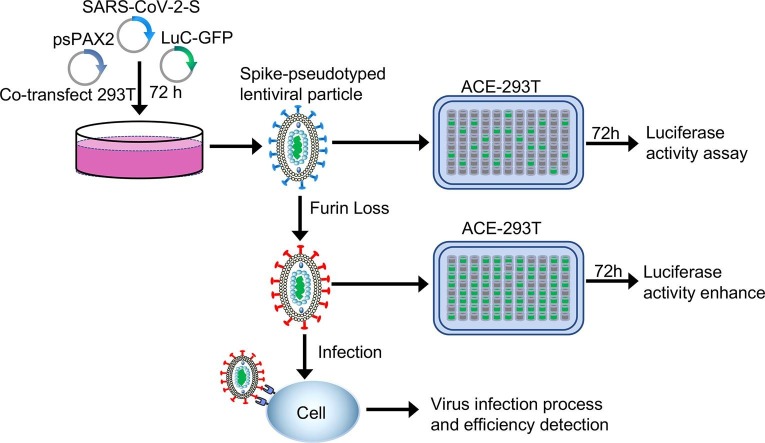
Graphical abstract
Keywords: COVID-19, SARS-CoV-2, Spike protein, Furin, Viral pseudotyping
Abbreviations: SARS-CoV-2, acute respiratory syndrome coronavirus 2; ACE2, angiotensin-converting enzyme 2; FCS, furin cleavage site; PVs, Pseudoviruses; PEI, polyetherimide; VLP, virus-like particle; RLU, relative luminescence signals; ER, endoplasmic reticulum; VSV-dG, G protein-deficient vesicular stomatitis virus
Abstract
Background
SARS-CoV-2 has a significant impact on healthcare systems all around the world. Due to its high pathogenicity, live SARS-CoV-2 must be handled under biosafety level 3 conditions. Pseudoviruses are useful virological tools because of their safety and versatility, but the low titer of these viruses remains a limitation for their more comprehensive applications.
Method
Here, we constructed a Luc/eGFP based on a pseudotyped lentiviral HIV-1 system to transduce SARS-CoV-2 S glycoprotein to detect cell entry properties and cellular tropism.
Results
The furin cleavage site deletion of the S protein removed (SFko) can help SARS-CoV-2 S to be cleaved during viral packaging to improve infection efficiency. The furin cleavage site in SARS-CoV-2-S mediates membrane fusion and SFko leads to an increased level of S protein and limits S1/S2 cleavage to enhance pseudovirus infection in cells. Full-length S (SFL) pseudotyped with N, M, and E helper packaging can effectively help SFL infect cells. Finally, pseudotyped SFko particles were successfully used to detect neutralizing antibodies in RBD protein-immunized mouse serum.
Conclusion
Overall, our study indicates a series of modifications that result in the production of relatively high-titer SARS-COV-2 pseudo-particles that may be suitable for the detection of neutralizing antibodies from COVID-19 patients.
1. Introduction
A large outbreak of severe acute respiratory disease (COVID-19) was characterized as a pandemic by the World Health Organization on March 11, 2020 (Zhu et al., 2020, Zhou et al., 2020). It is identified as an acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes lethality in humans and is classified as a β-coronavirus genus (Perlman and Netland, 2009). It has already been successfully demonstrated that it has a high affinity for the host human angiotensin-converting enzyme 2 (ACE2) receptor (Walls et al., 2020). SARS-CoV-2 encodes four structural proteins: the spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins (Kim et al., 2020); with S, E, and M being three integral membrane proteins. Understanding how SARS-CoV-2 infects human cells will provide insights into viral transmission and pathogenesis.
The S protein is a class I fusion protein comprised of two domains, S1 and S2. The surface unit S1 binds to a cellular receptor while the transmembrane unit S2 facilitates the fusion of the viral membrane with a cellular membrane (Hoffmann et al., 2020). Cleavage of the S1/S2 site results in the activation of a highly conserved the fusion peptide immediately downstream of the S2 site (Millet and Whittaker, 2015, Davidson et al., 2020, Andersen et al., 2020). Notably, SARS-CoV-2 in the presence of four amino acids, “682RRAR685”, results in a furin cleavage site (FCS) (Wrapp et al., 2020). This FCS has been present in all the peak proteins of clinical SARS-CoV-2 isolates and in some other highly pathogenic viruses (such as avian influenza A) (Luczo et al., 2015, Chu et al., 2020, Belouzard et al., 2009).
Pseudoviruses (PVs) are useful virological tools due to their safety and versatility. This method can “pseudotype” assemble the S protein of SARS-CoV-2 into a safer nonreplicating virus particle, replacing the endogenous protein of the virus particle itself, so that these virus particles rely on the S protein to enter cells (Temperton et al., 2005, Grehan et al., 2015). Recently, the use of HIV-based lentiviral particles (Ou et al., 2020), MLV-based retroviral particles (Pinto et al., 2020), and VSV-based SARS-CoV-2 pseudotyped virus particles (Letko et al., 2020, Xiong et al., 2020) have been reported. The results of these PV neutralization measurements have a good correlation with the use of live SARS-CoV-2 virus (Chen et al., 2020).
Here, we constructed a Luc and eGFP gene expressing PVs containing the wild-type or mutant S protein of SARS-CoV-2 in the envelope-defective HIV-1 backbone as an evaluation system in a preliminary in vitro. Meanwhile, using the SARS-CoV-2 S protein PV system, we focused on specific FCS to determine whether it is essential for SARS-CoV-2 S multibasic cleavage site processing and how it impacts both viral entry and cell-cell fusion. We confirmed that in cells infected with PVs, deletion of FCS can greatly increase infection efficiency.
2. Material and methods
2.1. Cell line and plasmids
Huh-7 (human hepatocarcinoma cells), Vero cells (African green monkey kidney cells), Calu-3 (human lung epithelial cells), and MCF-7 (human breast adenocarcinoma cells) were obtained from the American Type Culture Collection. HEK293T cells stably expressing ACE2 receptor (293 T-ACE2) were constructed using the lentiviral transduction method. The plasmid PCAGGS-spike coding full-length (1–1273) was kindly provided by Prof. Guangwen Lu (Sichuan University, China). The VSV-G-encoding plasmid lentiviral packaging plasmid psPAX2 (#12260, Addgene). The eGFP fluorescent protein or pCDH-lentiviral reporter plasmid that expresses luciferase and GFP was generously transformed. The plasmids coding membrane (M), envelope (E), and nucleocapsid (N) proteins were synthesized by General Biological System Co. Ltd (Anhui, China).
2.2. Animals
Using a fusion protein that contained the RBD linked to mouse IgG1 Fc fragment (designated RBD-mFc) as an immunogen, Bal/bc mice (the Center of Experimental Animals, Sichuan University) were immunized 2-week intervals between immunizations and three occasions later, blood collection was performed via a tail vein draw of ∼ 0.1 mL. Collection of serum to measure neutralizing antibody titers.
2.3. Production of SARS-CoV-2 S PVs or PVs with helper packaging and virus entry
Pseudovirions were produced by cotransfection of HEK293T cells with pLenti-eGFP/pCDH-Luc-GFP, psPAX2, and plasmids encoding either SFL, Sdel18, SG614, SFko, or VSV-G were (6:3:1) by using polyetherimide (PEI) reagent (Polysciences Inc, USA) based on the manufacturer’s instructions as the transfection reagent. After 6 h of transfection, the medium was changed to a fresh medium, and the viral supernatant was collected at 24 h, 48 h, and 72 h, respectively. Cell debris was removed by filtration, and HIV-1 lentiviral titers were detected using a p24 kit (Sino Biological Inc., Beijing, China). On the basis of pseudovirion packaging, helper plasmids (M, N, E) were added to the proportion of (5:2:1:5:5:1) during packaging. Different cells were seeded into 96-well plates and inoculated with 100 μl media containing pseudovirions. Approximately 48 h postinoculation, eGFP expression in the infected cells was determined by fluorescence microscopy and flow cytometry. Cell lysates were collected 48 h postinfection, and luciferase activity was analyzed to monitor PV integration by the ONE-Glo™ Luciferase Assay System (Promega, USA). The transduction efficiency was analyzed using a multifunctional enzyme labeling instrument (Thermo Fisher Scientific, Inc., USA) All experiments were performed in triplicate or more.
2.4. Pseudovirus titer assay by p24 ELISA kit
The 96-well strip plate is precoated with a monoclonal antibody specific for HIV-1 p24 / capsid protein p24. The pseudovirus supernatant from different mutants was in the same packaging conditions. Add the diluted test virus samples to each well and incubate for 2 h at room temperature. After washing, a horseradish peroxidase-conjugated anti- HIV-1 p24 / capsid protein p24 secondary antibody is added and incubated at room temperature for 1 h. A TMB substrate solution is added to the wells and color develops in proportion to the amount of HIV-1 p24 / capsid protein p24 bound in the initial step. The color development is stopped and the intensity of the color is measured at 450 nm.
2.5. Detection of the S protein of SARS-CoV-2 by western blot
Briefly, purified pseudovirion virus pellets of SFL, Sdel18, SG614, SFko, or VSV-G were passed through a 20% sucrose layer and cell lysates were resuspended in 30 μl RIPA buffer, and incubated on ice for 30 min with an interval vortex. Then the samples were boiled for 10 min and separated in a 10% SDS-PAGE gel and transferred to nitrocellulose filter membranes. After blocking with 5% milk, the membranes were blotted with SARS-CoV-2 (2019-nCoV) Spike Antibody, Rabbit PAb (1:2000) (Sinobiological Inc., China) and incubated in blocking solution overnight at 4℃, followed by incubation with horseradish peroxidase (HRP)-conjugated goat anti-rabbit secondary antibody (1:3000) (Proteintech, China) and visualization with Chemiluminescent Reagent (Bio-Rad, USA).
2.6. Flow cytometric analysis of S protein expression
Briefly, HEK293T cells were transfected with 2 μg of plasmids encoding SFL, Sdel18, SG614, and SFko using Lipofectamine 3000 (Invitrogen, USA) reagents. Forty hours later, the cells were washed with PBS and incubated with monoclonal mouse anti-SARS Spike glycoprotein antibody 1A9 (1:500) (Abcam, UK) for 1 h on ice, followed by Alexa Fluor 488-conjugated goat anti-mouse IgG (1:500) (Proteintech, China) for 1 h. After washing, the cells were analyzed by flow cytometry.
2.7. virus-like particle (VLP) production and expression
SARS-CoV-2 VLPs were produced by SARS-CoV-2 M, S, E co-transfection of 293 T cells with 70% cell density in six-well plates at a proportion of (8:1:1) using PEI as the transfection reagent. VLP culture supernatants were harvested 72 h post-transfection and cleared by 0.45 μm filtration. Then, the supernatants were concentrated using a 100 kD ultrafiltration tube (Millipore, USA) for further concentration and purification. boiled for 10 min, separated in a 10% SDS-PAGE gel, and transferred to nitrocellulose filter membranes for blotting.
2.8. Cell-cell fusion assay
Plasmids encoding SFL, Sdel18, SG614, SFko, and eGFP were over-expressed into 293 T cells as the effector cells and 293 T-ACE2 as the target cells. Effector cells were collected separately and added to 293 T-ACE2 cells at an effector: target ratio (E: T) of 1:1 (104 effectors:104 targets) in 96-well plate coculture for 4 to 24 h. Then the fusion was observed under a fluorescence microscope.
2.9. Analysis of SARS-CoV-2 S protein subcellular localization
Confocal microscopy analysis was performed to study the localization of the S protein. Briefly, 293 T cells overexpressing SFL, Sdel18, SG614, SFko, and 293 T-ACE2-GFP were cocultured in 24-well plates for 12 h, and SARS-CoV-2 S proteins were detected using monoclonal mouse anti-SARS Spike glycoprotein antibody 1A9 (1:500). Specific signals were visualized using Alexa Fluor 594-conjugated goat anti-mouse IgG (1:500) (Cell Signaling Technology, USA). For nuclear staining, cells were treated with DAPI for 5 min. Staining sections were analyzed using a laser scanning confocal microscope (Zeiss 880).
2.10. Indirect ELISA
The RBD protein with His tags and SARS-CoV-2 Spike S1 + S2 ECD-His Recombinant Protein (Sinobiological Inc., China) were separately diluted and added to antigen-coated wells (Thermo Scientific™) and incubated at 4 °C overnight. A dilution series (from 1:2 × 103 to 1:2 × 107) of mouse serum was added to the wells, followed by the addition of HRP-conjugated goat anti-mouse IgG (1:2000) (Beyotime, China). After washing, tetramethylbenzidine liquid substrate (TMB) (Sigma) was added to the plates, and the reaction was stopped with 2 M H2SO4, and the optical density at 450 nm (OD450nm) was measured using a multifunctional enzyme labeling instrument. The binding ability was determined using a four-parameter nonlinear regression curve fit (GraphPad Prism 7.0).
2.11. Detection neutralization effect of pseudotyped virus infection
PVs containing Sdel18, SG614, and SFko were incubated with the sera from the immunized mouse at various dilutions (1:100–1:10000) and unimmunized sera for 1 h at 37 °C. The mixture was added to 293 T-ACE2 cells to detect viral infectivity. After 48 h of infection, luciferase or eGFP expression in the infected cells was determined by fluorescence microscopy and the ONE-Glo™ Luciferase Assay System. The relative luciferase units (RLU) from the negative control wells were normalized and used to calculate the neutralization percentage for each concentration.
2.12. Statistical analysis
All statistical analyses were performed using GraphPad Prism version 7.0 (GraphPad Software, San Diego, CA, USA). All presented data are shown as the means ± SDs, which contain five replicates. Comparisons among multiple groups were performed using two-way ANOVA with Dunnett's posttest. Only p values of 0.05 or lower were considered statistically significant (p>0.05[ns, not significant], p ≤ 0.05[*], p ≤ 0.01[**], p ≤ 0.001[***]).
3. Results
3.1. Pseudovirion incorporation of SARS-CoV-2 S
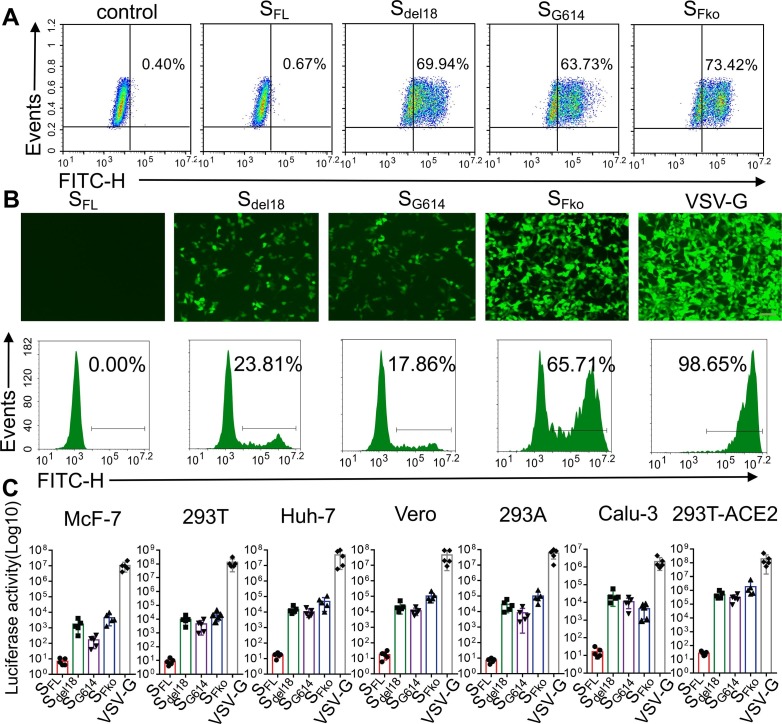
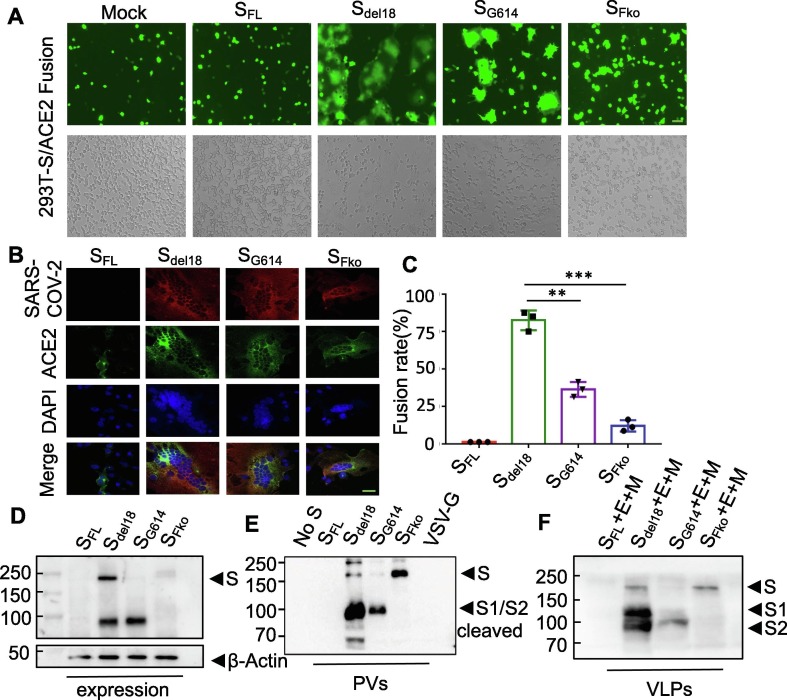
To construct a pseudovirus carrying the S protein of SARS-CoV-2, the S gene sequence was codon-optimized for expression and cloned into the eukaryotic expression plasmid pCAGGS (Yang et al., 2020) to generate the recombinant plasmid SARS-CoV-2 S full-length (SFL) (Fig. 1 A). Because truncation of the cytoplasmic tail of SARS Spike was shown to enhance the production of viral pseudotypes (Johnson et al., 2020), we also generated a SARS-COV-2 Spike subclone S with C-terminal 18 aa truncation (Sdel18) to generate SARS-CoV-2 S deleting an endoplasmic reticulum (ER)-retention signal from the cytoplasmic tail. On the basis of the variation of Sdel18, we mutated R682 to R685 to inhibit furin-mediated cleavage (SFko) to detect the cleavage state for cell infection impact. An aspartate(D) at position 614 mutated to glycine (G) (SG614) as a comparison (Fig. 1B). A pseudovirus system based on the lentivirus Packaging System was employed to produce the SARS-CoV-2 S protein (Fig. 1C).
Fig. 1.
Overall structure of the SARS-CoV-2 spike protein PVs system. A Schematic representation of the 1D structure of full-length coronavirus Spike protein. Segments of S1 and S2 include: NTD, N-terminal domain; RBD, receptor-binding domain; RRAR, S1/S2 Furin cleavage site; FP, fusion peptide; HR1, heptad repeat1; and HR2, heptad repeat2 are structural units in coronavirus S2 that function in membrane fusion; TM, transmembrane; CT, cytoplasmic tail. B Amino acid sequence alignment of the SARS-CoV-2 S mutant variants. Including Sdel18 with C-terminal 18 amino acid truncation, S proteins with aspartic acid (D) mutant to glycine (G) at residue 614 (SG614), and Furin cleavage “RRAR” deletion (SFko). The C-terminal 18 amino acids were lacking in all S mutant variants, compared with the SFL. C. The cell entry mechanism of SARS-CoV-2 PVs system.
3.2. FCS deletion can enhance PV infection
The different mutations for the expression of the SARS-CoV-2 S protein were monitored using flow detection and Western blotting. Compared with SFL, there were obvious expressions in these variants, while the maximum expression was 73.42% in SFko in HEK293T cells, there is no significant difference among Sdel18, SG614, and SFko (Fig. 2 A). Collect the PVs supernatant expressing the Luc and GFP genes pseudotyped with different mutant viruses, and add equal amounts into 293 T-ACE2 cells for 48 h. The fluorescence intensity of GFP was evaluated by fluorescence microscopy and flow cytometry. As a result, the efficiency of Sdel18 was 23.70%. The efficiency of SFko has increased drastically to 65.38%. Surprisingly, the efficiency of SG614 decreased instead of increasing as previously reported (Shi et al., 2020). Wild full-length S protein of SARS-CoV-2 was used as a negative control. VSV-G virus was used as a positive control (Fig. 2B). Different vectors, such as Plenti-eGFP, pCDH-GFP, pCDH-luciferase-GFP, and pCDH-luciferase-mCherry, were used for repeated verification. Different vectors had different viral packaging efficiencies, but the results consistently showed that the viral infection titer of SFko was significantly higher than that of Sdel18 (Supplementary Fig. 1). The cell type entry efficiency of PVs with different SARS-CoV-2 mutations further validates the results, further supporting the notion that ACE2 is the receptor and SFko-treated PVs demonstrated significant increases in cell entry efficiency. However, SFko-mediated furin deletion was inefficient in entering human lung Calu-3 cells (Fig. 2C).
Fig. 2.
Furin cleavage site deletion increases S pseudovirus entry. A. HEK293T cells transiently expressing SARS-CoV-2 S mutant variants were incubated with the anti-SARS-CoV-2 spike antibody on ice. Cells have analyzed the expressions by flow cytometry. B. The infection efficiency of SARS-CoV S different variants in the similar virus harvest supernatant titers (Scale bar = 100 µm). EGFP fluorescence intensity was detected using the FITC channel (527/32 filter) by flow cytometry (SFL-negative control, VSV-G-positive control). C. Transduction efficiency of different six types of cells was tested for S variants PVs. The y-axis showed the luciferase activity (Log10) value detected 48 h after infection with SARS-CoV S different variants. The experiments were repeated at least three times.
3.3. Neutralizing activities of SARS-CoV-2 S RBD-specific mouse serum
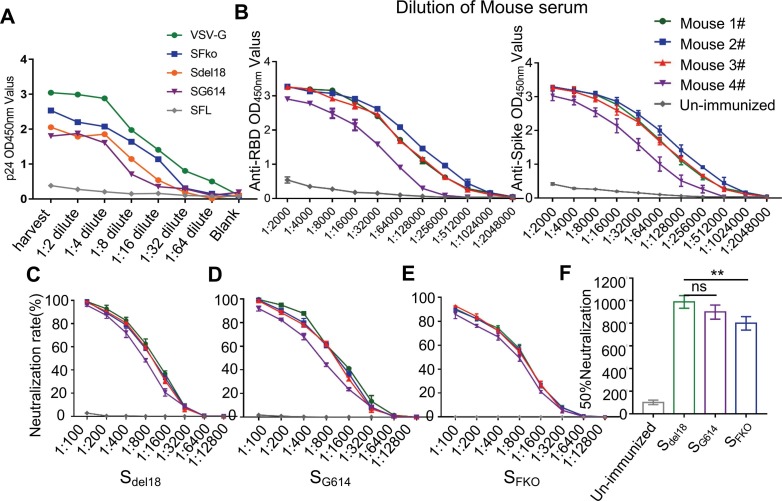
P24 ELISA assay results indicated that the HIV-1-based PVs pseudotyped with SFL, Sdel18, SG614, SFko, and VSV-G packaged by HEK293T under the same virus packaging conditions showed different titers. The full-length S protein had no p24 titration (Fig. 3 A). Indirect ELISA results indicated that RBD-mFc immune mouse serum simultaneously has the binding affinity of the RBD protein with His tags and SARS-CoV-2 S1 + S2 ECD-His Recombinant Protein (Fig. 3B). The neutralization activity of RBD-immunized mouse serum against S pseudovirus was detected in 1:5000 titer. Neutralizing activities are similar among Sdel18 and SG614 PVs. SFko showed weaker neutralization efficiency which means that the neutralization efficiency was only 90% at a ratio of 1:100 (Fig. 3C-E). The 50% neutralization was presented with titers of 1:1000 among Sdel18, SG614, and a titer of 1:800 with SFko (Fig. 3F). Collectively, the results indicate that the SFko-mediated convenient and enhanced pseudovirus system has been established to test the neutralizing activity of mouse sera against SARS-CoV-2.
Fig. 3.
Detection neutralization effect of RBD-mFc serum of SARS-CoV-2 pseudovirus infection. A Virus supernatants were analyzed by p24 ELISA for PVs-associated p24. Under the same packaging conditions, the p24 expression level of SFko is the highest when the PVs of different mutants (SFL-negative control, VSV-G-positive control). B. Determination of the binding affinity of immune serum with RBD-His protein and Spike S1 + S2 ECD-His recombinant protein using indirect ELISA. C-E. Neutralization potency was measured by using S variants pseudotyped virus luciferase neutralization assay. F. 50% neutralization was presented as the mean ± SD four mouse serum samples in each group (n = 4) (ns, no significance; p ≤ 0.01[**] indicate significant differences).
3.4. SARS-CoV-2-S needs cleavage at the S1/S2 site to mediate membrane fusion
The 293 T-S-GFP/293 T-ACE2 fusion showed that Sdel18 and SG614 promoted the transport of S to the target cell surface for fusion, while SFko, with deletion of the polybasic site in the S protein, showed negligible syncytia formation in cell culture, but not completely (Fig. 4 A). We further verified this fusion phenomenon in 293 T-S-GFP/293 T-ACE2 cells by confocal microscopy. Colocalization of SARS-CoV-2 (red) with ACE2 (green) and the nucleus (blue) during cell fusion by immunofluorescence analysis. When fusion occurred, cells with green fluorescent membranes and blue fluorescent nuclei were observed to fuse to form multinucleated syncytia and the adjacent cells grew and fused into pieces (Fig. 4B). The SFko group exhibited syncytia, but very few syncytia, 10% fusion rate (Fig. 4C). The Western blot results revealed that Sdel18 and SG614 in PVs during viral packaging and overexpression in 293 T cells were all cleaved, while the mutant SFko was no longer cleaved during viral packaging at 72 h post packaging (Fig. 4D-E). When the spike enters the cell, it is cleaved by the protease on the surface of the host cell, then the S1 protein falls off into the supernatant, and S2 fuses with the cell. This confirmed that the 100 kDa polypeptides corresponding to the S2 subunit were processed in the biosynthesis process to promote syncytia formation. The virus-like particles (VLPs) further confirm that there is indeed cleavage into two domains, S1 and S2, in SARS-CoV-2 S; in contrast, the mutant type SFko does not (Fig. 4F).
Fig. 4.
Furin cleavage in the S1/S2 Site of SARS-CoV-2 spike is required for efficient cell-cell fusion butisnot essential. A. 293 T-S-GFP/293 T-ACE2 mediated cell-cell fusion with the four variants. The effector cell was identified as co-expressing S and GFP proteins, 293 T-ACE2 was the target cell. Mock-fusion between target cells and 293 T/GFP effector cells without S-expression as control. (Scale bar = 100 µm). B. Detection of SARS-CoV-2 S subcellular localization in 293 T/293 T-ACE2-GFP cells fusion by confocal microscopy. Red represented high SARA-Cov-2 antibody binding. GFP (green) represented GFP over-expressed 293 T-ACE2 cells. Nuclei (blue) were counter-stained with DAPI. The nucleus was observed to fuse to form multinucleated syncytia and the adjacent cells grew and fused into pieces. (Scale bar = 20 µm). C. Statistical analysis of fusion rates mediated by wild-type or mutated S protein after co-culture for 4 h. Experiments were repeated at least three times independently, and the data are expressed as means ± SD. p ≤ 0.001[***] indicates significant differences; ns: no significance. D. Detection of S variant proteins in cells lysate by western blot. E. Furin-mediated S protein cleavage with PVs subjected to western blot analysis. F. The virus-like particles (VLPs) with S, M, E were subjected to western blot analysis. All experiments were biologically repeated at least three times.
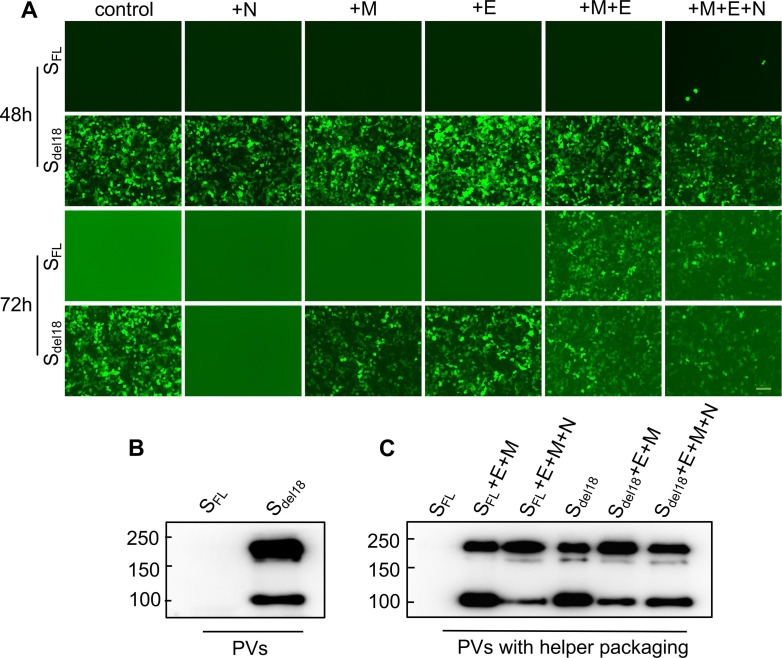
3.5. E, M, N helper packaging can effectively help SFL infect cells
N, E, and M of SARS-CoV-2 were added for helper packaging to explore the effect these proteins have on the packaging of the PVs. Virus supernatant harvested at 48 h, Sdel18 + N successfully infected 293 T-ACE2 cells. However, the virus supernatant harvested at 72 h cannot infect the cells. Notably, the virus was captured by SFL + M + E cells, and SFL + M + E + N cells successfully infected 293 T-ACE2 cells with the virus supernatant harvested at 72 h (Fig. 5 A). The incorporation of SARS-CoV-2 S on the surface of PVs was detected by using Western blotting with a SARS-CoV-2 S antibody. During virus maturation, it is cleaved into two subunits: S1, which binds to receptors in the host cell, and S2, which mediates membrane fusion. When the Sdel18 pseudovirus successfully enters the cell, the Sdel18 protein will be cleaved, it was observed at approximately 180 kDa and confirmed that the 100 kDa protein corresponded to the cleaved subunit (Fig. 5B). Similarly, we demonstrated that PVs of SFL with M, E, and N helper packaging were cleaved successfully. (Fig. 5C).
Fig. 5.
SFL pseudotyped with E, M,andN helper packaging can effectively help SFL infect cells. A. Fluorescence images of SFL PVs with helper packaging after 48 h and 72 h cell infection. 72 h packaging SFL PVs cannot infect 293 T-ACE2 while 72 h packaging SFL PVs with M, E or M, E, N helper packaging infect cells successfully. But 72 h packaging Sdel18 PVs with N helper packaging without infectivity of budding viruses (Scale bar = 100 µm). Experiments were done twice and one representative is shown. B. SARS-CoV-2 S protein PVs were cleaved successfully when infecting the cells triumphantly by western blot. uncleaved S protein, about 180 kDa; cleaved S protein, about 100 kDa. C. PVs with E, M, and N helper packaging in 72 h show similar results. The total amount of uncleaved and cleaved spike molecules was calibrated for SARS-CoV-2 PVs entry. Experiments were done twice and one is shown.
4. Discussion
Because of the COVID-19 pandemic, the novel coronavirus SARS-CoV-2 rose to shape scientific research in 2020, with its S protein being a predominant focus. Currently, attention has been focused specifically on the S protein, the key determinant for immunological responses and virus infection. The main purpose of this study was to improve the infection efficiency of S protein pseudoviruses. In fact, in the course of the study, research methods to increase the titer of PVs are constantly being reported (Yang et al., 2021).
Indeed, the incorporation of envelope glycoproteins into virions is an essential step in the retroviral replication cycle. The infectivity of lentiviral systems, such as HIV-1, can be adversely affected by the expression of abnormal heterologous glycoproteins with cytoplasmic tail deletions (Murakami and Freed, 2000). It is reported that the full-length spike has an endoplasmic reticulum (ER)-retention signal from the cytoplasmic tail (Ou et al., 2020). During virus packaging, the proteins are trapped in the endoplasmic reticulum, making it impossible to secrete. Considering the entire length of S protein is not in favor of lentivirus pseudotyping, most of the f pseudovirus systems deleted the last 18 amino acids, which may increase the levels of S on the cell surface, resulting in a higher titer of pseudovirus (Wang et al., 2022, Hu et al., 2020, Crawford et al., 2020). The results of virus infection and western blot both showed the SARS-CoV-2-S full-length construct fail to be expressed. Additionally, to improve the packaging efficiency of the PV, the packaging conditions and the ratio of packaging plasmids were optimized. At the same time, to elevate PV productivity, the packaging conditions and the proportion of packaging plasmids were optimized. Selection of vectors with the features desired to use should be made with consideration for lentiviral packaging proportions and the relationship between the viral titer and the sizes of the Luc and eGFP being packaged. It was also noteworthy that the human embryonic kidney cell line 293A showed a higher infection efficiency with SARS-CoV-2 S pseudovirions.
We also find that if the SFL PVs with M, E, and N proteins with helper packaging, they can successfully infect 293 T-ACE2 (Fig. S1). The M protein is responsible for directing most of the protein interactions required for the assembly of SARS-CoV-2. However, the M protein itself cannot form viral particles; when M and E proteins are co-expressed, virus-like particles (VLPs) are formed to produce the envelope of SARS-CoV-2. The cytoplasmic tails of some coronavirus S proteins contain an endoplasmic reticulum retrieval signal (ERRS) that can retrieve S proteins from the Golgi to the endoplasmic reticulum; this process is thought to accumulate S proteins at the CoV budding site, the ER-Golgi intermediate compartment (ERGIC), and to facilitate S protein incorporation into VLPs (Ujike et al., 2016, Nguyen Dzung and Hildreth James, 2000). The key cell entry mechanisms need to be further understood. This packaging protocol will help to study the assembly of SARS-CoV-2 virus particles and the interaction in these structural proteins. Moreover, taking into account that some SARS-CoV-2 S molecules were cleaved during PV packaging, we used the total amount of uncleaved and cleaved S molecules to calibrate SARS-CoV-2 PV entry.
We sought to determine the influence of related mutations on the PV packaging system for SARS-CoV-2 entry. A striking difference of the S protein of SARS-CoV-2 has an FCS that did not exist in SARS-CoV at its S1/S2 boundary encoding four amino acid residues “RRAR” (Coutard et al., 2020). It has been proven by Markus Hoffmann, an infection biologist at the German Primate Center, to cause the cleavage of the SARS-CoV-2 S protein and is a necessary step for the virus to enter the lung cells (Hoffmann et al., 2020). There are also some studies showing that the host cell protease furin can cleave the SARS-CoV-2 S protein at the S1/S2 site, but this is mainly based on the use of indirect furin inhibitor experiments (Shang et al., 2020, Xia et al., 2020). It has been reported that formation-specific furin inhibitors can suppress SARS-CoV-2 S-mediated syncytium formation and block SARS-CoV-2 entry and virus replication which are potential antiviral agents for SARS-CoV-2 infection and pathogenesis (Cheng et al., 2020). Many infections with coronavirus syncytium formation have been found to be associated with increased invasiveness (Park et al., 2016). We think that the full-length S protein structure is not conducive to the expression The results of plasmids encoding SFL, Sdel18, SG614 and SFko transient transfection 293 T to express show that the full-length S protein is not conducive to expression, while the expression of other mutants is significantly higher and there is no significant difference among them (Fig. 2A). Through the packaging of the SFko PVs, it was found that the deletion of the “RRAR” FCS can significantly enhance the infection efficiency of the S protein, such as Vero, Huh7, and other cells. However, the infection efficiency of Calu-3 cells was reduced but remains to be further studied. We initially discovered that FCS might be essential for the activation of SARS-CoV-2 S in human airway epithelial cells. We also found that FCS plays an important role in coronavirus S protein-mediated cell-cell fusion. While FCS will reduce the large lack of S protein-mediated cell fusion, it does not stop it (Buchrieser et al., 2020, Follis et al., 2006). S must be processed at the S1/S2 border to mediate cell-cell fusion and then the potential cleavage sites within the S2 subunit alter S processing at the S1/S2 border, mediating cell-cell fusion (Barrett et al., 2021).
Meanwhile, to detect the cell entry mechanisms of SARS-CoV-2 S on the surface of PVs, we evaluated their infectivity by performing Western blotting on the PVs. SG614 showed a decrease in S1/S2 cleaved shedding and a lower level of S protein in the virion. The S: S1/S2 cleaved ratio by western blot monitoring was markedly greater in Sdel18 compared to SG614, indicating that glycine at SG614 stabilizes the interaction between the S1 and S2 domains, limiting S1/S2 cleaved shedding (Yurkovetskiy et al., 2020). There was no S1/S2 cleaved shedding and increasing total S protein in SFko that the mutations show to enhance virus infection through related mechanisms that increase total S protein and limit S1/S2 cleaved shedding when incorporated into the virion (Zhang et al., 2020).
The fusogenic capacity of SFL, Sdel18, SG614, and SFko via an S-mediated cell-cell fusion assay in the absence of trypsin protease (Xia et al., 2020). We provide evidence that only S1/S2 cleavage occurs during packing biosynthesis of S, and deletion of the polybasic site in the S protein reduces syncytia formation in cell culture. These results suggest that FCS may play a role in SARS-CoV-2 S-mediated membrane fusion. Notably, this enhanced infectivity of SFko is not an artifact of titer normalization, as virus titers were determined in the supernatant, which was very similar to the similar packing condition. However, after binding the receptor in host cells, a nearby host protease cleaves S, which releases the fusion peptide, facilitating virus entry mechanisms that should be further explored. SARS-CoV-2 S protein entry into susceptible cells is a complex process that requires the concerted action of receptor-binding and proteolytic processing of the S protein to promote virus-cell fusion syncytium formation.
There is a limit that lentivirus-based SARS-CoV-2 pseudotyped viruses have a relatively low pseudoviral titer for which the relative luciferase units (RLU) is ∼ 106 and be applied only to hACE2-overexpressing cell lines (Crawford et al., 2020), while they cannot efficiently infect both Huh-7 and Vero cells, which were used to isolate live virus (Nie et al., 2020). We contribute to solving this problem with FCS deletion to enhance infection.
Taken together, we successfully built a robust pseudovirus packaging system to produce infectious SARS-CoV-2 PVs. In this PV system, we established FCS mutations, which are conducive to virus packaging and infection. This methodology is only suitable for virus neutralization, virus invasion cell mechanism research, and anti-spike inhibitor screening, but we do not know whether this system can simulate real viruses or assess their infectivity.
5. Conclusion
In conclusion, this study constructed a furin site deletion HIV-1 PVs evaluation system in a preliminary in vitro. The furin cleavage site deletion of the S protein can help SARS-CoV-2 S to be cleaved during viral packaging to improve infection efficiency. As a widely used pseudovirus neutralizing system, it can provide strong support for the production of relatively high-titer SARS-COV-2 pseudo-particles that may be suitable for the detection of neutralizing antibodies from COVID-19 patients.
CRediT authorship contribution statement
Zeng Wang: Conceptualization, Data curation, Formal analysis, Methodology, Investigation, Writing – original draft. Kunhong Zhong: Data curation, Formal analysis, Methodology, Investigation, Writing – review and editing. Guoqing Wang: Data curation, Formal analysis, Methodology, Investigation, Writing – review and editing. Qizhong Lu: Resources, Data curation, Methodology, Software. Hexian Li: Resources, Data curation, Methodology. Zhiguo Wu: Resources, Data curation, Methodology. Zongliang Zhang: Resources, Data curation, Methodology. Nian Yang: Resources, Data curation, Methodology. Meijun Zheng: Resources, Data curation, Methodology. Yuelong Wang: Funding acquisition. Chunlai Nie: Funding acquisition. Liangxue Zhou: Supervision, Funding acquisition, Writing – review and editing. Aiping Tong: Supervision, Funding acquisition, Writing – review and editing. All authors have read and approved the final manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
This study was supported by the National Natural Science Foundation of China (82073404 & 81773188 & 82002648), the Major Subject of the Science and Technology Department of Sichuan Province (2019YFS0110), the 1.3.5 project for disciplines of excellence, West China Hospital, Sichuan University (ZYJC18007) and the special research fund on COVID-19 of West China Hospital Sichuan University (HX-2019-nCoV-064 & HX-2019-nCoV-058).
Ethics approval
This study was approved by the Medical Ethics Committee of Hospital of West China Hospital of Sichuan University Biomedical Ethics Committee (20210535A) and was performed according to the Institutional Guidelines.
Edited by: Shitao Li
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gene.2022.147144.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., Chen H.-D., Chen J., Luo Y., Guo H., Jiang R.-D., Liu M.-Q., Chen Y., Shen X.-R., Wang X.i., Zheng X.-S., Zhao K., Chen Q.-J., Deng F., Liu L.-L., Yan B., Zhan F.-X., Wang Y.-Y., Xiao G.-F., Shi Z.-L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A., Park Y., Tortorici M., Wall A., McGuire A., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Lee J., Yang J., Kim J., Kim V., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78(4):779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millet J., Whittaker G. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12(1) doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen K., Rambaut A., Lipkin W., Holmes E., Garry R. The proximal origin of SARS-CoV-2. Nat. Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science (New York, N.Y.) 2020;367(6483):1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luczo J., Stambas J., Durr P., Michalski W., Bingham J. Molecular pathogenesis of H5 highly pathogenic avian influenza: the role of the haemagglutinin cleavage site motif. Rev. Med. Virol. 2015;25(6):406–430. doi: 10.1002/rmv.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu H., Chan J.-W., Yuen T.-T., Shuai H., Yuan S., Wang Y., Hu B., Yip C.-Y., Tsang J.-L., Huang X., Chai Y., Yang D., Hou Y., Chik K.-H., Zhang X.i., Fung A.-F., Tsoi H.-W., Cai J.-P., Chan W.-M., Ip J.D., Chu A.-H., Zhou J., Lung D.C., Kok K.-H., To K.-W., Tsang O.-Y., Chan K.-H., Yuen K.-Y. Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: an observational study. Lancet Microbe. 2020;1(1):e14–e23. doi: 10.1016/S2666-5247(20)30004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belouzard S., Chu V., Whittaker G. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. PNAS. 2009;106(14):5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperton N., Chan P., Simmons G., Zambon M., Tedder R., Takeuchi Y., et al. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg. Infect. Dis. 2005;11(3):411–416. doi: 10.3201/eid1103.040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grehan K., Ferrara F., Temperton N. An optimised method for the production of MERS-CoV spike expressing viral pseudotypes. MethodsX. 2015;2:379–384. doi: 10.1016/j.mex.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L.i., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q.i., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11(1) doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto, D., Park, Y., Beltramello, M., Walls, A., Tortorici, M., Bianchi, S., et al. Structural and functional analysis of a potent sarbecovirus neutralizing antibody. bioRxiv: Preprint Server Biol. 2020. doi: 10.1101/2020.04.07.023903.
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020;5(4):562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong H.-L., Wu Y.-T., Cao J.-L., Yang R., Liu Y.-X., Ma J., Qiao X.-Y., Yao X.-Y., Zhang B.-H., Zhang Y.-L., Hou W.-H., Shi Y., Xu J.-J., Zhang L., Wang S.-J., Fu B.-R., Yang T., Ge S.-X., Zhang J., Yuan Q., Huang B.-Y., Li Z.-Y., Zhang T.-Y., Xia N.-S. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerging Microbes Infect. 2020;9(1):2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z., Huang Q., Xu L., Tang J., Tian Q., Yao W., Hu L., Yan X., Zhou X., Wu Y., Deng K., Zhang Z., Qian Z., Chen Y., Ye L. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020;17(6):647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., Bao L., Mo F., Li X., Huang Y., Hong W., Yang Y., Zhao Y., Ye F., Lin S., Deng W., Chen H., Lei H., Zhang Z., Luo M., Gao H., Zheng Y., Gong Y., Jiang X., Xu Y., Lv Q.i., Li D., Wang M., Li F., Wang S., Wang G., Yu P., Qu Y., Yang L.i., Deng H., Tong A., Li J., Wang Z., Yang J., Shen G., Zhao Z., Li Y., Luo J., Liu H., Yu W., Yang M., Xu J., Wang J., Li H., Wang H., Kuang D., Lin P., Hu Z., Guo W., Cheng W., He Y., Song X., Chen C., Xue Z., Yao S., Chen L.u., Ma X., Chen S., Gou M., Huang W., Wang Y., Fan C., Tian Z., Shi M., Wang F.-S., Dai L., Wu M., Li G., Wang G., Peng Y., Qian Z., Huang C., Lau J.-N., Yang Z., Wei Y., Cen X., Peng X., Qin C., Zhang K., Lu G., Wei X. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586(7830):572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- Johnson M.C., Lyddon T.D., Suarez R., Salcedo B., LePique M., Graham M., Ricana C., Robinson C., Ritter D.G., Simon V. Optimized pseudotyping conditions for the SARS-COV-2 spike glycoprotein. J. Virol. 2020;94(21) doi: 10.1128/JVI.01062-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, P.Y., Plante, J., Liu, Y., Liu, J., Xia, H., Johnson, B., et al. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. Res Sq. 2020. doi: 10.21203/rs.3.rs-70482/v1.
- Yang P., Yang Y., Wu Y., Huang C., Ding Y., Wang X., Wang S. An optimized and robust SARS-CoV-2 pseudovirus system for viral entry research. J. Virol. Methods. 2021;295:114221. doi: 10.1016/j.jviromet.2021.114221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami T., Freed E. Genetic evidence for an interaction between human immunodeficiency virus type 1 matrix and alpha-helix 2 of the gp41 cytoplasmic tail. J. Virol. 2000;74(8):3548–3554. doi: 10.1128/jvi.74.8.3548-3554.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Liu L., Wang C., Wang Z., Duan X., Chen G., Zhou H., Shao H. Establishment of a pseudovirus neutralization assay based on SARS-CoV-2 S protein incorporated into lentiviral particles. Biosafety Health. 2022;4(1):38–44. doi: 10.1016/j.bsheal.2021.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Gao Q., He C., Huang A., Tang N., Wang K. Development of cell-based pseudovirus entry assay to identify potential viral entry inhibitors and neutralizing antibodies against SARS-CoV-2. Genes Diseases. 2020;7(4):551–557. doi: 10.1016/j.gendis.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., Pettie D., King N.P., Balazs A.B., Bloom J.D. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12(5):513. doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujike M., Huang C., Shirato K., Makino S., Taguchi F. The contribution of the cytoplasmic retrieval signal of severe acute respiratory syndrome coronavirus to intracellular accumulation of S proteins and incorporation of S protein into virus-like particles. J. Gen. Virol. 2016;97(8):1853–1864. doi: 10.1099/jgv.0.000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen Dzung H., Hildreth James E.K. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 2000;74(7):3264–3272. doi: 10.1128/JVI.74.7.3264-3272.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117(21):11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct. Target. Ther. 2020;5(1) doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y.-W., Chao T.-L., Li C.-L., Chiu M.-F., Kao H.-C., Wang S.-H., Pang Y.-H., Lin C.-H., Tsai Y.-M., Lee W.-H., Tao M.-H., Ho T.-C., Wu P.-Y., Jang L.-T., Chen P.-J., Chang S.-Y., Yeh S.-H. Furin inhibitors block SARS-CoV-2 spike protein cleavage to suppress virus production and cytopathic effects. Cell Rep. 2020;33(2):108254. doi: 10.1016/j.celrep.2020.108254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.-E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Gallagher T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. PNAS. 2016;113(43):12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchrieser J., Dufloo J., Hubert M., Monel B., Planas D., Rajah M.M., Planchais C., Porrot F., Guivel‐Benhassine F., Van der Werf S., Casartelli N., Mouquet H., Bruel T., Schwartz O. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020;39(23):e106267. doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Follis K., York J., Nunberg J. Furin cleavage of the SARS coronavirus spike glycoprotein enhances cell-cell fusion but does not affect virion entry. Virology. 2006;350(2):358–369. doi: 10.1016/j.virol.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett C.T., Neal H.E., Edmonds K., Moncman C.L., Thompson R., Branttie J.M., Boggs K.B., Wu C.-Y., Leung D.W., Dutch R.E. Effect of clinical isolate or cleavage site mutations in the SARS-CoV-2 spike protein on protein stability, cleavage, and cell-cell fusion. J. Biol. Chem. 2021;297(1):100902. doi: 10.1016/j.jbc.2021.100902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yurkovetskiy L., Wang X., Pascal K.E., Tomkins-Tinch C., Nyalile T.P., Wang Y., Baum A., Diehl W.E., Dauphin A., Carbone C., Veinotte K., Egri S.B., Schaffner S.F., Lemieux J.E., Munro J.B., Rafique A., Barve A., Sabeti P.C., Kyratsous C.A., Dudkina N.V., Shen K., Luban J. Structural and functional analysis of the D614G SARS-CoV-2 spike protein variant. Cell. 2020;183(3):739–751.e8. doi: 10.1016/j.cell.2020.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Jackson, C., Mou, H., Ojha, A., Rangarajan, E., Izard, T., et al., 2020. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv: Preprint Server Biol. doi: 10.1101/2020.06.12.148726.
- Crawford, K., Eguia, R., Dingens, A., Loes, A., Malone, K., Wolf, C., et al., 2020. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12(5). doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed]
- Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., Zhang L.i., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Fan C., Huang W., Xu M., Wang Y. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020;15(11):3699–3715. doi: 10.1038/s41596-020-0394-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.