Abstract
CYP3A4, CYP3A5, and CYP3A7, which are located in a multigene locus (CYP3A), play crucial roles in drug metabolism. To understand the highly variable hepatic expression of CYP3As, regulatory network analyses have focused on transcription factors (TFs). Since long non-coding RNAs (lncRNAs) likely contribute to such networks, we assessed the regulatory effects of both TFs and lncRNAs on CYP3A expression in the human liver and small intestine, main organs of CYP3A expression. Using weighted gene co-expression network analysis (WGCNA) of GTEx v8 RNA expression data and multiple stepwise regression analysis, we constructed TF-lncRNA-CYP3A co-expression networks. Multiple lncRNAs and TFs displayed robust associations with CYP3A expression that differed between liver and small intestines (LINC02499, HNF4A-AS1, AC027682.6, LOC102724153, and RP11-503C24.6), indicating that lncRNAs contribute to variance in CYP3A expression in both organs. Of these, HNF4A-AS1 had been experimentally demonstrated to affect CYP3A expression. Incorporating ncRNAs into CYP3A expression regulatory network revealed additional candidate TFs associated with CYP3A expression. These results serve as a guide for experimental studies on lncRNA-TF regulation of CYP3A expression in the liver and small intestines.
Keywords: cytochrome P450, WGCNA, transcription factor, long non-coding RNA, regulatory network
1. Introduction
As a key subfamily of cytochrome P450 (CYP) enzymes, CYP3A enzymes contribute to the metabolism of more than 50% of all marketed drugs [1]. Members of this subfamily include CYP3A4, CYP3A5, CYP3A7, and CYP3A43. CYP3A4 and CYP3A5 are the primary metabolizing enzymes of the CYP3A subfamily. CYP3A7 is primarily expressed in the fetus and newborn, but also exhibits relatively high expression in some adult livers [2]. The liver and small intestine are the main organs of drug metabolism and express high CYP3A levels, except for CYP3A43 [3,4], which is therefore not considered further here.
CYP3A expression displays substantial inter-individual variability in the human liver and small intestine [3,4], contributing to variable drug responses. Regulatory mechanisms at transcriptional and post-transcriptional levels appear to play the primary role in variable CYP3A expression [5]. It is critical to uncover the regulatory mechanism of CYP3A expression to improve the therapeutic effect and reduce the side effects of CYP3A-related drugs. By binding to cis-acting elements of CYP3As, multiple transcription factors (TFs) and their networks can regulate CYP3A expression, including progesterone X receptor (PXR; NR1I2), constitutive androgen receptor (CAR; NR1I3), hepatocyte nuclear factor 4 alpha (HNF4α; NR2A1), vitamin D receptor (VDR; NR1I1), glucocorticoid receptor (GR; NR3C1), Yin Yang 1 (YY1), and estrogen receptor α (ESR1) [3,6,7]. Evidence suggests that the CYP3A gene cluster represents a regulome that is characterized by interacting enhancer/suppressor domains. Collins et al. [8] discovered that some distal cis-acting regulatory elements can simultaneously affect CYP3A4, 3A5, and 3A7 expression because of chromatin looping. Therefore, combined analysis of CYP3As can identify overlapping regulators, addressing coordinate expression.
Published network studies on CYP3A expression analyzed only the associations between TFs and CYP3A expression in liver [5,7,9]. Whereas the pervasive role of non-coding RNAs (ncRNAs) is well established [10,11], any regulatory roles of ncRNAs in CYP3A regulation remain unexplored. Therefore, inclusion of ncRNAs in co-expression networks of TFs and ncRNAs with CYP3As has the potential to identify novel factors regulating CYP3A expression. Since CYP3A regulation may differ between the liver and small intestine [12], we included both tissues in the analysis to assess tissue-specific CYP3A regulation. NcRNAs can be divided into housekeeping ncRNAs and regulatory ncRNAs. Regulatory ncRNAs could be further classified as small non-coding RNAs (sncRNAs) (<200 nucleotides) and long non-coding RNAs (lncRNAs) (>200 nucleotides). The main classes of small ncRNAs are microRNAs (miRNAs), small interfering RNAs (siRNAs), and piwi-interacting RNAs (piRNAs). The lncRNAs, including cytoplasmic and nuclear lncRNAs, microRNA precursors, circular RNAs, and more, have diverse functions [13]. We focus on lncRNAs available in large databases generated with next generation RNAseq methodology.
Unlike methods that focus on single genes or a few genes [14], weighted gene co-expression network analysis (WGCNA) can transform gene expression data into co-expression modules, providing insight into signaling and regulatory networks [15]. In addition, WGCNA can identify RNAs with low abundance that may play important regulatory roles in biological responses [16]. Several studies have shown that WGCNA identifies modules and pathways and lncRNAs using gene clustering [9,17,18,19,20]. The Genotype-Tissue Expression (GTEx) database provides gene expression data across multiple human tissues [21]. After extracting GTEx v8 data of TFs and lncRNA expression in the liver and small intestine, we tested whether lncRNAs contribute to regulation of CYP3A expression, expanding previous CYP3A-TF networks with lncRNAs acting as potential network links and modulators.
2. Materials and Methods
2.1. GTEx v8 Data and Data Pre-Process
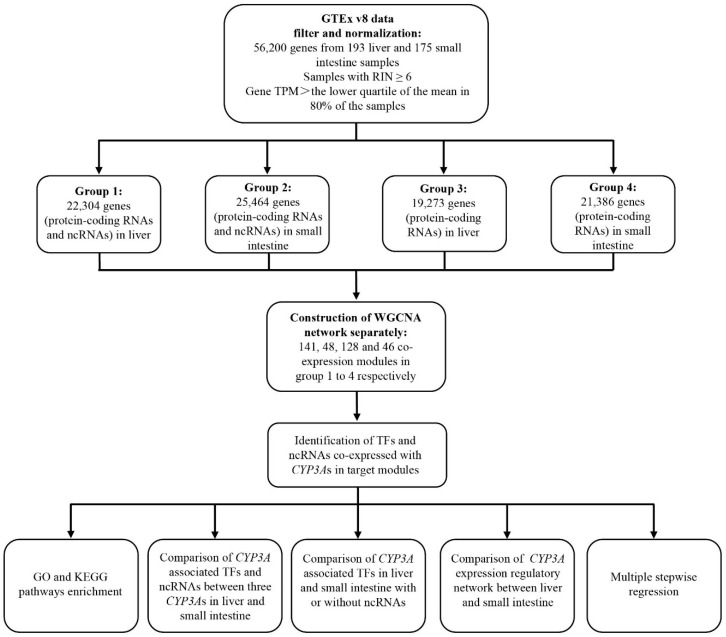
The GTEx v8 data dataset (https://gtexportal.org/home/ (accessed on 19 February 2020)) provides 56,200 genes for extraction from 193 liver and 175 small intestine samples [21]. Donor phenotype, RNAseq data (TPM, transcripts per million), and data processing information of samples were also obtained from the GTEx v8 dataset. Since most genes express multiple distinct transcripts, the sum of all transcripts TPM value of each gene was used as the expression value of the gene, and only the samples with RNA integrity number (RIN) ≥ 6 were selected. For each gene, we calculated the mean expression values for the liver and small intestine, and set the lower quartile of the means of all genes as the threshold. Next, genes with an expression value over the threshold (lower quartile of the mean) in more than 80% of the samples were selected, identifying 22,304 genes in the liver and 25,464 genes in the small intestines. Following the GTEx data processing method reported by Somekh et al. [22], we performed quantile normalization within each tissue, then transformed gene TPM to log2(TPM + 1), and adjusted for the effect of ischemic time by regressing the log2(TPM + 1) expression values with ischemic time. The annotations of gene types were obtained from Gencode v33 (https://www.gencodegenes.org/ (accessed on 14 March 2020)) [23]. Human transcription factors were downloaded from the animalTFDB 3.0 database (http://bioinfo.life.hust.edu.cn/HumanTFDB#!/ (accessed on 20 April 2020)) [24]. Genes encoding the RNAs were then divided into four groups: group 1 consisted of 22,304 genes in the liver; group 2 consisted of 25,464 genes in the small intestine; ncRNAs were then removed from group 1 to yield group 3 (19,273 genes) in the liver, and from group 2, generating group 4 (21,386 genes) in the small intestine. The demographics and gene types of the liver and small intestine samples in GTEx v8 dataset are shown in Table 1. The experimental approach is shown in Figure 1.
Table 1.
Demographics and gene types of the liver and small intestine samples in GTEx v8 dataset.
| Liver (Mean ± SD) | Small Intestine (Mean ± SD) | |
|---|---|---|
| Total number (n) | 193 | 175 |
| Age (years) | 54.48 ± 11.04 | 47.66 ± 13.63 |
| Sex (male/female) (n) | 131/62 | 109/66 |
| Race(White/Black/Asian/unknown) | 169/19/4/1 | 144/27/2/2 |
| Height (in) | 68.11 ± 3.74 | 67.57 ± 3.80 |
| Weight (lb) | 179.03 ± 36.15 | 179.62 ± 34.47 |
| BMI (kg/m2) | 26.97 ± 4.14 | 27.51 ± 3.98 |
| Liver diseases (Yes/No) | 3/190 | 3/172 |
| Gastrointestinal diseases (Yes/No) | 0/193 | 0/175 |
| Total number of genes (n) | 56,200 | |
| Protein-coding genes (n) a | 19,646 | |
| Pseudogenes (n) a | 14,897 | |
| To be Experimentally Confirmed (TEC) a | 1008 | |
| Unidentified genes (n) b | 497 | |
| NcRNAs (n) a,c | 20,152 | |
| Long non-coding RNAs (lncRNA) (n) a,d | 13,731 | |
| MicroRNAs precursors (miRNA) (n) a,d | 1576 | |
| Miscellaneous other RNAs (misc_RNA) (n) a,d | 2007 | |
| Small nuclear RNAs (snRNA) (n) a,d | 1864 | |
| Small nucleolar RNAs (snoRNA) (n) a,d | 847 | |
| Ribosomal RNAs (rRNA) (n) a,d | 51 | |
| Small Cajal body-specific RNAs (scaRNA) (n) a,d | 40 | |
| Mitochondrial transfer RNAs (Mt_tRNA) (n) a,d | 22 | |
| Mitochondrial ribosomal RNAs (Mt_rRNA) (n) a,d | 2 | |
| Ribozymes (n) a,d | 6 | |
| Small non-coding RNAs (sRNA) (n) a,d | 4 | |
| Small cytoplasmic RNA (scRNA) (n) a,d | 1 | |
| VaultRNA (vtRNA) (n) a,d | 1 | |
a The annotations of gene types were obtained from Gencode v33 (https://www.gencodegenes.org/ accessed on 7 July 2022). b Gene types cannot be obtained in Gencode v33 database. NcRNAs d are the further classification of ncRNAs. c GTEx: Genotype-Tissue Expression project. NcRNAs: non-coding RNAs. BMI: Body mass index.
Figure 1.
Study design. GTEx: Genotype-Tissue Expression project. RIN: RNA integrity number. TPM: transcripts per million. WGCNA: weighted gene co-expression network analysis. TFs: transcription factors. NcRNAs: non-coding RNAs.
2.2. Construction of WGCNA Network and Detection of Modules
The construction of the WGCNA network and detection of modules were conducted using the WGCNA R package [15]. The power known as soft threshold (β) for the four groups of datasets was pre-calculated using the pickSoftThreshold function. BlockwiseModules, an automatic block-wise network construction function for large datasets, was used to construct an unsigned co-expression network and detect modules. The correlation method used in WGCNA was biweight midcorrelation (bicor), as it is more robust than the Pearson correlation. Modules, termed target modules, in each dataset contained three target genes (CYP3A4, 3A5, and 3A7), and these were extracted for further analysis.
2.3. Function Enrichment Analysis in Target Modules
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses, based on hypergeometric distribution for genes in each target module with p value <0.05 and q value <0.05, were performed in R using the ClusterProfiler [25] R package.
2.4. Identification of TFs and ncRNAs Co-Expressed with CYP3As in Target Modules
For each module, WGCNA served to calculate the module eigengene E (ME), which is the first principal component of a given module and represents the gene expression profile of the module. For each gene, WGCNA calculated the correlation between gene expression value and ME as module membership (kME). The kME value represents the degree of membership of the gene to the module and is highly correlated with the connectivity of the gene. For a given module, the larger the |kME| of the gene, the more likely it is to be a hub gene [15]. In addition, WGCNA was used to calculate the weight value between two genes using the biweight midcorrelation method (bicor(A,B)). All gene pairs with a weight value >0.05 were retained. Amongst the four groups, the top 80 regulators (TFs and ncRNAs) were determined by selecting the highest weight values for each CYP3A, and these were chosen for further analysis. Their co-expression networks were performed using Cytoscape v.3.7.2 [26], and the positive and negative correlations were calculated using the bicor function in the WGCNA R package.
2.5. Functional Annotation of TFs and ncRNAs Co-Expressed with CYP3As in Target Modules
The ClusterProfiler [25] R package was used to annotate the top 80 regulators (TFs and ncRNAs) co-expressed with CYP3As along with the CYP3As in each target module from the four groups using the GO and KEGG function, animalTFDB 3.0 [24], miRBase (https://www.mirbase.org/ (accessed on 22 May 2020)) [27], and RNAcentral databases (https://rnacentral.org/ (accessed on 20 June 2020)) [28]. These were also combined with auxiliary annotations.
2.6. Multiple Stepwise Regression Analysis
A multiple stepwise regression analysis was performed using the SPSS software package (ver. 20.0; SPSS Inc., Chicago, IL, USA). The TPM values of the top 80 regulators (TFs and ncRNAs) co-expressed with CYP3As for each CYP3A were set as independent variables, and the TPM values of each CYP3A were set as dependent variables. A p value less than 0.05 was considered statistically significant.
The R scripts are available on GitHub: https://github.com/servicemanli/CYP3A_regulatory_network.git (accessed on 22 August 2022).
3. Results
3.1. Construction of the WGCNA Network and Expression Modules
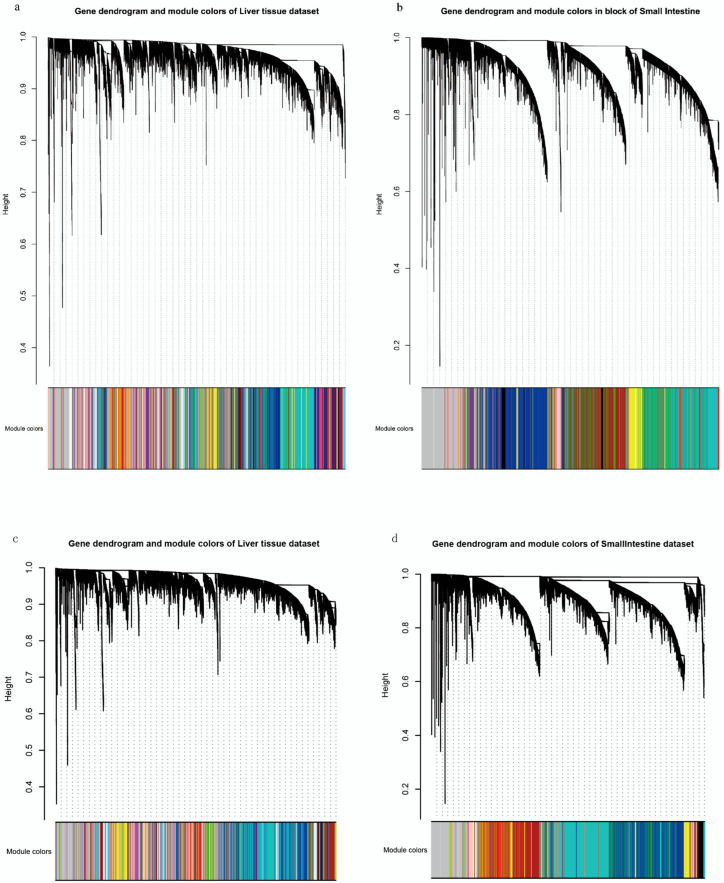
In this study, we applied WGCNA to explore CYP3A-TF networks with lncRNAs acting as potential network links and modulators, to determine whether CYP3As expression networks in the liver and small intestine are altered by the inclusion of ncRNAs. Thus, we used WGCNA to analyze and compare the following four groups of data extracted from GTEx v8: group 1 with all genes in the liver, group 2 with all genes in the small intestine, group 3 with protein-coding genes in the liver, and group 4 with protein-coding genes in the small intestine (ncRNAs were removed in group 3 and 4). Shown in Supplementary Figure S1, the lowest soft-thresholding powers were 4, 7, 4, and 7 for the four groups, respectively, for which the scale-free topology fit index (scale-free R2) reached 0.8. The appropriate soft-thresholding values emphasize strong gene–gene correlations. The correlation matrix was subsequently transformed into an adjacency matrix. Each adjacency matrix was normalized using a topological overlap measure (TOM). A dissimilarity matrix based on TOM was used to identify gene modules with a dynamic tree-cutting algorithm. Totals of 141, 48, 128, and 46 co-expression modules were identified in the four groups, respectively (Figure 2). In group 1, all three CYP3As were in the turquoise module, whereas in group 3, CYP3A5 and 3A7 were in the turquoise module and CYP3A4 was in the blue module. The number of genes within CYP3A-related modules also changed significantly from group 1 to group 3. These analyses revealed that liver modules containing CYP3As and genes related CYP3As changed after removal of the ncRNAs. The modules containing the CYP3As from group 1 to group 4 and the numbers of genes within the modules are listed in Table 2.
Figure 2.
Gene clustering tree and module identification. The top halves of the four plots represent the gene clustering tree constructed based on topological overlap, and the height on the y-axis represents distance between modules; the higher the height value, the less likely it is that the two modules are co-expressed. The lower half of each of the four plots represents the gene modules, and each colored row represents a color-coded module that contains a group of highly connected genes. (a) The first group, (b) second group, (c) third group and (d) fourth group.
Table 2.
Modules containing CYP3As identified from group 1 to 4 using weighted gene co-expression network analysis.
| Group | Tissue | Gene Type | Module | Module Size | CYP3A | Number of TFs | Number of ncRNAs |
|---|---|---|---|---|---|---|---|
| Group 1 | Liver | All genes | Turquoise | 3103 | CYP3A4, 3A5 and 3A7 | 161 | 390 lncRNAs and 5 miRNAs |
| Group 2 | Small intestine | All genes | Turquoise | 4866 | CYP3A4 and 3A5 | 339 | 658 lncRNAs and 3 miRNAs |
| Red | 1437 | CYP3A7 | 55 | 257 lncRNAs and 4 miRNAs | |||
| Group 3 | Liver | Protein coding genes | Turquoise | 3090 | CYP3A5 and 3A7 | 223 | NA |
| Blue | 1991 | CYP3A4 | 109 | NA | |||
| Group 4 | Small intestine | Protein coding genes | Blue | 4078 | CYP3A4 and 3A5 | 358 | NA |
| Green | 1809 | CYP3A7 | 83 | NA |
All genes in group 1 and 2 represent the genes that meet the criteria, including protein-coding genes and ncRNAs. The numbers for group 1 and 2 are 22,304 and 25,464, respectively. Protein-coding genes in group 3 and 4 represent the genes that meet the criteria, excluding ncRNAs. The numbers for group 3 and 4 are 19,273 and 21,386, respectively. NA represents not applicable. LncRNAs: Long non-coding RNAs. MiRNAs: microRNAs.
3.2. Identification of TFs and ncRNAs Co-Expressed with CYP3As in Target Modules
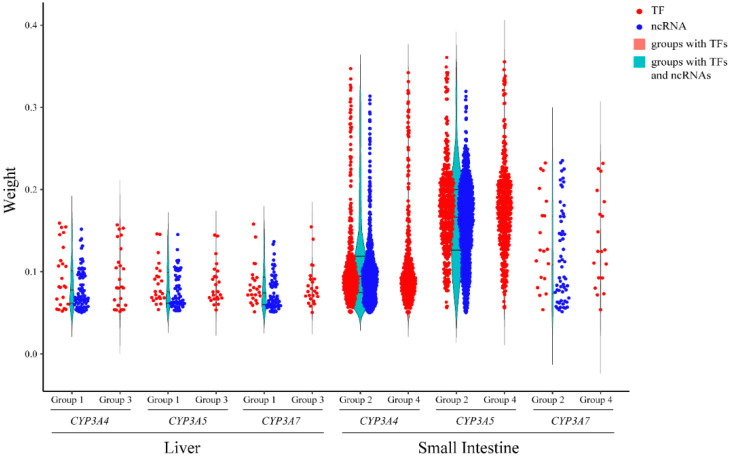
We next searched for ncRNAs and TFs in the same expression modules as CYP3As. For each CYP3A, the TFs and ncRNAs with weight values > 0.05 in the same module are shown in Supplementary Table S1. For group 1 and group 2 (liver and intestine + ncRNAs), the number of ncRNAs associated with each CYP3A exceeded the number of TFs (Supplementary Table S1). We compared the weight values of TFs and ncRNAs using an independent-sample t-test in groups 1 and 2, respectively. In group 1, for each CYP3A, there was no significant difference in the weight values between TFs and ncRNAs (data not shown). In group 2, for CYP3A4 and 3A5, the weight values of TFs were significantly higher than those of ncRNAs (CYP3A4: 0.12 ± 0.062 vs. 0.10 ± 0.046, p = 0.003; CYP3A5: 0.17 ± 0.057 vs. 0.16 ± 0.052, p = 3.10 × 10−4). For CYP3A7 in group 2, no significant difference was found in the weight values between TFs and ncRNAs (data not shown). These results show that multiple ncRNAs can reach weight values with CYP3As matching those of TFs in the liver. In the small intestine, TFs are more closely associated with CYP3A4 and 3A5 than ncRNAs.
We then ordered the potential regulators by weight and analyzed the associations between the top 80 potential regulators and each CYP3A expression from group 1 to 4 via biweight midcorrelation method. As shown in Supplementary Table S2, most of the potential regulators are significantly associated with CYP3A expression, even after Bonferroni correction. If there are multiple CYP3As in one module, the regulators often have similar associations between these CYP3As, whether positive or negative associations. Among the associations with CYP3As, some lncRNAs had lower p values compared to TFs, such as lncRNA LINC02499 with CYP3A5 (p = 5 × 10−23) in group 1; and lncRNA MIR194-2HG (miRNA precursor) with CYP3A5 (p = 1 × 10−98) and lncRNA EGFR-AS1 (anti-sense RNA) with CYP3A7 (p = 9 × 10−38) in group 2. These results indicate that lncRNAs are significantly associated with CYP3As in the liver and small intestine (Supplementary Table S2). In addition, we found that most lncRNAs are positively associated with CYP3A expression, including LINC02499, MIR194-2HG, and EGFR-AS1.
After removal of ncRNAs in the liver, the modules containing CYP3As were changed, and CYP3A-related TFs were changed. In group 1, the previously reported CYP3A-related TFs such as NR1I2, NR1I3, and ESR1 were in the turquoise module (same module as CYP3A4, 3A5 and 3A7) [7,29,30]. However, in group 3, NR1I2 and NR1I3 were in the turquoise module (same module as CYP3A5 and 3A7), whereas ESR1 was in blue module (same module as CYP3A4). In the small intestine, CYP3A-containing modules and CYP3A-related TFs were similar before and after the removal of ncRNAs. These results suggest that the removal of ncRNAs from the CYP3A alters inferred regulatory networks, especially in the liver.
3.3. Functional Enrichment Analysis in Target Modules
Next, we aimed to evaluate the functions of genes within the modules co-expressed with CYP3As. Functional enrichment analysis was performed on the seven modules containing CYP3As (turquoise module in group 1, turquoise and red modules in group 2, turquoise and blue modules in group 3, blue and green modules in group 4) by GO and KEGG analysis. Supplementary Figure S2 shows the most significantly enriched category for each module. The gene-enriched biological processes (BPs) of these modules were predominantly biometabolism and biosynthesis. The gene-enriched molecular functions (MFs) of this module were mainly enzyme activity, binding with other molecules, and substance-transporting activities. The module-enriched cellular components (CCs) included mitochondrial matrix and brush border membrane. The seven modules were enriched with multiple functional gene entries related to the CYP450 family (Supplementary Table S3). These functional enrichment results of module genes support the reliability of the WGCNA-constructed network and the identified modules.
3.4. Comparison of CYP3A Associated TFs in the Liver and Small Intestine with and without ncRNAs
We evaluated whether TFs and CYP3A regulatory networks were altered in the absence of ncRNAs. For each CYP3A, we compared the numbers of TFs associated with CYP3A with and without ncRNAs in the liver and small intestine.
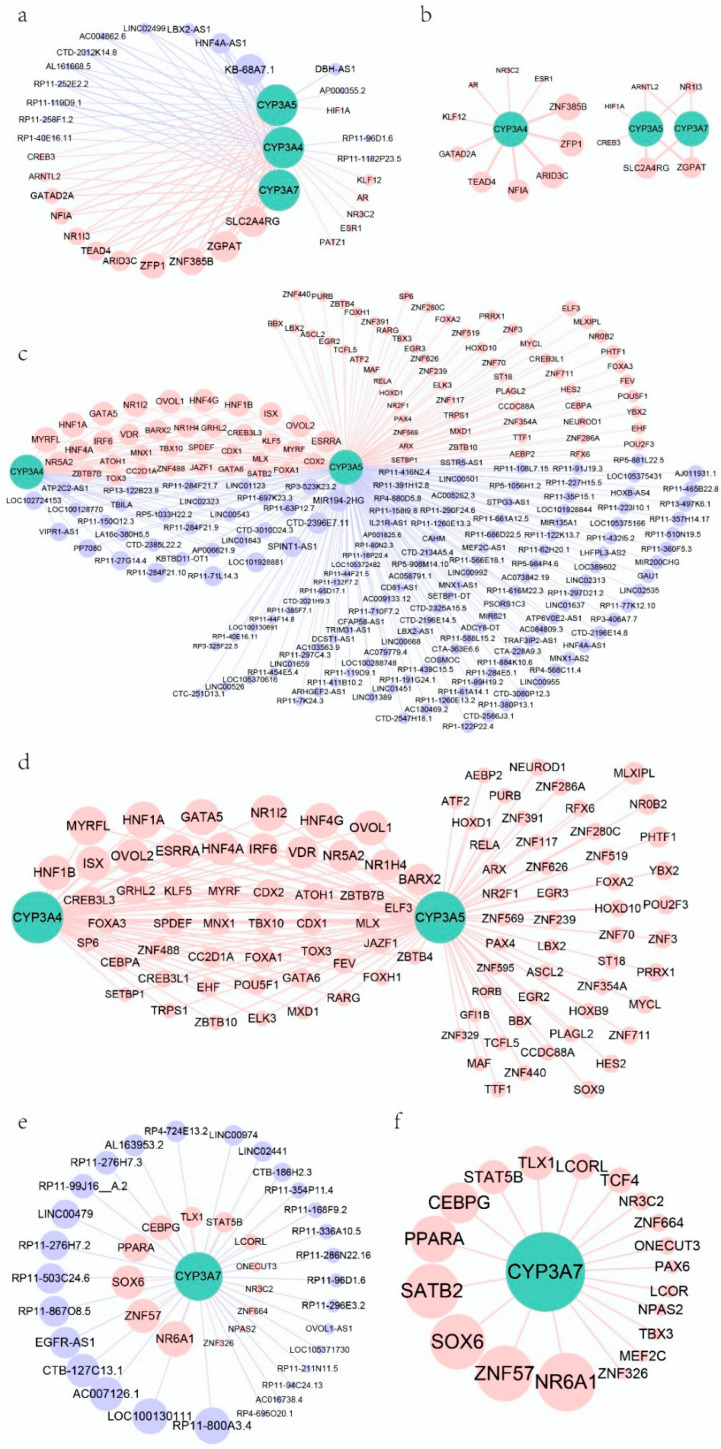
Focusing on liver expression with and without ncRNAs, most of the TFs associated with CYP3As overlapped with TFs found in group 1 when including ncRNAs (Supplementary Table S4 and Figure 3). Group 1 also had a subset of TFs that were not in group 3. Compared to the overlapping TFs, some of these TFs had higher weight values with CYP3As, such as ZGPAT (weight value with CYP3A4 was 0.16), ZNF385B (weight value with CYP3A5 was 0.14), and ARID3C (weight value with CYP3A5 was 0.14). We found that after removing ncRNAs, the numbers of TFs related to CYP3A4, 3A5, and 3A7 in the liver decreased. These results indicate that omission of ncRNAs in the liver can lead to missed relationships TFs associated with CYP3A expression. In addition, the regulatory networks for the three CYP3As in the liver differed between group 1 with ncRNAs and group 3 without ncRNAs (Figure 4a,b).
Figure 3.
Comparison of CYP3A-associated transcription factors (TFs) in liver and small intestine with and without ncRNAs. The red dots represent the overlapping TFs associated with each CYP3A in liver and small intestine with and without ncRNAs. The blue dots represent the ncRNAs in liver and small intestine (group 1 and group 2). The red violin plots represent all TFs associated with each CYP3A in liver and small intestine (group 3 and group 4), and the green violin plots represent all TFs and ncRNAs associated with each CYP3A in liver and small intestine (group 1 and group 2). The y-axis has the weight values calculated by WGCNA.
Figure 4.
The direct co-expression relationships among TFs, lncRNAs, and CYP3A4, 3A5, and 3A7. (a) The co-expression network of TFs, ncRNAs, and CYP3As in group 1 and all TFs and ncRNAs with weight values > 0.1. (b) The co-expression network in group 3 (weight value > 0.1). (c) The co-expression network of TFs, ncRNAs, CYP3A4, and 3A5 in group 2. (d) The co-expression network of TFs, CYP3A4, and 3A5 in group 4 (weight value > 0.2). (e) The co-expression network of TFs, ncRNAs, and CYP3A7 in group 2. (f) The co-expression network of TFs and CYP3A7 in group 4 (weight value > 0.1). The thicker the edges in (a,b), the larger the weight values. The larger the dots in (a–f), the larger the weight values. The size of dots represents weight value with CYP3A5 when there is more than one relationship among TFs, ncRNAs, and CYP3As. The red and blue dots represent TFs and ncRNAs, respectively.
In the small intestine, group 4 without ncRNAs, most TFs associated with CYP3A4 and 3A5, overlapped with TFs in group 3, including ncRNAs (Supplementary Table S4 and Figure 3). Similarly, the regulatory networks for CYP3A4 and 3A5 did not change substantially in small intestine upon removal of ncRNAs (Figure 4c,d).
3.5. Comparison of CYP3A-Associated TFs and ncRNAs between Three CYP3As in the Liver and Small Intestine
We next asked whether TFs and ncRNAs are shared between different CYP3As in the liver and small intestine. In group 1 and group 2 (liver and intestines + ncRNAs), the numbers of TFs and ncRNAs associated with each CYP3A were compared among the three CYP3As. In group 1, most TFs and ncRNAs overlapped between all three CYP3As (Supplementary Table S5 and Figure 5). In the small intestine, most TFs and ncRNAs overlapped only between CYP3A4 and 3A5 (Supplementary Table S5 and Figure 5). Since CYP3A7 resides in a different module, its regulatory mode appears to differ from those of CYP3A4 and 3A5. These results indicate that CYP3A4, 3A5, and 3A7 are under similar regulatory control in the liver. Meanwhile, CYP3A4 and 3A5 but not 3A7 are under similar regulatory control in the small intestine.
Figure 5.
Comparison of CYP3A-associated TFs and ncRNAs with three CYP3As in the liver and small intestine. The y-axis is for the weight values calculated by WGCNA. Red and blue dots, respectively, represent the overlapping TFs and ncRNAs associated with two different CYP3As in the liver or small intestine. The violin plots with same color represent the same CYP3A in the same tissue.
3.6. Comparison of CYP3A Expression Network between Liver and Small Intestines
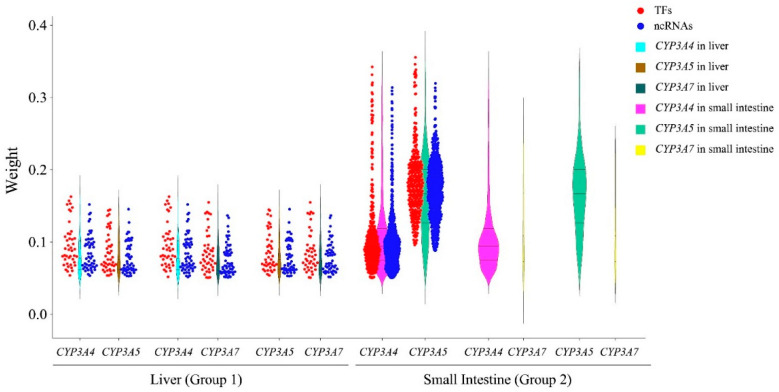
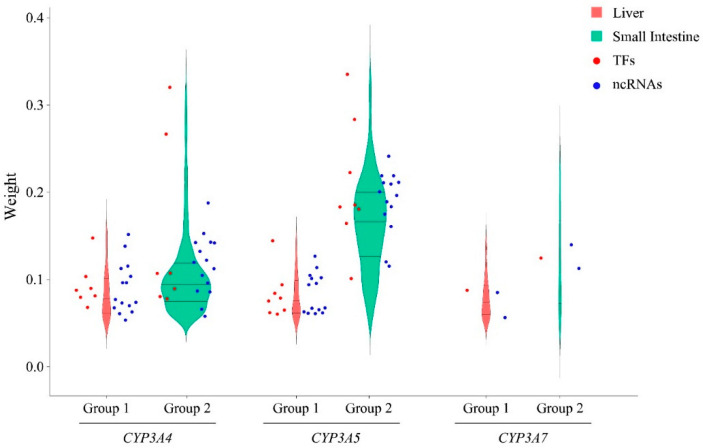
To compare CYP3A-associated TFs and ncRNAs in the liver and small intestine, TFs and ncRNAs associated with each CYP3A with a weight value greater than 0.05 from groups 1 and 2 were selected. As shown in Supplementary Table S6 and Figure 6, most TFs and ncRNAs differ between the liver and small intestines for each CYP3A. CYP3A4 and 3A5 are associated with more TFs and ncRNAs in the small intestines than in the liver. In addition, the average weight values of TFs and ncRNAs associated with CYP3As in the small intestine were higher than that in liver. These results indicate that different TFs and ncRNAs are involved in the regulatory modes of CYP3A expression in the liver and small intestine.
Figure 6.
Comparison of CYP3A expression regulatory networks between liver and small intestine. The y-axis displays the weight values calculated by WGCNA. Red and blue dots represent the overlapping TFs and ncRNAs associated with each CYP3A in the liver and small intestine, respectively. The red and green violin plots represent all TFs and ncRNAs associated with each CYP3A in the liver and small intestine, respectively.
3.7. Multiple Stepwise Regression Analysis for the Association with CYP3A4,5,7 RNA Expression Values
We reasoned that ncRNAs and TFs could explain part of the variance in CYP3As. To this end, for the liver and small intestine, the top 80 regulators (TFs and ncRNAs) of each CYP3A (Supplementary Table S1) were selected. Of the 80 regulators, all the ncRNAs were lncRNAs. The TPM values (level of expression) of the 80 regulators were set as independent variables, and TPM values of each CYP3A were set as dependent variables. Then, we conducted a multiple stepwise regression analysis to evaluate the contribution of regulators to CYP3A’s expression variance. In the liver, the TFs and lncRNAs in the final models explained 68.1%, 69.5%, and 44.5% of the expression variance in CYP3A4, 3A5, and 3A7, respectively (Table 3). The results reveal that the top significant variables, ESR1, LINC02499, and HNF4A-AS1 explained 36.4%, 48.5%, and 23.8% of the expression variance in CYP3A4, CYP3A5, and CYP3A7, respectively (Table 3). After the removal of the lncRNAs from the top 80 variables, the TFs in the final models explained 45.1%, 55.3%, and 26.0% of the expression variance in CYP3A4, 3A5, and 3A7, respectively. The most significant TFs, ESR1, NR1I2, and ZBTB47, explained 36.4%, 40.5%, and 21.5% of expression variance in CYP3A4, CYP3A5, and CYP3A7, respectively (Supplementary Table S7).
Table 3.
Transcriptional factors and non-coding RNAs contributing to CYP3A expression in the liver and the small intestine.
| Liver (n = 193) | |||||||
|---|---|---|---|---|---|---|---|
| Dependent Variable | Independent Variable | Type | R-Square | p-Value | R-Square Change | Beta | p-Value |
| CYP3A4 | 68.11% | 3.19 × 10−36 | |||||
| ESR1 | humanTF | 36.36% | 0.488 | 1.44 × 10−9 | |||
| DBH-AS1 | lncRNA | 0.80% | −0.462 | 1.23 × 10−6 | |||
| AL161668.4 | lncRNA | 2.17% | −0.411 | 3.74 × 10−6 | |||
| AC008537.3 | lncRNA | 1.94% | 0.334 | 3.88 × 10−6 | |||
| HNF4A-AS1 | lncRNA | 2.60% | 0.515 | 5.18 × 10−6 | |||
| AC027682.6 | lncRNA | 2.48% | −0.346 | 1.29 × 10−4 | |||
| HIF1A | humanTF | 0.86% | −0.262 | 1.84 × 10−3 | |||
| AC004160.2 | lncRNA | 3.95% | 0.186 | 2.75 × 10−3 | |||
| LINC02499 | lncRNA | 2.59% | −0.266 | 6.23 × 10−3 | |||
| CTD-2325A15.5 | lncRNA | 1.46% | −0.172 | 1.12 × 10−2 | |||
| AC122713.2 | lncRNA | 0.94% | −0.179 | 1.13 × 10−2 | |||
| ZNF385B | humanTF | 0.79% | 0.234 | 1.82 × 10−2 | |||
| AL359715.3 | lncRNA | 0.84% | 0.146 | 2.11 × 10−2 | |||
| HMGB3 | humanTF | 0.82% | −0.159 | 3.40 × 10−2 | |||
| ZGPAT | humanTF | 0.73% | 0.221 | 3.53 × 10−2 | |||
| CYP3A5 | 69.52% | 2.10 × 10−39 | |||||
| LINC02499 | lncRNA | 48.48% | 0.514 | 3.84 × 10−8 | |||
| AL161668.4 | lncRNA | 1.50% | −0.407 | 9.04 × 10−7 | |||
| HLF | humanTF | 2.48% | 0.302 | 2.16 × 10−4 | |||
| CTD-2325A15.5 | lncRNA | 1.77% | −0.242 | 2.49 × 10−4 | |||
| AL122035.2 | lncRNA | 0.71% | 0.211 | 9.84 × 10−4 | |||
| HMGB3 | humanTF | 1.19% | −0.214 | 2.67 × 10−3 | |||
| ZNF680 | humanTF | 0.86% | −0.203 | 3.44 × 10−3 | |||
| NFYC | humanTF | 0.74% | 0.174 | 8.33 × 10−3 | |||
| AC008537.3 | lncRNA | 0.99% | 0.179 | 1.11 × 10−2 | |||
| NR1I2 | humanTF | 0.41% | 0.187 | 1.86 × 10−2 | |||
| GCFC2 | humanTF | 2.01% | 0.157 | 2.45 × 10−2 | |||
| ZNF473 | humanTF | 0.62% | 0.146 | 2.48 × 10−2 | |||
| AC137056.1 | lncRNA | 0.47% | −0.142 | 3.26 × 10−2 | |||
| CYP3A7 | 44.52% | 1.64 × 10−19 | |||||
| HNF4A-AS1 | lncRNA | 23.84% | 0.875 | 1.84 × 10−8 | |||
| DCXR-DT | lncRNA | 4.53% | −0.447 | 3.24 × 10−5 | |||
| AL161668.4 | lncRNA | 2.32% | −0.431 | 8.09 × 10−5 | |||
| AL117382.3 | lncRNA | 1.52% | −0.267 | 1.83 × 10−2 | |||
| AL513327.1 | lncRNA | 1.98% | −0.169 | 2.49 × 10−2 | |||
| AC104809.1 | lncRNA | 1.48% | 0.193 | 2.88 × 10−2 | |||
| AC083841.1 | lncRNA | 1.44% | 0.186 | 3.04 × 10−2 | |||
| NR3C2 | humanTF | 2.48% | 0.154 | 7.77 × 10−2 | |||
| RP11-669E14.4 | lncRNA | 4.94% | 0.151 | 9.43 × 10−2 | |||
| Small Intestine (n = 175) | |||||||
| Dependent Variable | Independent Variable | Type | R-Square | p-Value | R-Square change | Beta | p-Value |
| CYP3A4 | 95.67% | 3.18 × 10−101 | |||||
| NR1I2 | humanTF | 1.46% | 0.937 | 1.05 × 10−13 | |||
| CREB3L3 | humanTF | 2.34% | 0.427 | 1.28 × 10−8 | |||
| LOC102724153 | lncRNA | 83.59% | 0.362 | 5.84 × 10−7 | |||
| FOXA3 | humanTF | 1.52% | −0.323 | 1.23 × 10−5 | |||
| BARX2 | humanTF | 0.91% | −0.293 | 2.24 × 10−5 | |||
| CTD-2547H18.1 | lncRNA | 0.63% | 0.207 | 1.03 × 10−4 | |||
| GATA6 | humanTF | 0.19% | 0.186 | 1.49 × 10−4 | |||
| LINC02323 | lncRNA | 3.60% | −0.198 | 1.95 × 10−4 | |||
| OVOL2 | humanTF | 0.21% | −0.298 | 3.63 × 10−4 | |||
| RP11-284F21.10 | lncRNA | 0.20% | −0.214 | 1.93 × 10−3 | |||
| SPINT1-AS1 | lncRNA | 0.17% | 0.209 | 3.36 × 10−3 | |||
| LINC02313 | lncRNA | 0.28% | 0.121 | 4.17 × 10−3 | |||
| LOC105375431 | lncRNA | 0.43% | −0.123 | 1.60 × 10−2 | |||
| MNX1-AS2 | lncRNA | 0.15% | −0.116 | 1.93 × 10−2 | |||
| CYP3A5 | 98.09% | 7.01 × 10−127 | |||||
| NR1I2 | humanTF | 0.64% | 0.771 | 2.67 × 10−22 | |||
| RP11-63P12.7 | lncRNA | 1.64% | 0.255 | 4.15 × 10−11 | |||
| MLXIPL | humanTF | 0.55% | 0.132 | 1.13 × 10−9 | |||
| HNF1B | humanTF | 0.28% | 0.409 | 4.42 × 10−9 | |||
| RP3-523K23.2 | lncRNA | 0.15% | −0.264 | 1.94 × 10−6 | |||
| ZBTB7B | humanTF | 0.20% | −0.190 | 1.25 × 10−5 | |||
| KBTBD11-OT1 | lncRNA | 0.14% | −0.137 | 1.29 × 10−5 | |||
| RP11-150O12.3 | lncRNA | 0.25% | −0.143 | 1.07 × 10−4 | |||
| RP11-284F21.9 | lncRNA | 0.30% | 0.127 | 7.77 × 10−4 | |||
| TBILA | lncRNA | 0.12% | 0.121 | 1.46 × 10−3 | |||
| TBX10 | humanTF | 0.38% | −0.107 | 3.69 × 10−3 | |||
| RP11-284F21.10 | lncRNA | 0.08% | −0.138 | 7.97 × 10−3 | |||
| LINC00543 | lncRNA | 0.35% | −0.093 | 8.63 × 10−3 | |||
| MNX1 | humanTF | 0.08% | 0.122 | 1.33 × 10−2 | |||
| RP13-497K6.1 | lncRNA | 0.11% | 0.050 | 1.52 × 10−2 | |||
| LOC100128770 | lncRNA | 0.05% | 0.074 | 4.77 × 10−2 | |||
| CYP3A7 | 80.38% | 4.53 × 10−50 | |||||
| RP11-94C24.13 | lncRNA | 6.16% | 0.470 | 5.77 × 10−9 | |||
| FOXD1 | humanTF | 2.04% | −0.347 | 3.14 × 10−7 | |||
| NR6A1 | humanTF | 0.91% | 0.367 | 2.38 × 10−4 | |||
| NR3C2 | humanTF | 2.01% | 0.307 | 2.40 × 10−4 | |||
| ONECUT3 | humanTF | 0.77% | −0.209 | 8.86 × 10−4 | |||
| RP11-503C24.6 | lncRNA | 62.50% | 0.345 | 9.47 × 10−4 | |||
| RP11-794G24.1 | lncRNA | 1.78% | −0.234 | 1.50 × 10−3 | |||
| ZNF664 | humanTF | 0.82% | −0.225 | 1.94 × 10−3 | |||
| RP11-459I19.1 | lncRNA | 0.76% | −0.212 | 2.21 × 10−3 | |||
| EGFR-AS1 | lncRNA | 0.60% | 0.310 | 5.12 × 10−3 | |||
| LOC105373958 | lncRNA | 0.65% | 0.180 | 5.81 × 10−3 | |||
| RAG1 | humanTF | 0.69% | 0.190 | 1.04 × 10−2 | |||
| RP11-211N11.5 | lncRNA | 0.70% | −0.164 | 1.78 × 10−2 | |||
Βeta: standardized coefficients.
In small intestine, TFs plus lncRNAs in the final models accounted for 95.7%, 98.1%, and 80.4% of the expression variance in CYP3A4, 3A5, and 3A7, respectively (Table 3). The results reveal that the significant variables LOC102724153 and RP11-503C24.6 explained 83.6% and 62.5% of the expression variance of CYP3A4 and CYP3A7, respectively (Table 3). For CYP3A5, NR1I2 was the most significant variable (p = 2.7 × 10−22) associated with CYP3A5 expression, and no other TFs or lncRNAs significantly accounted for the variance in CYP3A5 expression. After the removal of the lncRNAs from top 80 variables, the TFs in the final models explained 93.8%, 96.0%, and 69.3% of the expression variance in CYP3A4, 3A5, and 3A7, respectively. The significant TFs CREB3L3, NR1I2, and NR6A1 explained 83.3%, 92.7%, and 56.8% of expression variance in CYP3A4, CYP3A5, and CYP3A7, respectively (Supplementary Table S7).
For each CYP3A, the weight values between the two lncRNAs accounting for the highest proportion of CYP3A expression variance and the TFs within the module are listed in Supplementary Table S8.
We conclude that, after the removal of lncRNAs, the proportions of regulators explaining expression variance in CYP3A4, 3A5, and 3A7 in liver decreased by 23.0%, 14.2%, and 20.7%; and the decreases in regulators explaining expression variance in CYP3A4, 3A5, and 3A7 in the small intestine were only 1.9%, 2.1%, and 11.1%, respectively. The results indicate that lncRNAs had a greater regulatory effect on CYP3As in the liver than in the small intestine.
4. Discussion
In this study, we conducted a weighted gene co-expression network analysis to identify TFs and ncRNAs associated with CYP3As in the liver and small intestine based on GTEx v8 data. The expression data were analyzed with and without ncRNAs in both the liver and small intestine. Bioinformatic databases were applied to further prioritize TFs and ncRNAs using multiple stepwise regression analysis to identify TFs and ncRNAs contributing to CYP3A expression in the liver and small intestine. Networks obtained with CYP3As and TFs alone are similar to those published earlier, and a series of previously reported TFs related to CYP3A, such as ESR1, HNF4α, and NR1I2, were also identified here [5,7,9]. Including ncRNAs with the analysis yielded an expanded set of TFs and multiple significant lncRNAs affecting CYP3A expression.
Our results show that multiple ncRNAs have weight values with CYP3As similar to those of TFs in both liver and small intestine (Supplementary Table S1). Co-expression networks show that a series of lncRNAs and TFs are significantly associated with CYP3A expression even after Bonferroni correction in both tissues; some lncRNAs outranked TFs as potential regulatory factors (Supplementary Table S2). In the liver, the lncRNAs HNF4A-AS1, LINC02499, and RP11-669E14.4 showed the most significant associations with CYP3A4, 3A5, and 3A7, respectively (Supplementary Table S2). In the small intestine, LOC102724153, NR1I2 (a TF), and EGFR-AS1 ranked highest for CYP3A4, 3A5, and 3A7, respectively (Supplementary Table S2). Of these six top regulators, HNF4A-AS1 and NR1I2 were reported to be associated with CYP3A expression [29,31]. Experimental discovery of the role of lncRNA HNF4A-AS1 provides strong validation for the lncRNAs inferred by WGCNA [32,33,34]. These results reveal that multiple lncRNAs have robust associations with CYP3As similar to that of TFs in both liver and small intestine. Most of the significant lncRNAs were positively associated with CYP3A expression.
LncRNAs can recruit regulatory protein complexes to a gene to regulate its transcription [10]. In addition, the lncRNA transcripts can bind to and regulate proteins [35]. Comparing regulatory networks of CYP3A expression with and without ncRNAs showed significant changes in hepatic CYP3A networks. Inclusion of ncRNAs (Group 1) identified more TFs than present in networks without ncRNAs (Group 3) (Figure 3 and Figure 4, Supplementary Table S4). Most ncRNAs in the same module as CYP3As were lncRNAs. This result supports the hypothesis that lncRNAs indeed function as co-regulators in the liver.
A number of TFs regulating CYP3A expression had been identified, such as NR1I2, ESR1 and PPARA [5,7,9]. While these TFs were also identified here, our results indicate that adding lncRNAs to hepatic CYP3A regulatory networks not only reveals a role for lncRNAs but also identifies additional TFs candidates affecting CYP3A expression. In the small intestine, on the other hand, significant TFs in the CYP3A4 and 3A5 networks were similar with and without ncRNAs in the analysis (Figure 3 and Figure 4, Supplementary Table S4). In addition, the mean weight values of TFs for CYP3A4 and 3A5 were significantly higher than those of ncRNAs in the small intestine. These results suggest that ncRNAs may play a more prevalent role in the regulation of CYP3A4 and 3A5 expression in the liver than in the small intestine.
The multigene locus with CYP3A4, 3A5, and 3A7 contains overlapping promoter and enhancer regions as a result of chromatin looping [8]. Our results indicate that the three CYP3As share most significant TFs and ncRNAs in the liver with similar weight values (Figure 5). These results indicate that CYP3A4, 3A5, and 3A7 may have similar, likely overlapping regulatory modes. Collins et al. [8] and Wang et al. [7] reported enhancer sites associated with expression of more than one CYP3A gene and TFs affecting more than one CYP3A. On the other hand, a single regulator can have different targets for each individual CYP3A, resulting in co-expression with multiple CYP3As. In the small intestine, CYP3A4 and 3A5 shared most TFs and ncRNAs. For each regulator, the weight value with CYP3A5 was significantly higher than the weight value with CYP3A4 (Figure 5). This result suggests that the regulatory effects of TFs and ncRNAs on CYP3A5 were stronger than that of CYP3A4 in the small intestine. Considering that CYP3A7 is in different modules from CYP3A4 and 3A5, the TFs and ncRNAs associated with CYP3A7 differ from those of CYP3A4 and 3A5, suggesting different regulatory modes of CYP3A4, 3A5, and 3A7 in the small intestine. In addition, each CYP3A has indifferent and more significantly associated TFs and ncRNAs in the small intestine than the liver (Figure 6), demonstrating distinct co-expression patterns of CYP3As in the liver and small intestine.
We also performed multiple stepwise regression analysis to evaluate the contribution of each regulator to CYP3A expression variance. ESR1 was the most significant regulator affecting CYP3A4 expression in the liver (Table 3). In prior studies, Wang et al. [7] found that ESR1 is a master regulator in the hepatic expression of CYP3A4, and our results confirm this finding. LINC02499 and HNF4A-AS1 are the top-ranking lncRNAs affecting CYP3A5 and 3A7 expression, respectively (Table 3). Ma et al. [36] reported that LINC02499 expression was remarkably decreased in hepatocellular carcinoma (HCC) tissues compared to adjacent non-tumor tissues, and decreased LINC02499 was also significantly associated with poorer overall survival in the HCC cohort. The mechanisms by which LINC02499 regulates CYP3A5 expression in the liver still require further investigation. Chen et al. [31] reported that knockdown of HNF4A-AS1 increases mRNA expression of basal levels of P450s, including CYP3A4, in HepaRG cells. These results confirmed that HNF4A-AS1 participates in the regulatory network of CYP3As in the liver. In the small intestine, LOC102724153 and RP11-503C24.6 account for large proportions of the variance in CYP3A4 and 3A7 expression, respectively (Table 3). In addition, after removal of ncRNAs, the proportion of CYP3A expression variance explained by regulatory factors decreased in the liver more than in the small intestine (Table 3, Supplementary Table S7). These results suggest that lncRNAs account for a portion of variance in CYP3A expression in the liver, and more markedly so in the small intestines (Table 3). We also analyzed the weight values between the lncRNAs and TFs in the same modules. Some lncRNA–TF weight values exceeded those between lncRNAs and CYP3As (Supplementary Table S7), providing insight as to whether these lncRNAs might interact directly with CYP3As or indirectly through TFs.
Predicting CYP3A expression as a guide for clinical drug therapy more broadly must consider diverse factors, including genetic polymorphisms, induction by xenobiotics, and clinical factors [37,38]. For example, CYP3A5*3, the nonfunctional allele of CYP3A5, can account for a large portion of variance in CYP3A5 expression and pharmacokinetic variability of CYP3A5 substrates [39,40]. According to our results, lncRNAs should also be considered as important factors in CYP3A expression. By including lncRNAs, our study provides new insights into the CYP3A regulation that underlies the interindividual pharmacokinetic and pharmacodynamic variability of CYP3As. Well-defined regulatory networks may enhance clinical predictions of CYP3A related drug metabolism, with lncRNAs as additional biomarkers of toxicity or metabolism [34,41].
GTEx v8 provides extensive expression data of protein-coding genes and ncRNA genes in multiple tissues, enabling us to identify TFs and lncRNAs affecting CYP3A expression in the liver and small intestine. Our results revealed that several lncRNAs have robust associations with CYP3As expression in the liver and small intestine, and lncRNAs may play crucial roles in the CYP3A regulatory network in the small intestine, and even more so in the liver. Our study expanded on potential factors influencing CYP3A expression and highlighted a role of lncRNAs in contributing to tissue-specific CYP3A expression. Further experiments should be applied to verify the mechanism of lncRNAs significantly associated with CYP3A expression in regulating CYP3A expression.
Our study has several limitations. First, a series of miRNAs have been reported to affect CYP3A expression [42,43,44,45,46]. GTEx v8 provides only microRNA precursors but not mature miRNAs that require distinct assay methods and therefore could not be included in the network analysis. In our study, only three miRNA precursors were found to be co-expressed with CYP3A (Supplementary Table S1). Second, in multiple stepwise regression analysis, multi-collinearity existed among some independent variables, and the final model may have missed relevant independent variables. Last, all results were obtained using bioinformatic methods, and further experiments should be carried out to validate the results.
In summary, we used GTEx v8 datasets and WGCNA to generate a comprehensive catalog of regulator-associated CYP3A gene expression in the liver and small intestine.
Acknowledgments
Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health; and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The data used for the analyses described in this manuscript were obtained from the GTEx Analysis Release V8, dbGaP Accession phs000424.v8.p2.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/biomedicines10123061/s1. Figure S1: Network topology for different soft-thresholding powers. Figure S2: GO and KEGG enrichment analysis. Table S1: The TFs and ncRNAs co-expressed with each CYP3A in the different modules. Table S2: The correlation results between the top 80 TFs and ncRNAs by weight for each CYP3A and CYP3A in different modules from the four groups. Table S3: Results of GO and KEGG enrichment analysis of groups 1 to 4. Table S4: Comparison of CYP3A-associated TFs with and without ncRNAs in liver and small intestine. Table S5: Comparison of CYP3A-associated TFs and ncRNAs between three CYP3As in liver and small intestine. Table S6: Comparison of CYP3A-associated ncRNAs and TFs between liver and small intestine. Table S7: Transcriptional factors contributing to CYP3A expression level in the liver and the small intestine; Table S8: Weight values between two lncRNAs and TFs.
Author Contributions
Conceptualization and writing—original draft, L.L.; formal analysis and writing—original draft, H.H.; formal analysis and visualization, S.Z.; formal analysis, X.W.; conceptualization and revising draft, W.S.; conceptualization, D.W.; formal analysis, S.Y. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: the GTEx v8 dataset (https://gtexportal.org/home/ (accessed on 19 February 2020)).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by Science and Technology Projects in Guangzhou (202201011752), Southern Medical University Innovation Training Program for Undergraduate Students (S202112121123 and S202212121153) and Southern Medical University Scientific Research Enlightenment Program for Undergraduate Students.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hohmann N., Haefeli W.E., Mikus G. CYP3A activity: Towards dose adaptation to the individual. Expert Opin. Drug Metab. Toxicol. 2016;12:479–497. doi: 10.1517/17425255.2016.1163337. [DOI] [PubMed] [Google Scholar]
- 2.Sim S.C., Edwards R.J., Boobis A.R., Ingelman-Sundberg M. CYP3A7 Protein Expression Is High in a Fraction of Adult Human Livers and Partially Associated with the CYP3A7*1C Allele. Pharm. Genom. 2005;15:625–631. doi: 10.1097/01.fpc.0000171516.84139.89. [DOI] [PubMed] [Google Scholar]
- 3.Lolodi O., Wang Y.M., Wright W.C., Chen T. Differential Regulation of CYP3A4 and CYP3A5 and its Implication in Drug Discovery. Curr. Drug Metab. 2017;18:1095–1105. doi: 10.2174/1389200218666170531112038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He H., Nie Y.L., Li J.F., Meng X.G., Yang W.H., Chen Y.L., Wang S.J., Ma X., Kan Q.C., Zhang L.R. Developmental regulation of CYP3A4 and CYP3A7 in Chinese Han population. Drug Metab. Pharmacokinet. 2016;31:433–444. doi: 10.1016/j.dmpk.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 5.Yang X., Zhang B., Molony C., Chudin E., Hao K., Zhu J., Gaedigk A., Suver C., Zhong H., Leeder J.S., et al. Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res. 2010;20:1020–1036. doi: 10.1101/gr.103341.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith N.F., Mani S., Schuetz E.G., Yasuda K., Sissung T.M., Bates S.E., Figg W.D., Sparreboom A. Induction of CYP3A4 by vinblastine: Role of the nuclear receptor NR1I2. Ann. Pharmacother. 2010;44:1709–1717. doi: 10.1345/aph.1P354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang D., Lu R., Rempala G., Sadee W. Ligand-Free Estrogen Receptor α (ESR1) as Master Regulator for the Expression of CYP3A4 and Other Cytochrome P450 Enzymes in the Human Liver. Mol. Pharmacol. 2019;96:430–440. doi: 10.1124/mol.119.116897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins J.M., Wang D. Cis-acting regulatory elements regulating CYP3A4 transcription in human liver. Pharm. Genom. 2020;30:107–116. doi: 10.1097/FPC.0000000000000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong S., Han W., Hou C., Liu J., Wu L., Liu M., Liang Z., Lin H., Zhou L., Liu S., et al. Relation of Transcriptional Factors to the Expression and Activity of Cytochrome P450 and UDP-Glucuronosyltransferases 1A in Human Liver: Co-Expression Network Analysis. AAPS J. 2017;19:203–214. doi: 10.1208/s12248-016-9990-2. [DOI] [PubMed] [Google Scholar]
- 10.Long Y., Wang X., Youmans D.T., Cech T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017;3:eaao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin Y.S., Dowling A.L., Quigley S.D., Farin F.M., Zhang J., Lamba J., Schuetz E.G., Thummel K.E. Co-regulation of CYP3A4 and CYP3A5 and contribution to hepatic and intestinal midazolam metabolism. Mol. Pharmacol. 2002;62:162–172. doi: 10.1124/mol.62.1.162. [DOI] [PubMed] [Google Scholar]
- 13.Zhang P., Wu W., Chen Q., Chen M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019;16:20190027. doi: 10.1515/jib-2019-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He W., Wei D., Cai D., Chen S., Li S., Chen W. Altered Long Non-Coding RNA Transcriptomic Profiles in Ischemic Stroke. Hum. Gene Ther. 2018;29:719–732. doi: 10.1089/hum.2017.064. [DOI] [PubMed] [Google Scholar]
- 15.Langfelder P., Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pei G., Chen L., Zhang W. WGCNA Application to Proteomic and Metabolomic Data Analysis. Methods Enzymol. 2017;585:135–158. doi: 10.1016/bs.mie.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 17.Zheng S., Tansey W.P., Hiebert S.W., Zhao Z. Integrative network analysis identifies key genes and pathways in the progression of hepatitis C virus induced hepatocellular carcinoma. BMC Med. Genom. 2011;4:62. doi: 10.1186/1755-8794-4-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., Cui Y., Ding X., Liu S., Han B., Duan X., Zhang H., Sun T. Analysis of mRNA-lncRNA and mRNA-lncRNA-pathway co-expression networks based on WGCNA in developing pediatric sepsis. Bioengineered. 2021;12:1457–1470. doi: 10.1080/21655979.2021.1908029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin X., Wang P., Yang T., Li G., Teng X., Huang W., Yu H. Identification of key modules and genes associated with breast cancer prognosis using WGCNA and ceRNA network analysis. Aging. 2020;13:2519–2538. doi: 10.18632/aging.202285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yin L., Cai Z., Zhu B., Xu C. Identification of Key Pathways and Genes in the Dynamic Progression of HCC Based on WGCNA. Genes. 2018;9:92. doi: 10.3390/genes9020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., Shad S., Hasz R., Walters G., Garcia F., Young N., et al. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Somekh J., Shen-Orr S.S., Kohane I.S. Batch correction evaluation framework using a-priori gene-gene associations: Applied to the GTEx dataset. BMC Bioinform. 2019;20:268. doi: 10.1186/s12859-019-2855-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J., et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47:D766–D773. doi: 10.1093/nar/gky955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu H., Miao Y.R., Jia L.H., Yu Q.Y., Zhang Q., Guo A.Y. AnimalTFDB 3.0: A comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 2019;47:D33–D38. doi: 10.1093/nar/gky822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kozomara A., Birgaoanu M., Griffiths-Jones S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019;47:D155–D162. doi: 10.1093/nar/gky1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.RNAcentral Consortium RNAcentral 2021: Secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021;49:D212–D220. doi: 10.1093/nar/gkaa921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Banerjee M., Chen T. Differential Regulation of CYP3A4 Promoter Activity by a New Class of Natural Product Derivatives Binding to Pregnane X Receptor. Biochem. Pharmacol. 2013;86:824–835. doi: 10.1016/j.bcp.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu Q., Yao N., Wu J., Liu M., Liu F., Zhang H., Xiong Y., Xia C. Constitutive androstane receptor weakens the induction of panaxytriol on CYP3A4 by repressing the activation of pregnane X receptor. Biochem. Pharmacol. 2019;159:32–39. doi: 10.1016/j.bcp.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 31.Chen L., Bao Y., Piekos S.C., Zhu K., Zhang L., Zhong X.B. A Transcriptional Regulatory Network Containing Nuclear Receptors and Long Noncoding RNAs Controls Basal and Drug-Induced Expression of Cytochrome P450s in HepaRG Cells. Mol. Pharmacol. 2018;94:749–759. doi: 10.1124/mol.118.112235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen L., Wang P., Manautou J.E., Zhong X.B. Knockdown of Long Noncoding RNAs Hepatocyte Nuclear Factor 1α Antisense RNA 1 and Hepatocyte Nuclear Factor 4α Antisense RNA 1 Alters Susceptibility of Acetaminophen-Induced Cytotoxicity in HepaRG Cells. Mol. Pharmacol. 2020;97:278–286. doi: 10.1124/mol.119.118778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P., Chen S., Wang Y., Wang X., Yan L., Yang K., Zhong X.B., Han S., Zhang L. The Long Noncoding RNA Hepatocyte Nuclear Factor 4α Antisense RNA 1 Negatively Regulates Cytochrome P450 Enzymes in Huh7 Cells via Histone Modifications. Drug Metab. Dispos. 2021;49:361–368. doi: 10.1124/dmd.120.000316. [DOI] [PubMed] [Google Scholar]
- 34.Wang X., Yu Y., Wang P., Yang K., Wang Y., Yan L., Zhong X.B., Zhang L. Long Noncoding RNAs Hepatocyte Nuclear Factor 4A Antisense RNA 1 and Hepatocyte Nuclear Factor 1A Antisense RNA 1 Are Involved in Ritonavir-Induced Cytotoxicity in Hepatoma Cells. Drug Metab. Dispos. 2022;50:704–715. doi: 10.1124/dmd.121.000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Engreitz J.M., Haines J.E., Perez E.M., Munson G., Chen J., Kane M., McDonel P.E., Guttman M., Lander E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539:452–455. doi: 10.1038/nature20149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma X., Mo M., Tan H.J.J., Tan C., Zeng X., Zhang G., Huang D., Liang J., Liu S., Qiu X. LINC02499, a novel liver-specific long non-coding RNA with potential diagnostic and prognostic value, inhibits hepatocellular carcinoma cell proliferation, migration, and invasion. Hepatol. Res. 2020;50:726–740. doi: 10.1111/hepr.13491. [DOI] [PubMed] [Google Scholar]
- 37.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: Regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 38.Matthaei J., Bonat W.H., Kerb R., Tzvetkov M.V., Strube J., Brunke S., Sachse-Seeboth C., Sehrt D., Hofmann U., von Bornemann Hjelmborg J., et al. Inherited and Acquired Determinants of Hepatic CYP3A Activity in Humans. Front. Genet. 2020;11:944. doi: 10.3389/fgene.2020.00944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie H.G., Wood A.J., Kim R.B., Stein C.M., Wilkinson G.R. Genetic variability in CYP3A5 and its possible consequences. Pharmacogenomics. 2004;5:243–272. doi: 10.1517/phgs.5.3.243.29833. [DOI] [PubMed] [Google Scholar]
- 40.Chen D., Lu H., Sui W., Li L., Xu J., Yang T., Yang S., Zheng P., Chen Y., Chen J., et al. Functional CYP3A variants affecting tacrolimus trough blood concentrations in Chinese renal transplant recipients. Pharmacogenomics J. 2021;21:376–389. doi: 10.1038/s41397-021-00216-w. [DOI] [PubMed] [Google Scholar]
- 41.Yin J., Li F., Zhou Y., Mou M., Lu Y., Chen K., Xue J., Luo Y., Fu J., He X., et al. INTEDE: Interactome of drug-metabolizing enzymes. Nucleic Acids Res. 2021;49:D1233–D1243. doi: 10.1093/nar/gkaa755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wei Z., Jiang S., Zhang Y., Wang X., Peng X., Meng C., Liu Y., Wang H., Guo L., Qin S., et al. The effect of microRNAs in the regulation of human CYP3A4: A systematic study using a mathematical model. Sci. Rep. 2014;4:4283. doi: 10.1038/srep04283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan Y.Z., Gao W., Yu A.M. MicroRNAs regulate CYP3A4 expression via direct and indirect targeting. Drug Metab. Dispos. 2009;37:2112–2117. doi: 10.1124/dmd.109.027680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takagi S., Nakajima M., Kida K., Yamaura Y., Fukami T., Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J. Biol. Chem. 2010;285:4415–4422. doi: 10.1074/jbc.M109.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takagi S., Nakajima M., Mohri T., Yokoi T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J. Biol. Chem. 2008;283:9674–9680. doi: 10.1074/jbc.M709382200. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H.F., Zhu L.L., Yang X.B., Gao N., Fang Y., Wen Q., Qiao H.L. Variation in the expression of cytochrome P450-related miRNAs and transcriptional factors in human livers: Correlation with cytochrome P450 gene phenotypes. Toxicol. Appl. Pharmacol. 2021;412:115389. doi: 10.1016/j.taap.2020.115389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: the GTEx v8 dataset (https://gtexportal.org/home/ (accessed on 19 February 2020)).