Abstract
Determination of the full-length thyroid-stimulating hormone receptor (TSHR) structure by cryo-electron microscopy (cryo-EM) is described. The TSHR complexed with human monoclonal TSHR autoantibody K1-70™ (a powerful inhibitor of TSH action) was detergent solubilised, purified to homogeneity and analysed by cryo-EM. The structure (global resolution 3.3 Å) is a monomer with all three domains visible: leucine-rich domain (LRD), hinge region (HR) and transmembrane domain (TMD). The TSHR extracellular domain (ECD, composed of the LRD and HR) is positioned on top of the TMD extracellular surface. Extensive interactions between the TMD and ECD are observed in the structure, and their analysis provides an explanation of the effects of various TSHR mutations on TSHR constitutive activity and on ligand-induced activation. K1-70™ is seen to be well clear of the lipid bilayer. However, superimposition of M22™ (a human monoclonal TSHR autoantibody which is a powerful stimulator of the TSHR) on the cryo-EM structure shows that it would clash with the bilayer unless the TSHR HR rotates upwards as part of the M22™ binding process. This rotation could have an important role in TSHR stimulation by M22™ and as such provides an explanation as to why K1-70™ blocks the binding of TSH and M22™ without activating the receptor itself.
Keywords: TSHR, cryo-EM, autoantibodies, autoimmunity, structure
Introduction
The G-protein coupled receptor (GPCR) for thyroid-stimulating hormone (TSH) receptor (TSHR) has a key role in the regulation of thyroid function and is a major thyroid autoantigen (Rees Smith et al. 1988, Rapoport et al. 1998, Morshed et al. 2012, Furmaniak et al. 2015). In particular, TSHR autoantibodies (TRAbs) bind to the receptor in such a way as to mimic TSH action and cause the hyperthyroidism of Graves’ disease (Rees Smith et al. 1988, Rapoport et al. 1998, Furmaniak et al. 2022). TRAbs also act on TSHRs in the orbit and have an important role in the eye signs of Graves’ disease (Bahn 2010, Furmaniak et al. 2022).
TSH and TRAbs bind principally to the concave surface of the receptor’s leucine-rich-repeat domain (LRD, amino acids 22–279). The crystal structures of the human TSHR LRD (amino acids, 22–260; TSHR260) bound to the thyroid-stimulating human monoclonal autoantibody (MAB) M22™ (Sanders et al. 2003) and the blocking type MAB K1-70™ (Evans et al. 2010, Furmaniak et al. 2019) have been solved at 2.55 and 1.9 Å resolution, respectively (Sanders et al. 2007, 2011). Also the crystal structure of a thermostable ligand-free TSHR LRD has been solved at 2.83 Å resolution (Miller-Gallacher et al. 2019). Also, the crystal structure of the TSHR LRD (Sanders et al. 2007) and the crystal structure of the FSHR extracellular domain (ECD) (Jiang et al. 2012) allowed structural modelling of the full-length TSHR by different groups (Davies et al. 2014, Brüser et al. 2016, Schaarschmidt et al. 2016, Kleinau et al. 2017).
We now describe the structure of the full-length TSHR (amino acids 22–764) in complex with K1-70™ Fab determined by cryo-electron microscopy (cryo-EM).
Materials and methods
Preparation of TSHR–K1-70™ complexes
Full-length human TSHR (amino acids 22–764) was expressed in CHO-K1 cells as described previously (Oda et al. 1998). The cells were resuspended in 10 mmol/L Tris–HCl pH 7.5 supplemented with complete protease inhibitors (Roche), homogenised and centrifuged at 12,000 g for 30 min at 4°C to obtain a crude membrane preparation. This was suspended in 10 mmol/L Tris–HCl pH 7.5, 50 mmol/L NaCl, re-sedimented and re-suspended in the same buffer.
K1-70™ IgG was purified from heterohybridoma culture supernatants, Fab was prepared using mercuripapain (Sanders et al. 2011) and added to the TSHR membrane preparation. The mixture containing 10 μg/mL of K1-70™ Fab was then incubated for 1 h at 20°C followed by centrifugation at 12,000 g for 45 min at 4°C and the pellets were homogenised in 10 mmol/L Tris–HCl pH 7.5, 50 mmol/L NaCl, 2% lauryl maltose neopentyl glycol (LMNG), 0.2% cholesteryl hemisuccinate (CHS) containing 25 µg/mL K1-70™ Fab. After incubation for 2 h at 4°C, the homogenate was centrifuged at 90,000 g for 1 h at 4°C and the supernatant containing solubilised TSHR–K1-70™ Fab complex was stored in aliquots at −70°C.
Purification of TSHR–K1-70™ complexes
A mouse monoclonal TSHR antibody (14C4) that binds to a conformational epitope within amino acids 22–260 of the TSHR (but distinct from the K1-70™ binding site) was coupled to CNBr-activated Sepharose 4B (Sigma-Aldrich) and a 3 mL column was used to affinity purify solubilised TSHR–K1-70™ Fab complex. After washing with 6 column volumes of 50 mmol/L Tris–HCl pH 8.0,150 mmol/L NaCl, 0.2% LMNG and 0.02% CHS containing 2 µg/mL of K1-70™ Fab, the complex was eluted with 0.1 mmol/L sodium citrate pH 4.5, 0.2% LMNG and 0.02% CHS, 2 µg/mL K1-70™ Fab. One-millilitre fractions were collected and dialysed into 50 mmol/L Tris pH 8.0, 150 mmol/L NaCl, 0.02% LMNG and 0.002% CHS.
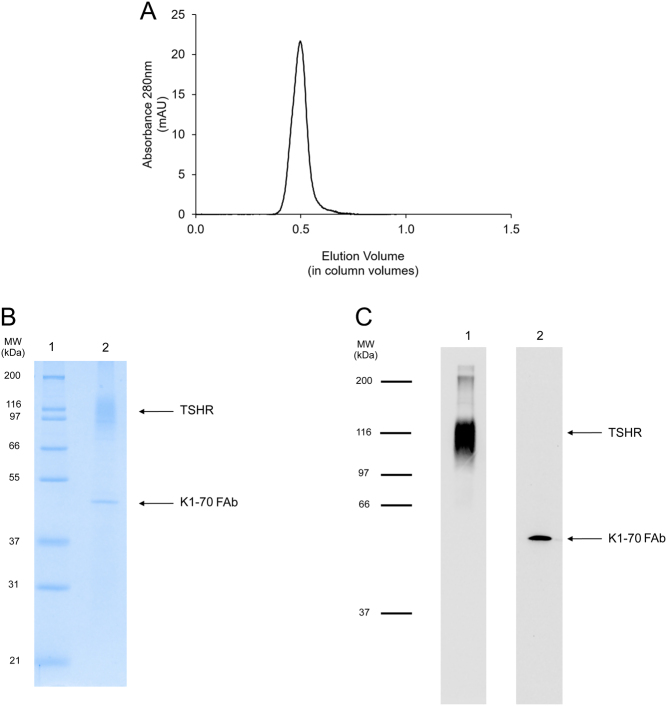
After dialysis, the complex was concentrated using a 10 kDa MWCO spin concentrator (Thermo Scientific) and run on a Superdex 200 XK 16-100 column (GE Healthcare) in 50 mmol/L Tris–HCl pH 8.0, 150 mmol/L NaCl, 0.02% LMNG and 0.002% CHS. The fractions containing the complex (Fig. 1) were collected and concentrated to 1 mg/mL for cryo-EM.
Figure 1.
(A) Analysis of purified TSHR–K1-70™ complex by size exclusion chromatography (Superdex 200 XK 16-100 column run in 50 mmol/L Tris–HCl pH 8.0, 150 mmol/L NaCl, 0.02% LMNG and 0.002% CHS). (B) Analysis of purified TSHR–K1-70™ complex by SDS-PAGE (12% acrylamide gel) under non-reducing conditions. The positions of the molecular weight standards are shown and the positions of the TSHR and the K1-70™ Fab are marked (lane 1: molecular weight standards; lane 2: purified TSHR–K1-70™ complex). (C) Western blotting analysis of purified TSHR–K1-70™ complex. Lane 1: blotting with mouse monoclonal TSHR antibody 8E2 labelled with horseradish peroxidase; lane 2: blotting with mouse anti-human IgG (Fab-specific) MAB labelled with horseradish peroxidase. See text for experimental details.
Analysis of TSHR–K1-70™ complexes
During purification of the complex, total protein concentration was determined by Bradford assay (Biorad). Also, the presence of K1-70™ Fab and the presence of the TSHR in various fractions were monitored. Briefly, 150 μL of TSHR–K1-70™ complex was incubated overnight at 4°C in ELISA plate wells coated with a mouse MAB which binds to the C-terminus of the TSHR. The wells were washed thrice with wash buffer (50 mmol/L NaCl, 10 mmol/L Tris–HCl pH 7.8, 0.05% LMNG) and 100 μL of either TSHR MAb 14C4 labelled with horse radish peroxidase (14C4-POD; RSR Ltd, Cardiff, UK) or anti-human Fab-specific antibody labelled with horse radish peroxidase (Sigma Aldrich) was added and the samples were incubated for 20 min at room temperature. The plate wells were then washed twice with wash buffer, once with water before the addition of 100 μL 3,3’,5,5’ tetramethylbenzidine substrate and a further 35 min incubation in the dark at room temperature. The reaction was then stopped by the addition of 50 μL of 0.5M H2SO4 and the absorbance was read at 450 nm using an ELISA plate reader.
The purity of the final complex was assessed by 12% non-reduced sodium dodecyl sulphate-PAGE (SDS-PAGE) (Laemmli 1970) and Western blotting (Birk & Koepsell 1987) with anti-human Fab-specific antibody labelled with peroxidase (Sigma Aldrich) to detect the presence of K1-70™ Fab and peroxidase-labelled mouse MAB to the N-terminus of the TSHR (8E2; Oda et al. 2000) to detect TSHR. The concentration of the purified complex was determined by optical density measurements at 280 nm using the extinction coefficient of 1.330 mL/mg/cm calculated from the amino acid sequence using ProtParam (https://www.expasy.org/resources/protparam).
Cryo-EM grid preparation and data collection
The purified complex (3 μL of 1 mg/mL) was applied to UltrAufoil R1.2/1.3 300 mesh grids and the grids glow discharged once per slide, the foil side being the most recently charged. A total of 12 cryo-TEM grids were prepared using Vitrobot Mark IV (Life Sciences). The data were collected from the UltrAufoil grid from undiluted complex (1 mg/mL) with 0 blotting force and 3 s blot time.
Data collection was performed on a Titan Krios 300kV (at Cryo-EM Facility, Department of Biochemistry, University of Cambridge, UK) using 130,000× magnification, corresponding to a pixel size of 0.652 Å/pixel. The data were collected with a Falcon 3 Direct Detector in electron counting mode. A total dose of 47.3 e/Å2 and 50 fractions were used for recording movies during each exposure which lasted 501.34 s and a defocus range from −2.4 to −1.0 µm with a defocus step of 0.2 µm.
A total of 8219 movies were collected during the data collection session. WARP (Tegunov & Cramer 2019) identified about 1,600,000 particles which were then passed into Cryo-SPARC processing for 3D reconstruction (Punjani et al. 2017).
Molecular replacement and refinement
The electron density of the TSHR–K1-70™ complex showed the TSHR bound to K1-70™ Fab. The crystal structure of the TSHR LRD in complex with K1-70™ Fab (Sanders et al. 2011; PDB-Id: 2xwt) and the model of TSHR (AlphaFold database: https://alphafold.ebi.ac.uk/; Tunyasuvunakool et al. 2021) were fitted into the electron density.
The structure was manually refined using COOT v0.9 (Emsley et al. 2010) as the first step. Then, the Refinement Cascade protocol within the Discovery Studio 2021 suite of programs (BIOVIA Dassault Systèmes 2021) was run to automatically refine the structure of the complex. Finally, minimisation was carried out with MODELLER (Sali & Blundell 1993, Webb & Sali 2016) followed by an automatic All-atom Refine protocol with COOT. This process was repeated three times.
Results
TSHR–K1-70™ complex
The peak fractions from the preparative size exclusion chromatography (SEC) of the TSHR–K1-70™ complex were pooled and concentrated to 1 mg/mL. This preparation bound to 14C4-POD and to anti-Fab-POD, confirming the presence of both TSHR and K1-70™ Fab components in the complex and analytical SEC of the preparation, indicated purity of >95% (Fig. 1A). On non-reduced SDS-PAGE, the complex separated into two components corresponding to TSHR (molecular weight 89–116 kDa) and K1-70™ Fab (47 kDa) (Fig. 1B). Western blotting analysis also confirmed the presence of both the TSHR and K1-70™ Fab in the complex (Fig. 1C).
Full-length human TSHR structure
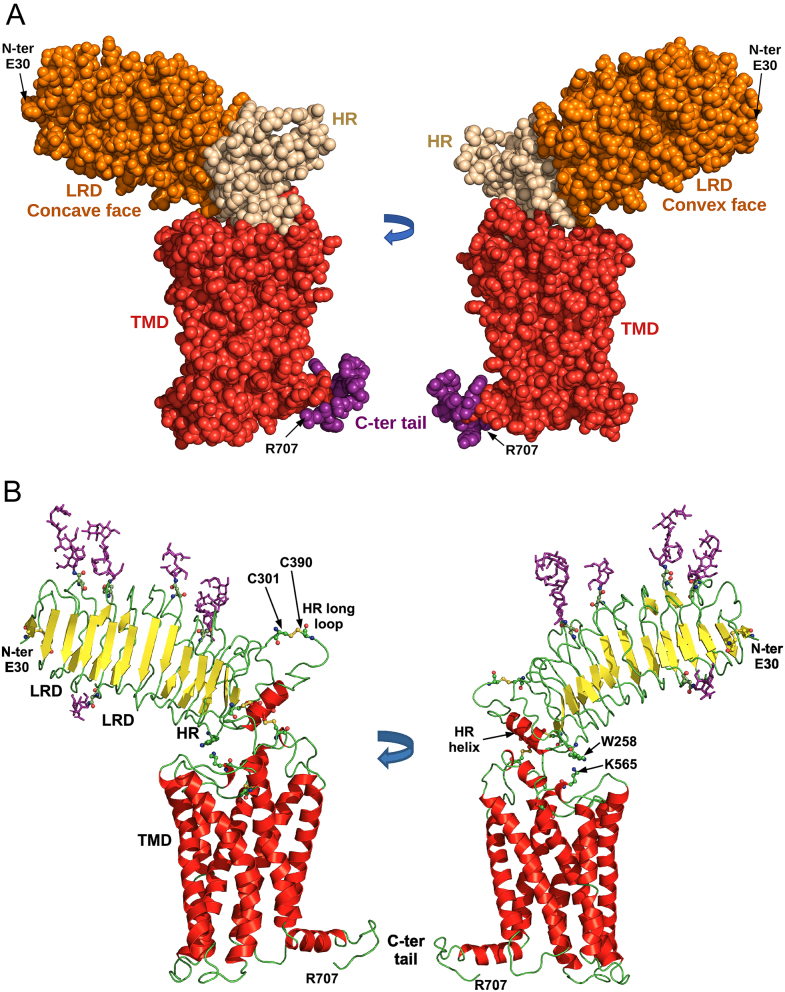
Processing of the cryo-EM data resulted in a final structure for the TSHR–K1-70™ Fab complex at 3.32 Å overall resolution (Supplementary Fig. 1 and Supplementary Table 1, see section on supplementary materials given at the end of this article). It consists of the TSHR bound to K1-70™ Fab with the electron density showing the TSHR structure from Glu30 to Arg707. This is composed of three domains, the ECD (amino acids 22–409) consisting of the LRD (amino acids 22–279) and hinge region (HR; amino acids 280–409), the TMD and the intracellular C-terminus (amino acids 410–764) (Fig. 2A and B). The structure of the ECD, formed by the LRD and HR, is that of a typical leucine-rich-repeat (LRR) structure with 11 repeats in the LRD and 1 repeat in the HR. Each LRR consists of a parallel β-strand on its concave surface while the N-terminal cap (N-cap) has an additional β-strand antiparallel to the β-strand of the first repeat (Supplementary Fig. 2A). The LRD and the HR form a continuum structure with the N-cap, with two disulphide bonds, and a C-terminal cap (C-cap) with three disulphide bonds and an alpha helix. One of the two N-terminal disulphide bonds between Cys31 and Cys41 is visible in the structure while the other one between Cys24 and Cys29 is not. All three intra-domain disulphide bonds which form the C-cap are visible in the structure between Cys283 and Cys398, Cys284 and Cys408, and Cys301 and Cys390. However, TSHR residues from Asn302 to Ile389, which form part of the unstructured long hinge loop, are not visible in the structure.
Figure 2.
Structural features of the full-length TSHR. (A) TSHR structure (amino acids E30 to R707) in two views rotated 180o. The structure is a space fill representation with the leucine-rich-repeat domain (LRD) in orange, the hinge region (HR) in beige and the transmembrane domain (TMD) in red with the intracellular C-terminus in purple. (B) TSHR structure in two views rotated 180o. The structure is in cartoon representation with the disulphide bonds and glycosylation sites in ball and stick representation with oxygen in red, nitrogen in blue and sulphur in yellow. The glycans are shown in purple. The LRD, HR and TMD are marked. Cys301 forms a disulphide bond with Cys390 and the residues in between (amino acids 302–389) are missing from the structure.
The cryo-EM structure shows five N-linked glycans in the LRD in all the positions expected from the sequence consensus for N-linked glycosylation and shown in the TSHR LRD crystal structures (amino acids Asn77, Asn99, Asn113, Asn177 and Asn198). A further glycosylation site, Asn302, is not visible in the cryo-EM structure (Fig. 2A, B and Supplementary Fig. 2A).
The structure of the TSHR TMD (Supplementary Fig. 2B) is that of a typical GPCR 7 helices TMD (Núñez Miguel et al. 2017a) with an 8th helix (TM8) parallel to the membrane. The long extracellular loop (ECL) 2 is disulphide bonded to transmembrane helix (TM) 3 through cysteine residues 569 and 494. The long ECL1 loop has an α-helix close to its C-terminus and proline kinks are seen in the structure of helices TM6 and TM7 which correspond to a change in direction of the helix axis. Helix TM4 is broken by the Gly–Gly sequence at residues 544 and 545 and a proline distortion is seen at the extracellular end of the helix.
Cation–π interactions are observed between LRD residue Trp258 (10th LRR) and TMD residue Lys565 of ECL2. Residues Lys261 and Tyr279, in the 10th and 11th LRR, respectively, and Ser281, Cys284 and Asn288 in the HR helix interact with residues in ECL1 (Table 1). Furthermore, residue E404 in the C-cap interacts with both ECL2 and the extracellular part of TMD helix 7.
Table 1.
Interactions between the TSHR ECD and TMD.
| TSHR ECD | TSHR TMD | Interaction types | ||
|---|---|---|---|---|
| Residue | Location | Residue | Location | |
| W258 | 10th Repeat | K565 | ECL2 | Cation–π |
| K261 | 10th Repeat | I486 | ECL1 | Induced dipole |
| T490 | ECL1 | Hydrogen bond, hydrophobic | ||
| Y279 | 11th Repeat | I486 | ECL1 | Hydrophobic |
| T490 | ECL1 | Polar | ||
| S281 | HR-helix | Y482 | ECL1 | Hydrogen bond, induced dipole |
| A485 | ECL1 | Hydrophobic | ||
| I486 | ECL1 | Induced dipole | ||
| C284 | HR-helix | Y482 | ECL1 | Hydrophobic |
| N288 | HR-helix | Y482 | ECL1 | Hydrogen bond, hydrophobic, induced dipole |
| E404 | C-ter cap | V566 | ECL2 | Hydrophobic, induced dipole |
| S567 | ECL2 | Induced dipole | ||
| I568 | ECL2 | Induced dipole | ||
| K660 | TM7 | Hydrophobic | ||
| V656 | TM7 | Induced dipole | ||
| F405 | P10 seg | Q489 | ECL1 | Hydrophobic, induced dipole |
| A564 | ECL2 | Induced dipole | ||
| K565 | ECL2 | Hydrophobic, induced dipole | ||
| V566 | ECL2 | Hydrophobic | ||
| S567 | ECL2 | Polar, hydrophobic, induced dipole | ||
| P407 | P10 seg | Y481 | ECL1 | Hydrophobic |
| C408 | P10 seg | Y481 | ECL1 | Hydrogen bond, induced dipole |
| Y482 | ECL1 | Hydrogen bond, induced dipole | ||
| E409 | P10 seg | T477 | ECL1 | Induced dipole |
| S479 | ECL1 | Polar, hydrophobic, induced dipole | ||
| E480 | ECL1 | Hydrogen bond, induced dipole | ||
| Y481 | ECL1 | Hydrogen bond, hydrophobic, induced dipole | ||
| D410 | P10 seg | R418 | TM1 | Salt bridge (2), ion pair (3), induced dipole, long-range charge–charge |
| D474 | TM2 | Polar, induced dipole | ||
| T477 | ECL1 | Polar, induced dipole | ||
| S479 | ECL1 | Induced dipole | ||
| I411 | P10 seg | V473 | TM2 | Hydrophobic |
| D474 | TM2 | Polar, hydrophobic, induced dipole | ||
| Y481 | ECL1 | Hydrophobic | ||
| S567 | ECL2 | Induced dipole | ||
| M412 | P10 seg | R418 | TM1 | Polar, hydrophobic, induced dipole |
| I568 | ECL2 | Hydrophobic | ||
| K660 | TM7 | Polar, induced dipole | ||
| V664 | TM7 | Induced dipole | ||
| G413 | P10 seg | K415 | N-ter | Polar, induced dipole |
| Y414 | P10 seg | F416 | TM1 | Hydrogen bond, aromatic, hydrophobic, induced dipole |
| L417 | TM1 | Polar, hydrophobic, induced dipole | ||
| S657 | TM7 | Hydrogen bond | ||
| I661 | TM7 | Hydrophobic | ||
ECLX, extracellular loop X; HR-helix, hinge region helix; P10 seg, P10 segment; TMX, transmembrane helix X.
The TSHR P10 peptide (Brüser et al. 2016) (amino acids 405–414 of the TSHR) is visible in the cryo-EM structure and forms the C-terminal half of the linker between the TSHR ECD and the TMD. The P10 peptide interacts with TMD residues in ECL1, ECL2, TM1, TM2 and TM7 (Table 1) forming two salt bridges, six hydrogen bonds, three ion pairs and several polar, aromatic, hydrophobic and induced dipole interactions. The intracellular C-terminal tail of the TSHR is visible up to amino acid 707 with the last 57 amino acids not visible in the structure.
A structural superimposition of the TSHR ECD residues 30–257, which are present in all four available TSHR structures (cryo-EM TSHR–K1-70™, TSHR260–K1-70™ (PDB code 2XWT), TSHR260–M22™ (PDB code 3G04) and the thermostable TSHR260–JMG55™ (Miller-Gallacher et al. 2019), gave low root-mean-square deviation (RMSD) values (0.51–0.75 Å; Supplementary Table 2). A superimposition of the hinge region (residues 261–301 and 390–411) of our cryo-EM structure of the TSHR with the hinge region of the AlphaFold TSHR model (Tunyasuvunakool et al. 2021) gave a high RMSD of 3.5 Å mainly due to a longer hinge helix in the AlphaFold models of all 3 glycoprotein hormone receptors (GPHRs).
K1-70™ structure
The K1-70™ Fab structure consists of K1-70™ heavy chain (HC) residues Gln1 to Ser229 and light chain (LC) residues Ser2 to Ala212 and is the structure of a typical Fab fragment.
TSHR–K1-70™ complex structure
K1-70™ Fab binding is principally to the concave surface of the TSHR LRD (Fig. 3 and Supplementary Fig. 4), which has no glycans attached. K1-70™ Fab HC and LC bind to the N-terminal repeats of the TSHR (Tables 2 and 3) from amino acid Glu35 in the TSHR N-cap to Lys183 in the 7th LRR. There is a mixture of an extensive hydrogen bonding and salt bridge network (20 hydrogen bonds and salt bridges), 5 ion pairs, 13 polar interactions and 13 hydrophobic/aromatic contacts. Differences in some of the interactions seen in the cryo-EM structure of the full-length TSHR bound to K1-70™ Fab compared to the crystal structure of the TSHR LRD in complex with K1-70™ Fab are most likely due to side chain flexibility in the interacting amino acids. There are no interactions between the K1-70™ Fab and either the HR or the TMD.
Figure 3.
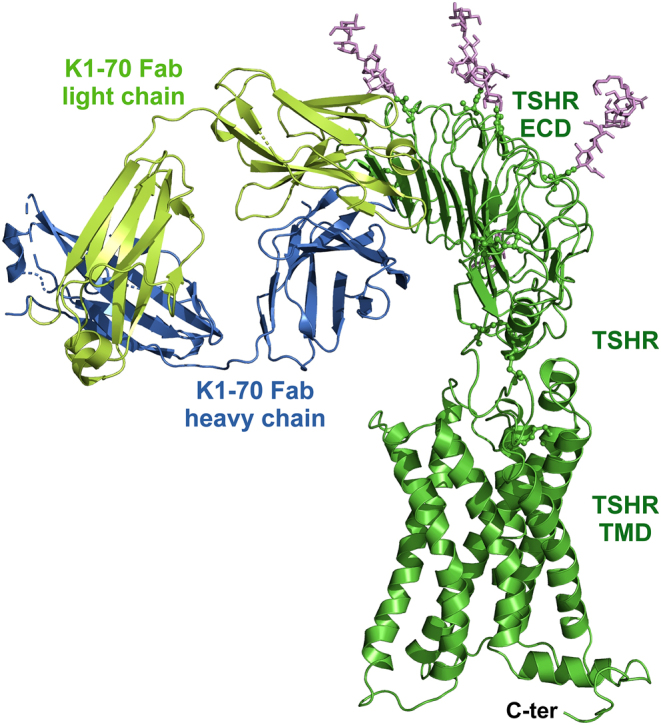
Structural features of the full-length TSHR in complex with the blocking monoclonal autoantibody K1-70™. The TSHR–K1-70™ Fab structure is in cartoon representation with the glycans shown in purple sticks. The K1-70™ heavy chain is shown in blue, the K1-70™ light chain in light green and the TSHR in green. The TSHR, extracellular domain (ECD), transmembrane domain (TMD) and intracellular C-terminus are marked.
Table 2.
Interactions between the TSHR LRD and K1-70 heavy chain.
| TSHR LRD | K1-70 Heavy chain residue | Interaction types | |
|---|---|---|---|
| Residue | Location | ||
| D36 | First repeat | R101 | Salt bridge, induced dipole, long-range charge–charge |
| R38 | First repeat | D96 | Salt bridge, ion pair (2), induced dipole, long-range charge–charge |
| R101 | Polar, induced dipole | ||
| Y102 | Hydrogen bond, polar, cation–π, induced dipole | ||
| K42 | First repeat | D31 | Hydrogen bond, salt bridge (2), induced dipole |
| K58 | Second repeat | D31 | Hydrogen bond, induced dipole |
| N32 | Induced dipole | ||
| D96 | Salt bridge, Hydrogen bond, induced dipole, long-range charge–charge | ||
| W97 | Cation–π | ||
| I60 | Second repeat | D31 | Hydrophobic, induced dipole |
| R80 | Third repeat | Y99 | Hydrogen bond, induced dipole |
| N100 | Hydrogen bond, polar, hydrophobic, induced dipole | ||
| Y82 | Third repeat | W97 | Aromatic, induced dipole |
| N100 | Hydrogen bond, polar, induced dipole | ||
| S84 | Third repeat | Y52 | Polar, induced dipole |
| W97 | Hydrophobic | ||
| H105 | Fourth repeat | N98 | Polar, induced dipole |
| E107 | Fourth repeat | R58 | Long-range charge–charge |
| W97 | Hydrogen bond, induced dipole | ||
| R109 | Fourth repeat | W33 | Cation–π |
| D54 | Salt bridge (2), induced dipole, long-range charge–charge | ||
| D56 | Salt bridge, ion pair (2), induced dipole, long-range charge–charge | ||
| R58 | Polar | ||
| W97 | Hydrophobic | ||
| N110 | Fourth repeat | D54 | Hydrogen bond, polar, hydrophobic, induced dipole |
| R112 | Fourth repeat | D54 | Long-range charge–charge |
| K129 | Fifth repeat | Y99 | Cation–π |
| F130 | Fifth repeat | Y99 | Aromatic |
| F134 | Fifth repeat | R58 | Cation–π |
| N135 | Fifth repeat | D54 | Polar, induced dipole |
| E157 | Sixth repeat | R58 | Salt bridge, ion pair, long-range charge–charge |
Table 3.
Interactions between the TSHR LRD and K1-70 light chain.
| TSHR LRD | K1-70 Light chain residue | Interaction types | |
|---|---|---|---|
| Residue | Location | ||
| E35 | N-ter cap | S56 | Hydrogen bond, hydrophobic, induced dipole |
| D36 | First repeat | Y49 | Polar, hydrophobic, induced dipole |
| P55 | Polar, induced dipole | ||
| S56 | Polar, induced dipole | ||
| R80 | Third repeat | Y32 | Cation–π |
| D50 | Long-range charge–charge | ||
| K102 | Fourth repeat | Y32 | Hydrogen bond |
| D50 | Long-range charge–charge | ||
| T104 | Fourth repeat | Y32 | Induced dipole |
| K129 | Fifth repeat | N31 | Induced dipole |
| S93 | Induced dipole | ||
| I152 | Sixth repeat | S27A | Induced dipole |
| G29 | Hydrophobic | ||
| S30 | Induced dipole | ||
| F153 | Sixth repeat | S27A | Hydrophobic, induced dipole |
| S93 | Hydrophobic, induced dipole | ||
| R94 | Cation–π, hydrophobic, induced dipole | ||
| I155 | Sixth repeat | S93 | Induced dipole |
| K183 | Seventh repeat | S93 | Polar |
Discussion
The TSHR LRD structure seen by cryo-EM is in good agreement with the three LRD structures determined previously by X-ray crystallography (Sanders et al. 2007, 2011, Miller-Gallacher et al. 2019). All four structures show the extensive concave surface of the LRD forming the binding site for K1-70™ Fab and M22™ Fab. Also, no glycans are seen on the concave surface of the LRD in any of the four structures and this would be expected from the assessment of the consensus sequence for N-linked glycosylation (Asn-Xxx-Ser/Thr, where Xxx is any amino acid except proline; Núñez Miguel et al. 2017b). The full-length TSHR used to determine the cryo-EM structure was not deglycosylated in the process (in contrast to the deglycosylation carried out during preparation of material for X-ray crystallography), and this provides further evidence of the absence of glycans on the concave surface of the TSHR LRD.
The cryo-EM structure of the TSHR has intact disulphide bonds at five of the six expected disulphide bond sites, confirming the disulphide bond arrangements at the N-terminus, the intra-domain disulphide bonding which forms the C-cap of the HR and the disulphide bond between the ECL2 and TM3 in the TMD. The disulphide bond between Cys301 and Cys390 in the HR long loop is clearly visible in the structure; however, residues 302–389 between these cysteine residues lack clear electron density in this region. This could be due to both flexibility of the long unstructured loop and cleavage of the TSHR into two subunits (A and B; Rees Smith et al. 1988), with the removal of approximately 50 amino acids (Chazenbalk et al. 1997, Oda et al. 2000). Part of the cleaved region (amino acids 316–366) which is not visible in our structure has been reported to bind to neutral TSHR antibodies (Morshed et al. 2012).
There are clear interactions between some TSHR ECD and TMD residues. Amino acid Trp258 in the 10th LRR interacts with Lys565 in ECL2 and Lys565 also interacts with F405 (P10 segment). Mutation of the amino acid tryptophan 258 of the TSHR ECD to alanine results in a decrease in the constitutive activity of the TSHR by approximately 50% (unpublished data) and decreased stimulating activity by TSHR stimulating human MABs such as M22™ and K1-18™ (Núñez Miguel et al. 2012). Furthermore, TSHR Trp258 is seen to form important interactions with M22™ in the crystal structure of the TSHR-M22™ complex (Sanders et al. 2007). Also, mutation of TSHR Lysine 565 to alanine has been reported to stop the constitutive activity of the TSHR and strongly impair hormone-induced signalling activity (Kleinau et al. 2007). The decrease in the constitutive activity of the TSHR when the interaction between Trp258 and Lys565 is disrupted by the introduction of alanine at either position 258 or 565 suggests an important role for this interaction in the constitutive activity of the TSHR. No interactions between K1-70™ Fab and TSHR Trp258 are seen in the cryo-EM or crystal structures (Sanders et al. 2011). This is consistent with the observation that K1-70™ has no effect on the constitutive activity of the TSHR despite being a powerful inhibitor of TSHR stimulation by TSH and by TRAbs. Furthermore, the luteinizing hormone (LH)/chorionic gonadotrophin (CGR) has no constitutive activity (Van Sande et al. 1995) and no interaction is seen between the equivalent residues, Tyr254 and Lys510, in the inactive LH/CGR cryo-EM structure (WT LH/CGR bound to the small molecule antagonist, compound 26) (Duan et al. 2021).
Mutations in Ser281 to alanine, asparagine, threonine or isoleucine are well documented for their ability to increase the constitutive activity of the TSHR. In the TSHR cryo-EM structure, Ser281 is involved in interactions with three residues of ECL1 including one hydrogen bond (Table 1). In addition, Ser281 also interacts with residues Cys283, Cys284 and Ala285 of the HR and Asn606 of the P10 segment. Disruption of some or all of these interactions leads to increased constitutive activity of the TSHR.
Amino acid substitutions in TSHR ECL1, ECL2 and ICL3 have previously been shown to increase the constitutive activity of the TSHR, with Ile486 (ECL1) and Ile568 (ECL2) showing strong activating activity (Parma et al. 1995). In our cryo-EM structure residues Lys261, Tyr297 and Ser281 in the 10th LRR, 11th LRR and HR helix all interact with Ile486 (ECL1). Furthermore, Glu404 (C-cap of HR) and Met412 (P10 region) interact with Ile568 (Table 1). Disruption of these interactions with amino acid mutations will weaken the binding of the ECD to the TMD and cause the observed increase in constitutive activity.
In the cryo-EM structure, the positioning of the TSHR HR on top of the TMD can be seen for the first time. The P10 region (amino acids 405–414) is visible in the structure and forms the C-terminal half of the linker between the TSHR ECD and the TMD. There are multiple interactions between nine of the ten amino acids in the P10 region with residues in ECL1 and ECL2 of the TMD as well as interactions with TM1, TM2 and TM7 (Table 1). Furthermore, the P10 region residue Cys408 is disulphide bonded to Cys284 of the HR. The presence of this network of interactions with the P10 region would be expected in the inactive conformation of the receptor. The P10 region is the most highly conserved among glycoprotein hormone receptors (Duan et al. 2021) and is known to undergo a conformational rearrangement during activation of some class B GPCRs (Zhang et al. 2017, 2018). It has been proposed that activation of the TSHR induces a conformational change which leads to the P10 region forming an intramolecular agonist that interacts with the TMD of the receptor (Brüser et al. 2016, Krause et al. 2020). These proposed conformational changes in the TMD would allow G proteins to bind to the intracellular side of the TSHR. The functionally important ionic lock (salt bridge between Arg519 and Asp619) which stabilises the inactive state of GPCRs (Katritch et al. 2012) is also present in the TSHR–K1-70™ structure.
The TSHR cryo-EM structure shows that Leu5123.43 in the TMD makes hydrophobic interactions with Ser508, Thr511, Thr513, Asp633 and Phe634 (superscripts refer to Ballesteros–Weinstein numbering; Ballesteros & Weinstein 1995). In addition, there are also interactions (hydrogen bond, polar and induced dipole) between Asp6336.44 and Asn6747.49. Residue Leu3.43 of the glycoprotein hormone receptor family is functionally important and is involved in a network of hydrophobic interactions in the inactive state. Furthermore, Asp6.44 which interacts with Asn7.49 in the inactive state of the GPHRs is Phe6.44 instead of Asp6.44 in most GPCRs (Tehan et al. 2014). Three naturally occurring TSHR Leu5123.43 polar mutants (Gln, Asn, Arg) that increase the constitutive activity of the receptor (Kosugi et al. 2000, Trülzsch et al. 2001, Nishihara et al. 2006) show reduced inter-helical packing. In addition, four naturally occurring TSHR Asp6336.44 mutants (Ala, Glu, His and Tyr) that increase the constitutive activity of the receptor (Porcellini et al. 1994, Russo et al. 1996, Parma et al. 1997) break or modify the interaction between Asp6336.44 and Asn6747.49. Mutational studies have shown that the interaction between Asp6.44 and Asn7.49 stabilises the inactive form of the TSHR TMD (Govaerts et al. 2001).
The complex of TSHR–K1-70™ Fab is seen as a monomer as is the case for the cryo-EM structure of LH/CGR in complex with hCG and G proteins (Duan et al. 2021). The LH/CGR complex is also described as being a rigid structure that would prevent receptor dimerisation (Duan et al. 2021) and this appears to be the case for TSHR complexed with K1-70™ Fab. However, there are reports that TSHR dimerisation and multimerisation occur (Urizar et al. 2005, Latif et al. 2015, Boutin et al. 2020).
A structural superimposition (Supplementary Fig. 5A, B and C) of the cryo-EM structure of the TSHR ECD with the crystal structure of the FSHR ECD (Fan & Hendrickson 2005) shows that they are similar to each other. Structural differences are observed in areas with amino acid insertions or deletions, in the N-terminal first two repeats, in the C-terminal cap and mostly in the flexible and unstructured long hinge loop.
A structural superimposition of the full-length TSHR and the full-length LH/CGR cryo-EM inactive structure (Supplementary Fig. 5D, E and F) determined byDuan et al. (2021) shows that the structures are similar. The functionally important linkers between the ECD and the TMD and the two long ECL1 and ECL2 loops are similar in both structures. The transmembrane helices show similar structures and relative positions except for TM6. There is a difference in the position of the extracellular end of TSHR TM6 which is displaced approximately 7 Å away from the centre of the helix bundle compared to the LH/CGR TM6 in the inactive structure. In contrast, there is no displacement of the intracellular end of TM6. The entire intracellular region of the TSHR TMD in our structure superimposes well with the equivalent region of the inactive LH/CGR structure. In both structures, access to the G-protein binding pocket is blocked by TM helix 6 which prevents the binding of G proteins and receptor activation. The inactive LH/CGR structure has the small molecule antagonist compound 26 bound within the TMD and this interacts with Ser586 and Met582 of the TM6 helix. In our structure of the TSHR bound to K1-70™, there is no antagonist in the allosteric binding pocket within the TMD for TM6 to interact with. It may be that the difference in the position of TM6 in the TSHR structure bound to K1-70™ and that seen in the LH/CGR is one of the reasons that the TSHR displays constitutive activity while LH/CGR does not (Van Sande et al. 1995).
Electron density is observed in the space in the TMD that is created by the displacement of TM6 of the TSHR in the TSHR–K1-70™ complex, but this does not fit with the structures of LMNG or CHS used in solubilisation and purification of the complex. Instead the electron density resembles that of a small phospholipid (Supplementary Fig. 3) which was most probably inside the receptor prior to solubilisation. Testing of potential phospholipid candidates for their ability to fit this electron density revealed inositol-1,4,5-triphosphate (IP3) as a good candidate which fits the electron density exactly. It is interesting to note that IP3 is a secondary signalling molecule produced by the activation of a Gq protein by GPCRs including the TSHR (Kleinau et al. 2010).
A structural superimposition of the full-length TSHR and the active LH/CGR structure (wild types LH/CGR in complex with hCG,Duan et al. 2021) shows an upward rotation of the ECD of the active LH/CGR by approximately 39o around the hinge region compared to the TSHR ECD (calculated using Ser281 Cα–Glu61 Cα vector from the TSHR and Ser277 Cα–Tyr58 Cα vector from the LH/CGR) and an upward displacement of approximately 2 Å between TSHR Ser281 and LH/CGR Ser277. A comparison of the TMDs of both receptors shows that TM2, TM4 and TM8 superimpose on top of each other, whereas the positions of TM1 and TM3 show a small difference. In contrast, the position of the extracellular end of LH/CGR TM5 differs by 2 Å and the extracellular ends of TM6 and TM7 by 3 Å towards the centre of the helix bundle compared to the TSHR. Furthermore the position of the intracellular end of TM6 in the active structure of LH/CGR relative to the TSHR is approximately 12 Å away from the centre of the helix bundle typical of the difference between the active and inactive states of GPCRs (Choe et al. 2011). These differences in the two structures suggest that our structure of the TSHR bound to the blocking autoantibody K1-70™ is not that of a fully active GPCR.
Analysis of the cryo-EM structures of the inactive and the active LH/CGR complexes (Duan et al. 2021) shows a rotation in the ECD of the receptor occurs when hCG binds in a so-called ‘push’ movement. Furthermore, in the case of FSH binding to the FSHR (Jiang et al. 2012), a sulfonated tyrosine residue surrounded by a negatively charged patch in the receptor’s long hinge loop has been shown to bind to the hormone, pulling the ECD upwards. In the case of hCG binding to LH/CGR, a similar ‘pull’ mechanism is suggested to occur (Duan et al. 2021). The upward movement of the LH/CGR ECD resulting from these ‘push’ and ‘pull’ effects has been proposed to cause receptor activation (Duan et al. 2021). Also, ‘push’ and ‘pull’ mechanisms similar to that proposed for hCG binding to LH/CGR may be involved in the hormone activation of the TSHR and FSHR.
In contrast, Tyr385 of the TSHR which has been shown to be required for stimulation of the TSHR by TSH (is not required for stimulation of the TSHR by autoantibodies or mouse monoclonal stimulating antibodies (Kosogi et al. 1991, Costagliola et al. 2002). Consequently, binding of M22™ and other stimulating antibodies to the TSHR does not appear to cause a corresponding ‘pull’" effect via the hinge loop through Tyr385.
Proposed mechanism of activation by M22
The crystal structure of TSHR260 in complex with the thyroid-stimulating human monoclonal TRAb M22™ shows that M22™ interacts with TSHR amino acids more C-terminal than those that interact with K1-70™ (Sanders et al. 2011). For example, TSHR Trp258 forms important interactions with the M22™ light chain, whereas Trp258 is not involved in K1-70™ binding.
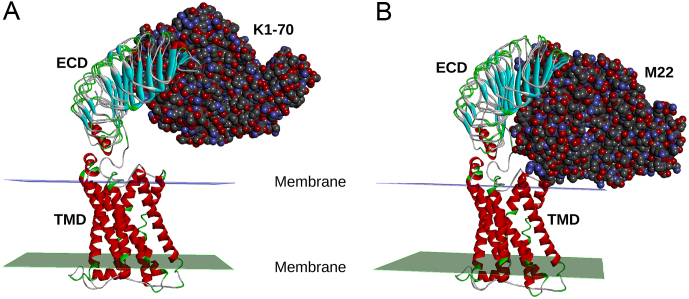
The implications of the differences in the TSHR LRD binding region of M22™ (more C-terminal) and K1-70™ (more N-terminal) can now be accessed using our cryo-EM full-length TSHR structure (Fig. 4). K1-70™ is well clear of the lipid bilayer in the TSHR–K1-70™ complex, whereas M22™ binding will result in a clash of the antibody light chain with the lipid bilayer unless the TSHR rotates upwards (i.e. away from the lipid bilayer) in the M22™ binding process. A similar ‘push’ process occurs when hCG binds to the LH/CG receptor (Duan et al. 2021) as mentioned earlier and stimulation of the TSHR by M22™ may also be caused by movements associated with such a ‘push’ process. This ‘push’ process would also explain the observations ofChazenbalk et al. (2002) who noted that thyroid-stimulating antibodies but not thyroid-blocking antibodies were partially sterically hindered when binding to the full-length TSHR compared to the TSHR ECD alone. One of their suggestions was that partial obstruction of the thyroid-stimulating autoantibody binding site may lead to a torsion effect on the TSHR ECD, on antibody binding, which may explain how thyroid-stimulating autoantibodies were able to activate the TSHR. Furthermore, Jiang et al. (2014) reported that their trimer model of full-length FSHR if applied to a TSHR trimer may explain the difference in the activities of M22™ (stimulating) and K1-70™ (blocking) autoantibodies, where M22™ but not K1-70™ would sterically clash with the TSHR hinge loop.
Figure 4.
Proposed mechanism of TSHR activation by M22™. The cryo-EM structure of the TSHR bound to the blocking autoantibody K1-70™ was superimposed with the crystal structure of the TSHR leucine-rich domain (LRD) in complex with K1-70™ (A) and with the crystal structure of the TSHR LRD in complex with the stimulating monoclonal autoantibody M22™ (B). K1-70™ does not contact the membrane while the stimulating antibody M22™ is seen to clash with the membrane. This clash between M22™ and the membrane would inhibit M22™ binding to the TSHR unless the receptor’s ECD rotates upwards as part of the M22™ binding process. This rotation could have an important role in TSHR stimulation by M22™.
The cryo-EM structure of the full-length TSHR in complex with the powerful blocking type monoclonal autoantibody K1-70™ (Figs 3 and 4) indicates that when K1-70™ binds to the TSHR LRD it does not form interactions with other parts of the receptor. This confirms that the ability of K1-70™ to prevent activation of the TSHR by TSH or by stimulating autoantibodies is due to the direct binding of K1-70™ to the same region of the TSHR LRD so that binding of other ligands is blocked. This understanding of the mechanism of action of K1-70™ is helpful in developing applications for its use in controlling activation of the TSHR in Graves’ disease (Furmaniak et al. 2022), in Graves’ orbitopathy (Furmaniak et al. 2022) and in thyroid cancer (Ryder et al. 2021).
Supplementary Material
Declaration of interest
RSR Ltd is a developer and manufacturer of in vitro medical diagnostics including kits for measuring autoantibodies to the TSHR. The work was carried out while all authors were employees of RSR Ltd.
Funding
The work was funded by RSR Ltd.
References
- Bahn RS.2010Graves’ ophthalmopathy. New England Journal of Medicine 362726–738. ( 10.1056/NEJMra0905750) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros JA, Weinstein H.1995Integrated methods for the construction of three dimensional models and computational probing of structure function relations in G protein-coupled receptors. Methods in Neurosciences 25366–428. ( 10.1016/S1043-9471(0580049-7) [DOI] [Google Scholar]
- BIOVIA Dassault Systèmes 2021Discovery Studio 2021. San Diego, CA, USA: BIOVIA Dassault Systèmes. [Google Scholar]
- Birk HW, Koepsell H.1987Reaction of monoclonal antibodies with plasma membrane proteins after binding on nitrocellulose: renaturation of antigenic sites and reduction of non-specific antibody binding. Analytical Biochemistry 16412–22. ( 10.1016/0003-2697(8790360-5) [DOI] [PubMed] [Google Scholar]
- Boutin A, Krieger CC, Marcus-Samuels B, Klubo-Gwiezdzinska J, Neumann S, Gershengorn MC.2020TSH receptor homodimerization in regulation of cAMP production in human thyrocytes in vitro. Frontiers in Endocrinology 11 276. ( 10.3389/fendo.2020.00276) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brüser A, Schulz A, Rothemund S, Ricken A, Calebiro D, Kleinau G, Schöneberg T.2016The activation mechanism of glycoprotein hormone receptors with implications in the cause and therapy of endocrine diseases. Journal of Biological Chemistry 291508–520. ( 10.1074/jbc.M115.701102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazenbalk GD, Tanaka K, Nagayama Y, Kakinuma A, Jaume JC, McLachlan SM, Rapoport B.1997Evidence that the thyrotropin receptor ectodomain contains not one, but two, cleavage sites. Endocrinology 1382893–2899. ( 10.1210/endo.138.7.5259) [DOI] [PubMed] [Google Scholar]
- Chazenbalk GD, Pichurin P, Chen CR, Latrofa F, Johnstone AP, McLachlan SM, Rapoport B.2002Thyroid-stimulating autoantibodies in Graves preferentially recognise free A subunit, not the thyrotropin holoreceptor. Journal of Clinical Investigation 110209–217. ( 10.1172/JCI15745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe HW, Kim YJ, Park JH, Morizumi T, Pa EF, Krauss N, Hofmann KP, Scheerer P, Ernst OP.2011Crystal structure of metarhodopsin II. Nature 471651–655. ( 10.1038/nature09789) [DOI] [PubMed] [Google Scholar]
- Costagliola S, Panneels V, Bonomi M, Koch J, Many MC, Smits G, Vassart G.2002Tyrosine sulfation is required for agonist recognition by glycoprotein hormone receptors. EMBO Journal 21504–513. ( 10.1093/emboj/21.4.504) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies TF, Ali MR, Latif R.2014Allosteric modulators hit the TSH receptor. Endocrinology 1551–5. ( 10.1210/en.2013-2079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Xu P, Cheng X, Mao C, Croll T, He X, Shi J, Luan X, Yin W, You Eet al. 2021Structures of full-length glycoprotein hormone receptor signalling complexes. Nature 598688–692. ( 10.1038/s41586-021-03924-2) [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K.2010Features and development of coot. Acta Crystallographica: Section D, Biological Crystallography 66486–501. ( 10.1107/S0907444910007493) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans M, Sanders J, Tagami T, Sanders P, Young S, Roberts E, Wilmot J, Hu X, Kabelis K, Clark Jet al. 2010Monoclonal autoantibodies to the TSH receptor, one with stimulating activity and one with blocking activity, obtained from the same blood sample. Clinical Endocrinology 73404–412. ( 10.1111/j.1365-2265.2010.03831.x) [DOI] [PubMed] [Google Scholar]
- Fan QR, Hendrickson WA.2005Structure of human follicle-stimulating hormone in complex with its receptor. Nature 433269–277. ( 10.1038/nature03206) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furmaniak J, Sanders J, Núñez Miguel R, Rees Smith B.2015Mechanisms of action of TSHR autoantibodies. Hormone and Metabolic Research 47735–752. ( 10.1055/s-0035-1559648) [DOI] [PubMed] [Google Scholar]
- Furmaniak J, Sanders J, Clark J, Wilmot J, Sanders P, Li Y, Rees Smith B.2019Preclinical studies on the toxicology, pharmacokinetics and safety of K1-70™ a human monoclonal autoantibody to the TSH receptor with TSH antagonist activity. Auto-Immunity Highlights 10 11. ( 10.1186/s13317-019-0121-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furmaniak J, Sanders J, Sanders P, Li Y, Rees Smith B.2022TSH receptor specific monoclonal autoantibody K1-70™ targeting of the TSH receptor in subjects with Graves’ disease and Graves’ orbitopathy – results from a phase I clinical trial. Clinical Endocrinology 96878–887. ( 10.1111/cen.14681) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govaerts C, Lefort A, Costagliola S, Wodak SJ, Ballesteros JA, Van Sande J, Pardo L, Vassart G.2001A conserved ASN in transmembrane helix 7 is an on/off switch in the activation of the thyrotropin receptor. Journal of Biological Chemistry 27622991–22999. ( 10.1074/jbc.M102244200) [DOI] [PubMed] [Google Scholar]
- Jiang X, Liu H, Chen X, Chen PH, Fischer D, Sriraman V, Yu HN, Arkinstall S, He X.2012Structure of follicle-stimulating hormone in complex with the entire ectodomain of its receptor. PNAS 10912491–12496. ( 10.1073/pnas.1206643109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Fischer D, Chen X, McKenna SD, Liu H, Sriraman V, Yu HN, Goutopoulos A, Arkinstall S, He X.2014Evidence for follicle-stimulating hormone receptor as a functional trimer. Journal of Biological Chemistry 28914273–14282. ( 10.1074/jbc.M114.549592) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC.2012Diversity and modularity of G protein-coupled receptor structures. Trends in Pharmacological Sciences 3317–27. ( 10.1016/j.tips.2011.09.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinau G, Claus M, Jaeschke H, Mueller S, Neumann S, Paschke R, Krause G.2007Contacts between extracellular loop two and transmembrane helix six determine basal activity of the thyroid-stimulating hormone receptor. Journal of Biological Chemistry 282518–525. ( 10.1074/jbc.M606176200) [DOI] [PubMed] [Google Scholar]
- Kleinau G, Jaeschke H, Worth CL, Mueller S, Gonzalez J, Paschke R, Krause G.2010Principles and determinants of G-protein coupling by the rhodopsin-like thyrotropin receptor. PLoS ONE 5 e9745. ( 10.1371/journal.pone.0009745) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinau G, Worth CL, Kreuchwig A, Biebermann H, Marcinkowski P, Scheerer P, Krause G.2017Structural-functional features of the thyrotropin receptor: a class A G-protein-coupled receptor at work. Frontiers in Endocrinology 8 86. ( 10.3389/fendo.2017.00086) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosogi S, Ban T, Akamizu T, Kohn LD.1991Further characterisation of a high affinity thyrotropin binding site on the rat thyrotropin receptor which is an epitope for blocking antibodies from idiopathic myxedema patients but not thyroid stimulating antibodies from Graves’ patients. Biochemical and Biophysical Research Communications 1801118–1124. ( 10.1016/S0006-291X(0581182-9) [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hai N, Okamoto H, Sugawa H, Mori T.2000A novel activating mutation in the thyrotropin receptor gene in an autonomously functioning thyroid nodule developed by a Japanese patient. European Journal of Endocrinology 143471–477. ( 10.1530/eje.0.1430471) [DOI] [PubMed] [Google Scholar]
- Krause G, Eckstein A, Schülein R.2020Modulating TSH receptor signaling for therapeutic benefit. European Thyroid Journal 966–77. ( 10.1159/000511871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK.1970Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680–685. ( 10.1038/227680a0) [DOI] [PubMed] [Google Scholar]
- Latif R, Ali MR, Mezei M, Davies TF.2015Transmembrane domains of attraction on the TSH receptor. Endocrinology 156488–498. ( 10.1210/en.2014-1509) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Gallacher J, Sanders P, Young S, Sullivan A, Baker S, Reddington SC, Clue M, Kabelis K, Clark J, Wilmot Jet al. 2019Crystal structure of a ligand-free stable TSH receptor leucine-rich repeat domain. Journal of Molecular Endocrinology 62117–128. ( 10.1530/JME-18-0213) [DOI] [PubMed] [Google Scholar]
- Morshed SA, Latif R, Davies TF.2012Delineating the autoimmune mechanisms in Graves’ disease. Immunologic Research 54191–203. ( 10.1007/s12026-012-8312-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara E, Fukata S, Hishinuma A, Kudo T, Ohye H, Ito M, Kubota S, Amino N, Kuma K, Miyauchi A.2006Sporadic congenital hyperthyroidism due to a germline mutation in the thyrotropin receptor gene (Leu 512 Gln) in a Japanese patient. Endocrine Journal 53735–740. ( 10.1507/endocrj.k06-090) [DOI] [PubMed] [Google Scholar]
- Núñez Miguel R, Sanders J, Sanders P, Young S, Clark J, Kabelis K, Wilmot J, Evans M, Roberts E, Hu Xet al. 2012Similarities and differences in interactions of thyroid stimulating and blocking autoantibodies with the TSH receptor. Journal of Molecular Endocrinology 49137–151. ( 10.1530/JME-12-0040) [DOI] [PubMed] [Google Scholar]
- Núñez Miguel R, Sanders J, Furmaniak J, Rees Smith BR.2017aStructure and activation of the TSH receptor transmembrane domain. Auto-Immunity Highlights 8 2. ( 10.1007/s13317-016-0090-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez Miguel R, Sanders J, Furmaniak J, Rees Smith B.2017bGlycosylation pattern analysis of glycoprotein hormones and their receptors. Journal of Molecular Endocrinology 5825–41. ( 10.1530/JME-16-0169) [DOI] [PubMed] [Google Scholar]
- Oda Y, Sanders J, Roberts S, Maruyama M, Kato R, Perez M, Petersen VB, Wedlock N, Furmaniak J, Rees Smith B.1998Binding characteristics of antibodies to the TSH receptor. Journal of Molecular Endocrinology 20233–244. ( 10.1677/jme.0.0200233) [DOI] [PubMed] [Google Scholar]
- Oda Y, Sanders J, Evans M, Kiddie A, Munkley A, James C, Richards T, Wills J, Furmaniak J, Smith BR.2000Epitope analysis of the human thyrotropin (TSH) receptor using monoclonal antibodies. Thyroid 101051–1059. ( 10.1089/thy.2000.10.1051) [DOI] [PubMed] [Google Scholar]
- Parma J, Van Sande J, Swillens S, Tonacchera M, Dumont J, Vassart G.1995Somatic mutations causing constitutive activity of the thyrotropin receptor are the major cause of hyperfunctioning thyroid adenomas: identification of additional mutations activating both the cyclic adenosine 3’,5’-monophosphate and inositol phosphate-Ca2+ cascades. Molecular Endocrinology 9725–733. ( 10.1210/mend.9.6.8592518) [DOI] [PubMed] [Google Scholar]
- Parma J, Duprez L, Van Sande J, Hermans J, Rocmans P, Van Vliet G, Costagliola S, Rodien P, Dumont JE, Vassart G.1997Diversity and prevalence of somatic mutations in the thyrotropin receptor and Gs alpha genes as a cause of toxic thyroid adenomas. Journal of Clinical Endocrinology and Metabolism 822695–2701. ( 10.1210/jcem.82.8.4144) [DOI] [PubMed] [Google Scholar]
- Porcellini A, Ciullo I, Laviola L, Amabile G, Fenzi G, Avvedimento VE.1994Novel mutations of thyrotropin receptor gene in thyroid hyperfunctioning adenomas. Rapid identification by fine needle aspiration biopsy. Journal of Clinical Endocrinology and Metabolism 79657–661. ( 10.1210/jcem.79.2.8045989) [DOI] [PubMed] [Google Scholar]
- Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA.2017cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature Methods 14290–296. ( 10.1038/nmeth.4169) [DOI] [PubMed] [Google Scholar]
- Rapoport B, Chazenbalk GD, Jaume JC, McLachlan SM.1998The thyrotropin (TSH) receptor: interaction with TSH and autoantibodies. Endocrine Reviews 19673–716. ( 10.1210/edrv.19.6.0352) [DOI] [PubMed] [Google Scholar]
- Rees Smith B, McLachlan SM, Furmaniak J.1988Autoantibodies to the thyrotropin receptor. Endocrine Reviews 9106–121. ( 10.1210/edrv-9-1-106) [DOI] [PubMed] [Google Scholar]
- Russo D, Arturi F, Suarez HG, Schlumberger M, Du Villard JA, Crocetti U, Filetti S.1996Thyrotropin receptor gene alterations in thyroid hyperfunctioning adenomas. Journal of Clinical Endocrinology and Metabolism 811548–1551. ( 10.1210/jcem.81.4.8636365) [DOI] [PubMed] [Google Scholar]
- Ryder M, Wentworth M, Algeciras-Schimnich A, Morris JC, Garrity J, Sanders J, Young S, Sanders P, Furmaniak J, Rees Smith B.2021Blocking the thyrotropin receptor with K1-70 in a patient with follicular thyroid cancer, Graves’ disease, and Graves’ ophthalmopathy. Thyroid 311597–1602. ( 10.1089/thy.2021.0053) [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL.1993Comparative protein modelling by satisfaction of spatial restraints. Journal of Molecular Biology 234779–815. ( 10.1006/jmbi.1993.1626) [DOI] [PubMed] [Google Scholar]
- Sanders J, Evans M, Premawardhana L, Depraetere H, Jeffreys J, Richards T, Furmaniak J, Rees Smith BR.2003Human monoclonal thyroid stimulating autoantibody. Lancet 362126–128. ( 10.1016/S0140-6736(0313866-4) [DOI] [PubMed] [Google Scholar]
- Sanders J, Chirgadze DY, Sanders P, Baker S, Sullivan A, Bhardwaja A, Bolton J, Reeve M, Nakatake N, Evans Met al. 2007Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody. Thyroid 17395–410. ( 10.1089/thy.2007.0034) [DOI] [PubMed] [Google Scholar]
- Sanders P, Young S, Sanders J, Kabelis K, Baker S, Sullivan A, Evans M, Clark J, Wilmot J, Hu Xet al. 2011Crystal structure of the TSH receptor bound to a blocking type TSHR autoantibody. Journal of Molecular Endocrinology 4681–99. ( 10.1530/JME-10-0127) [DOI] [PubMed] [Google Scholar]
- Schaarschmidt J, Nagel MBM, Huth S, Jaeschke H, Moretti R, Hintze V, Von Bergen M, Kalkhof S, Meiler J, Paschke R.2016Rearrangement of the extracellular domain/extracellular loop1 interface is critical for thyrotropin receptor activation. Journal of Biological Chemistry 29114095–14108. ( 10.1074/jbc.M115.709659) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegunov D, Cramer P.2019Real-time cryo-electron microscopy data preprocessing with warp. Nature Methods 161146–1152. ( 10.1038/s41592-019-0580-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehan BG, Bortolato A, Blaney FE, Weir MP, Mason JS.2014Unifying family A GPCR theories of activation. Pharmacology and Therapeutics 14351–60. ( 10.1016/j.pharmthera.2014.02.004) [DOI] [PubMed] [Google Scholar]
- Trülzsch B, Krohn K, Wonerow P, Chey S, Holzapfel HP, Ackermann F, Führer D, Paschke R.2001Detection of thyroid-stimulating hormone receptor and Gsalpha mutations: in 75 toxic thyroid nodules by denaturing gradient gel electrophoresis. Journal of Molecular Medicine 78684–691. ( 10.1007/s001090000170) [DOI] [PubMed] [Google Scholar]
- Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Žídek A, Bridgland A, Cowie A, Meyer C, Laydon Aet al. 2021Highly accurate protein structure prediction for the human proteome. Nature 596590–596. ( 10.1038/s41586-021-03828-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urizar E, Montanelli L, Loy T, Bonomi M, Swillens S, Gales C, Bouvier M, Smits G, Vassart G, Costagliola S.2005Glycoprotein hormone receptors: link between receptor homodimerization and negative cooperativity. EMBO Journal 241954–1964. ( 10.1038/sj.emboj.7600686) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sande J, Parma J, Tonacchera M, Swillens S, Dumont J, Vassart G.1995Somatic and germline mutations of the TSH receptor gene in thyroid diseases. Journal of Clinical Endocrinology and Metabolism 802577–2585. ( 10.1210/jcem.80.9.7673398) [DOI] [PubMed] [Google Scholar]
- Webb B, Sali A.2016Comparative protein structure modeling using modeller. Current Protocols in Bioinformatics 545.6.1–5.6.37. ( 10.1002/cpbi.3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Qiao A, Yang D, Yang L, Dai A, de Graaf C, Reedtz-Runge S, Dharmarajan V, Zhang H, Han GWet al. 2017Structure of the full-length glucagon class B G-protein-coupled receptor. Nature 546259–264. ( 10.1038/nature22363) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Qiao A, Yang L, Van Eps N, Frederiksen KS, Yang D, Dai A, Cai X, Zhang H, Yi Cet al. 2018Structure of the glucagon receptor in complex with a glucagon analogue. Nature 553106–110. ( 10.1038/nature25153) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a