Graphical abstract
Keywords: Brassica napus, Phosphoenolpyruvate, Plastid transporter, Seed oil content, Membrane lipid
Highlights
-
•
Knockout of BnaA08.PPT1 and BnaC08.PPT1 resulted retarded growth of B. napus.
-
•
BnaPPT1 is required for membrane lipid synthesis and chloroplast development in leaves.
-
•
Overexpression of BnaC08.PPT1 significantly elevated the seed oil content.
-
•
BnaPPT1 plays an important role in maintaining plant growth and seed oil accumulation in B. napus.
Abstract
Introduction
Phosphoenolpyruvate/phosphate translocator (PPT) transports phosphoenolpyruvate from the cytosol into the plastid for fatty acid (FA) and other metabolites biosynthesis.
Objectives
This study investigated PPTs’ functions in plant growth and seed oil biosynthesis in oilseed crop Brassica napus.
Methods
We created over-expression and mutant material of BnaPPT1. The plant development, oil content, lipids, metabolites and ultrastructure of seeds were compared to evaluate the gene function.
Results
The plastid membrane localized BnaPPT1 was found to be required for normal growth of B. napus. The plants grew slower with yellowish leaves in BnaA08.PPT1 and BnaC08.PPT1 double mutant plants. The results of chloroplast ultrastructural observation and lipid analysis show that BnaPPT1 plays an essential role in membrane lipid synthesis and chloroplast development in leaves, thereby affecting photosynthesis. Moreover, the analysis of primary metabolites and lipids in developing seeds showed that BnaPPT1 could impact seed glycolytic metabolism and lipid level. Knockout of BnaA08.PPT1 and BnaC08.PPT1 resulted in decreasing of the seed oil content by 2.2 to 9.1%, while overexpression of BnaC08.PPT1 significantly promoted the seed oil content by 2.1 to 3.3%.
Conclusion
Our results suggest that BnaPPT1 is necessary for plant chloroplast development, and it plays an important role in maintaining plant growth and promoting seed oil accumulation in B. napus.
Introduction
Phosphoenolpyruvate (PEP), an intermediate metabolites of glycolysis pathway, plays a crucial role in maintaining the development of plastids in plant cells. There are three pathways that provide PEP to plastid in plants. The first way is to catalyze pyruvate under the action of pyruvate orthophosphate dikinase (PPDK), thus providing PEP to plastid. However, existing studies have proved that this way mainly exists in C4 plants (such as maize), and there is no proof that C3 plants (such as Arabidopsis thaliana) can produce PEP through PPDK [1]. The second way is to catalyze 3-phosphoglycerate (3-PGA) of plastidic glycolysis by enolase (ENO). However, the activity of plastidic enolase is extremely low, which is not enough to meet the requirement of PEP for plastid development [2], [3]. The third way is that PEP can be supplied by importing from cytosol, which appears to be the main way due to the absence of PPDK and low activity of plastidic enolase in the C3 plants [3].
In plastid, PEP acts as a precursor substance mainly participates in the following metabolic pathways. Together with erythrose 4-phosphate (E-4-P), PEP enters the shikimate pathway, which produces essential aromatic amino acids and many other aromatic secondary metabolites [4]. Moreover, under the catalysis of pyruvate kinase (PK), PEP is converted into pyruvate, which enters the biosynthesis of fatty acids (FAs), isoprenoids, or branched-chain amino acids [4]. In Arabidopsis, plastidic PK is consisted of three subunits α, β1 and β2. Disruption of plastidic PK β1 subunit would cause a reduction in enzyme activity and 60 % reduction in seed oil content. The oil content of the pk mutant could be completely recovered by the expression of the β1 subunit-encoding gene and partially restored by the expression of the β2 subunit-encoding gene. Thus, PK is involved in the conversion process of photosynthate to oil, and its role reveals the preferred route from PEP to pyruvate, then to FAs pathway [5]. Phosphoenolpyruvate/phosphate translocator (PPT), connecting the glycolytic pathway in cytosol with the metabolic pathway in plastid, can transport PEP from cytosol into the plastid, and then participates the metabolic pathways [4], [6].
Previous studies about PPTs were mainly in Arabidopsis. Two PPT genes, AtPPT1 and AtPPT2 identified in Arabidopsis, share only 52 % identical amino acids [7], [8]. The expression of AtPPT1 is defective in a chlorophyll a/b-binding protein underexpressed 1 (cue1) mutant in Arabidopsis. The leaf of the mutants visibly exhibited a reticulate phenotype with dark green paraveinal regions and pale green interveinal regions [9], [10]. Furthermore, the biosynthesis of aromatics defected in cue1 mutant, and the reticulate phenotype could be rescued by feeding aromatic amino acids [10]. The phenotype of reticulate leaf could be completely recovered when AtPPT1 supplementation was performed, whereas the complementary plants of AtPPT2 only partially rescued the phenotype [7]. Compared to wild-type (WT), the size of mesophyll cells and chloroplasts were smaller in cue1, suggesting AtPPT1 might be involved in providing signals for mesophyll cell development [7], [9]. These results demonstrate that PEP provision to plastids by PPT is essential for plant growth and leaf development. However, the molecular mechanism of PPTs in plastid development is still unclear. Whether PPTs impact glycolysis and FA synthesis, thus affecting seed oil accumulation is not investigated.
In this study, we identified two PPT1 genes BnaA08.PPT1 and BnaC08.PPT1 in B. napus. They showed high expression levels in developing seeds, suggesting that PPT1 may play a role in seed development. Here, we found that the double mutant plants of BnaA08.PPT1 and BnaC08.PPT1 were defective in starch and plastid membrane lipid synthesis, leading to retarded leaf development. Moreover, the metabolism such as glycolysis, FA and lipid synthesis in mutant seed was also significantly altered. Overexpression (OE) of BnaC08.PPT1 driven by the promoter of the cauliflower mosaic virus 35S (CaMV-35S) exhibited a high level of metabolism of glycolysis and lipid synthesis. These results suggest that the BnaPPT1 plays an important role in plant chloroplast development and seed oil accumulation in B. napus.
Materials and methods
Plant growth condition and agronomic trait measurement
In this research, B. napus cultivar (cv.) Westar was employed for gene cloning and used as the transformation receptor. The seeds of OE, mutant, and WT were sown in the genetically modified field to set a plot experiment. The field have been approved by the local government and established by the supporting institution Huazhong Agricultural University, and iss specially used for planting genetically modified rapeseed. The plant grew under natural condition. The whole plant was harvested when the plants matured in the field and agronomic traits were measured. Totally, six to twelve replications were used for each line. The seeds were collected from each plant and desiccated in air, and the content of seed oil and protein was tested using a near-infrared reflectance spectroscope (Foss NIRSystems 5000) [11].
The growth chamber PGW40 (Conviron, Canada) was employed to observe the plant growth. OE, mutant, and WT seeds were random sown, which were set in the growth chamber under a condition with 16 h of light, 8 h of darkness, and a temperature of 22/20 °C. The environmental relative humidity was set at 60 % in the whole plant growth cycle. For the plant growth in the liquid media in the growth room, the condition is 16 h light/ 8 h dark, 22 / 20 °C. The seeds were germinated on water for 7 to 10-day and then explanted into the liquid media. The liquid media was updated every-one week. The formula of the liquid media as described previously [12].
Vector construction and plant transformation
To construct the OE vector of BnaPPT1, the CDS of BnaC08.PPT1 was cloned from Westar and ligated into p35S-FAST. The CRISPR/Cas9 vector was constructed as described previously [13]. Briefly, two sgRNAs in exons which could both target on BnaA08.PPT1 and BnaC08.PPT1 were designed by CRISPR-P [14]. The amplified product including two sgRNAs was linked into PKSE401 [15]. Then, the OE and CRISPR vectors were transformed into Agrobacterium tumefaciens GV3101, respectively. The method of transformation was described previously [16]. The OE plants were detected by PCR using specific primers. To detect the Cas9 edited plants, the PCR was performed using a pair of primers which specific to amplified Cas9 first. Subsequently, the PCR product of sgRNA target sequence was amplified and sent to sequence in Quintara (Wuhan) Biotechnology Co., ltd. The patterns of Cas9 edited mutation were analyzed using DSDecode [17]. All primers mentioned above were listed in Supplemental Table S1.
Subcellular localization
BnaC08.PPT1 was cloned and ligated into the GFP-tagged PMDC83 vector. The primers were listed in Supplementary Table S1. Protoplasts were isolated from 4-week-old leaves of Arabidopsis as follows. healthy, well-grown leaves were selected and cut into 0.5–1 mm in the solution of 0.4 M Mannitol. The cut leaves were immediately completely immersed in the enzyme solution (1 % Cellulase R10, 0.2 % Macerozyme R10, 0.4 M Mannitol, 20 mM KCl, 20 mM MES (PH 5.7), 10 mM CaCl2, 0.1 % BSA) and vacuumed in the dark for 5–30 min. Enzymolysis was carried out for 2–3 h on a 40 rpm shaker, protected from light. The enzymolysis reaction was terminated by adding equal volume of W5 Solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES (pH5.7)). Centrifugation was performed at 100 g at 4 ℃ for 3–8 min. The supernatant was carefully sucked out and discarded. Then 5–10 mL pre-cooled W5 Solution was used to gently resuspend the protoplast, and centrifugation was performed as before. MMG solution (0.4 M Mannitol, 15 mM MgCl2, 4 mM MES (pH5.7)) was used to resuspend protoplasts. The fused BnaC08.PPT1-GFP plasmid was transformed into the mesophyll protoplasts in 40 % PEG solution (V/V) as the protocol described previously [18]. Next, 12 h after infection, the fluorescence signal was observed using a Leica confocal microscope. The fluorescence of GFP was measured at 645–709 nm, and chloroplast autofluorescence was measured at 552 nm.
Quantitative real-time PCR
Total RNA was ectracted from 60-day leaves and 35 DAF (day after flowering) seeds using Transzol reagent (TransGen Biotech). The integrity and concentration of RNA were determined by gel electrophoresis and NanoDrop, respectively. Next, 2 µg RNA was used to synthesize cDNA by cDNA Synthesis Supermix (TransGen Biotech). The synthesized cDNA was used as the template to perform the quantitative real-time PCR (qPCR) using TransStart Green qPCR SuperMix (TransGen Biotech). BnaACTIN7 was used as the internal control [19]. The primers were listed in Supplementary Table S1.
Determination of chlorophyll and chlorophyll precursor contents
The content of the chlorophyll and chlorophyll precursor of leaves was measured when plants were three-week-old. The detailed procedures were performed as described previously [20].
Measurement of photosynthetic efficiency
The photosynthetic efficiency was measured using a portable photosynthesis system (LI-6800; Li-Cor) when plants were 80-day-old in the field. The measurement was performed in a sunning day, and the temperature is around 15–20 ℃. Each line measured 5 independent individual plants and the average value was calculated. The settings of leaf chamber environments were described as follows: 600 μmol s−1 flow rate, 60 % relative humidity, 400 μmol mol−1 CO2, 1000 rpm fan speed, 1200 μmol m−2 s−1 light intensity, 20 ℃.
Observation of cell ultrastructure
Transmission electron microscope (TEM) was used to observe the ultrastructure of chloroplasts in leaf and oil body in seed (inner cotyledon). 30-day leaves and mature seeds were cut and fixed in 2.5 % glutaraldehyde solution for overnight, washed by 0.1 M PBS buffer for 3 times, and then fixed in 2 % osmium solution for 2 h. After washing with 0.1 M PBS buffer for 3 times, gradient dehydration was performed in 50 %, 70 %, 90 % ethanol, 90 % ethanol/90 % acetone (1:1, V/V), 90 % acetone and 100 % acetone for 20 min for each step, respectively, and then the samples were embedded in resin. After 24 h of polymerization, samples were cut into ultra-thin sections (50–60 nm), followed by staining with 3 % uranium acetate and lead citrate for 2 h, and then added into copper mesh and observed under TEM (H-7650, HITACHI, Japan).
Metabolites extraction and analysis
The metabolites were extracted and determined by LC-MS/MS using a modified method described previously [21], [22]. Briefly, 10–15 mg of 35 DAF seeds were collected and ground with liquid nitrogen. A mix of 1.8 mL methanol:chloroform (7:3, v/v) containing 0.8 μg PIPES as internal standard was added and incubated for 2 h with intermittent mixing at −20 ℃. After adding 1.6 mL ddH2O, the samples were vortexed and centrifuged at 1000 rpm for 5 min, and then the upper phase was transferred into a new tube. Repeated the step above again. The upper extract was dried using nitrogen in water bath at 50 ℃, the extract was re-dissolved with 200 μL ddH2O and then filtered with 0.45 μM cellulose acetate centrifuge tube filters. The extract was diluted by 10 folds with ddH2O for metabolites analysis by LC-MS/MS (QTRAP 6500 plus). The instrumental parameters and the data analysis as described by Luo et al [21], [22].
Lipid extraction and analysis
10–15 mg of freeze-dried leaves or seeds were dropped into 6 mL isopropanol at 75 °C with 0.01 % BHT for 15 min, Cool and add 3 mL chloroform and 1.2 mL ddH2O, then the samples were vortexed and agitated at room temperature for more than 1 h, and then lipid extracts were transferred into a new glass screw-cap tube. 4 mL chloroform/methanol (2:1) with 0.01 % BHT was added followed by shaking for 30 min. This extraction procedure was repeated until the leaves or seeds become white. After adding 1 mL 1 M KCl, the combined extract was vortexed, and centrifuged to separate the phases, and then the upper phase was discard. The samples were washed with 2 mL ddH2O, and repeat the above step. The extracts were evaporated with N2, then re-dissolved in chloroform to a concentration of 10 mg/mL. The samples were prepared for analysis as follows: 210 μL 300 mM ammonium acetate–methanol (1:20, v:v), 60 μL 10 mg/ml lipid extracts, 3 μL foe each internal standard (PL, GL, DAG, TAG), then the volume was filled with chloroform to 300 μL. The lipids were measured and quantified by LC-MS/MS (QTRAP 6500 plus).
GC-FID analysis of seed fatty acid
FAs of WT, OE and CRISPR lines were analyzed by GC-FID (Gas chromatography-flame ionization detector). About 10–20 mg mature seeds for each line were crushed and used. Fatty acids were methylated and extracted according to the method described previously [23]. FAs were quantified using gas chromatography. The GC-FID parameters are described as following: oven temperature was maintained at 170 °C for 1 min and then increased to 210 °C with an increase rate of 3 °C/min. Finally, FAs were identified according to their retention times and peak areas were analyzed. FAs were calculated as nmol% [24].
Measurement of soluble sugar and starch content
The extraction and measurement of soluble sugar and starch content were performed as reported previously [20]. For the measurement of seed soluble sugar, 80–100 mg seeds were collected and added into 25 mL ddH2O, heating at 95 °C for 30 min. 50 μL of supernatant was added to 400 μL 2 mg/ml anthrone sulfuric acid, followed by heating at 95 °C for 5 min. The mix of heated supernatant and anthrone sulfuric acid was measured by spectrophotometer (HITACHI, UH5300) at wavelength of 625 nm. The concentration of soluble sugar was calculated according to the glucose standard curve. Content of soluble sugar (mg/g) = concentration × volume ÷ dry weight (DW).
The residue were dried and ground into powder under liquid nitrogen, which was ground into a homogenate after adding 5 mL 0.2 M KOH. Then the samples were transferred it to a tube heated in 95 °C for 30 min, and added 1 mL 1 M acetic acid, followed by centrifuge at 12000 rpm for 5 min. Finally, 50 μL of supernatant was used to perform the remaining steps as the measurement for soluble sugar. The concentration of hydrolysis product glucose was calculated according to the glucose standard curve. Content of starch (mg/g) = 0.9 × concentration × volume ÷ DW.
Results
Generation of PPT1 transgenic plants
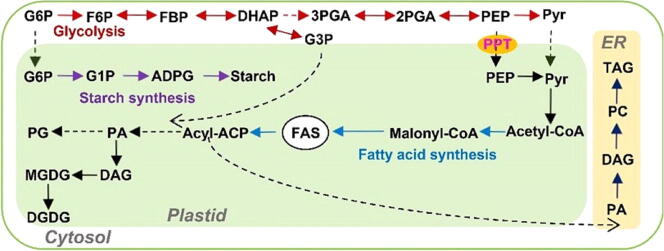
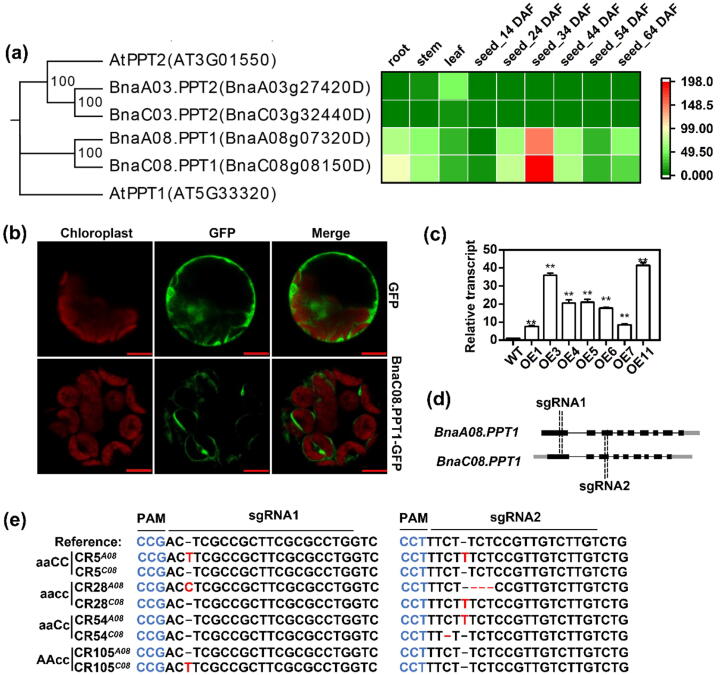
To investigate the function of PPT genes in B. napus, the amino acid sequences of Arabidopsis PPTs were used for blast analysis in the website of BnTIR (https://yanglab.hzau.edu.cn/BnTIR) [25]. The results show that there are two homologous copies of BnaPPT1 (BnaA08g07320D, designated as BnaA08.PPT1; BnaC08g08150D, designated as BnaC08.PPT1) and BnaPPT2 (BnaA03g07320D, designated as BnaA03.PPT2; and BnaC03g32440D, designated as BnaC03.PPT2), respectively (Fig. 1a). However, the data showed that the expression levels of BnaA03.PPT2 and BnaC03.PPT2 were very low in different tissues. BnaA08.PPT1 and BnaC08.PPT1 had higher expression in root, stem, and leaf, especially in developing seed (Fig. 1a). The expression level of BnaC08.PPT1 was higher in most tissues than that of BnaA08.PPT1, especially in developing seeds. To analyze the subcellular localization of BnaPPT1, BnaC08.PPT1 fused with GFP at its C-terminus was transiently expressed in the protoplasts of Arabidopsis. The green fluorescence signal was found to encapsulate the auto-fluorescence of chloroplast membrane, suggesting that BnaC08.PPT1 was localized at the chloroplast membrane (Fig. 1b). To explore the function of BnaPPT1 in B. napus, BnaC08.PPT1 was overexpressed in B. napus driven by 35S promoter. RT-qPCR data showed all the transgenic lines had higher expression of BnaC08.PPT1 than that in WT (Fig. 1c). The expression level of PPT1 in OE lines was increased by 8 to 41 folds compared to WT, among which OE1 and OE11 were selected for further study (Fig. 1c). Meantime, two sgRNAs were designed to target BnaA08.PPT1 and BnaC08.PPT1 at the same time, which were cloned into a CRISPR/Cas9 vector to generate Cas9-induced mutations of BnaPPT1 (Fig. 1d). Finally, four mutated genotypes were identified by sequencing. CR5 and CR105 were two homozygous mutant lines of BnaA08.PPT1 and BnaC08.PPT1, respectively. CR28 was a homozygous double mutant and CR54 was a heterozygous double mutant (Fig. 1e and Supplemental Fig. 1).
Fig. 1.
Generation of BnaPPT1 overexpression transgenic plants and knockout mutants. (a) Amino acid sequence alignment using MEGA7 and gene expression of PPTs in B. napus. At, Arabidopsis thaliana; Bn, B. napus; Gene expression data are from BnTIR (http://yanglab.hzau.edu.cn/). (b) BnaC08.PPT1 is a chloroplast membrane protein observed in Arabidopsis protoplast. Bars = 10 μm. (c) Relative expression level of BnaC08.PPT1 in overexpression lines by quantitative real-time PCR. Total RNA was extracted from 8-week-old leaves. Values are means ± SD (n = 3). ** indicates P < 0.01, based on a student t-test. (d) CRSIPR targets on BnaA08.PPT1 and BnaC08.PPT1. Two target sites are designed on the first and third exons respectively. (e) Acquisition of mutant lines by sequencing. CR5 and CR105 is a homozygous mutant line of BnaA08.PPT1 and BnaC08.PPT1 respectively. CR28 is a homozygous double mutant, and CR54 is a heterozygous double mutant. Mutation sites are indicated in red, and PAM sites are indicated in blue. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The alteration of plant growth in BnaPPT1 mutants
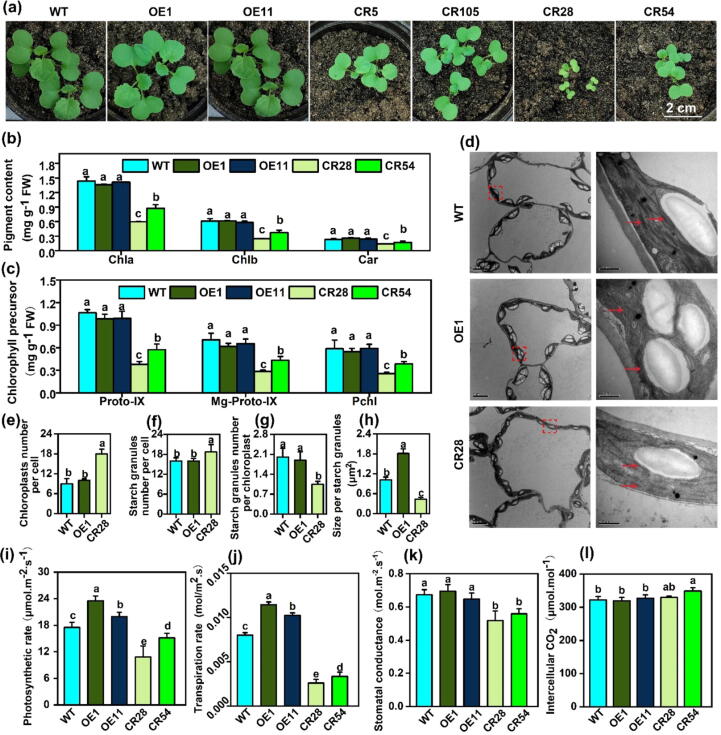
The seeds of transgenic lines including OE, mutant and WT were sown in the pots in growth chamber. Two weeks later, the seedlings of mutants were observed significantly smaller than WT, while there was no difference between OE and WT lines. Additionally, the mutant of CR28 was significantly smaller than other mutants, and the cotyledons of CR28 displayed a tendency to turn yellow (Fig. 2a). The seedlings size of CR5 and CR105 had no difference. However, the seedlings of CR54 were bigger than CR28, but smaller than CR5 and CR105 (Fig. 2a). CR28 and CR54 mutant plants showed retarded growth at different growth stages (Supplemental Fig. 2). We further evaluated the effect of BnaPPT1 on plant growth by growing these materials in the liquid media. Similarly, CR28 and CR54 mutant plants had shorter stem length, root length, less biomass than that of WT. However, these growth indexes had no difference between OE and WT plants (Supplemental Fig. 3). These data indicate that BnaA08.PPT1 and BnaC08.PPT1 have a key role in the maintenance of plant growth.
Fig. 2.
Disruption of BnaPPT1 genes affects plant growth of B. napus. (a) Growth phenotype of overexpression lines (OE1 and OE11) and mutants (CR5, CR105, CR28 and CR54). Pictures were taken from two-week-old plants. Bars = 2 cm. (b) Chlorophyll content in leaves. Samples were taken from three-week-old plants. Chla, Chlorophyll a; Chlb, Chlorophyll b; Car, Carotenoid. Values are means ± SD (n = 3). (c) Content of chlorophyll precursors from three-week-old plants. Values are means ± SD (n = 3). Proto-IX, protoporphyrin IX; Mg-Proto-IX, Mg protoporphyrin IX; Pchl, protochlorophyllide. (d) The transverse sections of leaves were observed under transmission electron microscope. Red arrows indicate the thylakoid grana. Samples were taken from leaves of 30-day-old plants. (e)-(h) Chloroplast number per cell, starch granule number per cell, starch granule number per chloroplast and size per starch granule. Values are means ± SD (n = 8–10). (i)-(l) Photosynthetic rate, transpiration rate, stomatal conductance and intercellular CO2 concentration in leaves. Data were collected from 80-day-old plants. Values are means ± SD (n = 5). Different letters represent significant differences at P < 0.05, based on an ANOVA analysis with Fisher LSD test. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Fig. 3.
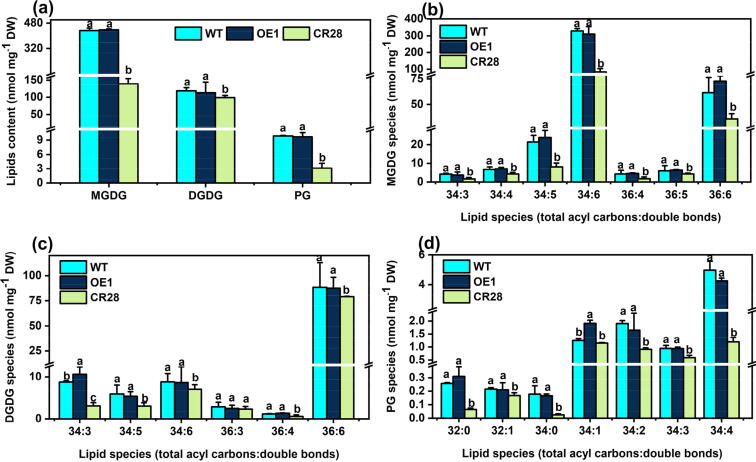
Content of lipids in leaves. (a) Content of total MGDG, DGDG and PG in leaves. (b) Content of MGDG species in leaves. (c) Content of DGDG species in leaves. (d) The species content of PG in leaves. Samples were taken from three-week-old plants. Values are means ± SD (n = 8). Different letters represent significant differences at P < 0.05, based on an ANOVA analysis with Fisher LSD test. MGDG, monogalactosyldiacylglycerol; DGDG, digalactoslydiacylglycerol; PG, phosphatidylglycerol.
To explore how BnaPPT1 impacts plant growth and the color of leaf, the metabolites related to chloroplast development were analyzed (Fig. 2b, 2c and Supplemental Fig. 4). Compared to WT, the content of chlorophyll a and b, and carotenoid in OE lines had no changes. However, the content of chlorophyll a was reduced by 30.5 %, 17.2 %, 58.4 % and 39.2 % in CR5, CR105, CR28 and CR54, respectively. Chlorophyll b was reduced by 19.8 %, 29.3 %, 59.6 % and 39.1 %, and carotenoid was reduced by 30.7 %, 63.3 %, 38.0 % and 27.3 % in CR5, CR105, CR28 and CR54, respectively (Fig. 2b and Supplemental Fig. 4a). The protoporphyrin IX (Proto-IX), Mg protoporphyrin IX (Mg-proto-IX) and protochlorophyllide (Pchl) are the precursors for chloroplast pigments synthesis [26]. The content of Proto-IX, Mg-proto-IX and Pchl was reduced by 64.5 %, 59.5 % and 56.4 % in CR28 than that in WT, and CR54 also showed decreased level of Proto-IX, Mg-proto-IX and Pchl (Fig. 2c and Supplemental Fig. 4b). However, the content of them in OE lines was equal to WT plants (Fig. 2c and Supplemental Fig. 4b). We analyzed the ultrastructure of chloroplast in the leaves of WT, mutant and OE lines using transmission electron microscope (TEM). The number of grana lamella was less in mutant plastids than that in OE and WT, and the density of grana was obvious less in mutant plastids (Fig. 2d). Compared to WT, CR28 had increased number of chloroplasts and starch granules in one cell, but the starch granules number per chloroplast and the size of starch were decreased in mutant CR28. There was no difference in the number of chloroplasts and starch granules in one cell, the starch granules number per chloroplast between OE1 and WT, but OE1 showed larger starch granules than WT (Fig. 2d-2 h). On the other hand, the results of the photosynthetic efficiency of 80-day-old plants in the field showed that OE lines had higher photosynthetic rate and transpiration rate than that in WT, but the mutants had lower photosynthetic rate, transpiration rate, and stomatal conductance compared to WT (Fig. 2i-2 k). However, there were no difference in stomatal conductance and intercellular CO2 concentration between OE and WT (Fig. 2k-2 l). In addition, the intercellular CO2 concentration was higher in CR54 than that in WT (Fig. 2l). These results suggest that BnaPPT1 plays an important role in chloroplast development and impacts plant photosynthesis.
Fig. 4.
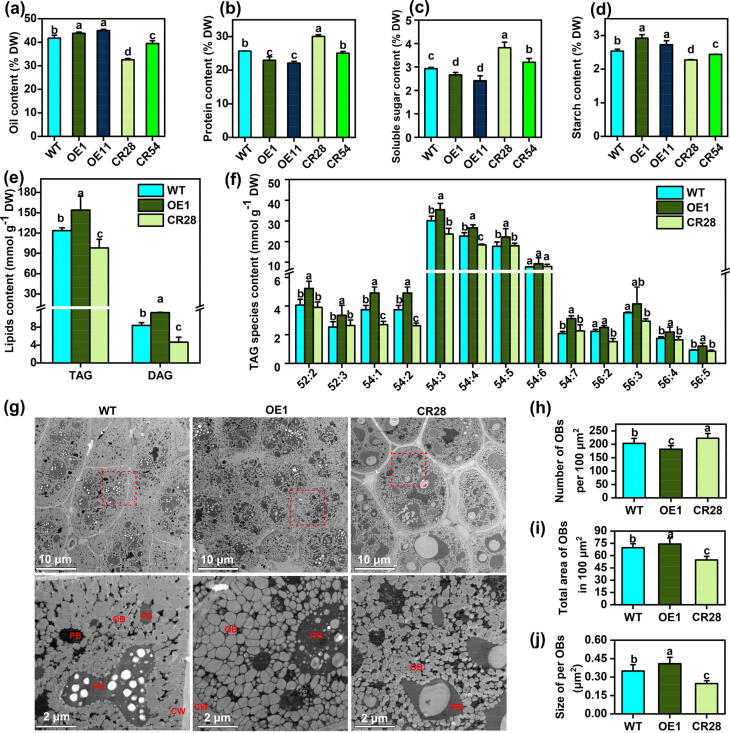
BnaPPT1 promotes seed oil accumulation in mature seeds. (a)-(d) Oil, protein, soluble sugar and starch content in mature seeds. Values are means ± SD (n = 4–6). (e) Contents of neutral lipids TAG and DAG. Values are means ± SD (n = 6). (f) Content of TAG species. Values are means (n = 6). (g) Cotyledon of mature seeds was examined by scanning transmission electron microscopy. (h)-(j) Number of OBs per 100 μm2, total area of OBs in 100 μm2 and size of per OBs in (g) Values are means ± SD (n = 12–16). CW, cell wall; OB, oil body; PB, protein body. Different letters represent significant differences at P < 0.05, based on an ANOVA analysis with Fisher LSD test.
To evaluate the impact of BnaPPT1 on agronomic traits of B. napus, WT, OE and mutant lines were sown in the field. During the whole plant growth period, the mutants displayed slower growth and yellowish leaf, while no visible different phenotype was found between OE and WT (Supplemental Fig. 5a and 5b). The agronomic traits including plant height, inflorescence length, number of effective branches, 1000 seeds weight, and plant yield etc. were investigated after plant harvest (Table 1). The results showed that the value of the most of the agronomic traits was significantly lower in mutants than that in WT, while there were no obvious differences between WT and OE lines (Table 1). In addition, the value of these traits was also significantly lower in CR28 than that in CR54 (Table 1). These results were consistence with the trends of plant growth in the growth chamber. Above results suggest that BnaPPT1 is necessary for maintaining plant growth, and the function of BnaA08.PPT1 and BnaC08.PPT1 is redundant.
Fig. 5.
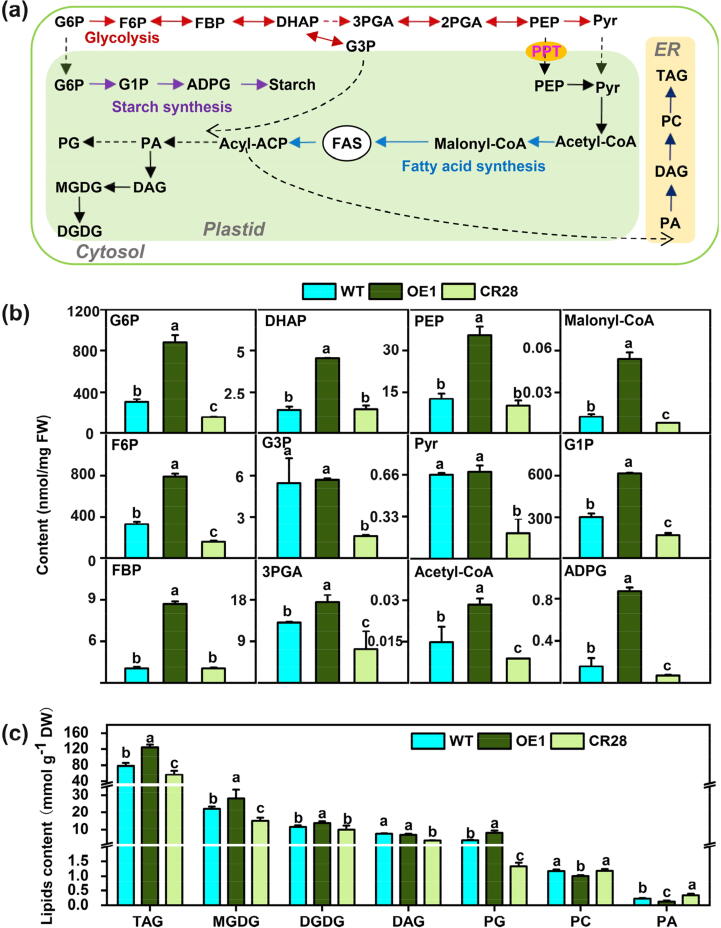
BnaPPT1 impacts glycolysis and lipids metabolism pathways in developing seeds. (a) Brief diagrams of glycolytic pathway, starch synthesis and lipids metabolism in plant. (b) The content of G6P, F6P, FBP, DHAP, G3P, 3PGA, PEP, Pyr, Acetyl-CoA, Malonyl-CoA, G1P and ADPG in seeds at 35 DAF. (c) Content of polar lipids in seeds at 35 DAF. Values are means ± SD (n = 6). Different letters represent significant differences at P < 0.05, based on an ANOVA analysis with Fisher LSD test. PPT, Phosphoenolpyruvate/phosphate translocator; G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; FBP, fructose-1,6-diphosphate; DHAP, dihydroxy-acetone-phosphate; G3P, glycerol-3-phosphate; 2-PGA, 2-phosphoglycerate; 3-PGA, 3-phosphoglycerate; PEP, phosphoenolpyruvate; Pyr, pyruvate; G1P, glucose-1-phosphate; ADPG, ADP-glucose. TAG, triacylglycerol; MGDG, monogalactosyldiacylglycerol; DGDG, digalactoslydiacylglycerol; DAG, diacylglycerol; PG, phosphatidylglycerol; PC, phospholipid choline; PA, phosphatidic acid.
Table 1.
Measurement of agronomic traits after plants were harvested.
| Plant height (cm) | Inflorescence length (cm) | Number of effective branches | Silique number of main inflorescence | Silique number per plant | Silique length (cm) | Seed number per silique | Thousand seed weight (g) | Yield per plant (g) | |
|---|---|---|---|---|---|---|---|---|---|
| WT | 92.9 ± 6.2a | 37.0 ± 1.4a | 7.0 ± 1.1a | 39.8 ± 4.2a | 164.8 ± 28.1a | 5.0 ± 0.2a | 20.2 ± 1.4a | 3.1 ± 0.3a | 6.1 ± 1.1a |
| OE1 | 92.1 ± 4.8a | 39.8 ± 3.0a | 7.3 ± 1.2a | 39.8 ± 3.0a | 161.8 ± 17.0a | 5.2 ± 0.2a | 19.6 ± 2.0a | 3.2 ± 0.1a | 7.2 ± 0.4a |
| OE11 | 87.9 ± 3.0a | 40.4 ± 2.7a | 7.3 ± 1.2a | 40.4 ± 2.7a | 171.0 ± 22.2a | 5.0 ± 0.1a | 18.2 ± 2.8 a | 3.1 ± 0.2a | 7.1 ± 2.2a |
| CR28 | 41.7 ± 1.5c | 10.5 ± 0.7c | 2.3 ± 0.5b | 15.7 ± 2.1c | 16.8 ± 2.2c | 4.6 ± 0.3b | 8.6 ± 0.9b | 2.8 ± 0.2b | 0.4 ± 0.2c |
| CR54 | 75.0 ± 7.9b | 29.6 ± 3.9b | 7.1 ± 1.0a | 34.0 ± 3.6b | 148.3 ± 15.1b | 5.2 ± 0.3a | 19.9 ± 2.4a | 3.0 ± 0.3a | 4.4 ± 0.3b |
Values are means ± SD (n = 6–12). Different letters represent significant differences at P < 0.05, based on an ANOVA analysis with Fisher LSD test.
Plastid membrane lipids level was significantly altered in BnaPPT1 mutant
Monogalactosyldiacyglycerol (MGDG), digalactosyldiacylglycerol (DGDG) and phosphatidylglycerol (PG) are main components of the chloroplast membrane [27]. We extracted the total lipids from OE1, CR28 mutant and WT leaf and then analyzed the lipids using liquid chromatography with tandem mass spectrometry (LC-MS/MS). The content of MGDG, DGDG and PG was decreased by 67.8 %, 68.5 % and 16.4 % in CR28 than that in WT, but there was no difference between OE1 and WT (Fig. 3a). These results were consistence with the trends of chlorophyll and chlorophyll precursor content among mutants, OE and WT lines. Compared to WT, the content of main species of MGDG-34:3 ∼ -34:6, MGDG-36:4 ∼ -36:6, DGDG-34:3, DGDG-36:6, PG-32:0 and PG-34:0 ∼ -34:4 was significantly reduced in the CR28 mutant (Fig. 3b-3d). However, most species of MGDG, DGDG and PG had comparable levels between OE1 and WT (Fig. 3b-3d). Besides, the FAs in leaves were measured (Supplemental Fig. 6). Compared to WT, there was no obvious difference in OE1. However, the content of C18:1 and C18:2 was increased, and C18:3 was reduced in CR28. These results imply that BnaPPT1-imported PEP is required for chloroplast membrane lipid synthesis.
Fig. 6.
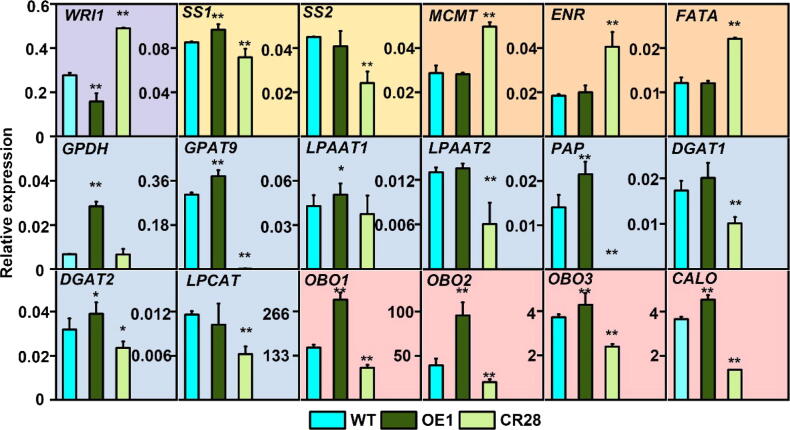
BnaPPT1 affects expression of lipid metabolism- and starch synthesis- related genes in developing seeds. Total RNA was extracted from 35 DAF seeds. Relative expression level was calculated by compared to actin’s expression level. * and ** indicates P < 0.05 and P < 0.01 respectively, based on a student t-test. WRI1, wrinkled1; SS1, starch synthase1; SS2, starch synthase2; MCMT, malonyltransferase; ENR, enoyl-ACP reductase; FATA, acyl-ACP thioesterase A; GPDH, glycerol-3-phosphate dehydrogenase; GPAT9, glycerol-3-phosphate acyltransferase 9; LPAAT1, lysophosphatidic acid acyltransferase 1; LPAAT2, lysophosphatidic acid acyltransferase 2; PAP, phosphatidate phosphatase; DGAT1, diacylglycerol acyltransferase 1; DGAT2, diacylglycerol acyltransferase 2; LPCAT, lysophosphatidylcholine acyltransferase; OBO1, oil body oleosin 1; OBO2, oil body oleosin 2; OBO3, oil body oleosin 3; CALO, Caleosin. Different colors represent different pathway genes. Purple, yellow, orange, blue and pink represent transcription factors, starch synthesis genes, fatty acid synthesis genes, TAG synthesis genes and oil body synthesis genes, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
BnaPPT1 affects seed oil accumulation
To evaluate the influence of BnaPPT1 on seed oil accumulation, several traits were investigated in mature seeds. The seed oil content (SOC) and protein content were determined by near infrared spectrometer (NIRS). The results showed that SOC was significantly increased from 41.7 % in WT to 43.8 % and 45.0 % in OE1 and OE11, respectively. However, SOC was decreased from 41.7 % in WT to 32.6 % and 39.5 % in CR28 and CR54, respectively (Fig. 4a). Furthermore, the FA composition in seeds was analyzed by gas chromatography-flame ionization detector (GC-FID). The results show that the content of C18:1 was slightly reduced and C18:2 was increased in CR28 and CR54 compared to WT. However, the content of all FAs had no changes in OE lines compared to WT (Supplemental Fig. 7). The protein content in OE lines was noticeably decreased by 2.8–3.5 %, while it was increased by 4.4 % in CR28 (Fig. 4b). Besides, the soluble sugar was decreased in OE lines but increased in mutants (Fig. 4c). Interestingly, the starch content was increased in OE lines but decreased in mutants (Fig. 4d). However, the content of seed oil, protein, soluble sugar, and starch had no difference between single mutants and WT (Supplemental Fig. 8). It suggests that BnaPPT1 can improve seed oil content, and the functions of BnaC08.PPT1 and BnaA08.PPT1 have redundancy to a certain extent.
TAG is the predominant lipid in mature seed, and DAG is the precursor for TAG. Total lipids were extracted from mature seeds and then analyzed using LC-MS/MS. The results showed that the content of TAG and DAG was increased by 24.6 % and 34.0 % in OE1, but decreased by 20.5 % and 44.6 % in CR28, as compared to that in WT (Fig. 4e). The content of TAG species −52:2, −52:3, −54:1 ∼ -54:5, −56:4, and −54:5 was increased in OE1, but the content of a few species like TAG-54:1, −54:2, and −54:4 was decreased in CR28 (Fig. 4f). The content of main species of DAG-36:2 ∼ -36:4 was significantly increased in OE1 compared to WT. However, the content of DAG-32:0, –32:1, −34:0 ∼ -34:2, −36:2, −36:3, and −36:5 was lower in CR28 than that in WT (Supplemental Fig. 9).
Oil bodies (OBs) are the place for TAG storage. The slices of seed cotyledon were made to analyze the parameters related to OBs by TEM. The density of OBs was obviously less in mutant than that in OE and WT (Fig. 4g, upper). In contrast, the ratio of other tissues such as protein bodies (PBs) was larger in CR28 than that in OE1 and WT (Fig. 4g, lower). The number of OBs and the size of OBs were quantified and compared among OE1, CR28 and WT. Compared to WT, OE1 had fewer OBs number in 100 μm2 while CR28 had more OBs. The total area of OBs in 100 μm2 and the size of per OB were larger in OE1 but smaller in CR28 (Fig. 4h-4j). Taken together, above results suggest that BnaPPT1 can affect seed oil accumulation.
BnaPPT1 affects glycolysis and lipids metabolism in developing seeds
To further explore how BnaPPT1 affects oil accumulation, metabolites in glycolysis, starch synthesis and lipid biosynthesis in developing seeds were determined at 35-day after flowering (DAF), when seed oil accumulation was rapid and BnaPPT1 had the highest expression level. A brief model of carbon influx into lipid and starch from glycolysis was shown in Fig. 5a. The content of the metabolites of glucose-6-phosphate (G6P), fructose-6-phosphate (F6P), fructose-1,6-diphosphate (FBP), dihydroxy-acetone-phosphate (DHAP), glycerol-3-phosphate (G3P), 3-phosphoglycerate (3PGA), PEP and Pyr (pyruvate) related to glycolysis was significantly lower in CR28 than that in WT (Fig. 5b). However, most of them was higher in OE1 than WT, except the content of G3P and pyruvate had no difference between OE1 and WT (Fig. 5b). Furthermore, the content of the precursors of starch synthesis, glucose-1-phosphate (G1P) and ADP-glucose (ADPG), was higher in OE1 than WT, but lower in CR28 (Fig. 5b). Acetyl-CoA and malonyl-CoA are the precursors for FA synthesis, the content of them was also higher in OE1 but lower in mutant compared to WT (Fig. 5b).
Lipid was also extracted from 35 DAF seeds. The results of lipidome analysis showed that TAG was increased by 45.4 % in OE1 but decreased by 22 % in CR28 compared to WT (Fig. 5c). The content of DAG was decreased by 49.8 % in CR28, while had no obvious difference in OE1 and WT. Moreover, compared to WT, the content of membrane lipids MGDG, DGDG and PG was noticeably increased in OE1 but decreased in CR28. The level of phosphatidylcholine (PC) was slightly decreased in OE1, while had no obvious difference between CR28 and WT (Fig. 5c). Interestingly, the content of PA was decreased by 45.6 % in OE1 but increased by 49.4 % in CR28 compared to WT (Fig. 5c).
The expression of some key genes related to glycolysis, starch and FA synthesis, and lipid metabolism was analyzed in 35 DAF seeds of OE1, CR28 and WT. The results showed that the expression level of starch synthesis-related genes starch synthase (SS1 and SS2) was decreased in CR28 compared to WT, while the expression of SS1 was increased in OE1 (Fig. 6). The expression of FA synthesis-related genes malonyltransferase (MCMT), enoyl-ACP reductase (ENR) and acyl-ACP thioesterase (FATA) was increased by 1.2, 0.8 and 0.7 folds in CR28 than that in WT, but showed no changes in OE1 and WT (Fig. 6). The expression of most genes related to TAG synthesis such as glycerol-3-phosphate dehydrogenase (GPDH), glycerol-3-phosphate acyltransferase 9 (GPAT9), lysophosphatidic acid acyltransferase 1 (LPAAT1), lysophosphatidic acid acyltransferase 2 (LPAAT2), phosphatidate phosphatase (PAP), diacylglycerol acyltransferase 1 (DGAT1), diacylglycerol acyltransferase 2 (DGAT2), and lysophosphatidylcholine acyltransferase (LPCAT) was lower in CR28 than that in WT, while they had higher expression in OE1 than WT except LPAAT2, DGAT1 and LPCAT (Fig. 6). Moreover, the gene expression of oil body oleosin 1 (OBO1), body oleosin 2 (OBO2), body oleosin 3 (OBO3) and Caleosin (CALO) which are related to oil storage was also determined. The data showed that all of them had higher expression in OE1 but lower in CR28 than WT (Fig. 6). It was worth noting that the expression of WRINKLED1 (WRI1), a vital transcription factor regulating FA biosynthesis and glycolytic pathway [28], [29], [30], [31], [32], [33], was decreased by 22.2 % in OE1 but elevated by 167.8 % in CR28 compared to WT (Fig. 6). These results indicate that manipulation of BnaPPT1 may affect the expression of key genes involved in glycolysis, oil biosynthesis and lipid droplet formation.
Discussion
In plants, there are many translocator related to transport the metabolites during glycolysis, such as PPT1, triose phosphate translocator (TPT), glucose 6-phosphate translocator (GPT) and xylulose 5-phosphate translocator (XPT) etc., which are belonged to plastidic phosphate translocator (pPT) family and play roles in transporting metabolites from cytosol into plastid [34]. Previous researches exhibited that all of these plastidic phosphate translocators had effect on plant growth and chloroplast development [9], [10], [35], [36], [37], [38], [39]. However, no further studies on how they affect plant growth and chloroplast development have been reported and their effect on seed oil accumulation has been rarely determined.
The function of PPT1 is transporting PEP from cytosol into the plastid [34]. Knockout of AtPPT1 caused the slower growth and yellowing leaf, and reduced the size of the mesophyll cells and mesophyll chloroplasts [9], [10]. These phenomena were consistency with our results in B. napus, suggesting the function of PPT1 was conserved in B. napus and Arabidopsis. However, how PPT1 affects the development of mesophyll cells and mesophyll chloroplasts is still unknown. In our study, the content of Chlorophyll a, b, carotenoid, proto-IX, Mg-Proto-IX, and Pchl was significantly lower in mutants than that in WT, but had no difference between OE and WT. Moreover, the content of them was obviously lower in homozygous double mutant than that in single or heterozygous double mutants, and the same trends were observed in plant growth and biomass accumulation. It suggests that PPT1 not only impacts chloroplast development but also impacts the synthesis of the precursors of chloroplast, leading to prevent plant growth, and the function of BnaC08.PPT1 and BnaA08.PPT1 is redundant to a certain extent. In addition, the number of chloroplasts in one cell was more in CR28 than that in WT and OE1, but the number of starch granules in one chloroplast was significantly less in CR28 than that in WT and OE1. Furthermore, the content of plastid membrane lipids containing MGDG, DGDG and PG [27] was also changed in CR28. Hence, we speculate that the development of chloroplast is abnormal in BnaPPT1 mutant, causing more chloroplasts are needed to maintain photosynthesis to keep the mutant alive. While, whether PPT1 impacts the synthesis of pigment and chlorophyll precursor first or impacting the development of chloroplast first is the key point which should be solved in future.
PEP is a precursor for pyruvate, which is an important initial metabolic substrate for many biochemical substances, such as FA and amino acid etc [40]. In plastid, one important role of pyruvate is to synthesize FA, which will be exported to endoplasmic reticulum (ER) to assemble into glycerol skeleton to form triacylglycerol (TAG) [41]. Previous studies showed that PK can convert PEP into pyruvate in plastid, and pyruvate transporter BILE ACID: SODIUM SYMPORTER FAMILY PROTEIN 2 (BASS2) can transport pyruvate from cytosol into plastid, thereby providing more substrates for the accumulation of seed oil [5], [42]. Although the previous studies showed that BnaPPT1 was one of the important plastidic transporters for channeling carbon intermediates PEP from the cytosol into the plastid in B. napus, the changes of FA and oil content in seed have not been reported [43], [44]. In this study, BnaPPT1 is highly expressed in seeds, which suggests that it may play a role in oil accumulation. The content of most intermediary metabolites in glycolysis, FA and TAG biosynthesis in 35 DAF seeds was increased in OE but decreased in mutant. Finally, the content of TAG and DAG was higher in OE but lower in mutants compared to WT. However, the content of pyruvate has no difference between WT and OE. Therefore, further analysis of the content of pyruvate and other primary metabolites in plastid of seeds needs to be measured.
A lot of transcription factors (TFs) and enzymes are involved in the biosynthesis processes of starch, FA and TAG [27], [45]. Among them, WRI1, a TF, which is the “Master Regulator” in plant oil biosynthesis [46]. Previous studies showed that the expression of WRI1 was positively related to FA and TAG synthesis, playing “push” and “pull” roles in FA synthesis [47], [48], [49]. During this process, WRI1 could also accelerate the process of glycolysis to supply more carbon resource to form the substrate for FA synthesis [33], [46]. The increased free FA will increase its toxic action for cells, which requires the enzymes involved in TAG synthesis to quickly assemble the FA onto the glycerol skeleton in the form of ester [31]. In our results, the changes in expression levels of most genes encoding these proteins among OE, mutants, and WT were basically as expected. Interestingly, the expression of MCMT, ENR, and FATA which are involved in FA synthesis genes was higher in mutants than that in WT, but no difference in OE and WT. Furthermore, the expression of WRI1 was much higher in mutants but lower in OE compared in WT. These results seem opposite to the previous studies [47], [48], [49]. Hence, we speculate that the defect in TAG metabolism in BnaPPT1 mutant seeds will form a feedback mechanism, which stimulates more WRI1 to be expressed to supply more carbon influx into FA biosynthesis pathway. Meanwhile, this feedback mechanism stimulates more enzymes related to FA synthesis to be expressed to rapidly synthesize FA to ensure TAG biosynthesis. However, the mutation of BnaPPT1 impedes the PEP to form pyruvate, leading to the initial substrate for FA synthesis is deficient, and then the TAG could not be synthesized normally.
Biotechnology has been broadly used to improve the oil content in oil crop seeds. Previous studies reported that SOC can be significantly improved by overexpressing transcription factors such as WRI1, LEC1, LEC2 [47], [50], [51], [52], GPDH [53], LPAAT [54], [55], DGAT1 [56], [57], oleosin protein-encoding gene OLEO1 [58] etc. However, the other traits including flowering, seed weight were also altered when the seed oil content was increased in these genes-overexpressed plants [58], [59]. Here, overexpression of BnaPPT1 could significantly improve the SOC in B. napus, but had no any other effect on the other agronomic traits. Thus, PPT1 is an important factor for improving the SOC in oilseed.
Compliance with Ethics Requirements
This article does not contain any studies with human or animal subjects.
CRediT authorship contribution statement
Shan Tang: Methodology, Investigation. Fei Peng: Investigation. Qingqing Tang: Investigation. Yunhao Liu: Investigation. Hui Xia: Investigation. Xuan Yao: Supervision, Writing-review & editing. Shaoping Lu: Supervision, Writing-review & editing. Liang Guo: Conceptualization, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by grants from the National Natural Science Foundation of China (31871658), China Postdoctoral Science Foundation (2021M691182), Hubei Hongshan Laboratory Research Funding (2021HSZD004) and Higher Education Discipline Innovation Project (B20051).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.07.008.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Aoyagi K., Bassham J.A. Pyruvate orthophosphate dikinase mRNA organ specificity in wheat and maize. Plant Physiol. 1984;76(1):278–280. doi: 10.1104/pp.76.1.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prabhakar V., Lottgert T., Gigolashvili T., Bell K., Flugge U.I., Hausler R.E. Molecular and functional characterization of the plastid-localized Phosphoenolpyruvate enolase (ENO1) from Arabidopsis thaliana. FEBS Lett. 2009;583(6):983–991. doi: 10.1016/j.febslet.2009.02.017. [DOI] [PubMed] [Google Scholar]
- 3.Flugge U.-I., Hausler R.E., Ludewig F., Gierth M. The role of transporters in supplying energy to plant plastids. J Exp Bot. 2011;62(7):2381–2392. doi: 10.1093/jxb/erq361. [DOI] [PubMed] [Google Scholar]
- 4.Prabhakar V., Löttgert T., Geimer S., Dörmann P., Krüger S., Vijayakumar V., et al. Phosphoenolpyruvate provision to plastids is essential for gametophyte and sporophyte development in Arabidopsis thaliana. Plant Cell. 2010;22(8):2594–2617. doi: 10.1105/tpc.109.073171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andre C, Froehlich JE, Moll MR, Benning C. A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. Plant Cell 2007;19(6):2006–22. [DOI] [PMC free article] [PubMed]
- 6.Fischer K., Kammerer B., Gutensohn M., Arbinger B., Weber A., Hausler R.E., et al. A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyruvate/phosphate antiporter. Plant Cell. 1997;9(3):453–462. doi: 10.1105/tpc.9.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knappe S., Lottgert T., Schneider A., Voll L., Flugge U.-I., Fischer K. Characterization of two functional phosphoenolpyruvate/phosphate translocator (PPT) genes in Arabidopsis–AtPPT1 may be involved in the provision of signals for correct mesophyll development. Plant J. 2003;36(3):411–420. doi: 10.1046/j.1365-313x.2003.01888.x. [DOI] [PubMed] [Google Scholar]
- 8.Knappe S., Flugge U.I., Fischer K. Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 2003;131(3):1178–1190. doi: 10.1104/pp.016519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H., Culligan K., Dixon R.A., Chory J. CUE1: a mesophyll cell-specific positive regulator of light-controlled gene expression in Arabidopsis. Plant Cell. 1995;7(10):1599–1610. doi: 10.1105/tpc.7.10.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Streatfield S.J., Weber A., Kinsman E.A., Häusler R.E., Li J., Post-Beittenmiller D., et al. The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell. 1999;11(9):1609–1621. doi: 10.1105/tpc.11.9.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gan L.S.X., Jin L., Wang G., Xiu J., Wei Z., Fu T. Establishment of math models of NIRS analysis for oil and protein contents in seed of Brassica napus. Scientia Agricultura Sinica. 2003;36(12):1609–1613. [Google Scholar]
- 12.Feigl G., Lehotai N., Molnár Á., Ördög A., Rodríguez-Ruiz M., Palma J.M., et al. Zinc induces distinct changes in the metabolism of reactive oxygen and nitrogen species (ROS and RNS) in the roots of two Brassica species with different sensitivity to zinc stress. Ann Bot. 2015;116(4):613–625. doi: 10.1093/aob/mcu246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xing H.-L., Dong L.i., Wang Z.-P., Zhang H.-Y., Han C.-Y., Liu B., et al. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14(1) doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lei Y., Lu L.i., Liu H.-Y., Li S., Xing F., Chen L.-L. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant. 2014;7(9):1494–1496. doi: 10.1093/mp/ssu044. [DOI] [PubMed] [Google Scholar]
- 15.Gao X., Yan P., Shen W., Li X., Zhou P., Li Y. Modular construction of plasmids by parallel assembly of linear vector components. Anal Biochem. 2013;437(2):172–177. doi: 10.1016/j.ab.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 16.Tang T., Yu X., Yang H., Gao Q.i., Ji H., Wang Y., et al. Development and validation of an effective CRISPR/Cas9 vector for efficiently isolating positive transformants and transgene-free mutants in a wide range of plant species. Front Plant Sci. 2018;9 doi: 10.3389/fpls.2018.01533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu W., Xie X., Ma X., Li J., Chen J., Liu Y.-G. DSDecode: a web-based tool for decoding of sequencing chromatograms for genotyping of targeted mutations. Mol Plant. 2015;8(9):1431–1433. doi: 10.1016/j.molp.2015.05.009. [DOI] [PubMed] [Google Scholar]
- 18.Yoo S.-D., Cho Y.-H., Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2(7):1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 19.Zhou Z., Dun X., Xia S., Shi D., Qin M., Yi B., et al. BnMs3 is required for tapetal differentiation and degradation, microspore separation, and pollen-wall biosynthesis in Brassica napus. J Exp Bot. 2012;63(5):2041–2058. doi: 10.1093/jxb/err405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong Y, Yuan S, Sun L, Wang X, Hong Y. Cytidinediphosphate-diacylglycerol synthase 5 is required for phospholipid homeostasis and is negatively involved in hyperosmotic stress tolerance. Plant J 2018;94(6):1038-50. [DOI] [PubMed]
- 21.Guo L., Ma F., Wei F., Fanella B., Allen D.K., Wang X. Cytosolic phosphorylating glyceraldehyde-3-phosphate dehydrogenases affect Arabidopsis cellular metabolism and promote seed oil accumulation. Plant Cell. 2014;26(7):3023–3035. doi: 10.1105/tpc.114.126946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo B., Groenke K., Takors R., Wandrey C., Oldiges M. Simultaneous determination of multiple intracellular metabolites in glycolysis, pentose phosphate pathway and tricarboxylic acid cycle by liquid chromatography-mass spectrometry. J Chromatogr A. 2007;1147(2):153–164. doi: 10.1016/j.chroma.2007.02.034. [DOI] [PubMed] [Google Scholar]
- 23.Lu S., Yao S., Wang G., Guo L., Zhou Y., Hong Y., et al. Phospholipase Dε enhances Brassica napus growth and seed production in response to nitrogen availability. Plant Biotechnol J. 2016;14(3):926–937. doi: 10.1111/pbi.12446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang S., Zhao H.u., Lu S., Yu L., Zhang G., Zhang Y., et al. Genome- and transcriptome-wide association studies provide insights into the genetic basis of natural variation of seed oil content in Brassica napus. Mol Plant. 2021;14(3):470–487. doi: 10.1016/j.molp.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 25.Liu D., Yu L., Wei L., Yu P., Wang J., Zhao H.u., et al. BnTIR: an online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnol J. 2021;19(10):1895–1897. doi: 10.1111/pbi.13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheminant S., Wild M., Bouvier F., Pelletier S., Renou J.-P., Erhardt M., et al. DELLAs regulate chlorophyll and carotenoid biosynthesis to prevent photooxidative damage during seedling deetiolation in Arabidopsis. Plant Cell. 2011;23(5):1849–1860. doi: 10.1105/tpc.111.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, et al. Acyl-lipid metabolism. The Arabidopsis book 2013;11:e0161. [DOI] [PMC free article] [PubMed]
- 28.Focks N., Benning C. wrinkled1: A novel, low-seed-oil mutant of Arabidopsis with a deficiency in the seed-specific regulation of carbohydrate metabolism. Plant Physiol. 1998;118(1):91–101. doi: 10.1104/pp.118.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baud S., Wuillème S., To A., Rochat C., Lepiniec Loïc. Role of WRINKLED1 in the transcriptional regulation of glycolytic and fatty acid biosynthetic genes in Arabidopsis. Plant J. 2009;60(6):933–947. doi: 10.1111/j.1365-313X.2009.04011.x. [DOI] [PubMed] [Google Scholar]
- 30.Adhikari N.D., Bates P.D., Browse J. WRINKLED1 rescues feedback inhibition of fatty acid synthesis in hydroxylase-expressing seeds. Plant Physio. 2016;171(1):179–191. doi: 10.1104/pp.15.01906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.To A., Joubès J., Barthole G., Lécureuil A., Scagnelli A., Jasinski S., et al. WRINKLED transcription factors orchestrate tissue-specific regulation of fatty acid biosynthesis in Arabidopsis. Plant Cell. 2012;24(12):5007–5023. doi: 10.1105/tpc.112.106120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kong Q, Ma W. WRINKLED1 as a novel 14-3-3 client: function of 14-3-3 proteins in plant lipid metabolism. Plant Signal Behav 2018;13(8):e1482176. [DOI] [PMC free article] [PubMed]
- 33.Kong Q., Yang Y., Guo L., Yuan L., Ma W. Molecular basis of plant oil biosynthesis: insights gained from studying the WRINKLED1 transcription factor. Front Plant Sci. 2020;11:24. doi: 10.3389/fpls.2020.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber A.P.M., Linka N. Connecting the plastid: transporters of the plastid envelope and their role in linking plastidial with cytosolic metabolism. Annu Rev Plant Biol. 2011;62(1):53–77. doi: 10.1146/annurev-arplant-042110-103903. [DOI] [PubMed] [Google Scholar]
- 35.Hilgers E.J.A., Staehr P., Flugge U.I., Hausler R.E. The xylulose 5-phosphate/phosphate translocator supports triose phosphate, but not phosphoenolpyruvate transport across the inner envelope membrane of plastids in Arabidopsis thaliana mutant plants. Front Plant Sci. 2018;9:1461. doi: 10.3389/fpls.2018.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hilgers EJA, Schottler MA, Mettler-Altmann T, Krueger S, Dormann P, Eicks M, et al. The combined loss of triose phosphate and xylulose 5-phosphate/phosphate translocators leads to severe growth retardation and impaired photosynthesis in Arabidopsis thaliana tpt/xpt double mutants. Front Plant Sci 2018;9:1331. [DOI] [PMC free article] [PubMed]
- 37.Kunz H.H., Häusler R.E., Fettke J., Herbst K., Niewiadomski P., Gierth M., et al. The role of plastidial glucose-6-phosphate/phosphate translocators in vegetative tissues of Arabidopsis thaliana mutants impaired in starch biosynthesis. Plant Biol. 2010;12:115–128. doi: 10.1111/j.1438-8677.2010.00349.x. [DOI] [PubMed] [Google Scholar]
- 38.Rolletschek H., Nguyen T.H., Häusler R.E., Rutten T., Göbel C., Feussner I., et al. Antisense inhibition of the plastidial glucose-6-phosphate/phosphate translocator in Vicia seeds shifts cellular differentiation and promotes protein storage. Plant J. 2007;51(3):468–484. doi: 10.1111/j.1365-313X.2007.03155.x. [DOI] [PubMed] [Google Scholar]
- 39.Schneider A., Häusler R.E., Kolukisaoglu Ü., Kunze R., Van Der Graaff E., Schwacke R., et al. An Arabidopsis thaliana knock-out mutant of the chloroplast triose phosphate/phosphate translocator is severely compromised only when starch synthesis, but not starch mobilisation is abolished. Plant J. 2002;32(5):685–699. doi: 10.1046/j.1365-313x.2002.01460.x. [DOI] [PubMed] [Google Scholar]
- 40.Schwender J., Ohlrogge J., Shachar-Hill Y. Understanding flux in plant metabolic networks. Curr Opin Plant Bio. 2004;7(3):309–317. doi: 10.1016/j.pbi.2004.03.016. [DOI] [PubMed] [Google Scholar]
- 41.Xu C., Shanklin J. Triacylglycerol metabolism, function, and accumulation in plant vegetative tissues. Annu Rev Plant Biol. 2016;67(1):179–206. doi: 10.1146/annurev-arplant-043015-111641. [DOI] [PubMed] [Google Scholar]
- 42.Lee E.J., Oh M., Hwang J.U., Li-Beisson Y., Nishida I., Lee Y. Seed-specific overexpression of the pyruvate transporter BASS2 increases oil content in Arabidopsis seeds. Front Plant Sci. 2017;8:194. doi: 10.3389/fpls.2017.00194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kubis S.E., Rawsthorne S. The role of plastidial transporters in developing embryos of oilseed rape (Brassica napus L.) for fatty acid synthesis. Biochem Soc Trans. 2000;28(6):665–666. [PubMed] [Google Scholar]
- 44.Kubis S.E., Pike M.J., Everett C.J., Hill L.M., Rawsthorne S. The import of phosphoenolpyruvate by plastids from developing embryos of oilseed rape, Brassica napus (L.), and its potential as a substrate for fatty acid synthesis. J Exp Bot. 2004;55(402):1455–1462. doi: 10.1093/jxb/erh157. [DOI] [PubMed] [Google Scholar]
- 45.Song J.-M., Zhang Y., Zhou Z.-W., Lu S., Ma W., Lu C., et al. Oil plant genomes: current state of the science. J Exp Bot. 2022;73(9):2859–2874. doi: 10.1093/jxb/erab472. [DOI] [PubMed] [Google Scholar]
- 46.Kong Q, Yuan L, Ma W. WRINKLED1, a “Master Regulator” in transcriptional control of plant oil biosynthesis. Plants (Basel) 2019;8(7). [DOI] [PMC free article] [PubMed]
- 47.Liu J., Hua W., Zhan G., Wei F., Wang X., Liu G., et al. Increasing seed mass and oil content in transgenic Arabidopsis by the overexpression of wri1-like gene from Brassica napus. Plant Physiol Bioc. 2010;48(1):9–15. doi: 10.1016/j.plaphy.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 48.Elahi N., Duncan R.W., Stasolla C. Decreased seed oil production in FUSCA3 Brassica napus mutant plants. Plant Physiol Bioc. 2015;96:222–230. doi: 10.1016/j.plaphy.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 49.Vanhercke T., El Tahchy A., Shrestha P., Zhou X.R., Singh S.P., Petrie J.R. Synergistic effect of WRI1 and DGAT1 coexpression on triacylglycerol biosynthesis in plants. FEBS Lett. 2013;587(4):364–369. doi: 10.1016/j.febslet.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 50.Tan H., Yang X., Zhang F., Zheng X., Qu C., Mu J., et al. Enhanced seed oil production in canola by conditional expression of Brassica napus LEAFY COTYLEDON1 and LEC1-LIKE in developing seeds. Plant Physiol. 2011;156(3):1577–1588. doi: 10.1104/pp.111.175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen B., Allen W.B., Zheng P., Li C., Glassman K., Ranch J., et al. Expression of ZmLEC1 and ZmWRI1 increases seed oil production in maize. Plant Physiol. 2010;153(3):980–987. doi: 10.1104/pp.110.157537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Santos Mendoza M, Dubreucq B, Miquel M, Caboche M, Lepiniec L. LEAFY COTYLEDON 2 activation is sufficient to trigger the accumulation of oil and seed specific mRNAs in Arabidopsis leaves. FEBS Lett 2005;579(21):4666–70. [DOI] [PubMed]
- 53.Vigeolas H., Waldeck P., Zank T., Geigenberger P. Increasing seed oil content in oil-seed rape (Brassica napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol J. 2007;5(3):431–441. doi: 10.1111/j.1467-7652.2007.00252.x. [DOI] [PubMed] [Google Scholar]
- 54.Woodfield H.K., Fenyk S., Wallington E., Bates R.E., Brown A., Guschina I.A., et al. Increase in lysophosphatidate acyltransferase activity in oilseed rape (Brassica napus) increases seed triacylglycerol content despite its low intrinsic flux control coefficient. New Phytol. 2019;224(2):700–711. doi: 10.1111/nph.16100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maisonneuve S., Bessoule J.-J., Lessire R., Delseny M., Roscoe T.J. Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis. Plant Physiol. 2010;152(2):670–684. doi: 10.1104/pp.109.148247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weselake R.J., Shah S., Tang M., Quant P.A., Snyder C.L., Furukawa-Stoffer T.L., et al. Metabolic control analysis is helpful for informed genetic manipulation of oilseed rape (Brassica napus) to increase seed oil content. J Exp Bot. 2008;59(13):3543–3549. doi: 10.1093/jxb/ern206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jako C., Kumar A., Wei Y., Zou J., Barton D.L., Giblin E.M., et al. Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol. 2001;126(2):861–874. doi: 10.1104/pp.126.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang D, Zhang H, Hu Z, Chu S, Yu K, Lv L, et al. Artificial selection on GmOLEO1 contributes to the increase in seed oil during soybean domestication. PLoS Genet 2019;15(7):e1008267. [DOI] [PMC free article] [PubMed]
- 59.Li Q, Shao J, Tang S, Shen Q, Wang T, Chen W, et al. Wrinkled1 accelerates flowering and regulates lipid homeostasis between oil accumulation and membrane lipid anabolism in Brassica napus. Front Plant Sci 2015;6:1015. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.