpG against the expected log10-transformed p-value in meta-analyses.
Abbreviations: PC, principal components; CpG, cytosine-phosphate-guanine; λ, inflation factor
We found 13 significant DMPs with almost half of the probes exhibiting more methylation at higher optimism levels using PC-correction (p-value<5.160×10−05). We present the top 13 DMPs ranked by p-values with their annotated genes and their between-study heterogeneity test results in Table 2. For example, the methylation level of cg24856537 (mapped to NOTCH4) increased 1.41×10−04 (%5-mC) (p-value=3.72×10−06), and the methylation level of cg03451670 (mapped to AGAP1) decreased 2.50×10−04 (%5-mC) (p-value=3.14×10−05), respectively, per 1% increase of maximum possible in optimism. Among the homogeneous CpGs (N=359,862), 9% had between-study heterogeneity. There were no significant DMPs in relation to optimism using the probe-correction threshold (p-value<1.389×10−07) (Figure S4).
Table 2.
The significant 13 DMPs ranked by p-value associated with optimism in site-by-site meta-analyses based on the homogeneous probes in both race/ethnicity and gender in WHI and NAS.
| CpG | Annotated gene a | Other close gene a | Chr | Position a | p-value | Difference in DNAm b | SE c | Heterogeneity I2 (%), p-value d |
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| cg23032421 | IL5RA | - | 3 | 3152038 | 2.68×10−06 | 2.12×10−04 | 4.52×10−05 | 0.00, 0.68 |
| cg24856537 | NOTCH4 | - | 6 | 32169929 | 3.72×10−06 | 1.41×10−04 | 3.05×10−05 | 19.52, 0.28 |
| cg18583565 | - | MGMT | 10 | 131567980 | 8.55×10−06 | −1.70×10−04 | 3.79×10−05 | 0.00, 0.78 |
| cg00702840 | - | MDH2 | 7 | 75703958 | 1.15×10−05 | −1.20×10−04 | 2.62×10−05 | 0.00, 0.78 |
| cg04219552 | - | DYSF | 2 | 72126504 | 1.52×10−05 | 9.17×10−05 | 2.12×10−05 | 0.00, 0.66 |
| cg05372549 | SRM | - | 1 | 11119861 | 2.45×10−05 | 2.29×10−05 | 5.42×10−06 | 0.00, 0.87 |
| cg15560586 | CSNK1G1 | - | 15 | 64648483 | 2.73×10−05 | −3.10×10−05 | 7.39×10−06 | 0.00, 0.54 |
| cg03451670 | AGAP1 | - | 2 | 236579469 | 3.14×10−05 | −2.50×10−04 | 5.91×10−05 | 0.00, 0.48 |
| cg05065507 | TACSTD2 | - | 1 | 59042931 | 3.24×10−05 | −3.10×10−04 | 7.44×10−05 | 0.00, 0.76 |
| cg06418871 | MARVELD2 | - | 5 | 68710831 | 3.26×10−05 | −2.60×10−04 | 6.28×10−05 | 0.00, 0.70 |
| cg24329557 | GCM2 | - | 6 | 10882326 | 3.45×10−05 | 2.53×10−04 | 6.11×10−05 | 0.00, 0.43 |
| cg27288968 | PAXBP1 | - | 21 | 34106765 | 3.76×10−05 | 1.37×10−04 | 3.32×10−05 | 0.00, 0.91 |
| cg16303846 | DOCK10 | - | 2 | 225907539 | 5.10×10−05 | −5.70×10−05 | 1.41×10−05 | 0.00, 0.95 |
Note: The models adjusted for age, race/ethnicity (WHI_AS315 and WHI_BAA23 only), education, smoking status, pack-years, body mass index, case status (WHI_BAA23 only), and methylation-related technical covariates.
Abbreviations: DMPs, differentially methylated probes; CpG, cytosine-phosphate-guanine; DNAm, DNA methylation; Chr, chromosome; SE, standard error; I2, I-squared.
The University of California Santa Cruz UCSC database (GRCh37/hg19)
Difference in DNAm level (% 5mC) per 1% increase of maximum possible in optimism
Standard error in DNAm level (% 5mC) per 1% increase of maximum possible in optimism
I2 < 50% and p-value of heterogeneity > 0.05 were considered as homogenous across sub-studies
Regional and pathway analyses
In the regional analyses, we identified 4 significant DMRs associated with optimism (see Table 3). The CpGs which were located in 4 DMRs and their associations with optimism are presented in Table S3. The 4 DMRs mapped to the genes of MARVELD2, MGMT, RHOU, and SIGLECL1.
Table 3.
Significant DMRs in regional analyses based on the meta-analyses on the homogeneous probes in both sex and race/ethnicity in WHI and NAS.*
| chr | Start a | End a | Sidak p-value | Number of CpGs | Covered CpGs a | Annotated Gene a | Other close gene a |
|---|---|---|---|---|---|---|---|
|
| |||||||
| 5 | 68710808 | 68711278 | 1.43×10−10 | 7 | cg02505827, cg05901765, cg06418871, cg06998965, cg12687157, cg17019292, cg18663063 | MARVELD2 | - |
| 10 | 131567747 | 131568314 | 3.07×10−05 | 6 | cg01079118, cg13742952, cg14308082, cg18583565, cg22768615, cg23889967 | - | MGMT |
| 1 | 228890801 | 228891037 | 4.34×10−05 | 5 | cg16061472, cg20462561, cg23012917, cg23926439, cg25138412 | - | RHOU |
| 19 | 51774377 | 51774667 | 1.98×10−3 | 3 | cg14724749, cg14884932, cg15497724 | - | SIGLECL1 |
Abbreviations: DMRs, differentially methylated regions; CpG, cytosine-phosphate-guanine; chr, chromosome.
We defined significant DMRs as one with ≥3 CpGs and Sidak p-value<0.05.
The University of California Santa Cruz UCSC database (GRCh37/hg19)
We identified two significant pathways in relation to optimism: “Signaling by Rho family GTPases” and “Axonal guidance signaling” (see Table 4).
Table 4.
The significantly differentially methylated pathways based on the meta-analyses on the homogeneous probes in both sex and race/ethnicity in WHI and NAS.*
| Pathway maps | p-value | N a | Genes |
|---|---|---|---|
|
| |||
| Signaling by Rho family GTPases | 0.01 | 4 | CDH13, SEPT9, MYL12B, ARHGEF7 |
| Axonal guidance signaling | 0.04 | 5 | COPS5, CXCR4, MYL12B, ARHGEF7, NTRK1 |
We defined significant pathways if p-value <0.05 and gene set contains≥4 genes.
The number of genes in each pathway
Sensitivity Analyses
In sensitivity analysis 1, we present the Manhattan and Q-Q plots for each of the three sub-studies in WHI_BAA23 (excluding cases from the analytic sample) (Figure S5, Supplemental Digital Content) and the random-effects meta-regression across seven sub-groups (Figure S6). In sensitivity analysis 2, we present the Manhattan and Q-Q plots for the fixed-effects meta-analyses (Figure S7). We then compared the effect sizes of the 13 significant DMPs (shown in Table 2) derived from our main analyses with the results obtained from the two sensitivity analyses (Figure S8). Effect sizes of these 13 CpG sites were highly consistent between the main analyses and the two sensitivity analyses (Table S4).
Replication Analyses
The first replication analyses showed that for all the 13 DMPs (shown in Table 2), estimates from meta-analyses were in the same direction as those from WHI_AS315, WHI_BAA23, and NAS. See Table S5 and Figure S9. The p-values from meta-analyses were more driven by WHI_BAA23. For example, the top 1 probe (cg23032421) had its p-value from BAA23 2.82×10−5. The second replication analyses found no replication across the three groups (WHI_AS315, WHI_BAA23, and NAS) (Table S6–S8). For example, there were 25 significant DMPs in WHI_BAA23. However, only two CpGs (cg16415104 for AS315 and cg23032421 for NAS) were statistically significantly associated with optimism in WHI_AS315 or NAS (Table S7). If we compared Table S5 with Tables S6–8, we found two probes (cg23032421 and cg04219552) in Table S5 were also identified in Table S7 but none of the probes in Table S5 were also identified in Tables S6 and S8.
Discussion
We conducted an EWAS of optimism in well-established cohorts of women (WHI) and men (NAS). An overwhelming majority of the CpGs (N= 359,862) were homogeneous across men and women and across racial/ethnic groups. Based on the 359,862 homogeneous CpGs, we identified 13 DMPs using the PC-correction approach. We further identified 4 statistically significant DMRs and 2 pathways associated with optimism. The annotated genes from DMPs and DMRs, and the pathways were related to psychiatric disorders, cardiovascular disease, neurodevelopmental and neurodegenerative disorders, and cancer. To our knowledge our results are novel as this is the first study to use an agnostic EWAS with optimism as an approach to understand the potential epigenetic mechanisms underlying observed relations between optimism and health.
Significant DMPs
Among the top 13 DMPs based on PC-correction, five (i.e., cg24856537, cg18583565, cg03451670, cg05065507, cg06418871) were annotated to genes (i.e., NOTCH4, MGMT, AGAP1 (also known as CENTG2), TACSTD2 (also known as TROP2), MARVELD2) that are associated with multiple health outcomes, including psychiatric disorders (42–44), neurodevelopmental disorders (45), cardiovascular disease (46), cancer (47) and hearing loss (48). For example, higher optimism was associated with increased DNAm at cg24856537 (body of NOTCH4) (p-value=3.72×10−06, β=1.41×10−04) in our study. The NOTCH4 gene maps to chromosome 6p21.32 and is expressed in endothelial cells and various tissues, including the nervous system (49). Several studies have identified NOTCH4 as a candidate gene involved in psychiatric disorders, such as schizophrenia (SCZ) (42) and bipolar disorder (44), and it plays an important role in heart development (46). Another example, an increase in methylation at cg03451670 (body of AGAP1) was negatively associated with optimism (p-value=3.14×10−05, β=−2.50×10−04). The AGAP1 gene maps to chromosome 2q37.2 and has been identified as a candidate gene for psychiatric disorders, such as SCZ (43) and autism (45).
In addition, cg18583565 is mapped to genes MGMT, which might be a risk factor for cognitive impairment (50) although the site is located to the gene body. Another probe (cg05065507) is mapped to the gene body of TACSTD2; overexpression of TACSTD2 (also known as TROP2) has been reported in multiple cancers (47). The probe of cg06418871 is mapped to the gene promoter of MARVELD2; mutations in this gene seem to be a significant cause of hearing loss (48).
Significant DMRs and pathways
We observed four significant DMRs annotating to four genes, among which three (i.e., MARVELD2, MGMT, SIGLECL1) have been associated with adverse health outcomes (51–53). The genes MARVELD2 and MGMT were also identified in the single CpG site analyses. The increased expression of gene SIGLECL1, which is located in chr19: 51774377–51774667, has been found in patients with dementia (54). The three CpGs (i.e., cg14724749, cg14884932, cg15497724) are mapped to either the island or N shore area of gene SIGLECL1 and their methylation level were all negatively (although not statistically) associated with optimism level in the single CpG site analyses (data not shown). We further identified two significant pathways with optimism. The pathway “Signaling by Rho family GTPases” has been implicated in a variety of cancers and other disorders including atherosclerosis (55); and “Axonal guidance signaling” pathway is involved in a neurodevelopmental and neurodegenerative disorders, such as autism, SCZ, Alzheimer’s and Parkinson’s disease (56).
These findings are concordant with previous epidemiological studies linking optimism to specific disease endpoints, such as cancer, cardiovascular disease, and neurodegenerative disorders (4, 5, 57, 58). Although underlying mechanisms have not been fully elucidated, plausible explanations include buffering against stress reactivity (33), greater likelihood of engaging in health-related behaviors (21), and better communication with doctors (59), among others. Furthermore, it is worth noting that findings of optimism with health outcomes are usually maintained after adjusting for health behaviors, suggesting biological mechanisms may be in play (4). DNAm impacts gene expression and gene regulation and multi-omics research will inform further biologic insights between optimism and health outcomes. Recently DNAm has been linked in the causal pathway between inflammation and chronic diseases including cardiovascular disease (60). One speculation regarding mechanisms is that optimism may buffer negative response to stressful events and the potential toxic neurobiological effects of stress caused by inflammation (61) and endothelial dysfunction (62). Further work is needed to ascertain how optimism may lead to disease-relevant epigenetic alterations.
Limitations and strengths
The current study has some limitations. Data from two sub-studies in WHI as well as NAS are cross-sectional, with optimism and DNAm assessed at a single point in time; hence we cannot draw causal conclusions concerning directionality of the observed associations. Our sub-studies primarily included older subjects, and DNAm may operate differently in younger individuals. While optimism is stable in adulthood, it is not immutable, and therefore, generalizability to other age groups remains uncertain (17). DNAm data pre-processing, quality control, and normalization differed by study, but this is unlikely to affect findings as only 9% heterogeneity was evident in the between-study heterogeneity test. In addition, given the modest correlation between two optimism measures (LOT-R and PSM-R) used across the cohorts, it is unclear if our findings reflect linkages with optimism per se, or with a shared element of having a positive orientation or positive affect more generally. Also important to note is that in the current study we considered optimism only; thus our findings are not comparable with those from studies examining EWAS in relation to other positive psychological factors (e.g., wellbeing) (18).
On the other hand, our study has a number of important strengths. This is the first study to investigate the associations between optimism and DNAm using EWAS. Second, we used well characterized cohorts for the EWAS. Third, we applied the number of independent degrees of freedom approach to account for the high correlation between CpGs (36). Fourth, we analyzed the associations between optimism and DNAm using single CpG site, regional and pathway levels to fully elucidate the genes and pathways potentially related to optimism. While effect sizes for associations with individual CpG sites were somewhat small, findings of associations with regions or pathways that contain multiple CpGs suggest that the overall effect may be larger. Further studies with larger sample sizes may facilitate clearer detection of small but important effects.
In summary, this study examined the associations between optimism and DNAm in older women and men from two cohorts. The annotated associated genes and pathways have been implicated in multiple pathophysiological processes also linked with optimism in several outcomes, including psychiatric disorders, cardiovascular disease, cancer, neurodevelopmental and neurodegenerative disorders. Further studies with other tissues and gene expression to understand functional relevance are needed to gain a more comprehensive assessment of epigenetic processes in relation to optimism and the causal pathway for human health and diseases.
Supplementary Material
Source of Funding
The authors acknowledge all participants in these studies. We also thank Dr. Zongli Xu from National Institute of Environmental Health Sciences for conducting pathway analyses using the Ingenuity Pathway Analysis (IPA) database. Epigenetic Mechanisms of PM-Mediated Cardiovascular Disease (WHI_AS315) was supported by NIH/National Institute of Environmental Health Sciences grant R01-ES020836. Integrative Genomics and Risk of Coronary Heart Disease and Related Phenotypes (WHI_BAA23) was supported by the National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services (HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, HHSN268201600004C), and the National Institute of Aging (1U01AG060908-01). Investigator effort for this study was supported by a grant from the National Institute of Aging (R01-AG53273). The Normative Aging Study is a research component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC) and is supported by the Veterans Affairs Cooperative Studies Program/Epidemiological Research Centers. The Normative Aging Study is also supported by grants from the National Institute of Aging (R01-AG018436, K99-AG055696, K08-AG048221) and National Institute of Environmental Health Sciences (R01-ES015172, 1R01-ES027747, R01-ES031259, R01-ES021733), and a Senior Research Career Scientist award from the Veterans Affairs Clinical Science R&D Service. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of funding bodies such as the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Departments of Health and Human Services and Veterans Affairs.
Acronyms:
- AA
African-American
- BMI
body mass index
- Chr
chromosome
- CpGs
cytosine-phosphate-guanines
- CHD
Coronary Heart Disease
- DMPs
differentially methylated probes
- DMRs
differently methylated regions
- DNAm
DNA methylation
- EWAS
epigenome-wide association study
- I 2
I-squared
- LME
linear mixed-effect
- NAS
Normative Aging Study
- PCs
principal components
- SD
standard deviation
- SE
standard error
- WHI
Women’s Health Initiative
- λ
inflation factor
Footnotes
Conflicts of Interest
Dr. DeMeo has received grant support from Bayer and honoraria from Novartis. The other authors report no financial relationships with commercial interests.
References
- 1.https://www.census.gov/newsroom/press-releases/2018/cb18-41-population-projections.html. 01/04/2022
- 2.Scheier MF, Carver CS, Bridges MW. Distinguishing optimism from neuroticism (and trait anxiety, self-mastery, and self-esteem): a reevaluation of the Life Orientation Test. J Pers Soc Psychol 1994. 67(6):1063–78. [DOI] [PubMed] [Google Scholar]
- 3.Malinchoc M, Offord KP, Colligan RC. PSM-R: Revised Optimism-Pessimism Scale for the MMPI-2 and MMPI. J Clin Psychol 1995. 51(2):205–14. [DOI] [PubMed] [Google Scholar]
- 4.Kubzansky LD, Huffman JC, Boehm JK, Hernandez R, Kim ES, Koga HK, et al. Positive psychological well-being and cardiovascular disease: JACC health promotion series. J Am Coll Cardiol 2018. 72(12):1382–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gruber-Baldini AL, Ye J, Anderson KE, Shulman LM. Effects of optimism/pessimism and locus of control on disability and quality of life in Parkinson’s disease. Parkinsonism Relat Disord 2009. 15(9):665–9. [DOI] [PubMed] [Google Scholar]
- 6.Scheier MF, Carver CS. Dispositional optimism and physical health: A long look back, a quick look forward. Am Psychol 2018. 73(9):1082–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robertson KD, Jones PA. DNA methylation: past, present and future directions. Carcinogenesis 2000;21(3):461–7. [DOI] [PubMed] [Google Scholar]
- 8.Dada O, Adanty C, Dai N, Zai C, Gerretsen P, Graff A, et al. Mediating effect of genome-wide DNA methylation on suicidal ideation induced by perceived stress. Psychiatr Genet. 2021;31(5):168–76. [DOI] [PubMed] [Google Scholar]
- 9.Kim D, Kubzansky LD, Baccarelli A, Sparrow D, Spiro A 3rd, Tarantini L, et al. Psychological factors and DNA methylation of genes related to immune/inflammatory system markers: the VA Normative Aging Study. BMJ Open. 2016;6(1):e009790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam LL, Emberly E, Fraser HB, Neumann SM, Chen E, Miller GE, et al. Factors underlying variable DNA methylation in a human community cohort. Proc Natl Acad Sci U S A. 2012;109 Suppl 2(Suppl 2):17253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S DNA methylation age of human tissues and cell types. Genome Biol 2013;14(10): R115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pepin ME, Ha CM, Crossman DK, Litovsky SH, Varambally S, Barchue JP, et al. Genome-wide DNA methylation encodes cardiac transcriptional reprogramming in human ischemic heart failure. Lab Invest 2019; 99(3):371–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang YP, Lei QY. Metabolic recoding of epigenetics in cancer. Cancer Commun (Lond) 2018. 38(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Os Jovanova, Nedeljkovic I, Spieler D, Walker RM, Liu C, Luciano M, et al. DNA Methylation Signatures of Depressive Symptoms in Middle-aged and Elderly Persons: Meta-analysis of Multiethnic Epigenome-wide Studies. JAMA psychiatry 2018. 75(9):949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C, Turecki G. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Molecular psychiatry 2015. 20(3):320–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakusic J, Schaufeli W, Claes S, Godderis L. Stress, burnout and depression: A systematic review on DNA methylation mechanisms. J Psychosom Res 2017. 92:34–44. [DOI] [PubMed] [Google Scholar]
- 17.Kim ES, Fong K, Lee L, Spiro A, Schwartz J, Whitsel E, et al. Optimism is not associated with two indicators of DNA methylation aging. Aging 2019. 11(14):4970–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baselmans BM, van Dongen J, Nivard MG, Lin BD, Zilhao NR, Boomsma DI, Bartels M. Epigenome-Wide Association Study of Wellbeing. Twin Res Hum Genet. 2015;18(6):710–9. [DOI] [PubMed] [Google Scholar]
- 19.Horvath S, Ritz BR. Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging 2015. 7(12):1130–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plomin R, Scheier MF, Bergeman CS, Pedersen NL, Nesselroade JR, McClearn GE. Optimism, pessimism and mental health: A twin/adoption analysis. Pers Individ Dif 1992. 13(8):921–30. [Google Scholar]
- 21.Carver CS, Scheier MF, Segerstrom SC. Optimism. Clin Psychol Rev 2010. 30(7):879–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malouff JM, Schutte NS. Can psychological interventions increase optimism? A meta-analysis. J Posit Psychol 2017. 12(6):594–604. [Google Scholar]
- 23.Eshun S Cultural variations in hopelessness, optimism, and suicidal ideation: A study of Ghana and US college samples. Cross-Cultural Research 1999. 33(3):227–38. [Google Scholar]
- 24.Study TWsHI. Design of the Women’s Health Initiative clinical trial and observational study. Control Clin Trials 1998;19(1):61–109. [DOI] [PubMed] [Google Scholar]
- 25.Hays J, Hunt JR, Hubbell FA, Anderson GL, Limacher M, Allen C, Rossouw J. The Women’s Health Initiative recruitment methods and results. Ann Epidemiol 2003. 13(9 Suppl): S18–77. [DOI] [PubMed] [Google Scholar]
- 26.Gondalia R, Baldassari A, Holliday KM, Justice AE, Méndez-Giráldez R, Stewart JD, et al. Methylome-wide association study provides evidence of particulate matter air pollution-associated DNA methylation. Environ Int 2019. 132:104723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang C, Koutrakis P, Gao X, Baccarelli A, Schwartz J. Associations of annual ambient PM2.5 components with DNAm PhenoAge acceleration in elderly men: The Normative Aging Study. Environ Pollut 2020. 258:113690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colligan RC, Offord KP, Malinchoc M, Schulman P, Seligman ME. CAVEing the MMPI for an optimism-pessimism scale: Seligman’s attributional model and the assessment of explantory style. J Clin Psychol 1994. 50(1):71–95. [DOI] [PubMed] [Google Scholar]
- 29.Kubzansky LD, Sparrow D, Vokonas P, Kawachi I. Is the glass half empty or half full? A prospective study of optimism and coronary heart disease in the normative aging study. Psychosom Med 2001. 63(6):910–6. [DOI] [PubMed] [Google Scholar]
- 30.Jane E Gillham AJS, Reivich KJ, Seligman ME. In: Chang EC, editor. Optimism and Pessimism: Implications for Theory, Research, and Practice. Washington, DC: American Psychological Association; 2001. p. 301–20. [Google Scholar]
- 31.Cohen P, Cohen J, Aiken LS, West SG. The problem of units and the circumstance for POMP. Multivariate Behav Res 1999. 34(3):315–46. [Google Scholar]
- 32.Heiss JA, Just AC. Identifying mislabeled and contaminated DNA methylation microarray data: an extended quality control toolset with examples from GEO. Clin Epigenetics 2018. 10:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Majeed NM, Tan JJ, Tov W, Hartanto A. Dispositional optimism as a buffer against emotional reactivity to daily stressors: A daily diary approach. J Res Pers 2021. 93:104105. [Google Scholar]
- 34.Kim ES, Hagan KA, Grodstein F, DeMeo DL, De Vivo I, Kubzansky LD. Optimism and cause-specific mortality: a prospective cohort study. Am J Epidemiol 2017. 185(1):21–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mansournia MA, Altman DG. Inverse probability weighting. BMJ 2016. 352:i189. [DOI] [PubMed] [Google Scholar]
- 36.Li M-X, Yeung JM, Cherny SS, Sham PC. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Human Genetics 2012;131(5):747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rapado-González Ó, Martínez-Reglero C, Salgado-Barreira Á, Muinelo-Romay L, Muinelo-Lorenzo J, López-López R, Díaz-Lagares Á, Suárez-Cunqueiro M. Salivary DNA Methylation as an Epigenetic Biomarker for Head and Neck Cancer. Part I: A Diagnostic Accuracy Meta-Analysis. J Pers Med 2021;11(6):568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mallik S, Odom GJ, Gao Z, Gomez L, Chen X, Wang L. An evaluation of supervised methods for identifying differentially methylated regions in Illumina methylation arrays. Brief Bioinform 2019. 20(6):2224–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Z, Xie C, Taylor JA, Niu L. ipDMR: Identification of differentially methylated regions with interval p-values. Bioinformatics 2021. 37(5):711–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Z, Sandler DP, Taylor JA. Blood DNA Methylation and Breast Cancer: A Prospective Case-Cohort Analysis in the Sister Study. J Natl Cancer Inst 2020. 112(1):87–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agha G, Mendelson MM, Ward-Caviness CK, Joehanes R, Huan T, Gondalia R, Salfat E, Brody J, Fiorito G, Bressler J, Chen B, Ligthart S, Guarrera S, Colicino E, Just A, Wahl S, Gieger C, Vandiver A, Tanaka T, Hernandez D, Pilling L, Singleton A, Sacerdote C, Krogh V, Panico S, Tumino R, Li Y, Zhang V, Stewart J, Floyd J, Wiggins K, Rotter J, Multhaup M, Bakulski K, Horvath S, Tsao P, Absher D, Vokonas P, Hirschhorn J, Fallin D, Liu C, Bandinelli S, Boerwinkle E, Dehghan A, Schwartz J, Psaty B, Feinberg A, Hou L, Ferrucci L, Sottodehnia N, Matullo G, Peters A, Fornage M, Assines T, Whitsel E, Levy D, Baccarelli A, Boerwinkle E, Dehghan A, Schwartz J, Psaty B, Feinberg A, Hou L, Ferrucci L, Sotoodehnia N, Matullo G, Peters A, Fornage M, Assimes T, Whitsel E, Levy D, Baccarelli A. Blood Leukocyte DNA Methylation Predicts Risk of Future Myocardial Infarction and Coronary Heart Disease. Circulation 2019. 140(8):645–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei J, Hemmings GP. The NOTCH4 locus is associated with susceptibility to schizophrenia. Nature Genetics 2000. 25(4):376–7. [DOI] [PubMed] [Google Scholar]
- 43.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe’er I, Dudbridge F, Holmans P, Whittemore A, Mowry B, Olincy A, Amin F, Cloninger C, Silverman J, Buccola N, Byerley W, Black D, Crowe R, Oksenberg J, Mirel D, Kendler K, Freedman R, Gejman P. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 2009. 460(7256):753–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dieset I, Djurovic S, Tesli M, Hope S, Mattingsdal M, Michelsen A, Inge Joa, Larsen T, Agartz I, Melle I, Rossberg J, Aukrust P, Andreassen O, Ueland T. Up-regulation of NOTCH4 gene expression in bipolar disorder. Am J Psychiatry 2012. 169(12):1292–300. [DOI] [PubMed] [Google Scholar]
- 45.Wassink TH, Piven J, Vieland VJ, Jenkins L, Frantz R, Bartlett CW, Goedken R, Childress D, Spence M, Smith M, Sheffield V. Evaluation of the chromosome 2q37.3 gene CENTG2 as an autism susceptibility gene. Am J Med Genet B Neuropsychiatr Genet 2005. 136b(1):36–44. [DOI] [PubMed] [Google Scholar]
- 46.Luxán G, D’Amato G, MacGrogan D, de la Pompa JL. Endocardial Notch Signaling in Cardiac Development and Disease. Circ Res 2016. 118(1):e1–e18. [DOI] [PubMed] [Google Scholar]
- 47.Shvartsur A, Bonavida B. Trop2 and its overexpression in cancers: regulation and clinical/therapeutic implications. Genes Cancer 2015. 6(3–4):84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mašindová I, Šoltýsová A, Varga L, Mátyás P, Ficek A, Hučková M, Sůrová M, Šafka-Brožková D, Anwar S, Judit Bene, Straka S, Janicsek I, Ahmed Z, Seeman P, Melegh B, Profant M, Klimeš I, Riazuddin S, Kádasi L, Gašperíková D. MARVELD2 (DFNB49) mutations in the hearing impaired Central European Roma population--prevalence, clinical impact and the common origin. PloS one 2015. 10(4):e0124232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murphy PA, Lu G, Shiah S, Bollen AW, Wang RA. Endothelial Notch signaling is upregulated in human brain arteriovenous malformations and a mouse model of the disease. Lab Invest 2009. 89(9):971–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Q, Xiao F, Qi F, Song X, Yu Y. Risk Factors for Cognitive Impairment in High-Grade Glioma Patients Treated with Postoperative Radiochemotherapy. Cancer Res Treat 2020. 52(2):586–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spaks A, Svirina D, Spaka I, Jaunalksne I, Breiva D, Tracums I, Krievins D. CXC chemokine ligand 4 (CXCL4) is predictor of tumour angiogenic activity and prognostic biomarker in non-small cell lung cancer (NSCLC) patients undergoing surgical treatment. Biomarkers 2016. 21(5):474–8. [DOI] [PubMed] [Google Scholar]
- 52.Ye C, Xu M, Lin M, Zhang Y, Zheng X, Sun Y, Deng Y, Pan J, Xu Z, Lu X, Chi P. Overexpression of FZD7 is associated with poor survival in patients with colon cancer. Pathol Res Pract 2019. 215(8):152478. [DOI] [PubMed] [Google Scholar]
- 53.Li G, Su Q, Liu H, Wang D, Zhang W, Lu Z, Chen Y, Huang X, Li W, Zhang C, He Y, Fu L, Bi J. Frizzled7 Promotes Epithelial-to-mesenchymal Transition and Stemness Via Activating Canonical Wnt/β-catenin Pathway in Gastric Cancer. Int J Biol Sci 2018. 14(3):280–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rendina A, Drongitis D, Donizetti A, Fucci L, Milan G, Tripodi F, Giustezza F, Postiglione A, Pappata S, Ferrari R, Bossu P, Angiolillo A, Costanzo A, Caiazza M, Vitale E. CD33 and SIGLECL1 Immunoglobulin Superfamily Involved in Dementia. J Neuropathol Exp Neurol 2020. 79(8):891–901. [DOI] [PubMed] [Google Scholar]
- 55.Hall A Rho family GTPases. Biochem Soc Trans 2012. 40(6):1378–82. [DOI] [PubMed] [Google Scholar]
- 56.Stoeckli ET. Understanding axon guidance: are we nearly there yet? Development 2018;145(10). [DOI] [PubMed] [Google Scholar]
- 57.Ciria-Suarez L, Calderon C, Fernández Montes A, Antoñanzas M, Hernández R, Rogado J, Pacheo-Barcia V, Ansensio-Martinez E, Palacin-Lois M, Jimenez-Fonseca P. Optimism and social support as contributing factors to spirituality in Cancer patients. Support Care Cancer 2021. 29(6):3367–73. [DOI] [PubMed] [Google Scholar]
- 58.Amonoo HL, Celano CM, Sadlonova M, Huffman JC. Is Optimism a Protective Factor for Cardiovascular Disease? Curr Cardiol Rep 2021. 23(11):158. [DOI] [PubMed] [Google Scholar]
- 59.Mannix MM, Feldman JM, Moody K. Optimism and health-related quality of life in adolescents with cancer. Child Care Health Dev 2009. 35(4):482–8. [DOI] [PubMed] [Google Scholar]
- 60.Wielscher M, Mandaviya PR, Kuehnel B, Joehanes R, Mustafa R, Robinson O, Zhang Y, Bodinier B, Walton E, Mishra P, Schlosser P, Wilson R, Tsai P, Palaniswamy S, Marioni RE, Fiorito G, Cugliari G, Karhunen V, Ghanbari M, Psaty B, Loh M, Bis J, Lehne B, Sotoodehnia N, Deary I, Chadeau-Hyam M, Brody J, Cardona A, Selvin E, Smith A, Miller A, Torres M, Marouli E, Gao X, Meurs J, Grafshindler J, Rathmann W, Koenig W, Peters A, Weninger W, Farlik M, Zhang T, Chen W, Xia Y, Teumer A, Nauck M, Grabe H, Doerr M, Lehtimaki T, Guan W, Milani L, Tanaka T, Fisher K, Waite L, Kasela S, Vineis P, Verweij N, Harst P, Lacoviello L, Sacerdote C, Panico S, Krogh V, Tumino R, Tzala E, Matullo G, Hurme M, Raitakari O, Colicino E, Baccarelli A, Kahonen M, Herzig KH, Li S, BIOS consortium, Conneely K, Kooner J, Kottgen A, Heijmans B, Deloukas P, Relton C, Ong K, Bell J, Boerwinkle E, Elliott P, Brenner H, Beekman M, Levy D, Waldenberger M, Chambers J, Dehghan A, Jarvelin M. DNA methylation signature of chronic low-grade inflammation and its role in cardio-respiratory diseases. Nat Commun 2022. 13(1):2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Black PH, Garbutt LD. Stress, inflammation and cardiovascular disease. J Psychosom Res 2002. 52(1):1–23. [DOI] [PubMed] [Google Scholar]
- 62.Ghiadoni L, Donald AE, Cropley M, Mullen MJ, Oakley G, Taylor M, O’Connor G, Betteridge J, Klein N, Steptoe A, Deanfield J. Mental stress induces transient endothelial dysfunction in humans. Circulation 2000. 102(20):2473–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


