Abstract
The spleen is a key immune-related organ that plays a role in communication between the brain and the immune system through the brain–spleen axis and brain–gut–microbiota axis. However, how the gut microbiota affects spleen and brain function remains unclear. Here, we investigated whether microbiome depletion induced by administration of an antibiotic cocktail (ABX) affects spleen and brain function. Treatment with ABX for 14 days resulted in a significant decrease in spleen weight and significant alterations in splenic functions, including the percentage of neutrophils, NK cells, macrophages, and CD8+ T cells. Furthermore, ABX treatment resulted in the depletion of a large portion of the gut microbiota. Untargeted metabolomics analysis showed that ABX treatment caused alterations in the levels of certain compounds in the plasma, spleen, and brain. Moreover, ABX treatment decreased the expression of microglia marker Iba1 in the cerebral cortex. Interestingly, correlations were found between the abundance of different microbiome components and metabolites in various tissues, as well as splenic cell populations and spleen weight. These findings suggest that ABX-induced microbiome depletion and altered metabolite levels may affect spleen and brain function through the gut–microbiota–spleen–brain axis.
Keywords: Brain-spleen axis, Brain-gut-microbiota axis, Metabolite, Microglia
Abbreviations: ABX, antibiotic cocktail; CNS, central nervous system; FDR, false discovery rate; GF, germ free; GFAP, glial fibrillary acidic protein; OPLS-DA, orthogonal partial least squares discriminant analysis; VIP, variable importance in projection
Highlights
-
•
Antibiotic cocktail (ABX) decreased spleen weight in mice.
-
•
Splenic cell compartments are associated with spleen weight.
-
•
Relative abundance of microbiota and metabolites was associated with spleen weight.
-
•
ABX-induced microbiome depletion caused reduction of microglia in the brain.
-
•
ABX-induced microbiome depletion can affect spleen–brain axis.
1. Introduction
Accumulating evidence demonstrates that the gut microbiota contributes to human health and disease. Dysbiosis (imbalance of the gut microbiota) is observed in patients with gastrointestinal (GI) disorders as well as patients with a variety of diseases, including metabolic, psychiatric, and neurological disorders (Aron-Wisnewsky and Clément, 2016; Cryan et al., 2020; Fan and Pedersen, 2021; Miyauchi et al., 2022; Sharon et al., 2016). Bidirectional communication takes place between the gut microbiota and the central nervous system (CNS) via neural, endocrine, immune, and humoral pathways, known as the brain–gut–microbiota axis (Chang et al., 2022; Cryan et al., 2019; Wei et al., 2022a, 2022b).
The spleen is a key immune-related organ that may mediate communication between the brain and the immune system through the brain–spleen axis (Bronte and Pittet, 2013; Lewis et al., 2019; Mebius and Kraal, 2005; Noble et al., 2018; Wei et al., 2022b). We previously demonstrated that the brain–spleen axis might play a role in stress-related psychiatric disorders such as depression (Hashimoto, 2020; Ma et al., 2022b; Wei et al., 2022a, 2022b; Yang et al., 2017; Zhang et al., 2020a, 2020b, 2021b).
The spleen contributes to brain–gut crosstalk and behavioral outcomes through the production of metabolites and immune factors (Buchmann Godinho et al., 2021; Rhee et al., 2009; Wei et al., 2021). Pathways involved in the gut–brain–spleen axis may include the cholinergic anti-inflammatory reflex and efficient activation of the vagus afferent fiber system by inflammatory intermediates (Borovikova et al., 2000; Huston et al., 2006; Rosas-Ballina et al., 2008). Previous studies suggest that a number of neuropsychiatric disorders, such as Alzheimer’s disease, Parkinson’s disease, schizophrenia, and depression, might be associated with the gut–brain–spleen axis (Breit et al., 2018; Buchmann Godinho et al., 2021; Mok et al., 2020). A dynamic, pathological crosstalk among the brain, gut, and spleen manifests as internal metabolite disturbances and CNS inflammation (Buchmann Godinho et al., 2021). However, whether microbiome depletion induces alterations in the metabolic profile and in the function of organs, such as the brain, spleen, and blood, remains unclear.
Germ-free (GF) mouse models are generally considered to be the gold standard for microbiota studies. Importantly, GF mice exhibit a broad range of developmental impairments, including development of the early immune system (Kennedy et al., 2018), and as such are not suitable for studying development-related conditions such as psychiatric and neurological disorders. In contrast, treating adult mice with a broad-spectrum antibiotic cocktail (ABX) allows study of the role that the microbiota plays in maintaining cell functionality and signaling pathways after development (Kennedy et al., 2018). Therefore, the aim of the present study was to investigate whether ABX-induced microbiome depletion affects spleen and brain function and the metabolites present in the blood, spleen, and brain. We performed 16S rRNA sequencing analysis to analyze gut microbiota and untargeted metabolomics analysis to analyze the metabolites in the blood, spleen, and brain.
2. Material and methods
2.1. Animals
Male C57BL/6 mice (8 weeks old, body weight 20–25 g) were purchased from Japan SLC Corporation (Hamamatsu, Shizuoka, Japan). All mice were carefully housed in clear polycarbonate cages (21 × 30 × 22.5 cm), 4–5 per cage, on an automatically managed light/dark cycle of 12 h/12 h (7:00 a.m. to 7:00 p.m.), with a constant and stable ambient temperature of 23 ± 1 °C and a relative humidity of 55 ± 5%. The experimental procedures used in this study were approved by the Animal Care and Use Committee of Chiba University Institution (License No. 4–237 and 4–407). Animals were promptly sacrificed by inducing deep anesthesia with inhaled isoflurane followed by skilled cervical dislocation. Animal suffering was minimized to the greatest extent possible.
2.2. Antibiotic cocktail therapy and sample collection
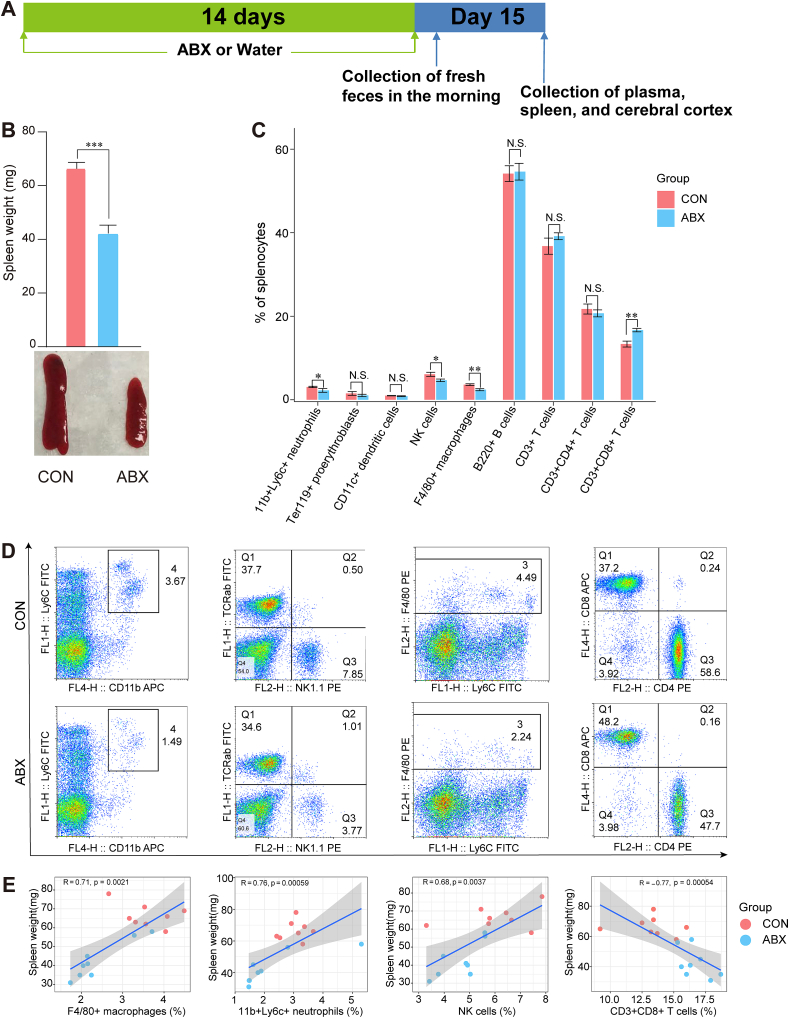
On days 1–14, mice were allowed ad libitum access to plain drinking water or drinking water containing the broad-spectrum antibiotic cocktail (ABX: ampicillin 1 g/L, FUJIFILM Wako Pure Chemical Corporation, Tokyo, Japan; neomycin sulfate 1 g/L, Sigma-Aldrich Co. Ltd, MO, USA; metronidazole 1 g/L, FUJIFILM Wako Pure Chemical Corporation, Tokyo, Japan) (Fig. 1A), as previously described (Hashimoto et al., 2022; Pu et al., 2019, 2021; Wang et al., 2020a, 2020b, 2021). Fresh mouse feces were collected on the morning of day 15 for 16S ribosomal RNA sequencing analysis as previously described (Hashimoto et al., 2022; Wan et al., 2022a, 2022b; Wang et al., 2022a, 2022b) (Fig. 1A). All mouse fecal samples were quickly frozen in liquid nitrogen after collection and then transferred to a −80 °C freezer for storage. Plasma, spleen, and cerebral cortex samples were also collected from each mouse on day 15 (Fig. 1A).
Fig. 1.
Effects of ABX on spleen weight and spleen cell types
A: Schedule of the experiment. ABX or water was injected as drinking water for 14 days. Fresh feces samples were collected in the morning of day 15. Subsequently, plasma, spleen, and cerebral cortex were collected in the same day. B: ABX induced a significant reduction in spleen weight in mice (Two-tailed unpaired Student’s T test, P < 0.0001). Representative photograph of spleen from the two groups. C: FACS analysis of spleen cell populations in all mice (Mann-Whitney test: 11b+Ly6c+ neutrophils, P = 0.0207; NK1.1+ NK cells, P = 0.0207; F4/80+ macrophages, P = 0.0070; CD3+CD8+ T cells, P = 0.002). D: Representative FACS dot plots of neutrophils, NK cells, macrophages, and CD8+ T cells. E: Spearman correlation analysis of spleen cell population and spleen weight in all mice. Values represent mean ± S.E.M. (CON: n = 8. ABX: n = 9). *P < 0.05, **P < 0.01, ***P < 0.001. N.S.: not significant.
After anesthetization with 5% isoflurane, cardiac blood was collected from each mouse using a 1-mL syringe and placed into a tube containing ethylenediamine-N,N,N′,N′-tetraacetic acid (EDTA) potassium salt dehydrate as an anticoagulant. Then, blood samples were centrifuged at 4 °C, and the supernatant (containing the plasma) was collected and stored at −80 °C. Half of the spleen samples were immediately subject to fluorescence activated cell sorting (FACS) analysis, while the remaining spleen, plasma, and left cerebral cortex samples were subjected to untargeted metabolomics analysis. The right cerebral cortex was used for western blot analysis.
2.3. FACS analysis of spleen samples
Single-cell suspensions were prepared from the spleen samples and stained for FACS (fluorescence activated cell sorting) analysis as previously described (Shinno-Hashimoto et al., 2022; Zhang et al., 2021b). The following antibodies were used for immunofluorescence staining: anti-CD3-FITC (x40, cat# 100305: BioLegnd, San Diego, CA), anti-CD4-allophycocyanin (x100, cat# 17-0042-82: eBioscience, San Diego, CA), anti-CD8a-allophycocyanin (x100, cat# 553035: BD Bioscience), anti-NK1.1-PE (x100, cat# 553165: BD Bioscience), anti-Ly6c-FITC (x100, cat# 553104: BD Bioscience), anti-CD11b-PE (x400 diluted using FACS buffer, cat# 553312: BD Bioscience, Franklin Lakes, NJ), anti-CD11c-PE (x100, cat# 557401: BD Bioscience), anti-Ter119-PE (x40, cat# 12-5921-83: eBioscience, San Diego, CA), anti-F4/80-PE (x40, cat# 12-4801-80: Invitrogen), and anti-B220-PE (x200, cat# 553309: BD Bioscience). The stained cells were analyzed using a FACSCanto II and FlowJo software (BD Bioscience).
2.4. Western blot analysis
Western blotting was carried out to assess the expression levels of ionized calcium binding adaptor molecule 1 for microglia (Iba1) and glial fibrillary acidic protein for astrocyte (GFAP) in the cerebral cortex, as previously described (Ma et al., 2022b; Wan et al., 2022b). The cerebral cortex tissue was ground in frozen Laemmli lysis buffer, the ground sample was centrifuged at 3000×g (RCF) for 10 min at 4 °C, and the supernatant was collected. The protein concentration in each sample was determined using a DC protein assay kit (Bio-Rad, Hercules, CA). Then, the protein samples were boiled for 10 min at 95 °C in loading buffer (125 mM Tris/HCl (pH 6.8), 20% glycerol, 0.1% bromophenol blue, 10% β-mercaptoethanol, 4% sodium dodecyl sulfate). The proteins were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (Mini-PROTEAN® TGX™ precast gels; Bio-Rad) and then electro-transferred to polyvinylidene difluoride (PVDF) membranes using a Trans Blot Mini Cell (Bio-Rad). The membranes were blocked with 5% skim milk powder dissolved in 0.1% Tween 20 in TBS solution for 60 min, followed by incubation with the corresponding primary antibodies (anti-Iba1 (1:1000, cat# 016–20001, 1 μg/mL, FUJIFILM, Tokyo, Japan), anti-GFAP (1:1000, cat# MA5-15086, Thermo Fisher Scientific, USA), and anti-β-actin (1:10,000; cat# A5441, Sigma-Aldrich Co., Ltd, St Louis, MO, USA)) overnight at 4 °C, followed by incubation with the corresponding secondary antibody (1:5000) for 1 h. Bands were visualized with an enhanced chemiluminescence (ECL) kit and quantified using a ChemiDoc™ Touch Imaging System (170–01,401; Bio-Rad Laboratories, Hercules, CA).
2.5. 16S rRNA sequencing analysis
DNA was extracted from mouse fecal samples using the NucleoSpin DNA Fecal Kit (REF: 740472.50, MACHEREY-NAGEL, Germany) following the manufacturer's protocol. The V3–V4 region of the 16S rRNA gene was amplified by polymerase chain reaction (PCR), and all PCR products were sequenced on a HiSeq2500 platform by BGI JAPAN K.K. (Kobe, Japan). The sequences were deposited with the National Center for Biotechnology Information (accession number: PRJNA887905).
We used the Observed species index, Chao index, Ace index, and Shannon index to measure α-diversity (species diversity within each sample). The Observed species index, Chao index, and ACE index reflect the species richness of the microbial community. The Shannon index reflects the species diversity of the community, which is influenced by species richness and species evenness. Principal component analysis (PCA) was performed to assess differences in β-diversity, or species complexity, among samples (Caporaso et al., 2010). Linear discriminant analysis (LDA) effect size (LEfSe) was employed using LEfSe Software to discover high-dimensional biomarkers and reveal taxonomic features (Segata et al., 2011).
PICRUSt2 (Douglas et al., 2020) software was applied to predict the functional composition of the metagenome. After predicting the functions of all of the samples, the Wilcox test was used to identify different functions among the separate groups.
2.6. Untargeted metabolomics analysis
Untargeted metabolomics analysis of the plasma samples was performed following the methods described in our previous reports (Hashimoto et al., 2022; Shinno-Hashimoto et al., 2022; Wang et al., 2022b; Wan et al., 2022a; Wan et al., 2022b; Yang et al., 2022a). The spleen and cerebral cortex samples were analyzed in a manner similar to that used for the plasma samples, with slight modifications to the metabolite extraction process. Briefly, 400 μL of methanol containing internal standards (25 μM N,N-diethyl-2-phenylacetamide, and d-camphor-10-sulfonic acid) and 400 μL of ultrapure water were added to the sample, and the sample was homogenized using BioMasher® II with PowerMasher® II to extract the metabolome. The samples were then centrifuged at 14,000 g for 5 min, and 500 μL of the supernatant was passed through an ultrafiltration filter to purify it. Then, the purified supernatant was transferred to Amicon® Ultra-0.5 3 kDa filter columns (Merck Millipore, Tokyo, Japan) and centrifuged at 14,000 × rpm for 1 h. The filtrates were transferred to glass vials for analysis. The details of the analysis are described in the supplementary material.
2.7. Statistical analysis
The data are expressed as the mean ± the standard error of the mean (S.E.M.). Student’s t-test and Wilcox test (Mann-Whitney test) were employed to analyze differences between two groups based on the data distribution. Regarding the β diversity analysis, differences between groups were statistically analyzed using analysis of similarity (ANOSIM). Analyses comparing multiple time points were assessed using repeated measures analysis of variance (ANOVA). For analysis of metabolites, we used orthogonal partial least squares discriminant analysis (OPLS-DA) as a multivariate analysis model implemented in SIMCA-P (V.14.0). Then, significant peaks were determined by combination of variable importance in projection (VIP) value > 1, Wilcox rank test P value < 0.05, and false discovery rate (FDR) < 0.3 between the two groups. P-values of less than 0.05 were considered statistically significant.
3. Results
3.1. Effects of ABX treatment on spleen weight and splenocyte population
The body weight of ABX-treated mice was lower than that of control mice, and then recovered gradually, consistent with previous reports (Fig. S1A) (Pu et al., 2019; Wang et al., 2020a, 2020b). Compared with the CON group, ABX treatment resulted in a significant reduction in spleen weight and in the ratio of spleen weight to body weight (Fig. 1B and Fig. S1B). To identify the cell populations associated with the ABX-induced decrease in spleen weight, we then evaluated the frequency of neutrophils (11b+Ly6c+), proerythroblasts (Ter119+), dendritic cells (CD11c+), natural killer (NK) cells (NK1.1+), macrophages (F4/80+), B cells (B220+), T cells (CD3+), CD4+ T cells (CD3+CD4+), and CD8+ T cells (CD3+CD8+) in the spleen using flow cytometry. Of note, the percentages of 11b+Ly6c+ neutrophils, NK1.1+ NK cells, and F4/80+ macrophages were significantly lower in the ABX group compared with the CON group (Fig. 1C and D). In contrast, the percentage of CD8+ T cells in the ABX group was higher than that in the CON group. There were no changes in the percentage of proerythrocytes, dendritic cells, B cells, CD4+ T cells or CD3+ T cells between the two groups (Fig. 1C and D).
Positive correlations were found between the percentage of splenic macrophages, neutrophils, NK cells, and spleen weight. In contrast, a negative correlation was found between the percentage of CD8+ T cells and spleen weight (Fig. 1E).
3.2. ABX-treated mice exhibit altered gut microbiota composition
To examine the effect of ABX on the mouse gut microbiota, we carried out high-throughput amplicon sequencing of 16S rRNA microbial genes from mice in the CON and ABX groups. To estimate the richness, evenness, and diversity of the gut microbiota of mice in the CON and ABX groups, α-diversity was estimated using the Shannon index, Chao index, Observed species index, and Ace index. We detected a significant reduction in α-diversity in the ABX group compared to the CON group (Fig. S2A). These results suggest that the within-individual bacterial diversity of ABX-treated mice differed from that of CON mice. PCA-based β-diversity analysis revealed distinct cluster separation between the CON and ABX groups (ANOSIM analysis, P = 0.001) (Fig. S2B), indicating differences in the individual microbial communities between the two groups.
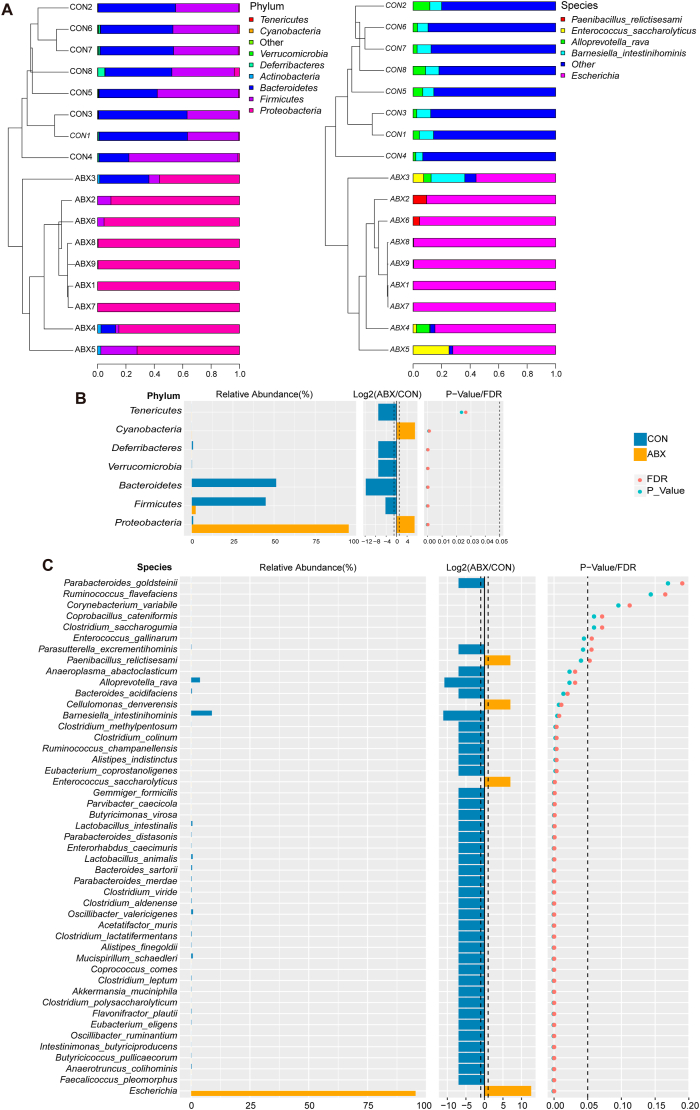
The microbial taxonomy and bacterial taxonomy of the gut microbiota were compared between the two groups at the phylum and species level (Fig. 2A). Comparing the percentages of bacteria at the phylum level, ABX eliminated or markedly reduced Tenericutes, Verrucomicrobia, Deferribacteres, Bacteroidetes, and Firmicutes levels compared with the CON group (Fig. 2A), while at the species level, the percentages of Alloprevotella rava and Barnesiella intestinihominis were considerably lower in the ABX group than in the CON group (Fig. 2A). Interestingly, ABX increased the percentages of the phyla Proteobacteria (88.7% in ABX and 1.1% in CON) and Actinobacteria (0.72% in ABX and 0.22% in CON) (Fig. 2A). At the species level, we observed an ABX-induced increase in the percentage of Escherichia (88.6% in ABX, 0.02% in CON), Enterococcus saccharolyticus (3.92% in ABX, 0% in CON), and Paenibacillus relictisesami (1.5% in ABX, 0% in CON) compared with the CON group (Fig. 2A).
Fig. 2.
Effects of ABX on the composition of gut microbiota
A: Bar chart of phylum-level bacterial community composition, with species level presenting the top five bacteria in abundance in all samples and the rest shown as "other". B: Differences of bacterial abundance at the phylum level in two groups (statistical results are presented in Table S1). C: Differences of bacterial abundance at the species level in two group of mice (statistical results are presented in Table S2).
Next, we investigated specific differences in the gut microbiota at different taxonomic levels in the ABX group compared with the CON group. At the phylum level, the ABX group had higher abundances of Proteobacteria and Cyanobacteria relative to the CON group; in fact, no Cyanobacteria were detected in the CON group (Fig. 2B, Table S1). In contrast, Bacteroidetes and Firmicutes were depleted in the ABX group in comparison with the CON group (Fig. 2B, Table S1). Three phyla were found to be completely absent in the ABX group, including Deferribacteres, Tenericutes, and Verrucomicrobia (Fig. 2B, Table S1). The abundance of 39 species was found to differ between the two groups (Fig. 2C, Table S2), of which 36 were significantly reduced or eliminated after ABX treatment (Fig. 2C, Table S2). Interestingly, the abundance of three species—Cellulomonas denverensis, Enterococcus saccharolyticus, and Escherichia—was dramatically higher in the ABX group than in the CON group (Fig. 2C, Table S2).
3.3. Microbial biomarkers and functional profiles
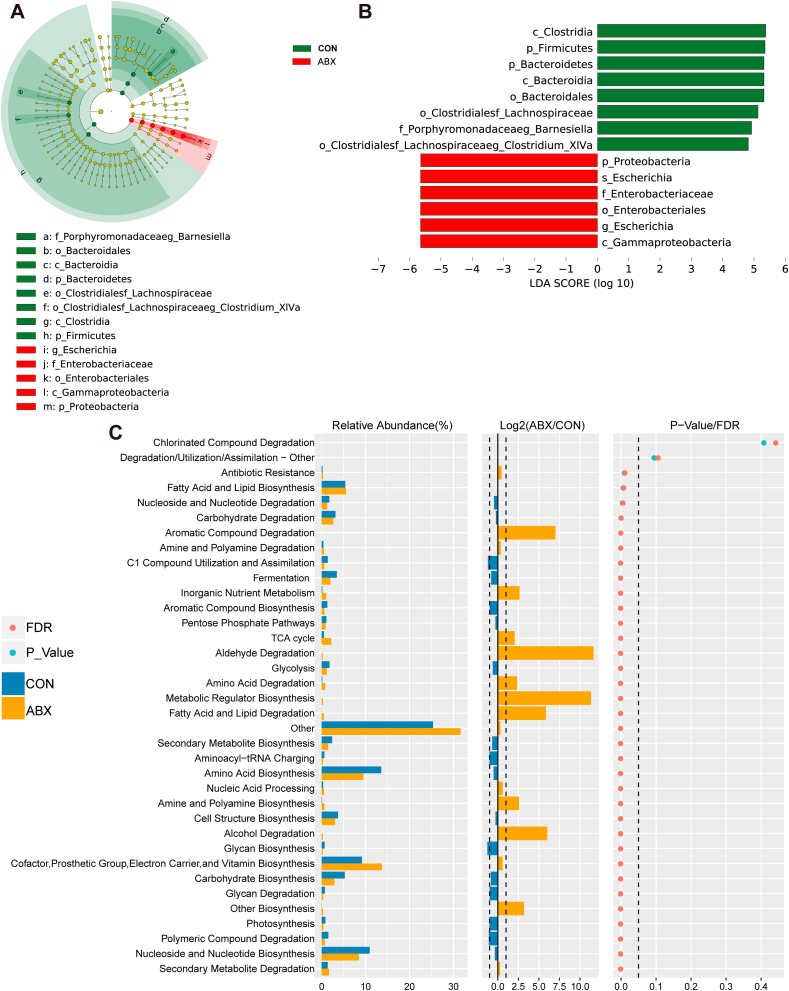
We employed LEfSe to determine taxonomic biomarkers that contributed to variations in the ABX-treated microbiome, with high strictness (LDA >4.8). As illustrated in the LEfSe-generated cladogram plot (Fig. 3A), the CON and ABX groups exhibited significantly different microbial community characteristics. Compared with the CON group, ABX group possessed significantly higher abundances of the class Gammaproteobacteria, the genus Escherichia, the order Enterobacteriales, the family Enterobacteriacea, the species Escherichia, and the phylum Proteobacteria. In contrast, the CON group contained higher abundances of the class Clostridia, the phylum Firmicutes, the phylum Bacteroidetes, the class Bacteroidia, the order Bacteroidales, the order Clostridialesf Lachnospiraceae, the family Porphyromonadaceaeg Barnesiella, and the order Clostridialesf Lachnospiraceaeg Clostridium XlVa (Fig. 3B).
Fig. 3.
Potential biomarkers defined by LEfSe, prediction of microbial function by PICRUSt2
A: Taxonomic cladogram generated by LEfSe analysis. B: LEfSe linear discriminant analysis (LDA) for taxa with P < 0.05 and scores >4.8. C: Significantly altered MetaCyc metabolic pathway between two groups predicted based on sequencing data of 16S rRNA gene (statistical results are presented in Table S3).
Next, PICRUSt2 was used to predict the metagenomes of the gut microbes. The resulting functional profiles highlighted 34 pathways that were significantly changed in ABX mice compared with CON mice (Fig. 3C). The categories of microbial functions that changed in response to treatment with ABX included biosynthesis and degradation of fatty acids and lipids, glycan biosynthesis and degradation, aminoacyl-tRNA charging, carbohydrate biosynthesis, TCA cycle, biosynthesis and degradation of amines and polyamines, and degradation of aldehydes (Fig. 3C, Table S3). Of these, 17 metabolic pathways (e.g., TCA cycle, inorganic nutrient metabolism, amino acid degradation, amine and polyamine biosynthesis, alcohol degradation, fatty acid and lipid degradation, aromatic compound degradation, and metabolic regulator biosynthesis, and aldehyde degradation, etc.) were significantly increased in the ABX group compared with the CON group (Fig. 3C, Table S3).
3.4. Splenic metabolomic profiles correlated with ABX-induced changes in percentage of splenic cell types, spleen weight, and the gut microbiota
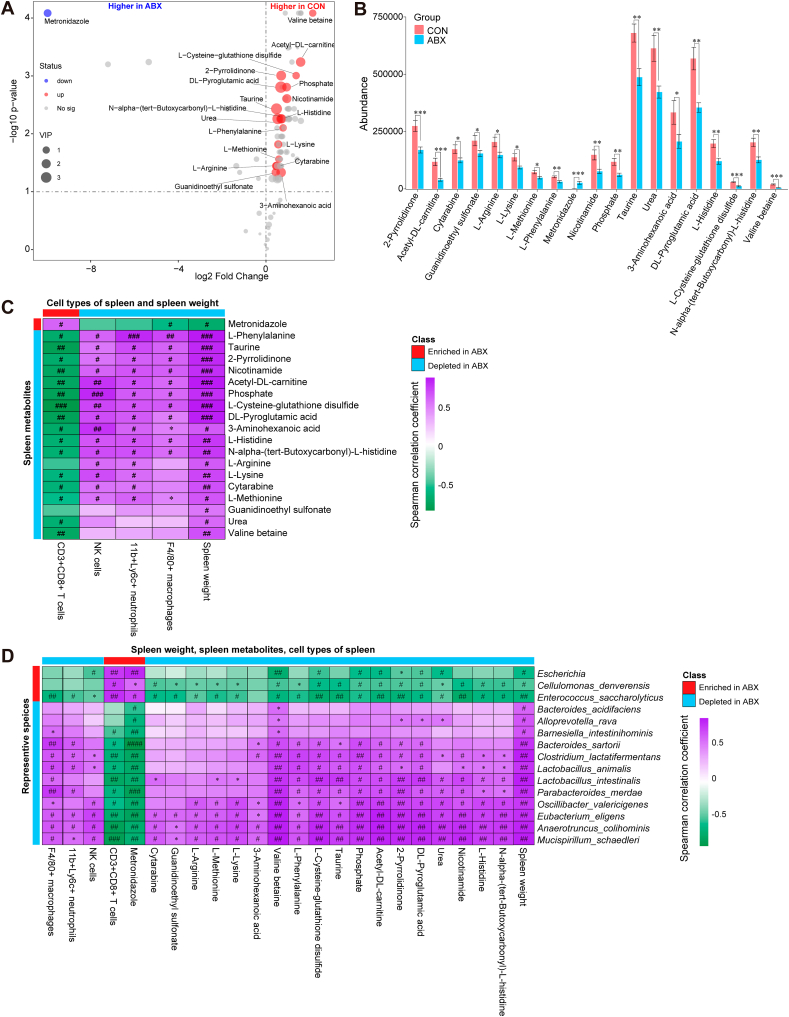
To further explore ABX-induced spleen weight loss, changes in the percentage of splenic cell populations, and metabolic changes, we performed untargeted metabolomic analysis of mouse spleen samples. After the initial screen, 99 metabolites from each group were selected for more detailed evaluation. We further carried out supervised data analysis using OPLS-DA to clarify metabolic variations. The OPLS-DA score plots for the CON and ABX groups were clearly separated, suggesting that the model was able to distinguish between metabolites from the CON group and from the ABX group (Fig. S3). A volcano plot was constructed by analyzing the P-values from the Mann-Whitney test and the variable importance (VIP) scores from the OPLS-DA. Metabolites matching the requirements of P < 0.05 (FDR-corrected P < 0.05) and VIP >1 were defined as differentially represented metabolites (Fig. 4A). Eighteen metabolites, including 2-pyrrolidinone, 3-aminohexanoic acid, acetyl-DL-carnitine, cytarabine, DL-pyroglutamic acid, guanidinoethyl sulfonate, L-histidine, L-arginine, L-cysteine-glutathione disulfide, L-lysine, L-methionine, L-phenylalanine, N-alpha-(tert-butoxycarbonyl)-L-histidine, nicotinamide, phosphate, taurine, urea, and valine betaine, exhibited significantly lower abundance in the spleens of mice in the ABX group than in the CON group (Fig. 4B, Table S4). In contrast metronidazole exhibited higher abundance in the spleens of ABX mice compared with the CON mice (Fig. 4B, Table S4). Taken together, these results suggest that, instead of being due solely to the decrease in spleen weight observed in ABX-treated mice, the reduction in specific metabolites in the spleen might result from long-term exposure to ABX.
Fig. 4.
Alterations in splenic metabolites between the two groups, and Spearman correlations in splenic metabolites, spleen cell populations, and gut microbes
A: Volcano plot showing splenic metabolite profiles between the CON and ABX groups. X-axis indicates the log2 transformed fold change in splenic metabolite abundance, and y-axis indicates the -log10 transformed P-value using the Wilcox test. Horizontal lines indicate P < 0.05. Red or blue dots indicate differential metabolites between the two groups. B: Differences in abundance of splenic differential metabolites between the two groups (statistical results are presented in Table S4). C: Spearman correlation analysis of splenic differential metabolites with percentage of splenic cell population or spleen weight. D: Spearman correlation analysis of differential species (average of abundance >0.1% in all samples) with splenic differential metabolites, percentage of splenic cell populations, or spleen weight. Values represent mean ± S.E.M. (CON: n = 8. ABX: n = 9). #P (FDR-corrected) < 0.05, ##P (FDR-corrected) < 0.01, ###P (FDR-corrected) < 0.001, ####P (FDR-corrected) < 0.0001. *P < 0.05, **P < 0.01, ***P < 0.001. N.S.: not significant.
The correlations among differentially represented spleen metabolites, percentage of spleen cell types, and spleen weight were analyzed for all mice (Fig. 4C). Spleen weight was positively correlated with the abundance of the 18 metabolites that were reduced in the ABX group and negatively correlated with the abundance of metronidazole in the ABX group (Fig. 4C). The abundance of 12 metabolites, including 3-aminohexanoic acid, L-methionine, L-histidine, N-alpha-(tert-butoxycarbonyl)-L-histidine, phosphate, acetyl-DL-carnitine, DL-pyroglutamic acid, L-cysteine-glutathione disulfide, L-phenylalanine, nicotinamide, 2-pyrrolidinone, and taurine, was positively associated with the percentage of several cell types in the splenic cell population, such as NK cells, macrophages, and neutrophils, but it was negatively associated with the percentage of CD8+ T cells (Fig. 4C). To determine the most unique and functionally critical taxa that differed in abundance between the ABX group and the CON group, correlation analyses were performed between the differentially represented species (>0.1% in mean abundance across all samples, henceforth referred to as differential species) and spleen weight, percentage of spleen cell populations, and spleen differential metabolites (Fig. 4D). Significant correlation was found between differential species and differential metabolites in the spleen and both the percentage of spleen cell populations and the spleen weight (Fig. 4D). Overall, the abundances of 10 species (Barnesiella intestinihominis, Bacteroides sartorii, Clostridium lactatifermentans, Lactobacillus animalis, Lactobacillus intestinalis, Parabacteroides merdae, Oscillibacter valericigenes, Eubacterium eligens, Anaerotruncus colihominis, Mucispirillum schaedleri) was negatively associated with the percentage of splenic CD8+ T cells and the abundance of the splenic metabolite metronidazole, but it was positively associated with the percentage of splenic macrophages and spleen weight (Fig. 4D). Metronidazole was one of the antibiotics used in this study. We found that the abundance of the ABX-depleted species Mucispirillum schaedleri, Anaerotruncus colihominis, and Lactobacillus intestinalis was negatively correlated with the percentage of CD8+ T cells in the ABX group, which in turn was negatively correlated with the concentration of L-cysteine-glutathione disulfide. Furthermore, L-cysteine-glutathione disulfide was positively correlated with spleen weight (Fig. S4).
3.5. Plasma metabolomic profiles correlated with ABX-induced changes in percentage of splenic cell types, spleen weight, and gut the microbiota
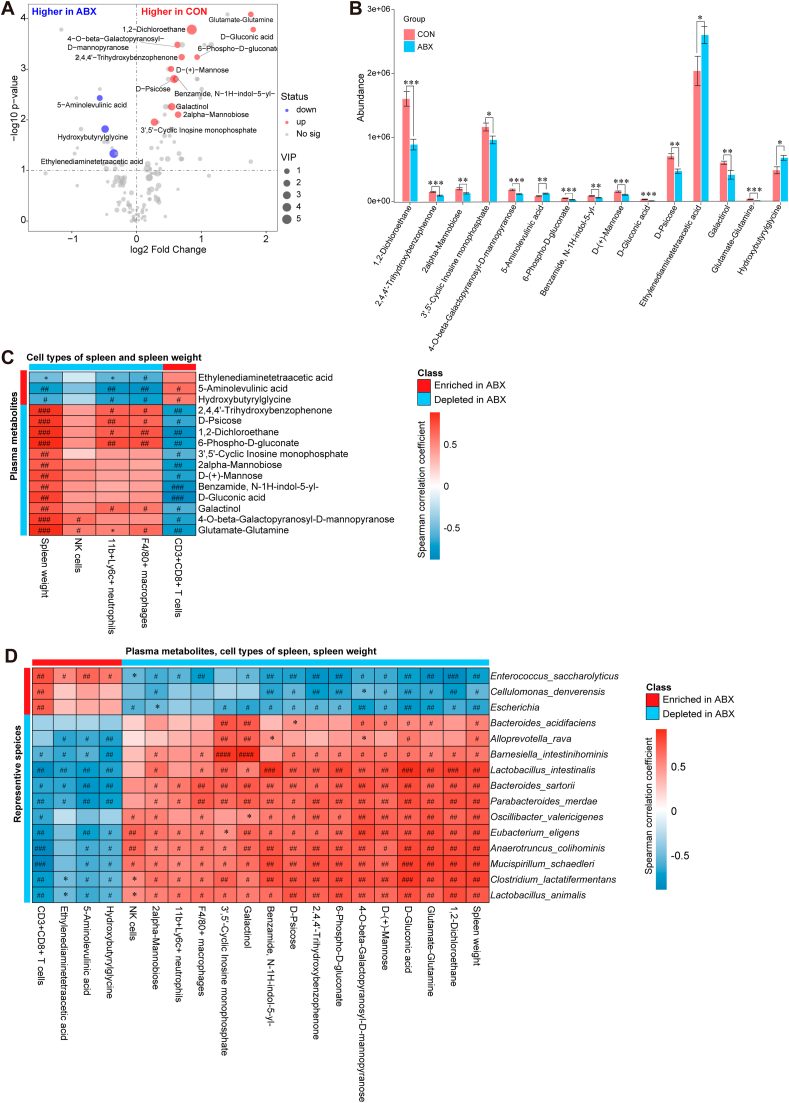
We further investigated whether plasma metabolomic biomarkers were associated with ABX-induced changes in the percentage of splenocyte types, spleen weight, and the gut microbiota. A total of 156 metabolite signatures were detected after preprocessing the detected signals. After 14 days of ABX treatment, the plasma metabolic profiles were significantly different between the CON and ABX groups. The OPLS-DA model shows that the metabolic profiles of the two groups were completely separated (Fig. S5). Using VIP >1 and P < 0.05 (FDR-corrected P < 0.05) as cut-offs, we identified 15 differentially represented metabolites (Fig. 5A). We identified 12 metabolites (1,2-dichloroethane, 2,4,4'-trihydroxybenzophenone, 2-alpha-mannobiose, 3',5'-cyclic inosine monophosphate, 4-O-beta-galactopyranosyl-D-mannopyranose, 6-phospho-D-gluconate, benzamide, N–1H-indol-5-yl-, D-(+)-mannose, D-gluconic acid, D-psicose, galactinol, and glutamate-glutamine) whose abundance was significantly lower in plasma from the ABX group compared with the CON group (Fig. 5B, Table S5). In contrast, we found a significantly higher abundance of three metabolites (5-aminolevulinic acid, ethylenediaminetetraacetic acid, hydroxybutyrylglycine) in plasma from the ABX group compared with the CON group (Fig. 5B, Table S5).
Fig. 5.
Alterations in plasma metabolites between the two groups, and Spearman correlations in plasma metabolites, spleen cell populations, and gut microbes
A: Volcano plot showing plasma metabolite profiles between the CON and ABX groups. X-axis indicates the log2 transformed fold change in plasma metabolite abundance, and y-axis indicates the -log10 transformed P-value using the Wilcox test. Horizontal lines indicate P < 0.05. Red or blue dots indicate differential metabolites between the two groups. B: Differences in abundance of plasma differential metabolites between the two groups (statistical results are presented in Table S5). C: Spearman correlation analysis of plasma differential metabolites with percentage of splenic cell population or spleen weight. D: Spearman correlation analysis of differential species (average of abundance >0.1% in all samples) with plasma differential metabolites, percentage of splenic cell populations, or spleen weight. Values represent mean ± S.E.M. (CON: n = 8. ABX: n = 9). #P (FDR-corrected) < 0.05, ##P (FDR-corrected) < 0.01, ###P (FDR-corrected) < 0.001, ####P (FDR-corrected) < 0.0001. *P < 0.05, **P < 0.01, ***P < 0.001. N.S.: not significant.
Spearman correlation analysis showed a positive correlation between all 12 plasma ABX-depleted metabolites and spleen weight, but a negative correlation between all 12 plasma ABX-depleted metabolites and splenic CD8+ T cell percentage (Fig. 5C). However, the two ABX-enriched plasma metabolites—5-aminolevulinic acid and hydroxybutyrylglycine—were positively correlated with splenic CD8+ T cell percentage and negatively correlated with the percentage of splenic neutrophils, splenic macrophages, and spleen weight (Fig. 5C). We also detected extensive correlations between differential species, plasma metabolites, splenic cell populations, and spleen weight (Fig. 5D, Fig. S6). A total of 10 ABX-depleted species (Barnesiella intestinihominis, Lactobacillus intestinalis, Bacteroides sartorii, Parabacteroides merdae, Oscillibacter valericigenes, Eubacterium eligens, Anaerotruncus colihominis, Mucispirillum schaedleri, Clostridium lactatifermentans, and Lactobacillus animalis) were significantly positively correlated with 12 ABX-depleted plasma metabolites, percentage of splenic cell populations (NK cells, neutrophils, macrophages), and spleen weight, whereas they were negatively correlated with two ABX-enriched plasma metabolites (5-aminolevulinic acid and hydroxybutyrylglycine) and percentage of splenic CD8+ T cells (Fig. 5D). These associations are shown in the correlation network diagram (Fig. S6).
3.6. Relationships between ABX-induced changes in the gut microbiota, splenic metabolites, plasma metabolites, cortical metabolites, and splenic function
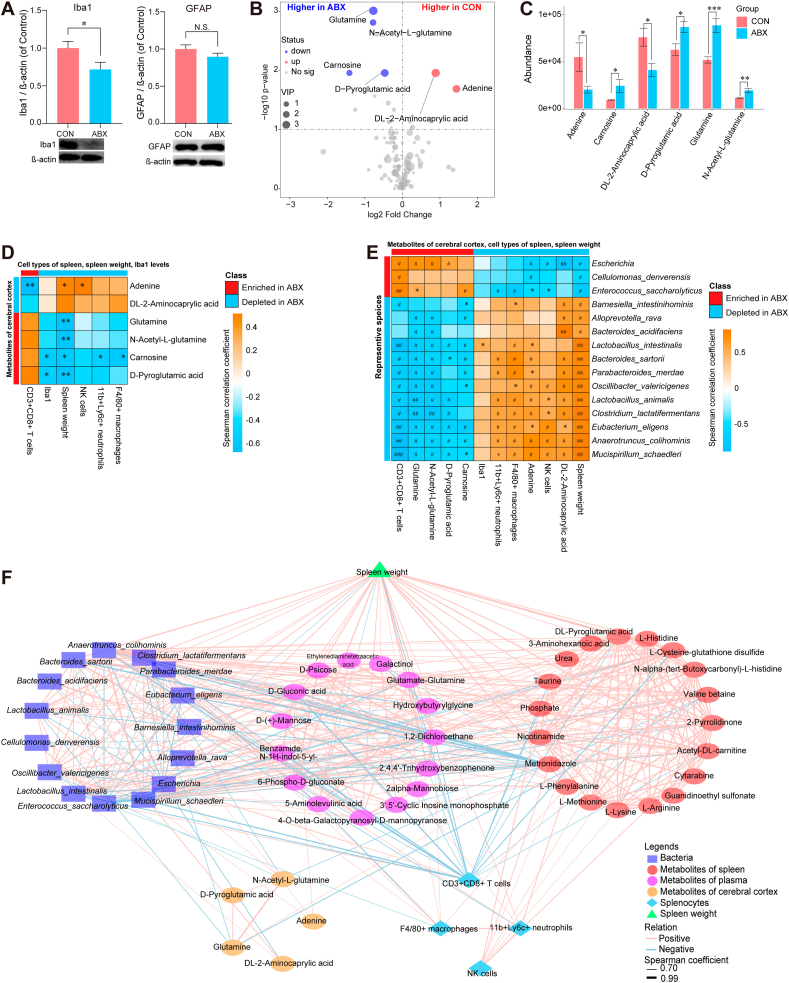
We measured the expression of the microglial marker Iba1 and the astrocytic marker GFAP in the cerebral cortex samples from the ABX and CON groups by western blotting. Mice treated with ABX showed reduced levels of Iba1 expression compared with CON mice; however, GFAP expression did not differ between the two groups (Fig. 6A).
Fig. 6.
Expression of Iba1 and GFAP in the cerebral cortex, altered metabolites in the cerebral cortex, and joint correlation analysis of microbial, multi-location metabolites, and splenic cell populations
A: Expression of Iba1 and GFAP in the cerebral cortex from two groups. B: Volcano plot showing cerebral cortical metabolite profiles between the CON and ABX groups. X-axis indicates the log2 transformed fold change in cerebral cortical metabolite abundance, and y-axis indicates the -log10 transformed P-value using the Wilcox test. Horizontal lines indicate P < 0.05. Red or blue dots indicate differential metabolites between the two groups. C: Differences in abundance of differential metabolites in the cerebral cortex between the two groups (statistical results are presented in Table S6). D: Spearman correlation analysis of differential metabolites in the cerebral cortex with percentage of splenic cell population, Iba1 expression in the cerebral cortex, or spleen weight. E: Spearman correlation analysis of differential species (average of abundance >0.1% in all samples) with differential metabolites in the cerebral cortex, Iba1 level in the cerebral cortex, percentage of splenic cell populations, or spleen weight. F: Joint correlation analysis of spleen weight, percentage of spleen cell population, representative metabolites of different tissues (spleen, plasma, cerebral cortex) and gut microbiota for all mice. Red lines denote positive correlations, blue lines denote negative correlations, and thicker lines represent larger correlation coefficients (Spearman correlation analysis, absolute value of correlation coefficient >0.7, FDR-corrected P < 0.05). Circles indicate metabolites, and different colors indicate originating from different types of tissues. Bacteria are indicated by squares, triangles indicate spleen weights, and diamonds indicate splenic cell populations. Values represent mean ± S.E.M. (CON: n = 8. ABX: n = 9). #P (FDR-corrected) < 0.05, ##P (FDR-corrected) < 0.01, ###P (FDR-corrected) < 0.001. *P < 0.05, **P < 0.01, ***P < 0.001. N.S.: not significant.
After data preprocessing, a total of 139 metabolic features were identified by untargeted metabolomics analysis of cerebral cortex samples from the ABX and CON groups. Subsequently, the OPLS-DA model was constructed to identify metabolites that contributed towards group differentiation. The metabolic profiles of the two groups as determined by the OPLS-DA model were somewhat separate (Fig. S7). Using the same procedures as that used for the spleen and plasma metabolomics analyses, six differential metabolites from the cerebral cortex were selected (P < 0.05, VIP >1, FDR-corrected P < 0.3) (Fig. 6B). Compared with the CON group, we identified four metabolites (carnosine, D-pyroglutamic acid, glutamine, and N-acetyl-L-glutamine) with elevated abundance in the ABX group and two metabolites (adenine and DL-2-aminocaprylic acid) with decreased abundance in the ABX group (Fig. 6C, Table S6).
We noticed that the level of Iba1 in the cerebral cortex was significantly and negatively correlated with the abundance of metabolites such as carnosine and D-pyroglutamic acid (Fig. 6D). Furthermore, spleen weight was negatively correlated with the four ABX-enriched metabolites in the cerebral cortex (Fig. 6D). Spearman rank correlation analysis showed extensive positive correlations between ABX-depleted species and ABX-depleted metabolites in the cerebral cortex, splenic cell populations (NK cells, neutrophils, and macrophages), and spleen weight. In contrast, significant negative correlations were found between ABX-depleted species and ABX-enriched metabolites, and splenic CD8+ T cells (Fig. 6E). The Iba1 level in the cerebral cortex was positively correlated with the abundance of Lactobacillus intestinalis (Fig. 6E). Similar correlations were found in the correlation network plots (Fig. S8).
To investigate the relationship between spleen weight, percentage of spleen cell populations, differential metabolites in the spleen, and the gut microbiota, we performed a joint analysis of the above data for all of the mice. Correlation network analysis revealed highly sophisticated patterns of interactions (463 correlations, Spearman rho ≥0.7, P < 0.05, FDR-corrected P < 0.05) between taxa (mean abundance >0.1% across all samples), splenic cell populations, spleen weight, and metabolites in the spleen, plasma, and cerebral cortex (Fig. 6F). Interestingly, the correlations were stronger between different tissues than within each tissue, suggesting strong functional associations between the brain, gut microbiota, plasma, and spleen. Of interest, the concentrations of five plasma metabolites (1,2-dichloroethane, D-gluconic acid, D-psicose, 6-phospho-D-gluconate, and 2,4,4'-trihydroxybenzophenone) were positively correlated with the abundance of Clostridium lactatifermentans and Lactobacillus animalis, which were negatively correlated with glutamine in the cerebral cortex. Plasma D-psicose was further positively correlated with the splenic metabolite L-phenylalanine, splenic neutrophil percentage, and spleen weight. Seven plasma metabolites, including D-gluconic acid, 1,2-dichloroethane, glutamate-glutamine, benzamide, N–1H-indol-5-yl-, 2,4,4'-trihydroxybenzophenone, 6-phospho-D-gluconate, and 4 -O-beta-galactopyranosyl-D-mannopyranose were positively correlated with the species Anaerotruncus colihominis and Mucispirillum schaedleri, which were further positively correlated with adenine in the cerebral cortex. Of note, the plasma D-gluconic acid level directly correlated with adenine in the cerebral cortex. The tight relationship between gut microbes, plasma metabolites, and cerebral cortical metabolites indicates that specific microbes may be involved in the metabolism of specific plasma and brain metabolites and regulate brain function. We observed correlations between plasma metabolites, splenic metabolites, and cerebral cortical metabolites. Some highly correlated features included the plasma metabolites glutamate-glutamine, 2,4,4'-trihydroxybenzophenone, and 1,2-dichloroethane, which correlated positively with the splenic metabolite valine-betaine; the latter further correlated with two metabolites (glutamine and N-acetyl-L-glutamine) in the cerebral cortex, spleen CD8+ T cell population, and spleen weight. We found that adenine in the cerebral cortex was positively correlated with the plasma metabolite D-gluconic acid, which was positively correlated with two splenic metabolites (phosphate and L-cysteine-glutathione disulfide); phosphate and L-cysteine-glutathione disulfide were further negatively correlated with splenic CD8+ T cell population and positively correlated with spleen weight.
4. Discussion
The main findings of this study are as follows. First, ABX treatment resulted in decreased spleen weight compared with the CON group. The percentage of cell types of spleen was significantly altered in the ABX group, including a decrease in the percentage of three cell types (NK cells, macrophages, and neutrophils) and, conversely, a significant increase in the percentage of CD8+ T cells. The percentage of these cell populations significantly correlated with spleen weight. Second, ABX treatment leads to depletion of a large proportion of the gut microbiota. The α- and β-diversity of the gut microbiota, as well as the composition of the gut microbiota, showed significant differences between the two groups. Third, LEfSe analysis determined that the class Gammaproteobacteria, the genus Escherichia, the phylum Proteobacteria, the order Enterobacteriales, and the family Enterobacteriacea are potential microbial markers of the ABX group. Predictive functional analysis of the gut microbiota revealed that ABX caused alterations in multiple metabolic pathways, such as those involved in biosynthesis and degradation of fatty acids and lipids, degradation of amines and polyamines, carbohydrate biosynthesis, and the TCA (tricarboxylic acid) cycle. Fourth, ABX treatment caused a reduction in the expression of the microglia marker Iba1 in the cerebral cortex. Fifth, untargeted metabolomics analysis of spleen, plasma, and cerebral cortex samples demonstrated that ABX treatment caused alterations in the levels of a number of compounds. There were strong correlations among these differential compounds, the level of Iba1 expression in the cerebral cortex, spleen cell populations, spleen weight, and the relative abundance of microbes. Finally, joint network analysis showed strong interconnections among gut microbes and metabolites (plasma, spleen, cerebral cortex), spleen cell populations, and spleen weight. Collectively, these findings suggest that ABX-induced microbiome depletion could affect spleen and brain function through the gut–microbiota–spleen–brain axis.
In this study, we found that the percentages of neutrophils, NK cells, and macrophages were significantly reduced in the spleens of ABX-treated mice, and that these cell types were positively correlated with spleen weight. We previously reported that spleen weight positively correlates with percentages of cell types (i.e., neutrophils, proerythroblasts, B cells, macrophages, and dendritic cells) in imiquimod-treated mice (Shinno-Hashimoto et al., 2022). Therefore, spleen weight is likely to be affected by changes in the abundance of specific splenic cellular components. In contrast, ABX treatment caused increase in CD8+ T cells in the spleen, and there was a negative correlation between CD8+ T cell percentage and spleen weight. Antibiotic treatment can cause abnormalities in the immune cell populations in the GI tract and spleen (Kennedy et al., 2018), indicating that the immune system responds to pathogenic and commensal microbial populations. Therefore, ABX-induced microbiome depletion may alter the cellular composition of the host spleen, resulting in a reduction in spleen weight.
Escherichia was the most predominant bacterium in ABX-treated mice, with a relative abundance of 88.6%. Previous reports have demonstrated that Escherichia coli (E. coli) strains are diverse, ranging from innocuous GI residents to multiple pathotypes capable of causing intestinal or extraintestinal disease, such as infantile diarrhea, hemorrhagic colitis, and more, related to antibiotic resistance (Croxen et al., 2013; Moyenuddin et al., 1989; Subramanian et al., 2009). Given the existence of various antibiotic-resistant E. coli strains, the high abundance of Escherichia in the gut of ABX-treated mice may indicate the presence of antibiotic-resistant strains. Collectively, it seems that antibiotic use drives changes in the composition of the intestinal microbiota, leading to an imbalance in physiological homeostasis and permitting the long-term growth and colonization of antibiotic-resistant microorganisms in the gut.
The 18 ABX-reduced splenic compounds were positively correlated with the proportion of spleen cell types such as NK cells, macrophages, and neutrophils, and negatively correlated with the proportion of splenic CD8+ T cells. Bacterial or parasitic infections are known to possibly affect splenic responses (Lewis et al., 2019). Although the current study does not provide strong evidence for this, these compounds may be involved in splenic-mediated immune responses. The differentially represented taxa between the two groups were significantly correlated with splenic metabolites and splenic cell populations. Collectively, these findings suggest that ABX-induced microbiome depletion modulates splenic function by altering metabolite expression, although further study is needed to confirm this.
In this study, we detected Enterococcus saccharolyticus (Hammad et al., 2014), a bacterium that is associated with antibiotic resistance, in the intestine of ABX-treated mice. Furthermore, there was a positive correlation between plasma levels of 5-aminolevulinic acid (5-ALA) and relative abundance of Enterococcus saccharolyticus, suggesting that Enterococcus saccharolyticus in the intestine may contribute to the production of 5-ALA. Microbial production of 5-ALA by photosynthetic bacteria has been demonstrated previously (Liu et al., 2014; Sasaki et al., 2002). Increasing evidence suggests that 5-ALA-induced apoptosis can be used to treat tumors (Huang et al., 2016; Liu et al., 2019; Mamalis et al., 2016; Stummer et al., 2006). We found that plasma levels of 5-ALA were significantly elevated in the ABX-treated mice, and negatively correlated with splenic neutrophils, macrophages, and spleen weight. Given the beneficial role of 5-ALA (Jiang et al., 2022), the increase in 5-ALA levels induced by ABX treatment may have a compensatory effect on impaired splenic functions. Taken together, our findings suggest that ABX-induced microbiome depletion could affect the synthesis and metabolism of a number of host metabolites that regulate splenic cell populations and spleen function via humoral circulation. Nonetheless, more detailed study is needed to elucidate the role of Enterococcus saccharolyticus in the production of 5-ALA, as well as the role of 5-ALA in the spleen.
Increasing evidence suggests that the gut microbiome plays a crucial role in microglial maturation and activation both in healthy individuals and in patients with CNS disease (Cook and Prinz, 2022; Erny and Prinz, 2020). Erny et al. (2015) demonstrated that GF mice have global defects in microglia and altered cell proportions, leading to an impaired innate immune response. Furthermore, microbiome-derived compounds are known to regulate microglial homeostasis in the brain (Erny et al., 2015). We previously reported that repeated administration of PLX5622 (a specific inhibitor of the colony-stimulating factor 1 receptor) affects the relative abundance of gut microbiota components, and that there are significant correlations between microglial markers in the brain and the relative abundance of gut microbiota components, suggesting microbiome-microglia crosstalk through the brain–gut axis (Yang et al., 2022b). Compared with CON mice, the ABX mice exhibited low expression of the microglia marker Iba1 in the cerebral cortex. Collectively, these findings suggest crosstalk between the gut microbiota and the microglia in the brain through the gut–microbiota–brain axis.
Carnosine, a dipeptide that is present in high concentrations in the brain, is a putative neurotransmitter (Caruso et al., 2019; Tiedje et al., 2010). A growing body of evidence shows the potentially beneficial effects of carnosine in psychiatric and neurological disorders (Schön et al., 2019). In this study, we found that carnosine was elevated in the cerebral cortex of ABX-treated mice, which is in keeping with its known neuroprotective effects in the brain. Furthermore, carnosine was negatively correlated with Iba1 expression in the cerebral cortex, consistent with the previous report that carnosine in the brain is mainly metabolized by microglia, oligodendrocytes, astrocytes, and macrophages (Caruso et al., 2019). Substantial evidence suggests that carnosine has anti-aggregatory, anti-oxidant, anti-inflammatory, and neuroprotective effects in the CNS (Caruso et al., 2019). Interestingly, we found that the level of carnosine in the cerebral cortex was negatively correlated with the proportion of splenic macrophages and neutrophils, suggesting a brain–spleen axis.
The joint analysis showed that the abundance of the probiotic bacterial strain Lactobacillus animalis (Sahoo et al., 2015) was correlated with plasma levels of D-psicose and negatively correlated with glutamine levels in the cerebral cortex. Glutamine is a by-product of ammonia metabolism that is capable of causing reactive oxidative stress in astrocytes and inducing the mitochondrial permeability transition, which causes neurotoxicity through the action of mitochondrial phosphate activated glutaminase (Albrecht and Norenberg, 2006). The anti-hyperlipidemic effect of D-psicose is related to its hypoglycemic, hypolipidemic, and antioxidant activities, which make it optimal for the prevention of diabetes and related health problems (Chung et al., 2012), further suggesting that plasma metabolism is regulated by intestinal microorganisms and affects the functional state of neuronal cells. Furthermore, plasma D-psicose levels were positively correlated with L-phenylalanine (an essential amino acid in the spleen) levels, which are positively correlated with the proportion of splenic neutrophils, and ultimately spleen weight. Collectively, our findings suggest that the gut–spleen–brain axis regulates the metabolic and functional state of neuronal cells via microbial-derived metabolites, and that the brain may modulate splenic cell types and splenic function.
We previously reported that ABX-induced depletion of the gut microbiome is associated with stress resilience in mice exposed to chronic social defeat stress (Wang et al., 2020b) and protects against MPTP-induced dopaminergic neurotoxicity in the brain (Pu et al., 2019) and LPS (lipopolysaccharide)-induced acute lung injury (Hashimoto et al., 2022). Furthermore, LPS-induced splenomegaly in mice was associated with higher levels of pro-inflammatory cytokines (Ma et al., 2022a, 2022b; Zhang et al., 2020b, 2021a), suggesting that an increase in spleen weight is associated with systemic inflammation. Considering the decreased spleen weight induced by treatment with ABX, ABX treatment possibly has anti-inflammatory effects in mice. Given the anti-inflammatory role of the gut–microbiota–spleen–brain axis, it is likely that ABX-induced microbiome depletion has anti-inflammatory effects in several animal models.
This study had one limitation, in that we divided the spleen to investigate the relationship between splenic cell types and metabolites in the spleen. However, the spleen is divided by function and structure into the red pulp, white pulp, and marginal zone, suggesting that the immune cells are differentially organized in these three structures (Lewis et al., 2019). The differences in these structures may affect the functions of splenic cells and metabolites, and further detailed study is needed to address this possibility.
5. Conclusion
The findings from the present study suggest that ABX-induced microbiome depletion causes a decrease in spleen weight and abnormal expression of metabolites in the blood, spleen, and brain. In addition to microbiome depletion, ABX treatment likely affects spleen and brain function through the gut–microbiota–spleen–brain axis.
Declaration of competing interest
Dr. Hashimoto is the inventor of filed patent applications on “The use of R-Ketamine in the treatment of psychiatric diseases”, “(S)-norketamine and salt thereof as pharmaceutical”, “R-Ketamine and derivative thereof as prophylactic or therapeutic agent for neurodegeneration disease or recognition function disorder”, “Preventive or therapeutic agent and pharmaceutical composition for inflammatory diseases or bone diseases”, “R-Ketamine and its derivatives as a preventive or therapeutic agent for a neurodevelopmental disorder”, and “Preventive or therapeutic agent and pharmaceutical composition for inflammatory diseases” by the Chiba University. Dr. Hashimoto also declares that he has received research support and consultant from Abbott, Boehringer Ingelheim, Daiichi-Sankyo, Meiji Seika Pharma, Seikagaku Corporation, Sumitomo-Pharma, Taisho, Otsuka, Murakami Farm and Perception Neuroscience. The other authors have no conflict of interest.
Acknowledgment
This study was supported by the grants from Japan Society for the Promotion of Science (to K.H., 21H00184 and 21H05612), JST OPERA Program Japan (to C.M JPMJOP1831), and unrestricted grant of Yamada Bee Company, Japan (to C.M). Ms. Xiayun Wan and Dr. Yong Yang were supported by the Academic Research & Innovation Management Organization of Chiba University (Chiba, Japan). Dr. Yong Yang was supported by the Japan China Sasakawa Medical Fellowship (Tokyo, Japan). We thank Emily Crow, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbih.2022.100573.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
The 16S rRNA sequencing data have been deposited to the NCBI Sequence Read Archive and are available at the accession number PRJNA887905.
References
- Albrecht J., Norenberg M.D. Glutamine: a Trojan horse in ammonia neurotoxicity. Hepatology (Baltimore, Md. 2006;44(4):788–794. doi: 10.1002/hep.21357. [DOI] [PubMed] [Google Scholar]
- Aron-Wisnewsky J., Clément K. The gut microbiome, diet, and links to cardiometabolic and chronic disorders. Nat. Rev. Nephrol. 2016;12(3):169–181. doi: 10.1038/nrneph.2015.191. [DOI] [PubMed] [Google Scholar]
- Borovikova L.V., Ivanova S., Zhang M., Yang H., Botchkina G.I., Watkins L.R., Wang H., Abumrad N., Eaton J.W., Tracey K.J. Vagus nerve stimulation attenuates the systemic inflammatory response to endotoxin. Nature. 2000;405(6785):458–462. doi: 10.1038/35013070. [DOI] [PubMed] [Google Scholar]
- Breit S., Kupferberg A., Rogler G., Hasler G. Vagus nerve as modulator of the brain-gut axis in psychiatric and inflammatory disorders. Front. Psychiatr. 2018;9:44. doi: 10.3389/fpsyt.2018.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronte V., Pittet M.J. The spleen in local and systemic regulation of immunity. Immunity. 2013;39(5):806–818. doi: 10.1016/j.immuni.2013.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchmann Godinho D., da Silva Fiorin F., Schneider Oliveira M., Furian A.F., Rechia Fighera M., Freire Royes L.F. The immunological influence of physical exercise on TBI-induced pathophysiology: crosstalk between the spleen, gut, and brain. Neurosci. Biobehav. Rev. 2021;130:15–30. doi: 10.1016/j.neubiorev.2021.08.006. [DOI] [PubMed] [Google Scholar]
- Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A., Kelley S.T., Knights D., Koenig J.E., Ley R.E., Lozupone C.A., McDonald D., Muegge B.D., Pirrung M., Reeder J., Sevinsky J.R., Turnbaugh P.J., Walters W.A., Widmann J., Yatsunenko T., Zaneveld J., Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso G., Caraci F., Jolivet R.B. Pivotal role of carnosine in the modulation of brain cells activity: multimodal mechanism of action and therapeutic potential in neurodegenerative disorders. Prog. Neurobiol. 2019;175:35–53. doi: 10.1016/j.pneurobio.2018.12.004. [DOI] [PubMed] [Google Scholar]
- Chang L., Wei Y., Hashimoto K. Brain-gut-microbiota axis in depression: a historical overview and future directions. Brain Res. Bull. 2022;182:44–56. doi: 10.1016/j.brainresbull.2022.02.004. [DOI] [PubMed] [Google Scholar]
- Chung M.Y., Oh D.K., Lee K.W. Hypoglycemic health benefits of D-psicose. J. Agric. Food Chem. 2012;60(4):863–869. doi: 10.1021/jf204050w. [DOI] [PubMed] [Google Scholar]
- Cook J., Prinz M. Regulation of microglial physiology by the microbiota. Gut Microb. 2022;14(1) doi: 10.1080/19490976.2022.2125739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxen M.A., Law R.J., Scholz R., Keeney K.M., Wlodarska M., Finlay B.B. Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 2013;26(4):822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cryan J.F., O'Riordan K.J., Sandhu K., Peterson V., Dinan T.G. The gut microbiome in neurological disorders. Lancet Neurol. 2020;19(2):179–194. doi: 10.1016/S1474-4422(19)30356-4. [DOI] [PubMed] [Google Scholar]
- Cryan J.F., O'Riordan K.J., Cowan C.S.M., Sandhu K.V., Bastiaanssen T.F.S., Boehme M., Codagnone M.G., Cussotto S., Fulling C., Golubeva A.V., Guzzetta K.E., Jaggar M., Long-Smith C.M., Lyte J.M., Martin J.A., Molinero-Perez A., Moloney G., Morelli E., Morillas E., O'Connor R., Cruz-Pereira J.S., Peterson V.L., Rea K., Ritz N.L., Sherwin E., Spichak S., Teichman E.M., van de Wouw M., Ventura-Silva A.P., Wallace-Fitzsimons S.E., Hyland N., Clarke G., Dinan T.G. The microbiota-gut-brain axis. Physiol. Rev. 2019;99(4):1877–2013. doi: 10.1152/physrev.00018.2018. [DOI] [PubMed] [Google Scholar]
- Douglas G.M., Maffei V.J., Zaneveld J.R., Yurgel S.N., Brown J.R., Taylor C.M., Huttenhower C., Langille M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38(6):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D., Hrabě de Angelis A.L., Jaitin D., Wieghofer P., Staszewski O., David E., Keren-Shaul H., Mahlakoiv T., Jakobshagen K., Buch T., Schwierzeck V., Utermöhlen O., Chun E., Garrett W.S., McCoy K.D., Diefenbach A., Staeheli P., Stecher B., Amit I., Prinz M. Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 2015;18(7):965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erny D., Prinz M. How microbiota shape microglial phenotypes and epigenetics. Glia. 2020;68(8):1655–1672. doi: 10.1002/glia.23822. [DOI] [PubMed] [Google Scholar]
- Fan Y., Pedersen O. Gut microbiota in human metabolic health and disease. Nat. Rev. Microbiol. 2021;19(1):55–71. doi: 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- Hammad A.M., Shimamoto T., Shimamoto T. Genetic characterization of antibiotic resistance and virulence factors in Enterococcus spp. from Japanese retail ready-to-eat raw fish. Food Microbiol. 2014;38:62–66. doi: 10.1016/j.fm.2013.08.010. [DOI] [PubMed] [Google Scholar]
- Hashimoto K. Molecular mechanisms of the rapid-acting and long-lasting antidepressant actions of (R)-ketamine. Biochem. Pharmacol. 2020;177 doi: 10.1016/j.bcp.2020.113935. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y., Eguchi A., Wei Y., Shinno-Hashimoto H., Fujita Y., Ishima T., Chang L., Mori C., Suzuki T., Hashimoto K. Antibiotic-induced microbiome depletion improves LPS-induced acute lung injury via gut-lung axis. Life Sci. 2022;307 doi: 10.1016/j.lfs.2022.120885. [DOI] [PubMed] [Google Scholar]
- Huang Q., Ou Y.S., Tao Y., Yin H., Tu P.H. Apoptosis and autophagy induced by pyropheophorbide-α methyl ester-mediated photodynamic therapy in human osteosarcoma MG-63 cells. Apoptosis. 2016;21(6):749–760. doi: 10.1007/s10495-016-1243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huston J.M., Ochani M., Rosas-Ballina M., Liao H., Ochani K., Pavlov V.A., Gallowitsch-Puerta M., Ashok M., Czura C.J., Foxwell B., Tracey K.J., Ulloa L. Splenectomy inactivates the cholinergic antiinflammatory pathway during lethal endotoxemia and polymicrobial sepsis. J. Exp. Med. 2006;203(7):1623–1628. doi: 10.1084/jem.20052362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M., Hong K., Mao Y., Ma H., Chen T., Wang Z. Natural 5-aminolevulinic acid: sources, biosynthesis, detection and applications. Front. Bioeng. Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.841443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy E.A., King K.Y., Baldridge M.T. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front. Physiol. 2018;9:1534. doi: 10.3389/fphys.2018.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis S.M., Williams A., Eisenbarth S.C. Structure and function of the immune system in the spleen. Sci. Immunol. 2019;4(33) doi: 10.1126/sciimmunol.aau6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Zhang G., Li X., Zhang J. Microbial production and application of 5-aminolevulic acid. Appl. Microbiol. Biotechnol. 2014;98:7349–7357. doi: 10.1007/s00253-014-5925-y. [DOI] [PubMed] [Google Scholar]
- Liu T., Ma X., Ouyang T., Chen H., Xiao Y., Huang Y., Liu J., Xu M. Efficacy of 5-aminolevulinic acid-based photodynamic therapy against keloid compromised by downregulation of SIRT1-SIRT3-SOD2-mROS dependent autophagy pathway. Redox Biol. 2019;20:195–203. doi: 10.1016/j.redox.2018.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Zhang J., Fujita Y., Qu Y., Shan J., Wan X., Wang X., Ishima T., Kobayashi K., Wang L., Hashimoto K. Nuclear factor of activated T cells 4 in the prefrontal cortex is required for prophylactic actions of (R)-ketamine. Transl. Psychiatry. 2022;12(1):27. doi: 10.1038/s41398-022-01803-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Zhang J., Fujita Y., Shinno-Hashimoto H., Shan J., Wan X., Qu Y., Chang L., Wang X., Hashimoto K. Effects of spleen nerve denervation on depression-like phenotype, systemic inflammation, and abnormal composition of gut microbiota in mice after administration of lipopolysaccharide: a role of brain-spleen axis. J. Affect. Disord. 2022;317:156–165. doi: 10.1016/j.jad.2022.08.087. [DOI] [PubMed] [Google Scholar]
- Mamalis A., Koo E., Sckisel G.D., Siegel D.M., Jagdeo J. Temperature-dependent impact of thermal aminolaevulinic acid photodynamic therapy on apoptosis and reactive oxygen species generation in human dermal fibroblasts. Br. J. Dermatol. 2016;175(3):512–519. doi: 10.1111/bjd.14509. [DOI] [PubMed] [Google Scholar]
- Mebius R.E., Kraal G. Structure and function of the spleen. Nat. Rev. Immunol. 2005;5(8):606–616. doi: 10.1038/nri1669. [DOI] [PubMed] [Google Scholar]
- Miyauchi E., Shimokawa C., Steimle A., Desai M.S., Ohno H. The impact of the gut microbiome on extra-intestinal autoimmune diseases. Nat. Rev. Immunol. 2022 doi: 10.1038/s41577-022-00727-y. online ahead of print. [DOI] [PubMed] [Google Scholar]
- Mok S.W., Wong V.K., Lo H.H., de Seabra Rodrigues Dias I.R., Leung E.L., Law B.Y., Liu L. Natural products-based polypharmacological modulation of the peripheral immune system for the treatment of neuropsychiatric disorders. Pharmacol. Ther. 2020;208 doi: 10.1016/j.pharmthera.2020.107480. [DOI] [PubMed] [Google Scholar]
- Moyenuddin M., Wachsmuth I.K., Moseley S.L., Bopp C.A., Blake P.A. Serotype, antimicrobial resistance, and adherence properties of Escherichia coli strains associated with outbreaks of diarrheal illness in children in the United States. J. Clin. Microbiol. 1989;27(10):2234–2239. doi: 10.1128/jcm.27.10.2234-2239.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble B.T., Brennan F.H., Popovich P.G. The spleen as a neuroimmune interface after spinal cord injury. J. Neuroimmunol. 2018;321:1–11. doi: 10.1016/j.jneuroim.2018.05.007. [DOI] [PubMed] [Google Scholar]
- Pu Y., Chang L., Qu Y., Wang S., Zhang K., Hashimoto K. Antibiotic-induced microbiome depletion protects against MPTP-induced dopaminergic neurotoxicity in the brain. Aging (Albany NY) 2019;11(17):6915–6929. doi: 10.18632/aging.102221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu Y., Tan Y., Qu Y., Chang L., Wang S., Wei Y., Wang X., Hashimoto K. A role of the subdiaphragmatic vagus nerve in depression-like phenotypes in mice after fecal microbiota transplantation from Chrna7 knock-out mice with depression-like phenotypes. Brain Behav. Immun. 2021;94:318–326. doi: 10.1016/j.bbi.2020.12.032. [DOI] [PubMed] [Google Scholar]
- Rhee S.H., Pothoulakis C., Mayer E.A. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat. Rev. Gastroenterol. Hepatol. 2009;6(5):306–314. doi: 10.1038/nrgastro.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas-Ballina M., Ochani M., Parrish W.R., Ochani K., Harris Y.T., Huston J.M., Chavan S., Tracey K.J. Splenic nerve is required for cholinergic antiinflammatory pathway control of TNF in endotoxemia. Proc. Natl. Acad. Sci. U.S.A. 2008;105(31):11008–11013. doi: 10.1073/pnas.0803237105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T.K., Jena P.K., Nagar N., Patel A.K., Seshadri S. In vitro evaluation of probiotic properties of lactic acid bacteria from the gut of Labeo rohita and Catla catla. Probiotics Antimicrob. Proteins. 2015;7(2):126–136. doi: 10.1007/s12602-015-9184-8. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Watanabe M., Tanaka T., Tanaka T. Biosynthesis, biotechnological production and applications of 5-aminolevulinic acid. Appl. Microbiol. Biotechnol. 2002;58:23–29. doi: 10.1007/s00253-001-0858-7. [DOI] [PubMed] [Google Scholar]
- Schön M., Mousa A., Berk M., Chia W.L., Ukropec J., Majid A., Ukropcová B., de Courten B. The potential of carnosine in brain-related disorders: a comprehensive review of current evidence. Nutrients. 2019;11(6):1196. doi: 10.3390/nu11061196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G., Sampson T.R., Geschwind D.H., Mazmanian S.K. The central nervous system and the gut microbiome. Cell. 2016;167(4):915–932. doi: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinno-Hashimoto H., Eguchi A., Sakamoto A., Wan X., Hashimoto Y., Fujita Y., Mori C., Hatano M., Matsue H., Hashimoto K. Effects of splenectomy on skin inflammation and psoriasis-like phenotype of imiquimod-treated mice. Sci. Rep. 2022;12(1) doi: 10.1038/s41598-022-18900-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stummer W., Pichlmeier U., Meinel T., Wiestler O.D., Zanella F., Reulen H.J., ALA-Glioma Study Group Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: a randomised controlled multicentre phase III trial. Lancet Oncol. 2006;7(5):392–401. doi: 10.1016/S1470-2045(06)70665-9. [DOI] [PubMed] [Google Scholar]
- Subramanian K., Selvakkumar C., Vinaykumar K.S., Goswami N., Meenakshisundaram S., Balakrishnan A., Lakshmi B.S. Tackling multiple antibiotic resistance in enteropathogenic Escherichia coli (EPEC) clinical isolates: a diarylheptanoid from Alpinia officinarum shows promising antibacterial and immunomodulatory activity against EPEC and its lipopolysaccharide-induced inflammation. Int. J. Antimicrob. Agents. 2009;33(3):244–250. doi: 10.1016/j.ijantimicag.2008.08.032. [DOI] [PubMed] [Google Scholar]
- Tiedje K.E., Stevens K., Barnes S., Weaver D.F. Beta-alanine as a small molecule neurotransmitter. Neurochem. Int. 2010;57(3):177–188. doi: 10.1016/j.neuint.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Wan X., Eguchi A., Fujita Y., Ma L., Wang X., Yang Y., Qu Y., Chang L., Zhang J., Mori C., Hashimoto K. Effects of (R)-ketamine on reduced bone mineral density in ovariectomized mice: a role of gut microbiota. Neuropharmacology. 2022;213 doi: 10.1016/j.neuropharm.2022.109139. [DOI] [PubMed] [Google Scholar]
- Wan X., Eguchi A., Qu Y., Yang Y., Chang L., Shan J., Mori C., Hashimoto K. Gut-microbiota-brain axis in the vulnerability to psychosis in adulthood after repeated cannabis exposure during adolescence. Eur. Arch. Psychiatr. Clin. Neurosci. 2022;272(7):1297–1309. doi: 10.1007/s00406-022-01437-1. [DOI] [PubMed] [Google Scholar]
- Wang S., Ishima T., Qu Y., Shan J., Chang L., Wei Y., Zhang J., Pu Y., Fujita Y., Tan Y., Wang X., Ma L., Wan X., Hammock B.D., Hashimoto K. Ingestion of Faecalibaculum rodentium causes depression-like phenotypes in resilient Ephx2 knock-out mice: a role of brain-gut-microbiota axis via the subdiaphragmatic vagus nerve. J. Affect. Disord. 2021;292:565–573. doi: 10.1016/j.jad.2021.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Ishima T., Zhang J., Qu Y., Chang L., Pu Y., Fujita Y., Tan Y., Wang X., Hashimoto K. Ingestion of Lactobacillus intestinalis and Lactobacillus reuteri causes depression- and anhedonia-like phenotypes in antibiotic-treated mice via the vagus nerve. J. Neuroinflammation. 2020;17(1):241. doi: 10.1186/s12974-020-01916-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Qu Y., Chang L., Pu Y., Zhang K., Hashimoto K. Antibiotic-induced microbiome depletion is associated with resilience in mice after chronic social defeat stress. J. Affect. Disord. 2020;260:448–457. doi: 10.1016/j.jad.2019.09.064. [DOI] [PubMed] [Google Scholar]
- Wei Y., Chang L., Ishima T., Wan X., Ma L., Wuyun G., Pu Y., Hashimoto K. Abnormalities of the composition of the gut microbiota and short-chain fatty acids in mice after splenectomy. Brain Behav. Immun. Health. 2021;11 doi: 10.1016/j.bbih.2021.100198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Chang L., Wan X., Tan Y., Qu Y., Shan J., Yang Y., Ma L., Hashimoto K. (R)-ketamine ameliorates demyelination and facilitates remyelination in cuprizone-treated mice: a role of gut-microbiota-brain axis. Neurobiol. Dis. 2022;165 doi: 10.1016/j.nbd.2022.105635. [DOI] [PubMed] [Google Scholar]
- Wang X., Eguchi A., yang Y., Chang L., Wan X., Shan J., Qu Y., Ma L., Mori C., Yang J., Hashimoto K. Key role of the gut–microbiota–brain axis via the subdiaphragmatic vagus nerve in demyelination of the cuprizone-treated mouse brain. Neurobiol. Dis. 2022;176 doi: 10.1016/j.nbd.2022.105951. [DOI] [PubMed] [Google Scholar]
- Wei Y., Chang L., Hashimoto K. Molecular mechanisms underlying the antidepressant actions of arketamine: beyond the NMDA receptor. Mol. Psychiatr. 2022;27(1):559–573. doi: 10.1038/s41380-021-01121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Wang T., Liao L., Fan X., Chang L., Hashimoto K. Brain-spleen axis in health and diseases: a review and future perspective. Brain Res. Bull. 2022;182:130–140. doi: 10.1016/j.brainresbull.2022.02.008. [DOI] [PubMed] [Google Scholar]
- Yang B., Ren Q., Zhang J.C., Chen Q.X., Hashimoto K. Altered expression of BDNF, BDNF pro-peptide and their precursor proBDNF in brain and liver tissues from psychiatric disorders: rethinking the brain-liver axis. Transl. Psychiatry. 2017;7(5) doi: 10.1038/tp.2017.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Eguchi A., Wan X., Chang L., Wang X., Qu Y., Mori C., Hashimoto K. A role of gut-microbiota-brain axis via subdiaphragmatic vagus nerve in depression-like phenotypes in Chrna7 knock-out mice. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2022;120 doi: 10.1016/j.pnpbp.2022.110652. [DOI] [PubMed] [Google Scholar]
- Yang Y., Ishima T., Wan X., Wei Y., Chang L., Zhang J., Qu Y., Hashimoto K. Microglial depletion and abnormalities in gut microbiota composition and short-chain fatty acids in mice after repeated administration of colony stimulating factor 1 receptor inhibitor PLX5622. Eur. Arch. Psychiatr. Clin. Neurosci. 2022;272(3):483–495. doi: 10.1007/s00406-021-01325-0. [DOI] [PubMed] [Google Scholar]
- Zhang J., Chang L., Pu Y., Hashimoto K. Abnormal expression of colony stimulating factor 1 receptor (CSF1R) and transcription factor PU.1 (SPI1) in the spleen from patients with major psychiatric disorders: a role of brain-spleen axis. J. Affect. Disord. 2020;272:110–115. doi: 10.1016/j.jad.2020.03.128. [DOI] [PubMed] [Google Scholar]
- Zhang J., Ma L., Chang L., Pu Y., Qu Y., Hashimoto K. A key role of the subdiaphragmatic vagus nerve in the depression-like phenotype and abnormal composition of gut microbiota in mice after lipopolysaccharide administration. Transl. Psychiatry. 2020;10(1):186. doi: 10.1038/s41398-020-00878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ma L., Wan X., Shan J., Qu Y., Hashimoto K. (R)-Ketamine attenuates LPS-induced endotoxin-derived delirium through inhibition of neuroinflammation. Psychopharmacology (Berl) 2021;238(10):2743–2753. doi: 10.1007/s00213-021-05889-6. [DOI] [PubMed] [Google Scholar]
- Zhang K., Sakamoto A., Chang L., Qu Y., Wang S., Pu Y., Tan Y., Wang X., Fujita Y., Ishima T., Hatano M., Hashimoto K. Splenic NKG2D confers resilience versus susceptibility in mice after chronic social defeat stress: beneficial effects of (R)-ketamine. Eur. Arch. Psychiatr. Clin. Neurosci. 2021;271(3):447–456. doi: 10.1007/s00406-019-01092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 16S rRNA sequencing data have been deposited to the NCBI Sequence Read Archive and are available at the accession number PRJNA887905.