Abstract
Background
Genetic variants within nearly 1000 loci are known to contribute to modulation of blood lipid levels. However, the biological pathways underlying these associations are frequently unknown, limiting understanding of these findings and hindering downstream translational efforts such as drug target discovery.
Results
To expand our understanding of the underlying biological pathways and mechanisms controlling blood lipid levels, we leverage a large multi-ancestry meta-analysis (N = 1,654,960) of blood lipids to prioritize putative causal genes for 2286 lipid associations using six gene prediction approaches. Using phenome-wide association (PheWAS) scans, we identify relationships of genetically predicted lipid levels to other diseases and conditions. We confirm known pleiotropic associations with cardiovascular phenotypes and determine novel associations, notably with cholelithiasis risk. We perform sex-stratified GWAS meta-analysis of lipid levels and show that 3–5% of autosomal lipid-associated loci demonstrate sex-biased effects. Finally, we report 21 novel lipid loci identified on the X chromosome. Many of the sex-biased autosomal and X chromosome lipid loci show pleiotropic associations with sex hormones, emphasizing the role of hormone regulation in lipid metabolism.
Conclusions
Taken together, our findings provide insights into the biological mechanisms through which associated variants lead to altered lipid levels and potentially cardiovascular disease risk.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13059-022-02837-1.
Keywords: Cholesterol, Lipids, Genetics, Genome-wide association study, GWAS
Background
Abnormal blood lipid levels are a major cause of cardiovascular disease [1], the leading cause of morbidity and mortality worldwide [2]. Conventional blood lipid measures, low-density lipoprotein cholesterol (LDL-C), total cholesterol (TC), triglyceride (TG), high-density lipoprotein cholesterol (HDL-C), and nonHDL-C (TC – HDL-C), are commonly used in clinical practice to identify individuals at high risk for cardiovascular events. Several treatments for reducing LDL-C, including statins, ezetimibe, and PCSK9 inhibitors [3], also reduce the risk of developing cardiovascular disease.
Genome-wide association studies (GWAS) for blood lipids have identified nearly 1000 associated genetic loci to date [4–23], including our recent multi-ancestry GWAS meta-analysis in 1.65 M individuals [24]. The latter focused on the gains from the multi-ancestry meta-analysis relative to the single-ancestry results, in terms of number of loci, fine-mapping, and polygenic score (PGS) transferability. However, a challenge in the field is that the underlying gene and biological pathways is often unknown for GWAS loci. Within lipid GWAS, prior fine-mapping studies combined with functional follow-up have successfully identified causal genes with high confidence for only a handful of associated GWAS loci, including SORT1 [25], TM6SF2 [12], and ANGPTL3 [26], among others. Highly sophisticated methods are emerging to prioritize causal genes in well-powered GWAS studies, such as the Data-driven Expression-Prioritized Integration for Complex Traits [27] (DEPICT) and the Polygenic Priority Score [28] (PoPS), that take into account genome-wide properties of associated loci and larger sets of associated loci are beneficial. These methods can be combined with algorithms that integrate expression data such as transcriptome-wide association studies (TWAS) and comprehensive experimental data sets such as mouse gene knockouts. Gene sets enriched for causal genes will enhance our ability to unravel the biological pathways underlying these associations and there is growing interest in using a combination of gene prioritization methods to provide compelling evidence for putative causal genes [29].
In parallel, the linkage of electronic health records with genetic data in large-scale population studies and patient biobanks allows for the systematic exploration of pleiotropy of lipid-associated alleles. While blood lipid levels have a well-documented causal effect on cardiovascular disease based on genetic association studies validated by randomized controlled trials [30–32], genetic pleiotropic associations might exist for other conditions. Unraveling such pleiotropy may yield new biological insights by revealing previously unrecognized connections between blood lipids and both cardiovascular and non-cardiovascular diseases. Phenome-wide association scans (PheWAS) adopt an agnostic approach to test for pleiotropic associations between genetic factors and a wide range of phenotypes [33]. Such knowledge may allow for the identification of lipid levels as novel diagnostic biomarkers, the repurposing of drugs, and the prevention of adverse drug events [34].
Finally, given empirical sex differences in blood lipid distributions, sex-specific genetic associations may yield novel biological insights. Pre-menopausal females have lower levels of LDL-C than same-age males, and HDL-C levels are higher among females of all ages compared to males [35]. Lipid levels also show a greater estimated heritability in females compared with males [36], especially for LDL-C and TC (> 1.3-fold difference). Sexual dimorphism in lipid levels may be partly explained by X chromosome variants. Evidence from human X-linked abnormalities (like Turner or Klinefelter syndromes) suggests an important role of this chromosome in lipid metabolism [37]. This is further corroborated by the lipid and atherosclerosis profiles in the Four Core Genotypes mouse model [38], which comprises XX and XY gonadal males and XX and XY gonadal females. GWAS studies have traditionally understudied the X chromosome due to technical and analytical difficulties. A recent high coverage whole X chromosome sequencing study [39] prioritized CHRDL1 as a candidate causal lipid gene, suggesting with larger sample sizes we may be able to discover additional variation on the X chromosome associated with lipid levels.
In this study, we first prioritize genes at GWAS lipid loci through multiple in silico gene prediction algorithms and experimental data sources using the latest Global Lipids Genetics Consortium multi-ancestry meta-analysis [24]. We then identify novel disease associations related to lipid levels through PheWAS in two large biobanks using PGSs. Finally, we perform sex-stratified meta-analysis to compare the associations between males and females to identify genetic loci with sex-specific associations and GWAS meta-analysis of the X chromosome, to better understand lipid level differences between the sexes. Together, our results highlight biological mechanisms through which lipid-associated variants lead to altered lipid levels.
Results
Identifying functional genes in lipid-associated loci
In a GWAS meta-analysis of blood lipid levels from 1.65 million individuals (Additional file 1: Table S1) at 91 million genotyped and imputed genetic variants, we observed a total of 2286 genome-wide significant index variants associated with lipid levels at 923 loci (± 500 kb regions). This corresponded to 416 index variants associated with LDL-C, 539 with HDL-C, 461 with TG, 487 with TC, and 383 with nonHDL-C. Uniquely, we observed 1750 variants associated with one or more lipid levels [24] (Additional file 2: Table S2).
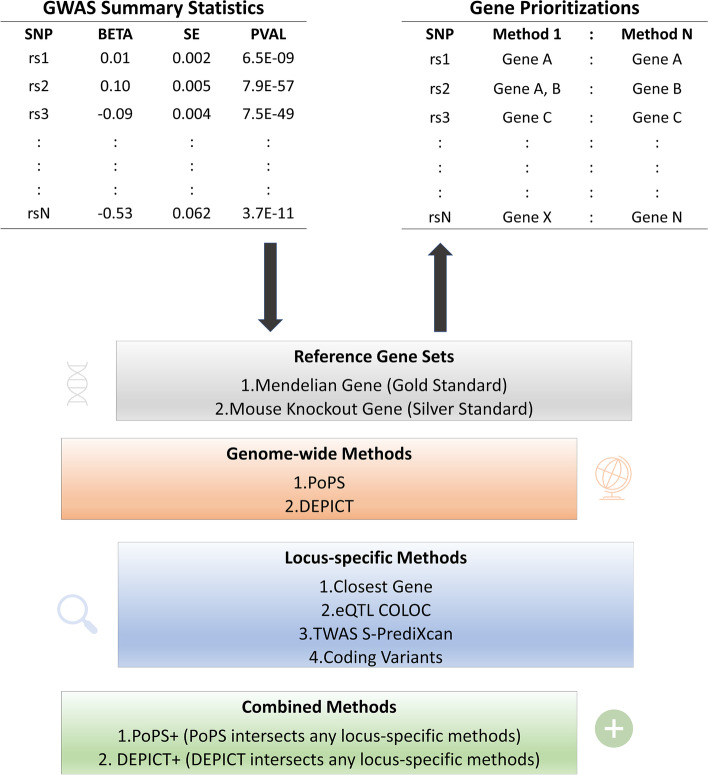
We employed six approaches to identify candidate functional genes for all 2286 lipid associations. Our prioritization approaches include four locus-specific methods that are based on local information around the indexed variant: (1) the closest gene to the index variant, (2) genes with lipid-associated protein-altering variants, (3) colocalized expression quantitative trait loci (eQTL) genes, and (4) nearby genes prioritized by transcript-wide association studies (TWAS). We also used two genome-wide methods that leveraged genome-wide similarities of features: (1) gene-level Polygenic Priority Score (PoPS) [28] and (2) Data-driven Expression-Prioritized Integration for Complex Traits (DEPICT) [27]. We further combined the two genome-wide methods with the locus-specific methods to increase the confidence in prioritized genes: (1) PoPS intersects with any locus-specific methods (PoPS +), and (2) DEPICT intersects with any locus-specific methods (DEPICT +) (Fig. 1). Since the genome-wide gene prioritization approaches can prioritize different genes for different lipid types at the same locus, we report the gene prioritization results for all 2286 lipid-variant associations (Additional file 2: Table S2, Additional file 3: Figure S1).
Fig. 1.
Schematic of multi-method candidate gene mapping of indexed variants associated with blood lipid levels. We defined indexed variants within the GLGC GWAS summary statistics and performed two similarity-based methods and four locus-based methods to prioritize genes for each of the indexed variants
We took the genes prioritized by PoPS + and performed text mining to determine whether previous biological evidence supported these genes as playing a role in lipid levels (Additional file 4: Table S3, S4). PoPS + leverages both locus-specific and genome-wide genetic signals to increase confidence level in prioritized genes [28]. In total, 882 out of 2286 lipid associations were assigned to one potential causal gene based on PoPS + . We identified a group of 466 unique genes among the 882 lipid associations. We determined that 31 out of the 466 PoPS + genes have over 1000 lipid-related publications, 91 PoPS + genes have 100–999 lipid-related publications, 321 PoPS + genes have 1–99 lipid-related publications, and 23 PoPS + genes had no lipid-related publications retrieved by the text mining algorithm. These 23 genes could indicate novel genes related to lipid levels for future work or be due to incorrect gene prioritization for a small fraction of index variants. (Additional file 5: Table S4). We then randomly selected 466 genes from 18,383 protein-coding genes using by the PoPS as the reference group to estimate the number of lipid-related publications we would expect to see by chance. A Mann–Whitney U test showed that there was a significant difference (W = 52,353, p-value < 2.2 x 10−16) between the set of genes identified by PoPS + compared to the reference set of 466 genes (Additional file 6: Figure S2). The median count of lipid-related publications was 19 for the PoPS + gene set compared with 2 lipid-related publications for genes in the reference set.
We performed a comprehensive lookup of all PoPS + prioritized lipid genes in the Therapeutic Target Database 2022 [24] and found 2092 drugs targeting at least one of our 102 PoPS + prioritized lipid genes observed in the database (Additional file 7: Table S5). Among those 102 PoPS + genes, we identify known drug target genes including PCSK9 druggable as subtilisin/kexin type 9 inhibitor, HMGCR druggable as HMG-CoA reductase inhibitor, PDE3A druggable as phosphodiesterase 3A inhibitor (CILOSTAZOL), and NR1H4 as a bile acid receptor FXR agonist (URSODIOL). We also identify several other potential drug targets [24] such as LIPG (lipase G) and NR1H3 (nuclear receptor subfamily 1 group H member 3), with relevant lipid biology. LIPG has phospholipase and triglyceride lipase activities and is a primary determinant of plasma HDL levels. NR1H3 has an important role in the regulation of cholesterol homeostasis, regulating cholesterol uptake through MYLIP-dependent ubiquitination of LDLR, VLDLR, and LRP8 that could be targeted as an LXR-alpha modulator.
Effects of protein-altering lipid alleles with protective effects on CAD, T2D, and NAFLD
Coronary artery disease (CAD), type 2 diabetes (T2D), and non-alcoholic fatty liver disease (NAFLD) are typically characterized by dyslipidemias. We examined protein-altering alleles with favorable lipid profiles for their associations with CAD, T2D, and NAFLD to identify potential cardiovascular drug targets without off-target liver or diabetes effects. Of the 2286 lipid associations, we observed 166 coding index variants. Eighteen coding variants with a protective lipid effect also had a protective effect for CAD or T2D (p-value < 0.001) and the lipid results colocalized with the CAD or T2D results, as appropriate, with a posterior probability of a shared causal variant > 0.8 (Table 1 and Additional file 8: Table S6). Six of these twenty variants had protective effects for both CAD and T2D, while nine were protective for CAD and three were protective for T2D (Table 1). Additionally, 269 noncoding alleles with a protective lipid effect also had a protective effect for CAD or T2D (p < 0.001; Additional file 8: Table S6).
Table 1.
Protective lipid coding alleles associated with CAD and/or T2D
| RSID | Trait | Coding variant | Effect allele (EAF) | Lipid effect | CAD OR (95% CI) |
CAD P-value |
T2D OR (95% CI) |
T2D P-value |
NAFLD OR (95% CI) |
NAFLD P-value |
|---|---|---|---|---|---|---|---|---|---|---|
| Protective lipid alleles associated with reduced risk of CAD and T2D | ||||||||||
| rs116843064 | TG |
ANGPTL4 p.Glu40Lys |
A (0.020) | − 0.238 |
0.87 (0.85,0.90) |
1.92x10−18 |
0.91 (0.86,0.95) |
2.30x10−5 |
0.99 (0.84,1.15) |
0.85 |
| rs1169288 | TC |
HNF1A p.Ile27Leu |
A (0.682) | − 0.035 |
0.97 (0.96,0.98) |
2.31x10−16 |
0.95 (0.94,0.96) |
7.30x10−13 |
0.95 (0.91,1.00) |
0.04 |
| rs2307111 | HDL-C |
POC5 p.His36Arg |
C (0.440) | 0.016 |
0.99 (0.98,0.99) |
9.09x10−05 |
0.95 (0.94,0.96) |
3.30x10−16 |
1.00 (0.96,1.05) |
0.85 |
| rs6480771 | HDL-C |
DUSP13 p.Ser111Gly |
T (0.531) | 0.008 |
0.99 (0.98,0.99) |
1.44x10−04 |
0.97 (0.96,0.99) |
4.40x10−05 |
0.92 (0.88,0.97) |
5.21x10−04 |
| rs35742417 | TG |
RREB1 p.Ser1554Tyr |
A (0.173) | − 0.012 |
0.98 (0.97,0.99) |
5.06x10 −04 |
0.96 (0.95,0.98) |
3.70 x10−06 |
0.98 (0.93,1.04) |
0.60 |
| rs72681869 | TG |
SOS2 p.Pro191Arg |
C (0.008) | − 0.053 |
0.93 (0.89,0.98) |
3.71x10−03 |
0.88 (0.82,0.94) |
3.90x10−04 |
0.87 (0.68,1.12) |
0.29 |
| Protective lipid alleles associated with reduced risk of CAD | ||||||||||
| rs7412 | LDL-C |
APOE p.Arg176Cys |
T (0.076) | − 0.517 |
0.90 (0.88,0.91) |
9.94x10−52 |
1.01 (0.98,1.03) |
0.55 |
1.01 (0.93,1.10) |
0.84 |
| rs11591147 | LDL-C |
PCSK9 p.Arg46Leu |
T (0.015) | − 0.434 |
0.80 (0.77,0.83) |
5.97x10−36 |
1.04 (0.99,1.09) |
0.16 |
1.05 (0.88,1.26) |
0.58 |
| rs11601507 | LDL-C |
TRIM5 p.Val112Phe |
C (0.926) | − 0.042 |
0.95 (0.94,0.96) |
2.80x10−12 |
0.99 (0.96,1.01) |
0.26 |
1.02 (0.93,1.11) |
0.72 |
| rs1132274 | HDL-C |
RRBP1 p.Arg891Leu |
C (0.827) | 0.017 |
0.97 (0.96,0.98) |
3.57x10−08 |
1.01 (0.99,1.02) |
0.43 |
1.00 (0.94,1.07) |
0.91 |
| rs4760 | HDL-C |
PLAUR p.Leu317Pro |
A (0.860) | 0.016 |
0.97 (0.96,0.98) |
7.31x10−07 |
0.99 (0.97,1.01) |
0.3 |
0.96 (0.91,1.03) |
0.26 |
| rs855791 | LDL-C |
TMPRSS6 p.Val736Ala |
G (0.578) | − 0.009 |
0.98 (0.97,0.99) |
1.08x10−06 |
1.00 (0.99,1.01) |
0.75 |
0.94 (0.9,0.98) |
4.83x10−03 |
| rs58542926 | TC |
TM6SF2 p.Glu167Lys |
T (0.073) | − 0.124 |
0.97 (0.95,0.98) |
4.02x10−06 |
1.10 (1.07,1.12) |
2.00x10−14 |
1.45 (1.33,1.58) |
1.05x10−16 |
| rs56196860 | HDL-C |
FKBP4 p.Asn197Lys |
A (0.027) | 0.031 |
0.95 (0.92,0.97) |
1.05x10-05 |
0.98 (0.94,1.02) |
0.33 |
1.03 (0.87,1.21) |
0.73 |
| rs72836561 | HDL-C |
CD300LG p.Arg82Cys |
C (0.971) | 0.187 | 0.95 (0.93,0.98) | 1.34x10−04 | 0.98 (0.95,1.02) | 0.4 | 0.98 (0.86,1.12) | 0.77 |
| Protective lipid alleles associated with reduced risk of T2D | ||||||||||
| rs1800961 | HDL-C |
HNF4A p.Thr139Ile |
C (0.969) | 0.134 |
0.99 (0.97,1.01) |
0.3751 |
0.85 (0.82,0.88) |
3.20x10− 20 |
1.02 (0.90,1.17) |
0.74 |
| rs1801253 | TG |
ADRB1 p.Gly389Arg |
C (0.732) | − 0.011 |
1.01 (1.00,1.02) |
8.76x10− 03 |
0.97 (0.96,0.98) |
1.9010x− 05 |
1.00 (0.95,1.06) |
0.85 |
| rs13107325 | HDL-C |
SLC39A8 p.Ala391Thr |
C (0.941) | 0.082 |
1.00 (0.98,1.01) |
0.8876 |
0.95 (0.93,0.98) |
3.30Ex10− 04 |
0.85 (0.78,0.93) |
1.85x10− 04 |
Driver tissues for lipid levels
We applied DESE (Driver-tissue Estimation by Selective Expression) [40] to estimate the driver tissues of five lipid traits using both gene-level and transcript-level eQTL summary statistics from GTEx v8 tissues [41]. We identified liver as the top-ranked tissue for HDL-C (gene-level p-value = 4.5 x 10−18, transcript-level p-value = 3.0 x 10−26), TC (gene-level p-value = 1.1 x 10−25, transcript-level p-value = 1.4 x 10−33), and nonHDL-C (gene-level p-value = 2.0 x 10−19, transcript-level p-value = 3.9 x 10−29) based on both gene-level and transcript-level selective expression (Additional file 9: Figure S3, Additional file 10: Table S7). For LDL-C, we identified the spleen as the top-ranked tissue using GTEx gene-level data (p-value = 8.3 x 10−20), while liver was ranked second (p-value = 4.8 x 10−17). However, when using GTEx transcript-level data, liver was the top-ranked tissue (p-value = 4.3 x 10−29) and second was whole blood (p-value = 4.3 x 10−20). The top tissue for TG according to both GTEx gene-level and transcript-level expression data was whole blood (gene-level p-value = 6.4 x 10−20, transcript-level p-value = 1.4 x 10−21). Spleen and liver were second according to GTEx gene-level and transcript-level expression data, respectively. The results were consistent with previous knowledge that the liver is a major tissue for lipid metabolism. Transcript-level selective expression provided more statistically significant results for the estimated driver tissues compared to the gene-level selective expression, as reported in the original [40].
Polygenic scores for lipid phenotypes and phenome-wide association scans
We have previously reported that a polygenic score (PGS) for LDL-C was most informative when generated from the multi-ancestry GWAS and that the multi-ancestry PGS performed equally well in European-ancestry Americans, African-ancestry Americans, and continental Africans [24]. Using a similar approach, we generated PGS for the other four lipid traits (“Methods”).
We next performed a phenome-wide association scan (PheWAS) for the multi-ancestry lipid PGS (LDL-C PGS previously reported [24]) to identify pleiotropic effects of lipids with other traits in the European subsets of the UK Biobank and the Million Veteran Program (MVP) cohorts. We compared the effect sizes from the PheWAS analysis between the UK Biobank and MVP per lipid PGS and observed a moderate correlation between the two datasets (Additional file 11: Figure S4). The correlation of the PGS effects on all phenotypes between the UK Biobank and MVP ranges from 0.12 for the HDL-C PGS to 0.39 for the TC PGS (Additional file 11: Figure S4). In general, correlations were stronger for the ICD-10-based phecodes (r2 of 0.42–0.52) compared to the biomarkers (r2 of 0.06–0.23) (Additional file 11: Figure S4), which may reflect differences in study populations due to varied environmental effects, prevalence of chronic health conditions, and sex distribution. Among PheWAS results with p-value ≤ 0.05 in the UK Biobank, the correlation was even higher for ICD-10-based phecodes (r2 of 0.52–0.76) but remained the same for the biomarkers (r2 of 0.07–0.22).
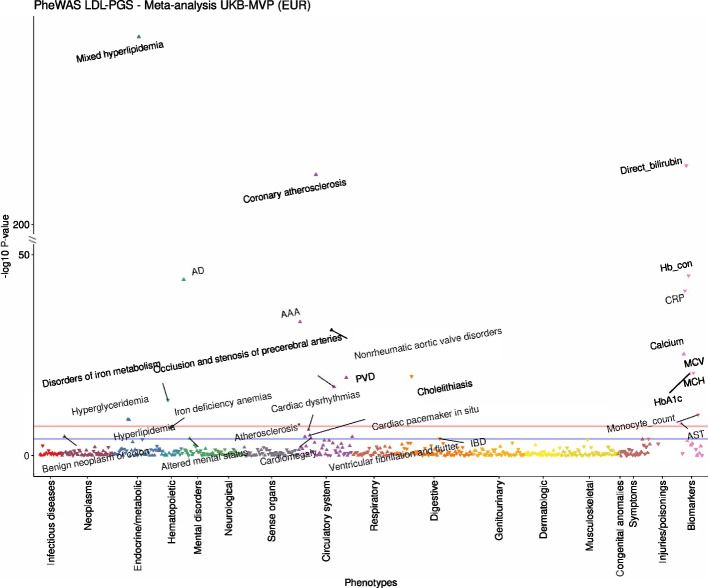
We meta-analyzed the results from the two cohorts to increase the power of the PheWAS, by matching ICD10-mapped phecodes and biomarkers. In the combined the UK Biobank-MVP PheWAS results, we detected 58 phenotypes associated with the LDL-C PGS at phenome-wide significance level (p-value ≤ 6.5 × 10−5, corrected for 773 phenotypes), 165 with the HDL-C PGS, 59 with the TC PGS, 166 with the TG PGS, and 78 with the nonHDL-C PGS (Fig. 2, Additional file 12: Table S8, Additional file 13: Figure S5, Additional file 14: Figure S6, Additional file 15: Figure S7, Additional file 16: Figure S8). As expected, multiple cardiovascular phenotypes related to atherosclerosis, including the expected coronary artery disease as well as aortic aneurysm and essential hypertension, were phenome-wide significantly associated with all five lipid PGSs, indicating increased risk of these diseases for individuals with genetically predicted increased LDL-C, TG, TC, or nonHDL-C or genetically predicted decreased HDL-C. A recent wide-ranging Mendelian randomization analysis confirmed the causal effect of circulating lipids, not only for coronary artery disease, but other cardiovascular outcomes [42]. Additionally, all lipid PGSs were also significantly associated with decreased levels of direct bilirubin (Additional file 12: Table S8, Fig. 2, Additional file 13: Figure S5, Additional file 14: Figure S6, Additional file 15: Figure S7, Additional file 16: Figure S8), indicating genetically predicted lower LDL-C increased levels of bilirubin (Fig. 2). Correspondingly, lipid PGSs were associated with lower risk for cholelithiasis (gallstones) with the opposite direction for TG PGS, indicating that extreme lowering of LDL-C may impact rates of cholelithiasis (Additional file 12: Table S8, Fig. 2, Additional file 13: Figure S5, Additional file 14: Figure S6, Additional file 15: Figure S7, Additional file 16: Figure S8). To further clarify whether this association might be driven by the ABCG8 gene alone, we excluded from the LDL-PGS all variants within the locus and tested the association between LDL-PGS and cholelithiasis in the UK Biobank. There was no attenuation of the observed association (OR = 0.94 and p-value = 7.94 × 10−17 without the ABCG8 locus vs. OR = 0.93 and p-value = 1.96 × 10−21).
Fig. 2.
PheWAS meta-analysis results for multi-ancestry LDL-C PGS in the UK Biobank and MVP. The blue horizontal line denotes phenome-wide significance (p-value ≤ 6.5 × 10−5, to account for multiple testing of 773 phenotypes) and the red line is genome-wide significance (p-value ≤ 5 × 10−8). Phenotypes have been pruned, so that the most significant one per correlated phenotype group (correlation coefficient > 0.2) is retained. Pairwise correlations were estimated with chi-square test and Cramer’s V for the dichotomous phenotypes and Pearson’s correlation for the continuous phenotypes. AAA: abdominal aortic aneurysm, AD: Alzheimer’s disease, AST: aspartate aminotransferase, Atherosclerosis*: atherosclerosis of native arteries of the extremities with intermittent claudication, Hb_con: hemoglobin concentration, IBD: irritable bowel disease, MCH: mean corpuscular hemoglobin, MCV: mean corpuscular volume, PVD: peripheral vascular disease
In the PheWAS analysis, we found that the TC and LDL-C PGS were significantly associated with increased levels of HbA1c (beta = 0.101 mmol/mol per SD PGS increase, p -value= 1.21 × 10−23 and beta = 0.095 mmol/mol per SD PGS increase, p-value = 4.37 × 10−21, respectively), while the HDL-C PGS was associated with decreased levels of HbA1c (beta = − 0.257 mmol/mol per SD PGS increase, p-value = 2.84 × 10−143) (Additional file 12: Table S8). Furthermore, genetically predicted increased LDL-C was significantly associated with decreased hemoglobin concentration (p-value = 1.92 × 10−45, similar significant associations for all other lipid PGSs with a reverse direction of effect for TG, Additional file 12: Table S8). As expected, genetically predicted increased LDL-C and TC were both associated with increased risk for Alzheimer’s disease [43] (OR = 1.33 per SD PGS increase, p-value = 1.74 × 10−44 and OR = 1.26 per SD PGS increase, p-value = 1.48 × 10−30, respectively; Additional file 12: Table S8). To further investigate how this association might be driven by the ApoE locus, we excluded all genetic variants overlapping this gene from the LDL-PGS and repeated the analysis in the UK Biobank. While the association between the LDL-PGS and the risk for Alzheimer’s disease was slightly attenuated after removing the ApoE locus (OR = 1.23 vs. 1.36 per SD PGS increase), the association remained significant (p-value = 2.51 × 10−21). Recent Mendelian randomization studies also provide evidence for the causal effect of lipids on risk for dementia [44] and Alzheimer’s disease [45]. The LDL-C and TC PGSs were also associated with increased aspartate aminotransferase levels (a liver enzyme), in accordance with other studies [46]. We also observed inverse associations between LDL-PGS (p-value = 1.43 × 10−14) and TC PGS (p-value = 8.34 × 10−14) with the risk of iron metabolism disorders (Additional file 12: Table S8).
To better understand the loci that drive the association between each of the lipid PGSs and cholelithiasis and cholecystitis, we interrogated the results from the single-variant PheWAS meta-analysis in the UK Biobank and MVP with all lipid multi-ancestry index variants (N = 1750 unique). We identified 22 genetic variants associated with cholelithiasis and/or cholecystitis at genome-wide significance. Genes prioritized for these index variants included genes already reported to be associated with gallstone disease [47] (CYP7A1, ABCG5/8), as well as additional genes (ABCB4, LRBA, HNF4A, NUCB1, GATA4), that may play also a role. Importantly, we found there was overlap (same index variant) between the previously published index variants for gallstone disease and our lipid index variants for these two loci (Additional file 17: Table S9).
Lipid loci show sex-specific effects
Sex-stratified analyses have the potential to identify loci missed by sex-combined analyses [48] as well as to detect loci exhibiting differential effects on lipids between sexes. First, we performed GWAS meta-analysis separately in each sex (Nmales = 749,391; Nfemales = 562,410), excluding loci discovered in the sex-combined analysis [24]. We identified twelve loci in females and four in males that reached genome-wide significance in the sex-stratified analysis (p-value < 5 × 10−8; Additional file 18: Table S10, Additional file 19: Table S11, Additional file 20: Table S12) but not in the sex-combined meta-analysis. As variants may show association to a single sex for reasons unrelated to biological sex differences, including differences in sample sizes between groups, we additionally tested for heterogeneity by sex for these variants in GLGC participating cohorts with close to equal number of males and females. Of the sixteen loci, eight showed significant sex-heterogeneity (p-value < 0.0031, Bonferroni-corrected threshold for sixteen tests). For example, the non-synonymous variant rs34372369 (EPHA1, p.Pro582Leu) is associated with nonHDL-C only in females (male p-value > 0.05) and shows significant sex-heterogeneity (p-value = 0.0012). This variant has been previously found to be linked with expression levels of the sex hormone-binding globulin gene (SHBG) more strongly in males than females [49], suggesting a possible reason for the difference in observed associations. We additionally sought to replicate the sex-heterogeneity results of these variants in 8 independent multi-ancestry cohorts (N = 311,639, 77% non-European ancestry, Additional file 21: Table S13). However, we did not detect significant differences in effect sizes between sexes for these variants after accounting for the number of tests (p-value > 0.0031, Additional file 22: Table S14), potentially due to the limited sample size or difference in ancestry makeup.
Second, we tested for a difference in the male- and female-specific effect sizes for each of the index variants identified from the sex-combined multi-ancestry meta-analysis. Of the 1750 unique index variants, 64 showed a significant difference in effect size by sex for one or more traits (Bonferroni correction for the number of index variants in each trait, Additional file 23: Table S15). These were evenly distributed across traits and more often had stronger effects in females than males (67%, Additional file 24: Figure S9). We tested for replication of the sex-specific differences in up to 311,120 participants from eight independent multi-ancestry cohorts not included in the original meta-analysis (Additional file 21: Table S13). Fifty-four of the 64 (84%) variants had effect size differences that were directionally consistent with the original meta-analysis (Additional file 25: Table S16). Of these, 10 had significantly different effect sizes (p-value < 7.8 × 10−4, Bonferroni correction for 64 variants) and 22 were nominally significant (p-value < 0.05). We attribute the low rate of replication to the small sample size and the differing proportions of ancestry groups within our replication samples, but we cannot dismiss the potential of false positives in the sex-specific discovery results.
We tested whether the observed sex differences could be caused by a higher frequency of cholesterol-lowering medications in males, potentially indicating an insufficient correction for pre-medication cholesterol levels. Among white British individuals in the UK Biobank, variants with significant sex differences had significantly higher effect size estimates on average after excluding individuals on medication (Additional file 26: Figure S10, Additional file 27: Table S17). However, of the 17 variants that exhibited a significant difference in effect size by sex in the UK Biobank alone, 15 remained significant after excluding individuals taking medications. Based on this observation, the observed differences did not appear to be driven solely, or even primarily, by differences in medication use between sexes. Furthermore, none of the identified sex-specific variants were associated with sex-participation bias [50] (Additional file 28: Table S18), indicating that differential study enrollment between sexes was unlikely to be the cause of the observed sex-specific lipid associations. We next investigated differences in environmental factors between sexes for these variants in the UK Biobank (Additional file 29: Table S19), including alcohol use [48], smoking status [48], body mass index (BMI) [51], and waist-hip ratio adjusted for BMI (WHRadjBMI) [51]. Twenty-two of the variants (34%) with differential effects on lipids by sex also exhibited a significant difference by sex for WHRadjBMI and one variant had a significant difference by sex for alcohol use (ADH1B p.His48Arg). The observed sex differences may therefore be partially attributed to pleiotropic associations with other traits.
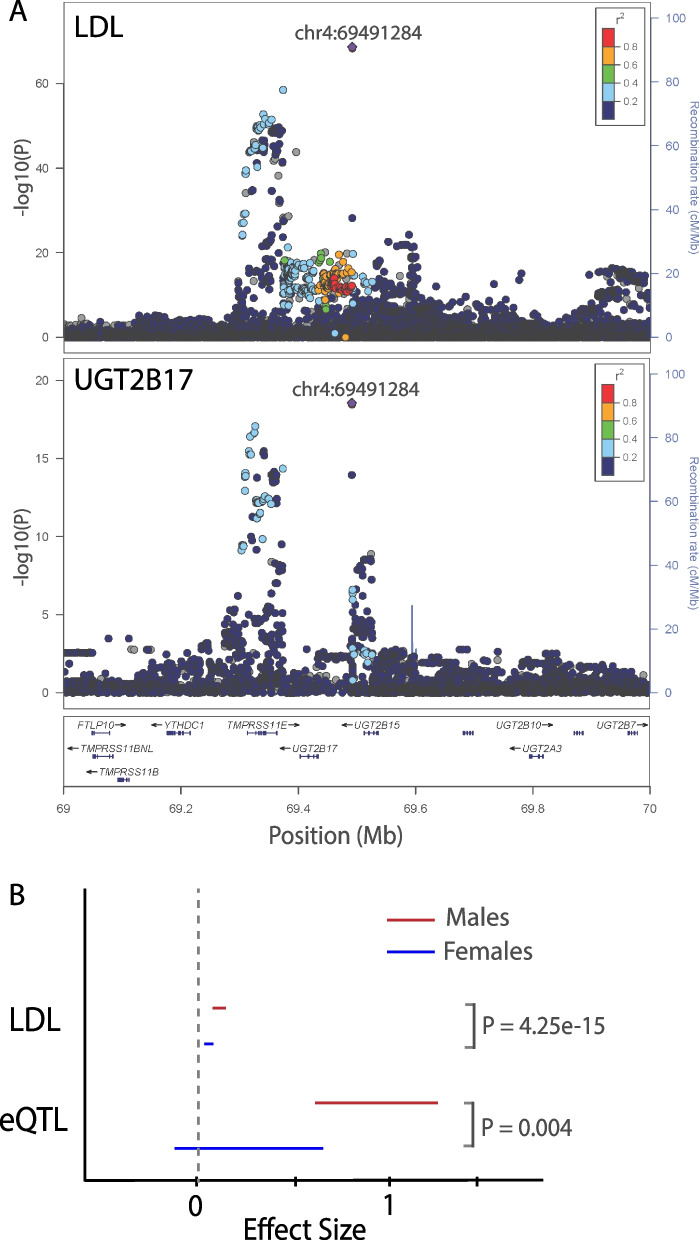
Finally, we annotated each locus that showed significant sex differences with regulatory information to identify biological mechanisms that could underlie this difference. Of the 64 lipid variants with significant sex-stratified associations, 14 colocalized (posterior probability of H4 > 0.8) with expression of 20 genes in lipid-related tissues (liver, adipose, or skeletal muscle; Additional file 30: Table S20). Eight of these loci also show a sex-biased eQTL effect in at least one tissue in the direction concordant with the observed sex specificity of the GWAS effect (Additional file 30: Table S20). Among these ten is CETP, a gene with strong prior evidence for association with lipids, and UGT2B17 [20] (Additional file 31: Supplementary Note, Fig. 3). The lead variant of UGT2B17, rs4860987, shows a significantly stronger effect of LDL-C in males (Betamale = 0.042, SEmale = 0.002, Betafemale = 0.016, SEfemale = 0.003, p-valuedifference = 4.2 × 10−15) and colocalizes with a male-specific liver eQTL associated with increased expression of UGT2B17. Common variants at this locus are in moderate LD (R2 = 0.51) with a common copy number variation (CNV), which may mediate the causal effect (Additional file 31: Supplementary Note). UGT2B17 plays a role in the metabolism of androgens [52], including testosterone, which is consistent with the observed pleiotropic relationship of this locus with testosterone in males (Additional file 30: Table S20). We note that the index variant in UGT2B17, rs4860987, did not show significant sex-specific effects in the replication cohorts, but this could be due to varying frequencies for the index variant between ancestry groups and the moderate LD to the causal CNV in the region. We observed that the combined frequency of rs4860987 across the replication studies was much lower (8%) compared with our combined frequency in the discovery (24%) due to differing proportions of ancestry groups and, along with the lower number of individuals (N = 218,437), led to a much-reduced power to replicate this sex-specific effect.
Fig. 3.
Sex specificity at the UGT2B17 Locus. A The association signal for LDL-C (top panel) colocalizes with the UGT2B17 eQTL signal in the liver (bottom panel). B The effect sizes of this variant on LDL-C and UGT2B17 expression are both significantly higher for males (red) compared to females (blue)
Lipid-associated loci on the X chromosome
Lastly, we meta-analyzed association statistics for 3.1 million X chromosomal variants, including PAR regions, across 1,238,180 individuals from multiple ancestry groups. We identified 28 index variants significantly associated with lipid levels (Additional file 32: Table S21), of which 21 have not been previously reported [20, 39, 53] (15 for HDL-C, 4 for LDL-C, 4 for TC, 5 for TG and 4 for nonHDL-C, Table 2). Among these 28 loci, two have index variants with a minor allele frequency (MAF) < 1% and three index variants are missense mutations (in genes ARSL, TSPAN6, and G6PD), all of which are novel. We validated the identified X chromosomal associations in up to 255,475 individuals from seven multi-ancestry cohorts (Additional file 21: Table S13). Twenty index variants were at least nominally associated (p-value < 0.05), with five reaching genome-wide significance in the replication cohorts alone (p-value < 5 × 10−8, Additional file 32: Table S21).
Table 2.
Novel X chromosome lipid-associated loci
| RSID | Position in chromosome X (hg19) | EA/NEA | Annotation (closest gene) |
Associated trait | EAF | N | Effect size (SE) from METAL |
Ancestry and GC corrected p-value from MR-MEGA | Sex difference p-value |
|---|---|---|---|---|---|---|---|---|---|
| rs35143646 | 2,856,155 | T/C | Missense (ARSL) | LDL-C | 0.6352 | 1,038,070 |
0.0115 (0.0014) |
1.83x10− 16 | 0.1791 |
| TC | 0.6449 | 1,100,310 |
0.0091 (0.0014) |
1.47x10− 14 | 0.08484 | ||||
| nonHDL-C | 0.6822 | 712,983 |
0.0112 (0.0017) |
2.08x10− 11 | 0.5294 | ||||
| rs5934507 | 8,917,206 | G/A | Intergenic (FAM9B) | HDL-C | 0.274 | 1,158,000 | − 0.0076 (0.0012) | 9.99x10− 10 | 5.83x10− 4 |
| rs191084933 | 14,133,208 | G/A | Intergenic (GEMIN8) | HDL-C | 0.0055 | 1,052,630 |
0.0694 (0.0101) |
5.20x10− 10 | 0.3272 |
| rs7888119 | 16,813,128 | T/C | Intronic (TXLNG) | HDL-C | 0.546 | 1,158,000 |
0.0114 (0.0012) |
1.47x10− 20 | 0.2212 |
| rs2230488 | 20,204,461 | T/G | Synonymous (RPS6KA3) | LDL-C | 0.1772 | 1,135,110 | − 0.0143 (0.0015) | 6.90x10− 18 | 0.9297 |
| TC | 0.1745 | 1,237,380 | − 0.0114 (0.0014) | 4.68x10− 12 | 0.8906 | ||||
| rs6527977 | 20,322,238 | A/C | Intergenic (RPS6KA3) | nonHDL-C | 0.1602 | 789,200 | − 0.0147 (0.0020) | 3.06x10− 10 | 0.7000 |
| rs12012576 | 21,813,178 | G/A | Intergenic (SMPX) | TG | 0.3103 | 1,160,340 | − 0.0072 (0.0012) | 4.88x10− 8 | 0.4558 |
| rs6609434 | 46,636,767 | C/A | Intergenic (SLC9A7) | HDL-C | 0.4959 | 767,051 |
0.0060 (0.0014) |
4.25x10− 8 | 0.9035 |
| rs113957181 | 49,848,600 | A/C | Intronic (CLCN5) | HDL-C | 0.0566 | 452,268 | − 0.0297 (0.0047) | 1.39x10− 9 | 0.1896 |
| rs782397956 | 53,993,589 | T/C | Intronic (PHF8) | HDL-C | 0.2050 | 621,342 | − 0.0009 (0.0021) | 2.65x10− 15 | 0.2585 |
| rs72305711 | 55,981,911 | T/TTA | Intronic (RP13-188A5.1) | HDL-C | 0.1111 | 534,967 | − 0.0167 (0.0033) | 2.22x10− 22 | 0.03202 |
| rs5914559 | 56,139,739 | G/T | Intergenic (KLF8) | TG | 0.6697 | 738,155 | − 0.0131 (0.0016) | 4.44x10− 16 | 0.3224 |
| rs5964416 | 64,368,487 | A/C | Intergenic (ZC4H2) | HDL-C | 0.0924 | 614,283 | − 0.0316 (0.0016) | 2.68x10− 11 | 0.1478 |
| rs5965342 | 66,204,144 | T/C | Intergenic (EDA2R) | HDL-C | 0.2037 | 745,721 | − 0.0212 (0.0020) | 1.20x10− 29 | 0.02050 |
| rs505520 | 66,258,914 | C/A | Intergenic (EDA2R) | TG | 0.2191 | 750,415 |
0.0217 (0.0019) |
2.91x10− 30 | 0.004371 |
| rs771540123 | 67,967,645 | A/G | Intergenic (STARD8) | nonHDL-C | 0.0027 | 15,311 |
− 0.0202 (0.115) |
1.61x10− 9 | NA |
| rs5937000 | 70,047,788 | C/T | Intronic (TEX11) | HDL-C | 0.4515 | 796,971 |
0.0075 (0.0013) |
9.55x10− 9 | 0.02603 |
| rs5938008 | 74,496,225 | C/T | Intronic (UPRT) | HDL-C | 0.9167 | 1,072,550 |
0.0174 (0.003) |
1.24x10− 8 | 0.7235 |
| rs1802288 | 99,890,204 | T/C | Missense (TSPAN6) | LDL-C | 0.1665 | 714,113 |
0.0159 (0.0019) |
1.42x10− 16 | 0.9642 |
| TC | 0.1684 | 753,479 |
0.0128 (0.0018) |
2.49x10− 10 | 0.8646 | ||||
| rs139144471 | 117,829,694 | G/T | Intergenic (DOCK11) | HDL-C | 0.0876 | 1,098,540 | − 0.0121 (0.0022) | 1.16x10− 8 | 0.1111 |
| rs6648533 | 122,804,678 | C/T | Intronic (THOC2) | HDL-C | 0.299 | 1,106,090 | − 0.0100 (0.0013) | 2.24x10− 12 | 0.4516 |
| rs5929738 | 135,265,287 | C/A | Intronic (FHL1) | HDL-C | 0.4679 | 970,330 |
0.0071 (0.0011) |
3.21x10− 10 | 0.4197 |
| rs5975692 | 135,266,089 | G/A | Intronic (FHL1) | TG | 0.465 | 1,138,740 | − 0.0084 (0.0011) | 7.87x10− 12 | 0.9215 |
| rs2070826 | 153,582,198 | C/T | Intronic (FLNA) | HDL-C | 0.1427 | 1,141,020 |
0.0170 (0.0017) |
6.65x10− 26 | 0.08671 |
| rs11593 | 153,627,145 | C/A | Intronic (RPL10) | TG | 0.1586 | 1,143,360 | − 0.0166 (0.0017) | 5.76x10− 21 | 0.8695 |
| rs7886627 | 153,679,609 | G/A | Intergenic (FAM50A) | nonHDL-C | 0.1214 | 771,706 | − 0.0147 (0.0022) | 9.26x10− 9 | 0.06819 |
| rs1050828 | 153,764,217 | T/C | Missense (G6PD) | LDL-C | 0.0113 | 744,968 | − 0.0514 (0.0061) | 2.54x10− 15 | 0.6533 |
| rs762517 | 153,764,734 | A/G | Intronic (G6PD) | TC | 0.0142 | 798,600 | − 0.0480 (0.0057) | 5.26x10− 16 | 0.7594 |
EA Effect allele, NEA Non-effect allele, EAF Effect allele frequency, N Number of samples, SE Standard error of the effect size, GC Genomic control, LDL-C Low-density lipoprotein cholesterol, TC Total cholesterol, HDL-C High-density lipoprotein cholesterol, TG Triglycerides
We additionally considered potential sex differences for the X chromosome variants. A missense variant in RENBP with MAF = 2.5% reached genome-wide significance only in males but was not significant in the sex-combined meta-analysis or in the female-only analysis (p-value = 4.59 × 10−8, 0.003 and 0.2, respectively). We also observe three X chromosome loci with significant heterogeneity in effects between sexes; however, these were not significant in the replication cohorts alone, possibly due to the lower sample size (Bonferroni correction for the number of index variants in each trait, Additional file 32: Table S21).
Using a PheWAS approach in the UK Biobank, we found four of the novel loci to have pleiotropic associations with body composition traits (FAM9B [HDL-C], EDA2R [HDL, TG], TSPAN6 [LDL-C, TC], and DOCK11 [HDL-C]), four variants with coronary atherosclerosis and ischemic heart disease, three with immune-related biomarkers (SLC9A7 [HDL-C], CLCN5 [HDL-C], THOC2 [HDL-C]), and two with blood clotting-related biomarkers (KLF8 [TG], TEX11 [HDL-C]) (Additional file 32: Table S21). Interestingly, two of the three sex-biased X chromosome variants demonstrate the most significant association with testosterone of all lipid X chromosome variants tested in the PheWAS (rs505520: beta/SE = − 0.089/0.007 nmol/L per TG-increasing allele and rs5934507: beta/SE = 0.237/0.006 nmol/L per HDL-increasing allele).
Discussion
In this study, we identify and prioritize likely candidate genes at lipid-associated loci discovered through a variety of approaches including multi-ancestry meta-analysis of autosomes [24] (~ 91 million variants) and the X chromosome (~ 3 million variants), as well as sex-specific meta-analyses using sample sizes ranging from 1.35 to 1.65 million individuals. We previously reported a comparison of multi-ancestry vs single-ancestry lipid findings using autosomal chromosomes and identified improvements in fine-mapping of credible sets and PGS performance, with slight differences in the number of identified loci by ancestry group [24]. Here, we add X chromosome and sex-specific discovery results. We also focus on lipid biology by prioritizing implicated genes, identifying novel phenotypes and diseases associated with genetically predicted lipid levels, and predicting candidate drug target genes.
Our results from this effort translate our GWAS findings for three complimentary research areas, helping us further elucidate the biological mechanisms underlying the lipid-associated genetic variants. We first sought to identify methods for prioritization of functional genes at GWAS loci by performing six gene prioritization methods. Lipids are an excellent exemplar phenotype for gene prioritization algorithms because of a wealth of GWAS loci (~ 1000), Mendelian dyslipidemia genes (21), and mouse dyslipidemia phenotypes observed in gene knockouts (740). While the gene prioritization approaches are not independent of each other, integrating several prioritization predictors provides higher confidence when attempting to characterize causal genes. Others have also highlighted the importance of such frameworks in different diseases [29, 54, 55].
We identify 466 unique genes by combining evidence from a global approach (PoPS) with local gene prioritization approaches. The vast majority of these genes had many lipid-related publications, suggesting the accuracy of our combined prioritization approach. Twenty-three PoPS + identified genes had no lipid-related publications, indicating they could be truly novel or possibly were incorrectly prioritized. Functional validation of the larger pool of prioritized genes, which will require highly parallel experimental methods, will help to further optimize bioinformatics algorithms to prioritize genes and is beyond the scope of this manuscript.
Our prioritization approach also indicates several genes as potential drug targets including PDE3A and NR1H4. PDE3A encodes the phosphodiesterase 3A gene and is predicted to be druggable as phosphodiesterase 3A inhibitor (CILOSTAZOL). Cilostazol has antiplatelet, anti-proliferative, vasodilatory, and ischemic-reperfusion protective properties [56] and has been previously suggested for the primary or secondary prevention of CAD [22]. NR1H4 encodes a bile acid receptor and regulates the expression of genes involved in bile acid synthesis and transport. The target gene is predicted to be druggable as a bile acid receptor FXR agonist (URSODIOL). Ursodiol is used to treat primary biliary cirrhosis and cholelithiasis and could be a potential candidate for drug repurposing.
We also identify eighteen coding variants where the protective lipid allele is also protective for CAD or T2D. Among these, PCSK9 is a well-documented drug target, not only for lipids but also for cardiovascular events [57–59]. In comparison to published studies [60], others find a non-significant increased risk for T2D [61] and an arguably stronger protective effect for CAD [62], for PCSK9 variant carriers. Our observation is consistent with the lack of excess T2D risk observed in PCSK9 inhibitor clinical trials [57–59, 63] and with strong protective effects for coronary heart disease [64]. Furthermore, these variants are potential therapeutic targets for protective lipid profiles and lowering risk of disease.
Our second goal was to identify diseases that may benefit from lipid-lowering as well as diseases or traits that may become problematic due to very low lipids. To accomplish this, we examined the association of genetically predicted lipid traits (using PGS) with 773 phenotypes in 478,556 individuals. We observed that genetically predicted increased LDL-C, TC, and HDL-C levels, or decreased TG levels, decrease the risk of cholelithiasis. Prior epidemiological studies have not consistently reported an association between lipid levels and risk of gallstones, with some studies showing that increased levels of LDL-C, TC, and TG and decreased levels of HDL-C predispose to the risk for cholelithiasis [65, 66], but others showing no association [67, 68]. Our results are corroborated by a recent Mendelian randomization meta-analysis study in the FinGen and UK Biobank cohorts [69]. The prioritized genes for the individual index lipid variants significantly associated with cholelithiasis in the PheWAS analysis include ABCG8, a hepatic cholesterol transporter, responsible for the efflux of cholesterol from the enterocytes to the lumen and from the hepatocytes into bile [70]. The lipid-decreasing allele of index variant in ABCG8, rs4245791, has been previously associated with high risk for gallstone disease [47] and high intestinal cholesterol absorption [71], possibly mediated by an increased expression of ABCG8 [72]. Furthermore, even after excluding ABCG5/8 variants from the LDL-PGS, the association with the risk of cholelithiasis was not attenuated. These PGS-PheWAS results suggest the existence of many other cholesterol transporters like ABCG8 that modify blood cholesterol levels perhaps in large part by facilitating an increased secretion of cholesterol into the biliary system, which in turn increases the risk of the formation of gallstones through the supersaturation of bile. We also observed that HbA1c levels were elevated among subjects with genetically predicted increased LDL-C and TC and with genetically predicted decreased HDL-C. Previous epidemiological studies have established associations between dyslipidemia (increased LDL-C, TC, TG, and decreased HDL-C levels) and increased HbA1c levels among subjects with T2D, as well as insulin-resistant subjects without diabetes [73, 74]. Our observations support a strong genetic basis to these associations and are in accordance with previous studies showing shared pathways between lipid biology, T2D, and HbA1c [75], as well as pleiotropic effects of blood red cell variants with lipid levels [76]. Mendelian randomization studies have shown that hemoglobin and LDL show bidirectional inverse relationships and hemoglobin effects on LDL are also mediated through Hb1Ac, implying that genetic variation influencing erythrocytic factors could also determine lipid levels and the opposite [77]. While most of our significant PheWAS findings could be confirmed via Mendelian randomization studies, we cannot exclude the possibility of spurious associations due to pleiotropy.
Lastly, we sought to expand the coverage of the genome and performed the most comprehensive GWAS of lipid levels to date by including assessment of 3 million variants on the X chromosome as well as explicitly testing for sex-specific effects across 23 chromosomes in 1.35 million individuals of diverse ancestries. We report 21 novel X chromosome loci, including an LDL-lowering locus involving a missense variant in G6PD, known to cause G6PD deficiency (p.V68M) [78]. The proposed mechanism is via the inhibition of the NADPH-dependent hydroxymethylglutaryl-CoA (HMG-CoA) reductase, resulting in decreased cholesterol biosynthesis, even though the protective effect of the G6PD deficiency on cardiovascular risk is debatable [79].
We also observed that approximately 3–5% of the genome-wide lipid index variants exhibited differential effects between sexes, which did not seem to be due to differential prevalence in the use of lipid medications or study selection bias. These findings may have important implications in the interpretation of lipid biology, the identification of novel drug targets, and possibly for more accurate prediction of blood cholesterol-related conditions. For example, the UGT2B17 locus, one of the ten sex-biased loci with corresponding sex-biased eQTL effect, is known to be implicated in androgen and drug metabolism [52]. A common CNV in the region, partially tagged by the lipid index variant, is associated with significant variations in expression levels between ethnic groups [80], which would explain lack of replication in the set of independent studies, and the deletion has been linked to testosterone-related decreased BMI levels [81], as well as decreased risk for osteoporosis in men [82].
Several of the reported sex-biased and X chromosome loci showed significant pleiotropic effects with sex hormone levels, including testosterone and SHBG, highlighting the role of hormone regulation in lipid metabolism [83]. In particular, we observe an overall inverse effect between the X chromosome lipid index variants and the sex hormone levels. Observational studies have long suggested a potential influence of the sex hormones on the risk for cardiovascular risk [84] but this hypothesis has not been consistently supported by recent Mendelian randomization studies, raising the issues of reverse causality [85, 86].
Conclusions
In conclusion, we leverage the power of a large multi-ancestry GWAS study to further our understanding of lipid metabolism and the impact on chronic diseases. We identify novel lipid loci on the X chromosome and autosomal loci with evident sex-biased lipid effects. We compare a range of gene prioritizing methods to identify causal genes, an approach applicable to studying other complex traits. We additionally further our understanding of lipid metabolism through a phenome-wide study that implicates a relationship between genetically predicted low cholesterol with risk of cholelithiasis.
Methods
Meta-analysis
Summary statistics for sex-combined autosomal analyses were previously published [24]. Following the same procedure, we carried out meta-analyses stratified by sex for 5 lipid traits (HDL-C, LDL-C, TG, nonHDL-C, and TC) for both the autosomes and chromosome X. The sample size for chromosome X (Total N = 1,238,180; males = 749,391; females = 562,410) was lower than available for autosomes as not all participating biobanks submitted results for chromosome X. Quality control of summary statistics from contributing cohorts was performed using EasyQC [87]. Prior to meta-analysis, we removed variants with low imputation info scores (r2 < 0.3), those with minor allele count < 3, and those with Hardy–Weinberg equilibrium p-value < 1 × 10−8. Variants on the X chromosome were filtered using the female imputation info scores and Hardy–Weinberg equilibrium p-values. Summary statistics were corrected by the genomic-control factor calculated from the median p-value of variants with minor allele frequency > 0.5%. For cohorts that contributed summary statistics imputed both on the Haplotype Reference Consortium (HRC) and 1000 Genomes Population v3 (1KGP3) panels, we generated a single file containing all possible variants, favoring those imputed from the HRC imputation panel due to generally higher imputation quality of these variants. Multi-ancestry meta-analysis was performed with MR-MEGA [88] with 5 principal components and using the inverse-variance weighted method in METAL to estimate effect sizes [89]. Independent loci were defined with physical distance > 500 kb or genetic distance > 0.25 cM, whichever one would result in a larger window, followed by a conditional analysis using rareGWAMA [90] as previously described [24], to identify index variants that were shadows of nearby, more-significant associations. Conditional analysis for chromosome X used a female-only UK Biobank LD reference (N = 21,510). In line with the analysis in the autosomes, a locus was identified as dependent if the effect size after conditioning on the most significant variant in the area was more than 1.43 times smaller than the original (95th percentile of the effect size ratios for chromosome X).
Differences in effect size between males and females were tested within each cohort using [91]:
and were then meta-analyzed across studies using METAL, to account for cohort-specific ascertainment (e.g., enrichment of cases for type 2 diabetes), or demographics, such as age.
Replication
We collected summary statistics from 8 cohorts across 6 ancestry groups, including African or African American, East Asian, European, Hispanic, Middle Eastern, and South Asian. Each cohort provided sex-stratified and X chromosome association results for the tested traits, as available. The difference in effect sizes between males and females was calculated within each cohort as described above and then meta-analyzed across studies using METAL. X chromosome association results were meta-analyzed using METAL with weighting by sample size.
Gene prioritization methods
Closest gene
We annotated the closest gene to the lipid multi-ancestry index variants [24] by identifying the closest gene transcript on either side (500 kb) of the index variant [92].
Colocalization with GTEx eQTLs
For each of the five lipid phenotypes, we first lifted over GWAS summary statistics from the multi-ancestry meta-analysis [24] to GRCh38 using the UCSC liftOver tool. Then, we defined a set of approximately independent windows across the genome within which colocalization with eQTLs was run. To define these, we first identified all genome-wide significant variants (p-value < 5e − 08) from the meta-analysis for each lipid trait and sorted them by significance, from most significant to least. Starting with the most significant variant, we aimed to define a window defining independent genetic signals; we define a variant’s window as a region within the greater of 500 kb or 0.25 cM on either side of this “sentinel variant.” Genetic distances were defined using reference maps from HapMap 3. All other genome-wide significant variants within this window were discarded from the list of sentinel variants, and similar windows were defined for the remaining genome-wide significant variants.
We ran an eQTL colocalization using GTEx v8 eQTL summary statistics within each of our defined windows for all lipid traits. For each of the 49 GTEx tissues, we first identified all genes within 1 Mb of the sentinel variant, and then restricted analysis to those genes with eQTLs (“eGenes”) in that tissue (FDR < 0.05). We used the R package “coloc” (run on R version 3.4.3, coloc version 3.2.1) [93] with default parameters to run colocalization between the GWAS signal and the eQTL signal for each of these cis-eGenes, using as input those variants in the defined window, i.e., all variants present in both datasets. A colocalization posterior probability of (PP3 + PP4) > 0.8 was used to identify loci with enough colocalization power, and PP4/PP3 > 0.9 was used to define those loci that show significant colocalization [94].
Transcriptome-wide association studies (TWAS)
For our transcriptome-wide association analysis (TWAS), we integrated the results of our GWAS with eQTL summary statistics from GTEx v8. The S-PrediXcan software [95] allows us to integrate these two datasets using only summary statistics from GWAS without needing individual-level genotype data. We used the multi-ancestry lipid GWAS summary statistics [24] and harmonized them with the GTEx summary statistics. Then we performed the TWAS using the eQTL models estimated on GTEx v8 expression data. For each of the 49 GTEx tissues, we identified “significant genes” those genes with p-values more significant than an FDR threshold of 0.05.
Genes with coding variants
We determine the coding variants within 99% credible sets and the coding variants in LD > 0.8 with variants in the 99% credible sets with the credible sets as defined here [24]. We define regions for construction of the credible sets as ± 500 kb around each index variant. We used Bayes factors (BFs) for each variant from the MR-MEGA output and generated the credible sets within each region by ranking all variants by BF and calculating the number of variants required to reach a cumulative probability of at least 99%. Additionally, we used previously established gene-based associations [96] to determine whether rare coding variation in a gene were associated with blood lipid levels (p < 0.001). We labeled a gene as having coding variants if any of these criteria were met.
DEPICT
We used Data-driven Expression-Prioritized Integration for Complex Traits (DEPICT, v1 beta version rel194 for 1 KG imputed GWAS) to prioritize genes at our index variants, on the assumption that truly associated genes share functional annotations [27]. Index variants [24] with p-value < 5 x 10−8 were retained as input. We implemented the DEPICT analysis with the default settings of 500 permutations for bias adjustment and 20 replications for FDR estimation. DEPICT prioritizes genes by calculating the similarity of a given gene to genes from other associated loci across 14,461 reconstituted gene sets and estimates the nominal gene prioritization p-value and the estimated false discovery rate of each gene. The prioritized genes at FDR < 0.05 were considered significant.
PoPS
We used the PoPS method to prioritize genes in the previously reported [24] multi-ancestry index variants for all lipid traits. The PoPS method [28] is a new gene prioritization method that identifies the causal genes by integrating GWAS summary statistics with gene expression, biological pathway, and predicted protein–protein interaction data. First, as part of the PoPS analysis, we used MAGMA to compute gene association statistics (z-scores) and gene–gene correlations from GWAS summary statistics and LD information from a multi-ancestry reference panel (1000 Genomes). Next, PoPS performs marginal feature selection by using MAGMA to perform enrichment analysis for each gene feature separately. The model is fitted by generalized least squares (GLS), and MAGMA results are used to perform marginal feature selection, retaining only features that pass a nominal significance threshold (p < 0.05). Then PoPS computes a joint enrichment of all selected features simultaneously in a leave one chromosome out (LOCO) framework. The gene features employed by PoPS are listed here: https://github.com/FinucaneLab/gene_features. Finally, PoPS computes polygenic priority scores for each gene by fitting a joint model for the enrichment of all selected features. The PoPS score for a gene is independent of the GWAS data on the chromosome where the gene is located. The PoPS analysis returned scores for a total of 18,383 genes per lipid trait. We only kept the top 20% genes among all 18,383 genes. We then annotated our index variants with the nearest ENSEMBL genes in a 500-kb window (either side) and selected the highest PoPS score gene in the locus as the prioritized one.
We performed the PoPS analysis on our lipid-specific multi-ancestry meta-analysis results, using all populations from 1000G as the reference for the LD information in MAGMA. As a sensitivity step, we also repeated the same analysis using only the European population from 1000G as the reference. We observed high concordance in the top two PoPS prioritized genes from both reference panels. In detail, the same 2119 genes (89%) were prioritized as the top genes from both panels, a further 203 genes were prioritized as a top gene with one panel and as the second top with the other and only 7 genes were completely mismatched between the two reference panels.
Monogenic genes
We annotated genes known to cause Mendelian lipid disorders based on proximity with identified GWAS loci [97, 98]. GWAS index variants within ± 500 kb of the transcription start and end positions from the USCS genome browser annotations were annotated as nearby known monogenic dyslipidemia genes.
Mouse knockout lipid phenotype silver set genes
Human gene symbols (9557 unique genes) were mapped to gene identifiers (HGNC) and their corresponding mouse ortholog genes were obtained using Ensembl (www.ensembl.org). Phenotype data for single-gene knockout mouse models were obtained from the International Mouse Phenotyping Consortium (IMPC) (www.mousephenotype.org) latest data release 12.0 (www.mousephenotype.org/data/release). The knockout mouse models were primarily produced by IMPC but also include some models that have been reported from the relevant literature and were curated by Mouse Genome Informatics (MGI) data release 6.16 (www.informatics.jax.org). For each mouse model, reported phenotypes were grouped using the mammalian phenotype ontology hierarchy into broad categories relevant to lipids: growth and body weight (MP:0001259), lipid homeostasis (MP:0002118), cholesterol homeostasis (MP:0005278), and lipid metabolism (MP:0013245). This resulted in mapping of human genes to significant phenotypes in animals.
For each of the multi-ancestry lipid index variant [24], we mapped the closest gene to the knockout mouse phenotypes and curated the set to only include mouse phenotypes strictly relating to lipid metabolism. That resulted in our silver set of 740 genes with mouse lipid phenotypes (Additional file 33: Table S22).
Overlap between methods
We standardized the gene names across different methods using the R/geneSynonym package, a wrapper to gene synonym information in ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/gene_info.gz. We also quantified how often the same gene was prioritized by multiple methods for each index variant and determined scores that ranged from 1 to 6 (S1-S6), based on the number of methods that prioritized the gene.
We integrated multiple gene prioritization methods to identify likely causal genes in the latest global lipid genetics consortium GWAS results. In total, we have implemented the 6 individual gene prioritization methods above that utilize the GWAS summary statistics from meta-analysis. Our gene prioritization methods can be placed into two broad categories, the locus-specific methods and the genome-wide methods. The locus-specific methods leverage local GWAS data by connecting the causal variants to the causal gene(s) using genomic distance, eQTLs, or protein-coding variants.
More specifically, there are four locus-specific methods that have been implemented including: (1) The closest protein-coding gene around the index variants based on the genomic distance, (2) eQTL colocalization using r COLOC package, (3) TWAS using S-PrediXcan, (4) coding variants which have been identified in 99% credible sets OR in LD > 0.8 with coding variants OR from gene-based tests (p < 0.001) of rare coding variants. For the eQTL and TWAS, we first used all the 49 GTEx tissues and then restricted to only 5 lipid-specific tissues: liver, adipose subcutaneous, adipose visceral, whole blood, and small intestine. In addition, two genome-wide methods were employed: (1) DEPICT (FDR < 0.05), (2) PoPS (Top 1 gene). It is reasonable to combine similarity-based methods with locus-based methods since they use two different sources of information.
To determine the relative performance of each prioritization method and their combined scores for lipid loci, we used 21 genes known to cause Mendelian dyslipidemias as a gold standard set (ABCA1, ABCG5, ANGPTL3, APOA5, APOB, APOE, CETP, CYP27A1, GPD1, GPIHBP1, LCAT, LDLR, LDLRAP1, LIPA, LIPC, LMF1, LPL, MTTP, PCSK9, SAR1B, SCARB1), and 740 mouse knockout genes causing lipid phenotypes as a silver standard set (Additional file 33: Table S22). We examined two metrics for each gene prioritization approach: (1) the proportion of prioritized genes in the gold/silver standard set, and (2) the proportion of correctly identified genes among all prioritized genes (Additional file 3: Figure S1). Note that out of the 2286 lipid associations, 97 fell within 500 kb of a Mendelian gene and 1280 within 500 kb of a mouse knockout gene with a lipid phenotype. We observed that the TWAS results yielded a high number of prioritized genes, but lead to a low proportion correctly identified. The TWAS approach had a much smaller proportion of genes correctly prioritized among all the prioritized genes, given it prioritized a total of 3511 genes, which was 3.5-fold greater than the other methods (~ 1000 genes). Notably, PoPS provided a similar proportion of correctly identified genes (78%) as of TWAS, while retained a high proportion of prioritized genes in the gold standard set (67%). Compared with PoPS, PoPS + (PoPS plus one of the local methods) slightly sacrificed the proportion of correctly identified genes from 78 to 71%, but improved the proportion of prioritized genes in the gold standard set from 67% to 79%. Overall, PoPS/PoPS + outperform other gene prioritization methods on both metrics for our gold (Additional file 3: Figure S1A) and silver (Additional file 3: Figure S1B) standard gene sets. We also assessed lipid-relevant tissue (liver, subcutaneous and visceral adipose, whole blood, and small intestine) expression QTLs (lipid eQTLs) and transcriptome-wide association (lipid TWAS) and found that the expression results from all tissues performed slightly better at recovering the reference gene sets compared with limiting to the lipid-relevant tissues (Additional file 3: Figure S1).
Text mining analysis
We retrieved the whole MEDLINE/PubMed titles and abstracts as of March 06, 2022, from National Library of Medicine (https://ftp.ncbi.nlm.nih.gov/pubmed/baseline/; https://ftp.ncbi.nlm.nih.gov/pubmed/updatefiles/). We then examined whether a list of genes prioritized by PoPS + and any one of the lipid-related keywords (lipid, lipids, triglyceride, triglycerides, fatty acid, cholesterol, dyslipidemias, hyperlipidemia, hypercholesteremia, diabetes, type 2 diabetes, type II diabetes, heart, cardiovascular, artery, coronary, coronary artery, coronary heart, atherosclerosis, peripheral vascular, PAD, stroke) occurred in the same abstract. We counted how many lipid-related publications that have a specific gene co-occurred with at least one lipid-related keyword. The same text mining approach was also implemented to a set of randomly selected genes from the 18,383 protein-coding genes used by the PoPS. We estimated the number of lipid-related publications we would expect to see by chance. A Mann–Whitney U test was performed to show whether there was a significant difference between the number of lipid-related publications of the PoPS + gene set and reference gene set.
Drug target mining analysis
To gain therapeutic insights from our gene prioritization results, we performed a lookup in Therapeutic Target Database (TTD) 2022 [99] (http://db.idrblab.net/ttd/). Specifically, we cross-referenced 466 unique lipid-associated genes prioritized by PoPS + (Additional file 2: Table S2) with 1563 genes corresponding to at least one drug (either under development or approved) with known clinical indication in TTD 2022. As a quality control for this lookup, we excluded all TTD entries related to drugs that were discontinued, terminated, or withdrawn from the market. The full lookup results are available in Additional file 8: Table S6.
Driver tissues for lipid levels
We performed phenotype-tissue association analysis using DESE (driver-tissue estimation by selective expression) [40]. DESE estimates the causal tissues by selective expression of phenotype-associated genes in GWAS. We used the GWAS summary statistics from the five lipid traits and the GTEx v8 normalized gene-level and transcript-level expression datasets as input. SNPs inside a gene and its ± 5 kb adjacent regions were first mapped to the gene, and then DESE ran iteratively to produce a list of driver tissues and the corresponding p-values of the associations. We used a Bonferroni-corrected significance threshold of 0.05/54 = 9.3 x 10−4.
PheWAS analysis
Construction of lipid PGSs
We had previously developed a multi-ancestry PGS for LDL-C that was demonstrated to perform well across multiple ancestry groups [24]. In a similar manner, we also generated PGS for HDL-C, nonHDL-C, TC, and triglycerides. First, multi-ancestry meta-analysis results were generated with METAL [89] after excluding individuals from the Michigan Genomics Initiative and the UK Biobank. The set of variants used to construct the PGS was limited to those that were well-imputed (R2 > 0.3) in MGI, UK Biobank, and MVP. Risk scores based on PRS-CS [100] or pruning and thresholding with Plink [101] across several r2 (0.1, 0.2), distance (250 kb, 500 kb), and p-value thresholds (5 × 10−10, 5 × 10−9, 5 × 10−8, 5 × 10−7, 5 × 10−6, 5 × 10−5, 5 × 10−4, 5 × 10−3, 0.05) were developed. For each trait, the single best score was selected based on the adjusted r2 calculated in the UK Biobank of the linear model for the lipid trait with the risk score and age, sex, batch, and PC1-4 as covariates. This corresponded to PRS-CS for HDL-C and nonHDL-C and pruning and thresholding for LDL-C (r2 = 0.1, p-value = 5 × 10−4, 500 kb), TG (r2 = 0.1, p-value = 5 × 10−3, 500 kb), and TC (r2 = 0.1, p-value = 5 × 10−4, 500 kb). The variance explained by the risk score among the UK Biobank participants was similar across traits (adjusted r2 of the full model-adjusted r2 of covariates: HDL-C = 0.13; LDL-C = 0.15; nonHDL-C = 0.14; TC = 0.14; TG = 0.10) and validated the ability of the risk score to predict genetically increased lipid levels.
PheWAS of lipid PGSs and index lipid variants in the UK Biobank and MVP
We used the European ancestry subset of individuals from the UK Biobank (408,886 samples) and the European samples from MVP (69,670 samples) to perform the PheWAS analysis.
We constructed a weighted PGS for each of the lipid traits, based on the corresponding genome-wide significant multi-ancestry index variants. We used the PheWAS package in R [102] to map ICD-10 codes from hospital records into clinically relevant phenotypes (phecodes) and to implement these association analyses, while adjusting for sex, age, 10 genetic principal components, and genotyping array (for the UK Biobank only) in each cohort. For the lipid-PGS PheWAS, each PGS was inverse normalized prior to analysis and lipid levels were corrected for statin use. The MVP samples used for the PheWAS analysis were not included in the GWAS meta-analysis [24].
Similarly, we extracted all multi-ancestry autosomal index variants for all lipid traits from the same European ancestry subset of the UK Biobank and MVP and performed a single-variant PheWAS association analysis per cohort. Additionally, we performed a single-variant PheWAS association analysis in the UK Biobank only with the sex-stratified and X chromosome index variants from the multi-ancestry analysis.
Meta-analysis of MVP and the UK Biobank PheWAS results
We combined, via meta-analysis, PheWAS lipid-specific PGS results for all intersecting phecodes and biomarkers between the UK Biobank and MVP (Europeans only) per lipid trait. We used ICD10-based phecodes and manually matched biomarkers to identify intersecting phenotypes between the two datasets. We restricted our meta-analysis to phenotypes that had at least 100 samples (total number for continuous traits or number of cases for binary traits) in each cohort. After the meta-analysis, we excluded phenotypes that had less than 500 combined samples (total number for continuous traits or number of cases for binary traits), to avoid reporting spurious results [103]. That resulted in a total of 773 phenotypes (739 phecodes and 34 biomarkers/measurements). We used both fixed and random effects model for the meta-analysis. We assessed heterogeneity using the p-value for Cochran’s q and set the level for significant heterogeneity at a Bonferroni threshold (p-value ≤ 6.5 × 10−5, to account for multiple testing of 773 phenotypes). We report the results from the fixed-effects model for the phenotypes with non-significant heterogeneity and the results from the random effects model for all others. Similarly, we meta-analyzed all index-variant PheWAS results between the UK Biobank and MVP and obtained results for 811 phenotypes and 1750 lipid multi-ancestry index variants, after excluding instances with a combined sample size < 500.
Lipid index variants with CAD, T2D, and NAFLD datasets
The GWAS meta-analysis results of CAD and T2D were acquired from MVP [62] and DIAGRAM Consortium [61], respectively. For variant rs1229984, the CAD result is from CARDIoGRAMPlusC4D meta-analysis [104], as it was not present in the MVP results. The NAFLD GWAS and meta-analysis was performed in the UK Biobank and Michigan Genomics Initiative (MGI). We determined the association of the lipid index variants with CAD, T2D, and NAFLD and aligned the alleles across all the traits to the LDL-lowering allele. We then highlighted the protective lipid coding alleles associated with CAD.
GWAS and meta-analysis of NAFLD in the UK Biobank and Michigan Genomics Initiative (MGI)
Individuals with NAFLD were identified using ICD-9 571.8 and ICD-10 K76.0. Individuals with hepatitis, liver cirrhosis, liver abscess, ascites, a liver transplant, hepatomegaly, jaundice, or with abnormal result of serum enzyme levels or a function study of the liver were excluded (exclusion phecodes 70.2, 70.3, 571.51, 571.6, 571.8, 571.81, 572, 573, 573.2, 573.3, 573.5, 573.7, 573.9) [105]. Analysis was performed using SAIGE v43.3 [106]. Analysis in the UK Biobank included white British individuals with batch, sex, birth year, and the first 4 genetic principal components as covariates. A total of 1122 cases and 399,900 controls were included in the analysis. Analysis in MGI included only European-ancestry participants with array version, sex, birth year, and the first 4 genetic principal components as covariates. A total of 2901 cases and 49,098 controls were analyzed. Meta-analysis was performed using METAL with weighting based on the effective sample size calculated as 4/((1/Ncases) + (1/Ncontrols)).
CAD/T2D colocalization analysis with lipid traits
We used R package coloc v3.2.1 [93] to perform summary statistics-based colocalization via a Bayesian approach and test whether the 5 lipid traits share common genetic causal variants with CAD or T2D. We first defined a window of ± 100 kb around each index variant [24]. Then for each window of the 10 pairs of traits, we ran colocalization with default parameters using those SNPs present in both datasets. A colocalization posterior probability of PP4 > 0.8 was used to define those loci that show significant colocalization.
Supplementary Information
Additional file 1: Table S1. Characteristics of contributing cohorts (as provided by each participating cohort).
Additional file 2: Table S2. Association results for the multi-ancestry index SNPs with the gene prioritization.
Additional file 3: Figure S1. Summary of prioritizing genes for A. Mendelian and B. mouse model genes separately by trait.
Additional file 4: Table S3. Text mining results for the PoPS+ prioritized genes.
Additional file 5: Table S4. Frequency of lipid-related publications for the PoPS+ prioritized genes.
Additional file 6: Figure S2. Frequency distribution of the lipid-related publications for both high confidence genes and the baseline genes.
Additional file 7: Table S5. Lookup of all prioritized lipid genes in the Therapeutic Target Database 2022.
Additional file 8: Table S6. Association of lipid index variants with CAD, T2D and NAFLD.
Additional file 9: Figure S3. Lipid traits – tissue/cell type associations estimated by DESE according to GTEx gene-level and GTEx transcript-level selective expression.
Additional file 10: Table S7. DESE phenotype-tissue association results using both GTEx gene-level and transcript-level selective expression.
Additional file 11: Figure S4. Comparison of PheWAS results in UKB and MVP for the LDL-C PGS, HDL-C PGS, TC PGS, TG PGS and nonHDL-C PGS.
Additional file 12: Table S8. PheWAS UKB-MVP meta-analysis results for each lipid PGS.
Additional file 13: Figure S5. PheWAS meta-analysis results for the trans-ethnic HDL-C PGS in UK Biobank and MVP.
Additional file 14: Figure S6. PheWAS meta-analysis results for the trans-ethnic TC PGS in UK Biobank and MVP.
Additional file 15: Figure S7. PheWAS meta-analysis results for the trans-ethnic TG PGS in UK Biobank and MVP.
Additional file 16: Figure S8. PheWAS meta-analysis results for the trans-ethnic nonHDL-C PGS in UK Biobank and MVP.
Additional file 17: Table S9. PheWAS UKB-MVP meta-analysis results for each index lipid variant at Bonferroni threshold for multiple testing p<=3.5e-8)
Additional file 18: Table S10. Lambda GC values across minor allele frequency bins for sex-specific meta-analyses.
Additional file 19: Table S11. Significant female-specific multi-ancestry meta-analysis results.
Additional file 20: Table S12. Significant male-specific multi-ancestry meta-analysis results.
Additional file 21: Table S13. Characteristics of replication cohorts (as provided by each participating cohort).
Additional file 22: Table S14. Test for difference in effects for index variants from sex-stratified meta-analysis.
Additional file 23: Table S15. Comparison of the sex-specific effects.
Additional file 24: Figure S9. Comparison of effect size estimates between males and females for index variants showing a significant difference in effect size between sexes.
Additional file 25: Table S16. Comparison of effect size estimates for sex-stratified analysis in the replication cohorts.
Additional file 26: Figure S10. Comparison of effect sizes for trans-ancestry index variants excluding cholesterol-lowering medication.
Additional file 27: Table S17. Sex-stratified effect sizes in UK Biobank considering all individuals or only those not on cholesterol lowering medications.
Additional file 28: Table S18. Sex-participation association of the variants with significant sex-specific lipid results.
Additional file 29: Table S19. Comparison of sex-stratified effect sizes in the UK Biobank for BMI, waist hip ratio adjusted for BMI, alcohol use, and smoking status.
Additional file 30: Table S20. Colocalization results for the sex-specific loci.
Additional file 31: Supplementary Note. Supplementary Note and Cohort Acknowledgments.
Additional file 32: Table S21. Significant X chromosome results in the sex-combined and sex-stratified analysis and replication.
Additional file 33: Table S22. Mouse genes with lipid phenotypes (silver set).
Acknowledgements
We thank Bethany Klunder for her administrative support of the Global Lipids Genetics Consortium. Study-specific acknowledgements are available in the Additional file 32: Supplementary Note.
Peer review information
Wenjing She was the primary editor of this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Review history
The review history is available as Additional file 34.
Authors’ contributions
S.Kanoni, S.E.G. Y.W., I.S., S.Ramdas, and Xiang.Zhu contributed equally to this work as co-first authors. All authors reviewed the manuscript. Consortium management: G.M.P., P.N., T.L.A., M.Boehnke, and C.J.W. Study design, interpretation of results, and drafting of the manuscript: S.Kanoni, S.E.G., Y.W., I.S., S.Ramdas, Xiang.Zhu, S.L.C., K.F.B., S.Vedantam, T.W.W., A.E.L., E.M., G.JM.Z., K-H.H.W., I.N., Y.V.S., A.P.M., M.Boehnke, C.D.B., P.N., P.D., C.J.W., T.L.A., and G.M.P. Primary analyses: S.Kanoni, S.E.G., Y.W., I.S., S.Ramdas, Xiang.Zhu, S.L.C., K.F.B., S.Vedantam, T.W.W., A.E.L. Individual study design, analysis, and oversight: S.Kanoni, S.E.G., Y.W., I.S., S.Ramdas, Xiang.Zhu, S.L.C., K.F.B., S.Vedantam, T.W.W., A.E.L., E.M., G.JM.Z., K-H.H.W., I.N., Q.H., D.K., A.T.Hilliard, Zeyuan.Wang, C.X., G.Thorleifsson, A.H., D.F.G., H.Holm, I.O., M.Y.H., S.Han, M.Akiyama, S.S., C.Terao, M.Kanai, W.Zhou, B.M.B., H.R., A.S.H., Y.V., J.A.P., E.A.R., T.Lingren, QP.F., I.J.K., A.N., J.Takayama, H.C.M., K.A.H., B.T., J.Haessler, F.G., Y.B., J.E.M., A.Campbell, K.L., I.Y.M., A.R., G.Hindy, J.D.F., W.Zhao, D.R.W., C.Turman, H.Huang, M.Graff, A.Choudhury, D.Sengupta, A.Mahajan, M.R.B., W.Zhang, K.Yu, E.M.S., A.Pandit, S.G., X.Y., J.Luan, J-H.Z., F.Matsuda, H-M.J.,.Yoon, C.M-G., A.Pitsillides, J.J.H., A.R.Wood, Y.J., Z.G., S.Haworth, R.E.M., J.F.C., M.Aadahl, A.A.B., J.Yao, A.Manichaikul, C-M.H., Y-J.H., H.R.W., J.R., J.B-J., L.L.K., A.G., M.S-L., R.N., P.Mauro, F.Matteo, A.F.McD., P.M-V., M.Wielscher, S.T., N.S., L.T.M., M.Munz, L.Z., J.Huang, B.Y., A.Poveda, A.K., C.Lamina, L.F., M.S., T.E..G., J.P.B., S.E.R., E.W.D., J.M.Z., J.S.M., C.Fuchsberger, H.Christensen, J.A.B., M.V-M., M.F.F., M.K.W., Zhe.Wang, M.H.P., M.Mangino, P.C., N.V., J.W.B., J.Engmann, N.L.T., A.V., R.C.S., K.S.L., N.R.Z., P.L., M.E.K., G.E.D., S.Huo, D.D.I., H.I., Jian.Yang, Jun.Liu, A.D., H.L.L., J.M., M.Frank, B.S., L.J.S., M.CG., C.W., M.Nakatochi, A.W., N.H–K., X.S., R.X., A.H-C., J.C.F-L., V.L., S.S.N., S.Bayyana, H.M.S., M.R.I., C.Oldmeadow, H-N.K., S.Ryu, P.RHJ.T., L.A., R.D., L.A.L., G.Prasad, L.L-M., M.P., J.Long, X.Li, E.T., F.T., C.N.S., A.Loukola, S.Bollepalli, S.C.W., Y.X.W., W.B.W., T.Nutile, D.R., Y.J.S., S.C., F.L., Jingyun.Yang, K.A.K., B.B., G.G.N., K.M., L.F.B., J.A.S., P.H., A-E.F., E.H., M.Lin, M.P.C., S.Vaccargiu, P.J.van der M., N.Pitkänen, B.E.C., S.W.van der L., K.N.C., S.W., A.R.B., A.P.D., A.A.A., J.Y.L., E.RB.P.,.A.N., H.S.C., M.Nethander, S.F-W., L.S., N.W.R., C.A.W., S-Y.L., J-S.W., C.C., L-P.L., K.N., G.C-P., H.Vestergaard, B.H., O.G., Q.C., M.O.O., J.van S., J.Liang, H.T., N.T., J.H.S., R.D.J., A.P.R., L.W.M., Z.C., L.Li, T.Kawaguchi, J.Thiery, J.C.B., L.J.L., Huaixing.Li, M.A.N., O.T.R., S.I., S.H.W., C.P.N., H.Campbell, S.J., T.Nabika, F.A-M., H.N., P.S.B., I.K., P.K., T.G., T.Katsuya, D.de K., Gert J.de B., E.K.K., H.H.H.A., M.A.I., Xiaofeng.Zhu, F.W.A., A.O.K., J.WJ.B., X-O.S., L.S.R., O.Pedersen, T.H., P.Mitchell, A.W.H., M.Kähönen, L.P., C.Bouchard, A.T., Y-D. I.C., C.E.P., T.A.M., W.L., A.F., C.Ohlsson, D.M., Y.S.C., H.Lee, J-M.Y., W–P.K., S.Y.R., J-T.W., I.M.H., K.J.S., M.E.Z., H.Völzke, G.Homuth, M.K.E., A.B.Z., O.Polasek, G.Pasterkamp, I.E.H., S.Redline, K.P., A.J.O., H.Snieder, G.B., R.S., H.Schmidt, S.Bandinelli, G.D., T.A.T., S.LR.K., P.A.P., N.K., M.B.S., G.G., C.A.B., B.J., P.K.J., D.A.B., P.L.De J., X.Lu, V.M., M.Brown, M.J.C., P.B.M., X.G., M.Ciullo, J.B.J., N.J.S., J.Kaprio, P.P., T.T-L., C.A.A-S., L.S.A., S.A.B., H. J.de S., A.R.Wickremasinghe, R.M.K., J-Y.W., W.Zeng, A.I.den H., D.B., A.Correa, J.G.W., L.Lind, C-K.H., A.E.N., Y.M.G., J.F.W., B.P., H–L.K., J.A., R.J.S., D.C.R., D.K.A., M.Walker, H.A.K., G.R.C., J.M.M., M.C.C., D.J., N.P.B., C.G.V., L.O., M.Fornage, E S.T., R.M.van D., T.Lehtimäki, N.C., M.Yokota, Jianjun.Liu, D.F.R., A.J.McK., F.Kee, K-H.J., M.I.McC., C.NA.P., V.V., C.H., E.S., C.M.van D., Z-B.J., J.Q., H.Hishigaki, X.Lin, W.M., V.G., J-C.T., G.L., L.M.t H., P.JM.E., S.M.D., M.Kumari, M.Kivimaki, P.van der H., T.D.S., R.J.F.L, M.A.P., E.J.P., M.Cruz, B.M.P., I.B., P.P.P., C.N.R., K.Christensen, S.Ripatti, E.W., H.Hakonarson, S.F.A.G., L.ALM.K., J.de G., M.Loeffler, F.Kronenberg, D.G., J.Erdmann, H.Schunkert, P.W.F., A.Linneberg, J.W.J., A.V.K., M.Männikkö, M-R.J., Z.K., C.Francesco, D.O.M–K., K.W.van D., H.W., D.P.S., N.G., P.S., N.Poulter, L-M.C., J.I.R., T.M.D., F.Karpe, M.J.N., N.J.T., C-Y.C., T-Y.W., C.C.K., Hengtong.Li, C.S., A.Peters, C.G., A.T.Hattersley, N.L.P., P.KE.M., D.I.B., A.HM.W., L.A.C., J.B.J.vanM, M.Ghanbari, P.G-L., W.H., YJ.K., Y.T., N.J.W.,.Langenberg, E.Z., J.Kuusisto, M.Laakso, E.I., G.A., J.C.C., J.S.K., P.S.de V., A.C.M., S.Hazelhurst, M.R., K.E.N., M.D., P.K., N.G.M., J.B.W., S.A., D.Saleheen, R.G.W., M.V.H., C.Black, B.H.S., A.B., A.E.J., J.E.B., P.M.R., D.I.C., C.K., G.Tamiya, M.Yamamoto, D.A.van H., R.C.T., W-Q.W., G.P.J., B.N., M.G.H., M.D.R., P.J., V.S., K.H., B.O.Å., M.Kubo, Y.K., Y.O., Y.M., B-J.K., U.T., K.S., J.Z., Y.E.C., Y-L.H., J.A.L., D.J.R., P.S.T., K-M.C., K.Cho, C.J.O'D., J.M.G., P.WF.W., T.M.F., J.N.H., S.Kathiresan, K.L.M., Y.V.S., A.P.M., M.Boehnke, C.D.B., P.N., P.D., C.J.W., T.L.A., and G.M.P. All author(s) read and approved the final manuscript.
Funding
GMP, PN, and CW are supported by NHLBI R01HL127564. GMP and PN are supported by R01HL142711. AG acknowledge support from the Wellcome Trust (201543/B/16/Z), European Union Seventh Framework Programme FP7/2007–2013 under grant agreement no. HEALTH-F2-2013–601456 (CVGenes@Target) & the TriPartite Immunometabolism Consortium [TrIC]-Novo Nordisk Foundation’s Grant number NNF15CC0018486. JMM is supported by American Diabetes Association Innovative and Clinical Translational Award 1–19-ICTS-068. SR was supported by the Academy of Finland Center of Excellence in Complex Disease Genetics (Grant No 312062), the Finnish Foundation for Cardiovascular Research, the Sigrid Juselius Foundation, and University of Helsinki HiLIFE Fellow and Grand Challenge grants. EW was supported by the Finnish innovation fund Sitra (EW) and Finska Läkaresällskapet. CNS was supported by American Heart Association Postdoctoral Fellowships 15POST24470131 and 17POST33650016. Charles N Rotimi is supported by Z01HG200362. Zhe Wang, Michael H Preuss, and Ruth JF Loos are supported by R01HL142302. NJT is a Wellcome Trust Investigator (202802/Z/16/Z), is the PI of the Avon Longitudinal Study of Parents and Children (MRC & WT 217065/Z/19/Z), is supported by the University of Bristol NIHR Biomedical Research Centre (BRC-1215–2001) and the MRC Integrative Epidemiology Unit (MC_UU_00011), and works within the CRUK Integrative Cancer Epidemiology Programme (C18281/A19169). Ruth E Mitchell is a member of the MRC Integrative Epidemiology Unit at the University of Bristol funded by the MRC (MC_UU_00011/1). Simon Haworth is supported by the UK National Institute for Health Research Academic Clinical Fellowship. Paul S. de Vries was supported by American Heart Association grant number 18CDA34110116. Julia Ramierz acknowledges support by the People Programme of the European Union’s Seventh Framework Programme grant n° 608765 and Marie Sklodowska-Curie grant n° 786833. Maria Sabater-Lleal is supported by a Miguel Servet contract from the ISCIII Spanish Health Institute (CP17/00142) and co-financed by the European Social Fund. Jian Yang is funded by the Westlake Education Foundation. Olga Giannakopoulou has received funding from the British Heart Foundation (BHF) (FS/14/66/3129). CHARGE Consortium cohorts were supported by R01HL105756. Study-specific acknowledgements are available in the Additional file 32: Supplementary Note. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
Availability of data and materials
The GWAS meta-analysis results (including both ancestry-specific and trans-ancestry analyses) and risk score weights are available at: http://csg.sph.umich.edu/willer/public/glgc-lipids2021 [107]. A web browser displaying the gene prioritization and PheWAS results is available at https://hugeamp.org:8000/research.html?pageid=GLGC_149 [108]. The optimized trans-ancestry polygenic score weights are deposited within the PGS Catalog (https://www.pgscatalog.org/publication/PGP000230/ [109] and https://www.pgscatalog.org/publication/PGP000366/ [110]. Scripts used for analysis and summary of results are available under the MIT license on this GitHub repository: https://github.com/Global-Lipids-Genetics [111]. The version of source code used in the manuscript is deposited in Zenodo: https://doi.org/10.5281/zenodo.7130299 [112].
Declarations
Ethics approval and consent to participate
The overall study was approved by the IRB of the Boston University Medical Center. Individual studies were approved by the appropriate institutional review boards (IRB) and informed consent was obtained from all participants.
Competing interests
Ioanna Ntalla is an employee and stock owner of Gilead Sciences since August 2019. Derek Klarin accepts consulting fees from Regeneron Pharmaceuticals. All deCODE-affiliated authors (Gudmar Thorleifsson, Anna Helgadottir, Daniel F Gudbjartsson, Hilma Holm, Unnur Thorsteinsdottir, Kari Stefansson) are employees of deCODE/Amgen Inc. As of January 2020, Anubha Mahajan is an employee of Genentech, and a holder of Roche stock. Markus Scholz receives funding from Pfizer Inc. for a project not related to this research. Marcus E Kleber is employed by SYNLAB MVZ Mannheim GmbH. Gabriel Cuellar-Partida contributed to this work while employed at The University of Queensland, but he is now an employee of 23andMe Inc. Mark J Caulfield is Chief Scientist for Genomics England, a UK Government company. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. Mark I McCarthy has served on advisory panels for Pfizer, NovoNordisk, and Zoe Global and has received honoraria from Merck, Pfizer, Novo Nordisk, and Eli Lilly and research funding from Abbvie, Astra Zeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, NovoNordisk, Pfizer, Roche, Sanofi Aventis, Servier, and Takeda. As of June 2019, Mark I McCarthy is an employee of Genentech, and a holder of Roche stock. Winfried März has received grants from Siemens Healthineers, grants and personal fees from Aegerion Pharmaceuticals, grants and personal fees from AMGEN, grants from Astrazeneca, grants and personal fees from Sanofi, grants and personal fees from Alexion Pharmaceuticals, grants and personal fees from BASF, grants and personal fees from Abbott Diagnostics, grants and personal fees from Numares AG, grants and personal fees from Berlin-Chemie, grants and personal fees from Akzea Therapeutics, grants from Bayer Vital GmbH, grants from bestbion dx GmbH, grants from Boehringer Ingelheim Pharma GmbH Co KG, grants from Immundiagnostik GmbH, grants from Merck Chemicals GmbH, grants from MSD Sharp and Dohme GmbH, grants from Novartis Pharma GmbH, grants from Olink Proteomics, other from Synlab Holding Deutschland GmbH, all outside the submitted work. Bruce M Psaty serves on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. Amit V Khera has served as a consultant to Sanofi, Medicines Company, Maze Pharmaceuticals, Navitor Pharmaceuticals, Verve Therapeutics, Amgen, and Color Genomics; received speaking fees from Illumina, the Novartis Institute for Biomedical Research; received sponsored research agreements from the Novartis Institute for Biomedical Research and IBM Research, and reports a patent related to a genetic risk predictor (20190017119). Dennis O Mook-Kanamori is a part-time clinical research consultant for Metabolon, Inc. Danish Saleheen has received support from the British Heart Foundation, Pfizer, Regeneron, Genentech, and Eli Lilly pharmaceuticals. Veikko Salomaa has received honoraria for consultations from Novo Nordisk and Sanofi and has ongoing research collaboration with Bayer Ltd, all unrelated to the present study. Sekar Kathiresan is an employee of Verve Therapeutics, and holds equity in Verve Therapeutics, Maze Therapeutics, Catabasis, and San Therapeutics. He is a member of the scientific advisory boards for Regeneron Genetics Center and Corvidia Therapeutics; he has served as a consultant for Acceleron, Eli Lilly, Novartis, Merck, Novo Nordisk, Novo Ventures, Ionis, Alnylam, Aegerion, Haug Partners, Noble Insights, Leerink Partners, Bayer Healthcare, Illumina, Color Genomics, MedGenome, Quest, and Medscape; he reports patents related to a method of identifying and treating a person having a predisposition to or afflicted with cardiometabolic disease (20180010185) and a genetics risk predictor (20190017119). Cristen J Willer’s spouse is employed by Regeneron.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Stavroula Kanoni, Sarah E. Graham, Yuxuan Wang, Ida Surakka, Shweta Ramdas, and Xiang Zhu contributed equally to this work.
Michael Boehnke, Christopher D. Brown, Pradeep Natarajan, Panos Deloukas, Cristen J. Willer, Themistocles L. Assimes, and Gina M. Peloso jointly supervised this work.
Contributor Information
Cristen J. Willer, Email: cristen@umich.edu
Gina M. Peloso, Email: gpeloso@bu.edu
References
- 1.Castelli WP, Anderson K, Wilson PW, Levy D. Lipids and risk of coronary heart disease. The Framingham Study. Ann Epidemiol. 1992;2:23–28. doi: 10.1016/1047-2797(92)90033-M. [DOI] [PubMed] [Google Scholar]
- 2.GBD Diseases and Injuries Collaborators: Global burden of 369 diseases and injuries in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet. 2019;2020(396):1204–1222. doi: 10.1016/S0140-6736(20)30925-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, Braun LT, de Ferranti S, Faiella-Tommasino J, Forman DE, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2019;139:e1082–e1143. doi: 10.1161/CIR.0000000000000625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 5.Kathiresan S, Manning AK, Demissie S, D'Agostino RB, Surti A, Guiducci C, Gianniny L, Burtt NP, Melander O, Orho-Melander M, et al. A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med Genet. 2007;8(Suppl 1):S17. doi: 10.1186/1471-2350-8-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 7.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asselbergs FW, Guo Y, van Iperen EP, Sivapalaratnam S, Tragante V, Lanktree MB, Lange LA, Almoguera B, Appelman YE, Barnard J, et al. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am J Hum Genet. 2012;91:823–838. doi: 10.1016/j.ajhg.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albrechtsen A, Grarup N, Li Y, Sparso T, Tian G, Cao H, Jiang T, Kim SY, Korneliussen T, Li Q, et al. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia. 2013;56:298–310. doi: 10.1007/s00125-012-2756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tachmazidou I, Dedoussis G, Southam L, Farmaki AE, Ritchie GR, Xifara DK, Matchan A, Hatzikotoulas K, Rayner NW, Chen Y, et al. A rare functional cardioprotective APOC3 variant has risen in frequency in distinct population isolates. Nat Commun. 2013;4:2872. doi: 10.1038/ncomms3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holmen OL, Zhang H, Fan Y, Hovelson DH, Schmidt EM, Zhou W, Guo Y, Zhang J, Langhammer A, Lochen ML, et al. Systematic evaluation of coding variation identifies a candidate causal variant in TM6SF2 influencing total cholesterol and myocardial infarction risk. Nat Genet. 2014;46:345–351. doi: 10.1038/ng.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO, Brody JA, Khetarpal SA, Crosby JR, Fornage M, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94:223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surakka I, Horikoshi M, Magi R, Sarin AP, Mahajan A, Lagou V, Marullo L, Ferreira T, Miraglio B, Timonen S, et al. The impact of low-frequency and rare variants on lipid levels. Nat Genet. 2015;47:589–597. doi: 10.1038/ng.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang CS, Zhang H, Cheung CY, Xu M, Ho JC, Zhou W, Cherny SS, Zhang Y, Holmen O, Au KW, et al. Exome-wide association analysis reveals novel coding sequence variants associated with lipid traits in Chinese. Nat Commun. 2015;6:10206. doi: 10.1038/ncomms10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Leeuwen EM, Karssen LC, Deelen J, Isaacs A, Medina-Gomez C, Mbarek H, Kanterakis A, Trompet S, Postmus I, Verweij N, et al. Genome of The Netherlands population-specific imputations identify an ABCA6 variant associated with cholesterol levels. Nat Commun. 2015;6:6065. doi: 10.1038/ncomms7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iotchkova V, Huang J, Morris JA, Jain D, Barbieri C, Walter K, Min JL, Chen L, Astle W, Cocca M, et al. Discovery and refinement of genetic loci associated with cardiometabolic risk using dense imputation maps. Nat Genet. 2016;48:1303–1312. doi: 10.1038/ng.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu DJ, Peloso GM, Yu H, Butterworth AS, Wang X, Mahajan A, Saleheen D, Emdin C, Alam D, Alves AC, et al. Exome-wide association study of plasma lipids in >300,000 individuals. Nat Genet. 2017;49:1758–1766. doi: 10.1038/ng.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu X, Peloso GM, Liu DJ, Wu Y, Zhang H, Zhou W, Li J, Tang CS, Dorajoo R, Li H, et al. Exome chip meta-analysis identifies novel loci and East Asian-specific coding variants that contribute to lipid levels and coronary artery disease. Nat Genet. 2017;49:1722–1730. doi: 10.1038/ng.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann TJ, Theusch E, Haldar T, Ranatunga DK, Jorgenson E, Medina MW, Kvale MN, Kwok PY, Schaefer C, Krauss RM, et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat Genet. 2018;50:401–413. doi: 10.1038/s41588-018-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 22.Klarin D, Damrauer SM, Cho K, Sun YV, Teslovich TM, Honerlaw J, Gagnon DR, DuVall SL, Li J, Peloso GM, et al. Genetics of blood lipids among ~300,000 multi-ethnic participants of the Million Veteran Program. Nat Genet. 2018;50:1514–1523. doi: 10.1038/s41588-018-0222-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spracklen CN, Chen P, Kim YJ, Wang X, Cai H, Li S, Long J, Wu Y, Wang YX, Takeuchi F. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum Mol Genet. 2018;27:1122. doi: 10.1093/hmg/ddx439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham SE, Clarke SL, Wu KH, Kanoni S, Zajac GJM, Ramdas S, Surakka I, Ntalla I, Vedantam S, Winkler TW, et al. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600:675–679. doi: 10.1038/s41586-021-04064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Musunuru K, Strong A, Frank-Kamenetsky M, Lee NE, Ahfeldt T, Sachs KV, Li X, Li H, Kuperwasser N, Ruda VM, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musunuru K, Pirruccello JP, Do R, Peloso GM, Guiducci C, Sougnez C, Garimella KV, Fisher S, Abreu J, Barry AJ, et al. Exome sequencing, ANGPTL3 mutations, and familial combined hypolipidemia. N Engl J Med. 2010;363:2220–2227. doi: 10.1056/NEJMoa1002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pers TH, Karjalainen JM, Chan Y, Westra HJ, Wood AR, Yang J, Lui JC, Vedantam S, Gustafsson S, Esko T, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weeks EM, Ulirsch JC, Cheng NY, Trippe BL, Fine RS, Miao J, Patwardhan TA, Kanai M, Nasser J, Fulco CP, et al: Leveraging polygenic enrichments of gene features to predict genes underlying complex traits and diseases. medRxiv 2020:2020.2009.2008.20190561. [DOI] [PMC free article] [PubMed]
- 29.Stanzick KJ, Li Y, Schlosser P, Gorski M, Wuttke M, Thomas LF, Rasheed H, Rowan BX, Graham SE, Vanderweff BR, et al. Discovery and prioritization of variants and genes for kidney function in >1.2 million individuals. Nature Communications. 2021;12:4350. doi: 10.1038/s41467-021-24491-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Emerging Risk Factors Collaboration. Di Angelantonio E, Sarwar N, Perry P, Kaptoge S, Ray KK, Thompson A, Wood AM, Lewington S, Sattar N, et al. Major lipids, apolipoproteins, and risk of vascular disease. JAMA. 2009;302:1993–2000. doi: 10.1001/jama.2009.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richardson TG, Sanderson E, Palmer TM, Ala-Korpela M, Ference BA, Davey Smith G, Holmes MV. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med. 2020;17:e1003062. doi: 10.1371/journal.pmed.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allara E, Morani G, Carter P, Gkatzionis A, Zuber V, Foley CN, Rees JMB, Mason AM, Bell S, Gill D, et al: Genetic determinants of lipids and cardiovascular disease outcomes. Circulation: Genomic Precision Med. 2019;12:e002711. [DOI] [PMC free article] [PubMed]
- 33.Veturi Y, Lucas A, Bradford Y, Hui D, Dudek S, Theusch E, Verma A, Miller JE, Kullo I, Hakonarson H, et al. A unified framework identifies new links between plasma lipids and diseases from electronic medical records across large-scale cohorts. Nat Genet. 2021;53:972–981. doi: 10.1038/s41588-021-00879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bush WS, Oetjens MT, Crawford DC. Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet. 2016;17:129–145. doi: 10.1038/nrg.2015.36. [DOI] [PubMed] [Google Scholar]
- 35.Abbott RD, Garrison RJ, Wilson PW, Epstein FH, Castelli WP, Feinleib M, LaRue C. Joint distribution of lipoprotein cholesterol classes. The Framingham study. Arteriosclerosis. 1983;3:260–272. doi: 10.1161/01.ATV.3.3.260. [DOI] [PubMed] [Google Scholar]
- 36.Flynn E, Tanigawa Y, Rodriguez F, Altman RB, Sinnott-Armstrong N, Rivas MA. Sex-specific genetic effects across biomarkers. Eur J Hum Genet. 2021;29:154–163. doi: 10.1038/s41431-020-00712-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zore T, Palafox M, Reue K. Sex differences in obesity, lipid metabolism, and inflammation-A role for the sex chromosomes? Mol Metab. 2018;15:35–44. doi: 10.1016/j.molmet.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.AlSiraj Y, Chen X, Thatcher SE, Temel RE, Cai L, Blalock E, Katz W, Ali HM, Petriello M, Deng P, et al. XX sex chromosome complement promotes atherosclerosis in mice. Nat Commun. 2019;10:2631. doi: 10.1038/s41467-019-10462-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Natarajan P, Pampana A, Graham SE, Ruotsalainen SE, Perry JA, de Vries PS, Broome JG, Pirruccello JP, Honigberg MC, Aragam K, et al. Chromosome Xq23 is associated with lower atherogenic lipid concentrations and favorable cardiometabolic indices. Nat Commun. 2021;12:2182. doi: 10.1038/s41467-021-22339-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang L, Xue C, Dai S, Chen S, Chen P, Sham PC, Wang H, Li M. DESE: estimating driver tissues by selective expression of genes associated with complex diseases or traits. Genome Biol. 2019;20:233. doi: 10.1186/s13059-019-1801-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allara E, Morani G, Carter P, Gkatzionis A, Zuber V, Foley CN, Rees JMB, Mason AM, Bell S, Gill D, et al. Genetic determinants of lipids and cardiovascular disease outcomes: a wide-angled Mendelian randomization investigation. Circ Genom Precis Med. 2019;12:e002711. doi: 10.1161/CIRCGEN.119.002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saiz-Vazquez O, Puente-Martinez A, Ubillos-Landa S, Pacheco-Bonrostro J, Santabarbara J. Cholesterol and Alzheimer's disease risk: a meta-meta-analysis. Brain Sci. 2020;10:386. [DOI] [PMC free article] [PubMed]
- 44.Zhang X, Tian Q, Liu D, Geng T, Xu X, Ge S, Zheng D, Wu L, Song M, Hou H, et al. Causal association of circulating cholesterol levels with dementia: a mendelian randomization meta-analysis. Transl Psychiatry. 2020;10:145. doi: 10.1038/s41398-020-0822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan JS, Hu MJ, Yang YM, Yang YJ. Genetic predisposition to low-density lipoprotein cholesterol may increase risks of both individual and familial Alzheimer's disease. Front Med (Lausanne) 2021;8:798334. doi: 10.3389/fmed.2021.798334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Deb S, Puthanveetil P, Sakharkar P. A population-based cross-sectional study of the association between liver enzymes and lipid levels. Int J Hepatol. 2018;2018:1286170. doi: 10.1155/2018/1286170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joshi AD, Andersson C, Buch S, Stender S, Noordam R, Weng LC, Weeke PE, Auer PL, Boehm B, Chen C, et al. Four susceptibility loci for gallstone disease identified in a meta-analysis of genome-wide association studies. Gastroenterology. 2016;151(351–363):e328. doi: 10.1053/j.gastro.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bernabeu E, Canela-Xandri O, Rawlik K, Talenti A, Prendergast J, Tenesa A. Sex differences in genetic architecture in the UK Biobank. Nat Genet. 2021;53:1283–1289. doi: 10.1038/s41588-021-00912-0. [DOI] [PubMed] [Google Scholar]
- 49.Ruth KS, Day FR, Tyrrell J, Thompson DJ, Wood AR, Mahajan A, Beaumont RN, Wittemans L, Martin S, Busch AS, et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat Med. 2020;26:252–258. doi: 10.1038/s41591-020-0751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pirastu N, Cordioli M, Nandakumar P, Mignogna G, Abdellaoui A, Hollis B, Kanai M, Rajagopal VM, Parolo PDB, Baya N, et al. Genetic analyses identify widespread sex-differential participation bias. Nat Genet. 2021;53:663–671. doi: 10.1038/s41588-021-00846-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM, Consortium G Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhatt DK, Basit A, Zhang H, Gaedigk A, Lee SB, Claw KG, Mehrotra A, Chaudhry AS, Pearce RE, Gaedigk R, et al. Hepatic abundance and activity of androgen- and drug-metabolizing enzyme UGT2B17 are associated with genotype, age, and sex. Drug Metab Dispos. 2018;46:888–896. doi: 10.1124/dmd.118.080952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nielsen JB, Rom O, Surakka I, Graham SE, Zhou W, Roychowdhury T, Fritsche LG, Gagliano Taliun SA, Sidore C, Liu Y, et al. Loss-of-function genomic variants highlight potential therapeutic targets for cardiovascular disease. Nat Commun. 2020;11:6417. doi: 10.1038/s41467-020-20086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aragam KG, Jiang T, Goel A, Kanoni S, Wolford BN, Weeks EM, Wang M, Hindy G, Zhou W, Grace C, et al: Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. medRxiv 2021:2021.2005.2024.21257377. [DOI] [PMC free article] [PubMed]
- 55.Votava JA, Parks BW. Cross-species data integration to prioritize causal genes in lipid metabolism. Curr Opin Lipidol. 2021;32:141–146. doi: 10.1097/MOL.0000000000000742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kherallah RY, Khawaja M, Olson M, Angiolillo D, Birnbaum Y. Cilostazol: a review of basic mechanisms and clinical uses. Cardiovasc Drugs Ther. 2022;36:777-92. [DOI] [PubMed]
- 57.Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, Kuder JF, Wang H, Liu T, Wasserman SM, et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017;376:1713–1722. doi: 10.1056/NEJMoa1615664. [DOI] [PubMed] [Google Scholar]
- 58.Schwartz GG, Steg PG, Szarek M, Bhatt DL, Bittner VA, Diaz R, Edelberg JM, Goodman SG, Hanotin C, Harrington RA, et al. Alirocumab and cardiovascular outcomes after acute coronary syndrome. N Engl J Med. 2018;379:2097–2107. doi: 10.1056/NEJMoa1801174. [DOI] [PubMed] [Google Scholar]
- 59.Ray KK, Wright RS, Kallend D, Koenig W, Leiter LA, Raal FJ, Bisch JA, Richardson T, Jaros M, Wijngaard PLJ, Kastelein JJP. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N Engl J Med. 2020;382:1507–1519. doi: 10.1056/NEJMoa1912387. [DOI] [PubMed] [Google Scholar]
- 60.Nelson CP, Lai FY, Nath M, Ye S, Webb TR, Schunkert H, Samani NJ. Genetic assessment of potential long-term on-target side effects of PCSK9 (Proprotein Convertase Subtilisin/Kexin Type 9) inhibitors. Circ Genom Precis Med. 2019;12:e002196. doi: 10.1161/CIRCGEN.118.002196. [DOI] [PubMed] [Google Scholar]
- 61.Mahajan A, Taliun D, Thurner M, Robertson NR, Torres JM, Rayner NW, Payne AJ, Steinthorsdottir V, Scott RA, Grarup N, et al. Fine-mapping type 2 diabetes loci to single-variant resolution using high-density imputation and islet-specific epigenome maps. Nat Genet. 2018;50:1505–1513. doi: 10.1038/s41588-018-0241-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Assimes T, Catherine T, Xiang Z, Austin H, Shoa C, Valerio N, Shining M, Huaying F, Bryan RG, Kyung Min L, et al. A large-scale multi-ethnic genome-wide association study of coronary artery disease. Nat Med. 2022;28:1679-92. [DOI] [PMC free article] [PubMed]
- 63.Ridker PM, Revkin J, Amarenco P, Brunell R, Curto M, Civeira F, Flather M, Glynn RJ, Gregoire J, Jukema JW, et al. Cardiovascular efficacy and safety of bococizumab in high-risk patients. N Engl J Med. 2017;376:1527–1539. doi: 10.1056/NEJMoa1701488. [DOI] [PubMed] [Google Scholar]
- 64.Hopewell JC, Malik R, Valdes-Marquez E, Worrall BB, Collins R. ISGC MCot: Differential effects of PCSK9 variants on risk of coronary disease and ischaemic stroke. Eur Heart J. 2018;39:354–359. doi: 10.1093/eurheartj/ehx373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayat S, Hassan Z, Changazi SH, Zahra A, Noman M. Zain Ul Abdin M, Javed H, Ans AH: Comparative analysis of serum lipid profiles in patients with and without gallstones: a prospective cross-sectional study. Ann Med Surg (Lond) 2019;42:11–13. doi: 10.1016/j.amsu.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, Shen S, Wang B, Ni X, Liu H, Ni X, Yu R, Suo T, Liu H. Serum lipid levels are the risk factors of gallbladder stones: a population-based study in China. Lipids Health Dis. 2020;19:50. doi: 10.1186/s12944-019-1184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gustafsson U, Sahlin S, Einarsson C. Biliary lipid composition in patients with cholesterol and pigment gallstones and gallstone-free subjects: deoxycholic acid does not contribute to formation of cholesterol gallstones. Eur J Clin Invest. 2000;30:1099–1106. doi: 10.1046/j.1365-2362.2000.00740.x. [DOI] [PubMed] [Google Scholar]
- 68.Weerakoon HT, Ranasinghe S, Navaratne A, Sivakanesan R, Galketiya KB, Rosairo S. Serum lipid concentrations in patients with cholesterol and pigment gallstones. BMC Res Notes. 2014;7:548. doi: 10.1186/1756-0500-7-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen L, Yang H, Li H, He C, Yang L, Lv G. Insights into modifiable risk factors of cholelithiasis: a Mendelian randomization study. Hepatology. 2022;75:785–796. doi: 10.1002/hep.32183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu XH, Qian K, Jiang N, Zheng XL, Cayabyab FS, Tang CK. ABCG5/ABCG8 in cholesterol excretion and atherosclerosis. Clin Chim Acta. 2014;428:82–88. doi: 10.1016/j.cca.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 71.Silbernagel G, Chapman MJ, Genser B, Kleber ME, Fauler G, Scharnagl H, Grammer TB, Boehm BO, Makela KM, Kahonen M, et al. High intestinal cholesterol absorption is associated with cardiovascular disease and risk alleles in ABCG8 and ABO: evidence from the LURIC and YFS cohorts and from a meta-analysis. J Am Coll Cardiol. 2013;62:291–299. doi: 10.1016/j.jacc.2013.01.100. [DOI] [PubMed] [Google Scholar]
- 72.Teupser D, Baber R, Ceglarek U, Scholz M, Illig T, Gieger C, Holdt LM, Leichtle A, Greiser KH, Huster D, et al. Genetic regulation of serum phytosterol levels and risk of coronary artery disease. Circulation Cardiovasc Genet. 2010;3:331–339. doi: 10.1161/CIRCGENETICS.109.907873. [DOI] [PubMed] [Google Scholar]
- 73.Artha I, Bhargah A, Dharmawan NK, Pande UW, Triyana KA, Mahariski PA, Yuwono J, Bhargah V, Prabawa IPY, Manuaba I, Rina IK. High level of individual lipid profile and lipid ratio as a predictive marker of poor glycemic control in type-2 diabetes mellitus. Vasc Health Risk Manag. 2019;15:149–157. doi: 10.2147/VHRM.S209830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hussain A, Ali I, Ijaz M, Rahim A. Correlation between hemoglobin A1c and serum lipid profile in Afghani patients with type 2 diabetes: hemoglobin A1c prognosticates dyslipidemia. Ther Adv Endocrinol Metab. 2017;8:51–57. doi: 10.1177/2042018817692296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen J, Spracklen CN, Marenne G, Varshney A, Corbin LJ, Luan J, Willems SM, Wu Y, Zhang X, Horikoshi M, et al. The trans-ancestral genomic architecture of glycemic traits. Nat Genet. 2021;53:840–860. doi: 10.1038/s41588-021-00852-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chami N, Chen MH, Slater AJ, Eicher JD, Evangelou E, Tajuddin SM, Love-Gregory L, Kacprowski T, Schick UM, Nomura A, et al. Exome genotyping identifies pleiotropic variants associated with red blood cell traits. Am J Hum Genet. 2016;99:8–21. doi: 10.1016/j.ajhg.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leong A, Chen J, Wheeler E, Hivert MF, Liu CT, Merino J, Dupuis J, Tai ES, Rotter JI, Florez JC, et al. Mendelian randomization analysis of hemoglobin A1c as a risk factor for coronary artery disease. Diabetes Care. 2019;42:1202–1208. doi: 10.2337/dc18-1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McDonagh EM, Thorn CF, Bautista JM, Youngster I, Altman RB, Klein TE. PharmGKB summary: very important pharmacogene information for G6PD. Pharmacogenet Genomics. 2012;22:219–228. doi: 10.1097/FPC.0b013e32834eb313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dore MP, Parodi G, Portoghese M, Pes GM. The controversial role of glucose-6-phosphate dehydrogenase deficiency on cardiovascular disease: a narrative review. Oxid Med Cell Longev. 2021;2021:5529256. doi: 10.1155/2021/5529256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spielman RS, Bastone LA, Burdick JT, Morley M, Ewens WJ, Cheung VG. Common genetic variants account for differences in gene expression among ethnic groups. Nat Genet. 2007;39:226–231. doi: 10.1038/ng1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhu AZ, Cox LS, Ahluwalia JS, Renner CC, Hatsukami DK, Benowitz NL, Tyndale RF. Genetic and phenotypic variation in UGT2B17, a testosterone-metabolizing enzyme, is associated with BMI in males. Pharmacogenet Genomics. 2015;25:263–269. doi: 10.1097/FPC.0000000000000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang TL, Chen XD, Guo Y, Lei SF, Wang JT, Zhou Q, Pan F, Chen Y, Zhang ZX, Dong SS, et al. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am J Hum Genet. 2008;83:663–674. doi: 10.1016/j.ajhg.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gencer B, Bonomi M, Adorni MP, Sirtori CR, Mach F, Ruscica M. Cardiovascular risk and testosterone - from subclinical atherosclerosis to lipoprotein function to heart failure. Rev Endocr Metab Disord. 2021;22:257–274. doi: 10.1007/s11154-021-09628-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Firtser S, Juonala M, Magnussen CG, Jula A, Loo BM, Marniemi J, Viikari JS, Toppari J, Perheentupa A, Hutri-Kahonen N, Raitakari OT. Relation of total and free testosterone and sex hormone-binding globulin with cardiovascular risk factors in men aged 24–45 years. The Cardiovascular Risk in Young Finns Study. Atherosclerosis. 2012;222:257–262. doi: 10.1016/j.atherosclerosis.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 85.Schooling CM, Luo S, Au Yeung SL, Thompson DJ, Karthikeyan S, Bolton TR, Mason AM, Ingelsson E, Burgess S. Genetic predictors of testosterone and their associations with cardiovascular disease and risk factors: a Mendelian randomization investigation. Int J Cardiol. 2018;267:171–176. doi: 10.1016/j.ijcard.2018.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Au Yeung SL, Cheng KK, Zhao J, Zhang W, Jiang C, Lam TH, Leung GM, Schooling CM. Genetically predicted 17beta-estradiol and cardiovascular risk factors in women: a Mendelian randomization analysis using young women in Hong Kong and older women in the Guangzhou Biobank Cohort Study. Ann Epidemiol. 2016;26:171–175. doi: 10.1016/j.annepidem.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 87.Winkler TW, Day FR, Croteau-Chonka DC, Wood AR, Locke AE, Magi R, Ferreira T, Fall T, Graff M, Justice AE, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Magi R, Horikoshi M, Sofer T, Mahajan A, Kitajima H, Franceschini N, McCarthy MI, Cogent-Kidney Consortium TDGC, Morris AP Trans-ethnic meta-regression of genome-wide association studies accounting for ancestry increases power for discovery and improves fine-mapping resolution. Hum Mol Genet. 2017;26:3639–3650. doi: 10.1093/hmg/ddx280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liu DJ, Peloso GM, Zhan X, Holmen OL, Zawistowski M, Feng S, Nikpay M, Auer PL, Goel A, Zhang H, et al. Meta-analysis of gene-level tests for rare variant association. Nat Genet. 2014;46:200–204. doi: 10.1038/ng.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Winkler TW, Justice AE, Cupples LA, Kronenberg F, Kutalik Z, Heid IM. consortium G: Approaches to detect genetic effects that differ between two strata in genome-wide meta-analyses: recommendations based on a systematic evaluation. PLoS ONE. 2017;12:e0181038. doi: 10.1371/journal.pone.0181038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Fauman EB, Hyde C: An optimal variant to gene distance window derived from an empirical definition of cis and trans protein QTLs. bioRxiv 2022:2022.2003.2007.483314. [DOI] [PMC free article] [PubMed]
- 93.Giambartolomei C, Vukcevic D, Schadt EE, Franke L, Hingorani AD, Wallace C, Plagnol V. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Caliskan M, Manduchi E, Rao HS, Segert JA, Beltrame MH, Trizzino M, Park Y, Baker SW, Chesi A, Johnson ME, et al. Genetic and epigenetic fine mapping of complex trait associated loci in the human liver. Am J Hum Genet. 2019;105:89–107. doi: 10.1016/j.ajhg.2019.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, Torstenson ES, Shah KP, Garcia T, Edwards TL, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 1825;2018:9. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hindy G, Dornbos P, Chaffin MD, Liu DJ, Wang M, Selvaraj MS, Zhang D, Park J, Aguilar-Salinas CA, Antonacci-Fulton L, et al. Rare coding variants in 35 genes associate with circulating lipid levels-a multi-ancestry analysis of 170,000 exomes. Am J Hum Genet. 2022;109:81–96. doi: 10.1016/j.ajhg.2021.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brown EE, Sturm AC, Cuchel M, Braun LT, Duell PB, Underberg JA, Jacobson TA, Hegele RA. Genetic testing in dyslipidemia: a scientific statement from the National Lipid Association. J Clin Lipidol. 2020;14:398–413. doi: 10.1016/j.jacl.2020.04.011. [DOI] [PubMed] [Google Scholar]
- 98.Hegele RA, Boren J, Ginsberg HN, Arca M, Averna M, Binder CJ, Calabresi L, Chapman MJ, Cuchel M, von Eckardstein A, et al. Rare dyslipidaemias, from phenotype to genotype to management: a European Atherosclerosis Society task force consensus statement. Lancet Diabetes Endocrinol. 2020;8:50–67. doi: 10.1016/S2213-8587(19)30264-5. [DOI] [PubMed] [Google Scholar]
- 99.Zhou Y, Zhang Y, Lian X, Li F, Wang C, Zhu F, Qiu Y, Chen Y. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022;50:D1398–D1407. doi: 10.1093/nar/gkab953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ge T, Chen CY, Ni Y, Feng YA, Smoller JW. Polygenic prediction via Bayesian regression and continuous shrinkage priors. Nat Commun. 2019;10:1776. doi: 10.1038/s41467-019-09718-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Denny JC, Ritchie MD, Basford MA, Pulley JM, Bastarache L, Brown-Gentry K, Wang D, Masys DR, Roden DM, Crawford DC. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Verma A, Bradford Y, Dudek S, Lucas AM, Verma SS, Pendergrass SA, Ritchie MD. A simulation study investigating power estimates in phenome-wide association studies. BMC Bioinformatics. 2018;19:120. doi: 10.1186/s12859-018-2135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nelson CP, Goel A, Butterworth AS, Kanoni S, Webb TR, Marouli E, Zeng L, Ntalla I, Lai FY, Hopewell JC, et al. Association analyses based on false discovery rate implicate new loci for coronary artery disease. Nat Genet. 2017;49:1385–1391. doi: 10.1038/ng.3913. [DOI] [PubMed] [Google Scholar]
- 105.Liu Z, Zhang Y, Graham S, Wang X, Cai D, Huang M, Pique-Regi R, Dong XC, Chen YE, Willer C, Liu W. Causal relationships between NAFLD, T2D and obesity have implications for disease subphenotyping. J Hepatol. 2020;73:263–276. doi: 10.1016/j.jhep.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou W, Nielsen JB, Fritsche LG, Dey R, Gabrielsen ME, Wolford BN, LeFaive J, VandeHaar P, Gagliano SA, Gifford A, et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Graham SE, Clarke SL, Wu KH, Kanoni S, Zajac GJM, Ramdas S, Surakka I, Ntalla I, Vedantam S, Winkler TW, et al: GLGC GWAS meta-analysis results and risk score weights repository: http://csg.sph.umich.edu/willer/public/glgc-lipids2021 2021.
- 108.Kanoni S GS, Wang Y, Surakka I, Ramdas S, Zhu X, Costanzo M, Jang D, Burtt NP, Willer CJ, Assimes TL, Peloso GM: A web browser displaying the gene prioritization and PheWAS results: https://hugeamp.org:8000/research.html?pageid=GLGC_149 2021.
- 109.Graham SE, Clarke SL, Wu KH, Kanoni S, Zajac GJM, Ramdas S, Surakka I, Ntalla I, Vedantam S, Winkler TW, et al: Optimized trans-ancestry polygenic score weights for LDL in the PGS Catalog : https://www.pgscatalog.org/publication/PGP000230/ 2021.
- 110.Kanoni S, Graham SE, Wang Y, Surakka I, Ramdas S, Zhu X, Clarke SL, Bhatti KF, Vedantam S, Winkler TW, et al: Optimized trans-ancestry polygenic score weights for HDL, TC, TG and non-HDL in the PGS Catalog: https://www.pgscatalog.org/publication/PGP000366/ 2022.
- 111.Kanoni S, Graham SE, Wang Y, Surakka I, Ramdas S, Zhu X, Clarke SL, Bhatti KF, Vedantam S, Winkler TW, et al: Implicating genes, pleiotropy and sexual dimorphism at blood lipid loci through multi-ancestry meta-analysis. Github. https://github.com/Global-Lipids-Genetics. 2022. [DOI] [PMC free article] [PubMed]
- 112.Kanoni S, Graham SE, Wang Y, Surakka I, Ramdas S, Zhu X, Clarke SL, Bhatti KF, Vedantam S, Winkler TW, et al. Implicating genes, pleiotropy and sexual dimorphism at blood lipid loci through multi-ancestry meta-analysis. 2022. Zenodo. 10.5281/zenodo.7130299. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Characteristics of contributing cohorts (as provided by each participating cohort).
Additional file 2: Table S2. Association results for the multi-ancestry index SNPs with the gene prioritization.
Additional file 3: Figure S1. Summary of prioritizing genes for A. Mendelian and B. mouse model genes separately by trait.
Additional file 4: Table S3. Text mining results for the PoPS+ prioritized genes.
Additional file 5: Table S4. Frequency of lipid-related publications for the PoPS+ prioritized genes.
Additional file 6: Figure S2. Frequency distribution of the lipid-related publications for both high confidence genes and the baseline genes.
Additional file 7: Table S5. Lookup of all prioritized lipid genes in the Therapeutic Target Database 2022.
Additional file 8: Table S6. Association of lipid index variants with CAD, T2D and NAFLD.
Additional file 9: Figure S3. Lipid traits – tissue/cell type associations estimated by DESE according to GTEx gene-level and GTEx transcript-level selective expression.
Additional file 10: Table S7. DESE phenotype-tissue association results using both GTEx gene-level and transcript-level selective expression.
Additional file 11: Figure S4. Comparison of PheWAS results in UKB and MVP for the LDL-C PGS, HDL-C PGS, TC PGS, TG PGS and nonHDL-C PGS.
Additional file 12: Table S8. PheWAS UKB-MVP meta-analysis results for each lipid PGS.
Additional file 13: Figure S5. PheWAS meta-analysis results for the trans-ethnic HDL-C PGS in UK Biobank and MVP.
Additional file 14: Figure S6. PheWAS meta-analysis results for the trans-ethnic TC PGS in UK Biobank and MVP.
Additional file 15: Figure S7. PheWAS meta-analysis results for the trans-ethnic TG PGS in UK Biobank and MVP.
Additional file 16: Figure S8. PheWAS meta-analysis results for the trans-ethnic nonHDL-C PGS in UK Biobank and MVP.
Additional file 17: Table S9. PheWAS UKB-MVP meta-analysis results for each index lipid variant at Bonferroni threshold for multiple testing p<=3.5e-8)
Additional file 18: Table S10. Lambda GC values across minor allele frequency bins for sex-specific meta-analyses.
Additional file 19: Table S11. Significant female-specific multi-ancestry meta-analysis results.
Additional file 20: Table S12. Significant male-specific multi-ancestry meta-analysis results.
Additional file 21: Table S13. Characteristics of replication cohorts (as provided by each participating cohort).
Additional file 22: Table S14. Test for difference in effects for index variants from sex-stratified meta-analysis.
Additional file 23: Table S15. Comparison of the sex-specific effects.
Additional file 24: Figure S9. Comparison of effect size estimates between males and females for index variants showing a significant difference in effect size between sexes.
Additional file 25: Table S16. Comparison of effect size estimates for sex-stratified analysis in the replication cohorts.
Additional file 26: Figure S10. Comparison of effect sizes for trans-ancestry index variants excluding cholesterol-lowering medication.
Additional file 27: Table S17. Sex-stratified effect sizes in UK Biobank considering all individuals or only those not on cholesterol lowering medications.
Additional file 28: Table S18. Sex-participation association of the variants with significant sex-specific lipid results.
Additional file 29: Table S19. Comparison of sex-stratified effect sizes in the UK Biobank for BMI, waist hip ratio adjusted for BMI, alcohol use, and smoking status.
Additional file 30: Table S20. Colocalization results for the sex-specific loci.
Additional file 31: Supplementary Note. Supplementary Note and Cohort Acknowledgments.
Additional file 32: Table S21. Significant X chromosome results in the sex-combined and sex-stratified analysis and replication.
Additional file 33: Table S22. Mouse genes with lipid phenotypes (silver set).
Data Availability Statement
The GWAS meta-analysis results (including both ancestry-specific and trans-ancestry analyses) and risk score weights are available at: http://csg.sph.umich.edu/willer/public/glgc-lipids2021 [107]. A web browser displaying the gene prioritization and PheWAS results is available at https://hugeamp.org:8000/research.html?pageid=GLGC_149 [108]. The optimized trans-ancestry polygenic score weights are deposited within the PGS Catalog (https://www.pgscatalog.org/publication/PGP000230/ [109] and https://www.pgscatalog.org/publication/PGP000366/ [110]. Scripts used for analysis and summary of results are available under the MIT license on this GitHub repository: https://github.com/Global-Lipids-Genetics [111]. The version of source code used in the manuscript is deposited in Zenodo: https://doi.org/10.5281/zenodo.7130299 [112].