Abstract
The transcription factor Yin Yang 1 (YY1) is ubiquitously expressed in mammalian cells, regulating the expression of a variety of genes involved in proliferation, differentiation, and apoptosis in a context-dependent manner. While it is well-established that global YY1 knockout (KO) leads to embryonic death in mice and YY1 deletion in neurons or oligodendrocytes induces impaired brain function, the role of astrocytic YY1 in the brain remains unknown. We investigated the role of astrocytic YY1 in the brain using a glial fibrillary acidic protein (GFAP)-specific YY1 conditional KO (YY1 cKO) mouse model to delete astrocytic YY1. Astrocytic YY1 cKO mice were tested for behavioral phenotypes, such as locomotor activity, coordination, and cognition, followed by an assessment of relevant biological pathways using RNA-sequencing analysis, immunoblotting, and immunohistochemistry in the cortex, midbrain, and cerebellum. YY1 cKO mice showed abnormal phenotype, movement deficits, and cognitive dysfunction. At the molecular level, astrocytic YY1 deletion altered the expression of genes associated with proliferation and differentiation, p53/caspase apoptotic pathways, oxidative stress response, and inflammatory signaling including NF-κB, STAT, and IRF in all regions. Astrocytic YY1 deletion significantly increased the expression of GFAP as astrocytic activation and Iba1 as microglial activation, indicating astrocytic YY1 deletion activated microglia as well. Accordingly, multiple inflammatory cytokines and chemokines including TNF-α and CXCL10 were elevated. Combined, these novel findings suggest that astrocytic YY1 is a critical transcription factor for normal brain development and locomotor activity, motor coordination, and cognition. Astrocytic YY1 is also essential for preventing pathological oxidative stress, apoptosis, and inflammation.
Keywords: yin yang 1, YY1, astrocytes, inflammation, chemokines, p53, RNA-sequencing
Introduction
Yin Yang 1 (YY1) is a ubiquitously expressed transcription factor, exerting multiple functions in the mammalian brain by acting either as an activator or a repressor for the transcription of its target genes (He & Casaccia-Bonnefil, 2008; Shi, Seto, Chang, & Shenk, 1991). YY1 controls protein activity in neural cell types of neurons, astrocytes, microglia, and oligodendrocytes, in addition to modifying DNA conformation (Galvin & Shi, 1997). The physiological function of YY1 in the central nervous system (CNS) is not completely understood, but the YY1 promoter is GC-rich and contains multiple Sp1 binding sites, similar to the promoter of a large subset of housekeeping and growth-regulating genes (Safrany & Perry, 1993; Yao et al., 1998), implicating that it might be involved in brain development. Indeed, it has been established that YY1 plays an essential role during embryogenesis (Donohoe et al., 1999), such as in the development of the neuroepithelium (Dong & Kwan, 2020) and maintaining the proliferation and survival of neural progenitor cells (Zurkirchen et al., 2019). YY1 also orchestrates chromatin looping during early neural lineage commitment (Beagan et al., 2017; Weintraub et al., 2017) and is considered a master regulator of a neural crest (NC) transcriptional program (Varum et al., 2019). Since NC is an embryonic stem cell population, giving rise to a plethora of cell types, including neurons and glia (Baggiolini et al., 2015; Dupin & Sommer, 2012), YY1’s activity during development could potentially determine the fate of many neural cell types in the brain.
The neural cell type-specific roles of YY1 in the brain remains to be established. In astrocytes, YY1 is highly expressed (Karki et al., 2014; Waters et al., 2019), but little is known as to whether astrocytic YY1 regulates brain function and astrocyte-specific functions. Studies have shown that YY1 is dispensable during the early stages of astrocyte development, but it is essential during late stages of brain development, which are governed by region-specific interactions with neurons, and for maintaining mature astrocytes in the adult brain (Mockenhaupt et al., 2021). Given that astrocytes reside throughout the entire CNS and perform critical functions (Farmer & Murai, 2017; Khakh & Sofroniew, 2015), the role of astrocytic YY1 in contributing to brain function is worthy of study.
Since it has been shown that YY1 is highly expressed in astrocytes (Karki et al., 2014; Rosas, Vargas, Lopez-Bayghen, & Ortega, 2007), it is important to identify the biological processes and pathways affected by the deletion of astrocytic YY1 in understanding its comprehensive role in astrocytes and CNS. YY1 function in developmental stages of the brain has been previously studied (Dong & Kwan, 2020; He et al., 2007; Pajarillo et al., 2020; Zurkirchen et al., 2019). Recently, it has been reported that astrocyte-specific YY1 deletion during the later stages of post-natal development in GFAP-controlled YY1 conditional knockout (YY1 cKO) mice impaired astrocytic maturation and motor function along with apoptosis, and increased the expression of many inflammatory genes (Mockenhaupt et al., 2021).
To investigate the role of astrocytic YY1 in the mouse brain, we have generated astrocyte GFAP-specific YY1 cKO mice. However, in addition to astrocytes, GFAP is expressed in radial glial cells and neural stem cells (Garcia, Doan, Imura, Bush, & Sofroniew, 2004; Mamber et al., 2012) thus YY1 deletion under the GFAP promoter control can shed light on the role of YY1, not only in astrocytes, but also in adult neurogenesis. It has been shown that GFAP co-expressed with nestin, a neural stem/progenitor cell marker at E18 in radial glial cells and neural progenitor cells of mouse embryonic SVZ (Suzuki & Goldman, 2003). In addition, GFAP is expressed in a brain region- and age-specific manner as it is expressed in the cortex from P0, but in the cerebellum only from P5 (Mamber et al., 2012) or P7 (Tao et al., 2011), suggesting that GFAP-dependent YY1 deletion may affect different brain regions in distinct manners.
Materials and methods
Animals
Eight weeks-old female B6.Cg-Tg(Gfap-cre)73.12Mvs/J (RRID:IMSR_JAX:012886) and male B6;129S4-Yy1tm2Yshi/J (RRID:IMSR_JAX:014649) mice were purchased from the Jackson Laboratory (Bar Harbor, ME) and housed in the Florida A&M University (FAMU) Animal Care Facility. Mice were group-housed and kept on a 12-h light and dark cycle with food and water available ad libitum. To generate GFAP-specific YY1 cKO mice, we used a cre-loxP system in which the B6;129S4-Yy1tm2Yshi/J (YY1-floxed or YY1flox/flox) uses a construct that introduces a floxed locus so that cell-type specific KO can be achieved through breeding with a Cre recombinase mouse. The first generation of GFAP-cre+/−:YY1flox/− mice were generated by crossing GFAP-cre+/− female with YY1flox/flox male mice. The GFAP-cre+/−:YY1flox/flox mice (GFAP-specific YY1 cKO) were generated by crossing GFAP-cre+/−:YY1flox/− female mice with YY1flox/flox male mice. A total of 38 mice with mixed sex (19 mice per group; control and YY1 cKO mice) were used in the study: for each group, 6 mice for survival, 7 mice for RNA-seq analysis, 3 mice for western blot, and 3 mice for immunohistochemistry experiments. All animal protocols were approved by the FAMU Institutional Animal Care and Use Committee (IACUC) for the care and use of laboratory animals. The weight of each mouse was measured weekly to assess overall growth.
Genotyping of mice
For genotyping, genomic DNA samples were obtained from tail snips of 4-5 week-old mice by incubating the tissue at 55 °C in the lysis buffer (0.1 M Tris-Cl, pH 8.8, 5mM EDTA, pH 8.0, 0,2% SDS, 0.2 M NaCl, 20 μg/ml proteinase K). The isolated DNA was subsequently genotyped using the following primer sets: GFAP-cre-F (5’- TCC ATA AAG GCC CTG ACA TC-3’) and GFAP-cre-R (5’-TGC GAA CCT CAT CAC TCG T-3’); YY1-loxP-F (5’- ACC TGG TCT ATC GAA AGG AAG CAC-3’) and YY1-loxP-R (5’- GCT TCG CCT ATT CCT CGC TCA TAA −3’). The PCR products were separated using agarose gel electrophoresis and visualized via blot imaging by the Bio-Rad ChemiDoc imaging system.
Open-field, rotarod, and novel object (NO) tests
Seven mice (mixed sex) per group (control and YY1 cKO) were randomly assigned for the following tests. Open-field test for locomotor activity and rotarod test for motor coordination were performed in 12-week-old mice as described in our previous study (Johnson et al., 2018). Briefly, locomotor activity was assessed in an open-field arena made of Plexiglas using Fusion SuperFlex software v6.25 (Omnitech Electronics, Columbus, OH). Each animal had an acclimation period at the same time of each day for 3 consecutive days at 12 weeks of age. Each mouse was placed in the center of the arena, followed by the measurement of locomotor activity for 30 min. The mean distance traveled, movement speed, and vertical activity were compared between groups.
For motor coordination, mice were trained for 3 consecutive days, with 1 session consisting of 3 trials separated by 5 min rest periods in the AccuRotor rotarod system (Omnitech Electronics). Each mouse was placed on the rod and the trial began when the rod started rotating, speed gradually increasing from 4 to 40 rpm up to 10 min by 0.1 revolution/sec. Latency to fall (the time for subjects to fall from the rotarod during a trial) was recorded with Fusion Software v6.3 for AccuRotor. Motor coordination for mice persisting on the rod for the entire trial duration was recorded for 650 sec. The mean duration for each group was used for comparisons.
For the NO recognition test, mice were acclimated in the arena for 3 consecutive days before the actual test. On the testing day, mice underwent a familiarization phase followed by a 5 min resting period and then the NO test. During familiarization, each mouse was exposed to two identical objects (familiar objects, FO) placed on the left and right back corner of the arena for a 10 min-period. During the NO test, one FO will be replaced with an item of different texture, shape, and color (novel object, NO) on the right back corner of the open-field arena. Mice were placed at the center of the arena for a 10 min period with FO and NO. The time spent exploring the NO and FO were recorded with the Fusion software. Time spent exploring the NO and the index to discriminate FO and NO were calculated and compared between groups (Bevins & Besheer, 2006). The time of novel object exploration and discrimination index values were compared between the control and YY1 cKO mice.
Dissection of the mouse brain
All dissections were performed within a 2 h interval (4:00 PM – 6:00 PM). The total time between decapitation and storage of brain samples into the liquid nitrogen was no more than 4 min. The mouse brain regions of cortex, midbrain, and cerebellum were used for the study as these regions are critically involved in regulation of movement and memory (Ackerman, 1992). After mouse brains were removed from the skull under anesthesia, frontal cortex, midbrain, and cerebellum were dissected according to the methods described previously (Li, 2011; Meyerhoff et al., 2021). The three brain regions were then rapidly frozen in liquid nitrogen before being stored at −80 °C.
RNA extraction and RNA-Seq library preparation
Following the manufacturer’s recommended procedures, total RNA was extracted from the cortex, midbrain, and cerebellum using a modified RNA extraction and purification protocol with TRIzol® Reagent (Life Technologies, Carlsbad, CA) and RNA Clean & Concentrator™−5 with Dnase I (Zymo Research, Irvine, CA). Seven animals (n=7, 4 males and 3 females) from the two groups (control and YY1 cKO) were used for a total of 42 Illumina sequencing libraries in the three brain regions. The library preparation failed for one YY1 cKO sample from the cortex, so 6 samples were used for this group.
RNA sample input was 400 ng total RNA (determined by Qubit RNA HS reagents, Thermo) with a RIN >7 (TapeStation 4200 High Sensitivity RNA ScreenTape, Agilent). Libraries were prepared with the Biomek 400 Automated Workstation (Beckman Coulter), using the NEBNext® Poly(A) mRNA Magnetic Isolation Module and NEBNext® Ultra II RNA Library Prep kit for Illumina (New England Biolabs, MA). A unique index (barcode) was added to each library to multiplex the libraries for the sequencing run. The multiplexed samples were quantified with qPCR (Kapa Biosystems, MO) and the average library size was determined with a Bioanalyzer high sensitivity DNA chip (Agilent Technologies, CA). The pooled libraries were sequenced with paired-end, 50 base pair reads on an Illumina NovaSeq 6000 located in the Translational Science Laboratory at the College of Medicine, Florida State University. The sequencing data were demultiplexed into individual sample data and adapter primer sequences were removed during the demultiplexing process.
RNA-seq data analysis
Each sequenced library was assessed for quality control using fastQC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc) and further analyzed using RNA-Seq Alignment version 1.1.0 (Illumina BaseSpace application). The reads were aligned with STAR aligner (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3530905/) to the mouse genome (genome release GRCm38/mm10), using default parameters, and counts for each gene were generated using Salmon (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5600148/). DESeq2 (Love, Huber, & Anders, 2014) software accounts for nonnormally distributed count data and differences in sequencing depth and was used to perform pairwise comparisons to determine statistically significant differentially expressed genes (DEG) using a False Discovery Rate, FDR, of <0.05 (Love, Huber, & Anders, 2014) . The WT versus YY1 cKO pairwise comparison included 7 replicates for each brain region and resulted in 3749 DEG in the cortex, 3201 DEG in the midbrain, and 6924 DEG in the cerebellum. These data were further analyzed using WebGestalt software and heatmap, volcano plots, principal component analysis (PCA), and other graphs were generated using R software, packages and Bioconductor open-source software (Wang, Duncan, Shi, & Zhang, 2013; B. Zhang, Kirov, & Snoddy). Briefly, the vegan package was used to create the heatmap. The ggplot2 package was employed for volcano plot and PCA. BioVenn was used to compare DEG lists of three brain regions and visualize into Venn diagram. Relevant genes and biological pathways for neurogenesis, apoptosis, oxidative stress, and proinflammatory cytokines were further assessed with the list of genes in the RT2 PCR Array Profilers (Qiagen). All data have been deposited and available in the Gene Expression Omnibus under the accession # GSE207014.
Perfusion and brain immunohistochemistry (IHC)
The IHC procedures were performed as described in our previous study with a slight modification (Pajarillo et al., 2020). Briefly, twenty-four hours after the last locomotor activity test, mice were processed for transcardial perfusion to fix brain tissues. Mice were perfused with 4% paraformaldehyde in PBS, and brains were soaked in 30% sucrose solution for 2 additional days. Afterward, tissue samples were immediately frozen in dry ice.
For immunohistochemistry, brain tissue was sliced for frozen coronal sections of cortex, midbrain, and cerebellum at 30 μm thickness in the HM525 NX Cryostat (Thermo Fisher). Tissue sections (3 mice/group) were prepared for IHC to assess multiple targets including YY1, Cre recombinase, GFAP, Iba1, NeuN, p53, caspase-3, CXCL10, and TNF-α in the three regions of the mouse brain. Tissue sections were incubated with PBST (1× PBS, 0.025% Triton X-100) then washed thrice with PBS, followed by incubation with blocking buffer (1× PBS, 10% normal goat serum, 1% bovine serum albumin) for 1 h at room temperature. Then, tissue sections on the slides were incubated with the primary antibody solution in a dark humidity chamber at 4°C overnight. Antibodies for YY1, Cre recombinase, GFAP, Iba1, NeuN, p53, CXCL10, and TNF-α (at 1:100 dilution) were diluted in blocking buffer. Antibodies for YY1 (sc-7341), p53 (sc-126), caspase-3 (sc-56053), CXCL10 (sc-374092), and TNF-α (sc-52746) were obtained from Santa Cruz Biotechnology and those for Iba1 (ab178846 or sc-28530), GFAP (ab4674), Cre recombinase (ab216262), and all secondary antibodies were from Abcam. NeuN (MAB377) antibody was from MilliporeSigma. After incubation, slides were washed with PBS thrice for 5 min in each wash, followed by incubation with secondary antibody solution conjugated with Alexa Fluor 405 (blue), 488 (green), 568 (magenta), or 647 (red) fluorescent dyes (1:1000 dilution) for 2 h at room temperature in the dark chamber. Immunostained tissue sections were washed thrice and then air-dried, followed by placing the mounting media between the coverslip and specimen. To verify the specificity of the immunohistological staining, negative controls that were incubated only with relevant secondary antibodies were used. Fluorescence intensity and localization of each protein target were assessed in the cortex, midbrain, and cerebellum with a Nikon Ts2R fluorescence microscope and Leica SPEII confocal microscope (Leica Microsystems, Inc).
shRNA lentiviral particles transduction
Human H4 astrocytes (HTB-14, ATCC) were grown in DMEM supplemented with 10% fetal bovine serum 100 U/ml of penicillin, and 100 μg/ml of streptomycin at 37 °C in a 95% air, 5% CO2 incubator. Following the manufacturer’s instruction, cells in 12-well plates (2.5 x 106 cells/well) were incubated with media containing 5 μg/ml of polybrene and 10 μl YY1 (sc-36863-V, Santa Cruz) or control shRNA (sc-108080, Santa Cruz) lentiviral particles (20 multiplicity of infection). After 24 h, lentiviral-containing medium was replaced with complete growth medium overnight, and cells were continuously grown in growth media with puromycin dihydrochloride (8 μg/ml) to select stable clones. Stable cells were used to assess the levels of proteins such as YY1, p53, active caspase-3, TNF-α, and CXCL10 in H4 cells.
Western blot
For protein analysis, protein samples (3 mice/group) were harvested from the three tissue regions collected for protein assay and immunoblotting. Brain tissue samples were homogenized in a radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors, followed by bicinchoninic acid assay. Equal amounts of proteins extracts were run on 10% SDS-PAGE, followed by immunoblotting analysis. Antibodies for YY1, Cre recombinase, p53, Bax, Bcl-2, caspase-3, CXCL10, TNF-α, and β-actin at 1:1000-1:2000 dilution were used, followed by horseradish peroxidase-conjugated secondary antibody (1:5000 dilution). Antibodies for YY1 (sc-7341), p53 (sc-126), caspase-3 (sc-56053), CXCL10 (sc-374092), TNF-α (sc-52746), Bax (sc-7480), Bcl-2 (sc-7382), and β-actin (sc-47778) were obtained from Santa Cruz Biotechnology and antibody for Cre recombinase (ab216262) were from Abcam. All blots were developed using a West Pico PLUS chemiluminescence substrate detection kit (Pierce, Rockford, IL), followed by blot imaging and quantification with the Bio-Rad ChemiDoc Imaging System and Image Laboratory Software version 5.2.1 (Bio-Rad), respectively.
Statistical analysis
Data were expressed as a mean ± S.D. Data analyses were conducted with GraphPad software (GraphPad, San Diego, CA). Bar graphs were generated to display a side-by-side comparison of the relative expression levels of selected genes and proteins between the two genotypes. Shapiro-Wilk normality test was used to determine whether the data-points had a normal distribution. For other comparisons except for RNA-seq results, statistical differences between two groups were determined by unpaired two-tailed Student’s t-test for the open-field, rotarod, novel object, relative gene and protein expression analyses. Statistical significance was set at p<0.05.
Results
Validation of YY1 gene deletion in the astrocytes of three mouse brain regions
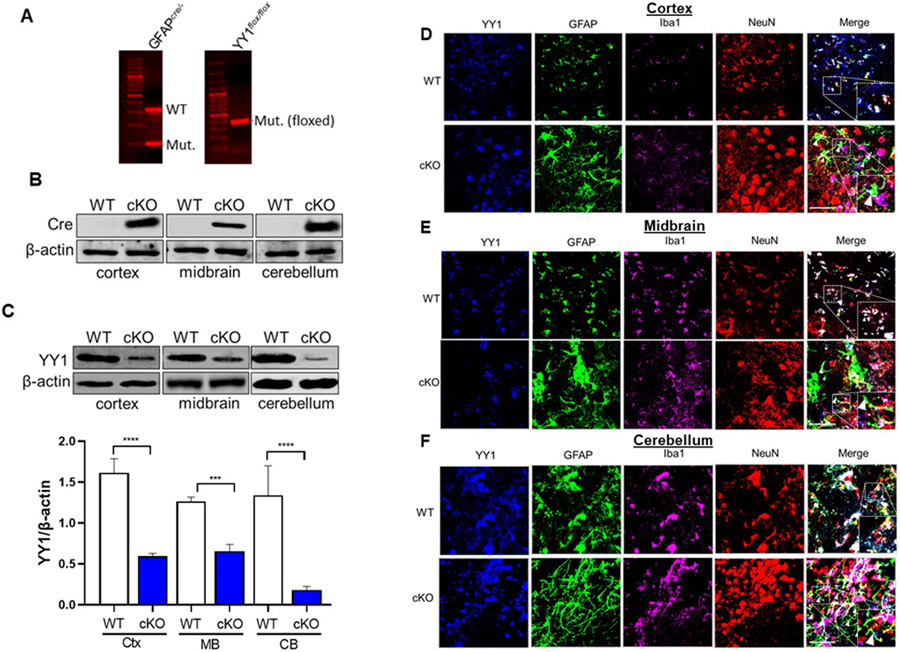
The transcription factor YY1 is a critical regulator of normal biological processes such as embryogenesis, differentiation, cell cycle, and proliferation (Verheul, van Hijfte, Perenthaler, & Barakat, 2020). Several studies have investigated the role of YY1 in neurons and oligodendrocytes (Dong & Kwan, 2020; He et al., 2007; Zurkirchen et al., 2019), but previous studies have been limited in characterizing YY1’s role in astrocyte function. To assess the role of YY1 in astrocyte function in vivo, we generated YY1 cKO mice by using a Cre/lox strategy, crossing YY1flox/flox (YY1-floxed) mice (Affar el et al., 2006) with a GFAP-cre line expressing the Cre recombinase from the astrocyte-specific GFAP promoter (Garcia et al., 2004). We confirmed DNA sequences for GFAP-cre and the YY1flox/flox (YY1 gene flanked with loxP sequences at exon 1) from mouse genomic DNA by PCR. Results showed that gfap-cre+/−/yy1flox/flox mice carry both the DNA sequences for GFAP-cre and YY1-floxed gene (Fig. 1A).
Figure 1. Expression of GFAP promoter-driven Cre recombinase deletes astrocytic YY1 in the brain.
(A) Genomic DNA from mouse tail snips were processed for genotyping by PCR, followed by agarose gel electrophoresis. GFAPcre/− and YY1-floxed (YY1flox/flox) mice show mutant DNA bands that are positive for inserting Cre recombinase and loxP sequences, respectively. YY1 cKO mice are positive for both GFAPcre/− and YY1-floxed sequences. (B-C) Brain tissue samples from YY1-floxed only (WT) and YY1 cKO (cKO) mice were assessed for the expression of Cre recombinase (B) and YY1 (C) by western blotting. (C) Immunoblotting image and quantification of relative YY1 protein levels were assessed in the cortex (Ctx), midbrain (MB), and cerebellum (CB). β-actin was used as a loading control for proteins. (D-F) Expression of YY1, Iba, GFAP, and NeuN in the cortex (D), midbrain (E), and cerebellum (F) of WT and YY1 cKO mice (×40 magnification with a confocal microscope, scale represent 50 μm). Insets show a higher magnification of a region from the imaging. Yellow arrowheads depict YY1 (blue) in astrocytes (GFAP, green), as well as in microglia (Iba1, magenta) and neurons (NeuN, red) of WT cortex, midbrain, and cerebellum, whereas white arrowheads indicate no YY1 expression in astrocytes of YY1 cKO mice.
Since our animal model of YY1 deletion is inherent to astrocytes, total levels of astrocytic YY1 in the brain cannot to be distinguished from other neural cell types. YY1 mRNA levels by RNA-seq analysis were significantly decreased in all three regions we assessed: 16.5% in cortex, 15.5% in midbrain and 46.5% in cerebellum (decreased the most in the cerebellum). YY1 protein levels were also mostly significantly decreased in the cerebellum, followed by the cortex, and midbrain in YY1 cKO mice (Fig. 1B). To confirm if astrocytic YY1 was deleted in the mouse brain, we first determined the expression of Cre recombinase in cortex, midbrain, and cerebellum. Results showed that all three regions expressed Cre recombinase protein in YY1 cKO, but not in WT (YY1flox/flox) mice (Fig. 1B). Astrocyte-specific YY1 deletion from these brain regions was also examined to confirm if the deletion is exclusive to GFAP+ astrocytes using antibodies against YY1 as well as cell type-specific markers for astrocytes (GFAP), microglia (Iba1), and neurons (NeuN) by western blot and IHC. The results showed that YY1 protein levels in the frontal cortex, midbrain, and cerebellum were significantly decreased in YY1 cKO mice, compared to WT mice. In addition, while imaging data in WT mice showed that YY1 is expressed in neurons, astrocytes, and microglia in all brain regions tested (Fig. 1D-F), astrocytic YY1 deletion did not show YY1 immunoreactivity in GFAP+ astrocytes, but YY1 was present in neurons and microglia in all three brain regions tested (Fig. 1D-F).
Impairment of development along with behavioral and cognitive deficits in YY1 cKO mice
YY1 is involved in neurodevelopment (Donohoe et al., 1999), but the specific role of astrocytic YY1 in development had not previously been studied. Thus, astrocytic YY1 cKO mice with age-matched control group (3 months old) for both sexes were used to investigate the effect of astrocytic YY1 deletion on growth and development. The astrocytic YY1 cKO mice generated by GFAP-specific YY1 deletion appeared to be normal at birth, but showed slower growth compared to WT mice (Fig. 2A), exhibiting lower body weight compared to that of WT at 12 weeks of age (Fig. 2B). Astrocytic YY1 cKO mice showed increased mortality from 3 months of age (data not shown), indicating that astrocytic YY1 is critical for normal postnatal development and survival.
Figure 2. GFAP promoter-driven YY1 deletion caused phenotypic abnormalities and impairment of locomotor activity, motor coordination, and recognition memory.
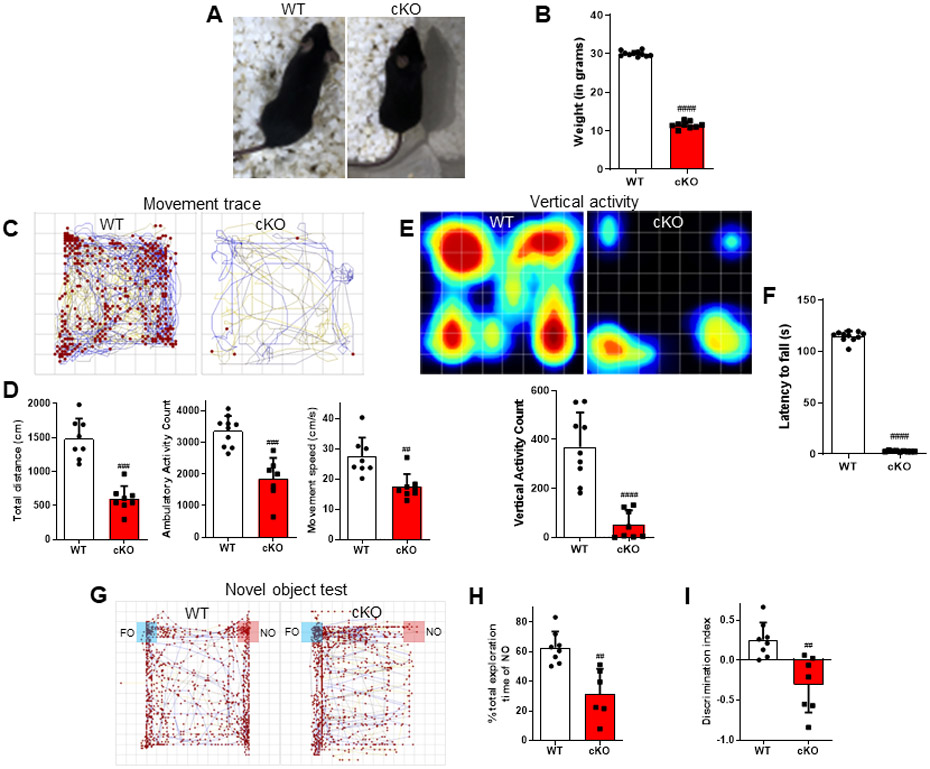
(A) Image showing differences in size between WT and YY1 cKO mice. (B) The body weight of mice in each group was measured prior to the experiments. (C-H) Locomotor activity, motor coordination, and recognition memory were analyzed. (C) The traces show a representative mouse movement. (D) Total distance traveled, ambulatory (walking) activity count, movement speed, (E) vertical activity, and (F) latency to fall were recorded and compared between groups. Total exploration of the novel object (G) and ability to discriminate between NO and FO were quantified and compared between groups. ##, p<0.01 ###, p<0.001 ####, p<0.0001 compared with the control (WT) (Student t-test; n = 6-7). Data are expressed as mean ± SD.
We also assessed the role of astrocytic YY1 in behavioral phenotypes in mice using the open-field test for locomotor activity, rotarod for motor coordination, and novel object recognition for memory. Results showed that astrocytic YY1 deletion decreased locomotor activity in YY1 cKO mice (Movie 1), displaying a shorter distance traveled compared to WT mice (Fig. 2C). Astrocytic YY1- deleted mice also exhibited significantly decreased ambulatory (walking) activity and movement speed (Fig. 2D) as well as reducing vertical activity compared to WT mice (Fig. 2E). In addition, YY1 cKO mice showed a significantly impeded motor coordination and ability to stay on the rod during the rotarod test (Fig. 2F).
The effect of astrocytic YY1 deletion on recognition memory function was examined using the novel object recognition test, which measures mice’s ability to discriminate novel object from the familiar object. Results showed that astrocytic YY1 deletion impaired memory function as shown by the reduced total exploring time on the novel object (Fig. 2G-H) and decreased ability of the mice to distinguish between familiar and novel objects (Fig. 2I).
Effect of astrocytic YY1 deletion in overall gene expression in the brain
Since astrocytic YY1 deletion induced significantly impaired phenotypes such as reduced locomotor activities and cognition function, we have further analyzed three brain regions that significantly affect movement and cognitive function, namely cortex, midbrain, and cerebellum. Studies have shown that YY1 controls cortex development by regulating metabolic pathways, protein translation, and apoptosis of neural progenitor cells at early stages of brain development (Zurkirchen et al., 2019). Studies have also shown that YY1 deletion in mid-hindbrain region produced abnormalities in brain development particularly in midbrain and cerebellum regions by disrupting cell cycle, Wnt signaling, and p53-induced cell death of neuroepithelial cells (Dong & Kwan, 2020). Recent studies have also shown that GFAP-specific YY1 deletion during end-stage embryonic development caused phenotype abnormalities and severe motor deficits, suggesting linkage to impaired cerebellar astrocyte maturation (Mockenhaupt et al., 2021).
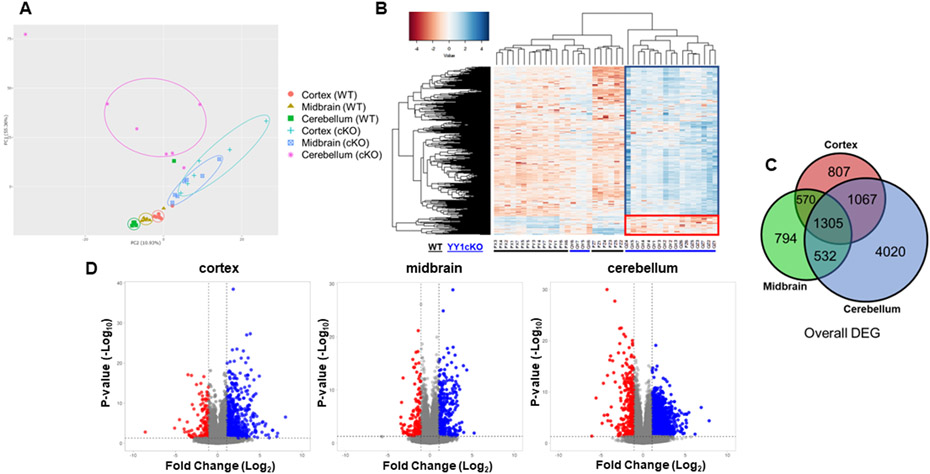
To understand the impact of astrocytic YY1 deletion in gene expression profiles of the brain, we performed RNA-sequencing analysis of frontal cortex, midbrain, and cerebellum from WT and YY1 cKO mice, relevant to altered phenotypes. PCA plots of each RNA-seq datasets from the cortex, midbrain, and cerebellum of WT and cKO mice showed that the first principal component (PC1) of 55.36% accounted for the largest separation of YY1 cKO from WT group (genotype) while the second principal component (PC2) of 10.93% accounted for the separation of samples by brain region (Fig. 3A). Hierarchical clustering of normalized gene expression data confirmed the distinct gene expression profile in the same regions (Fig. 3B). These results demonstrate that astrocytic YY1 deletion significantly altered overall gene expression in a brain-region specific manner. The overall transcriptomic analysis showed that astrocytic YY1 deletion induced higher numbers of upregulated genes than those of downregulated genes (Fig. 3B). The commonality and region-specificity of gene expression profile in the three regions were assessed by the overlap of DEG between the regions in YY1 cKO using a Venn diagram. Venn diagram showed that DEG was common to all or at least 2 brain regions as well as DEG of non-overlapping genes (Fig. 3C). The results revealed that 1305 DEG were common to the cortex, midbrain, and cerebellum (Fig. 3C).
Figure 3. RNA-seq analysis identified differentially expressed genes in the cortex, midbrain, and cerebellum of WT and YY1 cKO mice.
(A) Principal component analysis (PCA) plot of the individual RNA-seq data from cortex, midbrain, and cerebellum of WT and cKO mice. PCA plot showed brain region-specific and astrocytic YY1 deletion-specific differences in gene expression. (B) Heatmap of overall gene expression data between WT and cKO murine cortex, midbrain, and cerebellum. Low (red), no change (white), and high (blue) expression levels are depicted. (C) Venn diagram showed the number of differentially expressed genes (DEG) that are common (shared) in all brain regions and uniquely expressed in the cortex, midbrain, or cerebellum. (D) Volcano plots comparing all genes in the cortex, midbrain, and cerebellum identifies significantly up- or down-regulated in cKO mice. The significance threshold for fold change (log2, twofold change) and p-value (-log10) are shown as black lines on the x- and y-axis, respectively (using DESeq2 statistics, N = 6-7 mice per group).
Next, we analyzed expression levels of genes that were significantly altered by astrocytic YY1 deletion with a log2 fold-change threshold of ≥0.1 and an adjusted p-value <0.05 in the RNA-seq data set using DESeq2 statistics. The volcano plot demonstrated that higher number of genes were upregulated in the cerebellum (3728) followed by cortex (1959) and then the midbrain (1675) of YY1 cKO mice (Fig. 3D). Moreover, higher numbers of down-regulated genes were found in the cerebellum (3196), followed by the cortex (1790), and then the midbrain (1526) of YY1 cKO mice (Fig. 3D). Cerebellum had the highest number of DEG (6924) altered by astrocytic YY1 deletion, followed by cortex (3749) and then midbrain (3201) (Fig. 3D). To gain insight into the biological processes and molecular mechanisms altered by astrocytic YY1 deletion, we performed ontology enrichment of genes that were differentially expressed between WT and YY1 cKO in different brain regions. Regardless of the brain region, the most enriched categories that were upregulated in YY1 cKO appeared to be associated with immune response including “positive regulation of cytokine production” and “negative regulation of immune system process” (Suppl. Table 1). On the other hand, those that are downregulated in YY1 cKO are related to “CNS development,” “neurogenesis,” “apoptotic process,” “regulation of anatomical structure morphogenesis,” and “positive regulation of cellular biosynthetic process” (Suppl. Table 1). The enrichment of these sets of genes demonstrates that astrocytic YY1 deficiency exerted a significant impact on brain development, oxidative stress, cell death, and inflammation.
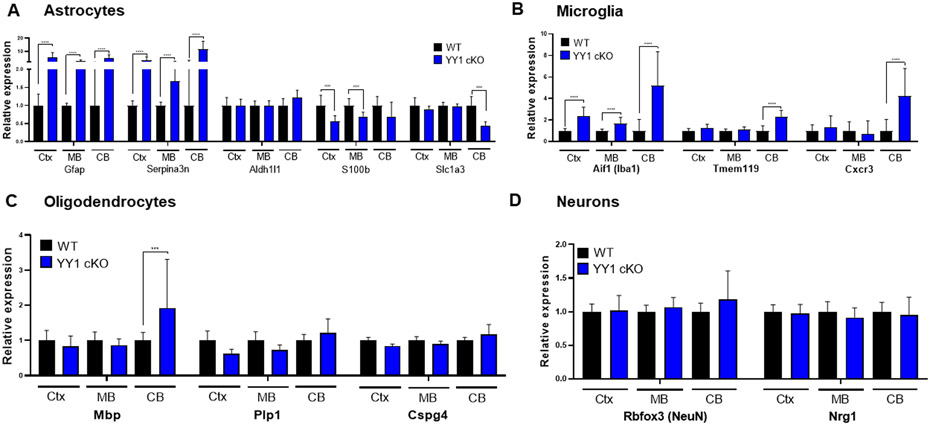
Since astrocytic YY1 deletion significantly impacted brain function, we tested if astrocytic YY1 deletion altered gene expressions in a neural cell type-dependent manner by examining expression levels of cell type-specific genes of astrocytes, microglia, oligodendrocytes, and neurons (Cahoy et al., 2008; Darmanis et al., 2015). Results showed that astrocytic YY1 deletion dramatically increased astrocytic genes Gfap and Serpina3n, but decreased S100b (S100β) in cortex and cerebellum, and reduced Slc1a3 (GLAST) in cerebellum (Fig. 4A). Moreover, astrocytic YY1 deletion greatly increased microglial specific markers Aif1 (Iba) in all regions, and cerebellum alone showed higher levels of Tmem119 and Cxcr3 in YY1 cKO mice, compared to WT control (Fig. 4B). We also determined if astrocytic YY1 deletion affected gene expression of oligodendrocyte and neuronal genes. Results showed that astrocytic YY1 deletion increased expression of the oligodendrocyte marker Mbp only in the cerebellum (Fig. 4C). Astrocytic YY1 did not impact neuronal markers Rbfox3 (NeuN) and Nrg1 (Fig. 4D). These results indicate that astrocytic YY1 deletion significantly activated not only astrocytes but also microglia as indicative of dramatic increases of Gfap and Iba1, respectively.
Figure 4. Astrocytic YY1 deletion altered gene expression of neural cell markers in the cortex, midbrain, and cerebellum.
(A-D) Bar graphs of common cell type-specific markers of astrocytes (A), microglia (B), oligodendrocytes (C), and neurons (D) in the cortex, midbrain, and cerebellum. ***, p<0.001; ****, p<0.0001; ####, p<0.0001 compared with the control (WT) (Student t-test, n = 6-7). Data are expressed as mean ± SD.
Astrocytic YY1 deletion impairs CNS development and neurogenesis
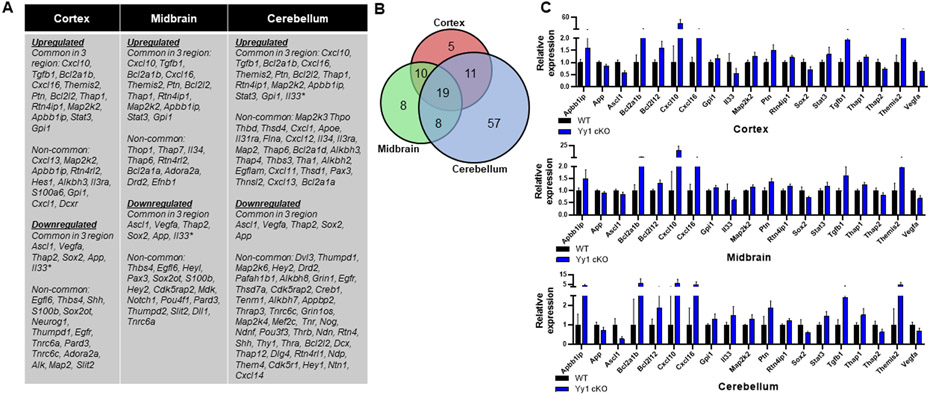
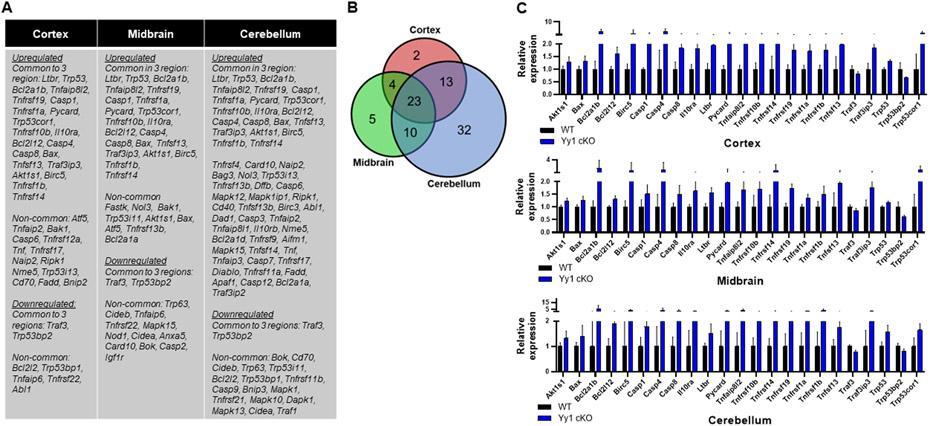
Since astrocytic YY1 deletion slowed growth and development as well as impairing movement and cognitive function, we sought to determine if genes associated with CNS development and neurogenesis were altered by astrocytic YY1 deletion in the three brain regions examined. Results revealed that higher numbers of genes associated with neurogenesis were altered in astrocytic YY1-deleted cerebellum compared to those of cortex and midbrain (Fig. 5A). Nineteen genes related to neurogenesis (Kumar et al., 2020) were commonly altered in all three regions of YY1 cKO mice (Fig. 5B). Astrocytic YY1 deletion significantly increased Apbb1ip, Cxcl10, Cxcl16, Stat3, and Tgfb1, but decreased App, Ascl1, Il33, Sox2, Thap2, and Vegfa in all three regions (Fig. 5C). The altered expression levels of neurogenesis-related genes by astrocytic YY1 deletion were highest in cerebellum (57), followed by midbrain (8), and then cortex (5).
Figure 5. Astrocytic YY1 deletion altered gene expression associated with neurogenesis.
(A) List of up- and down-regulated genes associated with neurogenesis that are common (shared) and non-common to the cortex, midbrain, and cerebellum. (B) Venn diagram showed the number of neurogenesis-related DEG that are common (shared) in all brain regions and uniquely expressed in the cortex, midbrain, or cerebellum. (C) Bar graphs of neurogenesis-related DEG common to all brain regions. (p<0.05 using Student t-test, n = 6-7). Data are expressed as mean ± SD.
Astrocytic YY1 deletion induces apoptosis
Since astrocytic YY1 deletion impaired neurogenesis and CNS development, we tested if apoptosis and synaptic pruning process-related genes (Jiang, Chen, & Zheng, 2021) were also altered in YY1 cKO mice. Results showed that astrocytic YY1 deletion altered significantly higher number of genes associated with apoptosis in cerebellum (78) (Fig. 6B), compared to cortex (42) and midbrain (42) (Fig. 6A). Twenty-three apoptosis-related genes were commonly altered in all three regions from astrocytic YY1 deficient genotype (Fig. 6C). Given that YY1 has been shown to directly regulate the p53 apoptotic pathway (Chen, Foreman, Sant'Angelo, & Krangel, 2016; Gronroos, Terentiev, Punga, & Ericsson, 2004), we tested whether the p53 pathway and apoptosis-associated genes were altered in the astrocytic YY1-deleted cortex, midbrain, and cerebellum. The results showed that astrocytic YY1 deletion increased mRNA levels of p53 and proapoptotic genes (Fig. 6C), indicating that astrocytic YY1 is critical for maintaining cell survival by regulating p53 levels in all brain regions. Further analysis revealed increased expression of proapoptotic genes including Bax, Casp4, and Casp8 in YY1 cKO mice, and decreased that of antiapoptotic genes such as Bcl2l12.
Figure 6. Astrocytic YY1 deletion altered gene expression associated with apoptosis in the cortex, midbrain, and cerebellum.
(A) List of up- and down-regulated genes associated with apoptosis that are common (shared) and non-common to the cortex, midbrain, and cerebellum. (B) Venn diagram showed the number of apoptosis-related DEG that are common (shared) in all brain regions and uniquely expressed in the cortex, midbrain, or cerebellum. (C) Bar graphs of apoptosis-related DEG common to all brain regions. (p<0.05 using Student t-test, n = 6-7). Data are expressed as mean ± SD.
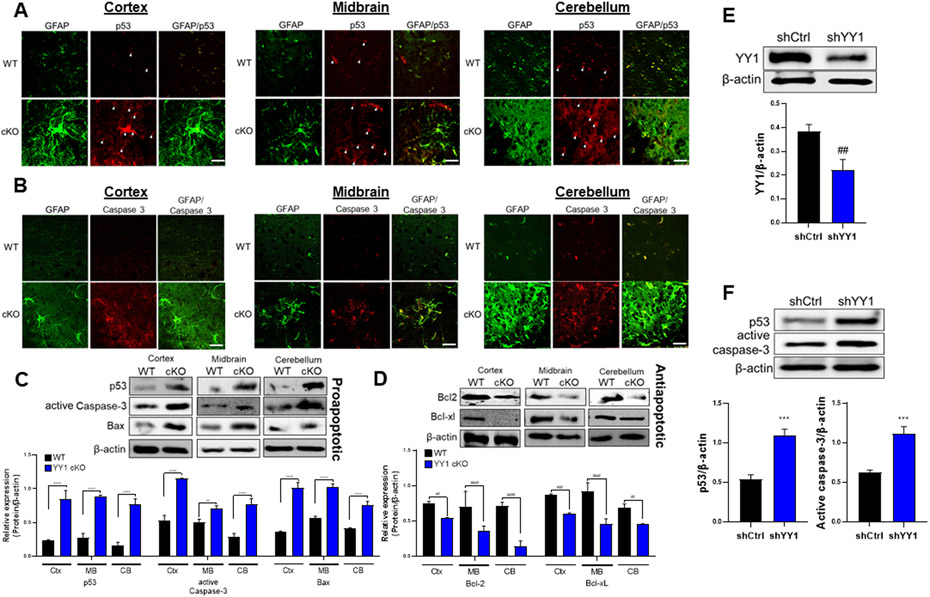
Since mRNA levels of apoptosis-related genes were significantly altered, we assessed if protein levels of apoptosis-related genes were also altered, using several selected proteins such as antiapoptotic proteins, Bcl-2 and Bcl-xL and proapoptotic proteins, p53, caspase-3, and Bax. Results showed that astrocytic YY1 deletion significantly decreased protein levels of Bcl-2 and Bcl-xL in cortex, midbrain, and cerebellum, while increasing protein levels of proapoptotic p53, active (or cleaved) caspase-3, and Bax in each region (Fig. 7A-D). We further tested if astrocytic YY1 deletion induced apoptosis via the p53-caspase-associated apoptotic pathway. Immunohistochemistry results showed that p53 and caspase-3 were expressed at lower levels in all neural cell types in the WT control group, while astrocytic YY1 deletion significantly increased fluorescence intensity levels of p53 (Fig. 7A) and caspase-3 (Fig. 7B) in astrocytes (GFAP+ cells) as well as non-astrocytes (GFAP− cells) in all regions examined. Immunoblotting results showed that protein levels of proapoptotic Bax, active caspase-3, and p53 were increased (Fig. 7C), whereas those of antiapoptotic Bcl2 and Bcl-xL were decreased in astrocytic YY1-deleted cortex, midbrain, and cerebellum (Fig. 7D). These results indicate that astrocytic YY1 has a critical role in preventing apoptosis in cortex, midbrain, and cerebellum, at least in part, via the p53 apoptotic pathway.
Figure 7. Deletion of astrocytic YY1 regulates brain apoptosis.
(A-B) Coronal sections of brain tissues were immunostained with GFAP and p53 or caspase-3. The expression of GFAP and p53/caspase-3 were shown as green and red fluorescence signals, respectively, in the cortex, midbrain, and cerebellum of the WT and YY1 cKO mouse brain (×40 magnification with a confocal microscope, scale represent 50 μm). (C-D) Cortex, midbrain, and cerebellum regions were processed for proapoptotic proteins (i.e., p53, active caspase-3, and Bax) (C) and antiapoptotic proteins (i.e., Bcl-2, Bcl-xL) (D) by western blotting. (E-F) Human H4 astrocytes were transduced with shRNA lentiviral particles for YY1 knockdown and control, followed by detection and quantification of the proteins YY1 (E), p53 and active caspase-3 (F). β-actin was used as a loading control for protein. **, p<0.01; ***, p<0.001; ****, p<0.0001; ##, p<0.01; ###, p<0.001; ####, p<0.0001 compared with the control (WT) (Student t-test, n = 3). Data are expressed as mean ± SD.
To determine if the role of astrocytic YY1 in p53-caspase-3 pathway from in vivo study can be recapitulated in vitro condition, we did knockdown YY1 in human H4 astrocytes by YY1 shRNA lentiviral particles transduction and assessed protein levels of p53 and caspase-3. YY1 shRNA significantly reduced YY1 protein levels in H4 astrocytes (Fig. 7E), confirming YY1 knockdown. Moreover, YY1 knockdown increased protein levels of p53 and active caspase-3 in astrocytes (Fig. 7F), indicating that YY1 is required to maintain normal levels of p53 and caspase-3 in astrocytes.
Astrocytic YY1 deletion induces oxidative stress
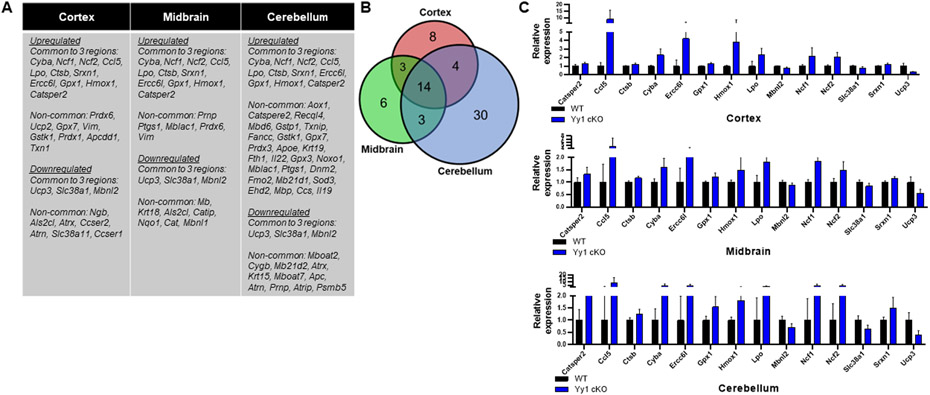
Since mitochondria play an important role in apoptosis and are the major sources of reactive oxygen species (ROS) which contribute to oxidative stress, we tested whether oxidative stress-related genes were altered by astrocytic YY1 deletion in all three brain regions. Results showed that cerebellum showed a higher number of altered oxidative stress-related genes compared to those in cortex and midbrain (Fig. 8A-C). Fourteen genes associated with oxidative stress were commonly dysregulated in all brain regions of astrocytic YY1 cKO mice (Fig. 8A-C). Astrocytic YY1 deletion significantly increased Ncf1, Ncf2, and chemokine ligand Ccl5 (RANTES), indicating increased ROS production and could contribute to oxidative damage in all brain regions examined. Several antioxidant genes such as heme oxygenase 1 (Hmox1) and glutathione peroxidase 1 (Gpx1) were increased in YY1 cKO mice (Fig. 8A,C). In addition, astrocytic YY1 deletion upregulated the lysosomal cysteine protease cathepsin B (Ctsb) (Fig. 8A,C), which is also involved in oxidative stress, as well as autophagy and lysosomal protein degradation.
Figure 8. Astrocytic YY1 deletion altered gene expression associated with oxidative stress in the cortex, midbrain, and cerebellum.
(A) List of up- and down-regulated genes associated with oxidative stress that are common (shared) and non-common to the cortex, midbrain, and cerebellum. (B) Venn diagram showed the number of oxidative stress-related DEG that are common (shared) in all brain regions and uniquely expressed in the cortex, midbrain, or cerebellum. (C) Bar graphs of oxidative stress-related DEG common to all brain regions. (p<0.05 using Student t-test, n = 6-7). Data are expressed as mean ± SD.
Astrocytic YY1 deletion induces inflammation
Studies have shown that YY1 directly regulates transcription of inflammatory genes (X. C. Zhang et al., 2018) or indirectly regulates inflammatory genes by activating signaling proteins (Xu et al., 2016; Zan et al., 2017). The present study also reveals that astrocytic YY1 deletion activated astrocytes to induce an inflammatory state, as shown by increased levels of GFAP and the enlargement of astrocytic cell body and processes (Fig. 10). Thus, we determined if astrocytic YY1 is involved in regulating specific inflammatory signaling pathways in cortex, midbrain, and cerebellum. Expression levels of several genes associated with proinflammatory cytokine production and signaling pathways including TNF-α and interleukins were increased by astrocytic YY1 deletion (Fig. 9A-C). Imaging data revealed that astrocytic YY1 deletion increased fluorescence intensities of TNF-α, GFAP as well as Iba1 in all brain regions (Fig. 10A). Astrocytic YY1 deletion also increased protein levels of TNF-α in cortex, midbrain, and the cerebellum (Fig. 10C). TNF-α was co-localized with GFAP, indicating that astrocytic YY1 is important for preventing production of proinflammatory TNF-α in these brain regions (Fig. 10A).
Figure 10. Deletion of astrocytic YY1 regulates proinflammatory cytokine TNF-α and chemokine CXCL10 in the brain.
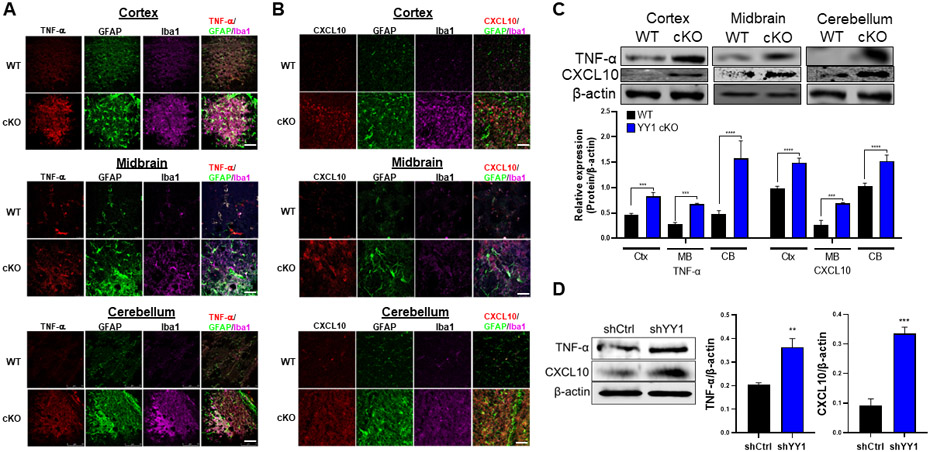
(A-B) Coronal sections of brain tissues were immunostained with GFAP, Iba1, TNF-α, and CXCL10. The expression of GFAP, Iba1, and TNF-α/CXCL10 were shown as green, magenta, and red fluorescence signals, respectively, in the cortex, midbrain, and cerebellum of the WT and YY1 cKO mouse brain (×40 magnification with a confocal microscope, scale represent 50 μm). (C-D) Cortex, midbrain, and cerebellum regions were processed for TNF-α and CXCL10 by western blotting. (E) Human H4 astrocytes were transduced with shRNA lentiviral particles for YY1 knockdown and control, followed by detection and quantification of the proteins TNF-α and CXCL10. β-actin was used as a loading control for protein. ***, p<0.001; ****, p<0.0001 compared with the control (WT) (Student t-test, n = 3). Data are expressed as mean ± SD.
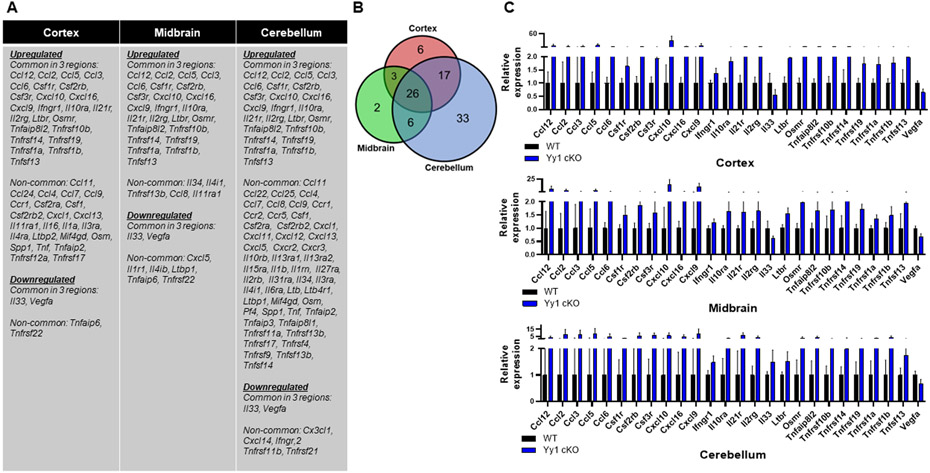
Figure 9. Astrocytic YY1 deletion altered gene expression associated with inflammation in the cortex, midbrain, and cerebellum.
(A) List of up- and down-regulated genes associated with inflammation that are common (shared) and non-common to the cortex, midbrain, and cerebellum. (B) Venn diagram showed the number of inflammation-related DEG that are common (shared) in all brain regions and uniquely expressed in the cortex, midbrain, or cerebellum. (C) Bar graphs of inflammation-related DEG common to all brain regions. (p<0.05 using Student t-test, n = 6-7). Data are expressed as mean ± SD.
Furthermore, astrocytic YY1 appears to be critically involved in preventing expression of variety of chemokines in cortex, midbrain, and cerebellum since astrocytic YY1 deletion greatly increased expression levels of Ccl12, Ccl2, Ccl3, Ccl4, Ccl5, Ccl6, Ccl9, Cxcl10, Cxcl11, Cxcl16, and Cxcl9 in all three regions (Fig. 9A,C). Some other chemokines were increased in specific brain regions (Fig. 9A). The cellular sources of these chemokines were assessed by immunohistochemical staining for CXCL10 as a representative chemokine in three brain regions as well as protein quantification by western blot (Fig. 10B,C). Results showed that astrocytic YY1 deletion increased expression of CXCL10 proteins in astrocytes, co-localizing with GFAP in cortex, midbrain, and cerebellum (Fig. 10B). To determine if modulatory effects of astrocytic YY1 deletion on inflammation in the mouse brain can occur in astrocyte cultures, we assessed the effects of astrocytic YY1 knockdown on the levels of TNF-α and CXCL10 in H4 astrocytes. The results showed that YY1 knockdown increased protein levels of TNF-α and CXCL10 in astrocytes, indicating that astrocytes are responsible for producing these proteins (Fig. 10D). We also evaluated if microglia are activated and expressed chemokines by astrocytic YY1 deletion since microglia are main cell types in the brain involved in inflammation. The results revealed that astrocytic YY1 deletion activated microglia by increasing Iba1 expression and co-localized with TNF-α and CXCL10 in cortex, midbrain, and cerebellum compared to the WT control (Fig. 10A,B).
Discussion
Our novel findings demonstrate for the first time that astrocytic YY1 plays a critical role in mouse brain development, function, and sustained health by preventing pathological conditions, such as inflammation, oxidative stress and apoptosis. The effects of astrocytic YY1 deletion were brain region-specific, with the cerebellum being the most affected, followed by the cortex and midbrain, and these effects correlated with YY1 expression levels. At the cellular level, astrocytic YY1 deletion activated not only astrocytes but also microglia by enhancing expression of GFAP and Iba1, as activation indicators of each cell type, respectively. Accordingly, many proinflammatory cytokines and chemokines were also largely increased in astrocytic YY1-deleted brain tissues. At the molecular level, YY1 target genes such as p53/caspase for apoptotic signaling were also involved in neurodegeneration and brain dysfunction in YY1 cKO mice. These findings demonstrate that astrocytic YY1 is essential for the maintenance of neurodevelopment, CNS function and health.
Studies have shown that global YY1 deficiency impaired growth and viability during embryonic development, leading to embryonic lethality (Affar el et al., 2006; Dong & Kwan, 2020). YY1 deletion in oligodendrocytes (He et al., 2007) and neural progenitor cells (Zurkirchen et al., 2019) also impaired myelination and brain development. As supported by the PCA plot and hierarchical clustering, YY1 deletion in astrocytes also caused abnormal gene expression profiles and is crucial for mouse growth as its deletion led to a >60% reduction in body weight (Fig. 2). In particular, astrocytic YY1 is essential for normal postnatal development during which GFAP expression levels are elevated (Mamber et al., 2012). Although further studies are necessary to dissect the role of astrocytic YY1 during the postnatal stages of mouse growth, our findings highlight that astrocytic YY1 is indispensable for normal growth, and brain function including locomotor activity and cognition. Interestingly, tyrosine hydroxylase (TH) mRNA levels were not changed, while those of dopamine transporter (DAT, Slc6a3) were significantly decreased in the midbrain, suggesting that dopaminergic neuronal function was compromised, potentially contributing to the impaired locomotor activity. One caveat of our mouse model for astrocytic YY1 deletion under GFAP control could also directly modulate neuronal functions since low levels of GFAP are also expressed in radial glial cells, which are key progenitor cells during late stages of neurogenesis (for review, see (Arellano, Morozov, Micali, & Rakic, 2021)).
Astrocytic YY1 deletion significantly altered numerous genes involved in neurogenesis, apoptosis, oxidative stress, and inflammation, suggesting that astrocytic YY1 plays a critical role in preventing cellular toxicity. YY1 regulates the gene expression related to a plethora of biological activities by either directly binding to the target genes or indirectly via YY1-dependent proteins and signaling pathways (for review, see (Verheul et al., 2020)). Our findings that YY1 expression levels were reduced most significantly in cerebellum, compared to other regions tested in YY1 cKO brain, parallel with changes in the RNA-Seq dataset suggest that astrocytic YY1 function is brain regions-specific, but mechanisms involved is unclear at present. Epigenetic regulation may play a role in the region-specific effects as astrocytic YY1 deletion modulated several histone deacetylases (HDAC) specific to each brain region with more pronounced reduction of HDAC subtypes in the cerebellum (Suppl. Table 2).
Astrocytic YY1 deletion modulated 19 neurogenesis genes common in the three regions. Particularly, decreased expression of Sox2, App, Ascl1, Thap2, and Vegfa (Fig. 5C) might be responsible for the impairment of neurogenesis. Sox2 maintains neural progenitor cells during development (Graham, Khudyakov, Ellis, & Pevny, 2003) and global YY1 deletion causes abnormal cell cycle progression along with reduction of neural progenitor cells population (Dong & Kwan, 2020; Zurkirchen et al., 2019), suggesting the role of astrocytic YY1 in neural development. Moreover, astrocytic YY1 deletion dramatically increased expression of the complement protein C1q and related genes in all three brain regions (Suppl. Table 3). C1q protein prunes the neuronal synapse during brain development in order to inhibit the formation of excessive dendrites and unnecessary synapses (Rupprecht, Rupprecht, & Rammes, 2021), suggesting that excessive pruning might have occurred in the astrocytic YY1-deleted brain.
Increased expression of genes associated with apoptotic process might have also contributed to abnormal brain function in astrocytic YY1-deleted mice since there were increased mRNA levels of 23 proapoptotic genes in all three regions (Fig. 6), including Bax, Casp8, and Trp53 (p53) with the highest increase in the cerebellum. Moreover, protein levels of p53, active caspase-3 and Bax were also significantly increased in all three brain regions by western blot, corroborating with higher immunofluorescence signals of p53 and caspase-3 in astrocytes (Fig. 7) in both in vivo and in vitro settings, supporting the role of YY1 in p53 and caspase-3 signaling. YY1 represses p53 protein by either directly binding to the P53 gene (Chen et al., 2016) or by post-transcriptional facilitation of p53’s interaction with Mdm2 (an E3 ubiquitin ligase) for p53 degradation (Gronroos et al., 2004), suggesting that astrocytic YY1 is critical in preventing the apoptotic pathway of p53/caspase-3. In addition, mRNA and protein levels of anti-apoptotic genes Bcl-2 and Bcl-xL were decreased in all three brain regions of astrocytic YY1 cKO.
Astrocytic YY1 deletion also altered the expression of genes associated with antioxidant defense and ROS production in the mouse brain with the highest alteration in the cerebellum, underscoring the importance of cerebellar astrocytic YY1 in protecting against oxidative stress. Among several oxidative stress-associated genes upregulated in all brain regions, regulatory subunits of NAD(P)H oxidase, namely Cyba, Ncf1 and Ncf2 are involved in ROS production in the mitochondria (Dvoriantchikova, Grant, Santos, Hernandez, & Ivanov, 2012), suggesting that the astrocytic YY1 deletion-induced oxidative stress is associated with mitochondrial dysfunction. This is supported by previous studies showing that astrocytic YY1 deletion induced mitochondrial dysfunction and oxidative stress in the diabetic (Song et al., 2020) and cerebral ischemic mouse model (W. Liu, Guo, & Zhao, 2018). Antioxidant genes such as Gpx1 and Hmox1 were also upregulated in astrocytic YY1 cKO mouse brain, suggesting that compensatory response to oxidative stress has also occurred in astrocytic YY1-deleted brain tissues.
Astrocytic YY1 appears critical in the prevention of inflammatory signal activation in the brain, as its deletion induced a dramatic increase of GFAP expression in all three regions. Although GFAP upregulation could be associated with inflammatory activation or protection of neurons (for review, see (Pekny, Wilhelmsson, & Pekna, 2014)), many inflammatory genes were also upregulated along with GFAP, suggesting that GFAP activation is associated with activation of inflammation in astrocytes (Colombo & Farina, 2016; Croitoru-Lamoury et al., 2003). Intriguingly, astrocytic YY1 deletion activated microglia and upregulated numerous proinflammatory cytokines and chemokines that are likely produced by microglia, as Aif1 (Iba1) mRNA levels and Iba1-immunostained cells were sharply increased in YY1 cKO mice (Fig. 4 and 10). Other microglial activation markers such as Cx3cr1, Tlr2, Ctss, and P2rx4 (Suppl. Table 4), and several toll-like receptors (TLR) (Suppl. Table 5) which are preferentially expressed in microglia (Trudler, Farfara, & Frenkel, 2010) were also significantly increased, suggesting that astrocytic YY1 deletion led to microglial activation, possibly via substances released from YY1-deleted astrocytes by astrocyte-microglia crosstalk (Linnerbauer, Wheeler, & Quintana, 2020).
Regardless of the neural cell type, dramatic increases of inflammatory mediators in the astrocytic YY1-deleted mouse brain could lead to neuronal insults as proinflammatory cytokines and chemokines contribute to a wide array of neurodegenerative disorders (Ramesh, MacLean, & Philipp, 2013), (Dhaiban, Al-Ani, Elemam, & Maghazachi, 2020; J. Q. Liu, Chu, Zhou, Zhang, & Chen, 2019; Lourenco et al., 2021). Increased expression levels of multiple chemokines including Cxcl10 may facilitate intercellular communication between glia and neurons (Banisadr, Rostene, Kitabgi, & Parsadaniantz, 2005). CXCL10 was highly upregulated in both astrocytes and microglia of all brain regions in astrocytic YY1 cKO mice as well as YY1-knockdown astrocytic cultures (Fig. 10), suggesting that astrocytes are responsible for the inflammatory phenotype in the brain (Mockenhaupt et al., 2021), at least in part, by YY1 deficiency in astrocytes. CXCL10 upregulation is linked to neurodegenerative diseases (Xia, Bacskai, Knowles, Qin, & Hyman, 2000) and induces neuronal injury by dysregulating mitochondrial function (Sui et al., 2006) and promoting a proinflammatory environment (Y. Jiang et al., 2021). TNF-α was also increased in microglia in AD model (Shippy, Watters, & Ulland, 2022), supporting the preventive role of astrocytic YY1 in neuroinflammation.
Although molecular mechanisms of astrocytic YY1 on chemokine regulation are unclear, YY1 may directly target several chemokine genes (Fig. 9), and interact with other transcription factors, such as NF-κB, signal transducer and activator of transcription (STAT) 1-3, interferon regulatory factor (IRF) (Xu et al., 2016; Zan et al., 2017) to modulate its downstream inflammatory target genes (Suppl. Tables 6-8). A previous study has shown that NF-κB directly binds to the promoter region of CXCL10 to increase CXCL10 in neurons and astrocytes (Pugazhenthi, Zhang, Bouchard, & Mahaffey, 2013), suggesting that NF-κB could be involved in CXCL10 expression.
Astrocytic YY1 deletion significantly altered pathways and biological processes in the brain, but further studies are required to get more insight into the molecular and cellular mechanisms by which astrocytic YY1 regulates other neural cell types such as microglia and neurons. Moreover, as our study used adult mouse brain samples (3 months of age), the transcriptional changes in astrocytic YY1 cKO during embryonic or postnatal development stages have yet to be determined.
Taken together, our novel findings demonstrate that astrocytic YY1 is essential for CNS development, movement and cognitive function. Astrocytic YY1 deletion induced a significant alteration in the expression of numerous genes in all brain regions tested, suggesting an indispensable role of astrocytic YY1 in brain function, including modulation of neurogenesis, apoptosis, oxidative stress, and inflammation. Our findings present potential mechanisms by which astrocytic YY1 modulate critical cellular and molecular functions, warranting further studies for understanding the role of astrocytic YY1 in the brain.
Supplementary Material
Movie 1. Mouse movement and behavior in WT (YY1 loxP mice) and GFAP-specific YY1 cKO mice. Mice (12-week-old) were placed in the cages and behavior recorded for 30 sec.
Main points.
Astrocytic YY1 deletion caused growth retardation and impairment of motor function and cognition in mice
Astrocytic YY1 is involved in regulation of development, apoptosis, oxidative stress, and inflammation in the mouse brain
Acknowledgments
This work was supported by National Institutes of Health Grants NIEHS R01 ES024756 (to EL), R01 ES031282 (to EL), and R01 ES10563 (to MA), NCI SC1 CA200519 (to DS) and NIMHD U54 MD007582. The content is solely the authors' responsibility and does not necessarily represent the official views of the National Institutes of Health. The data discussed in this article have been deposited in NCBI’s Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO series accession # GSE207014.
Footnotes
Conflict of interest statement
The authors declare no conflicts of interest with the contents of this article.
References
- Ackerman S (1992). In Discovering the Brain. Washington (DC). [PubMed] [Google Scholar]
- Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, … Shi Y (2006). Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol, 26(9), 3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arellano JI, Morozov YM, Micali N, & Rakic P (2021). Radial Glial Cells: New Views on Old Questions. Neurochem Res, 46(10), 2512–2524. doi: 10.1007/s11064-021-03296-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggiolini A, Varum S, Mateos JM, Bettosini D, John N, Bonalli M, … Sommer L (2015). Premigratory and migratory neural crest cells are multipotent in vivo. Cell Stem Cell, 16(3), 314–322. doi: 10.1016/j.stem.2015.02.017 [DOI] [PubMed] [Google Scholar]
- Banisadr G, Rostene W, Kitabgi P, & Parsadaniantz SM (2005). Chemokines and brain functions. Curr Drug Targets Inflamm Allergy, 4(3), 387–399. doi: 10.2174/1568010054022097 [DOI] [PubMed] [Google Scholar]
- Beagan JA, Duong MT, Titus KR, Zhou L, Cao Z, Ma J, … Phillips-Cremins JE (2017). YY1 and CTCF orchestrate a 3D chromatin looping switch during early neural lineage commitment. Genome Res, 27(7), 1139–1152. doi: 10.1101/gr.215160.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevins RA, & Besheer J (2006). Object recognition in rats and mice: a one-trial non-matching-to-sample learning task to study 'recognition memory'. Nat Protoc, 1(3), 1306–1311. doi: 10.1038/nprot.2006.205 [DOI] [PubMed] [Google Scholar]
- Cahoy JD, Emery B, Kaushal A, Foo LC, Zamanian JL, Christopherson KS, … Barres BA (2008). A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J Neurosci, 28(1), 264–278. doi: 10.1523/JNEUROSCI.4178-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Foreman DP, Sant'Angelo DB, & Krangel MS (2016). Yin Yang 1 Promotes Thymocyte Survival by Downregulating p53. J Immunol, 196(6), 2572–2582. doi: 10.4049/jimmunol.1501916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E, & Farina C (2016). Astrocytes: Key Regulators of Neuroinflammation. Trends Immunol, 37(9), 608–620. doi: 10.1016/j.it.2016.06.006 [DOI] [PubMed] [Google Scholar]
- Croitoru-Lamoury J, Guillemin GJ, Boussin FD, Mognetti B, Gigout LI, Cheret A, … Dormont D (2003). Expression of chemokines and their receptors in human and simian astrocytes: evidence for a central role of TNF alpha and IFN gamma in CXCR4 and CCR5 modulation. Glia, 41(4), 354–370. doi: 10.1002/glia.10181 [DOI] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, … Quake SR (2015). A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci U S A, 112(23), 7285–7290. doi: 10.1073/pnas.1507125112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhaiban S, Al-Ani M, Elemam NM, & Maghazachi AA (2020). Targeting Chemokines and Chemokine Receptors in Multiple Sclerosis and Experimental Autoimmune Encephalomyelitis. J Inflamm Res, 13, 619–633. doi: 10.2147/JIR.S270872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, & Kwan KM (2020). Yin Yang 1 is critical for mid-hindbrain neuroepithelium development and involved in cerebellar agenesis. Mol Brain, 13(1), 104. doi: 10.1186/s13041-020-00643-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, & Shi Y (1999). Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol, 19(10), 7237–7244. doi: 10.1128/MCB.19.10.7237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupin E, & Sommer L (2012). Neural crest progenitors and stem cells: from early development to adulthood. Dev Biol, 366(1), 83–95. doi: 10.1016/j.ydbio.2012.02.035 [DOI] [PubMed] [Google Scholar]
- Dvoriantchikova G, Grant J, Santos AR, Hernandez E, & Ivanov D (2012). Neuronal NAD(P)H oxidases contribute to ROS production and mediate RGC death after ischemia. Invest Ophthalmol Vis Sci, 53(6), 2823–2830. doi: 10.1167/iovs.12-9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer WT, & Murai K (2017). Resolving Astrocyte Heterogeneity in the CNS. Front Cell Neurosci, 11, 300. doi: 10.3389/fncel.2017.00300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvin KM, & Shi Y (1997). Multiple mechanisms of transcriptional repression by YY1. Mol Cell Biol, 17(7), 3723–3732. doi: 10.1128/MCB.17.7.3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia AD, Doan NB, Imura T, Bush TG, & Sofroniew MV (2004). GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat Neurosci, 7(11), 1233–1241. doi: 10.1038/nn1340 [DOI] [PubMed] [Google Scholar]
- Graham V, Khudyakov J, Ellis P, & Pevny L (2003). SOX2 functions to maintain neural progenitor identity. Neuron, 39(5), 749–765. doi: 10.1016/s0896-6273(03)00497-5 [DOI] [PubMed] [Google Scholar]
- Gronroos E, Terentiev AA, Punga T, & Ericsson J (2004). YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci U S A, 101(33), 12165–12170. doi: 10.1073/pnas.0402283101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, & Casaccia-Bonnefil P (2008). The Yin and Yang of YY1 in the nervous system. J Neurochem, 106(4), 1493–1502. doi: 10.1111/j.1471-4159.2008.05486.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, … Casaccia-Bonnefil P (2007). The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron, 55(2), 217–230. doi: 10.1016/j.neuron.2007.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Chen L, & Zheng S (2021). Global Reprogramming of Apoptosis-Related Genes during Brain Development. Cells, 10(11). doi: 10.3390/cells10112901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Huang F, Chai X, Yuan W, Ding H, & Wu X (2021). The role of IP-10 and its receptor CXCR3 in early pregnancy. Mol Immunol, 140, 59–69. doi: 10.1016/j.molimm.2021.09.013 [DOI] [PubMed] [Google Scholar]
- Johnson J Jr., Pajarillo E, Karki P, Kim J, Son DS, Aschner M, & Lee E (2018). Valproic acid attenuates manganese-induced reduction in expression of GLT-1 and GLAST with concomitant changes in murine dopaminergic neurotoxicity. Neurotoxicology, 67, 112–120. doi: 10.1016/j.neuro.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki P, Webb A, Smith K, Johnson J Jr., Lee K, Son DS, … Lee E (2014). Yin Yang 1 is a repressor of glutamate transporter EAAT2, and it mediates manganese-induced decrease of EAAT2 expression in astrocytes. Mol Cell Biol, 34(7), 1280–1289. doi: 10.1128/MCB.01176-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh BS, & Sofroniew MV (2015). Diversity of astrocyte functions and phenotypes in neural circuits. Nat Neurosci, 18(7), 942–952. doi: 10.1038/nn.4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Pareek V, Faiq MA, Kumar P, Kumari C, Singh HN, & Ghosh SK (2020). Transcriptomic analysis of the signature of neurogenesis in human hippocampus suggests restricted progenitor cell progression post-childhood. IBRO Rep, 9, 224–232. doi: 10.1016/j.ibror.2020.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KW (2011). Neuroproteomics. New York: Humana Press/Springer. [Google Scholar]
- Linnerbauer M, Wheeler MA, & Quintana FJ (2020). Astrocyte Crosstalk in CNS Inflammation. Neuron, 108(4), 608–622. doi: 10.1016/j.neuron.2020.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JQ, Chu SF, Zhou X, Zhang DY, & Chen NH (2019). Role of chemokines in Parkinson's disease. Brain Res Bull, 152, 11–18. doi: 10.1016/j.brainresbull.2019.05.020 [DOI] [PubMed] [Google Scholar]
- Liu W, Guo Q, & Zhao H (2018). Oxidative stress-elicited YY1 potentiates antioxidative response via enhancement of NRF2-driven transcriptional activity: A potential neuronal defensive mechanism against ischemia/reperfusion cerebral injury. Biomed Pharmacother, 108, 698–706. doi: 10.1016/j.biopha.2018.09.082 [DOI] [PubMed] [Google Scholar]
- Lourenco MV, Ribeiro FC, Santos LE, Beckman D, Melo HM, Sudo FK, … Ferreira ST (2021). Cerebrospinal Fluid Neurotransmitters, Cytokines, and Chemokines in Alzheimer's and Lewy Body Diseases. J Alzheimers Dis, 82(3), 1067–1074. doi: 10.3233/JAD-210147 [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15(12), 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamber C, Kamphuis W, Haring NL, Peprah N, Middeldorp J, & Hol EM (2012). GFAPdelta expression in glia of the developmental and adolescent mouse brain. PLoS One, 7(12), e52659. doi: 10.1371/journal.pone.0052659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhoff J, Muhie S, Chakraborty N, Naidu L, Sowe B, Hammamieh R, … Gautam A (2021). Microdissection of Mouse Brain into Functionally and Anatomically Different Regions. J Vis Exp(168). doi: 10.3791/61941 [DOI] [PubMed] [Google Scholar]
- Mockenhaupt K, Tyc KM, McQuiston A, Hariprashad A, Biswas DD, Gupta AS, … Kordula T (2021). Yin Yang 1 sets up the stage for cerebellar astrocyte maturation. bioRxiv, 2021.2005.2014.444129. doi: 10.1101/2021.05.14.444129 [DOI] [Google Scholar]
- Pajarillo E, Johnson J Jr., Rizor A, Nyarko-Danquah I, Adinew G, Bornhorst J, … Lee E (2020). Astrocyte-specific deletion of the transcription factor Yin Yang 1 in murine substantia nigra mitigates manganese-induced dopaminergic neurotoxicity. J Biol Chem, 295(46), 15662–15676. doi: 10.1074/jbc.RA120.015552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekny M, Wilhelmsson U, & Pekna M (2014). The dual role of astrocyte activation and reactive gliosis. Neurosci Lett, 565, 30–38. doi: 10.1016/j.neulet.2013.12.071 [DOI] [PubMed] [Google Scholar]
- Pugazhenthi S, Zhang Y, Bouchard R, & Mahaffey G (2013). Induction of an inflammatory loop by interleukin-1beta and tumor necrosis factor-alpha involves NF-kB and STAT-1 in differentiated human neuroprogenitor cells. PLoS One, 8(7), e69585. doi: 10.1371/journal.pone.0069585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh G, MacLean AG, & Philipp MT (2013). Cytokines and chemokines at the crossroads of neuroinflammation, neurodegeneration, and neuropathic pain. Mediators Inflamm, 2013, 480739. doi: 10.1155/2013/480739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas S, Vargas MA, Lopez-Bayghen E, & Ortega A (2007). Glutamate-dependent transcriptional regulation of GLAST/EAAT1: a role for YY1. J Neurochem, 101(4), 1134–1144. doi: 10.1111/j.1471-4159.2007.04517.x [DOI] [PubMed] [Google Scholar]
- Rupprecht C, Rupprecht R, & Rammes G (2021). C1q, a small molecule with high impact on brain development: putative role for aging processes and the occurrence of Alzheimer's disease. Eur Arch Psychiatry Clin Neurosci, 271(5), 809–812. doi: 10.1007/s00406-021-01273-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safrany G, & Perry RP (1993). Characterization of the mouse gene that encodes the delta/YY1/NF-E1/UCRBP transcription factor. Proc Natl Acad Sci U S A, 90(12), 5559–5563. doi: 10.1073/pnas.90.12.5559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Seto E, Chang LS, & Shenk T (1991). Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell, 67(2), 377–388. doi: 10.1016/0092-8674(91)90189-6 [DOI] [PubMed] [Google Scholar]
- Shippy DC, Watters JJ, & Ulland TK (2022). Transcriptional response of murine microglia in Alzheimer's disease and inflammation. BMC Genomics, 23(1), 183. doi: 10.1186/s12864-022-08417-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D, Yang Q, Jiang X, Shan A, Nan J, Lei Y, … Cao Y (2020). YY1 deficiency in beta-cells leads to mitochondrial dysfunction and diabetes in mice. Metabolism, 112, 154353. doi: 10.1016/j.metabol.2020.154353 [DOI] [PubMed] [Google Scholar]
- Sui Y, Stehno-Bittel L, Li S, Loganathan R, Dhillon NK, Pinson D, … Buch S (2006). CXCL10-induced cell death in neurons: role of calcium dysregulation. Eur J Neurosci, 23(4), 957–964. doi: 10.1111/j.1460-9568.2006.04631.x [DOI] [PubMed] [Google Scholar]
- Suzuki SO, & Goldman JE (2003). Multiple cell populations in the early postnatal subventricular zone take distinct migratory pathways: a dynamic study of glial and neuronal progenitor migration. J Neurosci, 23(10), 4240–4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J, Wu H, Lin Q, Wei W, Lu XH, Cantle JP, … Sun YE (2011). Deletion of astroglial Dicer causes non-cell-autonomous neuronal dysfunction and degeneration. J Neurosci, 31(22), 8306–8319. doi: 10.1523/JNEUROSCI.0567-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trudler D, Farfara D, & Frenkel D (2010). Toll-like receptors expression and signaling in glia cells in neuro-amyloidogenic diseases: towards future therapeutic application. Mediators Inflamm, 2010. doi: 10.1155/2010/497987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varum S, Baggiolini A, Zurkirchen L, Atak ZK, Cantu C, Marzorati E, … Sommer L (2019). Yin Yang 1 Orchestrates a Metabolic Program Required for Both Neural Crest Development and Melanoma Formation. Cell Stem Cell, 24(4), 637–653 e639. doi: 10.1016/j.stem.2019.03.011 [DOI] [PubMed] [Google Scholar]
- Verheul TCJ, van Hijfte L, Perenthaler E, & Barakat TS (2020). The Why of YY1: Mechanisms of Transcriptional Regulation by Yin Yang 1. Front Cell Dev Biol, 8, 592164. doi: 10.3389/fcell.2020.592164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, & Zhang B (2013). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res, 41(Web Server issue), W77–83. doi: 10.1093/nar/gkt439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters MR, Gupta AS, Mockenhaupt K, Brown LN, Biswas DD, & Kordula T (2019). RelB acts as a molecular switch driving chronic inflammation in glioblastoma multiforme. Oncogenesis, 8(6), 37. doi: 10.1038/s41389-019-0146-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub AS, Li CH, Zamudio AV, Sigova AA, Hannett NM, Day DS, … Young RA (2017). YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell, 171(7), 1573–1588 e1528. doi: 10.1016/j.cell.2017.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia MQ, Bacskai BJ, Knowles RB, Qin SX, & Hyman BT (2000). Expression of the chemokine receptor CXCR3 on neurons and the elevated expression of its ligand IP-10 in reactive astrocytes: in vitro ERK1/2 activation and role in Alzheimer's disease. J Neuroimmunol, 108(1-2), 227–235. doi: 10.1016/s0165-5728(00)00285-x [DOI] [PubMed] [Google Scholar]
- Xu HG, Liu L, Gao S, Jin R, Ren W, & Zhou GP (2016). Cloning and characterizing of the murine IRF-3 gene promoter region. Immunol Res, 64(4), 969–977. doi: 10.1007/s12026-015-8780-8 [DOI] [PubMed] [Google Scholar]
- Yao YL, Dupont BR, Ghosh S, Fang Y, Leach RJ, & Seto E (1998). Cloning, chromosomal localization and promoter analysis of the human transcription factor YY1. Nucleic Acids Res, 26(16), 3776–3783. doi: 10.1093/nar/26.16.3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zan J, Zhang H, Gu AP, Zhong KL, Lu MY, Bai XX, … Cai J (2017). Yin Yang 1 Dynamically Regulates Antiviral Innate Immune Responses During Viral Infection. Cell Physiol Biochem, 44(2), 607–617. doi: 10.1159/000485116 [DOI] [PubMed] [Google Scholar]
- Zhang B, Kirov S, & Snoddy J (2005). WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res, 33(Web Server issue), W741–748. doi: 10.1093/nar/gki475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Liang HF, Luo XD, Wang HJ, Gu AP, Zheng CY, … Cai J (2018). YY1 promotes IL-6 expression in LPS-stimulated BV2 microglial cells by interacting with p65 to promote transcriptional activation of IL-6. Biochem Biophys Res Commun, 502(2), 269–275. doi: 10.1016/j.bbrc.2018.05.159 [DOI] [PubMed] [Google Scholar]
- Zurkirchen L, Varum S, Giger S, Klug A, Hausel J, Bossart R, … Sommer L (2019). Yin Yang 1 sustains biosynthetic demands during brain development in a stage-specific manner. Nat Commun, 10(1), 2192. doi: 10.1038/s41467-019-09823-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie 1. Mouse movement and behavior in WT (YY1 loxP mice) and GFAP-specific YY1 cKO mice. Mice (12-week-old) were placed in the cages and behavior recorded for 30 sec.