Abstract
Background
The immunological changes underpinning acquisition of remission (also called sustained unresponsiveness) following food immunotherapy remain poorly defined. Limited access to effective therapies and biosamples from treatment responders has prevented progress. Probiotic peanut oral immunotherapy is highly effective at inducing remission, providing an opportunity to investigate immune changes.
Methods
Using a systems biology approach, we examined gene co‐expression network patterns in peanut‐specific CD4+ T cell responses before and after probiotic and peanut oral immunotherapy in subjects enrolled in the PPOIT‐001 randomized trial: Responders who attained remission (n = 16), placebo‐treated who remained allergic (n = 16).
Results
Acquisition of remission was associated with rewiring of gene network patterns, which was characterized by integration of T helper 2 and interferon signalling modules, markedly reduced T helper 2 gene connectivity, and shutdown in co‐expression activity between T helper 2 effectors and cell cycle regulators.
Conclusion
The immunological changes underlying remission following peanut oral immunotherapy are mediated by reprogramming of T helper 2‐associated gene networks in the CD4+ T cell compartment. Findings provide insight into immune mechanisms driving the acquisition of remission following oral immunotherapy, paving the way for the development of improved approaches to induce remission/sustained unresponsiveness in patients with food allergy.
Keywords: CD4+ T cells, food oral immunotherapy, network analysis, remission of allergy, sustained unresponsiveness
We examined longitudinal changes in gene co‐expression networks in peanut‐specific CD4+ T cells from participants with remission following oral immunotherapy (OIT) (n = 16) and placebo‐treated who remained allergic (n = 16). Remission was associated with emergence of a regulatory module dominated by type 1 interferon signalling and collapse of the allergic T helper 2 module, with Th2 genes incorporated within the regulatory module. Findings provide insight into immune mechanisms driving the acquisition of remission following oral immunotherapy.Abbreviations: CCNB, cyclin B; CDK1, cyclin dependent kinase 1; CEP55, centrosomal protein 55; DC, dendritic cell; IFN, interferon; IL, interleukin; PBMC, peripheral blood mononuclear cell; PPOIT, probiotic + peanut oral immunotherapy; Th2, T helper 2

1. INTRODUCTION
Food allergy is a global public health concern, affecting 10% of infants and 5%–8% of children. 1 , 2 There is currently no cure and management relies on avoidance of the eliciting food. The lifestyle restrictions imposed by allergen avoidance and unpredictability of accidental ingestion/reaction result in severely reduced quality of life and psychological distress for both patients and their families. A therapy that induces remission of allergy, also referred to as sustained unresponsiveness (SU), can improve quality of life for patients and prevent deaths. Understanding the immune mechanisms underpinning remission/SU, and particularly long‐lasting remission/SU, will facilitate the development of effective therapies that deliver long‐term solutions for patients.
Efforts to retrain the immune system from allergy towards tolerance have led to interest in oral immunotherapy (OIT) for the treatment of food allergy. Peanut OIT is highly effective at inducing desensitization and induces remission/SU in a limited subset (~30%) of treated patients. 3 , 4 Desensitization during OIT results in transient changes in immunoglobulin (Ig) E and IgG4 and basophil activation, which often regress after treatment cessation or even during ongoing maintenance dosing. 5 Changes in the allergen‐specific T cell response on the contrary are considered central to achieving lasting redirection of the allergic state to remission/SU. Th2 cells are essential for the generation of allergen‐specific IgE and the development of food allergy, 6 , 7 and remission/SU following OIT was associated with transition of Th2 subsets towards an anergic phenotype, 8 a decrease in pro‐allergic Th2A cells 9 and in some cases an induction of T regulatory (Treg) cells. 10 , 11 Nevertheless, the key immune factors driving these changes remain unknown.
Previous research into the immune mechanisms supporting acquisition of remission/SU has been hampered by limited access to patient cohorts that have successfully transitioned from allergy to remission/SU. Furthermore, previous mechanistic studies have examined selected genes or pathways which generates an oversimplified and incomplete picture of the underlying mechanisms. Because immune responses are determined by networks of proteins/genes working together in a coordinated fashion to mediate healthy or allergic responses, a more comprehensive investigation of the complex immune processes and their coordinated interrelationships that critically determine acquisition of remission/SU is expected to provide greater insight into key drivers of treatment success. 12 We have previously shown that a combination treatment of Probiotic and Peanut Oral Immunotherapy (PPOIT) is highly effective at inducing remission/SU that persisted to 4‐year post‐treatment, 13 providing a unique opportunity to investigate immune changes driving the shift from allergy to remission/SU. Here, we apply a systems biology approach to characterize the evolution of gene co‐expression patterns in CD4+ T cells from peanut‐allergic participants enrolled in the PPOIT‐001 randomized trial who transition from allergy to remission/SU following immunotherapy.
2. METHODS
2.1. PPOIT‐001 randomized trial design
Peanut‐allergic children (n = 62) aged 1–10 years were randomized 1:1 stratified by age (≤5, >5 years) and peanut skin prick test wheal size (≤10, >10 mm), to receive Probiotic Peanut Oral Immunotherapy (PPOIT) or placebo for 18 months. Remission/SU was defined as passing a double‐blind placebo‐controlled food challenge (DBPCFC; cumulative 3950 mg peanut protein) performed 2–6‐week post‐treatment; Allergy was defined as failing a DBPCFC at end‐of‐treatment. Ethics approval was attained from the Royal Children's Hospital, Melbourne, Australia (HREC 27086), and informed written consent was obtained from a parent/guardian.
2.2. Immunotherapy protocol
PPOIT comprised a daily dose of Lactobacillus rhamnosus CGMCC 1.3724 (2 × 1010 cfu) and peanut oral immunotherapy (2000 mg peanut protein maintenance) administered for 18 months.
2.3. Study samples and PBMC isolation
Blood samples from PPOIT‐responders who attained remission/SU (n = 16) and placebo‐treated subjects who remained allergic (n = 16), collected at study entry (T0), end‐of‐treatment (T1) and 3‐month post‐treatment (T3) were used (Figure 1). PBMC were isolated by density gradient centrifugation and cryopreserved. Briefly, 10 ml of fresh blood collected into sodium heparin vacutainer tubes (BD) were processed within 2 h. Plasma was removed (centrifuged 700 g 10 min) and the sample topped up to 10 ml with Transport medium (10 IU/ml preservative‐free heparin in RPMI‐1640 Medium). PBMC were isolated by density gradient centrifugation (400 g , 30 min, brake off). PBMCs were washed with RPMI (Thermo Fisher Scientific) containing 2% (v/v) heat‐inactivated fetal calf serum (HI‐FCS) and cryopreserved in 1 ml RPMI+HI‐FCS + 1 ml HI‐FCS containing 15% (v/v) dimethyl sulfoxide (DMSO), at a maximum density of 1 × 107/ml.
FIGURE 1.
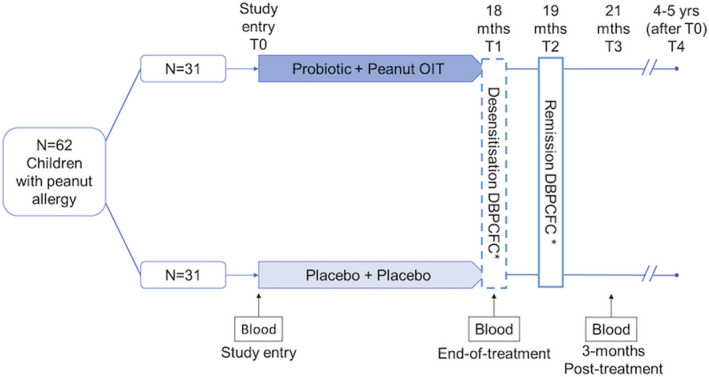
PPOIT‐001 randomized trial design. *DBPCFC, double‐blind placebo‐controlled food challenge. n = 62 children aged 1–10 years with peanut allergy were randomized 1:1 to receive probiotic and peanut oral immunotherapy (PPOIT) or placebo for 18 months. A double‐blind placebo‐controlled food challenge (DBPCFC; cumulative 4 g peanut protein) was performed at end‐of‐treatment (T1) and at 2–6‐week post‐treatment (T2). Subjects who failed the T1 DBPCFC were classified as allergic. Subjects who passed both the T1 and T2 DBPCFC were classified as having attained remission/SU. Blood samples were collected at various times. This study included n = 16 PPOIT‐treated children who attained remission/SU and n = 16 placebo‐treated children who remained allergic. PBMC collected from these subjects at baseline (T0), end‐of‐treatment (T1), and 3‐month post‐treatment (T3) underwent transcriptomic profiling
2.4. In vitro cell cultures and CD4+ T cell isolation
Cryopreserved PBMC were thawed and cultured for 48 h in the presence of endotoxin‐free crude peanut extract (CPE, 200 µg/ml) 14 or AIM‐V media (MED) alone. The CPE preparation was treated using the Pierce High Capacity Endotoxin Removal Spin Columns (Thermo Fisher Scientific). Cultured CD4+ T cells were isolated by positive selection using Dynabeads CD4 Positive Isolation Kit (ThermoFisher Scientific) and stored in TRIzol (ThermoFisher Scientific) until processed. Isolated CD4+ cells were analyzed for purity and shown to comprise >98% CD3+CD4+ T cells, with <1% CD14+ monocytes. Findings for CD4+ cells are therefore assumed to reflect changes in T cell transcriptional profiles. Total RNA was extracted using RNeasy Mini Kit (Qiagen) according to manufacturer's instructions. Quantity and quality of RNA was assessed by TapeStation (Agilent) and Qubit (RIN 8‐10). Gene expression profiling of CD4+ T cell total RNA was performed by the Ramaciotti Centre for Genomics (The University of New South Wales, Sydney NSW Australia) using Human Genome U219 arrays (Affymetrix).
2.5. Gene expression profiling of CD4+ cells
Raw microarray data were analyzed using open‐source statistical programming language R. A custom chip description file was used to annotate probe sets (hgu219hsentrezgcdf, version 24). Quality control of arrays was carried out with ArrayQualityMetrics, 15 and 5 probe sets were removed based on QC plots. Probe sets were background corrected, quantile normalized and summarized employing the robust multi‐array algorithm (RMA) 16 and filtered using the proportion of variation accounted for by the first principal component (PVAC > 4), 17 resulting in 3673 probe sets. Unwanted variation due to array processing was removed using ComBat 18 prior to downstream expression, network and gene pair correlation analysis.
2.6. Differential gene expression analysis of peanut‐specific CD4+ T cell responses
Differentially expressed genes associated with peanut stimulation were identified using a paired study design with limma and adjusted for multiple testing using Benjamini–Hochberg. In the limma model, participant ID was used as a covariate, as well as a group covariate categorizing by outcome group (Remission/SU or allergic), timepoint (T0 or T1 or T3) and cell culture condition (CPE or unstimulated/Media). The contrast matrix compared stimulated (CPE) to unstimulated (Media) cells, within each outcome group (remission/SU or allergic) at each timepoint. An adjusted p < 0.05 and an absolute log‐fold change >0.1 were deemed significant. Peanut‐specific responses were compared with media control at each time point (T0/T1/T3) in subjects with remission/SU and allergic controls.
2.7. Gene co‐expression network analysis
Gene co‐expression networks were constructed independently for subjects with remission/SU and allergy using weighted gene co‐expression network analysis (WGCNA). 19 Signed networks were constructed with the following parameters: SoftPower = 10, minimum module size = 20, deepsplit = 1 and a merge height = 0.1. The overall expression of network modules was summarized employing module eigengenes, representing the first principal component. A Wilcoxon signed‐rank test was utilized to test for differences in eigengene values comparing CPE stimulated versus Media control in each group, respectively.
To visualize the network wiring diagram, pairwise weighted gene‐gene correlations were exported from the adjacency matrix and reconstructed in Cytoscape 20 (version 3.6.1). The top 800 highest ranking edges (gene–gene connections) were imported, and the Cytoscape NetworkAnalyzer tool was employed with the following parameters: undirected edges, organic layout and map node size to degree: low node values to small size.
2.8. Differential gene correlation analysis
To systematically assess gene regulatory relationships across time points (changes between gene co‐expression pairs), differential gene correlation analysis (DGCA) 21 was employed across multiple conditions (remission/SU and allergic) and time points (T0/T1/T3). Pearson correlations, delta Z‐scores and delta Benjamini–Hochberg adjusted p‐values were computed.
2.9. Pathway analysis
Functional pathway analysis of network modules was carried out with Gene Ontology enrichment with clusterProfiler 22 and InnateDB pathway analysis 23 with Benjamini–Hochberg corrected p‐values. InnateDB pathway analysis uses data from 6 ontology databases: REACTOME, KEGG, PID NCI, NETPATH, PID BIOCARTA and INOH.
2.10. Upstream regulator analysis
Ingenuity Systems upstream regulator analysis 24 was employed to identify putative upstream molecular drivers of the network modules. Regulators with Benjamini & Hochberg corrected p ≤ 0.05 and absolute z‐scores > 2.0 were deemed significant.
3. RESULTS
3.1. Study population
This study profiled biospecimens from children aged 1–10 years in the PPOIT‐001 randomized trial. Following 18 months of treatment, 74% of PPOIT‐treated subjects achieved remission/SU compared with 3% of placebo‐treated patients. A 4‐year follow‐on study of the parent trial demonstrated that 70% of subjects with 2–6‐week SU at the end‐of‐treatment had persistent 8‐week SU at 4‐year post‐treatment. 13 N = 16 PPOIT‐treated patients who achieved remission/SU and n = 16 placebo‐treated patients who remained allergic were included in this study. Eight subjects examined in the remission group had long‐term challenge test to assess for persistent SU and 6 of the 8 were confirmed to have persistent remission, in line with the rate of persistent SU reported previously. Baseline characteristics of subjects included in this study are presented in Table 1. There were no differences in baseline characteristics between remission/SU and allergic groups.
TABLE 1.
Demographic characteristics at study entry
|
Remission/SU n = 16 |
Allergic n = 16 |
|
|---|---|---|
| Age (y) | ||
| Median (IQR) | 5.8 (4.2–8.7) | 6.3 (3.8–7.9) |
| Min–max | 2.1–9.5 | 2.1–10 |
| Weight (kg) | ||
| Mean (SD) | 23.9 (9.3) | 24.6 (10.2) |
| Male sex | ||
| n (%) | 12 (75%) | 11 (68.8%) |
| History of doctor diagnosed eczema (ever) | ||
| n (%) | 12 (75%) | 12 (75%) |
| If ever eczema, medication for eczema in last 12 months | ||
| n (% of ever eczema) | 9 (75%) | 10 (83.3%) |
| History of doctor‐diagnosed asthma (ever) | ||
| n (%) | 7 (43.8%) | 8 (50%) |
| If ever asthma, medication for asthma in last 12 months | ||
| n (% of asthma ever) | 6 (85.7%) | 8 (100%) |
| History of anaphylaxis to peanut* | ||
| n (%) | 5 (31.2%) | 6 (37.5%) |
| Peanut‐specific SPT wheal size (mm) | ||
| Mean (SD) | 16.6 (5.4) | 17.6 (7.3) |
| sIgE (kU/L) | ||
| Median (IQR) | 4.7 (0.9–56.5) | 11.7 (2.8–62) |
Abbreviations: IQR, interquartile range; SD, standard deviation; kU/L, kilo unit per liter.
*One missing in the placebo group.
3.2. Genomic profiling of peanut‐specific CD4+ T cell responses
Peripheral blood mononuclear cells (PBMC) were cultured in the presence or absence of CPE for 48 h, and gene expression was profiled using microarrays in CD4+ T cells isolated by positive immunomagnetic separation. Gene expression levels of endotoxin‐free crude peanut extract (CPE, 200 µg/ml) stimulated and unstimulated CD4+ T cells were compared to identify differentially expressed genes (DEGs). Prior to allergen immunotherapy (T0), there were 667 DEGs (205/462 upregulated/downregulated) in remission/SU subjects and 677 DEGs (214/463 upregulated/downregulated) in allergic subjects (Figure 2A). The DEGs had small log‐fold changes (LFC range: −0.91 to +0.53). The expression patterns of the top 50 upregulated/downregulated genes are illustrated in Figure S1 and Tables [Link], [Link] in the Online Repository. As expected, highly ranked DEGs were similar in both groups and included cardinal Th2 effectors (IL4, IL5, IL9 and IL13; Figure 2B), Th1 effectors (IFNG) and other cytokines (IL2, IL3, CSF2 and IL17F).
FIGURE 2.
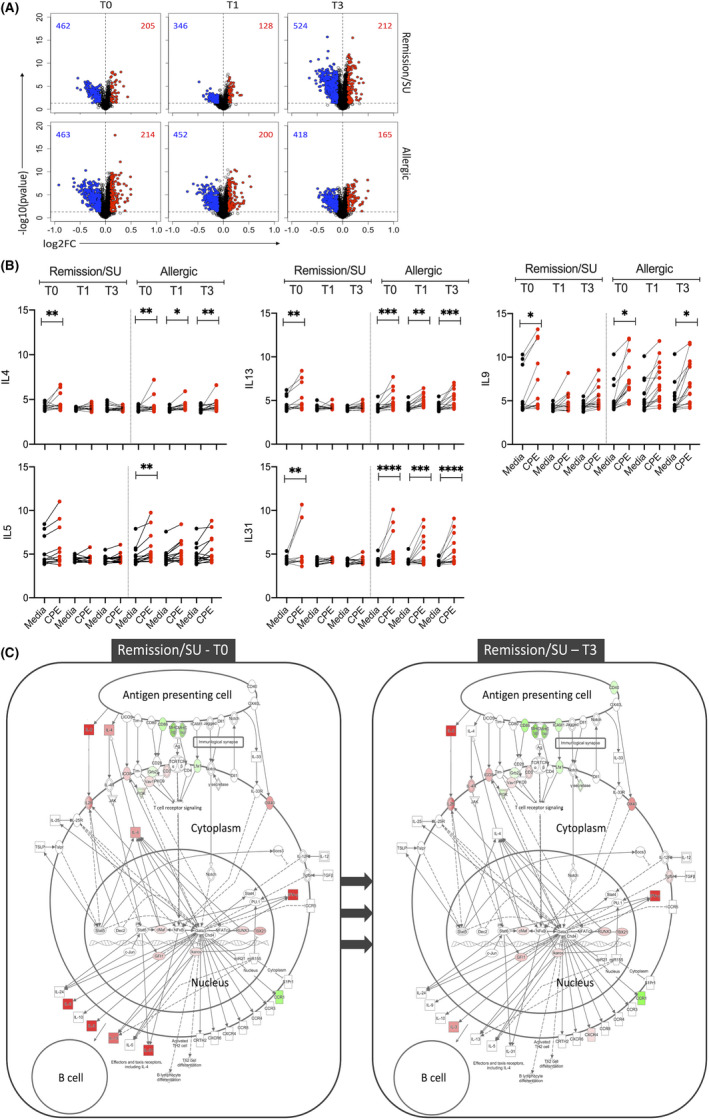
Differential expression analysis showing peanut‐induced gene expression patterns. Differentially expressed genes (DEG) were identified with limma, employing an FDR < 0.05 and absolute log‐fold change >0.1, at T0 (study entry), T1 (end‐of‐treatment) and T3 (3‐month post‐treatment) in remission/SU and allergic groups. (A) Volcano plots of peanut‐induced DEG in remission/sustained unresponsiveness (SU) and allergic, (B) Gene expression levels of cardinal Th2 genes (IL4, IL5, IL9, IL13 and IL31) with and without peanut stimulation (CPE/Media). Significance values were obtained from limma analysis comparing CPE stimulated versus Media control, adjusted *p < 0.05, **<0.01, ***<0.001, ****<0.0001, (C) Th2 DEG were projected onto the canonical Th2 pathway employing Ingenuity pathways diagram. The data show the cellular context of where the Th2 genes operate in remission/SU at T0/T3. Th2 genes were active prior to treatment (T0) and no longer differentially expressed at T3. Red colour: upregulated and green: downregulated comparing CPE versus media control
At end‐of‐treatment (T1), the number of DEGs in the remission/SU group reduced to 474 (128/346 upregulated/downregulated) whereas DEGs in the allergic group remained largely unchanged (652 DEGs; 200/452 upregulated/downregulated) (Figure 2A). Notably, at end‐of‐treatment (T1), differential expression of Th2 cytokines IL4, IL5, IL9, IL13 and IL31 was lost in the remission/SU subjects, whereas Th2 cytokines remained upregulated in allergic subjects (Figure 2B).
At 3‐month post‐treatment (T3), there were 736 DEGs in the remission/SU subjects (212/524 upregulated/downregulated) and 583 DEGs in allergic subjects (165/418 upregulated/downregulated) (Figure 2A). Similar to findings at end‐of‐treatment, Th2 cytokines (IL4, IL9, IL13, IL31, but not IL5) remained upregulated only in allergic subjects (Figure 2B). Although there appeared to be an increase in peanut‐stimulated DEG at T3 compared with T1 in the remission/SU group, formal statistical analysis comparing responses between T1 and T3 resulted in zero DEGs between the two visits. We conducted pathway analysis of unique DEG at T3 compared with T1 and found that the upregulated pathways at T3 were associated with cytokine‐cytokine receptor interactions/JAK‐STAT signalling (p < 2.4E‐03) and cell cycle (p < 2.8E‐06), and downregulated pathways were related to lysosome/phagosome (p < 4.6E‐07) and TLR signalling (p = 8.7E‐06).
We then projected the DEGs onto the canonical Th2 pathway employing Ingenuity to visualize the cellular context (Figure 2C) in the Th2 pathway and overlaid the gene expression levels from T0 to T3 in remission/SU. Th2 genes were upregulated prior to treatment (T0) and no longer differentially expressed at T3 in remission/SU. Despite the observed shifts in Th2 gene expression levels for remission/SU subjects compared with allergic subjects (described above), these differences were not statistically significant in between‐group comparisons at any time point (data not shown).
3.3. Gene co‐expression network patterns in CD4+ T cells are rewired during the transition from allergy to remission/SU
We have previously demonstrated in house dust mite (HDM)‐allergic patients that gene network patterns underlying CD4+ T cell responses to the HDM are rewired during HDM immunotherapy, 25 and we reasoned that a similar process may occur with transition from allergy to remission/SU following PPOIT treatment in peanut‐allergic subjects. To investigate this, we constructed separate weighted gene co‐expression networks for remission/SU and allergic subjects at each time point. At T0, the co‐expression networks comprised 10 modules for the remission/SU group and 7 modules for the allergic group. Gene Ontology enrichment 22 and pathway analyses demonstrated that the modules were enriched for biological pathways (Figure S2). We computed ranked expression levels and ranked network connectivity and then compared findings at T0 with subsequent time points in the remission/SU and allergic groups (Figure 3A). The data showed that ranked expression was tightly correlated in both groups, whereas ranked network connectivity was highly variable between groups, demonstrating that PPOIT‐induced changes in network connectivity (i.e. correlation patterns between genes) were much more striking compared with changes in gene expression levels.
FIGURE 3.
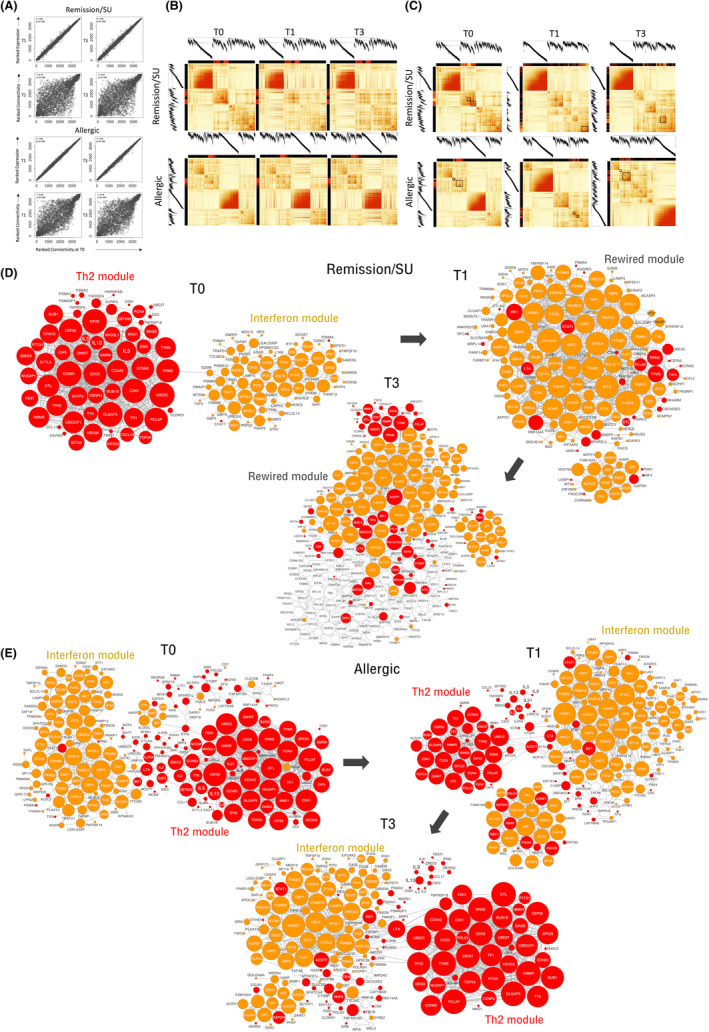
Correlation of CD4+ T cell gene networks at T0/T1 and T0/T3 in subjects who develop remission/SU and subjects who remain allergic. Co‐expression networks were constructed separately for subjects with remission/SU and allergic subjects. (A) Ranked expression levels and ranked connectivity in the remission/SU (upper panel) and allergic groups (lower panel), presented as correlation plots at T0/T1 and T0/T3, (B) Network correlation patterns at T1 and T3 were superimposed over the network structure from T0 in remission/SU and allergic subjects. The Th2‐associated module is highlighted with a red colour band, (C) network connectivity patterns in remission/SU highlighting Th2 (red colour bands) and IFN modules (yellow colour bands) showing discrete bands at T0 and merged/integrated bands at T1 and T3 (black box), network connectivity patterns in allergic subjects are shown in the second panel below, network wiring were reconstructed employing WGCNA and visualized in Cytoscape in (D) remission/SU and (E) allergic subjects; The top 800 edges/pairwise connections were used for network reconstruction. The node size reflects the number of connections. Red colour denotes Th2 community genes, and gold colour denotes interferon community genes
We next sought to visualize changes in network topology over the course of treatment. First in Figure 3B, we plotted the network structure at T0 and superimposed the intensity of the correlation patterns at each time point over the network structure at T0. The data showed that the intensity of the correlation patterns for the Th2‐associated module (identified in red within the border of the heatmap) was markedly decreased over time in the remission/SU group (upper panel), but remained unchanged in the allergic group (lower panel). Second in Figure 3C, we plotted the network structure for each time point to visualize how the modules are reprogrammed by the therapy. The data showed that in the SU group, the Th2 and IFN gene networks comprised discrete modules at T0, but became merged and integrated at T1 and T3; whereas in the allergic group, the Th2 and IFN response genes remained separated/discrete modules at all time points (Figure 3C). To visualize the gene‐level changes that were rewired during the course of PPOIT, we reconstructed the top‐ranking network edges/connectivity patterns from WGCNA using Cytoscape, focussing on the Th2 and interferon associated modules pre‐immunotherapy/T0. The data showed that in the remission/SU group (Figure 3D), individual genes from the Th2 and IFN modules were not connected and formed distinct modules at T0 (when subjects were allergic); however, with transition to remission/SU, genes from the Th2 and IFN modules became interconnected and there was a shift from Th2 dominance to IFN dominance at the T1 timepoint. Moreover, extensive pairwise connections between Th2 and interferon module genes were retained at T3 in remission/SU subjects (Figure S3). In contrast, this rewiring between Th2 and IFN genes was not observed in allergic subjects and Th2 dominance was sustained at T1 and T3, as evidenced in the network architecture (Figure 3E). This correlated with parallel changes in immunological parameters; the remission/SU group demonstrated an overall reduction in peanut sIgE and SPT and increase in peanut sIgG4 at end‐of‐treatment (T1) and 3‐months after treatment cessation (T3), whereas the allergic individuals did not (Table S7), as published previously. 26
We next summarized the overall expression of each module employing principal component analysis and plotted the module eigengenes for each group over the course of therapy (Figure S4A,B). Responses to in vitro peanut stimulation at T0 in both remission/SU and allergic groups included upregulated modules associated with Th2/cell cycle, type 1 interferons, ribosomal translation, spliceosome and TFGB receptor, and one downregulated module related to lysosome/phagosome. At T1 and T3, glycolysis was upregulated in both groups in response to peanut stimulation.
We performed upstream regulator analysis to identify putative molecular drivers of the gene expression patterns at each timepoint, focussing on Th2/cell cycle and IFN‐associated network modules in remission/SU and allergic groups (Table 2). The data showed that at T0 in both remission/SU and allergic, the Th2/cell cycle module was driven by activation of T cell‐associated drivers (CD3, CD28, TCR and IL2), inflammatory (TNF, IL1B, NfkB complex and IL15), growth factors/cell cycle related (ERBB2, MYC and EGF) and Th2 (IL4 and IL5). The IFN module was driven by Th1 effector IFNG and type 1 interferons, as well as inflammatory (TNF and IL1B) and Th2 (IL5) drivers (Table 2). At T1 in remission/SU, the top‐ranking drivers of the rewired network Th2/IFN module were IFNG and type 1 interferons. In contrast in the allergic group, the drivers remained unchanged and the Th2 module was regulated by Th2 pathways.
TABLE 2.
Activation Z‐scores of the upstream drivers of the Th2/cell cycle and IFN‐associated modules at T0/T1/T3 in remission/SU and allergic
| Remission/SU | Allergic | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Th2+ cell cycle | IFN | Th2+ cell cycle/IFN | Th2+ cell cycle/IFN | Th2+ cell cycle | IFN | |||||
| T0 | T0 | T1 | T3 | T0 | T1 | T3 | T0 | T1 | T3 | |
| Th1 | ||||||||||
| IFNG | 2.52 | 8.06 | 8.75 | 7.40 | 2.90 | ns | 7.31 | 2.70 | 8.84 | 4.40 |
| Th2 | ||||||||||
| IL4 | 5.56 | ns | ns | 2.19 | 5.86 | 4.02 | 3.66 | ns | ns | ns |
| IL5 | 4.70 | 4.37 a | 4.42 a | 4.26 a | 4.99 | 3.61 | 5.35 | ns | 3.69 | ns |
| IL9 | ns | ns | 2.11 | ns | N/A | 2.62 | 2.53 | ns | ns | ns |
| IL13 | 2.00 | ns | ns | ns | 3.10 | 2.45 | 2.56 | ns | ns | ns |
| Interferons type 1 and 3 | ||||||||||
| IFNA2 | 3.18 | 7.16 | 7.82 | 6.57 | 2.52 | ns | 6.07 | 2.54 | 7.77 | 4.28 |
| IRF7 | ns | 7.16 | 7.33 | 6.83 | ns | ns | 6.10 | 3.35 | 7.00 | 3.90 |
| STAT1 | ns | 6.13 | 6.88 | 6.61 | 2.11 | ns | 5.83 | 3.28 | 6.84 | 3.22 |
| IFNA group | 2.72 | 6.56 | 7.24 | 6.52 | ns | ns | 6.01 | 2.29 | 7.30 | 3.40 |
| IFNL1 | ns | 5.76 | 6.33 | 5.25 | ns | ns | 4.46 | 2.10 | 6.52 | 4.05 |
| T cell associated | ||||||||||
| CD3 complex | 7.04 | 2.92 | 5.91 | 5.64 | 6.25 | 5.31 | 6.74 | ns | 4.10 | ns |
| CD28 | 6.09 | ns | 5.10 | 5.48 | 5.79 | 4.53 | 5.59 | ns | 2.94 | ns |
| TCR complex | 4.85 | ns | 5.97 | 4.86 | 4.26 | 3.35 | 5.45 | ns | 3.34 | ns |
| IL2 | 5.11 | 3.64 | 5.40 | 5.44 | 5.11 | 4.16 | 5.78 | ns | 4.05 | ns |
| CSF2 | 6.39 | 2.95 | 5.84 | 6.17 | 5.62 | 4.95 | 6.83 | ns | 3.44 | ns |
| CD40LG | 4.64 | 2.91 | 5.22 | 3.99 | 3.77 | 2.74 | 5.32 | ns | 3.86 | ns |
| Growth factors/cell cycle | ||||||||||
| ERBB2 | 6.16 | ns | 3.83 | 3.72 | 6.06 | 4.29 | 4.84 | ns | ns | ns |
| MYC | 5.58 | ns | 2.89 | 2.79 | 6.27 | 3.48 | 4.50 | ns | ns | ns |
| EGF | 4.99 | ns | ns | 3.75 | 4.63 | 4.51 | 4.86 | ns | ns | ns |
| EGFR | 4.00 | ns | 2.59 | ns | 4.15 | 3.52 | 4.31 | ns | ns | ns |
| Inflammatory | ||||||||||
| TNF | 6.60 | 6.17 | 7.40 | 7.62 | 5.38 | 5.04 | 7.58 | ns | 6.26 | 3.32 |
| IL1B | 5.60 | 4.82 | 6.57 | 5.86 | 5.16 | 3.37 | 6.67 | 2.60 | 5.74 | 2.55 |
| NFkB complex | 5.48 | 4.35 | 5.63 | 5.56 | 5.29 | 3.61 | 6.51 | 2.17 | 4.73 | ns |
Putative regulators of the modules were identified at each timepoint T0/T1/T3 employing Upstream Regulator Analysis. Molecular drivers with Benjamini & Hochberg adjusted p < 0.05 and absolute Z‐scores >2.0 were deemed significant. Positive values indicate activation, and negative values indicate inhibition.
Bold text highlights z‐scores for Th1 INFG, type 1 and 3 interferons and Th2.
Abbreviation: ns, not significant.
Z‐score ranking for IL5 is > top 30.
3.4. Remission/SU is associated with rewiring of gene network patterns between Th2 effectors and cell cycle regulators
To systematically investigate changes in the wiring of the gene networks that underpin CD4+ T cell responses in the transition from allergy to remission/SU during therapy, we employed differential gene correlation analysis, which identifies differential correlation patterns between the respective responses at the level of individual gene pairs. The data showed that in remission/SU subjects, the top‐ranking differentially correlated gene pairs at T1 compared with T0 were associated with cell cycle regulation (CEP55, CCNB1, CCNB2 and CDK1) and Th2 effector function (IL9) (Figure 4A). Specifically, the differential correlation patterns (T1 versus T0, delta adj. p < 3.97E‐03) showed a significant decrease in correlation on acquisition of remission/SU which was sustained at T3 (T1 versus T3, delta adj. p not significant, Table S8). In addition, correlation patterns between Th2 effectors (IL9 paired with IL4, IL5 and IL13) were also decreased (Figure 4B, Table S8). In the allergic group, however, correlation patterns between Th2 and cell cycle genes and the Th2 effectors themselves were highly correlated at all time points (T0, T1 and T3).
FIGURE 4.
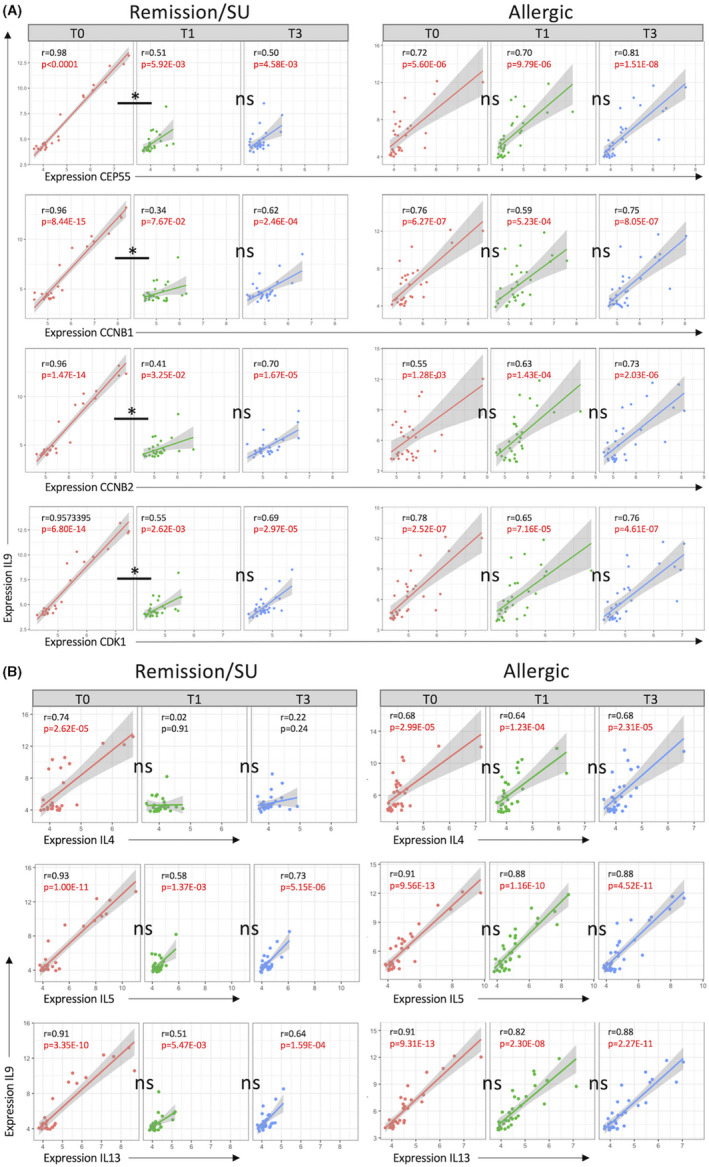
Remission/SU is associated with loss of correlation patterns between Th2 effectors and cell cycle/proliferation. Differential gene correlation analysis (DGCA) was employed to examine gene–gene relationships (network connectivity) across time in the remission/SU and allergic groups. Pearson correlations were computed at each time point (T0, T1 and T3) and differential delta z‐scores and Benjamini–Hochberg adjusted p‐values within and across time points. (A) Th2 effector and cell cycle/proliferation gene‐gene pairs and (B) differential gene‐gene correlation for Th2 gene pairs
4. DISCUSSION
The immunological changes that underpin the transition from peanut allergy to remission/SU remain largely unknown. We employed a systems biology approach to compare and contrast peanut‐specific CD4+ T cell responses in peanut‐allergic children enrolled in the PPOIT‐001 randomized trial who attained remission/SU following PPOIT treatment and who remained allergic following placebo treatment. At the gene expression level, acquisition of remission/SU was associated with loss of Th2 upregulation by end‐of‐treatment which persisted to 3‐month post‐treatment. In contrast, Th2 responses remained upregulated at all three time points in the allergic subjects. At the systems level, comparison of CD4+ T cell gene expression in remission/SU and allergic subjects revealed profound differences in network connectivity patterns whereas differences in gene expression levels were modest. For instance, the magnitude of the correlation patterns within the Th2 module was markedly decreased following acquisition of remission/SU. Moreover, upon attainment of remission/SU, the Th2 and IFN modules merged and the connectivity patterns at the gene level were integrated, and the Th2 gene network patterns that were initially dominant at T0 were replaced by IFN‐dominant patterns at T1/T3. This remodelling of connectivity patterns correlates with parallel shifts in immunological allergic parameters; peanut SPT and peanut sIgE and sIgG4, as published previously. 26 Finally, acquisition of remission/SU resulted in a dramatic reduction in the correlation between Th2 effectors and cell cycle regulators, suggesting that the CD4+ T cell responses were being driven towards an anergic phenotype. In summary, our findings demonstrate that acquisition of remission/SU is associated with reprogramming of Th2‐memory responses with diminished expression of Th2 effectors, integration of Th2 and IFN modules, and uncoupling of co‐expression between Th2 effectors and cell cycle regulators.
Rewiring of Th2 and IFN modules has previously been reported in the context of allergen‐driven CD4+ T cell responses during HDM immunotherapy, where a progressive integration was observed between these distinct gene modules during therapy which was associated with persistent attenuation of HDM‐associated symptoms that persisted after treatment cessation. 25 Notably, the fact that we observed a similar mechanism operating in this study is compelling, given the striking differences between the two studies. First, the PPOIT subjects were children whereas the HDM studies were performed in adults with severe allergic rhinitis. Second, HDM exposure is ubiquitous in the environment, whereas food allergens are avoided, resulting in large variations in allergen dose and frequency of exposure. Third, HDM immunotherapy was performed via the subcutaneous route, as opposed to orally in the case of PPOIT. Fourth, PPOIT therapy includes a probiotic whereas HDM SIT does not. Fifth, the CD4+ T cell responses to peanut were stimulated for 48 h, whereas corresponding HDM responses were studied after 24 h of stimulation. Given these major differences in study settings, the immune changes we have identified employing this systems‐level approach are likely to be fundamental to the basic mechanism of action of allergen‐specific immunotherapy, and comparisons between different therapies in future studies employing similar methodology may further elucidate the underlying processes.
The precise mechanisms that determine how IFN and Th2 responses counter‐regulate each other in the context of allergen immunotherapy are unknown. Previous studies have demonstrated that adding type I IFN to allergen stimulated PBMC abrogates the production of Th2 cytokines 27 and expression of the high‐affinity IgE receptor. 28 Moreover, in CD4+ T cells, type I interferon signalling suppresses the expression of gata‐3 during Th2 differentiation as well as in fully committed Th2 cells and promotes epigenetic silencing of non‐coding regions in the Th2 locus that controls Th2 cytokine expression. 29 Although we anticipated a role for T regulatory cells in the acquisition of remission/SU, we were not able to detect FOXP3 mRNA changes as the probe set did not pass our stringent controls to remove noisy probes (PVAC filter). It is important to acknowledge that our studies were based on profiling total CD4+ T cells, and therefore, we cannot determine whether the Th2 and IFN modules are operating in the same CD4+ T cell populations or in discrete subpopulations. In this context, it is noteworthy that single‐cell genomic studies of CD4+ T cell memory responses to HDM have identified novel subsets of T helper and T regulatory cells that express a type I interferon response signature, and these cells are preferentially expanded in subjects without allergy, suggesting they may play a key role in dampening allergic responses. 30
Our differential gene correlation analysis demonstrated that remission/SU was associated with markedly decreased correlation patterns between Th2 effectors and positive regulators of the cell cycle/proliferation, which was not observed for the allergic group. Notably, these changes are persistent out to 3‐month post‐treatment, demonstrating a lasting effect. The finding that arrest of Th2‐induced signalling occurred in parallel to a loss of cell cycle‐Th2 gene pair correlation suggests that pathogenic Th2 responses may be deleted or become more anergic, as described in other studies of OIT. 8 , 9 Our findings are consistent with an early single‐cell study of antigen‐specific CD4+ T cells from participants undergoing OIT, which showed that acquisition of remission/SU (defined as passing a food challenge 3‐month post‐treatment) was associated with antigen‐specific CD4+ T cells shifting towards an anergic phenotype (CD28−/CD38−/IFN‐γ/IL4‐/IL13‐/IL10‐). 8 Another study identified a novel terminally differentiated Th2 subset, Th2A, expressing receptors for IL‐25 (IL‐17RB), IL‐33 (IL1RL1) and thymic stromal lymphopoietin (CLRF2), which was upregulated in participants with food allergy. OIT led to a reduction in this Th2A subset, and this decrease correlated with treatment success. 9 More recently, a study described an allergic Th2 subset in peanut‐specific T cells using single‐cell sequencing with matching cell surface marker expression similar to the Th2A subset. 31 Our findings of lost correlation between Th2 and cell cycle genes may similarly reflect these mechanisms in which the allergic Th2A subset is subsequently lost in those who successfully gain remission of allergy/SU following OIT. It is evident that genes do not exist in isolation, but instead work together in co‐expression networks; ours is the first study to systematically assess CD4+ co‐expression patterns in peanut OIT, in a robust, well‐phenotyped cohort with high SU rates and post‐treatment follow‐up.
We acknowledge several limitations of our study. We employed microarray technology to profile peanut‐stimulated gene expression in total CD4+ T cells. Future studies could employ single‐cell RNA‐Seq on peanut‐allergen‐specific CD4+ T cells isolated using a series of activation markers (e.g. OX40/AIM assay) and obtain genomic profiles at single‐cell resolution. 30 A limitation is that isolated CD4+ cells included small numbers of monocytes, which are known producers of high quantities of interferons. Although we confirmed >98% purity of CD3+CD4+ T cells with our isolation protocol, monocytes may have contributed to the emergent type 1 IFN module either directly or by supporting cognate interactions with allergen‐specific T cells leading to activation of type 1 IFN signalling pathways. A further limitation is that the peanut extract likely contained residual amounts of endotoxin, despite use of an endotoxin removal kit, which may initiate non‐antigen‐specific interactions; however, any such contribution is expected to be minimal. Another limitation of the study is that we did not include a PPOIT‐treated non‐responder group to distinguish non‐remission/SU treatment effects. Additionally, we cannot make any inferences on the individual contributions of probiotic and peanut OIT because we did not include subjects who received peanut OIT (without adjuvant). We acknowledge an increased number of DEG at T3 relative to T1 in the remission group raising the possibility of slight regression in changes; however, several findings provide confidence that there is no material loss of key changes underpinning transition from allergy to remission between the T1 and T3 timepoints. Firstly, formal statistical analysis comparing responses at T1 and T3 did not reveal any significant DEG. Secondly, pathway analysis of unique DEG at T3 compared with T1 showed that neither the upregulated pathways nor downregulated pathways were related allergic or regulatory responses. Thirdly, Th2‐related pathways remained absent at T3 while type 1/2 interferon signalling pathways were similarly dominant at both T1 and T3, albeit with higher activation at T1. Finally, downregulation of cardinal Th2 genes and cell cycle genes at T1 compared with T0 was also retained at T3. Thus, while some of the changes associated with remission at T1 appear to regress at T3, our findings provide compelling evidence that the crucial rewiring of gene‐gene communications away from a Th2‐dominant module to an integrated regulatory module that engulfs the Th2 elements remain in place at T3. Notwithstanding these limitations, we present the first longitudinal analysis of immunological mechanisms that underpin the transition from peanut allergy to remission/SU, and additionally, for the first time, apply a systems biology approach to examine the complex interrelationships driving this process. Our findings pave the way for new approaches to monitor therapeutic responses during food immunotherapy and identify candidate pathways that can be targeted to improve treatment efficacy.
AUTHOR CONTRIBUTIONS
MLKT, AB and PH were involved in conceptualization and funding acquisition. SEA, MLKT, ACJ and AB were involved in methodology. SEA, ACJ, AB, DA were involved in investigation. SEA and ACJ were involved in visualization. MLKT and AB were involved in supervision. SEA, ACJ, AB and MLKT were involved in writing—original draft. SEA, MLKT, ACJ, AB, PH and DA were involved in writing—review and editing.
CONFLICT OF INTEREST
MLKT has received consultant fees from Pfizer; is an inventor on patents covering PPOIT; is an employee and scientific founder and holds share interest/options in Prota Therapeutics; is a member of the Medical Advisory Board of Anaphylaxis & Anaphylaxis Australia and past member of the International Union of Immunological Societies (IUIS, ended 2019), the International Expert Panel on Guidelines for Food Allergy in Schools (ended 2021) and of the Board of Directors and an expert committee of the World Allergy Organisation (WAO, both ended 2019); is a member of expert committees of the American Academy of Allergy Asthma and Immunology (AAAAI), Asia Pacific Association of Allergy Asthma and Clinical Immunology (APAAACI) and the Australasian Society of Clinical Immunology and Allergy (ASCIA). All other authors declare no competing interests.
Supporting information
Fig S1
Fig S2
Fig S3
Fig S4
Table S1‐S8
ACKNOWLEDGEMENTS
The parent trial was supported by the Food Allergy and Anaphylaxis Network (FAAN), the Murdoch Children’s Research Institute, Perpetual Philanthropy (grant ID 493), the CASS Foundation, the Financial Markets Foundation for Children and the National Health and Medical Research Council Australia (NHMRC project grant no. 1029690). Nestle Health Science provided the probiotic and placebo treatments. Anthony Bosco was supported by the Simon Lee Foundation and the NHMRC 1129996. The Murdoch Children's Research Institute is supported by the Victorian Government’s Operational Infrastructure Support Program. This trial was registered with the Australian New Zealand Clinical Trials Registry (ACTRN12608000594325, 25/11/2008). Funding organizations were not involved in any aspect of the study and did not contribute to study design, interpretation of results or writing of the manuscript. We would like to thank all the parents and participants who took part in the parent trial and the clinical team without whom the work would not have been possible. We also thank Helen Lescesen and Lakshi Starks for completing the cell culture and RNA isolation work and Wesley Burks for supplying the protocol used for crude peanut extract preparation. Open access publishing facilitated by The University of Melbourne, as part of the Wiley ‐ The University of Melbourne agreement via the Council of Australian University Librarians.
Ashley SE, Jones AC, Anderson D, Holt PG, Bosco A, Tang MLK. Remission of peanut allergy is associated with rewiring of allergen‐driven T helper 2‐related gene networks. Allergy. 2022;77:3015–3027. doi: 10.1111/all.15324
Sarah E. Ashley, Anya C. Jones contributed equally to this work. Anthony Bosco and Mimi LK Tang contributed equally to this work.
REFERENCES
- 1. Gupta RS, Warren CM, Smith BM, et al. The public health impact of parent‐reported childhood food allergies in the United States. Pediatrics. 2018;142(6):e20181235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Osborne NJ, Koplin JJ, Martin PE, et al. Prevalence of challenge‐proven IgE‐mediated food allergy using population‐based sampling and predetermined challenge criteria in infants. J Allergy Clin Immunol. 2011;127(3):668‐676.e2. [DOI] [PubMed] [Google Scholar]
- 3. Chinthrajah RS, Purington N, Andorf S, et al. Sustained outcomes in oral immunotherapy for peanut allergy (POISED study): a large, randomised, double‐blind, placebo‐controlled, phase 2 study. Lancet. 2019;394(10207):1437‐1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jones SM, Burks AW, Keet C, et al. Long‐term treatment with egg oral immunotherapy enhances sustained unresponsiveness that persists after cessation of therapy. J Allergy Clin Immunol. 2016;137(4):1117‐1127 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gorelik M, Narisety SD, Guerrerio AL, et al. Suppression of the immunologic response to peanut during immunotherapy is often transient. J Allergy Clin Immunol. 2015;135(5):1283‐1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martino DJ, Bosco A, McKenna KL, et al. T‐cell activation genes differentially expressed at birth in CD4+ T‐cells from children who develop IgE food allergy. Allergy. 2012;67(2):191‐200. [DOI] [PubMed] [Google Scholar]
- 7. Martino D, Neeland M, Dang T, et al. Epigenetic dysregulation of naive CD4+ T‐cell activation genes in childhood food allergy. Nat Commun. 2018;9(1):3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ryan JF, Hovde R, Glanville J, et al. Successful immunotherapy induces previously unidentified allergen‐specific CD4+ T‐cell subsets. Proc Natl Acad Sci USA. 2016;113(9):E1286‐E1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wambre E, Bajzik V, DeLong JH, et al. A phenotypically and functionally distinct human TH2 cell subpopulation is associated with allergic disorders. Sci Transl Med. 2017;9(401):eaam9171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kulis MD, Patil SU, Wambre E, et al. Immune mechanisms of oral immunotherapy. J Allergy Clin Immunol. 2018;141(2):491‐498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sampath V, Tupa D, Graham MT, et al. Deciphering the black box of food allergy mechanisms. Ann Allergy Asthma Immunol. 2017;118(1):21‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. de Jong E, Bosco A. Unlocking immune‐mediated disease mechanisms with transcriptomics. Biochem Soc Trans. 2021;49(2):705‐714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hsiao K‐C, Ponsonby A‐L, Axelrad C, et al. Long‐term clinical and immunological effects of probiotic and peanut oral immunotherapy after treatment cessation: 4‐year follow‐up of a randomised, double‐blind, placebo‐controlled trial. Lancet Child Adolesc Health. 2017;1(2):97‐105. [DOI] [PubMed] [Google Scholar]
- 14. Kim EH, Bird JA, Kulis M, et al. Sublingual immunotherapy for peanut allergy: clinical and immunologic evidence of desensitization. J Allergy Clin Immunol. 2011;127(3):640‐646.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kauffmann A, Gentleman R, Huber W. arrayQualityMetrics–a bioconductor package for quality assessment of microarray data. Bioinformatics. 2009;25(3):415‐416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249‐264. [DOI] [PubMed] [Google Scholar]
- 17. Lu J, Kerns RT, Peddada SD, et al. Principal component analysis‐based filtering improves detection for Affymetrix gene expression arrays. Nucleic Acids Res. 2011;39(13):e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high‐throughput experiments. Bioinformatics. 2012;28(6):882‐883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinform. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498‐2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McKenzie AT, Katsyv I, Song W‐M, et al. DGCA: a comprehensive R package for differential gene correlation analysis. BMC Syst Biol. 2016;10(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu G, Wang L‐G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284‐287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Breuer K, Foroushani AK, Laird MR, et al. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 2013;41(Database issue):D1228‐D1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krämer A, Green J, Pollard J, et al. Causal analysis approaches in ingenuity pathway analysis. Bioinformatics. 2014;30(4):523‐530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jones AC, Anderson D, Troy NM, et al. Rewiring of gene networks underlying mite allergen‐induced CD4+ Th‐cell responses during immunotherapy. Allergy. 2020;75(9):2330‐2341. [DOI] [PubMed] [Google Scholar]
- 26. Tang ML, Ponsonby AL, Orsini F, et al. Administration of a probiotic with peanut oral immunotherapy: a randomized trial. J Allergy Clin Immunol. 2015;135(3):737‐744.e8. [DOI] [PubMed] [Google Scholar]
- 27. Pritchard AL, White OJ, Burel JG, Upham JW. Innate interferons inhibit allergen and microbial specific T(H)2 responses. Immunol Cell Biol. 2012;90(10):974‐977. [DOI] [PubMed] [Google Scholar]
- 28. Subrata LS, Bizzintino J, Mamessier E, et al. Interactions between innate antiviral and atopic immunoinflammatory pathways precipitate and sustain asthma exacerbations in children. J Immunol. 2009;183(4):2793‐2800. [DOI] [PubMed] [Google Scholar]
- 29. Huber JP, Gonzales‐van Horn SR, Roybal KT, Gill MA, Farrar JD. IFN‐alpha suppresses GATA3 transcription from a distal exon and promotes H3K27 trimethylation of the CNS‐1 enhancer in human Th2 cells. J Immunol. 2014;192(12):5687‐5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seumois G, Ramírez‐Suástegui C, Schmiedel BJ, et al. Single‐cell transcriptomic analysis of allergen‐specific T cells in allergy and asthma. Sci Immunol. 2020;5(48):eaba6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chiang D, Chen X, Jones SM, et al. Single‐cell profiling of peanut‐responsive T cells in patients with peanut allergy reveals heterogeneous effector TH2 subsets. J Allergy Clin Immunol. 2018;141(6):2107‐2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2
Fig S3
Fig S4
Table S1‐S8


