Abstract
Scope
Epidemiological evidence suggests that a reduced risk of colorectal cancer (CRC) is correlated with high consumption of fruits and vegetables, which are major sources of fiber and phytochemicals, such as flavan‐3‐ols. However, it remains unknown how these phytochemicals and their specific gut‐related metabolites may alter cancer cell behavior.
Methods and results
A focused screening using native (poly)phenols and gut microbial metabolites (GMMs) on 3D HCT116 spheroids is carried out using a high‐throughput imaging approach. Dose–responses, IC50, and long‐term exposure are calculated for the most promising native (poly)phenols and GMMs. As a result, this research shows that (poly)phenol catabolites may play a key role in preventing cancer propagation. Indeed, µM concentration levels of (4R)‐5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone significantly decrease spheroid size at early stages of spheroid aggregation and gene expression of matrix metalloproteinases.
Conclusion
A chronic exposure to (4R)‐5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone may lead to a reduced CRC risk. Daily intake of monomeric, oligomeric, and polymeric flavan‐3‐ols may increase the colonic concentrations of this metabolite, and, in turn, this compound may act locally interacting with intestinal epithelial cells, precancerous and cancer cells.
Keywords: 3D spheroids, colorectal cancer, flavan‐3‐ols, gut microbial metabolites, high content imaging
This work explores the potential of native phytochemicals and gut microbial metabolites in promoting gut health. 3D HCT116 spheroids are first exposed to (poly)phenols and (poly)phenols metabolized by the gut microbiota. Secondly, by combining high‐throughput imaging techniques and transcriptional profiling, this research shows that a flavan‐3‐ol metabolite may play a pivotal role in promoting gut health.
1. Introduction
Diet‐gut microbiota interactions exhibit a substantial impact on intestinal epithelial cells (IECs).[ 1 ] Apart from the host genetics, environmental factors, dietary patterns, and microbiome dysbiosis, gut microbial metabolites (GMMs) play a pivotal role in the homeostasis[ 2 ] of IECs and progression of diseases, such as colorectal cancer (CRC).[ 3 ] CRC is one of the most commonly diagnosed malignancies and one of the most common causes of cancer death in Europe.[ 4,5 ] It is estimated that the global burden of CRC will increase by 60%, with over 2.2 million new cases by 2030,[ 4 ] due to an aging population and Western dietary patterns. Dietary patterns modulate the gut microbiota community and influence its function,[ 6 ] leading to changes in metabolite production.[ 7 , 8 , 9 , 10 , 11 ] Then, GMMs can quickly reach the gut epithelium, promoting gut health[ 12 ] or triggering gastrointestinal (GI) diseases.[ 13 ] The scientific literature has brilliantly explained the essential function of short‐chain fatty acids (SCFAs) released from the fermentation of dietary fibers. Acetic acid, propionic acid, and butyric acid have been linked to bacteria producers and their key roles in regulating host metabolism, immune system, cell proliferation, mucus production, and secretion.[ 14 ] By contrast, how gut microbiota interacts with bioactive phytochemicals, such as (poly)phenols,[ 15 ] species involved in the breakdown of (poly)phenols,[ 16 ] and specific GMMs that may alter cancer cell behavior have been scarcely studied.[ 17 , 18 ]
Evidence suggests that a reduced risk of CRC[ 19 , 20 , 21 ] is associated with high consumption of fruit and vegetables, which are major sources of fiber and phytochemicals, such as (poly)phenols.[ 22 , 23 , 24 ] After the digestion of fruit and vegetables, up to 90–95% of oligomeric (poly)phenols, mainly proanthocyanidins (PACs), are not absorbed by the small intestine[ 8 , 25 ] and, together with nondigestible polysaccharides, reach the colon almost intact,[ 26 ] where they interact with gut microbiota.[ 7 , 8 ] Firstly, PACs, among other (poly)phenols, and fiber undergo extensive microbial bioconversion, producing hydroxyphenyl‐γ‐valerolactones (PVLs), their derived hydroxyphenylvaleric acids (PVAs), other phenolic acids, and SCFAs.[ 26 , 27 ] Lastly, these GMMs may then act locally interacting with IECs,[ 28 ] but these catabolites can also either modulate the gut microbiota[ 15 ] or be absorbed and released into the bloodstream.[ 8 , 29 , 30 ] Dietary patterns are associated with health outcomes, however beneficial effects of GMMs, particularly those derived from flavan‐3‐ols, are often difficult to demonstrate in epidemiological and interventional studies targeting gut health because the gut microbiota underlies interindividual variability,[ 31 ] and then, metabolites occur locally and are rapidly absorbed.[ 28 ]
The intestinal epithelium is rapidly renewed. The continuous cellular turnover of the intestinal epithelium is conserved by Lgr5+ intestinal stem cells, which generate transit‐amplifying cells, and then differentiate into various IECs.[ 32 ] Intestinal stem cells have been linked with the origin of CRC,[ 33 ] a multistage process that progresses from normal cells to neoplastic transformation and tissue invasion.[ 34 ] Throughout this process, the extracellular matrix plays a pivotal role in CRC. The extracellular matrix provides an essential physical scaffolding and regulates important biochemical processes, mediating cell adhesion and cell signaling, thereby regulating processes as diverse as proliferation, differentiation, migration, and apoptosis.[ 35 ] This specialized niche is maintained by degrading enzymes released to rehabilitate the extracellular matrix and to reinstate tissue homeostasis. However, in cancer progression remodeling is seized control, and extracellular components and enzymes lead toward tumorigenesis. One important step in CRC invasion is the disassembly of the extracellular matrix and its constituents through enzymes such as matrix metalloproteinases (MMPs).[ 36 ] Dietary phenolic compounds have been shown to act at multiple key steps in carcinogenesis and inflammation, modulating key signal transduction pathways, as those mediated by NF‐κB and STAT, PI3K and COX9,[ 22 , 37 ] but also other linked to cancer propagation, such as MPPs.[ 38 ] However, the role of specific GMMs, such as PVLs, PVAs, and small phenolic acids in modulating these pathways and preventing GI diseases is limited.
Several strategies have been proposed to determine the local effects of native phytochemicals or GMMs on cancer cells of the GI tract. Immortalized human cancer cell lines grown as monolayers (2D) is the most popular model to study the bioactivity of food‐related compounds.[ 22 , 31 , 39 , 40 , 41 , 42 ] However, 2D cell models have serious drawbacks. For instance, they do not recapitulate the tumor cell microenvironment, lack dimensionality, and differ metabolically, genetically, and phenotypically from in vivo cells.[ 43 ] As an alternative, animal models have been used to translate and confirm hypotheses observed by 2D in vitro models.[ 44 , 45 ] Although animal models represent a powerful approach for bioactive assessment, they still present several limitations. First, the gut environment including the gut architecture, animal diet, and gut microbiota is nonhuman.[ 46 ] Secondly, there are high‐costs related to animal care and breeding, and ethical concerns to be considered.[ 41 ] During the last decade, 3D models have emerged as an alternative to animal models and 2D cell lines.[ 12 , 41 ] Tumor spheroids mitigate the disadvantages of the 2D cell models, gaining intrinsic properties and better mimicking the in vivo scenario, for instance, cell‐to‐cell and cell‐extracellular matrix interactions.[ 47 ] At the same time, protocols are simple, reproducible, and cost‐effective to reveal the role and dose–response of food‐related compounds in preventing CRC.
In this research work, we developed a high‐throughput imaging strategy to investigate the responses of 3D HCT116 spheroids to native (poly)phenols, such as flavan‐3‐ols, flavonols, flavones, dihydrochalcones, isoflavones, stilbenes, phenolic acids, and GMMs like PVLs, PVAs and small phenolic acids, as well as other bioactive compounds. As a first step, a focused screening was performed to investigate the ability of those compounds to affect 3D spheroid integrity (SI). Afterward, dose–response relationship was determined for the most promising native (poly)phenols and GMMs, and lastly, we investigated in depth the role of one PVL. This research shows that specific (poly)phenol catabolites may play a role in promoting gut health, and provides vital information for the future design of in vivo studies.
2. Results and Discussion
2.1. The Impact of Native (Poly)phenols and GMMs on Spheroid Integrity
2.1.1. Optimization of a High‐Throughput Imaging Workflow
Firstly, native phytochemicals, as they occur in plants, and released GMMs were selected. A detailed Table S1, it can be found in the Supporting Information. These native (poly)phenols were chosen because they can be found in many fruits that humans consume daily, such as apples, pears, stone fruits, cranberries, blueberries, grapes, and foodstuffs such as wine, tea, sorghum, lentils, nuts, and dark chocolate, among others.[ 26 , 30 ] Afterwards, as mentioned above, (poly)phenols are the substrate needed by the gut microbiota to release specific GMMs, such as PVLs, PVAs, and small phenolic acids. Secondly, a cell model closely recapitulating an in vivo tumor environment was used. To visualize cell morphology and quantify efficiently phenotypic changes by imaging techniques, this line was genetically modified to express an EGFP protein (HTC116 EGFP).[ 48 ] Then, cell adhesion, cell proliferation, and the capacity of spheroid formation were compared. Both 2D and 3D HCT116 WT and HCT116 EGFP responded equally to 5‐FU (Figure S1, Supporting Information). Lastly, the Z’ factor was determined to quantify the suitability of the high‐throughput screening. 3D HTC116 EGFP spheroids (n = 27/condition) were treated with 5‐FU (50 µM), chemotherapeutic agent used for colon cancer carcinoma, which was used as a positive control, and with a negative control, DMSO 0.05%, for 72 h to measure the Z’ factor parameter.[ 49 ] Considering that this parameter ranges from one (excellent assay) to below zero values (unworkable assay), a Z’ factor of 0.4 indicates a feasible assay to study 3D spheroid responses to (poly)phenols and gut‐related metabolites (Figure S2, Supporting Information).
2.1.2. A Screening of Phytochemicals and GMMs
At this point, the screening assay was performed evaluating the role of native phytochemicals and GMMs. As several studies claimed, the colonic concentration of (poly)phenols and GMMs may range from µM to mM.[ 28 , 31 ] However, it is difficult to quantify accurately in vivo the colonic concentration of these compounds since the gut microbiota leads concurrent biotransformations and the metabolized forms may rapidly be absorbed by enterocytes. Note that physiological concentrations ranging from 10 to 100 µM of various compounds studied in the research work exerted limited cytotoxicity on normal human colon cell lines.[ 50 , 51 ] The SI was evaluated on 3D HCT116 EGFP spheroids, which were immediately treated (50 mM), after seeding, for a period of 72 h from at least four independent experiments. Spheroids were then imaged, and the SI was calculated (Table S2, Supporting Information). 3D spheroids were initially treated at an early time point,[ 52 , 53 ] because we hypothesized that native (poly)phenols and GMMs play a role in prevention, modulating cancer cell growth, and propagation that occur early in spheroid formation. Only those native (poly)phenols and GMMs that were able to alter the SI more than 20% (SI inferior to 80%) were further evaluated (Table 1 and Figure S3, Supporting Information). The screening revealed that (+)‐catechin and (−)‐epicatechin did not alter the SI (Table S2, Supporting Information). By contrast, ursolic acid, pyrogallol, gallic acid, and flavan‐3‐ol monomers with three hydroxyl groups ((+)‐gallocatechin) and esterified forms with gallic acid ((+)‐gallocatechin‐3‐gallate (GCG) and (−)‐epigallocatechin‐3‐gallate (EGCG)) significantly affected the SI (Table 1). Various of these natural products exhibit cancer chemopreventive and chemotherapeutic activities. For example, ursolic acid has been used as natural product‐based nanoformulations targeting cancer cells,[ 54 ] and has also inhibited proliferation on cancer HCT116 cells.[ 55 ] A recent study has proven the EGCG mechanism of action on cancer cells. This green tea flavan‐3‐ol shields the murine double minute 2 (MDM2) binding site of p53‐N‐terminal domain, thus stabilizing p53 by inhibiting p53 ubiquitination and degradation, and, in turn, promotes apoptosis.[ 56 ] Besides, the gallic acid portion of the EGCG may interact with the cytosine active site on the DNA methyltransferase enzyme, which may be one of the reasons EGCG can also be an effective DNA methyltransferase inhibitor.[ 57 ] Although these phenolics and monomeric flavan‐3‐ols disturbed the SI and 3D spheroid propagation and, in a real‐life scenario, a small fraction of the ingested amount is rapidly absorbed in the upper GI tract,[ 58 , 59 ] the bulk of phenolics, (epi)(gallo)catechins, and (epi)(gallo)catechin‐3‐gallates arrive unmetabolized at the colonic region, where they are rapidly biotransformed by the gut microbiota.[ 60 , 61 ] Therefore, the expected low concentrations and the rapid biotransformation in the colonic region suggest that flavan‐3‐ol monomers do not directly play a key role in preventing CRC, as described elsewhere.[ 21 ] The same applies to PACs, such as, procyanidins B1 and B2, that arrive nearly intact at the distal GI tract and, in this study, did not influence the SI (Table S2, Supporting Information). Even if the direct repercussion on IECs of monomeric and oligomeric flavan‐3‐ols may be considered marginal, in a real‐life scenario, indirectly, they may be crucial in promoting GI health. Because the gut microbiota uses monomeric, dimeric, and oligomeric flavan‐3‐ols as substrate to produce PVLs, PVAs, and small phenolic acids.[ 28 , 62 , 63 ] The SI was notably affected by one PVL, (4R)‐5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone (44.7%), one PVA, 5‐(3′,4′‐dihydroxyphenyl)valeric acid (68.7%), and several small phenolic acids, such as benzene‐1,2‐diol (50.7%), 3′,4′‐dihydroxyphenylacetic acid (67.6%), and 4′‐hydroxy‐3′‐methoxycinnamic acid (also named ferulic acid) (80.1%), as it can be seen in Table 1. PVLs belong to an important class of flavan‐3‐ol microbial metabolites characterized by their high concentrations in different compartments. The nutrikinetics pattern of PVLs is defined by a steady concentration in plasma, and a delayed appearance in urine caused by a prolonged breakdown of PACs, (epi)(gallo)catechins, and (epi)(gallo)catechin‐3‐gallates in the colonic region.[ 8 ] For example, Brindani et al.[ 64 ] reported urinary concentrations of PVLs reaching 132 µM after 1 week of green tea supplementation. In another study, after a single intake of 400 g of Elstar apple the cumulative excretion of PVLs and PVAs resulted on 524 µM.[ 31 ] Taken all together, this evidence suggests that i) the gut microbiota plays a pivotal role shaping the chemical structure of flavan‐3‐ols, and ii) µM concentrations in the colonic region may affect cancer cell behavior.
Table 1.
Spheroid integrity (SI) was calculated as the average value and standard deviations (SD) for each polyphenol and gut‐related metabolite
| Compound | SI | SD |
|---|---|---|
| (+)‐Gallocatechin‐3‐gallate (GCG) | 1.0 | 1.1 |
| Benzene‐1,2,3‐triol (pyrogallol) | 1.5 | 1.6 |
| (−)‐Epigallocatechin‐3‐gallate (EGCG) | 1.6 | 0.6 |
| Ursolic acid | 1.9 | 2.5 |
| (+)‐Gallocatechin | 9.7 | 4.9 |
| 3,4,5‐Trihydroxybenzoic acid (gallic acid) | 34.0 | 19.1 |
| 5‐Fluorouracil | 39.0 | 10.8 |
| (4R)‐5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone | 44.7 | 22.3 |
| Benzene‐1,2‐diol (catechol or pyrocatechol) | 50.7 | 16.3 |
| Naringenin | 54.7 | 6.3 |
| Resveratrol | 55.6 | 27.7 |
| Luteolin | 58.9 | 28.2 |
| 2′‐Hydroxycinnamic acid (o‐coumaric acid) | 63.8 | 36.5 |
| 3‐(3′,4′‐Dihydroxyphenyl)propanoic acid (dihydrocaffeic acid) | 65.2 | 14.7 |
| 3′,4′‐Dihydroxyphenylacetic acid | 67.6 | 18.8 |
| 5‐(3′,4′‐Dihydroxyphenyl)valeric acid | 68.7 | 7.9 |
| Glycitein 7‐O‐glucoside (glycitin) | 76.1 | 10.1 |
| 4′‐Hydroxy‐3′‐methoxycinnamic acid (ferulic acid) | 80.1 | 13.4 |
3D HTC116 spheroids were treated at 50 µM from at least four independent experiments. After 72 h of exposure, the SI parameter was calculated as a percentage of the control (DMSO 0.05%).
2.2. Zooming in the Potential of PVLs and Small Phenolic Acids as Inhibitors of 3D HTC116 Spheroid Aggregation
The first broad screening demonstrated that low molecular phenolic compounds released by the colonic microbiota substantially affected 3D SI. This morphological parameter was remarkably affected by one PVL, 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone. For this reason, we decided to further evaluate the influence of the chemical structure. 3D HCT116 spheroids were treated with 50 µM of PVLs ((4R)‐5‐(3′‐hydroxyphenyl)‐γ‐valerolactone, (4S)‐5‐(4′‐hydroxyphenyl)‐γ‐valerolactone, (4R)‐5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone, and (4R)‐5‐(3′,5′‐dihydroxyphenyl)‐γ‐valerolactone), one phase II conjugate ((4R)‐5‐(3′‐hydroxyphenyl)‐γ‐valerolactone‐4′‐sulfate), and small phenolic acids (3‐hydroxybenzoic acid, 3′‐hydroxyphenylacetic acid, 4′‐hydroxyphenylacetic acid, and 3‐(3′‐hydroxy‐4′‐methoxyphenyl)propanoic acid (also named dihydro‐isoferulic acid)) (Figure 1 ).
Figure 1.
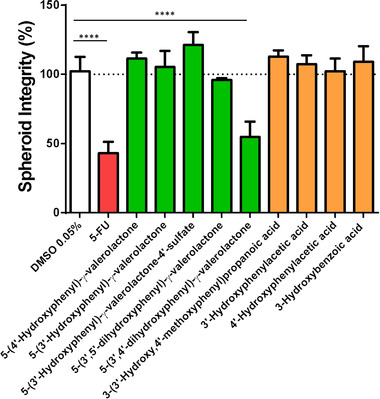
The impact of hydroxy‐phenyl‐γ‐valerolactones and small phenolic acids on 3D HCT116 EGFP spheroid integrity. Data are the mean ± SEM from at least four independent experiments. One‐way ANOVA followed by Holm–Sidak test (**** Adjusted p < 0.0001).
Following the proposed biotransformation pathway,[ 61 ] PACs and (epi)catechins are biotransformed releasing 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone.[ 28 , 30 ] As it can be seen in Figure 1, this GMM significantly affected SI (adjusted p < 0.0001), likewise the positive control 5‐FU, a drug commonly used in CRC treatments. Afterward, this GMM may be dehydroxylated to yield the analogous 5‐(3′‐hydroxyphenyl)‐γ‐valerolactone and 5‐(4′‐hydroxyphenyl)‐γ‐valerolactone derivatives. In this study, these colonic metabolites did not alter SI. Neither the sulfate conjugate 5‐(3′‐hydroxyphenyl)‐γ‐valerolactone‐4′‐sulfate did it (Figure 1). On the other hand, (epi)gallocatechins are colonic precursors of 5‐(3′,4′,5′‐trihydroxyphenyl)‐γ‐valerolactone, which is consecutively dehydroxylated to generate the analogous 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone and 5‐(3′,5′‐dihydroxyphenyl)‐γ‐valerolactone. The hydroxyl groups at the C3′ and C4′ positions of ring B highlighted a superior bioactivity (adjusted p < 0.0001) compared to C3′ and C5′ positions (Figure 1). Lastly, the small phenolic acids tested, end‐products of the proposed biotransformation pathway of PACs, did not modify the SI. However, if we compared these results with the previous screening (Table S2, Supporting Information), 3‐(3′,4′‐dihydroxyphenyl)propanoic acid and 3′,4′‐dihydroxyphenylacetic acid, containing two hydroxyl groups, were more effective lowering the SI than those small phenolics having exclusively one hydroxyl group (Figure 1 and Table S2, Supporting Information). These metabolites bearing the ortho‐dihydroxy substitution, are reported among the major circulating metabolites, deriving from the dietary intake of catechin, epicatechin and procyanidins, and flavan‐3‐ol oligomers.[ 28 ]
2.2.1. Native (Poly)phenols and GMMs: Half‐Maximal Inhibitory Concentrations (IC50)
Eleven compounds were further validated using the IC50. The IC50 calculates efficacy and indicates how much compound is needed to inhibit SI by half. Spheroid treatments were performed at early stages of aggregation (t = 0 h), and when spheroids were fully formed (t = 72 h). Phytochemical and GMM concentrations ranged from 0 to 100 µM, as these concentrations are in line with the expected amount of phenolic compounds in the intestinal lumen upon dietary (poly)phenol consumption.[ 65 , 66 ]
To further validate our research, butyric acid was first used as a positive control. Butyric acid is a SCFA present in a high concentration in the gut lumen. This GMM is produced by the fermentation of dietary fiber via the gut microbiota. Butyric acid is used by healthy colonocytes as an energy source and promotes proliferation.[ 14 ] However, this metabolite also inhibits cell proliferation and induces apoptosis on cancer cells.[ 9 , 67 ] Several studies have reported that butyric acid inhibited cell proliferation (250–10 000 µM) on different 2D immortalized cancer cell lines.[ 68 , 69 ] At this point, we investigated how much butyric acid was needed to inhibit SI by half on formed‐spheroids (IC50 = 670 µM) and on early stages of spheroid formation (IC50 = 712 µM), (Figure S4, Supporting Information). These responses were similar to those obtained from the 2D‐cultured HCT116 cells.[ 62 , 68 ] However, we noticed that higher concentrations of butyric acid were needed to effectively inhibit proliferation on HCT116 spheroids.[ 47 ] As expected, butyric acid was able to spill into the nucleus and act as a histone deacetylase inhibitor, increasing differentiation and apoptosis, and suppressing proliferation on spheroids in formation and formed spheroids.
At this stage, the IC50 was first calculated for flavan‐3‐ol monomers with three hydroxyl groups ((+)‐gallocatechin), esterified forms with gallic acid (EGCG and (−)‐epicatechin‐3‐gallate) and 3,4,5‐trihydroxybenzoic acid (aka gallic acid) on early stages of aggregation (t = 0 h) (Figure 2 ). (−)‐Epicatechin‐3‐gallate (IC50 = 70.3 µM), (+)‐gallocatechin (IC50 = 74.6 µM), and gallic acid (IC50 = 68.1 µM) as single agents highlighted similar efficacies to reduce SI by half. However, the gallic acid esterification with (−)‐epigallocatechin, EGCG (IC50 = 35.2 µM), showed a superior efficacy. Indeed, the effect was amplified two fold. When spheroids were fully formed (t = 72 h), we noticed that SI was to some extent altered, but fully formed spheroids tolerated higher concentration (Figure 2). GIC cells were also used to study how native (poly)phenols could alter SI and viability in other cell lines (Figure S5, Supporting Information). This experiment illustrated that the SI on 3D HCT116 (IC50 = 35.2 µM) and 3D GIC (IC50 = 22.6 µM) spheroids in aggregation was uniformly affected by EGCG. Lastly, 3D GICs spheroids were also employed to study whether quercetin, isorhamnetin, and a type II metabolite, quercetin‐3‐glucuronide, could alter SI since no effects of these compounds on 3D HTC116 were observed. In this sense, quercetin did not alter SI on GIC cells, while 100 µM of quercetin‐3‐glucuronide showed an inhibition by half of SI on GIC cells (Figure S6, Supporting Information). Several studies have investigated quercetin and luteolin claiming several cancer relevant phenotypes in GBM cells, GICs, and tumor‐organoids.[ 70 , 71 ]
Figure 2.
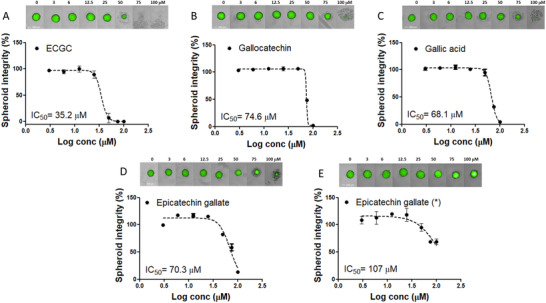
Seven‐point dose‐response curve of EGCG A), gallocatechin B), gallic acid C), and epicatechin gallate D) on HCT116 EGFP spheroids in aggregation and epicatechin gallate on formed spheroids E). Spheroids were exposed to increasing concentrations from 3 to 100 µM for 72 h. Spheroid integrity was quantified by the High Content Imaging System Operetta (Perkin Elmer) and expressed as a percentage of SI of vehicle‐treated controls (DMSO 0.05%). IC50 was calculated applying dose–response‐inhibition nonlinear regression analysis. Data are expressed as mean ± SEM, and all experiments were performed from at least four independent experiments. Spheroids were imaged on High Content Imaging System Operetta (Perkin Elmer) with 10XLWD objective in brightfield and green fluorescent channels (merged images) at different concentrations. Scale bars, 200 µm.
As a last step, we compared the efficacy of 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone and 5‐(3′,4′‐dihydroxyphenyl)valeric acid on formed spheroids and at the early stage of spheroid formation in reducing SI (Figure 3 ). The effectiveness of these GMMs was associated with prevention, since 3D SI was substantially affected at early stages. For example, a concentration of 78.2 µM of 5‐(3′,4′‐dihydroxyphenyl)valeric acid was needed to inhibit SI by half. By contrast, 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone showed 50% efficacy at 51 µM. When spheroids were fully formed, 5‐(3′,4′‐dihydroxyphenyl)valeric acid could alter SI at the highest concentration points, while 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone did not affect SI at the concentrations studied. Cells in the aggregation were sensitive to 5‐(3′,4′‐dihydroxyphenyl)valeric acid and 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone, the latter being more significant. Whereas the cells in compact spheroids were resistant to the range of physiological concentration studied. This evidence suggests the role of flavan‐3‐ol metabolites as preventive agents.
Figure 3.

Dose–response curves of 5‐(3′,4′‐dihydroxyphenyl)valeric acid (PVA) A) and 5‐(3′,4′‐ dihydroxyphenyl)‐γ‐valerolactone (PVL) B) on 3D HCT116 EGFP spheroids in aggregation and on formed spheroids C and D). The concentration tested for (3′,4′‐dihydroxyphenyl)‐γ‐valerolactone on spheroids in aggregation ranged from 30 to 100 µM for 72 h, while the remaining experiments ranged from 3 to 100 µM for 72 h. Spheroid integrity was quantified by High Content Imaging System Operetta (Perkin Elmer) and expressed as a percentage of the vehicle‐treated controls (DMSO 0.05%). IC50 was calculated applying dose–response‐inhibition nonlinear regression analysis. The data were calculated as the mean ± SEM from at least four independent experiments. Spheroids were imaged on the High Content Imaging System Operetta (Perkin Elmer) with 10XLWD objective in brightfield and green fluorescent channels (merged images) at different concentrations. Scale bars, 200 µm.
2.3. A Long Exposure to (4R)‐5‐(3′,4′‐dihydroxyphenyl)‐γ‐Valerolactone Affects Matrix Metalloproteinase Expression Levels on 3D HCT116 Spheroid
2.3.1. A Long‐Term Study Simulating a Chronic Exposure
The PVL, 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone, consistently affected the SI at early stages of aggregation, which may be associated with prevention, avoiding cancer cell propagation. For this reason, we continued evaluating the role of this PVL in‐depth. A long‐term exposure of 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone was first investigated, simulating a long‐term exposure in the colonic region. The efficacy to alter SI at different time points (48, 120, 168, and 216 h) was quantified, and the results revealed a time‐dependent efficacy (Figure 4 ). A fixed concentration of this compound had different effect on SI over time. For example, when 3D spheroids were treated at 30, 50, and 70 µM concentrations, statistical differences were noticed between 48 h time point versus 120, 168, and 216 h time points (Figure 4A,B). Spheroids treated with rising concentrations ranged from 30 to 100 µM and imaged at different time points also showed that the IC50 decreased gradually over time from 58.4 µM at 48 h to 49.9 at 120 h, becoming a stable response after 120 h treatment. These data suggest that a chronic exposure to 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone may lead to a reduction of CRC risk. A sustained daily intake of monomeric, oligomeric, and polymeric flavan‐3‐ols might increase the colonic concentrations of PVLs, and, in turn, PVLs may act locally interacting with IECs and precancerous and cancer cells. Nevertheless, interindividual differences in the formation of PVLs, PVAs, and phenolic acids have to be considered,[ 31 , 63 , 72 ] and the composition and function of the gut microbiota further investigated.
Figure 4.
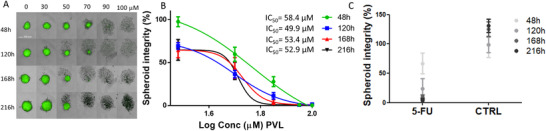
3D spheroid images at different concentration and time points A), and five‐point dose–response curves B) increasing concentration from 30 to 100 µM (5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone) on HCT116 EGFP spheroids in aggregation for 48, 120, 168, and 216 h. Two‐way ANOVA followed by Tukey multiple comparison test revealed a time‐dependent efficacy: 30 µM significant differences between 48 h time point versus 120, 168, and 216 h time points (p < 0,0001), 50 µM significant differences between 48 h time point versus 120 h (p < 0.0001), 168 h (p < 0.01), and 216 h (p < 0.0001) time points, and 70 µM significant differences between 48 h time point versus 120 h (p < 0.05), 168 h (p < 0.001), and 216 h (p < 0.0001) time points, and between 120 h time point versus 216 h time point (p < 0.05). The time‐dependent efficacy was also calculated using 5‐FU (10 µM) and DMSO at 0.05% C). In this case, the two‐way ANOVA followed by Tukey's multiple comparison tests revealed that 5‐FU significantly affected SI between 48 h time point versus 168 and 216 h time points (p < 0.05). SI was quantified by High Content Imaging System Operetta (Perkin Elmer) and expressed as a percentage of SI of vehicle‐treated controls at 48 h (DMSO 0.05%). IC50 was calculated applying dose–response‐inhibition nonlinear regression analysis. Data are expressed as mean ± SEM. All experiments were performed from at least four independent experiments. Spheroids were imaged on High Content Imaging System Operetta (Perkin Elmer) with 10XLWD objective in brightfield and green fluorescent channels (merged images) at different concentrations. Scale bars, 200 µm.
2.3.2. Matrix Metalloproteinases Expression Levels
GMMs may potentially reach the extracellular matrix, which provides an essential physical scaffolding,[ 35 ] and this niche is maintained by degrading enzymes, such as MMPs. However, in cancer propagation, these enzymes are hijacked.[ 36 ] For this reason, in this research work, we decided to study the gene expression of MMPs. Initially, the transcriptional profiling of MMP‐2, MMP‐7, and MMP‐9 target genes was evaluated on 2D HCT116 cell lines (Figure 5 ). Cells were treated with 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone at two concentration levels (30 and 100 µM). MMP‐2 and MMP‐9 gene expressions were not altered by raising concentrations of this flavan‐3‐ol metabolite. By contrast, a dose‐dependent response was noticed for MMP‐7 mRNA levels. 5‐(3′,4′‐Dihydroxyphenyl)‐γ‐valerolactone at 100 µM significantly affected gene expression levels of MMP‐7. Although the treatment did not statistically modify the MMP‐9 gene expression, a dose‐dependent response was noticed, and we decided to investigate ultimately it on 3D spheroids. 3D HCT116 spheroids were exposed to 50 µM of 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone using different time points (48 h as short‐term and 120 h as long‐term exposures) to study the gene expression of MMP‐7 and MMP‐9. Again, this metabolite did not modify the gene expression of MMP‐9. However, MMP‐7 expression was diminished after 48 h treatment. MMP‐7 binds to CRC cells via cell surface‐bound cholesterol sulfate and induces significant cell aggregation by cleaving cell‐surface protein,[ 73 ] which is required for cell migration.[ 74 ] Thus, we conjectured that SI depletion of 2D cells and 3D spheroids might be related to the downregulation of MMP‐7, and downstream associated pathways, which may promote apoptosis and inhibit proliferation. One of first attempts in vivo has demonstrated that the mouse microbial catabolism of PACs efficiently decreased the mRNA levels of the proliferation marker Cyclin D1 on cancer cells, and attenuated tumor growth in vivo.[ 39 ] However, more studies have to be carried out to reveal the specific mechanism using 3D intestinal organoids and tumoroids,[ 12 ] and ultimately validated in vivo.
Figure 5.
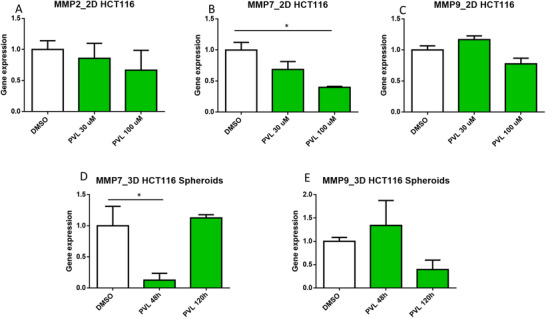
Transcriptional profiling of MMP2 A), MMP7 B), and MMP9 C) target genes in 2D HCT116 cells treated with DMSO 0.05% and 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone at 30 and 100 µM. After 72 h treatment, RNA was extracted to perform a qPCR. Gene expression was normalized to the reference gene RPLP0 and compared to the DMSO control. The transcriptional profiling of MMP7 D) and MMP9 E) target genes were evaluated at two different time points (48 and 120 h) on 3D HCT116 EGFP spheroids treated with DMSO 0.05% and 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone (50 µM). Gene expression was normalized to the reference gene RPLP0 and compared to the DMSO control. Data are the mean ± SEM from at least three independent experiments. One‐way ANOVA followed by Holm–Sidak test (* Adjusted p < 0.05).
3. Concluding Remarks
Dietary patterns and health outcomes are highly correlated, but there are still unresolved questions regarding the molecular and cellular mechanisms that underlie the link between diet and health. In this research work, we investigated the role of native phytochemical and GMMs in preventing CRC progression in vitro. To tackle this challenge, first, a screening of native phytochemicals and GMMs was performed demonstrating that a few (poly)phenol catabolites may play a role in preventing CRC. GMMs linked to the microbial catabolism of native dietary constituents were deeply evaluated because these compounds reach the gut epithelium and, so far, the local effects have been scarcely studied. The screening, IC50 and long‐term exposure studies proved that µM concentration levels of 5‐(3′,4′‐dihydroxyphenyl)‐γ‐valerolactone significantly decreased SI at early stages of 3D spheroid formation, whereas formed spheroids needed higher concentrations. The transcriptional profiling of MMP‐2, MMP‐7, and MMP‐9 was then investigated on 2D and 3D HTC116 cell lines treated with this epicatechin metabolite, revealing that SI depletion may be related to the downregulation of MMP‐7.
The type, quantity, and biological activity of GMMs produced in humans depend on the composition of gut microbiota. Thus, interindividual differences in the formation of PVLs, PVAs, and phenolic acids and spatial location have to be investigated in vitro and in vivo to corroborate this findings. Although 3D spheroids provided a superior in vitro model compared to 2D cell lines, future research works should consider the use of intestinal organoids and patient‐derived tumoroids, and ultimately validate relevant findings in vivo.
4. Experimental Section
Polyphenol Library and Reagents
A library of phytochemicals, highly focused on (poly)phenols and including native molecules and GMMs was created, selecting 54 compounds commercially available (Table S1, Supporting Information). (4R)‐5‐(3′‐Hydroxyphenyl)‐γ‐valerolactone, (4S)‐5‐(4′‐hydroxyphenyl)‐γ‐valerolactone, (4R)‐5‐(3′,5′‐dihydroxyphenyl)‐γ‐valerolactone, and (4R)‐5‐(3′‐hydroxyphenyl)‐γ‐valerolactone‐4′‐sulfate were synthesized in house at the University of Parma.[ 64 , 75 ] Compounds were first dissolved in dimethyl sulfoxide (DMSO) (CAS 67‐68‐5, Sigma‐Aldrich, Schnelldorf, Germany) at 10 mM concentration, and then were arrayed in three 96‐well plates using a robotic platform EVO200 (Tecan, Männedorf, Switzerland) and stored at −20 °C. Fluorouracil (5‐FU) (CAS 51‐21‐8, Sigma‐Aldrich, Schnelldorf, Germany), etoposide (CAS 33419‐42‐0, Sigma‐Aldrich), temozolomide (CAS 85622‐93‐1, Sigma‐Aldrich), and butyric acid (CAS 107‐92‐6, Sigma Aldrich) were used as positive controls. Nomenclature for phenolic compounds was reported according to the nomenclature proposed elsewhere.[ 76 ]
Cell Lines
The human colon adenocarcinoma HCT116 (p53+/+) cell line was kindly provided by Prof. Inga and Dr. Bisio (University of Trento, Italy). HCT116 EGFP cells were created from HCT116 (p53+/+) as described elsewhere.[ 77 ] Human glioblastoma stem cell lines (GICS) 030616 were facilitated by Dr. Galli (H. S. Raffaele, Milan, Italy). GICS 030616 DsRed reporter cell line was established by Dr. Leo, and Dr. Tarter (University of Trento, Italy). Briefly, a dsRed2 lentiviral construct was obtained by GATEWAY Cloning Technology recombining the pENTRdsRed2 N1 (cod.22523, Invitrogen, Waltham, MA, USA) and the pLenti CMV Puro Dest (cod. 17452, Invitrogen, Waltham, MA, USA) plasmids. After lentivirus packing and production, GICS 030616 were infected with 0.3 RTU of virus, amplified and FACS‐sortedGICS cells (FACS Aria II, BD Biosciences, NJ, USA).
High‐Content Screening
Clear round‐bottom ultralow attachment 96‐well plates (Corning, NY, USA) were used to seed HCT116 EGFP cells (2000 each well). Afterwards, plates were centrifugated for 8 min at 300 × g, forming aggregated cells, a loose spheroid. To assess assay suitability by high‐throughput screening (Z’ factor), immediately after seeding, the wells were supplemented with 0.05% DMSO or 50 µM 5‐FU (spheroid number = 27). With regards to the screening, controls (0.05% DMSO and 5‐FU), GMMs, and native polyphenols were added into the appropriate well, as single agents, at 50 µM from at least four independent experiments, immediately after the seeding. In all the cases, dispensing steps were performed by the Tecan Freedom EVO 200 robot (Tecan, Switzerland).
After 72 h of treatment, bright‐field and fluorescent spheroid images were automatically captured on the High Content Imaging System Operetta (PerkinElmer, Waltham, MA, USA) using 10× LWD objective. Images were processed by Harmony software (PerkinElmer, Waltham, MA, USA), different features, such as spheroids’ area, roundness, SER texture, as well as reporter fluorescent intensity were quantified in the green fluorescent channel (λ = 495 nm excitation/λ = 519 nm emission). The combination of these parameters was defined as SI. SI was calculated using the following equation: A sph (spheroid area) × R sph (spheroid roundness) × I sph (EGFP mean fluorescence intensity in the entire spheroid region). The effect of a compound on the spheroid was expressed as a percentage of the SI mean in relation to vehicle‐treated controls.
Cell Viability Assay
Cell growth, morphology, and attachment quality were monitored by xCELLigence real‐time cell analysis (RTCA) DP instrument (Agilent Technologies, CA, USA) by measuring impedance of electron flow caused by adherent cells. Cells were seeded into E‐plates 16 and impedance was continuously recorded every 15 min. A parameter called cell index (CI) was calculated and reported by the RTCA software. The CI was normalized to 24 h time point, which corresponded to the treatment or vehicle addition.
Spheroid Assay
HCT116 wild type (WT) and engineered (EGFP) were grown as a multicellular spheroid with Corning 96 well clear round bottom ultralow attachment microplate and GICs cells were grown as multicellular spheroids using InSphero GravityPLUS Hanging Drop System (InSphero AG, Schlieren, Switzerland) according to the manufacturer's instructions. The treatments were carried out in a single dose or dose range from at least four independent experiments immediately after cell seeding (spheroid in aggregation assay t = 0 h) or 3 days after seeding (formed spheroid assay t = 72 h) and imaged at different time points. Images of HCT116 EGFP spheroids were acquired and analyzed as reported in the section High‐Content Screening. For butyric acid treatment, the SI was calculated using features quantified in brightfield channel since this compound exhibited a strong green autofluorescence as the following equation: A sph × R sph × T sph (SER texture calculated for spheroid region). For HCT116 WT spheroids treated with 5‐FU, the SI was calculated as follow A sph × R sph in brightfield channel. The SI on GICs spheroid was calculated as A sph × R sph in the red channel (with λ = 535 nm excitation/λ = 615 nm emission). To evaluate apoptosis on GICS DsRed spheroids, 5 mL/spheroid of CellEvent Caspase‐3/7 Green (ThermoFisher) were added. Then, after 30 min of incubation, then spheroids were imaged in the green channel.
Quantitative Real‐Time PCR
Total RNA was extracted by pooling three spheroids for each treatment or by collecting cells growing as monolayers using QIAzol reagent (QIAGEN, Hilden, Germany) according to the manufacturer's instructions. Reverse transcription was performed using iScript Reverse Transcription Supermix (BioRad, CA, USA) on C1000 Thermal Cycler (BioRad, CA, USA). Quantitative real‐time PCR was performed using 2× qPCR Probe KAPA Biosystem following the manufacturer's instructions on CFX384 Real‐Time System (BioRad, CA, USA). All assays were performed from at least three independent experiments. Cycle quantification and relative expression measurement were performed using 2−∆∆Cq method implemented in the CFX Manager software (BioRad). Relative expression values of each target gene were normalized to ribosomal protein, large, P0 (RPLP0 level).
Statistical Methods
The IC50 was calculated applying a nonlinear regression model to the log (inhibitor) versus response curve (variable slope four parameters), and statistical analysis were performed with one‐way ANOVA followed by Holm–Sidak test, or two‐way ANOVA followed by Tukey's test as posthoc. All significance level of the data were analyzed by using GraphPad Prism Software version 6 (Graphpad Software, San Diego, CA, USA). Data are expressed as mean values ± SEM.
Conflict of Interest
The authors declare no conflict of interest.
Author Contributions
F.M. and A.Q. designed the study. P.G., M.P., V.S. conducted the experiments and performed the data analysis. P.G. and J.R. analyzed and interpreted data, and J.R. drafted the manuscript. C.C. synthetized and provided the tested PVLs. P.G., F.M., C.C., A.Q., P.M., D.D.R., and J.R. edited the manuscript. All authors critically read and approved the final version of the manuscript.
Supporting information
Supporting Information.
Acknowledgements
This research was conducted with the project “Apple fruit quality in the post‐genomic era, from breeding new genotypes to post‐harvest: nutrition and health,” funded by the AGER (Agribusiness and research) with grant no. 2010–2119. The funding body was not involved in the study design, sample collection, analysis and interpretation of the data, drafting the report, or the decision to submit the article. A few spelling mistakes were updated on November 3, 2022.
Mena P., Crozier A., Rubert J., Gatto P., Pancher M., Sidarovich V., Curti C., Mena P., Del Rio D., Quattrone A., Mattivi F., A Screening of Native (Poly)phenols and Gut‐Related Metabolites on 3D HCT116 Spheroids Reveals Gut Health Benefits of a Flavan‐3‐ol Metabolite. Mol. Nutr. Food Res. 2022, 66, 2101043. 10.1002/mnfr.202101043
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Calibasi‐Kocal G., Mashinchian O., Basbinar Y., Ellidokuz E., Cheng C.‐W., Yilmaz Ö. H., Trends Endocrinol. Metab. 2021, 32, 20. [DOI] [PubMed] [Google Scholar]
- 2. Nakamura A., Kurihara S., Takahashi D., Ohashi W., Nakamura Y., Kimura S., Onuki M., Kume A., Sasazawa Y., Furusawa Y., Obata Y., Fukuda S., Saiki S., Matsumoto M., Hase K., Nat. Commun. 2021, 12, 2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bouvard V., Loomis D., Guyton K. Z., Grosse Y., El Ghissassi F., Benbrahim‐Tallaa L., Guha N., Mattock H., Straif K., Lancet Oncol. 2015, 16, 1599. [DOI] [PubMed] [Google Scholar]
- 4. Arnold M., Sierra M. S., Laversanne M., Soerjomataram I., Jemal A., Bray F., Gut 2017, 66, 683. [DOI] [PubMed] [Google Scholar]
- 5. Torre L. A., Bray F., Siegel R. L., Ferlay J., Lortet‐Tieulent J., Jemal A., CA Cancer J. Clin. 2015, 65, 87. [DOI] [PubMed] [Google Scholar]
- 6. Shetty S. A., Smidt H., de Vos W. M., Curr. Opin. Biotechnol. 2019, 58, 146. [DOI] [PubMed] [Google Scholar]
- 7. Koutsos A., Lima M., Conterno L., Gasperotti M., Bianchi M., Fava F., Vrhovsek U., Lovegrove J., Tuohy K., Nutrients 2017, 9, 533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Trošt K., Ulaszewska M. M., Stanstrup J., Albanese D., De Filippo C., Tuohy K. M., Natella F., Scaccini C., Mattivi F., Food Res. Int. 2018, 112, 108. [DOI] [PubMed] [Google Scholar]
- 9. O'Keefe S. J. D., Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang J., Yu J., Protein Cell 2018, 9, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang L. S., Davies S. S., Genome Med. 2016, 8, 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rubert J., Schweiger P. J., Mattivi F., Tuohy K., Jensen K. B., Lunardi A., Trends Endocrinol. Metab. 2020, 31, 848. [DOI] [PubMed] [Google Scholar]
- 13. Thomas A. M., Manghi P., Asnicar F., Pasolli E., Armanini F., Zolfo M., Beghini F., Manara S., Karcher N., Pozzi C., Gandini S., Serrano D., Tarallo S., Francavilla A., Gallo G., Trompetto M., Ferrero G., Mizutani S., Shiroma H., Shiba S., Shibata T., Yachida S., Yamada T., Wirbel J., Schrotz‐King P., Ulrich C. M., Brenner H., Arumugam M., Bork P., Zeller G., et al., Nat. Med. 2019, 25, 667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Makki K., Deehan E. C., Walter J., Bäckhed F., Cell Host Microbe 2018, 23, 705. [DOI] [PubMed] [Google Scholar]
- 15. Braune A., Blaut M., Gut Microbes 2016, 7, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rowland I., Gibson G., Heinken A., Scott K., Swann J., Thiele I., Tuohy K., Eur. J. Nutr. 2018, 57, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown E. M., McDougall G. J., Stewart D., Pereira‐Caro G., González‐Barrio R., Allsopp P., Magee P., Crozier A., Rowland I., Gill C. I. R., PLoS One 2012, 7, e49740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. López de las Hazas M.‐C., Piñol C., Macià A., Motilva M.‐J., J. Agric. Food Chem. 2017, 65, 6467. [DOI] [PubMed] [Google Scholar]
- 19. Turati F., Rossi M., Pelucchi C., Levi F., Vecchia C. L.a, Br. J. Nutr. 2015, 113, S102. [DOI] [PubMed] [Google Scholar]
- 20. Nabavi S. F., Atanasov A. G., Khan H., Barreca D., Trombetta D., Testai L., Sureda A., Tejada S., Vacca R. A., Pittalà V., Gulei D., Berindan‐Neagoe I., Shirooie S., Nabavi S. M., Cancer Lett. 2018, 434, 101. [DOI] [PubMed] [Google Scholar]
- 21. Rossi M., Negri E., Parpinel M., Lagiou P., Bosetti C., Talamini R., Montella M., Giacosa A., Franceschi S., La Vecchia C., Cancer Causes Control 2010, 21, 243. [DOI] [PubMed] [Google Scholar]
- 22. Little C. H., Combet E., McMillan D. C., Horgan P. G., Roxburgh C. S. D., Crit. Rev. Food Sci. Nutr. 2017, 57, 2310. [DOI] [PubMed] [Google Scholar]
- 23. Langhans W., J. Agric. Food Chem. 2018, 66, 2287. [DOI] [PubMed] [Google Scholar]
- 24. Hou T. Y., Davidson L. A., Kim E., Fan Y.‐Y., Fuentes N. R., Triff K., Chapkin R. S., Annu. Rev. Nutr. 2016, 36, 543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Monagas M., Urpi‐Sarda M., Sánchez‐Patán F., Llorach R., Garrido I., Gómez‐Cordovés C., Andres‐Lacueva C., Bartolomé B., Food Funct. 2010, 1, 233. [DOI] [PubMed] [Google Scholar]
- 26. Ozdal T., Sela D. A., Xiao J., Boyacioglu D., Chen F., Capanoglu E., Nutrients 2016, 8, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lotti C., Rubert J., Fava F., Tuohy K., Mattivi F., Vrhovsek U., Anal. Bioanal. Chem. 2017, 409, 5555. [DOI] [PubMed] [Google Scholar]
- 28. Mena P., Bresciani L., Brindani N., Ludwig I. A., Pereira‐Caro G., Angelino D., Llorach R., Calani L., Brighenti F., Clifford M. N., Gill C. I. R., Crozier A., Curti C., Del Rio D., Nat. Prod. Rep. 2019, 36, 714. [DOI] [PubMed] [Google Scholar]
- 29. Favari C., Righetti L., Tassotti M., Gethings L. A., Martini D., Rosi A., Antonini M., Rubert J., Manach C., Dei Cas A., Bonadonna R., Brighenti F., Dall'Asta C., Mena P., Del Rio D., Mol. Nutr. Food Res. 2021, 65, 2000875. [DOI] [PubMed] [Google Scholar]
- 30. Tao W., Zhang Y., Shen X., Cao Y., Shi J., Ye X., Chen S., Compr. Rev. Food Sci. Food Saf. 2019, 18, 971. [DOI] [PubMed] [Google Scholar]
- 31. Anesi A., Mena P., Bub A., Ulaszewska M., Del Rio D., Kulling S. E., Mattivi F., Metabolites 2019, 9, 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clevers H., Cell 2013, 154, 274. [DOI] [PubMed] [Google Scholar]
- 33. Kemper K., Prasetyanti P. R., De Lau W., Rodermond H., Clevers H., Medema J. P., Stem Cells 2012, 30, 2378. [DOI] [PubMed] [Google Scholar]
- 34. La Vecchia S., Sebastián C., Semin. Cell Dev. Biol. 2020, 98, 63. [DOI] [PubMed] [Google Scholar]
- 35. Fares J., Fares M. Y., Khachfe H. H., Salhab H. A., Fares Y., Signal Transduct. Target. Ther. 2020, 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Said A. H., Raufman J. P., Xie G., Cancers (Basel) 2014, 6, 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shankar E., Kanwal R., Candamo M., Gupta S., Semin. Cancer Biol. 2016, 40–41, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Crascì L., Lauro M. R., Puglisi G., Panico A., Crit. Rev. Food Sci. Nutr. 2018, 58, 893. [DOI] [PubMed] [Google Scholar]
- 39. Ravindranathan P., Pasham D., Balaji U., Cardenas J., Gu J., Toden S., Goel A., Sci. Rep. 2018, 8, 13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu Y., Chen Y.‐G., Cells 2018, 7, 225.30469504 [Google Scholar]
- 41. Porter R. J., Murray G. I., McLean M. H., Br. J. Cancer 2020, 123, 1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Russo G. L., Tedesco I., Spagnuolo C., Russo M., Semin. Cancer Biol. 2017, 46, 1. [DOI] [PubMed] [Google Scholar]
- 43. Sayed I. M., El‐Hafeez A. A. A., Maity P. P., Das S., Ghosh P., Adv. Cancer Res. 2021, 151, 345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jang S., Sun J., Chen P., Lakshman S., Molokin A., Harnly J. M., Vinyard B. T., Urban J. F., Davis C. D., Solano‐Aguilar G., J. Nutr. 2015, 146, 673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chen T.‐Y., Ferruzzi M. G., Wu Q.‐L., Simon J. E., Talcott S. T., Wang J., Ho L., Todd G., Cooper B., Pasinetti G. M., Janle E. M., Mol. Nutr. Food Res. 2017, 61, 1700111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Walter J., Armet A. M., Finlay B. B., Shanahan F., Cell 2020, 180, 221. [DOI] [PubMed] [Google Scholar]
- 47. Han S. J., Kwon S., Kim K. S., Cancer Cell Int. 2021, 21, 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Boutros M., Heigwer F., Laufer C., Cell 2015, 163, 1314. [DOI] [PubMed] [Google Scholar]
- 49. Strovel J., Sittampalam S., Coussens N. P., Hughes M., Inglese J., Kurtz A., Andalibi A., Patton L., Austin C., Baltezor M., Beckloff M., Weingarten M., Weir S., Early Drug Discovery and Development Guidelines: For Academic Researchers, Collaborators, and Start‐up Companies 2004, pp. 661–669. [PubMed]
- 50. Dobani S., Latimer C., McDougall G. J., Allwood J. W., Pereira‐Caro G., Moreno‐Rojas J. M., Ternan N. G., Pourshahidi L. K., Lawther R., Tuohy K. M., Del Rio D., O'Connor G., Rowland I., Almutairi T. M., Crozier A., Gill C. I. R., Redox Biol. 2021, 40, 101862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ganesan T., Sinniah A., Chik Z., Alshawsh M. A., Nutrients 2020, 12, 2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Boylan K. L. M., Manion R. D., Shah H., Skubitz K. M., Skubitz A. P. N., Int. J. Mol. Sci. 2020, 21, 4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McLoughlin P., Roengvoraphoj M., Gissel C., Hescheler J., Certa U., Sachinidis A., Genes Cells 2004, 9, 661. [DOI] [PubMed] [Google Scholar]
- 54. Kashyap D., Tuli H. S., Yerer M. B., Sharma A., Sak K., Srivastava S., Pandey A., Garg V. K., Sethi G., Bishayee A., Semin. Cancer Biol. 2021, 69, 5. [DOI] [PubMed] [Google Scholar]
- 55. Kim K., Shin E., Jung J., Park J., Kim D., Shim B., Kim S.‐H., Int. J. Mol. Sci. 2018, 20, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao J., Blayney A., Liu X., Gandy L., Jin W., Yan L., Ha J.‐H., Canning A. J., Connelly M., Yang C., Liu X., Xiao Y., Cosgrove M. S., Solmaz S. R., Zhang Y., Ban D., Chen J., Loh S. N., Wang C., Nat. Commun. 2021, 12, 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weng Y.‐P., Hung P.‐F., Ku W.‐Y., Chang C.‐Y., Wu B.‐H., Wu M.‐H., Yao J.‐Y., Yang J.‐R., Lee C.‐H., Oncotarget 2018, 9, 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ottaviani J. I., Borges G., Momma T. Y., Spencer J. P. E., Keen C. L., Crozier A., Schroeter H., Sci. Rep. 2016, 6, 29034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wiese S., Esatbeyoglu T., Winterhalter P., Kruse H. P., Winkler S., Bub A., Kulling S. E., Mol. Nutr. Food Res. 2015, 59, 610. [DOI] [PubMed] [Google Scholar]
- 60. Zhang L., Wang Y., Li D., Ho C. T., Li J., Wan X., Food Funct. 2016, 7, 1273. [DOI] [PubMed] [Google Scholar]
- 61. Stoupi S., Williamson G., Drynan J. W., Barron D., Clifford M. N., Mole. Nutr. Food Res. 2010, 54, 747. [DOI] [PubMed] [Google Scholar]
- 62. Cires M. J., Wong X., Carrasco‐Pozo C., Gotteland M., Front. Nutr. 2017, 10.3389/fnut.2016.00057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liu C., Vervoort J., Beekmann K., Baccaro M., Kamelia L., Wesseling S., Rietjens I. M. C. M., J. Agric. Food Chem. 2020, 68, 14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Brindani N., Mena P., Calani L., Benzie I., Choi S.‐W. W., Brighenti F., Zanardi F., Curti C., Del Rio D., Mol. Nutr. Food Res. 2017, 61, 6. [DOI] [PubMed] [Google Scholar]
- 65. Bianchi M. G., Chiu M., Taurino G., Brighenti F., Del Rio D., Mena P., Bussolati O., Nutrients 2019, 11, 2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Del Rio D., Rodriguez‐Mateos A., Spencer J. P. E., Tognolini M., Borges G., Crozier A., Antioxid. Redox Signal 2013, 18, 1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tilg H., Adolph T. E., Gerner R. R., Moschen A. R., Cancer Cell 2018, 33, 954. [DOI] [PubMed] [Google Scholar]
- 68. Li Q., Cao L., Tian Y., Zhang P., Ding C., Lu W., Jia C., Shao C., Liu W., Wang D., Ye H., Hao H., Mol. Cell. Proteomics 2018, 17, 1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yuille S., Reichardt N., Panda S., Dunbar H., Mulder I. E., PLoS One 2018, 13, e0201073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yi C., Li G., Ivanov D. N., Wang Z., Velasco M. X., Hernández G., Kaundal S., Villarreal J., Gupta Y. K., Qiao M., Hubert C. G., Hart M. J., Penalva L. O. F., RNA Biol. 2018, 15, 1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tsiailanis A. D., Renziehausen A., Kiriakidi S., Vrettos E. I., Markopoulos G. S., Sayyad N., Hirmiz B., Aguilar M.‐I., Del Borgo M. P., Kolettas E., Widdop R. E., Mavromoustakos T., Crook T., Syed N., Tzakos A. G., Free Radical Biol. Med. 2020, 160, 391. [DOI] [PubMed] [Google Scholar]
- 72. Mena P., Ludwig I. A., Tomatis V. B., Acharjee A., Calani L., Rosi A., Brighenti F., Ray S., Griffin J. L., Bluck L. J., Del Rio D., Eur. J. Nutr. 2019, 58, 1529. [DOI] [PubMed] [Google Scholar]
- 73. Ishikawa T., Kimura Y., Hirano H., Higashi S., J. Biol. Chem. 2017, 292, 20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Basu S., Thorat R., Dalal S. N., PLoS One 2015, 10, e0123979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Curti C., Brindani N., Battistini L., Sartori A., Pelosi G., Mena P., Brighenti F., Zanardi F., Del Rio D., Adv. Synth. Catal. 2015, 357, 4082. [Google Scholar]
- 76. Kay C. D., Clifford M. N., Mena P., McDougall G. J., Andres‐Lacueva C., Cassidy A., Del Rio D., Kuhnert N., Manach C., Pereira‐Caro G., Rodriguez‐Mateos A., Scalbert A., Tomás‐Barberán F., Williamson G., Wishart D. S., Crozier A., Am. J. Clin. Nutr. 2020, 112, 1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sidarovich V., De Mariano M., Aveic S., Pancher M., Adami V., Gatto P., Pizzini S., Pasini L., Croce M., Parodi F., Cimmino F., Avitabile M., Emionite L., Cilli M., Ferrini S., Pagano A., Capasso M., Quattrone A., Tonini G. P., Longo L., Mol. Cancer Ther. 2018, 17, 1405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


