Abstract
Bipolar androgen therapy (BAT) is a novel treatment strategy for patients with metastatic castration-resistant prostate cancer (mCRPC). We retrospectively identified patients with mCRPC that achieved deep PSA responses to BAT. All patients with available next-generation molecular sequencing harbored pathogenic mutations in TP53 and/or a homologous recombination DNA repair gene.
Background:
Bipolar androgen therapy (BAT) is an emerging treatment strategy for men with metastatic castration resistant prostate cancer (mCRPC) whereby serum testosterone is cycled from supraphysiologic to near-castrate levels each month. BAT has been shown to induce clinical responses in a significant proportion of patients, some of which are extreme. We explored the clinical and molecular characteristics of patients with mCRPC who achieved deep responses to BAT.
Methods:
We conducted a retrospective analysis of patients with mCRPC treated with BAT at Johns Hopkins. We identified 22 of 114 (19%) patients with mCRPC who achieved ≥70% PSA reductions upon treatment with BAT. All patients were treated using 400 mg testosterone cypionate intramuscularly every 28 days, together with continuous LHRH agonist therapy. Clinical-grade DNA sequencing was obtained using commercially available assays.
Results:
Somatic next-generation sequencing was obtained for 15 of 22 (68%) patients. Of these 15 extreme PSA responders with sequencing data available, All 15 of 15 (100%) harbored a pathogenic mutation in TP53 and/or a homologous recombination DNA repair (HRD) gene. Among the subset of patients with measureable disease (N = 15), 10 patients (67%) achieved an objective response including one patient with a complete response. The median radiographic progression-free survival of these deep PSA responders was 11.3 months (95% CI, 7.9–25.0 months).
Conclusions:
We observed an enrichment of TP53 and HRD mutations in mCRPC patients with extreme PSA responses to BAT, with durability lasting about a year. These data support the hypothesis that BAT may most benefit patients with DNA repair-deficient mCRPC. Further studies of BAT in biomarker-selected mCRPC populations (ie, TP53/HRD-mutated) are warranted.
Keywords: Testosterone, DNA repair, Homologous recombination deficiency, Biomarker, Next-generation sequencing
Introduction
Bipolar androgen therapy (BAT) is emerging as a novel therapy for men with metastatic castration resistant prostate cancer (mCRPC). Intramuscular injections of testosterone are administered every 28 days, resulting in rapid oscillations of circulating testosterone concentrations from supraphysiologic levels to near-castrate levels.1 Men are concurrently maintained on an LHRH agonist/antagonist to suppress endogenous testosterone production from the testes. Several clinical trials have demonstrated the efficacy of BAT as a treatment option for mCRPC patients, with acceptable toxicity.2–6
The rapid cycling of testosterone levels in preclinical models has been shown to induce DNA damage and genomic rearrangements such as TMPRSS2-ERG, which may provide insight into the mechanism of action.7–9 Enzalutamide-resistant, patient-derived xenograft models showed that clinical responses to BAT were due, in part, to suppression of the DNA damage response transcriptome.10 In clinical practice, a durable response to BAT was observed in a patient with mCRPC who harbored an inactivating BRCA2 mutation.11 We hypothesized that patients who experience extreme clinical responses to BAT may have a predictive genomic signature. In this study, we interrogated the molecular profiles of tumors from mCRPC patients who achieved deep responses while on BAT.
Methods
We conducted a retrospective analysis of three IRB-approved clinical trials conducted at Johns Hopkins University (NCT02090114, NCT02286921, NCT03554317) between April 2014 and October 2019. Patients who achieved a 70% or greater decline in PSA while receiving BAT were included in the analysis, and were defined as extreme responders. All patients received intramuscular injections of 400mg testosterone cypionate every 28 days and were maintained on an LHRH agonist/antagonist. Patients were evaluated every 28 days with safety labs and PSA measurement. CT and bone scan imaging were obtained every 12 weeks. BAT was discontinued in the event of: 1. radiographic progression, 2. ≥25% rise in PSA from baseline for patients who did not have a decrease in PSA below baseline during BAT, or 3. per the treating physician’s discretion for patients who experienced a decrease in PSA from baseline. Clinical or radiographic progression was defined by RECIST 1.1 and Prostate Cancer Working Group 2 (PCWG2) guidelines.
Radiographic progression-free survival (rPFS) on BAT was defined as the time from initiation of BAT therapy to the time of investigator-assessed radiographic progression (defined by RECIST 1.1 and PCWG2). Clinical-grade targeted DNA sequencing was performed using commercially available assays (Personal Genome Diagnostics, Foundation One, Caris Life Sciences) on the primary tumor or circulating tumor DNA. These gene panels included homologous recombination DNA repair (HRD) genes: BRCA1, BRCA2, ATM, CHEK2, BARD1, ARID1A, NBN, RAD50, RAD51C, RAD51D, PALB2, MRE11, FANCA, FANCB, FANCC, FANCD2, FANCE, FANCF, FANCG, FANCI, FANCL, FANCM.
Results
In total, 114 patients with mCRPC were treated with BAT within a clinical trial at Johns Hopkins during this time period. 19% (N = 22/114) of these patients achieved a 70% or greater decline in PSA from baseline upon treatment with BAT. The clinical and pathologic characteristics of these patients are shown in Table 1. The majority of patients had Gleason 8 or higher disease (59.1%) and prior treatment with one or more novel AR-targeted therapies (81.8%). Most patients did not have prior treatment with a taxane chemotherapy (90.9%).
Table 1.
Baseline Patient Pathologic and Clinical Characteristics
| Baseline Characteristics | N = 22 |
|---|---|
| Age (years) | |
| Median | 70.5 |
| (Range) | (51–84) |
| Gleason sum at diagnosis, N (%) | |
| ≤7 | 9 (40.9%) |
| 8 | 2 (9.1%) |
| 9 | 8 (36.4%) |
| 10 | 3 (13.6%) |
| Baseline PSA (ng/mL) | |
| Median | 41.1 |
| (Range) | (2.4–366) |
| Lines of prior novel AR targeted therapy, N (%) | |
| 0 | 4 (18.2%) |
| 1 | 9 (40.9%) |
| 2 | 9 (40.9%) |
| Prior taxane chemotherapy, N (%) | |
| Yes | 2 (9.1%) |
| No | 20 (90.9%) |
| NGS mutation profile, N (%) | |
| TP53 and/or HRD | 15 (68.2%) |
| Unknown | 7 (31.8%) |
AR = androgen receptor; HRD = homologous recombination DNA repair gene; N = number of patients; NGS = next-generation sequencing; PSA = prostate-specific antigen.
Fifteen of 22 deep PSA responders (68.2%) had available somatic DNA sequencing data. Of these patients, 100% (N = 15/15) harbored a pathogenic mutation in TP53 and/or a HRD gene. BRCA2 and TP53 were the two most frequently mutated genes in this cohort (Table 2). ATM, ARID1A and BARD1 mutations were also observed. One patient was identified as having an inactivating ARID1A mutation without a concomitant TP53. A list of specific mutations is provided (Supplemental Table 1).
Table 2.
List of observed mutated genes in extreme responder cohort.
| Mutated Gene | N (%) |
|---|---|
| BRCA2 | 5 (33.3%) |
| TP53 | 4 (26.6%) |
| TP53, BRCA2 | 2 (13.3%) |
| TP53, BARD1 | 1 (6.7%) |
| BRCA2, ATM | 1 (6.7%) |
| ARID1A | 1 (6.7%) |
| ATM, RB1 | 1 (6.7%) |
The PSA response to BAT of these extreme responders is shown as a waterfall plot in Figure 1A. 59% (N = 13/22) of patients achieved a ≥90% decrease in PSA. Two patients (one BRCA2-mutated, one with unknown mutation status) had complete biochemical responses. In this cohort, 15 patients had measureable soft-tissue disease. The observed objective response rate was 66.7% (N = 10/15; Figure 1B). One patient harboring a BRCA2 mutation achieved a completed objective response. The median rPFS of the extreme responder cohort was estimated at 11.3 months (95% CI: 7.9–25.0 months) (Figure 2). Three patients were free from radiographic progression for over two years.
Figure 1.
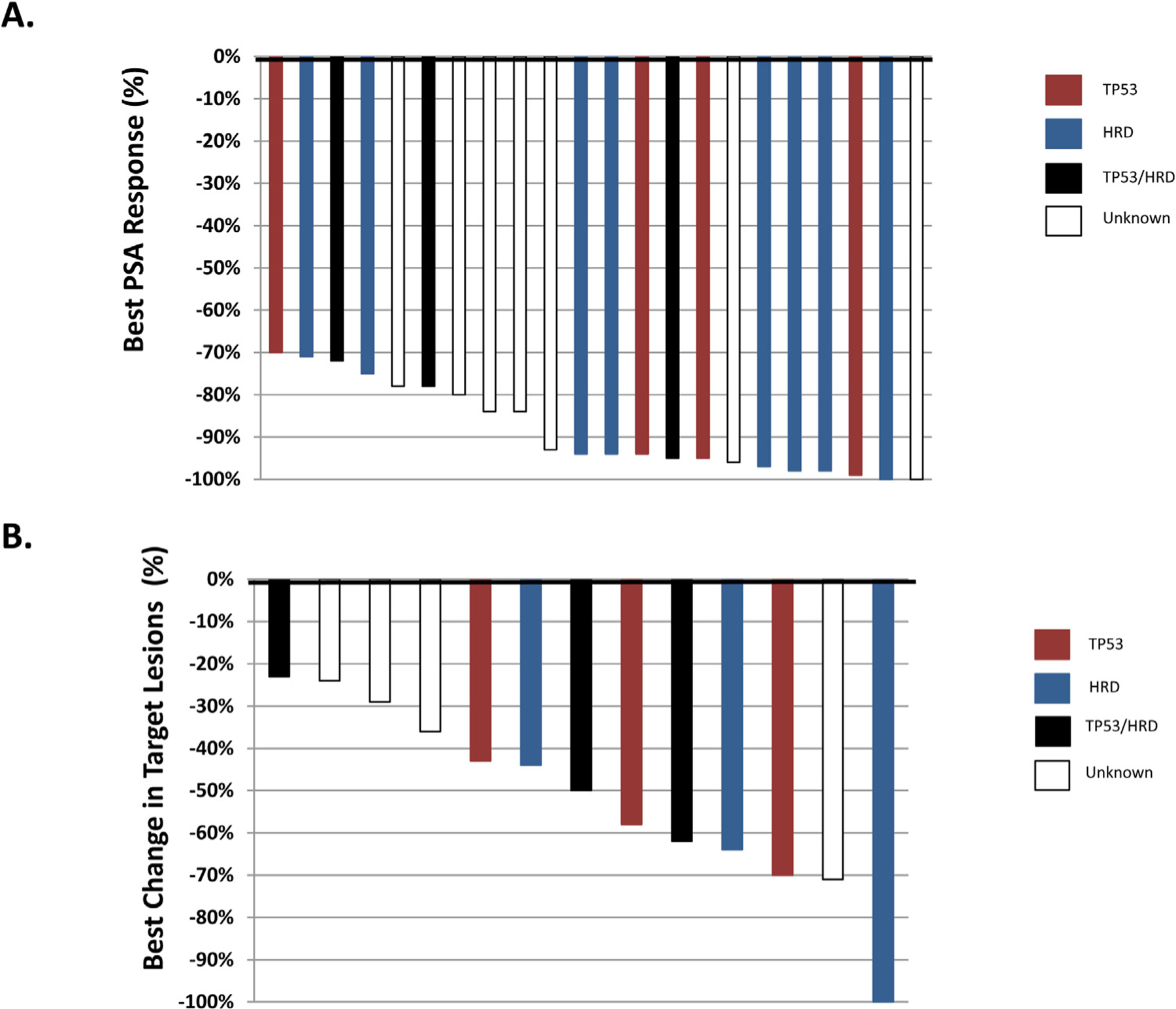
Best PSA and objective responses in mCRPC patients treated with BAT, stratified by mutational profile. (A) Twenty-two (22) mCRPC patients were observed to have a PSA decline of 70% or greater. Fifteen of the 15 patients with molecular testing information had a TP53 and/or HRD mutation (B) Best change in target lesions in 15 mCRPC patients with a deep PSA response on BAT is shown. Ten of 15 patients achieved an objective response on BAT. TP53 mutated – Red, HRD mutated – Blue, TP53/HRD mutated – Black, Unknown - White.
Figure 2.
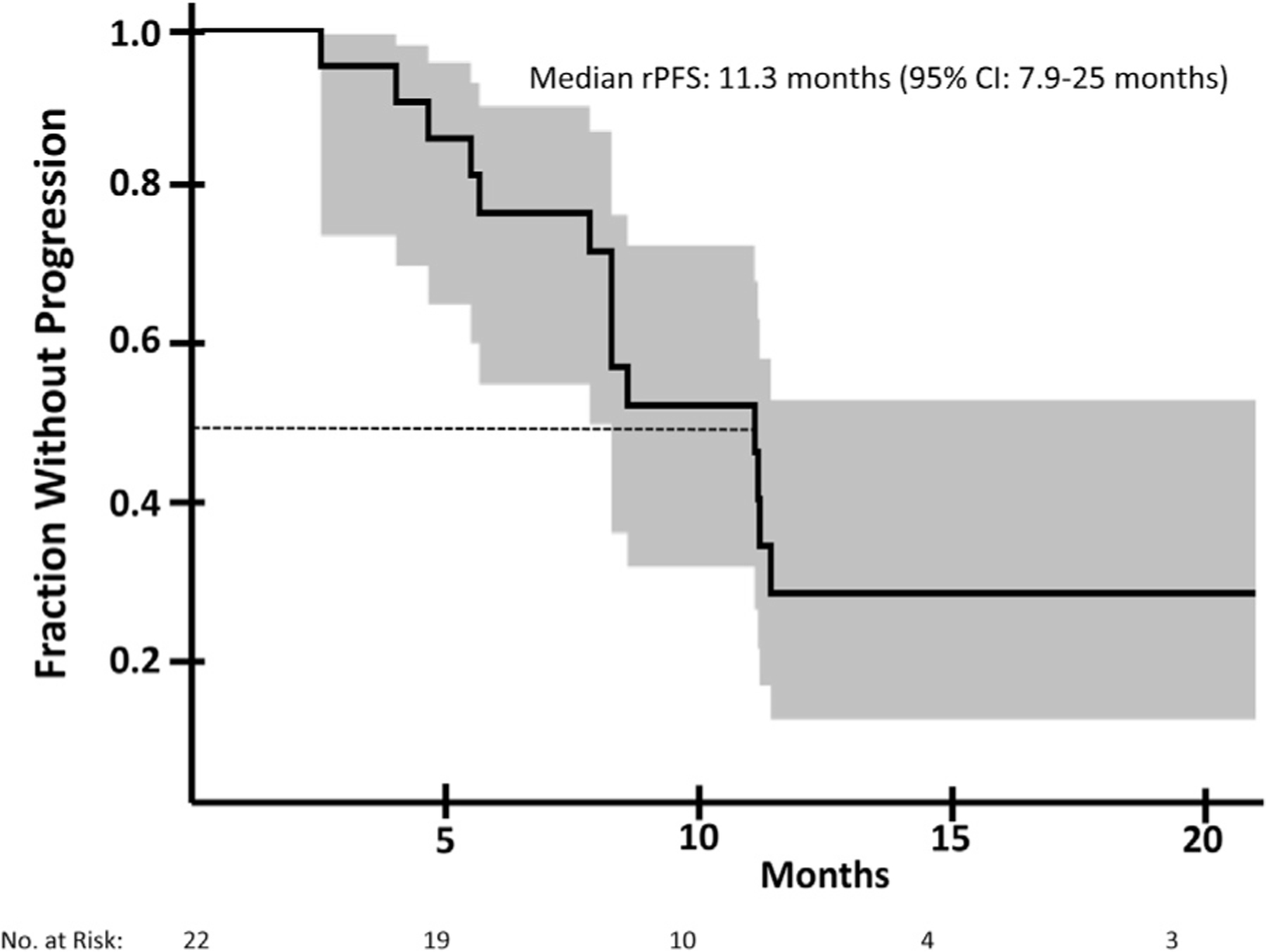
Radiographic progression-free survival in mCRPC patients with extreme clinical response to bipolar androgen therapy. Kaplan-Meier estimate of radiographic progression-free survival (rPFS) in mCRPC patients treated with BAT. The median rPFS is estimated at 11.3 months (95% confidence interval: 7.9–25.0 months). 95% confidence interval showed in shaded area.
Discussion
Preclinical data suggest that supraphysiologic levels of testosterone can engage the androgen receptor, resulting in:
inhibition of DNA relicensing during cell cycle progression
creation of double-stranded DNA breaks and
We speculated that an impaired underlying DNA repair mechanism may predispose mCRPC patients to extreme responses to BAT. We retrospectively identified those patients with mCRPC who achieved a PSA decline of 70% or more to BAT, in order to better characterize their clinical and molecular features. Interestingly, the majority of these extreme responders were identified as having inactivating BRCA2 and/or TP53 mutations. Somatic and germline mutations in BRCA2 and other HRD genes in men with metastatic prostate cancer are well documented, and have been associated with clinical benefit to PARP inhibitors in these patients.14 Studies also implicate ARID1A in maintaining genomic stability through protecting telomere cohesion as well as repair of double stranded DNA breaks, perhaps explaining the response to BAT observed in our ARID1A-altered patient.15,16 TP53 also has a key role in cellular DNA repair through its multiple functions as a master regulator.17 Our data suggest a preliminary association between impaired DNA repair response and favorable BAT activity. Prospective data of BAT in a biomarker-selected mCRPC population (ie, HRD or TP53 mutated) is warranted. Moreover, these data support investigation of combining BAT with PARP inhibitors, which may impair DNA repair in the absence of HRD mutations and induce synthetic lethality with BAT. Results of an ongoing trial studying this combination therapy are eagerly awaited (NCT03516812).
TP53 mutations in prostate cancer generally confer a poor prognosis due, in part, to poor responses to AR-targeted therapies.18 When observed in concert with PTEN or RB1 alterations, TP53 mutations may drive the development of AR-indifferent and/or neuroendocrine prostate cancer through a process called lineage plasticity.19 Given the dramatic response to BAT observed here in patients with TP53-mutated prostate cancer, we hypothesize that BAT may delay or reverse differentiation to the neuroendocrine phenotype. To this end, we are prospectively studying BAT in mCRPC patients who have at least two of the three following mutations: TP53, PTEN, RB1 (NCT02090114). A second trial (NCT03522064) is examining the role of BAT in mCRPC patients with HRD mutations. These data will be key to understanding the potential association between response to BAT and HRD and/or TP53 mutations.
There are several limitations to this analysis. First, this is a retrospective study of a small number of highly selected patients. Thus, prospective data are needed to confirm these findings, and such prospective trials are ongoing (NCT02090114, NCT03522064). Second, several patients in this analysis did not have molecular testing as part of their routine clinical care. It is possible that additional DNA sequencing would have shown less frequent TP53 and/or HRD mutations among these extreme responders. Third, this is a heterogeneous population with respect to prior therapies. Clinical response to BAT may have been affected by the number and type of prior treatments received, such as the number of AR-targeted therapies or exposure to chemotherapy. Fourth, we have not systematically examined the molecular features of BAT non-responders, nor have we compared clinical outcomes to BAT in patients with and without TP53 and/or HRD mutations. Thus, it is theoretically possible that the prevalence of these mutations may be just as high in the non-responding patients. However as a point of comparison, somatic mutations in HRD genes and/or TP53 occurred in only 247 of 429 patients (58%) with metastatic CRPC in the SU2C/PCF dream team cohort as determined through cBioPortal.20,21 Lastly, we do not know loss-of-heterozygosity status of the genes of interest in the study. Determining true biallelic vs. monoallelic inactivation would be helpful to determine the predictive nature of our findings.
These preliminary data suggest an enrichment of TP53 and HRD mutations among men with mCRPC achieving extreme responses to BAT, supporting prior preclinical studies.8 These retrospective data are now being confirmed in biomarker-selected prospective clinical trial that are likely to shed further light on the working hypothesis that supraphysiologic androgens may exert their effect on prostate cancer through AR-mediated DNA damage.
Clinical Practice Points
As BAT becomes more commonplace in the treatment of patients with metastatic castration-resistant prostate cancer (mCRPC), further study will be needed to identify a biomarker of response.
We know that the PSA response rate to BAT is 20–25% in a heavily pretreated population. Preclinical data suggest that impaired DNA repair may enrich for response to BAT. Preliminary clinical studies have also shown this to be the case.
In our study, all patients with mCRPC that achieved a deep PSA response to BAT harbored a pathogenic mutation in TP53 and/or a homologous recombination DNA repair gene. This finding may direct clinicians when considering BAT as a treatment option for patients with advanced mCRPC that are asymptomatic and have a specific molecular profile. Our study should also inform future clinical trials involving BAT.
Supplementary Material
Acknowledgments
The project was supported by the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins NIH grant P30 CA006973, R01 CA184012, Department of Defense (W81XWH1910724) and a PCF Young Investigator Award. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
Credit Author Statement
Mark C. Markowski – Conceptualization, Methodology, Formal Analysis, Writing – Original Draft; Sushant Kachhap – Methodology, Formal Analysis, Writing – Review and Editing; Angelo M. De Marzo – Methodology, Resources, Writing – Review and Editing, Supervision; Laura A. Sena – Methodology, Writing – Review and Editing; Jun Luo – Resources, Writing – Review and Editing, Supervision; Samuel R. Denmeade – Resources, Writing – Review and Editing, Supervision, Funding Acquisition; Emmanuel S. Antonarakis – Conceptualization, Methodology, Formal Analysis, Funding Acquisition, Writing – Review and Editing, Supervision, Funding Acquisition.
Prior Presentation of Work: 2021 Genitourinary Cancers Symposium (GU ASCO).
Disclosure
The authors have stated that they have no conflicts of interest.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.clgc.2021.08.001.
References
- 1.Denmeade SR, Isaacs JT. Bipolar androgen therapy: the rationale for rapid cycling of supraphysiologic androgen/ablation in men with castration resistant prostate cancer. Prostate 2010;70:1600–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schweizer MT, Antonarakis ES, Wang H, et al. Effect of bipolar androgen therapy for asymptomatic men with castration-resistant prostate cancer: results from a pilot clinical study. Sci Transl Med 2015;7 269ra262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schweizer MT, Wang H, Luber B, et al. Bipolar androgen therapy for men with androgen ablation naive prostate cancer: results from the phase II BATMAN study. Prostate 2016;76:1218–1226. [DOI] [PubMed] [Google Scholar]
- 4.Teply BA, Wang H, Luber B, et al. Bipolar androgen therapy in men with metastatic castration-resistant prostate cancer after progression on enzalutamide: an open-label, phase 2, multicohort study. Lancet Oncol 2018;19:76–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markowski MC, Wang H, Sullivan R, et al. A multicohort open-label phase II trial of bipolar androgen therapy in men with metastatic castration-resistant prostate cancer (RESTORE): a comparison of post-abiraterone versus post-enzalutamide cohorts. Eur Urol 2020;79:692–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denmeade SR, Wang H, Agarwal N, et al. TRANSFORMER: a randomized phase II study comparing bipolar androgen therapy versus enzalutamide in asymptomatic men with castration-resistant metastatic prostate cancer. J Clin Oncol 2021;39:1371–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haffner MC, Aryee MJ, Toubaji A, et al. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet 2010;42:668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bastus NC, Boyd LK, Mao X, et al. Androgen-induced TMPRSS2:ERG fusion in nonmalignant prostate epithelial cells. Cancer Res 2010;70:9544–9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chatterjee P, Schweizer MT, Lucas JM, et al. Supraphysiological androgens suppress prostate cancer growth through androgen receptor-mediated DNA damage. J Clin Invest 2019:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lam HM, Nguyen HM, Labrecque MP, et al. Durable response of enzalutamide-resistant prostate cancer to supraphysiological testosterone is associated with a multifaceted growth suppression and impaired DNA damage response transcriptomic program in patient-derived xenografts. Eur Urol 2020;77:144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teply BA, Kachhap S, Eisenberger MA, Denmeade SR. Extreme response to high-dose testosterone in BRCA2- and ATM-mutated prostate cancer. Eur Urol 2017;71:499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Litvinov IV, Vander Griend DJ, Antony L, et al. Androgen receptor as a licensing factor for DNA replication in androgen-sensitive prostate cancer cells. Proc Natl Acad Sci 2006;103:15085–15090 U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vander Griend DJ, Litvinov IV, Isaacs JT. Stabilizing androgen receptor in mitosis inhibits prostate cancer proliferation. Cell Cycle 2007;6:647–651. [DOI] [PubMed] [Google Scholar]
- 14.Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med 2015;373:1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao B, Lin J, Rong L, et al. ARID1A promotes genomic stability through protecting telomere cohesion. Nat Commun 2019;10:4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen J, Peng Y, Wei L, et al. ARID1A deficiency impairs the DNA damage checkpoint and sensitizes cells to PARP inhibitors. Cancer Discov 2015;5:752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res 1991;51:6304–6311. [PubMed] [Google Scholar]
- 18.Maughan BL, Guedes LB, Boucher K, et al. p53 status in the primary tumor predicts efficacy of subsequent abiraterone and enzalutamide in castration-resistant prostate cancer. Prostate Cancer Prostatic Dis 2018;21:260–268. [DOI] [PubMed] [Google Scholar]
- 19.Aparicio AM, Shen L, Tapia EL, et al. Combined tumor suppressor defects characterize clinically defined aggressive variant prostate cancers. Clin Cancer Res 2016;22:1520–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abida W, Cyrta J, Heller G, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


