Graphical abstract
Keywords: Oryza sativa, Domestication, Modern breeding, Novel functional variation selection, Standing functional variation selection
Highlights
-
•
We quantified the genome wide consequences of selection responsible for the subspecific and population differentiation of rice before and during domestication using a 3D differentiation model, which provided insights into the evolutionary process of rice.
-
•
We showed the presence of equivalent rich genomic diversity and subspecific/population differentiation within both Os (landraces) and Or of the similar geographic origins and sample sizes, providing strong evidence rejecting the single origin (domestication) model of Os from China.
-
•
Our results revealed the evolutionary dynamics of rice at the genomic level during domestication and modern breeding by comparing the genome wide selective signatures.
-
•
Our results, combined with all available evidence from previous studies, led us to a new hypothesis of multiple independent domestications of Os at different sites and timing over a long period of time in Asia with the earliest domestication events occurred in China.
-
•
Our results suggest the more efficient utilization of the rich diversity within Os by exploiting inter-subspecific and among population diversity in future rice improvement.
Abstract
Introduction
Rice, Oryza sativa L. (Os), is one of the oldest domesticated cereals that has also gone through extensive improvement in modern breeding.
Objectives
How rice was domesticated and impacted by modern breeding.
Methods
We performed comprehensive analyses of genomic sequences of 504 accessions of Os and 456 accessions of O. rufipogon/O. nivara (Or).
Results
The natural selection on Or before domestication and the natural and artificial selection during domestication together shaped the well-differentiated genomes of two subspecies, geng(j) (japonica) and xian(i) (indica), while breeding has made apparent genomic imprints between landrace and modern varieties of each subspecies, and also between primary modern and advanced modern varieties of xian(i). Selection during domestication and breeding left genome-wide selective signals covering ∼ 22.8 % and ∼ 8.6 % of the Os genome, significantly reduced within-population genomic diversity by ∼ 22 % in xian(i) and ∼ 53 % in geng(j) plus more pronounced subspecific differentiation. Only ∼ 10 % reduction in the total genomic diversity was observed between the Os and Or populations, indicating domestication did not suffer severe genetic bottleneck.
Conclusion
Our results revealed clear differentiation of the Or accessions into three large populations, two of which correspond to the well-differentiated Os subspecies, geng(j) and xian(i). Improved productivity and common changes in the same suit of adaptive traits in xian(i) and geng(j) during domestication and breeding resulted apparently from compensatory and convergent selections for different genes/alleles acting in the common KEGG terms and/or same gene families, and thus maintaining or even increasing the within population diversity and subspecific differentiation of Os, while more genes/alleles of novel function were selected during domestication than modern breeding. Our results supported the multiple independent domestication of Os in Asia and suggest the more efficient utilization of the rich diversity within Os by exploiting inter-subspecific and among population diversity in future rice improvement.
Introduction
Asian cultivated rice (Oryza sativa L.) (Os) is the staple food for more than half of the world population. As one of the oldest domesticated cereals, rice is well-known for its rich within species diversity, xian(i) (indica) - geng(j) (japonica) subspecific differentiation and subpopulation differentiation [1], [2], [3], [4]. Also, rice has gone through intensive improvement for decades since the green revolution in 1950s. Domestication practiced by ancient humans and breeding performed by modern breeders shared a common element, i.e. selection for increased productivity and adaptation to specific environments. However, the two processes differ in many aspects. The former (domestication) involved a long term of selection for increased productivity from O. rufipogon/O. nivara (Or) each consisting of largely homozygous individuals with limited introgression from other populations, while the latter (modern breeding) involves strong selection of relatively few generations for increased productivity from segregating populations of crosses involving two or more genetically different lines. Both processes, undoubtedly, have shaped the Os population architecture, but how both domestication and modern breeding influenced the organization of genomic diversity within and among different rice populations have not been fully characterized. The answer to this question will not only shed light on the genetic dynamics and process regarding the population differentiation during domestication and modern breeding of Os, but also provide important information for future rice improvement.
During the past decade, tremendous efforts have been taken to understand the impacts of domestication and modern breeding on the population structure of Os using DNA markers and cloned domestication genes [5], [6], [7], [8], [9], [10], [11]. While provided valuable information on the patterns of selection signatures in the rice genome and some ‘domestication’ genes, results from these studies remain inconclusive regarding the origin (domestication) of rice and impacts of domestication and breeding on the Os population structure due to small and/or biased sampling in the sequenced genomes and genes. It is now possible to address this issue with the availability of the genome sequence data of our newly sequenced 94 modern rice varieties and 410 representative rice varieties from 3,024 rice accessions of the 3,000 Rice Genome (3 K RG) panel [4], [12] with two reference genomes and wild rice accessions [9], [10].
In this study, we performed comprehensive analyses of the genome sequence data of a selected set of 504 Os accessions with balanced number of landraces and modern varieties (Tables S1-S2) and public sequence data of 456 wild rice accessions. Our results revealed many interesting features of the population architecture of rice resulting from domestication and modern breeding and shed new light on the genetic mechanisms of domestication and modern breeding with important implications for future improvement of this important crop.
Results
Summary information of sequenced O. Sativa genomes
The 504 Os varieties included 179 landraces and 325 modern varieties from 45 countries, including 94 newly sequenced modern varieties and 410 accessions from the 3KRG [4], [12] that consisted of 248 accessions from a global mini core collection [13] and 162 parental lines from the International Molecular Breeding Program [14], [15] (Table S1, Fig. S1). The genome sequence data of 504 accessions contained a total of 3.14 Tb clean bases, 2.93 Tb of which were mapped to the Nipponbare reference genome (here after called Nip_ref) with an average mapping depth of 15.5 X and an average mapping coverage of 91.2 %. When mapped to the 9311 reference genome (here after called 9311_ref), the 504 genomes had an average mapping depth 14.3 X and an average mapping coverage of 87.2 % (Table S3, Fig. S2). Using GATK [16], we discovered ∼ 12 M SNPs and ∼ 2 M InDels in the 504 sequenced genomes when calling on Nip_ref, which were denoted as Nip-SNPs and Nip-InDels. We discovered additional 1,563,657 SNPs and 299,844 InDels from the reads unmappable to Nip_ref calling on 9311_ref (denoted as Nip9311-SNPs and Nip9311-InDels, respectively). Approximately the same numbers of SNPs and Indels were identified when calling on 9311_ref (denoted as 9311-SNPs and 9311-InDels). Annotation of the SNPs identified 84,224 and 21,449 large-effect variants according to Nip_ref and 9311_ref, respectively (Table S4).
The public SNP set (7,970,357) of the 446 O. rufipogon accessions [10] were converted to the coordinate of MSU RGAP 7. SNPs of additional 10 wild accessions (including five O. rufipogon ones and five O. nivara ones) were called using reads of their re-sequencing data [9] and the same method as the 504 Os accessions.
Differentiation of O. Rufipogon and O. Sativa shaped by domestication
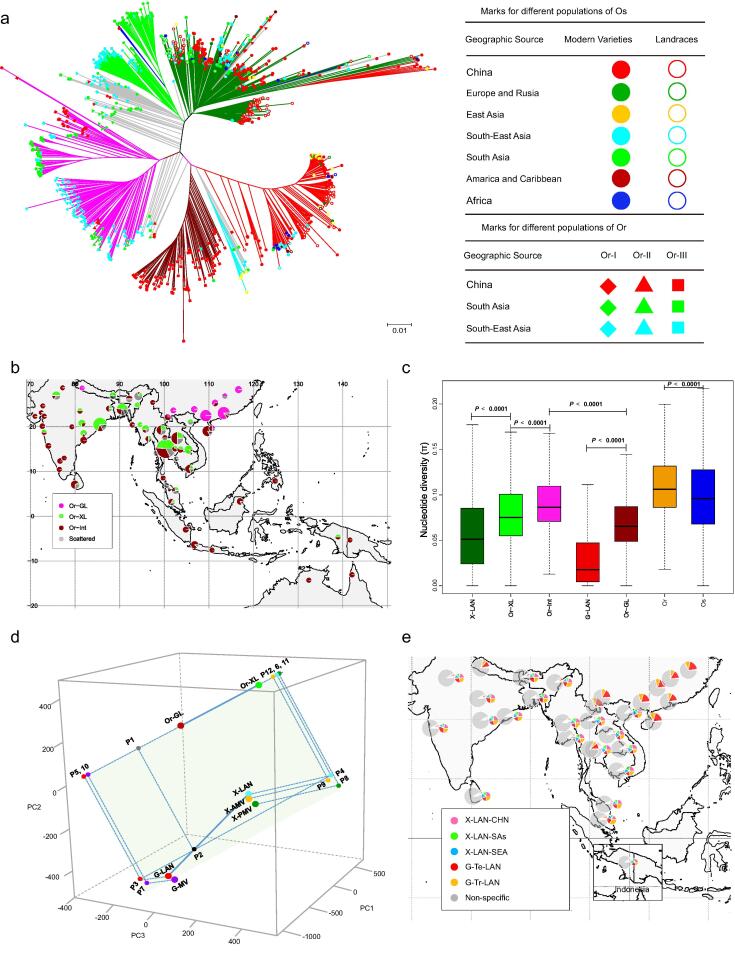
The phylogenetic tree classified the 504 Os and 456 Or accessions (Table S5) into several well-differentiated populations with the two Os subspecies, geng(j) (japonica) and xian(i) (indica) representing the direction of maximum diversity and the Or accessions locating between them (Fig. S3). The 456 Or accessions were classified into three distinct groups (Fig. 1a and Fig. S3). One group shows the closest relationship with xian, and thus is called xian-like Or (Or-XL), which consists of 116 (77 %) of the 150O. rufipogon I (Or-I) accessions from South Asia (SAs) and Southeast Asia (SEA). A second group shows the closest relationship to geng, and thus called geng-like Or (Or-GL), which comprises most Or-IIIa accessions (77 of 99) from China plus two accessions from SAs and SEA. The third group locates between Or-XL and Or-GL, and comprises most Or-IIIb and Or-II accessions plus some Or-IIIa accessions, and shows a wide geographical distribution, including all rice growing countries of Asia, and thus called Or-Int. Geographically, the proportion of Or-Int accessions decreased gradually from low to high latitudes, Or-XL accessions were distributed primarily in SAs and SEA, while Or-GL accessions were found primarily in China and a few in Nepal (Fig. 1b). The principal component (PC) analysis using SNPs was performed to further characterize the genetic relationships among Or and Os subpopulations. Or-GL, Or-XL, Or-Int, geng(j) landraces (G-LAN), modern geng(j) varieties (G-MV), xian(i) landraces (X-LAN), primary modern xian(i) varieties (X-PMV) and advanced modern xian(i) varieties (X-AMV) could be well distinguished in the PC plots constructed with top three PCs. Furthermore, PC1 could divided wild rice into two subgroups (Or-GL/japonica and Or-XL/indica) (Fig. S4). These results suggested that the xian-geng(j) subspecific differentiation predated far the Os domestication and the natural selection along with the changed ecological conditions during Or spreading from low to high latitude may have played important roles on the differentiation of Or before domestication.
Fig. 1.
Phylogeny, differentiation and diversity of/among O. sativa (Os) and O. rufipogon (Or) populations. (a) The phylogenetic tree of 960 rice accessions, including 504O. sativa accessions and 456O. rufipogon/O. nivara accessions, in which the colored lines indicate different types of Os accessions with red = geng(j), green = xian(i), blue = Aus, cyan = Aro, light green = Or-XL, dark red = Or-GL, pink = Or-Int, and grey = admixtures; Or-I, Or-II and Or-III in the panel representing the three major Or populations reported by Huang et al. [10]; (b) the geographic distribution of the three Or populations; (c) the genetic diversity estimates, θπ, of the two Os subspecific landraces (X-LAN and G-LAN) and three Or populations (Or-XL, Or-GL and Or-Int) inferred by the phylogenetic tree; (d) a 3-dimensional presentation of the differentiation among different Or and Os populations based on the principal component analysis of SNP variation, in which the ×, y and z coordinates were indicated by PC1, PC2 and PC3, the large points represent centers of the Or and Os populations; the distance between points along line Or-GL:Or-XL (i.e. the line through points Or-GL and Or-XL, similar for others) accounts for the natural selection consequences (NSC) of two corresponding or projected populations; the dot line P1:P2 is the common vertical line between line Or-GL:Or-XL and line G-LAN:X-LAN; P3, P4, P7, P8 and P9 are the projection points of populations G-LAN, X-LAN, G-MV, X-PMV and X-AMV to the plane consisting of lines Or-GL:Or-XL and P1:P2, thus the distance between each population and its projection point accounts for the subspecies-specific artificial selection consequence (ssASC); P5, P6, P10, P11 and P12 are the projection points of P3, P4, P7, P8 and P9 to line Or-GL:Or-XL, thus the distance between each point and its projection point accounts for the subspecies-common artificial selection consequence (scASC); (e) the genomic proportions of population-specific genomic blocks of various Os populations in different Or populations from the same geographic regions of 2°×2° of latitude by longitude, where the small pies are distributions for population-specific genomic blocks. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To understand how domestication affected population structures of Or and Os subpopulations, we calculated the nucleotide diversity (θπ) of different Or and Os populations, their differentiation statistics (PCA based 3D differentiation model and Fst) and genomic introgression (Fig. 1). The diversity (θπ) of the whole Os population reduced only by 10 % when compared to Or, in agreement with a previous report [10]. Surprisingly, geng(j) landraces (G-LAN) showed a sharp reduced genetic diversity by 53 % when compared to Or-GL, and xian(i) landraces (X-LAN) showed a significantly reduced diversity by 22 % when compared to Or-XL (Fig. 1c, Table S6). Based on estimated Fst statistics (Fig. S4a, Table S7), the differentiation between Or-XL and G-LAN was the strongest, followed by that between X-LAN and G-LAN, and between X-LAN and Or-GL, while the differentiations between Or-GL and G-LAN, and between Or-XL and X-LAN were very weak. This result revealed that domestication resulted in more pronounced subspecific differentiation. When the geographical differentiations between different Os xian(i) and Or-XL populations were examined, the differentiation between Chinese xian(i) landraces (X-LAN-CHN) and Or-CHN was the strongest, and the differentiation between xian(i) landraces of SEA and SAs (X-LAN-SEA and X-LAN-SAs) and Or-XL of Southeast Asia (Or-XL-SEA) was weaker than that between those from Or-XL of South Asia (Or-XL-SAs). In the phylogenetic trees of different Os and Or populations constructed based on the proportions of the total shared ‘core’ genome segments (Fig. S5), each of which was characterized by a set of nearly-fixed linked SNPs within 100-kb windows, the temperate geng(j) landraces and tropical geng(j) subpopulations (G-Te and G-Tr) shared more “core” genome segments and further grouped together with Or-GL. All xian(i) landraces from CHN, SEA and SAs clustered together, and Or-XL from SEA and SAs clustered together, while geng(j) landraces and Or-GL were separated from xian(i) landraces and Or-XL by Or-Int. Fig. 1e shows the proportions of subpopulation-specific core genome segments of different Os populations in Or-XL and Or-GL populations of different geographic origins. The geng-population specific core genomic blocks were detected almost exclusively in those Or-GL populations from South China with two exceptions, while those xian-population specific core genomic blocks were present widely in different Or populations from SAs and SEA.
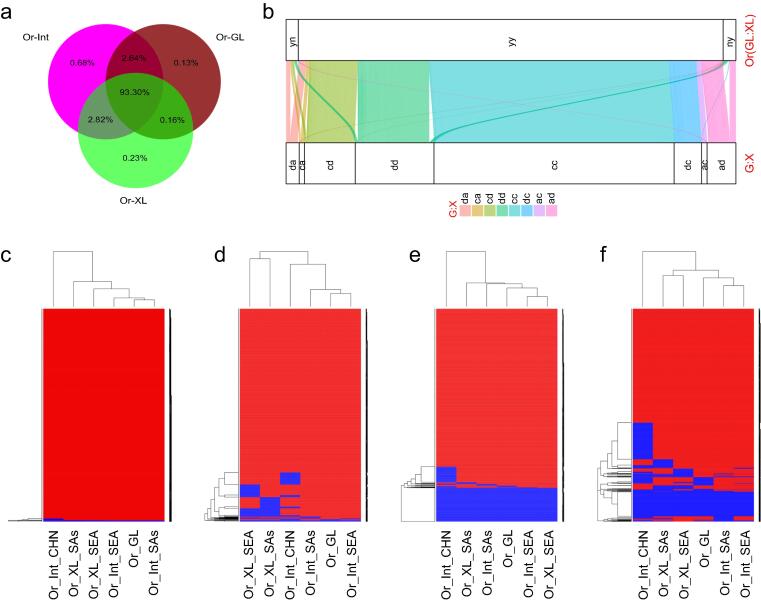
To look into the relationships in total genomic constitution between different Os and Or populations, we performed a comprehensive pan genome analysis using the pan genome data of all Os genes of the 3KRG and “map-to-pan” method [17]. Virtually all (99.4 %) of the 45,982 Os genes were detectable in Or-Int, and this portion was less in Or-XL (96.5 %) and Or-GL (96.2 %) (Fig. 2a). This result suggested that the Or-Int populations were more likely the common ancestral gene pool of Os, Or-GL and Or-XL. When looked at the distributions of the Os genes in Or-GL and Or-XL (Fig. 2b), we found that 80.4 % Or-GL specific genes were transmitted to Os (72.0 % to geng(j) and 34.8 % to xian), but so for only 69.7 % Or-XL specific genes (63.1 % to xian(i) and 40.4 % to geng). To infer the evolutionary relationship between Or subpopulations (Or-GL and Or-XL) and Os subspecies (geng(j) and xian), we examined the distributions of the core (with frequency ≥ 0.8) and distributed genes (with frequency less than 0.8) of Os populations (or subpopulations) in different Or subpopulations (Fig. 2c-f, Tables S8-S9). As expected, almost all 24,180 core genes of the two Os subspecies were shared by Or-GL and Or-XL (Fig. 2c), while almost all of the 5,114 genes core in geng(j) but distributed in xian(i) were present in very high frequencies in Or-GL, Or-Int-SEA and Or-Int-SAs accessions, but in much lower frequencies in Or-Int-CHN and Or-XL populations (Fig. 2d). In contrast, clustering using the 2,770 genes core in xian(i) but distributed in geng(j) could not separate Or-GL and Or-XL subpopulations except for Or-Int-CHN (Fig. 2e), while clustering using the 7,998 genes distributed in both subspecies classified all Or populations into two major groups, Or-Int-CHN and the other containing two subpopulations, Or-XL and Or-GL/Or-Int-SEA and Or-Int-SAs (Fig. 2f). Taking together, our pan genome analyses strongly suggested that geng(j) was most likely directly domesticated from Or-GL accessions, while xian(i) did not appear to have originated from any single ancestor Or population.
Fig. 2.
Pan-genome analysis between Oryza sativa (Os) and O. rufipogon (Or). (a) The proportions of Os genes detectable in different Or populations; (b) the distribution of genes of two subspecies, geng(j) (G) and xian(i) (X), in Or-GL and Or-XL, where a, c or d in the G:X column represent absence, core or distributed genes in the corresponding subspecies, y or n in the Or(GL:XL) column represent presence or absence in the corresponding Or populations; (c)-(f) the bi-clustering analysis for genes both core in two subspecies (24,180), those core in G and distributed in X (5,114), those distributed in G and core in X (2,770), and those both distributed in two subspecies (7,998) according to their presence (red) or absence (blue) in different Or populations. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The population structure of O. Sativa
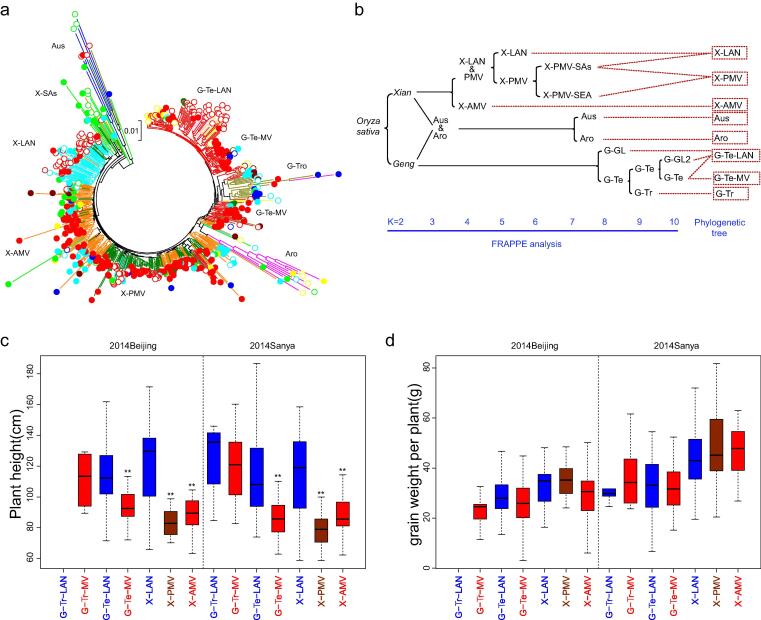
To better understand the Os population structure, we reconstructed the phylogenetic tree and performed the model based population structure inference for k = 2 to 13 using 567 k high quality SNPs, which resolved the 504 Os accessions into 8 well-differentiated subpopulations resulting apparently from domestication and breeding (Fig. 3a, Figs. S6-8). In the model-based population structure inference, when k = 2 to 5, the percentage of accessions with the maximum Q-values > 0.5 was higher than 95 %, and dropped to less than 95 % for k = 6 to 13 with a peak at k = 10 (Fig. S6). We thus systematically compared the population structures of the 504 Os accessions from k = 2 to 10 with their documented taxa and geographical origins (Table S1). When k = 2, all Os accessions were classified into two major subspecies, geng(j) and xian(i), with more admixture in xian(i) than geng(j) (Fig. 3b, Figs. S7a&8a). When k = 3, Aus and aromatic (Aus & Aro) accessions were differentiated from geng(j) and xian, forming a single cluster, and were separated from each other at k = 7. We found that ∼ 65 % of the Aro genome was from Aus and the remaining ∼ 35 % from geng(j) (Fig. S8b), suggesting Aro was originated from crosses between Aus and geng(j). Similar to the reported results by Civáň and Brown [18], introgression from xian(i) to Aus was detected (Figs. S7b-c&8b-c). When k = 4, the advanced modern xian(i) varieties (X-AMV) released after 1980 s were separated from X-LAN and the primary modern varieties (X-PMV) released since the Green Revolution (GR) from late 1950 s to 1980 s (Figs. S7&9, Table S1). Significant amounts of introgression from X-LAN and X-PMV into X-AMV were detected (Fig. S8, k = 4), implying that X-AMV were descended from X-LAN and X-PMV during recent breeding. At k = 5 and 6, X-LAN was separated from X-PMV, and X-LAN of China were separated from xian(i) accessions (landraces and modern varieties) of SEA and SAs. Those xian(i) accessions of SEA and SAs presented higher genetic mixture among LAN, PMV and AMV from k = 4 to 10. The temperate geng (G-Te) and tropical ones (G-Tr) were separated at k = 9 (Fig. 3b, Fig. S7h). When k = 10, the Os accessions were classified into ten subpopulations. Clearly, the subpopulation differentiation revealed at k = 4–10 resulted primarily from recent breeding efforts. Hereafter, we would focus on the nine Os subpopulations (X-LAN, X-PMV, X-AMV, G-LAN, G-Tr, G-Te-LAN, G-Te-MV, Aro, Aus) that have been consistently identified according to the results of FRAPPE, phylogenetic analyses and documented information (Fig. 3b, Fig. S7i&8i, Table S1).
Fig. 3.
The population structure and important agronomic phenotypes of O. sativa (Os). (a) The phylogenetic tree of 504 Os accessions, in which the marks are the same as Fig. 1a, and the colored lines represent different populations, which were inferred by population structure analysis of FRAPPE with k = 10 and labeled by symbols around the tree; (b) the step-by-step differentiation of different Os populations with increased k values; (c) and (d) the mean values and variation of plant height and grain weight per plant of different populations of xian(i) and geng(j) landraces and modern varieties, in which ** represents significant differences in mean trait values between the modern varieties and landraces.
Past breeding efforts have resulted in major changes in mean value and variation of the measured agronomic traits in the seven Os subpopulations (Fig. 3c&d, Fig. S10, Table S10). In xian, X-PMV showed an average ∼ 34 cm reduced height when compared to X-LAN, resulting clearly from GR. When compared to X-PMV, X-AMV showed significantly increased height by ∼ 9 cm, 13-day delayed heading and 4 more panicles/plant resulting from recent breeding efforts. A similar height reduction by ∼ 18 cm was observed in modern geng(j) varieties (G-MV) when compared to geng(j) landraces (G-LAN). Surprisingly, we did not detect consistent differences between modern varieties and landraces in both geng(j) and xian(i) for three main yield components. Major shift from breeding was the significant reduced trait variation in modern varieties for some measured traits, particularly for height and heading date, when compared with landraces of both geng(j) and xian.
Population differentiation attributed to natural and artificial selection
We then estimated the population differentiation to acclimate natural or artificial selection consequences (NSC and ASC) during modern breeding using the same 3D differentiation model (Fig. 1d, Fig. S4b, Table S11). As indicated above, the genome wide differentiation between the two Os subspecies could be primarily attributed to two parts of selection consequences (SC): natural selection on Or before domestication and natural plus artificial selection on Os during domestication. Using a 3D differentiation model constructed with the first three principle components of the SNP variation in all 960 Or and Os accessions (Fig. 1d), we were able to divide the total differentiation between or within the subspecies into different components: the static natural selection consequence (s-NSC), static subspecies-common artificial selection consequence (s-scASC), static subspecies-specific artificial selection consequence (s-ssASC) between populations within each subspecies, as well as the dynamic natural selection consequence (d-NSC), dynamic subspecies-common artificial selection consequence (d-scASC) and dynamic subspecies-specific artificial selection consequence (d-ssASC) within each population. The model shows that approximately 42 % and 58 % of the total xian-geng(j) subspecific differentiation were attributable to NSC before and during domestication, respectively (Fig. 1d, Fig. S4b, and Table S11). Furthermore, NSC of xian(i) (X-LAN from Or-XL) was weak, accounting only for 9.8 % of the total NSC between the two subspecies, while NSC of geng(j) (G-LAN from Or-GL) was much stronger, 4.9 times of that of xian(i) (indica). The s-scASC in geng(j) and xian(i) was aparently lower than s-ssASC of xian(i) (88 %) but much higher than s-ssASC of geng(j) (233 %) (Fig. 1d, Fig. S4b, and Table S11). The dynamic selection consequence within G-LAN and X-LAN in all three directions were apparently lower than the static selection consequence of G-LAN from Or-GL and X-LAN from Or-XL, respectively, implying the distinct differentiation of G-LAN from Or-GL and X-LAN from Or-XL (Fig. 1d, Fig. S4b, and Table S11). We noted that NSC of geng(j) was much higher than d-NSC within other populations (193 %), which could be attributed to the strong differentiation of the tropical, subtropical and temperate G-LAN populations within geng(j) (japonica).
Compared to domestication, s-SC from modern breeding was weaker in all three directions (s-NSC, s-scASC and s-ssASC) for both subspecies. Except for X-PMV from X-LAN and X-AMV from X-PMV, both s-NSC and s-ssASC were relatively small in both subspecies during modern breeding, when compared with s-scASC, indicating artificial selection in both xian(i) and geng(j) had resulted a greater level of common consequence. However, a higher d-NSC was observed in the G-MV population than in other populations (256 %), indicating modern geng(j) varieties had gone through a much stronger natural selection as compared to other populations.
Genome-wide selected blocks and selected genes resulted from domestication
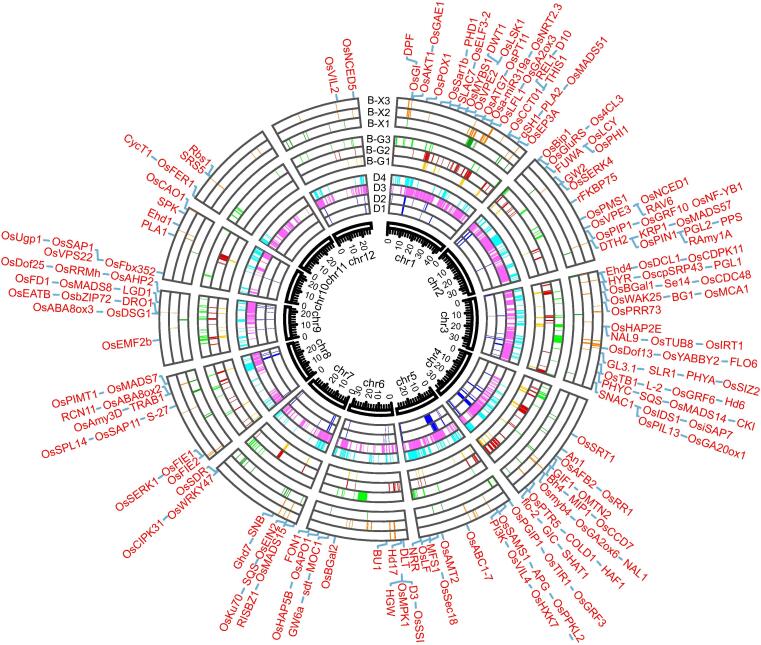
To gain insights into how domestication shaped the observed Os population structure described above, we performed a genome-wide search to detect SBs across the rice genome by comparing genome-wide values of Tajima's D of the Os landrace population and diversity reduction (θπ ratio) of the Os landrace populations relative to corresponding Or populations. We focused on the degree and difference between domestication and breeding, and the difference between the two subspecies. To achieve this, we used the same threshold of (θπ(5,1,8), or θπ(3,1,8) & D(-2,1,8) in detecting the domestication and breeding SBs of two subspecies rather than using the threshold of a top 5 % SBs in specific population, which would differ between the two subspecies [10]. According to our simulation by reshuffling alleles at each SNP site within either Os or Or, no blocks could be identified as SBs with this threshold, indicating it was a stringent one. Using this threshold, we identified 624 and 318 SBs attributable to domestication in G-LAN and X-LAN accessions using Nip-SNPs (Fig. 4, Figs. S11-13 and Tables S12-13). To investigate the domestication and breeding blocks at the subspecies level, we unified the above blocks or merged some of them if they overlap each other. The domestication selected blocks (d-SBs) in geng(j) (japonica) and xian(i) (indica) covered a total of 126.4 (34.0 %) and 44.5 Mb (11.6 %) of the two respective reference genomes, with only 179 d-SBs of 24.8 Mb shared between geng(j) and xian(i). The average size of the d-SBs detected in G-LAN was 203 kb, much larger than the average 140 kb of d-SBs detected in X-LAN (Fig. S12, Table S13). Further, to distinguish selected genes (SGs) from those ‘neutral’ genes due to genetic hitchhiking within the identified SBs, we calculated the diversity reduction and population based Ka/Ks ratios for all genes within each SB. We found a total of 14,202 and 5,163 domestication selected genes (d-SGs) in geng(j) and xian(i), respectively, with 3,115 d-SGs shared between the two subspecies (Table S14). In particular, 87.6 %, 67.6 % and 57.3 % of the SGs in geng, xian(i) and xian-geng(j) showed strong signals of diversity reduction (with θπ ratio > 5 or θπ > 3 and tajima’D < -2), while 32.4 %, 58.6 %, 67.7 % of them were apparently due to coding region selection (as measured by population based Ka/Ks), including either maintaining new advantageous mutants (thereafter called novel function selection, NFS) or eliminating new deleterious mutants (or enhancing standing function, thereafter called standing function selection, SFS). In the cases of coding region selection, 3,114 (19.2 %) of the d-SGs had gone through NFS, while 2,404 (14.8 %) cases of the d-SGs could be attributed to SFS. Surprisingly, only a very small proportion of the geng(j) (3.4 %) and xian(i) (1.9 %) d-SBs were attributable to genetic bottlenecks that either showed uniform low polymorphism or no genes within a SB showing coding region selection (Table S15). This result indicated that genetic bottleneck played little role during domestication of both geng(j) and xian(i).
Fig. 4.
Genome-wide selected blocks related to domestication and breeding, and cloned genes related to yield and domestication. The colored blocks in each layer from inner to outer represent respectively the domestication selected blocks detected from Oryza sativa (Os) relative to Oryza rufipogon (Or) (D1), Os landraces (LAN) relative to Or (D2), geng(j) LAN relative to geng-like Or (D3), xian(i) LAN relative to xian-like Or (D4), and the breeding selected blocks detected from geng(j) modern varieties (MV) relative to geng(j) LAN (B-G1), temperate geng(j) MV relative to temperate geng(j) LAN (B-G2), tropical geng(j) MV relative to tropical geng(j) LAN (B-G3), xian(i) primary modern varieties (PMV) relative to xian(i) LAN (B-X1), xian(i) advanced modern varieties (AMV) relative to xian(i) LAN (B-X2) and xian(i) AMV relative to PMV (B-X3); the outmost symbols are the cloned genes related to yield and domestication in the selected blocks.
Genome-wide selected blocks and selected genes attributable to breeding
We detected the breeding selected blocks (B-SBs) and breeding selected genes (B-SGs) using the same method by comparing the Os modern varieties to the Os landraces. Because of the well-known xian-geng(j) differentiation, we detected significantly more B-SBs in geng(j) populations when using Nip-SNPs, and so did for those in xian(i) populations when using 9311-SNPs (Fig. S11c&f). To overcome this bias, we compared all B-SBs detected in the xian(i) populations using 9311-SNPs with those in the geng(j) populations using Nip-SNPs, and compared all SGs within the identified B-SBs detected by using both Nip-SNPs and 9311-SNPs. Even using their respective reference genomes, we detected more B-SBs in geng(j) (3 1 4) than in xian(i) (2 4 6), covering 42.0 (11.3 %) and 22.7 (5.9 %) Mb the Nip_ref and 9311_ref, respectively. The average size of the detected B-SBs was 133.8 kb and 92.5 kb in geng(j) and xian, respectively, much smaller than their corresponding d-SBs (Fig. S11). Within the detected B-SBs, there were 5,265 and 2,589 B-SGs in geng(j) and xian, respectively, with only 360 B-SGs shared between the two subspecies (Fig. 4, Fig. S13, Tables S12-13). This was consistent with the expectation from largely independent breeding activities worldwide for geng(j) and xian(i). Interestingly, 736 (5.2 %) and 236 (4.6 %) of the detected d-SGs in geng(j) and xian(i) appeared to have resulted from “secondary domestication”, i.e. those genes had apparently gone through selection during domestication and further selection during modern breeding. Further, the majority of the detected d-SGs in xian(i) (10,544 or 74.2 %) and geng(j) (2,732 d-SGs or 52.9 %) were transmitted from landraces into modern varieties (Tables S14-S17). Of the geng-, xian- and geng-xian(i) shared B-SGs, 24.7 %, 32.0 % and 7.5 % were attributable to coding region selection, respectively. Different from the d-SGs, more of the detected B-SGs were attributable to SFS (1,672, 22.3 %) than to NFS (431, 5.8 %). We observed that 11.5 % and 12.3 % of the B-SBs in geng(j) and xian(i) showed uniform low polymorphism or 30.3 % and 21.1 % contained no selected genes (Table S15). This indicated that recent modern breeding has suffered a more severe genetic bottleneck than domestication.
Functionalities of the detected selected genes resulted from domestication and breeding
To gain insights into the functionalities of the detected selected genes resulting from domestication and breeding, we examined 1,957 cloned rice genes (885 of which are known to affect 22 traits of agronomic importance, https://www.ricedata.cn/gene) within the detected SBs. We found 509 (57.5 %) of the detected SGs were associated with important agronomic traits, which were thus called AI_SGs. This portion was higher than all cloned genes (50.2 %) and all annotated genes (43.9 %), suggesting the importance of these AI_SGs and cloned genes in domestication and modern breeding. Of the 509 AI_SGs, 72.7 % and 35.8 % were attributable to domestication and breeding, respectively (Fig. 4&5a-c, Figs. S13-14, Tables S12, S14, S16-S17). These cloned genes included 90 (64.4 %) of the detected d-SGs in xian(i), 330 (92.4 %) of the d-SGs in geng(j), and 50 (52.0 %) of the d-SGs shared. The cloned d-SGs under coding region selection included 75 of the d-SGs in geng, 46 of the d-SGs in xian(i) and 30 of the shared d-SGs, while d-SGs showing NFS included 97 d-SGs in geng, 22 d-SGs in xian(i) plus 13 of the d-SGs shared.
When the differentiations between different Or and Os landrace populations were examined using haplotypes at those domestication AI-SGs (Table S18, Fig. S15), the average differentiation index (DI = -log10p(chisq)/-log10p(chisq_asymptotic), significant when > 1) was the weakest (1.14) between G-Te and G-Tr, and the strongest between G-Te and xian(i) (5.58). Surprisingly, G-Tr showed apparently weak differentiation from G-Te, xian, Or-GL, Or-XL-SEA and Or-XL-SAs, suggesting G-Tr represents the most recent type that formed by introgression or fusion among Os subspecies and/or Or populations, or G-Tr is a reserved proto type of the domesticated Os. Furthermore, similarly strong differentiation of G-Te from Or-GL and that of xian(i) from Or-XL were observed at those d-SGs for seed shattering (5.14 and 5.59), grain size (3.88 and 3.34), grain quality (4.09 and 4.31), growth duration (4.45 and 4.24) and soil nutrient use efficiency (4.19 and 4.78). The strong differentiation between geng(j) and xian(i) (all > 5) at these d-SGs for all above traits except for seed shattering implies that parallel and convergent evolution had occurred in geng(j) and xian(i) during domestication. When the gene networks underlying growth duration were examined, we found that more new haplotypes at most downstream genes were strongly selected for promoting flowering under the long-day environments in geng, and the opposite was true in xian(i) where those standing haplotypes promoting flowering under short-day environments were strongly selected (Fig. 5d). Except for those common strong differentiated traits, geng(j) showed stronger differentiation from Or-GL in tillering and panicle architecture (4.39), while xian(i) did in photosynthesis (4.94), biomass (4.87) and plant height (4.61). These results indicated that xian(i) and geng(j) had independent evolutionary histories.
Fig. 5.
The scenario of agronomy-important cloned selected genes (SGs). (a) The proportion of all SGs in annotated genes and the proportion of agronomy-important cloned SGs relative to all cloned genes among different domestication or breeding events; (b) the portion of SGs subjecting to coding region selection relative to all SGs; (c) the portion of SGs subjecting to novel function selection (NFS) relative to SGs subjecting to coding region selection; (d) the regulation network of growth duration genes subjected to domestication and breeding, here, LD: long day condition, SD: short day condition, +: interaction, +p: phosphorylation, red frame: selected during geng(j) domestication, green solid or dot frame: selected during xian(i) domestication or breeding, orange filled: NFS, yellow filled: standing function selection (SFS), blank: allele-frequency-dependent selection, waved line: promoter; and (e) the cumulative percent of haplotypes with significant difference in growth duration, here, labels in the bracket indicate the types of SGs, such as B-G-Tr being the SGs during event of breeding for tropical geng(j) varieties. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To understand the functionalities of the detected B-SGs, we examined 182 AI_SGs in the B-SBs, and found that 163 of them were detected only in the B-SBs and the remaining 19 had gone through secondary domestication (Tables S14, S16-S17). Of these, 105 and 64 genes were specifically detected in the geng(j) and xian(i) B-SBs, respectively. Interestingly, the majority of the B-SGs were subspecies-specific and involved in disease resistance (66), plant height (28), heat tolerance (18), soil nutrient use efficiency (12), erect leaves (6), flood tolerance (4) and biomass (4) with only 13B-SGs shared between geng(j) and xian(i). This resulted from the largely independent breeding efforts in geng(j) and xian(i) in the past. In xian, 74 cloned genes were strongly selected during GR from X-LAN to X-PMV, at least 39 of them were reported to be associated with important agronomic traits. The post-GR breeding efforts revealed by comparisons between X-PMV and X-AMV were reflected primarily by strong selection on 44B-SGs. The majority of these genes are involved in stress tolerances. A third set of genes included 19 genes at which frequencies of the favorable alleles increased steadily from X-LAN to X-PMV then to X-AMV. The fourth set included 9 genes which had apparently gone through diversifying selection for GR as they showed low-diversity in X-LAN but high-diversity in X-PMV. Surprisingly, 33 of the 39 genes associated with GR were not SGs for X-AMV. Chi-tests of haplotype distribution among populations indicated that breeding in xian(i) did not result in strong differentiation for most genes associated with both GR and post-GR except for those B-SGs related to growth duration (DI = 3.32), pest resistance (DI = 2.84) and heat tolerance (DI = 1.92) during post-GR of xian(i) breeding efforts (Tables S14, S18). Taking together, genes selected strongly during GR in xian(i) are involved in soil nutrient use and photosynthesis efficiencies, population biomass accumulation etc., while the post-GR breeding acted apparently on genes for biotic and abiotic stress resistances/tolerances (Fig. 5e, Fig. S16, Tables S14, S16-S18).
In contrast, 79B-SGs were associated with breeding of the temperate geng(j) subpopulation (G-Te-LAN vs G-Te-MV), including 1 for seed shattering (SHAT1), 9 for grain size, 14 for cold tolerance, 10 for high temperature tolerance, 2 for flood resistance, 4 for erect leaves and 13 for plant height. SNK multiple comparisons indicated that G-Te-MV enriched more haplotypes with medium plant height, strong cold tolerance, low KGW and medium growth duration (Fig. 5e, Fig. S16), even though the differentiation between G-Te-LAN and G-Te-MV was very weak at these genes (Table S18). There were 62 AI-SGs in the detected B-SBs of tropical geng(j) (G-Tr-LAN vs G-Tr-MV), including 22 for disease resistance, 4 for pest resistance, 9 for high temperature tolerance, 3 for flood tolerance, 2 for erect leaves and 15 for plant height. G-Tr-MV had higher frequencies for haplotypes associated with medium plant, moderate cold tolerance, moderate disease resistance, more grain number and medium growth duration (Fig. 5e, Fig. S16), but the differentiation between G-Tr-LAN and G-Tr-MV at these genes was very weak (DI = 0.30–1.28) (Table S18). There were 23 AI-SGs shared between G-Te and G-Tr, which were mainly associated with seed germination, seed ripening, grain size, pest resistance, high temperature tolerance, flood tolerance and plant height. These results clearly indicated that past breeding efforts of temperate and tropical geng (G-Te and G-Tr) were largely independent from each other.
KEGG and gene family enrichment analyses indicated that genes in the SBs showed several interesting aspects regarding the differentiation between two subspecies during domestication or breeding, and between domestication and breeding in each subspecies (Fig. 6, Tables S19-S20). First, geng(j) and xian(i) shared a significant portion (52.6–63.8 %) or (21.2–34.2 %) of common KEGG terms that were selected during domestication or breeding, but the SGs in the same KEGG terms were very different between geng(j) and xian(i) with only 19.9–24.2 % or 4.9–7.9 % common SGs (Fig. 6a, Tables S19, S21,Fig. S17). There were 110 enriched KEGG terms in the SBs, including peroxisome, apoptosis, nitrogen metabolism, fatty acid metabolism, cell cycle, photosynthesis and so on. Taking peroxisome as an example, there were 78 genes of the peroxisome pathway in the SBs, eight of which were cloned, including OsACX1, OsCATA, GLO4, GLO1, GLO3, OsPEX5, OsCATC and OsACX3. These cloned genes were mainly involved in the stress tolerance/resistance. Thus, most, if not all, genes of this pathway were expected to be stress-related and should be the targets for future validation and application. Secondly, the detected SGs in geng(j) and xian(i) shared many common gene families (40 % on average), but different members of the same gene families were selected in geng(j) and xian(i) during domestication or breeding with only 6.6 % in common (Fig. 6b, Tables S20 & S22). This result indicated that it was the compensatory gene networks underlying important adaptive traits, instead of the same genes/alleles, that were the targets of selection during domestication and breeding of geng(j) and xian(i). In addition, we found that the detected SBs had more pronounced copy number variation than the unselected regions (Fig. S18, Table S23), suggesting gene duplication followed by functional diversification of duplicated genes may play an important role during the differentiation of Os populations and their adaptations to different environments.
Fig. 6.
Enrichment of pathways and gene families in different selected blocks. (a) The proportion of common terms with common genes or just common terms of pathway within two subspecies, xian(i) and geng(j) or two events (domestication and breeding), in which “To the minimum” or “To the maximum” means the proportion to the minimum or maximum number of enriched terms in the two compared types of selected blocks; and (b) the proportion of common gene families with common member genes or just common gene families within two subspecies xian(i) and geng(j) or two events (domestication and breeding).
Discussion
The differentiation of O. Rufipogon and O. Sativa and implication on the domestication of O. Sativa
How O. sativa was domesticated has been an issue of long debate. Results from recent molecular and genomic analyses suggested two models. The multiple-origin (domestication) model proposes that geng(j) and xian(i) were independently domesticated from divergent populations of their wild progenitors, O. rufipogon and O. nivara [19], in which xian(i) was first domesticated in the south range of Himalayas and eventually spread to other parts of the world, while geng(j) was first domesticated in the lower Yangtze River or South China and then spread to other places, or that geng(j) was first domesticated in China and then xian(i) originated from the introgression or crosses between geng(j) and O. rufipogon/O. nivara [10], [20]. In contrast, the single origin model proposes Os was first domesticated in the lower Yangtze Valley or Pearl River Valley of China about 8,200–10,000 years ago in which geng(j) was derived from xian(i) [21] or xian(i) derived from geng(j) [10], [22], [23], [24], and then spread towards north or south, and became diversified [25].
Here, we showed the organization of tremendous genome diversity and population differentiation of Os and its presumed ancestor Or populations. We also showed the pan genome relationships between the Os and Or populations and the genome-wide profile of selection signatures in the Os genome that shaped the population structure of Os during domestication. Several important results were obtained. Firstly, the sampled Or accessions were clearly differentiated into three major populations, Or-XL, Or-Int and Or-GL. Our classification of the Or and Os accessions was very similar to the phylogenetic tree from Huang et al. [10] with very few misclassifications even completely different Os accessions were used in the two studies. In fact, Or-XL consisted of 77 % of O. rufipogon I (Or-I) accessions from SAs and SEA; Or-GL comprised most Or-IIIa accessions from China plus two accessions from SAs and SEA, and Or-Int comprised most Or-IIIb and Or-II accessions and some Or-IIIa accessions. The major difference was apparently the misclassification of some Or-Int accessions, by Huang et al. [10], into Or-IIIb. With the largest pan genome, Or-Int accessions have the widest geographic distribution and the genetic potential to differentiate into either Or-XL and xian(i) or Or-GL and geng(j). Or-XL accessions are also widely distributed in SAs, SEA and south China (Fig. 1b), while Or-GL were distributed primarily in China and some mountainous areas of SAs and SEA. Second, domestication seemed to have resulted in little diversity reduction within Os as compared to Or, but caused significant reduction in within-subspecies (xian(i) or geng(j)) diversity and more pronounced subspecific and population differentiation, indicating minimum effects of genetic bottleneck during Os domestication. Third, domestication left tremendous selective signatures on large numbers of genes across a significant portion of the genomes of Os landraces, consistent with previous reports [7], [8], [9], [10], [11]. These results led us to an opinion, similar to that summarized by Larson et al. [26], during a long and complex process, under ancient gathers’ or farmers’ conscious or unconscious selection, the changes from wild rice to domesticated rice were systematic and more dramatic in morphological traits or characteristics related to improved productivity and their corresponding genes. However, the detected d-SBs and d-SGs in xian(i) and geng(j) were largely different with a very small portion of d-SGs shared resulting primarily from convergent selection during domestication. Fourth, as indicated by the 3D differentiation, approximately 42 % of the Os xian(i)-geng(j) subspecific differentiation could be attributed to the > half million-year natural selection causing the Or-GL and Or-XL differentiation before domestication [27], [28], while the remaining ∼ 58 % sub-specific differences could be attributed primarily to the combined effects of natural and artificial selection during the long process of domestication. During domestication and breeding, the predominant types of selection were pyramiding the old ‘advantageous’ alleles at large numbers of loci from ancestor Or accessions and removing the new deleterious variations (SFS) at many house-keeping genes, while new ‘advantageous’ alleles at relatively fewer loci for environmental-adaptations were apparently responsible for the spreading of rice to wider geographic areas. This was not surprising since the spreading of geng(j) from South and Yangtze River areas of China to other parts of the world by human was much faster than the natural spreading of Or populations, consistent with our observation that natural selection during geng(j) domestication was much stronger than that during xian(i) domestication. Thus, selection, instead of genetic drift, played a much more important role in the subspecific and population differentiation of Os. Fifth, although geng(j) and xian(i) shared some common KEGG terms selected during domestication, the SGs in their shared KEGG terms were very different between geng(j) and xian(i). Also, the detected SGs in geng(j) and xian(i) shared many common gene families, but different members of the same gene families were selected in geng(j) and xian(i) during domestication. Thus, evolution resulted from convergent selection actually acted on different genes/alleles in differentiated populations.
Thus, our results do not seem to support any single-origin models of Os because such models would have resulted in dramatically reduced genetic diversity within Os from the severe genetic bottleneck, which was not observed. Nor could the huge numbers of subspecies-specific genes/gene families, large SVs and SNPs [4] could arise in less than 4,000 y through any genetic mechanisms if xian(i) could have been derived from geng, and vice versa. The single origin (domestication) of either xian(i) or geng(j) in the current multiple-origin models [21], [22] does not seem to reconcile completely with our data based on the same argument and with available archaeological evidence [4], [29]. The tremendous genetic diversity and strong geographic differentiation within xian(i), plus the archaeological evidence of xian(i) cultivation for > 9,000 years in India (lower Ganges River) and > 10,000 years in China (lower Yangtze River) [23], [24], [30], [31], would suggest independent domestications of xian(i). The same argument may be held for the multiple and independent domestication events for the temperate and tropical geng(j) landraces in different geographic origins at different times, as only a few geng(j) landraces were sampled from SAs in this study. Otherwise, it would be difficult to imagine how the diverse indigenous xian(i) and geng(j) landraces of different geographic origins were evolved from few earlier domesticates of either xian(i) or geng(j) in the primary diversity center of O. sativa along the foothills of Himalayas from SA/SEA to Southwest China.
Taking together, we propose a hypothesis of multiple independent domestications of Os that seems to reconcile with all evidence. In this hypothesis, the ancestor of Os is inferred to be a perennial outcrossing wild rice type of A-genome that was widely distributed in both Asia and Africa long before domestication, from which perennial/annual types of O. rufipogon/O. nivara (Or-Int) were evolved and became spread across Asia and Africa. Then, as a result of their adaptations to different environments under natural selection, ancient types of annual Or-XL and Or-GL wild types became differentiated from largely outcrossing Or-Int types, with the former distributed widely in the tropic and sub-tropic areas of Asia and the latter in the sub-tropic and tropic mountainous areas of Asia. The environmental diversity along the Himalayas imposed diverse types of selection on the local Or populations, and thus became the centers of Or diversity and later Os diversity after domestication. Then, early domestication and eventual cultivation of Os were largely geographically independent local events [32], [33] and a long diffusion process that could have taken place for at least several thousand years repeatedly at different times and in different sites of SAs/SEA and China. The earliest domestication of xian(i) and temperate geng(j) from differentiated but sympatrically distributed Or-XL and Or-GL populations could have occurred almost simultaneously in the lower Yangtze River of China some 10,000 years ago [23], [24], [30], [31], [34]. The climate conditions in these areas have been suitable for cultivation of both xian(i) (as early and single crop) and geng(j) (as the second or late crop) until today. From there, then, the temperate geng(j) spread north to northeast China, Korea and Japan, and other parts of world, while the xian(i) types became the predominantly cultivated type in South China. Meanwhile, Aman (indica) was domesticated from differentiated Or-XL populations first in the Ganges River Valley of India and probably in other places of SAs and SEA at different but later times. Again, the ancient domestication events and following cultivation of locally adapted indigenous rice types in those regions are presumed to have occurred repeatedly for a long period of time in different sites, as suggested by the presence of many unique genes/gene families and large SVs in these populations that could not be easily explained by the limited introgression between populations [4]. Thus, this process, as summarized by Larson et al. [26], could have contributed significantly to early agriculture of Asia and to the greatest ancient civilizations in China and India, and reserved much of the genetic diversity of the ancestor Or populations in the thousands of Os landraces maintained in Genebanks worldwide. Clearly, this hypothesis should be validated in future with more balanced sampling, i.e. inclusion of more annual Or-XL accessions from China, more Or-GL and geng(j) landrace accessions from SAs/SEA, and with much deeper sequencing of the Or accessions. Also, more direct evidence for the multi-origin model of Os can be obtained by experimental validation of the genetic potential of the outcrossing Or-Int types to differentiate into either xian(i)or geng(j) types, even Aus and Bas types in future.
Impacts of modern breeding on the population structure of O. Sativa and implications
The impacts of modern breeding on the Os population structure revealed in this study have important implications for future rice improvement in order to achieve consistent and accelerated genetic gains. First, modern breeding has resulted in significant reduction in diversity within both geng(j) and xian(i) populations at both the phenotypic and genomic levels. In other words, the GR and post-GR breeding efforts in either xian(i) or geng(j) have been based on a narrow genetic basis. This provides an adequate explanation for the long-standing plateaus in raising yield potential of both inbred and hybrid cultivars of rice since 1990 s [35], [36], [37]. Thus, tremendous efforts are needed to broaden the genetic basis of the current breeding programs worldwide and this should be done by exploiting the rich diversity within Os accessions of different geographic origins and in particular from different subspecies, as proven in our large scale backcross breeding programs [38], [39], [40], [41], [42]. Second, as expected, the GR breeding was acting on very few loci for reduced height and early growth duration, while the post-GR breeding was apparently acting on loci affecting resistances to major biotic stresses and for slightly increased height/biomass. Third, the 509 genes in the SBs were of diverse functionalities, but their strong association with traits of agronomic importance implied that these genes should be the focuses of future functional genomic research and, once validated, target genes for future rice improvement by appropriate molecular breeding strategies. Fourth, pyramiding the old ‘advantageous’ alleles at large numbers of loci from ancestor Or accessions and sweeping away the new deleterious variations (SFS) were the predominant types of selection during both domestication and breeding and a large proportion of those genes under SFS were house-keeping ones. This is expected since advantageous alleles at most loci regulating resistances/tolerance to biotic and abiotic stresses would have been maintained in the Or populations under long-term natural selection before domestication. While, new ‘advantageous’ alleles at relatively few loci for environmental-adaptations such as growth duration and soil nutrient use efficiency were the target of selection during domestication [43], [44], [45], [46]. This suggested that genes with novel alleles under NFS could be more easily identified from landraces or wild rice to develop new varieties with wide-environment adaptability. Fifth, selection in geng(j) and xian(i) during domestication and breeding tended to act on different genes in the same pathways, or on different functional alleles of the same loci, or different members of the same gene families. This strongly suggested that xian(i) and geng(j) may have compensatory regulation and functional genetic networks affecting the same phenotypes for their adaptations to different environments. This can be well demonstrated by the global distribution of different Wx alleles controlling rice grain quality in the modern cultivars grown worldwide [47], in which alleles Wxa and Wxb were mainly distributed in temperate areas with Wxa rich in a mid-latitude area and Wxb more abundant in high-latitude areas, whereas alleles Wxlv and Wxin were mainly distributed in the tropical regions. This implied again that different Wx alleles had been selected in response to different cultural and/or environmental preferences around the world [48]. If so, xian(i) and geng(j) or even different populations within xian(i) and geng(j), should be valuable sources of useful genetic diversity for each other to improve complex traits, as clearly demonstrated by large-scale backcross breeding efforts [38], [41], [42], [49], [50]. This may also explain the reason why we usually detect so many different loci controlling the same traits when genetic mapping and allelic mining for complex traits using GWAS were practiced in different types of Os populations [4], [10], [51], [52], [53], [54], [55], [56], [57], [58], [59].
Finally, the free availability of the materials and their genome information of the diverse germplasm from 3KRG and the 94 modern varieties from this project will greatly facilitate more efficient discovery and exploitation of the rich diversity within Os and its wild progenitors for future rice improvement.
Methods
SNP identification of 504 accessions of cultivated rice (Oryza sativa) and 456 accessions of wild rice
Of the 504 sampled Os accessions, genomic sequence data of 410 accessions were from the 3KRG [4], [17], and additional 94 elite modern varieties were carefully selected to represent the major rice grown areas in China [60] (Table S1). The 410 accessions consisted of two parts: 248 from a global mini core collection [13] and 162 from the International Molecular Breeding Program [14], [15]. Among the 410 accessions, only 44 varieties were released in 1990 or later. So, we selected additional 94 elite modern varieties, 79 of which were released in 1990 or later. Together, the 138 varieties were the best representatives of modern rice varieties developed in China since 1990. These 94 varieties were sequenced using the same methods and platform as 3KRG, including the DNA extract, construction of sequencing library and sequencing. The clean reads of all 504 Os accessions were mapped to the Nipponpare reference genome (Nip_ref, MSU RGAP 7) [61] and to the 93–11 reference genome (9311_ref) [46]using BWA, and covered 371.7 Mb and 384.7 Mb of two reference genomes.
We downloaded the SNP set (7,970,357) of the 446 wild rice (O. rufipogon) accessions from the study [10]. According the local alignment using BWA, we converted the SNPs coordinate of IRGSP4 to MSU RGAP 7. We also downloaded the reads of 10 wild lines (five accessions of O. rufipogon and five accessions of O. nivara) from another rice re-sequencing study [9] and used the same method as the 504 Os accessions for genotype calling using MSU RGAP 7 as the reference. These sampling location of the wild rice collection takes full consideration of the habitat for extant wild rice, and represents an invaluable resource for studying genetic diversity of wild rice. Even though the sequencing price was very cheaper than before, it was very difficult to recollect and sequence representative wild rice materials. Although the sequencing depth of these wild rice was only about 2X, these materials were valuable resources for population genetic analysis [62].
Phylogenetic analysis and population structure
Similar to a recent study [63] where they successfully constructed the merged SNP set using the wild rice accessions with mean sequencing depth ∼ 2X [10] and the 3KRG rice accessions with the mean sequencing depth ∼ 14X [4] for the population structure and genome wide diversity scans, we combined the Nip-SNPs of the 504 Os accessions with the SNP data of publicly available 456 wild rice Or lines from the recent studies [9], [10] for the phylogenetic analysis. We randomly selected five sets of 1 M SNPs with the missing data rate of less than 50 % to construct the NJ-tree and distance matrix, all of which produced similar results. The neighbour-joining trees were then constructed using program Treebest (http://treesoft.sourceforge.net/treebest.shtml). The population structure of the all accessions was estimated by the model based population structure inference for k = 2 to 20 with 556 k high quality SNPs using ADMIXTURE tool with default parameters [64]. To reconstruct the phylogenetic tree of the 504 Os accessions using the genome information of both subspecies, we selected 567 k SNPs that combined 535 k Nip-SNPs (missing rate is 0) and 33 k Nip9311-SNPs (missing rate less than 0.2). The population structure of the 504 Os accessions was performed using FRAPPE [65].
The pan-genome analysis of the wild rice
We constructed the pan-genomes of the 504 Os and 446 Or accessions using the “map-to-pan” strategy, as described previously [4], and then compared the Or and Os pan-genomes accordingly. Because of the low sequencing depths of most Or accessions, we pooled all sequence data from the same Or populations (Or-Int, Or-XL and Or-GL) and then mapped to the Os pan-genome sequences and genes to determine the presence/absence of genes in Or-Int, Or-XL and Or-GL populations using the criteria for the presence of a gene when the gene body coverage was > 0.85 and its CDS coverage was > 0.95.
The construction of the 3D differentiation model
To estimate the contribution of natural and artificial selection to domestication and modern breeding, we constructed a 3D differentiation model using the first three principle coordinates of the SNP variation in all 960 Or and Os accessions (Fig. 1d, Fig. S4b, Table S11). In this model, the differentiation between Or-GL and Or-XL should be attributed to natural selection independent of domestication, and the differentiation between Os and Or populations (such as G-LAN vs Or-GL) or between two Os subpopulations (such as G-MV vs G-LAN) parellelling the line through Or-GL and Or-XL (we called line Or-GL:Or-XL) should be attributed to natural selection during domestication. The second direction parellelling line P1:P2 is vertical with natural selection and in the same differentiation direction for geng(j) from Or-GL and xian(i) from Or-XL during domestication and thus represents the differentiation of the subspecies common artifical selection. Line P1:P2 is the common vertical line between line Or-GL:Or-XL and line G-LAN:X-LAN and was inferred by the stepwise approximation according to the relationship among three sides of the right triangle, i.e. △ = c2 - a2 - b2 = 0. The third direction is vertical with both natural selection and subspecies common artical selection and respresents the subspecies specific artificial selection, which is in the opposite direction for geng(j) from Or-GL and xian(i) from Or-XL. Thus, the distance from each population (such as G-LAN) to its projection point (such as P3) on the plane containing line Or-GL:Or-XL and line P1:P2 measures the static subspecies specific artificial selection consequence (s-ssASC) (such as of G-LAN); the coordinates of the projecton point (such as xP3, yP3, zP3, and it is similar for the coordinates of the other points such as xOrGL, yOrGL, zOrGL for Or-GL) were inferred by the following algorithms,
And the distance from the above projection point (such as P3 of G-LAN) to its projection point (such as P5) on the line Or-GL:Or-XL measures the static subspecies common artificial selection consequence (s-scASC) (such as of G-LAN); the coordinates of the projecton point (such as xP5, yP5, zP5) are inferred by the following algorithms,
Where,
And the distance between populations or projection points on the line Or-GL:Or-XL (such as between Or-GL and projection point P5 (of G-LAN)) measures the static natural selection consequence (s-NSC). The standard deviation of the distance among individuals within one population (such as G-LAN) in each of the above three directions measures the ongoing selection within the population (such as G-LAN), and thus we have and call them dynamic natural selection consequence (d-NSC), the dynamic species-common artificial selection consequence (d-scASC) and the dynamic specific artifical selection consequence (d-ssASC) within each population.
Identification of genome wide non-neutral blocks (selected blocks, SBs)
Two criteria were used to identify the domestication and/or breeding blocks across the rice genome, which typically show loss of diversity or excess of rare alleles. The first criterion was θπ (5,1,8)[66], [67], i.e. the θπ ratio in a sliding window of 10 kb with a step of 5 kb of the compared population pair was ≥ 5, just one gap in each window allowed, and the minimum number of continued windows was ≥ 8. The other one was a criterion of θπ(3,1,8)[66], [67] and Tajima’s D (-2,1,8)[68], [69], [70], i.e. the θπ ratio in 10 kb-window was no less than 3 and simultaneously the D value in the 10 kb-window was no more than −2, with only one gap in each window allowed, and the minimum number of continued windows was no less than 8. Reshuffled alleles for each SNP site within either O. sativa or O. rufipogon indicated that no any block could be identified as non-neutral when using the above criteria, as suggested the low false positive rate in our selective sweep finding. And, to overcome the possible bias in calculating the nucleotide diversity (θπ), due to different sequencing depth between Os and Or, here we used SNP site number rather than window size to calculate these statistics when comparing Os and Or to detect the domestication SBs.
Identification of coding region selection and bottle neck effect using Ka/Ks
Because a dramatically increased or decreased Ka/Ks ratio in one gene represents the possible selection signature in its protein coding region, we call them the coding region selection. For each of those genes located in the d-SBs and B-SBs, we calculated and compared its population based Ka/Ks ratio in the landraces of either xian(i) (indica) or geng(j) (japonica) with 42 Or-Int accessions with sequencing depths > 4X using a c++ program ‘kaks_calculator2.0′ [71]. A gene with significantly increased or decreased Ka/Ks ratio would be considered under NFS or SFS, respectively. The high proportion of the house-keeping genes being identified as SFS validates our criteria to distinguish NFS and SFS. A selected block would be considered subjecting to apparent a bottleneck effect when the proportion of neutral genes within the block was equal or higher than 0.8 and no any gene under coding region selection was detected.
Gene flow or introgression between populations
To determine the gene flow or introgression between or among Os and/or Or subpopulations, we firstly detected the core (or population specific) genomic blocks with following steps (taking X-LAN as an example): (1) took the common SNP between the Nip_SNP of Os and those of Or; (2) calculated the frequency of major genotype of each site in X-LAN as PX-LAN, and called the site with PX-LAN >= 0.8 as X-LAN specific SNP; (3) denoted the X-LAN core (or specific) genomic blocks as one 10 kb window or the continued 10 kb windows with at least 80 % of the SNP being X-LAN specific. Then, we calculated the individual-average number and length of common core genomic blocks and the distributed core genomic blocks between subpopulations.
Availability of data and materials
The raw sequence data of 94 newly sequenced rice varieties reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2017) in BIG Data Center [72], Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA001912 that are publicly accessible at https://bigd.big.ac.cn/gsa. All the genomic data from 3KRG can be downloaded from https://aws.amazon.com/public-data-sets/3000-rice-genome/. All materials will provide for nonprofit research as required to ZKL, ZCL and HLZ.
CRediT authorship contribution statement
Xueqiang Wang: Methodology, Investigation, Validation, Data curation, Writing – review & editing. Wensheng Wang: Investigation, Validation, Data curation, Writing – review & editing. Shuaishuai Tai: Methodology, Investigation, Validation, Writing – review & editing. Min Li: Methodology, Investigation, Validation, Data curation. Qiang Gao: Methodology, Investigation, Validation. Zhiqiang Hu: Methodology, Data curation. Wushu Hu: Methodology, Data curation. Zhichao Wu: Data curation, Investigation. Xiaoyang Zhu: Methodology, Investigation. Jianyin Xie: Methodology, Investigation. Fengmei Li: Methodology. Zhifang Zhang: Investigation. Linran Zhi: Investigation. Fan Zhang: Investigation. Xiaoqian Ma: Methodology. Ming Yang: Investigation. Jiabao Xu: Investigation. Yanhong Li: Investigation. Wenzhuo Zhang: Investigation. Xiyu Yang: Investigation. Ying Chen: Investigation. Yan Zhao: Methodology, Investigation. Binying Fu: Funding acquisition, Investigation. Xiuqin Zhao: Resources, Investigation. Jinjie Li: Methodology, Investigation. Miao Wang: Investigation. Zhen Yue: Investigation. Xiaodong Fang: Investigation. Wei Zeng: Investigation. Ye Yin: Investigation. Gengyun Zhang: Funding acquisition, Investigation. Jianlong Xu: Methodology, Investigation. Hongliang Zhang: Conceptualization, Project administration, Supervision, Funding acquisition, Supervision, Validation, Resources, Writing – original draft, Writing – review & editing. Zichao Li: Conceptualization, Project administration, Supervision, Funding acquisition, Supervision, Validation, Resources, Writing – review & editing. Zhikang Li: Conceptualization, Project administration, Supervision, Funding acquisition, Supervision, Validation, Resources, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
This work was supported by grants to JLX and HLZ from the National Key Research and Development Program of China (2016YFD0100301, 2016YFD0100803), grants to ZKL from the Bill & Melinda Gates Foundation Project (grant no. OPP1130530), the Fundamental Research Funds for Central Non-Profit of CAAS (Y2017CG21) and the National Key Basic Research Program of China (948 Program) (2011-G2B), grants to ZCL from the National Key Technology Support Program of China (2015BAD02B01), grants to HLZ from the National Key Basic research Program of China (973 Program) (2010CB125904), grants to GYZ from State Key Laboratory of Agricultural Genomics (No.2011DQ782025), and grants to JLX from the Agricultural Science and Technology Innovation Program Cooperation and Innovation Mission (CAAS-XTCT2016001-7), the grants to BYF from CAAS Innovative Team Award and National Key Technology Support Program (2015BAD01B02).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.08.004.
Contributor Information
Hongliang Zhang, Email: zhangl@cau.edu.cn.
Zichao Li, Email: lizichao@cau.edu.cn.
Zhikang Li, Email: lizhikang@caas.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic Structure and Diversity in Oryza Sativa L. Genetics. 2005;169(3):1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang D, Zhang H, Wang M, Sun J, Qi Y, Wang F, et al. Genetic Structure and Differentiation of Oryza Sativa L. In China Revealed by Microsatellites. Theor Appl Genet. 2009;119(6):1105–1117. doi: 10.1007/s00122-009-1112-4. [DOI] [PubMed] [Google Scholar]
- 3.Zhang LB, Zhu Q, Wu ZQ, Ross‐Ibarra J, Gaut BS, Ge S, et al. Selection on Grain Shattering Genes and Rates of Rice Domestication. New Phytol. 2009;184(3):708–720. doi: 10.1111/j.1469-8137.2009.02984.x. [DOI] [PubMed] [Google Scholar]
- 4.Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, et al. Genomic Variation in 3,010 Diverse Accessions of Asian Cultivated Rice. Nature. 2018;557(7703):43–49. doi: 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma J, Bennetzen JL. Rapid Recent Growth and Divergence of Rice Nuclear Genomes. Proc Natl Acad Sci USA. 2004;101(34):12404–12410. doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu Q, Ge S. Phylogenetic Relationships among a-Genome Species of the Genus Oryza Revealed by Intron Sequences of Four Nuclear Genes. New Phytol. 2005;167:249–265. doi: 10.1111/j.1469-8137.2005.01406.x. [DOI] [PubMed] [Google Scholar]
- 7.McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, et al. Genomewide Snp Variation Reveals Relationships among Landraces and Modern Varieties of Rice. Proc Natl Acad Sci USA. 2009;106(30):12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao K, Wright M, Kimball J, Eizenga G, McClung A, Kovach M, et al. Genomic Diversity and Introgression in O. Sativa Reveal the Impact of Domestication and Breeding on the Rice Genome. Plos One 2010;5:e10780. [DOI] [PMC free article] [PubMed]
- 9.Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, et al. Resequencing 50 Accessions of Cultivated and Wild Rice Yields Markers for Identifying Agronomically Important Genes. Nat Biotechnol. 2012;30(1):105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 10.Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, et al. A Map of Rice Genome Variation Reveals the Origin of Cultivated Rice. Nature. 2012;490(7421):497–501. doi: 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie W, Wang G, Yuan M, Yao W, Lyu K, Zhao H, et al. Breeding Signatures of Rice Improvement Revealed by a Genomic Variation Map from a Large Germplasm Collection. Proc Natl Acad Sci U S A. 2015;112(39):E5411–E5419. doi: 10.1073/pnas.1515919112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Fu B, Gao Y, Wang W, Xu J, Zhang F. The 3,000 Rice Genomes Project. GigaScience. 2014;3:7. doi: 10.1186/2047-217X-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H, Zhang D, Wang M, Sun J, Qi Y, Li J, et al. A Core Collection and Mini Core Collection of Oryza Sativa L. In China Theor Appl Genet. 2011;122(1):49–61. doi: 10.1007/s00122-010-1421-7. [DOI] [PubMed] [Google Scholar]
- 14.Yu SB, Xu WJ, Vijayakumar CHM, Ali J, Fu BY, Xu JL, et al. Molecular Diversity and Multilocus Organization of the Parental Lines Used in the International Rice Molecular Breeding Program. Theor Appl Genet. 2003;108(1):131–140. doi: 10.1007/s00122-003-1400-3. [DOI] [PubMed] [Google Scholar]
- 15.Li Z, Rutger JN. Geographic Distribution and Multilocus Organization of Isozyme Variation of Rice (Oryza Sativa L.) Theor Appl Genet. 2000;101(3):379–387. [Google Scholar]
- 16.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A Mapreduce Framework for Analyzing Next-Generation DNA Sequencing Data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun C, Hu Z, Zheng T, Lu K, Zhao Y, Wang W, et al. Rpan: Rice Pan-Genome Browser for Approximately 3000 Rice Genomes. Nucleic Acids Res. 2017;45:597–605. doi: 10.1093/nar/gkw958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Civan P, Brown TA. Role of Genetic Introgression During the Evolution of Cultivated Rice (Oryza Sativa L.). BMC Evol Biol 2018;18:57. [DOI] [PMC free article] [PubMed]
- 19.Civan P, Craig H, Cox CJ, Brown TA. Three Geographically Separate Domestications of Asian Rice. Nat Plants. 2015;1:15164. doi: 10.1038/nplants.2015.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi JY, Platts AE, Fuller DQ, Hsing YI, Wing RA, Purugganan MD. The Rice Paradox: Multiple Origins but Single Domestication in Asian Rice. Mol Biol Evol. 2017;34:969–979. doi: 10.1093/molbev/msx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Molina J, Sikora M, Garud N, Flowers JM, Rubinstein S, Reynolds A, et al. Molecular Evidence for a Single Evolutionary Origin of Domesticated Rice. Proc Natl Acad Sci U S A. 2011;108(20):8351–8356. doi: 10.1073/pnas.1104686108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sweeney MT, Thomson MJ, Cho YG, Park YJ, Williamson SH, Bustamante CD, et al. Global Dissemination of a Single Mutation Conferring White Pericarp in Rice. PLoS Genet. 2007;3(8):e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y, Yang X, Huan X, Wang W, Ma Z, Li Z, et al. Rice Bulliform Phytoliths Reveal the Process of Rice Domestication in the Neolithic Lower Yangtze River Region. Quat Int. 2016;426:126–132. [Google Scholar]
- 24.Zuo X, Lu H, Jiang L, Zhang J, Yang X, Huan X, et al. Dating Rice Remains through Phytolith Carbon-14 Study Reveals Domestication at the Beginning of the Holocene. Proc Natl Acad Sci. 2017;114(25):6486–6491. doi: 10.1073/pnas.1704304114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F, Wang C, Li M, Cui Y, Shi Y, Wu Z, et al. The Landscape of Gene–Cds–Haplotype Diversity in Rice: Properties, Population Organization, Footprints of Domestication and Breeding, and Implications for Genetic Improvement. Molecular Plant. 2021;14(5):787–804. doi: 10.1016/j.molp.2021.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Larson G, Piperno DR, Allaby RG, Purugganan MD, Andersson L, Arroyo-Kalin M, et al. Current Perspectives and the Future of Domestication Studies. Proc Natl Acad Sci USA. 2014;111(17):6139–6146. doi: 10.1073/pnas.1323964111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein JC, Yu Y, Copetti D, Zwickl DJ, Wing RA. Genomes of 13 Domesticated and Wild Rice Relatives Highlight Genetic Conservation, Turnover and Innovation across the Genus Oryza. Nat Genet. 2018;50 doi: 10.1038/s41588-018-0040-0. [DOI] [PubMed] [Google Scholar]
- 28.Carpentier MC, Manfroi E, Wei FJ, Wu HP, Lasserre E, Llauro C, et al. Retrotranspositional Landscape of Asian Rice Revealed by 3000 Genomes. Nat Commun. 2019;10(1) doi: 10.1038/s41467-018-07974-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gross BL, Zhao Z. Archaeological and Genetic Insights into the Origins of Domesticated Rice. Proc Natl Acad Sci USA. 2014;111(17):6190–6197. doi: 10.1073/pnas.1308942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Lee GA, Leping J, Juzhong Z. Evidence for the Early Beginning (C. 9000 Cal. Bp) of Rice Domestication in China: A Response. The Holocene. 2007;17:1059–1068. [Google Scholar]
- 31.Fuller DQ, Allaby RG, Stevens C. Domestication as Innovation: The Entanglement of Techniques, Technology and Chance in the Domestication of Cereal Crops. World Archaeology. 2010;42(1):13–28. [Google Scholar]
- 32.Barigozzi C. Elsevier; Amsterdam: 1986. The Origin and Domestication of Cultivated Plants; pp. 21–34. [Google Scholar]
- 33.Harlan JR. Plant Domestication: Diffusion Origins and Diffusion. Developments in Agricultural and Managed Forest Ecology. 1986;16:21–34. [Google Scholar]
- 34.Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. Phylogeography of Asian Wild Rice, Oryza Rufipogon, Reveals Multiple Independent Domestications of Cultivated Rice, Oryza Sativa. Proc Natl Acad Sci USA. 2006;103(25):9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Tang Q, Zou Y, Li D, Qin J, Yang S, et al. Yield Potential and Radiation Use Efficiency of “Super” Hybrid Rice Grown under Subtropical Conditions. Field Crops Research. 2009;114(1):91–98. [Google Scholar]
- 36.Huang M, Zou YB, Jiang P, Xia B, Md I, Ao HJ. Relationship between Grain Yield and Yield Components in Super Hybrid Rice. Agricultural Sciences in China. 2011;10(10):1537–1544. [Google Scholar]
- 37.Jiang P, Xie X, Huang M, Zhou X, Zhang R, Chen J, et al. Potential Yield Increase of Hybrid Rice at Five Locations in Southern China. Rice. 2016;9(1) doi: 10.1186/s12284-016-0085-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ali AJ, Xu JL, Ismail AM, Fu BY, Vijaykumar CHM, Gao YM, et al. Hidden Diversity for Abiotic and Biotic Stress Tolerances in the Primary Gene Pool of Rice Revealed by a Large Backcross Breeding Program. Field Crops Research. 2006;97(1):66–76. [Google Scholar]
- 39.He YX, Zheng TQ, Hao XB, Wang LF, Gao YM, Hua ZT, et al. Yield Performances of Japonica Introgression Lines Selected for Drought Tolerance in a Bc Breeding Programme. Plant Breeding. 2010;129(2):167–175. [Google Scholar]
- 40.Meng L, Xu J, Li Z, Lin X, Sun H, Ma X, et al. Identification and Screening of Salt and Alkaline Tolerance in Rice Using Advanced Backcross Introgression Populations. Molecular Plant Breeding. 2010;18:72–80. [Google Scholar]
- 41.Li M, Wang WS, Pang YL, Domingo JR, Ali J, Xu JL, et al. Characterization of Salt-Induced Epigenetic Segregation by Genome-Wide Loss of Heterozygosity and Its Association with Salt Tolerance in Rice (Oryza Sativa L.). Front Plant Sci 2017;8:977. [DOI] [PMC free article] [PubMed]
- 42.Ali J, Aslam UM, Tariq R, Murugaiyan V, Schnable PS, Li D, et al. Exploiting the Genomic Diversity of Rice (Oryza Sativa L.): Snp-Typing in 11 Early-Backcross Introgression-Breeding Populations. Front Plant Sci 2018;9:849. [DOI] [PMC free article] [PubMed]
- 43.Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, et al. Natural Variation in Ghd7 Is an Important Regulator of Heading Date and Yield Potential in Rice. Nat Genet. 2008;40(6):761–767. doi: 10.1038/ng.143. [DOI] [PubMed] [Google Scholar]
- 44.Liu T, Liu H, Zhang H, Xing Y. Validation and Characterization of Ghd7.1, a Major Quantitative Trait Locus with Pleiotropic Effects on Spikelets Per Panicle, Plant Height, and Heading Date in Rice (Oryza Sativa L.) J Integr Plant Biol. 2013;55(10):917–927. doi: 10.1111/jipb.12070. [DOI] [PubMed] [Google Scholar]
- 45.Wu W, Zheng XM, Lu G, Zhong Z, Gao H, Chen L, et al. Association of Functional Nucleotide Polymorphisms at Dth2 with the Northward Expansion of Rice Cultivation in Asia. Proc Natl Acad Sci USA. 2013;110(8):2775–2780. doi: 10.1073/pnas.1213962110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao H, Jin M, Zheng XM, Chen J, Yuan D, Xin Y, et al. Days to Heading 7, a Major Quantitative Locus Determining Photoperiod Sensitivity and Regional Adaptation in Rice. Proc Natl Acad Sci USA. 2014;111(46):16337–16342. doi: 10.1073/pnas.1418204111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang C, Zhu J, Chen S, Fan X, Li Q, Lu Y, et al. Wx(Lv), the Ancestral Allele of Rice Waxy Gene. Mol Plant. 2019;12:1157–1166. doi: 10.1016/j.molp.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Calingacion M, Laborte A, Nelson A, Resurreccion A, Concepcion JC, Daygon VD, et al. Diversity of Global Rice Markets and the Science Required for Consumer-Targeted Rice Breeding. PLoS ONE. 2014;9(1):e85106. doi: 10.1371/journal.pone.0085106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng L, Lin X, Wang J, Chen K, Cui Y, Xu J, et al. Simultaneous Improvement in Cold Tolerance and Yield of Temperatejaponicarice (Oryza Sativa L.) by Introgression Breeding. Plant Breeding. 2013;132(6):604–612. [Google Scholar]
- 50.Cui Y, Li R, Li G, Zhang F, Zhu T, Zhang Q, et al. Hybrid Breeding of Rice Via Genomic Selection. Plant Biotechnol J. 2020;18(1):57–67. doi: 10.1111/pbi.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang X, Wei X, Sang T, Zhao Q, Feng Q, Zhao Y, et al. Genome-Wide Association Studies of 14 Agronomic Traits in Rice Landraces. Nat Genet. 2010;42(11):961–967. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 52.Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, et al. Genome-Wide Association Mapping Reveals a Rich Genetic Architecture of Complex Traits in Oryza Sativa. Nat Commun. 2011;2(1) doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang W, Guo Z, Huang C, Duan L, Chen G, Jiang N, et al. Combining High-Throughput Phenotyping and Genome-Wide Association Studies to Reveal Natural Genetic Variation in Rice. Nat Commun. 2014;5(1) doi: 10.1038/ncomms6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dong H, Zhao H, Xie W, Han Z, Li G, Yao W, et al. A Novel Tiller Angle Gene, Tac3, Together with Tac1 and D2 Largely Determine the Natural Variation of Tiller Angle in Rice Cultivars. PLoS Genet. 2016;12(11):e1006412. doi: 10.1371/journal.pgen.1006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magwa RA, Zhao H, Xing Y. Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.) BMC Genet. 2016;17(1) doi: 10.1186/s12863-016-0340-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yano K, Yamamoto E, Aya K, Takeuchi H, Lo PC, Hu L, et al. Genome-Wide Association Study Using Whole-Genome Sequencing Rapidly Identifies New Genes Influencing Agronomic Traits in Rice. Nat Genet. 2016;48(8):927–934. doi: 10.1038/ng.3596. [DOI] [PubMed] [Google Scholar]
- 57.Zhao Y, Zhang H, Xu J, Jiang C, Yin Z, Xiong H, et al. Loci and Natural Alleles Underlying Robust Roots and Adaptive Domestication of Upland Ecotype Rice in Aerobic Conditions. PLoS Genet. 2018;14(8):e1007521. doi: 10.1371/journal.pgen.1007521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao Y, Zhao W, Jiang C, Wang X, Xiong H, Todorovska EG, et al. Genetic Architecture and Candidate Genes for Deep-Sowing Tolerance in Rice Revealed by Non-Syn Gwas. Front Plant Sci. 2018;9 doi: 10.3389/fpls.2018.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao Y, Qiang C, Wang X, Chen Y, Deng J, Jiang C, et al. New Alleles for Chlorophyll Content and Stay-Green Traits Revealed by a Genome Wide Association Study in Rice (Oryza Sativa) Sci Rep. 2019;9(1) doi: 10.1038/s41598-019-39280-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu J, Xiong H, Zhu X, Zhang H, Li H, Miao J, et al. Oslg3 Contributing to Rice Grain Length and Yield Was Mined by Ho-Lamap. BMC Biol. 2017;15(1) doi: 10.1186/s12915-017-0365-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, et al. Improvement of the Oryza Sativa Nipponbare Reference Genome Using Next Generation Sequence and Optical Map Data. Rice. 2013;6(1) doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H, Vieira FG, Crawford JE, Chu C, Nielsen R. Asian Wild Rice Is a Hybrid Swarm with Extensive Gene Flow and Feralization from Domesticated Rice. Genome Res. 2017;27(6):1029–1038. doi: 10.1101/gr.204800.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Civan P, Ali S, Batista-Navarro R, Drosou K, Ihejieto C, Chakraborty D, et al. Origin of the Aromatic Group of Cultivated Rice (Oryza Sativa L.) Traced to the Indian Subcontinent. Genome Biol Evol. 2019;11:832–843. doi: 10.1093/gbe/evz039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alexander DH, Novembre J, Lange K. Fast Model-Based Estimation of Ancestry in Unrelated Individuals. Genome Res. 2009;19(9):1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tang H, Peng J, Wang P, Risch NJ. Estimation of Individual Admixture: Analytical and Study Design Considerations. Genet Epidemiol. 2005;28(4):289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 66.Watterson GA. On the Number of Segregating Sites in Genetical Models without Recombination. Theor Popul Biol. 1975;7(2):256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- 67.Tajima F. Evolutionary Relationship of DNA Sequences in Finite Populations. Genetics. 1983;105(2):437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tajima F. Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yongle, Li, Pradeep, Ruperao, Jacqueline, Batley, et al. Genome Analysis Identified Novel Candidate Genes for Ascochyta Blight Resistance in Chickpea Using Whole Genome Re-Sequencing Data. Frontiers in Plant Science 2017;8:359. [DOI] [PMC free article] [PubMed]
- 70.Thornton K. Libsequence: A C++ Class Library for Evolutionary Genetic Analysis. Bioinformatics. 2003;19(17):2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- 71.Wang D, Zhang Y, Zhang Z, Zhu J, Yu J. Kaks_Calculator 2.0: A Toolkit Incorporating Gamma-Series Methods and Sliding Window Strategies. Genomics, Proteomics & Bioinformatics. 2010;8(1):77–80. doi: 10.1016/S1672-0229(10)60008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Members BDC. Database Resources of the Big Data Center in 2019. Nucleic Acids Res. 2019;47:D8–D14. doi: 10.1093/nar/gky993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data of 94 newly sequenced rice varieties reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2017) in BIG Data Center [72], Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession number CRA001912 that are publicly accessible at https://bigd.big.ac.cn/gsa. All the genomic data from 3KRG can be downloaded from https://aws.amazon.com/public-data-sets/3000-rice-genome/. All materials will provide for nonprofit research as required to ZKL, ZCL and HLZ.
CRediT authorship contribution statement
Xueqiang Wang: Methodology, Investigation, Validation, Data curation, Writing – review & editing. Wensheng Wang: Investigation, Validation, Data curation, Writing – review & editing. Shuaishuai Tai: Methodology, Investigation, Validation, Writing – review & editing. Min Li: Methodology, Investigation, Validation, Data curation. Qiang Gao: Methodology, Investigation, Validation. Zhiqiang Hu: Methodology, Data curation. Wushu Hu: Methodology, Data curation. Zhichao Wu: Data curation, Investigation. Xiaoyang Zhu: Methodology, Investigation. Jianyin Xie: Methodology, Investigation. Fengmei Li: Methodology. Zhifang Zhang: Investigation. Linran Zhi: Investigation. Fan Zhang: Investigation. Xiaoqian Ma: Methodology. Ming Yang: Investigation. Jiabao Xu: Investigation. Yanhong Li: Investigation. Wenzhuo Zhang: Investigation. Xiyu Yang: Investigation. Ying Chen: Investigation. Yan Zhao: Methodology, Investigation. Binying Fu: Funding acquisition, Investigation. Xiuqin Zhao: Resources, Investigation. Jinjie Li: Methodology, Investigation. Miao Wang: Investigation. Zhen Yue: Investigation. Xiaodong Fang: Investigation. Wei Zeng: Investigation. Ye Yin: Investigation. Gengyun Zhang: Funding acquisition, Investigation. Jianlong Xu: Methodology, Investigation. Hongliang Zhang: Conceptualization, Project administration, Supervision, Funding acquisition, Supervision, Validation, Resources, Writing – original draft, Writing – review & editing. Zichao Li: Conceptualization, Project administration, Supervision, Funding acquisition, Supervision, Validation, Resources, Writing – review & editing. Zhikang Li: Conceptualization, Project administration, Supervision, Funding acquisition, Supervision, Validation, Resources, Writing – review & editing.