Abstract
Background
Arylacetamide deacetylase (AADAC) is a lipolytic enzyme involved in xenobiotic metabolism. The characterization in terms of activity and substrate preference has been limited to a few mammalian species. The potential role and catalytic activities of AADAC from other organisms are still poorly understood. Therefore, in this work, the physicochemical properties, proteomic analysis, and protein-protein interactions from Gnathostomata organisms were investigated.
Results
The analysis were performed with 142 orthologue sequences with ~ 48–100% identity with human AADAC. The catalytic motif HGG[A/G] tetrapeptide block was conserved through all AADAC orthologues. Four variations were found in the consensus pentapeptide GXSXG sequence (GDSAG, GESAG, GDSSG, and GSSSG), and a novel motif YXLXP was found. The prediction of N-glycosylation sites projected 4, 1, 6, and 4 different patterns for amphibians, birds, mammals, and reptiles, respectively. The transmembrane regions of AADAC orthologues were not conserved among groups, and variations in the number and orientation of the active site and C-terminal carboxyl were observed among the sequences studied. The protein-protein interaction of AADAC orthologues were related to cancer, lipid, and xenobiotic metabolism genes.
Conclusion
The findings from this computational analysis offer new insight into one of the main enzymes involved in xenobiotic metabolism from mammals, reptiles, amphibians, and birds and its potential use in medical and veterinarian biotechnological approaches.
Graphical Abstract
Supplementary Information
The online version contains supplementary material available at 10.1186/s43141-022-00443-z.
Keywords: Arylacetamide deacetylase, AADAC, Orthologues, Interactome, Computational analysis
Background
Arylacetamide deacetylase (AADAC) is a lipolytic enzyme involved mainly in the hydrolysis of esters and amides [1–3]. Its active site domain is a classic lipase/esterase GXSXG motif (catalytic triad Ser189, His373, and Asp343), sharing high sequence homology with the active site of hormone-sensitive lipases (HSL). Although AADAC was initially considered as an esterase, current findings suggest that AADAC could be classified as a lipase, due to the strong homology shared with HSL, the acceptance of water-insoluble substrates, and the observed inhibition by classic lipase inhibitors, such as E600 (diethyl-p-nitrophenyl phosphate) [4, 5].
AADAC strongly contributes to drug hydrolysis, resulting in xenobiotic detoxification or prodrug activation [1]. The catalytic activity of AADAC has been tested on the hydrolysis and deacetylation on a wide variety of prodrugs, such as flutamide, phenacetin, indiplon, ketoconazole, rifabutin, rifampicin, and rifapentine [2, 6–9]. In contrast to other transmembrane enzymes, such as carboxylesterase 1 (CES1), carboxylesterase 2 (CES2), and paraoxonase 1 (PON1), AADAC selectively prefers bulky and small acyl moieties, such as fluorescein diacetate, N-monoacetyldapsone, and propanil [1]. Similarly, AADAC has been linked to the hydrolysis and activation of abiraterone acetate, a prodrug for metastatic castration-resistant prostate cancer [10].
Although in humans, AADAC is primarily expressed in the lumen side of the endoplasmic reticulum (ER) of the liver and gastrointestinal tract (jejunum, small intestine, and colon) [11, 12], its expression has also been found in the brain, particularly in the Purkinje cell layer of the human cerebellum, hippocampus, corpus callosum, and caudate nucleus [13]; adrenal cortex/medulla, and pancreas [14]. Due to its vast location, the physiological effects linked to an upper or lower expression or inhibition of AADAC could be beneficial, e.g., the upregulation of AADAC expression contributed to a better prognosis for several types of cancer [15–19]. On the other hand, variants or deletions of the AADAC gene have been linked to increased susceptibility to suffer Tourette syndrome [13].
In drug development, the extrapolation of data from orthologues is used to estimate the pharmacological effects and toxicity of drug candidates [20]. Orthologues are genes that evolved from a common ancestor by speciation; therefore, orthologue proteins are likely to have similar biological roles [21]. This similarity allows the identification of enzymes from organisms other than model organisms, such as birds, reptiles, or amphibians. Some AADAC orthologues, such as mouse, dog, rat, and cynomolgus macaque AADAC have been studied for their substrate recognition and specificity [11, 12, 22–24]. For example, human AADAC can hydrolyze flutamide, phenacetin, indiplon, and rifampicin, whereas rat and mouse AADAC only hydrolyzes flutamide and phenacetin, and not rifampicin [11]. In contrast, dog AADAC only hydrolyzes phenacetin and not indiplon [12]. Differences in enzyme location are also observed among orthologue AADAC. For instance, in human and cynomolgus macaques, AADAC mRNA is found in the liver and small intestine, whereas in rats and mice, mRNA AADAC is expressed in the kidney [22].
Mice and rats are the preferred organisms for toxicological studies, and differences in substrate preference by human, rat, and mouse liver microsomes expressing AADAC have also been observed [2]. Human and rat liver microsomes could hydrolyze indiplon, but mouse liver microsomes could not, whereas human and mouse liver microsomes hydrolyzed phenacetin and ketoconazole. Interestingly, neither mouse nor rat liver microsomes had any activity on rifamycins, whereas human liver microsomes showed activity values from 20 to 60 pmol/min/mg.
Given the importance of human AADAC in lipid, endobiotic, and xenobiotic metabolism, the present work focused on the functional analysis of AADAC orthologues, human AADAC-protein interactions, and physicochemical properties of a series of AADAC orthologue sequences from mammals, reptiles, amphibians, and birds.
Methods
Collection of AADAC protein sequences
AADAC amino acid sequences were retrieved from GenBank and Ensembl databases through the Basic Local Alignment Search Tool (BLAST). Searches were performed using the annotated Human Arylacetamide Deacetylase amino acid sequence (GenBank accession no. NP_001077.2, and Ensembl accession no. ENSG00000114771), and InterProScan was used to confirm the protein domains [25]. The orthologue sequences were selected according to the following parameters: (i) sequences with lengths between 393 and 415 amino acids; (ii) sequences without gaps, unspecific amino acids, or truncated regions, (iii) sequences containing the α/β hydrolase-3 domain, and (iv) sequences belonging to mammals, amphibians, birds, and reptiles.
Phylogenetic analysis
The phylogenetic tree was constructed using an amino acid alignment of human AADAC orthologues with MUSCLE in Mega X Software (RRID:SCR_000667) [26] with the following settings: (i) hydrophobicity multiplier (1.20), (ii) max iterations (16), and (iii) cluster method (UPGMA), also, the distances were calculated using the Jones-Taylor-Thornton matrix (JTT), and the tree was created with the maximum likelihood method with 1000 bootstrap replicates. Finally, the Interactive Tree of Life (iTOL v6; https://itol.embl.de/) was employed to display the tree [27]. The Fitch (or similarity) matrix of the 142 orthologues was developed in RStudio software (v2022.07.2) using the R package SeqinR 1.0-2: Biological Sequences Retrieval and Analysis [28]. A protein alignment was used to calculate the pairwise distance matrix of aligned sequences using the function dist.alignment (x, matrix = similarity). The Numerical Taxonomy Multivariate Analysis System (NTSYSpc v2.02) software calculated the correlation matrix of amino acid sequences coding for the AADAC gene using the Dis/similarity module for qualitative data. Principal component analysis (PCA) was performed by plotting the 2-dimensional (2D) Dis/similarity matrix in NTSYSpc.
AADAC-protein interaction prediction
Potential protein-protein association networks for human AADAC were predicted with the STRING online database (RRID:SCR_005223) [29], whereas the protein-protein association for AADAC orthologues was predicted with the BioGRID database (https://thebiogrid.org/) (RRID:SCR_007393) [30]. For the STRING database (https://string-db.org/), the sequences of AADAC were selected according to the above criteria. For the interaction analysis in BioGRID, the target-interacting protein data from human AADAC, paraoxonase 1 (PON1), carboxylesterase 1 (CES1), carboxylesterase 2 (CES2), carboxylesterase 3 (CES3) were used.
Motif search
The lipase/esterase consensus motifs GXSXG and HGGG were inferred by sequence alignment with human AADAC and hormone-sensitive lipase (HSL, GenBank accession no. AAA69810.1). The logo plot representations were generated using Logomaker [31], and WebLogo [32]. The code that creates the Logomaker plots is available on GitHub at: https://github.com/jbkinney/logomaker/tree/master/logomaker.
To analyze the conserved motifs in AADAC orthologue proteins, 10 putative motifs between 6 and 50 amino acids were predicted with the MEME Suite 5.4.1 [33].
Modeling of three-dimensional AADAC structures and predicted physicochemical properties and transmembrane regions
Protein modeling of AADAC was performed with AlphaFold via the Google Colab platform [34], and visualized with UCSF Chimera software [35]. The physicochemical parameters of human AADAC orthologues were predicted with ProtParam using the EXPASY server (RRID:SCR_018087) [36]. The sequences in FASTA were used to analyze the molecular weight (MW), theoretical pI, the total number of negatively and positively charged residues, amino acid composition, extinction coefficients, instability index, aliphatic index, and grand average of hydropathicity (GRAVY). In addition, the transmembrane regions were inferred with TMHMM 2.0 (RRID:SCR_014935) [37], and glycosylations were predicted with NetOGlyc 4.0 (RRID:SCR_009026) [38]. One-way non-parametric ANOVA was used to evaluate the significative differences between orthologues characteristics using GraphPad Prism version 8.0.2 (RRID:SCR_002798).
Results
A total of 142 gene sequences from mammals (67), amphibians (4), birds (53), and reptiles (18) were collected from GenBank and confirmed with Ensembl database (Table 1) as orthologues. AADAC protein sequences showed ~ 48–100% identity across orthologues when human AADAC (NP_001077.2) was used as reference. In all cases, the α/β hydrolase domain was located between residues 107-376. Similarly, the amino acid residues of the catalytic triad of lipases/esterases (Ser, Asp/Glu, and His) were conserved among all the orthologues studied (Supplementary Figure S1).
Table 1.
Structural characteristics from AADAC orthologues
| Organism name | Taxonomy | Prediction method | Identity (%) | Motif | Active site (amino acid no.) | Carboxylesterase family | Transmembrane regions | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HGGG | GXSXG | Superfamily | S | D | H | ||||||
| Amblyraja radiata | Amphibian | Genome | 49.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 344 | 374 | IV | 1 |
| Geotrypetes seraphini | Amphibian | Genome | 55.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 344 | 373 | IV | 1 |
| Nanorana parkeri | Amphibian | Genome | 57.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Rhinatrema bivittatum | Amphibian | Genome | 57.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 372 | IV | 0 |
| Anas platyrhynchos | Birds | Genome | 58.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Anser cygnoides domesticus | Bird | Genome | 59.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Antrostomus carolinensis | Bird | Genome | 58.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Aptenodytes forsteri | Bird | Genome | 59.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Apteryx australis mantelli | Bird | Genome | 64.5 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Apteryx rowi | Bird | Genome | 64.5 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Aquila chrysaetos chrysaetos | Bird | Genome | 59.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Athene cunicularia | Bird | Genome | 59.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 342 | 373 | IV | 1 |
| Aythya fuligula | Bird | Genome | 59.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Balearica regulorum gibbericeps | Bird | Genome | 59.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Calidris pugnax | Bird | Genome | 62.5 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Calypte anna | Bird | Genome | 58.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Camarhynchus parvulus | Bird | Genome | 48 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 374 | IV | 0 |
| Catharus ustulatus | Bird | Genome | 53.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Charadrius vociferus | Bird | Genome | 59.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Chiroxiphia lanceolata | Bird | Genome | 57.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Corapipo altera | Bird | Genome | 57.1 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Corvus moneduloides | Bird | Genome | 54.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Coturnix japonica | Bird | Genome | 57.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Cuculus canorus | Bird | Genome | 61 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Cyanistes caeruleus | Bird | Genome | 55.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Cygnus atratus | Bird | Genome | 58.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Egretta garzetta | Bird | Genome | 60.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 190 | 344 | 374 | IV | 2 |
| Empidonax traillii | Bird | Genome | 56.1 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Eurypyga helias | Bird | Genome | 59.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Falco cherrug | Bird | Genome | 60 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Falco peregrinus | Bird | Genome | 60 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Fulmarus glacialis | Bird | Genome | 58.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Gallus gallus | Bird | Genome | 58.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Gavia stellata | Bird | Genome | 60 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Haliaeetus albicilla | Bird | Genome | 59.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Haliaeetus leucocephalus | Bird | Genome | 59.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Lepidothrix coronata | Bird | Genome | 57.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Lonchura striata domestica | Bird | Genome | 52.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Manacus vitellinus | Bird | Genome | 54.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 344 | 375 | IV | 0 |
| Meleagris gallopavo | Bird | Genome | 56.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Melopsittacus undulatus | Bird | Genome | 60.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Mesitornis unicolor | Bird | Genome | 58.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Molothrus ater | Bird | Genome | 51.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Neopelma chrysocephalum | Bird | Genome | 55.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Nestor notabilis | Bird | Genome | 60.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Numida meleagris | Bird | Genome | 58.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Opisthocomus hoazin | Bird | Genome | 59.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Parus major | Bird | Genome | 54.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Phalacrocorax carbo | Bird | Genome | 60.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Phasianus colchicus | Bird | Genome | 56.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Pipra filicauda | Bird | Genome | 56.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Pseudopodoces humilis | Bird | Genome | 55.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Pygoscelis adeliae | Bird | Genome | 59.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Strigops habroptila | Bird | Genome | 60 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Struthio camelus australis | Bird | Genome | 63.5 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Taeniopygia guttata | Bird | Genome | 52.9 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Tyto alba alba | Bird | Genome | 59.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Ailuropoda melanoleuca | Mammal | Genome | 74.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 190 | 344 | 374 | IV | 3 |
| Aotus nancymaae | Mammal | Genome | 88.5 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Arvicanthis niloticus | Mammal | Genome | 68.9 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | IV | 0 |
| Camelus bactrianus | Mammal | Genome | 74.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Camelus dromedarius | Mammal | Genome | 74.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Camelus ferus | Mammal | Genome | 74.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Canis lupus dingo | Mammal | Genome | 75.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 3 |
| Canis lupus familiaris | Mammal | Genome | 75.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 3 |
| Castor canadensis | Mammal | Genome | 74.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 3 |
| Chlorocebus sabaeus | Mammal | Genome | 93 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Chrysochloris asiatica | Mammal | Genome | 83.2 | HGGG | GGSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Colobus angolensis palliatus - revisar | Mammal | Genome | 94 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 346 | 376 | IV | 1 |
| Condylura cristata | Mammal | Genome | 79.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 344 | 374 | IV | 1 |
| Cricetulus griseus | Mammal | Genome | 67.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | IV | 1 |
| Desmodus rotundus | Mammal | Genome | 75.9 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Dipodomys ordii | Mammal | Genome | 66.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 3 |
| Elephantulus edwardii | Mammal | Genome | 74.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Equus asinus | Mammal | Genome | 83.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Erinaceus europaeus | Mammal | Genome | 75.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Galeopterus variegatus | Mammal | Genome | 77.9 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 2 |
| Gorilla gorilla gorilla | Mammal | Genome | 99 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Grammomys surdaster | Mammal | Genome | 68.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | IV | 1 |
| Halichoerus grypus | Mammal | Genome | 76.4 | HGGG | GESAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | VII | 3 |
| Homo sapiens | Mammal | Validated | 100 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Hylobates moloch | Mammal | Genome | 96.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Leptonychotes weddellii | Mammal | Genome | 74.2 | HGGG | GESAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | VII | 3 |
| Loxodonta africana | Mammal | Genome | 82.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Macaca mulatta | Mammal | Genome | 92.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Mandrillus leucophaeus | Mammal | Genome | 93.5 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Marmota flaviventris | Mammal | Genome | 68.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 183 | 337 | 366 | IV | 1 |
| Mastomys coucha | Mammal | Genome | 67.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 187 | 342 | 372 | IV | 3 |
| Meriones unguiculatus | Mammal | Genome | 70.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 187 | 342 | 372 | IV | 0 |
| Mesocricetus auratus | Mammal | Genome | 66.9 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | IV | 3 |
| Microcebus murinus | Mammal | Genome | 77.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Microtus ochrogaster | Mammal | Genome | 67.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 190 | 344 | 374 | IV | 0 |
| Miniopterus natalensis | Mammal | Genome | 77.4 | HGGG | GSSSG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | – | 1 |
| Mus caroli | Mammal | Genome | 68.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | IV | 1 |
| Mus musculus | Mammal | Validated | 69.9 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | IV | 0 |
| Mus pahari | Mammal | Genome | 69.2 | HGGG | GDSSG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | – | 2 |
| Myotis davidii | Mammal | Genome | 74.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 3 |
| Myotis lucifugus | Mammal | Genome | 75.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 2 |
| Myotis myotis | Mammal | Genome | 74.9 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 3 |
| Nannospalax galili | Mammal | Genome | 72.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Nomascus leucogenys | Mammal | Genome | 96.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Onychomys torridus | Mammal | Genome | 68.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Ornithorhynchus anatinus | Mammal | Genome | 67.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Pan paniscus | Mammal | Genome | 99.5 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Pan troglodytes | Mammal | Genome | 99.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Papio anubis | Mammal | Genome | 93.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Peromyscus leucopus | Mammal | Genome | 68.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | IV | 1 |
| Peromyscus maniculatus bairdii | Mammal | Genome | 68.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | IV | 1 |
| Phoca vitulina | Mammal | Genome | 76.2 | HGGG | GESAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | VII | 3 |
| Piliocolobus tephrosceles | Mammal | Genome | 93.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Pipistrellus kuhlii | Mammal | Genome | 74.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 2 |
| Pteropus alecto | Mammal | Genome | 69.9 | HGGG | GSSSG | Alpha/beta hydrolase 3 | 189 | 342 | 372 | – | 1 |
| Rattus norvegicus | Mammal | Transcriptome | 67.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | IV | 3 |
| Rattus rattus | Mammal | Genome | 67.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 188 | 342 | 372 | IV | 3 |
| Rhinopithecus bieti | Mammal | Genome | 92.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 372 | IV | 1 |
| Rhinopithecus roxellana | Mammal | Genome | 92.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Saimiri boliviensis boliviensis | Mammal | Genome | 89.5 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Sapajus apella | Mammal | Genome | 89.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Talpa occidentalis | Mammal | Genome | 82 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Theropithecus gelada | Mammal | Genome | 93.5 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Trachypithecus francoisi | Mammal | Genome | 92.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Urocitellus parryii | Mammal | Genome | 69.4 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 183 | 367 | 337 | IV | 3 |
| Vicugna pacos | Mammal | Genome | 75.2 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Vulpes vulpes | Mammal | Genome | 76.7 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 3 |
| Alligator sinensis | Reptiles | Genome | 62.0 | HGGG | GDSSG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | – | 1 |
| Anolis carolinensis | Reptiles | Genome | 61.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Chelonoidis abingdonii | Reptiles | Genome | 63.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Crocodylus porosus | Reptiles | Genome | 63 | HGGG | GDSSG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Gavialis gangeticus | Reptiles | Genome | 63 | HGGG | GDSSG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | – | 1 |
| Gekko japonicus | Reptiles | Genome | 60.1 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 190 | 344 | 374 | IV | 0 |
| Lacerta agilis | Reptiles | Genome | 60.5 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 372 | IV | 0 |
| Notechis scutatus | Reptiles | Genome | 58.1 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Pantherophis guttatus | Reptiles | Genome | 58.6 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Pelodiscus sinensis | Reptiles | Genome | 60.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 199 | 353 | 383 | IV | 0 |
| Podarcis muralis | Reptiles | Genome | 62.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 372 | IV | 0 |
| Pogona vitticeps | Reptiles | Genome | 58.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Protobothrops mucrosquamatus | Reptiles | Genome | 58.1 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Pseudonaja textilis | Reptiles | Genome | 58.8 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Python bivittatus | Reptiles | Genome | 59.1 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 1 |
| Thamnophis sirtalis | Reptiles | Genome | 58.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Trachemys scripta elegans | Reptiles | Genome | 63.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 373 | IV | 0 |
| Zootoca vivipara | Reptiles | Genome | 61.3 | HGGG | GDSAG | Alpha/beta hydrolase 3 | 189 | 343 | 372 | IV | 0 |
The phylogenetic relationships of AADAC orthologues are shown in Fig. 1. The avian taxon had a common ancestor from which descended two distinctive subgroups: (i) clades XII and XIII, and (ii) clades VIII to XI. Under these terms, it is inferred that birds have the lowest percentage of identity with human AADAC. The analysis shows a marked difference between AADAC from the selected organisms. In mammals, four distinctive clades were observed: in clade I; human AADAC shared a common ancestor with AADAC from Pan paniscus and Pan troglodytes; in clade II, mouse and rat AADAC were grouped; clade III belonged to dog AADAC, whereas camels and hedgehogs were grouped in clade IV. Amphibians, represented by clade V, and clade IV from mammals evolved from a common ancestor. Reptiles, divided into clades VI and VII, share a common ancestor. PCA shows the similarity between orthologs of different taxa (Fig. 2A, B). It can be observed that organisms of the same taxon share more significant similarities (Fig. 2B), comparable to that observed in the phylogenetic tree (Fig. 1).
Fig. 1.
Maximum Likelihood tree showing the phylogenetic relationships of AADAC orthologues. Mammal AADAC sequences are distributed in clades. Sequences from mammals are highlighted in blue, amphibians in lilac, reptiles in green, and birds in pink. Distances were calculated using the Jones-Taylor-Thornton matrix (JTT), and the tree was created with the Maximum Likelihood method with 1,000 bootstrap replicates
Fig. 2.
Principal component analysis. A The Fitch matrix of the 142 orthologues. B Correlation matrix of the amino acid sequences coding for the AADAC genes, the Dis/similarity matrix was created in NTSYSpc; amphibian organisms are represented in purple, avian organisms are represented in pink, mammalian organisms are represented in blue, and reptiles in green
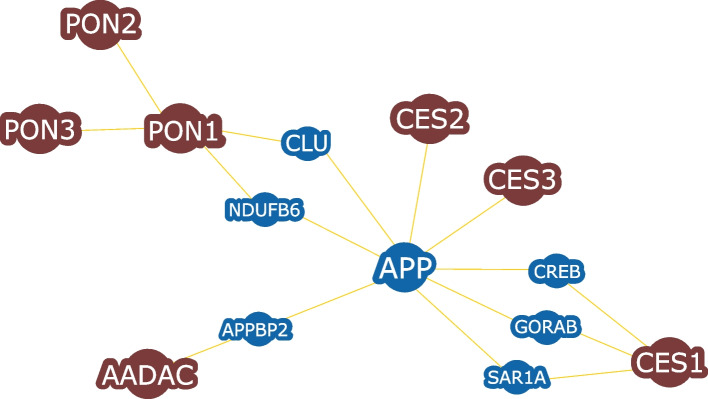
Human AADAC is closely related to three hydrolytic enzymes belonging to the carboxylesterase family: CES1, CES2, and CES3, which are ER-linked enzymes that contain the α/β hydrolase domain and are primarily known to be involved in phase I of drug or xenobiotic metabolism [39] . A single interactome to relate the genetic and physical interactions of CES1, CES2, PON1, and human AADAC proteins/genes was constructed (Fig. 3). The amyloid-beta precursor protein (APP) shows a chemical-protein interaction with CES2, AADAC, CES1, and PON1 (Fig. 3).
Fig. 3.
BioGRID-base human AADAC-protein interaction network
In humans, APP is a cell surface protein with signal transduction properties that control the viability, proliferation, migration, and aggressiveness of various cancer cells, such as breast cancer, ovarian cell adenocarcinoma, medulloblastoma, and neuroblastoma [32]. Additionally, APP interacted with Amyloid-Beta Precursor Protein Binding Protein 2 (APPBP2), which is involved in non-small cell lung cancer (NSCLC), a type of cancer accountable for ~ 80% of all lung carcinoma patients [33]. On the other hand, the interactions of 70 AADAC orthologues were analyzed using the STRING database (Supplementary Table S2). From the sequences studied, 45 AADAC orthologues were related to dihydrodiol dehydrogenases (DHDH), 57 sequences showed an interaction with the succinate receptor 1 (SUCNR1), and 44 interacted with ATP8B3. DHDH is involved in the detoxication of trans-dihydrodiol and carcinogenic metabolites of polycyclic aromatic hydrocarbons [40]; SUCNR1 is related to conditions such as hypertension, diabetes, and obesity [41], whereas ATP8B3 is involved in aminophospholipid transport in the liver [42].
The structural motif of the consensus pentapeptide GXSXG (Table 1 and Fig. 4A) was illustrated using consensus sequence logos grouped by amphibians, birds, mammals, and reptiles. Four different residues were found occupying the X position of the GXSXG sequence: GDSAG (92%), GESAG (2.1%), GDSSG (2.8%), and GSSSG (1.4%), while the HGG[G/A] tetrapeptide block was conserved through all AADAC orthologues, being the G/Gly residue following the HGG motif (HGGG) extensively conserved (99.2%) (Fig. 4B and Fig. 5C/motif 1). The search for structural motifs using the MEME-suite (motif-based sequence analysis tools) displayed 10 essential motifs for all AADAC sequences studied. Motifs 1, 3, 6, 8, and 9 were absent in star-nosed mole (Condylura cristata), arctic ground squirrel (Urocitellus parryii), yellow-bellied marmot (Marmota falviventris), chicken (Gallus gallus), gecko (Gekko japonicus), maiden ray (Amblyraja radiata), small tree finch (Camrahynchus parvulus), gaboon caecilian (Geotrypetes seraphini), and little egret (Egretta garzetta) (Fig. 5 and Supplementary Figure S2). Motif 7 (Fig. 5) corresponds to the transmembrane region, while motifs 1, 3, had glycosylation sites such as NVTV, NWSS, and NWSN. A possibly important region identified in motif 5 is the YXLXP sequence, where X represents the amino acids R/Arg or A/Ala present in all orthologues (Supplementary Figure S1, Fig. 5/motif 5), and forms a β-α loop in all orthologues sequences studied (Fig. 5A, B). The AADAC folding among organisms with 10 motifs was similar to human (Fig. 5C), and sequences with any missing motif showed structural differences, mainly in the formation of alpha helices and beta strands (Fig. 6D, F).
Fig. 4.
Conserved AADAC motifs using LogoMaker (A) and WebLogo (B and C). In all cases, the amino acid number indicated in the image is relative to the human AADAC sequence. A Logo showing the conserved GXSXG motif, which depicts the catalytic Ser, located in residues 113 to 126. B Logo showing the conserved HGGG motif, located in residues 113 to 126. C Logo showing the conserved transmembrane region of AADAC orthologues, located in residues 1 to 25
Fig. 5.
Conserved motifs in AADAC orthologues using MEME-suite. A Ten representative conserved motifs. Motif 1 (red), motif 2 (cyan), motif 3 (green), motif 4 (violet), motif 5 (orange), motif 6 (lime), motif 7 (blue), motif 8 (fuchsia), motif 9 (dark orange), and motif 10 (yellow). B The locations along the sequence of the 10 representative motifs depicted in A. The complete list is illustrated in Supplementary Figure S2. C Representation of all motifs of AADACs orthologues. The HGGG and GXSXG motifs are highlighted in gray. N and C, at the bottom of the images represent the amino terminal and carboxyl terminal
Fig. 6.
Tridimensional structural comparison of AADAC from orthologues. A ADDAC from Homo sapiens, the active site is shown in red, and the YXLXP motif in orange. B Active site of AADAC from Homo sapiens. The catalytic amino acids are represented by sticks. C Structural alignment of representative organisms from mammals (Macaca mulata in aqua), amphibian (Nanorana parkeri in pink), birds (Anas platyrhynchos in purple), reptiles (Trachemys scripta elegans in light green). D Structural alignment of mammalian AADAC showcasing the motif differences (Condylura cristata in grey color, Urocitellus parryii in salmon color and Marmota flaviventris in yellow color). E Structural alignment of birds (Camarhynchus parvulus in gold color, Gallus gallus in steel blue and Egretta garzetta in brown). F Structural alignment of amphibian and reptile organisms with motif differences (Amblyraja radiata in pink, Geotrypetes seraphini in aquamarine and Gekko japonicus in beige). The numbers indicate the structural motif related to MEME suite analysis (Fig. 3)
The physicochemical characterization shows that the molecular mass among orthologs remains between 44 and 46 kDa; however, for mammals, the mean is 45.8 kDa, while for birds and amphibians, it is 45.0 and 45.8 kDa, respectively. The number of negatively charged residues (Asp + Glu) was greater than the number of positively charged residues (Arg + Lys). The sum of hydropathy values of all amino acids divided by protein length showed similar tendencies between orthologues. The instability index is between 31 and 62 h, and some birds orthologues showed the most stable AADAC (Fig. 7). The prediction of N-glycosylation sites with NetNGlyc-4.0 server projected 4, 1, 6, and 4 different patterns for amphibians, bird, mammals, and reptiles, respectively.
Fig. 7.
Distribution plot depicting seven major predicted physicochemical properties of AADAC orthologues using the ProtParam Tool. A Molecular mass (kDa), B Percentage of negatively charged amino acids (Asp + Glu), C Percentage of positively charged amino acids (Arg + Lys), D Theoretical isoelectric point (pI), E Grand average of hydropathicity index (GRAVY), F Instability index, and G Aliphatic index. The complete computed physicochemical properties for each AADAC ortholog are depicted in Supplementary Table S1. One-way non-parametric ANOVA was used to evaluate significant differences between orthologues characteristics using GraphPad Prism version 8.0.2
The transmembrane regions of AADAC orthologues varied between 0 and 3. From 142 orthologue sequences, 48 organisms did not show transmembrane regions (33.8%), 73 organisms showed a single transmembrane region with the C-terminal oriented towards the lumen (51.4%), 5 organisms had 2 transmembrane regions with the active site towards the cytosol (3.5%), and 16 organisms showed 3 transmembrane regions with the C-terminal carboxyl oriented towards the ER lumen (11.2%) (Table 1).
Discussion
The phylogenetic history of human AADAC orthologues infers those common ancestral sequences evolved in the avian taxon, with speciation events occurring in a descending fashion in reptile, amphibian, and mammalian taxa, respectively.
Only nine AADAC orthologues showed interactions with carboxyl esterases (CES), belonging to red fox (Vulpes vulpes), Bolivian squirrel monkey (Samiri boliviensis), brown rat (Rattus norvegicus), house mouse (Mus musculus), Mus pahari, shrew-like mouse (Mus coroli), Mongolian gerbil (Meriones unguiculatus), one-humped camel (Camelus dromedarius), and human (Homo sapiens). It can be inferred that these nine AADAC may also be able to hydrolyze human AADAC substrates, such as xenobiotics.
The consensus sequence GDSAG found in mammals, birds, reptiles, and amphibians belongs to family IV of α/β hydrolases, also known as HSL family. The GESAG motif, found only in three orthologues, gray seal (Halichoerus grypus), common seal (Phoca vitulina), and weddell seal (Leptonychotes weddellii)), belonged to family VII of α/β hydrolases [43]. Three AADAC orthologues belonging to the order of Crocodilia (Alligator sinensis, Crocodylus porosus, and Gavialis gangeticus) and shrewmouse (Mus pahari) showed the GDSSG consensus sequence. The natal long-fingered bat (Miniopterus natalensis) and the black flying fox (Pteropus alecto) were the only AADAC orthologues showing the consensus pentapeptide GSSSG (Table 1 and Supplementary Figure S1).
The HGGG motif forms a loop close to the active site and directly participates in the catalytic hydrolysis by stabilizing the oxyanion intermediate of the reaction. The presence of multiple G/Gly residues confers greater flexibility to this loop. In contrast, some studies point that a change in the last G/Gly residue for an A/Ala decreases the enzyme activity up to 40% [44]. Pteropus alecto AADAC was the only orthologue to show an A/Ala residue placed at the last position (HGGA). The YXLXP motif has been previously reported in the telomeric protein SNM1B/Apollo (a member of the Metallo-β-lactamase/β-CASP family), which is involved in binding to TRF2 (telomeric repeat-binding factor 2) [45]; Similarly, SLX4 protein involved in DNA repair, contains a H/YXLXP motif and binds human TRF2. A similar site in the TRFH domain of TRF1 (around residue F142), FXLXP motif, serves as an anchor to TIN2 protein (TRF1-interacting protein 2) [46, 47].
Another essential structural region is the transmembrane region (Fig. 3C). The amino acids belonging to this region are responsible for anchoring the protein to the lipid bilayer. The most conserved amino acids among the studied orthologues were Y/Tyr, P/Pro, G/Gly, and K/Lys. In contrast, amino acids at positions 3, 5, 11, 12, 15, 21, and 23 showed increased variability among AADAC orthologues.
The predicted physicochemical characteristics of AADAC orthologues are resumed in the Supplementary Table S1. The calculated Molecular weight (MW) of AADAC orthologues is shown for amphibians, birds, mammals, and reptiles, and the minimum and maximum values are highlighted using a box and whisker plot (Fig. 7A). Human AADAC has been previously purified and characterized from liver tissue. Its MW was deduced at 45.671 kDa by gel electrophoresis [4], which is consistent with the MW value obtained by ProtParam for human AADAC (45.73 kDa), and similar to the average MW value calculated for mammals (45.68 kDa). The highest MW value belonged to AADAC from reptiles (46.77 kDa), while AADAC from birds had an average of 45.1 kDa, being the taxa with the lowest MW values.
The amount of Asp + Glu and Arg + Lys is related to the isoelectric point of the protein (Fig. 5B, C). Human AADAC orthologues showed isoelectric point (pI) values between 5.5 and 9.3 (Fig. 5D and Supplementary Table S1). The theoretical pI of human AADAC was 8.75, which differed from the pI calculated by in vivo experiments (9.36) [4]. Mammals showed pI values ranging from 6.0 to 9.3, followed by birds (5.6–9.2), reptiles (5.5–8.5), and amphibians (6.0–8.3).
The GRAVY value for a protein or a peptide is the sum of hydropathy values of all amino acids divided by the protein length [48]. Positive GRAVY values indicate hydrophobicity, whereas negative values mean hydrophilicity. The GRAVY value for human AADAC was found to be negative, implying a hydrophilic character, and similar GRAVY results were observed for mammals, amphibians, and reptiles AADAC (Fig. 7E). In contrast, birds were the only taxa showing positive GRAVY values, thus indicating hydrophobicity.
The instability index of a protein is calculated based on the presence of certain dipeptides. If the instability index value of a given protein is less than 40, it is considered stable under atmospheric conditions, and have a longer in vivo life span (~ 16 h), whereas proteins with an instability index > 40 show an in vivo half-life of less than 5 h [49]. The instability index value of human AADAC was 33.96 (Fig. 7E), which is considered a stable protein for in vivo experiments. In fact, human AADAC has already been expressed in sf9, DH10Bac, and E. coli cells, further purified and characterized for drug hydrolysis assays [6, 50, 51]. The average instability index values of AADAC orthologues ranged from 40 to 45 (Fig. 7F). Different model organisms for toxicological studies found in the mammalian taxon, such as mice, rats, and dogs, showed a protein instability index > 40, thus classifying them as in vivo unstable with a life span of fewer than five hours. Despite these values, mouse, rat, and dog AADAC microsomes have been successfully used in vitro to hydrolyze various prodrugs, confirming their role in the metabolism of the already known substrates of human AADAC [11, 12, 52].
For human AADAC, the high aliphatic index value was 99.15, which is considered thermostable. In general, all the AADAC orthologues studied showed aliphatic index values above 86.22, indicating a reliable degree of thermostability (Fig. 7G).
The potential N-glycosylation sites of amphibian, bird, mammal, and reptile AADAC were predicted with the NetNGlyc-4.0 server, and the results are detailed in Table 1. For N-glycosylation, an N-acetylglucosamine residue is linked via an amide bond to an asparagine (N/Asn) from the consensus sequence NX(S/T), where X can be any amino acid except proline (P/Pro). The presence of the consensus sequence is required for N-linked glycosylation; however, the potential sites might not be glycosylated [53]. For amphibians, the neuronal prediction gave two main glycosylation sites. All four amphibian sequences showed glycosylation sites at 78(NVTV/I). For bird AADAC, 39 sequences had one glycosylation site (78(NVTV), 73.6% of sequences), 12 sequences had two glycosylation sites (78(NVTV)-282(NWS[Q/R/V/L/N]) or 78(NVTV)-88(N[V/T/I][S/T]V), 22.6% of sequences), whereas the sequence of the bird Neopelma chrysocephalum showed no glycosylation site.
Mammalian AADAC showed six different glycosylation patterns. The most common pattern was 78(NV/ITV)-282(NWSS/A) (36 sequences in total, 53.7%), including AADAC sequences of Homo sapiens, Pan paniscus, Pan troglodytes, and Canis lupus familiaris. This is in agreement with the results reported in the literature [52], where their findings proved that N-glycosylation at the N282 residue of human AADAC was crucial for enzymatic activity by proper folding. The following observed patterns were 77(NVTV) (7 sequences, 10.4%, including Rattus rattus and Rattus norvegicus), and 282(NWSS) (5 sequences, 7.5%), and 77(NVTV)-281(NWSS) (4 sequences, 6%). Three glycosylation sites were found for four mammal sequences (78(NITV)-281(NWSS)-298(NRTY), 78(NVTV)-282(NWSS)-302(NGTS), 56(NHSM)-78(NVTV)-282(NWSS), and 55(NHSM)-77(NVTV)-281(NWSS), whereas 11 mammal sequences showed other glycosylation patterns. The AADAC sequence of Mus musculus had no N-glycosylation sites.
The prediction of N-glycosylation sites for reptile AADAC projected four different patterns. Six reptile sequences (33.3%) had the 282(NWS/N/R/H/K/E) site, 4 sequences (22.2%) had the 78(NVTV)-282(NWSN) site, 5 sequences (27.8%) showed different N-glycosylation patterns, and 3 sequences (16.7%) had no N-glycosylations.
In humans, AADAC is a type II transmembrane protein comprising a short, positively charged N-terminal region oriented towards the cytoplasm, a single transmembrane region, and a C-terminal tail oriented towards the lumen of the ER, containing the catalytic triad Ser189, Asp343, and His373 [5, 52, 54]. The lumen location of the catalytic triad was confirmed by the hydrolysis of D-13223, a substrate that is not observed in human liver cytosol [55].
The transmembrane regions of human AADAC and its orthologues were predicted with TMHMM (Table 1). TMHMM has a reliability of soluble and transmembrane proteins of 70–80% of assertiveness [37]. The server predicted a transmembrane helix located at residues 5–24 (Table 1), and an outer sequence of amino acid 25-399, which is consistent with the literature [54]. The active site of human AADAC faces the lumen site of the endoplasmic reticulum (RE), which implies different catalytic capacities compared to other carboxylesterases [5]. For example, substrates for cytosolic lipases, such as glycerolphospholipids or triglycerides stored in cytoplasmic lipid droplets, cannot cross the ER membrane and therefore are not hydrolyzed by AADAC [5].
All studied orthologues shared the same amino-terminal location at the cytosol, whereas the C-terminal was oriented towards the lumen of the RE.
Conclusions
Although AADAC is a ubiquitous protein spread among organisms from different taxa, drug hydrolysis assays and toxicological studies are limited to a few species. The predicted physicochemical characteristics of the studied 142 orthologues show similarity among the groups; however, the composition of amino acids as Arg + Lys and Asp + Glu ratios differed among the studied organisms, which generates differences between the isoelectric point. The orthologues were related to DHDH, ATP8B3, and P2RY1 genes, expressed in the lipid membrane, and directly related to skin and lung cancer genes (APP and APPBP2), suggesting that they may share mechanisms among these diseases. In addition, the comparative analysis of AADAC orthologues revealed that the sequence YXLXP, which has been involved as DNA repair mechanism, was shared by all the organisms studied. Other structural motifs that are fingerprints of lipolytic enzymes (GXSXG and HGGG) were found in all orthologs; however, according to the predicted 3D structures, missing motifs can generate conformational changes that can affect enzyme-substrate interaction.
The number of transmembrane regions varied among organisms and did not follow a pattern among mammals, amphibians, birds, or reptiles. Some substrate selectivities among the studied orthologues could be inferred by the phylogenetic tree constructed; however, further biochemical studies are mandatory to corroborate the results herein presented. This information, together with the analysis of the interactome, offers an overview of how these AADAC from the studied organisms could interact with various drugs tested mainly in primates or rodents. The collected data shown in this work broadens the knowledge of AADAC orthologues and could help to better select new model organisms as future candidates for detoxification, xenobiotic processing, and drug disposition studies.
Supplementary Information
Additional file 1: Figure S1. Amino acid sequence alignments for AADAC orthologues. The name of the orthologues is shown as an accession number on the left side. "⁎" shows identical residues for AADAC orthologues; ": "2 alternate residues. On grey color, principal identical residues referent to HGGG box, Consensus sequence (GXSXG), and YXLXP motif. Loops above the amino acid sequence indicate alpha-helix structures, and arrows indicate beta-sheet structures. Figure S2. The complete motifs analysis, using MEME-suite of AADAC orthologues. Table S1. Summary of the physicochemical characterization, transmembrane regions, and N-glycosylation sites predicted for AADAC orthologue. Table S2. The complete protein-protein interaction network analysis of AADAC orthologues using the STRING tool.
Acknowledgements
The authors would like to thank Dr. Mariana Díaz-Tenorio for her kind advice and helpful discussions.
Abbreviations
- AADAC
Arylacetamide deacetylase
- HSL
Hormone-sensitive lipases
- E600
Diethyl-p-nitrophenyl phosphate
- CES1
Carboxylesterase 1
- CES2
Carboxylesterase 2
- PON1
Paraoxonase 1
- BLAST
Basic Local Alignment Search Tool
- MW
Molecular weight
Authors’ contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by TD-V, CBR-O, and RBM-P. The first draft of the manuscript was written by TD-V, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript
Funding
This study was supported by PROFAPI-ITSON grant (DFP300320165). We gratefully recognize a postdoctoral fellowship for T.D.-V. from SEP-SES (23-007-C) postdoctoral grants.
Availability of data and materials
The manuscript has data included as electronic supplementary material.
Declarations
Ethics approval and consent to participate
Not applicable
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Fukami T, Kariya M, Kurokawa T, Iida A, Nakajima M. Comparison of substrate specificity among human arylacetamide deacetylase and carboxylesterases. Eur J Pharm Sci. 2015;78:47–53. doi: 10.1016/j.ejps.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Kisui F, Fukami T, Nakano M, Nakajima M. Strain and sex differences in drug hydrolase activities in rodent livers. Eur J Pharm Sci 2020;142:105143. https://doi.org/10.1016/j.ejps.2019.105143. [DOI] [PubMed]
- 3.Morikawa T, Fukami T, Gotoh-Saito S, Nakano M, Nakajima M. PPARα regulates the expression of human arylacetamide deacetylase involved in drug hydrolysis and lipid metabolism. Biochem Pharmacol 2022;199:115010. https://doi.org/10.1016/J.BCP.2022.115010. [DOI] [PubMed]
- 4.Probst MR, Beer M, Beer D, Jenö P, Meyer UA, Gasser R. Human liver arylacetamide deacetylase. Molecular cloning of a novel esterase involved in the metabolic activation of arylamine carcinogens with high sequence similarity to hormone-sensitive lipase. J Biol Chem. 1994;269:21650–21656. doi: 10.1016/S0021-9258(17)31855-0. [DOI] [PubMed] [Google Scholar]
- 5.Lo V, Erickson B, Thomason-Hughes M, Ko KWS, Dolinsky VW, Nelson R, et al. Arylacetamide deacetylase attenuates fatty-acid-induced triacylglycerol accumulation in rat hepatoma cells. J Lipid Res 2010;51:368–377. https://doi.org/10.1194/jlr.M000596. [DOI] [PMC free article] [PubMed]
- 6.Hirosawa K, Fukami T, Tashiro K, Sakai Y, Kisui F, Nakano M, et al. Role of human AADAC on hydrolysis of eslicarbazepine acetate and effects of AADAC genetic polymorphisms on hydrolase activity. Drug Metab Dispos 2021;49. https://doi.org/10.1124/dmd.120.000295. [DOI] [PubMed]
- 7.Probst MR, Jenö P, Meyer UA. Purification and characterization of a human liver arylacetamide deacetylase. Biochem Biophys Res Commun 1991;177:453–459. https://doi.org/10.1016/0006-291X(91)92005-5. [DOI] [PubMed]
- 8.Nagaoka M, Fukami T, Kisui F, Yamada T, Sakai Y, Tashiro K, et al. Arylacetamide deacetylase knockout mice are sensitive to ketoconazole-induced hepatotoxicity and adrenal insufficiency. Biochem Pharmacol 2022;195:114842. https://doi.org/10.1016/J.BCP.2021.114842. [DOI] [PubMed]
- 9.Phaisal W, Jantarabenjakul W, Wacharachaisurapol N, Tawan M, Puthanakit T, Wittayalertpanya S, et al. Pharmacokinetics of isoniazid and rifapentine in young pediatric patients with latent tuberculosis infection. Int J Infect Dis 2022;122:725–732. https://doi.org/10.1016/j.ijid.2022.07.040. [DOI] [PubMed]
- 10.Sakai Y, Fukami T, Nagaoka M, Hirosawa K, Ichida H, Sato R, et al. Arylacetamide deacetylase as a determinant of the hydrolysis and activation of abiraterone acetate in mice and humans. Life Sci 2021;284:119896. https://doi.org/10.1016/j.lfs.2021.119896. [DOI] [PubMed]
- 11.Kobayashi Y, Fukami T, Nakajima A, Watanabe A, Nakajima M, Yokoi T. Species differences in tissue distribution and enzyme activities of arylacetamide deacetylase in human, rat, and mouse. Drug Metab Dispos 2012;40:671–679. https://doi.org/10.1124/dmd.111.043067. [DOI] [PubMed]
- 12.Kurokawa T, Fukami T, Yoshida T, Nakajima M. Arylacetamide deacetylase is responsible for activation of prasugrel in human and dog. Drug Metab Dispos 2016;44:409–416. https://doi.org/10.1124/dmd.115.068221. [DOI] [PubMed]
- 13.Bertelsen B, Stefánsson H, Riff Jensen L, Melchior L, Mol Debes N, Groth C et al (2016) Association of AADAC Deletion and Gilles de la Tourette Syndrome in a Large European Cohort. 79. 10.1016/j.biopsych.2015.08.027 [DOI] [PubMed]
- 14.Trickett JI, Patel DD, Knight BL, Saggerson ED, Gibbons GF, Pease RJ. Characterization of the rodent genes for arylacetamide deacetylase, a putative microsomal lipase, and evidence for transcriptional regulation. J Biol Chem. 2001;276:39522–39532. doi: 10.1074/jbc.M101764200. [DOI] [PubMed] [Google Scholar]
- 15.Yuan L, Zheng W, Yang Z, Deng X, Song Z, Deng H. Association of the AADAC gene and Tourette syndrome in a Han Chinese cohort. Neurosci Lett. 2018;666:24–27. doi: 10.1016/j.neulet.2017.12.034. [DOI] [PubMed] [Google Scholar]
- 16.Pagliaroli L, Vereczkei A, Padmanabhuni SS, Tarnok Z, Farkas L, Nagy P, et al. Association of Genetic Variation in the 3’UTR of LHX6, IMMP2L, and AADAC With Tourette Syndrome. Front Neurol. 2020;11:803. doi: 10.3389/fneur.2020.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, Wu Z, Tian B. Five gene signatures were identified in the prediction of overall survival in resectable pancreatic cancer. BMC Surg. 2020;20:1–10. doi: 10.1186/s12893-020-00856-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Fang T, Wang Y, Yin X, Zhang L, Zhang X, et al. Impact of AADAC gene expression on prognosis in patients with Borrmann type III advanced gastric cancer. BMC Cancer. 2022;22:1–19. doi: 10.1186/S12885-022-09594-1/FIGURES/7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdel-Tawab MS, Fouad H, Othman AM, Eid RA, Mohammed MA, Hassan A, et al. Evaluation of gene expression of PLEKHS1, AADAC, and CDKN3 as novel genomic markers in gastric carcinoma. PLoS One. 2022;17:e0265184. doi: 10.1371/JOURNAL.PONE.0265184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verbruggen B, Gunnarsson L, Kristiansson E, Österlund T, Owen SF, Snape JR, et al. ECOdrug: a database connecting drugs and conservation of their targets across species. Nucleic Acids Res. 2018;46:D930. doi: 10.1093/NAR/GKX1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matsuya A, Sakate R, Kawahara Y, Koyanagi KO, Sato Y, Fujii Y, et al. Evola: Ortholog database of all human genes in H-InvDB with manual curation of phylogenetic trees. Nucleic Acids Res. 2008;36:D787–D792. doi: 10.1093/NAR/GKM878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uno Y, Hosokawa M, Imai T. Isolation and characterization of arylacetamide deacetylase in cynomolgus macaques. J Vet Med Sci. 2015;77:721–724. doi: 10.1292/jvms.14-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Honda S, Fukami T, Tsujiguchi T, Zhang Y, Nakano M, Nakajima M. Hydrolase activities of cynomolgus monkey liver microsomes and recombinant CES1, CES2, and AADAC. Eur J Pharm Sci. 2021;161:105807. doi: 10.1016/j.ejps.2021.105807. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida T, Fukami T, Kurokawa T, Gotoh S, Oda A, Nakajima M. Difference in substrate specificity of carboxylesterase and arylacetamide deacetylase between dogs and humans. European Journal of Pharmaceutical Sciences; 2018. [DOI] [PubMed] [Google Scholar]
- 25.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: Genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciccarelli FD, Doerks T, Von Mering C, Creevey CJ, Snel B, Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science (80- ) 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 28.Charif D, Lobry JR. SeqinR 1.0-2: a contributed package to the R project for statistical computing devoted to biological sequences retrieval and analysis. In: Bastolla U, Porto M, Roman HE, Vendruscolo M, editors. Struct. Approaches to Seq. Evol. Mol. Networks, Popul. Berlin, Heidelberg: Springer Berlin Heidelberg; 2007. pp. 207–232. [Google Scholar]
- 29.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oughtred R, Rust J, Chang C, Breitkreutz BJ, Stark C, Willems A, et al. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021;30:187–200. doi: 10.1002/pro.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tareen A, Kinney JB. Logomaker: beautiful sequence logos in Python. Bioinformatics. 2020;36:2272–2274. doi: 10.1093/BIOINFORMATICS/BTZ921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey TL, Johnson J, Grant CE, Noble WS. The MEME Suite. Nucleic Acids Res. 2015;43:W39–W49. doi: 10.1093/NAR/GKV416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nat Methods. 2022;196(19):679–682. doi: 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/JCC.20084. [DOI] [PubMed] [Google Scholar]
- 36.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD et al (2005) Protein identification and analysis tools on the ExPASy server. Proteomics Protoc Handb:571–607. 10.1385/1-59259-890-0:571
- 37.Möller S, Croning MDR, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/BIOINFORMATICS/17.7.646. [DOI] [PubMed] [Google Scholar]
- 38.Steentoft C, Vakhrushev SY, Joshi HJ, Kong Y, Vester-Christensen MB, Schjoldager KTBG, et al. Precision mapping of the human O-GalNAc glycoproteome through SimpleCell technology. EMBO J. 2013;32:1478–1488. doi: 10.1038/emboj.2013.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams ET, Ehsani ME, Wang X, Wang H, Qian YW, Wrighton SA, et al. Effect of buffer components and carrier solvents on in vitro activity of recombinant human carboxylesterases. J Pharmacol Toxicol Methods. 2008;57:138–144. doi: 10.1016/J.VASCN.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 40.Penning TM. Dihydrodiol dehydrogenase and its role in polycyclic aromatic hydrocarbon metabolism. Chem Biol Interact. 1993;89:1–34. doi: 10.1016/0009-2797(93)03203-7. [DOI] [PubMed] [Google Scholar]
- 41.Gilissen J, Jouret F, Pirotte B, Hanson J. Insight into SUCNR1 (GPR91) structure and function. Pharmacol Ther. 2016;159:56–65. doi: 10.1016/J.PHARMTHERA.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Molday LL, Hii T, Coleman JA, Wen T, Andersen JP, et al. Proteomic analysis and functional characterization of P4-ATPase phospholipid flippases from murine tissues. Sci Rep. 2018;8:1–14. doi: 10.1038/s41598-018-29108-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sood S, Sharma A, Sharma N, Kanwar SS (2018) Carboxylesterases: sources, characterization and broader applications. Insights Enzym Res 01. 10.21767/2573-4466.100002
- 44.Laurell H, Contreras JA, Castan I, Langin D, Holm C. Analysis of the psychrotolerant property of hormone-sensitive lipase through site-directed mutagenesis. Protein Eng. 2000;13:711–717. doi: 10.1093/protein/13.10.711. [DOI] [PubMed] [Google Scholar]
- 45.Higa M, Fujita M, Yoshida K. DNA replication origins and fork progression at mammalian telomeres. Genes. 2017;8:112. doi: 10.3390/genes8040112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martínez P, Blasco MA. Heart-breaking telomeres. Circ Res. 2018;123:787–802. doi: 10.1161/CIRCRESAHA.118.312202. [DOI] [PubMed] [Google Scholar]
- 47.De Lange T. Shelterin-mediated telomere protection. Annu Rev Genet. 2018;52:223–247. doi: 10.1146/annurev-genet-032918-021921. [DOI] [PubMed] [Google Scholar]
- 48.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 49.Guruprasad K, Reddy BVB, Pandit MW. Correlation between stability of a protein and its dipeptide composition: a novel approach for predicting in vivo stability of a protein from its primary sequence. Protein Eng Des Sel. 1990;4:155–161. doi: 10.1093/protein/4.2.155. [DOI] [PubMed] [Google Scholar]
- 50.Watanabe A, Fukami T, Takahashi S, Kobayashi Y, Nakagawa N, Nakajima M, et al. Arylacetamide deacetylase is a determinant enzyme for the difference in hydrolase activities of phenacetin and acetaminophen. Drug Metab Dispos. 2010;38:1532–1537. doi: 10.1124/dmd.110.033720. [DOI] [PubMed] [Google Scholar]
- 51.Mizoi K, Takahashi M, Sakai S, Ogihara T, Haba M, Hosokawa M. Structure-activity relationship of atorvastatin derivatives for metabolic activation by hydrolases. Xenobiotica. 2020;50:261–269. doi: 10.1080/00498254.2019.1625083. [DOI] [PubMed] [Google Scholar]
- 52.Muta K, Fukami T, Nakajima M, Yokoi T. N-Glycosylation during translation is essential for human arylacetamide deacetylase enzyme activity. Biochem Pharmacol. 2014;87:352–359. doi: 10.1016/j.bcp.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 53.An HJ, Froehlich JW, Lebrilla CB. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr Opin Chem Biol. 2009;13:421–426. doi: 10.1016/J.CBPA.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frick C, Atanasov AG, Arnold P, Ozols J, Odermatt A. Appropriate function of 11β-hydroxysteroid dehydrogenase type 1 in the endoplasmic reticulum lumen is dependent on its N-terminal region sharing similar topological determinants with 50-kDa esterase. J Biol Chem. 2004;279:31131–31138. doi: 10.1074/jbc.M313666200. [DOI] [PubMed] [Google Scholar]
- 55.Konishi K, Fukami T, Ogiso T, Nakajima M. In vitro approach to elucidate the relevance of carboxylesterase 2 and N-acetyltransferase 2 to flupirtine-induced liver injury. Biochem Pharmacol. 2018;155:242–251. doi: 10.1016/j.bcp.2018.07.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Amino acid sequence alignments for AADAC orthologues. The name of the orthologues is shown as an accession number on the left side. "⁎" shows identical residues for AADAC orthologues; ": "2 alternate residues. On grey color, principal identical residues referent to HGGG box, Consensus sequence (GXSXG), and YXLXP motif. Loops above the amino acid sequence indicate alpha-helix structures, and arrows indicate beta-sheet structures. Figure S2. The complete motifs analysis, using MEME-suite of AADAC orthologues. Table S1. Summary of the physicochemical characterization, transmembrane regions, and N-glycosylation sites predicted for AADAC orthologue. Table S2. The complete protein-protein interaction network analysis of AADAC orthologues using the STRING tool.
Data Availability Statement
The manuscript has data included as electronic supplementary material.